Abstract
While neurological and psychiatric disorders have historically been considered to reflect distinct pathogenic entities, recent findings suggest shared pathobiological mechanisms. However, the extent to which these heritable disorders share genetic influences remains unclear. Here, we performed a comprehensive analysis of GWAS data, involving nearly 1 million cases across ten neurological diseases and ten psychiatric disorders, to compare their common genetic risk and biological underpinnings. Using complementary statistical tools, we demonstrate widespread genetic overlap across the disorders, even in the absence of genetic correlations. This indicates that a large set of common variants impact risk of multiple neurological and psychiatric disorders, but with divergent effect sizes. Furthermore, biological interrogation revealed a range of biological processes associated with neurological diseases, while psychiatric disorders consistently implicated neuronal biology. Altogether, the study indicates that neurological and psychiatric disorders share key etiological aspects, which has important implications for disease classification, precision medicine, and clinical practice.
Introduction
Neurological and psychiatric disorders rank among the leading causes of disability and mortality worldwide1. Despite their shared link to the nervous system, the disorders have generally been considered to reflect distinct pathogenic entities, as emphasized by their separate classification in the International Classification of Diseases2. The clinical division was driven by progress in brain research during the 19th and 20th century3,4. While neurology laid claim on the disorders with demonstrable neuropathology, such as Alzheimer’s disease (ALZ), psychiatry focused on the mental disorders without recognizable pathology, such as schizophrenia (SCZ). However, findings in neuroscience over the past decades, combined with clinical and epidemiological observations, have challenged the validity of this clinical distinction3–8. Various therapeutic interventions are effective in both groups of disorders, for example transcranial magnetic stimulation in Parkinson’s disease (PD) and depression9 and anticonvulsants in epilepsy and bipolar disorder (BD)10. Neurological and psychiatric disorders also share clinical features, notably cognitive impairment, a key functional determinant2,11. Additionally, debilitating psychiatric symptoms such as hallucinations, delusions and mood disturbances are prominent across neurological diseases12–14, while classical neurological symptoms such as movement abnormalities are observed in psychiatric disorders15. Moreover, environmental risk factors such as pollutants increase risk of both neurological and psychiatric illnesses8, and epidemiological studies demonstrate high comorbidity between several neurological and psychiatric disorders14,16–18, including a higher incidence of dementia among individuals with psychotic disorders18. Furthermore, in vivo neuroimaging19 and postmortem20 investigations report systematic brain abnormalities in psychiatric disorders, indicating that mental disorders have a neural basis akin to neurological disease. Altogether, the existing clinical dichotomy inadequately reflects the interconnected nature of neurological and psychiatric disorders, emphasizing the need for a more unified clinical approach3–8. However, the extent to which these conditions share an etiological basis still remains largely unclear.
The significant heritability of neurological and psychiatric disorders indicates that genomic research could provide new insights into their etiology21. This could bridge the nosological gap by forming the basis for an etiology-driven approach to disease classification, reveal novel treatment targets, and inform the development of precision medicine approaches. In recent years, genome-wide association studies (GWAS) have identified multiple common genetic variants for neurological and psychiatric disorders. Two key findings have emerged: the conditions are polygenic and genetic overlap is ubiquitous22,23. Genetic overlap has mainly been assessed by estimating pairwise genetic correlations using tools such as linkage disequilibrium (LD) score regression (LDSC)24, demonstrating that the genetic risk of psychiatric disorders is highly intercorrelated25–28. On the contrary, there are fewer pairwise genetic correlations among neurological diseases25,29,30 and between neurological and psychiatric disorders25,31. Accordingly, neurological diseases have been considered to be genetically disparate from psychiatric disorders25, in line with their clinical distinction2. However, estimates of genetic correlation are sensitive to low GWAS power and do not provide a complete picture of the genetic relationship between complex human phenotypes22,32. Importantly, they may conceal genetic overlap involving a mixture of concordant and discordant effect directions33,34, and they do not account for differences in polygenicity33, which governs the extent to which phenotypes may share genetic variants. Moreover, recent analyses using LAVA34 and MiXeR33 have demonstrated extensive genetic overlap across complex human phenotypes irrespective of the genetic correlations, along with differences in their polygenic architetures22,27,33–35. Additionally, genetic analyses have identified overlapping common variants, rare variants and expression profiles between psychiatric and neurological disorders8,31,32,36–41, indicative of a partially shared pathobiological basis.
In the present study, we aimed to provide a novel genetic perspective on the clinical distinction between neurological and psychiatric disorders. To this end, we conducted a comprehensive cross-disorder analysis of recent large-scale GWAS datasets to characterize their shared genetic architecture. We applied novel statistical tools that capture distinct forms of genetic overlap and extensive follow up analyses to link the genomic findings to biological pathways and relevant tissue and cell types.
Results
Study design
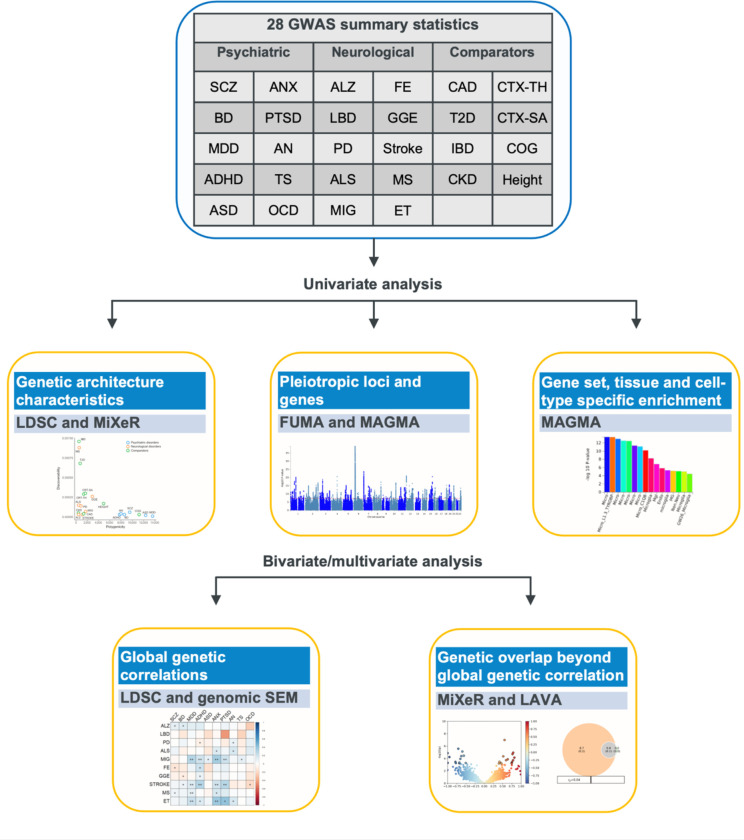
(Fig.1). We curated a collection of well-powered GWAS summary statistics, resulting in data on ten psychiatric disorders (attention-deficit/hyperactivity disorder (ADHD)42, anorexia nervosa (AN)43, autism spectrum disorder (ASD)44, anxiety disorders (ANX)45, BD46, major depressive disorder (MDD)47, obsessive-compulsive disorder (OCD)48, post-traumatic stress disorder (PTSD)49, SCZ50 and Tourette Syndrome (TS)51), and ten neurological diseases (ALZ52, amyotrophic lateral sclerosis (ALS)53, essential tremor (ET)54, Lewy body dementia (LBD)55, migraine (MIG)56, multiple sclerosis (MS)57, PD58–60, stroke61 and the epilepsy subtypes focal epilepsy (FE)62 and genetic generalized epilepsy (GGE)62). Additionally, we included GWAS data on brain-related traits (general cognitive ability (COG)63 and cortical surface area (CRT-SA) and thickness (CRT-TH)64), four somatic diseases (chronic kidney disease (CKD)65, coronary artery disease (CAD)66, inflammatory bowel disease (IBD)67 and Type 2 Diabetes (T2D)68) and height69 as comparators. All GWAS data were limited to participants of European ancestry to avoid bias due to differences in LD structure across ancestries. Ascertainment and diagnostic criteria are described in the Supplementary Note.
Fig. 1 |. Study design.
