Abstract
Chimeric antigen receptor (CAR) engineering of natural killer (NK) cells is promising, with early-phase clinical studies showing encouraging responses. However, the transcriptional signatures that control the fate of CAR-NK cells after infusion and factors that influence tumor control remain poorly understood. We performed single-cell RNA sequencing and mass cytometry to study the heterogeneity of CAR-NK cells and their in vivo evolution after adoptive transfer, from the phase of tumor control to relapse. Using a preclinical model of noncurative lymphoma and samples from a responder and a nonresponder patient treated with CAR19/IL-15 NK cells, we observed the emergence of NK cell clusters with distinct patterns of activation, function, and metabolic signature associated with different phases of in vivo evolution and tumor control. Interaction with the highly metabolically active tumor resulted in loss of metabolic fitness in NK cells that could be partly overcome by incorporation of IL-15 in the CAR construct.
Loss of metabolic fitness is a mechanism of resistance after CAR-NK cell therapy that could be overcome by IL-15.
INTRODUCTION
Chimeric antigen receptor (CAR) T cell therapy has emerged as a promising approach in which T cells are engineered to recognize a specific target on tumor cells. Several CAR T cell trials have shown durable remissions in patients with relapsed or refractory hematologic cancers (1–6), resulting in U.S. Food and Drug Administration (FDA) approvals of multiple CAR T cell products (7). Although these clinical responses are very encouraging, the potent systemic immune activation responsible for the efficacy of CAR T cell treatment may induce life-threatening toxicities (8).
NK cells are an attractive resource for immunotherapy. NK cells are innate cytotoxic lymphocytes that have intrinsic antitumor function and play an important role in surveillance and elimination of cancer (9). While T cell receptors recognize peptide antigens presented on human leukocyte antigen (HLA) molecules, NK cells can mediate cytotoxicity and induce death of target in an HLA-unrestricted manner. Also, unlike CAR T cells, which are most commonly derived from autologous donors, NK cells can be generated from a wide range of allogeneic sources without inducing graft-versus-host disease, allowing for the development of well-characterized cell products that can be made available off the shelf for clinical use (10). Cytokine release syndrome and neurotoxicity are less likely to occur with CAR-NK therapy (11), possibly due to CAR-NK cells having a different cytokine profile from CAR T cells (12).
In the clinical setting, although CAR T cell therapy has led to initial impressive results, between 30 and 50% of patients experienced relapse after having achieved complete remission (2, 4). Similarly, despite showing an excellent safety profile and encouraging responses, a subset of patients treated with CAR-NK cells developed progressive disease (11). Therefore, it is important to identify the underlying mechanisms responsible for disease relapse. Extensive studies of CAR T cell therapy describe resistance as often resulting from antigen loss or T cell dysfunction (13, 14). For CAR-NK cell therapy, however, the mechanisms driving resistance and disease relapse have not been fully elucidated.
Here, we investigated mechanisms that may be associated with resistance to CAR-NK cell therapy. To accomplish this, we designed an in vivo study to recapitulate the kinetics of CAR-NK cell activity in a noncurative CD19+ lymphoma model. We used single-cell transcriptomic and proteomic profiling to interrogate the heterogeneity in CAR-NK cells and their functional, phenotypic, and transcriptional evolution over time. Our results reveal a previously unidentified mechanism of tumor resistance to CAR-NK cell therapy characterized by transcriptional signatures indicating loss of metabolic fitness associated with NK cell dysfunction. The antitumor effect can be improved by armoring CAR19 NK cells with interleukin-15 (IL-15) to increase their metabolic fitness and effector function. Moreover, we show that administering a second dose of CAR19/IL-15 NK cells at the time of minimal measurable disease can result in enhanced tumor control in vivo. Finally, by comparing the transcriptional profiles of CAR-NK cells from a responder and a nonresponder patient after treatment with CAR19/IL-15 NK cells, we reproduce our observations that the ability of CAR-NK cells to clear the tumor is closely linked to their metabolic fitness.
RESULTS
Single-cell transcriptome profiling reveals distinct phenotypic, functional, and metabolic profile of NK cells after CAR engineering
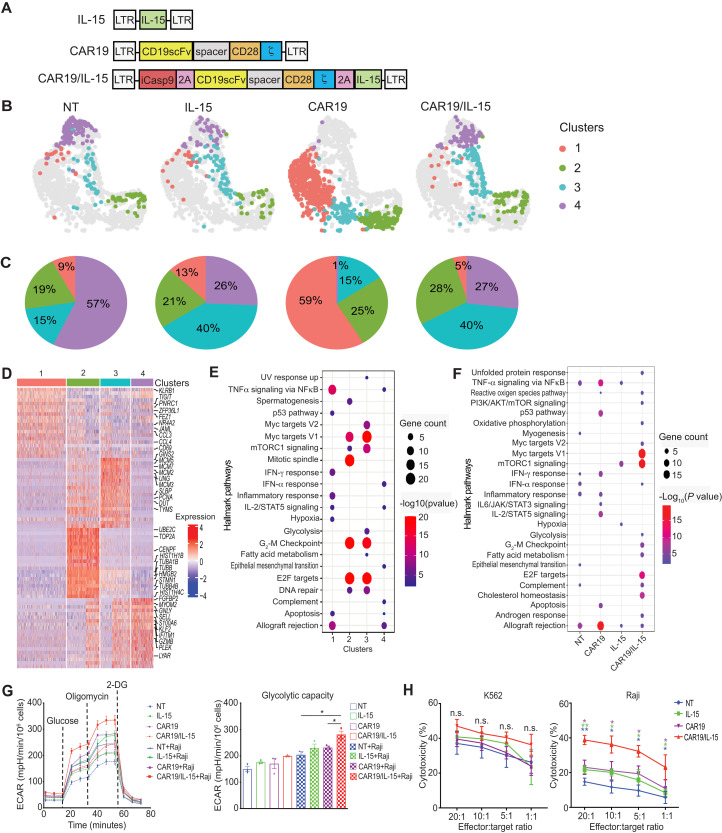
We previously showed that CAR-engineered cord blood (CB) NK cells displayed higher antitumor cytotoxicity in vitro and in vivo when engineered to coexpress a CAR and IL-15 (15). To further investigate how IL-15 modulates and/or synergizes with CAR-mediated signaling in NK cells, we genetically engineered NK cells to express either CAR19 alone, IL-15 alone, or CAR19 + IL-15 (herein referred to as CAR19/IL-15) (Fig. 1A). Nontransduced (NT) NK cells were used as control. After activation, transduction, and in vitro expansion (day 14 of culture), NK cells were collected for single-cell RNA sequencing (scRNA-seq). Upon completion of quality filtering, a total of 1613 cells were retained from four products for subsequent analysis (see Materials and Methods and table S1). Data visualization by uniform manifold approximation and projection (UMAP) (16) and unsupervised clustering identified four main clusters, each with a distinct cell population for the different engineered NK cell groups when compared to NT-NK cells (Fig. 1B). While cluster 3 comprised a large portion (40%) of IL-15 and CAR19/IL-15 NK cells, cluster 1 (59%) and cluster 4 (57%) were the most abundant clusters in CAR19 and NT-NK cells, respectively (Fig. 1, B and C). To further evaluate the transcriptomic profiles of the different engineered NK cells, we identified up-regulated differentially expressed genes (DEGs) and the corresponding gene-enriched pathways at both cluster and product levels (Fig. 1, D to F). We found that CAR19 NK cells were predominantly enriched in pathways associated with immune cell activation (cluster 1), while both IL-15 and CAR19/IL-15 NK cells showed a distinct increase in expression of genes associated with DNA replication, cell division, fatty acid metabolism, and glycolysis as well as mammalian target of rapamycin (mTOR) and c-Myc signaling (cluster 3) (Fig. 1E), thereby suggesting a role for IL-15 in NK cell proliferation as well as aerobic glycolysis. In addition to enrichment for immune-associated pathways, CAR19/IL-15 NK cells showed the strongest enrichment for metabolic pathways among the four products (Fig. 1F). We next studied the mitochondrial metabolism and glycolytic potential of engineered NK cells at baseline and following short-term stimulation with Raji lymphoma cells to investigate whether IL-15 signaling enhanced the metabolic potential of CAR-NK cells. CAR19 NK cells armed with IL-15 showed higher overall metabolic activity than CAR19 NK or IL-15 NK cells alone as indicated by substantially elevated extracellular acidification rate (ECAR) in a Seahorse extracellular flux assay (Fig. 1G).
Fig. 1. CAR-NK cells have a distinct single-cell transcriptome and metabolic profile.
(A) Schematic of vectors used to transduce CB-NK cells. (B) UMAP visualizing transcriptional clusters of NK cells across products. (C) Pie charts showing the relative proportions of the NK cell clusters. (D) Top up-regulated genes in each of the four NK cell clusters. Columns correspond to cells, and rows correspond to genes. Color represents Z-transformed expression. (E and F) Hallmark pathways enriched (Fisher’s test adjusted P value < 0.01) in genes up-regulated in NK cell clusters (E) or NK cell products (F). Color scale corresponds to −log10 transformed false discovery rate (FDR)–adjusted P values. (G) ECAR measured by Seahorse assays for NK cell products alone (dashed lines) or after coculture with Raji cells for 2 hours and then purified (solid lines). Bar graphs in the right panel summarize their glycolytic capacity. A representative graph of three independent experiments is shown. (H) 51Cr-release assay of NK cell products against K562 (left) and Raji (right) targets (n = 3 donors). Purple asterisks: CAR19/IL-15 versus CAR19 NK cells. Green asterisks: CAR19/IL-15 versus IL-15 NK cells. Blue asterisks: CAR19/IL-15 versus NT NK cells. The error bars represent mean values with SD. *P ≤ 0.05; **P ≤ 0.01; n.s., not significant. The statistical significance was determined by paired t test in (G) and (H).
Finally, we evaluated the antitumor function of the different NK cell products in response to K562 tumor cells (sensitive to NK cytotoxicity) and Raji lymphoma cells (resistant to NK cytotoxicity) using chromium (51Cr)–release assay. All NK cell groups were equally efficient at killing K562 targets (Fig. 1H, left panel); however, CAR19/IL-15 NK cells exerted substantially greater cytotoxicity against Raji cells when compared to CAR19, IL-15, or NT-NK cells, suggesting a synergistic effect between CAR-mediated activation and IL-15 signaling that contributed to potentiating NK cell response (Fig. 1H).
