Abstract
White adipose tissue (WAT) is a robust energy storage and endocrine organ critical for maintaining metabolic health as we age. Our aim was to identify cell-specific transcriptional aberrations that occur in WAT with aging. We leveraged full-length snRNA-Seq to characterize the cellular landscape of human subcutaneous WAT in a prospective cohort of 10 Younger (≤ 30 years) and 10 Older individuals (≥ 65 years) balanced for sex and body mass index (BMI). We highlight that aging WAT is associated with adipocyte hypertrophy, increased proportions of resident macrophages (M2), an upregulated innate immune response and senescence profiles in specific adipocyte populations, highlighting CXCL14 as a biomarker of this process. We also identify novel markers of pre-adipocytes and track their expression levels through pre-adipocyte differentiation. We propose that aging WAT is associated with low-grade inflammation that is managed by a foundation of innate immunity to preserve the metabolic health of the WAT.
Aging is associated with a progressive decline in physiological function leading to augmented human pathology and vulnerability to death1. White adipose tissue (WAT) functions as a robust energy store, an endocrine organ that governs whole-body metabolic homeostasis and a tissue that regulates immune modulation and regeneration2,3. Impairments in WAT function lead to unfavorable WAT redistribution towards central abdominal stores4, ectopic lipid accumulation and subsequent peripheral insulin resistance in organs such as skeletal muscle and liver5,6, and low-grade chronic systemic inflammation7,8. Elevated cellular senescence9–11, reduced progenitor proliferation, impaired adipogenic potential of the progenitor pool12 and immune cell infiltration13 have all been purported as factors contributing to age-associated decline in WAT function. These prior analyses, however, have largely been restricted to in vitro assessments or targeted histological and FACS approaches. Single cell/nuclei and spatial transcriptomics in human subcutaneous WAT is a rapidly growing area of research14–25 with aging recently being explored in a limited scope14. Our aim was to advance these findings at single nuclei resolution by analyzing nuclei-specific transcriptional differences in WAT between Younger and Older adults. In this study, for the first time, we prospectively obtained abdominal subcutaneous WAT biopsies for snRNA-Seq analyses from individuals who were metabolically normal and were not undergoing surgery. We leveraged our recent advancements in full-length snRNA-Seq in human WAT which yields superior gene detection capabilities compared to prior single cell/nuclei RNA-Seq platforms and technologies such as 10X Genomics24,26, combined with histological and in vitro assessments, to delineate nuclei-specific aberrations in human WAT with aging.
Results
To explore the effects of aging on WAT, we performed full-length snRNA-Seq analysis on abdominal subcutaneous WAT from 10 Older (≥ 65 years old) and 10 Younger (≤ 30 years old) individuals balanced for sex and BMI (Figure 1A). All individuals were metabolically healthy ≤ (Table 1, Supplementary Information 1). WAT biopsies were performed at Translational Research Institute (TRI) for the sole purpose of interrogating cell-specific aberrations with aging (see methods for processing details). The Older group had greater levels of systolic blood pressure, waist circumference, waist-to-hip-ratio, plasma cholesterol, very-low-density lipoprotein, triglycerides, thyroid stimulating hormone and aspartate transaminase vs. the Younger group (P < 0.05); however, all values were in the normal range. There were no differences in BMI, plasma glucose, plasma free fatty acids (FFAs), serum insulin, serum CRP, or adipose tissue insulin sensitivity measured by ADIPO-IR27,28 between the Older and Younger groups, indicative of comparable metabolic health, particularly for the WAT.
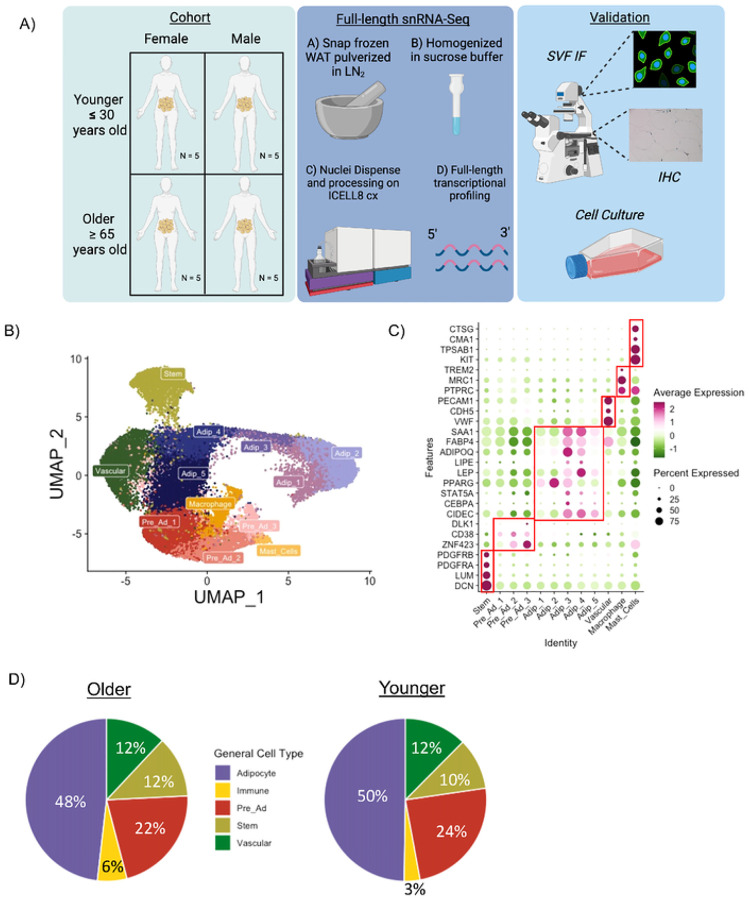
Figure 1. A single nuclei atlas of aging human abdominal subcutaneous white adipose tissue.
Schematic overview of study design and methods (A). UMAP of 25,736 nuclei from abdominal subcutaneous white adipose tissue (WAT) of 10 Younger and 10 Older participants (B). Dotplot of known marker genes for each cell population (C). Cell composition differences in the main cell population between Older and Younger participants (D).
Table 1.
| Clinical Variable | Older | Younger | P value |
|---|---|---|---|
| Age (years) | 74 ± 7 | 26 ± 3 | 1.19E −10 |
| Sex (F/M) | 5/5 | 5/5 | |
| Race (WH/BL/AI/UNK) | (8/1/0/1) | (7/2/1/0) | |
| Ethnicity (NH/H/UNK) | (5/1/4) | (2/5/3) | |
| BMI (kg/m2) | 29.38 ± 4.78 | 28.31 ± 4.80 | 0.623 |
| Waist (cm) | 102.69 ± 13.39 | 90.81 ± 8.07 | 0.0272 |
| Waist-to-hip Ratio | 0.96 ± 0.11 | 0.86 ± 0.08 | 0.0252 |
| SBP (mmHg) | 136 ± 13 | 122 ± 9 | 0.0256 |
| DBP (mmHg) | 78 ± 8 | 72 ± 7 | 0.109 |
| Glucose (mg/dL) | 97.50 ± 15.71 | 89.39 ± 7.23 | 0.151 |
| Insulin (μIU/mL) | 13.52 ± 7.36 | 11.34 ± 6.20 | 0.497 |
| HbAlc (%) | 5.63 ± 0.27 | 5.26 ± 0.54 | 0.0694 |
| Cholesterol (mg/dL) | 187.90 ± 43.51 | 151.20 ± 26.37 | 0.0349 |
| LDL (mg/dL) | 111.30 ± 42.58 | 80.60 ± 26.68 | 0.0692 |
| VLDL (mg/dL) | 21.10 ± 8.35 | 13.20 ± 6.32 | 0.0406 |
| HDL (mg/dL) | 55.50 ± 11.32 | 57.40 ± 12.20 | 0.722 |
| Non-HDL Cholesterol (mg/dL) | 132.40 ± 44.76 | 93.80 ± 24.53 | 0.0279 |
| Triglycerides (mg/dL) | 104.90 ± 41.95 | 66.10 ± 31.80 | 0.0316 |
| FFA (mmol/L) | 0.26 ± 0.14 | 0.34 ± 0.20 | 0.374 |
| ADIPO-IR | 0.56 ± 0.33 | 0.69 ± 0.52 | 0.522 |
| TSH (μIU/mL) | 2.59 ± 1.19 | 1.52 ± 0.78 | 0.0279 |
| ALT (units/L) | 20.00 ± 3.13 | 26.90 ± 28.47 | 0.361 |
| AST (units/L) | 26.00 ± 2.16 | 25.70 ± 19.56 | 0.0278 |
| CRP (mg/L) | 2.43 ± 1.61 | 2.68 ± 2.82 | 1 |
Abbreviations: ADIPO-IR, Adipose Tissue Insulin Resistance Index; AI, American Indian; ALT, Alanine Transaminase; AST, Aspartate Aminotransferase; BL, Black; BMI, Body Mass Index; CRP, C-Reactive Protein; DBP, Diastolic Blood Pressure; F, Female; FFA, Free-fatty Acid; H, Hispanic; HbA1c, Hemoglobin A1C; HDL, High-Density Lipoprotein; LDL, Low-Density Lipoprotein; M, Male; NH, Non-Hispanic; SBP, Systolic Blood Pressure; TSH, Thyroid Stimulating Hormone; WH, White; UNK, unknown;
Aging human WAT single nuclei atlas
Our dataset included 25,736 nuclei composed of 12 different clusters (Figure 1B) with an average of 5304 genes detected per nuclei. Known cell markers were used for initial annotation (Figure 1C) revealing populations of stem cells, pre-adipocytes, adipocytes, vascular cells, macrophages and mast cells. By transcriptionally profiling the nuclei isolated from WAT, we determined cell composition of adipocytes and non-adipocyte cells using an array of markers that are not limited to pre-specific cell markers. The largest cell type proportion in WAT is adipocytes (~50%) followed by pre-ads (22–24%), vascular cells (~12%), stem cells (10–12%) and immune cells (3–6%) (Figure 1D). Each participant had every cell type quantified except for mast cells (Extended Figure 1A-B). The Older group in comparison to the Younger group had double the proportion of immune cells (Figure 1D) in agreement with previous findings13. There were no major cell compositional differences between males and females (Extended Figure 1C).
