Abstract
Cell death coordinates repair programs following pathogen attack and tissue injury. However, aberrant cell death can interfere with such programs and cause organ failure. Cellular FLICE-like inhibitory protein (cFLIP) is a crucial regulator of cell death and a substrate of Caspase-8. However, the physiological role of cFLIP cleavage by Caspase-8 remains elusive. Here, we found an essential role for cFLIP cleavage in restraining cell death in different pathophysiological scenarios. Mice expressing a cleavage-resistant cFLIP mutant, CflipD377A, exhibited increased sensitivity to severe acute respiratory syndrome coronavirus (SARS-CoV)–induced lethality, impaired skin wound healing, and increased tissue damage caused by Sharpin deficiency. In vitro, abrogation of cFLIP cleavage sensitizes cells to tumor necrosis factor(TNF)–induced necroptosis and apoptosis by favoring complex-II formation. Mechanistically, the cell death–sensitizing effect of the D377A mutation depends on glutamine-469. These results reveal a crucial role for cFLIP cleavage in controlling the amplitude of cell death responses occurring upon tissue stress to ensure the execution of repair programs.
cFLIP cleavage acts as a molecular brake on cell death and is essential for the ability of tissues to overcome damage.
INTRODUCTION
Cell death is a fundamental biological process that ensures tissue homeostasis and orchestrates tissue remodeling following injury or infection. However, if on the one hand abrogation of cell death responses can prevent the activation of repair programs, on the other hand exacerbated cell death can lead to tissue failure (1–3). Therefore, the ability of tissues to control the extent of cell death under stress conditions is of fundamental importance for the activation of optimal repair programs. Tumor necrosis factor (TNF) is a proinflammatory cytokine that is produced in response to a large variety of stressors, including viral infection and injury, and it can initiate repair processes by inducing the expression of proinflammatory genes or by triggering cell death (1, 4, 5). The mechanisms regulating the decision between these two outcomes are of fundamental importance for the maintenance of tissue homeostasis and for the capacity of tissues to overcome damage (6, 7).
Binding of TNF to TNF receptor 1 (TNFR1) results in the formation of two spatially and temporally distinct complexes (8). A membrane-bound complex, called complex-I, is assembled on the intracellular death domain (DD)–containing portion of TNFR1. It is composed of adaptor proteins, such as TNFR1-associated death domain protein (TRADD) and TRAF2 (TNFR-associated factor 2); kinases, such as RIPK1 (receptor-interacting protein kinase 1), IKKα and IKKβ [inhibitor of nuclear factor κB (NF-κB) kinase subunits α and β], TAK1 (transforming growth factor β–activated kinase 1), TANK-binding kinase 1 (TBK1), and IKKε; and E3 ligases, such as cellular inhibitor of apoptosis protein 1 and 2 (cIAP1/2) and linear ubiquitin chain assembly complex (LUBAC) (9–14). The concerted action of phosphorylation and ubiquitination events ensures the correct assembly and stability of the complex, leading to the activation of genes required to mount an inflammatory response (1, 15, 16). Any perturbation of phosphorylation or ubiquitination processes leads to the formation of a secondary, cytoplasmic complex, referred to as complex-II (1, 14, 16–21). This complex consists of Fas associated via death domain (FADD), RIPK1, cellular FLICE-like inhibitory protein (cFLIPL), Caspase-8 and, depending on the cell types, RIPK3 (18, 22). Complex-II has cytotoxic activity and can trigger Caspase-8–mediated apoptosis and RIPK1/RIPK3/mixed lineage kinase domain like pseudokinase (MLKL)–mediated necroptosis (23). In immune cells, Caspase-8 activation can result in gasdermin D cleavage with the consequent induction of pyroptosis (24). Apart from TNFR1, other immune receptors including Toll-like receptor 3 (TLR3), TLR4, and type I and type II interferon (IFN) receptors have the potential to induce a complex-II–like cell death–inducing platform (25–27). The cFLIPL/Caspase-8 heterodimer acts as a molecular switch that controls the cell death outcomes of complex-II. While the cFLIPL/Caspase-8 heterodimer is required to suppress necroptosis, the Caspase-8/Caspase-8 homodimer formation is required for the activation of downstream Caspase-3 and Caspase-7 and the induction of apoptosis (28–30). Therefore, cFLIPL (here referred to as cFLIP) represents both an activator and an inhibitor of Caspase-8, because it is needed for the ability of the latter to suppress necroptosis, but at the same time, it prevents Caspase-8 full activation and apoptosis (28). cFLIP is a catalytically inactive homolog of Caspase-8, composed of tandem death effector domains (DED) and a caspase-like domain formed of a large and a small subunit (31). Apart from being the most direct, nonredundant regulator of Caspase-8 activity, cFLIP is also a Caspase-8 substrate. Caspase-8 can cleave cFLIP at aspartic acid-367 (D376) (D377 in mouse) (32). Different studies have addressed the function of the cleavage at D376 mainly using cell-free systems and hybrid protein expression (31, 33, 34). A recent report showed that mice bearing a noncleavable cFLIP mutant, where D371 and D377 were mutated to alanine, were born at the expected Mendelian ratio, and thymocytes derived from these double-mutant mice were more resistant to FasL-induced cell death than their wild-type (WT) counterparts (35). However, the biological role of the proteolytic event on cFLIP remains to be elucidated.
Here, we characterized the dynamics and function of cFLIP cleavage at D377 both in vivo and in vitro by generating a mutant mouse bearing a point mutation that abrogates the cleavage at D377 (CflipD377A mice). The D377A mutation renders mice more sensitive to the lethal effect of severe acute respiratory syndrome coronavirus (SARS-CoV) infection, exacerbates the phenotype of the Sharpincpdm mice, and impairs skin wound healing. Cells derived from the CflipD377A mice are substantiallyt more sensitive to TNF-induced necroptosis and apoptosis and exhibit increased complex-II formation. At the mechanistic level, we observed that glutamine-469 (Q469), a residue important for cFLIP heterodimerization with Caspase-8, is responsible for the cell death–sensitizing effect of the cFLIP-D377A mutant. Therefore, we report on the precise biological function of cFLIP cleavage in keeping cell death responses in check during pathogen attack, tissue injury, and tissue damage, to favor remodeling and repair.
RESULTS
cFLIP cleavage by Caspase-8 limits TNF cytotoxicity
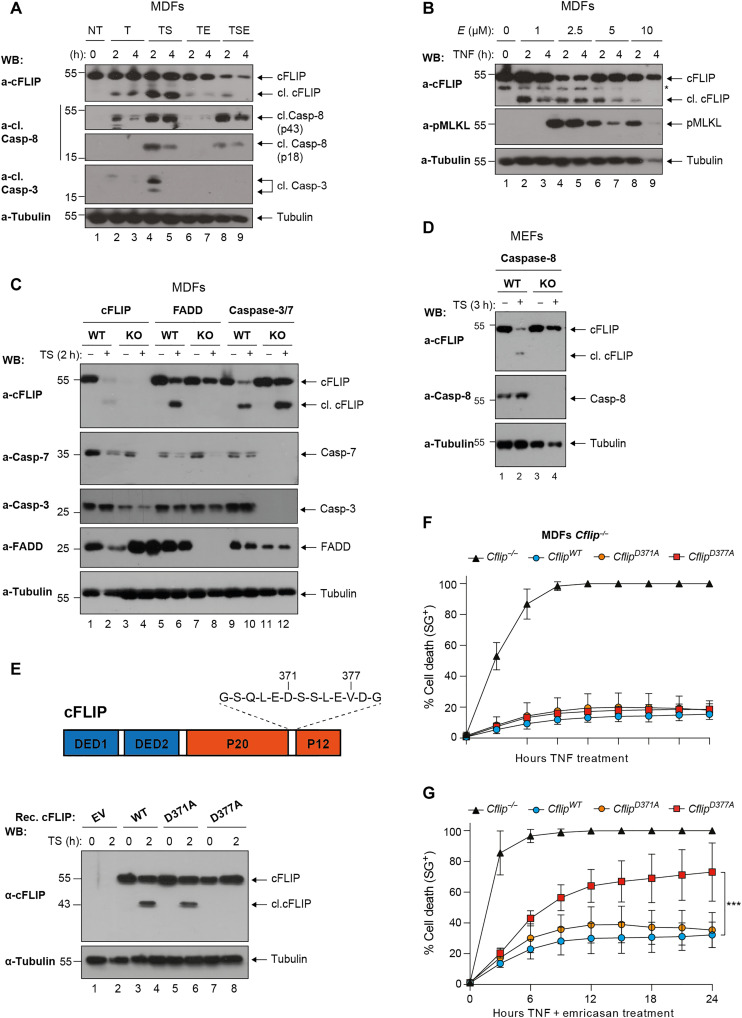
We first characterized the dynamics of cFLIP cleavage in TNF signaling in vitro. TNF treatment alone (T) or in combination with Smac mimetic (S) and/or emricasan (E) rapidly induced cleavage of cFLIP in mouse dermal fibroblasts (MDFs), as demonstrated by the appearance of the p43 fragment (Fig. 1A). Notably, cFLIP cleavage occurs in the absence of full Caspase-8 activation, as in the case of TNF-treatment alone, and despite the inhibition of Caspase-8 activity (Fig. 1, A and B). We next addressed the molecular requirements for cFLIP cleavage, and we found that FADD and Caspase-8, but not Caspase-3/7, are necessary for cFLIP to be cleaved (Fig. 1, C and D, and fig. S1, A to D). Because the co-deletion of Caspase-3 and Caspase-7 did not affect the cleavage event (Fig. 1C), we conclude that, upon TNF stimulation, the formation of a FADD/Caspase-8 complex is required for cFLIP cleavage and that Caspase-8 is most likely the only caspase able to cleave cFLIP in mouse cells. cFLIP bears an aspartic acid residue in close proximity to D377, the D371, which could potentially undergo Caspase-8–mediated cleavage (Fig. 1E) (35). To elucidate whether D371 can substitute for D377A, we reconstituted Cflip−/− MDFs with either WT cFLIP or mutant cFLIP bearing alanine substitutions in D371 or D377 (fig. S1E). We found that, whereas D371 is dispensable for cFLIP cleavage, D377 is the only cleavage site of cFLIP upon TNF signaling pathway activation (Fig. 1E and fig. S1F). Reexpression of the WT or mutant cFLIP variants could protect the Cflip−/− MDFs from TNF-induced cell death (Fig. 1F). Intriguingly, the cleavage-deficient D377A mutant cFLIP sensitized MDFs to TNF and emricasan (TE)–induced necroptosis (Fig. 1G). These findings indicate that cFLIP is cleaved by Caspase-8 following TNF signaling pathway stimulation at residue D377 and that this cleavage event has prosurvival functions.
Fig. 1. cFLIP is cleaved at D377 following TNFR1 activation.
(A) WT MDFs were treated with TNF (T: 100 ng/ml), TNF and Smac mimetic (TS, S:250 nM), TNF and emricasan (TE, E:1 μM) and TNF, Smac mimetic and emricasan (TSE) for the indicated time points. Cell lysates were analyzed by immunoblotting for the indicated specific antibodies (n = 2). WB, Western blot. (B) WT MDFs were treated with TNF (100 ng/ml) and increasing concentration of emricasan for the indicated time points. Cell lysates were analyzed by immunoblotting with the indicated specific antibodies (n = 3). The asterisk (*) indicates a nonspecific band. (C) MDFs and (D) mouse embryonic fibroblasts (MEFs) of the indicated genotypes were treated with TNF (100 ng/ml) and Smac mimetics (250 nM) for 2 hours, and cell lysates were analyzed by immunoblotting with the indicated specific antibodies (n = 2). KO, knockout. (E) Cartoon depicting cFLIP domain composition and D371 and D377 (top). Cflip−/− MDFs were reconstituted either with empty virus or virus expressing WT or the indicated mutant versions of cFLIP and treated with TNF (100 ng/ml) and Smac mimetic (250 nM) for 2 hours. Cell lysates were analyzed by immunoblotting with the indicated specific antibodies (n = 2). (F and G) Cells as in (E) were treated with TNF (10 ng/ml) alone (F) or in combination with emricasan (G) (1 μM), and cell death was measured over time by calculating the percentage of Sytox Green–positive cells (SG+) (n = 4). Data are presented as means ± SD; **P < 0.01 (n = 4).