Overview of the GWAS summary statistics and analyses performed in the study. Abbreviations psychiatric disorders: Attention-deficit/hyperactivity disorder (ADHD), anorexia nervosa (AN), autism spectrum disorder (ASD), anxiety disorders (ANX), bipolar disorder (BD), major depressive disorder (MDD), obsessive-compulsive disorder (OCD), post-traumatic stress disorder (PTSD), schizophrenia (SCZ), Tourette syndrome (TS); neurological diseases: Alzheimer’s disease (ALZ), amyotrophic lateral sclerosis (ALS), Essential Tremor (ET), Lewy body dementia (LBD), migraine (MIG), multiple sclerosis (MS), Parkinson disease (PD), focal epilepsy (FE), genetic generalized epilepsy (GGE); comparators: general cognitive ability (COG), total cortical surface area (CRT-SA) and average cortical thickness (CRT-TH), coronary artery disease (CAD), chronic kidney disease (CKD), inflammatory bowel disease (IBD) and Type 2 Diabetes (T2D); methods: linkage disequilibrium score regression (LDSC), genomic structural equation modeling (genomic SEM).
After data harmonization and pre-processing of the GWAS summary data, we conducted systematic cross-trait analyses and biological interrogation (Fig. 1). We first provide information on the characteristics of the genetic architecture distinguishing each phenotype. Next, we provide an overview of the overlapping genome-wide significant loci and implicated genes. Third, we present the patterns of global genetic correlations across the phenotypes, and clusters of inter-related brain disorders. Fourth, we provide estimates of genetic overlap beyond genetic correlation. Finally, we interrogate the implicated biological pathways, tissues and cell types across the included GWAS.
Individual genetic architecture characteristics.
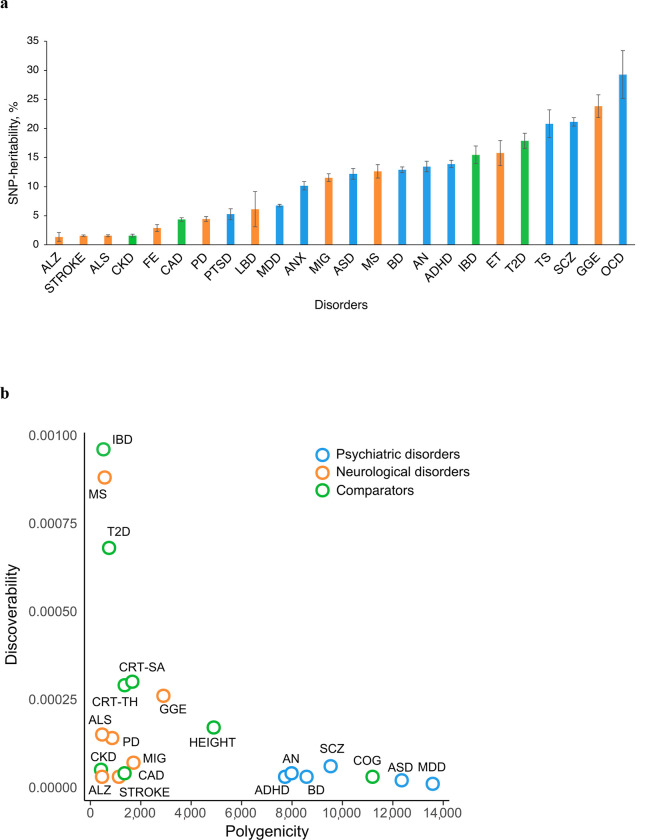
The genetic architecture of complex human phenotypes differs in terms of the heritability accounted for by single-nucleotide polymorphisms (SNP-heritability), the estimated number of SNPs influencing the phenotype (the polygenicity), and the variance of effect sizes across the associated SNPs (the discoverability)22,32. For each phenotype (trait or disorder), we estimated the SNP-heritability using LDSC70 (Table 1, Fig. 2a). On average, the estimated SNP-heritability on the liability scale was almost twice as large for psychiatric disorders (14.6%, range 5.3–29.3%) compared to neurological diseases (8.2%, range 1.4–23.8%). Regardless of disease category, however, illnesses with typical onset during childhood or adolescence had the highest estimated SNP-heritability, specifically OCD, GGE, SCZ and TS, all of putative neurodevelopmental origin. The average estimated SNP-heritability for non-brain related diseases was 9.8% (range 1.6–17.9%).
Table 1 |.
Overview of the GWAS contributing to the study
| Phenotypes | Abbreviation | Population prevalence | SNP-heritability (se.) | GWAS loci | Cases/controls | SNPs in dataset | PubMed ID |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Psychiatric disorders | |||||||
|
| |||||||
| Anorexia nervosa | AN | 0.009 | 0.135 (0.01) | 7 | 16,992/55,525 | 6,173,547 | 3130854543 |
| Anxiety disorders | ANX | 0.20 | 0.101 (0.007) | 2 | 31,977/82,114 | 4,757,986 | 3174869045 |
| Attention deficit hyperactivity disorder | ADHD | 0.05 | 0.139 (0.006) | 25 | 38,691/186,843 | 5,746,721 | 3670299742 |
| Autism spectrum disorder | ASD | 0.012 | 0.122 (0.009) | 2 | 18,381/27,969 | 6,783,844 | 3080455844 |
| Bipolar disorder | BD | 0.02 | 0.129 (0.005) | 57 | 41,917/371,549 | 6,449,398 | 3400209646 |
| Major depressive disorder | MDD | 0.15 | 0.068 (0.002) | 263 | 412,305/1,588,397 | 10,933,226 | 3404574447 |
| Obsessive-compulsive disorder | OCD | 0.025 | 0.293 (0.041) | 0 | 2,699/7,037 | 7,101,923 | 2876108348 |
| Post-traumatic stress disorder | PTSD | 0.30 | 0.053 (0.009) | 2 | 20,329/124,440 | 7,281,726 | 3159494949 |
| Schizophrenia | SCZ | 0.01 | 0.211 (0.007) | 173 | 53,386/77,258 | 6,493,147 | 3539658050 |
| Tourette syndrome | TS | 0.008 | 0.208 (0.024) | 1 | 4,819/9488 | 6,988,485 | 3081899051 |
|
| |||||||
| Neurological diseases | |||||||
|
| |||||||
| Alzheimer’s disease | ALZ | 0.05 | 0.014 (0.008) | 33 | 86,531/676,386 | 10 670 851 | 3449387052 |
| Amyotrophic lateral sclerosis | ALS | 0.0000625 | 0.016 (0.002) | 10 | 27,205/110,881 | 8 872 927 | 3487333553 |
| Essential tremor | ET | 0.01 | 0.158 (0.022) | 1 | 3,408/65,772 | 5 194 059 | 3498211354 |
| Focal epilepsy | FE | 0.003 | 0.029 (0.006) | 0 | 14,939/42,436 | 4 121 250 | medRxiv62 |
| Genetic generalized epilepsy | GGE | 0.002 | 0.238 (0.020) | 22 | 6,952/42,436 | 4 123 711 | medRxiv62 |
| Lewy body dementia | LBD | 0.001 | 0.061 (0.030) | 5 | 2,591/4,027 | 6 119 431 | 3358984155 |
| Migraine | MIG | 0.16 | 0.115 (0.007) | 35 | 48,975/540,381 | 8 484 427 | 3511568756 |
| Multiple sclerosis | MS | 0.002 | 0.126 (0.012) | 75 | 14,802/26,703 | 6 979 613 | 3160424457 |
| Parkinson’s disease | PD | 0.005 | 0.044 (0.004) | 52 | 53,858/846,380 | 8 955 805 | 2506400960, 2889205959, 3170189258 |
| Stroke | Stroke | 0.01 | 0.016 (0.001) | 24 | 73,652/1,234,808 | 6 372 181 | 3618079561 |
|
| |||||||
| Comparators | |||||||
|
| |||||||
| Cognitive ability | COG | - | 0.183 (0.006) | 201 | 269,867 | 8,002,023 | 2994208663 |
| Cortical surface area | CRT-SA | - | 0.383 (0.034) | 28 | 32,877 | 12,322,316 | 3387589164 |
| Cortical thickness | CRT-TH | - | 0.310 (0.025) | 26 | 32,877 | 12,322,316 | 3387589164 |
| Chronic kidney disease | CKD | 0.15 | 0.016 (0.002) | 21 | 41,395/439,303 | 7,767,542 | 3115216365 |
| Coronary artery disease | CAD | 0.082 | 0.044 (0.003) | 48 | 71,602/260,875 | 7,161,097 | 2871497566 |
| Inflammatory Bowel Disease | IBD | 0.0054 | 0.155 (0.015) | 117 | 25,042/34,915 | 7,969,489 | 2806790867 |
| Type 2 Diabetes | T2D | 0.10 | 0.179 (0.013) | 145 | 74,124/824,006 | 18,317,551 | 3029796968 |
| Height | Height | - | 0.371 (0.017) | 1405 | 4,080,687 | 1,265,438 | 3622439669 |
Overview of included GWAS summary datasets on psychiatric disorders, neurological diseases and comparators contributing to the study. The table displays the abbreviations used in Tables and Figures, the reported population prevalence used to estimate SNP-heritability on the liability scale for disorders, the estimated SNP-heritability calculated using LD score regression70 (observed scale for continuous traits), the total number of genome-wide significant loci after merging any physically overlapping lead SNPs (LD blocks <250 kb apart), the number of cases and controls or participants, the number of SNPs in the GWAS summary statistics and the associated PubMed IDs. Note that for PTSD, the prevalence estimate is based on the reported prevalence after trauma exposure, rather than the prevalence estimate in the whole population49.
