Summary
TET1-mediated active DNA demethylation is required for endogenous retrovirus (ERV) enhancer activation during human ES differentiation into definitive endoderm (DE) cells. Here we present a protocol for siRNA-mediated TET1 knockdown during this process to decipher TET1’s role in ERV activation and DE differentiation. We describe steps for inducing ES into DE cells. We then detail steps for knocking down TET1 during differentiation and for examining the effects of TET1 knockdown on LTR6B methylation, cell morphology, and gene expression.
For complete details on the use and execution of this protocol, please refer to Wu et al. (2022).1
Subject areas: Cell Culture, Genetics, Molecular Biology, Gene Expression, Cell Differentiation
Graphical abstract

Highlights
-
•
Induce differentiation of human ES cells into definitive endoderm (DE) cells
-
•
Two consecutive transfections with TET1 siRNA to enhance knockdown efficiency
-
•
Investigate the effects of TET1 knockdown on cell morphology and gene expression
-
•
Investigate the effects of TET1 knockdown on LTR6B methylation of DE cells
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
TET1-mediated active DNA demethylation is required for endogenous retrovirus (ERV) enhancer activation during human ES differentiation into definitive endoderm (DE) cells. Here we present a protocol for siRNA-mediated TET1 knockdown during this process to decipher TET1’s role in ERV activation and DE differentiation. We describe steps for inducing ES into DE cells. We then detail steps for knocking down TET1 during differentiation and for examining the effects of TET1 knockdown on LTR6B methylation, cell morphology, and gene expression.
Before you begin
All experiments involving the use of human pluripotent stem cells must obey relevant legal and ethical standards. The use of human embryonic stem cells (hESCs) of this study was approved by the Guangzhou Institute of Biomedicine and Health, Chinese Academy of Sciences, approval number: GIBH-IRB2022-019. In this protocol, we provide a detailed description of derivation of definitive endoderm cells from hESCs. Additionally, we explain how to perform siRNA knockdown for molecular mechanism exploration during this process.
Protocol overview
In this protocol, we provide a detailed description of how to knock down TET1 activities in the process of inducing DE cells using siRNAs, as well as methods to examine the resulting effects on CpG methylation and expression of LTR6B. References have previously described the functional roles of ERVs,2 including LTR6B, and the necessity of TET1-mediated demethylation for their activation during DE cell differentiation.1 To enhance knockdown effects, we perform two consecutive rounds of transfection, each co-transfecting 3 siRNAs targeting TET1. We introduce in detail the steps for DE cell induction and siRNA transfection, along with methods to examine changes in CpG methylation of LTR6B using bisulfite conversion sequencing and expression changes through qPCR. This protocol is easily adaptable for use in other cell lines or biological processes, as we have used it to knock down TET2 and TET3 during induction of DE cells, as well as other genes (for example, Setdb1, Dnmt1, Ythdc1, Mettl3) in the process of mouse ES differentiation to Epiblast stem cells. However, it is important to note that some parameters of specific steps, such as transfected concentration or whether additional rounds of transfection are necessary, should be carefully adjusted based on the efficiency of specific siRNAs or cell lines. To ensure biologically meaningful and reproducible results, proper attention must be taken throughout the experiment to maintain cells in a good state, avoid any contamination during cell culture, and avoid nuclease contamination in molecular experiments. It is recommended to perform most experiments in a Biosafety Cabinet and use high-quality reagents and materials.
Preparation primers and siRNAs
Timing: ∼1 week
-
1.
Order qPCR and Bisulfite PCR primers listed in the key resources table and dissolve them with nuclease-free water to a final concentration of 100 μM.
Note: Bisultife PCR primers were designed using the MethPrimer tool (https://www.urogene.org/methprimer/).3 It is usually recommended to designed 3–5 pairs of primers and tested their effectiveness prior to performing the BSP experiments.
-
2.
Order siRNAs for human TET1 list in the key resources table and dissolve them to a final concentration of 20 μM, using nuclease-free water.
Preparation of LB agar plate and LB liquid medium
Timing: ∼5 h
-
3.Preparation of LB agar plates with Ampicillin resistance.
-
a.Prepare pre-mixed powder consisting of 5.0 g of yeast extract, 10.0 g of tryptone, 10.0 g of NaCl, and 15.0 g of agar.
-
b.Add the powder to 1 L distilled water in an Erlenmeyer flask.
-
c.Sterilize for 20 min at 120°C using an autoclave.
-
d.Allow the agar medium to cool down to 50°C and add 1 mL 100 mg/mL of Ampicillin into the medium.
-
e.Mix, and pour the agar medium into 10 cm bacterial plates and allow it to cool and solidify.
-
f.Seal the plate with parafilm and store at 4°C.
-
a.
-
4.Prepare LB liquid media with or without Ampicillin.
-
a.To make 500 mL of liquid LB media, add 2.5 g yeast extract, 5.0 g tryptone and 5.0 g NaCl to 500 mL distilled water in a container.
-
b.Autoclave the container for 20 min.
-
c.Allow the media to cool to 15°C–25°C, add 500 μL of 100 mg/mL ampicillin stock to prepare LB media with ampicillin. Alternatively, skip this step to prepare LB media without ampicillin.
-
d.The medium should be stored at 4°C.
-
a.
Preparation of Matrigel
Timing: ∼6 h
-
5.Preparation of Matrigel.Note: These steps must be performed in a sterile environment, such as a biosafety cabinet. Matrigel must always be kept on ice.
-
a.Thaw the whole bottle of Matrigel from −20°C and place it on ice for 4–5 h to allow complete thawing.
-
b.Prepare 1.5 mL Eppendorf tubes, 200 μL and 1 mL pipette tips, and place them in the −80°C for pre-cooling. During the preparation process, place all the 1.5 mL Eppendort tubes on ice to ensure they remain chilled.Note: It is crucial that any item that will come in contact with the Matrigel be chilled. Matrigel will solidify and adhere to any item that is above 10°C.
-
c.Once the Matrigel has fully thawed, gently shake the bottle to ensure homogenization of the Matrigel.
