Abstract
Colorectal cancer (CRC) remains a leading cause of cancer-related deaths despite being the most preventable and treatable forms of cancer when caught early through screening. There is an unmet need for novel screening approaches with improved accuracy, less invasiveness, and reduced costs. In recent years, evidence has accumulated around particular biological events that happen during the adenoma-to-carcinoma transition, especially focusing on precancerous immune responses in the colonic crypt. Protein glycosylation plays a central role in driving those responses, and recently, numerous reports have been published on how aberrant protein glycosylation both in colonic tissue and on circulating glycoproteins reflects these precancerous developments. The complex field of glycosylation, which exceeds complexity of proteins by several orders of magnitude, can now be studied primarily because of the availability of new high-throughput technologies such as mass spectrometry and artificial intelligence-powered data processing. This has now opened new avenues for studying novel biomarkers for CRC screening. This review summarizes the early events taking place from the normal colon mucosa toward adenoma and adenocarcinoma formation and associated critical protein glycosylation phenomena, both on the tissue level and in the circulation. These insights will help establish an understanding in the interpretation of novel CRC detection modalities that involve high-throughput glycomics.
INTRODUCTION
Despite worldwide population-based screening efforts, colorectal cancer (CRC) remains a global public health issue with an estimated 1.9 million new cases and 935,000 deaths worldwide (1,2). Nearly 80% arise from adenomatous polyps, progressing slowly from a small-to-large adenoma, showing dysplastic changes that transform to carcinoma over many years (3). Successful CRC screening programs identify individuals for colonoscopy to remove precancerous polyps or diagnose CRC (Table 1). Predictions have estimated the number of new CRC cases to increase to 3.2 million in 2040, primarily driven by a shifting lifestyle factor (4,5). Therefore, there is an unmet need for novel screening approaches with improved accuracy, less invasiveness and patient discomfort, lower risk of complications, reduced costs, and improved access.
Table 1.
Performance characteristics of recommended colorectal cancer screening modalities
| Screening modality | Sensitivity | Specificity |
| Direct visualization tests | ||
| Colonoscopy | CRC: 0.92–0.99 (99) AA: 0.89–0.95 (100–102) |
CRC: 0.90 (99) AA: 0.89 (100–102) |
| CT colonography | CRC: 0.86–1.0 (103) AA:0.67–0.94 (100,102,104–108) |
CRC: Not reported (103) AA: 0.86–0.98 (100,102,104–108) |
| Flexible sigmoidoscopy (within reach) | CRC: 0.92–0.99 (99) AA: 0.90–0.92 (99) |
CRC: 0.92 (99) AA: not reported (99) |
| Stool-based tests | ||
| gFOBT | CRC: 0.5–0.75 (109,110) AA: 0.06–0.17 (109,110) |
CRC: 0.96–0.98 (109,110) AA: 0.96–0.99 (109,110) |
| FIT | CRC: 0.74 (110–118) AA: 0.23 (110–118) |
CRC: 0.94 (110–118) AA: 0.96 (110–118) |
| sDNA-FIT | CRC: 0.93 (115,118–120) AA: 0.43 (115,118–120) |
CRC: 0.85 (115,118–120) AA: 0.89 (115,118–120) |
AA, advanced adenomas; CEA, carcinoembryonic antigen; CT, colonography; CRC, colorectal cancer; FIT, fecal immunochemical test; gFOBT, guaiac-based fecal occult blood test; sDNA, stool DNA.
Studies are underway to evaluate minimally invasive blood tests (Table 2; see Supplementary Table 1, http://links.lww.com/CTG/A940), and most fall short for screening purposes because of low sensitivity and/or specificity. Liquid biopsy is a novel category aiming to detect circulating cancer, including circulating tumor cells, circulating tumor DNA, and cell-free DNA (6). Currently, there are 35 liquid biopsy CRC trials ongoing to help establish their clinical utility (7); however, a potential problem is the need for sufficient tumor volume, which challenges its suitability for screening purposes. Developing a blood-based screening test targeting precancerous events would be an appealing approach.
Table 2.
Colorectal cancer blood-based liquid biopsy biomarkers
| Biomarker target | Sensitivity | Specificity |
| Circulating tumor cells (CTCs)a | ||
| EpCAM enrichment | CRC: 0.86 (121) AA: 0.71 (121) |
CRC: 0.95 (121) AA: 0.85 (121) |
| Circulating tumor DNA (ctDNA) | ||
| cg10673833 (MYO1-G) | CRC: 0.84–0.87 (122,123) AA: 0.33 (123) |
CRC: 0.90–0.95 (122,123) AA: 0.67 (123) |
| VIM | CRC: 0.59–0.84 (124) AA: 0.45 (124) |
CRC: 0.93–0.95 (124) AA: 0.95 (124) |
| BCAT1/IKZF1 | CRC: 0.66 (125) AA: 0.06 (125) |
CRC: 0.94–0.95 (125) AA: 0.94–0.95 (125) |
| Cell-free DNA (cfDNA) | ||
| SEPT9 (mSEPT9) | CRC: 0.37–0.88 (126) AA: 0.23 (126) |
CRC: 0.77–0.99 (126) AA: 0.91 (126) |
| 11 methylation markersb | CRC: 0.84 (127) AA: 0.77 (127) |
CRC: 0.86 (127) AA: 0.83 (127) |
| SFRP1/SFRP2/SDC2/PRIMA1 | CRC: 0.92 (128) AA: 0.89 (128) |
CRC: 0.97 (128) AA: 0.87 (128) |
| Circulating microRNA (miRNA) markers | ||
| miR-760 | CRC: 0.80 (129) AA: 0.70 (129) |
CRC: 0.70 (129) AA: 0.62 (129) |
| miR-601/miR-760 | CRC: 0.83 (129) AA: 0.72 (129) |
CRC: 0.69 (129) AA: 0.62 (129) |
| miR-29a/miR-92a | CRC: 0.83 (130) AA: 0.73 (130) |
CRC: 0.85 (130) AA: 0.80 (130) |
| Exosomal long noncoding RNA (lncRNAs) markers | ||
| UCA1 | CRC: 1.0 (131) | CRC: 0.43 (131) |
| TUG1/UCA1 | CRC: 0.93 (131) | CRC: 0.64 (131) |
| SNHG11 | CRC: 0.93 (132) | CRC: 0.71 (132) |
| ZFAS1/SNHG11/LINC00909/LINC00654 | CRC: 0.92 (132) | CRC: 0.83 (132) |
| Exosomal circular RNA (circRNA) markers | ||
| circPanel (hsa_circ_0001900/hsa_circ_0001178/hsa_circ_0005927) | CRC: 0.68–0.73 (133) AA: 0.84 (133) |
CRC: 0.83–0.90 (133) AA: 0.70 (133) |
| hsa_circ_0007534 | CRC: 0.92 (134) | CRC: 0.52 (134) |
| hsa-circ-0004771 | CRC: 0.81 (135) | CRC: 0.83 (135) |
| Messenger RNA (mRNA markers) | ||
| hTERT | CRC: 0.98 (136) | CRC: 0.64 (136) |
| CK-19/CK-20/CEA/REG4/uPA/TIAM1 mRNA | CRC: 0.89 (137) | CRC: 0.88 (137) |
| CEA/CK20/survivin | CRC: 0.61 (138) | CRC: 0.76 (138) |
| Exosomal proteins markers | ||
| QSOX1 | CRC: 0.74–0.88 (139) | CRC: 0.83–0.94 (139) |
| CPNE3 | CRC: 0.68 (140) | CRC: 0.84 (140) |
| HSP60 | CRC: 0.63 (141) | CRC: 0.95 (141) |
| Other protein and cytological markers | ||
| CEA | CRC: 0.11 (142) AA: 0.05 (142) |
CRC: 0.98 (142) AA: 0.98 (142) |
| CA 19-9 | CRC: 0.08 (142) AA: 0.03 (142) |
CRC: 0.98 (142) AA: 0.98 (142) |
| TDGF 1/Cripto 1/CEA/ECM | CRC: 0.79–0.91 (143) AA: >0.60 (143) |
CRC: 0.81–0.87 (143) AA: not reported (143) |
| Combination of biological targets | ||
| CECs/cfDNA | CRC: 1.0 (144) AA: 0.73–0.83 (144) |
CRC: 0.89–0.91 (144) AA: 0.87–0.91 (144) |
| cfDNA and protein immunoassays | CRC: 0.91 (145) AA: 0.41 (145) |
CRC: 0.94 (145) AA: 0.90 (145) |
| cfDNA/ctDNA | CRC: 0.90–0.96 (146–148) AA: 0.20 (149) |
CRC: 0.94–0.97 (146–148) AA: not reported (149) |
AA, advanced adenomas; BCAT1, branched-chain amino acid transaminase 1; CA, carbohydrate antigen; CEA, carcinoembryonic antigen; CK, cytokeratin; CPNE3, exosomal Copine III; CRC, colorectal cancer; DNA, desoxyribonucleic acid; ECM, extracellular matrix; EpCAM, epithelial cell adhesion molecule; HSP60, heat-shock protein 60; hTERT, human telomerase reverse transcriptase; IKZF1, IKAROS, family zinc finger 1; LINC00, long intergenic noncoding RNA; PRIMA1, Proline-rich membrane anchor 1; REG4, regenerating islet-derived family, member 4; RNA, ribonucleic acid; QSOX1, quiescin sulfhydryl oxidase 1; SDC2, Syndecan 2; SEPT9, septin 9; SFRP, secreted frizzled-related protein; SNHG11, small nucleolar RNA, host; TDGF-1, teratocarcinoma-derived growth factor 1; TIAM1, T-cell lymphoma invasion and metastasis 1; TUG1, taurine upregulated gene 1; UCA1, urothelial cancer-associated 1; uPA, plasminogen activator urokinase; VIM, vimentin; ZFAS1, zinc finger antisense 1.
