Abstract
Many respiratory isolates of Pseudomonas aeruginosa from cystic fibrosis patients are mucoid (alginate producing) yet lack flagella. It was hypothesized that an alginate regulator inhibits flagellar gene expression. Mutations in algB, algR, and algT resulted in nonmucoid derivatives, yet algT mutants expressed flagella. AlgT-dependent control of flagellum synthesis occurred through inhibition of fliC but not rpoN transcription.
Pseudomonas aeruginosa causes a variety of acute infections, but the organism is also responsible for most of the life-threatening chronic respiratory tract infections in people with cystic fibrosis (CF). Although lungs of CF patients are colonized by motile, nonmucoid P. aeruginosa strains, during the course of chronic infection there appears to be a selection for certain phenotypes (8, 16). Among these are rough lipopolysaccharide structure, mucoidy, and loss of motility. The mucoid phenotype is due to the overproduction of the exopolysaccharide alginate, a virulence factor which provides a selective advantage to the bacteria (references 8 and 16 and references therein). Nonmotility is also rare in P. aeruginosa except among CF isolates (11, 13). The occurrence of these two phenotypes (alginate and lack of flagella) in many CF isolates prompted us to examine whether alginate and flagellum synthesis were coordinately regulated.
Evidence for coordinate regulation between alginate synthesis and flagellum expression.
A collection of mucoid and nonmucoid P. aeruginosa CF isolates (3) were cultured on L agar plates and scored for the mucoid phenotype (Luria broth [LB] contained the following [per liter]: 10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl; L agar contained 1.5% agar in LB). Flagellum expression was examined by transmission electron microscopy (TEM) and Western blotting using antiserum against flagella which had been purified by published techniques (14) from strain PAK (serotype A) or PAO1 (serotype B). All nonmucoid P. aeruginosa strains examined synthesized flagella, whereas all mucoid isolates lacked flagella (data not shown). The results confirm those reported elsewhere (11, 13) and suggest a correlation between alginate synthesis and lack of flagellum expression.
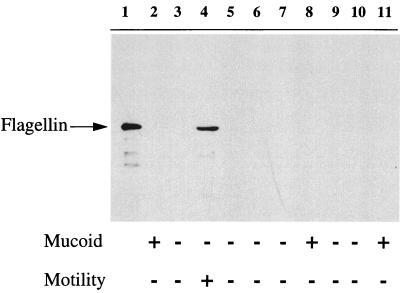
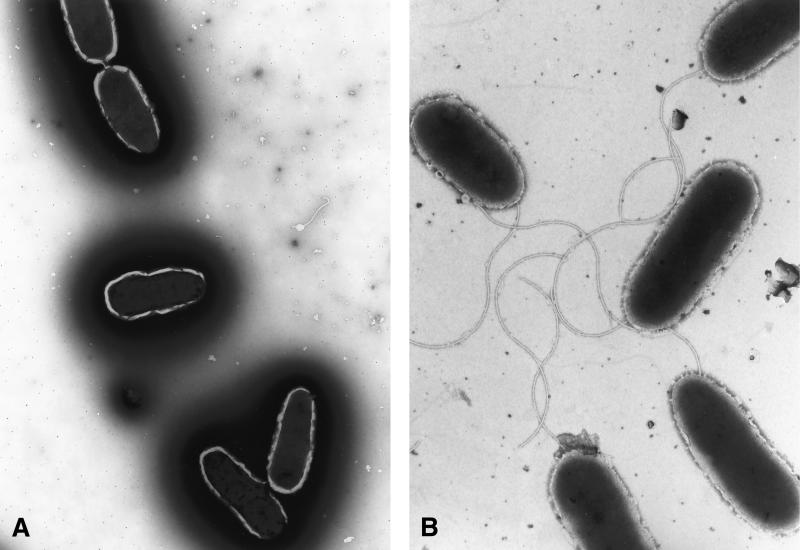
It was unclear if these mucoid CF isolates acquired flagellar gene mutations during selection in the lungs of CF patients. This mechanism has been proposed as an explanation for the high frequency of nonmotile variants in P. aeruginosa CF isolates (6, 13). We propose an alternative explanation. Since most mucoid CF isolates acquire mutations in mucA resulting in elevated levels of the alternative sigma factor ς22 (8), we reasoned that the inhibition of flagellum expression was due to increased expression of ς22. If this was true, inactivation of algT, encoding ς22, in a mucoid, nonmotile strain should restore flagellum synthesis. However, if mucoid CF isolates were to acquire a mutation in a flagellar gene(s), algT mutants should not be able to express flagella. To distinguish these, isogenic strains FRD1 (mucA22) (22) and FRD440 (mucA22 algT::Tn501) (22) were examined for flagellum expression by Western blotting of whole-cell extracts derived from cells cultured in LB lacking NaCl. While the mucoid strain FRD1 lacked flagella, the algT mutant expressed flagella and was motile (Fig. 1, compare lanes 2 and 4). Analysis of the representative AlgT+ strain P. aeruginosa FRD875 (mucA22 algD::xylE-aacC1) (21) by TEM (Fig. 2A), revealed that few, if any, bacteria were expressing a flagellum. However, the isogenic algT mutant clearly expressed a flagellum (Fig. 2B). Complementation studies with plasmid pJF15, which contains algT (7), revealed FRD440/pJF15 transconjugants were mucoid and lacked flagella (data not shown). This indicates the flagellum synthesis observed in FRD440 is due to the loss of the algT gene and not to polar effects on downstream genes.