-
d.Thoroughly wipe the Matrigel bottle with 75% alcohol and carefully remove the rubber stopper to open it. Immediately after opening, put the bottle back into the ice.
-
e.Using pre-chilled pipette tips, aliquot 280 μL of Matrigel into each pre-chilled EP tube. Secure the tubes with a sealing film and store them at −80°C.Note: Matrigel can only be aliquoted once and cannot be repeatedly frozen and thawed for use.
-
f.Thaw Matrigel tubes on ice for 1–2 h until fully liquefied.
-
g.Aliquot 25 mL of pre-chilled DMEM/F12 into a 50 mL centrifuge tube.
-
h.Using pre-chilled pipette tips aspirate 300–500 μL of DMEM/F12 into the Matrigel tube, and gently tap to mix evenly. Then transfer all the liquid back into the 50 mL centrifuge tube with DMEM/F12.
-
a.
Prepare mTeSR1 medium
Timing: ∼6 h
-
6.Prepare mTeSR1 complete medium (All operations were performed inside a biosafety cabinet).Note: All additives and medium should be thawed at 15°C–25°C or 8–12 h at 2°C–8°C and should not be thawed in a 37°C water bath.
-
a.Thaw 100 mL of mTeSR1 5× supplement at 15°C–25°C or 8–12 h in a 4°C refrigerator.Note: Once thawed, use immediately or aliquot and store at −20°C. The solution can be stored at −20°C for up to 3 months and should not be repeatedly thawed and refrozen.
-
b.Add the thawed mTeSR1 5× supplement to 400 mL of mTeSR1 basal medium and mix thoroughly.Note: mTeSR1 complete medium can be stored at 2°C–8°C for 2 weeks or aliquoted and stored at −20°C for up to 6 months.
-
a.
Prepare 0.5 mM EDTA
Timing: ∼4 h
-
7.EDTA was prepared as a 50 mM storage solution (100×) and diluted to a final concentration of 0.5 mM for use.
-
a.Weigh 1.4612 g of EDTA powder into a 100 mL beaker.
-
b.Add approximately 80 mL of deionized water and stir thoroughly.
-
c.Adjust the pH to 8.0 using NaOH.Note: EDTA powder will only dissolve completely at pH 8.0.
-
d.Add deionized water to bring the solution to a final volume of 100 mL.
-
e.Transfer the prepared EDTA stock solution to a 100 mL reagent bottle, seal with sterilization tape, and sterilize it at 120°C for 50 min.
-
f.Store the sterilized EDTA stock solution at 15°C–25°C. To prepare a working solution of 0.5 mM, add 50 μL of the stock solution to 50 mL of DPBS.
-
a.
Prepare Y27632
Timing: 1–2 h
-
8.Dissolve Y27632 powder in a solvent to prepare a 10 mM stock solution. For use, dilute the stock solution to a working concentration of 10 μM.Note: All operations were performed inside a biosafety cabinet.
-
a.Prepare 100 mL of sterilized water.
-
b.Prepare 32 EP tubes labeled with "10 mM Y27632″ in a biosafety cabinet.
-
c.Retrieve the tube containing 100 mg of Y27632 powder and centrifuge it for 5 min at 350 g.
-
d.Add 1 mL of sterilized water to the tube and gently vortex to obtain a suspension.
-
e.Transfer the suspension to a 50 mL centrifuge tube.
-
f.Add 1 mL of sterile water to wash the remaining powder and transfer the solution to the 50 mL centrifuge tube as well.
-
g.Add 29.2246 mL of sterile water to the 50 mL centrifuge tube and gently vortex to ensure homogeneous mixing. The total volume of the resulting solution is 31.2246 mL, and the concentration is 10 mM. Dispense the solution into EP tubes at 1 mL per tube and seal the tube openings. Store at −20°C.
-
a.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Competent cell DH5 alpha | TSINGKE | TSC-C01 |
| Chemicals, peptides, and recombinant proteins | ||
| Matrigel | Corning | 354277 |
| DMEM/F-12 | Gibco | C11330500BT |
| mTeSR1/5× Supplement | STEMCELL | 85851 |
| EDTA | Sigma | ED-500G |
| NaOH | Sigma | V800383 |
| Tris | Solarbio | 77-86-1 |
| Na2EDTA·2H2O | Sigma | E5134 |
| Acetic acid | Sigma | A6283 |
| Accutase | Sigma | A6964 |
| Fetal bovine serum | RMBIO | FBS-AB-5XM |
| Rock inhibitor Y-27632 | CHEMCD | C14357 |
| DPBS | GlgBio | EH80028 |
| Ampicillin | MD | A010 |
| Agarose | Solarbio | A8201 |
| Trypton | Solarbio | T8490 |
| Yeast extract | Solarbio | Y8020 |
| Sodium chloride | CHRON chemicals | 7647-14-5 |
| Agar | Solarbio | A8190 |
| Trypan blue dye | Sigma | T8154 |
| RPMI1640 | GIBCO | C11875500BT |
| Activin A | R&D | 338-AC |
| CHIR99021 | TOCRIS | 4423 |
| BSA | Solarbio | A8010 |
| ITS supplement | Gibco | 41400045 |
| Opti-MEM | Gibco | 31985070 |
| 75% ethanol | Ante | 2212134119 |
| Absolute ethanol | Sigma | E7148 |
| Chloroform | Sigma | 34854 |
| Isopropanol | Sigma | 278475 |
| Nuclease-free water | TIANGEN | RT121 |
| IPTG | Thermo Fisher | 15529019 |
| X-gal | Thermo Fisher | R0941 |
| Critical commercial assays | ||
| La Taq | TAKARA | Cat# RR52A |
| Trizol | Gene BioTech | TR118 |
| TIANamp Genomic DNA Kit | TIANGEN | Cat# DP403 |
| PrimeScript RT Master Mix | TAKARA | Cat# RR036A |
| ChamQ Universal SYBR qPCR Master Mix | Vazyme | Cat# Q311 |