IL-15 improves CAR-NK cell persistence and delays dysfunction in vivo
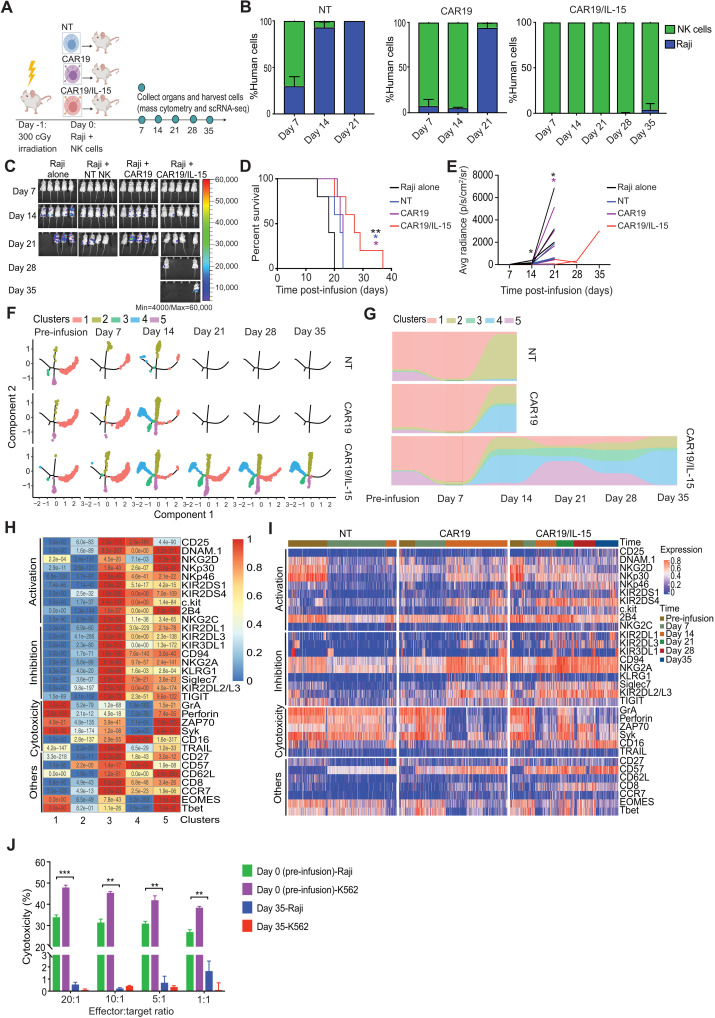
To determine how IL-15 enhances CAR-mediated function in vivo, we next compared the persistence and antitumor activity of CAR19, CAR19/IL-15, and NT-NK cells in a noncurative Raji lymphoma mouse model, as previously described (15). One cohort was kept for survival monitoring, while a parallel cohort was designated for collection of cells from bone marrow, spleen, liver, and blood every 7 days for a period of 35 days (Fig. 2A). As we have previously reported (15), mice bearing Raji lymphoma and treated with one dose of CAR19/IL-15 NK cells had notably lower tumor burden and improved survival compared to animals receiving NT-NK or CAR19 NK cells (Fig. 2, B to E, and table S2). However, although the response was superior in this group, mice treated with CAR19/IL-15 NK cells eventually succumbed to disease (Fig. 2, C to E).
Fig. 2. IL-15 promotes the persistence of CAR-NK cells in vivo.
(A) Schematic timeline of experiments (n = 13 to 15 mice per group; 5 mice were followed for survival and 8 to 10 were assigned for single-cell analyses; 2 mice per group were sacrificed at each time point). (B) Bar plots of NK and Raji cell percentages in samples collected at multiple time points from mice treated with NT, CAR19, or CAR19/IL-15 NK cells. (C) Bioluminescence imaging (n = 5 mice per group). Kaplan-Meier plots (D) showing mice survival and average radiance (E). Black asterisks: Raji alone versus CAR19/IL-15. Blue asterisks: NT versus CAR19/IL-15. Purple asterisks: CAR19 versus CAR19/IL-15. (F) Trajectory evolution of NK cell products from pre-infusion (day 0) to day 35 post-infusion (n = 8 mice per group, 2 mice analyzed at each time point). No data were available at days 21 to 35 in NT and CAR19 groups due to limited in vivo persistence. (G) Relative proportion of NK cell clusters. (H) Heatmap showing the average expression levels of the proteomic markers for the five clusters. P values in each square were calculated using unpaired t test by comparing the levels of marker abundance for cells in their cluster versus cells from all other cluster. (I) Heatmap of protein expression at different time points across products. (J) 51Cr-release assay of CAR19/IL-15 day 0 or 35 days after infusion against K562 or Raji. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. The P values were determined by log-rank (Mantel-Cox; D) and unpaired t test in (E) and (J).
The results from our in vivo and in vitro studies led to the hypothesis that, although IL-15 enhances the metabolic fitness and antitumor activity of CAR-NK cells, NK cells eventually become dysfunctional, thus allowing the tumor to relapse. To test this hypothesis, we applied high-parameter mass cytometry to analyze the functional phenotype of adoptively infused human NK cells collected from mouse tissues at various time points during the study (Fig. 2A; table S3 includes the list of antibodies). Using Monocle v2 (17), we identified trajectories consisting of five distinct NK cell clusters, which evolved dynamically over time in the three NK cell treatment groups (NT, CAR19, and CAR19/IL-15 NK cells) (Fig. 2, F to H, and fig. S1). Our findings revealed that the phenotypic profiles of the various NK products in the blood and tissue at the early (days 7 to 14), mid (day 21), and late time points (days 28 to 35) progressively diverged from that of pre-infusion (day 0). In the early time point, cluster 1, characterized by a mature phenotype (expressing high levels of Eomes and T-bet) and an activated, cytotoxic profile as shown by activation of ZAP70 and Syk kinases, and high expression of granzyme A (GrA) and perforin, was the dominant cluster for all treatment groups. However, after day 7, adoptively infused NK cells derived from different products followed different trajectories in vivo (Fig. 2, F and G). By day 14, NT-NK cells were rapidly dominated by cluster 2 (92.7%), characterized by reduced activation and function as shown by down-regulation of activating receptors such as DNAM1, NKG2D, NKp30, and NKp46, and low cytotoxic granule production (Fig. 2H), and were no longer detected after day 14. Consistent with this observation, NT-NK cells were unable to control tumor cells past day 7. CAR19 NK cells could control tumor growth until day 14 (Fig. 2B) and predominantly consisted of clusters 4 (52.6%) and 2 (37%). Cluster 4 was characterized not only by higher expression of inhibitory markers (NKG2A, Siglec7, and TIGIT) and lower expression of transcription factors (Eomes and T-bet) and cytolytic proteins (GrA, perforin) but also by up-regulation of cytokine receptors IL-2R (CD25), stem cell factor receptor (c-Kit), and coactivating receptors (2B4, CD16, DNAM1, and NKG2C) (Fig. 2, H and I), which is consistent with a previously activated phenotype and eventual exhaustion. CAR19/IL-15 NK cells were detected for much longer, with a sizable (12.5%) fraction of functional cluster 3, characterized by high expression of activation and inhibition markers along with modest expression of cytolytic proteins (GrA and perforin; Fig. 2H) and persistence of cluster 1 until day 28. In addition, between days 14 and 28, CAR19/IL-15 NK cells displayed the reemergence of cluster 5, characterized by high expression of not only activation markers (NKG2D, Nkp30, 2B4, and DNAM1), cytolytic proteins, and Eomes and T-bet but also inhibitory receptors (KIRs and NKG2A) (Fig. 2H). Consistent with the presence of multiple functional clusters (clusters 1, 3, and 5; Fig. 2, F to I) for much of the duration of the experiment, CAR19/IL-15 NK cells were able to control tumor growth for longer (Fig. 2B). However, these functional clusters were eventually dominated by nonfunctional clusters 2 and 4 (24% and 56%, respectively). These observations suggest that NK cells evolve in vivo following adoptive infusion, with different kinetics of activation and eventual exhaustion, with IL-15 coexpression resulting in longer persistence, and an activated profile associated with efficient control of the tumor that is then superseded by a dysfunctional phenotype that coincides with tumor progression.
To evaluate the potency of CAR19/IL-15 NK cells as they evolve in vivo, we sort-purified human NK cells from murine blood, liver, and spleen harvested on day 35 after infusion and compared their in vitro cytotoxicity against K562 or Raji targets to that of the pre-infusion product (day 0) by 51Cr-release assay. Day 35 CAR19/IL-15 NK cells displayed substantial impairment in cytotoxic function and were unable to kill K562 or Raji tumor cells (Fig. 2J), suggesting the acquisition of global NK cell dysfunction in CAR-NK cells over time. Together, these data support our hypothesis that IL-15 boosts the activity of CAR-engineered NK cells and can sustain NK cell function and persistence for a longer time. However, NK cells eventually become dysfunctional, resulting in tumor progression.
Coevolution of NK cells with tumor in vivo
Although IL-15 coexpression enhanced CAR-NK function and persistence, tumor relapse occurred in all treatment groups. To understand the mechanisms associated with treatment resistance, we sort-purified NK cells and Raji tumor cells from the bone marrow of animals at multiple time points after infusion and performed scRNA-seq to interrogate transcriptional changes at single-cell resolution in NK cells upon interaction with the tumor in vivo. To achieve unbiased comparison among groups, scRNA-seq data from multiple treatment groups and time points were pooled for downstream analyses. After removing low-quality cells, transcriptomes of 3447 NK cells (defined as NKG7 > 0 and CD19 = 0; see Materials and Methods) and 3818 tumor cells (defined as NKG7 = 0 and CD19 or MS4A1 > 0) were profiled (figs. S2 and S3).