Noncleavable cFLIP sensitizes to TNF-induced necroptosis and apoptosis
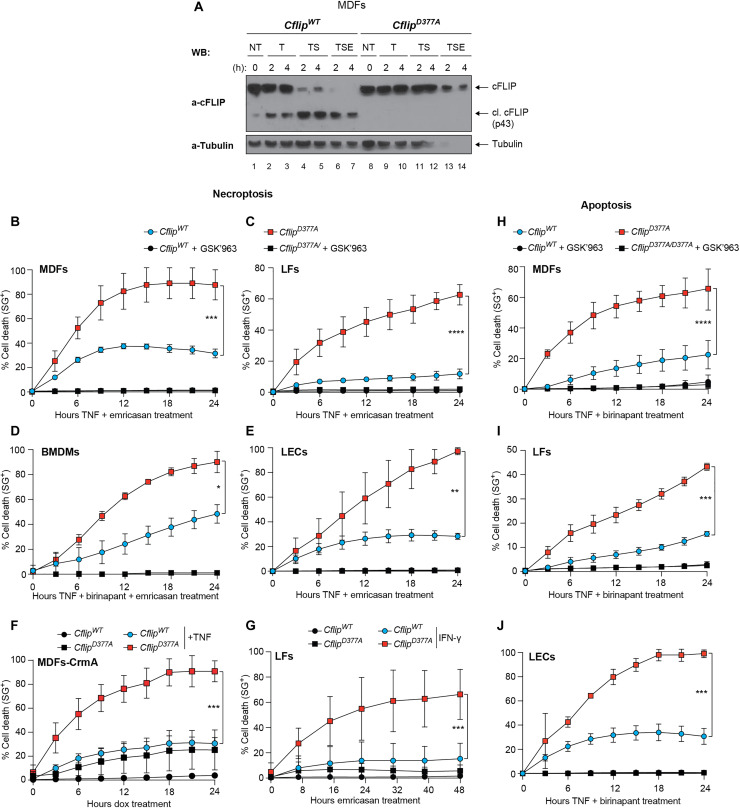
To study the physiological function of cFLIP cleavage at D377, we generated cFLIP cleavage–resistant mice, referred to as CflipD377A mice, where the D377 was mutated to alanine (fig. S2A). The CflipD377A mutant mice were weaned at the expected Mendelian ratio and do not exhibit any overt phenotype (fig. S2, B to D). These mice had an overtly normal immune system (fig. S2E). MDFs isolated from WT and cFLIP mutant mice had comparable levels of cFLIP, and in the D377A mutant cells, cFLIP could not undergo proteolytic cleavage following TNFR1 pathway activation (Fig. 2A). Next, we tested the role of cFLIP cleavage in TNF-induced cell death. While CflipD377A cells were not sensitive to TNF treatment alone (fig. S3A), MDFs, lung endothelial cells (LECs) and lung fibroblasts (LFs) were significantly more sensitive to TE- and bone marrow–derived macrophages (BMDMs) to TNF, emricasan, and Smac mimetic birinapant (TBE)–induced necroptosis, compared to their WT counterparts (Fig. 2, B to E). Consistent with the fact that RIPK1 kinase activity is required for necroptosis induction (36), the selective RIPK1 inhibitor GSK’963 suppressed TE- and TBE-mediated killing (Fig. 2, B to E). In addition, D377A mutant MDFs were also significantly more sensitive than WT MDFs to TNF treatment following expression of CrmA (37), a cowpox virus–encoded Caspase-8 inhibitor, via a doxycycline-induced construct (Fig. 2F and fig. S3B). Necroptosis can also be triggered by IFN-γ–induced Z-DNA-binding protein 1 (ZBP1) up-regulation, in combination with Caspase-8 inhibition (25). cFLIP mutant cells were also more sensitive than WT cells to IFN-γ and emricasan (IE)–induced necroptosis, in a TNF-independent manner (Fig. 2G and fig. S3C). cFLIP mutant cells were also more sensitive to TNF and birinapant (TB)– and TNF and cycloheximide (TC)–induced apoptosis (Fig. 2, H to J, and fig. S3D). TB-induced cell death was also RIPK1 kinase dependent, because the GSK’963 inhibitor could abolish cell death both in WT and mutant cells (Fig. 2, H to J). Last, the genotoxic drugs gemcitabine, paclitaxel, and doxorubicin, known to activate intrinsic apoptosis, killed WT and D377A mutant cells to the same extent (fig. S3, E to H). Together, these data indicate that cFLIP cleavage limits the ability of TNF to induce apoptosis and necroptosis in multiple cell types and also the ability of IFN-γ to trigger necroptosis, while it plays no role in cell death processes induced by genotoxins.
Fig. 2. Cleavage-resistant cFLIP mutant sensitizes to TNF-induced necroptosis and apoptosis.
(A) CflipWT and CflipD377A MDFs were treated as indicated [TNF (100 ng/ml), emricasan (1 μg/ml), and birinapant (250 nM)], and cell lysates were analyzed by immunoblotting using the indicated specific antibodies (n = 2). (B) WT and D377A mutant MDFs, (C) LFs, (D) BMDMs and (E) LECs were treated with TNF and emricasan (MDFs, LFs, and LECs) or TNF, birinapant, and emricasan (BMDMs) in the presence or not of the RIPK1-specific inhibitor GSK’963 (100 nM), and cell death was measured over time by calculating the percentage of Sytox Green–positive cells. For MDFs, TNF (1 ng/ml) and 1 μM emricasan (n = 6), for LFs, TNF (10 ng/ml) and 1 μM emricasan (n = 4) and for BMDMs and LECs, TNF (10 ng/ml), 250 nM birinapant, and 1 μM emricasan (n = 5 and n = 3, respectively). (F) CflipWT and CflipD377A MDFs stably expressing a doxycycline-inducible HA-tagged CrmA construct were treated for 48 hours with doxycycline (1 μg/ml) and then treated or not with TNF (100 ng/ml). Cell death was measured as in (B) to (E) (n = 3). (G) WT and D377A mutant LFs were pretreated with IFN-γ for 18 hours and then subjected to emricasan (1 μm) treatment. Cell death was measured as in (B) to (E) (n = 3). (H) WT and D377A mutant MDFs, (I) LFs, and (J) LECs were treated with TNF and birinapant in the presence or not of the RIPK1-specific inhibitor GSK’963 (100 nM), and cell death was measured over time by calculating the percentage of Sytox Green–positive cells. For MDFs, LFs, and LECs, TNF (10 ng/ml) and 250 nM birinapant, n = 4. Data are presented as means ± SD; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
The D377A mutation enhances complex-II formation, independently of NF-kB
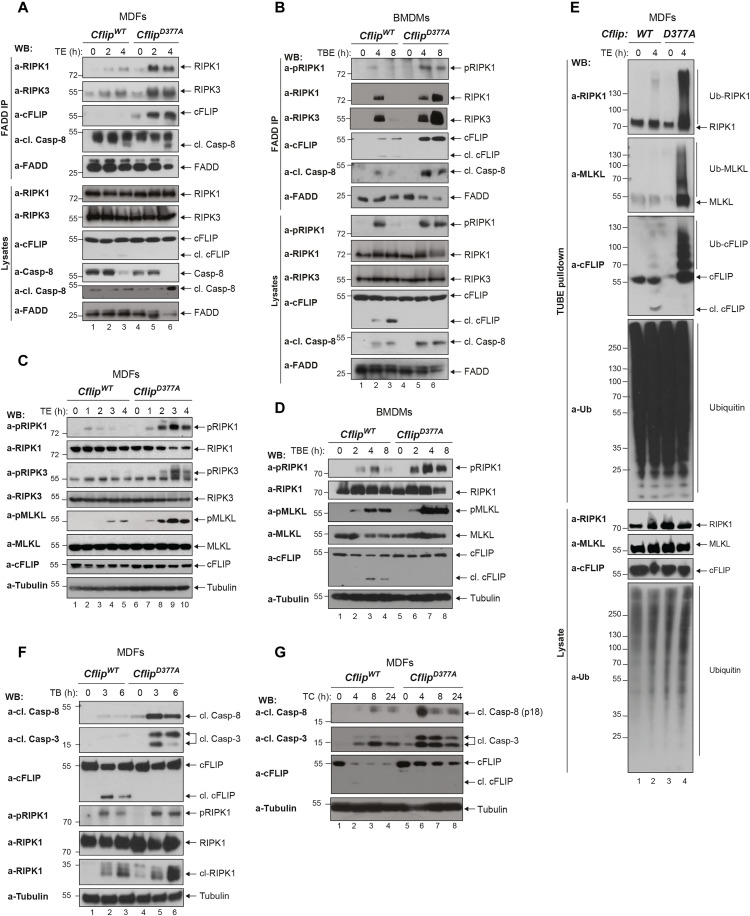
TNF-induced necroptosis is driven by the formation of the RIPK1 kinase activity–dependent complex-II and the consequent phosphorylation of RIPK3 and MLKL (23, 36). To ascertain how the abrogation of cFLIP cleavage sensitizes cells to necroptosis, we treated MDFs and BMDMs with TE or TBE, respectively, and immunoprecipitated complex-II via FADD. The subsequent immunoblotting analysis revealed increased association of RIPK1, RIPK3, Caspase-8, and cFLIP with FADD in the CflipD377A cells (Fig. 3, A and B). In addition, we observed significantly higher levels of phosphorylated RIPK1, RIPK3, and MLKL in cFLIP mutant cells treated with TE, TBE, or IE, or expressing CrmA (Fig. 3, C and D, and fig. S4, A and B). In addition to phosphorylation, RIPK1 and MLKL also undergo ubiquitin modification that promotes their killing activity during necroptosis (38). Consistent with CflipD377A cells being more sensitive to necroptosis, we observed markedly higher ubiquitination of RIPK1, MLKL, and cFLIP in the mutant cells compared to WT cells, upon TE treatment (Fig. 3E and fig. S4C). TNF-induced apoptosis is mediated by Caspase-8 activation, by means of autoproteolytic cleavage, which, in turn, activates the executioner caspases, Caspase-3 and Caspase-7 (39). Consistent with the observation that CflipD377A cells are more sensitive to apoptosis, we observed increased cleaved Caspase-8 and cleaved Caspase-3 in cFLIP mutant cells treated with TB or TC (Fig. 3, F and G). Last, the enhanced sensitivity to cell death imparted by the cFLIP mutation is not because of defects in activation of NF-κB or MAPKs (mitogen-activated protein kinases), since these signaling pathways were not affected by the D377A mutation (fig. S4, D and E). Recent studies reported on the transcriptional role of Caspase-8 downstream of TLRs (40, 41). Caspase-8 can cleave and inactivate N4BP1 to allow TLR-induced expression of proinflammatory cytokines (40). Therefore, we set up to understand whether cFLIP cleavage affects Caspase-8–mediated cytokine production. Western blot and reverse transcription quantitative polymerase chain reaction (qPCR) analysis of WT and CflipD377A BMDMs treated with LPS, to stimulate TLR4, or Poly I:C, to stimulate TLR3, revealed a highly similar N4BP1 cleavage pattern (fig. S4F) and cytokine mRNA expression levels (TNF and CCL2) (fig. S4G) between the two genotypes. Collectively, these data demonstrate that the D377A mutation promotes complex-II formation and its ability to induce MLKL-dependent necroptosis and Caspase-3–dependent apoptosis, while it does not seem to affect the transcriptional programs activated by TNFR1 and TLRs.
Fig. 3. The D377A mutation enhances complex-II formation, independently of NF-κB.
