Abstract
Reverse transcriptase PCR (RT-PCR) was used for polyribonucleotide assays with sera from deployed Persian Gulf War veterans with the Gulf War Syndrome and a cohort of nonmilitary controls. Sera from veterans contained polyribonucleotides (amplicons) that were obtained by RT-PCR and that ranged in size from 200 to ca. 2,000 bp. Sera from controls did not contain amplicons larger than 450 bp. DNA sequences were derived from two amplicons unique to veterans. These amplicons, which were 414 and 759 nucleotides, were unrelated to each other or to any sequence in gene bank databases. The amplicons contained short segments that were homologous to regions of chromosome 22q11.2, an antigen-responsive hot spot for genetic rearrangements. Many of these short amplicon segments occurred near, between, or in chromosome 22q11.2 Alu sequences. These results suggest that genetic alterations in the 22q11.2 region, possibly induced by exposures to environmental genotoxins during the Persian Gulf War, may have played a role in the pathogenesis of the Gulf War Syndrome. However, the data did not exclude the possibility that other chromosomes also may have been involved. Nonetheless, the detection of polyribonucleotides such as those reported here may have application to the laboratory diagnosis of chronic diseases that have a multifactorial etiology.
During the Persian Gulf War approximately 700,000 individuals were exposed to genotoxic hazardous materials (GHM) (42, 42a, 51). The GHMs to which these individuals were exposed included low-level chemical warfare agents, investigational drugs (including pyridostigmine bromide, which is used as a prophylactic agent against nerve agents), organophosphate, carbamate, and other pesticides and insect repellents. Other occupational and environmental contaminants included low levels of nuclear and electromagnetic radiation, toxic combustion products from oil-well fires, diesel exhaust products, and airborne particulates. A significant proportion of the Persian Gulf War veterans (GWVs) developed a pattern of symptomatic health disorders that have been referred to as Persian Gulf War-Related Illnesses (42). The pattern of illness is reasonably consistent: rash, fatigue, muscle and joint pain, headache, irritability, depression, unrefreshing sleep, gastrointestinal and respiratory disorders, and cognitive defects (22). These Gulf War Syndrome (GWS) disorders were recently defined as a clinical entity (16).
We elected to test for polyribonucleotides in the sera of GWVs on the basis of several considerations. Most GWVs received oral poliovirus vaccine before deployment to the Persian Gulf. Persistent enterovirus infection has been implicated in the chronic fatigue syndrome (18), one of the major health disorders of GWS. Clements et al. (8) reported that enterovirus-related sequences persisted in the sera of patients with the chronic fatigue syndrome. The availability of primers (14) to the nontranslated sequences of most enteroviruses and to the P2-P3 junction of oral polioviruses provided a means to test whether enterovirus sequences persisted in the sera of GWVs. We used a reverse transcriptase (RT) PCR (RT-PCR), described in this report, to detect amplicons (RT-PCR amplicons [RPAs]) in the sera tested. We report that amplicons that were 750 bp or larger occurred in the sera of GWVs but not in the sera of healthy nonmilitary controls. Two amplicons (of 414 and 714 bp) unique to GWVs were sequenced. They contained short segments homologous to regions of chromosome 22q11.2, a hot spot for genetic rearrangements and mutations.
MATERIALS AND METHODS
RT-PCR.
Sera from peripheral blood specimens were obtained after the provision of informed consent from 24 veterans with GWS (Rheumatology Clinic, Veterans Affairs, Northern California Health Care System, Martinez, Calif.) who had been deployed to the Persian Gulf approximately 5 years previously. The major signs and symptoms in the 24 GWVs with GWS were rash (n = 20), muscle and joint pain (n = 20), headache depression irritability (n = 19), gastrointestinal and respiratory disorders (n = 18), chronic fatigue syndrome (n = 17), posttraumatic stress disorder (n = 12), and cognitive losses (n = 6). Combinations of these symptoms occurred in all but one veteran. Blinded serum samples from 50 healthy nonmilitary subjects were obtained from life insurance applicants (Osborn Laboratories, Lenexa, Kans.). For