Fig. 2 |. Individual genetic architecture characteristics.
a, SNP-based heritability on the liability scale for all disorders estimated using LD score regression70. b, Polygenicity and discoverability of all phenotypes estimated using MiXeR71, excluding GWAS with poor model fit. For full univariate MiXeR results, see Supplementary Table 1.
Using MiXeR71, we estimated the polygenicity and discoverability for each phenotype (Fig. 2b; Supplementary Table 1), except for seven GWAS displaying poor model fit due to insufficient statistical power (ANX, PTSD, TS, OCD, FE, ET and LBD). The polygenicity estimates for all psychiatric disorders (7,725 (SD=349) – 13,582 (SD=387)) and COG (11,195 (SD=369)) exceeded those of neurological diseases (464 (SD=43) – 2,898 (SD=220)), somatic disorders (423 (SD=55) – 1,358 (SD=85)), height (4,894 (SD=90)) and cortical imaging measures (1,361 (SD=100) – 1,666 (SD=125)). For example, the least polygenic psychiatric disorder ADHD (7,725 (SD=349)) was estimated to be influenced by ~2.7 times more genetic variants than the most polygenic neurological disease GGE (2,898 (SD=220)). In line with prior work22,71, the most polygenic phenotypes were characterized by relatively low discoverability, indicating a larger fraction of trait-influencing variants with smaller effect sizes. In Supplementary Fig. 1, we present GWAS power plots displaying the estimated fraction of SNP-heritability explained by genome-wide significant SNPs as a function of sample size, demonstrating that the discovery trajectories for most of the GWAS are still in the early stages, except for height.
Overlapping genome-wide significant loci and genes.
We estimated the number and fraction of significantly associated loci and genes shared across the phenotype categories (Table 2) with results for each phenotypic pair provided in Supplementary Tables 2–3. For each GWAS, we identified genome-wide significant loci according to the FUMA protocol72. We subsequently grouped physically overlapping loci, resulting in a total number of 1,988 distinct loci. Of these, 441 loci were linked to psychiatric disorders and 227 loci to neurological diseases. In total, 41 loci were overlapping between psychiatric and neurological disorders, constituting 9.3% and 18.1% of the total number of loci linked to these categories, respectively. Additionally, we mapped GWAS associations to protein-coding genes using MAGMA73, yielding a total number of 7,829 distinct genes. Of these, 796 genes were linked to psychiatric disorders and 497 to neurological diseases. A total of 51 genes were shared between psychiatric and neurological disorders, constituting 6.4% and 10.3% of the total number of genes linked to these categories, respectively. As expected, the pleiotropy across genome-wide significant loci and genes were largely driven by GWAS power, warranting cautious interpretation of these results. Most of the pleiotropy for psychiatric disorders were observed for SCZ and MDD, while the neurological GWAS were more evenly powered.
Table 2 |.
Overview of pleiotropic loci and genes linked to psychiatric or neurological diseases at the genome-wide significant level
| Loci |
Genes |
|||
|---|---|---|---|---|
| Psychiatric disorders | Count | Fraction (%) | Count | Fraction (%) |
|
| ||||
| Total | 441 | 796 | ||
| Pleiotropic with | ||||
| Psychiatric disorders | 60 | 13.6% | 148 | 18.6% |
| Neurological diseases | 41 | 9.3% | 51 | 6.4% |
| Cognitive ability | 64 | 14.5% | 136 | 17.1% |
| Cortical thickness and surface area | 16 | 3.6% | 23 | 2.9% |
| Somatic disorders | 46 | 10.4% | 72 | 9.0% |
| Height | 162 | 36.7% | 463 | 58.2% |
|
| ||||
| Neurological diseases | ||||
|
| ||||
| Total | 227 | 497 | ||
| Pleiotropic with | ||||
| Psychiatric disorders | 41 | 18.1% | 51 | 10.3% |
| Neurological diseases | 16 | 7.0% | 16 | 3.2% |
| Cognitive ability | 19 | 8.4% | 35 | 7.0% |
| Cortical thickness and surface area | 10 | 4.4% | 19 | 3.8% |
| Somatic disorders | 53 | 23.3% | 58 | 11.7% |
| Height | 109 | 48.0% | 256 | 51.5% |
Count and fraction of pleiotropic genome-wide significant loci and genes linked to psychiatric and neurological disorders across the phenotype categories. After identifying genome-wide significant loci, physically overlapping loci were merged into grouped loci. Protein-coding genes were identified using MAGMA73. Across all phenotypes, 1,988 grouped loci and 7,829 genes were identified. The extended MHC-region (chr6: 25–37 Mb) was excluded from these analyses. Supplementary Tables 2–3 present the number of overlapping loci and genes for each pair of phenotypes.
Global genetic correlations.
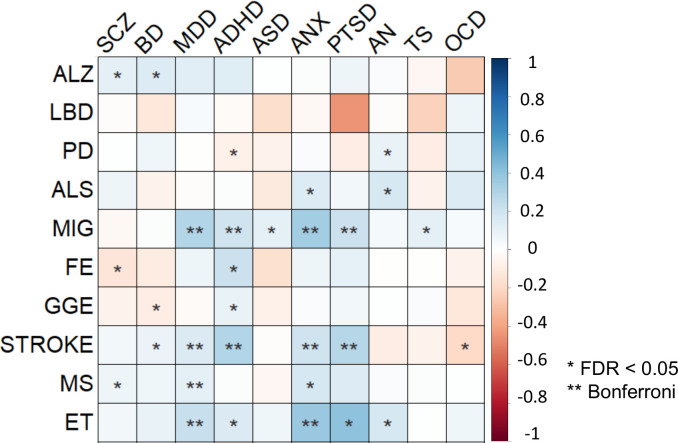
Using bivariate LDSC24, we estimated the global pairwise genetic correlations across all phenotypes (Supplementary Fig. 2; Supplementary Table 4). Our results corroborate prior findings of highly intercorrelated genetic risk among psychiatric disorders25–28,31. In total, 40 out of 45 genetic correlations among psychiatric disorders reached significance (FDR < 0.05). In comparison, 12 out of 45 correlations among neurological diseases reached significance (FDR < 0.05). As recently demonstrated29,30, the neurodegenerative disorders ALS, LBD, ALZ and PD formed a cluster of correlated disorders. Additionally, ET was correlated with both PD (rg=0.31, p=1.80×10−7) and MIG (rg=0.17, p=3.90×10−3), FE was correlated with stroke (rg=0.30, p=1.40×10−3), ALS (rg=0.32, p=7.10×10−3) and the other epilepsy subtype GGE (rg=0.61, p=8.04×10−17), while PD was negatively correlated with both MIG (rg=−0.08, p=1.40×10−2) and stroke (rg=−0.10, p=1.57×10−2).