(A) WT and D377A mutant MDFs and (B) BMDMs were treated with TNF (1 ng/ml) and emricasan (1 μg/ml) and TNF (1 ng/ml), birinapant (250 nM), and emricasan (1 μg/ml), respectively, for the indicated time points. Cell lysates were subjected to immunoprecipitation using a FADD-specific antibody. Immunocomplexes and cellular lysates were then analyzed by immunoblotting using the indicated specific antibodies (n = 3). (C) MDFs and (D) BMDMs were treated as in (A) and (B) for the indicated time points. Cell lysates were analyzed by immunoblotting using the indicated specific antibodies (n = 2). (E) MDFs were treated with TNF (10 ng/ml) and emricasan (1 μg/ml), and cell lysates were subjected to TUBE pull-down, followed by immunoblotting analysis with the indicated specific antibodies (n = 3). (F) WT and D377A mutant MDFs were treated with TNF (10 ng/ml) and birinapant (250 nM) or (G) TNF (10 ng/ml) and cycloheximide (1 μg/ml) for the indicated time points, and cell lysates were immunoblotted using the indicated specific antibodies (n = 2).
cFLIP cleavage arrests complex-II formation
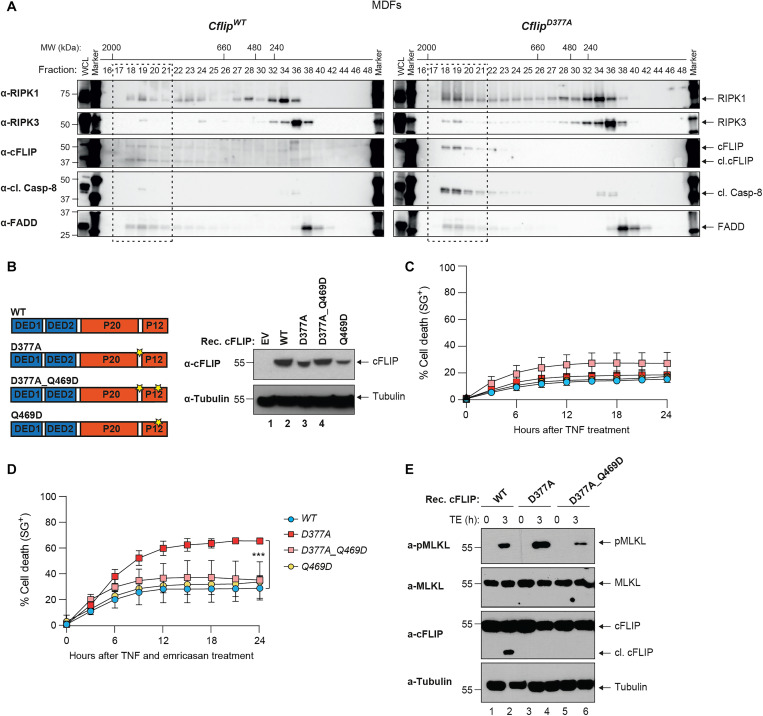
To mechanistically understand how cFLIP cleavage limits TNF-induced complex-II formation and cell death, we performed size exclusion chromatography. As expected, in WT cells treated with TBE, a small portion of RIPK1, RIPK3, cFLIP, and cleaved Caspase-8 co-eluted with FADD in fractions 17 to 21, which contain high molecular weight complexes of apparent molecular mass of ~2 MDa. Immunoprecipitation of FADD from pooled column fractions (17 to 21) further confirmed that these proteins are together in the same complex (fig. S5A). When gel filtration profiles of lysates from CflipWT and CflipD377A MDFs treated with TBE were compared, we observed increased abundance of RIPK1, RIPK3, cleaved Caspase-8, and cFLIP in fractions 17 to 21, corresponding to the high molecular weight complex, of CflipD377A lysates (Fig. 4A). However, the D377A mutation did not further increase the mass of the high molecular weight complex, because in both CflipWT and CflipD377A MDFs complex-II components co-eluted in the same fractions (17 to 21). This corroborated the results obtained with FADD immunoprecipitation (Fig. 3, A and B) and indicated that cleavage of cFLIP at position D377 limits the extent of complex-II formation. We next reasoned that the p12 domain of cFLIP, which is C-terminal to the D377A, might be important for the assembly and stability of complex-II. This fragment contains a residue, Q469, which was reported to be involved in the process of heterodimerization with Caspase-8 (Fig. 4B) (31). Thus, cFLIP cleavage could regulate the formation of cFLIP/Caspase-8 heterodimers and, as a consequence, the assembly, stabilization and killing potential of complex-II. To interrogate this possibility, we reconstituted Cflip−/− MDFs with different cFLIP mutant constructs (Fig. 4B). Notably, the Q469D mutation did not impair the ability of cFLIP to protect cells from the cytotoxic effect of TNF alone (Fig. 4C). If our hypothesis were correct, then the Q469D mutation would abrogate the pro–cell death effects of the D377A mutation. While D377A-reconstituted cells were significantly more sensitive to TE-induced cell death than WT-reconstituted cells, the Q469D mutation reverted the sensitizing effect of the D377A mutation (Fig. 4D). The levels of TE-induced cell death in D377A/Q469D-reconstituted cells were comparable to those observed in WT- and Q469D-reconstituted cells. Consistent with this, phosphorylated MLKL levels in WT- and D377A/Q469D-reconstituted cells were reduced when compared to D377A-reconstituted cells (Fig. 4E). Together, these findings indicate that the cell death–sensitizing ability of D377A depends on Q469. This supports a model whereby cFLIP cleavage represents a molecular mechanism that controls the availability of a cFLIP/Caspase-8 interaction surface, the Q469, to limit the extent to which complex-II can form and mitigate cell death responses.
Fig. 4. cFLIP cleavage counteracts complex-II formation.
(A) CflipWT and CflipD377A MDFs were treated with TNF (10 ng/ml), Smac mimetic (250 nM), and emricasan (1 μg/ml) for 4 hours, and lysates were separated on a Superose 6 size exclusion column. Aliquots from each fraction were retained and analyzed by immunoblotting with the indicated specific antibodies. (B) Cartoon depicting cFLIP domain composition and position of the D377 and Q469 residues (left). Immunoblotting analysis of Cflip−/− MDFs reconstituted with the indicated cFLIP constructs via lentiviral infection or with an empty lentivirus (EV). cFLIP- and tubulin-specific antibodies were used (right). (C and D) Cflip−/− MDFs reconstituted as in (B) were treated with TNF (10 ng/ml) (C) or TNF (1 ng/ml) and emricasan (1 μg/ml) (D) and cell death was measured over time by calculating the percentage of Sytox Green–positive cells (n = 3). (E) MDFs Cflip−/− MDFs reconstituted as in (C) and treated as in (D) for 3 hours. Cell lysates were analyzed by immunoblotting with the indicated specific antibodies.
Abrogation of cFLIP cleavage exacerbates the phenotype of Sharpin mutant mice
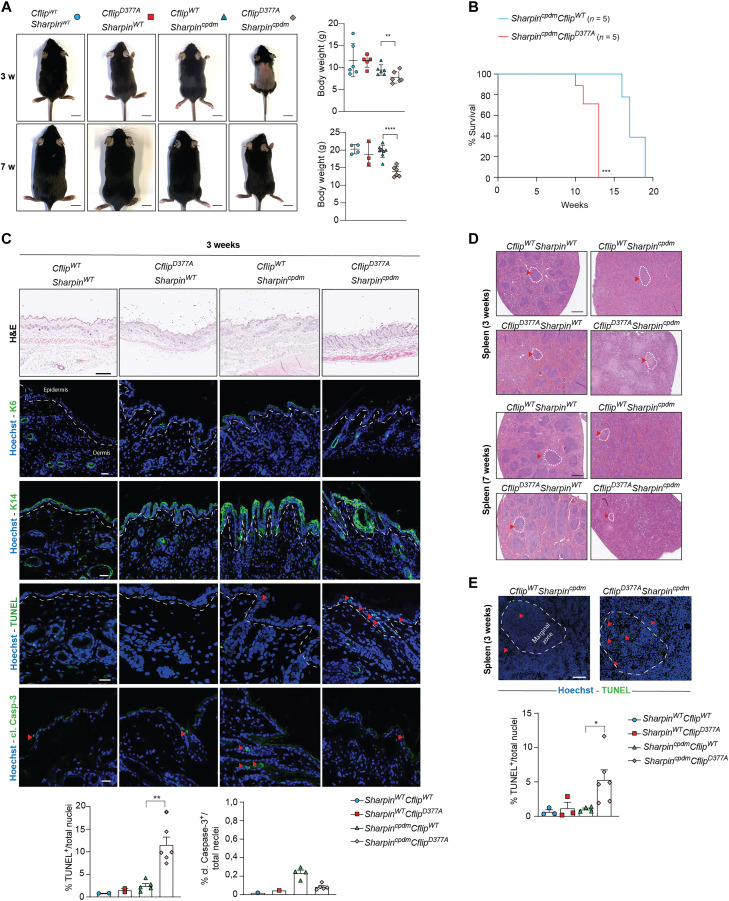
Next, we sought to investigate in a genetic model of TNF-induced, cell death–mediated tissue damage whether the cFLIP cleavage had cell death–limiting functions. Sharpin deficiency in mice causes a TNF-mediated, cell death–dependent multiorgan inflammation, referred to as chronic proliferative dermatitis mice (cpdm), characterized by skin dermatitis, lung and liver inflammation, loss of marginal zones in the spleen, and loss of Peyer’s patches (PP) (42–44). We observed that skin extracts of 7-week-old Sharpincpdm mice exhibited cleavage of cFLIP (fig. S6A). Because cFLIP cleavage at D377 limits TNF-induced complex-II–mediated cell death, we set up to investigate whether the loss of cFLIP cleavage would exacerbate the effects of Sharpin deletion in vivo. For this purpose, we generated CflipD377A/D377ASharpincpdm/cpdm double-mutant mice (CflipD377ASharpincpdm). CflipD377ASharpincpdm mice were born at the expected Mendelian ratio, yet they were runted and displayed significantly lower body weight than Sharpincpdm mice (Fig. 5A). Notably, the CflipD377ASharpincpdm mice developed inflammatory skin lesions and had to be euthanized significantly earlier than the Sharpincpdm mice because of these lesions (Fig. 5B). While 12-week-old Sharpincpdm did not show any evident sign of dermatitis, CflipD377ASharpincpdm mice exhibited damaged skin, increased epidermal thickness [hematoxylin and eosin (H&E)], and epidermal hyperplasia (K6), to an extent that was comparable to 18-week-old Sharpincpdm mice with clear signs of dermatitis (Fig. 5B and fig. S6B). Consistently with an earlier onset of dermatitis, the skin of 3-week-old CflipD377ASharpincpdm mice, although not visibly inflamed, had a significantly higher amount of terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL)–positive cells than the skin of the Sharpincpdm mice (Fig. 5C). Curiously, no increase in cleaved Caspase-3 was detected (Fig. 5C). TUNEL/K14 double staining showed that most dead cells in the skin of 3-week-old CflipD377ASharpincpdm mice were keratinocytes (fig. S6C). These findings therefore support a model where the D377A mutation accelerates the Sharpin deficiency–mediated cell death of keratinocytes and the onset of dermatitis. Another feature of Sharpin-deficient mice is splenomegaly and loss of marginal zones. Although 7-week-old Sharpincpdm mice exhibited splenomegaly, D377A mutation did not cause further spleen enlargement (fig. S6D). However, we observed a complete loss of marginal zones in CflipD377ASharpincpdm mice, while Sharpincpdm exhibited only minor spleen architecture alterations at 3 and 7 weeks of age (Fig. 5D). Consistent with this finding, we detected a significantly higher number of TUNEL-positive cells in the spleen of CflipD377ASharpincpdm mice compared to Sharpincpdm and control mice (Fig. 5E). Despite the loss of PPs (fig. S6E), histological analysis revealed that intestines of 7-week-old Sharpincpdm mice were normal and largely comparable to those of control mice (fig. S6F). On the contrary, intestines of CflipD377ASharpincpdm mice exhibited signs of damage, inflammation, and cell death, both in the small and large intestine, as shown by the histology score (fig. S6F), which correlates with a significant increase in the amount of TUNEL-positive cells (fig. S6G). Together, these findings indicate that the abrogation of cFLIP cleavage enhances the cell death–inducing potential of Sharpin deletion in vivo.
Fig. 5. Abrogation of cFLIP cleavage exacerbates the phenotype of Sharpin mutant mice.
