Abstract
Purpose
Polyamine modulating factor 1 binding protein (PMFBP1) acts as a scaffold protein for the maintenance of sperm structure. The aim of this study was further to identify the new role and molecular mechanism of PMFBP1 during mouse spermatogenesis.
Methods and Results
We identified a profile of proteins interacting with PMFBP1 by immunoprecipitation combined with mass spectrometry and demonstrated that class I histone deacetylases, particularly HDAC3 and chaperonin-containing TCP1 subunit 3 (CCT3), were potential interaction partners of PMFBP1 based on network analysis of protein-protein interactions and co-immunoprecipitation. Immunoblotting and immunochemistry assays showed that loss of Pmfbp1 would result in a decline in HDACs and change the proteomic profile of mouse testis, in which differently expressed proteins are associated with spermatogenesis and assembly of flagella, which was proved by proteomic analysis of testis tissue obtained from Pmfbp1−/− mice. After integrating with transcriptome data for Hdac3−/− and Sox30−/− round sperm obtained from a public database, RT-qPCR confirmed ring finger protein 151 (Rnf151) and ring finger protein 133 (Rnf133) were key downstream response factors of the Pmfbp1-Hdac axis affecting mouse spermatogenesis.
Conclusion
Taken together, this study indicates a previously unidentified molecular mechanism of PMFBP1 in spermatogenesis whereby PMFBP1 interacts with CCT3, affecting the expression of HDAC3, followed by the downregulation of RNF151 and RNF133, resulting in an abnormal phenotype of sperm beyond the headless sperm tails. These findings not only advance our understanding of the function of Pmfbp1 in mouse spermatogenesis but also provide a typical case for multi-omics analysis used in the functional annotation of specific genes.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-023-02874-0.
Keywords: PMFBP1, CCT3, Protein-protein interaction, HDAC3, Proteome, Spermatogenesis
Introduction
Spermatogenesis is a complex process that includes spermatogonial self-renewal and differentiation, meiosis, postmeiotic spermiogenesis, and finally the formation of mature spermatozoa [1, 2]. Abnormalities in these processes may lead to male reproductive disorders ranging from azoospermia to oligozoospermia, asthenozoospermia, and teratozoospermia. Acephalic spermatozoa syndrome (ASS) is a classical type of the most serious teratozoospermia, defined as headless spermatozoa predominance in the ejaculate. Partial ASS originates from a developmental disturbance of the sperm head-tail coupling apparatus (HTCA). Recently, significant progress in the discovery of genetic causes of ASS was established, and some key pathogenic genes were identified to play an important role in ASS including SUN5, PMFBP1, TSGA10, BRDT, DNAH6, and CEP112 [3–5].
Polyamine modulating factor 1 binding protein (PMFBP1) is the major pathogenic gene responsible for ASS, which is characterized by an abnormal head-tail connection and ejaculate of nearly 100% abnormal spermatozoa in semen, including a large number of headless sperm tails, a small number of tailless sperm heads, and a few sperm with an abnormal head-tail junction [3, 5–10]. Our previous research has shown that PMFBP1 acts as a scaffold protein in the sperm head-to-tail coupling apparatus (HTCA), which maintains the head-tail structural integrity by the formation of a sandwiched localized structure that cooperates with SUN5 and SPATA6, and that the absent of PMFBP1 leads to acephalic spermatozoa [3, 11]. Mutations in PMFBP1 (11%) and SUN5 (59%) have been found to account for approximately 70% of all cases of ASS, which suggests that defects in these genes could be relevant in the clinical diagnosis of this syndrome. Recently, Zhang et al. found that CENTLEIN could interact with PMFBP1 and SUN5 to participate in the assembly of the HTCA [5]. A comparison of the proteome of testicular sperm of the Pmfbp1 knockout mouse with that of the wild-type mouse has identified the profile of differentially expressed proteins (DEPs) in sperm [7], in which Pmfbp1 acts as a scaffold protein for the connection between HTCA and the nuclear membrane of sperm and maintains its head-tail structural integrity. PMFBP1 is initially expressed at the early round spermatid (RS) stage when the germ cells are undergoing transcriptome and proteome remodeling in preparation for the elongation of spermatids. The distribution of PMFBP1 was shifted to the implantation fossa during elongation of the cytoplasm, eventually becoming located at the junction of the head and mid-piece in mature sperm [7, 11]. Although PMFBP1 has been extensively studied in mature sperm, little is known about its functional and kinetic features before the elongation of RS during spermatogenesis.
With the advent of sequencing technology, it is now possible to obtain a comprehensive understanding of human health and disease at multiple levels, including the genome, epigenome, transcriptome, proteome, and metabolome [12–15]. Therefore, we sought to identify the functions of Pmfbp1 in mice by proteomic analysis. Using this method, we systematically characterized the network of protein-protein interactions (PPIs) in which Pmfbp1 is involved. First, the proteins that interact with Pmfbp1 were identified in mouse testis lysate using affinity purification mass spectrometry (AP-MS). The key proteins that potentially interact with PMFBP1 were identified by analysis of interactions within the PPI network and gene functional analysis. Combined with transcriptome analysis of data for Hdac3−/− and Sox−/− round sperm obtained from a public database, we identified the important downstream target genes of PMFBP1. Overall, this research has clarified the role of PMFBP1 in the formation of sperm, providing evidence from a typical case to increase our understanding of the mechanism of the PMFBP1 deficiency that results in asthenoteratozoospermia and male infertility by multi-omics analysis. Our findings will assist in the elucidation of the pathogenic mechanism of ASS and the development of novel clinical diagnostic and treatment methods.