FIG. 1.
Western blot analysis of flagellum expression in P. aeruginosa strains. Whole-cell extracts were prepared from P. aeruginosa strains by culturing cells in 10 ml of LB lacking NaCl at 37°C to an A580 of 0.4. The culture was centrifuged (5,000 × g for 10 min), and pellets were suspended in 2% of the original culture volume in fractionation buffer (10 mM Tris HCl [pH 8.0], 100 mM NaCl, 1 mM MgCl2). A 10-μl sample of this preparation was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting with anti-flagellum B antibodies. Polyclonal antiserum against flagella was elicited in New Zealand White rabbits (Covance) using flagella (0.75 mg) purified from sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Anti-flagellum antibodies were used in Western blots at a dilution of 1:25,000 with chemiluminescent reagents by procedures outlined by the manufacturer (Amersham), and film was exposed for 30 s prior to development. Lane 1, flagella B (250 ng) purified from PAO1. Lanes 2 through 11 contain extracts derived from strains FRD1 (mucA22), FRD875 (mucA22 algD::xylE aacC1), FRD440 (mucA22 algT::Tn501), FRD444 (mucA22 algB::Tn501), FRD810 (mucA22 algR::Ωstr), FRD831 (mucA22 ΔalgR::ΩaacC1), FRD1230 (mucA22 fliC::xylE aacC1), FRD1234 (mucA22 algT::Tn501 fliC::xylE aacC1), FRD1240 (mucA22 algT::Tn501 rpoN::xylE aacC1), and FRD1242 (mucA22 rpoN::xylE aacC1), respectively. The mucoid and motility phenotypes of the strains analyzed in each lane are depicted along the bottom. Motility assays were performed by inoculating a single colony into 0.3% L agar lacking sodium chloride. Following overnight growth at 37°C, motility was assessed qualitatively by examining colonies which spread beyond the point of inoculation (2).
FIG. 2.
TEM of P. aeruginosa strains. Magnification, ×18,090. (A) FRD875 (AlgT+mucA22 algD::xylE aacC1); (B) FRD440 (mucA22 algT::Tn501). TEM was performed by scraping individual colonies from plates cultured overnight and resuspending the samples in 15 μl of LB containing alpha lactalbumin carrier protein (500 μg/ml). Glow-discharged, Formvar-coated copper mesh grids were floated on this suspension for 2 min. The excess suspension was wicked off, and the grid was floated on a drop of 2% phosphotungstic acid for 1 min and wicked dry. TEM was performed on a Philips TEM 400 operated at 80 kV.
One potential explanation for the loss of flagellum expression in mucoid strains was that expression of alginate blocked the secretion of flagellin or the assembly of a functional flagellum. This was apparently not the case as a nonmucoid FRD1 derivative with an insertion in the algD gene (FRD875) remained nonmotile and lacked a flagellum (Fig. 1, lane 3; Fig. 2A). Taken together, these data suggest an inverse coordinate regulation between alginate synthesis and flagellum production.