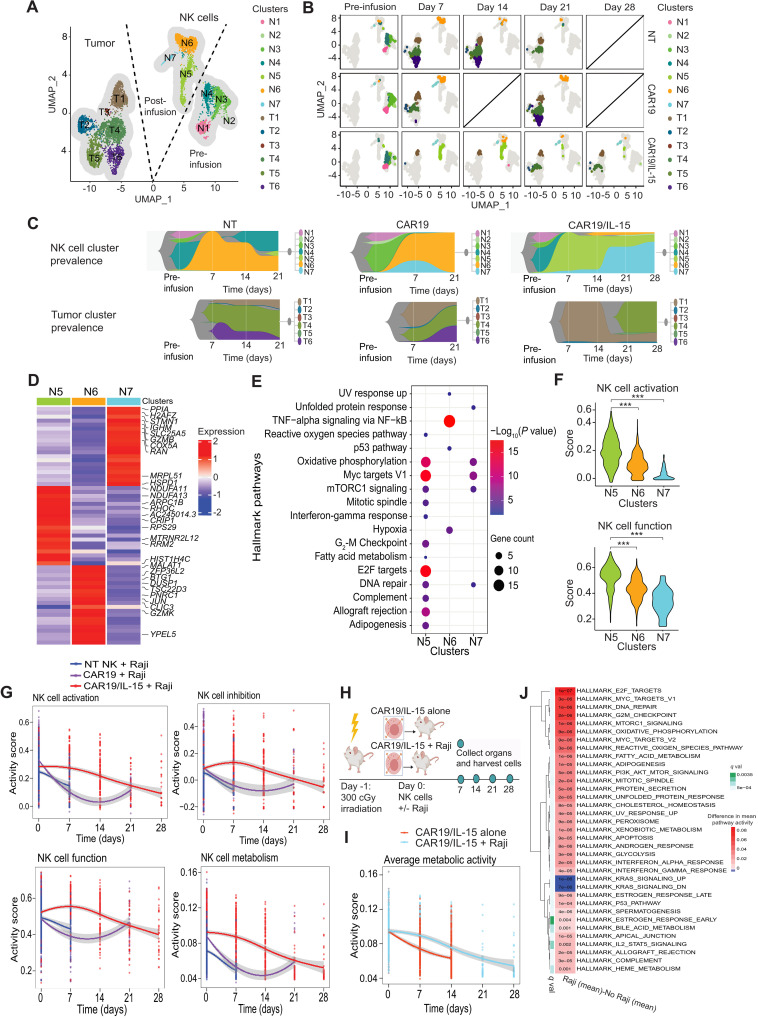
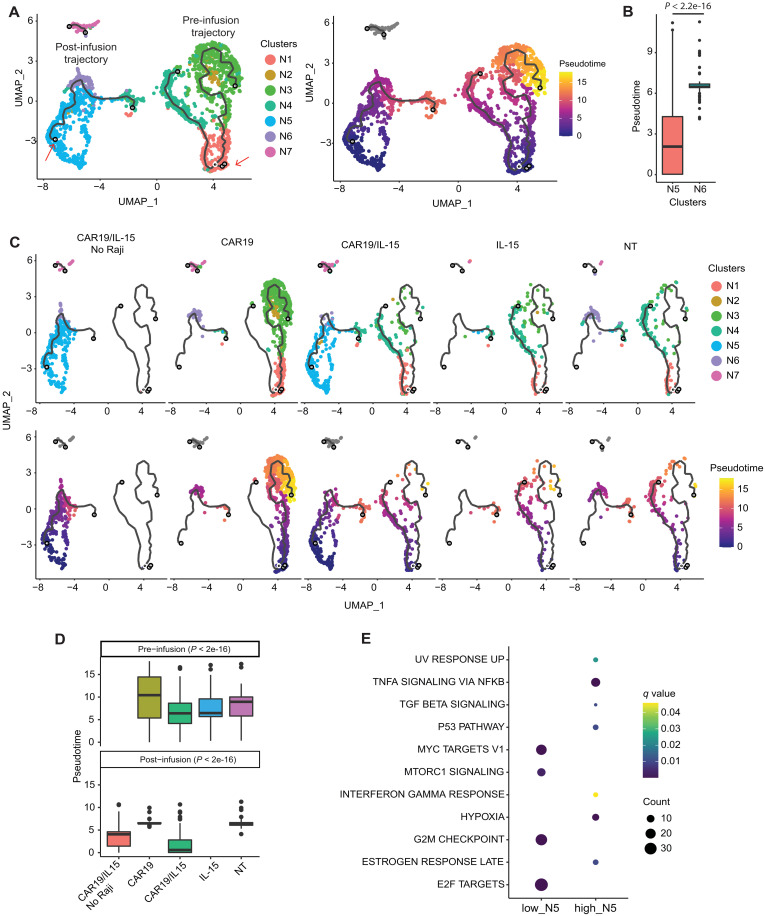
To study the distinct transcriptional states that exist in the different NK cell products and the tumor cells as they coevolve in vivo, we visualized the cells using UMAP analysis. Like the observation from mass cytometry data (Fig. 2F), pre-infusion NK cells differed from post-infusion NK cells with respect to their transcriptional profiles, suggesting that the products evolved in vivo after interacting with the tumor (Fig. 3A). We subsequently applied unsupervised clustering to analyze the evolution of both tumor and NK cells over the course of treatment. Our analysis showed NK cell cluster 5 (N5) as overwhelmingly predominant (>94%) in the CAR19/IL-15 NK group at day 7 after infusion and persisting during the 4 weeks of treatment (Fig. 3, B and C). As expected, tumor burden and heterogeneity in animals treated with CAR19/IL-15 NK cells were remarkably lower than that observed in CAR19 and NT-NK cell groups (Fig. 3B), with tumor cells initially predominantly confined to Raji tumor cluster 1 (T1). However, as the NK N5 cluster began to diminish at day 21, cluster N7 increased (Fig. 3C) and remained the predominant NK cell population through day 28 (>65%). Notably, this shift in NK transcriptional profile was accompanied by tumor relapse in these animals, with the emergence of tumor cluster 4 (T4) in CAR19/IL-15 animals (Fig. 3B). Tumor cells in cluster T1 were detected at all time points irrespective of the NK product the animals received, indicating that they might have inherent resistance to NK cell–mediated clearance (Fig 3, B and C).
Fig. 3. In vivo evolution of CAR-NK cells and Raji tumor.
(A) UMAP visualizing transcriptomic clusters of NK and Raji cells (n = 8 mice per group, 2 mice per group were sacrificed at each time point). (B) UMAP from (A) faceted by product and time point. Data are not available on day 14 (CAR19) or on day 28 (NT and CAR19) due to technical issues and limited in vivo persistence of NK cells. (C) Fish plots of NK and tumor cell clusters over time. (D) Z-transformed mean expression of top up-regulated genes in post-infusion clusters. (E) Hallmark pathway enriched in them (Fisher’s test adjusted P value < 0.01). (F) Violin plots of NK activation (top) and function (bottom) scores in post-infusion NK clusters. P value was calculated with Wilcoxon rank sum test. ***P < 0.01. (G) Trends in NK activation, inhibition, function, and metabolism across products before infusion (day 0) to day 28 after infusion. (H) Schematic of in vivo studies to test CAR19/IL-15 NK behavior in the presence or absence of tumor cells (n = 8 mice per group; 2 mice were sacrificed per time point per treatment group). (I) Pathway activity of the in vivo trend in NK cell metabolism for CAR19/IL-15 when exposed to Raji or not. (J) Heatmap of differential mean pathway activity (q < 0.01) between CAR19/IL-15 NK cells exposed or not to tumor. (G to I) The curves are the mean scores, and the shaded region is the 95% confidence interval of Student’s t-distribution at each data point.
To further evaluate the transcriptomic profiles of NK cells as they evolve in vivo over time, we conducted an analysis for DEGs (Fig. 3D) and the corresponding enriched pathways (Fig. 3E) for the different NK cell clusters. We found that compared to the other post-infusion NK cells, NK cells within the N5 cluster, which was prevalent in the first 3 weeks in CAR19/IL-15 NK-treated animals, mostly expressed hallmark pathways associated with immune function [allograft rejection and interferon-γ (IFN-γ) response], DNA replication, cell division, and metabolic activity such as oxidative phosphorylation (OXPHOS) and c-Myc signaling (Fig. 3E), with the top overexpressed genes associated with the mitochondrial electron transport chain (ETC) genes (NDUFA11 and NDUFA13) and ribosomal protein S9 (RPS29). N6, in comparison, was characterized by overexpression of genes in the tumor necrosis factor–α (TNF-α)–nuclear factor κB (NFκB) signaling pathway, although these NK cells also showed overexpression of the hypoxia pathways (Fig. 3E), which is associated with NK cell dysfunction (18) (Fig. 3D). Using single-sample gene set enrichment analysis (ssGSEA) (19) of the relevant gene sets (see Materials and Methods), we showed that the N6 and N7 clusters, which emerged at later time points in CAR19/IL-15 NK-treated animals and coincided with recurrence of tumor cells, had substantially lower activation and function scores when compared to N5 (Fig. 3F; P < 10−28), pointing to gradual exhaustion of NK cells in the CAR19/IL-15 group.
We also examined the dynamics of gene set activity scores for NK cell activation, inhibition, function, and metabolism in NT, CAR19, and CAR19/IL-15 NK cells over time (see Materials and Methods and Fig. 3G). Although the level of inhibition, activation, and function in CAR19/IL-15 NK cells started at a similar level to that observed for CAR19 NK cells on day 0 (pre-infusion), post-infusion CAR19/IL-15 cells showed consistently higher activity and were characterized by higher metabolic activity, with the temporal patterns closely mirroring NK activation and function. In contrast, NT-NK and CAR19 NK cells exhibited a high level of function early on, but they experienced a rapid decrease in activity and were no longer detectable beyond day 7 and day 21, respectively. In summary, these data demonstrate that CAR19/IL-15 NK cells maintained increased functional phenotype, metabolic fitness, and persistence, resulting in enhanced cell activity and antitumor response that decreased over time, which was associated with tumor recurrence.
