Abstract
KRAS/ERK pathway phosphorylates DICER1, causing its nuclear translocation, and phosphomimetic Dicer1 contributes to tumorigenesis in mice. Mechanisms through which phospho-DICER1 regulates tumor progression remain undefined. While DICER1 canonically regulates microRNAs (miRNA) and epithelial-to-mesenchymal transition (EMT), we found that phosphorylated nuclear DICER1 (phospho–nuclear DICER1) promotes late-stage tumor progression in mice with oncogenic Kras, independent of miRNAs and EMT. Instead, we observe that the murine AT2 tumor cells exhibit altered chromatin compaction, and cells from disorganized advanced tumors, but not localized tumors, express gastric genes. Collectively, this results in subpopulations of tumor cells transitioning from a restricted alveolar to a broader endodermal lineage state. In human LUADs, we observed expression of phospho–nuclear DICER1 in advanced tumors together with the expression of gastric genes. We define a multimeric chromatin-DICER1 complex composed of the Mediator complex subunit 12, CBX1, MACROH2A.1, and transcriptional regulators supporting the model that phospho–nuclear DICER1 leads to lineage reprogramming of AT2 tumor cells to mediate lung cancer progression.
Phosphorylated DICER1 reprograms alveolar tumor cell identity to promote late-stage progression of lung cancers.
INTRODUCTION
Lung cancer is the leading cause of cancer-related mortality worldwide primarily because of the formation of metastatic lesions, which induce multi-organ failure (1). Oncogenic gain-of-function mutations in the small guanosine triphosphatase (GTPase) KRAS account for 20 to 30% of human lung adenocarcinomas (LUADs) (2–6). The oncogenic mutations in the GTPase domain render KRAS locked in the guanosine triphosphate (GTP)–loaded and, thus, “active” state (7, 8) Notably, expression of oncogenic mutant Kras is sufficient to cause cell transformation and lung tumor formation in murine models (9). Molecularly, KRAS functions through the activation of the core kinase cascade RAF–mitogen-activated protein kinase kinase–ERK (extracellular signal–regulated kinase) to control a multitude of cellular behaviors (10). More specifically, active di-phosphorylated ERK, which is the final effector of the cascade, phosphorylates diverse substrates that, in turn, control cellular processes such as proliferation, migration, apoptosis, and differentiation, among others (11, 12). Thus, to understand the pathobiology of LUADs driven by the KRAS-ERK signaling pathway, we need to identify and define the substrates through which KRAS oncogenic mutations specifically drive tumor onset, progression, and invasion. However, unique targets of ERK that mediate only late stages of cancer progression and invasion remain undefined.
We identified DICER1 as a unique substrate of active ERK in worms, mice, and humans (13–15). ERK phosphorylates DICER1 at two serine residues in the ribonuclease IIIb (RNase IIIb) domain and the double-stranded RNA binding domain (13–16). In cancers, Dicer1 functions as a haploinsufficient tumor suppressor (17, 18). In this context, loss of one copy of Dicer1 in a Kras oncogenic murine background leads to an increase in tumor number and size. However, somatic mutations that result in the deletion or loss of Dicer1 are rare in cancers (19–21). Thus, the role of Dicer1 in cancer onset and progression is nuanced and indicates that Dicer1 function is context dependent. We hypothesized that phosphorylation of DICER1 likely provides one context for regulation of DICER1 activity and tested this function in cancer development. Genetically modified mouse models that carry the constitutive homozygous form of phosphorylated DICER1 (phospho-DICER1; phosphomimetic Dicer1; Dicer1S2D) wherein the phosphorylated serines at positions 1712 and 1836 are replaced with aspartic acid (13) are perinatal lethal, and the survivors display accelerated aging and hypermetabolism, suggesting that regulated phosphorylation of DICER1 is critical for normal mammalian development (13). In a cancer model, heterozygous Dicer1S2D allele together with the heterozygous oncogenic Kras G12D (KrasLA1) allele (9) leads to the generation of LUADs and tumor spread across the animal body (14).
Canonically, DICER1 functions in the cytoplasm as an RNase enzyme to generate small noncoding RNAs of 21 nucleotides in length, the microRNAs (miRNAs) (19, 22). Phosphomimetic DICER1, however, is nuclear in the primary lung tumors in the mouse model with KrasLA1, suggesting that phosphorylation and nuclear localization of DICER1 play an important role in tumor progression and spread in vivo (14). The discovery that phosphorylated nuclear DICER1 functions with a Kras oncogenic mutation to mediate tumor spread suggests that we have uncovered a specific signaling axis of KRAS-ERK-DICER1 cascade that may uniquely drive tumor spread. However, the function of phosphorylated nuclear DICER1 in tumor progression is currently unknown. Here, we investigated the cellular and molecular mechanisms through which phosphorylated nuclear DICER1 affects tumor progression in mice and humans.
We show that DICER1 is phosphorylated and nuclear in human LUADs. Using genetically modified mouse models, we observe that unlike loss of one copy of Dicer1, which regulates tumor onset in a KrasLA1/+ background, phosphomimetic DICER1 only regulates late stages of tumor progression. In addition and unexpectedly, phosphomimetic Dicer1 regulates late-stage tumor progression independent of epithelial-to-mesenchymal transition (EMT) and miRNA production. Instead, through a combination of single-cell RNA sequencing (scRNA-seq), in situ RNA hybridization, and immunofluorescence for validation of single-cell RNA analysis, Assay for Transposable-Accessible Chromatin (ATAC)-sequencing, and chromatin crosslinking–based immunoprecipitation proteomic analysis, we found that the phosphorylated nuclear DICER1 associates with an open chromatin state and binds to a unique chromatin complex composed of Mediator complex subunit 12, RNA polymerase fourth subunit, Chromobox Protein Homolog 1 (CBX1) chromatin remodeler, and various transcription factors, which collectively likely lead to the expression of gastric (endodermal) genes in lung tumor alveolar epithelial cells, among others. In addition, we observed that grade 3 human LUAD tumors with KRAS oncogenic mutations and phospho-DICER1 ectopically express gastric gene signature, suggesting that phosphorylation of DICER1 may play similar roles in cell reprogramming in mice and humans during progression of LUADs. We propose that the increased tumor cell plasticity because of lineage reprogramming may be the underlying mechanism through which phosphorylated nuclear DICER1 drives late-stage tumor progression.
RESULTS
DICER1 is nuclear and phosphorylated in human LUADs
To determine whether DICER1 is phosphorylated in human LUADs, we performed immunohistochemistry (IHC) analysis using anti-phospho-DICER1 and anti-phosphorylated ERK antibodies (Materials and Methods) on 88 LUAD tumors including 38 (~43%) samples with KRAS oncogenic mutations and 50 (~57%) without KRAS oncogenic mutations. KRAS oncogenic mutation status and baseline characteristics of the patient population are presented in Table 1 and tables S1 and S2. The quantification of positive antibody staining for phospho-DICER1 and active ERK was performed using the Fiji software and following Andy’s algorithm on the entire tumor section (Materials and Methods). Tumors with ≥1% but <30% of cells with phospho-DICER1 were classified as “low positive,” tumors with ≥30% but <70% of cells as “moderate positive,” ≥70% of cells as “high positive,” and <1% of cells with phospho-DICER1 as negative (Materials and Methods; Table 1 and fig. S1A). Tumors were simultaneously analyzed for phospho-ERK status following the same parameters as described for phospho-DICER1 (Materials and Methods; Table 1, tables S1 and S2, and fig. S1).
Table 1. Relationship between phospho-DICER1 (A), phospho-ERK (B), and clinicopathological features in LUADs.
LVI, lymph node invasion. There are a total of 88 samples in phospho-DICER1 staining and 87 samples in phospho-ERK staining. Sex data are missing from two cases; grade and LVI data are missing from six cases, and stage is missing from twelve cases. T4 only has one sample.
| A. Clinicopathology factors | Nuclear phospho-DICER1 expression | ||||
|---|---|---|---|---|---|
| Negative | Low | Moderate | High | ||
| N | N | N> | N | P* | |
| Sex | |||||
| Female | 0 | 19 | 22 | 5 | 0.6046 |
| Male | 0 | 17 | 21 | 2 | |
| Tumor size (T) | |||||
| T1 | 0 | 9 | 13 | 4 | 0.5589 |
| T2 | 0 | 19 | 19 | 2 | |
| T3–T4 | 0 | 6 | 9 | 1 | |
| Stage | |||||
| I | 0 | 21 | 20 | 4 | 0.839 |
| II | 0 | 8 | 11 | 2 | |
| III | 0 | 5 | 5 | 0 | |
| LVI | |||||
| Present | 0 | 8 | 12 | 2 | 0.7012 |
| Absent | 0 | 28 | 27 | 5 | |
| KRAS mutations | |||||
| Mutated | 0 | 9 | 25 | 4 | 0.0098 |
| Not Mutated | 0 | 28 | 19 | 3 | |
| B. Clinicopathology factors | Phospho-ERK expression | ||||
| Negative | Low | Moderate | High | ||
| N | N | N | N | P* | |
| Sex | |||||
| Female | 15 | 22 | 5 | 3 | |
| Male | 8 | 28 | 2 | 2 | |
| Tumor size (T) | |||||
| T1 | 6 | 13 | 3 | 4 | 0.1888 |
| T2 | 10 | 27 | 1 | 1 | |
| T3–T4 | 5 | 9 | 2 | 0 | |
| Stage | |||||
| I | 12 | 29 | 0 | 3 | 0.676 |
| II | 5 | 14 | 1 | 1 | |
| III | 4 | 6 | 0 | 0 | |
| LVI | |||||
| Present | 5 | 13 | 3 | 2 | 0.1048 |
| Absent | 16 | 38 | 2 | 2 | |
| KRAS mutations | |||||
| Mutated | 7 | 20 | 6 | 5 | 0.0034 |
| Not mutated | 16 | 32 | 1 | 0 | |
*Chi-square test.
Phospho-DICER1 was positive and nuclear in all LUAD tumors (Fig. 1, A and B). Tumors with KRAS oncogenic mutations correlated with moderate (65% of tumors) to high (10% of tumors) phospho-DICER1 positivity (Fig. 1B), whereas tumors with wild-type KRAS correlated with low (56% of tumors) phospho-DICER1 positivity (Fig. 1B). These data demonstrate that positive phospho-DICER1 status correlates with human LUADs bearing KRAS mutations.
Fig. 1. DICER1 is phosphorylated and nuclear in human LUADs.
(A) Representative images of human LUADs stained with anti–phospho-DICER1 (left) and anti–phospho-ERK (right) antibodies. 40×: Scale bars, 100 μm. Brown staining marks positive antibody signal; blue staining marks nuclei labeled by hematoxylin. Bottom: Magnification of tumor cells. The magnified black box highlights the tumor section stained with phospho-DICER1, and the green box highlights active-ERK staining. Scale bars, 10 μm. (B) Human LUADs with low, moderate, and high phospho-DICER1 signal plotted as percentages on the y axis and KRAS mutation status on the x axis. (C) Human LUADs with negative, low, moderate, and high phospho-ERK signal plotted by their KRAS mutation status. (D) Human LUADs with KRAS oncogenic mutations classified on the basis of the presence [LNI (+)] or absence [LNI (−)] of lymph node invasion and phospho-DICER1 nuclear signal. (E) Human LUAD tumors with KRAS oncogenic mutations classified on the basis of the presence [LNI (+)] or absence [LNI (−)] of lymph node invasion and phospho-ERK nuclear signal. “KRAS mutations” denotes samples with oncogenic KRAS mutations. “Non-KRAS” denotes samples without any mutations in the KRAS locus in these tumors. Samples are classified on the basis of the percentage of cells positive for phospho-DICER1 (pDICER1). Low (<30% positive cells), moderate (≥30% and <70% of positive cells), high (≥70% of positive cells). Chi-square, (B) **P = 0.0098 and (C) **P = 0.0034
Unlike phospho-DICER1 status, only 64 (73%) of the tumors were positive for phospho-ERK (Fig. 1, A and C). Of the tumors with KRAS oncogenic mutations that displayed positive phospho-ERK staining, we observed that 15% of the tumors showed moderate positivity, while 13% displayed a high level of phospho-ERK signal. Most of the LUAD tumors assayed displayed low phospho-ERK positive signal irrespective of their KRAS mutation profile. Unexpectedly, however, 18% of the tumors with KRAS mutations were negative for phospho-ERK signal (Fig. 1C). This was unexpected because it revealed that phospho-DICER1 and phospho-ERK do not perfectly overlap. This is likely due to a difference in the rate of dephosphorylation or the presence of different phosphatases that regulate each phosphorylation event of these two proteins.
To next determine whether the presence or absence of phospho-ERK and phospho-DICER1 correlated with tumor invasion, we evaluated tumor pathology and patient clinicopathologic features, and while we did not observe any correlations, we found that phospho-DICER1 is expressed in moderate to high levels in higher percentage in KRAS-mutated LUADs with invasion to the lymph node (Fig. 1D). A total of 22 LUAD tumors displayed lymph node invasion of which 13 tumors had mutations in KRAS and 9 tumors were wild type for KRAS (Fig. 1D and fig. S1B). Overall, we observed that a higher proportion of LUAD tumors with lymph node invasion and the presence of KRAS oncogenic mutations displayed either moderate or high levels of phospho-DICER1 signal. Unexpectedly, some tumors with KRAS oncogenic mutations and lymph node invasion still showed negative staining for phospho-ERK (Fig. 1E and fig. S1C), suggesting that phospho-ERK may be dynamic and a poorer prognostic marker for tumor progression or invasion compared to phospho-DICER1. Collectively, these analyses demonstrate that DICER1 is phosphorylated in human LUADs and correlates with tumor invasion and KRAS oncogenic mutations.