| 96-well white plate | Bio-Rad | HSP9601 |
| Lipofectamine™ RNAiMAX kit | Invitrogen | Cat# 13778150 |
| EpiTect Fast Bisulfite Conversion Kits | QIAGEN | Cat# 59824 |
| HiPure Gel DNA Mini Kit | MAGEN | Cat# D211102 |
| pMD™18-T Vector Cloning Kit | TAKARA | Cat# 6011 |
| Experimental models: Cell lines | ||
| Human: H1 hESC line | Provided by Jams A. Tomson, University of Wisconsin | WA01 |
| Oligonucleotides | ||
| NC-siRNA sense UUCUCCGAACGUGUCACGUdTdT |
This paper | N/A |
| NC-siRNA antisense ACGUGACACGUUCGGAGAAdTdT |
This paper | N/A |
| TET1-siRNA-1 sense GGAGUAAAGCACUCAGAAAdTdT |
This paper | N/A |
| TET1-siRNA-1 antisense UUUCUGAGUGCUUUACUCCdTdT |
This paper | N/A |
| TET1-siRNA-2 sense CGAAGCUACUGCAAAUCAAdTdT |
This paper | N/A |
| TET1-siRNA-2 antisense UUGAUUUGCAGUAGCUUCGdTdT |
This paper | N/A |
| TET1-siRNA-3 sense GAAGAGACAUUGAAUGAUAdTdT |
This paper | N/A |
| TET1-siRNA-3 aitisense UAUCAUUCAAUGUCUCUUCdTdT |
This paper | N/A |
| qPCR Primer TET1-qP-Forward ATCCTTGCTGTGTAGAGTTC |
This paper | N/A |
| qPCR Primer TET1-qP-Reverse AGTATTGGTGATATGCTTCCTAA |
This paper | N/A |
| qPCR Primer LTR6B-qP-Forward CCGAGAGAATTTTGAGCGTTAGC |
This paper | N/A |
| qPCR Primer LTR6B-qP-Reverse TGTTGTACCTGAGCGAGTTAGAG |
This paper | N/A |
| qPCR Primer FOXA2-qP-Forward ACAGCAGTCTTCTTCACC |
This paper | N/A |
| qPCR Primer FOXA2-qP-Reverse AGCAGGAGTCTACACAGTA |
This paper | N/A |
| qPCR Primer SOX17-qP-Forward ACCGCACGGAATTTGAAC |
This paper | N/A |
| qPCR Primer SOX17-qP-Reverse GCAGTAATATACCGCGGAGC |
This paper | N/A |
| qPCR Primer ACTIN-qP-Forward CCCAGAGCAAGAGAGG |
This paper | N/A |
| qPCR Primer ACTIN-qP-Reverse GTCCAGACGCAGGATG |
This paper | N/A |
| BSP primer LTR6B-BS-Forward: AAGAATGTAATTAATTGATAAG |
This paper | N/A |
| BSP primer LTR6B-BS-Reverse: CCCCRAAAAAAAAACTAAATT |
This paper | N/A |
| Primer M13F: GTAAAACGACGGCCAGT |
This paper | N/A |
| Primer M13R: CAGGAAACAGCTATGAC |
This paper | N/A |
| Recombinant DNA | ||
| pMD18-T vector | TAKARA | 6011 |
| Software and algorithms | ||
| QUMA | Kumaki et al.4 | http://quma.cdb.riken.jp/ |
| Graph Pad prism | GraphPad | https://www.graphpad.com/scientific- software/prism/ |
| SnapGene | Insightful Science | https://www.snapgene.com |
Materials and equipment
-
•
Prepare 50× TAE buffer: Add 242 g of Tris, 37.2 g of Na2EDTA·2H2O and 800 mL of deionized water to a 1 L beaker, stir well. Then add 57.1 mL of acetic acid and stir thoroughly. After that, add deionized water to bring the solution to a final volume of 1 L. This buffer can be stored at 15°C–25°C for 1 year.
TAE buffer (50×)
| Reagent | Final concentration | Amount |
|---|---|---|
| Tris | 2 M | 242 g |
| Na2EDTA·2H2O | 10 mM | 37.2 g |
| Acetic acid | 2 M | 57.1 mL |
| ddH2O | N/A | 1 L |
| Total | N/A | 1 L |
This buffer.
Step-by-step method details
Cell culture of hES before induction of hDE
Timing: ∼6 days
In this step, we describe how to passage and culture hES cells before inducing them into hDE cells. We also outline the conditions that must be met for successful hDE induction.
-
1.Recovery of hES cells.
-
a.Add 1 mL of diluted Matrigel per well of 6 well plate, let the plate sit at 37°C for 0.5–1 h.Note: Matrigel is used here as a substrate for cell attachment.
-
b.Turn on a water bath and maintain the temperature at 37°C.
-
c.Retrieve the cell cryovials from liquid nitrogen.
-
i.Roll the vial between gloved hands for 3–5 s to remove the frost.
-
ii.Immerse the cryovials to the 37°C water bath quickly.
-
i.
-
d.Gently swirl the cryovials, monitor the thaw process by holding the vial up to the light to assess the size of the ice crystals.Note: Do not submerge the cap of the vial in the water bath as this could contaminate the cells.
-
e.When only small ice crystal remains, thoroughly wipe the cryovials with 75% alcohol to sterilize.
-
f.Transfer the contents of the cryovials to a 15 mL centrifuge tube.
-
g.Slowly add 5 mL of mTeSR1 medium (containing 10 μM Y27632) to the cells in the 15 mL centrifuge tube. When adding the medium, gently rock the tube back and forth to ensure the osmotic pressure of the cells remains stable.Note: The additional of Y27632 in this step is necessary to improve cell survival since the cells were in a single-cell state. Usually, when repleting cells that have been digested into a single-cell state, the addition of Y27632 is required to improve cell viability. However, if cells are in a clonal state during repletion, Y27632 is no necessary. Additionally, when replacing the culture medium after cells have attached well to the substrate (for example, in step 1.m), Y27632 is also not required.
-
h.Centrifuge the cells at 250 g for 5 min.
-
i.Carefully aspirate the supernatant.Note: Remove as much supernatant as possible but be careful not to aspirate the cell pellet.