In total, 30 out of 100 genetic correlations between neurological and psychiatric disorders reached significance at FDR < 0.05 (rg range: −0.19 – 0.40; Fig. 3), adding further evidence that genetic risk transcends the categorical boundary between these disorders. We found that MIG, ET and stroke were positively correlated with several psychiatric disorders, in particular MDD, ADHD, ANX and PTSD. The same psychiatric disorders were also correlated with CAD, consistent with a connection between mental disorders and cardiovascular illness74. However, neither MIG or ET were significantly correlated with any somatic comparator or stroke, suggesting that their shared genetic effects with psychiatric disorders relate to other aspects. Furthermore, MS was significantly correlated with ANX (rg=0.17, p=6.00×10−4), MDD (rg=0.11, p=1.16×10−5) and SCZ (rg=0.07, p=1.02×10−2). All of these disorders were positively correlated with the immune-mediated disease IBD, indicating a common link to immunity. We also observed significant correlations between ALZ and both BD (rg=0.14, p=1.81×10−2) and SCZ (rg=0.11, p=1.14×10−2), in line with the comorbidity between dementia and psychosis12,18,31. Finally, we observed significant correlations between several comparators and psychiatric and neurological disorders, indicating body-wide effects of the involved genetic variants (Supplementary Note).
Fig. 3 |. Genetic correlations.
Global pairwise genetic correlations across neurological and psychiatric disorders estimated using linkage disequilibrium score regression24. One asterisk denotes statistical significance at FDR < 0.05, two asterisks denote statistical significance after Bonferroni correction. The color denotes the magnitude and direction of correlation.
Genetic covariance structure.
Applying genomic SEM75, we modeled the genetic covariance structure of neurological and psychiatric disorders. The model did not successfully converge for neurological diseases, likely due to insufficient correlation structure. We then leveraged a recently established factor model for psychiatric disorders28, which specified four latent factors that fit the data well (Supplementary Table 5). The first factor consisted of compulsive disorders (AN, OCD and TS), the second factor of psychotic disorders (SCZ and BD), the third factor was characterized by neurodevelopmental disorders (ASD and ADHD) as well as PTSD, MDD and TS, while the fourth factor consisted of internalizing disorders (MDD, ANX and PTSD). We then conducted confirmatory factor analyses (CFAs) and estimated whether any of the neurological diseases correlated with the psychiatric factors. After Bonferroni-correction (0.05/40 = 1.25×10−3), four neurological diseases (MIG, stroke, MS and ET) were found to significantly correlate with a psychiatric factor, indicating shared genetic covariance structure with psychiatric disorders. Specifically, MIG was positively correlated with the neurodevelopmental (rg=0.23, p=7.24×10−11) and internalizing (rg=0.31, p=2.98×10−31) factors, stroke was negatively correlated with the compulsive factor (rg=−0.20, p=6.95×10−4), but positively correlated with the neurodevelopmental (rg=0.28, p=5.66×10−11) and internalizing (rg=0.16, p=2.43×10−5) factors, while both MS (rg=0.15, p=2.12×10−6) and ET (rg=0.30, p=1.62×10−7) were positively correlated with the internalizing factor. No neurologic disease was significantly correlated with the psychotic factor.
Genetic overlap beyond global genetic correlations.
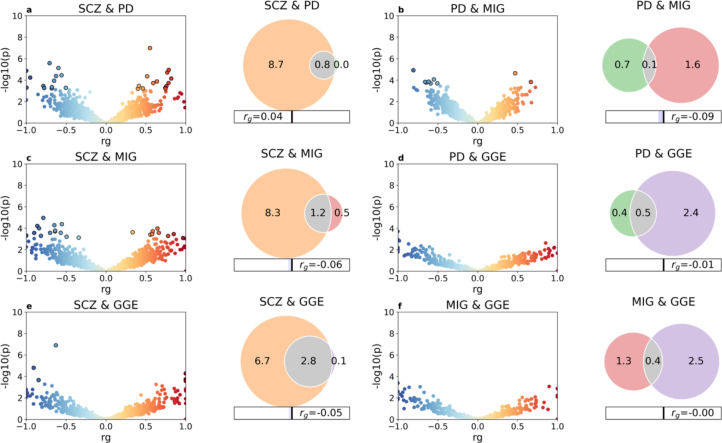
Using bivariate MiXeR33, we estimated the unique and overlapping genetic architectures between pairs of phenotypes (Supplementary Table 6). Unlike LDSC24, MiXeR can detect genetic overlap regardless of the global genetic correlations33. Corroborating recent work35, we found extensive genetic overlap across all psychiatric disorders, with a minor proportion of disorder-specific variants (Supplementary Fig. 3). MiXeR indicated varying degrees of genetic overlap between neurological diseases, with smaller proportions of shared risk compared to that observed among psychiatric disorders, suggesting that neurological diseases are more genetically distinct from each other. Despite disparate polygenicity estimates, we observed widespread genetic overlap between neurological and psychiatric disorders. This constituted a larger proportion of the genetic architectures of neurological diseases given their smaller polygenicity estimates relative to psychiatric disorders. As an example, MiXeR estimated pronounced genetic overlap between SCZ and neurological diseases PD, GGE and MIG, despite absent genetic correlations, indicative of a balanced mix of concordant and discordant effects among the shared variants (Fig. 4). Almost all genetic variants linked to PD and GGE and 70% of those linked to MIG were estimated to also influence risk of SCZ, while the overlap represented less than 30% of the SCZ variants.
Fig. 4 |. Genetic overlap beyond global genetic correlations.
LAVA local correlations and MiXeR-modeled genome-wide genetic overlap for selected disorders schizophrenia (SCZ), Parkinson’s disease (PD), migraine (MIG) and genetic generalized epilepsy (GGE). To the left, volcano plots of local genetic correlation coefficients (rho) against −log10 p-values for each pairwise analysis per locus estimated using LAVA34 (See Supplementary Table 7 for full results). Dots encircled in black represent significantly correlated loci after false discovery rate correction. To the right, Venn diagrams showing the number (in thousands) of shared and disorder-specific variants and the global genetic correlation (rg) estimated using MiXeR33 (See Supplementary Fig. 3 and Supplementary Table 6 for full results). The total polygenicity for each disorder represents the estimated number of variants required to explain 90% the SNP-based heritability.
Applying LAVA34, we calculated the local genetic correlations across 2,495 genomic regions between all pairs of phenotypes. We performed local genetic correlation tests at loci where both phenotypes had heritability estimates significantly different from zero, and corrected for multiple testing using FDR. Corroborating the MiXeR findings, LAVA estimated multiple significantly correlated loci across most pairs of phenotypes, including between neurological and psychiatric disorders (Supplementary Table 7). As observed for locus and gene pleiotropy at the genome-wide significance level (Supplementary Tables 2 and 3), the number of LAVA local correlations largely reflected GWAS power. Consistent with the MiXeR findings, LAVA estimated correlated loci between SCZ and PD (14 positively correlated and 13 negatively correlated loci), GGE (six positively correlated and six negatively correlated loci), and MIG (10 positively correlated and 15 negatively correlated loci), adding further support for a shared genetic basis (Fig. 4).
Tissue, cell-type and gene-set enrichment analyses.
Finally, we compared GWAS enrichment with specific tissues, cell types and gene sets (Table 4), leveraging RNA sequencing data from the Genotype-Tissue Expression (GTEx) project76, single-cell RNA sequencing datasets from the developing and adult human brain77, and predefined Gene Ontology gene sets implemented in FUMA72. We performed Bonferroni correction for the number of tested items in each analysis.
The analyses revealed a range of neural and somatic biological associations associated with neurological diseases. As previously shown52,57,77, ALZ and MS were both significantly associated with immune-enriched tissues, microglia and immunological pathways, implying a key role of the immune system. Additionally, ALZ was associated with amyloid-beta related processes. Risk genes for PD were significantly associated with various neurobiological processes, particularly concerning synaptic vesicles, and were specifically upregulated in the substantia nigra58, central to PD pathogenesis. Risk genes for GGE were significantly associated with both GABAergic and excitatory neurons62, in line with hyperexcitability being the main pathophysiological feature of epilepsy, but were not associated with any tissue or gene set. Stroke was significantly associated with one gene set only, ‘fibrinogen’, an established stroke risk factor involved in clot formation78. Risk genes for LBD were linked to lipid metabolism.
