Abstract
Activity-regulated cytoskeletal-associated protein (Arc, also known as activity-regulated gene 3.1 or Arg3.1) is induced in neurons in response to salient experience and neural activity and is necessary for activityinduced forms of synaptic plasticity, such as long-term potentiation (LTP) and long-term depression (LTD), cellular substrates of learning and memory. The bestcharacterized function of Arc is enhancement of the endocytic internalization of AMPA receptors in dendritic spines, a process associated with LTD. Arc has also been implicated in the proteolytic processing of amyloid precursor protein on the surface of endosomes. To mediate these activities, Arc must associate with cellular membranes, but it is unclear whether Arc binds directly to the lipid bilayer or requires protein–protein interactions for membrane recruitment. In this study, we show that Arc associates with pure phospholipid vesicles in vitro and undergoes palmitoylation in neurons, a modification that allows it to insert directly into the hydrophobic core of the bilayer. The palmitoylated cysteines are clustered in a motif, 94CLCRC98, located in the N-terminal half of the protein, which has not yet been structurally characterized. Expression of Arc with three mutated cysteines in that motif cannot support synaptic depression induced by the activity-dependent transcription factor, MEF2 (myocyte enhancer factor 2), in contrast to wild-type Arc. Thus, it appears that palmitoylation regulates at least a subset of Arc functions in synaptic plasticity.
Activity-regulated cytoskeleton-associated protein (Arc), also known as the product of the activity-regulated gene of 3.1 kb (Arg3.1), is rapidly induced in response to experience and neuronal activity and plays critical, though still poorly defined, roles in multiple forms of synaptic plasticity, learning, and memory (reviewed in refs 1 and 2). For example, Arc is essential for long-term depression (LTD) and synapse elimination, functioning, at least in part, by promoting the internalization of AMPA-type glutamate receptors (AMPARs).3-10 Arc may facilitate AMPAR endocytosis by directly interacting with two membrane remodeling proteins, endophilin and dynamin,3,11 and with clathrin adaptor protein, AP-2.12 Although Arc has been localized to membranes of the endocytic pathway, it is uncertain whether it binds to membranes directly or via interactions with membraneassociated proteins. Here we show that Arc can associate directly with membranes, without the intervention of endocytic or cytoskeletal proteins, by direct interaction with phospholipids. We further show that Arc can be palmitoylated on cysteines clustered in a motif, 94CLCRC98. Lastly, we demonstrate that mutation of these cysteines abolishes an Arc-dependent form of synaptic depression, pointing to a functional role for Arc palmitoylation in neurons.
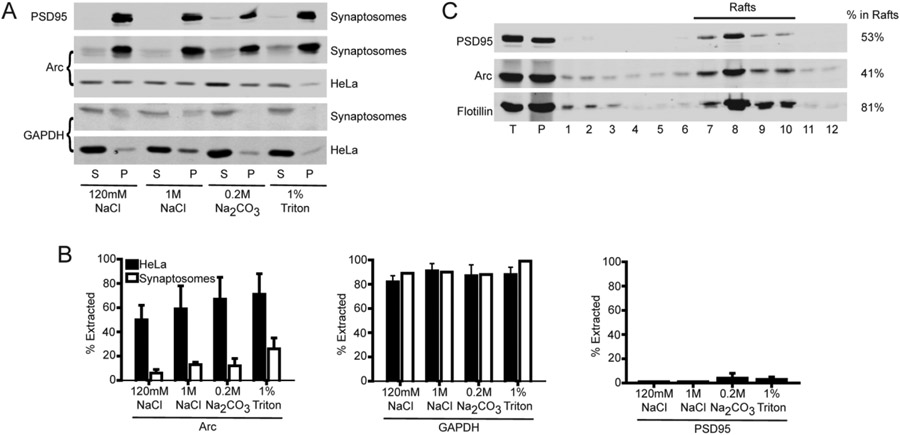
In their original report of the identification and partial characterization of Arc, Lyford et al.13 noted that the majority of Arc could not be solubilized from brain by 2 M KCl or 1% Triton X-100. We confirmed that only ~5–10% of Arc was extracted from rat brain synaptosomes using buffers that release peripheral membrane proteins [120 mM NaCl, 1 M NaCl, or 0.2% sodium carbonate (pH 10)] and only ~20% was extracted with 1% Triton X-100, which solubilizes integral membrane proteins (Figure 1A). In contrast, approximately half of myc-tagged Arc was solubilized from transfected HeLa cells by 120 mM NaCl, and ≤80% was solubilized by sequential treatment with 1 M NaCl, 0.1 M sodium carbonate (pH 10), and 1% Triton X-100 (Figure 1B). Thus, Arc apparently incorporates into insoluble complexes that are present in neurons but not in heterologous expression systems.14
Figure 1.