(A) Pictures (left) and body weight (right) of 3- and 7-week-old CflipWTSharpinWT, CflipD377ASharpinWT, CflipWTSharpincpdm, and CflipD377ASharpincpdm mice. Each symbol corresponds to one mouse. Scale bars, 1 cm. Data are presented as means ± SD; **P < 0.01, ****P < 0.0001. (B) Kaplan-Meier survival curve of CflipWTSharpincpdm and CflipD377ASharpincpdm mice. The mice were euthanized when the termination criteria, based on the severity of dermatitis, were reached. ***P < 0.001. (C) Representative skin sections of 3-week-old mice of the indicated genotypes stained with H&E, K6, K14, cleaved Caspase-3, and TUNEL. Nuclei were stained with Hoechst. White dashed lines separate the epidermis from the dermis. Red arrowheads indicate TUNEL- and cleaved Caspase-3–positive cells. The graphics represent the percentage of TUNEL- and cleaved Caspase-3–positive cells over the total number of cells. Data are presented as means ± SD; **P < 0.01. Scale bars 20 μm. (D) Representative images of spleen sections of 3- and 7-week-old mice of the indicated genotypes [as in (A)] stained with H&E. Scale bars, 500 μm. Dotted circles and red arrowheads indicate marginal zones. (E) Representative pictures of TUNEL staining of spleen sections of 3-weeks-old mice of the indicted genotypes (top) and the relative quantification (bottom) expressed as percentage of TUNEL-positive cells over the total number of cells. Hoechst stains nuclei. Red arrowheads indicate TUNEL-positive cells. Data are presented as means ± SD; *P < 0.05. Scale bars, 100 μm.
cFLIPD377A mice exhibit impaired skin wound healing
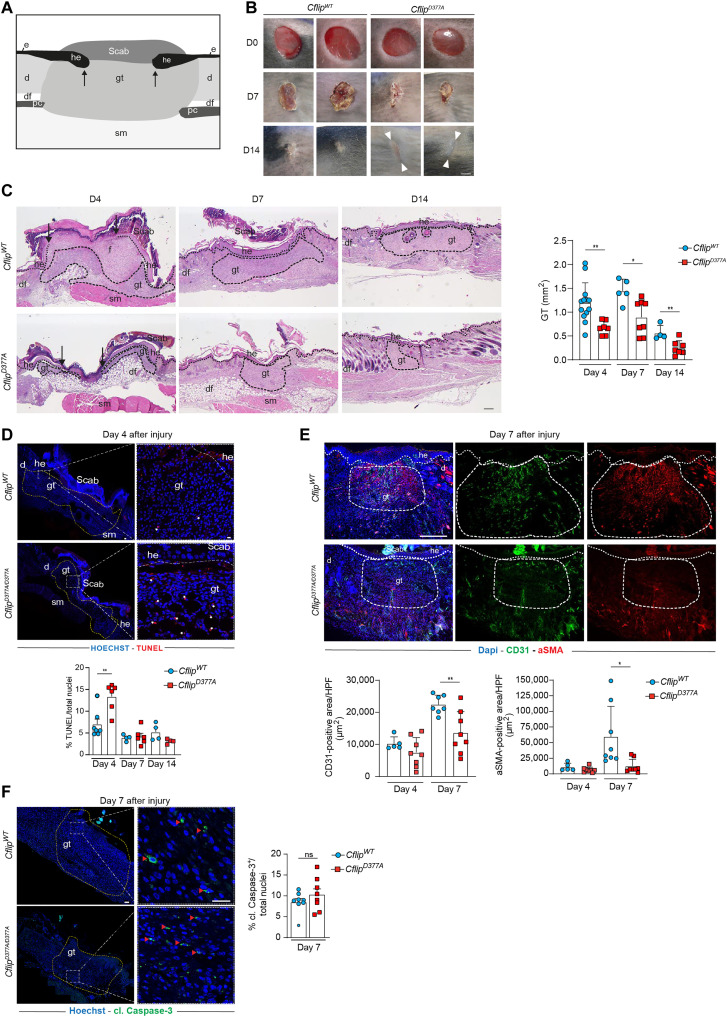
To further validate the role of cFLIP cleavage in protecting tissues from cell death–induced damage, we used a model of mouse full-thickness excisional skin injury. Because Caspase-8 was reported to play a crucial role in the wound healing response (45) and it was very recently shown that wound healing is a TNF- and cell death–dependent process (46), we wondered whether Caspase-8–mediated cFLIP cleavage could contribute to wound closure. The wound closure is characterized by the formation of the granulation tissue in the damaged area that is composed of different cell types whose concerted action promotes wound closure (Fig. 6A) (47, 48). Endothelial cells, myofibroblasts, and macrophages are the main components of the granulation tissue (48). Histomorphological analysis indicated a significant delay in the dynamics of wound closure in CflipD377A mice as compared to WT mice, as shown by reduced scar formation (Fig. 6B) and reduced granulation tissue area at days 4, 7, and 14 after injury (Fig. 6C). Consistent with a reduction in the granulation tissue, CflipD377A mice exhibited significantly higher levels of TUNEL-positive cells (Fig. 6D) at day 4 after injury, reduced wound vascularization (CD31 staining), and less myofibroblasts [α–smooth muscle actin (α-SMA) staining] at day 7 after injury (Fig. 6E). No difference in the percentage of cleaved Caspase-3–positive cells was detected (Fig. 6F). Thus, impairing cFLIP cleavage causes an increased cell death response that interferes with the wound healing response. We therefore conclude that cFLIP cleavage controls the extent of cell death occurring after wound injury to favor an optimal healing process.
Fig. 6. CflipD377A mice exhibit impaired skin wound healing.
(A) Cartoon depicting a skin wound at around 4 dpi (days post-injury), where the following skin areas are marked: d, dermis; df, dermal fat tissue; e, epidermis; gt, granulation tissue; he, hyperproliferative epithelium; pc, panniculus carnosus; sm, skeletal muscle. (B) Macroscopic pictures of wounds at 0, 7, and 14 dpi in CflipWT and CflipD377A mice. White arrowheads indicate scar tissue. Scale bar, 2 mm. (C) Representative H&E-stained wound sections (left) of CflipWT (top) and CflipD377A mice (bottom) at 4, 7, and 14 dpi. Scale bar, 200 μm. Arrows indicate the tips of the epithelial tongues. Quantitative analysis (right) of granulation tissue (gt) area of sections in (C) at 4, 7, and 14 dpi (n = 4 to 12 total wounds per genotype per dpi). (D) Representative TUNEL-stained wound sections (top) at 4 dpi of CflipWT mice and CflipD377A mice. Nuclei were stained with Hoechst (blue). Scale bars, 50 μm (left) and 20 μm (right). White asterisks indicate TUNEL-positive cells. Percentage of total TUNEL-positive cells (bottom) at 4, 7, and 14 dpi (n = 4 to 7 total wounds per genotype per dpi). (E) Representative CD31 (green) and α-SMA (red) immunostainings (top) at day 7 dpi of wound sections of CflipWT and CflipD377A mice. Scale bar, 200 μm. Percentage of total CD31- and α-SMA–positive area per HPF (high power field) in wound sections (bottom) at 4 and 7 dpi (n = 5 to 8 total wounds per genotype per dpi). (F) Representative cleaved Caspase-3–stained wound sections (left) at 7 dpi. Red arrowheads indicate cleaved Caspase-3–positive cells. Percentage of cleaved Caspase-3–positive cells (right) at 7 dpi (n = 4 to 7 total wounds per genotype per dpi). Data are presented as means ± SD; *P < 0.05 and **P < 0.01. ns, not significant.
cFLIPD377A mice are more sensitive to SARS-CoV–induced lethality
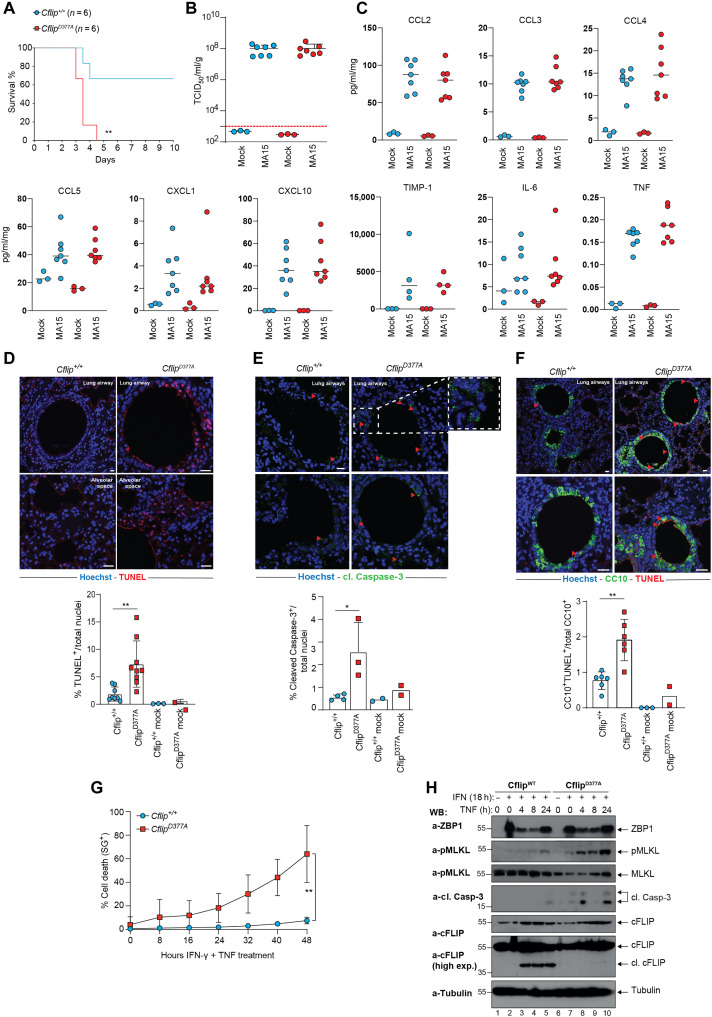
Different types of viruses, including coronaviruses, can induce cell death by triggering Caspase-8–dependent apoptosis or ZBP1/RIPK3-dependent necroptosis, directly in the infected cells or as a consequence of the hyperinflammatory state created by the immune system (49–51). Our observation that the D377A mutation sensitizes cells to both apoptosis and necroptosis prompted us to investigate the biological role of cFLIP cleavage in a model of SARS-CoV infection. We therefore infected WT and CflipD377A mice with a sublethal dose of a mouse-adapted strain of SARS-CoV-1 (MA15) (52), known to induce the production of TNF and other cytokines in the lung of infected mice (53, 54). Notably, CflipD377A mice were markedly more sensitive than WT mice to the lethal effect of the virus (Fig. 7A). However, we could not detect any difference in terms of viral titer between the two genotypes (Fig. 7B). We therefore reasoned that cFLIP cleavage is not involved in controlling viral replication. In addition, the levels of proinflammatory cytokines and chemokines between WT and mutant mice were largely comparable (Fig. 7C). When examining lung sections of infected mice, we detected a significantly higher number of TUNEL- and cleaved Caspase-3–positive cells in the lungs of infected CflipD377A mice compared to infected WT mice, both in the bronchioles and alveoli (Fig. 7, D and E). In addition, costaining with TUNEL and CC10, a marker of club cells that are present in the respiratory bronchioles, revealed a higher number of TUNEL-positive club cells in CflipD377A mice, indicating increased lung tissue damage (Fig. 7F). To explain the augmented cell death levels observed in the lungs of CflipD377A mice, we considered the possibility that the cFLIP mutant lung cells exhibited higher sensitivity to the cytokines produced during the viral response. TNF and IFN-γ are among the main produced cytokines during SARS-CoV infection. Therefore, we pretreated WT and D377A mutant LFs with IFN-γ before TNF treatment and found that cFLIP mutant LFs were significantly more sensitive than their WT counterparts to IFN-γ/TNF-induced cell death (Fig. 7G). Immunoblotting analysis revealed that D377A mutant LFs underwent necroptosis and apoptosis, as shown by the higher levels of phosphorylated MLKL and cleaved Casapse-3 (Fig. 7H). This suggested that cFLIP cleavage can control the killing activity of a cytotoxic complex induced by TNF and IFN-γ. Together, these findings show that cFLIP cleavage represents a mechanism that protects mice from SARS-CoV–induced lethality by limiting the extent of cytokine-induced cell death, apoptosis, and potentially necroptosis occurring in the lungs.
Fig. 7. CflipD377A mice are more sensitive to SARS-CoV–induced lethality.
(A) Survival curve of CflipWT and CflipD377A mice infected with SARS-Cov MA15 virus (n = 6). **P < 0.01. Viral titer (B) and cytokine levels (C) from lungs of CflipWT and CflipD377A mice at 3 dpi (n = 3 for mock infection and n = 7 for MA15 infection). (D) Representative TUNEL-stained lung sections of CflipWT mice and CflipD377A mice 3 dpi (top). Nuclei were stained with Hoechst (blue). Scale bars, 10 μm (top left) and 20 μm (top right and bottom). Percentage of total TUNEL-positive cells (bottom) (n = 8 for CflipWT-infected lungs, n = 10 CflipD377A-infected lungs, and n = 2–3 for mock infection). Data are presented as mean ± SD; **P < 0.01. (E) Representative pictures of lung sections of CflipWT and CflipD377A mice at 3 dpi, stained with cleaved Caspase-3 (top). Nuclei were stained with Hoechst (blue). Scale bar, 10 μm. Red arrowheads indicate cleaved Caspase-3–positive cells. Percentage of total cleaved Caspase-3–positive cells (bottom). Data are presented as means ± SD; *P < 0.05. (F) Representative CC10 (green)/TUNEL (red) double-stained lung sections of CflipWT mice and CflipD377A mice at 3 dpi (top). Nuclei were stained with Hoechst (blue). Scale bars, 10 μm (top) and 20 μm (bottom). Red arrowheads indicate TUNEL-positive/CC10-positive cells. Percentage of total CC10/TUNEL double-positive cells (bottom). (n = 6 for CflipWT infected lungs, n = 6 CflipD377A infected lungs, and n = 2 to 3 for mock infection). Data are presented as means ± SD; **P < 0.01. (G) CflipWT and CflipD377A LFs were pretreated with IFN-γ (100 nM) for 18 hours and then treated with TNF (100 ng/ml) for additional 24 hours. Cell death was measured over time by calculating the percentage of Sytox Green–positive cells (n = 3). (H) Total lysates of LFs treated like in (G) for the indicated times points were immunoblotted with the indicated specific antibodies (n = 2).
