Abstract
The value of computer-assisted image analysis has been shown in several studies. The performance of tools with artificial intelligence (AI), such as GestaltMatcher, is improved with the size and diversity of the training set, but properly labeled training data is currently the biggest bottleneck in developing next-generation phenotyping (NGP) applications. Therefore, we developed GestaltMatcher Database (GMDB) - a database for machine-readable medical image data that complies with the FAIR principles and improves the openness and accessibility of scientific findings in Medical Genetics.
An entry in GMDB consists of a medical image such as a portrait, X-ray, or fundoscopy, and machine-readable meta information such as a clinical feature encoded in HPO terminology or a disease-causing mutation reported in HGVS format. In the beginning, data was mainly collected by curators gathering images from the literature. Currently, clinicians and individuals recruited from patient support groups provide their previously unpublished data. For this patient-centered approach, we developed a digital consent form. GMDB is a modern publication medium for case reports that complements preprints, e.g., on medRxiv. To enable inter-cohort comparisons, we implemented a research feature in GMDB that computes the pairwise syndromic similarity between hand-picked cases.
Through a community-driven effort, we compiled an image collection of over 7,533 cases with 792 disorders in GMDB. Most of the data was collected from 2,058 publications. In addition, about 1,018 frontal images of 498 previously unpublished cases were obtained. The web interface enables gene- and phenotype-centered queries or infinite scrolls in the gallery. Digital consent has led to increasing adoption of the approach by patients. The research app within GMDB was used to generate syndromic similarity matrices to characterize two novel phenotypes (CSNK2B, PSMC3).
GMDB is the first FAIR database for NGP, where data are findable, accessible, interoperable, and reusable. It is a repository for medical images that cannot be included in medRxiv. That means GMDB connects clinicians with a shared interest in particular phenotypes and improves the performance of AI.
Introduction
Although next-generation sequencing (NGS) has significantly increased the diagnostic yield in rare diseases, it is still challenging to analyze a considerable amount of variants to reach the diagnosis, which is often called the “diagnostic odyssey”1. With the recent advance in artificial intelligence (AI), next-generation phenotyping (NGP) technology which can analyze the patients’ frontal images, has significantly progressed over the last few years2–14. Facial analysis applications such as DeepGestalt, which runs behind Face2Gene (https://www.face2gene.com), are increasingly used by geneticists and pediatricians to diagnose patients with facial dysmorphism9. The recent approach, GestaltMatcher11, can not only predict the ultra-rare and novel disorders but also delineate the facial phenotypes of the diseases15–22. Moreover, the facial analysis can be further integrated into the exome variants prioritization to facilitate the diagnosis23,24.
However, despite the increasing interest and technological advances, properly labeled training data is still the biggest bottleneck in developing NGP applications25. Furthermore, the existing data are often siloed, so curation is usually done repeatedly26. In recent years, principles have been developed to support the reuse of scientific data, thus meeting the increasing need to make data findable, accessible, interoperable, and reusable (FAIR) for humans and computers27. Nevertheless, most current databases, e.g., the London Medical Database (LMD) used to train Face2Gene’s DeepGestalt algorithm, are commercial and not freely accessible to the scientific community, thus building yet another data silo28, even though it has long been clear that patients with rare diseases benefit enormously from data sharing29. Due to the mentioned diagnostic odyssey, patients are increasingly interested in data sharing. A recent study showed that over 73.7% of surveyed parents are willing to allow the storage of their children’s facial images in a secure database that other researchers and doctors can access without general public access30. Therefore, the need and the will to improve the data situation are present, but these efforts often fail due to time-consuming recruitment processes. A faster and easier way for the patient recruiting process is needed.
Therefore, we proposed GestaltMatcher Database (GMDB; https://db.gestaltmatcher.org), a web framework that addresses the needs of human syndromologist first and yields data curation for AI as a by-product. On the one hand, it serves as a modern publication medium that enables updating case reports after the first publication and improves the accessibility from the former image databases such as LMD. With the implementation of digital consent, we also provide a more efficient way of patient recruitment. We have collected 9,863 frontal images from 7,533 patients with 792 different disorders, including 1,018 unpublished frontal images uploaded by 26 clinicians worldwide. At the same time, the data compiled through this community-driven effort was not only made available to other clinicians and scientists but could also be used to train and test AI, in accordance with the FAIR principles. With this incentive, we were able to curate thousands of case reports in a short time and to enhance collaboration amongst researchers all over the world. We envisioned that GMDB could be further connected with other databases with the protocols, such as Beacon31 and Phenopackets32.