Outcomes for CTC-based studies in CRC are frequently reported as detection rates, overall survival, and progression-free survival.
This panel contains cg00310855, cg01857475, cg01922936, cg11320449, cg11407741, cg11596863, cg15020425, cg22329423, cg24733262, cg25300584, cg26337020.
Key drivers for loss of colonic homeostasis and subsequent neoplasia formation include dysbiosis, (epi)genetic changes, and lack of proper immune surveillance intercalated with glycosylation changes. Aberrant protein glycosylation is a universal feature in various steps of malignant transformation and CRC development (8). In this review, we will summarize the sequence of events from normal colon mucosa to carcinoma and its correlations with glycoproteomic changes that might serve as potential biomarkers for CRC.
THE ADENOMA-TO-CARCINOMA SEQUENCE
(Epi)genetic changes during adenoma-to-carcinoma progression
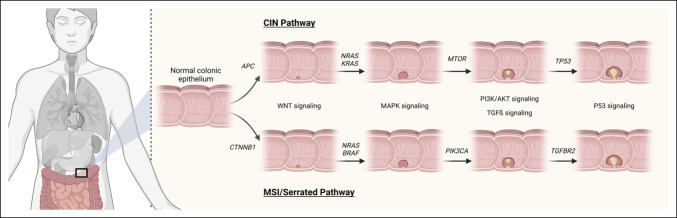
Chromosomal instability, microsatellite instability, and serrated neoplasia are 3 mechanisms that drive tumorigenesis in the adenoma-to-carcinoma pathway (Figure 1) (9). Conventional adenomas stem from biallelic mutations in the tumor suppressor gene adenomatous polyposis coli (APC) in the chromosomal instability pathway but can also occur due to hypermethylation of the APC promoter (9). Mutated APC triggers the activation of the WNT signaling pathway (10,11), as a result the oncoprotein, β-catenin, encoded by the CTNNB1 gene, accumulates in the cytosol because of ineffective phosphorylation (11). β-catenin translocates to the nucleus (11). Inactivation of APC leads to activation of the proto-oncogene, Kirsten rat sarcoma viral homolog (KRAS) (12). β-catenin is aided by additional signaling pathways mediated by KRAS to drive gene transcription responsible for tumor growth and invasion (13). KRAS activates the downstream Raf-MEK-ERK pathway and PI3K/AKT signaling through mTOR (14). The loss of heterozygosity at chromosome 18q can lead to loss of function of tumor suppressor genes in this location including SMAD2/4 which are part of transforming growth factor-β (TGF-β) pathway (15). Loss of TP53 occurs late in tumorigenesis and can coincide with the transition of a large adenoma to adenocarcinoma (12).
Figure 1.
Genetic alterations and signaling pathways associated with the polyposis to the colorectal cancer sequence. The 2 main pathways involved in the normal colonic epithelium to colorectal carcinoma pathway are the classic tubular adenomas to colorectal carcinoma pathway (upper half) and the serrated polyps to serrated colorectal cancer pathway (lower half). Adapted from Figure 3, Kuipers et al (159) and Figure 2, Davies et al (160). Created with BioRender.com. AKT, protein kinase B; APC, adenomatous polyposis coli; BRAF, v-raf murine sarcoma viral oncogene homolog B1; CIN, chromosomal instability; CTNNB1, catenin-β1; KRAS, Kirsten rat sarcoma viral oncogene homolog; MAPK, mitogen-activated protein kinase; MSI, microsatellite instability; mTOR, mammalian target of rapamycin; NRAS, neuroblastoma ras viral oncogene homolog; P53, tumor protein 53; PI3K, phosphatidylinositol 3‐kinase; PI3KCA, phosphatidylinositol-4; 5-bisphosphate 3-kinase catalytic subunit-α; TGFβ, transforming growth factor-β; TGFBR2, TGFβ receptor 2; TP53, tumor protein 53 gene; WNT, Wingless-related integration site.
The microsatellite instability (MSI) pathway leads to the formation of the hypermutable phenotype characterized by DNA base pair mutations (9). Epigenetic disruptions including MLH1 hypermethylation and germline mutations in the DNA mismatch repair gene form the MSI phenotype (9). The MSI pathway can predict a favorable outcome and is seen in all patients with Lynch syndrome (12). The MSI phenotype can lead to aberrant methylation of the CpG dinucleotides leading to CpG island methylator phenotype. An APC or BRAF mutation can be the initiating event of adenoma formation in the MSI pathway (12). TGF-β receptor 2 gene mutation causes cell proliferation and is mutated in more than 90% of MSI colorectal tumors (9). There are a plethora of genes disrupted by MSI, including those that regulate proliferation, DNA repair, and apoptosis (9).
An important feature of the serrated pathway is the activation of V600E in the v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) and subsequent MAPK pathway activation (9). Once acquiring a BRAF mutation, there are 2 ways serrated tumors form. One way is typically through sessile serrated adenomas when the pathway converges with the MSI pathway, which results in the MSI-H phenotype (10). Another way serrated tumors can form is through mutation of TP53, which leads to activation of other pathways including WNT, TGF-β, and epithelial-to-mesenchymal transition (9).
Role of host immunity in the adenoma-to-carcinoma sequence
The progression from normal tissue to carcinoma is accompanied by a series of phenotypic and functional changes of immune cells at the site of the lesion. CRC develops slowly from single crypt lesions to adenoma and carcinoma through a multistep process that is both controlled and shaped by the immune system (Figure 2). Transformed epithelial cells release danger signals, a group of molecules that are chemically unrelated but promote sterile inflammation. These damage-associated molecular patterns include proteins (e.g., high-mobility group box 1 protein, S100 proteins, and heat shock proteins) that are typically found inside the cell but are released under stress conditions or cell death (16). Damage-associated molecular patterns lead to the recruitment of immune cells that can eliminate damaged cells and induce regression of lesions (immunosurveillance) (17,18). However, premalignant cells develop diverse strategies to evade host control while increasing in size and progressing to a high degree of dysplasia. This state of relative dormancy (equilibrium), in which cancer expansion is kept at bay by the immune system, may extend for a very long time (19). A progressive shift of the immune landscape from an antitumor state to a protumor state, during which immune cells undergo marked phenotypic and functional changes, facilitates the progression of premalignant lesions to cancer (17,18). In this escape phase, the premalignant cells acquire the ability to escape immune control, undergo uncontrolled invasive growth, and develop into CRC.