Corroborating previous work26,28,32,42,46,47,50, GWAS on psychiatric disorders consistently implicated neuronal biology. Risk genes for ADHD, MDD and SCZ were all upregulated in brain tissue, implicated neurobiological processes and neuronal cell types. MDD was also associated with oligodendrocyte progenitor cells. BD risk genes were significantly associated with both GABAergic and excitatory neurons, but not with any tissue or gene set. ANX was significantly associated with several neurobiological pathways, while risk genes for AN were significantly downregulated in specific brain tissues.
Apart from COG, no comparator was significantly associated with neurons. COG and CRT-TH were the only comparators whose genes were significantly upregulated in brain tissue. Further results are described in the Supplementary Note. Full results are provided in Supplementary Tables 8 and 9 and Supplementary Figs. 4 and 5.
Discussion
In the present study, we elucidate the shared genetic architecture of neurological and psychiatric disorders, indicating that they partly share a genetic basis. Overall, the results represent a major advance in our understanding of the common genetic variation underlying brain-related disorders, suggesting that a large set of genetic variants influence a range of pathogenic processes, wherein disorder specificity is determined by the distribution of effect sizes. While the shared genomic components suggest that neurological and psychiatric disorders partially share molecular genetic mechanisms, a more central role of neuronal biology was implicated in psychiatric disorders, while neurological diseases implicated a larger variety of biological processes. Altogether, the findings are consistent with accumulating evidence indicating that neurological and psychiatric disorders share key etiological aspects, contrasting their clinical distinction.
To compare the genetic basis of neurological and psychiatric disorders, we analyzed GWAS summary data from 20 major disorders, representing the largest cross-disorder analysis on this subject to date (Table 1). Moreover, the application of statistical methods with different modelling assumptions and different techniques for measuring genetic overlap allowed us to interrogate their genetic relationship in a more comprehensive manner than previous work25. In the univariate analysis, psychiatric disorders were more polygenic than neurological diseases. Polygenicity indicates the number of additive genetic effects that may combine to yield increased trait susceptibility, providing a measure of genetic architecture complexity and possibly heterogeneity22,79. While both neurological and psychiatric disorders are multifactorial and clinically heterogenous, the higher levels of polygenicity of psychiatric disorders is consistent with a hypothesis that multiple causal pathways may converge on the same mental illness, while fewer causal pathways may underlie neurological diseases. Despite similar twin-based heritability estimates across neurological and psychiatric disorders21, the SNP-based heritability estimates appeared to negatively correlate with typical onset of illness, regardless of disease category. This contrasts the theoretical expectation that common genetic variants might explain more variance in late-onset disorders, given their weaker impact on reproductive fitness, thereby reducing selective pressure80. However, current methodology may inappropriately account for the effect of age and large-effect variants such as APOE variants, warranting cautious interpretation of SNP-heritability estimates for late-onset disorders81.
Expanding upon previous work based on less powerful GWAS25,31,36–40, we demonstrate widespread genetic correlations between neurological and psychiatric disorders, most of which were positive (Fig. 3). The results indicate that neurological and psychiatric disorders partly exist on genetic continua, providing new insights into their genetic relationship. Importantly, the shared genetic components may map onto overlapping biological aspects that could be targeted therapeutically. The pattern of correlations was not uniform across disorders, with clusters of disorders being more correlated with each other. Notably, both MIG and ET were positively correlated with several psychiatric disorders, in particular the internalizing disorders ANX, MDD and PTSD, consistent with their extensive psychiatric comorbidities16,17. On a cautious note, however, the GWAS on both MIG and ET were largely based on self-reports54,56. Although self-reported and clinically ascertained cases are shown to strongly correlate47,56,58, we cannot exclude the possibility that some self-reports were based on underlying mental illness with somatoform symptomatology. The findings nevertheless emphasize the interconnected nature of these disorders, and may motivate further trialing of psychotherapy or antidepressants, which show beneficial effects for MIG prevention82.
Beyond genetic correlations, we observed a more pervasive degree of genetic overlap across neurological and psychiatric disorders, involving a mixture of concordant and discordant effect sizes (Supplementary Fig. 3). As an example, MiXeR33 indicated that a pronounced fraction of the genetic risk underlying PD, GGE and MIG overlaps with SCZ, despite absence of global genetic correlations (Fig. 4). The findings align with the discovery of multiple correlated genomic regions between these disorders using LAVA34 (Fig. 4), and shared loci detected below the genome-wide significance level36,37,40. The emerging results indicate a substantial genetic basis shared across neurological and psychiatric disorders, in which multiple common genetic variants impact risk of several disorders, but with divergent effect sizes. Accordingly, a given genetic variant may influence numerous biological pathways, each of which may be differently involved in the pathogenesis underlying distinct brain disorders. This is consistent with recent findings of highly distributed genetic effects across brain morphological, cognitive and personality traits83–86, indicating that multiple genetic variants with small effects affect the fine-tuning and dynamic interplay across a range of neural and behavioral systems. From a clinical perspective, the findings are highly relevant to the potential implementation of genomic precision medicine in psychiatry and neurology. Integrating genomic data across multiple disorders in a multivariate prediction framework may aid in identifying individuals who are more likely to experience comorbid symptoms, either endogenously or due to adverse treatment effects, which is currently an unmet clinical need.
The study has some limitations. The analysis was restricted to individuals of European ancestry, given the lack of well-powered GWAS on other ancestries. Trans-ancestral follow-up studies are required to assess the generalizability of these results. The present analysis was based on common genetic variants, but rare variants likely impact the comorbidity between neurological and psychiatric disorders as well. For example, rare variants are jointly associated with epilepsy, SCZ and ASD32. Our study was limited by bias inherent to the original GWAS, including population stratification and ascertainment procedures. As noted above, misdiagnosis could affect the results, in particular with more common disorders like anxiety or depression. However, prior extensive simulations did not find that misdiagnosis could explain the magnitude of correlated risk across psychiatric disorders25,26. Comorbid illness may also bias the assessment of genetic overlap, warranting more deeply phenotyped cohorts to assess differential genetic overlap among clinical subtypes. The results may be affected by LD, whereby a causal variant may be correlated with multiple nearby variants, leading to spurious pleiotropy. To address this, statistical fine-mapping follow-up studies are needed. Finally, there was uneven power among the included GWAS, which limit the value of cross-disorder comparison at the present stage. This particularly affects the biological interpretation of the mapped genes, which only represent a minor fraction of the genetic risk architectures underlying these disorders. As GWAS samples get larger, cross-trait analyses based on more diverse datasets, additional disorders, and specific subtypes, should be conducted.
In conclusion, by leveraging recent large-scale GWAS datasets and novel statistical tools, we demonstrate that neurological and psychiatric disorders partly share genetic etiology and biological associations related to the brain. Incorporating these complex and interconnected illnesses into a more unified framework may help accelerate progress in these fields, lead to a more coherent and productive clinical approach3–8, and promote precision medicine implementation.
Methods
GWAS summary statistics.
We collated large-scale GWAS summary statistics based on available sample sizes and the quality of the phenotyping procedures (Table 1; See Supplementary Note for description of each GWAS dataset). All individuals included in the analysis were of European ancestry. Informed consent was obtained from all participants in the respective GWAS. The Regional Committee for Medical Research Ethics – Southeast Norway evaluated the current protocol and found that no additional institutional review board approval was necessary as no individual data were used. All GWAS datasets were derived from existing GWAS except the two datasets on total cortical surface area and average cortical thickness, which were generated from the UK Biobank under accession number 27412, after excluding all individuals with neurological and psychiatric disorders (Supplementary Note). For epilepsy, we chose to include its two main subtypes, focal epilepsy and genetic generalized epilepsy (GGE), rather than including the phenotype ‘all epilepsies combined’, due to the substantial differences in the genetic risk architectures underlying these two subtypes62, as emphasized by their differences in estimated SNP-heritability (2.9% vs 23.8%, respectively; Table 1). Before commencing analysis, all GWAS summary statistics underwent uniform quality control and were harmonized and preprocessed into a consistent file structure with a common reference for positions, rsIDs and effect alleles using the v1.6.0 cleansumstats pipeline87.
Genome-wide significant loci.
For each GWAS, we defined independently associated genomic loci using FUMA72. First, we identified independent significant SNPs with a genome-wide significant p-value (5×10−8) that were independent from each other at r2<0.60. LD r2 values were obtained from the 1000 Genomes Project European-ancestry haplotype reference panel88. The borders of the loci were defined by identifying all candidate SNPs in LD (r2≥0.6) with one of the independent significant SNPs in the locus. All loci less than 250kb apart were merged.