Results
Overview of GMDB
The aim of the GMDB is, on the one hand, to satisfy the needs of clinicians for a reference database and a more modern publication medium while also achieving data curation with crowdsourced labeling for machine learning. With this incentive, we have curated 9863 frontal images of 7,533 patients with 792 disorders (Figure 1 and Figure 2a). Our curators gathered images from 2,058 publications, and 1,018 frontal images from 498 cases that had not been published before were uploaded by clinicians or patients (Figure 2b).
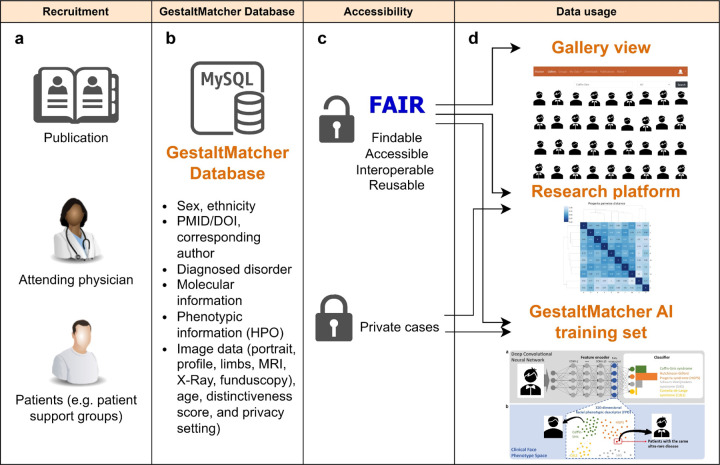
Figure 1:
Overview of GestaltMatcher database. a) GMDB recruited patients from the publication, clinicians, and patient support group; b) GMDB stored the patient metadata such as sex, ethnicity, age, phenotypic and genomic information; c) GMDB provided two types of data: public and private patients; d) Public patients can be accessible by searching in gallery view. For example, users can search for “Coffin-Siris syndrome” in the gallery and visualize the patient images of Coffin-Siris syndrome. Both public and private patients can be used in the research platform for the patients’ similarity analysis and the training of GestaltMatcher approach.
Figure 2:
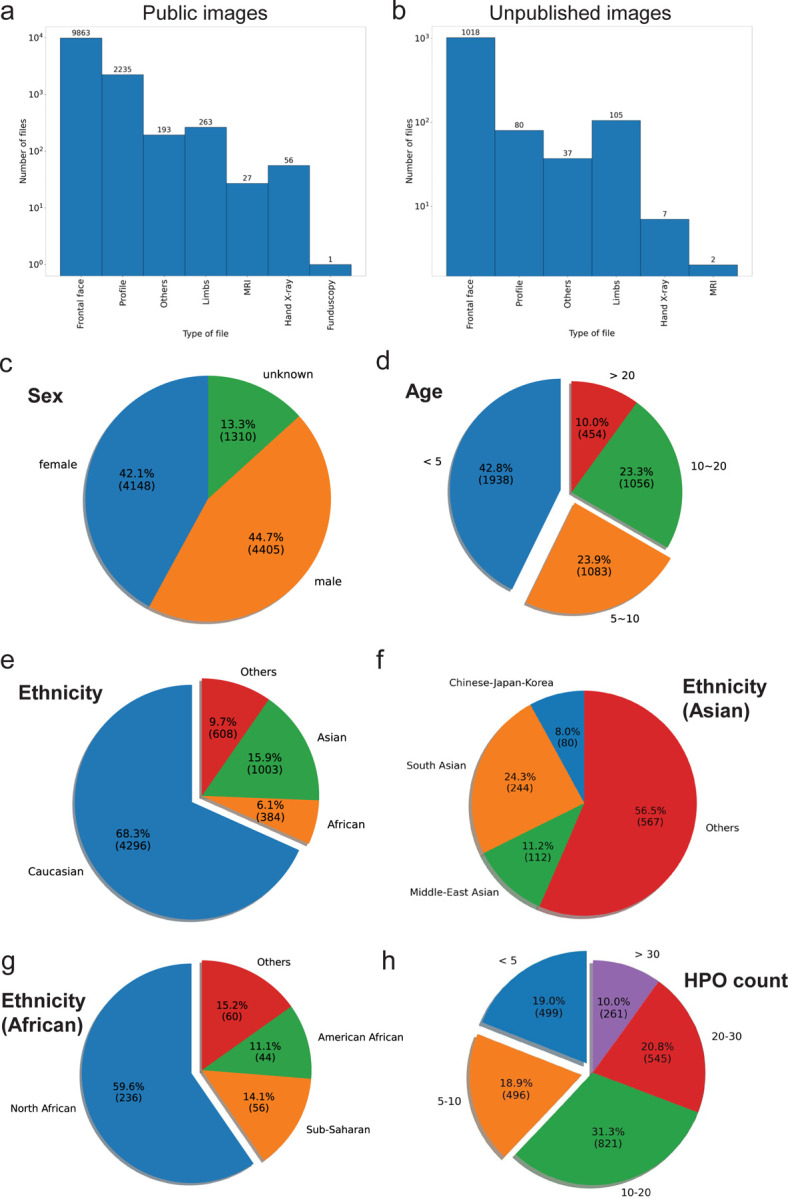
Overall statistic of data in GMDB. a) distribution of the public images (images from publication and unpublished image); b) distribution of the unpublished images; c) distribution of age; d) distribution of age; e) distribution of ethnicity; f) distribution of asian ethnicity; g) distribution of african ethnicity; h) distribution of the counts of HPO for each image.
The male patients are slightly more than the female patients (2.6% higher). The age distribution is imbalanced (Figure 2c). 42.8% of images were taken below five years old, and 23.9% were taken between five to ten years old (Figure 2d). The distribution of ethnic backgrounds is also imbalanced. Most images (68.3%) are from Caucasian patients (Figure 2e). The other ethnic groups are under-represented (Asian: 15.9% and African: 6.1%). When we look into the composition of Asian patients, the majority of images have no specific ethnicity labels, and the combination of Chinese, Japanese, and Korean patients is only 8% of the total Asian patients (Figure 2e). African patients are mainly recruited from North Africa, especially Egypt (Figure 2f). We also stored the clinical description encoded in HPO terminology. For the images with HPO terms, 62.1% of images have more than ten HPO terms (Figure 2g).
Reference Database and modern publication medium