Figure 2.
Mechanisms of tumor initiation and promotion in colorectal cancer influence the pool of secreted proteins. Tumor-initiating events that transform normal intestinal epithelial cells by spontaneous mutations, environmental mutagens, and bacteria. Cytokines produced by innate and adaptive immune cells, stromal compartment, and tumor cells exhibit pleiotropic roles, which depend on the developmental stage of the tumor. Cytokines and other factors produced in the tumor might enter circulation and influence the composition of glycoproteins secreted by the liver and by circulating blood cells. Created with BioRender.com.
The adenoma microenvironment contains immune cells that engage in complex interactions with premalignant cells and exhibit both antitumor and protumor functions (19–21). These immune cells change in number and function in the transition from adenoma to carcinoma. For instance, an increased population of T lymphocytes, including CD8+ cytotoxic T lymphocytes (CTLs) and CD4+ T helper cells, has been reported in adenoma tissues, mostly distributed in the stromal region and infiltrated into the adenomatous epithelium (22–24). CTLs can recognize and eliminate transformed cells, whereas CD4+ T helper cells play critical roles in regulating host immune response and promoting CTL function (25). Strikingly, CD4+ T cells progressively increase across the normal mucosa-adenoma-carcinoma sequence, whereas CTLs increase in premalignant adenoma and decrease in the transition to CRC (22,26). The antitumor capacity of CTLs can be further affected by exhaustion and by suppression driven by regulatory T cells (Tregs). T-cell exhaustion is a dysfunctional state that occurs in response to chronic antigen stimulation and is characterized by reduced cytokine production and increased expression of inhibitory receptors; it is detected only in CRC and not in the premalignant state (27). Similarly, Tregs are significantly increased in adenoma tissues compared with control tissues and are even higher in CRC tissues (28,29).
High T-helper 17 (TH17) cell activity contributes to the development of malignant lesions (30). TH17 cells are the dominant source of interleukin-17A (IL-17A), a cytokine that stimulates proliferation, survival, and mobilization of stromal fibroblasts through the upregulation of the TGF-β receptor that enhances the expression of profibrotic genes. The density of IL-17A-positive TH17 cells is increased in colorectal adenoma tissues (31,32). Given that TGF-β is also an upstream stimulator of TH17 cell differentiation and increased expression levels of both TGF-β and IL-17A occur in patients with CRC, TGF-β may work in synergy with TH17/IL-17A on the progression of CRC (33).
Myeloid cells are also found in premalignant lesions and change in number during the transition from normal tissue to carcinoma. Tumor infiltration by CD68+ macrophages increases progressively as colorectal adenomas arise, grow, and become dysplastic; this trend continues in the early stages of CRC (22). Progression from adenoma to cancer is accompanied by the emergence of mechanisms that suppress the generation, maturation, and function of dendritic cells to escape immune recognition and elimination, resulting in lower antigen presentation and lower numbers of these cells in both adenoma and CRC tissues (34).
In summary, the progression from normal tissue to adenoma and carcinoma is accompanied by a series of phenotypic and functional changes of the immune cells in the site of the lesion, supported by alterations in associated cytokine networks. We propose that cytokines, metabolites, and other factors produced in the lesion may travel to the periphery and exert global effects on the liver and circulating leukocytes.
ABERRANT GLYCOSYLATION DURING THE ADENOMA-TO-CARCINOMA SEQUENCE
Introduction to protein glycosylation
Protein glycosylation is the most abundant and complex post-translational modification common to all eukaryotic cells and involves an enzymatic process in which carbohydrate structures (glycans) are attached to proteins, mainly through nitrogen (N) and oxygen (O) linkages (35). It is estimated that 85% of serum proteins are glycosylated (36) and that specific glycosylation determines a wide range of physiological events, including protein folding and trafficking, cell-cell and cell-matrix interactions, cellular differentiations, and the immune response (37–42). Glycosylation significantly amplifies the proteome by producing diverse protein isoforms that demonstrate a variety of properties and functions (43–45). Figure 3 shows a schematic overview of N-linked and O-linked protein glycosylation.
Figure 3.
A schematic overview of N-linked and O-linked glycosylation. Glycosylation is a post-translational modification where sugar moieties are attached to proteins, affecting protein structure, conformation, and function. N-glycosylation and O-glycosylation are the 2 main types of glycosylation. Glycosylation changes have been associated with physiological and pathological events. The analysis helps identify novel biomarkers for diagnosis, prognosis, and disease monitoring. Created with BioRender.com.
Aberrant glycosylation is described as a hallmark of cancer and a critical feature during malignant transformation and tumor development (46,47). Aberrant N-glycan structures in cancer cells are associated with malignant transformation, tumor invasiveness, and metastatic disease (48–52). Importantly, aberrant glycosylation of cancer cells greatly influences the interaction between cancer cells and the immune system, particularly immunosurveillance and immunoediting. The aberrant glycosylation observed during cancer development includes increased branching of N-glycans, higher density of O-glycans, incomplete synthesis of glycans, neosynthesis, increased sialylation, and increased fucosylation.
Cellular and tissue glycosylation during CRC development
The dynamically changing glycoproteomic profiles in the colonic crypt result from the concordant enzymatic activity of glycosyltransferases and glycosidases, orchestrating the cellular transformations from normal mucosa to adenoma and ultimately to CRC. During this transition, predominantly N-linked and O-linked glycans are added to or removed from tissue proteins, thereby constantly changing the glycocalyx of cellular surfaces and mediating the local immune responses in the crypt. One example of an environmental trigger influencing local glycosylation is red meat consumption (53). This leads to aberrant cell surface glycosylation due to metabolic incorporation of the red meat-derived sialic acid (N-glycolyl neuraminic acid [Neu5Gc]) (54). Humans are unable to synthesize Neu5Gc because of a mutation in the Cmah gene (55,56); however, dietary intake of Neu5Gc from red meat leads to its incorporation in glycoconjugates (57,58). Such Neu5Gc-containing glycans are targeted by naturally circulating anti-Neu5Gc antibodies, which are present in all humans (59,60). This leads to an inflammatory response that has been shown to be associated with CRC risk in 2 large independent human cohorts (54,61,62).
During malignant transformation, changing glycoproteomic crypt profiles specifically influence the host immune response. For instance, increased branching of N-glycans is used by transforming cells to escape immune recognition, instructing the creation of immunosuppressive networks through inhibition of interferon gamma (53). In addition, mutations in the guanosine diphosphate-mannose-4,6-dehydratase gene that is involved in the synthesis of fucosylated proteins have been identified in human CRC tissue (63). Cells deficient in GDP-mannose-4, 6-dehydratase, therefore lacking fucosylation, evade natural killer cell-induced apoptosis through the death receptors TNF-related apoptosis-inducing ligand (63). In a large clustered regularly interspaced short palindromic repeats screening, it was demonstrated that the enzyme B3GNT2, responsible for the synthesis of polylactosaminylated N-glycans, can glycosylate various cell surface receptors on tumor cells to prevent subsequent killing by the cytotoxic T cells (64). Furthermore, CRC cells express high levels of sialic acid-containing glycans that are known engagers of the immune-suppressing receptors called Siglecs (sialic acid-binding immunoglobulin-like lectins) (65). Siglec/sialoglycan interaction can compromise dendritic cell activation and antigen presentation, tumor-associated macrophage polarization, and CD8 T-cell antitumor effector functions (66–68). Not surprisingly, many studies have pointed to the blocking of Siglec/sialoglycan interaction as a new potential immune checkpoint blockade therapy during cancer (69). Finally, recent work has provided an insight into the mechanism underlying the low response of INF-y therapy in patients with CRC. Krug et al (70) have elucidated the role of bisecting N-glycans on INFyRa, demonstrating that a decrease in INFyRa bisection leads to receptor degradation and resistance to INF-y therapy, which could be salvaged by pharmacological reconstitution of bisection through administration of all-trans retinoic acid.