Materials and Methods
Animals
C57BL/6 Pmfbp1−/− mice were generated by CRISPR/Cas9 targeting exons 3 and 10 of the Pmfbp1 gene, which has been described in detail elsewhere [3]. The primers used for genotyping were as follows: WT-FP, 5′-GATGAGTAATAACAGCCCAGG-3′ and WT-RP, 5′-CTTGATGCATGCGCAGTTAG-3′ for the identification of wild-type alleles; and KO-FP, 5′-GATCTCAGACTCCCTCGTCAACTC-3′ and KO-RP, 5′-GGAGTGGGCAGGATTGAAAG-3′ for the identification of knockout alleles. All experimental procedures involving mice were conducted according to the guidelines of the Laboratory Animal Ethical Commission of the Chinese Academy of Sciences and the “Instructive Notions with Respect to Caring for Laboratory Animals” established by the Ministry of Science and Technology of the People’s Republic of China. All experimental procedures were authorized by the Committee for Animal Research, Anhui Medical University, China.
Construction of Recombinant Plasmids
Total RNA was isolated from testis tissue harvested from 21-day-old mice or a human cell line HeLa using TRIzol reagent (Cat# 15596026; Invitrogen, Waltham, MA, USA). Reverse transcription was then performed using a PrimerScript RT reagent kit (Cat# RR037B; Takara Bio Co., Ltd., Kusatsu City, Shiga Prefecture, Japan) to gain the complementary DNA as the template. The primers for polymerase chain reaction (PCR) amplification and vector are listed in Supplementary Table 1. The cDNA was digested with restriction enzymes (Cat# R0101/R3138/R0146; New England Biolabs, Inc., Ipswich, MA, USA) and connected to a linearized vector (Ordinary DNA Product Purification Kit, Cat# DP204-02; Tiangen Biotech, Beijing, China) using a T4 DNA ligase kit (Cat# 2011A; Takara Bio). Parts of recombinant plasmids were constructed by homologous recombination using a ClonExpress II One Step Cloning Kit (Cat# C112; Vazyme, Nanjing, China).
Cell Culture and Transfection
Human embryonic kidney cells 293T (HEK293T; American Type Culture Collection, Manassas, VA, USA) and human cervical cells (HeLa; American Type Culture Collection) were cultured in Dulbecco’s Modified Eagle’s Medium with 4.5 g/L D-glucose (Cat# A4192101; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% of fetal bovine serum (Cat# C04001; VivaCell, Shanghai, China) and 1% of penicillin-streptomycin (Cat# P1400; Solarbio, Beijing, China). When the density of cells was about 70%, the plasmids were transfected into cells using Lipofectamine 2000 (Cat# 11668019; Thermo Fisher Scientific).
Affinity Purification and Mass Spectrometry (AP-MS Assays)
Protein purification and mass spectrometry were performed as previously described [16, 17]. First, pCDNA3.1-3xflag-Pmfbp1 and the empty vector were respectively transfected into HEK293T cells (2 × 107). After 24 h, the cells were collected and lysed in RIPA buffer containing protease inhibitor and phosphatase inhibitor (Cat# C510006; Sangon Biotech, Shanghai, China). FLAG-Pmfbp1 was extracted with anti-flag-M2 Affinity Gel (Cat# A2220; Millipore, Sigma-Aldrich, St. Louis, MO, USA) and incubated with whole protein extracts from 21-day-old mouse testis tissue. The potential binding protein of Pmfbp1 was dissolved in 1 × Laemmli sample loading buffer (0.1 M Tris-Cl, pH 6.8, 1% sodium dodecyl sulfate, 5% glycerol, 0.5% 2-mercaptoethanol, and 2.5 mg of bromophenol blue) by boiling for 5 min at 95 °C. The eluates were assessed with silver staining using a Fast Silver Stain Kit (Cat# P0017S-1; Beyotime Biotechnology, Shanghai, China). The target-stained protein bands were excised from the gel and then enzymatically digested into peptides, which were purified for analysis by mass spectrometry using an Orbitrap Elite ETD hybrid mass spectrometer (Thermo Fisher Scientific), as mentioned earlier.
The raw mass spectrometry data were processed using MaxQuant (v.1.5.3.8) with the following settings. Trypsin was the proteolytic enzyme, and up to two missed cleavages were allowed with a minimum peptide length of seven amino acids. Proteins or gene-specific peptides with q values ≤ 0.01 were used for the identification and quantification of proteins. All proteomic data were then searched against the mouse proteome downloaded from the UniProt database with mass error tolerances of 15 ppm and 0.05 Da. The peptide false discovery rate was set to < 0.05. The identified proteins were subjected to pathway enrichment analyses using the Metascape database. Further bioinformatics analysis of the data was carried out using Perseus (v.1.6.5.0), STRING, Metascape, R (v.4.1.1), Cytoscape, and the CORUM databases [18, 19]. The STRING database was used to predict the interactions between identified proteins, and the Biological General Repository for Interaction Data (BioGRID, http://thebiogrid.org/) and Search Tool for the Retrieval of Interacting Gene/Protein database (STRING, http://string-db.org/) were used to compile the protein interaction network [20, 21]. The Cytoscape software platform (version 3.8.2) was used to visualize high-confidence PPIs.
