Abstract
The Bradyrhizobium japonicum ftsH gene was cloned by using a set of widely applicable degenerated oligonucleotides. Western blot experiments indicated that the FtsH protein was produced under standard growth conditions and that it was not heat inducible. Attempts to delete the ftsH gene in B. japonicum failed, suggesting a pivotal cellular function of this gene. The expression of B. japonicum ftsH in an ftsH-negative Escherichia coli strain significantly enhanced the fitness of this mutant and reduced the steady-state level of ς32.
The Escherichia coli ftsH (filamentation temperature-sensitive) gene encodes a 70-kDa membrane-anchored ATP-dependent metalloprotease (24). FtsH belongs to the AAA family (ATPases associated with diverse cellular activities), members of which are found universally in prokaryotes and in mitochondria and chloroplasts of eukaryotes. The presence of ftsH even in the minimal genome of Mycoplasma genitalium (7) is indicative of its important cellular function(s). A regulatory function of FtsH in E. coli is the degradation of the heat shock sigma factor ς32 under normal growth conditions, in which only minute amounts of this alternative sigma factor are required (10, 24). FtsH has opposing effects on the stability or activity of ς32 and ς54. While the first is degraded by FtsH, the latter gains functionality by the action of FtsH through an unknown mechanism (6). Another task of FtsH, the capacity to degrade the λ cII and cIII proteins, led to its alternative designation, HflB, reflecting the high frequency of lysogeny phenotype of an hflB mutant (11, 21). Up-regulated expression of lipopolysaccharides due to stabilization of the committed deacetylase in lipopolysaccharide biosynthesis is assumed to be the reason for the lethality of an E. coli ftsH mutant (17). FtsH also degrades integral membrane proteins such as SecY, a subunit of the protein translocase, and subunit α of the F1F0 ATPase complex (1, 2). Two N-terminal transmembrane regions of FtsH mediate multimer formation of the protein, resulting in the appearance of ring-shaped structures (5, 21). There is accumulating evidence that FtsH serves other functions in addition to being a protease. It participates in the assembly of proteins into the membrane and in the translocation of exported proteins (3, 4). The multifunctionality of FtsH is manifested by its requirement in mRNA turnover. Its putative role in mRNA decay has provoked yet another designation, namely, MrsC for mRNA stability (8, 27).
Bradyrhizobium japonicum, the nitrogen-fixing root nodule symbiont of soybean, is an interesting organism in this context because it encodes three ς32-type RpoH factors (14) and two ς54-type RpoN proteins (12) which could be potential targets of FtsH. In order to determine whether B. japonicum FtsH is involved in the regulation of responses to heat shock and nitrogen limitation, we set out to characterize the corresponding gene and its product.
Cloning of the B. japonicum ftsH gene.
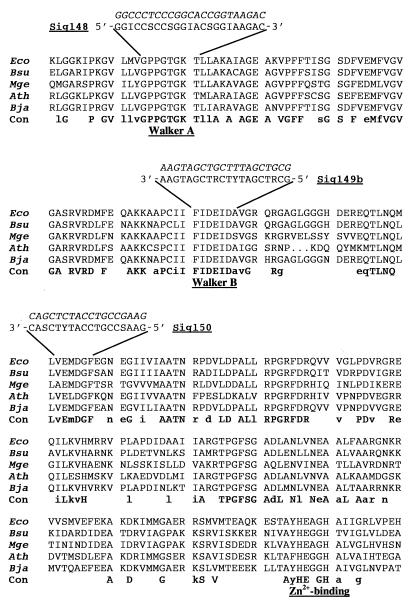
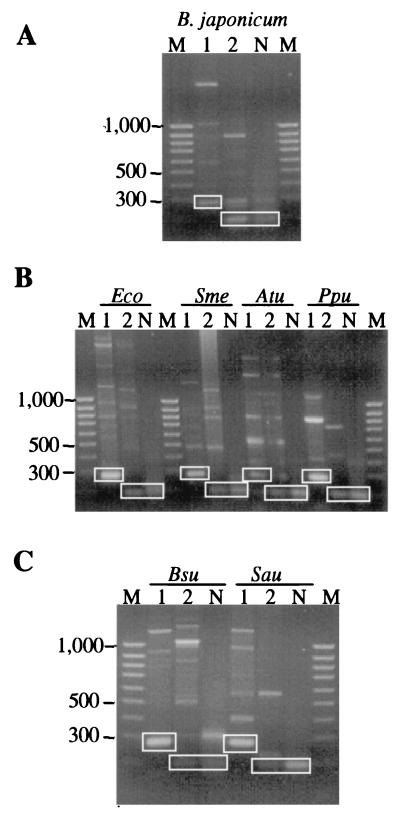
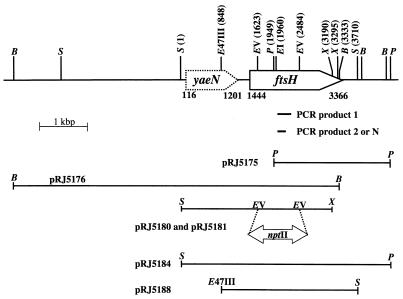