We next asked if CAR19/IL-15 NK cells require antigen-mediated activation from the tumor to persist and function. Thus, we performed an in vivo study to test CAR19/IL-15 NK cell behavior in the presence or absence of tumor cells. Animals were sacrificed every 7 days, and cells from bone marrow were harvested and submitted for scRNA-seq (Fig. 3H). Our findings showed that CAR19/IL-15 NK cells displayed prolonged persistence (fig. S4) and higher metabolic activity in the presence of tumor compared to the no tumor group (at days 7 and 14) (Fig. 3I). These data suggest that tumor cells provide CAR19/IL-15 NK cells with the signal to drive their proliferation and survival. We then compared hallmark pathway activity, inferred from gene expression using ssGSEA (see Materials and Methods), from CAR19/IL-15 NK cells exposed to tumor in vivo versus CAR19/IL-15 NK cells alone. Our findings confirmed that metabolic pathways, including OXPHOS and fatty acid metabolism, were up-regulated in CAR-NK cells exposed to tumor cells (Fig. 3J). Together, these data suggest that, upon initially engaging tumor cells, NK cells are activated and exhibit increased function and metabolism. Nevertheless, over time, NK cells can become dysfunctional and their antitumor response declines.
Tumor resistance to CAR-NK cell treatment secondary to loss of NK cell metabolic fitness
To understand the mechanisms by which tumor cells evade clearance by NK cells, we first studied the composition of tumor cells at the level of cell clusters identified from scRNA-seq across available time points from mice treated with different NK cell products (Fig. 3, A to C). CAR19/IL-15 NK cells rapidly eliminated Raji lymphoma clusters T2, T3, T4, T5, and T6, unlike CAR19 and NT-NK cell groups. There was also heterogeneity in the sensitivity of different tumor cells to CAR19/IL-15 NK cell therapy, with cluster T1 demonstrating primary resistance, while cluster T4 was initially eliminated but reappeared at later time points as N5 NK cells decreased, suggesting that N5 NK cells could keep these tumor cells in check initially but failed to completely clear the tumor (Fig. 3, B and C). These observations suggested that the different engineering strategies endowed NK cells with different potency against different tumor cell populations but that, over time, tumors develop mechanisms of resistance to evade NK cell immune surveillance.
We next sought to investigate the biological processes driving this shift in tumor resistance to NK cell cytotoxicity. To this end, we performed comparative analysis of pathway activity between the NK-resistant tumor clusters (T1 and T4) and the NK-sensitive tumor clusters (T2, T5, and T6). T2 and T6 clusters showed activation in the IFN response pathway, TNF-α, and IL-6/Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) signaling, indicating activation of proinflammatory signaling (fig. S5), while NK-resistant tumor clusters showed activation of pathways associated with proliferation and metabolism (MYC targets, G2-M checkpoint, mTOR signaling, OXPHOS, and fatty acid metabolism), suggesting that tumor cells with higher metabolic activity may evade NK cell recognition (fig. S5), likely by depleting the nutrient pool available in the tumor microenvironment (TME), resulting in the NK cell dysfunction observed over time (Figs. 2, F to J, and 3, F and G).
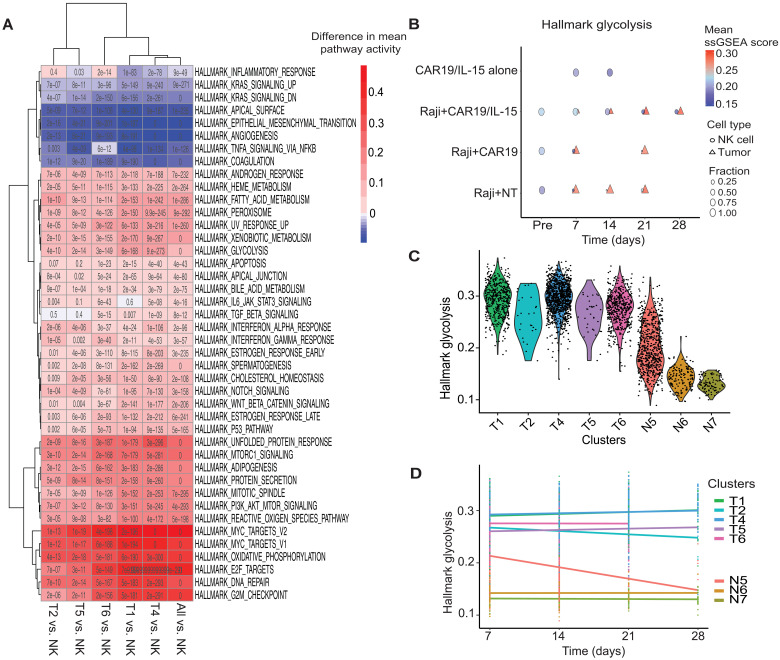
Thus, we next asked if resistance to CAR-NK cell therapy can be explained by relative differences in metabolic fitness between NK and tumor cells. Functional measurements of glycolysis confirmed that Raji tumor cells are substantially more metabolically active than NK cells (fig. S6A). Thus, we quantified and compared the activity of hallmark pathways between tumor and NK cells in our Raji lymphoma model (Fig. 4A; see Materials and Methods). Previous studies (20, 21) have reported that both glycolysis and fatty acid metabolism play a critical role in tumor cell proliferation. Therefore, we focused our analysis on these pathways along with OXPHOS. All three pathways showed higher activity in tumor cells across all NK products and time points where both tumor and NK cells were detected in vivo (Fig. 4B and fig. S6B). Additionally, at the level of DEGs, tumor cells expressed higher levels of metabolic enzymes like LDHA and LDHB relative to NK cells [q < 0.01, log2 fold change (FC) > 1]. We also observed overexpression of NDUFAB1, a subunit of NADH (reduced form of nicotinamide adenine dinucleotide) dehydrogenase, the largest complex in the ETC, as well as multiple mitochondrial ribosomal proteins (fig. S6C).
Fig. 4. Metabolic activity of CAR-NK cells and tumor cells in vivo.
(A) Heatmap of difference in pathway activity between indicated tumor and post-infusion NK cell clusters. Only pathways significantly dysregulated in at least one comparison are reported (q < 0.01; q values are reported in each cell). (B) Dot plots comparing pathway activity of glycolysis for tumor and NK cells in each product before infusion (Pre, day 0) and at days 7, 14, 21, and 28 after NK cell infusion. Significance of difference in pathway activity between NK and tumor cells at each time point, across products, was tested using Wilcoxon rank sum test and FDR-corrected (table S6). The color of a symbol indicates the mean pathway activity, and the size indicates the relative fraction of the cells. (C) Violin plots of glycolysis pathway activity across tumor and NK cell clusters. (D) Glycolysis pathway activity of the NK and tumor cell clusters across time. Linear regression was performed for each cell cluster. The slopes of the regression lines and P values are reported in table S7.
We next focused our metabolic pathway analysis on individual clusters. While all tumor clusters showed higher glycolysis, fatty acid metabolism, and OXPHOS pathway activity relative to NK cells, the two resistant tumor clusters T1 and T4 had the highest pathway activity levels (Fig. 4C and fig. S6, C and D). Last, we analyzed the dynamics of metabolic pathway activity for each cluster as tumor cells reemerged over time (Fig. 4D and fig. S6E; see Materials and Methods). All tumor clusters showed high metabolic activity at day 7, which stayed relatively stable over time. We also examined the activity of various metabolic pathways in NK cell clusters (Fig. 4C and fig. S6D) and found that they closely mirrored their in vivo functional activity (Fig. 3F). Nonfunctional NK cell clusters (N6 and N7) showed the lowest glycolytic activity, which remained relatively stable over time. The functional NK cell cluster N5, which was predominant in the CAR19/IL-15 NK cell group, showed the closest level of glycolytic activity to tumor cells initially, which decreased progressively over time (Fig. 4D). Similar trends were also observed for fatty acid metabolism and OXPHOS (fig. S6E).
Together, these findings indicate that Raji cells generally show greater metabolic fitness relative to NK cells. CAR19/IL-15 NK cells were characterized by higher metabolic fitness, although this dramatically decreased over time along with their antitumor activity. These data show a correlation between higher NK cell metabolic fitness and function. It also demonstrates that despite having higher metabolic activity relative to other NK cell products, CAR19/IL-15 NK cells lose metabolic fitness and their antitumor function over time in the presence of highly metabolic tumor cells, likely due to their inability to compete with the tumor cells for nutrient in the TME over time.
NK cells show a continuum of gene expression states
While clustering is a systematic approach to stratify a cell population, it does not always capture the extent of cell heterogeneity. We observed clear differences in NK cell metabolic fitness and antitumor function among the different NK cell clusters and within the functional cluster N5 (fig. S7, A and B). To better characterize this heterogeneity, we used Monocle 3 to construct a trajectory, which pseudo-temporally orders NK cells (see Materials and Methods). We identified two trajectories each composed of predominantly pre-infusion (pre-infusion trajectory) or post-infusion (post-infusion trajectory) cells (Fig. 5A). The post-infusion trajectory captures a transition of functional NK cells (with high metabolic fitness and antitumor activity) to nonfunctional NK cells. The post-infusion trajectory is rooted in functional N5 cells and progresses to terminally dysfunctional N6 cells (Fig. 5, A and B, and fig. S7, A to C). Thus, the pseudotime of the post-infusion trajectory describes an NK cell dysfunction axis.
Fig. 5. Trajectory analysis of NK cells.
(A) Trajectory of NK cells: colored by cluster membership (left) and pseudotime (right). The red arrows indicate the root nodes for the pre- and post-infusion trajectories (see Materials and Methods) relative to which pseudotime is computed. (B) Boxplot of pseudotime of NK clusters N5 and N6 that form the post-infusion trajectory; P value reported was computed using Wilcoxon rank sum test. (C) Trajectory plots split by NK cell product. (D) Violin plots showing the distribution pseudotime in pre- and post-infusion NK cells by groups. Differences in pseudo-time between groups in both pre- and post-infusion cells was significantly different [analysis of variance (ANOVA), P < 2 × 10−16 by a pairwise Tukey’s test reported in fig. S8]. (E) Hallmark pathways enriched in genes overexpressed in the low and high N5 cells (P < 0.05 and q < 0.1, respectively).
These differences were also reflected at the product level (Fig. 5, C and D). While pseudotime between cells from different products showed minor differences before infusion (fig. S7D, right), the differences were dramatically amplified after infusion (fig. S7D, left). CAR19/IL-15 NK cells showed an enrichment in cells closer to the beginning of the post-infusion trajectory and a depletion in cells further along the dysfunction axis (consistent with enrichment of N5 in this group) (Fig. 5, B and C).