Co-Immunoprecipitation
The plasmids for expressing CCT3, HDAC1, HDAC2, HDAC3, HDAC4, and HDAC5 with FLAG or GFP tag were co-transfected into HEK293T cells (1 × 105 cells/well) with Lipofectamine 2000 (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol. Cells were collected 24 h after transfection, washed in ice-cold phosphate-buffered saline, and lysed with 1 mL of lysis buffer (50 mM HEPES pH 8.0, 5 mM EDTA, 150 mM NaCl, 50 mM NaF, 0.5% IGEPAL, 1 mM dithiothreitol, 1 mM PMSF, 1.5 mM Na3VO4, 1× protease inhibitor cocktail) per well on ice. The clarified cell lysates were incubated with FLAG-M2 resin (Sigma) for 2 h. For endogenous co-immunoprecipitation, the testis extracts were collected to incubate with specific antibody-coupled Strep-Tactin® Sepharose® resin (50% suspension, IBA Lifesciences GmbH, Göttingen, Germany) on a rotation wheel at 4 °C for 2 h. Finally, the binding fraction was washed with lysis buffer 3 times before being resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Western Blot
The testis tissues from Pmfbp1−/− and Pmfbp1+/+ mice were lysed with M-PER reagent (Cat# 78505; Thermo Fisher Scientific) containing phenylmethanesulfonyl fluoride and a protease inhibitor cocktail. The concentrations of the extracted proteins were determined using a BCA protein assay kit (Cat# CW0014S; Beijing CoWin Biotech Co., Ltd., Beijing, China). Tissue lysate (30 μg/lane) was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred onto polyvinylidene difluoride membranes. After blocking with 5% bovine serum albumin, the membranes were successively incubated with specific primary antibodies and horseradish peroxide-conjugated secondary antibodies. The primary antibodies used are listed in Supplementary Table 2. Signals were detected using a Pro-Light horseradish peroxidase kit (PA112 Tiangen Biotech). Positive bands were detected using a FluorChem E System (UVP, Uplands, CA, USA) and normalized to GAPDH or tubulin expression.
Immunofluorescence Staining
The pCDNA3.1-3xflag-PMFBP1 plasmid was transiently transfected into HeLa cells (1 × 106 cells/well) cultured on cover glasses (Cat# 12-545-80; Fisherbrand, Thermo Fisher Scientific) using Lipofectamine 2000 transfection reagent (Cat# 11668030; Thermo Fisher Scientific) following the manufacturer’s instructions. After 24 h, the cells were fixed in 4% paraformaldehyde (Cat# E672002; Sangon Biotech) for 5 min, then permeabilized with 0.1% Triton X-100 (Cat# A110694; Sangon Biotech) for 1 min after transfection for 24 h [22]. After blocking with 1% bovine serum albumin (Sigma-Aldrich, Santa Clara, CA, USA) at room temperature for 1 h, the cells were incubated overnight with specific antibodies (Supplementary Table 2) at 4 °C, followed by fluorescent-labeled secondary antibodies (Cat# A21422/A11008/A31572/A21202; Thermo Fisher Scientific) for 1 h. DAPI (Cat# D9542; Sigma-Aldrich) was used to label the nuclei. Images were obtained and processed by a laser confocal microscope (LSM 800; Zeiss, Oberkochen, Germany) and analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Testis tissues from 21-day-old Pmfbp1−/− and Pmfbp1+/+ mice were fixed in 4% paraformaldehyde (Cat# BC-B-002; Biochannel, Nanjing, China) overnight at 4 °C and then dehydrated in a 20% sucrose-phosphate-buffered saline solution for 24 h and embedded in OCT compound (Cat# 4583; Sakura Finetek Inc., Torrance, CA, USA). Next, the testis tissues were sectioned into 5-μm slices using a freezing microtome (Leica Biosystems, Wetzlar, Germany). Frozen sections of the testis samples were then fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with blocking buffer (10% bovine serum albumin and 1% Triton X-100 in 1× phosphate-buffered saline) for 30 min at room temperature. Sections were stained as above.
Proteomic Analysis
Testes from three 21-day-old Pmfbp1−/−mice and three 21-day-old wild-type mice were ground into a powder with liquid nitrogen for the extraction of protein. Next, the powder was dissolved in lysis buffer (7 M urea, 2 M thiourea, 4% w/v CHAPS, 65 mM dithiothreitol) containing a protease inhibitor cocktail and phenylmethanesulfonyl fluoride. After sonication, the supernatant was collected by centrifugation, and the protein concentration was determined using a BCA protein assay kit (Cat# CW0014S, Beijing CoWin Biotech). Each sample corresponding to 240 g was reduced using 10 mM dithiothreitol for 30 min at 56°C for 1 h and alkylated with 55 mM iodoacetamide for 45 min in the dark at room temperature. Next, 100 mM triethylammonium bicarbonate was used to dilute the protein sample to urea with a concentration of less than 2 M. Trypsin was added to the mixture at a trypsin-to-protein ratio of 1:100 and kept at 37 °C overnight. Finally, 0.1% trifluoroacetic acid was added to stop the digestion reaction. After trypsin digestion and purification, the peptides were reconstituted in 0.5 M triethylammonium bicarbonate and processed using the TMT Tagging kit (Cat#90060; Thermo Fisher Scientific) according to the manufacturer’s instructions. The peptides were then subjected to LC-MS/MS identification and quantification on a Q Exactive™ Plus system (Thermo Fisher Scientific) coupled online to an ultraperformance liquid chromatography system.