A wide range of observations have further solidified tissue glycosylation alterations to be a hallmark of CRC development, including increased branching of N-glycans, higher density of O-glycans, incomplete glycan synthesis, glycan neo-synthesis, increased sialylation, and increased fucosylation (8). A major N-glycan branching structure related to cancer formation is the complex β1,6-GlcNAc-branched N-glycan (51). The aberrant overexpression is catalyzed by the N-acetylglucosaminyltransferase V (GnT-V) enzyme, implicated in inducing pro-malignant, pro-invasive, and pro-metastatic cancer phenotypes (48,49,71). Interestingly, upregulation of GnT-V is implicated early in colon adenoma progression and is detectable in immunohistochemical staining of aberrant crypt loci, the earliest morphological lesions visible in colonic carcinogenesis (72). Mechanistically, it has been shown that GnT-V promotes colon cancer stem cell self-renewal and tumorigenesis potential through regulation of Wnt/β-catenin signaling (72). The WNT/beta-catenin pathway is also known to interact with the P13K/AKT pathway to promote CRC progression (73). The WNT pathway is also known to interact with the Notch pathway through Jagged1 (74) to promote stem cell renewal and differentiation. Similarly, the WNT pathway interacts with the TGF-beta (75), MAPK/ERK (76), Hedgehog (77), and ND-KappaB pathway (78), leading to various hallmarks of cancer. Removal of these GnT-V-synthesized glycans renders CRC cells more susceptible to the immune system and improves cancer immunotherapy (79). GnT-V has also been implicated in metastasis development by modifying proteins involved in extracellular matrix degradation, cell adhesion, and cell motility, helping the spread of metastatic cancer cells (80,81). One of the best described epitopes in metastatic disease are sialyl Lewis antigens, namely sialyl Lewis X and A, which are key in aiding tumor extravasation through the endothelium by interactions with E-selectin (82–84).
Changes in serum glycosylation during CRC development
The adenoma-to-carcinoma sequence is associated with dynamically changing glycosylation profiles in colonic crypts and associated host immune responses that, in turn, influence systemic immune responses, including glycosylated acute-phase proteins and plasma-derived antibodies. For purposes of detection, immunoassays for circulatory glycoproteins have been tried with mixed results, including carcinoembryonic antigen, CA19-9, and CA-125 (85,86). Although aiding in staging evaluation and monitoring after treatment, they lack sensitivity and specificity for proper screening. Table 3 summarizes the most relevant markers to date.
Table 3.
Immunoassays that measure glycosylation-related changes in CRC
| Assay | GlycoEpitope | Changes in CRC | Clinical utility |
| CEA (150–152) | CD66 family of glycoproteins | Elevated in certain cases of CRC | Limited or no utility |
| CA19-9 (153–155) | Sialyl-LewisA | Elevated in metastatic CRC | Limited or no utility |
| CA-125 (156–158) | Heavily O-glycosylated MUC16 | Unclear or mixed results | May be useful for prognostication |
CEA, carcinoembryonic antigen; CRC, colorectal cancer; MUC16, Mucin-16.
Although glycosylation immunoassays have limitations, mass spectrometry (MS) has opened new avenues. Owing to the simplicity of acquiring blood, numerous studies have evaluated aberrant glycosylation, especially N-glycosylation as disease biomarkers (87). A study of the N-glycome in patients with CRC demonstrated a strong association with branched and poly-LacNAc elongated N-glycans, normalizing after treatment (88). Additional serum studies on N-glycan profiling demonstrated that increased fucosylation and sialylation on complement C3 and kininogen-1 may aid in CRC detection (89). Studying IgG N glycome profiles in CRC, differentially expressed levels of N-glycosylation were shown for normal individuals compared to those with CRC (90). Moreover, using matrix-assisted laser desorption/ionization-MS in patient sera, a positive correlation between multiantennary, sialylated N-glycans was shown during cancer progression while biantennary core-fucosylated N-glycans correlated negatively (91). Interestingly, the authors not only showed accuracy for rightly classifying CRC but also for advanced adenomas (AA), which could potentially introduce a paradigm shift for CRC screening. This was recently addressed in a study using ultra performance liquid chromatography and demonstrating distinct serum IgG N-glycan profiles for AA and CRC (92). Another group studying 34 serum glycans reported that 9 of both hybrid-type and multiantennary glycans were differentially expressed between CRC and AA (93). Not only glycosylated proteins but also glycan-binding proteins such as galectin-2, galectin-3, galectin-4, and galectin-8 are found to be elevated in the serum from patients with CRC. For instance, high levels of galectin-2 positively correlate with high CRC mortality (94). This study demonstrated that galectins can promote the adhesion of cancer cells to the blood vascular endothelium, therefore contributing to cancer metastases (94).
In summary, this suggests that glycosylation events associated with CRC development can be readily observed at the periphery through aberrant glycosylation observed on circulating glycoproteins. The question remains on how early does developing colonic neoplasia result in aberrant serum glycosylation profiles, mainly composed of liver-secreted glycoproteins and B-cell-derived antibodies. This will need to be addressed, but there seems to be sufficient evidence to justify larger prospective studies on the use of blood-based glycan profiles for the purpose of AA/CRC detection and screening. It also remains to be seen whether the glycoproteomic changes in circulation differ by the pathway of CRC development and/or by individual mutations present in these lesions. Nevertheless, it is an important pursuit particularly in the context of biomarkers and surrogate end points for personalized therapies.
CONCLUSION
To improve CRC screening, there is a need to develop biomarkers that are associated with precancerous and early cancerous events in the colonic crypt. We reviewed the early events taking place during the adenoma-to-carcinoma sequence and introduced critical protein glycosylation phenomena associated with the changing crypt microenvironment. The host immune response plays a central role in orchestrating the process from single crypt lesions to carcinoma with aberrant protein glycosylation being a key factor, both locally and on circulating glycoproteins. It is not until recently that the enormously complex field of glycosylation can be studied, primarily because of the availability of the appropriate tools (95). Machine learning models (e.g., Support Vector Machines and Random Forest) can decipher the vast amount of complex data coming from, for instance, liquid chromatography-MS (96–98). Large-scale prospective clinical studies are currently underway to evaluate the clinical validity of glycosylation markers and determine their clinical utility and become an additional tool in the armamentarium for CRC screening.
CONFLICTS OF INTEREST
Guarantor of the article: Daniel W. Hommes, MD, PhD.
Specific author contributions: D.C., D.W.H.: study planning; D.C., C.G., F.S., C.D., T.C., F.S., D.W.H.: data collection and interpretation; D.C., C.G., F.S., C.D., T.C., F.S., D.W.H.: manuscript drafting.
Financial support: None to report.
Potential competing interests: All authors are employed at InterVenn BioSciences, a life sciences company that specializes in glycomic analysis and biomarker development and holds multiple patents in this field.
Supplementary Material
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A940
Dharini Chandrasekar and Christina Guerrier contributed equally to this work.
Contributor Information
Dharini Chandrasekar, Email: dharini.chandrasekar@venn.bio.
Christina Guerrier, Email: christina.guerrier@venn.bio.
Frederico Alisson-Silva, Email: fred.silva@venn.bio.
Chirag Dhar, Email: chirag.dhar@venn.bio.
Tomislav Caval, Email: tomislav.caval@venn.bio.