Phosphomimetic Dicer1 causes late-stage tumor progression
Dicer1 functions as a haploinsufficient tumor suppressor because loss of one copy of Dicer1 in a Kras oncogenic murine background results in increased tumor size and number as early as 12 weeks in tumorigenesis (17, 18). Thus, to determine whether phosphorylation of DICER1 contributes to tumor initiation or late-stage progression in an oncogenic Kras background, we used the phosphomimetic Dicer1 murine model (Dicer1S2D) (13). In the Dicer1S2D model, serines at positions 1712 and 1836 are replaced by aspartic acid at the endogenous locus to mimic constitutive phosphorylation. Homozygous Dicer1S2D mice develop a detrimental aging phenotype, resulting in early lethality hindering the study of tumors in these animals (13). Thus, we crossed the heterozygous mutant Dicer1S2D with a heterozygous oncogenic Kras LA1 murine model (9) and assayed KrasLA1/+ and KrasLA1/+;Dicer1S2D/+ animals for tumor generation. Animals were evaluated for tumor number, size, and grade at multiple time points. Dicer1S2D/+ mice are phenotypically wild-type and do not develop tumors as described previously (13, 14). KrasLA1/+ mice developed multifocal lung adenomas and adenocarcinomas as described (9).
Previously, we had observed that Dicer1S2D/+ allele together with the KrasLA1/+ results in mice with multiple LUADs and tumor spread throughout the body of the animal (14). However, we do not know whether DICER1 is phosphorylated in wild-type adult lungs and whether the increase in tumor burden in the double mutant mice was due to a difference in DICER1 phosphorylation level within the tumor cells. Thus, we performed an IHC with phospho-DICER1 antibody and assessed both wild-type adult lungs and lung tumors from KrasLA1/+ and KrasLA1/+;Dicer1S2D/+ animals. We observed that in wild-type lungs, phospho-DICER1 is absent in the alveolar cells but expressed in few of the epithelial cells in the bronchiole structures [arrows in Fig. 2A (top)]. However, unlike in wild-type lungs, where alveolar cells do not express phospho-DICER1, the alveolar type 2 (AT2) tumor cells from both KrasLA1/+ and KrasLA1/+;Dicer1S2D/+ animals express phosphorylated nuclear DICER1 (Fig. 2A, bottom). AT2 cells have been shown to be the cell type for tumor origin in the KrasLA1/+ model (9). Thus, this observation demonstrates that DICER1 is aberrantly phosphorylated in alveolar cells in the KrasLA1/+ tumors, consistent with it being phosphorylated downstream to the KRAS/ERK signaling axis. Furthermore, we observed a higher intensity of phospho-DICER1 staining of the lung tumor cells of KrasLA1/+;Dicer1S2D/+ mice when compared to tumor cells of the KrasLA1/+ mice (Fig. 2A, bottom). The increased intensity of staining is likely because of the presence of one copy of phosphomimetic DICER1, wherein the aspartic acid is a true phosphomimic because it is detected by the phosphorylated antibody as shown previously (15). Together these IHC data suggest that (i) the KRAS/ERK cascade is active in the lung tumors of both KrasLA1/+;Dicer1S2D/+ and KrasLA1/+ mice, leading to the phosphorylation of DICER1, and that (ii) phosphomimetic Dicer1 allele mimics ERK-dependent phosphorylation and nuclear translocation of DICER1, resulting in nuclear accumulation of DICER1 in the lung tumor cells of KrasLA1/+;Dicer1S2D/+ mice.
Fig. 2. Phosphomimetic Dicer1 regulates tumor invasion in mice.
(A) Representative images of IHC staining with anti–phospho-DICER1 antibody in brown in murine wild-type (WT) lung tissue, KrasLA1/+ lung tumor, and KrasLA1/+;Dicer1S2D/+ lung tumor. Scale bars, 50 μm. (B) Number of LUADs from KrasLA1/+ (green, n = 4) and KrasLA1/+;Dicer1S2D/+ (orange, n = 5) animals at 24 weeks of age. *P = 0.03. (C) Representative hematoxylin and eosin (H&E) image of the LUADs from KrasLA1/+;Dicer1S2D/+ mice at 34 weeks of age. The nuclei are marked by purple hematoxylin staining. Left: 10× magnification of a lung tumor with a thymic lymphoma, marked by yellow asterisk. Right: A higher magnification of the yellow box highlighting the invasive LUAD marked by white asterisk (from left). Blue asterisk indicates infiltrating lymphocytes. Encircled in white dashed lines are LUAD cells that are invading nearby bronchial structures. Scale bar, 100 μm (D) Representative images of gross morphology of a lung and a heart from KrasLA1/+ and KrasLA1/+;Dicer1S2D/+ mice at 34 weeks of age. Yellow arrowheads point to lung tumors. Encircled in yellow is a tumor lesion spreading into and compressing the heart. Scale bar, 1000 μm.
We next performed a tumor watch to determine whether the presence of phosphomimetic Dicer1 allele affected tumor development or progression in the KrasLA1/+ model. We observed that at 6 and 12 weeks of age, KrasLA1/+ and KrasLA1/+;Dicer1S2D/+ mice developed numerous hyperplastic lesions and adenomas (fig.S2A) with no significant difference in the number, size, and histological grade (fig. S2, B and C), unlike what was observed with loss of one copy of Dicer1 in a Kras oncogenic background (17). At 24 weeks of age, the KrasLA1/+;Dicer1S2D/+ double mutant mice developed an increase in the number of LUADs with increased cellular atypia and invasion of adjacent bronchial or vessel structures when compared to KrasLA1/+ mice (Fig. 2, B and C, and fig. S2D). Most of the adenocarcinomas were organized into papillary-like structures with a few solid and lepidic adenocarcinomas. At 34 weeks of age, the KrasLA1/+;Dicer1S2D/+ mice displayed tumors across the animal body including lymph node and heart and thymic lymphomas, in addition to the primary lung adenomas and adenocarcinomas, as previously described (14) (Fig. 2D). KrasLA1/+ single mutant mice displayed no spread of tumors at these time points. Together, our data indicate that the oncogenic mutation in Kras is responsible for tumor initiation and the presence of phosphomimetic Dicer1 leads to late-stage tumor progression and invasion. These data suggest that the function of DICER1 phosphorylation in mediating tumor progression is distinct from the loss of one copy of Dicer1 (9) wherein the latter leads to an enhancement of tumor onset and accelerates early stages of tumor development. Thus, together with the genetic analysis, we conclude that constitutive phosphorylation of DICER1 downstream of the KRAS oncogenic pathway promotes late-stage tumor progression and leads to the formation of invasive tumors in vivo.
The presence of phospho-DICER1 does not regulate EMT to mediate tumor invasion
To determine the cellular mechanism through which the presence of phosphomimetic Dicer1 leads to an increased number of adenocarcinomas and spread of tumors, we evaluated EMT as a mechanism for tumor progression (Materials and Methods). Lung tumors from KrasLA1/+;Dicer1S2D/+ and KrasLA1/+ genotypes (1 year old, n = 5 per genotype) were assayed using immunofluorescence for epithelial and mesenchymal markers (Materials and Methods). Pro-SFPC, which marks mature AT2 cell, was used to stain and identify the AT2 cells (fig. S3A, magenta staining). We then assayed for vimentin expression within the AT2-positive tumor cells (fig. S3A, white staining). We hypothesized that the tumor epithelial cells undergoing EMT will display AT2 alveolar markers and coexpress the vimentin mesenchymal marker. However, we observed that AT2 cells did not exhibit vimentin signal in any of the tumors assayed (fig. S3A). Instead, AT2 tumor cells displayed a dual pattern for E-cadherin expression (fig. S3A, green staining), where some (i) exhibited high levels of E-cadherin staining, consistent with their epithelial nature, and some (ii) exhibited low levels of E-cadherin signal (fig. S3A). This variation in E-cadherin staining suggests that the AT2 tumor cell population is heterogeneous with regard to E-cadherin expression. However, even in AT2 tumor cells with low E-cadherin expression, we did not observe any vimentin expression (fig. S3A). In addition, we observed that the lung tumors from KrasLA1/+;Dicer1S2D/+ and KrasLA1/+ mice displayed similar percentages of vimentin-positive cells irrespective of their E-cadherin status (fig. S3B′). These data demonstrate that the tumor cells are not transitioning from an epithelial (E-cadherin) to a mesenchymal (vimentin positive) state and tumor cells that display E-cadherin expression are mutually exclusive from cells that display vimentin expression (fig. S3B″). In general, cells that express vimentin are likely immune and fibroblast cells. Thus, given the lack of vimentin staining in AT2 cells and no difference in the number of vimentin-positive cells between both the genotypes, we conclude that phosphomimetic DICER1-induced tumor progression is independent of EMT.
The presence of phospho-DICER1 results in the expression of gastrointestinal genes in lung tumors
During the EMT analysis, we observed a heterogeneity in E-cadherin expression within the AT2 tumor cell populations. Thus, we hypothesized that phosphomimetic Dicer1 likely generates subclonal populations of cells that contribute to tumor progression or invasion. To determine whether phosphomimetic Dicer1 leads to the generation of subclonal populations of tumor cells that contribute to tumor invasion in the KrasLA1/+;Dicer1S2D/+ relative to KrasLA1/+ animals, we performed scRNA-seq on the lung tumors from both the genotypes at 39 weeks of age (Materials and Methods). We isolated and sequenced 27 lung tumors (20,214 cells) from two KrasLA1/+ mice and 23 lung tumors (18,754 cells) from two KrasLA1/+;Dicer1S2D/+ mice (Materials and Methods). In addition, we sequenced lung tissue (7288 cells) from a Dicer1S2D/+ mouse, which is morphologically like wild-type, to validate the expression of canonical lung markers (fig. S4A). The single-cell sequencing identified a total of 18,816 tumor cells from both tumor models and 6783 cells from Dicer1S2D/+ mouse. Together, these cells clustered into more than 23 types of cells (fig. S4A). We broadly grouped these clusters into three major cell populations named epithelial, immune [which include natural killer (NK) cells, T cells and B cells, macrophages, etc.], and mesenchymal cells (includes endothelial cells, fibroblasts, etc.; figs. S4, B and C, S5, and S6). Because our goal was to define the population of tumor cells that may be heterogeneous and contribute to tumor progression, we focused only on the epithelial cells from this point forward because we assume that these are the tumor cells.
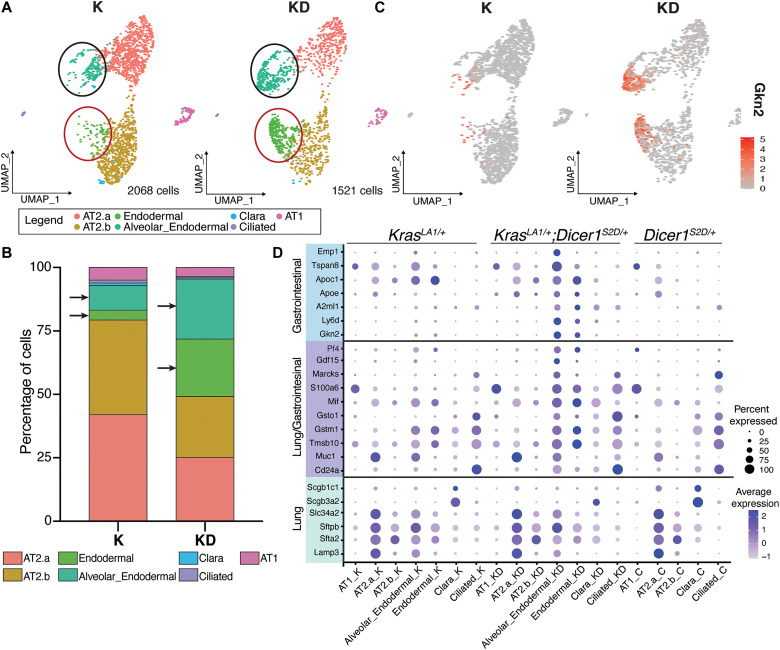
The epithelial cells were identified as those positive for Epcam, Cdh1, Sfta2, Lamp3, Scl34a2, Rtkn2, Pdpn, Hopx, Ager, Scgb1a1, Scgb3a2, and Foxj1 in both the tumor genotypes and the wild-type animals. In the absence of a tumor, however, the number of epithelial cells is very few in an adult mouse lung (figs. S4, A and B, and S6A), defined largely by alveolar cells, bronchial cells, Clara, and Club cells. Compared to a wild-type lung, however, tumors display a large expansion of epithelial cells. To determine the types of cells that were expanded in the lung tumors, we assayed for genes that were expressed in each cell, defined them by known categories, and used these categories as markers for each cell type. Using these markers, the tumor epithelial cells were then subclustered (Materials and Methods and figs. S6 to S8). These subclusters are AT1, AT2, Clara, and ciliated cells (Fig. 3A and fig. S6B). We observed, and as shown previously (9), that the largest population of tumor cells was the AT2 cell population. Within the AT2 cell population, we observed at least four distinct subpopulations, categorized as such by their gene expression. We defined these four populations as (i) those positive for Cdh1 (E-cadherin) and (ii) those negative for Cdh1 (three–fold change difference; fig. S8A), consistent with the immunofluorescence analysis (above). Because these two populations were a variant of AT2 cells, we named them AT2.a and AT2.b, respectively (Fig. 3, A and B, and figs. S7 and S8). (iii) The third subcluster of epithelial cells expressed AT2 cell markers but, unexpectedly, also expressed gastrointestinal genes including Gkn2, Ly6d, Apoe, A2ml1, and Emp1 (Fig. 3, A to D, and figs. S7 and S8). Expression of the gastrointestinal genes is unexpected because gastric genes are normally not expressed in the lung. Because these tumor cells express broad signatures of endodermal genes, AT2 cells and Cdh1, we named this subcluster as Alveolar_ Endodermal cells. (iv) The final subcluster of tumor epithelial cells displayed low expression of AT2 cell markers and no expression of Cdh1 or Lamp3 but still expressed endodermal genes. This subcluster was named “Endodermal cells” (Fig. 3, A to D, and figs. S7 and S8).