Unbiased annotation of cell types
We performed an unbiased annotation of the cell clusters by running DGE analysis on each cluster to identify uncommon cell type markers. The top 5 DEGs for each cluster are highlighted in Figure 2A (Known cell markers of each cluster [i.e. PDGFRA, CD38, LEP, PPARG, PECAM1, MRC1 and TPSAB1] are included as a reference for the main cell types. As anticipated, stem cells had the highest expression of DCN, LUM and COL1A2, vascular cells had the highest expression of VWF, macrophages had the highest expression of HLA-DRA and LIPA, and mast cells had the highest expression of KIT. Classical markers of adipocytes ADIPOQ and PLIN1 were upregulated to varying degrees in all five adipocyte clusters. Classical pre-adipocyte markers such as ZNF423, CD38 and DLK1 were not in top DEGs for each pre-adipocyte population; but rather, uncommon markers such as CTNNA2, RBFOX3, PTPRD and NRXN3 were highly expressed in our pre-adipocyte populations. Previous snRNA-Seq data sets in mouse and human WAT have identified pre-adipocyte populations within a subset of progenitor cells, usually marked by ICAM1 or CD3620,23,29–32; however, these previously identified pre-adipocytes still express known stem cells markers such as PDGFRA, COL4A1, COL1A120,29,31,32 which should diminish upon commitment to the adipogenic lineage33. Therefore, these previously identified pre-adipocyte populations may not be committed pre-adipocytes. In contrast, our pre-adipocyte populations are transcriptionally distinct from our stem cell pool and have reduced expression of stem cell markers (PDGFRA, LUM, DCN; Figure 2A).
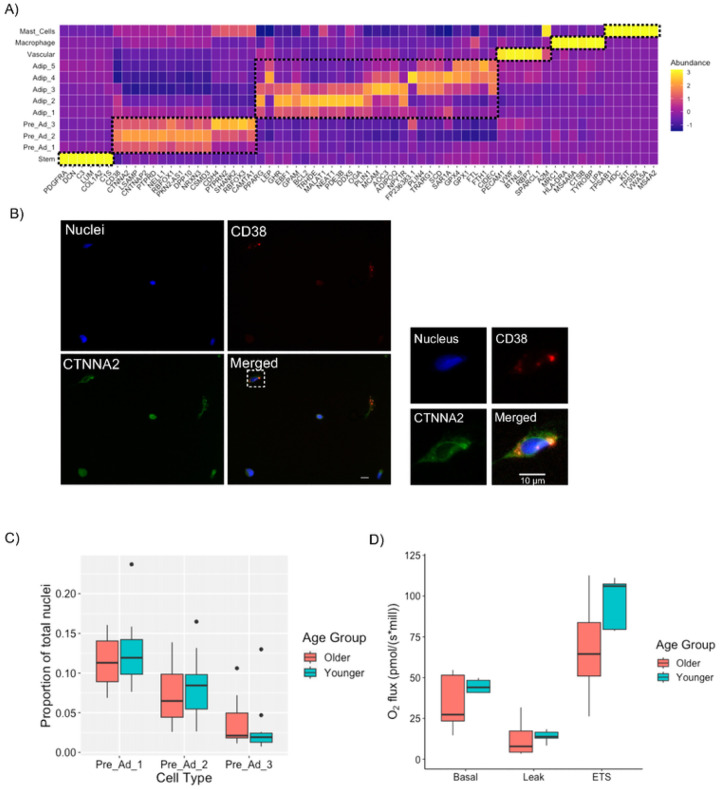
Figure 2. Full-length snRNA-seq reveals uncommon markers of pre-adipocytes.
Heatmap of Top 5 upregulated genes in each cell population with known marker genes included as a reference (A). Immunofluorescence staining of SVF obtained from subcutaneous adipose tissue with putative marker CD38 and novel marker CTNNA2. Images obtained at 40x magnification (B). Cell composition of the different pre-adipocyte populations in relation to the total number of nuclei in the Older and Younger groups (C). Pre-Adipocyte respiratory capacity measured with high resolution respirometry from Younger (n = 5) and Older (n = 5) Male participants (D).
To validate our cluster annotations in the pre-adipocyte populations, we compared gene expression profiles to previously published literature. We recently published full-length scRNA-Seq on the stromal vascular fraction (SVF) of abdominal subcutaneous WAT and transcriptionally profiled the commitment of pre-adipocytes from pluripotent stem cells (Whytock et al., 2022). We integrated our current dataset (annotated as BA) with the scRNA-Seq SVF and ran pairwise correlation analysis of the gene expression profiles between the identified cell clusters. Gene expression profiles from our defined cell populations correlated strongly with each other from both data sets, including our pre-adipocyte populations (black dashed box, Extended Figure 2A). Recently, Emont et al.,20 published a single cell atlas of WAT from various mouse and human fat pads and depots, respectively. We filtered the Emont data set to only include human abdominal subcutaneous WAT and compared gene expression profiles of major cell clusters against our data set. Gene expression profiles from major cell populations highly correlated with each other between the two data sets (Extended Figure 2B). Interestingly, our pre-adipocyte populations correlated most strongly with the lymphatic endothelial cell (EC) cluster in the Emont data set (black dashed box, Extended Figure 2B). All three of our pre-adipocyte clusters did not express upregulated endothelial cell makers (PECAM1, VWF, CDH5) or the specific lymphatic EC marker LYVE1. We are therefore confident that our pre-adipocyte populations are not lymphatic ECs and are indeed pre-adipocytes based on expression of expected markers (CD38, ZNF423 and DLK1)34–36. Both Emont et al.20 and a recent meta-analysis of human scRNA-Seq data25 had more defined immune cell populations (NK/T/B cells) in comparison to our data set, which is likely driven by the manner in which we procure our abdominal subcutaneous WAT samples. Ours24 are the only human single cell/nuclei RNA-Seq data generated from subcutaneous WAT obtained via lipoaspiration of the superficial subcutaneous depot rather than during surgery, the latter of which likely induces tissue inflammation.
Pre-Adipocytes
The top five DEGs for each of the three pre-adipocyte clusters (Figure 2A) have established roles in cell development including cell-to-cell adhesion, proliferation and differentiation which have primarily been shown in synapse/neuronal cells37–49. It is therefore plausible that these novel genes play important yet understated roles in pre-adipocyte proliferation, commitment, and differentiation, particularly in humans. CTNNA2 was recently identified as a gene with a single nucleotide polymorphism in polyadenylation signals upregulated in mouse lines for fatness50. PRPTD has been identified in pre-adipocyte populations, but there is conflicting evidence on whether it contributes to or downregulates adipogenesis51,52. NELL1 inhibits adipogenesis in 3T3-L1 and adipose-derived stem cell lines53. CDH4 is among differentially expressed genes in pre-adipocytes compared to bone-resident progenitor cells and has been identified as negative regulator of UCP154.
To characterize these pre-adipocyte populations further, transcription factor (TF) enrichment analysis was performed on the top DEGs (logfc >0.5) from all pre-adipocytes using ChEA355 (Supplementary Table 2). Top ranking TFs include: 1) SATB2 which interacts with and increases transcriptional activity of pre-adipocyte marker ZNF42356; 2) ZNF365 which increases in expression during differentiation in pre-adipocytes57; and 3) CTCF a multifactorial protein required during adipogenesis58. Therefore, our pre-adipocyte populations express genes that can be transcriptionally regulated by known TFs during pre-adipocyte differentiation.
Cadherin-Associated Protein Alpha 2 (CTNNA2) was among one of the highest expressed novel pre-adipocyte genes and was ubiquitously expressed across the three pre-adipocyte populations. We validated this particular pre-adipocyte marker at the protein level using immunofluorescence staining of SVF cells collected from abdominal subcutaneous WAT (Figure 2B). We co-stained CTNNA2 with known pre-adipocyte marker CD3836. CTNNA2 protein was present on the majority of SVF cells but to varying degrees. CTNNA2+ cells with the highest CTNNA2 abundance were also CD38+ positive cells, confirming cells with high CTNNA2 expression are pre-adipocytes (Figure 2B).
DGE and pathway analysis between the three pre-adipocyte populations (Supplemental Table 3) revelated Pre-Ad 1 had an upregulation of genes encoding ribosomal proteins (RPS6, RPL19) indicative of greater cell proliferation59; whereas, Pre-Ad2 and 3 had an upregulation of gens related to synaptic and cell-to-cell signaling that was more pronounced in Pre-Ad 3. There were no proportional differences among the pre-adipocyte populations between the Older and Younger groups (Figure 2C). Mitochondrial capacity is essential for preadipocyte proliferation and differentiation60 and can be blunted in WAT of metabolically unhealthy individuals61,62. Differences in mitochondrial capacity are more pronounced in pre-adipocytes compared to in vitro differentiated adipocytes63, as putative differences between groups can be diminished following induction with an adipogenic cocktail. Using high-resolution respirometry, we observed no differences between the Older and Younger groups in basal, leak nor maximal (uncoupled) respiration (electron transport system, ETS) of cultured pre-adipocytes (Figure 2D), suggesting that mitochondrial capacity in WAT is not attenuated with aging.
Cell sub-clustering
We resolved adipocyte and pre-adipocyte heterogeneity in our initial clustering analyses. To resolve our remaining cell types further, we performed additional sub-clustering on our macrophage, stem cell and vascular populations. Macrophages sub-clustered into two distinct clusters (Supplementary Figure 1A). Macrophage_0 had markers of resident macrophages (MRC1, PDGFC); whereas Macrophage_1 had markers of lipid-associated macrophages (LIPA, TREM2, Supplementary Figure 1B)64. The increased proportion of immune cells in the Older group was due to a greater proportion of resident type macrophages (Macrophage_0) compared to the Younger group (Supplementary Figure 1C). There were no proportional differences in Macrophage_1 between the Older and Younger groups (Supplementary Figure 1C).
