Abstract
Everted membrane vesicles of Pseudomonas aeruginosa PAO1 harboring plasmid pCRO616, expressing the ChrA chromate resistance protein, accumulated four times more 51CrO42− than vesicles from plasmidless cells, indicating that a chromate efflux system functions in the resistant strain. Chromate uptake showed saturation kinetics with an apparent Km of 0.12 mM chromate and a Vmax of 0.5 nmol of chromate/min per mg of protein. Uptake of chromate by vesicles was dependent on NADH oxidation and was abolished by energy inhibitors and by the chromate analog sulfate. The mechanism of resistance to chromate determined by ChrA appears to be based on the active efflux of chromate driven by the membrane potential.
Plasmid-determined resistance to chromate ions has been found in the genera Streptococcus (5), Pseudomonas (1, 2, 17), and Alcaligenes (9). Molecular analysis of chromate resistance determinants from plasmid pUM505 from Pseudomonas aeruginosa (3) and plasmid pMOL28 from Alcaligenes eutrophus (10) revealed that the deduced product of the chrA gene, the hydrophobic protein ChrA (416 and 401 amino acid residues, respectively), was responsible for the resistance phenotype. Chromate tolerance conferred by the ChrA protein was associated with reduced accumulation of 51CrO42− in both P. aeruginosa and A. eutrophus, and it was hypothesized that ChrA was involved in the extrusion of chromate ions (3, 10). Nevertheless, direct evidence for efflux was missing. Here we show that everted membrane vesicles from chromate-resistant P. aeruginosa cells expressing the ChrA protein accumulate more 51CrO42− than vesicles prepared from a plasmidless chromate-sensitive derivative.
Bacterial strains and chromate susceptibility.
Chromate-sensitive P. aeruginosa PAO1 and chromate-resistant PAO1(pCRO616), which contains the chrA gene from plasmid pUM505 (2) cloned in the pKT230 vector, have been previously described (3). Cells were grown in nutrient broth (Bioxon, Mexico City, México) and incubated at 37°C with shaking. The presence of plasmid pCRO616 allowed strain PAO1 to tolerate about six times more chromate than the plasmid-less derivative or the isogenic strain bearing the pKT230 cloning vector (data not shown).
Accumulation of 51CrO42− by whole cells.
Cultures of PAO1(pCRO616) accumulated about 2.5 times less chromate than the sensitive PAO1 strain after an 8-h incubation in the presence of 10 μM chromate (data not shown). Initial rates of chromate uptake were determined with suspensions of exponential-phase cells (0.3 mg/ml [dry weight]) in 0.1 M phosphate buffer (pH 7.0) with 50 μM 51CrO42− (New England Nuclear Corp., Boston, Mass.; specific activity, 400 to 1,200 mCi/mg). Aliquots (0.1 ml) were filtered through 0.45-μm-pore-diameter membranes (Millipore Corp., Bedford, Mass.) and washed twice with 5 ml of distilled water. The radioactivity on the filters was quantified in a Packard Multi-Prias gamma radiation counter. Cell suspensions from PAO1(pCRO616) showed a decreased initial rate of 51CrO42− uptake compared with that from the plasmidless strain (data not shown). Resistance to chromate was also related to diminished chromate accumulation in Pseudomonas ambigua (6), Pseudomonas fluorescens (11), and Enterobacter cloacae (12), although the precise mechanism of resistance was not elucidated in these bacteria. Decreased chromate uptake by resistant cells may be caused either by an efflux system, by a blockage in chromate uptake, or by both processes. To distinguish between these two possibilities, the uptake of chromate by inside-out membrane vesicles was measured. The properties of ChrA suggested that it might function as a membrane transporter involved in the extrusion of chromate (3).
Uptake of 51CrO42− by vesicles.