Fig. 3. Enrichment of gastrointestinal genes in lung tumor cells with phosphomimetic Dicer1.
(A) Uniform manifold approximation and projection (UMAP) graphs of the subclusters of lung tumor epithelial cells identified from scRNA-seq analysis of KrasLA1/+ (K, n = 2) and KrasLA1/+;Dicer1S2D/+ (KD, n = 2) mice. Each dot represents a cell. The cells were subclustered on the basis of the expression of genes as AT2a (pink), AT2b (brown), Endodermal (green), and Alveolar-Endodermal (dark green). The black and red circles around tumor cells highlight AT2 cells that express gastrointestinal genes. (B) Percentages of the various cell clusters identified in KrasLA1/+ (K) and KrasLA1/+;Dicer1S2D/+ (KD) lung tumors. Arrows point to Alveolar_Endodermal and Endodermal clusters. (C) UMAP showing the relative expression for Gkn2 gene in red within the tumor epithelial cell cluster from KrasLA1/+ (K) and KrasLA1/+;Dicer1S2D/+ (KD). (D) Dot plot showing gene expression of lung and gastrointestinal gene signatures. The size of the dots corresponds to the percentage of cells (on the X axis) that express the genes displayed on the y axis. The color density corresponds to the fold expression of each gene on the Y axis. K labels lung tumor cells from KrasLA1/+; KD labels lung tumor cells from KrasLA1/+;Dicer1S2D/+, and C labels the control lung cells from Dicer1S2D/+.
Together, these data reveal that the lung tumors from KrasLA1/+;Dicer1S2D/+ mice bear tumor epithelial cell populations composed of 25% of AT2.a cells, 24% of AT2.b cells, 23% Alveolar_ Endodermal, and 22% Endodermal clusters cells (Fig. 3B). In contrast, in the KrasLA1/+ single mutant lung tumors, over 42 and 37% of the tumor epithelial cells are composed of AT2.a and AT2.b tumor cells, respectively, whereas Alveolar_ Endodermal and Endodermal clusters represent only 9.7% and 3.9%, respectively (Fig. 3B). The enrichment in the Alveolar_ Endodermal and Endodermal clusters in the KrasLA1/+;Dicer1S2D/+ relative to the KrasLA1/+ tumors suggests that phosphorylated nuclear DICER1 in the lung tumors leads to ectopic expression of gastric and other endodermal genes, which creates subclonal populations of tumor cells that alter tumor cell plasticity and likely drive invasion.
The presence of phospho-DICER1 in murine lung tumors alters AT2 identity
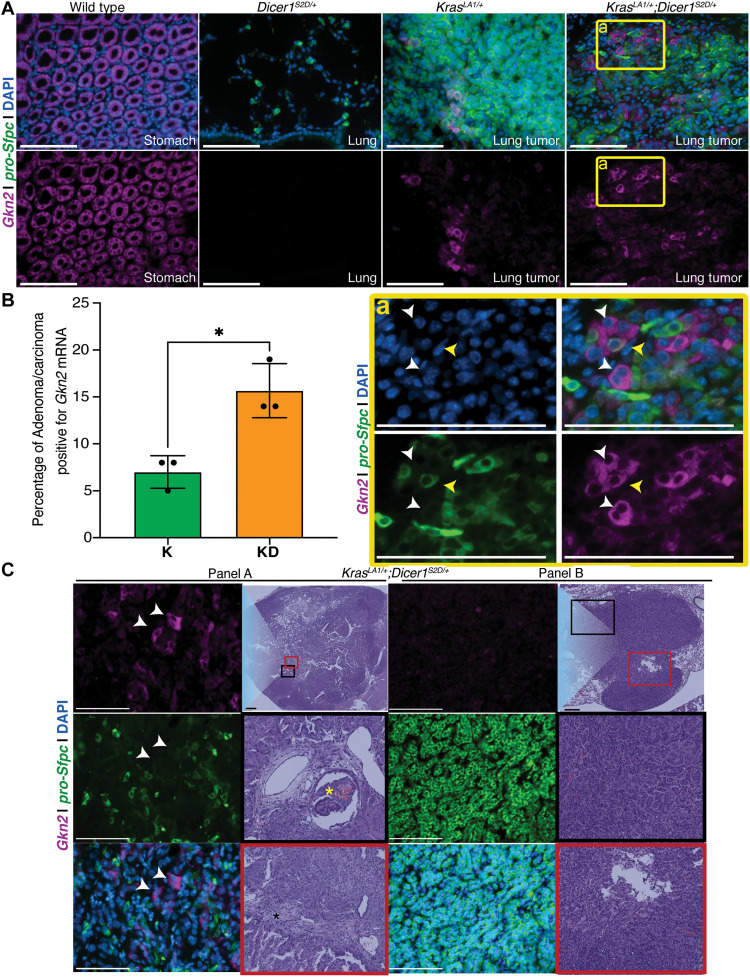
To validate the expression of gastric genes identified from the scRNA-seq analysis (above), we performed hairpin chain reaction (HCR)–RNA–fluorescent in situ hybridization (FISH) (Molecular Instruments; Materials and Methods) and immunofluorescence staining on lung tumors from KrasLA1/+ and KrasLA1/+;Dicer1S2D/+ mice (Materials and Methods). We tested for gastrokine-2 (Gkn2) expression because (i) it is up-regulated in the KrasLA1/+;Dicer1S2D/+ lung tumor cells, (ii) Gkn2 is expressed in the wild-type stomach but not lungs (Fig. 4A and fig. S9), and (iii) a subset of poorly characterized human lung cancers known as mucinous adenocarcinoma display GKN1 expression (23). Although, the mechanism for the latter remains unknown. We first tested murine wild-type stomach and lung tissue with Gkn2 and pro-Surfactant RNA (a marker for mature AT2 cells) probes to assess the specificity of staining. We observed that the wild-type stomach expressed Gkn2 and did not express pro-Surfactant, as expected (Fig. 4A); similarly, Dicer1S2D/+ lung (which is morphologically like wild type) does not express Gkn2 but expresses pro-Surfactant in the AT2 cells (Fig. 4A), as expected. Staining of the lung tumors from KrasLA1/+;Dicer1S2D/+ and KrasLA1/+animals reveals that as found by the scRNA-seq analysis, Gkn2 mRNA and protein are expressed in AT2 tumor epithelial cells (assayed with the pro-Surfactant marker) from both KrasLA1/+ and KrasLA1/+;Dicer1S2D/+ animals (Fig. 4A and fig. S9). Furthermore, the number of lung tumors with populations of epithelial cells that express Gkn2 was significantly higher in the KrasLA1/+;Dicer1S2D/+ relative to the KrasLA1/+ mice (Materials and Methods; Fig. 4B). These data suggest that the presence of phosphomimetic Dicer1 results in an increased incidence of AT2 cells displaying endodermal gene expression because the cells are potentially undergoing lineage reprogramming.
Fig. 4. Phosphomimetic Dicer1 leads to expression of gastrointestinal genes in AT2 tumor cells.
(A) Wild-type stomach tissue and normal Dicer1S2D/+ lung tissue mouse stained with Gkn2 RNA (magenta), pro-Sfpc RNA (green), and nuclei [4′,6-diamidino-2-phenylindole (DAPI) in blue] as positive controls for the expression of Gkn2 (stomach) and pro-Sfpc (lung). Gkn2 signal is negative in normal lung tissue, and pro-Sfpc is negative in normal stomach tissue, demonstrating the specificity of the probes. Lung tumor sections from KrasLA1/+ and KrasLA1/+;Dicer1S2D/+ mice reveal staining for both Gkn2 and pro-Sfpc. (a) is the magnified inset from (A) from KrasLA1/+;Dicer1S2D/+ animal. Yellow arrowheads point to cells that coexpress Gkn2 and pro-Sfpc (alveolar-endodermal). White arrowheads point to cells that only express Gkn2 (endodermal). (B) Percentage of lung tumors with epithelial cells positive for Gkn2 mRNA in KrasLA1/+ (K, N = 3 mice, n = 45 lung tumors) and KrasLA1/+;Dicer1S2D/+ (KD, N = 3 mice, n = 44 lung tumors) mice. (C) LUAD from KrasLA1/+;Dicer1S2D/+ animal stained with Gkn2 mRNA (magenta), pro-Sfpc (green), and DAPI (blue) on the left column and H&E staining on right column. Panel A represents a disorganized tumor. H&E for Panel A (top) is 10× magnification of tumor. Black and red inset boxes are magnified to highlight the disorganization in the tumor. Yellow asterisk marks invasion to bronchial structure; black asterisk marks desmoplasia. Fluorescent RNA hybridization images (63×) show tumor cells that express Gkn2 mRNA (white arrow) and do not express pro-Sfpc. Panel B represents an organized tumor. H&E for Panel B (top) is 10× magnification of tumor. Black and red inset boxes are magnified as insets on the bottom to highlight the organized and localized nature of the tumor. Fluorescent RNA hybridization images (63×) show tumor cells with no expression for Gkn2 but strong expression for pro-Sfpc. White scale bar, 50 μm; black scale bar, 100 μm
More detailed analysis of the AT2 tumor cells with the pro-Surfactant and Gkn2 markers further revealed subpopulations of tumor cells, which either coexpressed Gkn2 with pro-Surfactant [Fig. 4A (inset), yellow arrow heads] or subpopulations that had reduced or no expression of pro-Surfactant, a marker for mature AT2 cells [Fig. 4A (inset), white head arrows] (23, 24). These data lead us to interpret that even within the same tumor, there exists a subclonal population of tumor epithelial cells that loses their AT2 marker and, thus, AT2 cell identity while gaining the expression of gastric genes, suggesting that the cells may be transitioning from alveolar to alveolar-endodermal and endodermal identity. If this was the case, we next wondered whether the degree of expression of alveolar-endodermal pattern or endodermal pattern in tumors was related in any manner to the state of tumor differentiation.
To assess whether the presence of alveolar-endodermal pattern or endodermal pattern in tumors was related in any manner to the state of tumor differentiation, we assessed tumors from KrasLA1/+;Dicer1S2D/+ mice that were either well organized and thus well differentiated or poorly organized and thus poorly differentiated (or more dedifferentiated). Using histology, we defined poorly organized or dedifferentiated as tumors with features of desmoplasia, secretion of mucus, disorganized cells with variable nuclear size and shape, and invasion to bronchial structures (Fig. 4C, Panel A). Well-organized and differentiated tumors were defined as tumors exhibiting either papillary, solid, or lepidic type organization (Fig. 4C, Panel B). We then assessed these tumors for Gkn2 and pro-Surfactant expression. We observed that the well-organized tumors did not display Gkn2 but displayed pro-Surfactant, whereas tumors (sometimes from the same lung) that were poorly differentiated expressed Gkn2 (white arrow heads, Fig. 4C, Panel A) and did not express pro-Surfactant. Together, these data suggest that the presence of phosphomimetic Dicer1 in the lung tumors results in lineage reprogramming of the AT2 cell identity and causes the onset of gastric-like cell fate together likely leading to a poorly differentiated tumor in the KrasLA1/+;Dicer1S2D/+ mice.
Human grade II LUAD tumors express phospho-DICER1 and gastrokine 2
Next, we assessed whether human LUADs with oncogenic KRAS mutations and phospho-DICER1 expression also expressed GKN2 RNA. We assayed 16 tumors that had KRAS oncogenic mutations for GKN2 RNA (table S3). Of these, five have low phospho-DICER1, nine have moderate phospho-DICER1, and two have high phospho-DICER1. We observed the presence of GKN2-positive tumor cells in one poorly differentiated grade 3 stage IIB LUAD (Fig. 5) with 50% (moderate) positive signal for nuclear phospho-DICER1 (Fig. 5). We also observed tumors with low and high phospho-DICER1 positivity that did not display GKN2 expression (fig. S10). While these dataset are small, they, however, allow us to hypothesize that the presence of phospho-DICER1 associates with increased tumor plasticity and disease grade in human cancers. These data suggest that phosphorylation of DICER1 should be more carefully monitored in the future.
Fig. 5. Human LUAD grade III, stage IIB tumor with KRAS mutation, and phosphorylated nuclear DICER1 expresses GKN2 mRNA.