DISCUSSION
cFLIP is an essential, nonredundant cell death suppressor required to maintain tissue integrity (55–60). Its cell death inhibitory functions are due to the fact that it heterodimerizes with Caspase-8 and modulates its activity. Apart from being a modulator of Caspase-8 activity, cFLIP is also a Caspase-8 substrate, cleaved at position D377 (32). However, despite earlier studies on cFLIP proteolysis at D377 (31, 33, 34, 61), the biological relevance of this cleavage event remained to be elucidated. Here, we generated mutant mice carrying a noncleavable cFLIP version, the CflipD377A, which were born at the expected Mendelian ratio. It was previously shown that cFLIP is required for Caspase-8–mediated suppression of necroptosis. Our results indicate that the cleavage of cFLIP is dispensable for the ability of Caspase-8 to suppress necroptosis. When we analyzed different D377A mutant mouse cells, including BMDMs and LECs, we found that the abrogation of cFLIP cleavage sensitizes them to TNF-mediated apoptosis and necroptosis (induced by TNF and Smac mimetic as well as TNF and emricasan, respectively) by enhancing complex-II formation. The D377A mutation sensitizes cells not only to RIPK1-dependent cell death (apoptosis and necroptosis) but also to RIPK1-independent apoptosis (TNF and cycloheximide) and ZBP1-dependent necroptosis (IFN-γ and emricasan). Notably, in a previous study, a mutant mouse expressing noncleavable cFLIP was generated, where both D377 and the nearby D371 were mutated (35). Here, we show that the D377A mutation is sufficient to render cFLIP resistant to Caspase-8–mediated cleavage.
In vivo, the CflipD377A mice were more susceptible to SARS-CoV–mediated lethality than WT mice, and this correlated with significantly higher levels of lung epithelial cell death, while no difference in viral titer or cytokine production was observed in lung extracts. Therefore, we envisage a scenario whereby the D377A mutation sensitizes lung cells to cell death induced by the overproduction of cytokines following lung infection, referred to as cytokine storm (62), which include TNF and IFN-γ. Therefore, the increased cell death observed in the lungs of mutant mice would not be the trigger but the consequence of the cytokine storm and the link between this last and the more severe lung pathology. Consistent with this scenario, we found that LFs from the CflipD377A mouse are significantly more sensitive than WT cells to TNF/IFN-γ–induced cell death. Because IFN-γ induces the up-regulation of ZBP1 that can, in turn, trigger the assembly of a complex composed of RIPK1/RIPK3/Caspase-8/cFLIP (63), it is tempting to speculate that cFLIP cleavage can regulate the activity of this ZBP1-mediated cytotoxic complex. Recent reports showed that both Caspase-8–mediated apoptosis and MLKL-driven necroptosis contribute to disease severity following SARS-CoV infection (41, 50, 51, 62). We detected both cleaved Caspase-3– and TUNEL-positive cells in the lungs of the infected CflipD377A mice. In light of the fact that D377A mutant LFs treated with TNF/IFN-γ exhibited features of both apoptosis (cleaved Caspase-3) and necroptosis (phosphorylated MLKL) (Fig. 7, F to H), we hypothesize that noncleavable cFLIP can promote not only apoptosis but also necroptosis in the lungs of SARS-CoV–infected mice. In addition, CflipD377A mice exhibited impaired wound healing following skin excision, as a consequence of increased cell death observed in the granulation tissue. Again, this would be consistent with a model whereby D377A mutation renders granulation tissue cells more sensitive to death induced by the cytokines, including TNF, produced during the wound healing processes likely by macrophages. To further provide understanding of the biological role of D377A mutation in vivo, we crossed the Sharpincpdm mice, which carry an inactivating mutation on Sharpin that causes TNF-dependent cell death–mediated systemic inflammatory syndrome (44), with the CflipD377A mouse. The resulting SharpincpdmCflipD377A mice were runted; showed increased cell death levels in skin, spleen, and intestine in the first weeks after birth; and developed dermatitis significantly earlier than the Sharpincpdm mice. At the mechanistic level, we were able to show that the cleavage of cFLIP controls the extent of complex-II formation by regulating cFLIP/Caspase-8 heterodimerization via Q469. Two pieces of evidence support this model: (i) In the D377A mutant cells stimulated with TNF, birinapant, and emricasan, there is a higher abundance of complex-II components co-eluting in the ~2 MDa gel filtration fractions. This suggests that the D377A mutation favors the assembly of complex-II. (ii) The Q469D mutation abrogates the cell death–sensitizing effects of the D377A mutation. This indicates that, in the absence of cFLIP cleavage, Q469 favors complex-II assembly and stability most probably by promoting cFLIP/Caspase-8 heterodimerization. Caspase-8 can, in turn, recruit more FADD molecules via DED-mediated interaction, which, in turn, can recruit more RIPK1 molecules via DD-mediated interaction. Therefore, the Caspase-8–mediated cleavage of cFLIP at position D377 represents a mechanism that counterbalances complex-II formation to keep its killing activity in check. Previous studies have reported conflicting results regarding the role of cFLIP cleavage on Caspase-8 activity, mainly using cell-free system assays containing recombinant or hybrid cFLIP and Caspase-8 (31, 33, 34). Such systems do not recapitulate the complexity of cell death–inducing platforms, such as TNF-induced complex-II, which need to be assembled to activate Caspase-8. Therefore, in light of our findings, we believe that cFLIP cleavage does not affect Caspase-8 activity per se, but rather acts indirectly by regulating the dynamics of a complex to which Caspase-8 is recruited and at which it is activated. Notably, Caspase-8 can also cleave other substrates in TNF-induced complex-II, such as RIPK1. While RIPK1 cleavage is a decisive event to prevent TNF from killing and the absence of cleavage results in mouse embryonic lethality and multiorgan inflammation in human (35, 64, 65), cFLIP cleavage is decisive in limiting TNF cytotoxicity once the TNF-mediated cell death arm is triggered. As a consequence, cFLIP cleavage becomes highly relevant under pathological conditions, where the extent of cell death have to be tightly regulated to prevent the detrimental effects of aberrant cell death. This highlights the different layers of regulation that complex-II is subjected to. The ability of tissues to overcome stress-induced damage lies also in their capacity to tightly control the amplitude of cell death responses, by modulating the formation and activity of cell death–inducing complexes. If in the absence of cell death tissue repair programs cannot be initiated, excessive cell death can lead to hyperinflammatory responses that are, in turn, detrimental for the tissue. Different types of insults, such as viral and bacterial infections or mechanical tissue damage, are known to activate a cFLIP/Caspase-8/RIPK1–containing cell death complex. The results presented here reveal that the cleavage of cFLIP is a required regulatory module in the intricated process that controls the activity of this cytotoxic complex, thereby ensuring optimal cell death responses and the consequent activation of tissue repair programs.
MATERIALS AND METHODS
Mice generation
The CflipD377A mutant mice were generated by the MAGEC laboratory, Walter and Eliza Hall Institute of Medical Research (WEHI) on a C57BL/6J background. To generate these mice, Cas9 mRNA (20 ng/μl), single guide RNA (10 ng/μl; TTGATGGCCCATCTACCTCC), and oligo donor (40 ng/μl; GCCAAAGCTCTTTTTTATTCAGAACTATGAGTCGTTAGGTAGCCAGTTGGAAGATAGCAGTCTGGAGGTAGCTGGGCCATCAATAAAAAATGTGGACTCTAAGCCCCTGCAACCCAGACACTGCACAACTCA) were injected into the cytoplasm of fertilized one-cell stage embryos generated from WT C57BL/6J breeders. Twenty-four hours later, two-cell stage embryos were transferred into the uteri of pseudo-pregnant female mice. Viable offspring were genotyped by next-generation sequencing. Targeted animals were backcrossed twice to WT C57BL/6J to eliminate off-target mutations.
Cell lines
Immortalized MDFs and LFs, Platinum-E, and human embryotic kidney 293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin under 10% CO2. LECs were seeded on 0.1% gelatine-coated wells and cultured in a 1:1 mixture of Endothelial Cell Growth Medium 2 (EGM2) (PromoCell) and fully supplemented DMEM (Merck) [containing 20% fetal calf serum, glucose (4 g/liter), 2 mM glutamine, 1% penicillin (100 U/ml)–streptomycin (100 μg/ml), sodium pyruvate 1% (1 mM), Hepes (20 mM), and 1% nonessential amino acids]. Vero E6 cells were supplied by J. Vergara from the Centro de Investigación en Sanidad Animal IRTA-CReSA (Barcelona, Spain). Vero E6 cells were grown in DMEM (Sigma-Aldrich) supplemented with 10% FBS (Sigma-Aldrich), 2 mM GlutaMAX (Gibco), penicillin (100 U/ml; Sigma-Aldrich), streptomycin (100 μg/ml; Sigma-Aldrich), amphotericin B (0.25 μg/ml; Sigma-Aldrich), 1% nonessential amino acids (Gibco), and 25 mM Hepes (Biowest).
Isolation of primary cells and immortalization
MDFs were generated from the tail skin of 8- to 10-week-old mice. Once separated from the tail, the skin was minced with a scalpel and digested in 3 ml of trypsin (Sigma-Aldrich, T4049) for 1 hour at 37°C. Seven milliliters of DMEM was then added, the tube was manually shaken several times, and the digested tail was then filtered through a 100 μM cell strainer. Cells were washed once with phosphate-buffered saline (PBS) and seeded on a 6 cm plate. Two to 3 days later, they were infected with a SV40 Large T antigen expressing retrovirus for immortalization. LFs were generated from whole lungs of 8- to 10-week-old mice. Once separated from non-lung tissue, the lungs were cut in very small pieces and digested in 1.5 ml of collagenase type 2 (Worthington, 44N15307B) for 1 hour at 37°C and 500 rpm. One digested lung was seeded on two 15 cm dishes containing 20 ml of DMEM. After 2 days, the medium was changed and cells were grown until confluence. LFs were immortalized using an SV40 Large T antigen–encoding retrovirus.
LECs were isolated from the lungs of 8- to 12-week-old mice. Lungs were briefly washed in PBS, minced, and digested in 1 ml of 0.5% collagenase type II (Merck Millipore, C2-22) for 45 min at 37°C. The cell solution was then filtered through a 70 μm cell strainer and separated with magnetic beads (mouse CD31, Miltenyi Biotec) according to the manufacturer’s protocol. CD31+ endothelial cells were seeded on gelatin-coated wells and cultured in the endothelial cell medium mentioned above. After the first passage, cells were resorted using the same magnetic beads. Isolated primary endothelial cells were analyzed using the anti-CD31 antibody (Ab) to distinguish endothelial cells from other cell types. To generate BMDMs, bone marrow cells from tibia and femur of 2-month-old mice were seeded in noncoated petri dishes and cultured for 6 days in DMEM + 10% FBS + 20% (v/v) L929 mouse fibroblast–conditioned medium. Isolated cells were routinely tested negative for mycoplasma contamination by PCR.
Reagents and Abs
The following reagents were used in this study: birinapant (MedChem Express, HY-16591), emricasan (Absource Diagnostics, S7775-0005), GSK’963 (SeleckChem, S8642), IFN-γ (PeproTech, 315-05-100), mouse TNF-α (Enzo Life Sciences, ALX-522-009-C050), and Sytox Green (Thermo Fisher Scientific, S7020). The following antibiotics were used: blasticidine (InvivoGen, ant-bl-1), hygromycin-B (InvivoGen, ant-hg-1/5), neomycin (InvivoGen, ant-gn-5), and puromycin (InvivoGen, ant-pr-1). The following Abs were used: Caspase-7 rabbit Ab (Cell Signaling Technology, 12827), CC10 (Santa Cruz Biotechnology, sc365992), CD31 (BD Pharmingen, 550274), cFLIP rabbit Ab (Cell Signaling Technology, 56343), cleaved Caspase-3 rabbit Ab (Cell Signaling Technology, 9661), cleaved Caspase-8 rabbit Ab (Cell Signaling Technology, 9429S), SMA-Cy3 conjugated (Sigma-Aldrich, C6198), FADD mouse Ab (Millipore, 05-486 /1F7), FADD rabbit Ab (Abcam, ab124812), MLKL rat Ab (EMD Millipore, MABC604), phospho-MLKL rabbit Ab (Abcam, ab196436), phospho-RIPK1 rabbit Ab (Cell Signaling Technology, 31122), Phospho-RIPK3 rabbit Ab (Cell Signaling Technology, 57220), RIPK1 mouse Ab (BD Bioscience, 610459), RIPK1 rabbit Ab (Cell Signaling Technology, 3493), RIPK3 rabbit Ab (ProSci Inc., 2283), tubulin mouse Ab (Thermo Fisher Scientific/Sigma-Aldrich, T9026/047M4789V), ubiquitin mouse Ab (Santa Cruz Biotechnology, SC-8017), and ZBP1 mouse Ab (AdipoGen, AG-20B-0010-C100).