We next focused on the heterogeneity within N5, which was readily captured based on the post-infusion trajectory. NK cells in N5 themselves show a gradation in their functional indicators along the pseudotime dysfunction axis (fig. S7, A to C). N5 cells further along the dysfunction axis were enriched at later experimental time points, when tumor cells reemerged, indicating acquisition of their dysfunction (fig. S7E). To further illustrate this gradation in dysfunction, we compared the top (high_N5) and bottom (low_N5) 30% of N5 NK cells based on their pseudotime (fig. S7F). The high_N5 cells were characterized by low proliferation and NK function (fig. S7G). Functional analysis of DEGs also indicated that low N5 cells showed high expression of genes involved in cell cycle/proliferation (MYC targets, E2F targets, and G2-M checkpoint) and metabolism-associated pathways (MTORC1 signaling). In contrast, high N5 cells were characterized by overexpression of genes in the transforming growth factor–β (TGF-β) and hypoxia pathways, along with those in the inflammatory pathways (IFN-γ signaling and TNF-α signaling) (Fig. 5E). These data quantitatively demonstrate a continuous evolution of NK cells and support the effect of IL-15 in maintaining metabolic fitness and antitumor activity and delaying NK cell dysfunction in an in vivo TME.
Improvement of CAR-NK cell treatment by administering two infusions
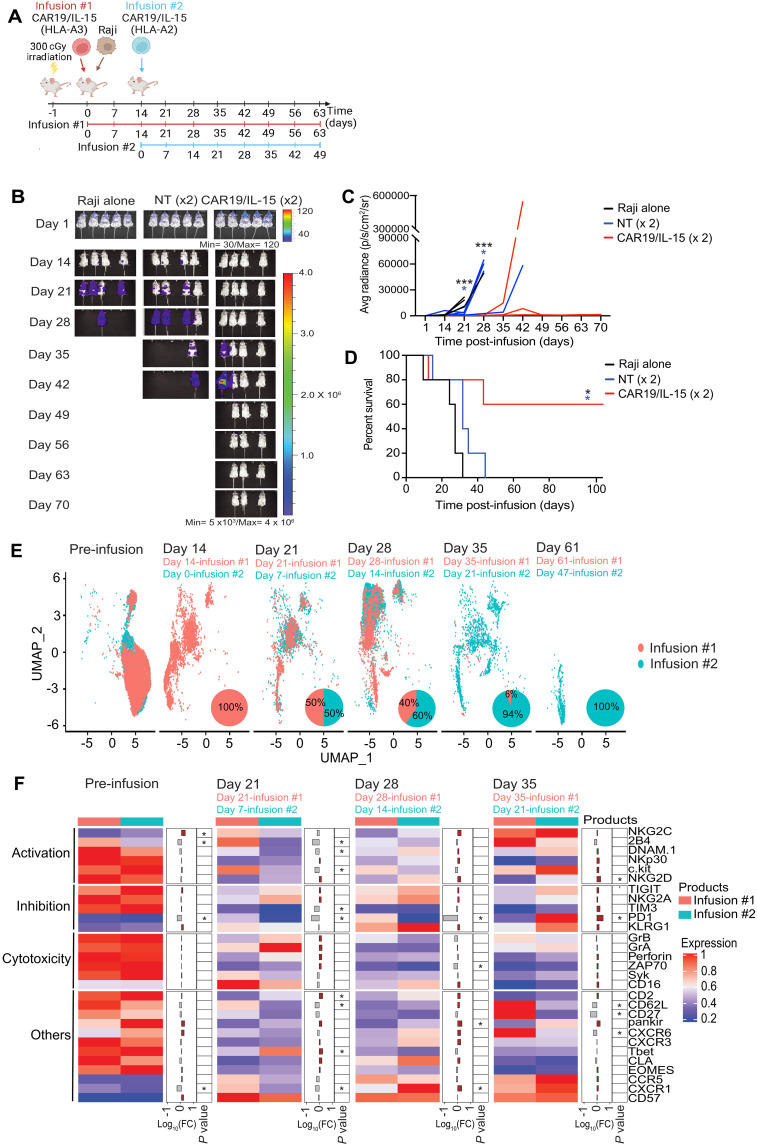
Given our findings that the activation, metabolic fitness, and function of CAR19/IL-15 NK cells decreased over time, we hypothesized that a second infusion would bolster the population of functional NK cells at later time points and consolidate the response, resulting in long-term tumor eradication. Thus, we treated mice bearing Raji lymphoma tumors with two infusions of CAR19/IL-15 NK cells 2 weeks apart (days 0 and 14) (Fig. 6A). We generated CAR-NK cells from two donors with different human leukocyte antigen (HLA) types (donor #1-HLA-A3 and donor #2-HLA-A2) to facilitate their simultaneous in vivo tracking and profiling using a mass cytometry panel that included antibodies against HLA-A3 and HLA-A2 (Fig. 6A, fig. S8, and table S4). Mice treated with two infusions of CAR19/IL-15 achieved 60% survival rate with long-term cures, whereas mice treated with double infusion of NT-NK (Fig. 6, B to D) or a single infusion experienced limited benefit (Fig. 2, C to E). To elucidate the mechanism underlying the improved survival in mice treated with two doses of CAR19/IL-15 NK cells, we studied the in vivo expansion and persistence of CAR-NK cells derived from each donor at various time points throughout treatment. We observed that the proportion of NK cells from the first infusion (HLA-A3) was highest at day 14 (100%), but steadily decreased over time and was almost undetectable at day 35 (6%; Fig. 6E). NK cells from the second donor (HLA-A2) administered 2 weeks after the first infusion were the predominant source of cells at day 35 and persisted until day 61 of the experiment (Fig. 6E). We next compared the phenotype of CAR-NK cells derived from the two infusions given 2 weeks apart by gating on HLA-A3– versus HLA-A2–expressing NK cells. At each respective time point after their infusion (days 21 and 28 for infusion #1, which corresponded to days 7 and 14 of the experiment for infusion #2), NK cells from the first and second infusion clustered together and expressed similar levels of markers of NK function, activation, and inhibition (Fig. 6F and fig. S9). The overall CAR-NK cell pool at day 61 derived solely from infusion #2. Phenotypically, these cells were characterized not only by expression of activation receptors (such as DNAM1 and NKG2D), activation-related kinases (Syk and ZAP70), and cytotoxic granules but also by higher expression of exhaustion markers (PD-1, TIM3, and TIGIT), consistent with a highly activated profile with evidence of exhaustion (Fig. 6F and fig. S9). These observations indicate that while early after infusion NK cells from the first and second infusion are phenotypically similar, administration of a second infusion of CAR19/IL-15 NK cells during a period of low tumor burden may increase the population of functional NK cells at later time points and, thus, tip the balance of competition in favor of NK cells with enhanced antitumor function in vivo.
Fig. 6. Improvement of CAR-NK cell antitumor activity following two infusions of CAR19/IL-15 NK cells.
(A) Schematic of the in vivo studies testing the two infusion treatment protocols [n = 15 mice per group; 5 mice were followed for survival and 10 mice were assigned for single-cell analyses; 2 mice per group were sacrificed at each time point (days 14, 21, 28, 35, and 61)]. (B) Bioluminescent imaging (n = 5 mice per group). The average radiance (C) and Kaplan-Meier plots showing the probability of survival (D) for the three groups of mice. *P ≤ 0.05 and **P ≤ 0.01. Black asterisks: Raji alone versus CAR19/IL-15. Blue asterisk: NT versus CAR19/IL-15. P values were determined by log-rank (Mantel-Cox, C) and unpaired t test (D). (E) UMAP plot showing the cell population change over time from mass cytometry analysis of the CAR19/IL-15 NK cells derived from the first donor infusion (HLA-A3+; red) and the second donor infusion (HLA-A2+; blue) (n = 10 mice per group; n = 2 mice per time point). Pie charts show the relative proportions of the first donor infusion (red) and the second donor infusion (blue). (F) Heatmap showing the mass cytometry proteomic marker abundance level in the first and the second infusion at different time points. The middle panel shows the FC between the first and second infusion. Brown bars represent higher expression in the second infusion; gray bars indicate higher expression in the first infusion. *P ≤ 0.05 and log10(FC) > 0.25.
Transcriptional profiling of CAR-NK cells and B cells in patients after CAR19/IL-15 NK cell immunotherapy
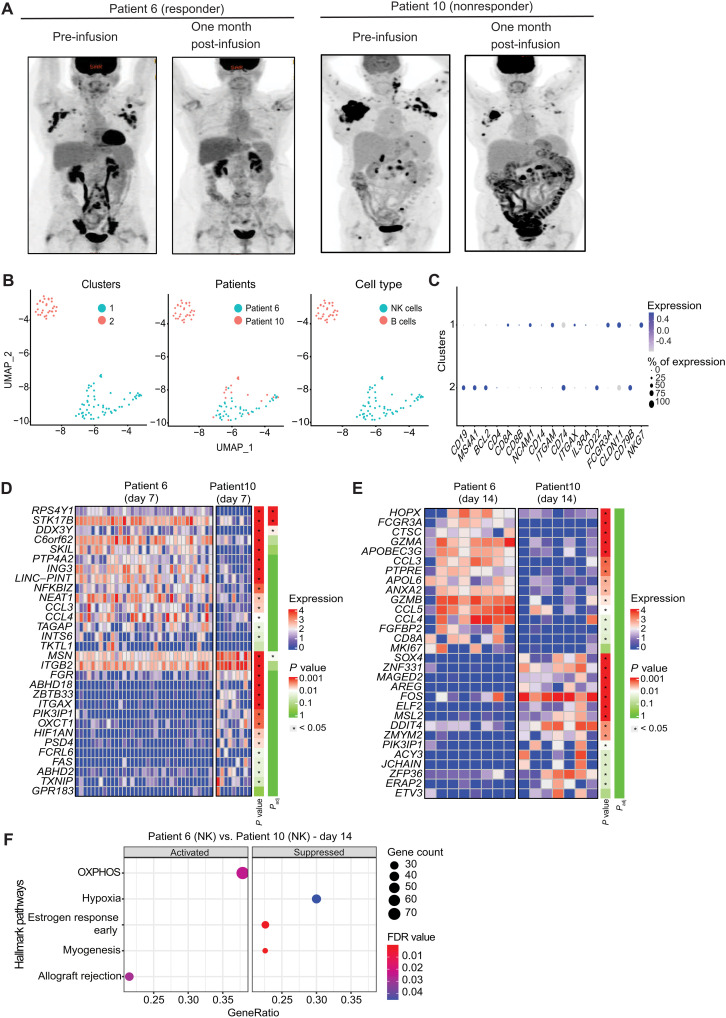
To evaluate whether the findings of our preclinical study hold true in patients treated with CAR19/IL-15 NK cells, we isolated and profiled CAR-NK cells and B cells from peripheral blood collected at multiple time points from two patients diagnosed with CD19+ relapse/refractory hematological malignancies and treated with CAR19/IL-15 NK cells (11). Each patient received one infusion of CAR-NK cells. Patient 6 was a 59-year-old female with accelerated chronic lymphocytic leukemia (CLL) who achieved a complete response (CR) 30 days after CAR-NK cell infusion (Fig. 7A). Patient 10 was a 61-year-old female with multiply relapsed transformed follicular lymphoma who failed to respond to CAR-NK cell infusion (Fig. 7A) (11). Single-cell fluorescence-activated cell sorting (FACS)–purified B cells and NK cells were sequenced using SMART-seq2 (see Materials and Methods), and after processing and quality control, 90 cells were retained (table S5). Two clusters were identified, and their identity was assessed based on expression of marker genes and reference transcriptome-based cell type annotations (Fig. 7, B and C, and fig. S10; see Materials and Methods). While cluster 1 includes cells with an NK cell signature from patients 6 and 10, cluster 2 is a B cell cluster defined based on expression of CD19 and MS4A1 derived solely from patient 10. Most cells from the NK cell cluster 1 and B cell cluster 2 are from days 7 and 14 after CAR-NK infusion, respectively.
