Abstract
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern such as Omicron hampered efforts in controlling the ongoing coronavirus disease 2019 pandemic due to their ability to escape neutralizing antibodies induced by vaccination or prior infection, highlighting the need to develop broad-spectrum vaccines and therapeutics. Most human monoclonal antibodies (mAbs) reported to date have not demonstrated true pan-sarbecovirus neutralizing breadth especially against animal sarbecoviruses. Here, we report the isolation and characterization of highly potent mAbs targeting the receptor binding domain (RBD) of huACE2-dependent sarbecovirus from a SARS-CoV survivor vaccinated with BNT162b2. Among the six mAbs identified, one (E7) showed better huACE2-dependent sarbecovirus neutralizing potency and breadth than any other mAbs reported to date. Mutagenesis and cryo–electron microscopy studies indicate that these mAbs have a unique RBD contact footprint and that E7 binds to a quaternary structure–dependent epitope.
A potent, broad-spectrum human monoclonal antibody capable of neutralizing all ACE2-binding sarbecoviruses is identified.
INTRODUCTION
In the past two decades, there have been three major outbreaks caused by zoonotic coronaviruses: severe acute respiratory syndrome (SARS-CoV) in 2003 (1), Middle East respiratory syndrome (MERS-CoV) in 2012 (2), and the most recent and ongoing pandemic caused by SARS-CoV-2 (3). Since the emergence of SARS-CoV-2, additional genetically closely related viruses, defined as Clade-1b viruses hereafter, have been found in humans, especially the variants of concern (VOCs), and in animals such as bats and pangolins (4–6). SARS-CoV-2 shares approximately 70% genome sequence identity with SARS-CoV (7). Similar to Clade-1b viruses, many other Clade-1a viruses have been found over the years, which share a close genetic relatedness to SARS-CoV. It is well documented that neutralizing antibodies (NAbs) and T cell–mediated immunity both play important roles in preventing severe coronavirus disease 2019 (COVID-19), with NAbs playing a more important role as a first line of defense against infection (8, 9). NAbs can be induced either through natural infection or vaccination. Most SARS-CoV-2 vaccines that are currently licensed for use in humans were developed against the Spike (S) protein of the ancestral Wuhan strain (10). The emergence and dominance of VOCs have posed a substantial threat and challenge as some VOCs, especially Omicron, have evolved to escape NAb immunity in either infected or vaccinated individuals regardless of the type of vaccines received, number of booster vaccinations received, or hybrid immunity status achieved from both infection and vaccination (11).
Rapid development of therapeutic monoclonal antibodies (mAbs) has been one of the cornerstones of the global response to COVID-19, with the first human clinical trial started just 5 months after the SARS-CoV-2 genome became publicly available (12). Passive immunization using therapeutic mAbs remains an important part of pandemic response and containment as they can play a key role in treating patients with severe disease, especially in the vulnerable populations (such as immunocompromised patients) or preventing onward transmission by ring-fence application in targeted high-risk populations. Unfortunately, all therapeutic mAbs currently licensed for COVID-19 prophylaxis or treatment have lost their effectiveness against the newly emergent VOCs (13–16). Therefore, there is an urgency to develop therapeutic mAbs that can be VOC agnostic or have a broad spectrum against different Clade-1a or Clade-1b viruses in the subgenus Sarbecovirus. NAbs targeting different regions of the S protein of SARS-CoV-2, such as N-terminal domain (NTD), S2 region, and receptor binding domain (RBD), have been identified from multiple studies (17–20). From a depletion study using polyclonal human sera vaccinated with mRNA-1273 or BNT162b2 depleted of either RBD or NTD antibodies, it was demonstrated that first, nearly all neutralizing activity is due to RBD-targeting NAbs, and second, vaccination-elicited neutralization breadth against variants is almost completely accounted for by antibodies targeting RBD rather than NTD (21). Hence, it is not a coincidence that all currently approved therapeutic mAbs are targeting RBD and most highly potent published NAbs are those targeting RBD (22). Broad-spectrum mAbs have been isolated and reported from a variety of sources including patients with SARS (23), patients with COVID-19 (24, 25), and animals immunized with different combinations of antigens (26). Previously, we reported that broad and potent pan-sarbecovirus human neutralizing antisera were generated in BNT162b2-vaccinated SARS survivors (27). Here, we report the isolation and characterization of highly potent pan huACE2-dependent sarbecovirus neutralizing mAbs from one of the donors in this cohort.
RESULTS
Isolation of broadly huACE2-dependent sarbecovirus neutralizing mAbs from a BNT162b2-vaccinated SARS survivor donor
SARS survivors who received the BNT162b2 mRNA vaccine generated broad NAbs against 10 huACE2-dependent sarbecoviruses in Clade-1a and Clade-1b, including multiple VOCs of SARS-CoV-2 and animal sarbecoviruses (27). We confirmed the neutralization capacity of one donor’s (SS6V) sera to SARS-CoV and SARS-CoV-2 before and after vaccination using a surrogate virus neutralization test (sVNT, see Materials and Methods) (table S1). The peripheral blood mononuclear cells (PBMCs) at day 23 after the first dose of the BNT162b2 vaccine were used for B cell enrichment and isolation. CD19+ B cells that were positive for binding to SARS-CoV (SC1+) and SARS-CoV-2 (SC2+) RBD tetramers (fig. S1) were sorted, and their antibody genes were amplified and cloned. In total, 19 paired heavy and light kappa fragments were recovered, and the mAbs were expressed in vitro. Majority (17 of 19) were from the double-positive (SC1+SC2+) B cells, while two from SC2+ and none from SC1+ single-positive B cells were recovered. We were unable to recover any paired heavy and light lambda fragments.
For control and comparative studies, we also included five published mAbs—S309 (Sotrovimab, GSK) (23), CR3022 (28), S2X259 (VirBiotech) (25), S2K146 (VirBiotech) (29), and LyCoV-1404 (Bebtelovimab, Eli Lilly) (30)—in this study. The two mAbs cloned from SC2+ single-positive B cells showed minimal reactivity to SARS-CoV RBD (table S2). All 17 mAbs recovered from SC1+SC2+ B cells showed binding to both SARS-CoV and SARS-CoV-2 RBDs, but their neutralization capacities varied across different mAbs (table S2). The six most potent neutralizers (B5, E7, E11, F1, F4, and F5) were selected for large-scale production and further characterization. The most potent mAbs (E7, F1, and F5) use a unique heavy- and light-chain gene family combination that has not been previously reported (fig. S2). All three mAbs use IGHV4-59/IGHJ4 heavy chains and IGKV2-28/IGKJ5 light chains, and the high sequence homology (>90%) suggests that the B cells most likely originated from the same clonal family, and the slight variations of the heavy and light chains occurred through somatic hypermutation and affinity maturation.
Potency, breadth, and functionality of huACE2-dependent sarbecovirus mAbs
We next performed an 18-plex sVNT based on RBDs derived from SARS-CoV-2 ancestral virus and its variants (Alpha, Beta, Delta, Delta Plus, Gamma, Lambda, and Mu); animal sarbecoviruses BANAL-52, BANAL-236, GD-1, RaTG13, GX-P5L, RsYN2018B, LYRa11, RsSHC014, and WIV-1; and SARS-CoV (Fig. 1, A and B). E7, F1, and F5 mAbs exhibited an ultrapotent ability to neutralize all 18 sarbecoviruses with sVNT median inhibitory concentration (IC50) ranging from 10.44 to 120.30 ng/ml (0.070 to 0.802 nM). Although B5, E11, and F4 showed broad-spectrum neutralizing activity, they were less effective against several of the viruses tested, i.e., E11 on Lambda, F4 on Lambda, GX-P5L, and RsSHC014. The IC50 of B5, E11, and F4 was higher than 1000 ng/ml (6.67 nM) for certain Clade-1b sarbecoviruses and ≥200 ng/ml (1.33 nM) for Clade-1a sarbecoviruses. Immune escape was also observed for the five comparator mAbs against various sarbecoviruses, i.e., S309 by Lambda and WIV-1, CR3022 by most sarbecoviruses, LyCoV-1404 by two Clade-1b zoonotic sarbecoviruses (RaTG13 and GX-P5L) and all the Clade-1a sarbecoviruses (WIV-1, RsSHC014, RsYN2018B, LYRa11, and SARS-CoV), and S2K146 by RsSHC014. S2X259 displayed the broadest neutralizing activities against all huACE2-dependent sarbecoviruses among all the comparator mAbs with IC50 ranging between 47.39 and 370.50 ng/ml (0.316 to 2.47 nM).
Fig. 1. Determination of breadth of neutralization of mAbs against 18 RBDs in Clade-1a and Clade-1b sarbecoviruses.
(A) In the multiplex-sVNT test, mAbs were serially diluted fourfold for six points and tested against 18 uniquely coded streptavidin Luminex beads coated with biotinylated RBD from different sarbecoviruses. huACE2-phycoerythrin was then added to test for the ability of the mAbs to inhibit RBD binding to ACE2. Percent inhibition was calculated as a fraction of signal obtained from the test reaction against the signal obtained from negative control reactions whereby no inhibitory agents were added. (B) Summary of IC50 (in nanograms per milliliter) of all 11 tested mAbs against 18 sarbecoviruses. The less potent the mAb, the darker is the red color. “X” refers to no neutralization observed at the highest concentration (10,000 ng/ml) tested. (C) Pseudovirus neutralization IC50 for mAbs against eight sarbecoviruses. Each data point is computed by performing fourfold serial dilution eight points with three technical replicates per dilution and calculating the mean of three biological replicates. Data points at 10,000 ng/ml represent no neutralization observed at the highest concentration tested.
We then performed functional characterizations on three of the most potent mAbs (E7, F1, and F5) and benchmarked them against the five control mAbs (S309, CR3022, S2X259, S2K146, and LyCoV-1404). The ability of the mAbs to neutralize different sarbecoviruses was assessed using eight spike-pseudotyped reporter viruses, which included SARS-CoV-2 ancestral and four VOCs (Alpha, Delta, Beta, and Gamma), two animal sarbecoviruses (GX-P5L and WIV-1), and SARS-CoV. We observed that E7, F1, and F5 retained highly potent neutralizing activity against all the eight pseudoviruses with a relative IC50 of less than 10 ng/ml (0.067 nM) (Fig. 1C). S309 has an IC50 of 100 to 1000 ng/ml (0.667 to 6.67 nM) against SARS-CoV-2 VOCs and animal sarbecoviruses. S2K146, being an ACE2-mimicking mAb, displayed the highest potency of all the control mAbs tested. CR3022 was only able to neutralize some Clade-1a sarbecoviruses (WIV-1 and SARS-CoV) and none of the Clade-1b sarbecoviruses, while LyCoV-1404 was able to neutralize all the SARS-CoV-2 variants tested but not against zoonotic sarbecoviruses and SARS-CoV.
Unique epitope footprints of the six cross-reactive ACE2-dependent sarbecovirus neutralizing mAbs
To determine the impact of individual RBD amino acid mutations on the binding of different mAbs and to map the epitope footprint of each mAb on RBD in relation to known ACE2 interface residues, we used deep mutational scanning (DMS) of a yeast-display RBD mutant library (fig. S3) (31). In general, we could classify the mAbs into three disparate epitope profiles consisting of B5 and E11 in one group; F4 in its unique group; and E7, F1, and F5 mAbs in the third group (Fig. 2). The epitope profiles of B5, E11, F4, E7, F1, and F5 mAbs are markedly different from those of the four comparator mAbs (S309, CR3022, S2X259, and S2K146) (Fig. 2A and fig. S4). Most mAbs have epitopes that are composed of key residues clustered within a narrow region, except for F4, as its epitope footprint covers a wider region spanning residues from G339-T345 and N440-N448. Figure 2B shows the alignment of 20 RBDs in Clade-1a and Clade-1b sarbecoviruses in relation to the binding epitopes of the mAbs. The mAbs target highly conserved regions across most of the huACE2-dependent sarbecoviruses, explaining their broadly neutralizing potency. The binding epitopes of the mAbs on RBD are also modeled in a three-dimensional (3D) structure (Fig. 2C), which reflects the three distinct epitope profiles observed.