SARS-CoV infection
SARS-CoV-1 MA15 was provided by S. Zúñiga and L. Enjuanes (CNB-CSIC, Madrid, Spain). For the virus titration in lungs, previously weighed portions were homogenized in 500 μl of DMEM with a GentleMACS dissociator (Miltenyi), with centrifugation of 1500 rpm × 5 min, and the supernatant was taken. Virus titration was determined by median tissue culture infectious dose (TCID50)/ml assay performing serial dilutions and calculated using the Ramakrishan newly proposed method formula (66).
Mice were infected intranasally with 106 TCID50/ml in a total volume of 40 μl of PBS after isoflurane anesthesia. Mice were weighed daily and reached humanitarian end point with a 25% weight loss. A clinical score was generated if needed, following different components: mouse appearance, level of consciousness, activity, response to stimuli, eye appearance, and frequency and quality of respiration, and mice reached humanitarian end point when the clinical score reached 21, when the respiratory characteristics were higher than 3, or when weight loss is greater than 25% of the initial weight. Animals were kept under standard conditions of temperature, humidity, and light at the Research Center on Encephalopathies and Emerging Diseases of the University of Zaragoza. Animal experimentation was approved by the Animal Experimentation Ethics Committee of the University of Zaragoza (number: PI44/20).
Excisional punch injury
Mice were anesthetized by intraperitoneal injection of 100 mg/kg body weight of Ketavet (Pfizer) and 10 mg/kg body weight of Rompun 2% (Bayer). The back skin was shaved using an electric shaver and disinfected with 70% ethanol. Full-thickness punch biopsies were created on the back using a standard biopsy puncher (Stiefel). For histological analysis, wounds were excised at different times after injury and processed following as described in (47). The tissue was either fixed for 2 hours in Roti Histofix or embedded in O.C.T. compound (Thermo Fisher Scientific) and stored at −80°C.
Retroviral production and infection
Replication-incompetent retroviral particles were generated in Platinum E cells. The cells were seeded in a 10 cm dish at 2,500,000 cells per dish in 10 ml of culturing medium. The following day, 1 ml of Opti-MEM I reduced serum medium containing 10 μg of plasmid of interest and 30 μl of polyethylenimine (1 mg/ml) was added to the medium and incubated overnight. The medium was changed after 18 hours of incubation. The supernatant containing retroviral particles was collected 2 days posttransfection. One day before the infection of target cells with retroviral particles, 80,000 cells per well were seed in six-well plate. On the day of infection, 3 ml of supernatant-containing retroviral particles and polybrene (1 μg/ml) were added to the respective wells of six-well plate and kept for 72 hours. To selectively expand the infected cells, a 7-day selection was performed using the respective antibiotics depending on the resistance cassette contained in the retroviral plasmid, i.e., puromycin (4 μg/ml), hygromycin (200 μg/ml), blasticidine (6 μg/ml), and neomycin (1 mg/ml).
Cell culture, constructs, and transfection
The SV40 Large T antigen and all the mouse cFLIP WT and mutant coding sequences (D371A, D377A, D371A/D377A, and D377A/Q469D) were in the pBABE retroviral vectors, carrying puromycin, hygromycin, neomycin, or blasticidin resistance. Cflip−/− MDFs were generated from immortalized Cflipf/f MDFs infected with a Cre-expressing retrovirus, followed by antibiotic selection. All the reconstitutions of Cflip−/− MDFs were done by infection with retroviruses expressing the different cFLIP sequences, followed by antibiotic selection.
Mouse cFLIP WT was amplified from mouse cDNA and cloned into pBABE using Bam HI and Eco RI restriction enzymes. All the cFLIP mutants were generated by site-directed mutagenesis and clones into pBABE using Bam HI and Eco RI restriction enzymes.
Complex-II purification
Cells were seeded in 10 cm dishes and treated as indicated using media containing mouse TNF (100 ng/ml) and emricasan (1 μM). Cells were lysed in 1% Triton X-100 lysis buffer [30 mM tris-HCl (pH 7.4), 120 mM NaCl, 2 mM EDTA, 2 mM KCl, 1% Triton X-100 supplemented with protease inhibitors, and 10 mM PR619] on ice. Cell lysates were rotated at 4°C for 20 min and then centrifuged at 4°C at 14,000 rpm for 15 min. Twenty microliters of protein G Sepharose (Sigma-Aldrich), previously blocked for 1 hour with lysis buffer containing 1% bovine serum albumin, was bound with FADD antibody (1.5 mg antibody/mg protein lysate) and was rotated with cleared protein lysates for 4 hours at 4°C. Three times washes in lysis buffer were performed, and immunocomplexes were eluted by boiling in 60 μl of 1× SDS Laemmli buffer.
Tube assay
Cells were lysed in DISC lysis buffer [20 mM tris-HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, and 10% glycerol] supplemented with protease inhibitors, 1 mM dithiothreitol (DTT), PR619 (10 mM), and Glutathione S-Transferase (GST)–Tandem Ubiquitin Binding Entities (TUBE) (50 mg/ml; 50 mg of TUBE/mg protein lysate). Cell lysates were rotated at 4°C for 20 min and then clarified at 4°C at 14,000 rpm for 10 min. Twenty-five microliters of GST beads was added, and pull downs were performed overnight. Beads were washed three times in wash buffer [50 mM tris (pH 7.5), 150 mM NaCl, 0.1% Triton X-100, and 5% glycerol], and pulled down proteins eluted by boiling in 50 μl of 1× SDS Laemmli buffer.
Immunostaining
Freshly isolated organs were fixed with 4% paraformaldehyde overnight, washed with PBS for 24 hours, and embedded in paraffin (lung, spleen, and skin) or cryopreserved (skin). Paraffin blocks and cryopreserved tissues were sectioned into 3 μm thick consecutive thick slices. After standard rehydration, a short antigen-retrieval with 1× sodium citrate (pH 6.0) (Sigma-Aldrich, C9999) was performed in a microwave for 5 min at 80% power. Tissue sections were then permeabilized with 0.2% (v/v) Triton X-100 in the Animal Free Blocker and Diluent (Vector Lab, SPS035) at room temperature (RT) for 10 min. Next, the sections were stained for the following antibodies [diluted in 0.2% (v/v) Triton X-100 in Animal Free Blocker and Diluent] overnight at 4°C: CC10 (1:50; Santa Cruz Biotechnology, sc365992), α-SMA–Cy3–conjugated (1:50; C6198, Sigma-Aldrich), and CD31 (1:50; 550274 BD Pharmingen). Note that, in CD31-stained cryosections, antigen retrieval was not performed. Primary antibodies were then visualized by secondary antibodies conjugated to Alexa Fluor 488 (1:200; Thermo Fisher Scientific, A-11008) diluted in 0.2% (v/v) Triton X-100 in Animal Free Blocker and Diluent containing 1:1000 Hoechst 33342 for nuclei staining at RT for 1 hour. For the detection of late PCD+ cells, the ApopTag Red In Situ Apoptosis Detection Kit (Merck S7165) was used. Briefly, sample sections were washed twice with PBS and treated with equilibration buffer for 10 s, incubated with the working strength terminal deoxynucleotidyl transferase enzyme for 1 hour at 37°C, followed by 10 min of stop buffer, and subsequent 30 min of rhodamine antibody solution at RT. Slides were quickly washed with PBS and mounted with ProLong Gold Antifade Mountant (Thermo Fisher Scientific, P36934). Last, images were acquired with a confocal fluorescence microscope (Stellaris 5 LIAchroic inverse).
Intestine histological score
Formalin-fixed and paraffin-embedded intestinal Swiss rolls were sectioned (3 μm) and stained with H&E. Histological evaluation was performed using the scoring system as described in (67). Briefly, histopathology scores are composed of four parameters: epithelial hyperplasia, epithelial injury, tissue inflammation, and epithelial cell death. Histological subscores for each parameter: 0, absent; 1, mild; 2, moderate; and 3, severe. An “area factor” for the fraction of affected tissue was assigned and multiplied with the respective parameter score (1 = 0 to 25%; 2 = 25 to 50%; 3 = 50 to 75%; and 4 = 75 to 100%). Each area was scored individually and multiplied with the correlating area factor. Total histology score was calculated as a sum of all parameter scores multiplied with their area factors. Maximum score was 48. Evaluation was performed in a blinded fashion.
Cell death analysis
Cells were seeded at 8000 cells (MDFs and LFs), 10,000 cells (LECs), and 50,000 cells (BMDMs) per well of a 96-well plate the day before the experiment. The following day, they were treated as indicated in the figure legends in the presence of 5 μM Sytox Green. Live uptake of Sytox Green by dead cells was monitored every hour over a period of 24 or 48 hours via an IncuCyte S3. The percentage of dead cells was calculated by using the Basic Analyzer of the IncuCyte 2020B software and the metric Sytox Object count per well normalized to the area confluence. The positive control (cells treated with TNF, Smac mimetic, and emricasan) was taken as 100% death, and with this, the percentage of cell death over time was calculated.
Protein expression and purification
The pGEX-GST-TUBE construct was transformed into BL21 (DE3) cells and cultured in LB medium at 37°C. Protein expression was induced overnight at 18°C with 0.1 mM isopropyl-β-d-thiogalactopyranoside when OD600 (optical density at 600 nm) reached 0.6. Cells were centrifuged at 4500 rpm for 20 min, resuspended in lysis buffer (PBS + 300 mM NaCl and 1 mM DTT supplemented with protease inhibitors), and sonicated four times for 30 s at maximum amplitude. Lysates were then spun for 30 min, 4°C, 4500 rpm, and cleared lysates were added to GST beads O.N. in rotation at 4°C. Beads were then washed and GST-TUBE eluted using elution buffer [50 mM tris-HCl (pH 8.5), 150 mM NaCl, 1 mM DTT, and glutathione (6 mg/ml)] for two consecutive times. The elution product was dialyzed using a Slide-A-Lyzer cassette in TBS buffer [50 mM tris (pH 7.5) and 150 mM NaC]/1 mM DTT and stored at −80°C.
Gel filtration
Cellular lysates were separated on a Superose 6 HR 10/30 size exclusion column and an AKTA purifier protein purification system (GE Healthcare), essentially as described previously (68). Aliquots from each fraction were retained for Western blotting, and fractions 12 to 16, 17 to 21, 22 to 26, 27 to 31, 32 to 36, 37 to 41, 42 to 46, and 47 to 51 were pooled and used for immunoprecipitation experiments.
Reverse transcription qPCR
RNA samples were isolated using the commercially available RNA isolaton kit (Zymo Research) according to the manufacturer’s instructions. cDNA was synthesized using the MMLV reverse transcriptase (Promega) according to the manufacturer’s instruction. qPCR was performed using the ORA qPCR Green ROX L Mix (HighQu) according to the manufacturer’s instruction and a real-time PCR thermocycler (Bio-Rad). The amount of mRNA detected was normalized to actin mRNA values. Primer sequences are the following: actin forward: ATGGTGGGAATGGGTCAGAAGGAC, actin reverse: CATTGTAGAAGGTGTGGTGC, TNF forward: CATCTTCTCAAAATTCGAGTGACAA, TNF reverse: TGGGAGTAGACAAGGTACAACCC, CCL2 forward: CCACTCACCTGCTGCTACTCAT, and CCL2 reverse: TGGTGATCCTCTTGTAGCTCTCC.
Statistical analysis
The number of independent experiments for each dataset is stipulated in the respective figure legends. Comparisons were performed with Student’s t test, repeated-measure analysis of variance (ANOVA), and log-rank Mantel-Cox test (Fig. 7A), whose values are represented in the figures as*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001 using Prism v.8.2 (GraphPad).
Acknowledgments
We would like to thank all members of the Liccardi, Kashkar, Pasparakis, and Peltzer laboratories for sharing reagents and for helpful discussions. We thank L. Kurian for invaluable advice during the preparation of the manuscript and for sharing reagents and space. We also thank the CECAD imaging facility for the support with image acquisition.
