ABSTRACT
Dimethylsulfoniopropionate (DMSP) and related organic sulfur compounds play key roles in global sulfur cycling. Bacteria have been found to be important DMSP producers in seawater and surface sediments of the aphotic Mariana Trench (MT). However, detailed bacterial DMSP cycling in the Mariana Trench subseafloor remains largely unknown. Here, the bacterial DMSP-cycling potential in a Mariana Trench sediment core (7.5 m in length) obtained at a 10,816-m water depth was investigated using culture-dependent and -independent methods. The DMSP content fluctuated along the sediment depth and reached the highest concentration at 15 to 18 cm below the seafloor (cmbsf). dsyB was the dominant known DMSP synthetic gene, existing in 0.36 to 1.19% of the bacteria, and was identified in the metagenome-assembled genomes (MAGs) of previously unknown bacterial DMSP synthetic groups such as Acidimicrobiia, Phycisphaerae, and Hydrogenedentia. dddP, dmdA, and dddX were the major DMSP catabolic genes. The DMSP catabolic activities of DddP and DddX retrieved from Anaerolineales MAGs were confirmed by heterologous expression, indicating that such anaerobic bacteria might participate in DMSP catabolism. Moreover, genes involved in methanethiol (MeSH) production from methylmercaptopropionate (MMPA) and dimethyl sulfide (DMS), MeSH oxidation, and DMS production were highly abundant, suggesting active conversions between different organic sulfur compounds. Finally, most culturable DMSP synthetic and catabolic isolates possessed no known DMSP synthetic and catabolic genes, and actinomycetes could be important groups involved in both DMSP synthesis and catabolism in Mariana Trench sediment. This study extends the current understanding of DMSP cycling in Mariana Trench sediment and highlights the need to uncover novel DMSP metabolic genes/pathways in extreme environments.
IMPORTANCE Dimethylsulfoniopropionate (DMSP) is an abundant organosulfur molecule in the ocean and is the precursor for the climate-active volatile gas dimethyl sulfide. Previous studies focused mainly on bacterial DMSP cycling in seawater, coastal sediment, and surface trench sediment samples, but DMSP metabolism in the Mariana Trench (MT) subseafloor sediments remains unknown. Here, we describe the DMSP content and metabolic bacterial groups in the subseafloor of the MT sediment. We found that the tendency for vertical variation of the DMSP content in the MT was distinct from that of the continent shelf sediment. Although dsyB and dddP were the dominant DMSP synthetic and catabolic genes in the MT sediment, respectively, both metagenomic and culture methods revealed multiple previously unknown DMSP metabolic bacterial groups, especially anaerobic bacteria and actinomycetes. The active conversion of DMSP, DMS, and methanethiol may also occur in the MT sediments. These results provide novel insights for understanding DMSP cycling in the MT.
KEYWORDS: DMSP, deep sediment, Mariana Trench, bacteria
INTRODUCTION
Dimethylsulfoniopropionate (DMSP) is an abundant organic sulfur compound in the global ocean, with an estimated level of biogenic production of 109 tons annually (1, 2). Marine algae, corals, plants, and heterotrophic bacteria can produce DMSP, which serves as a key nutrient (3), a signaling molecule (4, 5), or an antistress compound (6–9) in diverse organisms. Most importantly, bacterial cleavage of DMSP is the predominant source of dimethyl sulfide (DMS), which potentially plays a vital role in the regulation of the Earth’s climate (10). Once released into the atmosphere, the oxidation products of DMS aggregate into aerosols that can affect cloud formation, resulting in the backscatter of solar radiation and affecting the global temperature (11, 12). Another important volatile sulfur compound produced by DMSP demethylation is methanethiol (MeSH) (13), which can be converted to DMS by diverse bacteria (14).
The biosynthesis of DMSP is initiated from methionine (Met) through three known pathways, namely, the transamination pathway (15–19), the methylation pathway (20–26), and the decarboxylation pathway (27). In the well-studied transamination pathway, three key methylthiohydroxybutyrate (MTHB) S-methyltransferase enzymes were identified from different organisms: DsyB in some Alphaproteobacteria (15), DSYB in green algae and corals (16), and TpMMT in the diatom Thalassiosira pseudonana (28). Homologs of DsyB are widespread in many marine Alphaproteobacteria (predominantly Rhodobacterales), some Gammaproteobacteria, and Actinobacteria (9, 15, 25). For the methylation pathway, only one key Met methyltransferase, MmtN, was identified in Alphaproteobacteria, Gammaproteobacteria, and some Gram-positive bacteria (25, 29, 30). However, bacteria possessing mmtN are commonly less abundant than those with dsyB in seawater and sediments (25). The decarboxylation pathway was found in the heterotrophic microalga Crypthecodinium cohnii (30), but the key DMSP biosynthetic enzyme in this pathway remains unknown.
DMSP can be catabolized through the cleavage and demethylation pathways. In the cleavage pathway, DMS is produced as a coproduct with acrylate, hydroxypropionate, or acryloyl-CoA. Several bacterial DMSP lyases (DddD, DddL, DddP, DddQ, DddY, DddW, DddK, and DddX) and an algal DMSP lyase (Alma1) have been identified in this pathway (10, 31–36). Among these bacterial DMSP lyases, DddP is the most abundant environmental DMSP lyase and is often found in the marine Roseobacter clade (MRC) (9, 37). However, the major pathway for the degradation of DMSP is the demethylation pathway, and it is estimated that 50 to 90% of DMSP is metabolized by marine bacteria in this way (38). The key demethylase DmdA first converts DMSP to methylmercaptopropionate (MMPA), which is subsequently degraded by the MMPA-CoA ligase DmdB, the MMPA-CoA dehydrogenase DmdC, the methylthioacryloyl-CoA (MTA-CoA) hydratase DmdD, or the acrylate utilization hydratase AcuH to generate MeSH in diverse bacteria (39). DmdA has been reported to be prevalent in the MRC and the SAR11 clade (40, 41), which are considered the main bacterial groups for DMSP catabolism in the oceans (42).
The conversion between other organic sulfur compounds related to DMSP/DMS is also important and highly active in marine environments. The transformation of MeSH to DMS contributes to DMS production in both marine and terrestrial environments (14, 43), catalyzed by the key S-adenosylmethionine (SAM)-dependent methyltransferase MddA (14). On the other hand, MeSH can be oxidized to formaldehyde by MTO found in Thiobacillus, Rhodococcus, and Hyphomicrobium strains (37, 43). DMS can also be produced through the reduction of dimethyl sulfoxide (DMSO) by the bacterial DMSO reductase DorA (44). Conversely, DMS oxidation to DMSO represents a major sink of DMS in surface seawater (45). The multicomponent monooxygenase DsoABCDEF (46), the DMS dehydrogenase DdhAB (47), and the flavin-containing trimethylamine (TMA) monooxygenase Tmm (48) are responsible for DMS oxidation. In addition, the two-component DMS monooxygenase DmoAB can oxidize DMS to MeSH (49). The megL gene could encode a Met gamma lyase (MegL) that cleaves Met into MeSH (50).
The DMSP concentration in surface marine sediments can be up to 3 orders of magnitude higher than that in most seawater samples, but these levels were reported to decrease with sediment depth and oxygen availability (25, 41, 51). Microbial cycling of DMSP in sediment has been revealed recently, and bacteria are considered the key DMSP producers in these aphotic sediments, with dsyB being the dominant DMSP biosynthetic gene (9, 25, 37). The microbial DMSP catabolic potential seems to vary between different sediments (52). In surface salt marsh sediments, DMSP lyase genes (dddD, dddL, and dddP) were far more abundant than dsyB (25), but the abundances of dmdA and dddP were higher than that of dsyB in Mariana Trench (MT) deep-sea sediment, where dsyB far outnumbered both dmdA and dddP (9). In the hydrothermal sediment of Bohai Sea and Yellow Sea sediment (BYSS) samples, the abundance of DMSP degradation genes was low, and only the dddP gene was detected (37). For other genes involved in DMSP cycling, mddA was found mainly in Rhodopseudomonas and Thioalkalivibrio in salt marsh sediment (43) as well as in Desulfospira, Thioalkalivibrio, Crocosphaera, and Pseudomonas in BYSS samples (37). dmoA was predicted to be found mainly in Alcaligenes and Pseudomonas within BYSS samples (37). These results indicate that active DMSP cycling may occur in sediment environments.
The Mariana Trench is the deepest site in the ocean, with a depth of 11,000 m at the Challenger Deep (53). Despite the extreme environmental conditions in the Mariana Trench, there is still a high prokaryotic biomass (approximately 2.01 μg C g−1) and high activity in the sediments of this region (54). Bacterial DMSP synthesis and catabolism were examined in seawater (0 to 10,500 m) and surface sediment samples from the Challenger Deep of the Mariana Trench (9), demonstrating that bacteria are important DMSP producers in marine aphotic environments and that DMSP may be involved in hydrostatic pressure protection. However, the DMSP concentration, DMSP-cycling microbes, and their importance in deep Mariana Trench subseafloor sediments remain unexplored. In this study, we obtained a sediment core (~8 m) from a 10,816-m depth in the Challenger Deep and investigated the detailed vertical distribution of the DMSP content. The abundance of bacterial genes involved in DMSP cycling was analyzed by quantitative PCR (qPCR) and metagenome sequencing. Culture-dependent and -independent methods were employed to identify previously unknown DMSP metabolic bacteria and genes in Challenger Deep sediments.
RESULTS
DMSP content in the Mariana Trench deep sediment.
The total DMSP concentrations in the sediment samples from the Challenger Deep (10,816 m) ranged from 3.26 ± 0.37 to 9.66 ± 0.76 nmol g−1 wet sediment (Fig. 1) and varied with sediment depth. The DMSP concentration in the surface sediment samples was 6.35 ± 0.09 nmol g−1, whereas higher concentrations of DMSP were quantified at 15 to 18 cm below the seafloor (cmbsf) (9.66 ± 0.76 nmol g−1) and 37 to 40 cmbsf (7.90 ± 0.63 nmol g−1). The DMSP concentration was lower at 40 to 96 cmbsf (3.55 to 5.47 nmol g−1) than at the above layers and fluctuated between 96 cmbsf and 200 cmbsf, with a higher value at ~150 cmbsf (6.80 ± 0.31 nmol g−1). In the deep sediment below 250 cmbsf, the concentration of DMSP reduced gradually from 5.45 ± 0.18 nmol g−1 to 3.26 ± 0.37 nmol g−1. Generally, DMSP in the Challenger Deep sediment was present at levels similar to those reported previously for the surface sediment of the Mariana Trench (3.15 to 6.14 nmol g−1 wet sediment) (9) and lower than those in marine surface sediment (11.25 to 20.90 nmol g−1 wet sediment) (52). However, the variations in DMSP concentrations along the sediment depth were more fluctuant in the Challenger Deep than in other subseafloor sediments (52).