the most part, the subjects were matched by age, sex, and race. They ranged in age from 26 to 56 years. All sera were separated from clots immediately after blood was drawn and were used for RT-PCR within 48 h. To prevent cross contamination, separate facilities dedicated to specimen processing, PCR amplification, and amplicon detection were used. RNA from 0.25 ml of the sample was extracted in a laminar flow hood with 0.75 ml of TRIZOL LS reagent (Gibco BRL, Gaithersburg, Md.). RNA was precipitated with 10 μg of RNase-free glycogen as a carrier. Both methods were performed as specified by the manufacturer. Precipitated RNA was washed once with 70% ethanol by centrifugation at 4°C, resuspended in 10 μl of RNase-free distilled water, and added to 17 μl of the RT mixture (GeneAmp RNA PCR kit; Perkin-Elmer, Norwalk, Conn.) containing MgCl2 (5 mM), 1× PCR Buffer II, RNase inhibitor (2.5 U), murine leukemia virus RT (2.5 U), random hexamer primers (2.5 μM), and 1 mM each dATP, dGTP, dCTP, and dTTP. Poliovirus Sabin type 1 RNA (National Institute for Biological Standards and Control, Hertfordshire, United Kingdom) was used as a positive control. RT was omitted from the reaction mixture for the negative control. The RT mixture was incubated for 10 min at 22°C, 30 min at 42°C, and 5 min at 95°C with a Perkin-Elmer thermocycler. The RT mixture was then added to the top of a hot-start PCR, with a melted Ampliwax bead (Perkin-Elmer) used as the barrier. The 70 μl of the top PCR mixture contained 1× PCR Buffer II and Amplitaq (2.5 U). The 30 μl of the bottom PCR mixture contained 1× PCR Buffer II, 2 mM MgCl2, and the appropriate primer pairs (15 μM). Primers from the enteroviral nontranslated region (primer PG01 [5′-AAGCACTTCTGTTTCC-3′] and primer PG02 [5′-CATTCAGGGGCCGGAGGA-3′]) and the poliovirus viral protein region (P2-P3 junction of poliovirus types 1 and 2; primer PG03 [5′-GAAATGTGTAAGAACTGTCA-3′] and primer PG04 [5′-GTAACAATGTTTCTTTTAGCC-3′]) were used as primer pairs or in a multiplex combination. After 35 cycles of amplification (1 min at 94°C, 2 min at 48°C, and 1 min at 72°C), 8 μl of the PCR mixture was electrophoresed with a precast 6% polyacrylamide gel in TBE buffer (45 mM Tris, 45 mM boric acid, 1 mM EDTA) (NOVEX, San Diego, Calif.) for 30 min at 200 V. The gels were stained for 20 min in a 0.5-μg/ml ethidium bromide solution and were photographed under UV light.
Cloning and sequencing.
Sera from three different veterans were processed on three different days. The PCR products were run on and excised from a 2% NuSieve GTG low-melting-point agarose gel (FMC BioProducts, Rockland, Maine). The bands were blunt-end cloned with the Prime PCR Cloner Kit (5 PRIME-3 PRIME, Inc., Boulder, Colo.) according to the manufacturer’s specifications. Sequence analysis was performed with the automated sequencer from ABI PRISM (Operon Technologies, Inc., Alameda, Calif.).
Statistical analysis.
A 2-by-2 contingency analysis (see Table 1) was done by using Graphpad InStat software (Graphpad Program Software, San Diego, Calif.).
TABLE 1.
Occurrence of polyribonucleotide bands in sera from GWVs and nonmilitary controls
| Banda | Band size (bp) | No. (%) positive
|
P valueb | |
|---|---|---|---|---|
| GWVs (n = 24) | Nonmilitary controls (n = 50) | |||
| EV NTR | 297c | 14 (58) | 21 (42) | 0.22 |
| Polio P2/P3 | 565d | 10 (42) | 11 (22) | 0.10 |
| 200 | 2 (8) | 15 (30) | 0.043 | |
| Non-EV | 350 | 4 (17) | 0 (0) | 0.0092 |
| 450 | 17 (71) | 19 (38) | 0.0125 | |
| 750 | 12 (50) | 0 (0) | <0.0001 | |
EV NTR, enterovirus nontranslated region; EV, enterovirus; Non-EV, nonenterovirus.
See Materials and Methods for description of statistical analysis.
The PG01-PG02 primer pair detects a 297-bp band from the nontranslated region of a majority of enteroviruses.
The PG03-PG04 primer pair detects a 565-bp band of the P2-P3 junction of the oral poliovirus vaccine strains, Sabin types 1 and 2.
GenBank Search.
All GenBank and EMBL searches were done with the DNASTAR Lasergene CD-ROM and software (release 103, November 1997; DNASTAR, Madison, Wis.). Homology searches were performed with 2 through 6 k-tuples with window sizes of 11 to 100 nucleotides (nt). Homology searches for 14 nt or higher were done by starting with position 1 and continuing through to the last 14-nt segment of each amplicon. All sequences with 100% homology were recorded and are presented in Table 2.
TABLE 2.