Fig. 2. Complete mapping of mutations reducing each mAb binding using yeast-displayed RBD and DMS to determine escapability of mAbs.
(A) Complete mapping of mutations reducing mAb binding using yeast-displayed SARS-CoV-2 (Wuhan-Hu-1) RBD and DMS. Line height (y axis) indicates each mutation’s strength of escape from antibody binding at the position corresponding to the RBD (x axis). S2X259 data are from Tortorici et al. (25), and S2K146 data are from Park et al. (29). (B) Alignment of RBDs from sarbecoviruses with mutations that differ from ancestral SARS-CoV-2 highlighted in black. Three distinct binding profiles of mAbs are observed, which are different from the comparator mAbs used here. Binding epitopes of broadly cross-reactive sarbecovirus mAbs (E7, F1, and F5) target a highly conserved region in the RBD as highlighted in green. (C) Illustration of binding epitopes of mAbs on the 3D structure of RBD in red.
Affinity of pan huACE2-dependent sarbecovirus neutralizing mAbs
The affinity of mAbs’ binding to RBDs of SARS-CoV-2 mutants was determined using an RBD mutant array. The RBD array contains 38 most prevalent RBD mutants compiled from the RBD surveillance databank Global Initiative on Sharing All Influenza Data (GISAID). Some RBDs contain single-point mutations, while other RBDs contain multiple-point mutations corresponding to variants of interest (VOIs)/VOCs (i.e., Beta, Gamma, Kappa, and Delta), and the whole SARS-CoV-2 S1 subunit consisted of RBD and NTD. E7, F1, and F5 mAbs bind to all RBD mutants strongly with a relative half maximal effective concentration (EC50) of less than 25 nM (Fig. 3A). CR3022, an mAb that was isolated from a person infected with SARS-CoV (32), had the lowest binding affinity to all the SARS-CoV-2 RBD mutants. This was expected as multiple studies have reported that CR3022 binds more stably and with higher affinity to SARS-CoV than SARS-CoV-2 due to one or some key residues in SARS-CoV (33, 34), which were different in SARS-CoV-2.
Fig. 3. Binding affinity, neutralization potency, and bio-layer interferometry affinity of mAbs.
Heatmap of mAbs binding to 39 SARS-CoV-2 global RBD variants with the range of relative EC50 (in nanomolar) shown in (A) and blocking of ACE2 interaction and relative IC50 (in nanomolar) are shown in (B). Bio-layer interferometry affinity measurements with immobilized mAbs and SARS-CoV RBD (C) and SARS-CoV-2 RBD (D) serially diluted in solution. Representative binding curves of seven different RBD concentrations from 0.75 to 50 nM to immobilized mAbs are plotted and overlaid to a 1:1 kinetic model. Corresponding mean ± SEM Kd values (in molar) are indicated.
The same RBD variant array was also used to examine whether the mAbs can block RBD-ACE2 engagement in a multiplex competition format (Fig. 3B) (35). E7, F1, F5, and S2X259 were able to disrupt all tested RBD-ACE2 binding at a relative IC50 of 15 nM or less, consistent with the 18-plex sVNT data in Fig. 1A. We also observed that some mAbs, i.e., S309 and CR3022, performed poorly in the RBD-ACE2 inhibition assay despite having moderate to good binding activity to wild-type and early VOCs of SARS-CoV-2, as demonstrated in Fig. 3A and other studies (36, 37). This suggests that binding ability does not always equate to neutralizing ability. The observations using the RBD mutation array to study antibody inhibition of ACE2 engagement are in slight contrast to the 18-plex sVNT data whereby some neutralization by S309 and CR3022 were observed against some or most of the sarbecoviruses tested. However, we would like to emphasize that both assay formats have distinct differences. For instance, biotinylated RBD proteins were displayed in an upright manner on the Luminex beads for the 18-plex sVNT, while the direct conjugation used for the RBD variant array may result in heterogeneous access of the antibodies to the target neutralization epitopes. This could likely explain the increase in overall sensitivity of the 18-plex sVNT.
The affinities of mAbs to SARS-CoV or SARS-CoV-2 RBD proteins were also measured using biolayer interferometry (Fig. 3, C and D). E7 and F1 showed a higher affinity to SARS-CoV than to SARS-CoV-2, while F5 displayed a similar affinity to both. S309 and CR3022 exhibited stronger binding to SARS-CoV than SARS-CoV-2, while the reverse was observed for S2X259. These findings are consistent with the fact that S309 and CR3022 were derived from patients with SARS, whereas S2X259 was derived from a patient with COVID-19.
Cryo-EM of E7 Fab fragment in complex with SARS-CoV-2 S protein trimer
We then determined the structure of the E7 Fab fragment complexed with recombinant Wuhan-Hu-1 SARS-CoV-2 S protein trimer by single-particle cryo–electron microscopy (cryo-EM). Asymmetric (C1) 3D classification of the cryo-EM data yielded a single, homogeneous S trimer class with bound Fab E7. Although C1 nonuniform refinement of this class in cryoSPARC (38) produced a map at a global resolution of 3.12 Å (Fig. 4A and fig. S5D), the local resolution of the Fab:S protein interacting interface is poorer (4 to 5 Å) (fig. S5F). Subsequent local refinement of this region yielded a map with an overall resolution of 4.03 Å (fig. S5E) (Fig. 4, B to E) and an improved local resolution map (3.5 to 4.0 Å) of the Fab:S protein interface (fig. S5G).
Fig. 4. The cryo-EM structure of Fab E7 complexed with recombinant SARS-CoV-2 S protein trimer.
(A) Nonuniform refinement produced a 3.12 Å map, shown without B-factor sharpening for clarity, with Fab E7 bound to two RBDs, one in the up conformation and the other in the down conformation. Color key for atomic model: Fab light chain, yellow; Fab heavy chain, orange; S protomers are shown in red, blue, and green with corresponding RBDs in magenta, light blue, and teal, respectively. (B) Masked local refinement of the boxed region in (A) improved the Fab-RBD density markedly, which was resolved to 4.03 Å. (C) The epitope-paratope interfaces involving the down RBD and up RBD are boxed with red and blue dotted lines, respectively. (D) Zoomed-in view of the epitope-paratope interface for the down RBD. (E) Zoomed-in view of the epitope-paratope interface for the up RBD. Side chains of residues forming the epitope are shown as cyan sticks, with those of the paratope shown in green. (F) Open book representation of the interface between the epitope and paratope, with labeled residues and color scheme as in (E).
Our structure shows that E7 binds to a quaternary epitope across an “up” RBD and a neighboring “down” RBD. Most of its interaction is on the up RBD (Fig. 4E). On the up RBD, a large part of the E7 epitope overlaps with the class 1 mAbs epitopes, while the remaining parts overlap with class 4 mAb epitopes (39). On the down RBD, E7 binds to two residues that are located within the previously described class 2 mAb epitopes. 3D variability analysis in cryoSPARC did not reveal any appreciable flexibility in this E7:2-RBD interaction. Table S4 shows the summary of the likely interactions between Fab and RBDs. The major epitope on the up RBD likely forms extensive interactions with all three complementarity-determining region (CDR) loops of the heavy chain (CDRH1 to CDRH3). In agreement with our DMS analysis, the up RBD residues at 408 to 414 are likely involved in extensive polar interactions with the E7 heavy chain (Fig. 4E and fig. S7A). We hypothesize that R408 forms hydrogen bonds with CDRH3 T104, and both R408 and Q414 can form polar interactions with CDRH3 E103. The carbonyl oxygen of R408 is close enough to the Q414 side chain to form an intra-RBD hydrogen bond, while P412, G413, and Q414 on the same RBD also interact via hydrogen bonds with the backbone and side chains of N32, Q31, and N33 from light-chain CDR1 (CDRL1). Mutation of A411 (fig. S4) likely disrupts this network of polar interactions (Fig. 4E). Residues Y421, F456, and Y473 on the same RBM also likely form hydrophobic interactions with E7 residues I29 on CDRH1 and Y52-I53 on CDRH2, while the backbone oxygen of R457 is within hydrogen bonding range of S54 CDRH2.
The down RBD contains a smaller portion of the E7 epitope and only interacts with the E7 light chain (Fig. 4E). Down RBD residue Q493 likely forms hydrogen bonds with non-CDR residues S72 and D75, whereas F486 likely forms a hydrophobic interaction with CDRL1 Y35. A multiple sequence alignment and analysis of charges/hydrophobicity of the RBD from SARS-CoV-2 variants, SARS-CoV, and animal ACE2-binding sarbecoviruses reveals that most of the residues on both parts of the E7 epitope are strongly conserved, with most of the residues on the up RBD portion of the epitope showing absolute sequence conservation (figs. S5, S6, and S7B).
Ability to neutralize Omicron viruses of different sublineages
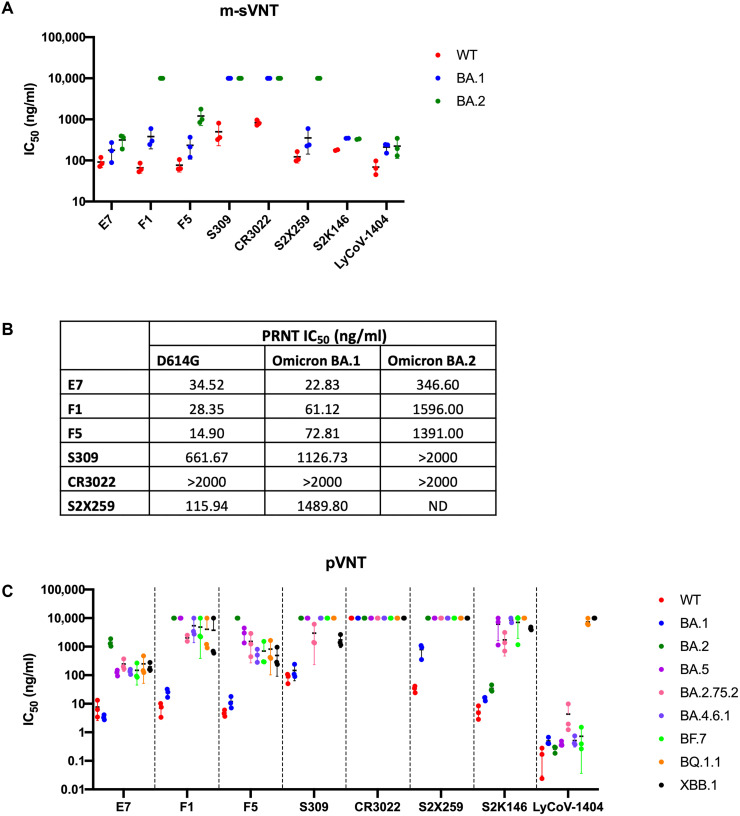
During the final stage of the current study, the Omicron VOC (B.1.1.529) emerged and was found to contain additional mutations in circulating new subvariants within just a few months of discovery. To assess the breadth of neutralizing ability of our newly identified mAbs against the Omicron subvariants, we conducted potency testing of the mAbs using multiplex sVNT (m-sVNT), plaque reduction neutralization tests (PRNTs), and pseudovirus neutralization tests (pVNTs). E7 and F5 retained potent activity on both Omicron BA.1 and BA.2, with IC50 ranging between 177 and 315 ng/ml (1.18 to 2.10 nM) on m-sVNT (Fig. 5A). In authentic virus neutralization tests, all three mAbs potently neutralize Omicron BA.1 with PRNT IC50 ranging from 3.38 to 72.81 ng/ml (0.023 to 0.49 nM). However, only E7 retained neutralizing activity against Omicron BA.2 with IC50 of 346.6 ng/ml (2.31 nM) by PRNT (Fig. 5B).