Funding: This work was funded by the following: Center for Molecular Medicine Cologne (CMMC) Junior Research Group program, Deutsche Forschungsgemainschaft (DFG; German Research Foundation, project no AN 1717/1-1), Jürgen Manchot Foundation, Collaborative Research Centre SFB1403 (ID 414786233), Collaborative Research Centre SFB1530 (ID 455784452), and Network Cancer Targeting (CANTAR). Work at the J.P. laboratory is funded by the following: FEDER (Fondo Europeo de Desarrollo Regional, Gobierno de Aragón, Group B29_17R), grant PID2020-113963RBI00 by MICIN/AEI and CIBER de Enfermedades Infecciosas, and Juan de la Cierva postdoc contract (MICIN/AEI). Work at the H.W. laboratory is funded by the following: Wellcome Trust Investigator Award (214342/Z/18/Z), Medical Research Council Grant (MR/S00811X/1), Cancer Research UK Programme Grant (A17341), Collaborative Research Centre SFB1403 (Project A10, ID 414786233), Collaborative Research Centre SFB1399 (Project C06), Collaborative Research Centre SFB1530 (Project A03, ID 455784452), and Alexander von Humboldt Foundation. Work at the M.M. laboratory is funded by the UKRI Medical Research Council (grant number MC_UU_00025/4).
Author contributions: A.A. supervised the study. A.A. and K.M.L. conceptualized and designed the experiments. K.M.L., D.P.S., M.Z., C.C.F., and A.A. performed and analyzed most of the experiments. K.M.L., M.Z., and M.P. performed all the histological experiments and analysis. I.L., K.N., E.J., and N.I. helped in performing some of the experiments in Figs. 3 and 4. P.M. provided the CflipD377A mouse. A.A., K.M.L., I.U.-M., D.d.M., M.A., J.P., M.C.A., and H.W. designed, performed, and helped with the analysis of the SARS-CoV infection model. A.C., E.S., and M.M. designed, performed, and analyzed the gel filtration experiments. M.P. and S.E. performed the wound healing experiment and helped with the analysis of the results. M.B., X.S., W.T., and A.L. helped with infection models. M.I. and X.H. performed the FACS-based immune analysis. N.P. contributed intellectually and provided guidance with the Sharpincpdm model. The figures were prepared by A.A., K.M.L., D.P.S., and M.Z. A.A. and K.M.L. wrote, edited, and revised the manuscript. All authors assisted with data interpretation and manuscript editing.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S6
REFERENCES AND NOTES
- 1.A. Annibaldi, P. Meier, Checkpoints in TNF-induced cell death: Implications in inflammation and cancer. Trends Mol. Med. 24, 49–65 (2018). [DOI] [PubMed] [Google Scholar]
- 2.J. Zinngrebe, E. Rieser, L. Taraborrelli, N. Peltzer, T. Hartwig, H. Ren, I. Kovács, C. Endres, P. Draber, M. Darding, S. von Karstedt, J. Lemke, B. Dome, M. Bergmann, B. J. Ferguson, H. Walczak, LUBAC deficiency perturbs TLR3 signaling to cause immunodeficiency and autoinflammation. J. Exp. Med. 213, 2671–2689 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.T. Oltean, E. van San, T. Divert, T. vanden Berghe, X. Saelens, J. Maelfait, N. Takahashi, P. Vandenabeele, Viral dosing of influenza A infection reveals involvement of RIPK3 and FADD, but not MLKL. Cell Death Dis. 12, 471 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.F. Balkwill, Tumour necrosis factor and cancer. Nat. Rev. Cancer 9, 361–371 (2009). [DOI] [PubMed] [Google Scholar]
- 5.G. D. Kalliolias, L. B. Ivashkiv, TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 12, 49–62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.N. Peltzer, H. Walczak, Cell death and inflammation—A vital but dangerous liaison. Trends Immunol. 40, 387–402 (2019). [DOI] [PubMed] [Google Scholar]
- 7.A. Annibaldi, H. Walczak, Death receptors and their ligands in inflammatory disease and cancer. Cold Spring Harb. Perspect. Biol. 12, a036384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O. Micheau, J. Tschopp, Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 (2003). [DOI] [PubMed] [Google Scholar]
- 9.M. A. Kelliher, S. Grimm, Y. Ishida, F. Kuo, B. Z. Stanger, P. Leder, The death domain kinase RIP mediates the TNF-induced NF-kappaB signal. Immunity 8, 297–303 (1998). [DOI] [PubMed] [Google Scholar]
- 10.M. Rothe, M. G. Pan, W. J. Henzel, T. M. Ayres, D. V. Goeddel, The TNFR2-TRAF signaling complex contains two novel proteins related to baculoviral inhibitor of apoptosis proteins. Cell 83, 1243–1252 (1995). [DOI] [PubMed] [Google Scholar]
- 11.A. Annibaldi, S. Wicky John, T. Vanden Berghe, K. N. Swatek, J. Ruan, G. Liccardi, K. Bianchi, P. R. Elliott, S. M. Choi, S. Van Coillie, J. Bertin, H. Wu, D. Komander, P. Vandenabeele, J. Silke, P. Meier, Ubiquitin-mediated regulation of RIPK1 kinase activity independent of IKK and MK2. Mol. Cell 69, 566–580.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.T. L. Haas, C. H. Emmerich, B. Gerlach, A. C. Schmukle, S. M. Cordier, E. Rieser, R. Feltham, J. Vince, U. Warnken, T. Wenger, R. Koschny, D. Komander, J. Silke, H. Walczak, Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36, 831–844 (2009). [DOI] [PubMed] [Google Scholar]
- 13.S. Rahighi, F. Ikeda, M. Kawasaki, M. Akutsu, N. Suzuki, R. Kato, T. Kensche, T. Uejima, S. Bloor, D. Komander, F. Randow, S. Wakatsuki, I. Dikic, Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136, 1098–1109 (2009). [DOI] [PubMed] [Google Scholar]
- 14.E. Lafont, P. Draber, E. Rieser, M. Reichert, S. Kupka, D. de Miguel, H. Draberova, A. von Mässenhausen, A. Bhamra, S. Henderson, K. Wojdyla, A. Chalk, S. Surinova, A. Linkermann, H. Walczak, TBK1 and IKKε prevent TNF-induced cell death by RIPK1 phosphorylation. Nat. Cell Biol. 20, 1389–1399 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.I. Jaco, A. Annibaldi, N. Lalaoui, R. Wilson, T. Tenev, L. Laurien, C. Kim, K. Jamal, S. Wicky John, G. Liccardi, D. Chau, J. M. Murphy, G. Brumatti, R. Feltham, M. Pasparakis, J. Silke, P. Meier, MK2 phosphorylates RIPK1 to prevent TNF-induced cell death. Mol. Cell 66, 698–710.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. T. Ting, M. J. M. Bertrand, More to life than NF-κB in TNFR1 signaling. Trends Immunol. 37, 535–545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.N. Peltzer, M. Darding, H. Walczak, Holding RIPK1 on the ubiquitin leash in TNFR1 signaling. Trends Cell Biol. 26, 445–461 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Y. Dondelinger, S. Jouan-Lanhouet, T. Divert, E. Theatre, J. Bertin, P. J. Gough, P. Giansanti, A. J. R. Heck, E. Dejardin, P. Vandenabeele, M. J. M. Bertrand, NF-κB-independent role of IKKα/IKKβ in preventing RIPK1 kinase-dependent apoptotic and necroptotic cell death during TNF signaling. Mol. Cell 60, 63–76 (2015). [DOI] [PubMed] [Google Scholar]
- 19.N. Peltzer, E. Rieser, L. Taraborrelli, P. Draber, M. Darding, B. Pernaute, Y. Shimizu, A. Sarr, H. Draberova, A. Montinaro, J. P. Martinez-Barbera, J. Silke, T. A. Rodriguez, H. Walczak, HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell Rep. 9, 153–165 (2014). [DOI] [PubMed] [Google Scholar]
- 20.M. Moulin, H. Anderton, A. K. Voss, T. Thomas, W. W.-L. Wong, A. Bankovacki, R. Feltham, D. Chau, W. D. Cook, J. Silke, D. L. Vaux, IAPs limit activation of RIP kinases by TNF receptor 1 during development. EMBO J. 31, 1679–1691 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Y. Dondelinger, T. Delanghe, D. Rojas-Rivera, D. Priem, T. Delvaeye, I. Bruggeman, F. Van Herreweghe, P. Vandenabeele, M. J. M. Bertrand, MK2 phosphorylation of RIPK1 regulates TNF-mediated cell death. Nat. Cell Biol. 19, 1237–1247 (2017). [DOI] [PubMed] [Google Scholar]
- 22.L. Wang, F. Du, X. Wang, TNF-alpha induces two distinct caspase-8 activation pathways. Cell 133, 693–703 (2008). [DOI] [PubMed] [Google Scholar]
- 23.G. Liccardi, A. Annibaldi, MLKL post-translational modifications: Road signs to infection, inflammation and unknown destinations. Cell Death Differ. 30, 269–278 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K. W. Chen, B. Demarco, R. Heilig, K. Shkarina, A. Boettcher, C. J. Farady, P. Pelczar, P. Broz, Extrinsic and intrinsic apoptosis activate pannexin-1 to drive NLRP3 inflammasome assembly. EMBO J. 38, e101638 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.H. Jiao, L. Wachsmuth, S. Kumari, R. Schwarzer, J. Lin, R. O. Eren, A. Fisher, R. Lane, G. R. Young, G. Kassiotis, W. J. Kaiser, M. Pasparakis, Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 580, 391–395 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M. Feoktistova, P. Geserick, B. Kellert, D. P. Dimitrova, C. Langlais, M. Hupe, K. Cain, M. MacFarlane, G. Häcker, M. Leverkus, cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol. Cell 43, 449–463 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.H. I. Muendlein, D. Jetton, W. M. Connolly, K. P. Eidell, Z. Magri, I. Smirnova, A. Poltorak, cFLIP(L) protects macrophages from LPS-induced pyroptosis via inhibition of complex II formation. Science 367, 1379–1384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A. Oberst, C. P. Dillon, R. Weinlich, L. L. McCormick, P. Fitzgerald, C. Pop, R. Hakem, G. S. Salvesen, D. R. Green, Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 471, 363–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. Thome, P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schröter, C. Scaffidi, P. H. Krammer, M. E. Peter, J. Tschopp, Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386, 517–521 (1997). [DOI] [PubMed] [Google Scholar]
- 30.O. Micheau, M. Thome, P. Schneider, N. Holler, J. Tschopp, D. W. Nicholson, C. Briand, M. G. Grütter, The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J. Biol. Chem. 277, 45162–45171 (2002). [DOI] [PubMed] [Google Scholar]
- 31.M. A. Hughes, I. R. Powley, R. Jukes-Jones, S. Horn, M. Feoktistova, L. Fairall, J. W. R. Schwabe, M. Leverkus, K. Cain, M. MacFarlane, Co-operative and hierarchical binding of c-FLIP and caspase-8: A unified model defines how c-FLIP isoforms differentially control cell fate. Mol. Cell 61, 834–849 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.T. Kataoka, J. Tschopp, N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol. Cell. Biol. 24, 2627–2636 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.C. Pop, A. Oberst, M. Drag, B. J. van Raam, S. J. Riedl, D. R. Green, G. S. Salvesen, FLIP(L) induces caspase 8 activity in the absence of interdomain caspase 8 cleavage and alters substrate specificity. Biochem. J. 433, 447–457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J. W. Yu, P. D. Jeffrey, Y. Shi, Mechanism of procaspase-8 activation by c-FLIPL. Proc. Natl. Acad. Sci. U.S.A. 106, 8169–8174 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.K. Newton, K. E. Wickliffe, D. L. Dugger, A. Maltzman, M. Roose-Girma, M. Dohse, L. Kőműves, J. D. Webster, V. M. Dixit, Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature 574, 428–431 (2019). [DOI] [PubMed] [Google Scholar]