Association of Arc with lipid rafts. (A) Solubility of Arc in HeLa cells and synaptosomes. Synaptosomes or PNS from myc-Arc-expressing HeLa cells were processed as described in Experimental Procedures in the Supporting Information. Arc, GAPDH (cytosolic marker), and PSD95 (PSD marker) in extracts were detected by immunoblotting. Representative blots of the supernatant (S) and pellet (P). (B) Quantification of extracted (soluble) proteins. (C) Triton-insoluble pellets from purified synaptosomes fractionated on a discontinuous sucrose gradient. Fractions were collected from the bottom of the centrifuge tubes (i.e., decreasing density with increasing fraction number). T (total) represents 2% of the sample loaded on the gradient (12 μL of 600 μL); 5% of each gradient fraction (20 μL of 400 μL) was loaded on the gel. Quantification (percent in rafts) was performed by immunoblotting with indicated antibodies as described in Experimental Procedures in the Supporting Information. Quantification of three experiments gave 50, 53.6, and 54% of Arc and 36, 47.8, and 40% of PSD95 fractionating with rafts. Flotillin-1 was used as a raft marker (81%).
Neurons contain two overlapping detergent-insoluble structures: postsynaptic densities (PSDs) and membrane rafts.14 Rafts are cholesterol- and sphingolipid-rich membrane microdomains that serve as platforms for signaling complexes and endocytosis.15 Following density gradient centrifugation of brain or synaptosome homogenates, PSD- and raft-associated proteins are obtained in the pellets and low-buoyant density fractions, respectively. In prior studies, Arc was recovered from both PSD and raft fractions.16,17 We confirmed that ~50% of synaptosomal Arc that is not extracted by cold Triton X-100 associates with membrane rafts (Figure 1C). Similar results were obtained upon fractionation of homogenates from crude synaptosomes (Figure S1).
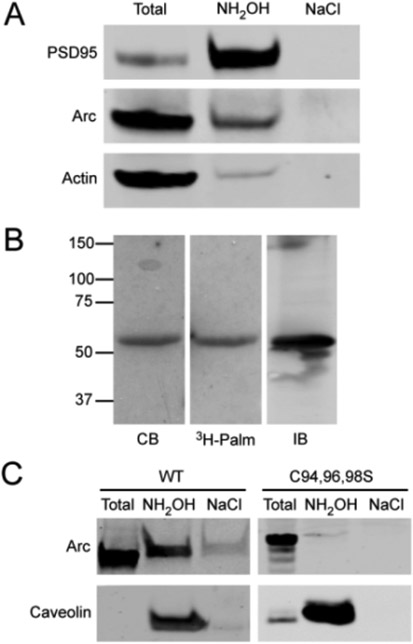
Membrane rafts are enriched in palmitoylated proteins. Palmitoylation is a reversible form of lipidation (typically on cysteines) that anchors peripheral proteins into membranes and regulates protein stability, interactions, and intracellular trafficking.18 Numerous postsynaptic proteins are tethered to the bilayer via palmitoylation,19 including several that have been implicated in the regulation of AMPA receptor trafficking (e.g., PSD95, GRIP1, and PICK1). Therefore, we tested whether Arc can undergo palmitoylation in cells. Using two experimental approaches, [3H]palmitate labeling and affinity capture on thiol-binding resin [known as Acyl-RAC (see Experimental Procedures in the Supporting Information)], we detected palmitoylation of endogenous Arc in mouse brain synaptosomes (Figure 2A) and whole mouse brain homogenates (Figure