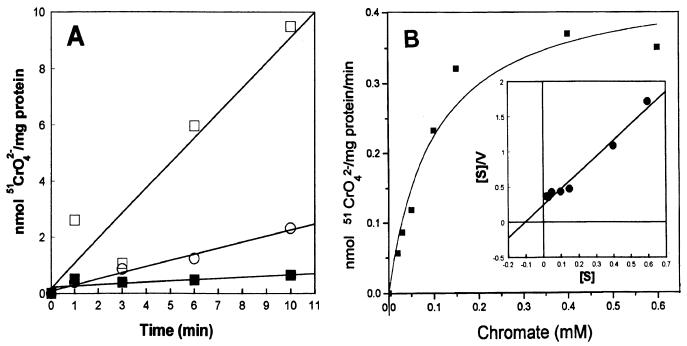
Everted membrane vesicles from Pseudomonas strains were prepared as follows. Cultures (1 liter) grown for 16 to 18 h at 37°C with shaking were harvested (12,000 × g, 15 min, 4°C), and then the cells were washed twice in 0.1 M phosphate buffer, suspended in 20 to 25 ml of the same buffer containing 1 mM dithiothreitol, and disrupted by passage twice through an Aminco French pressure cell at 12,000 lb/in2. Intact cells were removed by repeating the centrifugation step. Membrane vesicles were recovered by centrifugation at 200,000 × g for 45 min at 4°C, suspended in SHE buffer (250 mM sucrose, 10 mM HEPES, 1 mM EGTA [pH 7.4]), and stored at −70°C until used. The activity of inside-out vesicles was assessed as reported by Rottenberg and Moreno-Sánchez (13). Protein was determined by a modification of the method of Lowry et al. (8) by using bovine serum albumin as a standard. Transport assays were carried out at 25°C in 0.1 M phosphate buffer (pH 7.0) containing 2 mM NADH, 5 mM MgCl2, and 200 μM 51CrO42− and were initiated by the addition of membrane vesicles (0.15-mg/ml final concentration). Samples (0.4 ml) were filtered through 0.22-μm-pore-diameter Millipore membranes and processed as described above. Chromate uptake by vesicles from PAO1(pCRO616) was four times higher than that of its chromate-sensitive derivative (Fig. 1A). These data support the notion that a chromate efflux system is present in the plasmid-containing chromate-resistant strain. In experiments with whole cells, 20-fold excess sulfate, an analog of chromate, inhibited 50 to 60% 51CrO42− uptake by PAO1 (data not shown). Apparently, chromate uptake by PAO1(pCRO616) was unaffected by excess sulfate, since this strain accumulated an amount of chromate similar to that accumulated by the sensitive one in the presence of 1 mM sulfate (data not shown). Hence, these data suggested that chromate efflux was not functioning in the presence of high concentrations of sulfate. Accordingly, 51CrO42− uptake by membrane vesicles of PAO1(pCRO616) was severely inhibited by sulfate (Table 1). Thus, it appears that sulfate competes with chromate for extrusion by the ChrA protein. This is not surprising, since it has been established that chromate is a competitive inhibitor of sulfate transport in Pseudomonas (3, 11).
FIG. 1.
(A) Chromate uptake by everted membrane vesicles of PAO1 (○) and PAO1(pCRO616) (□). Membrane vesicles (0.15 mg/ml) were suspended in phosphate buffer with 200 μM 51CrO42−. After incubation at 25°C in the presence of 2 mM NADH, 51CrO42− uptake was measured. Uptake by vesicles from PAO1(pCRO616) was assayed in the absence of NADH (■). (B) Kinetics of chromate uptake by vesicles of PAO1(pCRO616). The uptake conditions were as described above, except that various concentrations of chromate were tested and aliquots were filtered after a 10-min incubation. The solid line represents the best fit to the Michaelis-Menten (hyperbolic) equation. The inset shows a linear transformation of the Michaelis-Menten equation by using the Hanes-Woolf plot (14). Data are representative of three assays in duplicate.
TABLE 1.
Effect of energy inhibitors on chromate uptake by everted membrane vesicles
| Treatment | Initial rate of chromate uptake (nmol of 51CrO42−/mg of protein/min)a |
|---|---|
| +NADH | 1.59 ± 0.10 |
| No NADH | 0.52 ± 0.06 |
| +4 mM NaCN | 0.34 ± 0.20 |
| +50 μM HQNO | 0.54 ± 0.26 |
| +20 mM NaN3 | 0.60 ± 0.26 |
| +50 μM Carbonyl cyanide m-chlorophenylhydrazone | 0.66 ± 0.10 |
| +5 μM Nigericin | 0.55 ± 0.20 |
| +1 mM Na2SO4 | 0.48 ± 0.13 |
Chromate uptake by vesicles from PAO1(pCRO616) was assayed in phosphate buffer with 2 mM NADH and 400 μM chromate, as described in the text. The values are the mean ± standard deviation of three assays in duplicate.
Kinetics of chromate uptake.
For the determination of kinetics constants, 51CrO42− uptake by membrane vesicles from strain PAO1(pCRO616) was measured with various concentrations of chromate after a 10-min incubation. 51CrO42− was taken up by the vesicles of PAO1(pCRO616) according to substrate saturation kinetics (Fig. 1B). An apparent Km of 0.12 ± 0.05 (n = 3) mM chromate and a Vmax of 0.5 ± 0.23 (n = 3) nmol of chromate/min per mg of protein were calculated (Fig. 1B). This Km value is similar to that of 0.14 mM reported for the accumulation of arsenite by everted membrane vesicles of Escherichia coli cells expressing the ArsB protein from plasmid R773 (7). ChrA and ArsB are hydrophobic proteins of about 400 amino acid residues, with no significant identity at the amino acid sequence level, but with similar amino acid composition and hydropathy profiles (4). Both proteins seem to be involved in similar anion extrusion systems.
Energetics of chromate uptake.