Segment homologies among GWS RPAs and human chromosome 22q11.2
| RPA, sequence, and position | GenBank accession no. | 22q11.2 GenBank sequence | Segment location | No. of other 100% matches |
|---|---|---|---|---|
| 759-nt RPA | ||||
| 15mers | ||||
| 59–73 | HSU07000 | 809–823 | Between two Alu regions | 0 human, 0 nonhuman |
| 83–97 | HSCN37F10 | 36332–36346 | Between MIR and Alu | 0 human, 0 nonhuman |
| 711–725 | HS322B1 | 45987–46001 | Between AluSx and MIR | 1 human, 0 nonhuman |
| 14mers | ||||
| 11–24 | D86998 | 23151–23164 | Between V2–8 and V1–3 | 2 human, 2 nonhuman |
| 136–149 | U30597 | 227965–227978 | Between two Alu regions | 0 human, 8 nonhuman |
| 194–207 | HSE78G1 | 29369–29382 | Between two Alu regions | 0 human, 3 nonhuman |
| 343–356 | HSN44A4 | 1460–1473 | In AluY | 9 human, 25 nonhuman |
| 377–390 (GWV 1) | HSN38E12 | 19903–19916 | Between AluSx and repeat region | 2 human, 3 nonhuman |
| 377–390 (GWV 2 and 3) | HSF4G12 | 39069–39082 | Between two repeat regions | 18 human, 22 nonhuman |
| 462–475 | HSN20A6 | 17205–17218 | Near flanking repeat region | 4 human, 1 nonhuman |
| 551–564 | AC000068 | 14136–14149 | No description | 0 human, 2 nonhuman |
| 634–647 | D87021 | 17508–17521 | Between V2–7 and V2–6 light-chain genes | 15 human, 7 nonhuman |
| 414-nt RPA | ||||
| 15mers | ||||
| 190–204 | AC002475 | 1301–1315 | No description | 23 human, 1 nonhuman |
| 310–324 | HSN74G7 | 10657–10671 | Between Alu repeat and repeat region | 3 human, 0 nonhuman |
| 14mers | ||||
| 136–149 | HS65B7 | 4097–4110 | Inside MIR repeat | 4 human, 0 nonhuman |
| 155–168 | HSE78G1 | 35878–35891 | Between repeat regions | 18 human, 2 nonhuman |
| 270–283 | HSE146D10 | 522–535 | Between repeat regions | 4 human, 2 nonhuman |
| 395–408 | HSE116C6 | 9265–9278 | Between Alu and repeat region | 1 human, 14 nonhuman |
Sequences from a survey of consensus sequences with 100% homology to the designated RPAs from sera from GWVs with GWS (GWS RPAs) were divided into human and nonhuman categories according to the GenBank definition of the entry. MIR, mammalian-wide interspersed repeat.
Nucleotide sequence accession numbers.
The sequences of the 414- and 759-nt sequences derived from sera from patients with GWS were placed in the GenBank database under accession nos. AF100637 and AF100636, respectively.
RESULTS
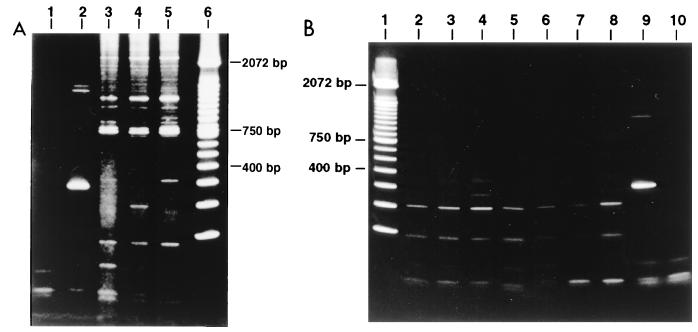
Sera from 24 deployed GWVs and 50 serum samples from healthy nonmilitary controls were tested for RPAs. Figure 1A shows the presence of multiple bands in the sera from GWVs. The pattern was typical for most veterans, i.e., the occurrence of several bands in the 300- to 750-bp regions accompanied by discrete bands with sequences longer than 2,000 bp. These band patterns were detected by RT-PCR but not by direct PCR, implicating the presence of RNAs and not DNAs in the sera. Figure 1B shows a representative gel in which sera from seven healthy nonmilitary controls were tested. Only a few distinct bands were found. There were no bands larger than 450 bp. The results for 24 veterans and 50 healthy controls (Table 1) indicate the differences in the occurrence of RPAs in the two cohorts.
FIG. 1.
Nucleotide bands (amplicons) in sera from GWVs and nonmilitary controls. (A) Results for representative samples from three different veterans. Lane 1, poliovirus without RT as a negative control; lane 2, poliovirus-positive control; lane 3, serum from veteran 1; lane 4, serum from veteran 2; lane 5, serum from veteran 3; lane 6, 100-bp ladder. (B) Results for representative samples from seven different nonmilitary controls. Lane 1, 100-bp ladder; lanes 2 to 8, sera from seven healthy controls, respectively; lane 9, poliovirus-positive control; lane 10, poliovirus without RT as a negative control.
Two bands in the gel regions of ca. 400 and 750 bp that occurred only in the sera of GWVs were isolated, cloned, and sequenced. Figure 2 presents the consensus sequence data for isolates from three different veterans. Each of the 414- and 759-nt sequences from the three different isolates had approximately 99% homology. The 414 and 759-nt GWS sequences contained several initiation and stop codons (open reading frames) that could code for small polypeptides. Neither the 414- nor 759-nt sequences had direct homologies to sequences in GenBank. In analogous studies with sera from approximately 30 patients with active multiple myeloma (13), we detected RPAs that were related to chromosome 22q11.2. We therefore elected to search the chromosome 22q11.2 database for homologies to the 414- and 759-nt sequences. Several short segments of 15 nt (15mer) and 14 nt (14mer) were found. Table 2 shows that three 15mer and eight 14mer segments of the 759-nt sequence had 100% homology to sequences in chromosome 22q11.2. One 14mer segment, from positions 377 to 390 (Fig. 2 and Table 2), was identical for GWVs 2 and 3 but differed by two nucleotides for GWV 1. The 14mer from GWVs 2 and 3 had 100% homology with a segment in the sequence with GenBank accession no. HSF4G12. The 14mer from GWV 1 had 100% homology with a segment in the sequence with GenBank accession no. HSN38E12. The gene sequences from GenBank accession nos. HSF4G12 and HSN38E12 are both located on chromosome 22q11.2. Six of 11 RPA segments were located either within an Alu region (12), between Alu and other repeat regions, or as segments flanking an Alu region. Five 759-nt segments occurred only in the chromosome 22q11.2 region. Two 14mers of the 759-nt sequence were located proximal to the immunoglobulin lambda light-chain variable-region genes. For the 414-nt sequence, there were two 15mer and four 14mer segments that also had 100% homology within the 22q11.2 region. However, these six segments also occurred at sites on other chromosomes. Interestingly, unique 15mer segments were not found in any chromosomal region other than 22q11.2.