To evaluate locus pleiotropy, we used the procedure previously applied by Watanabe et al. (2019)22. After identifying genome-wide significant loci for each phenotype, we grouped any physically overlapping loci across all phenotypes. A grouped locus could therefore contain more than one independent locus for a given phenotype if several loci were combined (i.e., loci A and C could both overlap with locus B but not with each other, but they would be grouped into one locus resulting in a continuous genomic region). Each grouped locus was then assigned to their specific phenotypes and the following categories: psychiatric disorders, neurological diseases, COG, cortical MRI measures (CRT-SA and CRT-TH), somatic diseases and height. We then determined the number and fraction of grouped loci shared across categories and between all pairs of phenotypes. The extended MHC-region (chr6: 25–37 Mb) was excluded from this analysis due to its complex LD structure.
MAGMA gene, gene-property and gene-set analysis.
For each GWAS dataset, we identified significantly associated protein-coding genes and gene-sets using MAGMA (v1.08)73 as implemented in FUMA72 with default settings, using the SNP-wise mean model and the European 1000 Genomes reference cohort phase 3 as reference panel. The input SNPs were mapped to 20,260 protein-coding genes, excluding the extended MHC-region (chr6: 25–37 Mb). Gene boundaries were expanded to 35 kb upstream and 10 kb downstream to include probable regulatory regions outside the transcribed region89. Genes were considered significant if the p-value was less than 0.05 after Bonferroni correction for the number of tested genes (0.05/20,260 = 2.47×10−6). MAGMA calculates an association p-value for each gene based on the aggregate of all SNPs mapped to each gene, accounting for gene-size, number of SNPs in a gene and LD between markers. We then carried out competitive gene-set analysis based on the identified genes in each phenotype. Specifically, we focused on the Gene Ontology gene set terms: biological processes (7,350 gene sets), cellular components (1,001 gene sets) and molecular functions (1,645 gene sets) obtained from MsigDB version 7.090. Gene sets were considered significant if the p-value was <0.05 after Bonferroni correction for the number of tested gene sets in each category (0.05/7,350 = 6,80×10−6, 0.05/1,001 = 5,00×10−5, 0.05/1,645 = 3.04×10−5, respectively).
Based on the gene-based results above, we carried out tissue specific expression analysis in 54 adult tissue types based on RNA sequencing data GTEx v.876 implemented in FUMA72. Tissues were considered significant if the P value was less than 0.05 after Bonferroni correction for 54 tissues. For cell type specificity analysis, we tested for enrichment in 24 single-cell RNA sequencing data sets from the developing and adult human brain available in FUMA using MAGMA gene-property analysis77. The specific datasets were: Allen_Human_LGN_level191, Allen_Human_LGN_level291, Allen_Human_MTG_level191, Allen_Human_MTG_level291, DroNc_Human_Hippocampus92, GSE104276_Human_Prefrontal_cortex_all_ages93, GSE104276_Human_Prefrontal_cortex_per_ages93, GSE67835_Human_Cortex94, Linnarsson_GSE101601_Human_Temporal_cortex95, Linnarsson_GSE76381_Human_Midbrain96, PsychENCODE_Developmental97 PsychENCODE_Adult97, and GSE168408_Human_Prefrontal_Cortex datasets from level 1 to 2, spanning six developmental stages: fetal, neonatal, infancy, childhood, adolescence and adult98. In the cell type specific analysis, systematic stepwise conditional analysis was performed within datasets to ensure that complex batch effects did not lead to false positives, as well as Bonferroni correction for multiple testing of 379 cell types (0.05/379 = 1.30×10−4).
All statistical tests conducted using MAGMA were one sided. We did not perform additional correction for multiple testing across the 28 phenotypes, since the aim of analysis was not to determine which of the phenotypes a specific gene, gene-set, tissue or cell type was associated with, but to explore group level patterns of shared associations across the phenotypes.
SNP-heritability and global genetic correlations.
Using LDSC70, we estimated the SNP-based heritability in the liability scale for each disorder, using reported population prevalence estimates (Table 1), and the SNP-based heritability on the observed scale for the continuous traits. LDSC distinguishes confounding from polygenicity by regressing the association statistics of SNPs on their LD scores70. All analyses were based on HapMap 3 SNPs only, with the MHC region (chr6: 25–34 Mb) excluded. Precalculated LD scores from the European 1000 Genomes reference cohort were used (https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2). Additionally, we used the bivariate extension of LDSC24 to estimate the global genetic correlations, i.e. the covariance in the SNP-heritability, between all pairs of phenotypes. Adjusting for the number of traits tested, we applied both the FDR method of Benjamini-Yekutieli99 given the dependence between the tests and Bonferroni-correction.
Genomic SEM.
Genomic SEM75 models the multivariate genetic architecture across traits and may uncover broad latent factors underlying genetic correlations, reveal clusters of correlated traits, and determine how latent factors correlate with each other. Genomic SEM analysis typically involves two steps, exploratory factor analysis (EFA) and CFA. While EFA identifies the most appropriate number of latent factors, assigning factors to specific traits given sufficiently high loadings to those factors, the resulting model is validated using CFA. In the present study, we first aimed to conduct a joint genomic SEM model of psychiatric and neurological disorders. However, the model did not successfully converge for neurological diseases, likely due to insufficient correlation structure across the neurological diseases. Instead, we applied a recently established factor model for psychiatric disorders28, and conducted separate CFAs for each individual neurological disease and the group of psychiatric disorders and estimated whether any of the neurological diseases significantly correlated with the psychiatric factors. We performed Bonferroni correction for the number of tested items (N=10*4, testing loadings for 10 neurological diseases onto 4 psychiatric factors). The CFA was conducted using all available variants, without partitioning genetic variants into even and odd chromosomes, as the analysis only included the CFA stage using a set of predefined models. The goodness of fit was evaluated through standard metric (AIC, CFI, SRMR, presented in Supplementary Table 5), showing appropriate model fit.
Univariate and bivariate MiXeR analysis.
We first applied univariate MiXeR71 analysis to each GWAS summary dataset to estimate the proportion of causally associated genetic variants from a reference panel (the polygenicity) and the variance of effect size per causal variant (the discoverability) using maximum likelihood estimation, and the GWAS sample size necessary to discover genetic variants that explain 90% of SNP-heritability of each phenotype. We applied a threshold of 90% SNP-heritability to avoid extrapolating model parameters into variants with infinitesimally small effects. MiXeR is based on a Gaussian mixture model, assuming that a given GWAS summary dataset can be modeled as a “mixture” of pre-defined components with causal and non-causal variants, each with its own Gaussian (normal) distribution. MiXeR incorporates the effects of LD structure, minor allele frequency, GWAS sample size, genomic inflation due to cryptic relatedness, and sample overlap (in the bivariate extension). Before analysis, the MHC region was excluded from all GWAS, while the chromosome 19 was in addition excluded from ALZ due to the strong effects of the APOE region52 and complicated LD that biases the estimates of polygenicity.
Informed by the model parameters from univariate MiXeR for each phenotype, MiXeR constructs a bivariate mixture model for pairs of phenotypes, in which a mixture of four bivariate Gaussian components is modeled: variants influencing one phenotype only, variants influencing both phenotypes, and variants that are not associated with either phenotype. Bivariate MiXeR estimates the polygenicity of the shared component irrespective of effect directions and correlation of effect sizes. Additionally, MiXeR estimates the genetic correlation of shared variants, and the global genetic correlation. Model fit is evaluated by calculating the difference between the Akaike information criterion (AIC) for best-fitting MiXeR estimates and reference models. Positive AIC differences are interpreted as evidence that the best-fitting MiXeR estimates are distinguishable from the reference model. For univariate MiXeR, an “infinitesimal model” in which all variants are assumed to be ‘causal’ is used as the reference. For bivariate MiXeR, AIC differences are calculated by comparing the best-fitting model to minimum possible overlap, constrained by rg, and maximum possible overlap, constrained by the polygenicity of the least polygenic trait. We provide conditional Q-Q plots and log-likelihood plots to visualize the stability of the fitness procedure.