-
j.Re-suspend the pellet by adding 1 mL of mTeSR1 medium (containing 10 μM Y27632) and pipetting gently several times.
-
k.Aspirate Matrigel from the 6 well plate. Then:
-
i.Slowly add all the 1 mL of the hES cell suspension to the well.
-
ii.Add an addition 1 mL mTeSR1 medium (containing 10 μM Y27632) to bring the total amount of medium to 2 mL.
-
i.
-
l.Carefully slide the plate forward-to-back and side-to-side to ensure even cell distribution, then place the plate into the 37°C incubator.
-
m.Observe the cell growth under the microscope daily and replace 2 mL of fresh mTeSR1 medium (without Y27632) for each well.
-
a.
-
2.hES culture and passaging.
-
a.After hES culture for 4–7 days, we observed cell morphology under a microscope to determine when to passage and amplify the cells.
-
i.The clone boundaries are clear, the nuclear material is uniform, the nucleoli are prominent, the center appears bright lipid droplets, and there is no fusion between clones.
-
ii.Confluence rate reach to 85%.Note: If the passaging is performed too early, the cells may not adhere well, resulting in a lower yield and increase risk of differentiation. And if the passaging is performed too late, the cells may undergo excessive growth leading to spontaneous differentiation.
-
i.
-
b.Pre-warm DMEM/F12 medium, 0.5 mM EDTA, and mTeSR1 medium at 15°C–25°C.
-
c.Add 1 mL of diluted Matrigel per well of 6 well plate, let the plate sit at 37°C for 0.5–1 h.
-
d.Before digesting the cells, observe the cells under microscope and take pictures of the cells.
-
e.Aspirate the supernatant, and 1 mL of DMEM/F12 medium to each well to wash, aspirate, and add 1 mL of 0.5 mM EDTA. Incubate at 37°C for 5 min.
-
f.Observe digestion of the cells in each well under the microscope. When bright gaps between cells are visible, aspirate the EDTA and add 1 mL of mTeSR1 medium.
-
g.Gently pipette the cells in an up-to-down, left-to-right sequence to dislodge the cells from the plate, avoiding forceful pipetting that may lead to the cells being dislodged in too small clusters.
-
h.Aspirate the Matrigel from each well of the Matrigel plate (step c.), add 1.5 mL of mTeSR1 medium, and label the cell line name, passage number, and date of passaging.
-
i.Slowly pipette the cell suspension into the wells at an appropriate ratio (usually 1: 3 to1: 20).
-
i.Gently slide the plate forward-to-back and side-to-side to ensure even cell distribution.
-
ii.Then gently slide the plate once before placing it in the incubate and leaving it undisturbed.
-
iii.Culture at 37°C with 5% CO2.
-
i.
-
j.Observe the cell growth under the microscope daily and replace 2 mL of fresh medium for each well.
-
k.When hES cells are growing well and not differentiated, induction of definitive endoderm (DE) cells can be initiated.
-
a.
Inducing ES differentiate to DE cells
Timing: ∼6 days
The procedures below describe how to differentiate hES cells into hDE cells, isolate RNA and perform qPCR assays to evaluate the efficiency of differentiation.
-
3.Preparations before hDE cells induction.
-
a.Pre-warm DMEM/F12 medium, 0.5 mM EDTA, and mTeSR1 medium at 15°C–25°C.
-
b.Add 0.5 mL of diluted Matrigel per well of 12 well plate, let the plate sit at 37°C for 0.5–1 h.
-
c.Aspirate the supernatant (hES cells in 6-well plate of Step 2), and 1 mL of DMEM/F12 medium to each well to wash, aspirate, and add 1 mL of accutase. Incubate at 37°C for 7 min.
-
d.While the hES cells are incubated with accutase, aspirate the Matrigel from each well of the Matrigel plate (step c.), add 1 mL of mTeSR1 medium (containing 10 μM Y27632).
-
e.Observe digestion of the cells in each well under the microscope. When bright gaps between cells are visible, add 1 mL mTeSR1 medium (containing 10 μM Y27632) and gently pipette the cells to generate single-cell suspension.
-
f.Transfer the cell suspension to a 15 mL centrifuge tube and centrifuge at 250 g for 5 min.
-
g.Carefully aspirate the supernatant and add 1 mL mTeSR1 medium (contain 10 μM Y27632), gently pipette to suspend the cells.
-
h.Aspirate 20 μL of the cell suspension and add 20 μL of trypan blue dye to calculate the number of viable cells.
-
i.Slowly pipette the cell suspension into each well at a density of 400,000 cells per well of 12-well plate.
-
i.Slide the plate gently forward-to-back to side-to-side to ensure an even distribution of cells.
-
ii.Place the plate in the incubator, culture at 37°C with 5% CO2.
-
i.
-
j.Prepare DE induction medium as followed recipes:
-
i.D1 medium: RPMI1640 base medium + Activin A (final concentration: 100 ng/mL) + CHIR99021 (final concentration: 3 μM) + BSA (final concentration: 5 mg/mL).
-
ii.D2 medium: RPMI1640 base medium + Activin A (final concentration:100 ng/mL) + ITS supplement (final concentration: 0.1%) + BSA (final concentration: 5 mg/mL).
-
iii.D3 medium: RPMI1640 base medium + Activin A (final concentration: 100 ng/mL) + ITS supplement (final concentration: 1%) + BSA (final concentration: 5 mg/mL)
-
i.
-
a.
-
4.The induction of DE.
-
a.The next day (denoted as D0), confirm cell growth under the microscope, replace the medium in each well with 1 mL of D1 induction medium.
-
b.On Day 1 (24 h later), remove the D1 induction medium from each well, and add 1 mL of D2 induction medium.
-
c.On Day 2 (another 24 h later), remove the D2 induction medium from each well, and add 1 mL of D3 induction medium.
-
d.On Day 3 (an additional 24 h later), the induction process is complete, proceed to next step.
-
a.
-
5.Examine Cell morphology and collect cell samples.
-
a.Examine Cell Morphology under microscopy (Figure 1).