FIG. 2.
Sequences of the 759-nt (A) and 414-nt (B) RPAs derived from the sera from three different GWVs. Boxed sequences denote 22q11.2 homologies (Table 2). Enteroviral primers are underlined.
DISCUSSION
The pattern of RPAs (polyribonucleotides) found in sera from GWVs was distinct from that found in sera from the nonmilitary cohort. Moreover, RPAs larger than 450 bp did not occur in the sera from healthy controls. The frequencies of occurrence of RPAs homologous to the poliovirus P2-P3 junction sequences and the enteroviral nontranslated region were not significantly different in the two groups (Table 1). The gels shown in Fig. 1 disclosed many bands larger than 2,000 bp in the sera of GWVs. No attempt was made to resolve or to characterize them at the molecular level. Such studies are in progress. Analysis of the 414- and 759-nt sequences showed that they are not related and that the 414-nt sequence is not a degradation product of the 759-nt sequence.
In attempting to understand the pathogenesis of GWS, the challenge has been to explain the diversity of the signs and symptoms typical of the disorder. A traditional approach of invoking a single cause is not applicable because it fails to accommodate three basic considerations. First, the etiology of the disease is multifactorial (49). Thus, different groups of signs and symptoms very likely have different causes. Second, exposure to environmental genotoxins during the Persian Gulf War likely caused an interaction among causative factors, thus affecting expression of signs and symptoms in given individuals. Third, and consistent with multifactorial diseases in general, the genetic and physiologic diversity of the affected population is in accord with the spectrum of disease expression seen. These concepts are known to be relevant to a number of chronic multifactorial diseases such as rheumatoid arthritis, systemic lupus erythematosus, multiple sclerosis, and insulin-dependent diabetes mellitus. For such diseases it has been essential to identify the individual causative factors, to weigh the contributions of each to the overall clinicopathologic picture, to determine how they interact in various population groups, and to evaluate the effects of different environmental influences.
The notions outlined above reflect our approach to an analysis of GWS. First, we sought to determine whether enterovirus infection could be a contributory factor in the pathogenesis of GWS. Molecular studies that have used PCR technologies have indicated persistent enterovirus infection in myalgia and myositis (54), dermatomyositis and polymyositis (5, 43), neuromuscular disease (28, 37), and the chronic fatigue syndrome (18). The signs and symptoms of these disorders are common in GWS. Moreover, enterovirus infection is known to cause a variety of immunologic and autoimmune disorders (9, 17). Immunologic disorders appear to make up an important component of the signs and symptoms of GWS. Studies of immunologic abnormalities in GWS, similar to those done for the chronic fatigue syndrome (4), appear to offer an important approach in an analysis of the pathogenesis of the disease.
To the best of our knowledge this is the first report of the occurrence of nonviral RPAs in the sera of subjects with a multifactorial chronic disease. We consider four central questions: (i) the possible origin(s) of the polyribonucleotides (amplicons) found in sera, (ii) the possible role(s) of chromosome 22q11.2 in the pathogenesis of the GWS, (iii) whether environmental genotoxins may have played a role in its pathogenesis, and (iv) the possible diagnostic value of detecting RPAs in the sera of patients with chronic diseases.
Identification of the possible origin(s) of the RPAs in sera is an important consideration. Since the occurrence of nonenterovirus RPAs in the sera of GWVs and controls was unexpected, we were concerned that they might have been PCR artifacts. Specific steps had been taken to minimize this possibility (see Materials and Methods). Two separate lines of evidence indicate that the RPAs described here were not artifactual in origin: (i) we developed a non-PCR, total RNA assay that independently confirmed that RNA species occur in the sera of patients with chronic diseases; and (ii) studies of approximately 30 patients with active multiple myeloma and 152 healthy controls by the described RT-PCR assay disclosed the occurrence of unique RPAs, e.g., GenBank accession no. AF018254, in test sera. Accordingly, our data suggest that individual chronic diseases may be characterized by the consistent occurrence of unique RPAs in the sera of patients with the individual chronic diseases.
An explanation of how polyribonucleotides could persist in the sera without being degraded is also needed. A reasonable account comes from the work of Wieczorek et al. (52), who reported that RNAs in the sera of patients with a variety of malignancies persisted as RNase-resistant RNA-proteolipid complexes. Salmon and Seligmann (45) referred to the occurrence of RNAs in the sera of patients with multiple myeloma. We recently confirmed and extended these findings (13). We detected a 705-bp segment homologous to the flanking region of the peroxisome proliferator-activated receptor exon 4 sequence located on chromosome 22q11.2. We are testing whether RPAs found in sera were derived from diverse tissue and cellular origins. These experiments are based on the clinical observation that immunologic abnormalities appear to be commonplace in GWS. In addition, Koga et al. (25) reported that uninfected thymocytes from healthy humans contained elevated amounts of heterodisperse RNA. Such heterodisperse RNA may be released into the circulation as a result of thymocyte apoptosis. Presumably, such RNAs would be protected from RNase degradation because of a physical association with cellular debris, as described by Wieczorek et al. (52). This hypothesis takes into consideration the evident immunologic dyscrasias that are observed in patients with GWS and that presumably occur because of underlying disorders in immune regulation.