Stem cells sub-clustered into three stem cell populations (Supplementary Figure 2A). Stem_0 had upregulation of pre-adipocyte markers compared to other stem cell populations (LSAMP, PDE4D) indicative of committing to the adipogenic lineage. Stem_1 was defined by nascent stem cell markers (DCN, LUM, COL1A1) and Stem_2 had upregulation of genes associated with lipid metabolism (PLIN4, G0S2; Supplementary Figure 2B). There was a trend towards Stem_0 having a higher proportion in the Older group when expressed as a proportion of total nuclei (Supplementary Figure 2C). There were, however, no differences in the proportions of Stem_1 or Stem_2 between the Older and Younger groups.
Vascular cells subclustered into four cell populations (Supplementary Figure 3A). Vascular_0 had upregulation of lipid metabolism genes (CIDEC, G0S2, PLIN4) defining a fatty-acid handling endothelial phenotype as previously described21,24. Vascular_1 had markers of pre-ads (DPP10, RBFOX1, PTRPD) indicative of being a vascular originating progenitor cell65,66. Vascular_2 had upregulation of classical markers of endothelial cells (PECAM1, VWF). Vascular_3 had markers of pericytes (PDGFRB, NOTCH3; Supplementary Figure 3B). There were also no differences in proportion of the each vascular subcluster between Older and Younger groups (Supplementary Figure 3C).
Adipocyte Heterogeneity
We identified five distinct adipocyte populations and sought to identify characteristics of each cluster. We performed DGE analysis comparing each adipocyte cluster to the remaining adipocyte clusters (Supplementary Table 4). We characterized Adipocyte 1 as “Nascent” adipocytes due to an upregulation of genes observed in pre-adipocyte populations (PTRPD, DPP10, NRXN3) and lower expression of classical adipocyte markers (ADIPOQ and LEP; Figure 3A). Adipocyte 2 had an upregulation of genes associated with adipocyte growth (PPARG, MALAT1 and NEAT1)67,68 in addition to insulin signaling genes (INSR and IRS1) and PDE3B which suppresses lipolysis69 and was therefore classified as “Immature” adipocytes (Figure 3A). Adipocyte 3 had upregulation of lipid synthesis genes (GPAM, SCD, DGAT), lipolytic genes (PNPLA2) and the classic adipokine ADIPOQ (Figure 3A). These features are characteristic of a “Mature” adipocyte carrying out routine adipocyte functions. Adipocyte 4 had upregulation of oxidative (ATP5F1B, ATP5MF and COX5A) and glutathione peroxidase (GPX1,3–4) genes but not typical browning or beiging markers and was therefore classified as “Oxidative”. Adipocyte 5 was defined as a “Remodeling” adipocyte due to upregulation of lysosomal proteases, cathepsins (CTSD and CTSS), complement factor (C3) and chemokines (CXCL12 and CXCL14). There were comparable adipocyte proportions between the Older and Younger groups (Figure 3B). When divided by sex, males tended to have a higher proportion of Adipocyte 5 (Remodeling) and a lower proportion of Adipocyte 4 (Oxidative) compared to females (Extended Figure 3A).
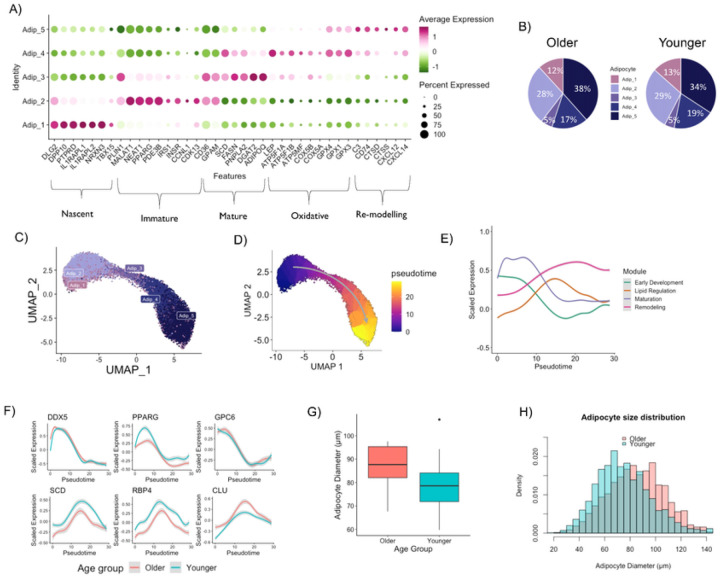
Figure 3. Adipocyte Heterogeneity in WAT.
Gene markers defining different adipocyte populations in WAT (A). Adipocyte composition differences between older and younger participants (B). Re-clustering of the adipocyte populations (C). Pseudotime trajectory of the five adipocyte populations (D). Smoothed expression dynamics for aggregated module scores throughout the adipocyte pseudotime (E). Expression patterns of key and novel genes throughout the adipocyte pseudotime (F). Mean adipocyte diameter in older and younger groups (G). Histogram of adipocyte size distribution split by age (H).
The adipocyte continuum
We assessed whether the different adipocyte populations were end-fate phenotypes with cell-specific biological functions or rather a continuum of biological processes through time and the impact of age. We performed pseudotime analysis on the adipocyte populations after re-clustering (Figure 3C), which revealed a clear one-dimensional trajectory initiating at Adipocyte 1 (Nascent) and terminating at Adipocyte 5 (Remodeling) (Figure 3D). The pseudotime visually displayed a similar trajectory when split into Older and Younger groups (Extended Figure 3B). The Older group had a lower proportion of nuclei in the first pseudotemporal bin (<5 pseudotime score; 30% vs. 34%) and higher proportion of nuclei in the last psuedotemporal bin (>20 pseudotime score; 39% vs. 35%) compared to the Younger group (Extended Figure 3C), indicating a shift towards later stages of the adipocyte continuum in the Older group. Unsupervised hierarchical gene clustering identified four modules of genes (Supplementary Table 5) whose patterns changed throughout the pseudotime (Figure 3E, Supplementary Figure 4A), with similar patterns observed when split by age (Extended Figure 3D, Supplementary Figure 4B-C). The module analysis revealed that adipocyte evolution initiates with early development, followed by maturation and lipid regulation and ends with remodeling (Figure 3E, Supplementary Figure 4). We therefore conclude that the observed adipocyte heterogeneity (i.e. five distinct adipocyte populations) is due to different developmental stages throughout time rather than end-fate phenotypes with cell-specific functions. Importantly, the biological processes of the adipocyte continuum that explain adipocyte heterogeneity are largely retained with aging; however, the Older group has more adipocytes in the later stages of remodeling, and the Younger group has more adipocytes in the early developmental stage. While the composition and trajectory of the modules were similar between groups (Supplementary Figure 4), some of the key temporally-regulated genes from each module showed disparity along the pseudotime with aging (Figure 3F, Supplementary Figure 5). Lipid homeostasis genes (e.g. PPARG, SRSF6, SCD, RBP4) had elevated expression in the Younger group, whereas remodeling genes (e.g. CLU, CFD) were elevated in the Older group (Figure 3F, Supplementary Figure 5). Taken together, our data indicate that the Younger group have a greater potential to develop adipocytes with more metabolically active properties, whereas the Older group have more adipocytes actively remodeling.
Adipocyte Size
Differences in adipocyte size were determined by histology70 with an average of 158 adipocytes measured per participant. Average adipocyte diameter (μm) was not statistically different between the Older and Younger groups (Figure 3G). Distribution frequency analyses revealed that the Older group displayed a greater frequency of larger adipocytes (>90 μm) and a reduced frequency of smaller adipocytes (<70 μm) compared to the Younger group (Figure 3H, Extended Figure 3E).
Adipokines
We quantified key adipokines (adiponectin, leptin and resistin) in the plasma and conditioned media (CM) of WAT explants (Extended Figure 3F). There was a trend towards leptin concentrations being higher in the CM from the Younger Group compared to the Older Group (P = 0.09, Extended Figure 3F) and no differences between groups for CM adiponectin and resistin. Plasma resistin concentrations were higher in the Older Group compared to the Younger group (P < 0.05) with no differences plasma adiponectin and leptin Extended Figure 3F). Adipokine concentrations in plasma and CM had expected correlations with adipocyte size and BMI (Supplementary Figure 6).
Pre-adipocyte differentiation
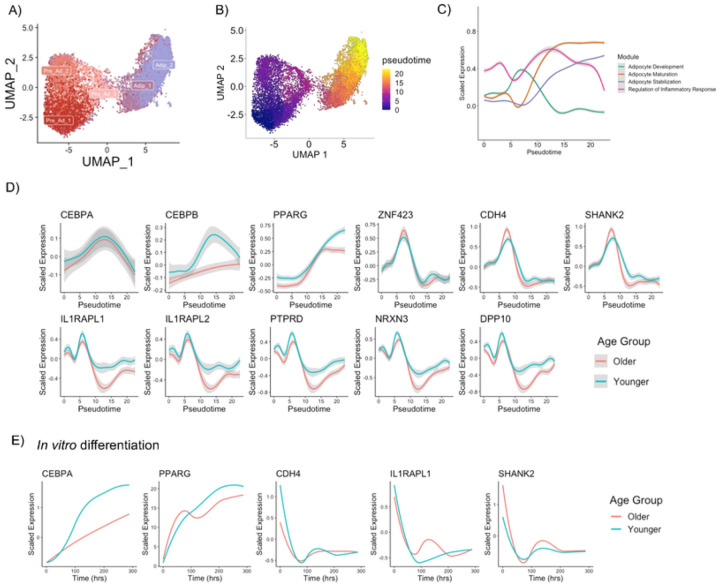
We next sought to track pre-adipocyte differentiation in vivo using a pseudotime trajectory. We re-clustered the pre-adipocyte populations with Adipocytes 1 (Nascent) and 2 (Immature) which are in the earlier stages of adipocyte development (Figure 4A). The pseudotime trajectory initiates in Pre-ad 1, branches off to Pre-ad 2 or 3, with Pre-ad 3 then converting towards Adipocyte 1 and Adipocyte 2 (Figure 4B). Similar pseudotime trajectories and similar proportions of nuclei in each pseudotemporal bin were observed between Older and Younger groups (Extended Figure 4A-B). Unsupervised hierarchical gene clustering identified four modules of genes (Supplementary Table 6) whose patterns changed throughout the pre-adipocyte differentiation pseudotime (Figure 4C, Supplementary Figure 7A), with similar patterns observed when split by age (Extended Figure 4C, Supplementary Figure 7B-C). The module analysis revelated that pre-adipocyte differentiation initiates with adipocyte development, transitions with adipocyte maturation and stabilization and has a fluctuation of inflammatory responses throughout (Figure 4C, Supplementary Figure 7). To validate our pre-adipocyte differentiation pseudotime, we plotted known markers of pre-adipocyte differentiation (CEPBA, CEBPB & PPARG; Figure 4D). As expected, early regulators of pre-adipocyte differentiation CEBPA and CEBPB increased during the early phases of the pseudotime (Figure 4D), while PPARG continues to increase throughout the pseudotime (Figure 4D). Both CEBPB and PPARG had a blunted response in the Older group (Figure 4D). Key genes from Module 1–3 showed little disparity with aging when transitioning along the pseudotime (Supplementary Figure 8). Due to module 4 expressing genes that align with commitment of pre-adipocytes to the adipogenic lineage, we mapped these novel markers along the pseudotime (Figure 4D). Known committed pre-adipocyte marker ZNF423 was used as reference. Several of these genesdisplayed a similar pattern to ZNF423 exhibiting a rapid increase at pseudotime score 7–8 corresponding to pre-adipocyte commitment (Figure 4D). Interestingly, the expression of the genes declined as the pseudotime transitioned to a score of 10–15 which corresponds to a Nascent adipocyte being formed and where a rapid increase in PPARG is observed (Figure 4D). This decline was dampened in the Younger group which retained elevated expression of several of these genes (IL1RAPL1, IL1RAPL2, PTPRD, NRXN3 & DPP10) as the pseudotime progressed (Figure 4D). We propose that previously assigned neuron/synapse genes play an essential role in forming committed pre-adipocytes and early adipocyte development and the elevated expression in the Younger group indicates a more poised state for differentiation during early development.