In finding the correct diagnosis for a rare genetic disease, comparing the patient’s face with images of other patients with a known diagnosis can sometimes be helpful. A literature search is often tedious due to data siloing and takes much time, which is rarely given in the everyday clinical routine. The “Gallery view” feature in the GMDB makes it easy to search for the candidate gene or disease and visualize all the images relevant to the search, collected by our curators from the literature and the previously unpublished cases, at a glance. Moreover, the user can search for HPO terms33 or even for PubMed ID (PMID) and DOI for a specific publication. A GMDB user can also easily find his or her cases using a personal reference. This search allows clinicians to compare their patients to the images in the database and get an overview of the heterogeneity of the individual facial characteristics of many diseases. In this context, having many patients of various ethnicities due to our numerous international collaborations is also helpful; these patients are historically underrepresented in other resources34.
In addition to its function as a reference database, the GMDB can also be seen as a new publication medium that shows the most recently published cases in the GMDB. Unlike conventional publication media, which are usually static, a case in the GMDB can be updated at any time, for example, after a follow-up patient consultation in which new symptoms were reported or after new laboratory results were obtained.
Facilitating the NGP development by open database
To address the idea of FAIR principles, we make the dataset of “publicly” uploaded patients in GMDB available to other researchers. They can then use the labeled images and the collected data to train and test their AI model. For example, the disorder prediction tool can be helpful to clinicians in finding a diagnosis, and it is possible to get suggestions for suitable syndromes and the GestaltMatcher score (https://www.gestaltmatcher.org/api.html). Brand et al. describe how the results of analysis with the GestaltMatcher disorder prediction tool helped to solve a typical phenotype of Koolen-de Vries syndrome (KdVS) with an unusual disease-causing mutation, by revealing a high gestalt-score for KdVS17. Moreover, the direct matching of patients via similarity is possible. Thus, not only matches with already solved cases can lead to a diagnosis. For instance, Marbach et al. describe two patients with a previously unknown genetic disorder caused by the same de novo mutation in LEMD2. GestaltMatcher was used to demonstrate the similarity of the two cases to each other, which supports the assumption that they represent a new phenotype35.