Fig. 7. Patient response to one infusion of CAR-NK cell therapy.
(A) Positron emission tomography (PET) scans from two patients with lymphoid malignancies before (Pre-infusion) and 1 month after receiving one dose of CAR19/IL-15 NK cell infusion. (B) UMAP showing the distribution of two transcriptomic clusters of cells from patient scRNA-seq data. Cluster 1 contained 60 NK cells, including 17 from patient 10 and 43 from patient 6. Cluster 2 contained 30 B cells from patient 10. (C) Dot plot showing the mRNA expression levels for the two patient clusters. The size of the dots represents the percentage of cells expressing the gene, and the color represents its average expression. (D and E) Heatmaps of scRNA-seq data showing the top 15 and bottom 15 genes ranked by log FC between patients 6 and 10 at days 7 and 14 after NK cell infusion. The color bar represents the significance of both P values and adjusted P values. The asterisks were added if P value/adjusted P value < 0.05. (F) Dot plots showing significantly activated (left panel) and suppressed (right panel) hallmark pathways in the NK cells in patient 6 at day 14 versus those in patient 10 at day 14 (see Materials and Methods).
We next compared the transcriptomic profile of CAR-NK cells in patient 6 to patient 10 at days 7 and 14 after infusion (see Materials and Methods). A caveat of this analysis was the low cell number, resulting in most genes failing to reach statistical significance after correcting for multiple hypothesis testing. We therefore used the log2 FC for each gene to look for pathway enrichment using GSEA and focused our gene-level analysis on the top 15 up- and down-regulated genes (see Materials and Methods). Differences between patient 6 and patient 10 at day 7 were minor, with no differentially expressed pathways. However, at the gene level, patient 6 NK cells showed higher expression of genes associated with NK cell activation and chemokine activity like NFKBIZ, CCL3, and CCL4 (Fig. 7D). These differences were more pronounced at day 14, with patient 6 NK cells showing higher expression of cytolytic genes (GZMB and GZMA) as well as multiple chemokine genes (CCL3, CCL4, and CCL5) associated with improved NK cell cytolytic function (Fig. 7E). Day 14 pathway analysis revealed notable differences, with patient 6 NK cells showing up-regulation of OXPHOS as well as allograft rejection (Fig. 7F), while patient 10 NK cells showed up-regulation of hypoxia signaling, which we also observed in the nonfunctional NK cluster N6 (Fig. 7D). Together, these data suggest that CAR-NK cells in the responder patient are likely to have improved metabolism and higher expression of immune-associated genes relative to the nonresponder.
DISCUSSION
The unprecedented success with CAR T cell therapy trials has led to FDA approval of multiple CAR T cell products for the treatment of several hematologic cancers (1, 3–5). However, the highly personalized nature and the potential for severe toxicities constitute notable limitations to CAR T cell therapy. CAR-NK cells have emerged as a viable alternative strategy due to their favorable safety profile and inherent antitumor potential. Initial studies have shown promising efficacy of CAR-NK cells in the clinical setting (11, 22, 23), but the mechanisms driving NK cell evolution and function in patients and factors dictating relapse have not yet been elucidated. Our study provides previously unidentified insights into the phenotypic and functional changes that occur in NK cells in the context of tumor challenge and describes a mechanism through which loss of metabolic fitness leads to NK cell dysfunction and, consequently, enables tumor relapse.
Robust NK cell activation is required to develop an effective antitumor response. To investigate the ability of CAR and IL-15 to modulate NK activation and enhance NK cell potency and persistence, we generated NK cells expressing either CAR19 alone, IL-15 alone, or CAR19 and IL-15 (CAR19/IL-15). In the latter, CAR-NK cells were engineered to secrete IL-15, which then activates NK cells via a positive feedback loop. IL-15 is essential for NK cell development and controls both the homeostasis and the peripheral activation of NK cells (24). Here, we showed that compared to NT-NK cells and NK cells expressing CAR19 only, armoring CAR19 NK cells with IL-15 increased their proliferation rate and effector function with improved tumor control and survival in vivo. We performed scRNA-seq and mass cytometry to investigate the differences in CAR19 NK cells that may be associated with IL-15 production, and as expected, CAR-mediated activation of NK cells led to enhanced antitumor activity. However, robust multifaceted metabolic activation occurred only in CAR19/IL-15 NK cells, with activation of the metabolic checkpoint kinase mTOR (24), which controls glycolysis by promoting the expression of transcription factors such as Myc (25). We observed up-regulation of Myc and confirmed that NK cells transduced with CAR19/IL-15 are functionally more metabolically fit, with improved glycolytic activity as measured by the ECAR compared to controls. Additionally, our results suggest that IL-15 plays a role in the persistence and survival of CAR-NK cells and longer-lasting antitumor response, as mice treated with CAR19/IL-15 NK cells were able to control tumor growth substantially longer than the CAR19 NK cell–treated group. A limitation of our scRNA-seq and mass cytometry analyses was the low numbers of NK and Raji cells isolated from the harvested organs of mice, especially at the later time points due to limited persistence of the NK cells. We therefore focused our analysis on describing differences between clusters of NK cells, which merged all cells with similar transcriptomes, for higher statistical power. Further, when analyzing trends across time, we made use of regression analysis to take advantage of the full distribution of the data. Additionally, we based our conclusions of metabolic pathway dysregulation in NK cells on coordinated changes in gene expression within these pathways rather than on changes in specific metabolites.
Although mechanisms of relapse after CAR-T therapy have been well described (10, 11), factors influencing progression after CAR-NK cell therapy have not. Here, we show (Fig. 3, I and J, and fig. S4) that following challenge with tumor antigens, CAR19/IL-15 NK cells become more metabolically active and proliferate, but in the absence of tumor, CAR19/IL-15 NK cells failed to proliferate and persist, pointing to the importance of CAR engagement and signaling in driving NK cell persistence in vivo. We next evaluated the transcriptomic profile of CAR19/IL-15 NK cells and Raji cells over time in vivo and identified patterns of in vivo evolution in both tumor and NK cells following treatment. In all treatment groups, tumor cells progressed toward a resistant phenotype, with preferential emergence of NK cell–resistant clones with higher levels of glycolysis and fatty acid metabolism. However, the CAR-NK cells appeared to evolve into a dysfunctional phenotype, characterized by emergence of clusters with low metabolic fitness and effector function, as confirmed by impaired cytotoxic activity in killing assays against K562 targets ex vivo. On the basis of the observation that, as the tumor progresses, NK cells acquire an increasingly dysfunctional phenotype, we hypothesized that loss of metabolic fitness by CAR-NK cells may be an underlying mechanism leading to NK cell exhaustion and loss of potency and, consequently, disease relapse.
Since tumor burden with chronic antigen exposure has been shown to drive CAR T cell exhaustion and thereby impair their antitumor activity (26), we adapted the treatment protocol to include two infusions of CAR-NK cells, 14 days apart, with administration of the second dose of CAR19/IL-15 NK cells at the time of minimal disease volume. Survival rates in the two-infusion setting reached 60%, a value notably higher than what had been achieved with a single dose of CAR-NK cells, with higher persistence and activation profile of CAR19/IL-15 NK cells from the second infusion compared to the first. We hypothesize that the second dose of CAR19/IL-15 NK cells shifted the balance of activity toward NK cells at the time of low tumor burden. These data suggest that successful treatment with CAR-NK cells may require multiple infusions, especially for the difficult to treat and highly metabolically active tumors. An important limitation of our study is the use of an immunocompromised mouse model, which limits a more comprehensive analysis of the immune microenvironment and the interaction of different immune subsets with the tumor.
In the clinic, CAR19/IL-15 NK cell therapy has achieved promising response (11); however, some patients fail to respond, and little is known about the underlying transcriptional programs in CAR19/IL-15 NK cells that govern a robust antitumor activity. To evaluate whether the findings in our preclinical study hold true in patients treated with CAR19/IL-15 NK cells, we also analyzed the in vivo evolution of adoptively infused CAR19/IL-15 NK cells in two patients with lymphoid malignancies treated on our clinical trial of CAR-NK cell therapy (NCT03056339). Although constrained by the low cell number, transcriptomic data suggest that the ability of NK cells to clear tumors is closely tied to their metabolic fitness.
Together, our data point to a previously unidentified mechanism of tumor relapse after CAR-NK cell therapy, characterized by loss of metabolic fitness that leads to an exhausted, dysfunctional phenotype. We demonstrated through in vivo studies that carefully designed therapeutic intervention can substantially minimize NK cell exhaustion, sustain the antitumor response, and improve survival. This knowledge is especially relevant as we seek to transition previously unknown therapies to clinical trials, where appropriately designed protocols are critical to achieving successful outcomes.