None of the RPA sequence data disclosed homologies to enterovirus or poliovirus sequences. Since only a fraction of the RPAs observed in gels were sequenced, we do not exclude the possibility that some of them were enterovirus related. We assume that the RPAs that were sequenced are direct transcripts of recombinant sequences, although direct experimental proof is still required. Both the 414- and 759-nt RPAs, which were found only in the sera from the three GWVs tested, had short 14mers or 15mers (Table 2) that were 100% homologous to chromosome 22q11.2 segments. These findings suggest that abnormalities in chromosome 22q11.2 are involved, either directly or indirectly, in the pathogenesis of GWS. This does not mean that chromosomal regions other than 22q11.2 are not involved. Nonetheless, it appears that the GWS may be added to the list of diseases in which abnormalities in chromosome 22q11.2 are involved. These include the recently defined chromosome 22q11.2 deletion syndrome (46, 48), juvenile rheumatoid arthritis-like polyarthritis (47), idiopathic thrombocytopenic purpura (29), and hypoparathyroidism (3). In fact, deletion from chromosome 22q11 is the most common microdeletion (36). Interestingly, up to 60% of subjects (36, 53) with such deletions suffer from behavioral or psychiatric disorders. Also of note, chromosome 22 appears to be involved in the so-called Goldenhar complex (21, 24), a birth defect possibly associated with GWS (19). The mechanisms involved in embryonic development and 22q deletion disorders are now being defined at the molecular level (33).
The occurrence of hot spots for genetic deletions, translocations (6), and rearrangements, e.g., immunoglobulin lambda light chains (15, 44), in chromosome 22q11.2 is recognized widely. Such hot spots may be particularly sensitive to adverse genotoxic effects of environmental GHMs encountered during service in the Persian Gulf War. Studies with animal models (2) suggest that combined or multiple exposures to GHMs may have a synergistic genotoxic effect, thus causing some of the symptoms seen in GWS.
The juxtaposition of the detected RPA sequences with Alu sequences in chromosome 22q11.2 also may be relevant to the pathogenesis of GWS. The contemporary notion that Alu sequences are “junk DNA” is not consistent with the accumulating evidence that Alu sequences become transcriptionally active when cells are exposed to physiologic insults such as infection with DNA viruses (10, 40) or human immunodeficiency virus type 1 (23, 25) or when cells are induced to express heat shock proteins (7). Liu et al. (32) reported that cells stressed by exposure to cycloheximide or puromycin “rapidly and transiently increased the abundance of Alu RNA.” We postulate that the expression of RNAs of Alu sequences, their flanking regions, and their recombinants in response to GHMs may be a supplemental mechanism for detoxification of GHMs (11, 38). Such Alu-Alu recombinants are generated by both extrachromosomal and chromosomal genetic mechanisms (20, 27, 31, 35, 39, 41). In addition, Makalowski et al. (34) described the role of Alu sequences in generating diverse proteins. Such diverse proteins may also contribute to autoimmune reactivities in patients with GWS and possibly other chronic disorders.
The possible roles of the detected RPAs in the pathogenesis of GWS are unknown. Nonetheless, their occurrence makes available markers that can be studied for possible pathophysiologic effects. The biological activities of such molecules can be significant. Krieg (26) reported that specific CpG Alu-rich DNA (30) sequences in the plasma of patients with systemic lupus erythematosus may play an important role in the pathophysiology of the disease. Interestingly, chromosome 22 is rich in CpG islands. In addition, Abken et al. (1) reported that novel mouse cytoplasmic DNA sequences immortalized human lymphocytes in vitro. Such studies provide a paradigm for GWS.
The patterns of the occurrence of RPAs in the sera of GWVs and healthy controls are sufficiently distinct to suggest possible future diagnostic applications. Sufficiently large numbers of subjects need to be studied (50) to determine the sensitivities and specificities of such tests. Our studies of patients with active multiple myeloma (13) suggest that patients with individual chronic multifactorial diseases may have unique RPAs in their sera. Validated tests for such putative surrogate markers may aid in the diagnosis of such diseases or in the evaluation of responses to therapeutic modalities.
ACKNOWLEDGMENTS
This work was supported by donations to the Chronic Illness Research Foundation.
We thank Miger Ornopia and Atefeh Farvard for excellent technical assistance. We thank Bradford Saget for reviewing the manuscript and for helpful suggestions.