The raw MS/MS data were processed using the MaxQuant software (version 1.6.17.0) with default settings. The resulting sequences were searched against the UniProt mouse proteome database, including a list of common contaminants. Carbamidomethylation (C) was considered a fixed modification, whereas oxidation (M), acetylation (protein N-term), and GlyGly (K) were specified as variable modifications. The false discovery rates at the protein and peptide–spectrum match levels were adjusted to < 1%, and the minimum score for modified peptides was set to > 40. The minimal peptide length was set to 7 amino acids, and the match-between runs option was enabled. Protein groups flagged as “reverse,” “potential contaminant,” or “only identified by site” were removed. All downstream analyses of the resulting output were performed using R (version 4.1.1), including heat maps, hierarchical clustering, and principal component analysis (implemented in DESeq2). Next, two-sided Student’s t-tests were used to compare the differential protein abundance between samples. The ratio thresholds for the upregulation and downregulation of proteins were set to > 1.50 and < 0.67, respectively, with a P-value < 0.05. The altered proteins were further analyzed by bioinformatics methods. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to annotate the protein pathway. DEPs were used to perform Gene Ontology (GO) enrichment by ClueGo [23]. GO terms with an adjusted P-value < 0.05 were selected for visualization. A PPI network of DEPs was constructed using the STRING database (version 11.5) [24]. Interactions that had a confidence score ≥ 0.7 were collected. The final data were visualized, and the key proteins in the interaction network were identified using Cytoscape software (version 3.8.2) [25].
Transcriptome Analysis
The transcriptomic data for purified RS from control and Stra8-cre/Hdac3fl/− mice or Sox30 knockout mice at postnatal day 21 (P21) or postnatal day 23 (P23) were respectively downloaded from the Gene Expression Omnibus database (P21 RS from Stra8-cre/Hdac3fl/– mice, number GSE153065; P23 RS from Sox30−/− mice, number GSE113073) and the National Omics Data Encyclopedia (P21 RS from Sox30−/− mice, accession number OEP000012). The ChIP-seq data in this study were generated from Sox30−/− RS using the Hdac3 antibody and wild-type mouse RS using the Sox30 antibody and downloaded from the Gene Expression Omnibus (GSE153065) database. The overlap between P21 Hdac3−/− RS, P23 Sox30−/− RS, P21 Sox30−/− RS, and the wild-type RS stages was obtained using the Draw Venn Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/). Pathway enrichment analysis of the overlapping genes was carried out using the Metascape database. The ChIP-seq data were visualized using Integrative Genomics Viewer software (version 2.5.0).
Quantitative Real-Time PCR
The isolated RNA sequencing samples from the testes of three Pmfbp1−/− mice and three wild-type mice were used to perform quantitative real-time PCR analysis. The total RNA was reverse-transcribed to single-stranded complementary DNA using the PrimerScript RT reagent kit with gDNA Eraser (Cat# RR037A; Takara Bio) according to the manufacturer’s instructions. Quantitative real-time PCR was performed on Quantitative real-time PCR (ABI, StepOnePlus) using SYBR® Premix Ex Taq™ II kits (Cat# RR820A; Takara Bio) under the following cycling conditions: 94 °C for 5 min followed by 40 cycles at 94 °C for 15 s and 60°C for 30 s. Relative expression was calculated using the 2−ΔΔCT method; all reactions were performed in triplicate; and each treatment had three biological repeats. GAPDH was used to compare the relative levels of mRNA. The primer sequences are shown in Supplementary Table 3.
Results
CCT3 May Be the Critical Protein that Interacts with PMFBP1 in Spermatogenesis
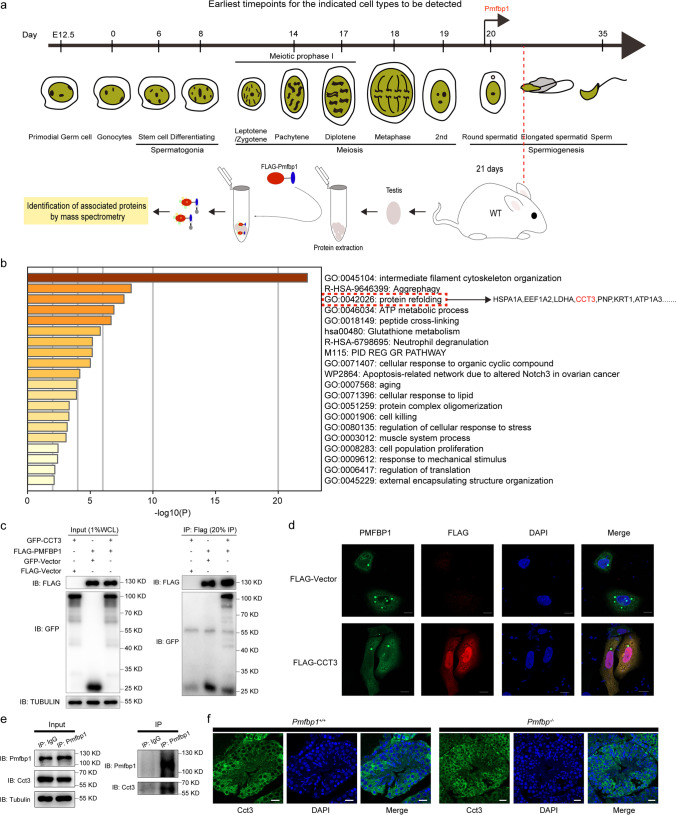
The AP-MS approach was used to systematically investigate the proteins that interact with PMFBP1 in an attempt to gain further insight into the molecular mechanisms of male infertility caused by PMFBP1 deficiency. As shown in Fig. 1a, mouse Pmfbp1 proteins fused with the Flag-tag were overexpressed in HEK293T cells, after which purified FLAG-Pmfbp1 was incubated with testis lysate from 21-day-old mice. The tagged Pmfbp1 proteins were affinity purified in biological triplicate and analyzed by mass spectrometry to identify co-purifying proteins. Seventy-four proteins were identified with high confidence (Supplementary Table 4). Functional enrichment analysis showed that significant GO-enriched terms were mostly related