Fig. 5. Neutralization assays to determine the potency of mAbs against Omicron subvariants.
Determination of neutralization ability of mAbs against wild-type (WT) D614 and Omicron sublineages using m-sVNTs (A), authentic live virus neutralization tests (B), and pVNTs (C). Neutralization assays were performed in biological triplicates, and means of IC50 ± SD are indicated in solid lines for m-sVNT and pVNT, respectively. Data points at 10,000 ng/ml represent no neutralization observed at the highest concentration tested. For authentic plaque reduction live virus neutralization test, starting concentration of mAbs at 2000 ng/ml twofold serially diluted six points to 62.5 ng/ml was used. >2000 ng/ml refers to no neutralization observed at the highest concentration, while ND refers to not done.
We also made pseudoviruses against eight Omicron sublineages—BA.1, BA.2, BA.5, BA.2.75.2, BA.4.6.1, BF.7, BQ.1.1, and XBB.1—to study the effect of additional mutations on the ability of the virus to escape NAbs (Fig. 5C). Although E7 displayed reduced potency to BA.2 with about 300-fold increase in IC50 compared to ancestral SARS-CoV-2 and BA.1 to 1400 ng/ml (9.3 nM), there was an approximately 10-fold improvement in potency of E7 from BA.2 compared to BA.5 with an IC50 of 123 ng/ml (0.8 nM). The newer Omicron subvariants, such as BQ.1.1 and XBB.1, were not observed to be more resistant to E7 as the IC50 stabilized at 134 to 248 ng/ml (0.89 to 1.65 nM), suggesting that despite additional mutations acquired by the circulating SARS-CoV-2 strains, they are still sensitive to E7. The discrepancies in neutralization potency to the BA.2 variant measured by the biochemical m-sVNT assay compared to the live virus assays (PRNT and pVNT) suggest that there may be synergistic effects between RBD mutations and non-RBD mutations of the whole S protein, which leads to more potent neutralization escape in the live virus assays.
DISCUSSION
In the 2 years since the World Health Organization declared COVID-19 a pandemic, we have witnessed the emergence of multiple VOCs resulting in increased transmissibility and/or immune escape, suggesting a continued evolution of the virus in the human population to become fitter in the current landscape of highly vaccinated and infected people. These included Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and the latest Omicron (B.1.1.529) variants. These variants contain mutations at key amino acid positions 417, 439, 452, 478, 484, and 501 of the RBD, which are found to severely affect the functionality of current vaccines and therapeutic mAbs (15, 40–42). Thus, the timely identification and characterization of broadly neutralizing mAbs is extremely important to fight or prevent future emergence of SARS-CoV-2 VOCs. Multiple studies have demonstrated that cross-clade and multi-variant NAbs do exist from both natural infection and vaccination (23, 43–45), but they are not immunodominant and represent a very small proportion of the total antibody repertoire.
Previously, we reported that BNT162b2-vaccinated SARS survivors were able to produce high levels of pan-huACE2–dependent sarbecovirus NAbs, which were not seen in any other cohorts, including those who received the BNT162b2 vaccination after SARS-CoV-2 infection (27). In our current study, we set out to isolate and characterize these highly cross-reactive huACE2-dependent sarbecovirus neutralizing mAbs from a BNT162b2-vaccinated SARS survivor who displayed the best quality of NAbs present in his/her serum. It was clear that these cross-clade ACE2-dependent sarbecovirus NAbs were only induced upon vaccination as there was minimal cross-neutralization of prevaccinated serum on ancestral SARS-CoV-2 virus (table S1). To further investigate the specificity of the NAbs from this donor, we isolated specific B cells that were double positive for staining by both SARS-CoV and SARS-CoV-2 RBD tetramers. From a very small pool of 19 paired heavy and light kappa productive B cell gene sequences, the majority already demonstrated both SARS-CoV and SARS-CoV-2 neutralizing capabilities in varying degrees. This shows that the peripheral circulating B cells in this individual are producing much more cross-clade NAbs than reported from previous studies using donors with SARS-CoV-2 infection or vaccination. In addition to the higher proportion of broad-spectrum NAbs in this donor, we have also observed much higher potency and breadth for the top three mAbs obtained in this study compared to other published mAbs. The three mAbs maintained strong neutralization capability across most SARS-CoV-2 VOCs, VOIs, and Clade-1a sarbecoviruses, as assayed using different virus neutralization assay platforms (sVNT, pVNT, and PRNT). Note that all three mAbs used a unique combination of heavy- and light-chain gene classes, which have not been previously reported for sarbecovirus-specific mAbs. The heavy- and light-chain sequences of E7, F1, and F5 shared a high degree of similarity of more than 90%. Comparison of E7’s CDRs and its paratope residues to those in F1 and F5 (fig. S8) shows slightly lower similarities. For E7 paratope residues, equivalent residues in both F1 and F5 have 84.6% similarity, whereas for their CDRs, F1 and F5 are 80.4 and 85.7% similar to E7, respectively. It is highly possible that the epitopes of E7, F1, and F5 overlap at least in part, as suggested by the high degree of similarity in DMS profiles. The clonal variations between the three mAbs affected susceptibilities to mutations at the virus structural interface, which changed the individual mAbs’ affinities, thus resulting in differences in neutralization abilities against the newer Omicron subvariants.
The DMS maps provide very good predictions as to whether a variant escapes mAb binding (31). For example, the G339 mutation, which is present in Omicron BA.1, came up as a site of escape for F4, suggesting that F4 will suffer some loss in binding and/or neutralization potency for this variant. This was correlated when F4 demonstrated no neutralization activity to BA.1 in our m-sVNT assay (fig. S9A). One key point from DMS is that although certain mutations enabled the virus to escape from binding by an mAb, the mutant might suffer secondary deleterious effects due to reduced biochemical folding stability in the RBD or loss in viral fitness. A notable example is BA.5, which had a reversion from R493 that was present in BA.1 and BA.2 back to Q493 (like in ancestral SARS-CoV-2). This unexpectedly resulted in partial restoration of the neutralizing capability of E7 against BA.5 compared to BA.2. To elucidate the reason for the recovery of E7 neutralization against BA.5, we performed pVNT using BA.2 + L452R and BA.2 + F486V single mutants. The IC50 for E7 remained high, similar to the IC50 for BA.2 at ~1000 ng/ml (fig. S9B). We thus hypothesize that the major resistance point of E7 is at S408 with a secondary minor resistance point at R493 (both mutations found in BA.2). For BA.1, which contained R408 (the same as ancestral SARS-CoV-2) and R493 (found in BA.2), the potency of E7 was comparable to IC50 against ancestral SARS-CoV-2 at ~10 ng/ml. On the other hand, for BA.5, which contained S408 (found in BA.2) and Q493 (the same as ancestral SARS-CoV-2), the reversion of R493 back to wild-type Q493 seemed to lead to a partial suppression of the major resistance mutation S408, an effect termed epistasis (46). The cryo-EM results support this hypothesis. Both F486V and R493Q mutations (fig. S7) affect the down RBD portion of the E7 epitope. F486, in particular, is involved in stabilizing the interface between the up and down RBDs in previously reported Delta and BA.1 spike structures, and antibodies targeting this residue may be able to overcome the tighter packing of Omicron variant RBDs relative to previous VOCs (47). Therefore, although the major portion of the E7 epitope lies on the up RBD, cobinding to the neighboring down RBD has a significant effect on E7’s overall binding affinity and neutralization. R408 occupies a central position within a network of hydrogen bonds between the up RBD and both chains of E7, and mutation of R408 to serine would disrupt these interactions. However, the quaternary interaction between two RBDs and Fab E7 is stabilized by both main- and side-chain interactions between Q493 on the down RBD and the E7 light chain, and this quaternary mode of binding masks the otherwise destabilizing effect of S408 in BA.5. Note that no new mutants containing R493 were sequenced after the BA.2 infection wave passed, suggesting that Q493 may be necessary for viral fitness, and E7 should continue to stay relevant in this pandemic.
Our cryo-EM structure of Fab E7 complexed with the SARS-CoV-2 S trimer suggests that the potency of E7 in neutralizing and binding across sarbecoviruses is due to its unique binding to a quaternary structure epitope consisting of highly conserved residues across the RBMs of one up RBD and the closest neighboring down RBD. Superposition of the Fab:S trimeric spike structures with existing structures of uncomplexed Wuhan-Hu-1, Delta, and Omicron BA.1 SARS-CoV-2 S trimers in the one RBD up and one RBD down conformation shows that the E7 epitope is accessible for antibody binding, but upon binding, E7 will have to induce a slight rotation of the down RBD relative to the up RBD (fig. S10B) to form the final structure. In addition to the presence of the major resistance mutation R408S on the E7 epitope on BA.2, the tighter packing of the BA.2 spike in the 3-RBD-down position may also contribute to the reduced ability of E7 to neutralize BA.2 relative to BA.1 (48, 49). We hypothesize that there could be two modes of neutralization by E7, namely, by inhibition of binding to the ACE2 receptor and by inhibition of fusion. By occupying a large portion of the RBM on the up RBD (which is required for receptor binding), E7 Fab might sterically hinder receptor binding (fig. S10A). Fusion of the virus to host membrane requires the S protein trimer to undergo massive conformational changes. The ability of E7 to bind across two RBDs could lock neighboring S proteins within a trimer together, thus preventing this conformational change and the subsequent fusion process. The ability of E7 to block these two important sarbecovirus entry processes might therefore explain the high potency of this antibody.
The classification system by Barnes et al. (39) divides anti–SARS-CoV-2 RBD antibodies into four epitope classes (fig. S11A) from classes 1 to 4. Class 1 and 2 epitopes are located adjacent to each other, and both overlap with the ACE2 binding site (fig. S11, A and B). Class 1 antibodies only target an up RBD, whereas class 2 antibodies can recognize the RBD in either the up or down conformation. Class 3 and 4 antibodies both bind to epitopes further away from the ACE2 binding site, with class 3 antibodies targeting the base of the RBD below the class 2 epitope and class 4 antibodies targeting a cryptic epitope located below the class 1 epitope. However, some class 3 and 4 antibodies do have epitopes that share some overlap with the ACE2 binding site, such as ADG-2 (50) and S2X259 (25). The class 4 epitope is exposed only in the up RBD conformation (fig. S11A). E7 antibody binds to a unique quaternary structure dependent epitope—across two neighboring RBDs, specifically to one in down and one in up conformation (fig. S12). The down RBD portion of the E7 epitope is more consistent with a class 2 epitope, while the up RBD portion of the E7 epitope spans both class 1 and 4 epitope residues (fig. S11, C and D).