-
b.Cell collections.
-
i.discard the medium and wash the cells with 2 mL of 1× PBS.
-
ii.Detach the cells from the surface using 1 mL of EDTA at 37°C for 5 min.
-
iii.Use a pipette tip to pipette the sample up and down several times to disperse the cells, and then transfer it to a 1.5 mL EP tube.
-
iv.Centrifugation at 250 g for 5 min, and then discard the EDTA solution carefully with pipette.
-
i.
-
c.Add different reagents depending on the intended use.
-
i.For RNA isolation, add 1 mL of Trizol to each cell sample and pipette vigorously several times to lyse the cells. Perform RNA isolation immediately or store at –80°C.
-
ii.For DNA isolation, add 200 μL of buffer GA from the kit TIANamp Genomic DNA Kit, perform DNA isolation intermittently or stored at –20°C.
-
i.
-
a.
-
6.RNA isolation.
-
a.Add 200 μL Chloroform (If the samples are stored at −80°C, they should be taken out from the freezer and fully thawed at 15°C–25°C before adding chloroform).
-
b.Mix the sample thoroughly by repeated inversion, incubate for 2 min at 15°C–25°C.
-
c.Repeat the above step for three more times.
-
d.Centrifugate at 12000 rpm (∼ 13000 g) for 15 min at 4°C.
-
e.During centrifugation, prepare a new EP tube containing 400 μL of isopropanol for each sample and place them on ice.
-
f.Carefully pipette 400 μL of the top transparent liquid (aqueous phase) and transfer it to the EP tube containing pre-chilled isopropanol.Note: Take care not to aspirate the middle interphase and lower organic phase.
-
g.Mix it thoroughly by repeated inversion for 15 s, place it on ice and incubate for 10 min.
-
h.Centrifugate at 12000 rpm (∼13000 g) for 10 min at 4°C.
-
i.Discard the supernatant, be careful not to discard the precipitate.
-
j.Add 700 μL of 75% ethanol, which has been freshly prepared and pre-cooled on ice, to the precipitate. Invert the mixture several times to thoroughly wash the precipitate.
-
k.Centrifugate at 12000 rpm (about 13000 g) for 5 min at 4°C, then discard the supernatant.
-
l.Centrifugate at 12000 rpm (about 13000 g) for 3 min, and then Carefully remove the residual ethanol completely using a micropipette.
-
m.Allow the precipitate to air-dry for 10 min at 15°C–25°C.
-
n.Dissolve the precipitate in an appropriate amount of RNase-free water, then incubate at 55°C for 10 min.
-
o.Measure the concentration of isolated RNA in a NanoDrop;
-
p.Assess the quality of isolated RNA by gel electrophoresis. The RNA can be stored at −80°C for several months.
-
a.
-
7.Synthesis of cDNA.
-
a.Synthesis cDNA using the kit PrimeScript RT Master Mix, following manufacture’s instruction (https://www.takarabio.com/documents/User%20Manual/RR036A_e.v2008Da.pdf) (Table 1).Note: During the preparation of the mix, the RNA should be kept on ice.
-
b.Perform the reverse-transcription reaction after gently mixing the reaction solution. Star the cDNA synthesis program as shown in Table 2.
-
a.
-
8.Examine the hDE inducing efficiency through qRT-PCR.
-
a.Perform qPCR experiments using the “ChamQ Universal SYBR qPCR Master Mix” kit from Vazyme, following manufacture’s instruction https://www.vazyme.com/product/167.html.
- b.
-
c.Perform qPCR program shown below.We use the CFX Connect Real-Time PCR Detection System (Bio-rad) for qRT-PCR, to detect the expression of FOXA2 and SOX17, two marker genes of hDE cells,5 as well as LTR6B, a transposable element that is specifically expressed in DE cells. The primers used in this study is provided in the resource table. More applicable primers of most of genes can be search in PrimerBank6 (https://pga.mgh.harvard.edu/primerbank).
-
i.In a 96-well white plate, pipet 7.5 μL of the qRT-PCR mix into each well.
-
ii.Pipet 2.5 μL of primer mix to each well.
-
iii.Place the 96-well plate in a centrifuge equipped with a suited rotor.
-
iv.Centrifuge at 4000 rpm for 2 min at 4°C.
-
v.Load the 96-well plate into the Real-Time PCR Detection System.
-
vi.Star the PCR program as shown in Table 5.
-
i.
-
d.Calculate relative expression.
-
i.The relative expression of FOXA2, SOX17 and LTR6B in relation to the endogenous reference gene β-actin was calculated using the “ΔCt” method (references).
-
ii.The relative expression was determined as follows:This method gives the mRNA transcript level of FOXA2 or SOX 17 in DE cells relative to in ES cells (Figure 2).
-
i.
-
a.
Figure 1.
Cell morphology during the 4-day induction of hESC to hDE cells under microscopy
(A‒D) four panels showing cell morphology on days 0–3 of induction, respectively.
Table 1.
cDNA synthesis reaction
| Reagent | Amount |
|---|---|
| Total RNA | 500 ng |
| 5× RT Master Mix | 2 μL |
| RNase free H2O | Up to 10 μL |
| Total | 10 μL |
Table 2.
cDNA synthesis program
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Reverse transcription | 37°C | 15 min | 1 |
| Heat inactivation | 85°C | 5 s | 1 |
| Hold | 4°C | forever | |
Table 3.
QRT-PCR mix
| Reagent | Amount |
|---|---|
| 2 × ChamQ Universal SYBR qPCR Master Mix | 5 μL |
| Template cDNA | 2.5 μL |
| Total | 7.5 μL |
Table 4.