Estimating local genetic correlations using LAVA.
For all pairs of phenotypes, we applied LAVA (v1.3.8) to estimate local genetic correlations across 2,495 semi-independent genetic loci of approximately equal size (~1 Mb). LAVA accounts for potential sample overlap using LDSC70. After computing local SNP-heritability estimates for each phenotype, we conducted pairwise local genetic correlation analysis for all loci with local SNP-heritability significantly different from zero. We applied FDR correction to account for multiple comparisons. The statistical tests conducted were all two sided.
Supplementary Material
Table 3 |.
Summary of tissue and cell type specificity analyses
| Tissue specific associations | Human brain cell type associations | |
|---|---|---|
|
| ||
| Neurological diseases | ||
|
| ||
| ALZ | Upregulated in the spleen, terminal ileum of the small intestine and whole blood | Microglia |
| GGE | - | GABAergic and excitatory neurons |
| MS | Upregulated in EBV-transformed lymphocytes, the spleen, whole blood and the terminal ileum of the small intestine | Microglia and endothelial cells |
| PD | Upregulated in the substantia nigra | - |
|
| ||
| Psychiatric disorders | ||
| ADHD | Upregulated in the cerebral cortex | GABAergic and excitatory neurons |
| AN | Downregulated in the putamen and hippocampus | - |
| BD | - | GABAergic and excitatory neurons |
| MDD | Upregulated in multiple brain tissues | GABAergic neurons and oligodendrocyte progenitor cells |
| SCZ | Upregulated in multiple brain tissues | GABAergic and excitatory neurons |
| Comparators | ||
|
| ||
| COG | Upregulated in multiple brain tissues Downregulated in the renal cortex | GABAergic and excitatory neurons |
| CAD | - | Vascular cells |
| CRT-SA | - | Oligodendrocytes, astrocytes, stem cells, and microglia |
| CRT-TH | Upregulated in the cerebellar hemisphere | - |
| IBD | Upregulated in the spleen and whole blood Downregulated in multiple central nervous tissues | Microglia and endothelial cells |
| Height | Up- and downregulated in multiple tissues body-wide | Vascular cells, endothelial cells, astrocytes, oligodendrocytes, and microglia |
| T2D | Downregulated in multiple central nervous tissues | Vascular cells, endothelial cells, astrocytes, oligodendrocytes, and microglia |
Summary of tissue and cell type specific analyses using FUMA72,77, only significant associations after Bonferroni correction are described. Tissue analysis was based on GTEx data76, while cell type analysis was based on 24 single-cell RNA sequencing data sets from the developing and adult human brain77. For full results, see Supplementary Results, Supplementary Figures 4–5, Supplementary Tables 8–9.
Acknowledgments
We thank the research participants, employees and researchers of the UK Biobank, 23andMe, Inc., MVP, iPSYCH and the many consortia for making this research possible. This research has been conducted using the UK Biobank Resource under Application Number 27412. Moreover, the research has been conducted using and data from MVP, under dbGap accession number phs001672. This work was partly performed on the TSD (Services for Sensitive Data) facilities, owned by the University of Oslo, operated and developed by the TSD service group at the University of Oslo, IT-Department (USIT). Computations were also performed on resources provided by UNINETT Sigma2—the National Infrastructure for High Performance Computing and Data Storage in Norway. We gratefully acknowledge support from the American National Institutes of Health (NS057198, EB000790, 1R01MH124839, R01MH120219, RF1AG073593), the Research Council of Norway (RCN) (229129, 213837, 324252, 300309, 273291, 223273, 248980, 326813), the South-East Norway Regional Health Authority (2019–108, 2022–073), KG Jebsen Stiftelsen (SKGJ-MED-021), EAA grant (#EEA-RO-NO-2018–0573). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 847776 and 964874 and 801133 (Marie Sklodowska-Curie grant agreement).
Footnotes
Competing interests
O.A.A. has received speaker’s honorarium from Lundbeck, Sunovion and Janssen and is a consultant for Cortechs.ai. A.M.D. is a founder of and holds equity interest in CorTechs Labs and serves on its scientific advisory board. He is also a member of the Scientific Advisory Board of Healthlytix and receives research funding from General Electric Healthcare (GEHC). The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. Remaining authors have nothing to disclose.
Code availability
Cleansumstats pipeline (https://github.com/BioPsyk/cleansumstats)
FUMA (https://fuma.ctglab.nl/)
Genomic SEM (https://github.com/MichelNivard/GenomicSEM)
LAVA (https://github.com/josefin-werme/LAVA)
LDSC (https://github.com/bulik/ldsc)
MAGMA (https://ctg.cncr.nl/software/magma)
MiXeR (https://github.com/precimed/mixer)
PLINK (https://www.cog-genomics.org/plink/2.0/)
Regenie (https://rgcgithub.github.io/regenie)
Data availability
All data are publicly available or available on request.
References
- 1.Whiteford H.A., Ferrari A.J., Degenhardt L., Feigin V. & Vos T. The global burden of mental, neurological and substance use disorders: an analysis from the Global Burden of Disease Study 2010. PLoS One 10, e0116820 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. International Classification of Diseases for Mortality and Morbidity Statistics (Eleventh Revision). (2018). [Google Scholar]
- 3.Kandel E.R. A new intellectual framework for psychiatry. Am J Psychiatry 155, 457–69 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Price B.H., Adams R.D. & Coyle J.T. Neurology and psychiatry: closing the great divide. Neurology 54, 8–14 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Keshavan M.S., Price B.H. & Martin J.B. The Convergence of Neurology and Psychiatry: The Importance of Cross-Disciplinary Education. JAMA 324, 554–555 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Insel T.R. & Quirion R. Psychiatry as a clinical neuroscience discipline. JAMA 294, 2221–4 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White P.D., Rickards H. & Zeman A.Z. Time to end the distinction between mental and neurological illnesses. BMJ 344, e3454 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Ibanez A. & Zimmer E.R. Time to synergize mental health with brain health. Nature Mental Health 1, 441–443 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lefaucheur J.P. et al. Evidence-based guidelines on the therapeutic use of repetitive transcranial magnetic stimulation (rTMS): An update (2014–2018). Clin Neurophysiol 131, 474–528 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Huhn M. et al. Efficacy of pharmacotherapy and psychotherapy for adult psychiatric disorders: a systematic overview of meta-analyses. JAMA Psychiatry 71, 706–15 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Millan M.J. et al. Cognitive dysfunction in psychiatric disorders: characteristics, causes and the quest for improved therapy. Nat Rev Drug Discov 11, 141–68 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Ismail Z. et al. Psychosis in Alzheimer disease - mechanisms, genetics and therapeutic opportunities. Nat Rev Neurol 18, 131–144 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ffytche D.H. et al. The psychosis spectrum in Parkinson disease. Nat Rev Neurol 13, 81–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaitatzis A., Trimble M.R. & Sander J.W. The psychiatric comorbidity of epilepsy. Acta Neurol Scand 110, 207–20 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Peralta V. & Cuesta M.J. Motor Abnormalities: From Neurodevelopmental to Neurodegenerative Through “Functional” (Neuro)Psychiatric Disorders. Schizophr Bull 43, 956–971 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minen M.T. et al. Migraine and its psychiatric comorbidities. J Neurol Neurosurg Psychiatry 87, 741–9 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Shanker V. Essential tremor: diagnosis and management. BMJ 366, l4485 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Richmond-Rakerd L.S., D’Souza S., Milne B.J., Caspi A. & Moffitt T.E. Longitudinal Associations of Mental Disorders With Dementia: 30-Year Analysis of 1.7 Million New Zealand Citizens. JAMA Psychiatry 79, 333–340 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson P.M. et al. ENIGMA and global neuroscience: A decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl Psychiatry 10, 100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung E. et al. Alterations in brain synaptic proteins and mRNAs in mood disorders: a systematic review and meta-analysis of postmortem brain studies. Mol Psychiatry 27, 1362–1372 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Polderman T.J. et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47, 702–9 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Watanabe K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat Genet 51, 1339–1348 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Abdellaoui A., Yengo L., Verweij K.J.H. & Visscher P.M. 15 years of GWAS discovery: Realizing the promise. Am J Hum Genet 110, 179–194 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulik-Sullivan B. et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 47, 1236–41 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brainstorm C. et al. Analysis of shared heritability in common disorders of the brain. Science 360(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cross-Disorder Group of the Psychiatric Genomics Consortium. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 179, 1469–1482 e11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero C. et al. Exploring the genetic overlap between twelve psychiatric disorders. Nat Genet 54, 1795–1802 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Grotzinger A.D. et al. Genetic architecture of 11 major psychiatric disorders at biobehavioral, functional genomic and molecular genetic levels of analysis. Nat Genet 54, 548–559 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wightman D.P. et al. The genetic overlap between Alzheimer’s disease, amyotrophic lateral sclerosis, Lewy body dementia, and Parkinson’s disease. Neurobiology of Aging (2023). [DOI] [PubMed] [Google Scholar]
- 30.Qiao J. et al. Genetic correlation and gene-based pleiotropy analysis for four major neurodegenerative diseases with summary statistics. Neurobiol Aging 124, 117–128 (2023). [DOI] [PubMed] [Google Scholar]