FtsH-like proteins from different bacteria display a number of conserved motifs and domains (20), among them the highly conserved ATP-binding Walker A and Walker B motifs. Degenerated oligonucleotides based on these motifs and on an additional conserved sequence further downstream were designed in accordance with the B. japonicum codon frequency table (19) (Fig. 1). Internal ftsH fragments were amplified by touchdown PCR. The annealing temperature was gradually lowered by 2°C from 50 to 42°C every second cycle. Amplification was completed after 30 cycles at an annealing temperature of 48°C. Amplification with total DNA of B. japonicum and the primers Sig148 and Sig150 resulted in the expected 280-bp fragment (PCR product 1; Fig. 2A and 3). Furthermore, a 190-bp fragment was amplified with Sig148 and Sig149b (PCR product 2). This short product was also obtained when the isolated 280-bp fragment served as the template for a nested PCR with Sig148 and Sig149b (PCR product N). The amplification products were cloned into pUC18, and sequencing of the inserts clearly revealed the internal sequence of an ftsH-like gene. The 280-bp ftsH fragment was then used as a hybridization probe to isolate the complete ftsH gene region of B. japonicum (Fig. 3). The presence of only a single band per digest in a Southern hybridization experiment suggested that the B. japonicum genome contains only a single copy of the ftsH gene (data not shown). Two fragments, a 2.1-kbp PstI fragment and a 6.9-kbp BamHI fragment, were cloned into pUC18 and designated pRJ5175 and pRJ5176, respectively (Fig. 3). Sequencing confirmed that the inserts carried large and overlapping portions of the B. japonicum ftsH gene. In order to obtain the complete ftsH gene on a single plasmid, pRJ5184 was constructed (Fig. 3) by ligating a 2.4-kbp EcoRI fragment from pRJ5175 into a 4.8-kbp EcoRI fragment of pRJ5177, a subclone of pRJ5176.
FIG. 1.
Alignment of the deduced amino acid sequences of an internal region of several AAA-type proteins. Only the amino acids corresponding to E. coli FtsH from position 179 to position 428 are shown. FtsH proteins from E. coli (Eco) (25), B. subtilis (Bsu) (16), M. genitalium (Mge) (7), and A. thaliana (Ath) (13) are listed and compared with B. japonicum (Bja) FtsH (this work). The conserved Walker motifs and the putative Zn2+ binding site comprising two conserved histidines are indicated. Residues that are identical in all five proteins are defined by capital letters in the consensus (Con) sequence line. Amino acids present in four of the five proteins are indicated by lowercase letters. The sequences of degenerated oligonucleotides used to amplify internal ftsH fragments are listed above the corresponding amino acid sequence. The code for mixed nucleotides is as follows: S is C or G, R is G or A, Y is C or T, and I is inosine. The true nucleotide sequence obtained by sequencing of the B. japonicum ftsH gene is provided in italic letters above the oligonucleotides.
FIG. 2.
PCR amplification of internal ftsH fragments from various bacteria. Total DNAs of B. japonicum (A); the gram-negative bacteria E. coli (Eco), S. meliloti (Sme), A. tumefaciens (Atu), and P. putida (Ppu) (B); and the gram-positive bacteria B. subtilis (Bsu) and S. aureus (Sau) (C) were subjected to touchdown PCR. An aliquot of the PCR mixture was separated on 1.5% (wt/vol) agarose gels. Amplification products obtained with the primer pair Sig148-Sig150 are in lanes 1, and products obtained with Sig148-Sig149b by using either total DNA or isolated product 1 as the template are in lanes 2 or N, respectively. Fragments corresponding to the expected sizes are boxed. The lengths (in base pairs) of representative bands from the 100-bp ladder (lanes M) are indicated on the left.
FIG. 3.