S2) and of recombinant Arc in HeLa cells (Figure 2B,C). Using these methods, we cannot quantitatively access the percentage of Arc that is palmitoylated in these systems. However, if we assume the identical efficiency of resin binding in the Acyl-RAC procedure, we estimate that the extent of Arc palmitoylation was between 5.0 and 8.0% of that of PSD95. CSS-Palm version 4.0 (http://csspalm.biocuckoo.org)20 identified the 94CLCRC98 motif as the likeliest palmitoylation site in Arc. Indeed, mutation of the three cysteines in this motif prevented Arc palmitoylation in HeLa cells (Figure 2C). Although a relatively small percentage of Arc was palmitoylated in mouse brain synaptosomes, the specific palmitoyl-Arc pool may play important roles in the neuron. For example, <1% of PICK1 is palmitoylated in neurons, yet palmitoylated PICK1 is required for induction of long-term depression in the cerebellum.21
Figure 2.
Palmitoylation of Arc. (A) Palmitoylation of Arc in synaptosomes detected by Acyl-RAC (see Experimental Procedures in the Supporting Information). PSD95 and actin are shown as positive and negative controls. (B) Incorporation of [3H]palmitate into myc-Arc expressed in HeLa cells: lane 1, Coomassie blue (CB)-stained gel of the anti-myc immunoprecipitate; lane 2, corresponding autoradiogram; lane 3, immunoblot of the anti-Arc immunoprecipitate. (C) Palmitoylation of myc-ArcWT or ArcC94,96,98S expressed in HeLa cells determined by Acyl-RAC. Caveolin 1 was the positive control. “Total” represents 1/15 of the input.
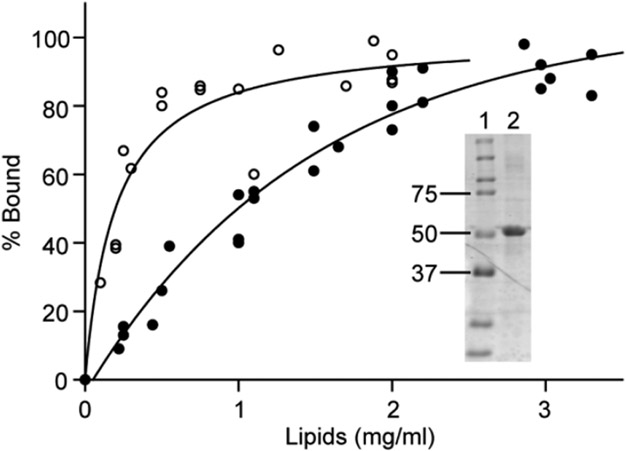
Protein acyltransferases (PATs), which catalyze protein palmitoylation, are integral membrane proteins. Therefore, palmitoylated proteins must associate with membranes, either by direct binding to lipids or through interactions with membrane proteins, prior to undergoing acylation. Using liposome co-sedimentation assays, we found that bacterially expressed (and, hence, nonpalmitoylated) Arc can interact directly with membrane lipids. As shown in Figure 3, ~50% of Arc co-sedimented with small unilamellar vesicles (SUVs) composed of 1 mg/mL phosphatidylcholine (PC). Arc bound with an approximately 4-fold higher affinity to vesicles, composed of a mixture of lipids found in brain membranes22 (Figure 3).
Figure 3.
Direct lipid binding by Arc. Purified Arc (4 μM) was incubated with liposomes made either from a brain lipid mixture22 (엯) or from PC (●) for 15 min at 25 °C in 20 mM Hepes (pH 7.4) and 100 mM NaCl. Each data point represents the result of a separate experiment (three experiments for brain lipid mixture and six experiments for PC). The inset shows the molecular weight standards (lane 1) and a CB-stained gel of representative Arc preparation used for binding analysis (lane 2).