Representative images of a human LUAD classified as grade III and stage IIB stained with IHC for phospho-DICER1 (top left). Phospho-DICER1 localizes as brown nuclear signal; blue signal is hematoxylin. Sections from the tumor assayed with fluorescent RNA hybridization images for GKN2 mRNA in magenta and pro-SFPC mRNA in green reveal colocalization (yellow arrows) between GKN2 and pro-SFPC, suggesting that the tumor cells are expressing alveolar-endodermal state. Blue staining is DAPI. White scale bar, 50 μm; Black scale bar, 100 μm
Phosphorylated nuclear DICER1 does not regulate miRNA production to control gene expression
DICER1 is an RNase enzyme known for its canonical role in the generation of mature miRNAs for posttranscriptional regulation of genes (19). To determine whether phosphorylation of DICER1 in lung tumors affects the production of miRNAs and, consequently mRNA expression, we assayed for mature miRNAs only from the lung tumors obtained from KrasLA1/+ (n = 4) and KrasLA1/+;Dicer1S2D/+ (n = 5) mice using the NanoString nCounter mouse miRNA expression array (Materials and Methods). Overall, we observed that 97.5% of the miRNAs were unchanged in the lung tumors between the two genotypes (n = 577 miRNAs; fig. S11A). To determine whether the remaining 2.5% of miRNAs that showed either up-regulation or down-regulation in the KrasLA1/+;Dicer1S2D/+ mice (cut off FC, ≥2 or ≤−1.5; none showed ≤−2) were known oncogenic or tumor suppressor miRNAs, we focused on each individual miRNA (fig. S11B). We found that miR-434-3p, miR-148b, miR-15b, miR-322, and miR-150 miRNAs were significantly up-regulated in the KrasLA1/+;Dicer1S2D/+ when compared to the KrasLA1/+ mice (fig. S11B), while miR-148a, miR-1937, miR-30a, and miR-429 were significantly down-regulated in the KrasLA1/+;Dicer1S2D/+ animals when compared to the KrasLA1/+ mice (fig. S11B). However, from published literature, we did not identify any correlation in function or similarity of seed sequences of these deregulated miRNAs (fig. S11C). For example, in our dataset, miR-148b, miR-15b, and miR-150 are up-regulated in the KrasLA1/+;Dicer1S2D/+ tumor when compared to the KrasLA1/+ tumor, yet in literature, while miR-15b and miR-150 are thought to be up-regulated in lung cancer cell lines, miR-148b is down-regulated (25–30). Similarly, miR-148a and miR-30a and miR-429 were down-regulated in KrasLA1/+;Dicer1S2D/+ when compared to KrasLA1/+ tumors, yet in literature, miR-429 up-regulation was observed in lung cancer cell lines (31–34). Together, given the minimal change in miRNA expression between the two genotypes, lack of any seed sequence conservation (fig. S11C) and lack of correlation between these miRNAs and cancer progression, we suggest that phosphorylation of DICER1 does not significantly affect miRNA production in lung tumors.
The presence of phosphorylated nuclear DICER1 does not deregulate NKX2.1 expression in lung tumors
Before this work, studies have reported (23, 35, 36) that loss of lineage transcription factor NKX2.1 together with oncogenic activation of Kras led to expression of endodermal genes in the lung tumor cells (23, 35, 36). As we did not observe a change in miRNA profiles in the tumors, we questioned whether the presence of phosphorylated nuclear DICER1 was causing the expression of endodermal genes due to a loss of NKX2.1. We performed an IHC analysis (Materials and Methods) to assay for NKX2.1 protein in the tumor cells from both KrasLA1/+;Dicer1S2D/+ and KrasLA1/+ animals. We observed that in wild-type adult lungs, NKX2.1 expresses in alveolar cells and is nuclear in nature (fig. S12A), as expected (37). We also observed that NKX2.1 is expressed in the nucleus in the AT2 tumor cells from both KrasLA1/+;Dicer1S2D/+ and KrasLA1/+ animals (fig. S12B) at similar levels, suggesting that in the tumors with phosphomimetic DICER1, expression of endodermal genes is not due to loss of NKX2.1 expression.
Phosphorylated nuclear DICER1 regulates the chromatin state to control gene expression and tumor progression
Since the first discovery of nuclear phosphorylated DICER1 in 2014, numerous studies have explored the noncanonical functions that DICER1 may be performing in the nucleus. Some of these implicate its role in controlling chromosome segregation and DNA damage (38, 39) through regulating chromatin remodelers (40). As we did not observe a change in miRNA abundance in mouse tumors (above), we assessed the noncanonical functions of nuclear phospho-DICER1. To determine whether the presence of nuclear phospho-DICER1 correlates with altered chromatin compaction, which may inform the altered gene expression in the tumors, we performed a global ATAC-sequencing analysis on lung tumors from KrasLA1/+;Dicer1S2D/+ and KrasLA1/+ animals (Materials and Methods).
We identified 43,137 and 42,524 peaks, on average, corresponding to open chromatin globally, across multiple genes, in the lung tumors from KrasLA1/+ and KrasLA1/+;Dicer1S2D/+ mice, respectively. While the peak distribution seems similar between the two genotypes (fig. S13, A to C, and table S4), we observed that the presence of phosphorylated nuclear DICER1 in the lung tumors correlates with increased changes in chromatin compaction in the KrasLA1/+;Dicer1S2D/+ tumors relative to the KrasLA1/+ tumors at several loci throughout the genome (Fig. 6, A to F, and fig. S13D). While we observed global changes to the chromatin accessibility in the KrasLA1/+;Dicer1S2D/+ tumors, we focused on the loci for endodermal genes that we had identified above to determine whether there was a change in chromatin status at these loci. We observed that the lung tumors from the KrasLA1/+;Dicer1S2D/+ mice display peaks corresponding to open chromatin at the beginning of the Gkn2 genomic locus, which are absent from the KrasLA1/+ lung tumors and the wild-type lung (Fig. 6A). Similarly, we observed peaks corresponding to open chromatin in the lung tumors of KrasLA1/+;Dicer1S2D/+ mice at other gastrointestinal genes such as A2ml1, Hnf4a, and Ctse locus, which were also absent in the KrasLA1/+ lung tumors and wild-type lung (Fig. 6, B to D). The state of the open chromatin at loci from gastrointestinal genes in the tumors with phosphomimetic Dicer1 supports the observed increase in gene expression of the gastrointestinal genes in KrasLA1/+;Dicer1S2D/+ lung tumors (Figs. 3 and 4 and figs. S7 and S8).
Fig. 6. Phosphomimetic Dicer1 affects chromatin compaction in the lung tumors.
(A to F) Representative histogram showing ATAC-sequencing results for KrasLA1/+ (blue) and KrasLA1/+; Dicer1S2D/+ (red) lung tumors and wild-type mouse lung (black) at the Gkn2 (A), A2ml1 (B), Hnf4a (C), Ctse (D), intergenic genomic region in chromosome 5 (E), and Nm1 (F) genomic locus. ATAC-sequencing from wild-type mouse lung was retrieved from ENCODE Consortium 3 database. Green bars show the Merged Regions. Orange bars show individual Intervals for each sample as described in Materials and Methods. For each gene, exons are represented as rectangular bars and introns as lines with arrows (pointing the direction of transcription). Regulatory elements in the mouse genome that are identified by the ENCODE Registry of candidate cis-Regulatory Elements (cCREs) database are shown at the bottom of each panel. Highlighted in light green are regions where peaks were present in the KrasLA1/+;Dicer1S2D/+ lung tumors and absent in the KrasLA1/+ lung tumors. Highlighted in light yellow are regions absent in the KrasLA1/+;Dicer1S2D/+ lung tumors and present in the KrasLA1/+ lung tumors.
In addition to specific gastrointestinal loci with open chromatin, we also observed peaks at Hnf4a, Ctse, and several intergenic regions with signatures of distal enhancers (Fig. 6, B to E). These data suggest that the presence of phosphorylated nuclear DICER1 also correlates with open chromatin state in regions with regulatory elements such as enhancers, which, in turn, leads to changes in gene expression. Last, we observed peaks in the KrasLA1/+ lung tumors that were low or absent in the KrasLA1/+;Dicer1S2D/+ lung tumors, such as in Nrn1, Pax2, and Fezf1 loci (Fig. 6F and fig. S13D). The absence of peaks or low peak signal in KrasLA1/+;Dicer1S2D/+ lung tumors relative to KrasLA1/+ animals suggests a closed chromatin state in the presence of phosphomimetic or phosphorylated nuclear DICER1. Further analysis of the regions that displayed closed chromatin in the KrasLA1/+;Dicer1S2D/+ revealed that these genomic regions bear CpG islands. These data suggest that these regions may be obscured from DNA methylation in the closed conformation, further causing a change in gene expression. Together, these data demonstrate that the presence of phosphomimetic Dicer1 in the lung tumors bearing Kras oncogenic mutation likely causes an alteration in chromatin compaction in regions with regulatory elements, which, in turn, likely leads to the ectopic expression of specific gastrointestinal genes in the tumor epithelial cells, resulting in a tumor with increased lineage plasticity as a mechanism for tumor progression.
Phosphorylated nuclear DICER1 interacts with a distinct chromatin-nuclear complex in LUAD cells
We observed that phospho-DICER1 correlates with altered chromatin compaction in the mouse lung tumors; thus, we next asked whether phospho-DICER1 interacts with any nuclear proteins that directly regulate chromatin compaction and transcriptional onset. However, because the mouse lung tumors are heterogeneous in cell composition and we failed to derive cell lines from the lung tumors from mice, we used human cell LUAD cell lines to assay for nuclear DICER1 interactors. We obtained eight human LUAD cell lines and one human lung carcinoid cell line (table S5) with gain-of-function mutations in KRAS, NRAS, or EGFR or loss-of-function mutations in P53. We screened these cell lines for expression of nuclear DICER1 using immunofluorescence staining. We used an antibody against total DICER1 for this analysis because we wanted to determine the expression of DICER1 in both cytoplasmic and nuclear compartments because if any cell lines displayed no nuclear DICER1, they should display cytoplasmic DICER1.
We observed that three LUAD cell lines [H441 (KRAS G12V), H23 (KRAS G12D), and H647 (KRAS G13D)] displayed clear DICER1 enrichment in the nuclear compartment as measured by pixel intensity in Fiji (Fig. 7, A and B; Materials and Methods). H1650 (KRAS wild type), H2073 (KRAS wild type), SK-LU-1 (KRAS G12D), and H1229 (KRAS wild type) displayed heterogeneous staining for nuclear DICER1 with some cells showing DICER1 enriched in the nucleus and other cells with similar pixel intensity of DICER1 expression in cytoplasm and nucleus (Fig. 7B). Unexpectedly, we observed that H727 (KRAS G12V) carcinoid lung cancer cells, despite having an oncogenic mutation in KRAS, displays DICER1 almost exclusively in the cytoplasm. This suggested to us that in some contexts DICER1 may not be phosphorylated by KRAS signaling axis. However, given that H727 was not an adenocarcinoma but a carcinoid (which are neuroendocrine in nature) and presented DICER1 almost exclusively in the cytoplasm yet has oncogenic mutations in KRAS provided a good control for us to compare to H23, which has KRAS G12D mutations and nuclear enrichment of DICER1 to identify specific chromatin complex of phosphorylated nuclear DICER1 in LUADs.
Fig. 7. Chromatin nuclear complex of phospho-DICER1 in human LUAD cells.
(A) Representative immunofluorescence image of H23 and H727 human lung cancer cell lines stained with DICER1 (magenta) and DNA (white, DNA). H23 displays nuclear enrichment for DICER1; H727 is cytoplasmic. Scale bars, 20 μm. (B) Human LUAD cell lines stained with DICER1 and DAPI quantified for percent enrichment of DICER1 in the nucleus (black) or cytoplasm (blue) or both the nucleus and the cytoplasm (pink). Quantification was done by measuring the pixel intensity in each compartment (Materials and Methods). (C) ChIP–mass spectrometry with DICER1 from nuclear extracts of H23 and H727 identified 147 proteins for H23 of which 95 are associated with nuclear function (orange bar) and 51 with cytoplasmic function (blue bar). Fourteen proteins were identified for H727; 3, with nuclear enrichment (orange bar); and 11, with cytoplasmic enrichment (blue bar). (D) Chip–mass spectrometry with DICER1 from nuclear extract with H23 and H727 measured as percentage of proteins identified to be nuclear or cytoplasmic. (E) Table listing the DICER1-ChIP interactors with known chromatin remodeling and transcriptional functions. (F) Western blot of CBX1 following ChIP with DICER1 shows the presence of CBX1 with the DICER1 chromatin complex. (G) Immunofluorescence analysis of H23 showing colocalization between MED12L (green), DICER1 (magenta), and DNA (blue, DAPI) in H23. Scale bars, 5 μm. (H) Model: Phosphorylated nuclear DICER1 interact with a multimeric protein complex in the chromatin of lung tumor cells, resulting in transcription of genes that are otherwise not transcribed.
To determine whether phospho-DICER1 interacts with other proteins at the chromatin of human LUADs cells, we used LUAD cell lines H23 (KRAS G12D, nuclear DICER1) and the carcinoid H727 (KRAS G12V, cytoplasmic DICER1) and performed chromatin cross-link–based proteomic analysis. We cross-linked H23 and H727 cells, extracted their nuclei, and performed a DICER1 immunoprecipitation, followed by mass spectrometry analysis (Materials and Methods). We identified 147 interactors of DICER1 from the H23 cells and 14 interactors DICER1 from H727 cells by mass spectrometry (table S6). Consistent with the nuclear expression of DICER1 in H23 cells, we observed that of the 147 DICER1- interactors identified in H23 cells, 95 proteins are associated with nuclear functions and 51 proteins are associated with cytoplasmic function (Fig. 7, C and D). Conversely, from the H727 immunoprecipitation analysis, we observed that of the 14 interactors, 3 proteins were associated with nuclear functions and 11 were associated with cytoplasmic function (Fig. 7, C and D). These data demonstrate that nuclear fraction of DICER1 is enriched at the chromatin with a unique fraction of nuclear proteins.
To determine the nature of these interacting proteins, we performed a Gene Ontology (GO) enrichment analysis for molecular functions and observed that most of the DICER1 interactors function in DNA binding and/or RNA binding, including ribosomal RNA binding (fig. S14A), consistent with DICER1 role in being an RNA binding protein. In addition, we also identified proteins with known roles regulating chromatin compaction and transcription, which belonged to categories of (i) DNA helicase activity, (ii) single-stranded DNA binding, (iii) promoter-specific chromatin binding proteins, (iv) adenosine triphosphate–dependent activity on DNA or RNA, (v) transcriptional co-regulators or co-activator activity, (vi) transcription factor binding, and (vii) chromatin binding enriched. Particularly, we observed proteins such as MED12L and POLR2D, which form part of the RNA polymerase II transcription complex and are directly involved with transcription (41–45). We also observed proteins such as the histone variant MACROH2A.1 and the chromodomain protein CBX1 (also known as HP1-β) both of which have known functions in condensation and decondensation of chromatin (46–48).