Physical map of the B. japonicum ftsH gene region. Numbers indicate the nucleotide positions of open reading frames and recognition sites of restriction enzymes as follows: B, BamHI; EI, EcoRI; EV, EcoRV; E47III, Eco47III; P, PstI; S, SalI; X, XhoI. The positions of PCR products 1, 2, and N with respect to the ftsH gene are indicated. The inserts of relevant plasmids harboring the ftsH gene region are given with their corresponding plasmid designations. Plasmids pRJ5175, pRJ5176, and pRJ5184 contain pUC18 (15) as a vector. The inserts of pRJ5180 and pRJ5181 were cloned into pSUP202pol3, a derivative of pSUP202 (22). Plasmid pRJ5188 is based on pBAD18-Cm (9).
Analysis of the ftsH gene region.
The deduced ftsH gene product displays significant overall sequence similarity to other known proteases of the AAA family. B. japonicum FtsH shares 57.7, 50.6, and 53.4% positionally identical amino acids with its homologs from E. coli, Bacillus subtilis, and Arabidopsis thaliana, respectively. Two putative transmembrane regions (residues 7 to 25 and 102 to 125) can be predicted as a potential membrane anchor. A particularly high degree of conservation is evident in the regions that had served for the design of degenerated oligonucleotides (Fig. 1). This finding prompted us to test whether the primers would also yield specific amplification products with chromosomal DNAs from other bacteria. Total DNAs from a variety of prokaryotes were subjected to PCR amplifications using the Sig148-Sig149b and Sig148-Sig150 primer pairs and to the nested PCR as described for B. japonicum. Appropriate amplification products of the expected sizes were obtained with DNAs from the gram-negative bacteria E. coli, Sinorhizobium meliloti, Agrobacterium tumefaciens, and Pseudomonas putida (Fig. 2B) and from the gram-positive bacteria B. subtilis and Staphylococcus aureus (Fig. 2C). In all cases, additional PCR products longer than the putative ftsH fragments were observed, most likely because the primers Sig148 and Sig149b were designed against the Walker motifs. These highly conserved sequences occur not only in FtsH but in many other nucleotide-binding proteins (26).
An open reading frame coding for a protein with an unknown function similar to E. coli YaeN (MesJ) (18) is located upstream of B. japonicum ftsH (Fig. 3). The gene is oriented in the same direction as ftsH, and the two genes might form an operon (see below).
Expression of the B. japonicum ftsH gene.
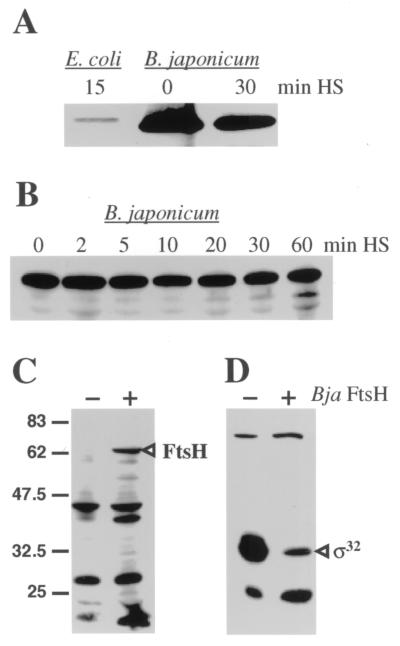
Western blot analysis of B. japonicum extracts with antiserum raised against E. coli FtsH revealed a protein band of approximately 70 kDa. Consistent with its calculated molecular mass of 69.9 kDa, the B. japonicum protein migrated slightly faster than the 70.7-kDa E. coli FtsH protein (Fig. 4A). In contrast to E. coli FtsH, which is a ς32-dependent heat shock protein (10), B. japonicum FtsH appeared to be constitutively expressed at 30°C without being heat inducible. A refined kinetic evaluation showed that the cellular level of B. japonicum FtsH did not change significantly over a period of 1 h after a temperature upshift from 30 to 43°C (Fig. 4B). This is unprecedented, as in all of the bacteria tested so far, ftsH expression was induced by a temperature upshift and by other stress conditions (20). Immunodetection of B. japonicum FtsH produced from plasmid pRJ5188 in ftsH-negative E. coli AR3291 served as a control to demonstrate that the antiserum used indeed recognizes the rhizobial protein (Fig. 4C).
FIG. 4.