to cytoplasmic proteins involved in protein folding, intermediate organization of the filament cytoskeleton, and the ATP metabolic process (Fig. 1b). Among these, Cct3 was significantly enriched in the protein folding pathway. A previous study indicated that CCT3, a component of the chaperonin-containing T-complex (TRiC) involved in the assembly of the BBSome complex, which functions as a coat complex required for sorting of specific membrane proteins to the primary cilia, may have functions in spermatogenesis and ciliogenesis [26, 27]. Therefore, we performed a co-immunoprecipitation assay to confirm an interaction between CCT3 and PMFBP1 (Fig. 1c). The immunofluorescence analysis also suggested the co-localization of CCT3 and PMFBP1 in the cytoplasm (Fig. 1d). Of note was that overexpression of CCT3 markedly diminished the aggregation “hot spot(s)” induced by PMFBP1 overexpression alone, which was expected from its protein folding function. In the mouse testis, Cct3 also interacted with Pmfbp1 (Fig. 1e). There was no significant difference in the expression between Pmfbp1 knockout mice and wild-type mice (Fig. 1f).
Fig. 1.
CCT3 interacts with PMFBP1. a Label-free mass spectrometry analysis of proteins that interact with Pmfbp1 in the testis of a 21-day-old mouse. Graphic representation of Pmfbp1 expression during spermatogenesis. The recombinant plasmid of the mouse Pmfbp1 gene was transfected into HEK293T cells. After 24 h, the total cell proteins were extracted for incubation with Flag-probe antibody agarose beads; these bead-bound proteins were incubated as bait with whole protein extracts from the testis of a 21-day-old mouse. The proteins that interacted with the Pmfbp1 protein were obtained for label-free mass spectrometry analysis. b Bar graph showing pathway enrichment of 74 Pmfbp1 interacting proteins identified by the affinity purification mass spectrometry approach via the Metascape database. Only the significant Gene Ontology terms (P < 0.05) are shown in rows. Terms with the prefix “GO” are from the Gene Ontology Consortium; the prefix “R-HSA” from the Reactome; the prefix “WP” from the WikiPathways, and the prefix “hsa” from the KEGG database. c The immunoprecipitation experiment was carried out using FLAG-M2 beads, and the lysates of HEK293T cells were transfected with FLAG-PMFBP1 and GFP-CCT3. The isolated proteins were then analyzed by western blotting with anti-FLAG and anti-GFP antibodies, respectively. This experiment was repeated three times, and representative blots are presented. d Immunofluorescent staining indicates the subcellular localization of PMFBP1 and CCT3. The upper panel of HeLa cells was transfected with the pEGFP-N2-PMFBP1 plasmid, and the lower panel was transfected with pEGFP-N2-PMFBP1 and pCDNA3.1-CMV-3×flag-CCT3 plasmids. Subcellular localization of CCT3 (red) was probed with an anti-FLAG antibody. Cell nuclei were counterstained with DAPI. Scale bar: 10 μm. e A co-immunoprecipitation assay confirmed the interaction between endogenous mouse Cct3 and Pmfbp1 in the mouse testis. The anti-IgG or anti-Pmfbp1 antibody and protein A/G beads were used for immunoprecipitation, and anti-Cct3 and anti-Pmfbp1 antibodies were used for western blot analysis. f Immunofluorescent staining of Cct3 (green) and DAPI (nucleus, blue) on frozen sections from P21 Pmfbp1−/− and wild-type mouse testes. Scale bar, 20 μm. CCT3, chaperonin-containing TCP1 subunit 3; GFP, green fluorescent protein; KEGG, Kyoto Encyclopedia of Genes and Genomes; P21, postnatal day 21; PMFBP1, polyamine modulating factor 1 binding protein
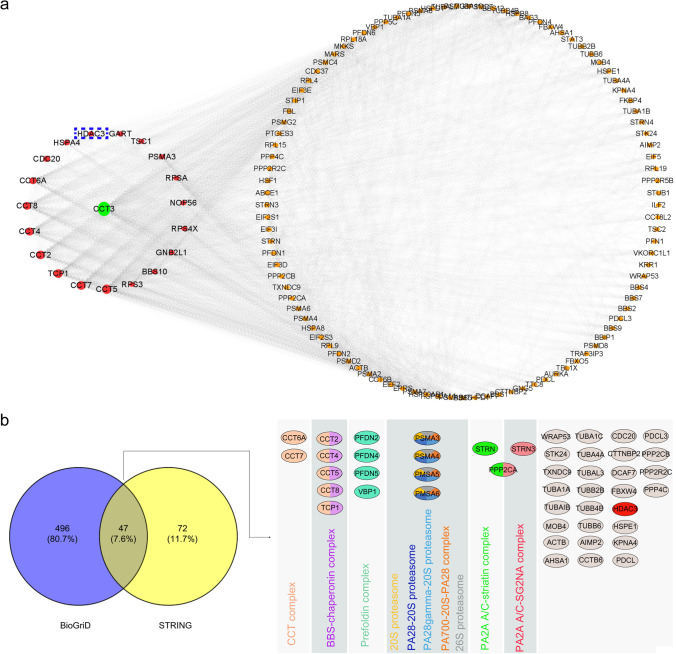
In order to further elucidate the function of the PMFBP1-CCT3 complex, the potential interacted proteins were predicted using BioGRID and STRING analyses (Fig. 2a, Figure S1). In total, 543 and 119 high-confidence PPIs were identified, respectively, including 47 proteins that intersected (Fig. 2b). The bioinformatics analysis from the CORUM database also indicated that CCT3 and its associated proteins including HDAC3 could form several important functional protein complexes, including a CCT complex, a BBS-chaperonin complex, and a prefoldin complex, which have been demonstrated to be critical for regulating ciliogenesis and protein folding [27] (Fig. 2b). Due to the fact that HDAC3 can interact with SMRT to regulate gene transcription after priming activation of the cellular chaperone [28], our results serve as a reminder that PMFBP1 may have an important role in spermatogenesis by interacting with HDAC3 in the presence of the cellular chaperone CCT3.