Primer mix
| Reagent | Amount |
|---|---|
| Primer 1 | 0.4 μL |
| Primer 2 | 0.4 μL |
| dd H2O | 1.7 μL |
| Total | 2.5 μL |
Table 5.
qPCR program
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Pre-Incubation | 95°C | 2 min | 1 |
| Denaturation | 95°C | 10 s | 1 |
| Annealing and Extension | 60°C | 20 s | 39 |
| Melting Curve | 70°C | 5 s | 1 |
| Melting Curve | Increase continuously to 95°C | 5 min | Measurement fluorescence every 0.1°C |
| Hold | 4°C | Forever | |
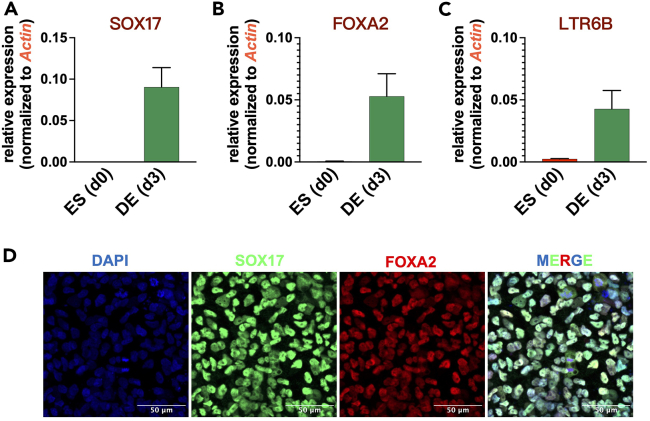
Figure 2.
qPCR results and immunofluorescence staining showing expression of hDE marker genes and LTR6B after induction of DE cells
(A‒C) Relative expression of SOX17 (A), FOXA2 (B) and LTR6B (C) was measured using qPCR with triple biological replicates. The relative expression levels of each gene were calculated using delta Ct method and normalized to endogenous reference gene β-Actin, presented as mean ± SEM.
(D) Immunostaining of SOX17 and FOXA2 at the DE stage (at D3).
siRNA knockdown of TET1 during induction of DE cells
Timing: ∼6 days
In below steps, we utilized siRNA to suppress TET1 expression during DE induction. Briefly, we induced DE cells as described in step 3–4 and performed two rounds of siRNA transfection to enhance the repression effect. The first round was performed simultaneously when cell seeding, and the second at D0, 24 h after the first transfection.
-
9.Transfection of siRNA of hTET1, using NC siRNA as control.
-
a.Dissolve siRNA with RNAse-free water to a concentration of 20 μM.
-
b.SiRNA is transfected using Lipofectamine™ RNAiMAX kit from ThermoFisher, following the manufacturer’s protocol https://www.thermofisher.cn/cn/zh/home/life-science/cell-culture/transfection/rnai-transfection/rnai-transfection-protocols.html.
-
c.For each round of transfection, prepare transfection mix shown below (Lipid dilution shown in Table 6 and siRNA dilution shown in Table 7).Add the siRNA dilution to the lipid solution (1:1) and incubate for 5 min at 15°C–25°C.
-
d.The first round of transfection was performed when cell seeding. Add the siRNA-lipid mixture to cells.
-
e.8 h later, aspirate the medium containing siRNA, replace with fresh mTeSR1 medium (contain 10 μM Y27632).
-
f.16 h later, replace the medium with D1 induction medium and performed the second round of transfection simultaneously.
-
g.8 h later, aspirate the medium containing siRNA-lipid mixture, replace with fresh D1 induction medium.
-
h.After culturing the cells for 16 h, replace the D1 medium with D2 induction medium.
-
i.Culture the cells for another 24 h before replacing the D2 medium with D3 medium.
-
ii.Allow the cells to continue to culture for an additional 24 h to complete the DE induction process.
-
i.
-
i.Check the cell morphology under microscope to confirm the ES cells have been successfully induced to DE cells (Figure 3) and collect cell samples according to step 4.
-
a.
Table 6.
Lipid dilution (for each 12-well)
| Reagent | Amount |
|---|---|
| Opti-MEM medium | 75 μL |
| Lipofectamine® RNAiMAX Reagent | 4.5 μL |
Table 7.
SiRNA dilution
| Reagent | Amount |
|---|---|
| Opti-MEM medium | 75 μL |
| TET1 siRNA-1 ∗ | 2.5 μL |
| TET1 siRNA-2 | 2.5 μL |
| TET1 siRNA-3 | 2.5 μL |
For NC siRNA (negative control), add 7.5 μL directly.
Figure 3.
Cell morphology during the 4-day induction of hDE cells under conditions of transfection of siNC (Negative Control) and siTET1 (TET1 siRNA)
(A‒D) four panels showing cell morphology on days 0–3 of induction with siNC transfection.
(E‒H) four panels showing cell morphology on days 0–3 of induction with siTET1 transfection.
(I and J) Immunostaining of SOX17 and FOXA2 at the DE stage (at D3) of siNC (I) and siTET1 (J).
Examine knockdown efficiency using qPCR
Timing: ∼2 days
-
10.
Perform RNA isolation and cDNA synthesis according to step 5 and step 6, respectively.
-
11.
Perform qRT-PCR according to step 7 to examine efficiency of siRNA knockdown and evaluate its impact on LTR6B expression and DE induction (Figure 4).
Figure 4.
qPCR results showing relative expression of genes in hDE cells under condition of siTET1 transfection
(A) relative expression of TET1, showing efficient knockdown of TET1 by siRNA.
(B) Relative expression of LTR6B.
(C and D) relative expression of hDE marker genes, SOX17(C) and FOXA2 (D). The relative expression levels of each gene were calculated using delta Ct method and normalized to endogenous reference gene β-Actin, presented as mean ± SEM. qPCR analysis was conducted using triple biological replicate. (∗, p < 0.05, ∗∗, p < 0.01, ∗∗∗<0.001, student’s t test).
Detect changes of CpG methylation of LTR6B
Timing: ∼5 days
In these steps, we introduce in detail procedures for isolating genomic DNA, performing bisulfite sequencing PCR (BSP) to examine the effects of TET1 knockdown on DNA methylation of LTR6B.
-
12.Genomic DNA isolation.
-
a.We isolated genomic DNA using TIANamp Genomic DNA Kit, following the manufacturer’s protocol https://en.tiangen.com/upload/file/20220509/20220509165835_34395.pdf.
-
b.Measure the concentration of genomic DNA in a NanoDrop;
-
a.