Previous studies have reported broadly NAbs against SARS-CoV-2 VOCs and sarbecovirus clades. Examples of these antibodies are the class 1 antibody S2K146; class 2 antibody C002; class 4 antibodies DH1047, C118, and C022; and ADG-2 antibody that binds between the class 1 and 4 sites (fig. S12) (29, 45, 50, 51). Comparison of the up RBD portion of the E7 epitope to the class 4 epitopes of DH1047, C118, and C022 showed limited overlaps (fig. S12), with only the conserved residue R408 shared between the C118, C022, and E7 epitopes. The S2K146 (class 1) and ADG-2 (between class 1 and 4) epitopes have larger overlaps with the up RBD portion of the E7 epitope. Although DH1047, C022, and C118 epitopes do not overlap with the ACE2 binding site, they inhibit ACE2 binding by steric hindrance, whereas E7, ADG-2, and S2K146 antibodies directly compete with ACE2 for binding. All these published antibodies engage a single RBD, and multiple copies of their Fabs can bind to an S trimer simultaneously. Although S2K146 and C002 are also able to interact with two RBDs within one spike, this is not their exclusive mode of binding. Binding of S2K146 and C002 is dependent only on the class 1 or class 2 epitope on a single RBD, and it is also used in their quaternary mode of binding. Quaternary binding of one copy of Fab S2K146 or C002 also does not preclude binding of additional copies to the same S protein. In the case of C002, quaternary binding also does not depend on a particular RBD conformation due to the accessibility of the core class 2 epitope in both the up and down RBD conformations. In contrast, Fab E7 exclusively binds to a quaternary epitope, which locks a neighboring pair of RBDs in a one-up plus one-down conformation, and therefore, only one Fab E7 can bind to one S protein trimer.
We also compared the E7 epitope to two other antibodies (P2B-1A1 and P5A-1B9) from the same VH4-59 germ line (52). Although they share the same germ line, the epitopes bound by all these antibodies are very different—antibodies P2B-1A1 and P5A-1B9 bind class 1 and 2 epitopes, respectively, and interaction with a single RBD is sufficient for their binding; although similar to S2K146 and C002, P5A-1B9 also shows the ability to interact with two RBDs at one time (52). This differs from E7, which has an epitope that spans across two RBDs and appears to exclusively bind to RBDs in a quaternary configuration (figs. S11 and S13). Note that C118 and C022 with strong pan-sarbecovirus neutralization activity, as well as S2X259, which only exhibits strong neutralization activity against variants before BA.2, also target the residue R408 but otherwise have nonquaternary epitopes mainly consisting of residues on a different part of the RBD showing strong sequence conservation across sarbecoviruses (25). Previously reported antibodies with a quaternary epitope on the SARS-CoV-2 RBD generally bind to two down RBDs to trap the S trimer in its closed state (25, 53, 54). A notable exception is the antibody N3-1 from a synthetic SARS-CoV-2 convalescent repertoire, which has a down RBD epitope similar to E7 involving F486 and Q493 but with different residues forming the up RBD portion of its epitope similar to the S2X259 epitope (55).
In conclusion, this study has demonstrated that cross-clade infection of SARS-CoV followed by vaccination with SARS-CoV-2 mRNA vaccines was able to induce potent, pan huACE2–dependent sarbecovirus NAbs both at the polyclonal serum level and at the single B cell/mAb level. This study’s findings will be extremely useful in guiding the development of future pan-sarbecovirus therapeutic antibodies and vaccines. This is critical not only for mitigating the current COVID-19 pandemic but also for preventing and responding to future coronavirus outbreaks.
MATERIALS AND METHODS
Single B cell receptor reconstruction into mAb expression plasmid
A 51-year-old SARS survivor (SS6V), who was vaccinated with BNT162b2 in January to February 2021, was recruited into an Institutional Review Board–approved study under the reference numbers NHG DSRB E 2020/00091 and NUS LH-20-006EC with informed consent. For peripheral blood processing, plasma from day 23 after the first vaccination dose was collected, followed by PBMC extraction using standard Ficoll-Paque density gradient centrifugation (GE Healthcare) and cryopreserved for long-term storage. For sorting, PBMCs were thawed, first stained for SARS-CoV RBD (SC1) and SARS-CoV-2 RBD (SC2) tetramers, and then surface stained with LIVE/DEAD Fixable aqua dead cell strain (Invitrogen), anti–human CD3–fluorescein isothiocyanate (FITC), anti–human CD14-FITC, anti–human CD56-FITC, anti–human CD19–phycoerythrin (PE)–Cy5, anti–human CD27-allophycocyanin (APC)–Cy7, and anti–human CD38-BV786. SC1+SC2+, SC1+, and SC2+ of viable CD19+ B cells were single cell sorted into 96-well polymerase chain reaction (PCR) plates (Axygen) prefilled with 10 μl per well of reverse transcription (RT)–PCR catch buffer containing 10 mM tris (pH 8.0) and 10 U of RNasin ribonuclease inhibitor (Promega) using BD FACSAria III (BD Biosciences) equipped with 355, 405, 488, 561, and 640 nm lasers (27, 56). The plates were then flash-frozen on dry ice and kept at −80°C until use.
RT was next performed using the Qiagen OneStep RT-PCR kit on each plate of sorted B cells. The primers used were listed in this paper (56). Nested PCR was done using Q5 polymerase (NEB), and the wells containing corresponding heavy and light chains were purified for sequencing. After analyzing the sequences, specific primers were then used to amplify for individual gene families, and the PCR products were cloned into pTRIOZ expression vector (Invivogen). pTRIOZ constructs were transfected into human embryonic kidney (HEK) 293 cells using Fugene6 (Promega), and the supernatant was harvested to check for small-scale efficacy screening using SARS-CoV and SARS-CoV-2 RBD binding enzyme-linked immunosorbent assay (ELISA) and sVNT. For binding ELISA, 100 ng of protein was coated onto Maxisorp plates (Nunc) overnight at 4°C. After blocking with OptEIA blocking buffer (BD), 50 μl of each supernatant was added neat per well and incubated at 37°C for 1 hour. Goat anti–human immunoglobulin (IgG)–horseradish peroxidase (HRP) (1:5000; Bethyl) diluted in OptEIA was then added, which will convert tetramethylbenzidine substrate into a colorimetric readout for quantification using Cytation 5 reader (BioTek). For sVNT, a commercially available kit for SARS-CoV-2 (cPass, Genscript) was used, and the manufacturer’s protocol was followed (57). The same kit was used to measure the amount of NAbs against SARS-CoV by substituting the SARS-CoV-2 RBD-HRP reagent with 6 ng per well of SARS-CoV RBD-HRP (Genscript). The best mAbs and control mAbs from the initial binding and neutralization screening were then batch produced by transfection into EXPI293 cells for large-scale expression and purified using protein G agarose beads (Millipore) for downstream characterizations.
Multiplex sVNT
Serum samples were tested with a newly developed m-sVNT assay (27). Briefly, AviTag-biotinylated RBD proteins from ancestral SARS-CoV-2 and SARS-CoV, nine VOCs/VOIs (Alpha, Delta, Beta, Gamma, Delta Plus, Lambda, Mu, Omicron BA.1, and Omicron BA.2), and nine zoonotic sarbecoviruses (BANAL-52, BANAL-236, GD-1, RaTG13, GX-P5L, Rs2018B, LYRa11, RsSHC014, and WIV-1) were coated on a MagPlex Avidin microsphere (Luminex) at 5 μg/1 million beads. The RBD-coated microspheres (600 beads per antigen) were preincubated with mAbs at a starting concentration of 10,000 ng/ml serially diluted fourfold for 15 min at 37°C with 250 rpm agitation. After 15 min incubation, 50 μl of PE-conjugated huACE2 (2 μg/ml; GenScript) were added to the well and incubated for 15 min at 37°C with agitation, followed by two phosphate-buffered saline (PBS)–1% bovine serum albumin washes. The final readings were acquired using the MAGPIX system (Luminex Corporation).
RBD mutant multiplex bead array
SARS-CoV-2 RBD mutants were purified and coupled onto magnetic multiplex beads (Luminex Corporation) as previously described and mixed in a bead cocktail (35). Bead cocktails were dispensed into each well of a black, clear-bottom, 384-well plate (Greiner), and mAbs at a starting concentration of 80 nM serially diluted were next added to the bead cocktail and incubated on a plate shaker for 2 hours at room temperature. The binding affinities of mAbs to different SARS-CoV-2 RBD mutants were detected using anti–human IgG-PE (Southern Biotech), and the mean fluorescence intensity (MFI) of the PE reporter was acquired on a FLEXMAP 3D machine (Luminex Corporation). To measure the competition of mAb-RBD binding by ACE2, bead cocktail and serially diluted mAbs were first mixed as described above, followed by the addition of Avi-tagged biotinylated ACE2. The plate was incubated on a plate shaker for 2 hours at room temperature. ACE2 binding was detected with streptavidin, R-PE conjugate (S866, Thermo Fisher Scientific) for 1 hour, followed by the addition of the R-PE Biotin-XX conjugate (P811, Thermo Fisher Scientific) for another 1 hour. The binding of ACE2 was detected as the MFI of the PE reporter using FLEXMAP 3D machine (Luminex Corporation).
Deep mutational scanning
The construction of a SARS-CoV-2 RBD single mutant yeast display library and DMS has been described previously (31, 58). Briefly, single-point mutations of 19 possible amino acids were introduced into all 201 positions of SARS-CoV-2 RBD (Wuhan-Hu-1) using PCR mutagenesis and constructed into a yeast display library. The mutagenized library was then used to screen against mAbs to determine the effect of each mutation on binding or expression to study the mutational constrain and antibody binding epitopes on RBD. Yeast libraries were induced for surface expression, labeled with primary (monoclonal) antibodies at a concentration corresponding to the EC90 of binding curves for each mAb to yeast-displayed RBD for 1 hour at room temperature, and then labeled with 1:200 anti–human IgG-PE antibody (Jackson ImmunoResearch, 109-115-098) and 1:100 anti-Myc antibody (Immunology Consultants Laboratory, CYMC-45F) after washing off unbound antibodies. Stained yeast cells were next sorted on BD FACSAria II to select cells with reduced antibody binding (fig. S2B), recovered overnight for plasmid extraction, and sequenced on Illumina NextSeq from pre-sort and antibody-escaped sorted populations. Sequencing counts before and after selection were used to calculate the “escape fraction” for each library variant using the code posted on Zenodo (https://zenodo.org/account/settings/github/repository/jbloomlab/SARS-CoV-2-RBD_MAP_NUS-mAbs (DOI: 10.5281/zenodo.7956405).
Bio-layer interferometry
The affinities of the antibodies binding to SARS-CoV or SARS-CoV-2 RBD protein were measured by bio-layer interferometry using an Octet RED96e (ForteBio) with anti–human IgG Fc capture biosensors (AHC biosensors, ForteBio). The tip of anti–human Fc biosensors was first hydrated in tris buffer containing 20 mM tris (pH 8.0), 150 mM NaCl, and 0.025% Tween-20 for 1 hour at room temperature before the experiment. The antibodies E7, F1, F5, S309, CR3022, and S2X259 IgGs and the recombinant SARS-CoV and SARS-CoV-2 S1-RBD proteins were also diluted in tris buffer. The RBD proteins were twofold serially diluted with final concentrations ranging from 0.78 to 50 nM, whereas antibody IgGs were diluted to 10 μg/ml. Two hundred microliters of solutions (buffers, RBD proteins, and antibody IgGs) were transferred into 96-well flat bottom black plates (Corning). The plate was shaken at 1000 rpm and incubated at a temperature of 30°C in the Octet RED96e throughout the experiment. The experiments were started with baseline measurement of each biosensor tip in tris buffer, and then the tips were transferred into wells containing the antibodies for antibody binding to the biosensor tips until achieving ~0.5 to 0.6 nm response. Following the antibody capture, the tips were dipped into tris buffer for 1 min to remove excess antibodies, and this was continued with a 200 s baseline measurement in other wells containing tris buffer. The association step was performed on the serially diluted RBD protein for 120 or 150 s, and then the tips were dipped back into the tris buffer that was used in the previous baseline measurement for a 900 s dissociation step. After the dissociation step, the biosensors tips were regenerated in 10 mM glycine (pH 2.0) to remove bound IgG. Baseline subtraction was performed with tips dipped into the tris buffer without the presence of RBD protein. The experiment was repeated for at least two times for each combination of antibody and SARS-CoV or SARS-CoV-2 RBD protein. The 1:1 kinetic model fit overlay was applied to calculate the binding affinity of each antibody.