REFERENCES
- 1.Abken H, Hegger R, Butzler C, Willecke K. Short DNA sequences from the cytoplasm of mouse tumor cells induce immortalization of human lymphocytes in vitro. Proc Natl Acad Sci USA. 1993;90:6518–6522. doi: 10.1073/pnas.90.14.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou-Donia M B, Wilmarth K R, Jensen K F, Oehme F W, Kurt T L. Neurotoxicity resulting from coexposure to pyridostigmine bromide, DEET and permethrin: implications of Gulf War chemical exposures. J Toxicol Environ Health. 1996;48:35–56. doi: 10.1080/009841096161456. [DOI] [PubMed] [Google Scholar]
- 3.Adachi M, Tachibana K, Masuno M, Makita Y, Maesaka H, Okada T, Hizukuri K, Imaizumi K, Kuroki Y, Kurahashi H, Suwa S. Clinical characteristics of children with hypoparathyroidism due to 22q11.2 microdeletion. Eur J Pediatr. 1998;157:34–38. doi: 10.1007/s004310050762. [DOI] [PubMed] [Google Scholar]
- 4.Barker W, Fujimura S F, Fadem M B, Landay A L, Levy J A. Immunologic abnormalities associated with the chronic fatigue syndrome. Clin Infect Dis. 1994;18:S136–S141. doi: 10.1093/clinids/18.supplement_1.s136. [DOI] [PubMed] [Google Scholar]
- 5.Bowles N E, Sewry C A, Dubowitz V, Archard L C. Dermatomyositis, polymyositis, and coxsackie-B-virus infection. Lancet. 1987;i:1004–1007. doi: 10.1016/s0140-6736(87)92271-9. [DOI] [PubMed] [Google Scholar]
- 6.Calasanz M J, Cigudosa J C, Odero M D, Ferreira C, Ardanaz M T, Fraile A, Carrasco J L, Sole F, Cuesta B, Gullon A. Cytogenetic analysis of 280 patients with multiple myeloma and related disorders: primary breakpoints and clinical correlations. Genes Chromosomes Cancer. 1997;18:84–93. [PubMed] [Google Scholar]
- 7.Chu W M, Ballard R, Carpick B W, Williams B R G, Schmid C W. Potential Alu function: regulation of the activity of double-stranded RNA-activated kinase PKR. Mol Cell Biol. 1998;18:58–68. doi: 10.1128/mcb.18.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clements G B, McGarry F, Nairn C, Galbraith D N. Detection of enterovirus-specific RNA in serum: the relationship to chronic fatigue. J Med Virol. 1995;45:156–161. doi: 10.1002/jmv.1890450208. [DOI] [PubMed] [Google Scholar]
- 9.Craighead J E, Huber S A, Sriram S. Animal models of picornavirus-induced autoimmune disease: their possible relevance to human disease. Lab Invest. 1990;63:432–446. [PubMed] [Google Scholar]
- 10.Darlix J L, Khandjian E, Weil R. Nature and origin of the RNA associated with simian virus 40 large tumor antigen. Proc Natl Acad Sci USA. 1984;81:5425–5429. doi: 10.1073/pnas.81.17.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies H G, Richter R J, Keifer M, Broomfield C A, Sowalla J, Furlong C E. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat Genet. 1996;14:334–336. doi: 10.1038/ng1196-334. [DOI] [PubMed] [Google Scholar]
- 12.Deininger P L. SINEs: short interspersed repeated DNA elements in higher eucaryotes. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 619–636. [Google Scholar]
- 13.Durie, B. G. M., W. H. Murphy, and H. B. Urnovitz. GenBank accession no. AF018254. Unpublished data.
- 14.Egger D, Pasamontes L, Ostermayer M, Bienz K. Reverse transcription multiplex PCR for differentiation between polio- and enteroviruses from clinical and environmental samples. J Clin Microbiol. 1995;33:1442–1447. doi: 10.1128/jcm.33.6.1442-1447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frippiat J P, Williams S C, Tomlinson I M, Cook G P, Cherif D, Le Paslier D, Collins J E, Dunham I, Winter G, Lefranc M P. Organization of the human immunoglobulin lambda light-chain locus on chromosome 22q11.2. Hum Mol Genet. 1995;4:983–991. doi: 10.1093/hmg/4.6.983. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda K, Nisenbaum R, Stewart G, Thompson W W, Robin L, Washko R M, Noah D L, Barrett D H, Randall B, Herwaldt B L, Mawle A C, Reeves W C. Chronic multisymptom illness affecting Air Force veterans of the Gulf War. JAMA. 1998;280:981–988. doi: 10.1001/jama.280.11.981. [DOI] [PubMed] [Google Scholar]
- 17.Garzelli C, Basolo F, Matteucci D, Prabhakar B S, Toniolo A. Picornavirus-induced immunosuppression. In: Specter S, Bendinelli M, Friedman H, editors. Virus-induced immunosuppression. New York, N.Y: Plenum Press; 1989. pp. 193–200. [Google Scholar]
- 18.Gow J W, Behan W M H. Amplification and identification of enteroviral sequences in the postviral fatigue syndrome. Br Med Bull. 1991;47:872–885. doi: 10.1093/oxfordjournals.bmb.a072517. [DOI] [PubMed] [Google Scholar]