Figure 4. Pre-adipocyte Differentiation.
Re-clustering of pre-adipocyte and early development adipocyte populations (A). Pseudotime trajectory of pre-adipocyte differentiation from pre-adipocytes to adipocytes (B). Smoothed expression dynamics for aggregated module score throughout the pre-adipocyte differentiation pseudotime (C). Expression patterns of key and novel genes throughout pre-adipocyte differentiation pseudotime (D). Expression patterns of key and novel genes through pre-adipocyte differentiation in vitro from Older (N = 4) and Younger (N= 3) individuals (E).
To further validate a subset of these novel pre-adipocyte markers, we tracked the expression levels during differentiation in vitro. Cultured pre-adipocytes derived from digested human subcutaneous abdominal WAT were differentiated using an established adipogenic cocktail71 and cells were harvested for RT-qPCR at time-points Day 0, Day 0 + 4hrs, Day 2, Day 4, Day 9 and Day 12 (Extended Figure 4D). To match the in vivo pseudotime trajectory, Days were converted to hours and gene expression patterns were modelled throughout the differentiation period. As anticipated, known adipogenesis markers CEBPA and PPARG increased with differentiation (Figure 4E). Novel pre-adipocyte markers from Module 4, CDH4, SHANK2 and IL1RAPL1 had their highest expression levels at time point 0 before the differentiation cocktail was added and when they would still be classified as pre-adipocytes. This aligns with a pseudotime score 7–8 corresponding to pre-adipocyte commitment in vivo. CDH4, SHANK2 and IL1RAPL1 expression rapidly decreased upon addition of the differentiation cocktail coinciding with the cells rapidly transitioning to adipocytes, with no discernable differences between the Older and Younger groups. Our data confirm that these are pre-adipocyte markers are temporally regulated during pre-adipocyte differentiation. We posit, however, that nuances in gene expression levels between groups are better detected using an in vivo pseudotime approach rather than artificial induction in vitro.
Cell-Type transcriptional differences between Older and Younger groups
Random Forest Classification
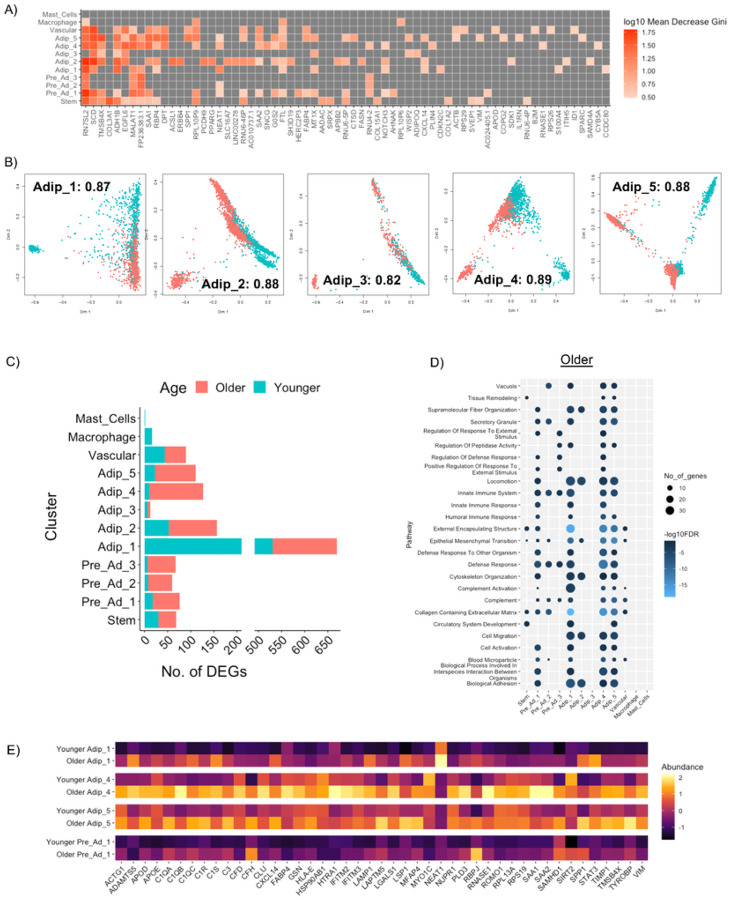
Random forest classification72 was performed on each cluster to determine the genes most important in classifying an Older and Younger cell for that specific cluster (Supplementary Table 7). The top genes based on a Mean Decrease Gini above 3 are shown in Figure 5A. Out of the 12 separate clusters, the five Adipocyte clusters, Pre-adipocyte 1, Vascular cells and Stem cells showed the highest accuracy (> 0.8) of random forest classification (Figure 5B, Extended Figure 5A), meaning that cells from Older and Younger individuals can be classified more accurately if selected at random from these clusters. Among the top genes contributing to this high accuracy that were also upregulated in Older individuals were DPT and CXCL14. DPT regulates extracellular matrix (ECM) formation and is upregulated in WAT in obesity-induced T2D73. Treatment of isolated adipocytes with DPT increases gene expression of ECM regulators (COL6A3, ELN, MMP9, TNMD) and inflammatory markers (IL6, IL8, TNF)73. CXCL14 is notable for its chemotactic properties74 but has recently been identified as a mediator of macrophage communication in brown adipose tissue75, as well as a regulator of insulin-mediated glucose uptake in 3T3-L1 adipocytes76. Together, these genes indicate an enhanced immune response in cells from the Older group. Among the top genes contributing to high accuracy that were also upregulated in Younger individuals were genes related to lipid metabolism (SCD, FASN, G0S2, ACSL1 and ADH1B) and adipogenesis (PPARG), indicative of active routine regulatory processes typical in WAT. Macrophages and Mast Cells had the lowest accuracy scores suggesting lower transcriptional diversity between Younger and Older groups for these cell types (Extended Figure 5A).
Figure 5. Transcriptional differences between old and young cell populations.
Top genes contributing to random forest classification for Older and Younger groups based on each cell cluster (A). Multi-dimensional scaling plots using top genes that contribute to random forest classification accuracy for each adipocyte cluster showing differences between the Older and Younger group. The number included is the accuracy score for that clusters (B). The number of DEGs between Older and Younger for each cluster (C). Top pathways upregulated in the Older group for each cluster (D). Heatmap of genes contributing to immune response and complement activation highlight differences between the Older and Younger Pre_Ad_1, Adip_1, Adip_4 and Adip_5 (E).
Differential gene expression
We performed DGE analysis comparing Older and Younger groups for each cell cluster (Supplementary Table 8). The Older group had the highest number of upregulated genes among the cell clusters (Figure 5C) except for Adipocyte 1 (Nascent adipocyte). Pathway analysis revealed the main pathways upregulated in the Older group were related to innate immune response and complement activation and were observed to the greatest extent in in Pre-Ad 1, Adipocyte 1 (Nascent adipocyte), Adipocyte 4 (Oxidative adipocyte) and Adipocyte 5 (Remodeling adipocyte) (Figure 5D). Genes contributing to immune response and complement activation pathways are shown in Figure 5E. In addition to bona fide immune cells, adipocytes themselves are now recognized as key innate immune cells within adipose tissue77. The data suggests that in the Older group, these pre-adipocyte and adipocyte populations are initiating an innate immune response via alternative complement pathway activation. As part of the innate immune response, these cells undergo remodeling of ECM (ADAMTS5, VIM, HTRA1, MFAP4, SPP1, TIMP1), phospholipid membrane (PLD3), lipids (APOD, APOE,) actin cytoskeleton (ACTG1, GSN, LSP1, MYO1C) and carbohydrate binding structure (LGALS1, RNASE1) that is in part mediated by lysosomal activity (LSAM1, LAPTMS) and ROS regulation (ROMO1). In the Younger group, the main pathways upregulated were related to neurogenesis and synapse organization in Adipocyte 1 (Extended Figure 5B). These genes have previously been attributed to the development of neurons; however, given their upregulation in Adipocyte 1 (Nascent) and in agreement with our pre-adipocyte differentiation pseudotime, we again reason that these genes are early regulators of pre-adipocyte differentiation.