Several other research groups have already achieved promising results by sharing the data. For example, the AI Bone2Gene uses our dataset of hand x-rays to determine bone age and in the future should also learn to reliably predict genetic diseases that manifest on the bone based on the image data36.
Worldwide collaboration
The FAIR database “GMDB” aims to reduce the time to diagnosis for patients with rare genetic diseases by acting as a reference database and as a training set for AI. By matching individual patients instead of classifying diseases, this approach enables clinicians to network based on the phenotype of their patients and enables collaborations around the world.
We currently collaborate with clinicians in Europe, Asia, Africa, and America who have published and analyzed their patients in the database. This globalization can also help to ensure that all ethnic groups are represented in the database and AI training sets, and thus also improve the diagnostic rate in ethnic minorities. At the same time, the community-driven effort worldwide also rapidly increases the number of patients in the GMDB.
The patient-centered approach also allows patients from patient support groups to contribute. We have now established collaborations with seven patient support groups, from which many patients have volunteered to share their photos in the database to shorten the time to diagnosis for others.
Research platform
Beyond the function of the disorder prediction tool, entire patient cohorts can also be analyzed within the Research platform in the GMDB. This research platform can therefore meet three known needs of clinician-scientists in genetics. First, it is possible to quantify the similarity of the individual patients in the cohort by generating a similarity matrix. This approach can detect clusters and see if, for example, cases with an identical variant cluster together. For example, Ebstein et al. showed that facial dysmorphism is more heterogeneous among their PSMC3 patient cohort, but more similarities were found in patients with identical variants22. Using this method in the research platform, similarity has already been quantified in 15 cohorts, of which seven have already been published15,16,18–22.
Discussion
GMDB is a web framework designed to empower the scientific community to compile and query labeled data for deep learning applications in the field of next-generation phenotyping. At the same time, it is also a modern publication organ that can be used as a searchable reference source by clinicians and researchers all over the world.
The ultimate goal is to drive research in rare genetic disorders and shorten the time to diagnosis of these disorders. This is achieved, in part, through GMDB function as a modern publication medium. It displays the latest submitted case reports in the Gallery view, which offers endless scrolling through the gallery. This feature allows phenotypes to be compared, but it can also be used to train residents and students to better and more quickly recognize these disorders based on facial dysmorphisms.
What is remarkable about this form of publication are the possibilities it offers. The previous static publication system is being replaced by a modern, more dynamic approach. Dynamic means updating a case at any time after a further patient consultation or new findings is possible. Often, specific symptoms of a disease do not all appear at the same time but develop over time. When describing a new phenotype, it is possible that further symptoms, which may also be relevant for patient surveillance, occur after publication. These cases can be better represented by a dynamic publication organ such as the GMDB.
It is well known that ethnicity can be a significant bias in detecting rare dysmorphic disorders - not only for AI but also for clinicians. Lumaka et al. also showed that European clinicians are worse at recognizing dysmorphic features in patients of African origin than in patients of European origin37. In the GMDB, clinicians get images of patients’ diseases of all ethnicities available in the database at a glance, and there is no need for a time-consuming literature search anymore. If the database is fed with images of many different ethnicities, the GestaltMatcher AI would also improve its performance for these ethnicities, as Lumaka et al. were also able to show using the example of Face2Gene37.
The research platform and the disorder prediction tool also aim to shorten the time to diagnosis. They offer the possibility to get suggestions for the underlying disorders, for direct matching of patients - with already diagnosed patients, and for matching with still unsolved patients. Tools for matching patients on genotype level with sequencing data already exist (e.g., GeneMatcher38 and DECIPHER). They are connected through the MatchMakerExchange Network API39,40. In the GMDB research platform, it is now possible to collect cohorts based on phenotype and to examine them to see whether the same or a related genetic mechanism underlies the disorder. Thus, the GMDB can also be seen as a photo version of GeneMatcher and will become a node of the MME network38,41.