To validate the chromatin immunoprecipitation (ChIP)/mass spectrometric results, we performed Western blot analysis for MED12L, CBX1, and MACROH2A.1 using H23 nuclear extracts that were chromatin–cross-linked and immunoprecipitated with DICER1. CBX1 was immunoprecipitated with DICER1 from the chromatin–cross-linked samples (Fig. 7F), demonstrating that the complex identified is a DICER1 chromatin nuclear complex. Unfortunately, however, the antibodies for MED12L and MACROH2A.1 did not work on the Western blot analysis in our hands, so we performed an immunofluorescence analysis using confocal imaging with AiryScan2.0 to assess whether MED12L and MACROH2A.1 was present with nuclear DICER1 in H23 samples. We observed that phosphorylated nuclear DICER1 [magenta in Fig. 7G and fig. S14 (C and D)] colocalizes with MED12L (green in Fig. 7G and fig. S14B) in H23 samples on the chromatin [4′,6-diamidino-2-phenylindole (DAPI); Fig. 7G and fig. S14B]. Similarly, we observed that MACROH2A.1 also colocalized with phosphorylated nuclear DICER1 on the chromatin of H23 cells (fig. S14, C and D).
Together, these data identify the first chromatin–cross-linked complex of nuclear phospho-DICER1 in LUADs and suggests that, in the nucleus, DICER1 likely forms a multimeric complex at the chromatin composed of proteins directly involved in regulating chromatin compaction and transcription and mediates multiple functions such as promoting transcription and regulating chromatin condensation (Fig. 7H). Because phosphorylated nuclear DICER1 is not present in wild-type adult lung alveolar cells, these data lead to the model that aberrant phosphorylation and expression of DICER1 in the nucleus results in aberrant expression of oncogenic genes that affect tumor cell plasticity and regulate tumor progression (Fig. 8).
Fig. 8. Phosphorylated nuclear DICER1 regulates late-stage tumor progression through reprogramming the chromatin and leading to increased cell plasticity.
In normal differentiated adult alveolar cells, DICER1 is not phosphorylated and does not localize to the nucleus. In transformed alveolar cells with KRAS oncogenic signal, DICER1 is phosphorylated and localized to the nucleus, where it binds to a chromatin remodeling complex consisting of the Mediator complex and chromatin remodelers, which result in unwinding of the chromatin and onset of aberrant gene transcription, resulting in relieving of lineage restriction and causing increased tumor cell plasticity.
DISCUSSION
Our work reports on the role of DICER1 phosphorylation in mediating tumor progression and spread, in a manner that is distinct from DICER1’s function as a haploinsufficient tumor suppressor and describes the unique chromatin complex that phosphorylated nuclear DICER1 likely regulates to mediate this function. Below, we discuss the role of phosphorylated nuclear DICER1 in regulating cellular reprogramming and lineage plasticity as a mechanism for cancer progression, how phosphorylated nuclear DICER1 functions independently of miRNA production, and the potential use of phospho-DICER1 antibody as a prognostic marker for detection of malignant human cancers.
The presence of phosphorylated nuclear DICER1 causes reprograming of tumor cell identity and tumor progression and spread
Canonically, DICER1 functions in the cell cytoplasm to process small noncoding RNAs (the miRNAs and small interfering RNAs) and regulate gene expression posttranscriptionally (19). Loss of one copy of DICER1, in this context, was shown to cooperate with the Kras oncogenic mutation G12D and lead to tumor onset and initiation (17, 18). The mechanism through which DICER1 functions to control tumor onset was proposed to be through the regulation of miRNA production (16). As a haploinsufficient tumor suppressor, DICER1 has also been shown to mediate metastasis through the regulation of miRNAs (49). In these situations, the aberrantly generated miRNAs regulate EMT to mediate tumor spread. Here, we uncovered a unique role for phospho-DICER1 in regulating only late-stage tumor progression in a manner independent of EMT. We find that phosphorylated nuclear DICER1 causes reprogramming of the lung tumor epithelial alveolar cells to an expanded endodermal state, leading to the expression of gastric genes. We propose a model wherein oncogenic mutations in Kras boosts alveolar proliferation and initiates the tumor and then phosphorylation of DICER1 through the KRAS-ERK signaling axis results in tumor progression and spread by altering the identity of AT2 cells (Fig. 8). We show that phosphorylated nuclear DICER1 causes the AT2 cells to assume either an “intermediate stage,” where the tumor cells express signatures for both alveolar and gastrointestinal genes, or an “advanced stage,” where the tumor cells lose the mature alveolar markers and only express the endodermal markers (Figs. 3 and 4). We did not observe any expression of phospho-DICER1 in mature adult alveolar cells from normal lungs, unlike the lung tumors from both KrasLA1/+;Dicer1S2D/+ and KrasLA1/+ mice, which express phospho-DICER1 (Fig. 2A). These data suggest that the aberrant expression of phosphorylated nuclear DICER1 in AT2 tumor cells likely results in de-repressing the differentiation program, which is laid down during embryonic lung development. During embryonic development lungs originate from the embryonic foregut during endoderm formation through lineage restriction; the embryonic foregut also generates esophagus, stomach, liver, and the pancreas (24, 50). Thus, it is likely that constitutive phosphorylation of DICER1 relieves the restriction of lung alveolar state, leading to dedifferentiation of the AT2 tumor cells to foregut progenitor state resulting in aberrant expression of endodermal genes. In addition, both Dicer1 and Kras are essential for normal lung development in mice (51). Loss of Dicer1 results in defective branching morphogenesis and alveolar differentiation (51), and KRAS promotes lung branching (52–55). Thus together, these observations suggest that KRAS-ERK-DICER1 signaling axis may play a central role during normal lung development and it is likely that this role is dysregulated during tumorigenesis.
Notably, Nkx2.1, a transcription factor important for alveolar differentiation when deleted from an oncogenic Kras LUAD in a genetically modified mouse model also results in ectopic expression of gastrointestinal genes (including gastrokine-1) and down-regulation of alveolar markers such as surfactant (pro-Sfpc) (23, 35, 36, 58). Similarly, coupling the loss of Nkx2.1 with overexpression of transcription factor Sox2 in murine models promoted the formation of squamous lung cancer with features of esophageal differentiation (56). Thus, suggesting that a change in cell fate is a common mechanism in a subset of lung cancer (57–59). However, loss of Nkx2.1 in a Kras wild-type lung failed to express gastrokine-1, suggesting that KRAS may activate specific signaling pathway that augment changes to the differentiation state, in a mechanism independent of, or redundant with, Nkx2.1 function. We observed that Nkx2.1 is nuclear in expression in the lung tumors from both Kras oncogenic mutants and tumors with phosphomimetic DICER1 (fig. S12), leading us to propose that phosphorylation of DICER1 may function in parallel with programs such as Nkx2.1 to regulate alveolar cell identity and result in lung tumors with enhanced plasticity as a mechanism for late-stage tumor progression and invasion.
The presence of phosphorylated nuclear DICER1 causes altered chromatin compaction and increased lineage plasticity of tumor cells
Unlike its canonical function in the cytoplasm, wherein DICER1 regulates miRNA production, to drive tumor initiation and metastasis in cancers (16, 49), we show that phosphomimetic DICER1 drives tumor progression in a mechanism independent of miRNA function (fig. S11). Instead, we find that the presence of phosphomimetic DICER1 in the lung tumors with Kras oncogenic mutations correlates with global chromatin changes (Fig. 6 and fig. S13). These changes lead to opening and altering of chromatin compaction in areas overlapping with regulatory elements in the genome. In support of this, we identified regions around gastrokine-2 that are “open” in the presence of phosphomimetic DICER1 relative to wild-type lungs (Fig. 6), suggesting that an aberrant gene expression program is driven in this instance by changes to the chromatin landscape.
To determine the mechanism through which phosphorylated nuclear DICER1 may regulate the chromatin landscape, we used a combination of human LUAD cell lines and ChIP/mass spectrometric method to identify a unique chromatin-DICER1 complex with previously unidentified protein interactors that affect chromatin remodeling. Our data lead to the model that once phosphorylated, DICER1 translocates to the nucleus where it can form a multimeric complex with proteins such as Mediator complex 12, CBX1, and transcription factors which can directly affect chromatin compaction and transcription (Fig. 7). We also identified nuclear RNA binding proteins in the complex, suggesting that the complex is likely RNA dependent, consistent with DICER1’s role in RNA binding. These data lead us to propose that in the nucleus, phospho-DICER1 together with the chromatin complex leads to the expression of gastrointestinal genes in alveolar tumor cells and changes to cellular identity and enhanced tumor plasticity (Fig. 8).
In support of these observations, DICER1 has been shown to function with the bromodomain transcription co-activator BRD4 to modulate genome stability and transcription in mouse embryonic stem (ES) cells (40). Gutbrod et al. (40) show that loss of Dicer1 in mouse ES cells leads to defects in proliferation and alterations in transcription of centromeric satellite repeats, defects that can be rescued by loss of Brd4 or inhibiting RNA polymerase II. In yeast, DICER1 have been shown to act on RNA substrates to release RNA polymerase II from sites where the transcription and replication machinery may collate (40, 60). Furthermore, Chitale et al. (38) demonstrated that DICER1 can interact with a chromatin remodeling protein Zuotin-Related Factor 1 to modulate chromatin decondensation during DNA damage response. Collectively, our work adds to these studies, which support a role for DICER1 in chromatin reprograming.
Together, because DICER1’s function as a haploinsufficient tumor suppressor occurs through loss of miRNA production (17) while phosphorylation of DICER1 promotes late-stage of tumor progression in a mechanism that is independent of the miRNAs, through noncanonical functions, such as chromatin compaction and transcription through the unique chromatin complex. These data provide new directions to the field and the identity of the molecular partners that phospho-DICER1 binds to will provide avenues for detailed mechanistic studies, especially in the context of LUADs.
Phosphorylated nuclear DICER1 as a prognostic marker for aggressive LUADs
Cancer development can be divided into two main phases, an initiating phase driven by mutations or genomic alterations that start the tumor formation and a progression phase driven and sustained by further genomic, metabolic, or epigenetic changes that drive metastasis (61, 62). Twenty to 30% of LUADs are initiated because of alveolar cells acquiring somatic oncogenic mutations in KRAS (2–5). As the tumor progresses, cells accumulate additional epigenetic and molecular alterations that modulate cellular behavior and, in some cases, cellular identity. While early detection of mutations, which lead to tumor onset are important to screen for in cancer prevention, not all mutations lead to cancer progression and often appear seemingly dormant for years before causing obvious drastic effects. Thus, it is critical to identify the genetic or epigenetic changes that lead to tumor progression and metastasis early enough such that patients can be stratified for effective therapeutic options. Here, we show that phosphorylated nuclear DICER1 correlates with aggressive human LUADs bearing KRAS oncogenic mutations. Experimentally, in the mouse models, we demonstrate that phosphorylated nuclear DICER1 drives late-stage tumor progression by affecting alveolar identity, a phenomenon we then observe in human LUADs as well. We show that phospho-DICER1 is more frequently detected by IHC than active ERK in human LUADs, and in some tumors, phospho-ERK was absent yet phospho-DICER1 was present, and the tumors were aggressive and invasive in nature. Collectively, these data suggest that phosphorylated nuclear DICER1 may represent a potential biomarker for early detection of metastatic cancers and opens the field to develop ways to block DICER1 phosphorylation and nuclear translocation as a means for therapy.
MATERIALS AND METHODS
Nomenclatures used in the study
The following nomenclatures were used: KRAS for human gene, Kras for mouse gene, and KRAS for mouse and human protein; Dicer1 for mouse gene and DICER1 for mouse and human protein; GKN2 for human gene, Gkn2 for mouse gene, and GKN2 for mouse and human protein; and Nkx2.1 for mouse gene and NKX2.1 for mouse protein.
Mice (Mus musculus) breeding, maintenance, screening, and genotyping
The following genotypes of mice were used in the study: Dicer1S2D/+ (13), KrasLA1/+ (9), and KrasLA1/+;Dicer1S2D/+ (13). Mice were maintained in >94% C57BL/J6 background. All mouse studies were conducted in compliance with the institutional animal care and use committee protocol (IACUC). Dicer1S2D/+ animals were crossed to KrasLA1/+ animals to generate a cohort of Dicer1S2D/+, KrasLA1/+, and KrasLA1/+;Dicer1S2D/+ animals. Pups were weaned at 3 weeks of age, ear tagged, and tail snipped for genotyping. Tails were digested in lysis buffer [1 M tris (pH 8.0), 5 M NaCl, 0.5 M EDTA (pH 8.0), and 10% SDS] with proteinase K (20 mg/ml) at 55°C overnight. The mice were genotyped each time through tail DNA purification and polymerase chain reaction (PCR)–based Sanger sequencing. PCR primers used to perform the genotype analysis are presented in the Supplementary Materials. Tumor watch, histopathology, and other tumor analyses were performed at indicated time points. Moribund animals were euthanized, and their tissues collected for pathology. Animals that were euthanized because of non–tumor-related issues (fighting wounds, dermatitis, and others) were not included in the analyses. The number of mice analyzed is stated in each figure. Unless specified, mice of both genders were used. No power analysis was used to determine the sample size.
Human LUAD tumor tissue microarrays
Tumor microarrays (TMAs) with de-identified patient information were obtained from Discovery Life Sciences (Powell, OH) and TriStar Technology Group LLC (Washington, DC). Eighty-eight LUAD samples from untreated patients were used. No patient or human subjects were recruited for this study. Samples were excluded in any of these instances: (i) Samples were damaged during sectioning and embedding. (ii) The core was missing more than 50% of the tissue. (iii) Key tumor histopathology information was missing, and (iv) the KRAS mutation profile was missing. Investigators were blinded for mutation profile and patient clinicopathology information during marker quantification and classification. Patient clinicopathology features are presented in table S1 and Table 1.