Flavio Schwarz, Email: flavio.schwartz@venn.bio.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209–49. [DOI] [PubMed] [Google Scholar]
- 2.Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol 2021;14(10):101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klos CL, Dharmarajan S. Polyp genetics. Clin Colon Rectal Surg 2016;29(04):289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keum N, Giovannucci E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat Rev Gastroenterol Hepatol 2019;16(12):713–32. [DOI] [PubMed] [Google Scholar]
- 5.Murphy N, Moreno V, Hughes DJ, et al. Lifestyle and dietary environmental factors in colorectal cancer susceptibility. Mol Aspects Med 2019;69:2–9. [DOI] [PubMed] [Google Scholar]
- 6.Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J Clin Oncol 2018;36(16):1631–41. [DOI] [PubMed] [Google Scholar]
- 7.Broccard SP, Kasbi AA, Bagaria SP, et al. Liquid biopsies for colorectal cancer: A narrative review of ongoing clinical trials and the current use of this technology at a comprehensive cancer center. J Gastrointest Oncol 2022;13(1):438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holst S, Wuhrer M, Rombouts Y. Glycosylation characteristics of colorectal cancer. Adv Cancer Res 2015;126:203–56. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen LH, Goel A, Chung DC. Pathways of colorectal carcinogenesis. Gastroenterology 2020;158(2):291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen B, Scurrah CR, McKinley ET, et al. Differential pre-malignant programs and microenvironment chart distinct paths to malignancy in human colorectal polyps. Cell 2021;184(26):6262–80.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao H, Ming T, Tang S, et al. Wnt signaling in colorectal cancer: Pathogenic role and therapeutic target. Mol Cancer 2022;21(1):144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Palma FDE, D'Argenio V, Pol J, et al. The molecular hallmarks of the serrated pathway in colorectal cancer. Cancers 2019;11(7):1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schatoff EM, Leach BI, Dow LE. Wnt signaling and colorectal cancer. Curr Colorectal Cancer Rep 2017;13(2):101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lam KK, Tang CL, Tan E, et al. KRAS mutation-independent downregulation of MAPK/PI3K signaling in colorectal cancer. Mol Oncol 2022;16(5):1171–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koveitypour Z, Panahi F, Vakilian M, et al. Signaling pathways involved in colorectal cancer progression. Cell Biosci 2019;9(1):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez C, Huebener P, Schwabe RF. Damage-associated molecular patterns in cancer: A double-edged sword. Oncogene 2016;35(46):5931–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlsson M, Lindberg K, Karlén P, et al. Evidence for immunosurveillance in intestinal premalignant lesions. Scand J Immunol 2010;71(5):362–8. [DOI] [PubMed] [Google Scholar]
- 18.Chow MT, Möller A, Smyth MJ. Inflammation and immune surveillance in cancer. Semin Cancer Biol 2012;22(1):23–32. [DOI] [PubMed] [Google Scholar]
- 19.Banner BF, Sonmez-Alpan E, Yousem SA. An immunophenotypic study of the inflammatory cell populations in colon adenomas and carcinomas. Mod Pathol 1993;6(3):295–301. [PubMed] [Google Scholar]
- 20.Cui G, Yuan A, Vonen B, et al. Progressive cellular response in the lamina propria of the colorectal adenoma-carcinoma sequence. Histopathology 2009;54(5):550–60. [DOI] [PubMed] [Google Scholar]
- 21.Whiteside TL. The role of immune cells in the tumor microenvironment. Cancer Treat Res 2006;130:103–24. [DOI] [PubMed] [Google Scholar]
- 22.Maglietta A, Maglietta R, Staiano T, et al. The immune landscapes of polypoid and nonpolypoid precancerous colorectal lesions. PLoS One 2016;11(7):e0159373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dock J, Ramirez CM, Hultin L, et al. Distinct aging profiles of CD8+ T cells in blood versus gastrointestinal mucosal compartments. PLoS One 2017;12(8):e0182498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preza GC, Yang OO, Elliott J, et al. T lymphocyte density and distribution in human colorectal mucosa, and inefficiency of current cell isolation protocols. PLoS One 2015;10(4):e0122723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4+ T cells in cancer immunotherapy—new insights into old paradigms. Cancer Gene Ther 2021;28(1-2):5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Hu X, Zimmerman M, et al. TNFα cooperates with IFN-γ to repress Bcl-xL expression to sensitize metastatic colon carcinoma cells to TRAIL-mediated apoptosis. PLoS One 2011;6(1):e16241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Becker WR, Nevins SA, Chen DC, et al. Single-cell analyses reveal a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat Genet 2021;54(7):985–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Cao X. Immunosuppressive cells in tumor immune escape and metastasis. J Mol Med Berl Ger 2016;94(5):509–22. [DOI] [PubMed] [Google Scholar]
- 29.Hua W, Yuan A, Zheng W, et al. Accumulation of FoxP3+ T regulatory cells in the tumor microenvironment of human colorectal adenomas. Pathol Res Pract 2016;212(2):106–12. [DOI] [PubMed] [Google Scholar]
- 30.Cui G. TH9, TH17, and TH22 cell subsets and their main cytokine products in the pathogenesis of colorectal cancer. Front Oncol 2019;9:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui G, Yang H, Zhao J, et al. Elevated proinflammatory cytokine IL-17A in the adjacent tissues along the adenoma-carcinoma sequence. Pathol Oncol Res 2015;21(1):139–46. [DOI] [PubMed] [Google Scholar]
- 32.Cui G, Yuan A, Goll R, et al. IL-17A in the tumor microenvironment of the human colorectal adenoma-carcinoma sequence. Scand J Gastroenterol 2012;47(11):1304–12. [DOI] [PubMed] [Google Scholar]
- 33.Cui G, Li Z, Florholmen J, et al. Dynamic stromal cellular reaction throughout human colorectal adenoma-carcinoma sequence: A role of TH17/IL-17A. Biomed Pharmacother 2021;140:111761. [DOI] [PubMed] [Google Scholar]
- 34.Yuan A, Steigen SE, Goll R, et al. Dendritic cell infiltration pattern along the colorectal adenoma-carcinoma sequence. APMIS 2008;116(6):445–56. [PubMed] [Google Scholar]
- 35.Ohtsubo K, Marth JD. Glycosylation in cellular mechanisms of health and disease. Cell 2006;126(5):855–67. [DOI] [PubMed] [Google Scholar]
- 36.Schjoldager KT, Narimatsu Y, Joshi HJ, et al. Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol 2020;21(12):729–49. [DOI] [PubMed] [Google Scholar]
- 37.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta 1999;1473(1):4–8. [DOI] [PubMed] [Google Scholar]
- 38.Crocker PR, Feizi T. Carbohydrate recognition systems: Functional triads in cell-cell interactions. Curr Opin Struct Biol 1996;6(5):679–91. [DOI] [PubMed] [Google Scholar]
- 39.Feizi T. Carbohydrate-mediated recognition systems in innate immunity. Immunol Rev 2000;173(1):79–88. [DOI] [PubMed] [Google Scholar]
- 40.Gabius HJ, Siebert HC, André S, et al. Chemical biology of the sugar code. Chembiochem Eur J Chem Biol 2004;5(6):740–64. [DOI] [PubMed] [Google Scholar]
- 41.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science 2001;291(5512):2364–9. [DOI] [PubMed] [Google Scholar]
- 42.Karlsson KA. Meaning and therapeutic potential of microbial recognition of host glycoconjugates. Mol Microbiol 1998;29(1):1–11. [DOI] [PubMed] [Google Scholar]
- 43.Aebersold R, Agar JN, Amster IJ, et al. How many human proteoforms are there? Nat Chem Biol 2018;14(3):206–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varki A. Biological roles of glycans. Glycobiology 2017;27(1):3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spiro RG. Protein glycosylation: Nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 2002;12(4):43R–56R. [DOI] [PubMed] [Google Scholar]
- 46.Fuster MM, Esko JD. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat Rev Cancer 2005;5(7):526–42. [DOI] [PubMed] [Google Scholar]
- 47.Pinho SS, Reis CA. Glycosylation in cancer: Mechanisms and clinical implications. Nat Rev Cancer 2015;15(9):540–55. [DOI] [PubMed] [Google Scholar]
- 48.Pinho SS, Reis CA, Paredes J, et al. The role of N-acetylglucosaminyltransferase III and V in the post-transcriptional modifications of E-cadherin. Hum Mol Genet 2009;18(14):2599–608. [DOI] [PubMed] [Google Scholar]