Senescence
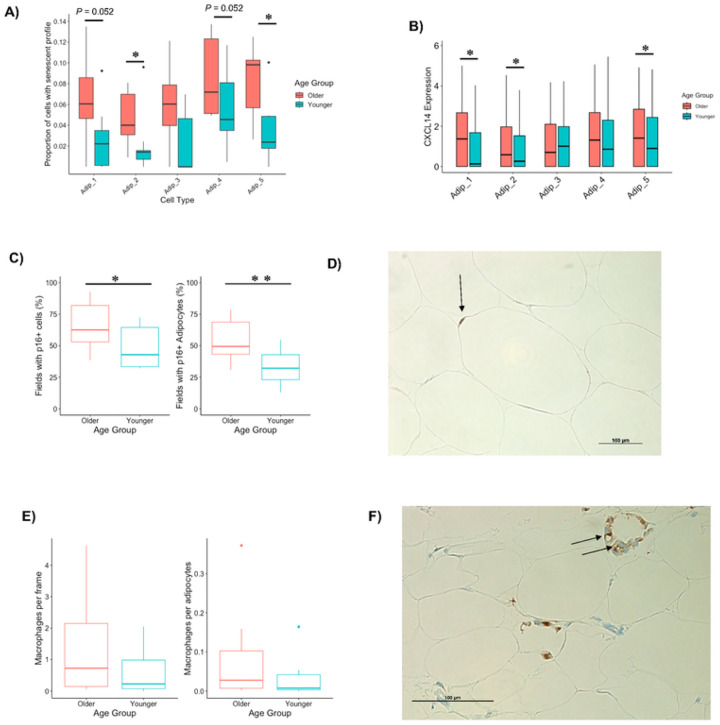
Cellular senescence is caused by accumulation of DNA damage78 and other cellular stressors79–81, leading to enlarged cells that are resistant to regulated cell death82 and secrete chemokines, cytokines, growth factors, and matrix metalloproteinases83 that can negatively impact the function of other cells in the microenvironment11,84. Senescence typically occurs in proliferating cell types but has also been found in terminally differentiated cells85,86 including adipocytes9,87. Senescent cells activate innate immunity to assist in the removal of nearby dying cells10,88. Given that the Older group has larger (ie, hypertrophic) adipocytes, upregulated innate immunity specifically in adipocyte populations and increased proportions of adipocytes in the end-stage of the adipocyte pseudotime, we reasoned that the Older group would have increased levels of senescence, particularly among their adipocytes. p16 (CDKN2A) and p21 (CDKN1A) are canonical markers of senescence but are unreliable in detecting senescence at the transcriptional level due to very low expression. To measure senescence profiles in our snRNA-Seq data set, we performed GSEA on each nucleus using the recently published SenMayo gene set which identifies the transcriptional profile of senescent cells with a combination of 125 genes89 (Extended Figure 6A). In line with Saul et al.89, we determined that the top 10% of cells could be classified as those with a senescent profile. The Older group had a significantly greater proportion of cells with a senescent profile in Adipocyte clusters 2, 5 and Pre-Adipocyte 1 and a trend towards a greater proportion of cells with a senescent profile in Adipocyte clusters 1 and 4 (Figure 6A, Extended Figure 6B). It was notable from our data set, that the only chemokine or cytokine upregulated in clusters that have an increased proportion of cells with a senescent profile was CXCL14 (Figure 6B). This gene was also included as high importance in distinguishing between Older and Younger individuals in our prior random forest classification (Figure 5A).
Figure 6. Markers of senescence in aging WAT.
The proportion of nuclei with a high senescence profile (top 10%) in adipocyte populations between Older and Younger participants (A). CXCL14 expression (B). The percentage of 20x magnification fields that had cells and adipocytes with p16 staining between Older and Younger participants (C). Representative image of p16 cell staining from an Older individual with p16 in and adipocyte highlighted (D). The number of macrophages per frame and per adipocytes (E). Representative image of macrophage crown like structure in the the Older group (F). *, P < 0.05; **, P < 0.01.
To confirm Older individuals had a greater proportion of cells with a senescent profile, we performed immunohistochemistry staining for the classic senescent marker p16. Abundance of p16+ cells was quantified as the percentage of 1) total fields (20x magnification) analyzed containing p16+ cells9 and 2) p16+ adipocytes. The Older group had significantly more fields containing p16+ cells than the Younger group (Figure 6C–D). The percentage of fields containing p16+ adipocytes was also greater among Older individuals (Figure 6C), suggesting the increased senescence profile in the Older group is driven by adipocytes and aligns with the snRNA-seq data (Figure 6A–B).
Given that CXCL14 may mediate macrophage infiltration90 and that our snRNA-seq data show a greater number of resident macrophages in the Older group, we quantified macrophage content via immunohistochemistry using a pan-macrophage marker CD6891. The numbers of macrophages quantified with CD68 per frame and per adipocyte were not statistically significant between the Older and Younger groups (Figure 6E). Three participants had crown-like structures of macrophages (Figure 6F), which form around dying or damaged adipocytes92. All of these individuals were male and two of them were from the Older group. Obesity is associated with macrophage infiltration91. In agreement, we noted a significant and positive correlation between a marker of abdominal obesity, waist-hip-ratio (WHR), and the number of macrophages per adipocytes (Extended Figure 6C). WHR was greater in the Older group (Table 1), and this was driven specifically by Older males (Extended Figure 6D). We therefore reason that macrophage infiltration occurs in Older males with higher WHR that can lead to crown-like structures developing around damaged adipocytes, perhaps as a means to maintain healthy WAT.
Discussion
We present a single nuclei atlas of aging human abdominal subcutaneous WAT in the largest prospective cohort study to date using cutting-edge full-length transcriptional profiling. We highlight new markers of pre-adipocyte populations that are temporally regulated during pre-adipocyte differentiation. We show that different adipocyte populations do not become distinct end-fate phenotypes with specific functions, but rather each adipocyte transitions through a developmental state along a continuum.
In the context of healthy aging, the main findings were that specific adipocyte populations in the Older group have an increased expression of genes related to innate immune response and an increased proportion of adipocytes with a senescent profile. Phenotypically, the Older group had a shift towards larger adipocytes which have been associated with hypoxia, oxidative stress and inflammation93,94. Resident-like (M2) macrophages are fundamental in providing an initial defense mechanism during the innate immune response95 and in agreement, the Older Group had increased proportions of resident-like macrophages. These differences exist despite the Older group having comparable whole-body and WAT metabolic health to the Younger group. We therefore propose that aging WAT endures very low-grade inflammation insults that are managed through their innate immunity foundation to preserve the metabolic health of the adipose tissue. CXCL14 was the only chemokine or cytokine upregulated in these specific adipocyte populations in the Older group. The role of CXCL14 in WAT has been confounded by a lack of consensus identifying and validating its receptor; however, it was recently proposed that CXCL14 synergizes with low concentrations of CXCL13 and CCL19/CCL21 during in vitro chemotaxis with immune cells expressing receptors CXCR5 and CCR796. CXCL14 is known to recruit M2 macrophages75 and its adipose tissue expression and circulating concentrations are reduced with metabolic abnormalities such as obesity, PCOS and T2D90,97, suggesting it may be a therapeutic candidate that mediates low-grade inflammation to maintain metabolic health of the tissue. Future research is needed to examine if the innate immunity foundation that preserves metabolic health in WAT is impaired in an unhealthy human aging model that is paired with metabolic and cardiovascular disease, as well as impairments in physical and physiological function.
Methods
Experimental model and subject details
Younger (≤ 30 years old) and Older (≥ 65 years old) individuals were recruited to the Translational Research Institute at AdventHealth to participate in the study. Full inclusion/exclusion criteria can be found in Supplementary information 1. Briefly, all participants were free from metabolic and infectious disease, were not taking medication related to diabetes or inflammation and had not had major surgery within the last 4 weeks. Five participants were taking 1 or more medications to treat the following conditions; hypertension, hyperlipidemia, urinary retention, thyroid, anxiety and osteoporosis. All participants were weight-stable for at least 3 months prior to the assessments and adipose biopsy. The study was approved by AdventHealth Institutional Review Board and carried out in accordance with the Declaration of Helsinki. Participants provided written informed consent to partake in the study.
Abdominal subcutaneous WAT biopsies were performed following an overnight fast using the tumescent lidocaine approach with a Mercedes aspiration cannula98. Following removal of excess blood and connective tissue, the sample was cleaned with PBS. A portion (~100mg) was immediately snap frozen for subsequent nuclei isolation for snRNA-seq. A portion (~20mg) was fixed in 10% Formalin for 24 hours and stored in 70% EtOH for subsequent histological analyses. A portion (~100mg) was used as an explant to collect conditioned media. The remaining tissue was stored in Medium 199 containing 25mM HEPES at room temperature prior to digestion of adipose derived progenitor cells and in vitro experiments.
Nuclei isolation from whole white adipose tissue
Nuclei was isolated from frozen WAT as previously detailed26. WAT was pulverized under linked nitrogen before being homogenized in 2mL of homogenization buffer (5mM MgCl2, 25mM Tris Buffer pH 8.0, 25mM KCL, 250mM sucrose, 1μm DDT, 1 x protease inhibitor, 0.2 U/μL SUPERase. In RNase Inhibitor (Thermofisher Scientific) in nuclease-free water) with a glasscol homogenizer. Following addition of Triton-X100 (0.1% v/v) the homogenate was incubated on ice with regular vortexing. Samples were then filtered through a 100μm and 40μm strainer (BD Falcon), centrifuged at 2,700g for 10 min at 4°C, resuspended in homogenization buffer and recentrifuged again at 2,700g for 10 min at 4°C. The pellet was then re-suspended in 1mL nuclei isolation medium (5mM MgCl2, 25mM Tris Buffer pH 8.0, 25mM KCL, 1 mM EDTA, 0.2 U/μL Ribolock RNAase inhibitor, 1% BSA in nuclease-free water) before being centrifuged at 2,700g for 10 min at 4°C. The sample was re-suspended in 500μL nuclei isolation medium before being filtered 10 x with a 25g syringe. Nuclei was stained with Hoechst 33342 (ReadyProbes Cell Viability Imaging Kit, Thermofisher Scientific) and counted with a countess II automated cell counter (Thermofisher Scientific).