51CrO42− uptake by membrane vesicles of strain PAO1(pCRO616) was much lower in the absence of NADH (Fig. 1A and Table 1), suggesting that a transmembrane H+ gradient is required for chromate transport. Accordingly, the respiratory chain inhibitors NaCN, HQNO, and NaN3 were tested and significantly lowered chromate uptake by vesicles (Table 1); notably, cyanide diminished uptake by almost 80%. Treatment of vesicles with ionophores also caused a significant inhibition of chromate uptake (Table 1); nigericin abolished chromate uptake by 65%. These data suggest that ChrA functions as a secondary transport system using the membrane potential as an energy source for the extrusion of chromate ions. Moreover, the addition of 50 mM lactic acid, which decreased the pH of assays from neutral to about 5.0, caused an increase in 51CrO42− uptake by vesicles of PAO1(pCRO616) (data not shown), also implying the involvement of protons in chromate transport. Arsenite extrusion by vesicles from E. coli expressing the ArsB protein also decreased at alkaline pH (7).
Whereas several bacterial determinants for the active efflux of diverse inorganic cations have been described (16), efflux of chromate by the ChrA protein represents the second example of an inorganic anion translocation system reported in bacteria, the first one being the already mentioned ars operon for the extrusion of arsenite (15).
Acknowledgments
We thank Eréndira Vargas, Irais Sánchez, and Concepción Bravo for technical help.
This work was supported by grants from Coordinación de Investigación Científica (UMSNH) to C.C. and CONACYT (25274-M) to R.M.-S. A.H.A. was supported by a fellowship from CONACYT.
REFERENCES
- 1.Bopp L H, Chakrabarty A M, Ehrlich H L. Chromate resistance plasmid in Pseudomonas fluorescens. J Bacteriol. 1983;155:1105–1109. doi: 10.1128/jb.155.3.1105-1109.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cervantes C, Ohtake H. Plasmid-determined resistance to chromate in Pseudomonas aeruginosa. FEMS Microbiol Lett. 1988;56:173–176. [Google Scholar]
- 3.Cervantes C, Ohtake H, Chu L, Misra T K, Silver S. Cloning, nucleotide sequence, and expression of the chromate resistance determinant of Pseudomonas aeruginosa plasmid pUM505. J Bacteriol. 1990;172:287–291. doi: 10.1128/jb.172.1.287-291.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cervantes C, Silver S. Plasmid chromate resistance and chromate reduction. Plasmid. 1992;27:65–71. doi: 10.1016/0147-619x(92)90007-w. [DOI] [PubMed] [Google Scholar]
- 5.Efstathiou J D, McKay L L. Inorganic salts resistance associated with a lactose-fermenting plasmid in Streptococcus lactis. J Bacteriol. 1977;130:257–265. doi: 10.1128/jb.130.1.257-265.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horitsu H, Futo S, Ozawa K, Kawai K. Comparison of characteristics of hexavalent-chromium tolerant bacterium, Pseudomonas ambigua G-1, and its hexavalent chromium-sensitive mutant. Agric Biol Chem. 1983;47:2907–2908. [Google Scholar]
- 7.Kuroda M, Dey S, Sanders O I, Rosen B P. Alternate energy coupling of ArsB, the membrane subunit of the ArsA anion-translocating ATPase. J Biol Chem. 1997;272:326–331. doi: 10.1074/jbc.272.1.326. [DOI] [PubMed] [Google Scholar]
- 8.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 9.Nies A, Nies D H, Silver S. Cloning and expression of plasmid genes encoding resistances to chromate and cobalt in Alcaligenes eutrophus. J Bacteriol. 1989;171:5065–5070. doi: 10.1128/jb.171.9.5065-5070.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nies A, Nies D H, Silver S. Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J Biol Chem. 1990;265:5648–5653. [PubMed] [Google Scholar]
- 11.Ohtake H, Cervantes C, Silver S. Decreased chromate uptake in Pseudomonas fluorescens carrying a chromate resistance plasmid. J Bacteriol. 1987;169:3853–3856. doi: 10.1128/jb.169.8.3853-3856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohtake H, Komori K, Cervantes C, Toda K. Chromate-resistance in a chromate-reducing strain of Enterobacter cloacae. FEMS Microbiol Lett. 1990;67:85–88. doi: 10.1016/0378-1097(90)90173-n. [DOI] [PubMed] [Google Scholar]
- 13.Rottenberg H, Moreno-Sánchez R. The proton pumping activity of H+-ATPases: an improved fluorescence assay. Biochim Biophys Acta. 1993;1183:161–170. doi: 10.1016/0005-2728(93)90014-7. [DOI] [PubMed] [Google Scholar]
- 14.Segel I H. Enzyme kinetics. New York, N.Y: John Wiley and Sons; 1975. pp. 210–212. [Google Scholar]
- 15.Silver S, Ji G, Bröer S, Dey S, Dou D, Rosen B P. Orphan enzyme or patriarch of a new tribe: the arsenic resistance ATPase of bacterial plasmids. Mol Microbiol. 1993;8:637–642. doi: 10.1111/j.1365-2958.1993.tb01607.x. [DOI] [PubMed] [Google Scholar]
- 16.Silver S, Phung L T. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 17.Summers A O, Jacoby G A. Plasmid-determined resistance to boron and chromium compounds in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1978;13:637–640. doi: 10.1128/aac.13.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]