- 49.Carvalho S, Catarino TA, Dias AM, et al. Preventing E-cadherin aberrant N-glycosylation at Asn-554 improves its critical function in gastric cancer. Oncogene 2016;35(13):1619–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinho SS, Carvalho S, Marcos-Pinto R, et al. Gastric cancer: Adding glycosylation to the equation. Trends Mol Med 2013;19(11):664–76. [DOI] [PubMed] [Google Scholar]
- 51.Taniguchi N, Kizuka Y. Glycans and cancer: Role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv Cancer Res 2015;126:11–51. [DOI] [PubMed] [Google Scholar]
- 52.Rodrigues JG, Balmaña M, Macedo JA, et al. Glycosylation in cancer: Selected roles in tumour progression, immune modulation and metastasis. Cell Immunol 2018;333:46–57. [DOI] [PubMed] [Google Scholar]
- 53.Bouvard V, Loomis D, Guyton KZ, et al. Carcinogenicity of consumption of red and processed meat. Lancet Oncol 2015;16(16):1599–600. [DOI] [PubMed] [Google Scholar]
- 54.Samraj AN, Läubli H, Varki N, et al. Involvement of a non-human sialic Acid in human cancer. Front Oncol 2014;4:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chou HH, Takematsu H, Diaz S, et al. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci U S A 1998;95(20):11751–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayakawa T, Aki I, Varki A, et al. Fixation of the human-specific CMP-N-acetylneuraminic acid hydroxylase pseudogene and implications of haplotype diversity for human evolution. Genetics 2006;172(2):1139–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alisson-Silva F, Kawanishi K, Varki A. Human risk of diseases associated with red meat intake: Analysis of current theories and proposed role for metabolic incorporation of a non-human sialic acid. Mol Aspects Med 2016;51:16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dhar C, Sasmal A, Varki A. From “serum sickness” to “Xenosialitis”: Past, present, and future significance of the non-human sialic acid Neu5Gc. Front Immunol 2019;10:807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higashi H, Hirabayashi Y, Fukui Y, et al. Characterization of N-glycolylneuraminic acid-containing gangliosides as tumor-associated Hanganutziu-Deicher antigen in human colon cancer. Cancer Res 1985;45(8):3796–802. [PubMed] [Google Scholar]
- 60.Padler-Karavani V, Yu H, Cao H, et al. Diversity in specificity, abundance, and composition of anti-Neu5Gc antibodies in normal humans: Potential implications for disease. Glycobiology 2008;18(10):818–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samraj AN, Bertrand KA, Luben R, et al. Polyclonal human antibodies against glycans bearing red meat-derived non-human sialic acid N-glycolylneuraminic acid are stable, reproducible, complex and vary between individuals: Total antibody levels are associated with colorectal cancer risk. PLoS One 2018;13(6):e0197464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bashir S, Fezeu LK, Leviatan Ben-Arye S, et al. Association between Neu5Gc carbohydrate and serum antibodies against it provides the molecular link to cancer: French NutriNet-santé study. BMC Med 2020;18(1):262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moriwaki K, Shinzaki S, Miyoshi E. GDP-mannose-4,6-dehydratase (GMDS) deficiency renders colon cancer cells resistant to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor- and CD95-mediated apoptosis by inhibiting complex II formation. J Biol Chem 2011;286(50):43123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joung J, Kirchgatterer PC, Singh A, et al. CRISPR activation screen identifies BCL-2 proteins and B3GNT2 as drivers of cancer resistance to T cell-mediated cytotoxicity. Nat Commun 2022;13(1):1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rodrigues E, Macauley MS. Hypersialylation in cancer: Modulation of inflammation and therapeutic opportunities. Cancers 2018;10(6):207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, Manni M, Bärenwaldt A, et al. Siglec receptors modulate dendritic cell activation and antigen presentation to T cells in cancer. Front Cell Dev Biol 2022;10:828916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stanczak MA, Rodrigues Mantuano N, Kirchhammer N, et al. Targeting cancer glycosylation repolarizes tumor-associated macrophages allowing effective immune checkpoint blockade. Sci Transl Med 2022;14(669):eabj1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stanczak MA, Siddiqui SS, Trefny MP, et al. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J Clin Invest 2018;128(11):4912–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stanczak MA, Läubli H. Siglec receptors as new immune checkpoints in cancer. Mol Aspects Med 2023;90:101112. [DOI] [PubMed] [Google Scholar]
- 70.Krug J, Rodrian G, Petter K, et al. N-Glycosylation regulates intrinsic IFN-γ resistance in colorectal cancer: Implications for immunotherapy. Gastroenterology 2023;164:392–406.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leite-Gomes E, Dias AM, Azevedo CM, et al. Bringing to light the risk of colorectal cancer in inflammatory bowel disease: Mucosal glycosylation as a key player. Inflamm Bowel Dis 2022;28(6):947–62. [DOI] [PubMed] [Google Scholar]
- 72.Guo H, Nagy T, Pierce M. Post-translational glycoprotein modifications regulate colon cancer stem cells and colon adenoma progression in Apcmin/+ mice through altered Wnt receptor signaling. J Biol Chem 2014;289(45):31534–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fleming-de-Moraes CD, Rocha MR, Tessmann JW, et al. Crosstalk between PI3K/Akt and Wnt/β-catenin pathways promote colorectal cancer progression regardless of mutational status. Cancer Biol Ther 2022;23(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodilla V, Villanueva A, Obrador-Hevia A, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci U S A 2009;106(15):6315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peng X, Luo Z, Kang Q, et al. FOXQ1 mediates the crosstalk between TGF-β and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol Ther 2015;16(7):1099–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cheruku HR, Mohamedali A, Cantor DI, et al. Transforming growth factor-β, MAPK and Wnt signaling interactions in colorectal cancer. EuPA Open Proteomics 2015;8:104–15. [Google Scholar]
- 77.Song L, Li ZY, Liu WP, et al. Crosstalk between Wnt/β-catenin and Hedgehog/Gli signaling pathways in colon cancer and implications for therapy. Cancer Biol Ther 2015;16(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma B, Hottiger MO. Crosstalk between wnt/β-catenin and NF-κB signaling pathway during inflammation. Front Immunol 2016;7:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silva MC, Fernandes Â, Oliveira M, et al. Glycans as immune checkpoints: Removal of branched N-glycans enhances immune recognition preventing cancer progression. Cancer Immunol Res 2020;8(11):1407–25. [DOI] [PubMed] [Google Scholar]
- 80.Demetriou M, Nabi IR, Coppolino M, et al. Reduced contact-inhibition and substratum adhesion in epithelial cells expressing GlcNAc-transferase V. J Cell Biol 1995;130(2):383–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Murata K, Miyoshi E, Kameyama M, et al. Expression of N-acetylglucosaminyltransferase V in colorectal cancer correlates with metastasis and poor prognosis. Clin Cancer Res 2000;6(5):1772–7. [PubMed] [Google Scholar]
- 82.St Hill CA, Farooqui M, Mitcheltree G, et al. The high affinity selectin glycan ligand C2-O-sLex and mRNA transcripts of the core 2 β-1,6-N-acetylglusaminyltransferase (C2GnT1) gene are highly expressed in human colorectal adenocarcinomas. BMC Cancer 2009;9(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.St Hill CA, Baharo-Hassan D, Farooqui M. C2-O-sLeX glycoproteins are E-selectin ligands that regulate invasion of human colon and hepatic carcinoma cells. PLoS One 2011;6(1):e16281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trinchera M, Malagolini N, Chiricolo M, et al. The biosynthesis of the selectin-ligand sialyl Lewis x in colorectal cancer tissues is regulated by fucosyltransferase VI and can be inhibited by an RNA interference-based approach. Int J Biochem Cell Biol 2011;43(1):130–9. [DOI] [PubMed] [Google Scholar]