MATERIALS AND METHODS
CAR retroviral vector production
The retroviral vectors encoding iC9.CAR19.CD28-zeta-2A-IL-15 (CAR19/IL-15), CAR19.CD28-zeta (CAR19), and IL-15 (Fig. 1A) were generated as previously described (27). Transient retroviral supernatant was produced, collected, and used for transduction of NK cells (28).
NK cell cytotoxicity assays
Chromium-release assay was performed as previously described (29). Details of this assays are provided in Supplementary Methods.
Xenograft lymphoma model
All animal studies were approved by the Institutional Animal Care and Use Committee. NOD/SCID IL-2Rγnull (NSG) mice (aged 8 to 10 weeks old; The Jackson Laboratory, Bar Harbor, ME) were irradiated with 300 cGy 1 day before intravenous injection with FFLuc-labeled Raji cells (2 × 105 per mouse) on day 0. Mice were randomized, and recipients with similar tumor burdens were distributed evenly across the treatment and control groups before NK cell infusion. Additional details of the in vivo studies are included in Supplementary Methods.
Single-cell RNA sequencing
Detailed protocols on scRNA-seq data processing, unsupervised clustering and dimension reduction, differential gene expression analysis, trajectory analysis, pathway activity in NK cells and tumor cells, gene set activity score calculation, GSEA, and cell type annotation are described in Supplementary Methods.
Mass cytometry
Two distinct panels of metal-tagged antibodies were used to (i) characterize the heterogeneity and phenotypic evolution of CAR-NK cells over time and (ii) to simultaneously profile NK cells from two donors with different HLA types across the duration of the experiments. See Supplementary Methods and supplementary tables for more details.
Acknowledgments
We thank Y. Tan for assisting with analytical support.
Funding: This work was supported, in part, by the generous philanthropic contributions to The University of Texas MD Anderson Cancer Center Moon Shots Program and The Sally Cooper Murray endowment and by grants (1 R01 CA211044-01, 5 P01CA148600-03, and U01CA247760) from the National Institutes of Health (NIH), the Cancer Prevention and Research Institute of Texas (grants RP180466 and RP180248), the Leukemia Specialized Program of Research Excellence (SPORE) Grant (P50CA100632), the Human Cell Atlas Seed Network Grant (CZF2019-02425 and CZF2019-002432 to K.C.) from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation, and a grant (P30 CA016672) from the NIH to the MD Anderson Cancer Center Flow Cytometry and Cellular Imaging Core Facility that assisted with the mass cytometry studies.
Author contributions: Conceptualization: D.M., S.M., R.E.C., E.J.S., K.C., and K.R. Methodology: L.L., V.M., J.D., Y.H., P.P.B., Q.M., S.L., R.B., M.K., M.D., K.C., K.R., Y.L., S.A., and P.L. Investigation: L.L., V.M., J.D., Y.H., P.P.B., Q.M., S.L., R.B., M.C., M.D., E.L., M.S., Y.L., S.A., P.L., J.G.L., T.V., J.F., V.N., M.K., H.R. and B.K. Funding acquisition: K.C. and K.R. Project administration: K.C. and K.R. Supervision: K.R. and K.C. Writing—original draft: L.L., L.M.-F., T.J.L., and K.R. Writing—review and editing: L.L., L.M.-F., T.J.L., K.R., and J.S.M.
Competing interests: R.B., E.L., P.P.B., D.M., M.D., P.L., R.E.C., E.J.S., K.R., and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Takeda Pharmaceutical. D.M., R.B., E.L., E.J.S., K.R., and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Affimed GmbH. K.R. participates on the Scientific Advisory Board for GemoAb, AvengeBio, Virogin Biotech, GSK, Bayer, Navan Technologies, and Caribou Biosciences. K.R. is the scientific founder of Syena. E.J.S. has served on the Scientific Advisory Board for Adaptimmune, Axio, Celaid, FibroBiologics, Navan Technologies, New York Blood Center, and Novartis. S.M. has received fees via his institution for presentations (Novartis, Celgene/BMS, Kite/Gilead, Janssen, Pfizer) and for services as head and/or member of DSMBs (Miltenyi, Immunicum/Mendus) in the field of advanced cell and gene therapy. S.M. is the founder of SWECARNET, Sweden’s national CAR T cell network. The remaining authors declare that they have no competing interests.
Data and materials availability: The scRNA-seq data are publicly available in Gene Expression Omnibus (GEO) repository at accession number GSE190976. The CyTOF data are publicly available in the Flow Repository at accession number FR-FCM-Z625. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S10
Tables S1 to S7
References
REFERENCES AND NOTES
- 1.S. S. Neelapu, F. L. Locke, N. L. Bartlett, L. J. Lekakis, D. B. Miklos, C. A. Jacobson, I. Braunschweig, O. O. Oluwole, T. Siddiqi, Y. Lin, J. M. Timmerman, P. J. Stiff, J. W. Friedberg, I. W. Flinn, A. Goy, B. T. Hill, M. R. Smith, A. Deol, U. Farooq, P. McSweeney, J. Munoz, I. Avivi, J. E. Castro, J. R. Westin, J. C. Chavez, A. Ghobadi, K. V. Komanduri, R. Levy, E. D. Jacobsen, T. E. Witzig, P. Reagan, A. Bot, J. Rossi, L. Navale, Y. Jiang, J. Aycock, M. Elias, D. Chang, J. Wiezorek, W. Y. Go, Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 377, 2531–2544 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.S. J. Schuster, M. R. Bishop, C. S. Tam, E. K. Waller, P. Borchmann, J. P. McGuirk, U. Jäger, S. Jaglowski, C. Andreadis, J. R. Westin, I. Fleury, V. Bachanova, S. R. Foley, P. J. Ho, S. Mielke, J. M. Magenau, H. Holte, S. Pantano, L. B. Pacaud, R. Awasthi, J. Chu, Ö. Anak, G. Salles, R. T. Maziarz, Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med. 380, 45–56 (2019). [DOI] [PubMed] [Google Scholar]
- 3.S. L. Maude, T. W. Laetsch, J. Buechner, S. Rives, M. Boyer, H. Bittencourt, P. Bader, M. R. Verneris, H. E. Stefanski, G. D. Myers, M. Qayed, B. de Moerloose, H. Hiramatsu, K. Schlis, K. L. Davis, P. L. Martin, E. R. Nemecek, G. A. Yanik, C. Peters, A. Baruchel, N. Boissel, F. Mechinaud, A. Balduzzi, J. Krueger, C. H. June, B. L. Levine, P. Wood, T. Taran, M. Leung, K. T. Mueller, Y. Zhang, K. Sen, D. Lebwohl, M. A. Pulsipher, S. A. Grupp, Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med. 378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J. H. Park, I. Rivière, M. Gonen, X. Wang, B. Sénéchal, K. J. Curran, C. Sauter, Y. Wang, B. Santomasso, E. Mead, M. Roshal, P. Maslak, M. Davila, R. J. Brentjens, M. Sadelain, Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 378, 449–459 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D. L. Porter, B. L. Levine, M. Kalos, A. Bagg, C. H. June, Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N. C. Munshi, L. D. Anderson Jr., N. Shah, D. Madduri, J. Berdeja, S. Lonial, N. Raje, Y. Lin, D. Siegel, A. Oriol, P. Moreau, I. Yakoub-Agha, M. Delforge, M. Cavo, H. Einsele, H. Goldschmidt, K. Weisel, A. Rambaldi, D. Reece, F. Petrocca, M. Massaro, J. N. Connarn, S. Kaiser, P. Patel, L. Huang, T. B. Campbell, K. Hege, J. San-Miguel, Idecabtagene vicleucel in relapsed and refractory multiple myeloma. N. Engl. J. Med. 384, 705–716 (2021). [DOI] [PubMed] [Google Scholar]
- 7.A. Mullard, FDA approves fourth CAR-T cell therapy. Nat. Rev. Drug Discov. 20, 166 (2021). [DOI] [PubMed] [Google Scholar]
- 8.S. S. Neelapu, S. Tummala, P. Kebriaei, W. Wierda, F. L. Locke, Y. Lin, N. Jain, N. Daver, A. M. Gulbis, S. Adkins, K. Rezvani, P. Hwu, E. J. Shpall, Toxicity management after chimeric antigen receptor T cell therapy: One size does not fit ‘ALL’. Nat. Rev. Clin. Oncol. 15, 218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. Chiossone, P. Y. Dumas, M. Vienne, E. Vivier, Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 18, 671–688 (2018). [DOI] [PubMed] [Google Scholar]
- 10.M. Daher, K. Rezvani, Outlook for new CAR-based therapies with a focus on CAR NK cells: What lies beyond CAR-engineered T cells in the race against cancer. Cancer Discov. 11, 45–58 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.E. Liu, D. Marin, P. Banerjee, H. A. Macapinlac, P. Thompson, R. Basar, L. Nassif Kerbauy, B. Overman, P. Thall, M. Kaplan, V. Nandivada, I. Kaur, A. Nunez Cortes, K. Cao, M. Daher, C. Hosing, E. N. Cohen, P. Kebriaei, R. Mehta, S. Neelapu, Y. Nieto, M. Wang, W. Wierda, M. Keating, R. Champlin, E. J. Shpall, K. Rezvani, Use of CAR-transduced natural killer cells in CD19-positive lymphoid tumors. N. Engl. J. Med. 382, 545–553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.H. Klingemann, Are natural killer cells superior CAR drivers? Onco. Targets Ther. 3, e28147 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.E. J. Orlando, X. Han, C. Tribouley, P. A. Wood, R. J. Leary, M. Riester, J. E. Levine, M. Qayed, S. A. Grupp, M. Boyer, B. de Moerloose, E. R. Nemecek, H. Bittencourt, H. Hiramatsu, J. Buechner, S. M. Davies, M. R. Verneris, K. Nguyen, J. L. Brogdon, H. Bitter, M. Morrissey, P. Pierog, S. Pantano, J. A. Engelman, W. Winckler, Genetic mechanisms of target antigen loss in CAR19 therapy of acute lymphoblastic leukemia. Nat. Med. 24, 1504–1506 (2018). [DOI] [PubMed] [Google Scholar]