Isolation of human adipose-derived progenitor cells
Abdominal subcutaneous WAT was minced and digested with collagenase (type 1, 1 mg/ml, 3% BSA in HBSS) (Worthington Type I Collagenase cat# S004196) using 1 g/ 2ml ratio, for 30 – 60 min at 37°C with shaking (100rpm), as described previously71. The mixture was passed through a 250 μm mesh, rinsed with HBSS, centrifuged at 200g for 5 minutes, and floating mature adipocytes were removed. Progenitor cells were span at 500g for 5 minutes and cell pellet was treated with erythrocyte lysis buffer (RBC lysis buffer, cat# 420301). Cells were re-pelleted with centrifugation, resuspended with alpha Dulbecco’s modified Eagle’s Medium (αMEM) + GlutaMax (Gibco, cat# 32561–037) supplemented with 10% fetal bovine serum (FBS), 100U/ml penicillin and 100μg/ml streptomycin, filtered through 100μm cell strainer, and then plated and culture in a humid atmosphere with 5% CO2. After 4 hrs, non-adherent cells were removed, and media was replaced. Adherent cells were allowed to reach 80% confluence with medium changed every other day and then either subculture or frozen (media supplemented with 10% DMSO and 16% FBS) for cryopreservation.
Single-nuclei RNA-Seq
Single-nuclei suspension (40K/mL) was aliquoted into 8 wells of a 384-well source plate (Takara Bio USA, San Jose, CA) and dispensed using an iCELL8 MultiSample NanoDispenser (Takara Bio USA) onto an iCELL8® 350v Chip (Takara Bio USA. Following dispense, the chip nanowells were imaged using the iCELL8 Imaging Station to identify nanowells containing a single nucleus, with only these nanowells being subjected for downstream dispenses. After imaging, the chip was subjected to freeze-thaw to lyse the nuclei, followed by a 3 min incubation at 72°C to denature the RNA. Selected nanowells were subjected to first-strand cDNA synthesis initiated by oligo dT primer (SMART-Seq iCELL8 CDS), followed by template switching with template switching oligo (SMART-Seq iCELL8 oligonucleotide) for 2nd strand cDNA synthesis, before unbiased amplification of full-length cDNA. Tagment DNA enzyme 1 (TDE1, Illumina, San Diego, CA) was used to tagment full-length cDNA before amplification with forward (i5) and reverse (i7) indexing primers. Each single nucleus was indexed by a unique combination of 1 of 72 forward and 1 of 72 reverse indexing primers allowing for downstream identification. Collected cDNA was purified twice using a 1:1 proportion of AMPure XP beads (Beckman Coulter, Brea, CA). cDNA was further amplified according to manufacturer’s instructions and purified again at a 1:1 proportion of AMPure XP beads. The resultant cDNA library was assessed for concentration by fluorometer (Qubit, Thermofisher Scientific) and quality by electrophoresis (Agilent Bioanalyzer high sensitivity DNA chips). Libraries were sequenced with Illumina HiSeq 4000 at an average sequencing depth of 270 M per library. This equated to an average 118,568 barcoded reads per nuclei.
Bioinformatic analyses
Initial analyses of the single nuclei libraries were performed using CogentAP™ Analysis Pipeline (Takara Bio, USA). GRCh38 was used as the genome reference. Cell and gene filtering was performed in R package scran and scuttle99. Nuclei were initially filtered if they had; < 500 genes, > 20% mitochondrial reads or if cell complexity was < 0.65. Following initial filtering, outliers were removed based on 3 median absolute deviations of log total counts. A minimum threshold of 0.1 was applied to filter out low expressed genes. All 20 samples were integrated and normalized in Seurat with SCTransform using 5000 highly variable protein coding genes100,101. Dimensional reduction to obtain integration anchors was achieved with reciprocal PCA. Once integrated, significant principal components were used to perform unsupervised K-nearest neighbor (KNN) graph-based clustering. Visualization was achieved with uniform manifold approximation and projection (UMAP). Differential gene expression analysis for cell clusters was performed using a Wilcoxon rank sum test with Seurat’s “FindMarkers” function with a FDR cut off of < 0.05, log2 FC > 0.25 or < −0.25 and expressed in >25% of nuclei in that cluster. Differential gene expression analysis between the Older and Younger group was performed using a Wilcoxon rank sum test with Seurat’s “FindMarkers” function with a FDR cut off of < 0.05, log2 FC > 0.1 or < −0.1 and expressed in >10% of nuclei in that cluster. A hypergeometric test was used to assess over-representation of upregulated genes (log2 FC < 0.25) in the Older and Younger group for each cell type using R package HypeR102 querying datasets of; gene ontology, KEGG, Reactome and Hallmark103–107. Significance was set at an FDR of 0.05. Genes detected in each cluster was used as a background reference. Senescent enrichment score was generated using R Package Ucell and function enrichIt()108, with the SenMayo list of genes used as the gene set89.
Pseudotime trajectory
Monocle 3 was used for trajectory analysis109–113. After analyzing the dynamic biological changes of each nucleus, an individual position of every single nucleus is plotted in a learned trajectory. Based on the clustered annotation and marker genes, we identified the root of the given trajectory and ordered nuclei along the pseudotime according to their developmental progress. For adipocyte lifecycle we identified this root as Adipocyte 1 and for the adipocyte differentiation we identified the roots as Pre-Ad 1 and 2. The nuclei were split into 5 pseudotemporal bins based on their pseudotime score (<5, 5–10, 10–15, 15–20, >20). Temporally regulated genes were identified by performing DGE (logfc > 0.8 & FDR < 0.05) for each bin against the first and last pseudotemporal bin as previously described114,115. Unsupervised hierarchical gene clustering was performed by K-means clustering using R package complex heatmap116 to identify modules of genes that change throughout the pseudotime. A hypergeometric test was used to assess over-representation of genes within a module using R package HypeR102 querying data set of gene ontology106,107. Significance was set at an FDR of 0.05. Aggregated gene expression for each module was calculated using the AddModuleScore() function in Seurat, allowing aggregated module gene expression to be plotted throughout the pseudotime.
Pairwise correlation analysis
We compared gene expression profiles between the current data set and previously published data sets. Initially samples were integrated together using Seurat as described above. Barcodes of cells/nuclei comprising the main cell types for each analysis were used to define the main cell populations. Correlation analyses were performed among these different cell populations.
Random Forest
Random Forest classification was used to rank genes on their importance in distinguishing between the Older and Younger group for each cell type72. The cell types were randomly split into a training (80%) and testing set (20%) to complete and evaluate modelling. Random Forest modelling was computed on each cell type to predict nuclei that originated from the Older and Younger groups. Accuracy scores for each cell type was computed and mean decrease gini was used to measure variable importance in the classification. Multi-dimensional scaling plots were generated using the top genes that contribute to the random forest classification accuracy for each cell-type.
Pre-adipocyte differentiation in vitro
Pre-Adipocytes derived from Younger (N = 3) and Older (N = 4) individuals were thawed from −80°C at a low passage, seeded and differentiated as previously described71. Briefly, 2-day post-confluency cells were induced to differentiate in DMEM/Ham’s F12 (1:1) containing isobutylmethylxanthine (0.5 mm), dexamethasone (100 nm), insulin (100 nm), T3 (2 nm), and transferrin (10 μg/liter) for 6 days and then maintained in DMEM/F12 containing only insulin (10 nm) and dexamethasone (10 nm) until day 12. Cells were harvested at; Day 0, 4 hrs, Day 2, Day 4, Day 9 and Day 12. RNA was extracted using Rneasy kit (Qiagen) and cDNA was generated using reverse transcriptase. Target Genes and housekeeping gene (PPIA) were measured by RT-qPCR using a ViiA7 sequence detection system (Life technologies) and SyberGreen technology suitable for relative gene expression quantification using the following parameters: one cycle of 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute.
Immunofluorescence staining of stromovascular fraction (SVF)
A cover slip was added in each well of a 24-well tissue culture plate. SVF were seed at a density of 140K/well and kept in culture overnight in αMEM supplemented with 10% FBS. Non-adherent cells were washed with PBS. The remaining adherent cells (ADSCs, endothelial cells and preadipocytes) were fixed with 4% paraformaldehyde for 30min. The plate was washed 3 times with PBS and kept at 4°C until IF staining was performed. Cells were incubated with 0.1M glycine in PBS for 15 minutes before being permeabilized for 5 min with PBS containing 1% NP40 and 0.05% saponin. Cells were blocked for 30 minutes using blocking solution (5% NGS in 0.1M PB). Cells were stained with primary antibodies targeting CTNNA2 (LSBio, #LS-C669704–50, dilution 1:50) and CD38 (abcam, #ab23518, dilution 1:200) overnight at 4°C, washed 6 times for 5 min with PBS and then incubated with secondary alexa fluor antibodies (Thermofisher; GARIgG 488 (#A32731), GAMIgG 633 (#A21052) for 45 min in the dark at room temperature. Following another 6 × 5 min PBS washes, cover slips were mounted onto slides with ProLong Gold Antifade mountant containing DAPI (Thermofisher, #P36941). Images were captured using a Nikon eclipse Ti microscope (Nikon Technologies, California) at 40x magnification.
Adipose Tissue Histology
Adipose tissue stored in 70% EtOH was embedded in paraffin and sectioned at 5 μm. Slides were stained with H&E to determine adipocyte sizing. Immunohistochemical detection of CD68 (Atlas Antibodies, AMAb90873) for macrophages and p16 (Enzo Life Science, ENZ-ABS664) with the avidin-biotin peroxidase method. Images were captured using a Nikon eclipse Ti microscope (Nikon Technologies, California) at 20x magnification.