- 85.Thirunavukarasu P, Sukumar S, Sathaiah M, et al. C-Stage in colon cancer: Implications of carcinoembryonic antigen biomarker in staging, prognosis, and management. J Natl Cancer Inst 2011;103(8):689–97. [DOI] [PubMed] [Google Scholar]
- 86.Drake PM, Cho W, Li B, et al. Sweetening the pot: Adding glycosylation to the biomarker discovery equation. Clin Chem 2010;56(2):223–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trbojević-Akmačić I, Lageveen-Kammeijer GSM, Heijs B, et al. High-throughput glycomic methods. Chem Rev 2022;122(20):15865–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.de Vroome SW, Holst S, Girondo MR, et al. Serum N-glycome alterations in colorectal cancer associate with survival. Oncotarget 2018;9(55):30610–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Qiu Y, Patwa TH, Xu L, et al. Plasma glycoprotein profiling for colorectal cancer biomarker identification by lectin glycoarray and lectin blot. J Proteome Res 2008;7(4):1693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu S, Cheng L, Fu Y, et al. Characterization of IgG N-glycome profile in colorectal cancer progression by MALDI-TOF-MS. J Proteomics 2018;181:225–37. [DOI] [PubMed] [Google Scholar]
- 91.Pan Y, Zhang L, Zhang R, et al. Screening and diagnosis of colorectal cancer and advanced adenoma by Bionic Glycome method and machine learning. Am J Cancer Res 2021;11(6):3002–20. [PMC free article] [PubMed] [Google Scholar]
- 92.Gu Y, Duan B, Sha J, et al. Serum IgG N-glycans enable early detection and early relapse prediction of colorectal cancer. Int J Cancer 2023;152(3):536–47. [DOI] [PubMed] [Google Scholar]
- 93.Takei D, Harada K, Nouso K, et al. Clinical utility of a serum glycome analysis in patients with colorectal cancer. J Gastroenterol Hepatol 2022;37(4):727–33. [DOI] [PubMed] [Google Scholar]
- 94.Barrow H, Guo X, Wandall HH, et al. Serum galectin-2, -4, and -8 are greatly increased in colon and breast cancer patients and promote cancer cell adhesion to blood vascular endothelium. Clin Cancer Res 2011;17(22):7035–46. [DOI] [PubMed] [Google Scholar]
- 95.de Haan N, Pučić-Baković M, Novokmet M, et al. Developments and perspectives in high-throughput protein glycomics: Enabling the analysis of thousands of samples. Glycobiology 2022;32(8):651–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pickering C, Zhou B, Xu G, et al. Differential peripheral blood glycoprotein profiles in symptomatic and asymptomatic COVID-19. Viruses 2022;14(3):553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ramachandran P, Xu G, Huang HH, et al. Serum glycoprotein markers in nonalcoholic steatohepatitis and hepatocellular carcinoma. J Proteome Res 2022;21(4):1083–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li Q, Kailemia MJ, Merleev AA, et al. Site-specific glycosylation quantitation of 50 serum glycoproteins enhanced by predictive glycopeptidomics for improved disease biomarker discovery. Anal Chem 2019;91(8):5433–45. [DOI] [PubMed] [Google Scholar]
- 99.Zauber AG, Lansdorp-Vogelaar I, Knudsen AB, et al. Evaluating test strategies for colorectal cancer screening: A decision analysis for the U.S. Preventive Services Task Force. Ann Intern Med 2008;149(9):659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Johnson CD, Fletcher JG, MacCarty RL, et al. Effect of slice thickness and primary 2D versus 3D virtual dissection on colorectal lesion detection at CT colonography in 452 asymptomatic adults. AJR Am J Roentgenol 2007;189(3):672–80. [DOI] [PubMed] [Google Scholar]
- 101.Pickhardt PJ, Puckett ML, Mysliwiec PA. Computed tomographic virtual colonoscopy to screen for colorectal neoplasia in asymptomatic adults. N Engl J Med 2003;349(23):2191–200. [DOI] [PubMed] [Google Scholar]
- 102.Zalis ME, Blake MA, Cai W, et al. Diagnostic accuracy of laxative-free computed tomographic colonography for detection of adenomatous polyps in asymptomatic adults: A prospective evaluation. Ann Intern Med 2012;156(10):692–702. [DOI] [PubMed] [Google Scholar]
- 103.US Preventive Services Task Force, Davidson KW, Barry MJ, Mangione CM, et al. Screening for colorectal cancer: US preventive Services Task Force recommendation statement. JAMA 2021;325(19):1965–77. [DOI] [PubMed] [Google Scholar]
- 104.Graser A, Stieber P, Nagel D, et al. Comparison of CT colonography, colonoscopy, sigmoidoscopy and faecal occult blood tests for the detection of advanced adenoma in an average risk population. Gut 2009;58(2):241–8. [DOI] [PubMed] [Google Scholar]
- 105.Johnson CD, Chen MH, Toledano AY, et al. Accuracy of CT colonography for detection of large adenomas and cancers. N Engl J Med 2008;359(12):1207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim YS, Kim N, Kim SH, et al. The efficacy of intravenous contrast-enhanced 16-raw multidetector CT colonography for detecting patients with colorectal polyps in an asymptomatic population in Korea. J Clin Gastroenterol 2008;42(7):791–8. [DOI] [PubMed] [Google Scholar]
- 107.Lefere P, Silva C, Gryspeerdt S, et al. Teleradiology based CT colonography to screen a population group of a remote island; at average risk for colorectal cancer. Eur J Radiol 2013;82(6):e262–267. [DOI] [PubMed] [Google Scholar]
- 108.Macari M, Bini EJ, Jacobs SL, et al. Colorectal polyps and cancers in asymptomatic average-risk patients: Evaluation with CT colonography. Radiology 2004;230(3):629–36. [DOI] [PubMed] [Google Scholar]
- 109.Ahlquist DA, Sargent DJ, Loprinzi CL, et al. Stool DNA and occult blood testing for screen detection of colorectal neoplasia. Ann Intern Med 2008;149(7):441–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shapiro JA, Bobo JK, Church TR, et al. A comparison of fecal immunochemical and high-sensitivity guaiac tests for colorectal cancer screening. Am J Gastroenterol 2017;112(11):1728–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Brenner H, Tao S. Superior diagnostic performance of faecal immunochemical tests for haemoglobin in a head-to-head comparison with guaiac based faecal occult blood test among 2235 participants of screening colonoscopy. Eur J Cancer Oxf Engl 2013;49(14):3049–54. [DOI] [PubMed] [Google Scholar]
- 112.Chiu HM, Ching JYL, Wu KC, et al. A risk-scoring system combined with a fecal immunochemical test is effective in screening high-risk subjects for early colonoscopy to detect advanced colorectal neoplasms. Gastroenterology 2016;150(3):617–25.e3. [DOI] [PubMed] [Google Scholar]
- 113.de Wijkerslooth TR, Stoop EM, Bossuyt PM, et al. Immunochemical fecal occult blood testing is equally sensitive for proximal and distal advanced neoplasia. Am J Gastroenterol 2012;107(10):1570–8. [DOI] [PubMed] [Google Scholar]
- 114.Hernandez V, Cubiella J, Gonzalez-Mao MC, et al. Fecal immunochemical test accuracy in average-risk colorectal cancer screening. World J Gastroenterol 2014;20(4):1038–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370(14):1287–97. [DOI] [PubMed] [Google Scholar]
- 116.Kim NH, Park JH, Park DI, et al. The fecal immunochemical test has high accuracy for detecting advanced colorectal neoplasia before age 50. Dig Liver Dis 2017;49(5):557–61. [DOI] [PubMed] [Google Scholar]
- 117.Park DI, Ryu S, Kim YH, et al. Comparison of guaiac-based and quantitative immunochemical fecal occult blood testing in a population at average risk undergoing colorectal cancer screening. Am J Gastroenterol 2010;105(9):2017–25. [DOI] [PubMed] [Google Scholar]
- 118.Redwood DG, Asay ED, Blake ID, et al. Stool DNA testing for screening detection of colorectal neoplasia in Alaska native people. Mayo Clin Proc 2016;91(1):61–70. [DOI] [PubMed] [Google Scholar]
- 119.Cooper GS, Markowitz SD, Chen Z, et al. Performance of multitarget stool DNA testing in African American patients. Cancer 2018;124(19):3876–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bosch LJW, Melotte V, Mongera S, et al. Multitarget stool DNA test performance in an average-risk colorectal cancer screening population. Am J Gastroenterol 2019;114(12):1909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsai WS, You JF, Hung HY, et al. Novel circulating tumor cell assay for detection of colorectal adenomas and cancer. Clin Transl Gastroenterol 2019;10(10):e00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lin WH, Xiao J, Ye ZY, et al. Circulating tumor DNA methylation marker MYO1-G for diagnosis and monitoring of colorectal cancer. Clin Epigenetics 2021;13(1):232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Luo H, Zhao Q, Wei W, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med 2020;12(524):eaax7533. [DOI] [PubMed] [Google Scholar]