Fig. 2.
Protein-protein interactions in the CCT3 network. a Protein-protein interactions in the CCT3 network (green, CCT3; red, top 19 proteins interacting with CCT3; yellow, other potentially common targets of CCT3) were generated by Cytoscape (version 3.8) (http://cytoscape.org/). b Venn diagram showing overlap of the genes identified by the BioGriD (blue) and STRING (yellow) databases. The g:Profiler based on information from the CORUM database was used to determine the specific cellular protein complexes (right panel) involved by the overlap of 47 proteins. Half of the prey belongs to different protein complexes that are highlighted by color coding. CCT3, chaperonin-containing TCP1 subunit 3; PMFBP1, polyamine modulating factor 1 binding protein
Pmfbp1 Interacts with Hdac3 and Affects its Expression in the Mouse Testis
In order to evaluate the predicted association between PMFBP1 and its potential binding protein, co-immunoprecipitation experiments were carried out, which revealed that HDAC1, HDAC2, and HDAC3 could specifically interact with PMFBP1, and the highest affinity was between HDAC3 and PMFBP1 (Fig. 3a, Figure S2). The endogenous co-immunoprecipitation analysis using P21 mouse testicular lysate also showed that Hdac3 and Pmfbp1 could pull down with each other (Fig. 3b and c). Immunofluorescence assays were performed to examine the distribution and expression of Hdacs in the mouse testis in response to the Pmfbp1 deficiency. Expression of Hdac1, Hdac2, and Hdac3 tended to decrease in Pmfbp1 knockout mice (Fig. 3d–f). Moreover, western blot analysis also revealed that expression levels of Hdac1, Hdac2, and Hdac3 were significantly lower in the Pmfbp1−/− mouse testis than in the wild-type mouse testis (Fig. 3g). This suggests that PMFBP1 may be involved in TRiC-mediated folding of HDAC3, consequently regulating the expression of HDAC3 and affecting spermatogenesis in late meiosis and early RS stages.
Fig. 3.
PMFBP1 interacts with HDAC3 and affects its expression level. a A co-immunoprecipitation assay confirmed the interaction of exogenous human GFP-PMFBP1 and FLAG-HDAC1/2/3/4/5 in HEK293T cells. FLAG-M2 beads were used for immunoprecipitation and anti-GFP and anti-FLAG antibodies for western blot analysis. The co-immunoprecipitation assay confirmed the interaction between endogenous mouse Pmfbp1 and Hdac3 in the mouse testis (b, c). Anti-Pmfbp1 and anti-Hdac3 antibodies were used for western blot analysis. (d–f) Hdac1, Hdac2, and Hdac3 expression in Pmfbp1−/− and wild-type mouse testes at P21. Hdac1, Hdac2, and Hdac3 expressions are shown in green and the nuclei in blue. Scale bar, 20 μm. g Western blot analysis of the Pmfbp1 and Hdac1/2/3 in Pmfbp1−/− and wild-type mouse testes at P21. GAPDH is shown as the loading control. The bands in the left panel were quantified by ImageJ software and show that the abundance of Hdac1, Hdac2, and Hdac3 proteins in the Pmfbp1−/− mouse was significantly lower than that in the wild-type animal. The expression level was set as 1 in the wild-type samples. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t-test). GFP, green fluorescent protein; HDAC3, histone deacetylase 3; PMFBP1, polyamine modulating factor 1 binding protein; P21, postnatal day 21
The Pmfbp1-Hdac3 Complex May Be the Key Factor Involved in Spermatogenesis in Mice
Proteins are major biological macromolecules that function in various histogeneses and life processes. To characterize the novel molecular mechanism of PMFBP1, we isolated the testes of three wild-types and three Pmfbp1−/− mice to observe the proteomic changes at P21 in view of the fact that Pmfbp1 has been expressed by this time and considering the difficulty involved in collecting intact sperm from the epididymis of a Pmfbp1−/− mouse. The amount of protein in these samples was analyzed by label-free quantification based on LC-MS/MS. A total of 7315 proteins were identified, with at least two unique peptides at a false discovery rate of < 1%. After quantification, 138 proteins were screened out as DEPs with a fold change of > 1.5 and a P-value of < 0.05, among which 125 were downregulated and only 13 were upregulated in Pmfbp1−/− mice when compared with the wild-type controls (Fig. 4a, Supplementary Table 5). Further bioinformatics analysis performed on these 138 DEPs showed that the terms annotated by GO were involved in spermatogenesis, the assembly of flagella, and ATP-dependent microtubule motor activity (Fig. 4b). As expected, the highly enriched proteins were associated with the assembly of cilia and spermatogenesis. KEGG pathway enrichment analysis demonstrated that many dysregulated proteins were involved in metabolism-related pathways, including AMP-activated protein kinase signaling, peroxisome proliferator-activated receptor signaling, pyruvate metabolism, and glycolysis/gluconeogenesis signaling related to spermatogenesis (Supplementary Table 6).