FIG 1.
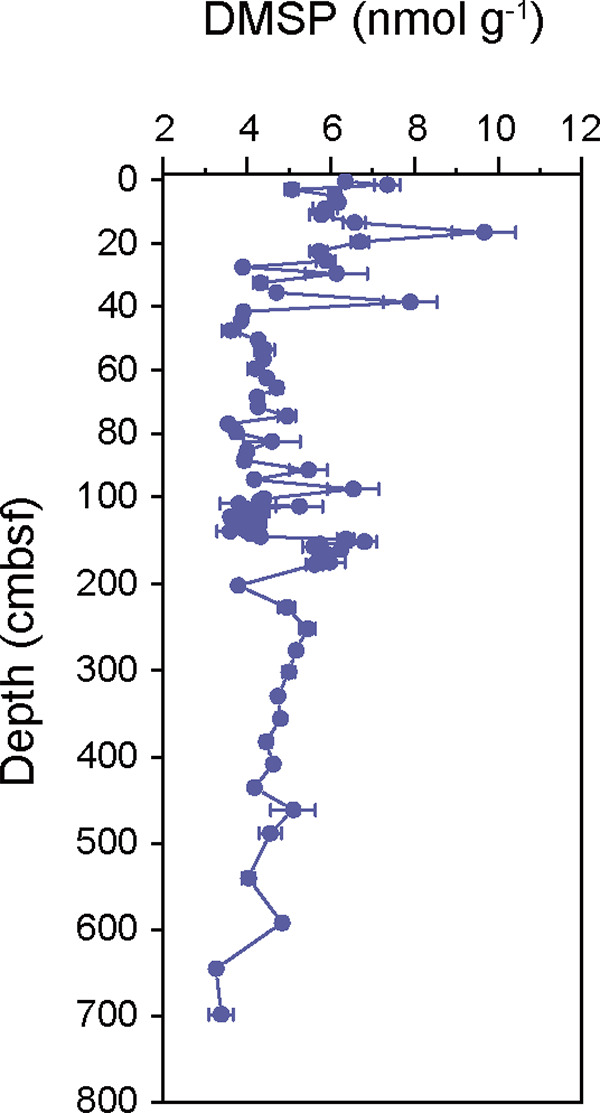
DMSP concentrations in the deep sediments from the Mariana Trench. Data are presented as means ± standard deviations (SD).
Vertical distribution of DMSP synthesis genes.
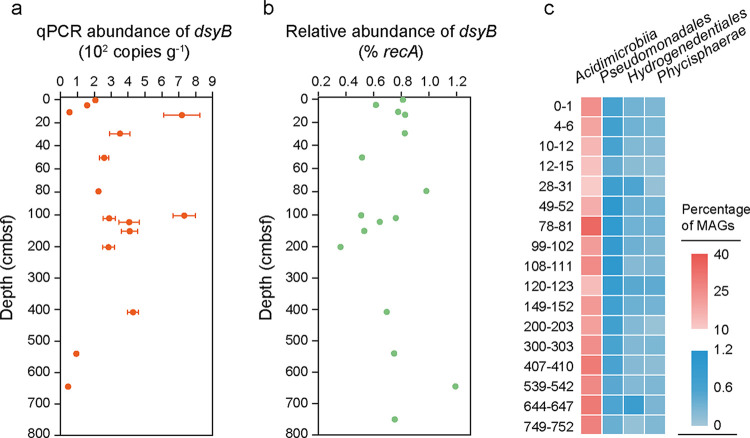
To quantify the abundance of bacterial DMSP biosynthetic genes, we performed qPCR analysis on the DMSP biosynthetic genes dsyB and mmtN. The abundance of the major bacterial DMSP biosynthetic gene dsyB ranged between 0.45 × 102 copies g−1 and 7.31 × 102 copies g−1 (Fig. 2a; see also Table S1 in the supplemental material). Corresponding to the DMSP content, dsyB was highly abundant at 12 to 15 cmbsf (3.59 × 102 copies g−1) and 99 to 102 cmbsf (3.66 × 102 copies g−1). The abundance of dsyB in samples from below 500 cmbsf was extremely low (<0.1 × 102 copies g−1). The mmtN gene could not be detected by qPCR at all depths of the Challenger Deep sediment (Table S1).
FIG 2.
Abundance of the DMSP synthesis gene dsyB in Mariana Trench sediment. (a) Absolute abundance of dsyB determined by qPCR. Data are presented as means ± SD. (b) Relative abundance of dsyB in metagenomes. (c) Relative abundance of MAGs with dsyB and their taxonomy information.
In the metagenomes from different depths, six protein homologs of DsyB were obtained, while homologs of MmtN (Fig. 2b, Table S2, and Fig. S1) were not found, which was consistent with the qPCR results, suggesting that bacteria synthesizing DMSP through the Met methylation pathway were scarce in these sediment samples. DsyB homologs were retrieved from all depths, and higher percentages of bacteria with DsyB existed at 12 to 15 cmbsf (0.83%), 28 to 31 cmbsf (0.83%), 78 to 81 cmbsf (0.98%), and 644 to 647 cmbsf (1.19%). Twelve high-quality metagenome-assembled genomes (MAGs) with dsyB (completeness of >50% and contamination of <10%) were obtained (Fig. 2c, Table S3, and Fig. S1), which were annotated as Acidimicrobiia (8/12), Gammaproteobacteria (Pseudomonadales order) (2/12), Phycisphaerae (1/12), as well as Hydrogenedentia (Hydrogenedentiales order) (1/12). Among these MAGs, only a few bacteria belonging to Gammaproteobacteria were previously reported to possess dsyB (25); no DsyB homologs were reported in the acidophilic actinomycetes Acidimicrobiia (55) as well as anaerobic Phycisphaerae (Planctomycetota) (56, 57) and Hydrogenedentia (58) bacteria. Acidimicrobiia MAGs harboring dsyB were present at high abundances across all depths, accounting for 1.25% to 3.09% in the sediment environment, and showed the highest abundance at 78 to 81 cmbsf (3.09%).
Vertical distribution of DMSP catabolism genes.
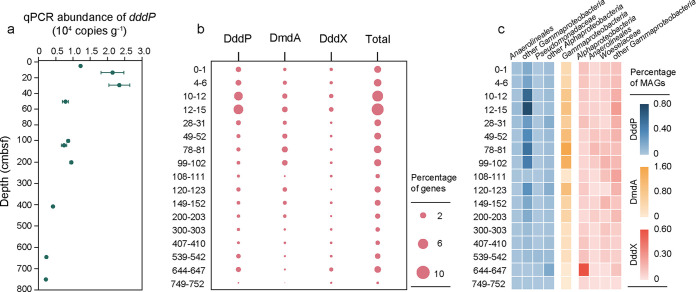
Quantification of the major DMSP catabolic genes dddP and dmdA (C2/D1 clades) showed that only dddP could be detected by qPCR analysis (Fig. 3a and Table S1), indicating that dddP might be the most abundant DMSP catabolic gene in these sediments. The abundance of dddP was highest in shallow sediment at 28 to 31 cmbsf (2.34 × 104 copies g−1) and was reduced to approximately 0.23 × 104 copies g−1 below 600 cmbsf. Although the key gene dmdA has been reported to be the dominant DMSP catabolic gene, with a higher abundance than those of the ddd genes (9), the levels of both the C2 and D1 clades of dmdA genes were below the qPCR detection threshold (Table S1).
FIG 3.
Abundance of DMSP catabolic genes in Mariana Trench sediment. (a) Absolute abundance of dddP determined by qPCR. Data are presented as means ± SD. (b) Relative abundances of dddP, dddX, and dmdA in metagenomes. (c) Relative abundance of MAGs with dddP, dddX, and dmdA and their taxonomy information.
Correspondingly, 17 protein homologs of DddP, 5 of DmdA, and 11 of DddX were retrieved from the metagenomes of the Challenger Deep sediment samples, but homologs of DddD, DddQ, DddL, DddK, DddW, DddY, and Alma1 were not found (Fig. 3b, Table S2, and Fig. S2 to S4). These homologs of DddP, DddX, and DmdA were distributed at all depths, and these DMSP catabolic genes were also more abundant at depths with higher DMSP concentrations, especially in samples at 10 to 12 cmbsf and 12 to 15 cmbsf. The percentage of bacteria with DddP reached 5.37% at 12 to 15 cmbsf, higher than those of DmdA (1.74%) and DddX (1.10%). In the deep subseafloor, the percentage of bacteria with DddP and DddX increased at 649 to 752 cmbsf (1.40% for DddP and 0.86% for DddX), while at 49 to 102 cmbsf, the relative abundance of DmdA (1.36% to 1.94%) seemed to be higher than those of DddP (0.72% to 0.85%) and DddX (0.32% to 0.35%). Further phylogenetic analysis indicated that the dmdA genes in the metagenomes clustered with dmdA subclades A, B, and E (Fig. S3), and this possibly explained why no dmdA was detected with primers for C2 and D1. However, dddP remained the most abundant DMSP catabolic gene in the shallow sediment above 15 cmbsf.
In total, 29 MAGs were annotated as possessing at least one DMSP catabolic gene (Table S3 and Fig. S2 to S4). Most MAGs with dddP belonged to Pseudomonadaceae and other Gammaproteobacteria as well as some Alphaproteobacteria (Fig. 3c). Surprisingly, one DddP homolog and three DddX homologs were found in anaerobic Anaerolineales MAGs (Table S3), indicating that such anaerobic bacteria may be involved in DMSP catabolism in Challenger Deep sediment. Homologs of DddX were also identified in MAGs from Woeseiaceae and other Gammaproteobacteria. The relative abundance of DddX-containing MAGs from Alphaproteobacteria was extremely high at 644 to 647 cmbsf (Fig. 3c). MAGs with DmdA were all from Gammaproteobacteria, with the highest abundance at 78 to 81 cmbsf (17.81%).
Genes involved in other DMS/MeSH production and oxidation processes.