Due to the interoperability of the data in the GMDB, it is possible to connect it in the MME network and may also be used for other tools. For instance, the phenotype also plays a particular role in interpreting genomic variants. It is also possible to link the GMDB with, for example, PEDIA, an AI-based approach that uses portrait images to interpret clinical exome data, improving the performance of bioinformatics pipelines for exome analysis23.
This database also addresses the problem of preprint servers like medRxiv. So far, all medical photography has to be removed because their board cannot check whether appropriate consent has been obtained. That also means the photos in case reports cannot be made accessible, and GMDB can be used as a repository for such data.
There are also several limitations of this database. Thus, the curation of data by different clinicians and even patients can also lead to different quality of image data. A standardized portrait image without irritating confounding factors, such as different facial expressions, camera angles, or even patients’ items such as glasses, can lead to a distortion of the information for the AI. An example is the problem we have encountered with images from previously published papers, which often use black bars over the patient’s eyes to prevent identification. Moreover, it is not possible to form a completely balanced data set, not only because the disorders vary so much in frequency. Other biases, such as the age and ethnicity of the patients shown in the images, can also affect the performance of AI. To positively influence these limiting factors, an attempt can be made to build up a data set that is as diverse as possible and collect images from patients of different age and ethnicity groups. Due to the dynamic form of publication, follow-up over the years is always possible.
However, the ever-growing numbers of images and cases in the database show how the GMDB can foster global collaboration and finally stop the data siloing by making data findable, accessible, reusable, and interoperable not only for humans but also for computers.
Methods
Curation process:
The curation process can be roughly subdivided into three phases. First, we started having medical students annotate cases from the literature, mainly by searching Pubmed and google scholar for publications with images of patients with facial dysmorphism and monogenic molecular diagnosis.
Second, we started to recruit solved patients from patient support groups. As we aimed to develop a patient-centered platform and strengthen patient autonomy, we collected feedback from the recruited patients during this phase to provide patients with a user-friendly experience. Patients are allowed to upload images and findings autonomously and access their data at any time. To facilitate the retrospective recruitment of patients, we have also implemented digital consent, which allows patients to decide under which conditions they consent to store their data in the database and enables direct signature online. We also further developed this feature in close cooperation with the German Smith-Magenis Syndrome patient organization Sirius e.V. to cover the patients’ requests precisely.
In the last phase, we expanded our database through international collaborations with clinicians from different continents. In the beginning, we also focused on the already solved but not yet published patients to improve the AI’s performance. However, as we progressed, more clinicians shared their unsolved cases with the scientific community. With the increasing number of database users, we also received feedback that other genetic mechanisms should be represented in the database.
Implementation and stored data
We first built an online platform with Ruby on Rails to allow users to input images and other patient data. For the back end, we set up a database by MySQL to store the patient data. An entry in GMDB consisted of a medical image such as a portrait, X-ray, or fundoscopy and machine-readable meta information such as age, ethnicity, and sex. We further collected the clinical features encoded in HPO terminology, the disease-causing mutation reported in HGVS format or ISCN nomenclature, and the test method and zygosity42–44. Despite the facial photograph, no other patient identifying information is stored in the GMDB. The diagnosed disorders will be stored based on the disorder list from OMIM45. The clinician can choose whether the patient was clinically diagnosed or molecularly diagnosed. It is also possible to add potential differential diagnoses. For cases curated from the literature, we collected the DOI and PMID as well as the contact details of the corresponding author. We then clarify whether reuse is possible while respecting intellectual property rights. The privacy settings of the patient, uploaded documents (laboratory findings or letters from the doctor), images from the unaffected family members, and information about the relationship between the cases were all stored.
Clinicians are also asked to provide their expert opinion about the distinctiveness of a phenotype: They have to score whether the medical imaging data was supportive (1), important (2), or key (3) in establishing the clinical diagnosis. Computer scientists can then use this information for the interpretation of the performance of their AIs.
GMDB started focusing on facial portraits of patients with rare monogenic diseases and is currently mainly populated by those cases, but not limited. Later in the curation process, we also annotated cytogenetic disorders with facial dysmorphism. Additional data under curation are X-rays documenting skeletal malformations and photos of the fundus of the eye documenting retinal diseases.