Pseudovirus neutralization test
SARS-CoV-2 Wuhan-hu-1 (ancestral), Alpha, Delta, Beta, Gamma, Omicron BA.1, Omicron BA.2, Omicron BA.5, Omicron BA.2.75.2, Omicron BA.4.6.1, BF.7, BQ.1.1, XBB.1, GX-P5L, WIV-1, and SARS-CoV full-length spike pseudotyped viruses were produced and packaged as previously described with minor modifications (57). Briefly, 5 million HEK293T cells were transfected with 20 μg of pCAGGS spike plasmid using FuGENE6 (Promega). At 24 hours after transfection, cells were incubated with vesicular stomatitis virus (VSV) ∆G luc seed virus (at multiplicity of infection of 5) for 2 hours. Following two PBS washes, the infected cells were replenished with complete growth media supplemented with 1:5000 diluted anti–VSV-glycoprotein mAb (clone 8GF11, Kerafast). At 24 hours after infection, pseudoviruses were harvested by centrifugation at 2000g for 5 min. For the pVNT assay, 3 × 106 RLU of pseudoviruses were preincubated with fourfold serially diluted mAb at a starting concentration of 20 μg/ml into a final volume of 50 μl for 1 hour at 37°C, followed by infection of huACE2 stably-expressing A549 cells. At 20 to 24 hours after infection, an equal volume of ONE-Glo luciferase substrate (Promega) was added, and the luminescence signal was measured using the Cytation 5 microplate reader (BioTek) with Gen5 software version 3.10.
Plaque reduction neutralization test
mAbs were serially diluted twofold using Dulbecco’s Modified Eagle’s Medium (DMEM) containing 2% fetal bovine serum (FBS) at a starting concentration of 2 μg/ml. SARS-CoV-2 virus (ancestral or Omicron BA.1 and BA.2 strains) was then diluted to 500 plaque-forming units/ml and mixed with the diluted mAbs and incubated at 37°C for 1 hour for the mAbs and viruses to bind. After 1 hour, the mAbs-virus mixture was added to the Vero E6 TMPRSS2 cell monolayer and incubated for a further 1 hour at 37°C. The inoculum was then decontaminated. The cells were replenished with plaque medium (DMEM supplemented with 2% FBS and 1% agarose) and incubated at 37°C for 3 days. Plaques were fixed and stained with 10% buffered formalin and 1% crystal violet, respectively.
SARS-CoV-2 S protein trimer purification
Lyophilized recombinant Wuhan-Hu-1 SARS-CoV-2 S protein trimer (GenBank accession code: QHD43416.1) (Acro Biosystems; catalog number: SPN-C52H9) was reconstituted according to the manufacturer’s instructions and subjected to gel filtration chromatography on a Superdex 200 Increase 300/100 GL column (GE Healthcare) using a buffer containing 50 mM tris (pH 7.40) and 300 mM NaCl. Fractions corresponding to the single, large peak seen at the approximate molecular weight of the S trimer were pooled and concentrated to 2.02 mg/ml S protein, as measured by Nanodrop (Thermo Fisher Scientific), using an Amicon centrifugal filter with a 100 kDa cutoff (Merck Millipore). The purity and size of the resulting purified S trimers were verified by SDS–polyacrylamide gel electrophoresis under denaturing conditions and dynamic light scattering, respectively.
Cryo-EM sample preparation
Purified SARS-CoV-2 S protein trimer was mixed with purified Fab E7 at a 1:3.6 molar ratio of S trimer:Fab, so that the final concentration of the S protein protomer in the solution was 0.5 mg/ml. UltrAuFoil 1.2/1.3 300-mesh gold grids (Ted Pella Inc.) were glow discharged at 5 mA for 60 s immediately before sample application. A total of 2.1 μl of the S protein–Fab E7 mixture was applied to a glow-discharged grid in a Vitrobot Mk IV (Thermo Fisher Scientific) operated at 4°C and 100% humidity. The sample was blotted off for 4 s with blot force 4 and 0.5 s drain time, immediately plunged into liquid ethane, and kept at liquid nitrogen temperature until it was ready to be imaged.
Cryo-EM imaging
Single-particle cryo-EM image acquisition was performed using SerialEM (59) on a Titan Krios transmission electron microscope (Thermo Fisher Scientific) operated at 300 kV and equipped with a K3 direct electron detector and GIF post-column energy filter (Gatan Inc.). A total of 4846 micrographs were acquired in counting mode at a nominal magnification of 81,000, corresponding to 1.105 Å per pixel on the camera, using beam image shift in SerialEM to target holes in a 3-by-3 multishot pattern after each stage movement. Micrographs were collected at a dose rate of 6.64 e−/Å2 s−1 with an exposure of 31.6 e−/Å2 s−1 per movie, fractionated into 25 frames, and with the energy filter operated in zero loss mode with a slit width of 20 eV.
Cryo-EM image processing
The movies were sorted into nine optics groups based on beam image shift values for processing in RELION (60). Movie frame alignment was performed in RELION, and contrast transfer function (CTF) parameters were determined using CTFFIND4 (61). Particle picking was performed by training the default BoxNet2 model in Warp (62), by manually picking all particles in 10 randomly selected micrographs, and using the resulting model to pick particles in all micrographs. Particles were extracted and binned by a factor of 4 for initial 2D classification, followed by manual selection of good 2D classes and extraction of particles from data binned by a factor of 2 for a second iteration of 2D classification. Good 2D classes were manually selected again; particles from these classes were extracted from unbinned images and then imported into cryoSPARC (38) for further processing.
Four initial 3D models were generated ab initio using cryoSPARC, of which one good model resembling an S trimer was identified and selected for use. A second 3D model was produced by scaling and low-pass filtering an existing S trimer map (EMDB accession code: EMD-11145) to 20 Å. Two copies of each 3D model were then used for a heterogeneous 3D refinement of the unbinned particles in cryoSPARC with four classes and without imposition of symmetry. Two good classes were manually selected and subjected to a second iteration of asymmetric (C1) heterogeneous refinement using the best class volume from the first iteration as a 3D model. Three classes were then manually selected and used to produce a 3D template by ab initio reconstruction in cryoSPARC with a single class. The selected particles and template were then used for C1 nonuniform refinement of the entire Fab–S trimer complex in cryoSPARC, resulting in a map at 3.69 Å resolution, as measured by gold standard Fourier shell correlation. Per-particle defocus, beam tilt, anisotropic magnification, and trefoil aberrations were refined in cryoSPARC, and a second iteration of C1 nonuniform refinement was performed to improve the resolution of the map to 3.32 Å. Polished particles were then imported into RELION for Bayesian polishing (63) and then reimported into cryoSPARC to repeat the CTF refinement steps described above with tetrafoil and spherical aberration refinement enabled. A third iteration of C1 nonuniform refinement resulted in a map at 3.12 Å resolution.
As the Fab-RBD complex density was poorer in quality than the rest of the map, the Fab and interacting RBD densities were manually identified and used to create a mask in UCSF Chimera (64). One iteration of C1 local refinement of this region was then performed using this mask in cryoSPARC, resulting in a 4.03 Å map with much more clearly resolved protein densities in this region.
Molecular model building
The S protein portion of the model was derived from an existing molecular model of the SARS-CoV-2 S protein trimer with one RBD in the up conformation [Protein Data Bank (PDB) accession code: 6XKL]. This model was docked into the nonuniform reconstruction of the spike-Fab complex, and individual chains were initially fit as rigid bodies using UCSF Chimera. The relevant residues in the fit S protein model were then mutated using Coot (65) to match the sequence of the commercial S protein preparation described above.
The sequence of the variable region of the Fab E7 heavy and light chains was used to generate an initial homology model using SWISS-MODEL (66), using the PDB models 7N3D and 5BK5 as templates for the heavy and light chains, respectively. The initial model for the Fab constant region was derived from PDB model 6N16, and the PDB atomic coordinates for the constant and variable region models were manually joined to produce a single model containing the heavy and light chains.
The Fab E7 and spike models were then combined into a single PDB model. The model for the spike region was subjected to real space refinement using Phenix (67) into the nonuniform refined map, and the same was done with the model for Fab E7 using the locally refined map of this region. Real space refinement was run using the default parameters in Phenix, with noncrystallographic symmetry (NCS) constraints and NCS refinement disabled. The fit of the spike and Fab-RBD models into the respective maps was manually improved in an iterative manner using Coot, and subsequent runs of real space refinement were performed in Phenix to improve the model quality. Possible epitope-paratope contacts were determined by first using the NCONT program from the CCP4 software package to identify pairs of Cα atoms in the final atomic model between (i) either chain of Fab E7 and (ii) either of the two RBDs in the binding site within <8.0 Å from each other. The amino acid residues in the model corresponding to these Cα pairs were then manually inspected in UCSF Chimera, UCSF ChimeraX (68), and Coot to determine which pairs of residues were able to interact, with a criterion of <3.5 Å used to filter possible hydrogen bond pairs.
Multiple sequence alignment
Full-length surface glycoprotein amino acid or nucleotide FASTA sequences were downloaded from UniProt, National Center for Biotechnology Information GenBank, and GISAID for a panel of SARS-CoV-2 variants, SARS-CoV, and zoonotic sarbecoviruses, with accession numbers listed in table S5. Downloaded nucleotide sequences were translated using Expasy Translate to obtain full-length S protein sequences. These full-length S protein sequences were used for a multiple sequence alignment using the default parameters in Clustal Omega (69) and rendered for interpretation using ESPript (70).
Data analysis
Data analysis was performed using GraphPad Prism 9 software to calculate IC50 for the mAbs.
Acknowledgments
We thank donor SS6V for the generous donation of blood sample for this study. We thank K. S. Tan for providing the ancestral (NUH-007) and Omicron (BA.1) viruses for the PRNT assays.
Funding: The work conducted at Duke-NUS was supported in part by Singapore National Research Foundation grant NRF2016NRF-NSFC002-013 (L.-F.W.), National Medical Research Council Singapore grant STPRG-FY19-001 (L.-F.W.), National Medical Research Council Singapore grant COVID19RF-003 (L.-F.W.), National Medical Research Council Singapore grant COVID19RF-060 (L.-F.W.), National Medical Research Council Singapore grant MOH-000535/MOH-OFYIRG19nov-0002 (C.W.T.), and National Medical Research Council Singapore grant OFLCG19May-0034 (L.-F.W.). The work conducted at University of Melbourne was supported in part by National Health and Medical Research Center Investigator grant GNT 2008092 (A.W.C.). The work conducted at Fred Hutchinson Cancer Research Centre was supported in part by the Damon Runyon Cancer Research Foundation (T.N.S.) and NIAIH/NIH grant K99AI166250 (T.N.S.).