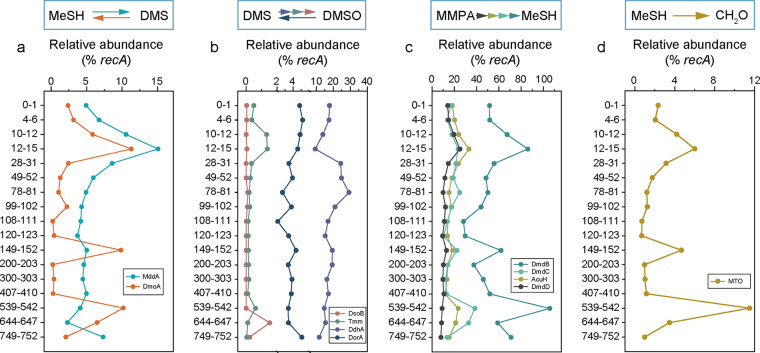
To better depict DMSP/DMS cycling in Challenger Deep sediment, we examined the relative abundances of other genes involved in these processes in metagenomes. In total, 51 mddA, 56 dmoA, 7 tmm, 178 ddhA, and 58 dorA homologs were obtained. For conversion between DMS and MeSH, the MeSH-dependent DMS-producing gene mddA was identified in 2.29 to 15.06% of bacteria and reached the highest abundance at 12 to 15 cmbsf (Fig. 4a). Bacteria with the DMS monooxygenase enzyme DmoA were generally less abundant than bacteria with MddA, accounting for 0.23 to 11.30% of the sediment metagenomes. However, the percentages of bacteria with DmoA were significantly higher at 12 to 15 cmbsf (11.30%), 149 to 152 cmbsf (9.83%), and 539 to 542 cmbsf (10.18%). Fifty-two MAGs harboring mddA were retrieved from Bacteroidota (1/52), Planctomycetota (4/52), Proteobacteria (6/52), Gemmatimonadota (14/52), Marinisomatota (16/52), Myxococcota (7/52), and Hydrogenedentota (2/52) (Table S4), which were not reported in previous studies except for those from Proteobacteria. DmoA was distributed in MAGs of Acinetobacter (4/28), Microbacterium (2/28), and Pseudomonas (1/28), as reported in a previous study on marine sediment samples (37). Additionally, DmoA homologs were also identified in MAGs from Burkholderiaceae (11/28), Dehalococcoidia (8/28), and Brevibacillus (2/28), which have not been reported previously.
FIG 4.
Abundance of genes involved in other processes of DMS/DMSP cycling in Mariana Trench sediment. (a) Relative abundances of the MeSH transmethylation gene mddA and the DMS oxidation gene dmoA. (b) Relative abundances of DMS oxidation-related genes (dsoB, tmm, and ddhA) and the DMSO reduction gene dorA. (c) Relative abundances of genes involved in the conversion of MMPA to MeSH (acuH, dmdB, dmdC, and dmdD). (d) Relative abundance of the MeSH oxidation gene mtoX.
For conversion between DMS and DMSO (Fig. 4b), ddhA was the dominant DMS oxidation gene throughout the sediment, compared with the other DMS oxidation genes tmm and dsoB. The relative abundance of ddhA ranged from 9.43% to 29.22% of bacteria, which increased at 28 to 81 cmbsf (>24.38%). The percentages of bacteria with Tmm were higher at 10 to 15 cmbsf (1.32%), 12 to 15 cmbsf (1.37%), and 539 to 542 cmbsf (0.60%). Only one dsoB homolog was found in the sediment metagenomes and reached the highest relative abundance of 1.52% at 644 to 647 cmbsf. A total of 146 MAGs, mainly from Actinobacteriota, Bacteroidota, Chloroflexota, Gemmatimonadota, Hydrogenedentota, Marinisomatota, Planctomycetota, and Proteobacteria, harbored ddhA, and more than one ddhA hit was found in 57 (out of 146) MAGs. A total of 3 (out of 1,350) MAGs harbored DsoB, which were retrieved only from Acinetobacter pittii, while Tmm was identified in only two Gammaproteobacteria MAGs. The DMSO reduction gene dorA was found in approximately 5% of bacteria in the sediment metagenomes, with a lower proportion than that of ddhA. Moreover, DorA predominantly existed in MAGs of Dehalococcoidia (17/31), Acidimicrobiia (5/31), Gammaproteobacteria (4/31), Rhodospirillales (1/31), and Anaerohalosphaeraceae (1/31).
Similar to mddA and dmoA, the MMPA-dependent MeSH-producing genes dmdB, dmdC, dmdD, and acuH and the MeSH oxidation gene mtoX reached higher abundances at 12 to 15 cmbsf, 149 to 152 cmbsf, and 539 to 542 cmbsf. The abundance of DmdB ranged from 28.12% to 105.75% (Fig. 4c) and was higher at 12 to 15 cmbsf (85.97%), 149 to 152 cmbsf (61.93%), and 539 to 542 cmbsf (105.75%). AcuH (33.04%) and DmdD (24.95%) were more abundant at 12 to 15 cmbsf, while the abundance of DmdC (38.35%) was higher at 539 to 542 cmbsf. Alternately, the abundance of MTO (Fig. 4d) was higher at 12 to 15 cmbsf (6.00%), 149 to 152 cmbsf (4.66%), and 539 to 542 cmbsf (11.50%). A total of 651 (out of 1,350) MAGs harbored DmdB, DmdC, DmdD, and AcuH, while 51 MAGs were found to harbor MTO (Table S4). Among them, DmdB was identified in 411 MAGs, which were from Acidimicrobiia (108 MAGs), Chloroflexota (77), Gammaproteobacteria (77), Gemmatimonadetes (33), Alphaproteobacteria (23), Myxococcota (16), Desulfobacterales (14), Elusimicrobiota (13), Thermodesulfovibrionia (11), Planctomycetota (9), Hydrogenedentiales (6), Acidobacteriota (3), Brevibacillus agri (2), “Candidatus Omnitrphota” (1), and unknown bacteria (18). A total of 33 (out of 51) MAGs possessing MTO were from Gammaproteobacteria, including Pseudomonadales (15 MAGs), Cupriavidus gilardii (10), Woeseiaceae (3), and other Gammaproteobacteria (5). Additionally, other MAGs with MTO were from Myxococcota (8/51), Thermodesulfovibrionia (6/51), Alphaproteobacteria (2/51), Elusimicrobiota (1/51), and Anaerolineales (1/51).
Isolates involved in DMSP cycling in surface sediment.
Surface sediment samples (0 to 1 cmbsf) were used to enrich and isolate DMSP-producing and -catabolizing bacteria. In total, 69 strains were obtained from enrichment cultures with Met, and 156 strains were obtained from enrichment cultures with DMSP. We further examined the DMSP synthetic and catabolic activities of these isolates as well as their ability to produce DMS or MeSH independent of DMSP.
Ten isolates, belonging to Alphaproteobacteria (3 isolates), Gammaproteobacteria (1), Actinomycetia (5), and Bacilli (1), could use Met to produce DMSP through the transamination pathway, accounting for 19.2% of all representative strains. At the genus level, these isolates were from Alcanivorax (1 isolate), Qipengyuania (3), Dietzia (3), Brachybacterium (2), and Mammaliicoccus (1). Among these DMSP-producing isolates, Qipengyuania citrea CHJ019 and Brachybacterium paraconglomeratum CHJ016 showed the highest DMSP concentrations in their cultures (>10 nmol DMSP/mL) (Table S5). So far, none of the culturable actinomycetes and bacillus isolates have been reported to produce DMSP in previous studies. Although multiple isolates from Qipengyuania (which used to be classified as Erythrobacter) have been found to produce DMSP, no known DMSP biosynthesis genes were identified in these isolates (9, 52). Indeed, dsyB and mmtN were not detected in these 10 strains by PCR amplification, suggesting that there may be previously unknown DMSP-producing genes in these isolates.
Moreover, all 10 of these isolates could produce MeSH and DMS when Met was added, indicating the existence of MegL and Mdd enzymes in these bacteria (14). In addition to the above-mentioned DMSP-producing isolates, Glutamicibacter mysorens CHJ159, Brachybacterium ginsengisoli CHJ148, and Arthrobacter crystallopoietes CHJ137 could also metabolize MeSH to produce DMS and may possess the MeSH transmethylation pathway (Table S5). This MeSH-dependent DMS-producing pathway was reported previously in diverse actinomycetes as well as in proteobacteria (37, 43).
For DMSP catabolism, 39 isolates were found to degrade DMSP to DMS, while 9 of them could also degrade DMSP to MeSH. These isolates, belonging to Alphaproteobacteria (9 isolates), Gammaproteobacteria (5), Actinomycetia (20), Bacilli (4), and Sphingobacteriia (1), exhibited varied levels of DMSP catabolism activity (Table S6). Surprisingly, more than one-half of these culturable DMSP catabolic isolates (51.3%) producing DMS were from Actinomycetia, including Glutamicibacter (9 isolates), Microbacterium (3), Brachybacterium (4), Agrococcus (1), Arthrobacter (1), Corynebacterium (1), and Brevibacterium (1) at the genus level. However, the DMSP catabolic gene in these actinomycetes remained unknown (34). DMSP catabolic isolates from Alphaproteobacteria accounted for 23%, including those from Qipengyuania (4 isolates), Tritonibacter (2), Aliihoeflea (1), Paracoccus (1), and Erythrobacter (1). DMSP catabolic isolates from Gammaproteobacteria were Psychrobacter (2 isolates), Pseudomonas (1), Alcanivorax (1), and Luteimonas (1). Additionally, two Bacilli isolates, from Staphylococcus and Mammaliicoccus, were also found to catabolize DMSP. DMSP catabolic isolates in Bacteroidetes have been identified in Mariana Trench samples, as described previously by Zheng et al. (9), but those from Sphingobacteriia were reported for the first time in this study. Isolates that could degrade DMSP to MeSH were mainly from Alphaproteobacteria (3 isolates), Gammaproteobacteria (1), Actinomycetia (2), Bacilli (2), and Sphingobacteriia (1). None of these DMSP catabolic isolates harbored dddP, dddD, dddL, and dmdA subclades (C2/D1) when detected by PCR. Although it could also be possible that the degenerate primers of known DMSP metabolic genes may not fully cover all divergent homologous genes in these strains, previously unknown DMSP catabolic and biosynthetic genes may exist in Mariana Trench sediments.