The GMDB architecture
The user interface of the database web service offers three main functions: The Gallery search, data download for developing NGP approaches, and the Research platform.
For diagnosing rare diseases, the face, if it shows dysmorphic features, can give clues to the underlying disease. In the case of very well-known and distinct phenotypes, experienced physicians can rely on their knowledge. Especially in the case of ultra-rare diseases this is often impossible because the physician has not seen a patient before, especially in the case of ultra-rare diseases. Here, sharing patients in publications can expand a physician’s wealth of experience. For this reason, GMDB can also be seen as a novel publication medium. The most recently published cases are displayed in the gallery view. Each case is initially represented only by the portrait image of the patient. After hovering over the image with the mouse, the affected gene and the disease are also displayed. However, the clinician can open the individual data sheet of the patient and thus also view all collected data of the patient. A clinician publishing a case can also select which uploaded images will be displayed in the Gallery.
To enable the analysis of patient cohorts with GM AI, we have also integrated a research platform into GMDB. This allows quantifying and comparing the similarities of all patients stored in the GMDB (from the literature or those publicly uploaded to the GMDB). Patients can be easily selected from the Gallery or “My Patients” section, and additional patients can be added to the research. As a result of the analysis, the Research platform provides a pairwise distance matrix, a pairwise rank matrix, and a tSNE projection46.
Data Governance and ELSI
The GestaltMatcher Database (GMDB) is hosted physically in the university hospital of Bonn and guarded by Arbeitsgemeinschaft für Gen-Diagnostik e.V. (AGD) which is a non-profit organization for genomic research. The service is free of charge for the users and is financed by membership fees of the AGD.
The GMDB is only accessible to the scientific community. To protect the patient data, the database underlies strict access control. Registration is only possible after receiving an invitation link from an existing user.
Beyond the initial purpose of GMDB as a search engine for medical imaging data for rare disorders, the framework is, in principle, also suitable for contributing health data to research as a referenceable micro-contribution. Clinicians who have already obtained appropriate informed consent to publish images can use this consent to upload their cases to GMDB. However, a case will only be visible to other users if he or she confirms this again by checking a checkbox and the patient’s image was set to be “public”.
“Public” means that the case is displayed in the Gallery and is also available to other users for further similarity comparisons and can be used for training and testing the GM AI and other AIs. A publication in the GMDB is comparable to a publication in a medical journal. So, when the case was not published before, appropriate consent for publishing the patient in GMDB is acquired. A “private” case, on the other hand, is only visible to the uploading clinician and to the patient himself. The data is included in the training set of the GMDB but is not available to other researchers. This means the case is not displayed in the Gallery and cannot be used to train other AIs. It is also possible to add cases from the existing literature. For this purpose, a PubMed ID or a DOI must be given and the paper’s corresponding author. In this way, these cases are also made reusable in the Research platform.
However, it is usually time-consuming to acquire consent in paper form. Therefore, we also implemented patient-centered digital consent. Patients can directly sign the consent in the database, for example, during a doctor’s visit on a tablet, or they can sign from home by receiving a safe invitation link via email. After creating a password, with this link, they can access their data at any time without having access to the other cases in the GMDB. The consent enables the patient to decide whether their case will only be used for training purposes of the AI or if they are also willing to show their facial image in the gallery and make it public in GMDB and sharable and visible to the database users.
After application with an IRB-approved research proposal, the dataset can be downloaded by other scientists for the training of other AIs.
Advisory Board
All the applications for acquiring the access to download GMDB public data for developing NGP approach will be reviewed by the advisory board consisted of the following coauthors: Koen Devriendt, Shahida Moosa, Christian Netzer, Martin Mücke, Christian Schaaf, Alain VERLOES, Christoffer Nellåker, Markus M. Nöthen, Gholson J. Lyon, Aleksandra Jezela-Stanek, and Karen W. Gripp. Once the majority of the board agree with the application, the applicant will be granted to the download access.
Acknowledgment
This research was supported [in part] by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health.