- 14.J. A. Fraietta, S. F. Lacey, E. J. Orlando, I. Pruteanu-Malinici, M. Gohil, S. Lundh, A. C. Boesteanu, Y. Wang, R. S. O’Connor, W. T. Hwang, E. Pequignot, D. E. Ambrose, C. Zhang, N. Wilcox, F. Bedoya, C. Dorfmeier, F. Chen, L. Tian, H. Parakandi, M. Gupta, R. M. Young, F. B. Johnson, I. Kulikovskaya, L. Liu, J. Xu, S. H. Kassim, M. M. Davis, B. L. Levine, N. V. Frey, D. L. Siegel, A. C. Huang, E. J. Wherry, H. Bitter, J. L. Brogdon, D. L. Porter, C. H. June, J. J. Melenhorst, Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat. Med. 24, 563–571 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.E. Liu, Y. Tong, G. Dotti, H. Shaim, B. Savoldo, M. Mukherjee, J. Orange, X. Wan, X. Lu, A. Reynolds, M. Gagea, P. Banerjee, R. Cai, M. H. Bdaiwi, R. Basar, M. Muftuoglu, L. Li, D. Marin, W. Wierda, M. Keating, R. Champlin, E. Shpall, K. Rezvani, Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 32, 520–531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.E. Becht, L. M. Innes, J. Healy, C.-A. Dutertre, I. W. H. Kwok, L. G. Ng, F. Ginhoux, E. W. Newell, Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 17.C. Trapnell, D. Cacchiarelli, J. Grimsby, P. Pokharel, S. Li, M. Morse, N. J. Lennon, K. J. Livak, T. S. Mikkelsen, J. L. Rinn, The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.K. Solocinski, M. R. Padget, K. P. Fabian, B. Wolfson, F. Cecchi, T. Hembrough, S. C. Benz, S. Rabizadeh, P. Soon-Shiong, J. Schlom, J. W. Hodge, Overcoming hypoxia-induced functional suppression of NK cells. J. Immunother. Cancer 8, e000246 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.S. Hänzelmann, R. Castelo, J. Guinney, GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.F. Rohrig, A. Schulze, The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 (2016). [DOI] [PubMed] [Google Scholar]
- 21.W. Li, M. Xu, Y. Li, Z. Huang, J. Zhou, Q. Zhao, K. le, F. Dong, C. Wan, P. Yi, Comprehensive analysis of the association between tumor glycolysis and immune/inflammation function in breast cancer. J. Transl. Med. 18, 92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.X. Tang, L. Yang, Z. Li, A. P. Nalin, H. Dai, T. Xu, J. Yin, F. You, M. Zhu, W. Shen, G. Chen, X. Zhu, D. Wu, J. Yu, First-in-man clinical trial of CAR NK-92 cells: Safety test of CD33-CAR NK-92 cells in patients with relapsed and refractory acute myeloid leukemia. Am. J. Cancer Res. 8, 1083–1089 (2018). [PMC free article] [PubMed] [Google Scholar]
- 23.V. Bachanova, A. Ghobadi, K. Patel, J. H. Park, I. W. Flinn, P. Shah, C. Wong, C. Bickers, P. Szabo, L. Wong, B. Valamehr, S. Atwal, Y. W. Chu, R. Elstrom, P. Strati, Safety and efficacy of FT596, a first-in-class, multi-antigen targeted, off-the-shelf, iPSC-derived CD19 CAR NK cell therapy in relapsed/refractory B-cell lymphoma. Blood 138, 823 (2021). [Google Scholar]
- 24.A. Marcais, J. Cherfils-Vicini, C. Viant, S. Degouve, S. Viel, A. Fenis, J. Rabilloud, K. Mayol, A. Tavares, J. Bienvenu, Y.-G. Gangloff, E. Gilson, E. Vivier, T. Walzer, The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 15, 749–757 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.C. Magaway, E. Kim, E. Jacinto, Targeting mTOR and metabolism in cancer: Lessons and innovations. Cell 8, 1584 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. H. Long, W. M. Haso, J. F. Shern, K. M. Wanhainen, M. Murgai, M. Ingaramo, J. P. Smith, A. J. Walker, M. E. Kohler, V. R. Venkateshwara, R. N. Kaplan, G. H. Patterson, T. J. Fry, R. J. Orentas, C. L. Mackall, 4-1BB costimulation ameliorates T cell exhaustion induced by tonic signaling of chimeric antigen receptors. Nat. Med. 21, 581–590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.V. Hoyos, B. Savoldo, C. Quintarelli, A. Mahendravada, M. Zhang, J. Vera, H. E. Heslop, C. M. Rooney, M. K. Brenner, G. Dotti, Engineering CD19-specific T lymphocytes with interleukin-15 and a suicide gene to enhance their anti-lymphoma/leukemia effects and safety. Leukemia 24, 1160–1170 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. Vera, B. Savoldo, S. Vigouroux, E. Biagi, M. Pule, C. Rossig, J. Wu, H. E. Heslop, C. M. Rooney, M. K. Brenner, G. Dotti, T lymphocytes redirected against the κ light chain of human immunoglobulin efficiently kill mature B lymphocyte-derived malignant cells. Blood 108, 3890–3897 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. Daher, R. Basar, E. Gokdemir, N. Baran, N. Uprety, A. K. Nunez Cortes, M. Mendt, L. N. Kerbauy, P. P. Banerjee, M. Shanley, N. Imahashi, L. Li, F. L. W. I. Lim, M. Fathi, A. Rezvan, V. Mohanty, Y. Shen, H. Shaim, J. Lu, G. Ozcan, E. Ensley, M. Kaplan, V. Nandivada, M. Bdiwi, S. Acharya, Y. Xi, X. Wan, D. Mak, E. Liu, X. R. Jiang, S. Ang, L. Muniz-Feliciano, Y. Li, J. Wang, S. Kordasti, N. Petrov, N. Varadarajan, D. Marin, L. Brunetti, R. J. Skinner, S. Lyu, L. Silva, R. Turk, M. S. Schubert, G. R. Rettig, M. S. McNeill, G. Kurgan, M. A. Behlke, H. Li, N. W. Fowlkes, K. Chen, M. Konopleva, R. E. Champlin, E. J. Shpall, K. Rezvani, Targeting a cytokine checkpoint enhances the fitness of armored cord blood CAR-NK cells. Blood 137, 624–636 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.E. Liu, S. O. T. Ang, L. Kerbauy, R. Basar, I. Kaur, M. Kaplan, L. Li, Y. Tong, M. Daher, E. L. Ensley, N. Uprety, A. K. Nunez Cortes, R. Z. Yang, Y. Li, H. Shaim, F. Reyes Silva, P. Lin, V. Mohanty, S. Acharya, M. Shanley, L. Muniz-Feliciano, P. P. Banerjee, K. Chen, R. E. Champlin, E. J. Shpall, K. Rezvani, GMP-compliant universal antigen presenting cells (uAPC) promote the metabolic fitness and antitumor activity of armored cord blood CAR-NK cells. Front. Immunol. 12, 626098 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.C. S. McGinnis, L. M. Murrow, Z. J. Gartner, DoubletFinder: Doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337. e324 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. Picelli, O. R. Faridani, Å. K. Björklund, G. Winberg, S. Sagasser, R. Sandberg, Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 33.T. Stuart, A. Butler, P. Hoffman, C. Hafemeister, E. Papalexi, W. M. Mauck 3rd, Y. Hao, M. Stoeckius, P. Smibert, R. Satija, Comprehensive integration of single-cell data. Cell 177, 1888–1902. e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Z. Gu, R. Eils, M. Schlesner, Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 35.M. V. Kuleshov, M. R. Jones, A. D. Rouillard, N. F. Fernandez, Q. Duan, Z. Wang, S. Koplev, S. L. Jenkins, K. M. Jagodnik, A. Lachmann, M. G. McDermott, C. D. Monteiro, G. W. Gundersen, A. Ma'ayan, Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44, W90–W97 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D. Venet, J. E. Dumont, V. Detours, Most random gene expression signatures are significantly associated with breast cancer outcome. PLOS Comput. Biol. 7, e1002240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. Cao, M. Spielmann, X. Qiu, X. Huang, D. M. Ibrahim, A. J. Hill, F. Zhang, S. Mundlos, L. Christiansen, F. J. Steemers, C. Trapnell, J. Shendure, The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.T. Wu, E. Hu, S. Xu, M. Chen, P. Guo, Z. Dai, T. Feng, L. Zhou, W. Tang, L. Zhan, X. Fu, S. Liu, X. Bo, G. Yu, clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.M. Kanehisa, S. Goto, KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. Subramanian, P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, J. P. Mesirov, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.G. Finak, A. McDavid, M. Yajima, J. Deng, V. Gersuk, A. K. Shalek, C. K. Slichter, H. W. Miller, M. J. McElrath, M. Prlic, P. S. Linsley, R. Gottardo, MAST: A flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.G. Yu, L.-G. Wang, Y. Han, Q.-Y. He, clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D. Aran, A. P. Looney, L. Liu, E. Wu, V. Fong, A. Hsu, S. Chak, R. P. Naikawadi, P. J. Wolters, A. R. Abate, A. J. Butte, M. Bhattacharya, Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Y. Lu, K. Li, Y. Hu, X. Wang, Expression of immune related genes and possible regulatory mechanisms in alzheimer's disease. Front. Immunol. 12, 768966 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.L. Li, H. Chen, D. Marin, Y. Xi, Q. Miao, J. Lv, P. P. Banerjee, H. Shaim, M. Daher, R. Basar, N. Imahashi, J. Jimenez, B. Hu, R. S. Mehta, L. N. Kerbauy, M. Kaplan, M. Mendt, G. Ozcan, E. Gokdemir, M. Hernandez Sanabria, Y. Li, K. Chen, J. Wang, L. Muniz-Feliciano, W. L. Zhao, R. E. Champlin, E. J. Shpall, K. Rezvani, A novel immature natural killer cell subpopulation predicts relapse after cord blood transplantation. Blood Adv. 3, 4117–4130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.H. Chen, M. C. Lau, M. T. Wong, E. W. Newell, M. Poidinger, J. Chen, Cytofkit: A bioconductor package for an integrated mass cytometry data analysis pipeline. PLOS Comput. Biol. 12, e1005112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S10
Tables S1 to S7
References