Enzyme activity of the DMSP metabolic proteins from MAGs.
To confirm whether the annotated DMSP metabolic enzymes from MAGs were active, we synthesized and heterologously expressed representative sequences to detect their activity, including homologs of dsyB (4 sequences), dddP (2 sequences), and dddX (1 sequence) (Table S7). Four DsyB sequences (690 bp to 1,014 bp) were retrieved from MAGs annotated as belonging to Acidimicrobiia, Gammaproteobacteria, Hydrogenedentia (Hydrogenedentiales order), and Phycisphaerae. However, none of the heterologously expressed dsyB genes showed DMSP synthetic activity in Escherichia coli BL21. This result indicated that these putative DsyB proteins may not be active MTHB S-methyltransferases. However, it is also possible that this may be due to the lack of an appropriate cofactor for the DsyB protein to be active in E. coli, as described previously by Li et al. (59). Further experiments are needed to confirm the activity of these putative DsyB proteins. For DMSP catabolism, two dddP sequences (1,161 bp and 1,305 bp) were chosen from Anaerolineales and Pseudomonadales MAGs, respectively, and one dddX sequence (2,127 bp) was chosen from an Anaerolineales MAG. The overexpressed DddP protein from Pseudomonas showed the highest DMSP catabolic enzyme activity (270,773.45 ± 18,002.42 nmol/mg protein) (Fig. S5). Surprisingly, we confirmed that both DddX and DddP from the Anaerolineales MAG can degrade DMSP to DMS, with activities of 138.91 ± 25.23 nmol/mg protein and 70.95 ± 4.17 nmol/mg protein, respectively.
DISCUSSION
As an important compound involved in global sulfur cycling, the process for the metabolism of DMSP in marine sediments has gradually been revealed in recent years. Although photosynthetic eukaryotes were thought to be dominant DMSP producers in the photic zones, bacteria were demonstrated to be significant contributors of DMSP in diverse marine sediments (25, 37, 52) as well as in the aphotic waters of Earth’s deepest ocean site: the Challenger Deep (9). Our previous study of the Mariana Trench revealed that bacteria are key DMSP producers in deep seawater and sediment, and a physiological function for DMSP in hydrostatic pressure protection has been proposed (9). However, the detailed and complete bacterial DMSP-cycling process through the depth profile of the Challenger Deep sediment remains to be further explored.
The DMSP concentrations in the surface sediment of the Mariana Trench at 5,000- to 10,500-m water depths varied between 3.15 and 6.14 nmol g−1 wet sediment, as reported previously by Zheng et al. (9). In the current study, the DMSP content in the surface sediment at 10,816 m is even slightly higher (6.35 ± 0.09 nmol g−1). Indeed, combined with the results of Zheng et al. (9), we suggest that the DMSP concentrations in the surface sediment are not correlated with the water depth in the Mariana Trench. In addition, the DMSP concentrations in hadal Mariana Trench surface sediment were lower than those in the South China Sea (SCS) continent slope sediment (11.25 to 20.90 nmol g−1) (60) and even lower than those in coastal sediment (~100 nmol g−1) (25), ferruginous sediment (~100 nmol g−1), and sulfidic sediment (~600 nmol g−1) (61). Most importantly, we found that the depth profile of the DMSP content from the surface to the subseafloor sediments showed distinct variation tendencies compared with those of the SCS sediment. There was no dramatic reduction in the DMSP concentrations in the top 30 cmbsf, and two extremely high DMSP concentrations were detected at 15 to 18 cmbsf and 37 to 40 cmbsf, which were even higher than those in the surface sediment. Such fluctuations in the DMSP content may indicate a nonhomogeneous process of sedimentation possibly affected by special topography and frequent tectonic activities (62). Notably, although the surface DMSP concentration in the Mariana Trench sediment is lower than that in the SCS sediment, the DMSP concentration in the deep seafloor (~700 cmbsf) of the Mariana Trench (3.26 ± 0.37 nmol g−1) is even higher than that in the SCS subseafloor sediment (0.56 to 2.08 nmol g−1). Such depth profiles of the DMSP content in the deep Mariana Trench sediment suggested that there might be active microbial cycling processes for DMSP and other organic sulfur compounds. Although some DMSP in the sediment is expected to arise from sinking particles of dead algae and/or fecal pellets, it seems that bacterial DMSP synthesis is likely an important source of DMSP considering the high DMSP turnover rates in photic seawater (9).
For DMSP biosynthesis, dsyB was the only gene detected by qPCR and metagenomic sequencing. This is reasonable since the abundance of mmtN was consistently lower than that of dsyB in diverse sediments (9, 52). However, as the dominant DMSP biosynthetic gene, the abundance of dsyB at 10,816 m in this study (102 copies g−1) was lower than those in the Mariana Trench surface sediment at 5,000 to 10,910 m (103 to 105 copies g−1) (9). Corresponding to the DMSP concentrations, peaks of dsyB abundance were observed at several depths (12 to 15 cmbsf and 99 to 102 cmbsf), and there seemed to be no obvious decline in either the absolute or relative abundance of dsyB along sediment depths. The metagenomic analysis predicted that approximately 1% of bacteria contain dsyB in deep sediment samples, which is comparable to that in Stiffkey salt marshes (~1% of bacteria) (25). MAGs containing dsyB in these samples were from Gammaproteobacteria (Pseudomonadales order), Acidimicrobiia, Hydrogenedentia (Hydrogenedentiales order), and Phycisphaerae, which are distinct from the commonly known dsyB-containing bacteria belonging to Alphaproteobacteria (9, 25). DsyB was also previously reported in some bacteria of the Gammaproteobacteria (all were unassigned), Betaproteobacteria, and Actinobacteria in coastal sediments, such as Flammeovirgaceae, Actinomycetospora, and Ponticoccus (25). However, anaerobic bacteria of the Acidimicrobiia (55), Phycisphaerae (56, 57), and Hydrogenedentia (Hydrogenedentiales order) (58), which were commonly found in deep anoxic sediment, have never been reported to possess dsyB or produce DMSP. Although we attempted to heterogeneously express these dsyB homologs from the above-mentioned MAGs, no DMSP-producing activity was detected in the recombinant E. coli strains. This result indicated that these putative DsyB proteins may not be active MTHB S-methyltransferases; however, this may also be due to the lack of certain cofactors in the E. coli strain, as indicated previously by Li et al. (59). Further experiments are required to confirm the activity of the DsyB homologs found in these MAGs. Moreover, culturable isolates from the Alcanivorax, Qipengyuania, Dietzia, Brachybacterium, and Mammaliicoccus genera were considered potential DMSP producers without dsyB or mmtN homologs. These isolates might contain previously unknown DMSP biosynthetic genes and pathways, and they could also possibly be the dominant DMSP producers instead of those bacteria with dsyB in Mariana Trench sediment.
For DMSP catabolism, 0.33 to 8.22% of the total bacteria in Mariana Trench sediment were predicted to contain at least one DMSP catabolic gene. The DMSP lyase DddP, instead of the demethylase DmdA, was the dominant enzyme, and higher abundances of DddP were found at ~20 and 40 cmbsf, corresponding to the DMSP content. dddP was identified in MAGs from Alphaproteobacteria, Gammaproteobacteria, and Anaerolineae, and MAGs from these groups with dddP were also retrieved from seawater of the Mariana Trench (9). dmdA genes were reported in Alphaproteobacteria, such as in the Changjiang Estuary (63), the Sanriku Coastal Region of Japan (41), and Eastern China Marginal Seas seawater and sediment (37, 64). However, MAGs containing dmdA genes in Mariana Trench deep sediment were mainly from Gammaproteobacteria. Furthermore, the dmdA genes in this study were clustered in dmdA subclades A, B, and E, while the dmdA genes in seawater and other sediments were mainly from subclades D1 and C2 (41, 65). For other ddd genes, only dddX was identified in the metagenomes and was predicted to be present in 0.12 to 1.10% of bacteria, which was identified in an Anaerolineales MAG. Both DddX and DddP retrieved from an Anaerolineales MAG were shown to be active in DMSP catabolism, indicating for the first time that anaerobes from Anaerolineales might be responsible for DMSP degradation in deep sediment. It should also be noted that Actinomycetia isolates accounted for a large proportion (51%) of the culturable DMSP catabolic bacteria, suggesting that actinobacteria can be important DMSP catabolic groups in Mariana Trench deep sediment. Although the DMSP catabolic capacity of many other Gram-positive actinobacteria has been previously reported (66), the DMSP catabolic gene and pathway remain unknown in these actinobacteria. Overall, we found that the known DMSP catabolic genes in deep Mariana Trench sediment were possibly from distinct bacterial groups compared with those in the seawater and other sediment environments, and potential previously unknown DMSP catabolic genes in actinomycetes and some anaerobic bacterial groups might be important for DMSP catabolism in such sediments. However, it should be noted that degenerate PCR may not fully cover the homologous genes of known DMSP metabolic enzymes in the obtained culturable isolates, considering the high level of divergence of these genes. Further experiments such as genome sequencing may be needed to confirm the existence of previously unknown active DMSP metabolic genes in these isolates.
For other genes involved in DMSP cycling, we found that the relative abundances of several genes were extremely high, especially the dmdB gene, encoding the MMPA-CoA ligase, as well as dmdCD and acuH, which are involved in the conversion of MMPA (resulting from DMSP catabolism by dmdA) to MeSH. A similarly high dmdB abundance was observed in Eastern China Marginal Seas sediments (37). Interestingly, we found that the relative abundances of genes related to MeSH production from MMPA and DMS, as well as MeSH oxidation to formaldehyde, consistently reached peaks at 12 to 15 cmbsf, 149 to 152 cmbsf, and 539 to 542 cmbsf, indicating that there might be active production and oxidation of MeSH at these depths for an unknown reason. The abundance of mddA, involved in the transformation of MeSH to DMS, was also the highest at 12 to 15 cmbsf. Therefore, except for DMSP biosynthesis and catabolism, active transformation between MeSH, DMS, and other compounds may occur in the deep sediment. Indeed, we found that many culturable isolates, including all of the DMSP-producing isolates, could use Met to produce DMS and MeSH and could use MeSH to produce DMS.