IHC of human tumors and mouse tissue
TMAs with 88 LUAD samples from untreated patients were used. IHC was performed using a monoclonal anti–phospho-DICER1 (1:200) (14, 15) and rabbit anti–phospho-ERK (1:100; Thr202/Tyr204; Cell Signaling Technology, #9101S), diluted in 30% normal goat serum (NGS; Thermo Fisher Scientific, 16210-064). Anti–phospho-DICER1 (1852) antibody were generated in house and tested for specificity as previously described (15). The slides were baked for 1 hour at 60°C, deparaffinized by immersing in Histo-Clear solution (National Diagnostics, HS-200) for 5 min twice, followed by a descending gradient of alcohol washes. Antigen retrieval was performed using 1× sodium citrate buffer (citric acid: BDH, catalog no. 10081; NaOH: BDH, catalog no. 30167) by HIER in 2100 Retriever machine (EM Sciences, #62700-100). Sections were blocked in 30% NGS for 1 hour at room temperature. After primary antibody incubation, sections were incubated in 1% hydrogen peroxide (H2O2) in methanol for endogenous peroxidase blocking and then in MACH2 Universal polymer-HRP (Biocare Medical LLC, #M2U522H). Signal was developed with 3,3′-diaminobenzidine (DAB) (SIGMAFAST 3,3′-Diaminobenzidine tablets; Sigma-Aldrich, D4293-50SET). Images were taken using Nikon Eclipse Ti2 equipped with Nikon DS-Ri2 color camera at MD Anderson Cancer Center, Department of Genetics Microscope core.
Quantification of antibody staining was performed using Fiji software and following Andy’s algorithm on the entire tumor section (63). Fiji program was used to select the nucleus of each cell and perform total cell count. Color deconvolution was then applied to each image to specifically identify cells with DAB brown pigmentation. The percentage of positively stained cells was calculated as follows: sum of all the DAB positive cells divided by the total number of nuclei per sample. LUAD samples that showed ≥1% but <30% of cells with phospho-DICER1 or phospho-ERK were classified as “low positive”; tumors with ≥30% but <70% of cells were classified as “moderate positive”; tumors with ≥70% of cells were classified as “high positive,” and <1% of cells with phospho-DICER1 or phospho-ERK as negative. Following quantification, KRAS mutation profile was unblinded and then correlated to tumor positivity for each marker.
IHC staining for phospho-DICER1 and NKX2.1 in mouse lung tissue was performed as following the same protocol as above. Five-micrometer sections of wild-type lung tissue (n = 2), KrasLA1/+ (n = 3), and KrasLA1/+;Dicer1S2D/+ (n = 3) mice were used. For NKX2.1 staining, anti-NKX2.1 (1:500; Santa Cruz Biotechnology, cs-13040) in 30% NGS was used. Images were captured on an EVOS M7000 (Invitrogen, AMF7000) microscope at 40× magnification.
Immunofluorescence staining on mouse tissues
Tumor tissues and indicated organs analyzed were harvested and fixed in 10% neutral buffered formalin. Right heart ventricle was injected with phosphate-buffered saline (PBS; pH 7.4) to perfuse the tissues. Lungs were inflated using formalin through a cannulated trachea. Fixed tissues were paraffin-embedded by the MD Anderson Department of Veterinary Medicine and Surgery Histology Laboratory. Five-micrometer sections were used for hematoxylin and eosin (H&E) staining for each of the histological analysis presented. Unstained sections were used for immunofluorescence staining. The slides were baked for 1 hour at 60°C, deparaffinized by immersing in Histo-Clear solution (National Diagnostics, HS-200) for 5 min twice, followed by a descending gradient of alcohol washes. Antigen retrieved by HIER as described above. Sections were blocked in 30% NGS plus goat anti-mouse immunoglobulin G(IgG; AffiniPure Fab Fragment Goat anti-Mouse IgG, Jackson ImmunoResearch, #115-007-003) for 1 hour at room temperature. The following primary antibodies were diluted in 30% NGS and incubated overnight at 4°C: mouse anti–E-cadherin (1:100; BD, #610182), rabbit anti–pro-SFPC (1:250; Millipore, AB3789), guinea pig anti-vimentin (1:50; Progen, #GP59), and rabbit anti-GKN2 (1:1000; Abcam, #ab188866). For GKN2 antibody, tris-EDTA [10 mM tris base (Sigma-Aldrich, T1503), 1 mM EDTA (Sigma-Aldrich, E5134), 0.05% Tween 20 (Fisher, BP337; pH 9.0)] was used for antigen retrieving. Samples were then washed in PBS with 1% Tween 20 (PBST). The secondary antibodies were used at 1:500 dilution in PBST: goat anti-mouse Alexa Fluor 488 (Invitrogen, #a11001), Donkey anti-rabbit Alexa Fluor 594 (Invitrogen, #a21207) and Goat anti-guinea pig Cy5 (Abcam, #ab102372). When using anti-IgG Alexa Fluor 488 antibody, lung tissue autofluorescence was quenched with Vector TrueVIEW (Vector Laboratories, SP-8400) following the manufacturer’s instructions. A 1:1000 dilution of DAPI (2 μg/ml) in 1× PBST was used for nuclear costaining, and slides were mounted in VECTASHIELD (Vector Laboratories, #H-100). Images were acquired using Zeiss Axio Imager M2 equipped with AxioCam MRm camera at 40× magnification (Zeiss, Thornwood, NY). Images for GKN2 staining were captured on an EVOS M7000 (Invitrogen, AMF7000) microscope at 40× magnification.
Cell dissociation for scRNA-seq
Lung tumor cell dissociation was performed following previously published protocol (64). Briefly, KrasLA1/+ (n = 2), KrasLA1/+:Dicer1S2D/+ (n = 2), and Dicer1S2D/+ (n = 1) mice of 39 weeks of age were anesthetized using Avertin (Sigma-Aldrich, T4802); right heart ventricle was perfused with 1× PBS without calcium and magnesium (Thermo Fisher Scientific, #100100023). Lungs were removed from each mouse and placed in Leibovitz media (Gibco, #21083-023). Lung tumors were identified and collected into a 1.7-ml tube under the dissecting microscope. A total of five tubes were collected, one tube from each mouse described above. Then, tumors or tissues were minced with forceps and digested in the Leibovitz media with collagenase type I (2 mg/ml; Worthington CLS-1, LS004197), deoxyribonuclease I (0.5 mg/ml; Worthington D, LS002007), and elastase (2 mg/ml; Worthington ESL, LS002294) for 30 min at 37°C. Digestion was stopped with 20% fetal bovine serum (FBS; Invitrogen 10082-139). Cells were filtered using a 70-μm cell strainer (Falcon, 352350) on ice and transferred to a 2-ml tube. Red blood cells were lysed using red blood cells lysis buffer (Miltenyi Biotec, 130-094-183), and the lysate was centrifuged for 1 min at 5000 rpm. Supernatant was removed, and cells in the pellet were washed twice with 1 ml of ice-cold 1× Leibovitz plus 10% FBS. The cellular suspension was then filtered with a 40-μm cell strainer (Falcon, #352340) and centrifuged, and cells were resuspended in ice-cold 1× Leibovitz plus 10% FBS. Cell viability was evaluated using Trypan blue and counted with a hemocytometer. Samples with more than 70% of viable cells were submitted for scRNA-seq at MD Anderson Cancer Center CPRIT Single Cell Genomics Core.
scRNA-seq and bioinformatic analysis
After cell dissociation and counting (as described above), samples were centrifuged and resuspended in 1× PBS with 0.04% BSA for single-cell suspensions and processed for the Chromium Single Cell Gene Expression Solution Platform (10x Chromium) with 3’v3 Library following the manufacturer’s instructions. Ten thousand cells from each sample cell suspension were sequence with Illumina NovaSeq6000 S1 with a 2 × 150 paired-end configuration and 400 million and 500 million reads per sample. A total of five lanes were used, each corresponding to a mouse.
FASTQC files were returned to the MD Anderson Cancer Center CPRIT Single Cell Genomics Core for demultiplexing, alignment to the mouse genome mm10-3.0.0, and unique molecular identifier (UMI) collapsing using the Cell Ranger v3.1.0. The cellranger aggr pipeline was used to combine the counts to produce one single feature-barcode matrix for further analysis. UMI counts were further analyzed in R v4.1.0 using Seurat v4.0.5 package. Low-quality cells were filtered out if they (i) had lower than 300 UMI, (ii) the transcript number was higher than 5000, and (iii) mitochondrial transcript fractions were lower than 15%. UMI counts were normalized and scaled using NormalizeData, ScaleData, and FindVariableFeatures functions. For each of these functions, the default settings in Seurat v4.0.5 were used which included “LogNormalize” method for NormalizeData function, and the “vst” method for FindVariableFeatures functions. Principle components analysis was performed, and the number of dimensions was estimated using the JackStraw and ElbowPlot function. Cell clusters were identified using a shared nearest neighbor–based algorithm using the FindNeighbors and FindClusters functions. Uniform manifold approximation and projection (UMAP) rendering was performed to visualize clusters. Major cell classes were identified on the basis of enrichment for Ptprc (immune cells); Col1a1, Col3a1, or Cdh5 (mesenchymal cells, including endothelial cells); and Epcam, Cdh1, Sfta2, Lamp3, and Rtkn2 (epithelial cells). In addition, other markers were used including gene signatures for T cells (Cd3e, Cd3d, and Cd2), B cells (Ms4a1, Cd79a, and Cd19), macrophages (Cd14, Cd68, and Cd163), NK cells (Nkg7 and Ctsw), monocyte cells (Aif1), dendritic (Samhd1 and Axl), fibroblast (Fbln1 and Lum), and endothelial cells (Vwf and Lyve1; figs. S4 to S6). Furthermore, automatic annotation with scCATCH v 2.1 package was performed to validate identified cell type. Cells that belong to a major class were subset using the subset function. Each subset was reclustered, and specific cell types were identified on the basis of gene expression using the FindAllMarkers functions. The significance of the difference in expressed genes between groups of cells was determined using Wilcoxon rank sum test. Only genes expressed in at least 25% of the cells per cluster and with a threshold of a log fold change of 0.25 were evaluated. Clusters were visualized using UMAP, as described above. Then, for the purpose of this study, we focused in further analyzing the epithelial population. The epithelial cells were identified and annotated on the basis of the gene expression of Sfta2, Lamp3, Scl34a2, Rtkn2, Pdpn, Hopx, Ager, Foxj1, Scgb1a1, and Muc5ac (figs. S6 to S7). Furthermore, FindAllMarkers functions was used to identified differentially expressed genes for each of the identity classes in the epithelial population following the same parameters as before (figs. S6 to S7). Heatmaps, volcano plots, and feature plots were generated using the DoHeatMap, EnhancedVolcano v1.10.0, and FeaturePlot functions and packages. KrasLA1/+ (K, n = 2), KrasLA1/+;Dicer1S2D/+ (KD, n = 2), and Dicer1S2D/+ (D, n = 1) mice were processed in parallel experimentally and computationally; hence, they are spatially comparable in UMAP plots.
Hairpin chain reaction–RNA–fluorescent in situ hybridization
HCR-RNA-FISH was performed following Molecular Instruments FFPE tissue section protocol with minor modifications (65). Twenty RNA probes were used per gene that was analyzed. Slides were baked for 1 hour at 60°C and deparaffinized by immersing in Histo-Clear solution (National Diagnostics, HS-200) twice for 5 min each, followed by a descending gradient of alcohol washes. Slides were incubated with 1× tris-EDTA buffer (pH 9.0) in the 2100 retriever machine for 30 min. Then, samples were cooled to 45°C by adding water every 5 min. This step was completed in a total of 20 min. Slides were washed in water, followed by 1× PBS rinse. Proteinase K step was eliminated from the recommended protocol to maintain higher tissue morphology and integrity. The signal was developed by incubating the samples in 200 μl of HCR hybridization buffer inside the humidified chamber at 37°C for 10 min, followed by 0.4 pmol of mouse Gkn2-B3 probe and 0.4 pmol of mouse pro-Sfpc B1 probe in 100 μl of HCR hybridization buffer at 37°C overnight. For human tumors, 0.4 pmol of human GKN2-B3 probe and 0.4 pmol of human SFPC-B1 probe were used. Slides were washed in a gradient of HCR probe wash buffer plus 5× SSCT solution (Sigma-Aldrich, S6639). For amplification, slides were incubated with 200 μl of HCR amplification buffer for 30 min at room temperature. HCR amplification buffer (100 μl) with 6 pmol of snap-cooled hairpin B3-h1-594, hairpin B3-h2-594, hairpin B1-h1-647, and hairpin B1-h2-647 each, and DAPI (2 μg/ml) was then added on top of the tissue and incubated overnight in a dark humidified chamber at room temperature. Last, excess hairpins were removed with multiple 5× SSCT washes at room temperature. Slides were mounted with VECTASHIELD (Vector Laboratories, #H-100) and imaged in EVOS M700 as described in immunostaining quantification section below.