Adipocyte size was quantified using ImageJ software. An average of 158 adipocytes were measured per participant from 6 frames. Each adipocyte was manually outlined to create an ROI, then the area was calculated by the ImageJ software. Adipocytes that were below 200μm2 and above 16,000μm2 were removed as they typically represent artefacts from processing70. Abundance of p16+ cells were quantified as the percentage of total fields (20x magnification) analyzed containing p16+ cells9 and p16+ adipocytes over an average of 26 frames per participant. Head-and-neck cancer tissue was used as a positive control for the presence of p16. The presence of CD68-positive cells was counted per frame with an average of 18 frames per participant and adjusted for the number of adipocytes per frame for each participant. Presence of crown-like structures for each frame was also noted. Tonsil tissue was used as a positive control for the presence of CD68.
Measurement of mitochondrial capacity (oxygen consumption rates)
Oxygen consumption (mitochondrial capacity) rates were performed via high resolution respirometry using the Oxygraph-2K (Oroboros Instruments, Innsbruck, Austria). Cultured pre-adipocytes (passages 3 to 6) derived from Younger (N = 5) and Older (N = 5) male individuals were plated in in 6 well/plate with 62,500 cells/well in αMEM supplemented with 10% FBS for 48 hrs to reach 80% confluency. Cells were detached from plates using StemPro Accutase (Gibco, cat#A11105–01)), centrifuged and resuspend in DMEM (Gibco, cat#11885–084)) medium. 3 wells of cells were used per chamber of the Oxygraph. Basal respiration was performed in intact cells. Addition of 1 μg/mL of oligomycin (Sigma-Aldrich, cat # 04876–5MG) and 0.2 mM of mitochondrial uncoupler (FCCP, Sigma-Aldrich, cat# C2920) allowed for measurements of Leak and maximal capacity of the electron transport system (ETS), respectively. Oxygen flux was normalized to cell number.
Human adipose tissue conditioned media
Adipose tissue was incubated in M199 media (Gibco, cat# 11043–023) containing 25 mM Hepes using a 20:1 ratio (μl:mg) in a 50 mL falcon tube and cultured in a water bath shaker (100 rpm) at 37°C for 3 hrs. After that period, tissue was collected by using a 100 μm cell strainer and conditioned media was centrifuged at 500 g for 5 min, then aliquoted and frozen at −80°C. Adipokine; leptin, adiponectin and resistin were measured by ELISA using reagents from the R&D systems, Inc. (cat# DLP00, cat# DRP300 and cat# DRSN00).
Blood metabolites
Fasting blood samples were collected for measurements of comprehensive metabolic panel, HbA1c (%), insulin, FFA and CRP and analyzed in the clinical chemistry laboratory at AdventHealth using standard assays. Adipose tissue insulin resistance index (ADIPO-IR) was calculated by multiplying plasma FFA (mmol/L) by serum insulin (pmol/L)27,28.
Statistical testing
For comparisons of clinical and phenotypical data between the Older and Younger group, an unpaired t-test was used to detect differences in normally distributed data and Mann-Whitney U test was used for non-normally distributed data. A two-way ANOVA was used when comparing variables for age and sex with post-hoc Tukey HSD.
Acknowledgements
The authors would like to acknowledge Meghan Hopf for help with the histology analyses, River Goodpaster and Reichelle Yeo for assisting with adipokine measurements and Nicolas Laurent for assistance with respiration analyses. We would like to thank the Clinical Research Unit and Laboratory at the Translational Research Institute for assisting in conducting this study and the volunteers for participation. The study was partially funded by James Vath, PhD and Huseyin Mehmet, PhD and was supported by an administrative supplement to grant #U01 AR071133. Figure 1A was generated with Biorender.com.
Footnotes
Code availability
No new code was generated during the production of this manuscript.
Supplementary Files
Contributor Information
Lauren Sparks, Translational Research Institute, AdventHealth.
Katie Whytock, Translational Research Institute, AdventHealth.
Adeline Divoux, Translational Research Institute, AdventHealth.
Yifei Sun, Icahn School of Medicine at Mount Sinai.
Maria Pino, Translational Research Institute, AdventHealth.
Gongxin Yu, Translational Research Institute, AdventHealth.
Steven Smith, Translational Research Institute, AdventHealth.
Martin Walsh, Icahn School of Medicine at Mount Sinai.
Data Availability
snRNA-Seq data generated during this study have been deposited under GSE235529.
References
- 1.López-Otín C., Blasco M. A., Partridge L., Serrano M. & Kroemer G. The hallmarks of aging. Cell 153, 1194–217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer A. K. & Kirkland J. L. Aging and adipose tissue: potential interventions for diabetes and regenerative medicine. Exp Gerontol 86, 97–105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodpaster B. H. & Sparks L. M. Metabolic Flexibility in Health and Disease. Cell Metab 25, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuk J. L., Saunders T. J., Davidson L. E. & Ross R. Age-related changes in total and regional fat distribution. Ageing Res Rev 8, 339–48 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Shulman G. I. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N Engl J Med 371, 1131–41 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Borén J., Taskinen M.-R., Olofsson S.-O. & Levin M. Ectopic lipid storage and insulin resistance: a harmful relationship. J Intern Med 274, 25–40 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Wu D. et al. Aging up-regulates expression of inflammatory mediators in mouse adipose tissue. J Immunol 179, 4829–39 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Starr M. E., Evers B. M. & Saito H. Age-associated increase in cytokine production during systemic inflammation: adipose tissue as a major source of IL-6. J Gerontol A Biol Sci Med Sci 64, 723–30 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Justice J. N. et al. Cellular Senescence Biomarker p16INK4a+ Cell Burden in Thigh Adipose is Associated With Poor Physical Function in Older Women. J Gerontol A Biol Sci Med Sci 73, 939–945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tchkonia T. et al. Fat tissue, aging, and cellular senescence. Aging Cell 9, 667–84 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu M. et al. Targeting senescent cells enhances adipogenesis and metabolic function in old age. Elife 4, e12997 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caso G. et al. Peripheral fat loss and decline in adipogenesis in older humans. Metabolism 62, 337–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trim W. V et al. Divergent immunometabolic changes in adipose tissue and skeletal muscle with ageing in healthy humans. J Physiol 600, 921–947 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Zhou W. et al. High-resolution aging niche of human adipose tissues. Signal Transduct Target Ther 8, 105 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angueira A. R. et al. Defining the lineage of thermogenic perivascular adipose tissue. Nat Metab 3, 469–484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karunakaran D. et al. RIPK1 gene variants associate with obesity in humans and can be therapeutically silenced to reduce obesity in mice. Nat Metab 2, 1113–1125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaitin D. A. et al. Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell 178, 686–698.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hildreth A. D. et al. Single-cell sequencing of human white adipose tissue identifies new cell states in health and obesity. Nat Immunol 22, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun W. et al. snRNA-seq reveals a subpopulation of adipocytes that regulates thermogenesis. Nature 587, 98–102 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Emont M. P. et al. A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vijay J. et al. Single-cell analysis of human adipose tissue identifies depot- and disease-specific cell types. Nat Metab 2, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acosta J. R. et al. Single cell transcriptomics suggest that human adipocyte progenitor cells constitute a homogeneous cell population. Stem Cell Res Ther 8, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merrick D. et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science (1979) 364, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whytock K. L. et al. Single cell full-length transcriptome of human subcutaneous adipose tissue reveals unique and heterogeneous cell populations. iScience 25, 104772 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massier L. et al. An integrated single cell and spatial transcriptomic map of human white adipose tissue. Nat Commun 14, 1438 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whytock K. L. et al. Isolation of nuclei from frozen human subcutaneous adipose tissue for full-length single-nuclei transcriptional profiling. STAR Protoc 4, 102054 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Søndergaard E., Espinosa De Ycaza A. E., Morgan-Bathke M. & Jensen M. D. How to Measure Adipose Tissue Insulin Sensitivity. J Clin Endocrinol Metab 102, 1193–1199 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Groop L. C. et al. Glucose and free fatty acid metabolism in non-insulin-dependent diabetes mellitus. Evidence for multiple sites of insulin resistance. J Clin Invest 84, 205–13 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burl R. B. et al. Deconstructing Adipogenesis Induced by β3-Adrenergic Receptor Activation with Single-Cell Expression Profiling. Cell Metab 28, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hepler C. et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sárvári A. K. et al. Plasticity of Epididymal Adipose Tissue in Response to Diet-Induced Obesity at Single-Nucleus Resolution. Cell Metab 33, (2021). [DOI] [PubMed] [Google Scholar]
- 32.Schwalie P. C. et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 559, (2018). [DOI] [PubMed] [Google Scholar]
- 33.Cawthorn W. P., Scheller E. L. & MacDougald O. A. Adipose tissue stem cells meet pre-adipocyte commitment: going back to the future. J Lipid Res 53, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta R. K. et al. Transcriptional control of preadipocyte determination by Zfp423. Nature 464, 619–23 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sul H. S., Smas C., Mei B. & Zhou L. Function of pref-1 as an inhibitor of adipocyte differentiation. Int J Obes Relat Metab Disord 24 Suppl 4, S15–9 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Carrière A. et al. Identification of the ectoenzyme CD38 as a marker of committed preadipocytes. Int J Obes (Lond) 41, 1539–1546 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Henrich K.-O. et al. CAMTA1, a 1p36 tumor suppressor candidate, inhibits growth and activates differentiation programs in neuroblastoma cells. Cancer Res 71, 3142–51 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Jacobs E. Z. et al. Expanding the molecular spectrum and the neurological phenotype related to CAMTA1 variants. Clin Genet 99, 259–268 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Kim K. K., Nam J., Mukouyama Y.-S. & Kawamoto S. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J Cell Biol 200, 443–58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Unsicker C. et al. SHANK2 mutations impair apoptosis, proliferation and neurite outgrowth during early neuronal differentiation in SH-SY5Y cells. Sci Rep 11, 2128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengelaub C. A., Navrazhina K., Ross J. B., Halberg N. & Tavazoie S. F. PTPRN2 and PLCβ1 promote metastatic breast cancer cell migration through PI(4,5)P2-dependent actin remodeling. EMBO J 35, 62–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ceresa D. et al. Cdh4 Down-Regulation Impairs in Vivo Infiltration and Malignancy in Patients Derived Glioblastoma Cells. Int J Mol Sci 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song W. et al. Putative complement control protein CSMD3 dysfunction impairs synaptogenesis and induces neurodevelopmental disorders. Brain Behav Immun 102, 237–250 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Danno S. et al. PKN2 is essential for mouse embryonic development and proliferation of mouse fibroblasts. Genes to Cells 22, 220–236 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Li C. et al. Neural EGFL like 1 as a potential pro-chondrogenic, anti-inflammatory dual-functional disease-modifying osteoarthritis drug. Biomaterials 226, 119541 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y. & Fisher G. J. Receptor type protein tyrosine phosphatases (RPTPs) - roles in signal transduction and human disease. J Cell Commun Signal 6, 125–38 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.St George-Hyslop F., Kivisild T. & Livesey F. J. The role of contactin-associated protein-like 2 in neurodevelopmental disease and human cerebral cortex evolution. Front Mol Neurosci 15, 1017144 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knudsen K. A., Soler A. P., Johnson K. R. & Wheelock M. J. Interaction of alpha-actinin with the cadherin/catenin cell-cell adhesion complex via alpha-catenin. J Cell Biol 130, 67–77 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bill B. R., Lowe J. K., Dybuncio C. T. & Fogel B. L. Orchestration of neurodevelopmental programs by RBFOX1: implications for autism spectrum disorder. Int Rev Neurobiol 113, 251–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Šimon M. et al. Genome-wide screening for genetic variants in polyadenylation signal (PAS) sites in mouse selection lines for fatness and leanness. Mamm Genome 34, 12–31 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Parrillo L. et al. Altered PTPRD DNA methylation associates with restricted adipogenesis in healthy first-degree relatives of Type 2 diabetes subjects. Epigenomics 12, 873–888 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Chen M. et al. Metastasis suppressor 1 interacts with protein tyrosine phosphatase receptor-δ to regulate adipogenesis. FASEB J 37, e22857 (2023). [DOI] [PubMed] [Google Scholar]