-
13.Bisulfite conversion:
-
a.We perform bisulfite conversion of genomic DNA using EpiTect Fast Bisulfite Conversion Kits, following the manufacture’s protocol: https://www.qiagen.com/us/resources/resourcedetail?id=12eec91a-0b70-4421-b0bf-2fdd616e3df9&lang=en and https://www.qiagen.com/us/resources/resourcedetail?id=e072eb69-54a4-430c-9648-ca92ad8c534e&lang=en. For each sample, in total 1 μg genomic DNA was used for conversion.
-
a.
- 14.
-
15.Gel electrophoresis and Purification of PCR products.
-
a.Load the PCR products along with a nucleic acid stain (e.g., SYBR Gold) and a tracking dye such as Bromophenol Blue. A suited DNA Ladder is also required.
-
b.Perform electrophoresis in 1× TAE buffer at 120 V, for 30 min.
-
c.Visualize DNA fragments on a UV gel imaging system.
-
d.Excise the DNA fragments with a size of 263 bp, using a clean razor blade.
-
e.Transfer the gel slices into a clean 1.5 mL EP tube.
-
f.Purify DNA from the gel pieces using a HiPure Gel DNA Mini Kit, following the manufacture’s protocols https://www.magen-tec.com/products/info.aspx?itemid=126#i5.
-
a.
-
16.Ligation of Purified PCR product into pMD18-T vector.
-
a.We perform TA cloning using pMD™18-T Vector Cloning Kit (https://www.takarabiomed.com.cn/ProductShow.aspx?m=20141220150817403017&productID=20141227130215047304#) from Takara Biomedical Technology.
-
b.Set up ligation reaction given in Table 10.
-
c.Incubate at 16°C for 4 h.
-
a.
-
17.Transformation of the ligation reaction.
-
a.Turn on a water bath and set the temperature to 42°C.
-
b.Thaw a required amount of E. coli competent cells (DH5α) on ice. For each transformation, 30 μL of competent cells are used.
-
c.Add all the ligation reaction in step 15 to the 30 μL competent cells kept on ice.
-
d.Gently tap the tube, and then incubate on ice for 30 min.
-
e.Place the competent cells in the water bath at 42°C and heat shocked for 45 s.
-
f.Immediately place the heat-shocked competent cells on ice and kept for 3 min.
-
g.Add 200 μL of pre-warmed LB liquid medium without any antibiotic, incubate the cells in a shaking incubator at 37°C for 5 min.
-
h.When the E. coli cells were incubated, Spread a 7 μL of IPTG and 40 μL X-gal evenly on the surface of the LB agar plate.
-
i.Plate the transformation mixes onto the LB agar plates with ampicillin antibiotics using a sterile loop.
-
j.Incubate at 37°C for ∼16 h in a bacterial incubator.
-
k.Check the growth of bacterial colonies on the agar plates. Most of the colonies are expected to be white, and some colonies might be blue formed by non-recombinant cells.
-
a.
-
18.Sequence the Colony.
-
a.Prepare sterile EP tubes, add 500 μL LB liquid medium with ampicillin antibiotic to each tube.
-
b.Pick a white colony using a sterile 10 μL pipette tip, transfer it into the LB liquid medium in the EP tube, agitate it for several times and discard the tips. Pick at least 20 colonies for each agar plate.Note: Each tube should only contain one single colony.Optional: To decrease the occurrence of false positives, it is recommended to perform colony PCR concurrently with colony picking for incubation.
-
c.Incubate these tubes in a shaker at 37°C in 200 rpm, for ∼ 16 h.
-
d.Perform Sange Sequencing using a universal primer M13F or M13R (sequences are shown in the key resources table) at a commercial sequencing facility.
-
a.
-
19.
Analyze sequencing results and calculate CpG methylation ratio (Figure 5).
Table 8.
PCR mix
| Reagent | Amount |
|---|---|
| 10× La Mix | 5 μL |
| dNTPs | 4 μL |
| Primer 1 | 2 μL |
| Primer 2 | 2 μL |
| Template DNA | 2 μL |
| La Taq | 0.4 μL |
| ddH2O | 34.4 μL |
| Total | 50 μL |
Table 9.
Bisulfite PCR condition
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Initial Denaturation | 95°C | 3 min | 1 |
| Denaturation | 95°C | 20 s | 35 cycles |
| Annealing | 55°C | 1 min | |
| Extension | 72°C | 30 s | |
| Final extension | 72°C | 3 min | 1 |
| Hold | 4°C | forever | |
Table 10.
pMD18-T ligation mix
| Reagent | Amount |
|---|---|
| Solution I | 5 μL |
| PMD18-T vector | 0.5 μL |
| Bisulfite converted DNA | 4.5 μL |
| Total | 10 μL |
Figure 5.
Bisulfite sequencing analysis of LTR6B DNA methylation in hESC, and hDE cells under transfection of siNC or siTET1
(A‒C) Bisulfite sequencing results of a region on LTR6B of hESC (A), hDE with siNC transfection (B), and hDE with siTET1 transfection (C). Each row stands for a sequenced DNA template molecule from a single colony. Methylated CpG sites are shown as solid circles, whereas open circles indicate unmethylated CpG sites.
(D) Barplot sumarise of CpG methylation rate in (A‒C).
We analyze the bisulfite sequence to calculate the CpG methylation using a web-based tool QUMA (http://quma.cdb.riken.jp/), following the user’s manual (http://quma.cdb.riken.jp/files/quma_manual_e.pdf).4
Optional: Other tools such as BiQ analyzer can be also used to analyze bisulfite sequence data.7
Expected outcomes
This protocol enables the induction of definitive endoderm (DE) cells from human embryonic stem (ES) cells, characterized by distinct cellular morphology and up-regulation of specific marker genes, such as FOXA2 and SOX17. During DE cell induction, siRNA-mediated downregulation of TET1 impairs demethylation of LTR6B, resulting in the subsequent downregulation of LTR6B and the majority of DEERVGs (DE-enhancer ERV-adjacent DE genes).1 The TET1 knockdown with siRNA approach can be applied in other scenarios where transient or conditional TET1 silencing is required. Moreover, this protocol is suitable for investigating the molecular mechanisms involved in driving ES towards DE cells, for example, identifying essential genes involved in this process.