Quantification of HCR-RNA-FISH
Immunofluorescence images for pro-Sfpc and Gkn2 RNA were captured using the EVOS M7000 (Invitrogen, AMF700). First, the whole lung sections were outlined and mapped with the 2× and 10× objective; then, the tumors were identified and marked for further analysis using EVOS microscope software. Images of the tumors were acquired upon zooming in on the tissue with the 40× objective. KrasLA1/+ (K, n = 3), KrasLA1/+;Dicer1S2D/+ (KD, n = 3), and Dicer1S2D/+ (D, n = 2) and wild type (n = 1) mice were analyzed simultaneously. For each lung section, tumors with cells expressing Gkn2 mRNA were counted as positive. To calculate the percentage of positive tumors per mouse, Gkn2-positive tumors were added and then divided by the total number of tumors identified in each lung section. The same quantification was performed for all lung sections. In addition, 16 human KRAS mutants LUADs were analyzed. For this, a block containing human LUAD TMA was sectioned and stained as described following the HCR-RNA-FISH Molecular Instruments FFPE tissue sections protocol (as described above). Then, GKN2 mRNA immunostaining results were compared to phospho-DICER1 positivity (as described as part of the LUADs screening).
ATAC-sequencing analysis
ATAC-sequencing and bioinformatic analysis were performed by Active Motif Inc. (Carlsbad, CA, USA). A total of 40 to 50 mg of lung tumors were dissected from euthanized mice, snap-frozen in liquid nitrogen, and submitted to Active Motif for ATAC-sequencing. At Active Motif, samples were processed and put through paired-end 42–base pair (bp) sequencing reads format using Illumina NovaSeq 6000 platform. Reads were mapped to mouse mm10 reference genome using BWA (v0.7.12) algorithm and saved as BAM files. Reads that passed Illumina’s purity filter, aligned with no more than two mismatches, and mapped uniquely to the genome were used for further analysis. In addition, duplicate reads (“PCR duplicates”) were removed. For comparative analysis, normalization was achieved by reducing the number of alignments per sample to match the number of alignments in the sample with a fewer number of alignments. Reduction was done through random sampling. Genomic regions with a high level of transposition/tagging events were determined using the MACS3 (v3.0.0) peak calling algorithm (66), with a cutoff of P = 1 × 10−7. To identify the density of transposition events along the genome (signal used to generate histograms), the genome was divided into 32-bp bins, and the number of fragments in each bin was determined. The histograms showing the density of each signal (peaks) was stored in a bigWig file and visualized using the USCC Genome browser. BigWig files were generated using deepTools (v3.5.1). A FRIP (fraction of reads in peaks) value of 10% or higher was used to define good data quality, and false peaks were defined within the ENCODE blacklist (67) and removed from analysis. Genomic regions identified by MACS3 in each individual sample were marked by an Interval bar (orange bars below each sample histogram). Last, because the overlapping Intervals (orange bars) do not have the same length, to be able to compare genomic regions between samples, intervals from all the samples were collected and grouped into “Merged Regions” (green bars on top of histograms). The Merged Regions defined the start coordinate of the most upstream region and the end coordinate of the most downstream region. After defining the Intervals and Merged Regions, their peak fragment density and their genomic locations along with their proximities to genes or other genomic features were presented in Excel spreadsheets (Supplementary Materials).
Immunofluorescence analysis of human LUADs
We assessed the presence of nuclear DICER1 in the following lung cancer cell lines: A459, H441, H23, H1299, H647, SK-LU-1, H1650, H2073, and H727 using immunofluorescence. The cells were cultured in RPMI 1640 with 10% FBS at 37°C with 5% CO2. Each cell line was split and seeded onto a four-chamber slide with 500 ml of cultured media at a density of 1 × 104 cells. After 32 to 36 hours, the culture medium was removed, and the cells were rinsed three times with 1× PBS before being fixed with 400 μl of 3% paraformaldehyde in phosphate buffer (pH 7.4) for 15 min at 37°C. After fixation, the cells were permeabilized with 400 μl of 0.1% Triton X-100 in 1× PBS for 5 min at room temperature, then washed three times with 1× PBS with 0.1% Tween 20 (PBS-T), and blocked with 500 μl of 30% NGS in PBS-T for 60 min at room temperature. The cells were then incubated with Monoclonal Mouse anti-Human DICER1 (LSBio, catalog no. LS-C338518) diluted (1:100) in 30% NGS overnight at 4°C. In addition, the following other antibodies were used: rabbit anti-MED12L (1:100; Cell Signaling Technology, catalog no. 4529) and rabbit anti-MacroH2A.1 (1:250; Cell Signaling Technology, catalog no. 8551). After washing the cells in PBS-T three times, they were incubated in secondary Donkey anti-rabbit Alexa Fluor 594 (Invitrogen, #a21207) antibodies at 1:500 dilution and DAPI (2 μg/ml; 1:1000) in 30% NGS. Last, the slides were mounted in VECTASHIELD (Vector Laboratories, #H-100) and imaged using a Zeiss Axio confocal with AiryScan2.0 microscope with a 63× magnification.
To quantify the immunofluorescence staining, we used Fiji software. First, we identified the nucleus of each cell and counted them. Then, we measured the pixel intensity in the nuclear compartment and compared it to the cytoplasm pixel intensity for each cell, per optical field. For quantification, we divided the nuclear intensity by the cytoplasmic intensity. Cells with nuclear intensity ≥1.5 were considered as having nuclear enrichment for DICER1, while cells with ≤0.5 were considered as having DICER1 enriched in the cytoplasm.
Chromatin cross-linking and immunoprecipitation for mass spectrometry analysis
ChIP–mass spectrometry analysis was performed on H23 and H727 cells. The cells were grown until they reached 70% confluence and then rinsed with 1× PBS and fixed with 1% formaldehyde (pH 7.4; Sigma-Aldrich, catalog no. F8775) for 10 min at 37°C. After fixation, 0.5 M glycine was added to stop the fixation; then, cells were washed with 1× PBS and trypsinized with 0.25% trypsin-EDTA for 5 min. Cells were lysed in detergent-free lysis buffer with protease inhibitor [5 mM PIPES (pH 8.0) and 85 mM KCl], scraped from the flask, and centrifuged. The resulting pellet was resuspended and incubated in lysis buffer with detergent [5 mM Pipes (pH 8.0), 85 mM KCl, 0.5% NP-40, and 0.5% Triton X-100] and protease inhibitors for 35 min at 4°C. The nuclei were pelleted by passing the lysate through a 25-gauge needle and centrifugation. The pellet was then lysed in nuclear lysis buffer [50 mM tris-HCl (pH 8.0), 0.5% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS] and sonicated at 40 A and five pulses with 10 s on and 30 s off. The nuclei lysate was then centrifuged, and the supernatant was used for ChIP.
To perform ChIP, protein A beads were prewashed in PBS followed by ChIP blocking buffer [0.01% SDS, 1% Triton X-100, 1.2 mM EDTA, 16.7 mM tris-HCl (pH 8.0), 16.7 mM NaCl, and BSA (1 mg/ml)] diluted in BSA and protease inhibitor. Prewashed protein A beads were then incubated with 1 mg of nuclei lysate for 1 hour at 4°C, followed by 10 μg of DICER1 antibody (NOVUS) at 4°C overnight. The next day, the beads were washed once with low-salt immune complex wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 tris-HCl (pH 8.0), and 150 mM NaCl], then three times with high-salt immune complex wash buffer [0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 tris-HCl (pH 8.0), and 500 mM NaCl], once with lithium chloride immune complex wash buffer [LiCl; 0.25 M LiCl, 1% IGEPAL-CA630, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM tris-Cl (pH 8.1)], and twice with TE buffer [10 mM tris-Cl (pH 8.1) and 1 mM EDTA]. Samples were then sent for mass spectrometry analysis at Harvard Medical School Taplin Mass Spectrometry Facility. To validate DICER1 protein interactors, Western blot and coimmunoprecipitation assay were performed using the anti-CBX1 (Anti-Heterochromatin Protein-1 β) antibody (Sigma-Aldrich, MAB3448).
GO enrichment analysis
Unique protein interactors identified in mass spectrometry analysis of H727 and H23 were analyzed for GO enrichment in http://geneontology.org. Graphs were generated following ShinyGO 0.77 (68) software (http://bioinformatics.sdstate.edu/go/).
Quantification and statistics analysis
Image quantifications are described here for individual experiments. Student’s t test, chi-square tests, and any other statistical analysis of data were done using GraphPad Prism v.9.2.0 software, unless otherwise noted. P values less than 0.05 were considered statistically significant. Scatter or bar plot graphed in GraphPad Prism displayed the SD from the mean value. At least three mice per genotype were used in each experiment, including males and females, unless otherwise noted. Criteria for exclusion are described in experimental model and subject details. The “n” values and statistical test for each experiment are denoted in the corresponding figure panels or graphs. Models were generated using Adobe Illustrator.
Acknowledgments
We thank G. Lozano for providing the mice, expertise in analysis, and critical comments on the manuscript. We thank N. Navin and J. Chen for help with scRNA-seq protocols and analysis. We thank E. Whitley for performing mouse pathology examination and diagnosis. We thank R. Behringer for help during the COVID-19 pandemic laboratory shutdown. We thank N. Copeland, N. Jenkins, and A. Kalia for critical comments on the manuscript. We thank members of the Arur Lab for critical discussions during this study. We thank the University of Texas MD Anderson Cancer Center, MD Anderson Cancer Center Department of Genetics Microscope Core, MD Anderson Cancer Center Advanced Technology Genomic Core (ATGC) (supported by NCI grant CA016672), and MD Anderson Cancer Center CPRIT Single Cell Genomics Core (supported by grant RP180684).
Funding: This work was supported by NCI R37 CA214609 (to D.L.G.) and NCI grant CA016672 (ATGC), and RP180684 CPRIT Grant (Single Cell Core). S.A. is an Andrew Sabin Family Foundation Fellow at University of Texas MD Anderson Cancer Center.
Author contributions: R.A.R.-C. and S.A designed the research, analyzed the data, and wrote the manuscript. R.A.R.-C., S.-Y.C., and J.S. performed key research experiments and contributed figures. S.T.K. and D.L.G. provided key experimental materials and read and edited the manuscripts. All authors read and approved the paper.
Competing interests: S.A. is the inventor of the phospho-DICER1 antibody used in this study; a patent application filed by University of Texas systems is pending. The authors declare that they have no other competing interests.