Immunodetection of FtsH protein in E. coli and B. japonicum extracts. E. coli C600 or B. japonicum 110spc4 cells were grown to mid-exponential phase at 30°C. After a reference sample had been taken, the culture was shifted to 43°C and samples were collected at the indicated time points after heat shock (HS). Crude cell extracts were prepared, separated on a sodium dodecyl sulfate–12% polyacrylamide gel, and subjected to Western blot analysis using anti-E. coli FtsH serum (U. Jenal, Basel, Switzerland; original source, P. Bouloc, Paris, France; 1,000-fold dilution). Only the area around 70 kDa is shown. Panels A and B represent two independent experiments with different sampling periods. A 10-fold-diluted E. coli extract was loaded in panel A in order to avoid a strong signal due to the use of a homologous antiserum. For the experiments shown in panels C and D, E. coli AR3291/pBAD18-Cm and AR3291/pRJ5188 were grown at 30°C in LB medium with arabinose (0.1% [wt/vol]) to mid-exponential phase and samples were subjected to Western blot analyses. The absence (−) or presence (+) of B. japonicum (Bja) FtsH expressed from pRJ5188 is indicated. Bands representing B. japonicum FtsH in panel C and E. coli ς32 in panel D are labeled. The sigma factor was detected by the use of anti-E. coli ς32 serum (B. Bukau, Freiburg, Germany; 3,000-fold dilution). The molecular masses of marker proteins are indicated in kilodaltons on the left.
The relative abundance of FtsH in B. japonicum under normal growth conditions suggested an important cellular function of this protein. In line with this assumption is the fact that repeated attempts to delete a large internal fragment of the ftsH gene by marker replacement mutagenesis using plasmids pRJ5180 and pRJ5181 (Fig. 3) failed. The occurrence of kanamycin- and tetracycline-resistant colonies demonstrated that the constructs containing a mutated ftsH gene had been mobilized and cointegrated into B. japonicum. However, tetracycline-sensitive colonies resulting from double homologous recombination could not be recovered.
Primer extension experiments performed to identify a promoter in the intergenic region between yaeN and ftsH or upstream of yaeN were unsuccessful, indicating that ftsH might be part of an extended operon. This proposition is supported by two additional observations: (i) the presence of a putative rho-independent transcription terminator downstream of ftsH (nucleotides 3410 to 3438) but not between yaeN and ftsH or in the sequenced region upstream of yaeN and (ii) the strict arabinose dependence of FtsH production from pRJ5188 in our complementation experiments (see below).
Functional expression of B. japonicum ftsH in an E. coli ΔftsH mutant.
Since the ftsH gene could not be eliminated in B. japonicum, we started an initial characterization of the gene and its product in an E. coli ftsH mutant strain. ΔftsH E. coli AR3291 is a viable ftsH null mutant with a suppressor mutation in sfhC (fabZ) that allows cells to survive (17, 23). The mutant was transformed with either pBAD18-Cm or pRJ5188 bearing B. japonicum ftsH on the same replicon (Fig. 3). The lack of FtsH in the strain containing the pBAD vector without an insert resulted in a concomitant, high level of ς32 protein (Fig. 4C and D). More importantly, the presence of B. japonicum FtsH in E. coli AR3291/pRJ5188 was paralleled by a significantly reduced level of the sigma factor. This result suggests that B. japonicum FtsH has the inherent capacity to degrade ς32 protein.
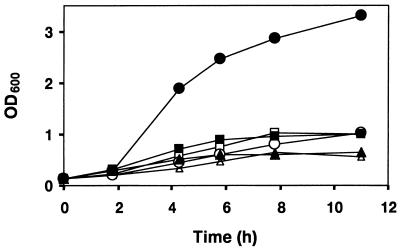
When E. coli AR3291/pBAD18-Cm and AR3291/pRJ5188 were grown at 37°C in the absence of the inducer arabinose, they showed a clear growth defect (Fig. 5). The growth curves of the two strains were almost indistinguishable in Luria-Bertani (LB) medium or in LB medium with glucose. Growth was significantly improved when expression of B. japonicum ftsH was induced by the addition of arabinose (closed circles in Fig. 5). We conclude from these growth experiments that B. japonicum ftsH is able to confer a general growth advantage upon the E. coli ftsH mutant, indicating that the heterologous protein can compensate for the loss of at least some of the important FtsH functions in E. coli AR3291. One of these functions might be to balance the cellular level of ς32. We also tested whether B. japonicum FtsH would be able to promote growth of the E. coli ftsH mutant under conditions that require active ς54. The expression of B. japonicum ftsH did not alleviate the growth defect of the mutant under nitrogen-limiting conditions (data not shown). Moreover, B. japonicum FtsH was unable to assist in ς54-dependent transcription of a pspA-lacZ fusion in E. coli (data not shown). Modulation of ς54 has only been demonstrated for the E. coli AAA protease (6). Apparently, many features of the multifunctional FtsH protein are not understood and remain to be explored.