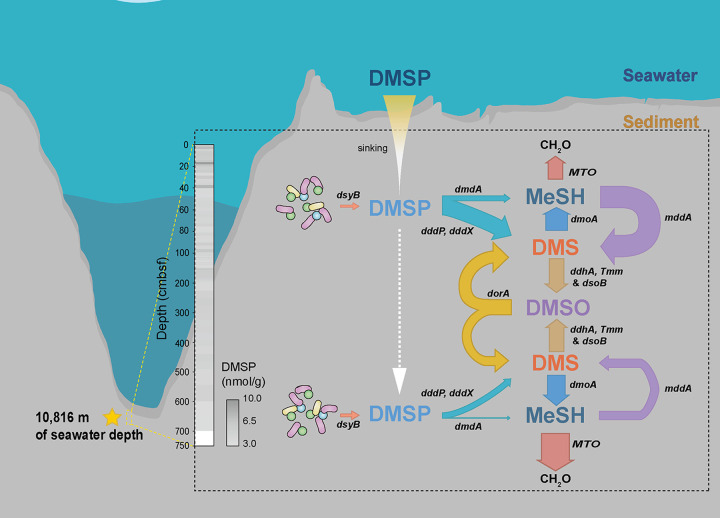
The whole DMSP-cycling processes in the deep sediment of the Mariana Trench are depicted in Fig. 5. In this study, we discuss the similarities and differences in bacterial DMSP metabolism in the Mariana Trench compared with other sediment environments. It seems that under extreme physical-chemical conditions, such as the low temperature and extremely high hydrostatic pressure in the trench sediment, there might be active but distinct bacterial groups involved in DMSP cycling. Further research is needed to reveal the potential previously unknown DMSP metabolic genes and pathways in these sediments, and the collection of more deep-sediment cores may help to provide a comprehensive understanding of DMSP metabolism in the Mariana Trench.
FIG 5.
Proposed bacterial DMSP cycling in Mariana Trench subseafloor sediment. The thickness of the arrows indicates the relative abundance of genes involved in DMSP cycling. The arrowheads indicate the flow directions of organic sulfur compounds.
Conclusion.
In this study, we outlined the complete bacterial DMSP-cycling processes in the deep sediment of the Mariana Trench. Both the DMSP contents and the DMSP metabolic gene levels fluctuated with the sediment depths. dsyB was the dominant DMSP biosynthesis gene, and dddP was the most abundant DMSP catabolic gene. Importantly, special bacterial groups in such deep sediments were involved in the DMSP biosynthetic pathway, such as Acidimicrobiia, Phycisphaerae, and Hydrogenedentia. Similarly, homologs of DddP and DddX were found in anaerobic Anaerolineales, and their DMSP catabolic activities were confirmed for the first time. More than one-half of the culturable bacteria involved in DMSP catabolism were actinomycetes, suggesting the significance of the potential novel DMSP catabolic gene in actinomycetes for DMSP catabolism in deep sediment. Moreover, highly abundant genes that participate in the conversion of MeSH, DMS, and other related organic compounds were identified.
MATERIALS AND METHODS
Sample collection and DNA preparation.
A sediment gravity core was collected from the Challenger Deep of the Mariana Trench (11°19.904′, 142°12.083′) during a cruise in July 2020 conducted on the R/V Dong Fang Hong 3. The depth of seawater was 10,816 m at the sampling site. The length of the sediment core reached approximately 750 cmbsf, and the core was sliced with a stainless steel cutter for subsampling. A total of 81 samples were obtained along the sediment core. The samples were transferred to sterilized plastic tubes and stored at −80°C before DNA extraction. Metagenomic DNA was extracted from 10 to 12 g (wet weight) of representative sediment samples from 17 depths, as previously described by Zhou et al. (67). Briefly, each 1-g sample was washed with 3.3 mL of extraction buffer (100 mM Tris-HCl [pH 8.0], 100 mM sodium EDTA [pH 8.0], 100 mM sodium phosphate [pH 8.0], 1.5 M NaCl, 1% cetyltrimethylammonium bromide [CTAB]) and then centrifuged at 6,000 × g for 20 min at room temperature. The concentrated biomass was ground in liquid nitrogen, 122 μg proteinase K and 0.37 mL 20% SDS were added, and the mixture was incubated at 37°C for 30 min and at 65°C for 2 h, respectively. After phenol-chloroform extraction (ratio of 24:1, 15 volumes), DNA was precipitated with 0.6 volumes of isopropanol, washed with 70% ethanol, and air dried. Finally, the PowerSoil DNA isolation kit (MoBio Lab) was used for DNA purification to improve the DNA quality and satisfy the sequencing quality threshold.
Quantification of DMSP in sediment.
The DMSP content was measured in samples from 82 depths, as described previously by Zheng et al. (9). One hundred microliters of purified water and 100 μL of NaOH (10 M) were added to 0.1-g samples placed into 2-mL vials, and the samples were then stored in the dark at room temperature for 24 h. The DMS released from DMSP cleavage was quantified by a purge-and-trap gas chromatography (GC) system or GC autoinjection using a flame photometric detector (Agilent 7890B GC system fitted with a 7693A autosampler) and an HP-INNOWax 30-m by 0.320-mm capillary column (Agilent Technologies, J&W Scientific). An eight-point calibration curve of DMS standards was used (60), and the detection limit for headspace DMS was 0.015 nmol. All experiments described here were performed using three biological replicates.
Quantitative PCR.
The abundance of genes involved in DMSP synthesis and degradation was quantified using qPCR in 17 samples from different depths, including dsyB, mmtN, as well as dddP and dmdA subclades (C2 and D1) (11, 40). The primers (5′ to 3′) used in this assay and their annealing temperatures are displayed in Table 1 (25, 40, 68, 69). A qPCR standard was prepared, and standard melt curves were conducted as described in previous reports (11, 40). The PCRs were conducted as follows: an initial denaturation step at 95°C for 3 min and then 35 cycles of 95°C for 30 s, the corresponding annealing temperature for 30 s, and 72°C for 30 s. A melt curve was run after PCR as follows: denaturation at 95°C for 1 min and 0.5°C increments from the annealing temperature with signal collection. Each sample was run with negative controls conducted in triplicate using a QuantStudio 5 system (Thermo Fisher Scientific). A 10-fold serial dilution for PCR amplification of plasmids containing each target gene fragment was used to construct the standard curves. The amplification curves exhibited clear linear relationships (R2 > 0.999) and yielded an amplification efficiency of 0.90 for each gene.
TABLE 1.
Primers and amplification conditions for qPCR detection and target gene amplicon sequencing for bacteria
| Target gene | Primer | Sequence (5′–3′) | Amplicon length (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| dsyB | dsyBF | CATGGGSTCSAAGGCSCTKTT | 246 | 58 | 25 |
| dsyBR | GCAGRTARTCGCCGAAATCGTA | ||||
| mmtN | mmtNF | CCGAGGTGGTCATGAAYTTYGG | 301 | 54 | 25 |
| mmtNR | GGATCACGCACACYTCRTGRTA | ||||
| dddP | 874F | AAYGAAATWGTTGCCTTTGA | 97 | 41 | 68 |
| 971R | GCATDGCRTAAATCATATC | ||||
| dmdA (C2) | 291F | AGATGAAAATGCTGGAATGATAAATG | 191 | 50 | 68 |
| 482R | AAATCTTCAGACTTTGGACCTTG | 40 | |||
| dmdA (D1) | 268F | AGATGTTATTATTGTCCAATAATTGATG | 89 | 49 | 68 |
| 356R | ATCCACCATCTATCTTCAGCTA | 40 | |||
| dddD | dddDF | ACCAACGTCATTGCAGGACC | 63 | 56 | 69 |
| dddDR | TGTGCGTGTTCTTCCGGTG | ||||
| dddL | dddLF | CTGGGAATACGGCTACGAGA | 239 | 53 | 69 |
| dddLR | GTTCAAGATCAGCGATCCGG |
Metagenomic sequencing and binning.
A total of 17 sediment DNA samples were used for the metagenomic analysis. Fifteen DNA samples (0 to 542 cmbsf, each about 1 μg) were sent to BGI Tech Solutions (Beijing Liuhe) Co., Ltd., and two DNA samples (644 to 647 cmbsf [0.4 μg] and 749 to 752 cmbsf [0.2 μg]) were sent to Majorbio BiFo-Pharm Technology Co., Ltd., Shanghai, China, for metagenomic sequencing. Libraries were prepared without any amplification step for each sample. Metagenomic shotgun sequencing was performed on the Illumina HiSeq X-Ten platform, with 2× 150-bp paired-end reads.
All of the raw reads containing >10% undefined bases and >40% low-quality bases and that had >15 bases matching the adapters were removed. The construction of genes was performed using MetaWrap-megahit (70), and gene prediction was performed by prodigal with default parameters (71). A total of 15,802,294 sequences were clustered at 95% identity into a set of 40,154,822 nonredundant sequences using cd-hit-est with the command cd-hit-est -c 0.95 -n 10 -d 0 -M 0 -T 1 -aS 0.9, and sequences of <100 bp were discarded (72). However, to retrieve more high-quality MAGs, the construction of MAGs was performed using MetaWrap-metaspades, -assembly, and -reassemble_bins with default parameters (70), and protein prediction for MAGs was performed using prokka with default parameters (73). MAGs with >50% completeness and <10% contamination were retained for further metagenomic analysis. The completion and contamination of each MAG were evaluated by using CheckM version 1.0.12 (74). The taxonomy of these MAGs was annotated by using GTDB-Tk version 1.6.0 (75).
Bioinformatic analyses of genes involved in DMS/DMSP cycling.
To explore the genetic potential for DMSP/DMS cycling, hidden Markov model (HMM)-based searches for homologs in metagenome data sets and MAGs were performed using HMMER 3.3.2 (76). Ratified MmtN, DsyB, DddD, DddP, DddK, DddQ, DddW, DddL, DddY, DddX, DmdA, Alma1, MddA, DmoA (the catalytic subunit of the DMS monooxygenase DmoAB), DorA, Tmm, DsoB (a key catalytic subunit of the monooxygenase DsoABCDEF), DdhA (the catalytic subunit of the DMS dehydrogenase DdhABC), DmdB, DmdC, DmdD, and MTO protein sequences were obtained from the National Center for Biotechnology Information (NCBI) database. HMMs were created for each enzyme using protein sequences that have been biochemically or structurally characterized, and the cutoff values used were selected based on established stringency cutoff values from previous reports (see Table S8 in the supplemental material) (10, 15, 16, 25, 33–36, 39, 48, 77–80). Separate cutoff E values were confirmed by BLAST analysis between functionally verified protein sequences (37). However, homologs of DmdD, DsoB, and DmoA in metagenomes were obtained using BLASTp with an E value cutoff of 10−30 and an identity cutoff of 40% (39, 49). All of the predicted DMSP/DMS cycling proteins were placed into phylogenetic trees to further identify functional homologs. Phylogenetic trees were constructed using IQ-TREE version 1.6.12 (81) and included nonfunctional sequences used as outgroups. Also, these amino acid sequences were aligned using MAFFT version 7 with default settings (82, 83). Sequences that clustered with nonfunctional sequences were removed. In addition, the DmdA sequences in the metagenomic analysis were used to build the neighbor-joining phylogenetic tree, as described previously by Varaljay et al. (40).