- 124.Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat Biotechnol 2009;27(9):858–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pedersen SK, Symonds EL, Baker RT, et al. Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer 2015;15(1):654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nian J, Sun X, Ming S, et al. Diagnostic accuracy of methylated SEPT9 for blood-based colorectal cancer detection: A systematic review and meta-analysis. Clin Transl Gastroenterol 2017;8(1):e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wu X, Zhang Y, Hu T, et al. A novel cell-free DNA methylation-based model improves the early detection of colorectal cancer. Mol Oncol 2021;15(10):2702–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barták BK, Kalmár A, Péterfia B, et al. Colorectal adenoma and cancer detection based on altered methylation pattern of SFRP1, SFRP2, SDC2, and PRIMA1 in plasma samples. Epigenetics 2017;12(9):751–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang Q, Huang Z, Ni S, et al. Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS One 2012;7(9):e44398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang Z, Huang D, Ni S, et al. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer 2010;127(1):118–26. [DOI] [PubMed] [Google Scholar]
- 131.Barbagallo C, Brex D, Caponnetto A, et al. LncRNA UCA1, upregulated in CRC biopsies and downregulated in serum exosomes, controls mRNA expression by RNA-RNA interactions. Mol Ther Nucleic Acids 2018;12:229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Xu W, Zhou G, Wang H, et al. Circulating lncRNA SNHG11 as a novel biomarker for early diagnosis and prognosis of colorectal cancer. Int J Cancer 2020;146(10):2901–12. [DOI] [PubMed] [Google Scholar]
- 133.Li J, Song Y, Wang J, et al. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer detection. Am J Transl Res 2020;12(11):7395–403. [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang W, Yang S, Liu Y, et al. Hsa_circ_0007534 as a blood-based marker for the diagnosis of colorectal cancer and its prognostic value. Int J Clin Exp Pathol 2018;11(3):1399–406. [PMC free article] [PubMed] [Google Scholar]
- 135.Pan B, Qin J, Liu X, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet 2019;10:1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lledó SM, Garcia-Granero E, Dasí F, et al. Real time quantification in plasma of human telomerase reverse transcriptase (hTERT) mRNA in patients with colorectal cancer. Colorectal Dis 2004;6(4):236–42. [DOI] [PubMed] [Google Scholar]
- 137.Yeh CS, Wang JY, Wu CH, et al. Molecular detection of circulating cancer cells in the peripheral blood of patients with colorectal cancer by using membrane array with a multiple mRNA marker panel. Int J Oncol 2006;28(2):411–20. [PubMed] [Google Scholar]
- 138.Shen C, Hu L, Xia L, et al. Quantitative real-time RT-PCR detection for survivin, CK20 and CEA in peripheral blood of colorectal cancer patients. Jpn J Clin Oncol 2008;38(11):770–6. [DOI] [PubMed] [Google Scholar]
- 139.Ganig N, Baenke F, Thepkaysone ML, et al. Proteomic analyses of fibroblast- and serum-derived exosomes identify QSOX1 as a marker for non-invasive detection of colorectal cancer. Cancers 2021;13(6):1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sun B, Li Y, Zhou Y, et al. Circulating exosomal CPNE3 as a diagnostic and prognostic biomarker for colorectal cancer. J Cell Physiol 2019;234(2):1416–25. [DOI] [PubMed] [Google Scholar]
- 141.Vocka M, Langer D, Fryba V, et al. Novel serum markers HSP60, CHI3L1, and IGFBP-2 in metastatic colorectal cancer. Oncol Lett 2019;18(6):6284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim NH, Lee MY, Park JH, et al. Serum CEA and CA 19-9 levels are associated with the presence and severity of colorectal neoplasia. Yonsei Med J 2017;58(5):918–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Non-Clinical Verification and Clinical Validation of BeScreened-CRC, a Blood-Based in Vitro Diagnostic Multivariate Index Assay for the Detection of Colorectal Cancer in Screening Non-compliant Patients. Beacon Biomedical, Inc; (https://static1.squarespace.com/static/5b8832f8f2e6b1941b7c53ac/t/5df286f293135176b05b5edb/1576175349526/BeScreened-CRC+White+Paper_2017_+201901R1.pdf) (2017). Accessed December 7, 2022. [Google Scholar]
- 144.Friedland S, Watson D, Pan J, et al. Development and clinical validation of a blood test for early detection of colorectal adenomas and cancer for screening and postpolypectomy surveillance. Gastro Hep Adv 2022;1(2):223–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lin J, Ariazi E, Dzamba M, et al. Evaluation of a sensitive blood test for the detection of colorectal advanced adenomas in a prospective cohort using a multiomics approach. J Clin Oncol 2021;39(3Suppl l):43. [Google Scholar]
- 146.Lee J, Kim HC, Kim ST, et al. Multimodal circulating tumor DNA (ctDNA) colorectal neoplasia detection assay for asymptomatic and early-stage colorectal cancer (CRC). J Clin Oncol 2021;39(15 Suppl l):3536. [Google Scholar]
- 147.Westesson O, Axelrod H, Dean J, et al. Abstract 2316: Integrated genomic and epigenomic cell-free DNA (cfDNA) analysis for the detection of early-stage colorectal cancer. Cancer Res 2020;80(16 Suppl):2316. [Google Scholar]
- 148.Kim HC, Kim ST, He Y, et al. S130 multimodal circulating tumor DNA (ctDNA) blood-based colorectal cancer (CRC) screening test demonstrates clinically meaningful sensitivity across multiple clinical parameters. Off J Am Coll Gastroenterol ACG 2021;116(1):S56. [Google Scholar]
- 149.Guardant Health, Inc. Shield detects CRC with high accuracy. Guardant health blood-based screening (https://bloodbasedscreening.com/crc-screening/) Accessed December 7, 2022.
- 150.Goldstein MJ, Mitchell EP. Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer Invest 2005;23(4):338–51. [DOI] [PubMed] [Google Scholar]
- 151.Wu S, Gu W. Association of T stage and serum CEA levels in determining survival of rectal cancer. Front Med 2019;6:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Björkman K, Jalkanen S, Salmi M, et al. A prognostic model for colorectal cancer based on CEA and a 48-multiplex serum biomarker panel. Sci Rep 2021;11(1):4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vukobrat-Bijedic Z, Husic-Selimovic A, Sofic A, et al. Cancer antigens (CEA and CA 19-9) as markers of advanced stage of colorectal carcinoma. Med Arch Sarajevo Bosnia Herzeg 2013;67(6):397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lakemeyer L, Sander S, Wittau M, et al. Diagnostic and prognostic value of CEA and CA19-9 in colorectal cancer. Dis Basel Switz 2021;9(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shibutani M, Maeda K, Nagahara H, et al. Significance of CEA and CA19-9 combination as a prognostic indicator and for recurrence monitoring in patients with stage II colorectal cancer. Anticancer Res 2014;34(7):3753–8. [PubMed] [Google Scholar]
- 156.Björkman K, Mustonen H, Kaprio T, et al. CA125: A superior prognostic biomarker for colorectal cancer compared to CEA, CA19-9 or CA242. Tumour Biol 2021;43(1):57–70. [DOI] [PubMed] [Google Scholar]
- 157.Huang CJ, Jiang JK, Chang SC, et al. Serum CA125 concentration as a predictor of peritoneal dissemination of colorectal cancer in men and women. Medicine (Baltimore) 2016;95(47):e5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gao Y, Wang J, Zhou Y, et al. Evaluation of serum CEA, CA19-9, CA72-4, CA125 and ferritin as diagnostic markers and factors of clinical parameters for colorectal cancer. Sci Rep 2018;8(1):2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kuipers EJ, Grady WM, Lieberman D, et al. Colorectal cancer. Nat Rev Dis Primer 2015;1:15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Davies RJ, Miller R, Coleman N. Colorectal cancer screening: Prospects for molecular stool analysis. Nat Rev Cancer 2005;5(3):199–209. [DOI] [PubMed] [Google Scholar]