FIG. 5.
Growth complementation of an E. coli ΔftsH mutant by B. japonicum ftsH. E. coli AR3291 transformed with pBAD18-Cm (open symbols) or with pRJ5188 carrying B. japonicum ftsH (closed symbols) was grown at 37°C in either LB medium (triangles), LB medium with glucose (0.02% [wt/vol]; squares), or LB medium with arabinose (0.1% [wt/vol]; circles). OD600, optical density at 600 nm.
Nucleotide sequence accession number.
The nucleotide sequence of the B. japonicum ftsH gene region between the SalI sites at positions 1 and 3710 in Fig. 3 has been deposited in the EMBL, GenBank, and DDBJ databases under accession no. AJ243808.
Acknowledgments
We are grateful to Amos Oppenheim for providing strains, Urs Jenal and Bernd Bukau for antisera, Hans-Martin Fischer for plasmid pSUP202pol3, Manuel Carmona for the pspA-lacZ fusion on plasmid pJD31, and Michael Kertesz and Ines Kullik for chromosomal DNA samples. Dino Bertani is acknowledged for performing the PCRs shown in Fig. 2.
This work was supported by grants from the Federal Institute of Technology, Zürich, Switzerland, and the Swiss National Foundation for Scientific Research.
REFERENCES
- 1.Akiyama Y, Kihara A, Ito K. Subunit of a proton ATPase F0 sector is a substrate of the FtsH protease in Escherichia coli. FEBS Lett. 1996;399:26–28. doi: 10.1016/s0014-5793(96)01283-5. [DOI] [PubMed] [Google Scholar]
- 2.Akiyama Y, Kihara A, Tokuda H, Ito K. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J Biol Chem. 1996;271:31196–31201. doi: 10.1074/jbc.271.49.31196. [DOI] [PubMed] [Google Scholar]
- 3.Akiyama Y, Ogura T, Ito K. Involvement of FtsH in protein assembly into and through the membrane. I. Mutations that reduce retention of a cytoplasmic reporter. J Biol Chem. 1994;269:5218–5224. [PubMed] [Google Scholar]
- 4.Akiyama Y, Shirai Y, Ito K. Involvement of FtsH in protein assembly into and through the membrane. II. Dominant mutations affecting FtsH functions. J Biol Chem. 1994;269:5225–5229. [PubMed] [Google Scholar]
- 5.Akiyama Y, Yoshihisa T, Ito K. FtsH, a membrane-bound ATPase, forms a complex in the cytoplasmic membrane of Escherichia coli. J Biol Chem. 1995;270:23485–23490. doi: 10.1074/jbc.270.40.23485. [DOI] [PubMed] [Google Scholar]
- 6.Carmona M, de Lorenzo V. Involvement of the FtsH (HflB) protease in the activity of ς54 promoters. Mol Microbiol. 1999;31:261–270. doi: 10.1046/j.1365-2958.1999.01169.x. [DOI] [PubMed] [Google Scholar]
- 7.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 8.Granger L L, O’Hara E B, Wang R F, Meffen F V, Armstrong K, Vancey S D, Babitzke P, Kushner S R. The Escherichia coli mrsC gene is required for cell growth and mRNA decay. J Bacteriol. 1998;180:1920–1928. doi: 10.1128/jb.180.7.1920-1928.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herman C, Thévenet D, D’Ari R, Bouloc P. Degradation of ς32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci USA. 1995;92:3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herman C, Thévenet D, D’Ari R, Bouloc P. The HflB protease of Escherichia coli degrades its inhibitor λcIII. J Bacteriol. 1997;179:358–363. doi: 10.1128/jb.179.2.358-363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kullik I, Fritsche S, Knobel H, Sanjuan J, Hennecke H, Fischer H M. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the ς54 gene (rpoN) J Bacteriol. 1991;173:1125–1138. doi: 10.1128/jb.173.3.1125-1138.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindahl M, Tabak S, Cseke L, Pichersky E, Andersson B, Adam Z. Identification, characterization and molecular cloning of a homologue of the bacterial FtsH protease in chloroplasts of high plants. J Biol Chem. 1996;271:29329–29334. doi: 10.1074/jbc.271.46.29329. [DOI] [PubMed] [Google Scholar]