As previously described by Zheng et al. (9), to compare the counts of bacterial cells among samples, the percentages of bacteria harboring genes were normalized using the single-copy housekeeping gene recA. The HMM profile for RecA was downloaded from FunGene (http://fungene.cme.msu.edu/), and sequences with an E value of ≤10−50 were retained. The percentage of cells containing a particular gene of interest was calculated as (gene homologs × 100)/recA. Their RPKM (reads per kilobase per million mapped reads) values were confirmed using BWA-MEM (bwa version 0.7.17-r1188, using default settings) and samtools version 1.10 (84). Reads with coverages of <80% were filtered and discarded using coverm filter version 0.4.0 (Ben Woodcroft, CMR, QUT, UK).
Heterologous expression and activity of potential DMSP metabolic enzymes.
To further validate the activity of the environmental sequences retrieved from these marine metagenomes predicted in MAGs, four DsyB sequences, two DddP sequences, and one DddX sequence (Table S7) were chosen for further experiments. These candidate sequences were synthesized by Sangon Biotech (Shanghai, China), cloned, and overexpressed using the pET24a plasmid in E. coli BL21(DE3) to test their activity. The recombinant E. coli strains were incubated in LB medium at 37°C for 3 h, and 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added before culturing for 20 h at 16°C. For DMSP catabolic proteins, 200 μL of an E. coli culture with 0.1 mM DMSP was mixed in a sealing vial, and the reaction mixture was incubated at 37°C at 170 rpm. After 24 h, DMS was measured to calculate the enzyme activities monitored by GC (see the section on DMSP concentration measurement, above). In addition, 200 μL of the culture was centrifuged to collect the cells, and the cell pellet was then resuspended in 1 mL Tris-HCl buffer (50 mM; pH 7) and sonicated (3 times for 10 s each) using a Scientz JY92-IIN ultrasonic homogenizer. The DMSP catabolic activity was normalized to the cellular protein content estimated by the Bradford method (Bio-Rad). E. coli BL21 containing the original pET24a plasmid was used as a negative control. For DMSP biosynthetic proteins, a 20-μL E. coli culture was mixed with 180 μL of LB medium containing 0.05 mM MTHB and 0.2 mM IPTG in a sealing vial at 37°C at 170 rpm for 12 h. Subsequently, 100 μL of 10 M NaOH was added to each sealing vial, and the reaction mixture was incubated at 28°C at 170 rpm for 2 h. As described above, the DMS concentration was then measured to calculate the enzyme activities monitored by a GC assay (see the section on DMSP concentration measurement, above). The DMSP biosynthetic activity was normalized to the cellular protein content, as mentioned above. All experiments described here were performed using three biological replicates.
Bacterial isolation and identification of DMSP-producing and -degrading isolates.
Culturable DMSP-producing and -degrading bacteria were isolated from the enrichment culture of a sample from 0 to 1 cmbsf. For the enrichment experiment, minimal marine basal medium (MBM) (salinity of 35 PSU [practical salinity units]) (85) supplemented with a mixed carbon source and either 0.5 mM l-Met (for DMSP-producing bacteria) or 0.5 mM DMSP (for DMSP-degrading bacteria) was used. The mixed carbon source contained succinate, glucose, pyruvate, sucrose, and glycerol (with final concentrations of 2 mM each). The enrichment process was carried out at 28°C for over 2 months, with supplementation with fresh medium each week. The resulting enriched cultures were serially diluted and spread onto marine agar (MA) (1 g of yeast extract, 5 g of peptone, and 0.01 g of ferric phosphate [pH 7.6] per L of seawater) plates and incubated at 28°C for 5 to 7 days. A single colony was picked and subsequently purified three times by streaking. The 16S rRNA genes of isolates were amplified using the primer set B8F (5′-AGAGTTTGATCCTGGCTCAG-3′) and B1510R (5′-GGTTACCTTGTTACGACTT-3′) (86) and sequenced. Taxonomy information identifying these cultivated strains was obtained from the Ezbiocloud server (http://www.ezbiocloud.net/identify).
All of these obtained isolates were tested for their DMSP production or catabolism capacity. For DMSP biosynthesis, bacterial isolates were cultivated in MBM supplemented with 0.5 mM l-Met at 28°C for 24 h. Two hundred microliters of the culture and 100 μL of NaOH (10 M) were mixed in 2-mL vials. The vials were crimped immediately and incubated at 170 rpm for 2 h in the dark, and headspace DMS was monitored by GC as described above. For DMSP catabolism, bacterial isolates were cultivated in MBM supplemented with 0.5 mM DMSP at 28°C for 24 h. The resulting DMS and MeSH were monitored directly by GC. To detect the ability to produce DMS directly from l-Met or MeSH, bacterial isolates were cultivated in MBM supplemented with 0.5 mM l-Met or MeSH at 28°C for 24 h in sealed vials, and the resulting DMS was monitored by GC. All experiments were conducted with three biological replicates. Degenerate primers of several known DMSP biosynthetic and catabolic genes (Table S7) were used to detect these genes in the obtained DMSP metabolic isolates by PCR. The PCR system and amplification conditions were described previously by Williams et al. (25).
Data availability.
The metagenome sequences and MAGs from the current study have been submitted to the NCBI under BioProject accession numbers PRJNA957232 and PRJNA957236, respectively.
ACKNOWLEDGMENTS
We acknowledge all of the scientists and crew members on the R/V Dong Fang Hong 3 for their assistance with sampling during the cruise. We also thank Ruihong Guo, Jinyan Wang, and Jian Jin for their help in the experiments.
This work was funded by the National Natural Science Foundation of China (92251303), and the Project of Laoshan Laboratory (LSKJ202203201 and LSKJ202203206).
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Xiao-Hua Zhang, Email: xhzhang@ouc.edu.cn.
Knut Rudi, Norwegian University of Life Sciences.
REFERENCES
- 1.Gali M, Devred E, Levasseur M, Royer SJ, Babin M. 2015. A remote sensing algorithm for planktonic dimethylsulfoniopropionate (DMSP) and an analysis of global patterns. Remote Sens Environ 171:171–184. doi: 10.1016/j.rse.2015.10.012. [DOI] [Google Scholar]
- 2.Ksionzek KB, Lechtenfeld OJ, McCallister SL, Schmitt-Kopplin P, Geuer JK, Geibert W, Koch BP. 2016. Dissolved organic sulfur in the ocean: biogeochemistry of a petagram inventory. Science 354:456–459. doi: 10.1126/science.aaf7796. [DOI] [PubMed] [Google Scholar]
- 3.Seymour JR, Simo R, Ahmed T, Stocker R. 2010. Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329:342–345. doi: 10.1126/science.1188418. [DOI] [PubMed] [Google Scholar]
- 4.Damm E, Nomura D, Martin A, Dieckmann GS, Meiners KM. 2016. DMSP and DMS cycling within Antarctic Sea ice during the winter-spring transition. Deep Sea Res 2 Top Stud Oceanogr 131:150–159. doi: 10.1016/j.dsr2.2015.12.015. [DOI] [Google Scholar]
- 5.Seyedsayamdost MR, Case RJ, Kolter R, Clardy J. 2011. The Jekyll-and-Hyde chemistry of Phaeobacter gallaeciensis. Nat Chem 3:331–335. doi: 10.1038/nchem.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raina JB, Tapiolas DM, Foret S, Lutz A, Abrego D, Ceh J, Seneca FO, Clode PL, Bourne DG, Willis BL, Motti CA. 2013. DMSP biosynthesis by an animal and its role in coral thermal stress response. Nature 502:677–680. doi: 10.1038/nature12677. [DOI] [PubMed] [Google Scholar]
- 7.Sunda W, Kieber DJ, Kiene RP, Huntsman S. 2002. An antioxidant function for DMSP and DMS in marine algae. Nature 418:317–320. doi: 10.1038/nature00851. [DOI] [PubMed] [Google Scholar]
- 8.Thume K, Gebser B, Chen L, Meyer N, Kieber DJ, Pohnert G. 2018. The metabolite dimethylsulfoxonium propionate extends the marine organosulfur cycle. Nature 563:412–415. doi: 10.1038/s41586-018-0675-0. [DOI] [PubMed] [Google Scholar]
- 9.Zheng Y, Wang J, Zhou S, Zhang Y, Liu J, Xue C-X, Williams BT, Zhao X, Zhao L, Zhu X-Y, Sun C, Zhang H-H, Xiao T, Yang G-P, Todd JD, Zhang X-H. 2020. Bacteria are important dimethylsulfoniopropionate producers in marine aphotic and high-pressure environments. Nat Commun 11:4658. doi: 10.1038/s41467-020-18434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curson ARJ, Todd JD, Sullivan MJ, Johnston AWB. 2011. Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Microbiol 9:849–859. doi: 10.1038/nrmicro2653. [DOI] [PubMed] [Google Scholar]
- 11.Todd JD, Rogers R, Li YG, Wexler M, Bond PL, Sun L, Curson ARJ, Malin G, Steinke M, Johnston AWB. 2007. Structural and regulatory genes required to make the gas dimethyl sulfide in bacteria. Science 315:666–669. doi: 10.1126/science.1135370. [DOI] [PubMed] [Google Scholar]
- 12.Vallina SM, Simo R. 2007. Strong relationship between DMS and the solar radiation dose over the global surface ocean. Science 315:506–508. doi: 10.1126/science.1133680. [DOI] [PubMed] [Google Scholar]