Data and materials availability: Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, S.A. (sarur@mdanderson.org). This study did not generate new unique reagents. scRNA-seq, NanoString array, and ATAC-sequencing data are deposited at Gene Expression Omnibus (https://ncbi.nlm.nih.gov/geo/) with accession number GSE228968 and are available as of the date of publication. Original code for scRNA-seq is the Supplementary Materials. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Methods
Figs. S1 to S14
Tables S1, S3, and S5
Legends for tables S2, S4, and S6
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Tables S2, S4, and S6
Data S1
REFERENCES AND NOTES
- 1.H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, F. Bray, Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.I. A. Prior, P. D. Lewis, C. Mattos, A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.B. El Osta, M. Behera, S. Kim, L. D. Berry, G. Sica, R. N. Pillai, T. K. Owonikoko, M. G. Kris, B. E. Johnson, D. J. Kwiatkowski, L. M. Sholl, D. L. Aisner, P. A. Bunn, F. R. Khuri, S. S. Ramalingam, Characteristics and outcomes of patients with metastatic KRAS-mutant lung adenocarcinomas: The Lung Cancer Mutation Consortium experience. J. Thorac. Oncol. 14, 876–889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.S. Dogan, R. Shen, D. C. Ang, M. L. Johnson, S. P. D'Angelo, P. K. Paik, E. B. Brzostowski, G. J. Riely, M. G. Kris, M. F. Zakowski, M. Ladanyi, Molecular epidemiology of EGFR and KRAS mutations in 3,026 lung adenocarcinomas: Higher susceptibility of women to smoking-related KRAS-mutant cancers. Clin. Cancer Res. 18, 6169–6177 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.I. Ferrer, J. Zugazagoitia, S. Herbertz, W. John, L. Paz-Ares, G. Schmid-Bindert, KRAS-mutant non-small cell lung cancer: From biology to therapy. Lung Cancer 124, 53–64 (2018). [DOI] [PubMed] [Google Scholar]
- 6.S. Vicent, J. M. López-Picazo, G. Toledo, M. D. Lozano, W. Torre, C. Garcia-Corchón, C. Quero, J. C. Soria, S. Martín-Algarra, R. G. Manzano, L. M. Montuenga, ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br. J. Cancer 90, 1047–1052 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.M. Malumbres, M. Barbacid, RAS oncogenes: The first 30 years. Nat. Rev. Cancer 3, 459–465 (2003). [DOI] [PubMed] [Google Scholar]
- 8.M. S. Miller, L. D. Miller, RAS mutations and oncogenesis: Not all RAS mutations are created equally. Front. Genet. 2, 100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.L. Johnson, K. Mercer, D. Greenbaum, R. T. Bronson, D. Crowley, D. A. Tuveson, T. Jacks, Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 (2001). [DOI] [PubMed] [Google Scholar]
- 10.L. Chang, M. Karin, Mammalian MAP kinase signalling cascades. Nature 410, 37–40 (2001). [DOI] [PubMed] [Google Scholar]
- 11.H. Lavoie, J. Gagnon, M. Therrien, ERK signalling: A master regulator of cell behaviour, life and fate. Nat. Rev. Mol. Cell Biol. 21, 607–632 (2020). [DOI] [PubMed] [Google Scholar]
- 12.S. Arur, M. Ohmachi, S. Nayak, M. Hayes, A. Miranda, A. Hay, A. Golden, T. Schedl, Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc. Natl. Acad. Sci. U.S.A. 106, 4776–4781 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.N. K. Aryal, V. Pant, A. R. Wasylishen, J. Parker-Thornburg, L. Baseler, A. K. El-Naggar, B. Liu, A. Kalia, G. Lozano, S. Arur, Constitutive Dicer1 phosphorylation accelerates metabolism and aging in vivo. Proc. Natl. Acad. Sci. U.S.A. 116, 960–969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.N. K. Aryal, V. Pant, A. R. Wasylishen, B. J. Rimel, L. Baseler, A. K. El-Naggar, D. G. Mutch, P. J. Goodfellow, S. Arur, G. Lozano, Dicer1 phosphomimetic promotes tumor progression and dissemination. Cancer Res. 79, 2662–2668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.M. Drake, T. Furuta, K. M. Suen, G. Gonzalez, B. Liu, A. Kalia, J. E. Ladbury, A. Z. Fire, J. B. Skeath, S. Arur, A requirement for ERK-dependent Dicer phosphorylation in coordinating oocyte-to-embryo transition in C. elegans. Dev. Cell 31, 614–628 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. Arur, Context-dependent regulation of Dicer activity and small RNA production: Implications to oocyte-to-embryo transition. Worm 4, e1086062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. S. Kumar, R. E. Pester, C. Y. Chen, K. Lane, C. Chin, J. Lu, D. G. Kirsch, T. R. Golub, T. Jacks, Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 23, 2700–2704 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.M. S. Kumar, J. Lu, K. L. Mercer, T. R. Golub, T. Jacks, Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39, 673–677 (2007). [DOI] [PubMed] [Google Scholar]
- 19.W. D. Foulkes, J. R. Priest, T. F. Duchaine, DICER1: Mutations, microRNAs and mechanisms. Nat. Rev. Cancer 14, 662–672 (2014). [DOI] [PubMed] [Google Scholar]
- 20.J. C. Robertson, C. L. Jorcyk, J. T. Oxford, DICER1 syndrome: DICER1 mutations in rare cancers. Cancers (Basel) 10, 143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.J. Vedanayagam, W. K. Chatila, B. A. Aksoy, S. Majumdar, A. J. Skanderup, E. Demir, N. Schultz, C. Sander, E. C. Lai, Cancer-associated mutations in DICER1 RNase IIIa and IIIb domains exert similar effects on miRNA biogenesis. Nat. Commun. 10, 3682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.M. Ha, V. N. Kim, Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15, 509–524 (2014). [DOI] [PubMed] [Google Scholar]
- 23.E. L. Snyder, H. Watanabe, M. Magendantz, S. Hoersch, T. A. Chen, D. G. Wang, D. Crowley, C. A. Whittaker, M. Meyerson, S. Kimura, T. Jacks, Nkx2-1 represses a latent gastric differentiation program in lung adenocarcinoma. Mol. Cell 50, 185–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Herriges, E. E. Morrisey, Lung development: Orchestrating the generation and regeneration of a complex organ. Development 141, 502–513 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.L. Lu, Q. Liu, P. Wang, Y. Wu, X. Liu, C. Weng, X. Fang, B. Li, X. Cao, H. Mao, L. Wang, M. Guan, W. Wang, G. Liu, MicroRNA-148b regulates tumor growth of non-small cell lung cancer through targeting MAPK/JNK pathway. BMC Cancer 19, 209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.J. Wang, S. Yao, Y. Diao, Y. Geng, Y. Bi, G. Liu, miR-15b enhances the proliferation and migration of lung adenocarcinoma by targeting BCL2. Thorac Cancer 11, 1396–1405 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Z. Wu, W. Li, J. Li, Y. Zhang, X. Zhang, Y. Xu, Y. Hu, Q. Li, Q. Sun, Z. Ma, Higher expression of miR-150-5p promotes tumorigenesis by suppressing LKB1 in non-small cell lung cancer. Pathol. Res. Pract. 216, 153145 (2020). [DOI] [PubMed] [Google Scholar]
- 28.H. Li, R. Ouyang, Z. Wang, W. Zhou, H. Chen, Y. Jiang, Y. Zhang, H. Li, M. Liao, W. Wang, M. Ye, Z. Ding, X. Feng, J. Liu, B. Zhang, MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci. Rep. 6, 39001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.K. Jiang, M. Shen, Y. Chen, W. Xu, miR‑150 promotes the proliferation and migration of non‑small cell lung cancer cells by regulating the SIRT2/JMJD2A signaling pathway. Oncol. Rep. 40, 943–951 (2018). [DOI] [PubMed] [Google Scholar]
- 30.N. Zhang, X. Wei, L. Xu, miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett. 587, 2346–2351 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Q. Xie, Z. Yu, Y. Lu, J. Fan, Y. Ni, L. Ma, microRNA-148a-3p inhibited the proliferation and epithelial-mesenchymal transition progression of non-small-cell lung cancer via modulating Ras/MAPK/Erk signaling. J. Cell. Physiol. 234, 12786–12799 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Y. Guan, Z. Rao, C. Chen, miR-30a suppresses lung cancer progression by targeting SIRT1. Oncotarget 9, 4924–4934 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R. Tang, L. Liang, D. Luo, Z. Feng, Q. Huang, R. He, T. Gan, L. Yang, G. Chen, Downregulation of MiR-30a is associated with poor prognosis in lung cancer. Med. Sci. Monit. 21, 2514–2520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Y. Lang, S. Xu, J. Ma, J. Wu, S. Jin, S. Cao, Y. Yu, MicroRNA-429 induces tumorigenesis of human non-small cell lung cancer cells and targets multiple tumor suppressor genes. Biochem. Biophys. Res. Commun. 450, 154–159 (2014). [DOI] [PubMed] [Google Scholar]
- 35.Y. Maeda, T. Tsuchiya, H. Hao, D. H. Tompkins, Y. Xu, M. L. Mucenski, L. Du, A. R. Keiser, T. Fukazawa, Y. Naomoto, T. Nagayasu, J. A. Whitsett, Kras(G12D) and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J. Clin. Invest. 122, 4388–4400 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.G. Orstad, G. Fort, T. J. Parnell, A. Jones, C. Stubben, B. Lohman, K. L. Gillis, W. Orellana, R. Tariq, O. Klingbeil, K. Kaestner, C. R. Vakoc, B. T. Spike, E. L. Snyder, FoxA1 and FoxA2 control growth and cellular identity in NKX2-1-positive lung adenocarcinoma. Dev. Cell 57, 1866–1882.e10 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.V. Boggaram, Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung. Clin. Sci. (Lond.) 116, 27–35 (2009). [DOI] [PubMed] [Google Scholar]
- 38.S. Chitale, H. Richly, DICER and ZRF1 contribute to chromatin decondensation during nucleotide excision repair. Nucleic Acids Res. 45, 5901–5912 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.K. Burger, M. Schlackow, M. Potts, S. Hester, S. Mohammed, M. Gullerova, Nuclear phosphorylated Dicer processes double-stranded RNA in response to DNA damage. J. Cell Biol. 216, 2373–2389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.M. J. Gutbrod, B. Roche, J. I. Steinberg, A. A. Lakhani, K. Chang, A. J. Schorn, R. A. Martienssen, Dicer promotes genome stability via the bromodomain transcriptional co-activator BRD4. Nat. Commun. 13, 1001 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.B. L. Allen, D. J. Taatjes, The Mediator complex: A central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16, 155–166 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R. C. Conaway, J. W. Conaway, Origins and activity of the Mediator complex. Semin. Cell Dev. Biol. 22, 729–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R. D. Kornberg, Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30, 235–239 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Y. Zhao, K. K. C. Li, K. P. Ng, C. H. Ng, K. A. W. Lee, The RNA Pol II sub-complex hsRpb4/7 is required for viability of multiple human cell lines. Protein Cell 3, 846–854 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.M. Maeta, M. Kataoka, Y. Nishiya, K. Ogino, M. Kashima, H. Hirata, RNA polymerase II subunit D is essential for zebrafish development. Sci. Rep. 10, 13213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.S. S. Baksh, R. E. Pratt, J. Gomez, V. J. Dzau, C. P. Hodgkinson, A novel Cbx1, PurB, and Sp3 complex mediates long-term silencing of tissue- and lineage-specific genes. J. Biol. Chem. 298, 102053 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M. Buschbeck, I. Uribesalgo, I. Wibowo, P. Rué, D. Martin, A. Gutierrez, L. Morey, R. Guigó, H. López-Schier, L. Di Croce, The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat. Struct. Mol. Biol. 16, 1074–1079 (2009). [DOI] [PubMed] [Google Scholar]
- 48.M. J. Gamble, K. M. Frizzell, C. Yang, R. Krishnakumar, W. L. Kraus, The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev. 24, 21–32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.X. Su, D. Chakravarti, M. S. Cho, L. Liu, Y. J. Gi, Y. L. Lin, M. L. Leung, A. El-Naggar, C. J. Creighton, M. B. Suraokar, I. Wistuba, E. R. Flores, TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature 467, 986–990 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.A. M. Zorn, J. M. Wells, Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 25, 221–251 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.K. S. Harris, Z. Zhang, M. T. McManus, B. D. Harfe, X. Sun, Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 2208–2213 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.L. De Moerlooze, B. Spencer-Dene, J. M. Revest, M. Hajihosseini, I. Rosewell, C. Dickson, An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development 127, 483–492 (2000). [DOI] [PubMed] [Google Scholar]
- 53.K. Sekine, H. Ohuchi, M. Fujiwara, M. Yamasaki, T. Yoshizawa, T. Sato, N. Yagishita, D. Matsui, Y. Koga, N. Itoh, S. Kato, Fgf10 is essential for limb and lung formation. Nat. Genet. 21, 138–141 (1999). [DOI] [PubMed] [Google Scholar]
- 54.N. Tang, W. F. Marshall, M. McMahon, R. J. Metzger, G. R. Martin, Control of mitotic spindle angle by the RAS-regulated ERK1/2 pathway determines lung tube shape. Science 333, 342–345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.D. R. Chang, D. Martinez Alanis, R. K. Miller, H. Ji, H. Akiyama, P. D. McCrea, J. Chen, Lung epithelial branching program antagonizes alveolar differentiation. Proc. Natl. Acad. Sci. U.S.A. 110, 18042–18051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.P. R. Tata, R. D. Chow, S. V. Saladi, A. Tata, A. Konkimalla, A. Bara, D. Montoro, L. P. Hariri, A. R. Shih, M. Mino-Kenudson, H. Mou, S. Kimura, L. W. Ellisen, J. Rajagopal, Developmental history provides a roadmap for the emergence of tumor plasticity. Dev. Cell 44, 679–693.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.N. D. Marjanovic, M. Hofree, J. E. Chan, D. Canner, K. Wu, M. Trakala, G. G. Hartmann, O. C. Smith, J. Y. Kim, K. V. Evans, A. Hudson, O. Ashenberg, C. B. M. Porter, A. Bejnood, A. Subramanian, K. Pitter, Y. Yan, T. Delorey, D. R. Phillips, N. Shah, O. Chaudhary, A. Tsankov, T. Hollmann, N. Rekhtman, P. P. Massion, J. T. Poirier, L. Mazutis, R. Li, J. H. Lee, A. Amon, C. M. Rudin, T. Jacks, A. Regev, T. Tammela, Emergence of a high-plasticity cell state during lung cancer evolution. Cancer Cell 38, 229–246.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.D. Yang, M. G. Jones, S. Naranjo, W. M. Rideout 3rd, K. H. J. Min, R. Ho, W. Wu, J. M. Replogle, J. L. Page, J. J. Quinn, F. Horns, X. Qiu, M. Z. Chen, W. A. Freed-Pastor, C. S. McGinnis, D. M. Patterson, Z. J. Gartner, E. D. Chow, T. G. Bivona, M. M. Chan, N. Yosef, T. Jacks, J. S. Weissman, Lineage tracing reveals the phylodynamics, plasticity, and paths of tumor evolution. Cell 185, 1905–1923.e25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.L. M. LaFave, V. K. Kartha, S. Ma, K. Meli, I. Del Priore, C. Lareau, S. Naranjo, P. M. K. Westcott, F. M. Duarte, V. Sankar, Z. Chiang, A. Brack, T. Law, H. Hauck, A. Okimoto, A. Regev, J. D. Buenrostro, T. Jacks, Epigenomic state transitions characterize tumor progression in mouse lung adenocarcinoma. Cancer Cell 38, 212–228.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.S. E. Castel, J. Ren, S. Bhattacharjee, A. Y. Chang, M. Sanchez, A. Valbuena, F. Antequera, R. A. Martienssen, Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell 159, 572–583 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R. Kanwal, S. Gupta, Epigenetic modifications in cancer. Clin. Genet. 81, 303–311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.F. Crispo, V. Condelli, S. Lepore, T. Notarangelo, A. Sgambato, F. Esposito, F. Maddalena, M. Landriscina, Metabolic dysregulations and epigenetics: A bidirectional interplay that drives tumor progression. Cell 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.A. M. K. Law, J. X. M. Yin, L. Castillo, A. I. J. Young, C. Piggin, S. Rogers, C. E. Caldon, A. Burgess, E. K. A. Millar, S. A. O'Toole, D. Gallego-Ortega, C. J. Ormandy, S. R. Oakes, Andy’s algorithms: New automated digital image analysis pipelines for FIJI. Sci. Rep. 7, 15717 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.D. R. Little, K. N. Gerner-Mauro, P. Flodby, E. D. Crandall, Z. Borok, H. Akiyama, S. Kimura, E. J. Ostrin, J. Chen, Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2-1. Proc. Natl. Acad. Sci. U.S.A. 116, 20545–20555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.H. M. T. Choi, M. Schwarzkopf, M. E. Fornace, A. Acharya, G. Artavanis, J. Stegmaier, A. Cunha, N. A. Pierce, Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Y. Zhang, T. Liu, C. A. Meyer, J. Eeckhoute, D. S. Johnson, B. E. Bernstein, C. Nusbaum, R. M. Myers, M. Brown, W. Li, X. S. Liu, Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.ENCODE Project Consortium , An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.S. X. Ge, D. Jung, R. Yao, ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Methods
Figs. S1 to S14
Tables S1, S3, and S5
Legends for tables S2, S4, and S6
Legend for data S1
Tables S2, S4, and S6
Data S1