- 53.James A. W. et al. A new function of Nell-1 protein in repressing adipogenic differentiation. Biochem Biophys Res Commun 411, 126–31 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ambrosi T. H. et al. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell 20, 771–784.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keenan A. B. et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res 47, W212–W224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Addison W. N. et al. Zfp423 Regulates Skeletal Muscle Regeneration and Proliferation. Mol Cell Biol 39, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao X. et al. Transcriptome Analysis of Landrace Pig Subcutaneous Preadipocytes during Adipogenic Differentiation. Genes (Basel) 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen Y. et al. Cooperation of ATF4 and CTCF promotes adipogenesis through transcriptional regulation. Cell Biol Toxicol 38, 741–763 (2022). [DOI] [PubMed] [Google Scholar]
- 59.Ceci M., Fazi F. & Romano N. The role of RNA-binding and ribosomal proteins as specific RNA translation regulators in cellular differentiation and carcinogenesis. Biochim Biophys Acta Mol Basis Dis 1867, 166046 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Wilson-Fritch L. et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol 23, 1085–94 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yin X. et al. Adipocyte mitochondrial function is reduced in human obesity independent of fat cell size. J Clin Endocrinol Metab 99, E209–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Böhm A. et al. Increased mitochondrial respiration of adipocytes from metabolically unhealthy obese compared to healthy obese individuals. Sci Rep 10, 12407 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keuper M. et al. Preadipocytes of obese humans display gender-specific bioenergetic responses to glucose and insulin. Mol Metab 20, 28–37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strieder-Barboza C. et al. Single-nuclei Transcriptome of Human AT Reveals Metabolically Distinct Depot-Specific Adipose Progenitor Subpopulations. BioRxiv (2022). [Google Scholar]
- 65.Tang W. et al. White fat progenitor cells reside in the adipose vasculature. Science 322, 583–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iyama K., Ohzono K. & Usuku G. Electron microscopical studies on the genesis of white adipocytes: differentiation of immature pericytes into adipocytes in transplanted preadipose tissue. Virchows Arch B Cell Pathol Incl Mol Pathol 31, 143–55 (1979). [DOI] [PubMed] [Google Scholar]
- 67.Farmer S. R. Regulation of PPARgamma activity during adipogenesis. Int J Obes (Lond) 29 Suppl 1, S13–6 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Han J. et al. The long noncoding RNA MALAT1 modulates adipose loss in cancer-associated cachexia by suppressing adipogenesis through PPAR-γ. Nutr Metab (Lond) 18, 27 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DiPilato L. M. et al. The Role of PDE3B Phosphorylation in the Inhibition of Lipolysis by Insulin. Mol Cell Biol 35, 2752–60 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Honecker J. et al. A distribution-centered approach for analyzing human adipocyte size estimates and their association with obesity-related traits and mitochondrial function. Int J Obes (Lond) 45, 2108–2117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee M.-J., Wu Y. & Fried S. K. A modified protocol to maximize differentiation of human preadipocytes and improve metabolic phenotypes. Obesity (Silver Spring) 20, 2334–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Toth R. et al. Random forest-based modelling to detect biomarkers for prostate cancer progression. Clin Epigenetics 11, 148 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Unamuno X. et al. Dermatopontin, A Novel Adipokine Promoting Adipose Tissue Extracellular Matrix Remodelling and inflammation in Obesity. J Clin Med 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu J., Chatterjee M., Schmid H., Beck S. & Gawaz M. CXCL14 as an emerging immune and inflammatory modulator. J Inflamm (Lond) 13, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cereijo R. et al. CXCL14, a Brown Adipokine that Mediates Brown-Fat-to-Macrophage Communication in Thermogenic Adaptation. Cell Metab 28, 750–763.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 76.Takahashi M. et al. CXCL14 enhances insulin-dependent glucose uptake in adipocytes and is related to high-fat diet-induced obesity. Biochem Biophys Res Commun 364, 1037–42 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Blaszczak A. M., Jalilvand A. & Hsueh W. A. Adipocytes, Innate Immunity and Obesity: A Mini-Review. Front Immunol 12, 650768 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campisi J. & d’Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol 8, 729–40 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Zhu Y., Armstrong J. L., Tchkonia T. & Kirkland J. L. Cellular senescence and the senescent secretory phenotype in age-related chronic diseases. Curr Opin Clin Nutr Metab Care 17, 324–8 (2014). [DOI] [PubMed] [Google Scholar]
- 80.Swanson E. C., Manning B., Zhang H. & Lawrence J. B. Higher-order unfolding of satellite heterochromatin is a consistent and early event in cell senescence. J Cell Biol 203, 929–42 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tchkonia T., Zhu Y., van Deursen J., Campisi J. & Kirkland J. L. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J Clin Invest 123, 966–72 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang E. Senescent human fibroblasts resist programmed cell death, and failure to suppress bcl2 is involved. Cancer Res 55, 2284–92 (1995). [PubMed] [Google Scholar]
- 83.Coppé J.-P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6, 2853–68 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu M. et al. JAK inhibition alleviates the cellular senescence-associated secretory phenotype and frailty in old age. Proc Natl Acad Sci U S A 112, E6301–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Farr J. N. et al. Identification of Senescent Cells in the Bone Microenvironment. J Bone Miner Res 31, 1920–1929 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jurk D. et al. Postmitotic neurons develop a p21-dependent senescence-like phenotype driven by a DNA damage response. Aging Cell 11, 996–1004 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Q. et al. Obesity and hyperinsulinemia drive adipocytes to activate a cell cycle program and senesce. Nat Med 27, 1941–1953 (2021). [DOI] [PubMed] [Google Scholar]
- 88.Xue W. et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–60 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Saul D. et al. A new gene set identifies senescent cells and predicts senescence-associated pathways across tissues. Nat Commun 13, 4827 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cereijo R. et al. The chemokine CXCL14 is negatively associated with obesity and concomitant type-2 diabetes in humans. Int J Obes (Lond) 45, 706–710 (2021). [DOI] [PubMed] [Google Scholar]
- 91.Weisberg S. P. et al. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112, 1796–808 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cinti S. et al. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J Lipid Res 46, (2005). [DOI] [PubMed] [Google Scholar]
- 93.Haczeyni F., Bell-Anderson K. S. & Farrell G. C. Causes and mechanisms of adipocyte enlargement and adipose expansion. Obes Rev 19, 406–420 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Muir L. A. et al. Adipose tissue fibrosis, hypertrophy, and hyperplasia: Correlations with diabetes in human obesity. Obesity (Silver Spring) 24, 597–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Solana R. et al. Innate immunosenescence: effect of aging on cells and receptors of the innate immune system in humans. Semin Immunol 24, 331–41 (2012). [DOI] [PubMed] [Google Scholar]
- 96.Kouzeli A. et al. CXCL14 Preferentially Synergizes With Homeostatic Chemokine Receptor Systems. Front Immunol 11, 561404 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.García-Beltran C. et al. Reduced circulating levels of chemokine CXCL14 in adolescent girls with polycystic ovary syndrome: normalization after insulin sensitization. BMJ Open Diabetes Res Care 8, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Divoux A. et al. Differential open chromatin profile and transcriptomic signature define depot-specific human subcutaneous preadipocytes: primary outcomes. Clin Epigenetics 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lun A. T. L., McCarthy D. J. & Marioni J. C. A step-by-step work ow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hao Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hafemeister C. & Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Federico A. & Monti S. hypeR: an R package for geneset enrichment workflows. Bioinformatics 36, 1307–1308 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liberzon A. et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gillespie M. et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res 50, D687–D692 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kanehisa M. & Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gene Ontology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49, D325–D334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ashburner M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–9 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Andreatta M. & Carmona S. J. UCell: Robust and scalable single-cell gene signature scoring. Comput Struct Biotechnol J 19, 3796–3798 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cao J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Levine J. H. et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qiu X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Traag V. A., Waltman L. & van Eck N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep 9, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Trapnell C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gupta A., Shamsi F., Patti M. E., Tseng Y.-H. & Streets A. Mapping the temporal transcriptional landscape of human white and brown adipogenesis using single-nuclei RNA-seq. bioRxiv (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Steier Z. et al. Single-cell multi-omic analysis of thymocyte development reveals drivers of CD4/CD8 lineage commitment. bioRxiv (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gu Z., Eils R. & Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–9 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
snRNA-Seq data generated during this study have been deposited under GSE235529.