- 13.Sievert SM, Kiene RP, Schulz-Vogt HN. 2007. The sulfur cycle. Oceanography 20:117–123. doi: 10.5670/oceanog.2007.55. [DOI] [Google Scholar]
- 14.Carrion O, Pratscher J, Curson ARJ, Williams BT, Rostant WG, Murrell JC, Todd JD. 2017. Methanethiol-dependent dimethylsulfide production in soil environments. ISME J 11:2379–2390. doi: 10.1038/ismej.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curson ARJ, Liu J, Bermejo Martinez A, Green RT, Chan Y, Carrion O, Williams BT, Zhang S-H, Yang G-P, Bulman Page PC, Zhang X-H, Todd JD. 2017. Dimethylsulfoniopropionate biosynthesis in marine bacteria and identification of the key gene in this process. Nat Microbiol 2:17009. doi: 10.1038/nmicrobiol.2017.9. [DOI] [PubMed] [Google Scholar]
- 16.Curson ARJ, Williams BT, Pinchbeck BJ, Sims LP, Martinez AB, Rivera PPL, Kumaresan D, Mercade E, Spurgin LG, Carrion O, Moxon S, Cattolico RA, Kuzhiumparambil U, Guagliardo P, Clode PL, Raina J-B, Todd JD. 2018. DSYB catalyses the key step of dimethylsulfoniopropionate biosynthesis in many phytoplankton. Nat Microbiol 3:430–439. doi: 10.1038/s41564-018-0119-5. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X-H, Liu J, Liu J, Yang G, Xue C-X, Curson ARJ, Todd JD. 2019. Biogenic production of DMSP and its degradation to DMS—their roles in the global sulfur cycle. Sci China Life Sci 62:1296–1319. doi: 10.1007/s11427-018-9524-y. [DOI] [PubMed] [Google Scholar]
- 18.Gage DA, Rhodes D, Nolte KD, Hicks WA, Leustek T, Cooper AJ, Hanson AD. 1997. A new route for synthesis of dimethylsulphoniopropionate in marine algae. Nature 387:891–894. doi: 10.1038/43160. [DOI] [PubMed] [Google Scholar]
- 19.Summers PS, Nolte KD, Cooper AJL, Borgeas H, Leustek T, Rhodes D, Hanson AD. 1998. Identification and stereospecificity of the first three enzymes of 3-dimethylsulfoniopropionate biosynthesis in a chlorophyte alga. Plant Physiol 116:369–378. doi: 10.1104/pp.116.1.369. [DOI] [Google Scholar]
- 20.James F, Paquet L, Sparace SA, Gage DA, Hanson AD. 1995. Evidence implicating dimethylsulfoniopropionaldehyde as an intermediate in dimethylsulfoniopropionate biosynthesis. Plant Physiol 108:1439–1448. doi: 10.1104/pp.108.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kocsis MG, Nolte KD, Rhodes D, Shen T-L, Gage DA, Hanson AD. 1998. Dimethylsulfoniopropionate biosynthesis in Spartina alterniflora: evidence that S-methylmethionine and dimethylsulfoniopropylamine are intermediates. Plant Physiol 117:273–281. doi: 10.1104/pp.117.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kocsis MG, Hanson AD. 2000. Biochemical evidence for two novel enzymes in the biosynthesis of 3-dimethylsulfoniopropionate in Spartina alterniflora. Plant Physiol 123:1153–1161. doi: 10.1104/pp.123.3.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otte ML, Wilson G, Morris JT, Moran BM. 2004. Dimethylsulphoniopropionate (DMSP) and related compounds in higher plants. J Exp Bot 55:1919–1925. doi: 10.1093/jxb/erh178. [DOI] [PubMed] [Google Scholar]
- 24.Lyon BR, Lee PA, Bennett JM, DiTullio GR, Janech MG. 2011. Proteomic analysis of a sea-ice diatom: salinity acclimation provides new insight into the dimethylsulfoniopropionate production pathway. Plant Physiol 157:1926–1941. doi: 10.1104/pp.111.185025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams BT, Cowles K, Bermejo Martinez A, Curson ARJ, Zheng Y, Liu J, Newton-Payne S, Hind AJ, Li C-Y, Rivera PPL, Carrion O, Liu J, Spurgin LG, Brearley CA, Mackenzie BW, Pinchbeck BJ, Peng M, Pratscher J, Zhang X-H, Zhang Y-Z, Murrell JC, Todd JD. 2019. Bacteria are important dimethylsulfoniopropionate producers in coastal sediments. Nat Microbiol 4:1815–1825. doi: 10.1038/s41564-019-0527-1. [DOI] [PubMed] [Google Scholar]
- 26.Hanson AD, Rivoal J, Paquet L, Gage DA. 1994. Biosynthesis of 3-dimethylsulfoniopropionate in Wollastonia biflora (L.) DC. (evidence that S-methylmethionine is an intermediate). Plant Physiol 105:103–110. doi: 10.1104/pp.105.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kiene RP. 1996. Biological and environmental chemistry of DMSP and related sulfonium compounds. Plenum Press, New York, NY. [Google Scholar]
- 28.Kageyama H, Tanaka Y, Shibata A, Waditee-Sirisattha R, Takabe T. 2018. Dimethylsulfoniopropionate biosynthesis in a diatom Thalassiosira pseudonana: identification of a gene encoding MTHB-methyltransferase. Arch Biochem Biophys 645:100–106. doi: 10.1016/j.abb.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Liao C, Seebeck FP. 2019. In vitro reconstitution of bacterial DMSP biosynthesis. Angew Chem Int Ed Engl 58:3553–3556. doi: 10.1002/anie.201814662. [DOI] [PubMed] [Google Scholar]
- 30.Uchida A, Ooguri T, Ishida T, Kitaguchi H, Ishida Y. 1996. Biosynthesis of dimethylsulfoniopropionate in Crypthecodinium cohnii (Dinophyceae), p 97–107. In Kiene RP, Visscher PT, Keller MD, Kirst GO (ed), Biological and environmental chemistry of DMSP and related sulfonium compounds. Springer, Boston, MA. [Google Scholar]
- 31.Todd JD, Curson ARJ, Dupont CL, Nicholson P, Johnston AWB. 2009. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some ascomycete fungi. Environ Microbiol 11:1376–1385. doi: 10.1111/j.1462-2920.2009.01864.x. [DOI] [PubMed] [Google Scholar]
- 32.Todd JD, Curson AR, Kirkwood M, Sullivan MJ, Green RT, Johnston AW. 2011. DddQ, a novel, cupin-containing, dimethylsulfoniopropionate lyase in marine roseobacters and in uncultured marine bacteria. Environ Microbiol 13:427–438. doi: 10.1111/j.1462-2920.2010.02348.x. [DOI] [PubMed] [Google Scholar]
- 33.Curson ARJ, Sullivan MJ, Todd JD, Johnston AWB. 2011. DddY, a periplasmic dimethylsulfoniopropionate lyase found in taxonomically diverse species of Proteobacteria. ISME J 5:1191–1200. doi: 10.1038/ismej.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C-Y, Wang X-J, Chen X-L, Sheng Q, Zhang S, Wang P, Quareshy M, Rihtman B, Shao X, Gao C, Li F, Li S, Zhang W, Zhang X-H, Yang G-P, Todd JD, Chen Y, Zhang Y-Z. 2021. A novel ATP dependent dimethylsulfoniopropionate lyase in bacteria that releases dimethyl sulfide and acryloyl-CoA. Elife 10:e64045. doi: 10.7554/eLife.64045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alcolombri U, Ben-Dor S, Feldmesser E, Levin Y, Tawfik DS, Vardi A. 2015. Identification of the algal dimethyl sulfide-releasing enzyme: a missing link in the marine sulfur cycle. Science 348:1466–1469. doi: 10.1126/science.aab1586. [DOI] [PubMed] [Google Scholar]
- 36.Schnicker NJ, De Silva SM, Todd JD, Dey M. 2017. Structural and biochemical insights into dimethylsulfoniopropionate cleavage by cofactor-bound DddK from the prolific marine bacterium Pelagibacter. Biochemistry 56:2873–2885. doi: 10.1021/acs.biochem.7b00099. [DOI] [PubMed] [Google Scholar]
- 37.Song D, Zhang Y, Liu J, Zhong H, Zheng Y, Zhou S, Yu M, Todd JD, Zhang X-H. 2020. Metagenomic insights into the cycling of dimethylsulfoniopropionate and related molecules in the Eastern China Marginal Seas. Front Microbiol 11:157. doi: 10.3389/fmicb.2020.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HO. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 39.Reisch CR, Stoudemayer MJ, Varaljay VA, Amster IJ, Moran MA, Whitman WB. 2011. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 473:208–211. doi: 10.1038/nature10078. [DOI] [PubMed] [Google Scholar]
- 40.Varaljay VA, Howard EC, Sun S, Moran MA. 2010. Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl Environ Microbiol 76:609–617. doi: 10.1128/AEM.01258-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui Y, Suzuki S, Omori Y, Wong S-K, Ijichi M, Kaneko R, Kameyama S, Tanimoto H, Hamasaki K. 2015. Abundance and distribution of dimethylsulfoniopropionate degradation genes and the corresponding bacterial community structure at dimethyl sulfide hot spots in the tropical and subtropical Pacific Ocean. Appl Environ Microbiol 81:4184–4194. doi: 10.1128/AEM.03873-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Howard EC, Sun S, Reisch CR, del Valle DA, Burgmann H, Kiene RP, Moran MA. 2011. Changes in dimethylsulfoniopropionate demethylase gene assemblages in response to an induced phytoplankton bloom. Appl Environ Microbiol 77:524–531. doi: 10.1128/AEM.01457-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrion O, Pratscher J, Richa K, Rostant WG, Farhan Ul Haque M, Murrell JC, Todd JD. 2019. Methanethiol and dimethylsulfide cycling in Stiffkey saltmarsh. Front Microbiol 10:1040. doi: 10.3389/fmicb.2019.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bray RC, Adams B, Smith AT, Richards RL, Lowe DJ, Bailey S. 2001. Reactions of dimethylsulfoxide reductase in the presence of dimethyl sulfide and the structure of the dimethyl sulfide-modified enzyme. Biochemistry 40:9810–9820. doi: 10.1021/bi010559r. [DOI] [PubMed] [Google Scholar]
- 45.Lidbury I, Krober E, Zhang Z, Zhu Y, Murrell JC, Chen Y, Schafer H. 2016. A mechanism for bacterial transformation of dimethylsulfide to dimethylsulfoxide: a missing link in the marine organic sulfur cycle. Environ Microbiol 18:2754–2766. doi: 10.1111/1462-2920.13354. [DOI] [PubMed] [Google Scholar]