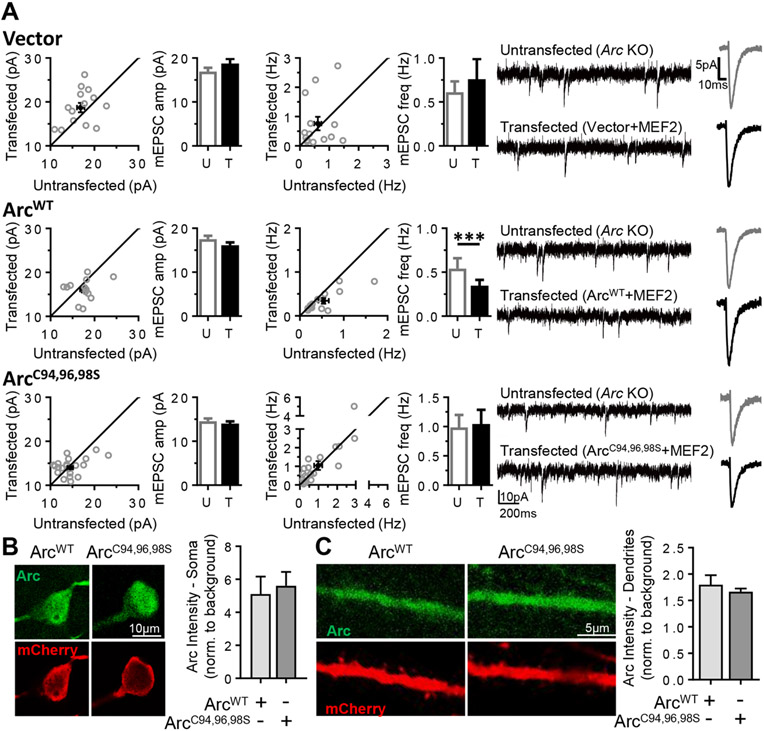
The Arc gene is induced in response to neural activity, which requires the activity-dependent transcription factor myocyte enhancer factor 2 (MEF2).23 We previously demonstrated that acute expression of constitutively active MEF2 (MEF2-VP16) induces Arc and depresses AMPAR-mediated synaptic transmission onto hippocampal CA1 neurons, an effect that requires Arc in the postsynaptic neuron.7 Here we tested the requirement for Arc palmitoylation in MEF2-induced depression of synaptic transmission measured as a decrease in the frequency of (m) excitatory postsynaptic currents (mEPSCs). Organotypic hippocampal slice cultures from Arc knockout (KO) mice were sparsely transfected biolistically with MEF2-VP16 and a fluorescent MEF2 transcriptional reporter (MRE-GFP) to identify transfected, MEF2-activated neurons.7 Synaptic transmission (mEPSCs) was measured simultaneously onto MEF2-transfected and neighboring untransfected CA1 neurons with whole cell voltage-clamp recordings. MEF2 transfection with an empty vector (pcDNA) of Arc KO neurons had no effect on mEPSC frequency or amplitude (ref 7, Figure 4A, and Figure S5 and Table S1). Cotransfection of Arc cDNA (ArcWT) and MEF2 into Arc KO neurons rescued MEF2-induced depression of mEPSC frequency but did not affect mEPSC amplitudes, as previously reported (ref 7, Figure 4A, and Figure S5 and Table S1). In contrast, co-transfection of Arc KO neurons with MEF2 and nonpalmitoylatable ArcC94,96,98S failed to rescue MEF2-induced depression of the mEPSC frequency (Figure 4C and Figure S5 and Table S1), suggesting that Arc palmitoylation is necessary for depression of synaptic transmission. Expression of ArcWT7 or ArcC94,96,98S alone, without MEF2, had no effect on mEPSC frequency (Table S1). The kinetics of mEPSCs were unaffected by any transfection condition (Table S1). The immunohistochemistry of Arc KO slice cultures co-transfected with either ArcWT or ArcC94,96,98S and mCherry (to fill dendrites) revealed that both forms of Arc were similarly expressed in the soma and dendrites (Figure 4B,C), indicating that the inability of ArcC94,96,98S to rescue MEF2-induced synaptic depression is likely not due to a deficit in expression.