Footnotes
Data Availability
All data are available at https://www.gestaltmatcher.org/
References
- 1.Baird P. A., Anderson T. W., Newcombe H. B. & Lowry R. B. Genetic disorders in children and young adults: A population study. Am. J. Hum. Genet. 42, 677–693 (1988). [PMC free article] [PubMed] [Google Scholar]
- 2.Ferry Q. et al. Diagnostically relevant facial gestalt information from ordinary photos. Elife 3, e02020 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuru K., Niranjan M., Tunca Y., Osvank E. & Azim T. Biomedical visual data analysis to build an intelligent diagnostic decision support system in medical genetics. Artif. Intell. Med. 62, 105–118 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Cerrolaza J. J. et al. Identification of dysmorphic syndromes using landmark-specific local texture descriptors. in 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI) 1080–1083 (2016). doi: 10.1109/ISBI.2016.7493453. [DOI] [Google Scholar]
- 5.Wang K. & Luo J. Detecting Visually Observable Disease Symptoms from Faces. EURASIP J. Bioinform. Syst. Biol. 2016, 13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dudding-Byth T. et al. Computer face-matching technology using two-dimensional photographs accurately matches the facial gestalt of unrelated individuals with the same syndromic form of intellectual disability. BMC Biotechnol. 17, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla P., Gupta T., Saini A., Singh P. & Balasubramanian R. A Deep Learning Frame-Work for Recognizing Developmental Disorders. in 2017 IEEE Winter Conference on Applications of Computer Vision (WACV) 705–714 (2017). doi: 10.1109/WACV.2017.84. [DOI] [Google Scholar]
- 8.Liehr T. et al. Next generation phenotyping in Emanuel and Pallister-Killian syndrome using computer-aided facial dysmorphology analysis of 2D photos. Clin. Genet. 93, 378–381 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Gurovich Y. et al. Identifying facial phenotypes of genetic disorders using deep learning. Nature Medicine 25, 60–64 (2019). [DOI] [PubMed] [Google Scholar]
- 10.van der Donk R. et al. Next-generation phenotyping using computer vision algorithms in rare genomic neurodevelopmental disorders. Genet. Med. 21, 1719–1725 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsieh T.-C. et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat. Genet. 54, 349–357 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duong D. et al. Neural Networks for Classification and Image Generation of Aging in Genetic Syndromes. Front. Genet. 13, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sümer Ö. et al. Few-Shot Meta Learning for Recognizing Facial Phenotypes of Genetic Disorders. arXiv [cs.CV] (2022). [DOI] [PubMed]
- 14.Hustinx A. et al. Improving deep facial phenotyping for ultra-rare disorder verification using model ensembles. in 2023 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) (IEEE, 2023). doi: 10.1109/wacv56688.2023.00499. [DOI] [Google Scholar]
- 15.Asif M. et al. De novo variants of CSNK2B cause a new intellectual disability-craniodigital syndrome by disrupting the canonical Wnt signaling pathway. Human Genetics and Genomics Advances 100111 (2022) doi: 10.1016/j.xhgg.2022.100111. [DOI] [PMC free article] [PubMed]
- 16.Guo L. et al. KBG syndrome: videoconferencing and use of artificial intelligence driven facial phenotyping in 25 new patients. Eur. J. Hum. Genet. 1–11 (2022) doi: 10.1038/s41431-022-01171-1. [DOI] [PMC free article] [PubMed]
- 17.Brand F. et al. Next-generation phenotyping contributing to the identification of a 4.7 kb deletion in KANSL1 causing Koolen-de Vries syndrome. Hum. Mutat. 43, 1659–1665 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Kampmeier A. et al. PHIP-associated Chung-Jansen syndrome: Report of 23 new individuals. Frontiers in Cell and Developmental Biology 10, (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aerden M. et al. The neurodevelopmental and facial phenotype in individuals with a TRIP12 variant. Eur. J. Hum. Genet. (2023) doi: 10.1038/s41431-023-01307-x. [DOI] [PMC free article] [PubMed]
- 20.Averdunk L. et al. Biallelic variants in CRIPT cause a Rothmund-Thomson-like syndrome with increased cellular senescence. Genet. Med. 100836 (2023) doi: 10.1016/j.gim.2023.100836. [DOI] [PubMed]
- 21.Lyon G. J. et al. Expanding the phenotypic spectrum of NAA10-related neurodevelopmental syndrome and NAA15-related neurodevelopmental syndrome. Eur. J. Hum. Genet. (2023) doi: 10.1038/s41431-023-01368-y. [DOI] [PMC free article] [PubMed]