Mutations in algB or algR do not affect flagellum expression.
ς22 directs the expression of several alginate transcriptional regulators (8, 22). These include algB and algR, encoding response regulators (8, 12). To determine if the ς22-dependent inhibition of flagellum synthesis was mediated through algB or algR, whole-cell extracts of isogenic algB and algR mutants were analyzed for flagellum expression and motility (Fig. 1). The algB mutant FRD444 (22) did not express flagella (Fig. 1, lane 5) and was nonmotile. Likewise, two algR mutants, FRD810 (22) and FRD831 (12), lacked motility and failed to synthesize flagella (Fig. 1, lanes 6 and 7, respectively). This suggests that the ς22-mediated inhibition of flagellum synthesis does not require the ς22-dependent algB or algR gene products.
Overexpression of ς22 in a motile P. aeruginosa isolate inhibits flagellum expression and motility.
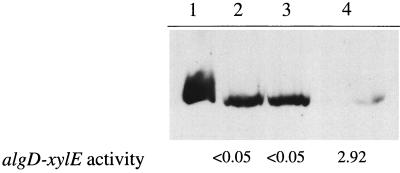
A prediction from the results above is that overexpression of ς22 in a motile P. aeruginosa strain should inhibit flagellum synthesis. This was shown to be the case in the experiment whose results are depicted in Fig. 3. The full-length algT gene was cloned by PCR amplification of P. aeruginosa FRD1 genomic DNA into the expression vector pMMB503EH (M. Bagdasarian), resulting in pWG21. The primers algT2 (5′-CGGGATCCTCAGGCTTCTCGCAACAAAGG-3′) and algT3 (5′-GGAATTCGAAGAGGAGCTTTCATG-3′) were used for PCR amplification with Taq polymerase by previously outlined conditions (3). The algT gene in pWG21 was sequenced, and the sequence was found to be identical to that published previously (4). Plasmid pWG21 or pMMB503EH was introduced into the motile strain P. aeruginosa WFPA14. WFPA14 was generated by allelic exchange of PAO1 wild-type algD with an algD::xylE aacC1 cassette from pDJW530 (21) by previously described techniques (12, 21). WFPA14 was chosen because the algD::xylE fusion provides a convenient screen for ς22 activity. A Western blot of whole-cell extracts demonstrated that expression of ς22 in WFPA14 inhibited flagellum expression (Fig. 3, compare lanes 3 and 4). ς22 expressed from pWG21 was active, since an increase in algD::xylE levels was observed (Fig. 3) (XylE assays were determined as described previously [21]). ς22-mediated inhibition of flagellum synthesis was also observed in the parental, motile strain PAO1 as well as in the serotype A strain PAK (data not shown).
FIG. 3.
Expression of ς22 in the nonmucoid, motile isolate PAO1 results in the inhibition of flagellum expression. Whole-cell extracts of PAO1-derived cells were analyzed on a Western blot probed with anti-flagellum B antibodies. Lane 1, a sample of purified flagella B (250 ng). Lanes 2 through 4 contain whole-cell extracts derived from strains WFPA14 (algD::xylE aacC1), WFPA14/pMMB503EH (vector), and WFPA14/pWG21 (algT), respectively. The algD::xylE activities (nanomoles of 2-hydroxymuconic semialdehyde/min/A540 of culture) of strains analyzed are indicated at the bottom of each lane.
The ς22-mediated inhibition of flagellum expression occurs through fliC transcription.