Figure 4.
Arc palmitoylation is required for MEF2-dependent synapse depression. (A) Simultaneous dual-patch mEPSC recordings from untransfected (U) Arc KO neurons and neighboring Arc KO neurons transfected (T) with MEF2-VP16 and MRE-mCherry with a vector (top), the coding region of ArcWT (middle), or a nonpalmitoylatable Arc mutant (ArcC94,96,98S, bottom). The dot plots show individual cell pairs in gray with means ± the standard error of the mean in black. Bar graphs represent data from 13–22 cell pairs. Representative mEPSC traces and averaged mEPSC waveforms for untransfected (gray) and transfected (black) neurons are shown at the right. (B and C) Representative immunohistochemistry images for Arc and mCherry (transfection marker) indicate that ArcWT and ArcC94,96,98S are similarly expressed in the (B) soma and (C) dendrites of transfected Arc KO neurons (n = 8–14, for ArcWT and ArcC94,96,98S, respectively). Bar graphs of fluorescence intensity normalized to background are shown as means ± the standard error of the mean.
In summary, we have shown that Arc undergoes palmitoylation in neurons and that mutation of the palmitoylated cysteines interferes with the plasticity of excitatory synaptic transmission. Unlike some proteins that require palmitoylation for dendritic localization (e.g., PSD95 and SynDig1),24,25 a nonpalmitoylatable Arc is still transported into the dendrite. Our electrophysiological studies demonstrate that the palmitoylated cysteines are critical for MEF2-dependent synaptic weakening. Although the requirement for AMPAR endocytosis has not been demonstrated for MEF2-induced synaptic depression, LTD-associated decreases in mEPSC frequency require Arc26 and are linked with AMPAR endocytosis.3,9 This suggests a role for Arc palmitoylation in the regulation of AMPAR endocytosis, perhaps by clustering of endocytic components in lipid rafts.
Although mutation of the palmitoylatable cysteines disrupts synaptic plasticity, it is possible that these mutations affect plasticity in a palmitoylation-independent manner, for example, by altering the conformation of Arc [although major changes in secondary structure were not detected by circular dichroism (Figure S4)] or its protein interactions. For example, the palmitoylation motif, 94CLCRC98, resides within a larger motif, 90SIKACLCRCQE100, which was previously identified as the endophilin-binding determinant in Arc.3 Deletion of this endophilin-binding motif inhibits Arc-mediated LTD,3 APP processing,17 and dendritic spine remodeling,27 raising the possibility that its deletion interferes with Arc activity because of the reduced level of endophilin binding, the absence of palmitoylation, or both. Unfortunately, using pull-down assays with purified proteins, we were unable to detect any significant endophilin interaction, even with ArcWT (Figure S3). Chowdhury et al.3 reported that Arc binds less tightly to fulllength endophilin than it does to an N-terminal truncation mutant. Perhaps endophilin adopts open, binding competent and closed, binding incompetent conformations in cells but is exclusively in the closed conformation in vitro. We plan to explore this issue further, using fluorescence approaches to compare the interactions of ArcWT and ArcC94,96,98S with endophilins in living cells.
Supplementary Material
ACKNOWLEDGMENTS
The authors kindly thank B. Walker and M. Tomasek for technical assistance and Dr. J. R. Gibson and members of the Huber laboratory and Dr. Leonel Malacrida (Laboratory of Fluorescence Dynamics, Irvine, CA) for valuable discussion.
Funding
This research was supported by National Institutes of Health Grants HD052731 (K.M.H.) and MH110223 (J.P.A. and K.M.H.).
ABBREVIATIONS
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- APP
amyloid precursor protein
- PNS
postnuclear supernatant
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00959.