- 19.Haley R W. Point: bias from the “healthy-warrior effect” and unequal follow-up in three governmental studies of health effects in the Gulf War. Am J Epidemiol. 1998;148:315–323. doi: 10.1093/oxfordjournals.aje.a009645. [DOI] [PubMed] [Google Scholar]
- 20.Harteveld K L, Losekoot M, Fodde R, Giordano P C, Bernini L F. The involvement of Alu repeats in recombination events at the alpha-globin gene cluster: characterization of two alphazero-thalassaemia deletion breakpoints. Hum Genet. 1997;99:528–534. doi: 10.1007/s004390050401. [DOI] [PubMed] [Google Scholar]
- 21.Herman G E, Greenberg F, Ledbetter D H. Multiple congenital anomaly/mental retardation (MCA/MR) syndrome with Goldenhar complex due to a terminal del(22q) Am J Med Genet. 1988;29:909–915. doi: 10.1002/ajmg.1320290423. [DOI] [PubMed] [Google Scholar]
- 22.The Iowa Persian Gulf Study Group. Self-reported illness and health status among Gulf War veterans. A population-based study. JAMA. 1997;277:238–245. [PubMed] [Google Scholar]
- 23.Jang K L, Collins M K L, Latchman D S. The HIV tat protein increases the transcription of human Alu repeated sequences by increasing the activity of the cellular transcription factor TFIIIC. J Acquired Immune Defic Syndr. 1992;5:1142–1147. [PubMed] [Google Scholar]
- 24.Kobrynski L, Chitayat D, Zahed L, McGregor D, Rochon L, Brownstein S, Vekemans M, Albert D L. Trisomy 22 and facioauriculovertebral (Goldenhar) sequence. Am J Med Genet. 1993;46:68–71. doi: 10.1002/ajmg.1320460111. [DOI] [PubMed] [Google Scholar]
- 25.Koga Y, Lindstrom E, Fenyo E M, Wigzell H, Mak T W. High levels of heterodisperse RNAs accumulate in T cells infected with HIV and in normal thymocytes. Proc Natl Acad Sci USA. 1988;85:4521–4525. doi: 10.1073/pnas.85.12.4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krieg A M. CpG DNA: a pathogenic factor in systemic lupus erythematosus? J Clin Immunol. 1995;15:284–292. doi: 10.1007/BF01541318. [DOI] [PubMed] [Google Scholar]
- 27.Lehrman M A, Goldstein J L, Russell D W, Brown M S. Duplication of seven exons in LDL receptor gene caused by Alu-Alu recombination in a subject with familial hypercholesterolemia. Cell. 1987;48:827–835. doi: 10.1016/0092-8674(87)90079-1. [DOI] [PubMed] [Google Scholar]
- 28.Leparc-Goffart L, Julien J, Fuchs F, Janatova I, Aymard M, Kopecka H. Evidence of presence of poliovirus genomic sequences in cerebrospinal fluid from patients with postpolio syndrome. J Clin Microbiol. 1996;34:2023–2026. doi: 10.1128/jcm.34.8.2023-2026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levy A, Michel G, Lemerrer M, Philip N. Idiopathic thrombocytopenic purpura in two mothers of children with DiGeorge sequence: a new component manifestation of deletion 22q11? Am J Med Genet. 1997;69:356–359. doi: 10.1002/(sici)1096-8628(19970414)69:4<356::aid-ajmg4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 30.Li J Z, Steinman C R. Plasma DNA in systemic lupus erythematosus. Characterization of cloned base sequences. Arthritis Rheum. 1989;32:726–733. doi: 10.1002/anr.1780320610. [DOI] [PubMed] [Google Scholar]
- 31.Li L, Bray P F. Homologous recombination among three intragene Alu sequences causes an inversion-deletion resulting in the hereditary bleeding disorder Glanzmann thrombasthenia. Am J Hum Genet. 1993;53:140–149. [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W-M, Chu W-M, Choudary P V, Schmid C W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lorain S, Quivy J P, Monier-Gavelle F, Scamps C, Lécluse Y, Almouzni G, Lipinski M. Core histones and HIRIP3, a novel histone-binding protein, directly interact with WD repeat protein HIRA. Mol Cell Biol. 1998;18:5546–5556. doi: 10.1128/mcb.18.9.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makalowski W, Mitchell G A, Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 35.Marcus S, Hellgren D, Lambert B, Fallstrom S P, Wahlstrom J. Duplication in the hypoxanthine phosphoribosyl-transferase gene caused by Alu-Alu recombination in a patient with Lesch Nyhan syndrome. Hum Genet. 1993;90:477–482. doi: 10.1007/BF00217444. [DOI] [PubMed] [Google Scholar]
- 36.McCandless S E, Scott J A, Robin N H. Deletion 22q11: a newly recognized cause of behavioral and psychiatric disorders. Arch Pediatr Adolesc Med. 1998;152:481–484. doi: 10.1001/archpedi.152.5.481. [DOI] [PubMed] [Google Scholar]
- 37.Muir P, Nicholson F, Sharief M K, Thompson E J, Cairns N J, Lantos P, Spencer G T, Kaminski H J, Banatvala J E. Evidence for persistent enterovirus infection of the central nervous system in patients with previous paralytic poliomyelitis. Ann N Y Acad Sci. 1995;753:219–232. doi: 10.1111/j.1749-6632.1995.tb27548.x. [DOI] [PubMed] [Google Scholar]