To determine if ς22-mediated inhibition of flagellum expression occurs through transcription of one or more flagellar genes, strains with fliC::xylE aacC1 or rpoN::xylE aacC1 operon fusions were constructed in P. aeruginosa FRD1 (mucA22) and FRD440 (mucA22 algT::Tn501). We chose rpoN and fliC since these genes represent early and late markers of flagellar gene expression, respectively (2, 20). Both the rpoN gene, which encodes the alternative sigma factor, ς54, as well as fliC, encoding the flagellar subunit protein flagellin, are essential for flagellum synthesis and motility (6, 20). A portion of fliC was obtained by PCR amplification of PAO1 genomic DNA with the primers fliC1 (5′-GCCTGCAGATCTCCAAC-3′) and fliC2 (5′-GCAGCTGGTTGGCCTGG-3′). The PCR fragment was cloned into pUC18 (23), resulting in pDJW567. The presence of fliC in pDJW567 was verified by DNA sequencing. The fliC gene of pDJW567 was subcloned into the gene replacement vector pEX100T (9) to generate pWG26. This plasmid was digested with AgeI, which cleaves within the fliC coding sequence, the ends were treated with Klenow fragment, and a 2.2-kb SmaI xylE aacC1 fragment from pX1918 (18) was inserted. The resulting plasmid, pDJW600, was used for generating chromosomal fliC::xylE aacC1 insertions by techniques outlined previously (12, 21). To create rpoN::xylE aacC1 operon fusions, a 10-kb fragment containing rpoN was obtained by BamHI digestion of pKI10 (10) and cloned into the gene replacement vector pDJW525 (12). The resulting plasmid, pWG23, was digested with NsiI, which cleaves within rpoN, and the 2.4-kb xylE aacC1 PstI cassette from pX1918GT (18) was inserted. The resulting plasmid, pWG24, was used for generating chromosomal rpoN::xylE aacC1 insertions.
Transcription levels of rpoN were similar in strains harboring either the wild-type or the algT::Tn501 allele, decreasing by only about 20% from 8.1 to 6.4 U of XylE in the algT mutant. By contrast, expression of the fliC::xylE operon fusion was increased by about 100-fold in the algT mutant from 0.24 to 22.4 U of XylE. This suggests that the ς22-dependent control of flagellum synthesis and motility occurs through inhibition of fliC transcription. As expected, stains with chromosomal fliC::xylE aacC1 or rpoN::xylE aacC1 insertions did not express any flagella detectable by Western blotting and were nonmotile (Fig. 1, lanes 8 through 11).
Published data (6, 8, 15, 16, 22) as well as data discussed in this paper can be summarized by a working model for explaining the coordinate control of flagellum synthesis with alginate. CF patients are colonized by motile P. aeruginosa strains, and there is evidence that flagella play a critical role in the early events of colonization of the lungs of CF patients and abiotic surfaces (2, 6, 15). Following or coincidental with this initial attachment, type IV fimbriae and twitching motility are utilized to form tighter association with the epithelium and to initiate microcolony formation. In the lungs of CF patients, impaired activity of mucocilliary clearance, decreased defensin activity, decreased bacterial uptake and desquamation, and increased expression of ganglioside surface receptors for P. aeruginosa favor progression of the infection. Concurrently with these processes airway inflammation occurs. Because of this, and the fact that they are convenient ligands for phagocytic cells, flagella become detrimental to P. aeruginosa. In CF, a strong selection is imposed on the organisms and mucA mutants predominate. Loss of the anti-sigma factor MucA function results in increased expression of the AlgT (ς22) regulon. The algT gene modulates a hierarchy of gene expression, leading to expression of the alginate operon. Induction of the ς22 regulon also appears to inhibit flagellum synthesis and motility.
The negative control of flagellum expression by ς22 could theoretically occur at any point in the flagellar hierarchy which includes the alternative sigma factors rpoN and fliA as well as several positive transcriptional regulators (1, 17, 19, 20). The ς22-mediated inhibition occurs through transcriptional control of fliC but is independent of rpoN. Since most sigma factors lack the ability to bind DNA without being complexed with core RNA polymerase (5), it is unlikely that ς22 directly represses flagellar gene expression. Instead, ς22 probably controls the expression of a negative effector of flagellum synthesis. Understanding the specific point in the flagellar genetic pathway in which ς22-dependent inhibition occurs is a focus of future studies. These experiments will provide insights into the pathogenesis of P. aeruginosa in chronic lung infections of CF patients and yield information regarding the natural role of the ς22 regulon in P. aeruginosa.
Acknowledgments
This work was supported by Public Health Service grant 5R01 HL58334 (D.J.W.) from the National Heart, Lung, and Blood Institute.
We are grateful to S. Lory and N. Baker for advice and for providing antiserum and strains. Assistance in TEM was provided by K. Grant and G. Jerome of the Micromed electron microscopy facility in the Department of Pathology at WFUBMC. We are also indebted to G. O’Toole, C. Whitchurch, and J. Mattick for helpful discussions. The DNA Synthesis Core Laboratory of the Cancer Center of Wake Forest University provided oligonucleotides. E. Jung of the DNA Sequencing Core Lab performed DNA sequencing. Both facilities are supported in part by NIH grant CA-12197.