Author contributions: Conceptualization: L.-F.W. Methodology: W.N.C., C.W.T., F.Z., G.F., A.W.K.T., S.L., B.Y., D.L., M.C., A.W.C., T.N.S., A.J.G., J.D.B., M.P., and J.B. Investigation: W.N.C., C.W.T., F.Z., A.Y.-Y.Y., W.C.Y., B.L.L., W.R.S., S.C., J.Z., M.S.S.K., G.F., A.W.K.T., T.-S.N., B.Y., E.L., T.N.S., A.J.G., S.C., and J.D.B. Visualization: W.N.C., C.W.T., A.W.K.T., G.F., E.L., A.W.C., T.N.S., and A.J.G. Funding acquisition: L.-F.W., S.L., D.L., A.W.C., T.N.S., J.D.B., M.P., G.G.A., and P.N.P. Project administration: L.-F.W. Supervision: L.-F.W., S.L., D.L., A.W.C., J.D.B., M.P., G.G.A., and P.N.P. Writing—original draft: W.N.C., C.W.T., F.Z., L.-F.W., A.W.K.T., and S.L. Writing—review and editing: All authors.
Competing interests: L.-F.W., W.N.C., C.W.T., and F.Z. are listed as inventors on patent number WO 2022/245288 A1, which discloses the subject matter described here. The patent is currently in filed stage, and the name of the organization filing the patent is National University of Singapore. J.D.B. consults for Apriori Bio and Flagship Labs 77 and is an inventor on patent number US 2021/0363661 A1, which is a patent related to deep mutational scanning currently in examination stage by Fred Hutchinson Cancer Research Centre and University of Washington. T.N.S. consults for Apriori Bio on deep mutational scanning. The remaining authors declare that they have no competing interests.
Data and materials availability: All data, code, and materials used in the analysis are available to any researcher for purposes of reproducing or extending the analysis. The monoclonal antibody materials can be provided by the corresponding authors pending scientific review and a completed material transfer agreement. Requests for the materials should be submitted to the corresponding author. Monoclonal antibody sequences reported in this study have been deposited in GenBank under the accession numbers OP019051 to OP019088. Raw sequencing data from DMS selections are available from the NCBI SRA, BioProject PRJNA639956, and BioSample SAMN27770976. The full computational pipeline for analyzing DMS sequencing data is available on Zenodo: https://zenodo.org/record/7956405 (DOI: 10.5281/zenodo.7956405). The cryo-EM density maps for the SARS-CoV-2 spike bound to Fab E7 are deposited in the Electron Microscopy Data Bank with accession codes EMD-33650 (whole spike bound to Fab E7) and EMD-33651 (local reconstruction of Fab E7 bound to two RBDs), with corresponding atomic models available in the Protein Data Bank with accession codes 7Y71 and 7Y72, respectively. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 to S6
REFERENCES AND NOTES
- 1.W. Li, Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, L. F. Wang, Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679 (2005). [DOI] [PubMed] [Google Scholar]
- 2.A. M. Zaki, S. van Boheemen, T. M. Bestebroer, A. D. Osterhaus, R. A. Fouchier, Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 (2012). [DOI] [PubMed] [Google Scholar]
- 3.P. Zhou, X. L. Yang, X. G. Wang, B. Hu, L. Zhang, W. Zhang, H. R. Si, Y. Zhu, B. Li, C. L. Huang, H. D. Chen, J. Chen, Y. Luo, H. Guo, R. D. Jiang, M. Q. Liu, Y. Chen, X. R. Shen, X. Wang, X. S. Zheng, K. Zhao, Q. J. Chen, F. Deng, L. L. Liu, B. Yan, F. X. Zhan, Y. Y. Wang, G. F. Xiao, Z. L. Shi, A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D. Delaune, V. Hul, E. A. Karlsson, A. Hassanin, T. P. Ou, A. Baidaliuk, F. Gambaro, M. Prot, V. T. Tu, S. Chea, L. Keatts, J. Mazet, C. K. Johnson, P. Buchy, P. Dussart, T. Goldstein, E. Simon-Loriere, V. Duong, A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat. Commun. 12, 6563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.T. T.-Y. Lam, N. Jia, Y.-W. Zhang, M. H.-H. Shum, J.-F. Jiang, H.-C. Zhu, Y.-G. Tong, Y.-X. Shi, X.-B. Ni, Y.-S. Liao, W.-J. Li, B.-G. Jiang, W. Wei, T.-T. Yuan, K. Zheng, X.-M. Cui, J. Li, G.-Q. Pei, X. Qiang, W. Y.-M. Cheung, L.-F. Li, F.-F. Sun, S. Qin, J.-C. Huang, G. M. Leung, E. C. Holmes, Y.-L. Hu, Y. Guan, W.-C. Cao, Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583, 282–285 (2020). [DOI] [PubMed] [Google Scholar]
- 6.S. Wacharapluesadee, C. W. Tan, P. Maneeorn, P. Duengkae, F. Zhu, Y. Joyjinda, T. Kaewpom, W. N. Chia, W. Ampoot, B. L. Lim, K. Worachotsueptrakun, V. C. Chen, N. Sirichan, C. Ruchisrisarod, A. Rodpan, K. Noradechanon, T. Phaichana, N. Jantarat, B. Thongnumchaima, C. Tu, G. Crameri, M. M. Stokes, T. Hemachudha, L. F. Wang, Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 12, 972 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.B. Hu, H. Guo, P. Zhou, Z. L. Shi, Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 19, 141–154 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.B. Israelow, T. Mao, J. Klein, E. Song, B. Menasche, S. B. Omer, A. Iwasaki, Adaptive immune determinants of viral clearance and protection in mouse models of SARS-CoV-2. Sci. Immunol. 6, eabl4509 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A. Sette, S. Crotty, Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 184, 861–880 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.F. Krammer, SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020). [DOI] [PubMed] [Google Scholar]
- 11.D. Planas, N. Saunders, P. Maes, F. Guivel-Benhassine, C. Planchais, J. Buchrieser, W. H. Bolland, F. Porrot, I. Staropoli, F. Lemoine, H. Pere, D. Veyer, J. Puech, J. Rodary, G. Baele, S. Dellicour, J. Raymenants, S. Gorissen, C. Geenen, B. Vanmechelen, T. Wawina-Bokalanga, J. Marti-Carreras, L. Cuypers, A. Seve, L. Hocqueloux, T. Prazuck, F. A. Rey, E. Simon-Loriere, T. Bruel, H. Mouquet, E. Andre, O. Schwartz, Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 602, 671–675 (2022). [DOI] [PubMed] [Google Scholar]
- 12.M. Tuccori, S. Ferraro, I. Convertino, E. Cappello, G. Valdiserra, C. Blandizzi, F. Maggi, D. Focosi, Anti-SARS-CoV-2 neutralizing monoclonal antibodies: Clinical pipeline. MAbs 12, 1854149 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Y. Cao, J. Wang, F. Jian, T. Xiao, W. Song, A. Yisimayi, W. Huang, Q. Li, P. Wang, R. An, J. Wang, Y. Wang, X. Niu, S. Yang, H. Liang, H. Sun, T. Li, Y. Yu, Q. Cui, S. Liu, X. Yang, S. Du, Z. Zhang, X. Hao, F. Shao, R. Jin, X. Wang, J. Xiao, Y. Wang, X. S. Xie, Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 602, 657–663 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Y. Cao, A. Yisimayi, F. Jian, W. Song, T. Xiao, L. Wang, S. Du, J. Wang, Q. Li, X. Chen, Y. Yu, P. Wang, Z. Zhang, P. Liu, R. An, X. Hao, Y. Wang, J. Wang, R. Feng, H. Sun, L. Zhao, W. Zhang, D. Zhao, J. Zheng, L. Yu, C. Li, N. Zhang, R. Wang, X. Niu, S. Yang, X. Song, Y. Chai, Y. Hu, Y. Shi, L. Zheng, Z. Li, Q. Gu, F. Shao, W. Huang, R. Jin, Z. Shen, Y. Wang, X. Wang, J. Xiao, X. S. Xie, BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature 608, 593–602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.S. Iketani, L. Liu, Y. Guo, L. Liu, J. F. Chan, Y. Huang, M. Wang, Y. Luo, J. Yu, H. Chu, K. K. Chik, T. T. Yuen, M. T. Yin, M. E. Sobieszczyk, Y. Huang, K. Y. Yuen, H. H. Wang, Z. Sheng, D. D. Ho, Antibody evasion properties of SARS-CoV-2 Omicron sublineages. Nature 604, 553–556 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.E. Takashita, S. Yamayoshi, V. Simon, H. van Bakel, E. M. Sordillo, A. Pekosz, S. Fukushi, T. Suzuki, K. Maeda, P. Halfmann, Y. Sakai-Tagawa, M. Ito, S. Watanabe, M. Imai, H. Hasegawa, Y. Kawaoka, Efficacy of antibodies and antiviral drugs against Omicron BA.2.12.1, BA.4, and BA.5 subvariants. N. Engl. J. Med. 387, 468–470 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. S. Low, J. Jerak, M. A. Tortorici, M. McCallum, D. Pinto, A. Cassotta, M. Foglierini, F. Mele, R. Abdelnabi, B. Weynand, J. Noack, M. Montiel-Ruiz, S. Bianchi, F. Benigni, N. Sprugasci, A. Joshi, J. E. Bowen, C. Stewart, M. Rexhepaj, A. C. Walls, D. Jarrossay, D. Morone, P. Paparoditis, C. Garzoni, P. Ferrari, A. Ceschi, J. Neyts, L. A. Purcell, G. Snell, D. Corti, A. Lanzavecchia, D. Veesler, F. Sallusto, ACE2-binding exposes the SARS-CoV-2 fusion peptide to broadly neutralizing coronavirus antibodies. Science 377, 735–742 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R. Otsubo, T. Minamitani, K. Kobiyama, J. Fujita, T. Ito, S. Ueno, I. Anzai, H. Tanino, H. Aoyama, Y. Matsuura, K. Namba, K. I. Imadome, K. J. Ishii, K. Tsumoto, W. Kamitani, T. Yasui, Human antibody recognition and neutralization mode on the NTD and RBD domains of SARS-CoV-2 spike protein. Sci. Rep. 12, 20120 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D. Pinto, M. M. Sauer, N. Czudnochowski, J. S. Low, M. A. Tortorici, M. P. Housley, J. Noack, A. C. Walls, J. E. Bowen, B. Guarino, L. E. Rosen, J. di Iulio, J. Jerak, H. Kaiser, S. Islam, S. Jaconi, N. Sprugasci, K. Culap, R. Abdelnabi, C. Foo, L. Coelmont, I. Bartha, S. Bianchi, C. Silacci-Fregni, J. Bassi, R. Marzi, E. Vetti, A. Cassotta, A. Ceschi, P. Ferrari, P. E. Cippa, O. Giannini, S. Ceruti, C. Garzoni, A. Riva, F. Benigni, E. Cameroni, L. Piccoli, M. S. Pizzuto, M. Smithey, D. Hong, A. Telenti, F. A. Lempp, J. Neyts, C. Havenar-Daughton, A. Lanzavecchia, F. Sallusto, G. Snell, H. W. Virgin, M. Beltramello, D. Corti, D. Veesler, Broad betacoronavirus neutralization by a stem helix-specific human antibody. Science 373, 1109–1116 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Y. Chen, X. Zhao, H. Zhou, H. Zhu, S. Jiang, P. Wang, Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses. Nat. Rev. Immunol. 