- 46.Horinouchi M, Yoshida T, Nojiri H, Yamane H, Omori T. 1999. Polypeptide requirement of multicomponent monooxygenase DsoABCDEF for dimethyl sulfide oxidizing activity. Biosci Biotechnol Biochem 63:1765–1771. doi: 10.1271/bbb.63.1765. [DOI] [PubMed] [Google Scholar]
- 47.McDevitt CA, Hanson GR, Noble CJ, Cheesman MR, McEwan AG. 2002. Characterization of the redox centers in dimethyl sulfide dehydrogenase from Rhodovulum sulfidophilum. Biochemistry 41:15234–15244. doi: 10.1021/bi026221u. [DOI] [PubMed] [Google Scholar]
- 48.Chen Y, Patel NA, Crombie A, Scrivens JH, Murrell JC. 2011. Bacterial flavin-containing monooxygenase is trimethylamine monooxygenase. Proc Natl Acad Sci USA 108:17791–17796. doi: 10.1073/pnas.1112928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boden R, Murrell JC, Schafer H. 2011. Dimethylsulfide is an energy source for the heterotrophic marine bacterium Sagittula stellata. FEMS Microbiol Lett 322:188–193. doi: 10.1111/j.1574-6968.2011.02349.x. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka H, Esaki N, Yamamoto T, Soda K. 1976. Purification and properties of methioninase from Pseudomonas ovalis. FEBS Lett 66:307–311. doi: 10.1016/0014-5793(76)80528-5. [DOI] [PubMed] [Google Scholar]
- 51.Zhuang G-C, Lin Y-S, Bowles MW, Heuer VB, Lever MA, Elvert M, Hinrichs K-U. 2017. Distribution and isotopic composition of trimethylamine, dimethylsulfide and dimethylsulfoniopropionate in marine sediments. Mar Chem 196:35–46. doi: 10.1016/j.marchem.2017.07.007. [DOI] [Google Scholar]
- 52.Zhang Y, Sun K, Sun C, Shi X, Todd JD, Zhang X-H. 2021. Dimethylsulfoniopropionate biosynthetic bacteria in the subseafloor sediments of the South China Sea. Front Microbiol 12:731524. doi: 10.3389/fmicb.2021.731524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang C, Xie Q, Wang D, Shu Y, Xu H, Xiao J, Zu T, Long T, Zhang T. 2018. Seasonal variability of water characteristics in the Challenger Deep observed by four cruises. Sci Rep 8:11791. doi: 10.1038/s41598-018-30176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manea E, Dell’Anno A, Rastelli E, Tangherlini M, Nunoura T, Nomaki H, Danovaro R, Corinaldesi C. 2019. Viral infections boost prokaryotic biomass production and organic C cycling in hadal trench sediments. Front Microbiol 10:1952. doi: 10.3389/fmicb.2019.01952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Itoh T, Yamanoi K, Kudo T, Ohkuma M, Takashina T. 2011. Aciditerrimonas ferrireducens gen. nov., sp. nov., an iron-reducing thermoacidophilic actinobacterium isolated from a solfataric field. Int J Syst Evol Microbiol 61:1281–1285. doi: 10.1099/ijs.0.023044-0. [DOI] [PubMed] [Google Scholar]
- 56.Fukunaga Y, Kurahashi M, Sakiyama Y, Ohuchi M, Yokota A, Harayama S. 2009. Phycisphaera mikurensis gen. nov., sp. nov., isolated from a marine alga, and proposal of Phycisphaeraceae fam. nov., Phycisphaerales ord. nov. and Phycisphaerae classis nov. in the phylum Planctomycetes. J Gen Appl Microbiol 55:267–275. doi: 10.2323/jgam.55.267. [DOI] [PubMed] [Google Scholar]
- 57.Lage OM, van Niftrik L, Jogler C, Devos DP. 2019. Planctomycetes, p 614–626. In Schmidt TM (ed), Encyclopedia of microbiology, 4th ed. Academic Press, Oxford, United Kingdom. doi: 10.1016/B978-0-12-809633-8.90689-7. [DOI] [Google Scholar]
- 58.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 59.Li C-Y, Crack JC, Newton-Payne S, Murphy ARJ, Chen X-L, Pinchbeck BJ, Zhou S, Williams BT, Peng M, Zhang X-H, Chen Y, Le Brun NE, Todd JD, Zhang Y-Z. 2022. Mechanistic insights into the key marine dimethylsulfoniopropionate synthesis enzyme DsyB/DSYB. Mlife 1:114–130. doi: 10.1002/mlf2.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu J, Zhang Y, Liu J, Zhong H, Williams BT, Zheng Y, Curson ARJ, Sun C, Sun H, Song D, Mackenzie BW, Bermejo Martinez A, Todd JD, Zhang X-H. 2021. Bacterial dimethylsulfoniopropionate biosynthesis in the East China Sea. Microorganisms 9:657. doi: 10.3390/microorganisms9030657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilkening JV, Turchyn AV, Redeker KR, Mills JV, Antler G, Carrión O, Todd JD. 2019. The production and fate of volatile organosulfur compounds in sulfidic and ferruginous sediment. J Geophys Res Biogeosci 124:3390–3402. doi: 10.1029/2019JG005248. [DOI] [Google Scholar]
- 62.Ichino MC, Clark MR, Drazen JC, Jamieson A, Jones DOB, Martin AP, Rowden AA, Shank TM, Yancey PH, Ruhl HA. 2015. The distribution of benthic biomass in hadal trenches: a modelling approach to investigate the effect of vertical and lateral organic matter transport to the seafloor. Deep Sea Res 1 Oceanogr Res Pap 100:21–33. doi: 10.1016/j.dsr.2015.01.010. [DOI] [Google Scholar]
- 63.Sun H, Liu J, Tan S, Zheng Y, Wang X, Liang J, Todd JD, Zhang X-H. 2021. Spatiotemporal distribution of bacterial dimethylsulfoniopropionate producing and catabolic genes in the Changjiang Estuary. Environ Microbiol 23:7073–7092. doi: 10.1111/1462-2920.15813. [DOI] [PubMed] [Google Scholar]
- 64.Sun H, Zhang Y, Tan S, Zheng Y, Zhou S, Ma Q-Y, Yang G-P, Todd JD, Zhang X-H. 2020. DMSP-producing bacteria are more abundant in the surface microlayer than subsurface seawater of the East China Sea. Microb Ecol 80:350–365. doi: 10.1007/s00248-020-01507-8. [DOI] [PubMed] [Google Scholar]
- 65.Varaljay VA, Gifford SM, Wilson ST, Sharma S, Karl DM, Moran MA. 2012. Bacterial dimethylsulfoniopropionate degradation genes in the oligotrophic North Pacific subtropical gyre. Appl Environ Microbiol 78:2775–2782. doi: 10.1128/AEM.07559-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu J, Liu J, Zhang S-H, Liang J, Lin H, Song D, Yang G-P, Todd JD, Zhang X-H. 2018. Novel insights into bacterial dimethylsulfoniopropionate catabolism in the East China Sea. Front Microbiol 9:3206. doi: 10.3389/fmicb.2018.03206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou J, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Levine NM, Varaljay VA, Toole DA, Dacey JW, Doney SC, Moran MA. 2012. Environmental, biochemical and genetic drivers of DMSP degradation and DMS production in the Sargasso Sea. Environ Microbiol 14:1210–1223. doi: 10.1111/j.1462-2920.2012.02700.x. [DOI] [PubMed] [Google Scholar]
- 69.Raina JB, Tapiolas D, Willis BL, Bourne DG. 2009. Coral-associated bacteria and their role in the biogeochemical cycling of sulfur. Appl Environ Microbiol 75:3492–3501. doi: 10.1128/AEM.02567-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Uritskiy GV, DiRuggiero J, Taylor J. 2018. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 6:158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 73.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 74.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 76.Kudo T, Kobiyama A, Rashid J, Reza MS, Yamada Y, Ikeda Y, Ikeda D, Mizusawa N, Ikeo K, Sato S, Ogata T, Jimbo M, Kaga S, Watanabe S, Naiki K, Kaga Y, Segawa S, Mineta K, Bajic V, Gojobori T, Watabe S. 2018. Seasonal changes in the abundance of bacterial genes related to dimethylsulfoniopropionate catabolism in seawater from Ofunato Bay revealed by metagenomic analysis. Gene 665:174–184. doi: 10.1016/j.gene.2018.04.072. [DOI] [PubMed] [Google Scholar]
- 77.Teng Z-J, Qin Q-L, Zhang W, Li J, Fu H-H, Wang P, Lan M, Luo G, He J, McMinn A, Wang M, Chen X-L, Zhang Y-Z, Chen Y, Li C-Y. 2021. Biogeographic traits of dimethyl sulfide and dimethylsulfoniopropionate cycling in polar oceans. Microbiome 9:207. doi: 10.1186/s40168-021-01153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carrion O, Curson ARJ, Kumaresan D, Fu Y, Lang AS, Mercade E, Todd JD. 2015. A novel pathway producing dimethylsulphide in bacteria is widespread in soil environments. Nat Commun 6:6579. doi: 10.1038/ncomms7579. [DOI] [PubMed] [Google Scholar]
- 79.Eyice O, Myronova N, Pol A, Carrion O, Todd JD, Smith TJ, Gurman SJ, Cuthbertson A, Mazard S, Mennink-Kersten MA, Bugg TDH, Andersson KK, Johnston AWB, Op den Camp HJM, Schafer H. 2018. Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME J 12:145–160. doi: 10.1038/ismej.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cao H-Y, Wang P, Xu F, Li P-Y, Xie B-B, Qin Q-L, Zhang Y-Z, Li C-Y, Chen X-L. 2017. Molecular insight into the acryloyl-CoA hydration by AcuH for acrylate detoxification in dimethylsulfoniopropionate-catabolizing bacteria. Front Microbiol 8:2034. doi: 10.3389/fmicb.2017.02034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Starr MP. 1981. The prokaryotes: a handbook on habitats, isolation, and identification of bacteria. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 86.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.00251-23-s0001.docx, DOCX file, 2.9 MB (2.9MB, docx)
Supplemental material. Download aem.00251-23-s0002.xlsx, XLSX file, 0.04 MB (37.6KB, xlsx)
Data Availability Statement
The metagenome sequences and MAGs from the current study have been submitted to the NCBI under BioProject accession numbers PRJNA957232 and PRJNA957236, respectively.