Fig. 4.
Pmfbp1 may cooperate with Hdac3 to affect gene expression profiling in the P21 mouse testis. a Heat map representing differentially regulated proteins (fold-change >1.5, P-value < 0.05, two-tailed Student’s t-test, false discovery rate < 0.05) in P21 mouse testis tissue after knockout of Pmfbp1. The color gradient represents the changes (log2 scale) from the most downregulated (dark blue) to the most upregulated (red) genes. Each row is a gene, and each column represents replicates (average of n = 3 replicates). b Histogram showing the Gene Ontology enrichment analysis results for 138 DEPs between the control group and the Pmfbp1−/− mouse. The tassel size corresponds to the −log10 P-value of the enrichment while the color to the genes belonging to each category. (c) Venn diagram of DEGs and DEPs. In total, 5126 DEGs and 138 DEPs were respectively identified in Hdac−/− and Pmfbp1−/− mouse testes at P21. Seventy overlapping target genes were found between the two sets. d Quantitative polymerase chain reaction analysis of key gene expression in P21 wild-type and Pmfbp1−/− testes (n = 3 each). Data are shown as the mean ± standard deviation. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t-test). e Western blot showing that the Efhc1, Spag6, Tekt4, Dnajb13, Nme5, and Odf1 proteins involved in mouse spermatogenesis were downregulated in Pmfbp1−/− mice. The P21 testis lysate was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. α-Tubulin was used as the loading control. The right panel shows the quantified western blot bands for Pmfbp1−/− and Pmfbp1+/+ mice by ImageJ software (right). The expression level of the wild-type samples was set at 1. f The indicated proteins in sperm samples from a patient with PMFBP1 deficiency were immunodetected using specific antibodies. α-Tubulin was used for the loading control. Bands for the patient and a normal subject were quantified by ImageJ software (right). The expression level of the wild-type samples was set at 1. BP, biological process; CC, cellular component; MF, molecular function; DEGs, differentially expressed genes; DEPs, differentially expressed proteins; HDAC3, histone deacetylase 3; PMFBP1, polyamine modulating factor 1 binding protein; P21, postnatal day 21
To evaluate the significance of the interaction between PMFBP1 and HDAC3, we also compared the gene expression profile of the mouse testis between Hdac3 knockout mice and Pmfbp1−/− mice. We analyzed the differentially expressed genes (DEGs) that were common to the proteome of the Pmfbp1−/− testis and the transcriptomic data for Stra8-cre/Hdac3fl/− RS in mice at P21 (the latter was downloaded from the Gene Expression Omnibus database [GSE153065] [29]), which showed that there were 70 genes in common (Fig. 4c). Quantitative real-time PCR proved that there were significant differences in the mRNA expression of some common genes in Pmfbp1−/− mice, including Ccdc13, Cep41, Spag6, Ccdc65, Rsph6a, Dnajb13, Tekt1, Tekt2, and Tekt4 (Fig. 4d), all of which are involved in the formation of sperm. Western blotting and immunofluorescence assays using mouse testis at P21 also indicated that several other DEPs, including Efhc1, Spag6, Tekt4, Dnajb13, Nme5, Odf1, Nme5, and Spag19, were present at the significantly lower relative abundance in the Pmfbp1−/− mouse testis than in the wild-type mouse testis (Fig. 4e, Figure S3). SPAG6, TEKT4, EFHC1, and NME5 were also found to be consistently downregulated in the sperm of a sterile patient with c.2725C>T [p.Arg909*] and c.2092delG [p.Ala698 Profs*7] mutations in the PMFBP1 gene [3] (Fig. 4f). These findings suggest that PMFBP1 may affect spermatogenesis by regulating the expression level of HDAC3 and the function of PMFBP1 is highly conserved in both mice and humans.