- 14.Narberhaus F, Krummenacher P, Fischer H M, Hennecke H. Three disparately regulated genes for ς32-like transcription factors in Bradyrhizobium japonicum. Mol Microbiol. 1997;24:93–104. doi: 10.1046/j.1365-2958.1997.3141685.x. [DOI] [PubMed] [Google Scholar]
- 15.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligonucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 16.Ogasawara N, Nakai S, Yoshikawa H. Systematic sequencing of the 180 kilobase region of the Bacillus subtilis chromosome containing the replication origin. DNA Res. 1994;1:1–14. doi: 10.1093/dnares/1.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Ogura T, Inoue K, Tatsuta T, Suzaki T, Karata K, Young K, Su L H, Fierke C A, Jackman J E, Raetz C R H, Coleman J, Tomoyasu T, Matsuzawa H. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the AAA protease FtsH (HflB) in Escherichia coli. Mol Microbiol. 1999;31:833–844. doi: 10.1046/j.1365-2958.1999.01221.x. [DOI] [PubMed] [Google Scholar]
- 18.Pichoff S, Alibaud L, Guédant A, Castanié M P, Bouché J P. An Escherichia coli gene (yaeO) suppresses temperature-sensitive mutations in essential genes by modulating Rho-dependent transcription termination. Mol Microbiol. 1998;29:859–869. doi: 10.1046/j.1365-2958.1998.00981.x. [DOI] [PubMed] [Google Scholar]
- 19.Ramseier T M, Göttfert M. Codon usage and G + C content in Bradyrhizobium japonicum genes are not uniform. Arch Microbiol. 1991;156:270–276. doi: 10.1007/BF00262997. [DOI] [PubMed] [Google Scholar]
- 20.Schumann W. FtsH—a single-chain charonin? FEMS Microbiol Rev. 1999;23:1–11. doi: 10.1111/j.1574-6976.1999.tb00389.x. [DOI] [PubMed] [Google Scholar]
- 21.Shotland Y, Koby S, Teff D, Mansur N, Oren D A, Tatematsu K, Tomoyasu T, Kessel M, Bukau B, Ogura T, Oppenheim A B. Proteolysis of the phage λ CII regulatory protein by FtsH (HflB) of Escherichia coli. Mol Microbiol. 1997;24:1303–1310. doi: 10.1046/j.1365-2958.1997.4231796.x. [DOI] [PubMed] [Google Scholar]
- 22.Simon R, Priefer U, Pühler A. Vector plasmids for in vivo and in vitro manipulation of Gram-negative bacteria. In: Pühler A, editor. Molecular genetics of the bacteria-plant interaction. Heidelberg, Germany: Springer Verlag; 1983. pp. 98–106. [Google Scholar]
- 23.Tatsuta T, Tomoyasu T, Bukau B, Kitagawa M, Mori H, Karata K, Ogura T. Heat shock regulation in the ftsH null mutant of Escherichia coli: dissection of stability and activity control mechanisms of ς32in vivo. Mol Microbiol. 1998;30:583–593. doi: 10.1046/j.1365-2958.1998.01091.x. [DOI] [PubMed] [Google Scholar]
- 24.Tomoyasu T, Gamer J, Bukau B, Kanemori M, Mori H, Rutman A J, Oppenheim A B, Yura T, Yamanaka K, Niki H, Hiraga S, Ogura T. Escherichia coli FtsH is a membrane bound, ATP-dependent protease which degrades the heat-shock transcription factor ς32. EMBO J. 1995;14:2551–2560. doi: 10.1002/j.1460-2075.1995.tb07253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomoyasu T, Yuki T, Morimura S, Mori H, Yamanaka K, Niki H, Hiraga S, Ogura T. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol. 1993;175:1344–1351. doi: 10.1128/jb.175.5.1344-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker J E, Saraste M, Runswick M J, Kelly D J. Distantly related sequences in the alpha and beta subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R F, O’Hara E B, Aldea M, Bargmann C I, Gromley H, Kushner S R. Escherichia coli mrsC is an allele of hflB, encoding a membrane-associated ATPase and protease that is required for mRNA decay. J Bacteriol. 1998;180:1929–1938. doi: 10.1128/jb.180.7.1929-1938.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]