Experimental Procedures, Figures S1–S5, and Table S1 (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Korb E, and Finkbeiner S (2011) Trends Neurosci. 34, 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Shepherd JD, and Bear MF (2011) Nat. Neurosci 14, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, and Worley PF (2006) Neuron 52, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, and Worley PF (2006) Neuron 52, 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bosl MR, Lipp HP, Grant SGN, Bliss TVP, Wolfer DP, and Kuhl D (2006) Neuron 52, 437–444. [DOI] [PubMed] [Google Scholar]
- (6).Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, and Worley PF (2008) Neuron 59, 70–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wilkerson JR, Tsai NP, Maksimova MA, Wu H, Cabalo NP, Loerwald KW, Dictenberg JB, Gibson JR, and Huber KM (2014) Cell Rep. 7, 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, and Cline HT (2006) Neuron 52, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, and Huber KM (2008) Neuron 59, 84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chen TJ, Wang DC, Hung HS, and Ho HF (2014) Cell. Mol. Life Sci 71, 4069–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Byers CE, Barylko B, Ross JA, Southworth DR, James NG, Taylor CA 4th, Wang L, Collins KA, Estrada A, Waung M, Tassin TC, Huber KM, Jameson DM, and Albanesi JP (2015) Biochim. Biophys. Acta, Gen. Subj 1850, 1310–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).DaSilva LL, Wall MJ, P de Almeida L, Wauters SC, Januario YC, Muller J, and Correa SA (2016) eNeuro 24, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, and Worley PF (1995) Neuron 14, 433–445. [DOI] [PubMed] [Google Scholar]
- (14).Suzuki T. (2002) Neurosci. Res 44, 1–9. [DOI] [PubMed] [Google Scholar]
- (15).Simons K, and Sampaio JL (2011) Cold Spring Harbor Perspect. Biol 3, a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Delint-Ramirez I, Fernández E, Bayés A, Kicsi E, Komiyama NH, and Grant SG (2010) J. Neurosci 30, 8162–8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wu J, Petralia RS, Kurushima H, Patel H, Jung MY, Volk L, Chowdhury S, Shepherd JD, Dehoff M, Li Y, Kuhl D, Huganir RL, Price DL, Scannevin R, Troncoso JC, Wong PC, and Worley PF (2011) Cell 147, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Linder ME, and Deschenes RJ (2007) Nat. Rev. Mol. Cell Biol 8, 74–84. [DOI] [PubMed] [Google Scholar]
- (19).Fukata Y, and Fukata M (2010) Nat. Rev. Neurosci 11, 161–175. [DOI] [PubMed] [Google Scholar]
- (20).Zhou F, Xue Y, Yao X, and Xu Y (2006) Bioinformatics 22, 894–896. [DOI] [PubMed] [Google Scholar]
- (21).Thomas GM, Hayashi T, Huganir RL, and Linden DJ (2013) J. Neurosci 33, 15401–15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Folch J, Lees M, and Sloane Stanley GH (1957) J. Biol. Chem 226, 496–509. [PubMed] [Google Scholar]
- (23).Kawashima T, Okuno H, Nonaka M, Adachi-Morishima A, Kyo N, Okamura M, Takemoto-Kimura S, Worley PF, and Bito H (2009) Proc. Natl. Acad. Sci. U. S. A 106, 316–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).El-Husseini AE-D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, and Bredt DS (2002) Cell 108, 849–863. [DOI] [PubMed] [Google Scholar]
- (25).Kaur I, Yarov-Yarovoy V, Kirk LM, Plambeck KE, Barragan EV, Ontiveros ES, and Díaz E (2016) J. Neurosci 36, 7562–7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Jakkamsetti V, Tsai NP, Gross C, Molinaro G, Collins KA, Nicoletti F, Wang KH, Osten P, Bassell GJ, Gibson JR, and Huber KM (2013) Neuron 80, 72–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Peebles CL, Yoo J, Thwin MT, Palop JJ, Noebels JL, and Finkbeiner S (2010) Proc. Natl. Acad. Sci. U. S. A 107, 18173–18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.