23, 189–199 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.J. E. Bowen, Y. J. Park, C. Stewart, J. T. Brown, W. K. Sharkey, A. C. Walls, A. Joshi, K. R. Sprouse, M. McCallum, M. A. Tortorici, N. M. Franko, J. K. Logue, I. G. Mazzitelli, A. W. Nguyen, R. P. Silva, Y. Huang, J. S. Low, J. Jerak, S. W. Tiles, K. Ahmed, A. Shariq, J. M. Dan, Z. Zhang, D. Weiskopf, A. Sette, G. Snell, C. M. Posavad, N. T. Iqbal, J. Geffner, A. Bandera, A. Gori, F. Sallusto, J. A. Maynard, S. Crotty, W. C. Van Voorhis, C. Simmerling, R. Grifantini, H. Y. Chu, D. Corti, D. Veesler, SARS-CoV-2 spike conformation determines plasma neutralizing activity elicited by a wide panel of human vaccines. Sci. Immunol. 7, eadf1421 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D. Corti, L. A. Purcell, G. Snell, D. Veesler, Tackling COVID-19 with neutralizing monoclonal antibodies. Cell 184, 3086–3108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D. Pinto, Y. J. Park, M. Beltramello, A. C. Walls, M. A. Tortorici, S. Bianchi, S. Jaconi, K. Culap, F. Zatta, A. De Marco, A. Peter, B. Guarino, R. Spreafico, E. Cameroni, J. B. Case, R. E. Chen, C. Havenar-Daughton, G. Snell, A. Telenti, H. W. Virgin, A. Lanzavecchia, M. S. Diamond, K. Fink, D. Veesler, D. Corti, Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). [DOI] [PubMed] [Google Scholar]
- 24.T. N. Starr, N. Czudnochowski, Z. Liu, F. Zatta, Y. J. Park, A. Addetia, D. Pinto, M. Beltramello, P. Hernandez, A. J. Greaney, R. Marzi, W. G. Glass, I. Zhang, A. S. Dingens, J. E. Bowen, M. A. Tortorici, A. C. Walls, J. A. Wojcechowskyj, A. De Marco, L. E. Rosen, J. Zhou, M. Montiel-Ruiz, H. Kaiser, J. R. Dillen, H. Tucker, J. Bassi, C. Silacci-Fregni, M. P. Housley, J. di Iulio, G. Lombardo, M. Agostini, N. Sprugasci, K. Culap, S. Jaconi, M. Meury, E. Dellota Jr., R. Abdelnabi, S. C. Foo, E. Cameroni, S. Stumpf, T. I. Croll, J. C. Nix, C. Havenar-Daughton, L. Piccoli, F. Benigni, J. Neyts, A. Telenti, F. A. Lempp, M. S. Pizzuto, J. D. Chodera, C. M. Hebner, H. W. Virgin, S. P. J. Whelan, D. Veesler, D. Corti, J. D. Bloom, G. Snell, SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature 597, 97–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. A. Tortorici, N. Czudnochowski, T. N. Starr, R. Marzi, A. C. Walls, F. Zatta, J. E. Bowen, S. Jaconi, J. Di Iulio, Z. Wang, A. De Marco, S. K. Zepeda, D. Pinto, Z. Liu, M. Beltramello, I. Bartha, M. P. Housley, F. A. Lempp, L. E. Rosen, E. Dellota Jr., H. Kaiser, M. Montiel-Ruiz, J. Zhou, A. Addetia, B. Guarino, K. Culap, N. Sprugasci, C. Saliba, E. Vetti, I. Giacchetto-Sasselli, C. S. Fregni, R. Abdelnabi, S.-Y. C. Foo, C. Havenar-Daughton, M. A. Schmid, F. Benigni, E. Cameroni, J. Neyts, A. Telenti, H. W. Virgin, S. P. J. Whelan, G. Snell, J. D. Bloom, D. Corti, D. Veesler, M. S. Pizzuto, Broad sarbecovirus neutralization by a human monoclonal antibody. Nature 597, 103–108 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. A. Cohen, P. N. P. Gnanapragasam, Y. E. Lee, P. R. Hoffman, S. Ou, L. M. Kakutani, J. R. Keeffe, H. J. Wu, M. Howarth, A. P. West, C. O. Barnes, M. C. Nussenzweig, P. J. Bjorkman, Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science 371, 735–741 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.C.-W. Tan, W.-N. Chia, B. E. Young, F. Zhu, B.-L. Lim, W.-R. Sia, T.-L. Thein, M. I.-C. Chen, Y.-S. Leo, D. C. Lye, L.-F. Wang, Pan-sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N. Engl. J. Med. 385, 1401–1406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.M. Yuan, H. Liu, N. C. Wu, C. D. Lee, X. Zhu, F. Zhao, D. Huang, W. Yu, Y. Hua, H. Tien, T. F. Rogers, E. Landais, D. Sok, J. G. Jardine, D. R. Burton, I. A. Wilson, Structural basis of a shared antibody response to SARS-CoV-2. Science 369, 1119–1123 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Y.-J. Park, A. De Marco, T. N. Starr, Z. Liu, D. Pinto, A. C. Walls, F. Zatta, S. K. Zepeda, J. E. Bowen, K. R. Sprouse, A. Joshi, M. Giurdanella, B. Guarino, J. Noack, R. Abdelnabi, S.-Y. C. Foo, L. E. Rosen, F. A. Lempp, F. Benigni, G. Snell, J. Neyts, S. P. J. Whelan, H. W. Virgin, J. D. Bloom, D. Corti, M. S. Pizzuto, D. Veesler, Antibody-mediated broad sarbecovirus neutralization through ACE2 molecular mimicry. Science 375, 449–454 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.K. Westendorf, S. Zentelis, L. Wang, D. Foster, P. Vaillancourt, M. Wiggin, E. Lovett, R. van der Lee, J. Hendle, A. Pustilnik, J. M. Sauder, L. Kraft, Y. Hwang, R. W. Siegel, J. Chen, B. A. Heinz, R. E. Higgs, N. L. Kallewaard, K. Jepson, R. Goya, M. A. Smith, D. W. Collins, D. Pellacani, P. Xiang, V. de Puyraimond, M. Ricicova, L. Devorkin, C. Pritchard, A. O'Neill, K. Dalal, P. Panwar, H. Dhupar, F. A. Garces, C. A. Cohen, J. M. Dye, K. E. Huie, C. V. Badger, D. Kobasa, J. Audet, J. J. Freitas, S. Hassanali, I. Hughes, L. Munoz, H. C. Palma, B. Ramamurthy, R. W. Cross, T. W. Geisbert, V. Menachery, K. Lokugamage, V. Borisevich, I. Lanz, L. Anderson, P. Sipahimalani, K. S. Corbett, E. S. Yang, Y. Zhang, W. Shi, T. Zhou, M. Choe, J. Misasi, P. D. Kwong, N. J. Sullivan, B. S. Graham, T. L. Fernandez, C. L. Hansen, E. Falconer, J. R. Mascola, B. E. Jones, B. C. Barnhart, LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 39, 110812 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.T. N. Starr, A. J. Greaney, S. K. Hilton, D. Ellis, K. H. D. Crawford, A. S. Dingens, M. J. Navarro, J. E. Bowen, M. A. Tortorici, A. C. Walls, N. P. King, D. Veesler, J. D. Bloom, Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182, 1295–1310.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. ter Meulen, E. N. van den Brink, L. L. Poon, W. E. Marissen, C. S. Leung, F. Cox, C. Y. Cheung, A. Q. Bakker, J. A. Bogaards, E. van Deventer, W. Preiser, H. W. Doerr, V. T. Chow, J. de Kruif, J. S. Peiris, J. Goudsmit, Human monoclonal antibody combination against SARS coronavirus: Synergy and coverage of escape mutants. PLOS Med. 3, e237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.N. C. Wu, M. Yuan, S. Bangaru, D. Huang, X. Zhu, C.-C. D. Lee, H. L. Turner, L. Peng, L. Yang, D. R. Burton, D. Nemazee, A. B. Ward, I. A. Wilson, A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody. PLOS Pathog. 16, e1009089 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.W. Yu, X. Wu, J. Ren, X. Zhang, Y. Wang, C. Li, W. Xu, J. Li, G. Li, W. Zheng, H. Liao, X. Yuan, Mechanistic insights to the binding of antibody CR3022 against RBD from SARS-CoV and HCoV-19/SARS-CoV-2: A computational study. Comb. Chem. High Throughput Screen. 24, 1069–1082 (2021). [DOI] [PubMed] [Google Scholar]
- 35.E. Lopez, E. R. Haycroft, A. Adair, F. L. Mordant, M. T. O'Neill, P. Pymm, S. J. Redmond, W. S. Lee, N. A. Gherardin, A. K. Wheatley, J. A. Juno, K. J. Selva, S. K. Davis, S. L. Grimley, L. Harty, D. F. Purcell, K. Subbarao, D. I. Godfrey, S. J. Kent, W. H. Tham, A. W. Chung, Simultaneous evaluation of antibodies that inhibit SARS-CoV-2 variants via multiplex assay. JCI Insight 6, e150012 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A.-L. Mader, L. Tydykov, V. Gluck, M. Bertok, T. Weidlich, C. Gottwald, A. Stefl, M. Vogel, A. Plentz, J. Kostler, B. Salzberger, J. J. Wenzel, H. H. Niller, J. Jantsch, R. Wagner, B. Schmidt, T. Gluck, A. Gessner, D. Peterhoff, Omicron's binding to sotrovimab, casirivimab, imdevimab, CR3022, and sera from previously infected or vaccinated individuals. iScience 25, 104076 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.M. Yuan, X. Zhu, W. T. He, P. Zhou, C. I. Kaku, T. Capozzola, C. Y. Zhu, X. Yu, H. Liu, W. Yu, Y. Hua, H. Tien, L. Peng, G. Song, C. A. Cottrell, W. R. Schief, D. Nemazee, L. M. Walker, R. Andrabi, D. R. Burton, I. A. Wilson, A broad and potent neutralization epitope in SARS-related coronaviruses. Proc. Natl. Acad. Sci. U.S.A. 119, e2205784119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.A. Punjani, J. L. Rubinstein, D. J. Fleet, M. A. Brubaker, cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 39.C. O. Barnes, C. A. Jette, M. E. Abernathy, K. A. Dam, S. R. Esswein, H. B. Gristick, A. G. Malyutin, N. G. Sharaf, K. E. Huey-Tubman, Y. E. Lee, D. F. Robbiani, M. C. Nussenzweig, A. P. West Jr., P. J. Bjorkman, SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.W. Dejnirattisai, J. Huo, D. Zhou, J. Zahradnik, P. Supasa, C. Liu, H. M. E. Duyvesteyn, H. M. Ginn, A. J. Mentzer, A. Tuekprakhon, R. Nutalai, B. Wang, A. Dijokaite, S. Khan, O. Avinoam, M. Bahar, D. Skelly, S. Adele, S. A. Johnson, A. Amini, T. G. Ritter, C. Mason, C. Dold, D. Pan, S. Assadi, A. Bellass, N. Omo-Dare, D. Koeckerling, A. Flaxman, D. Jenkin, P. K. Aley, M. Voysey, S. A. C. Clemens, F. G. Naveca, V. Nascimento, F. Nascimento, C. F. da Costa, P. C. Resende, A. Pauvolid-Correa, M. M. Siqueira, V. Baillie, N. Serafin, G. Kwatra, K. Da Silva, S. A. Madhi, M. C. Nunes, T. Malik, P. J. M. Openshaw, J. K. Baillie, M. G. Semple, A. R. Townsend, K. A. Huang, T. K. Tan, M. W. Carroll, P. Klenerman, E. Barnes, S. J. Dunachie, B. Constantinides, H. Webster, D. Crook, A. J. Pollard, T. Lambe, O. Consortium, I. C. Consortium, N. G. Paterson, M. A. Williams, D. R. Hall, E. E. Fry, J. Mongkolsapaya, J. Ren, G. Schreiber, D. I. Stuart, G. R. Screaton, SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell 185, 467–484.e15 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.A. Rossler, L. Knabl, D. von Laer, J. Kimpel, Neutralization profile after recovery from SARS-CoV-2 Omicron infection. N. Engl. J. Med. 386, 1764–1766 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.P. Wang, M. S. Nair, L. Liu, S. Iketani, Y. Luo, Y. Guo, M. Wang, J. Yu, B. Zhang, P. D. Kwong, B. S. Graham, J. R. Mascola, J. Y. Chang, M. T. Yin, M. Sobieszczyk, C. A. Kyratsous, L. Shapiro, Z. Sheng, Y. Huang, D. D. Ho, Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593, 130–135 (2021). [DOI] [PubMed] [Google Scholar]