- 31.Wingo T.S. et al. Shared mechanisms across the major psychiatric and neurodegenerative diseases. Nat Commun 13, 4314 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andreassen O.A., Hindley G.F.L., Frei O. & Smeland O.B. New insights from the last decade of research in psychiatric genetics: discoveries, challenges and clinical implications. World Psychiatry 22, 4–24 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frei O. et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat Commun 10, 2417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werme J., van der Sluis S., Posthuma D. & de Leeuw C.A. An integrated framework for local genetic correlation analysis. Nat Genet 54, 274–282 (2022). [DOI] [PubMed] [Google Scholar]
- 35.Hindley G. et al. Charting the Landscape of Genetic Overlap Between Mental Disorders and Related Traits Beyond Genetic Correlation. Am J Psychiatry 179, 833–843 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bahrami S. et al. Dissecting the shared genetic basis of migraine and mental disorders using novel statistical tools. Brain 145, 142–153 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smeland O.B. et al. Genome-wide Association Analysis of Parkinson’s Disease and Schizophrenia Reveals Shared Genetic Architecture and Identifies Novel Risk Loci. Biol Psychiatry 89, 227–235 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pickrell J.K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat Genet 48, 709–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahangari M. et al. Genome-wide analysis of schizophrenia and multiple sclerosis identifies shared genomic loci with mixed direction of effects. Brain Behav Immun 104, 183–190 (2022). [DOI] [PubMed] [Google Scholar]
- 40.Karadag N. et al. Identification of novel genomic risk loci shared between common epilepsies and psychiatric disorders. Brain (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeighami Y. et al. A comparison of anatomic and cellular transcriptome structures across 40 human brain diseases. PLoS Biol 21, e3002058 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demontis D. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat Genet (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson H.J. et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat Genet 51, 1207–1214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grove J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat Genet 51, 431–444 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purves K.L. et al. A major role for common genetic variation in anxiety disorders. Mol Psychiatry 25, 3292–3303 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullins N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet 53, 817–829 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levey D.F. et al. Bi-ancestral depression GWAS in the Million Veteran Program and meta-analysis in >1.2 million individuals highlight new therapeutic directions. Nat Neurosci 24, 954–963 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.International Obsessive Compulsive Disorder Foundation Genetics, C. & Studies, O.C.D.C.G.A. Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol Psychiatry 23, 1181–1188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nievergelt C.M. et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun 10, 4558 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trubetskoy V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu D. et al. Interrogating the Genetic Determinants of Tourette’s Syndrome and Other Tic Disorders Through Genome-Wide Association Studies. Am J Psychiatry 176, 217–227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wightman D.P. et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat Genet 53, 1276–1282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Rheenen W. et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat Genet 53, 1636–1648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao C. et al. Association of Essential Tremor With Novel Risk Loci: A Genome-Wide Association Study and Meta-analysis. JAMA Neurol 79, 185–193 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chia R. et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet 53, 294–303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hautakangas H. et al. Genome-wide analysis of 102,084 migraine cases identifies 123 risk loci and subtype-specific risk alleles. Nat Genet 54, 152–160 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.International Multiple Sclerosis Genetics, C. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nalls M.A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol 18, 1091–1102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang D. et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet 49, 1511–1516 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nalls M.A. et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet 46, 989–93 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mishra A. et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature 611, 115–123 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berkovic S.F., Cavalleri G.L. & Koeleman B.P.C. Genome-wide meta-analysis of over 29,000 people with epilepsy reveals 26 loci and subtype-specific genetic architecture. medRxiv, 2022.06.08.22276120 (2022). [Google Scholar]
- 63.Savage J.E. et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet 50, 912–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith S.M. et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci 24, 737–745 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wuttke M. et al. A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet 51, 957–972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nelson C.P. et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 49, 1385–1391 (2017). [DOI] [PubMed] [Google Scholar]
- 67.de Lange K.M. et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 49, 256–261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mahajan A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet 50, 1505–1513 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yengo L. et al. A saturated map of common genetic variants associated with human height. Nature 610, 704–712 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bulik-Sullivan B.K. et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47, 291–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holland D. et al. Beyond SNP heritability: Polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLoS Genet 16, e1008612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe K., Taskesen E., van Bochoven A. & Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 8, 1826 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Leeuw C.A., Mooij J.M., Heskes T. & Posthuma D. MAGMA: generalized geneset analysis of GWAS data. PLoS Comput Biol 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Colton C.W. & Manderscheid R.W. Congruencies in increased mortality rates, years of potential life lost, and causes of death among public mental health clients in eight states. Prev Chronic Dis 3, A42 (2006). [PMC free article] [PubMed] [Google Scholar]
- 75.Grotzinger A.D. et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav 3, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45, 580–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe K., Umicevic Mirkov M., de Leeuw C.A., van den Heuvel M.P. & Posthuma D. Genetic mapping of cell type specificity for complex traits. Nat Commun 10, 3222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wilhelmsen L. et al. Fibrinogen as a risk factor for stroke and myocardial infarction. N Engl J Med 311, 501–5 (1984). [DOI] [PubMed] [Google Scholar]
- 79.Smeland O.B., Frei O., Dale A.M. & Andreassen O.A. The polygenic architecture of schizophrenia - rethinking pathogenesis and nosology. Nat Rev Neurol 16, 366–379 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Zuk O. et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A 111, E455–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lambert J.C., Ramirez A., Grenier-Boley B. & Bellenguez C. Step by step: towards a better understanding of the genetic architecture of Alzheimer’s disease. Mol Psychiatry (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ashina M. et al. Migraine: integrated approaches to clinical management and emerging treatments. Lancet 397, 1505–1518 (2021). [DOI] [PubMed] [Google Scholar]
- 83.van der Meer D. et al. Understanding the genetic determinants of the brain with MOSTest. Nat Commun 11, 3512 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van der Meer D. et al. The genetic architecture of human cortical folding. Sci Adv 7, eabj9446 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bahrami S. et al. Distributed genetic architecture across the hippocampal formation implies common neuropathology across brain disorders. Nat Commun 13, 3436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hindley G. et al. Multivariate genetic analysis of personality and cognitive traits reveals abundant pleiotropy. Nat Hum Behav (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gadin J.R., Zetterberg R., Meijsen J. & Schork A.J. Cleansumstats: Converting GWAS sumstats to a common format to facilitate downstream applications. (Zenodo, 2023). [Google Scholar]
- 88.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maston G.A., Evans S.K. & Green M.R. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet 7, 29–59 (2006). [DOI] [PubMed] [Google Scholar]
- 90.Liberzon A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–40 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hodge R.D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Habib N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhong S. et al. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature 555, 524–528 (2018). [DOI] [PubMed] [Google Scholar]
- 94.Darmanis S. et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A 112, 7285–90 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hochgerner H. et al. STRT-seq-2i: dual-index 5’ single cell and nucleus RNA-seq on an addressable microwell array. Sci Rep 7, 16327 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.La Manno G. et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580 e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 362(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Herring C.A. et al. Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution. Cell 185, 4428–4447 e28 (2022). [DOI] [PubMed] [Google Scholar]
- 99.Benjamini Y. & Yekutieli D. The control of the false discovery rate in multiple testing under dependency. The Annals of Statistics 29, 1165–1188 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are publicly available or available on request.