Limitations
Our current protocol employs siRNA to knock down TET1 expression during ES to DE cell induction. Our test results demonstrate that the effects of siRNA are relatively short-lived, lasting approximately two days. Therefore, we perform two consecutive rounds of siRNA transfections to enhance the knockdown efficiency. However, multiple transfections may have subtle impacts on cellular states. Additionally, our protocol involves co-transfection of multiple siRNAs, which may bring risk of off-targeting.8 Therefore, we must exercise caution when conducting functional studies. Notably, our test results indicate that using one of the three siRNAs can generate significant knockdown effects, albeit weaker than those observed with co-transfection of all three siRNAs.
Troubleshooting
Problem 1
RNA degrades during the process of isolation (Step 5).
Potential solution
RNA is an unstable molecule that can easily degrade due to the presence of RNase, which can be found in various environmental contexts, including on skin, hair, saliva, and microorganisms. If RNA degradation is observed during isolation, the following tips can be used to improve RNA quality.
-
•
Once the cells are collected, add Trizol as soon as possible and pipette thoroughly to lysis cells immediately.
-
•
Designate a dedicated RNase-free area in the laboratory for RNA extraction and related experiments.
-
•
Thoroughly clean the experimental environment using decontamination solution (such as RNaseZap™ from ThermoFisher) before RNA isolation.
-
•
After touching skin (e.g., your face), door handles, and common object surfaces, change gloves.
Problem 2
qRT-PCR results present large variations among biological replicates (Step 7 and 10).
Potential solution
The large variation indicates the insufficient reliability of the data, which may be caused by RNA degradation, inaccurate concentration measurement, and random errors during the operation process. The following improvements can be made.
-
•
Prevent RNA degradation (problem 1).
-
•
Measure RNA concentration accurately.
-
•
Avoid leaving any residual reagent on the tip when using a pipette.
-
•
When loading samples onto the 96-well-white plate, keep the plate and samples on ice and avoid exposure to light.
Problem 3
No obvious knockdown effects were found after siRNA transfection (Step 8–10).
Potential solution
Poor knockdown efficiency may be attributed to low transfection efficiency or suboptimal cellular conditions. We may co-transfect a negative control with 5′-Fluorescein CE Phosphoramidite (6-Fam) modification with TET1 siRNA, which allow us to examine transfection efficiency under a fluorescence microscope. Alternatively, another group of cells can be transfected with GAPDH-siRNA as a positive control. Before the first round of transfection, it is necessary to ensure the healthy status of ES cells. The cells should present typical ES morphology, i.e., small round shape, high nucleus-to-cytoplasm ratio, less cytoplasm, etc.
Problem 4
Bisulfite sequencing PCR do not work (step 13).
Potential solution
DNA can become unstable after bisulfite conversion, so it is important to perform PCR soon after conversion. If this is not possible, the converted DNA should be stored at −20°C. Other factors that can lead to PCR failure include insufficient annealing time and low Taq enzyme activity. To address these issues, it may be necessary to use a new Taq enzyme and increase the annealing time.
Problem 5
An excessive number of false positive colonies were observed during the analysis of the bisulfite sequencing data (Step 18).
Potential solution
The primary reason for this issue is likely that the PCR products were not adequately purified during step 14. As a result, some PCR products from non-specific amplification may have remained in the final reaction during pMD18-T ligation. To address this problem, it is recommended to excise the target DNA fragment with greater care and to verify the fragment by electrophoresis after purification. Additionally, performing colony PCR to confirm the size of the products before Sanger sequencing is advisable.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jiangping He (he_jiangping@gzlab.ac.cn).
Materials availability
This study did not generate new unique reagents or cell lines. All reagents listed here can be found in the main article.1
Acknowledgments
This work was supported by the National Key R&D Program of China (2020YFA0112403, 2021YFA1102200, and 2019YFA0110200); the Frontier Science Research Program of the CAS (ZDBS-LY-SM007); the National Natural Science Foundation of China (32200461); the Science and Technology Planning Project of Guangdong Province, China (2020B1212060052); Plan on enhancing scientific research in GMU; Guangzhou Core Medical Disciplines Project (2021-2023); and the Shenzhen Innovation Committee of Science and Technology (ZDSYS20200811144002008) to the Shenzhen Key Laboratory of Gene Regulation and Systems Biology.
Author contributions
J.H. conceived and designed the project. Z.L. and Y.L. performed the main experiments. Z.L. and J.H. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Zhongqi Liufu, Email: liufzhq@gmail.com.
Jiangping He, Email: he_jiangping@gzlab.ac.cn.
Data and code availability
All data supporting the findings of this study are available within the paper. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Wu F., Liufu Z., Liu Y., Guo L., Wu J., Cao S., Qin Y., Guo N., Fu Y., Liu H., et al. Species-specific rewiring of definitive endoderm developmental gene activation via endogenous retroviruses through TET1-mediated demethylation. Cell Rep. 2022;41:111791. doi: 10.1016/j.celrep.2022.111791. [DOI] [PubMed] [Google Scholar]
- 2.Senft A.D., Macfarlan T.S. Transposable elements shape the evolution of mammalian development. Nat. Rev. Genet. 2021;22:691–711. doi: 10.1038/s41576-021-00385-1. [DOI] [PubMed] [Google Scholar]
- 3.Li L.-C., Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 4.Kumaki Y., Oda M., Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q., Hutchins A.P., Chen Y., Li S., Shan Y., Liao B., Zheng D., Shi X., Li Y., Chan W.-Y., et al. A sequential EMT-MET mechanism drives the differentiation of human embryonic stem cells towards hepatocytes. Nat. Commun. 2017;8 doi: 10.1038/ncomms15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang X., Spandidos A., Wang H., Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bock C., Reither S., Mikeska T., Paulsen M., Walter J., Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 8.Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data supporting the findings of this study are available within the paper. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.