- 36.A. Polykratis, N. Hermance, M. Zelic, J. Roderick, C. Kim, T. M. van, T. H. Lee, F. K. M. Chan, M. Pasparakis, M. A. Kelliher, Cutting edge: RIPK1 kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J. Immunol. 193, 1539–1543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Q. Zhou, S. Snipas, K. Orth, M. Muzio, V. M. Dixit, G. S. Salvesen, Target protease specificity of the viral serpin CrmA. Analysis of five caspases. J. Biol. Chem. 272, 7797–7800 (1997). [DOI] [PubMed] [Google Scholar]
- 38.L. R. Garcia, T. Tenev, R. Newman, R. O. Haich, G. Liccardi, S. W. John, A. Annibaldi, L. Yu, M. Pardo, S. N. Young, C. Fitzgibbon, W. Fernando, N. Guppy, H. Kim, L. Y. Liang, I. S. Lucet, A. Kueh, I. Roxanis, P. Gazinska, M. Sims, T. Smyth, G. Ward, J. Bertin, A. M. Beal, B. Geddes, J. S. Choudhary, J. M. Murphy, K. Aurelia Ball, J. W. Upton, P. Meier, Ubiquitylation of MLKL at lysine 219 positively regulates necroptosis-induced tissue injury and pathogen clearance. Nat. Commun. 12, 3364 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.A. Oberst, C. Pop, A. G. Tremblay, V. Blais, J.-B. Denault, G. S. Salvesen, D. R. Green, Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J. Biol. Chem. 285, 16632–16642 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. D. Gitlin, K. Heger, A. F. Schubert, R. Reja, D. Yan, V. C. Pham, E. Suto, J. Zhang, Y. C. Kwon, E. C. Freund, J. Kang, A. Pham, R. Caothien, N. Bacarro, T. Hinkle, M. Xu, B. S. McKenzie, B. Haley, W. P. Lee, J. R. Lill, M. Roose-Girma, M. Dohse, J. D. Webster, K. Newton, V. M. Dixit, Integration of innate immune signalling by caspase-8 cleavage of N4BP1. Nature 587, 275–280 (2020). [DOI] [PubMed] [Google Scholar]
- 41.D. S. Simpson, J. Pang, A. Weir, I. Y. Kong, M. Fritsch, M. Rashidi, J. P. Cooney, K. C. Davidson, M. Speir, T. M. Djajawi, S. Hughes, L. Mackiewicz, M. Dayton, H. Anderton, M. Doerflinger, Y. Deng, A. S. Huang, S. A. Conos, H. Tye, S. H. Chow, A. Rahman, R. S. Norton, T. Naderer, S. E. Nicholson, G. Burgio, S. M. Man, J. R. Groom, M. J. Herold, E. D. Hawkins, K. E. Lawlor, A. Strasser, J. Silke, M. Pellegrini, H. Kashkar, R. Feltham, J. E. Vince, Interferon-γ primes macrophages for pathogen ligand-induced killing via a caspase-8 and mitochondrial cell death pathway. Immunity 55, 423–441.e9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H. HogenEsch, S. Janke, D. Boggess, J. P. Sundberg, Absence of Peyer's patches and abnormal lymphoid architecture in chronic proliferative dermatitis (cpdm/cpdm) mice. J. Immunol. 162, 3890–3896 (1999). [PubMed] [Google Scholar]
- 43.H. HogenEsch, M. J. Gijbels, E. Offerman, J. van Hooft, D. W. van Bekkum, C. Zurcher, A spontaneous mutation characterized by chronic proliferative dermatitis in C57BL mice. Am. J. Pathol. 143, 972–982 (1993). [PMC free article] [PubMed] [Google Scholar]
- 44.B. Gerlach, S. M. Cordier, A. C. Schmukle, C. H. Emmerich, E. Rieser, T. L. Haas, A. I. Webb, J. A. Rickard, H. Anderton, W. W.-L. Wong, U. Nachbur, L. Gangoda, U. Warnken, A. W. Purcell, J. Silke, H. Walczak, Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596 (2011). [DOI] [PubMed] [Google Scholar]
- 45.P. Lee, D.-J. Lee, C. Chan, S.-W. Chen, I. Ch’en, C. Jamora, Dynamic expression of epidermal caspase 8 simulates a wound healing response. Nature 458, 519–523 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.L. Injarabian, S. Willenborg, D. Welcker, M. Pasparakis, H. Kashkar, S. A. Eming, FADD- and RIPK3-mediated cell death ensures timely clearance of wound macrophages and promotes wound healing. bioRxiv 2023.2003.2029.534669 [Preprint]. 29 March 2023. 10.1101/2023.03.29.534669 [DOI]
- 47.S. Willenborg, D. E. Sanin, A. Jais, X. Ding, T. Ulas, J. Nüchel, M. Popović, T. MacVicar, T. Langer, J. L. Schultze, A. Gerbaulet, A. Roers, E. J. Pearce, J. C. Brüning, A. Trifunovic, S. A. Eming, Mitochondrial metabolism coordinates stage-specific repair processes in macrophages during wound healing. Cell Metab. 33, 2398–2414.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 48.S. Willenborg, S. A. Eming, Cellular networks in wound healing. Science 362, 891–892 (2018). [DOI] [PubMed] [Google Scholar]
- 49.T. Zhang, C. Yin, D. F. Boyd, G. Quarato, J. P. Ingram, M. Shubina, K. B. Ragan, T. Ishizuka, J. C. Crawford, B. Tummers, D. A. Rodriguez, J. Xue, S. Peri, W. J. Kaiser, C. B. López, Y. Xu, J. W. Upton, P. G. Thomas, D. R. Green, S. Balachandran, Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell 180, 1115–1129.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R. Karki, S. Lee, R. Mall, N. Pandian, Y. Wang, B. R. Sharma, R. S. Malireddi, D. Yang, S. Trifkovic, J. A. Steele, J. P. Connelly, G. Vishwanath, M. Sasikala, D. N. Reddy, P. Vogel, S. M. Pruett-Miller, R. Webby, C. B. Jonsson, T.-D. Kanneganti, ZBP1-dependent inflammatory cell death, PANoptosis, and cytokine storm disrupt IFN therapeutic efficacy during coronavirus infection. Sci. Immunol. 7, eabo6294 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.S. Li, Y. Zhang, Z. Guan, H. Li, M. Ye, X. Chen, J. Shen, Y. Zhou, Z.-L. Shi, P. Zhou, K. Peng, SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 5, 235 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.A. Roberts, D. Deming, C. D. Paddock, A. Cheng, B. Yount, L. Vogel, B. D. Herman, T. Sheahan, M. Heise, G. L. Genrich, S. R. Zaki, R. Baric, K. Subbarao, A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLOS Pathog. 3, e5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.J. A. Regla-Nava, J. L. Nieto-Torres, J. M. Jimenez-Guardeño, R. Fernandez-Delgado, C. Fett, C. Castaño-Rodríguez, S. Perlman, L. Enjuanes, M. L. DeDiego, Severe acute respiratory syndrome coronaviruses with mutations in the E protein are attenuated and promising vaccine candidates. J. Virol. 89, 3870–3887 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.J. A. Regla-Nava, J. M. Jimenez-Guardeño, J. L. Nieto-Torres, T. M. Gallagher, L. Enjuanes, M. L. DeDiego, The replication of a mouse adapted SARS-CoV in a mouse cell line stably expressing the murine SARS-CoV receptor mACE2 efficiently induces the expression of proinflammatory cytokines. J. Virol. Methods 193, 639–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.H. I. Muendlein, A. Poltorak, Flipping the switch from inflammation to cell death. Trends Immunol. 41, 648–651 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.W. C. Yeh, A. Itie, A. J. Elia, M. Ng, H. B. Shu, A. Wakeham, C. Mirtsos, N. Suzuki, M. Bonnard, D. V. Goeddel, T. W. Mak, Requirement for Casper (c-FLIP) in regulation of death receptor-induced apoptosis and embryonic development. Immunity 12, 633–642 (2000). [DOI] [PubMed] [Google Scholar]
- 57.D. Panayotova-Dimitrova, M. Feoktistova, M. Ploesser, B. Kellert, M. Hupe, S. Horn, R. Makarov, F. Jensen, S. Porubsky, A. Schmieder, A. C. Zenclussen, A. Marx, A. Kerstan, P. Geserick, Y.-W. He, M. Leverkus, cFLIP regulates skin homeostasis and protects against TNF-induced keratinocyte apoptosis. Cell Rep. 5, 397–408 (2013). [DOI] [PubMed] [Google Scholar]
- 58.R. Weinlich, A. Oberst, C. P. Dillon, L. J. Janke, S. Milasta, J. R. Lukens, D. A. Rodriguez, P. Gurung, C. Savage, T. D. Kanneganti, D. R. Green, Protective roles for caspase-8 and cFLIP in adult homeostasis. Cell Rep. 5, 340–348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.N. Wittkopf, C. Günther, E. Martini, G. He, K. Amann, Y.–. W. He, M. Schuchmann, M. F. Neurath, C. Becker, Cellular FLICE-like inhibitory protein secures intestinal epithelial cell survival and immune homeostasis by regulating caspase-8. Gastroenterology 145, 1369–1379 (2013). [DOI] [PubMed] [Google Scholar]
- 60.X. Piao, S. Komazawa-Sakon, T. Nishina, M. Koike, J.-H. Piao, H. Ehlken, H. Kurihara, M. Hara, N. Van Rooijen, G. Schütz, M. Ohmuraya, Y. Uchiyama, H. Yagita, K. Okumura, Y.-W. He, H. Nakano, c-FLIP maintains tissue homeostasis by preventing apoptosis and programmed necrosis. Sci. Signal. 5, ra93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.K. M. Boatright, C. Deis, J. B. Denault, D. P. Sutherlin, G. S. Salvesen, Activation of caspases-8 and -10 by FLIP(L). Biochem. J. 382, 651–657 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.R. Karki, B. R. Sharma, S. Tuladhar, E. P. Williams, L. Zalduondo, P. Samir, M. Zheng, B. Sundaram, B. Banoth, R. K. S. Malireddi, P. Schreiner, G. Neale, P. Vogel, R. Webby, C. B. Jonsson, T.-D. Kanneganti, Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell 184, 149–168.e17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.R. Peng, C. K. Wang, X. Wang-Kan, M. Idorn, M. Kjær, F. Y. Zhou, B. K. Fiil, F. Timmermann, S. L. Orozco, J. McCarthy, C. S. Leung, X. Lu, K. Bagola, J. Rehwinkel, A. Oberst, J. Maelfait, S. R. Paludan, M. Gyrd-Hansen, Human ZBP1 induces cell death-independent inflammatory signaling via RIPK3 and RIPK1. EMBO Rep. 23, e55839 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.N. Lalaoui, S. E. Boyden, H. Oda, G. M. Wood, D. L. Stone, D. Chau, L. Liu, M. Stoffels, T. Kratina, K. E. Lawlor, K. J. M. Zaal, P. M. Hoffmann, N. Etemadi, K. Shield-Artin, C. Biben, W. L. Tsai, M. D. Blake, H. S. Kuehn, D. Yang, H. Anderton, N. Silke, L. Wachsmuth, L. Zheng, N. S. Moura, D. B. Beck, G. Gutierrez-Cruz, A. K. Ombrello, G. P. Pinto-Patarroyo, A. J. Kueh, M. J. Herold, C. Hall, H. Wang, J. J. Chae, N. I. Dmitrieva, M. McKenzie, A. Light, B. K. Barham, A. Jones, T. M. Romeo, Q. Zhou, I. Aksentijevich, J. C. Mullikin, A. J. Gross, A. K. Shum, E. D. Hawkins, S. L. Masters, M. J. Lenardo, M. Boehm, S. D. Rosenzweig, M. Pasparakis, A. K. Voss, M. Gadina, D. L. Kastner, J. Silke, Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 577, 103–108 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.P. Tao, J. Sun, Z. Wu, S. Wang, J. Wang, W. Li, H. Pan, R. Bai, J. Zhang, Y. Wang, P. Y. Lee, W. Ying, Q. Zhou, J. Hou, W. Wang, B. Sun, M. Yang, D. Liu, R. Fang, H. Han, Z. Yang, X. Huang, H. Li, N. Deuitch, Y. Zhang, D. Dissanayake, K. Haude, K. McWalter, C. Roadhouse, J. J. MacKenzie, R. M. Laxer, I. Aksentijevich, X. Yu, X. Wang, J. Yuan, Q. Zhou, A dominant autoinflammatory disease caused by non-cleavable variants of RIPK1. Nature 577, 109–114 (2020). [DOI] [PubMed] [Google Scholar]
- 66.M. A. Ramakrishnan, Determination of 50% endpoint titer using a simple formula. World J. Virol. 5, 85–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R. Schwarzer, H. Jiao, L. Wachsmuth, A. Tresch, M. Pasparakis, FADD and caspase-8 regulate gut homeostasis and inflammation by controlling MLKL- and GSDMD-mediated death of intestinal epithelial cells. Immunity 52, 978–993.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 68.T. Tenev, K. Bianchi, M. Darding, M. Broemer, C. Langlais, F. Wallberg, A. Zachariou, J. Lopez, M. MacFarlane, K. Cain, P. Meier, The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell 43, 432–448 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6