REFERENCES
- 1.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J Bacteriol. 1997;179:5574–5581. doi: 10.1128/jb.179.17.5574-5581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arora S K, Ritchings B W, Almira E C, Lory S, Ramphal R. The Pseudomonas aeruginosa flagellar cap protein, FliD, is responsible for mucin adhesion. Infect Immun. 1998;66:1000–1007. doi: 10.1128/iai.66.3.1000-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baynham P J, Wozniak D J. Identification and characterization of AlgZ, an AlgT-dependent DNA binding protein required for Pseudomonas aeruginosa algD transcription. Mol Microbiol. 1996;22:97–108. doi: 10.1111/j.1365-2958.1996.tb02659.x. [DOI] [PubMed] [Google Scholar]
- 4.Devries C A, Ohman D E. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternative sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dombroski A J, Walter W A, Gross C A. Amino-terminal amino acids modulate ς-factor DNA-binding activity. Genes Dev. 1993;7:2446–2455. doi: 10.1101/gad.7.12a.2446. [DOI] [PubMed] [Google Scholar]
- 6.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Role of flagella in pathogenesis of Pseudomonas aeruginosa pulmonary infection. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flynn J L, Ohman D E. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J Bacteriol. 1988;170:1452–1460. doi: 10.1128/jb.170.4.1452-1460.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoang T T, Karkhoff-Schweizer R R, Kutchma A J, Schweizer H. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: applications for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 10.Ishimoto K S, Lory S. Formation of pilin in Pseudomonas aeruginosa requires the alternative ς factor (RpoN) of RNA polymerase. Proc Natl Acad Sci USA. 1989;86:1954–1957. doi: 10.1073/pnas.86.6.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luzar M A, Thomassen M J, Montie T C. Flagella and motility alterations in Pseudomonas aeruginosa strains from patients with cystic fibrosis: relationship to patient clinical conditions. Infect Immun. 1985;50:577–582. doi: 10.1128/iai.50.2.577-582.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma S, Selvaraj U, Ohman D E, Quarless R, Hassett D J, Wozniak D J. Phosphorylation-independent activity of the response regulators AlgB and AlgR in promoting alginate biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1998;180:956–968. doi: 10.1128/jb.180.4.956-968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahenthiralingam E, Campbell M E, Speert D P. Nonmotility and phagocytic resistance of Pseudomonas aeruginosa isolates from chronically colonized patients with cystic fibrosis. Infect Immun. 1994;62:596–605. doi: 10.1128/iai.62.2.596-605.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montie T C, Craven R C, Holder I A. Flagellar preparations from Pseudomonas aeruginosa: isolation and characterization. Infect Immun. 1982;35:281–288. doi: 10.1128/iai.35.1.281-288.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;29:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 16.Pier G B. Pseudomonas aeruginosa: a key problem in cystic fibrosis. ASM News. 1998;6:339–347. [Google Scholar]
- 17.Ritchings B W, Almira E C, Lory S, Ramphal R. Cloning and phenotypic characterization of fleS and fleR, new response regulators of Pseudomonas aeruginosa which regulate motility and adhesion to mucin. Infect Immun. 1995;63:4868–4876. doi: 10.1128/iai.63.12.4868-4876.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schweizer H P, Hoang T T. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene. 1995;158:15–22. doi: 10.1016/0378-1119(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 19.Starnbach M N, Lory S. The fliA (rpoA) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol Microbiol. 1992;6:459–469. doi: 10.1111/j.1365-2958.1992.tb01490.x. [DOI] [PubMed] [Google Scholar]
- 20.Totten P A, Lara J C, Lory S. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J Bacteriol. 1990;172:389–396. doi: 10.1128/jb.172.1.389-396.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woolwine S, Wozniak D J. Identification of an Escherichia coli pepA homolog and its involvement in suppression of the algB phenotype in mucoid Pseudomonas aeruginosa. J Bacteriol. 1999;181:107–116. doi: 10.1128/jb.181.1.107-116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wozniak D J, Ohman D E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanisch-Perron C, Vieira J, Messing J. Improved M13 cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]