- 43.F. Bertoglio, V. Fuhner, M. Ruschig, P. A. Heine, L. Abassi, T. Klunemann, U. Rand, D. Meier, N. Langreder, S. Steinke, R. Ballmann, K. T. Schneider, K. D. R. Roth, P. Kuhn, P. Riese, D. Schackermann, J. Korn, A. Koch, M. Z. Chaudhry, K. Eschke, Y. Kim, S. Zock-Emmenthal, M. Becker, M. Scholz, G. Moreira, E. V. Wenzel, G. Russo, H. S. P. Garritsen, S. Casu, A. Gerstner, G. Roth, J. Adler, J. Trimpert, A. Hermann, T. Schirrmann, S. Dubel, A. Frenzel, J. Van den Heuvel, L. Cicin-Sain, M. Schubert, M. Hust, A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations. Cell Rep. 36, 109433 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.L. Wang, T. Zhou, Y. Zhang, E. S. Yang, C. A. Schramm, W. Shi, A. Pegu, O. K. Oloniniyi, A. R. Henry, S. Darko, S. R. Narpala, C. Hatcher, D. R. Martinez, Y. Tsybovsky, E. Phung, O. M. Abiona, A. Antia, E. M. Cale, L. A. Chang, M. Choe, K. S. Corbett, R. L. Davis, A. T. DiPiazza, I. J. Gordon, S. H. Hait, T. Hermanus, P. Kgagudi, F. Laboune, K. Leung, T. Liu, R. D. Mason, A. F. Nazzari, L. Novik, S. O'Connell, S. O'Dell, A. S. Olia, S. D. Schmidt, T. Stephens, C. D. Stringham, C. A. Talana, I. T. Teng, D. A. Wagner, A. T. Widge, B. Zhang, M. Roederer, J. E. Ledgerwood, T. J. Ruckwardt, M. R. Gaudinski, P. L. Moore, N. A. Doria-Rose, R. S. Baric, B. S. Graham, A. B. McDermott, D. C. Douek, P. D. Kwong, J. R. Mascola, N. J. Sullivan, J. Misasi, Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants. Science 373, eabh1766 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.D. R. Martinez, A. Schafer, S. Gobeil, D. Li, G. De la Cruz, R. Parks, X. Lu, M. Barr, V. Stalls, K. Janowska, E. Beaudoin, K. Manne, K. Mansouri, R. J. Edwards, K. Cronin, B. Yount, K. Anasti, S. A. Montgomery, J. Tang, H. Golding, S. Shen, T. Zhou, P. D. Kwong, B. S. Graham, J. R. Mascola, D. C. Montefiori, S. M. Alam, G. Sempowski, G. D. Sempowski, S. Khurana, K. Wiehe, K. O. Saunders, P. Acharya, B. F. Haynes, R. S. Baric, A broadly cross-reactive antibody neutralizes and protects against sarbecovirus challenge in mice. Sci. Transl. Med. 14, eabj7125 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.L. Witte, V. A. Baharani, F. Schmidt, Z. Wang, A. Cho, R. Raspe, C. Guzman-Cardozo, F. Muecksch, M. Canis, D. J. Park, C. Gaebler, M. Caskey, M. C. Nussenzweig, T. Hatziioannou, P. D. Bieniasz, Epistasis lowers the genetic barrier to SARS-CoV-2 neutralizing antibody escape. Nat. Commun. 14, 302 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Z. Cui, P. Liu, N. Wang, L. Wang, K. Fan, Q. Zhu, K. Wang, R. Chen, R. Feng, Z. Jia, M. Yang, G. Xu, B. Zhu, W. Fu, T. Chu, L. Feng, Y. Wang, X. Pei, P. Yang, X. S. Xie, L. Cao, Y. Cao, X. Wang, Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron. Cell 185, 860–871.e13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.V. Stalls, J. Lindenberger, S. M. Gobeil, R. Henderson, R. Parks, M. Barr, M. Deyton, M. Martin, K. Janowska, X. Huang, A. May, M. Speakman, E. Beaudoin, B. Kraft, X. Lu, R. J. Edwards, A. Eaton, D. C. Montefiori, W. B. Williams, K. O. Saunders, K. Wiehe, B. F. Haynes, P. Acharya, Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike. Cell Rep. 39, 111009 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.W. Yin, Y. Xu, P. Xu, X. Cao, C. Wu, C. Gu, X. He, X. Wang, S. Huang, Q. Yuan, K. Wu, W. Hu, Z. Huang, J. Liu, Z. Wang, F. Jia, K. Xia, P. Liu, X. Wang, B. Song, J. Zheng, H. Jiang, X. Cheng, Y. Jiang, S. J. Deng, H. E. Xu, Structures of the Omicron spike trimer with ACE2 and an anti-Omicron antibody. Science 375, 1048–1053 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.C. G. Rappazzo, L. V. Tse, C. I. Kaku, D. Wrapp, M. Sakharkar, D. Huang, L. M. Deveau, T. J. Yockachonis, A. S. Herbert, M. B. Battles, C. M. O'Brien, M. E. Brown, J. C. Geoghegan, J. Belk, L. Peng, L. Yang, Y. Hou, T. D. Scobey, D. R. Burton, D. Nemazee, J. M. Dye, J. E. Voss, B. M. Gunn, J. S. McLellan, R. S. Baric, L. E. Gralinski, L. M. Walker, Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Science 371, 823–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.C. A. Jette, A. A. Cohen, P. N. P. Gnanapragasam, F. Muecksch, Y. E. Lee, K. E. Huey-Tubman, F. Schmidt, T. Hatziioannou, P. D. Bieniasz, M. C. Nussenzweig, A. P. West, J. R. Keeffe, P. J. Bjorkman, C. O. Barnes, Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies. Cell Rep. 36, 109760 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.R. Yan, R. Wang, B. Ju, J. Yu, Y. Zhang, N. Liu, J. Wang, Q. Zhang, P. Chen, B. Zhou, Y. Li, Y. Shen, S. Zhang, L. Tian, Y. Guo, L. Xia, X. Zhong, L. Cheng, X. Ge, J. Zhao, H. W. Wang, X. Wang, Z. Zhang, L. Zhang, Q. Zhou, Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies. Cell Res. 31, 517–525 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.L. Liu, P. Wang, M. S. Nair, J. Yu, M. Rapp, Q. Wang, Y. Luo, J. F.-W. Chan, V. Sahi, A. Figueroa, X. V. Guo, G. Cerutti, J. Bimela, J. Gorman, T. Zhou, Z. Chen, K.-Y. Yuen, P. D. Kwong, J. G. Sodroski, M. T. Yin, Z. Sheng, Y. Huang, L. Shapiro, D. D. Ho, Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Z. Liu, W. Xu, Z. Chen, W. Fu, W. Zhan, Y. Gao, J. Zhou, Y. Zhou, J. Wu, Q. Wang, X. Zhang, A. Hao, W. Wu, Q. Zhang, Y. Li, K. Fan, R. Chen, Q. Jiang, C. T. Mayer, T. Schoofs, Y. Xie, S. Jiang, Y. Wen, Z. Yuan, K. Wang, L. Lu, L. Sun, Q. Wang, An ultrapotent pan-β-coronavirus lineage B (β-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope. Protein Cell 13, 655–675 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.J. Goike, C. L. Hsieh, A. Horton, E. C. Gardner, F. Bartzoka, N. Wang, K. Javanmardi, A. Herbert, S. Abbassi, R. Renberg, M. J. Johanson, J. A. Cardona, T. Segall-Shapiro, L. Zhou, R. H. Nissly, A. Gontu, M. Byrom, A. C. Maranhao, A. M. Battenhouse, V. Gejji, L. Soto-Sierra, E. R. Foster, S. L. Woodard, Z. L. Nikolov, J. Lavinder, W. N. Voss, A. Annapareddy, G. C. Ippolito, A. D. Ellington, E. M. Marcotte, I. J. Finkelstein, R. A. Hughes, J. M. Musser, S. V. Kuchipudi, V. Kapur, G. Georgiou, J. M. Dye, D. R. Boutz, J. S. McLellan, J. D. Gollihar, Synthetic repertoires derived from convalescent COVID-19 patients enable discovery of SARS-CoV-2 neutralizing antibodies and a novel quaternary binding modality. bioRxiv 438849 [Preprint] 9 April 2021. 10.1101/2021.04.07.438849. [DOI]
- 56.K. Smith, L. Garman, J. Wrammert, N. Y. Zheng, J. D. Capra, R. Ahmed, P. C. Wilson, Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat. Protoc. 4, 372–384 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.C. W. Tan, W. N. Chia, X. Qin, P. Liu, M. I. C. Chen, C. Tiu, Z. Hu, V. C.-W. Chen, B. E. Young, W. R. Sia, Y.-J. Tan, R. Foo, Y. Yi, D. C. Lye, D. E. Anderson, L.-F. Wang, A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 38, 1073–1078 (2020). [DOI] [PubMed] [Google Scholar]
- 58.A. J. Greaney, T. N. Starr, P. Gilchuk, S. J. Zost, E. Binshtein, A. N. Loes, S. K. Hilton, J. Huddleston, R. Eguia, K. H. D. Crawford, A. S. Dingens, R. S. Nargi, R. E. Sutton, N. Suryadevara, P. W. Rothlauf, Z. Liu, S. P. J. Whelan, R. H. Carnahan, J. E. Crowe Jr., J. D. Bloom, Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe 29, 44–57.e9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.D. N. Mastronarde, Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 60.S. H. W. Scheres, RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.A. Rohou, N. Grigorieff, CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D. Tegunov, P. Cramer, Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.J. Zivanov, T. Nakane, S. H. W. Scheres, A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin, UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 65.P. Emsley, B. Lohkamp, W. G. Scott, K. Cowtan, Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.A. Waterhouse, M. Bertoni, S. Bienert, G. Studer, G. Tauriello, R. Gumienny, F. T. Heer, T. A. P. de Beer, C. Rempfer, L. Bordoli, R. Lepore, T. Schwede, SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D. Liebschner, P. V. Afonine, M. L. Baker, G. Bunkoczi, V. B. Chen, T. I. Croll, B. Hintze, L. W. Hung, S. Jain, A. J. McCoy, N. W. Moriarty, R. D. Oeffner, B. K. Poon, M. G. Prisant, R. J. Read, J. S. Richardson, D. C. Richardson, M. D. Sammito, O. V. Sobolev, D. H. Stockwell, T. C. Terwilliger, A. G. Urzhumtsev, L. L. Videau, C. J. Williams, P. D. Adams, Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.T. D. Goddard, C. C. Huang, E. C. Meng, E. F. Pettersen, G. S. Couch, J. H. Morris, T. E. Ferrin, UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.F. Sievers, A. Wilm, D. Dineen, T. J. Gibson, K. Karplus, W. Li, R. Lopez, H. McWilliam, M. Remmert, J. Soding, J. D. Thompson, D. G. Higgins, Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.X. Robert, P. Gouet, Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 to S6