- 38.Nebert D W. Polymorphisms in drug-metabolizing enzymes: what is their clinical relevance and why do they exist? Am J Human Genet. 1997;60:265–271. [PMC free article] [PubMed] [Google Scholar]
- 39.Nystrom-Lahti M, Kristo P, Nicolaides N C, Chang S Y, Aaltonen L A, Moisio A L, Jarvinen H J, Mecklin J P, Kinzler K W, Vogelstein B, de la Chapelle A, Peltomaki P. Founding mutations and Alu-mediated recombination in hereditary colon cancer. Nat Med. 1995;1:1203–1206. doi: 10.1038/nm1195-1203. [DOI] [PubMed] [Google Scholar]
- 40.Panning B, Smiley J R. Activation of expression of multiple subfamilies of human Alu elements by adenovirus type 5 and herpes simplex virus type 1. J Mol Biol. 1995;248:513–524. doi: 10.1006/jmbi.1995.0239. [DOI] [PubMed] [Google Scholar]
- 41.Pousi B, Hautala T, Heikkinen J, Pajunen L, Kivirikko K I, Myllyla R. Alu-Alu recombination results in a duplication of seven exons in the lysyl hydroxylase gene in a patient with the type VI variant of Ehlers-Danlos syndrome. Am J Hum Genet. 1994;55:899–906. [PMC free article] [PubMed] [Google Scholar]
- 42.Presidential Advisory Committee on Gulf War Veteran’s Illnesses. Final report. U.S. Washington, D.C: Government Printing Office; 1996. [Google Scholar]
- 42a.Presidential Advisory Committee on Gulf War Veteran’s Illnesses. Special report. U.S. Washington, D.C: Government Printing Office; 1997. [Google Scholar]
- 43.Rosenberg N L, Rotbart H A, Abzug M J, Ringel S P, Levin M J. Evidence for a novel picornavirus in human dermatomyositis. Ann Neurol. 1989;26:204–209. doi: 10.1002/ana.410260204. [DOI] [PubMed] [Google Scholar]
- 44.Roth D B, Craig N L. VDJ recombination: a transposase goes to work. Cell. 1998;94:411–414. doi: 10.1016/s0092-8674(00)81580-9. [DOI] [PubMed] [Google Scholar]
- 45.Salmon S E, Seligmann M. B-cell neoplasia in man. Lancet. 1974;ii:1230–1233. doi: 10.1016/s0140-6736(74)90748-x. [DOI] [PubMed] [Google Scholar]
- 46.Smith C A, Driscoll D A, Emanuel B S, McDonald-McGinn D M, Zackai E H, Sullivan K E. Increased prevalence of immunoglobulin a deficiency in patients with the chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/Velocardiofacial syndrome) Clin Diagn Lab Immunol. 1998;5:415–417. doi: 10.1128/cdli.5.3.415-417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan K E, McDonald-McGinn D M, Driscoll D A, Zmijewski C M, Ellabban A S, Reed L, Emanuel B S, Zackai E H, Athreya B H, Keenan G. Juvenile rheumatoid arthritis-like polyarthritis in chromosome 22q11.2 deletion syndrome (DiGeorge anomalad/velocardiofacial syndrome/conotruncal anomaly face syndrome) Arthritis Rheum. 1997;40:430–436. doi: 10.1002/art.1780400307. [DOI] [PubMed] [Google Scholar]
- 48.Thomas J A, Graham J M., Jr Chromosome 22q11 deletion syndrome: an update and review for the primary pediatrician. Clin Pediatr. 1997;36:253–266. doi: 10.1177/000992289703600502. [DOI] [PubMed] [Google Scholar]
- 49.Urnovitz H B, Murphy W H. Human endogenous retroviruses: nature, occurrence, and clinical implications in human disease. Clin Microbiol Rev. 1996;9:72–99. doi: 10.1128/cmr.9.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urnovitz H B, Sturge J C, Gottfried T D. Increased sensitivity of HIV-1 antibody detection. Nat Med. 1997;3:1258. doi: 10.1038/nm1197-1258. [DOI] [PubMed] [Google Scholar]
- 51.U.S. Senate (J. J. Tuite, principal investigator) United States dual-use exports to Iraq and their impact on the health of the Persian Gulf War veterans. 1994. pp. 229–243. , 362–379. In Senate hearing document no. 103–900 and appendices. U.S. Government Printing Office, Washington, D.C. [Google Scholar]
- 52.Wieczorek A J, Rhyner C, Block L H. Isolation and characterization of an RNA-proteolipid complex associated with the malignant state in humans. Proc Natl Acad Sci USA. 1985;82:3455–3459. doi: 10.1073/pnas.82.10.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan W, Jacobsen L K, Krasnewich D M, Guan X Y, Lenane M C, Paul S P, Dalwadi H N, Zhang H, Long R T, Kumra S, Martinm B M, Scambler P J, Trent J M, Sidransky E, Ginns E I, Rapoport J L. Chromosome 22q11.2 interstitial deletions among childhood-onset schizophrenics and “multidimensionally impaired.”. Am J Med Genet. 1998;81:41–43. [PubMed] [Google Scholar]
- 54.Yousef G E, Isenberg D A, Mowbray J F. Detection of enterovirus specific RNA sequences in muscle biopsy specimens from patients with adult onset myositis. Ann Rheum Dis. 1990;49:310–315. doi: 10.1136/ard.49.5.310. [DOI] [PMC free article] [PubMed] [Google Scholar]