- 22.Ebstein F. et al. PSMC3 proteasome subunit variants are associated with neurodevelopmental delay and type I interferon production. Sci. Transl. Med. 15, eabo3189 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsieh T. C. et al. PEDIA: prioritization of exome data by image analysis. Genet. Med. 21, 2807–2814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt A. et al. Next-generation phenotyping integrated in a national framework for patients with ultra-rare disorders improves genetic diagnostics and yields new molecular findings. medRxiv (2023) doi: 10.1101/2023.04.19.23288824. [DOI] [PMC free article] [PubMed]
- 25.Hennekam R. C. M. & Biesecker L. G. Next-generation sequencing demands next-generation phenotyping. Hum. Mutat. 33, 884–886 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nellåker C. et al. Enabling Global Clinical Collaborations on Identifiable Patient Data: The Minerva Initiative. Front. Genet. 10, 611 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilkinson M. D. et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 3, 160018 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winter R. M. & Baraitser M. The London Dysmorphology Database. J. Med. Genet. 24, 509–510 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boycott K. M. et al. International Cooperation to Enable the Diagnosis of All Rare Genetic Diseases. Am. J. Hum. Genet. 100, 695–705 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoeman L., Honey E. M., Malherbe H. & Coetzee V. Parents’ perspectives on the use of children’s facial images for research and diagnosis: a survey. J. Community Genet. 13, 641–654 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiume M. et al. Federated discovery and sharing of genomic data using Beacons. Nat. Biotechnol. 37, 220–224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacobsen J. O. B. et al. The GA4GH Phenopacket schema defines a computable representation of clinical data. Nat. Biotechnol. 40, 817–820 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Köhler S. et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 49, D1207–D1217 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruszka P., Tekendo-Ngongang C. & Muenke M. Diversity and dysmorphology. Curr. Opin. Pediatr. 31, 702–707 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Marbach F. et al. The Discovery of a LEMD2-Associated Nuclear Envelopathy with Early Progeroid Appearance Suggests Advanced Applications for AI-Driven Facial Phenotyping. Am. J. Hum. Genet. 104, 749–757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rassmann S. et al. Deeplasia: prior-free deep learning for pediatric bone age assessment robust to skeletal dysplasias. bioRxiv (2023) doi: 10.1101/2023.03.07.23286906. [DOI] [PMC free article] [PubMed]
- 37.Lumaka A. et al. Facial dysmorphism is influenced by ethnic background of the patient and of the evaluator. Clin. Genet. 92, 166–171 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Sobreira N., Schiettecatte F., Valle D. & Hamosh A. GeneMatcher: A Matching Tool for Connecting Investigators with an Interest in the Same Gene. Hum. Mutat. 36, 928–930 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buske O. J. et al. The Matchmaker Exchange API: automating patient matching through the exchange of structured phenotypic and genotypic profiles. Hum. Mutat. 36, 922–927 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wohler E. et al. PhenoDB, GeneMatcher and VariantMatcher, tools for analysis and sharing of sequence data. Orphanet J. Rare Dis. 16, 365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Philippakis A. A. et al. The Matchmaker Exchange: A Platform for Rare Disease Gene Discovery. Hum. Mutat. 36, 915–921 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.den Dunnen J. T. et al. HGVS Recommendations for the Description of Sequence Variants: 2016 Update. Hum. Mutat. 37, 564–569 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Robinson P. N. et al. The Human Phenotype Ontology: A Tool for Annotating and Analyzing Human Hereditary Disease. Am. J. Hum. Genet. 610–615 (2008) doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed]
- 44.Stevens-Kroef M., Simons A., Rack K. & Hastings R. J. Cytogenetic Nomenclature and Reporting. in Cancer Cytogenetics: Methods and Protocols (ed. Wan T. S. K.) 303–309 (Springer New York, 2017). doi: 10.1007/978-1-4939-6703-2_24. [DOI] [PubMed] [Google Scholar]
- 45.Boyadjiev S. A. & Jabs E. W. Online Mendelian Inheritance in Man (OMIM) as a knowledgebase for human developmental disorders. Clin. Genet. 57, 253–266 (2000). [DOI] [PubMed] [Google Scholar]
- 46.van der Maaten L. & Hinton G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008). [Google Scholar]