Rnf151 and Rnf133 are Potential Critical Response Factor Downstream of the Pmfbp1-Hdac3 Axis with an Effect on Spermatogenesis in Mice
A previous study identified that Hdac3 could coordinate with Sox30 to form a transcriptional program in a deacetylase-independent manner during the meiotic-to-postmeiotic transition in spermatogenesis [29]. To further test the correlation of PMFBP1 with HDAC3 and identify the key factor downstream of the PMFBP1-HDAC3 axis, we analyzed the DEGs in common between the transcriptomic data for round sperm from Sox30 knockout mice and Stra8-cre/Hdac3fl/– mice. The published sequencing data for these two types of mice were downloaded from the Gene Expression Omnibus database (P21 RS from Stra8-cre/Hdac3fl/− mice, number GSE153065; P23 RS from Sox30−/− mice, number GSE113073) and the National Omics Data Encyclopedia (P21 RS from Sox30−/− mice, accession number OEP000012) [29–31]. Transcriptome profiling revealed a considerable overlap of 193 downregulated genes between P21 Sox30−/− RS, P23 Sox30−/− RS, and P21 Stra8-cre/Hdac3fl/− RS (Fig. 5a). Pathway enrichment analyses according to the Metascape database indicated that these downregulated genes were related to biological processes, including spermatogenesis (i.e., Rnf133 and Rnf151), fertilization, and the movement of cilia (Fig. 5a). Rnf151 and Rnf133 are two putative RING family E3 ubiquitin ligases, which have already been confirmed to play a critical role during spermiogenesis [32]. The alignments of downloaded ChIP-seq data for Rnf151 and Rnf133 using Integrative Genomics Viewer (IGV) software showed that Hdac3-Sox30 heterodimer binding to the promoter of Rnf151 and that of Rnf133 was significantly lower in the testes of mice lacking Sox30 than in wild-type mice (Fig. 5b). The result of quantitative real-time PCR also showed that Rnf151 and Rnf133 transcription levels were significantly lower in Pmfbp1−/− mice than in wild-type mice (Fig. 5c). It was worth mentioning that a droplet of cytoplasm is retained outside the flagellum in the sperm of Pmfbp1−/− mice [3, 7], and a similar abnormality has been observed in Rnf133 knockout sperm [32]. These observations suggest that Rnf133 is a critical response factor downstream of the Pmfbp1-Hdac3 axis and provide further biological evidence for Hdac3 being the key effector via which Pmfbp1 regulates spermatogenesis in mice.
Fig. 5.
Rnf151 and Rnf133 may be the response factor downstream of the Pmfbp1-Hdac3 axis in mouse spermatogenesis. a Venn diagram showing the overlapping downregulated genes among P21 Hdac3−/− round spermatids, P23 Sox30−/− round spermatids, and P21 Sox30−/− round spermatids compared with wild-type round spermatids. The 193 overlapping genes in these three groups were subjected to gene functional analysis using the Metascape database. The most significantly enriched pathway of these genes is “GO:0007283 spermatogenesis,” which harbors Rnf133 and Rnf151. b The Hdac3 ChIP-seq and Sox30 ChIP-seq reads in wild-type and Sox30 knockout testes at P20–P21 are aligned to the genomic sequence of Rnf151 (left) and Rnf133 (right) in the mouse reference genome. (C) Pmfbp1, Rnf151, and Rnf133 mRNA levels in testis tissue from Pmfbp1−/− (n = 3) and wild-type (n = 3) mice at P21. Values were determined by quantitative real-time polymerase chain reaction and are expressed as the mean ± standard error of triplicate experiments after normalization to BACT mRNA levels. The expression level of the wild-type samples was set at 1. The data are shown as the mean ± standard deviation. *P < 0.05, **P < 0.01, and ***P < 0.001 (Student’s t-test). GO, Gene Ontology; HDAC3, histone deacetylase 3; PMFBP1, polyamine modulating factor 1 binding protein; P20, postnatal day 20; P21, postnatal day 21; P23, postnatal day 23; Rnf, ring finger protein
So, we propose that Pmfbp1 could regulate the abundance of Hdac3 in spermatogenesis by cooperating with its interacting protein Cct3 and finally act on the downstream target genes. Furthermore, downregulated Rnf133 induces retention of the droplet of cytoplasm outside sperm, resulting in an abnormal phenotype beyond the headless sperm tails in Pmfbp1−/− mice.
Discussion
PMFBP1 is a testis-specific protein that has been identified to participate in the formation of normal sperm in humans and mice and to locate at the sperm head-to-tail connecting piece cooperating with SUN5 and SPATA6 [3, 7]. Although many studies of PMFBP1 in ASS have been reported, the molecular mechanisms underlying some of the phenotypic defects have not been well explained, such as disorganization of the mitochondrial sheath, a defect of the microtubule doublets in the axoneme, and the residue of a cytoplasmic droplet without distribution of PMFBP1. Our present study, which included mass spectrometry and proteomic data integrated with an analysis of publicly available data, provides further evidence for the key interaction protein Pmfbp1 during mouse spermatogenesis. Our three main findings are as follows. First, label-free mass spectrometry and co-immunoprecipitation revealed that CCT3, PMFBP1, and HDAC3 can form a functional protein complex in testicular cells. Second, proteomic analysis confirmed a general downregulation of proteins in Pmfbp1-defective mice; these proteins include Efhc1, Spag6, Tekt4, Dnajb13, Nme5, and Odf1, all of which are involved in spermatogenesis, assembly of flagella, and ATP-dependent microtubule motor activity. More than half of the DEPs were consistent with the transcriptional profile of Hdac3 knockout mice, and abnormal expression of key genes always leads to morphological abnormality in sperm, such as the abnormal “9 + 2” structure of the sperm flagellum [33]. Third, Hdac3 coordinates with Sox30 to achieve a transcriptional program during the meiotic-to-postmeiotic transition in spermatogenesis [32]. The results of our bioinformatics analysis show that there are binding sites for Hdac3-Sox30 on the promoters of the RING family E3 ubiquitin ligases Rnf151 and Rnf133 based on public transcriptomes and ChIP-seq data. Our real-time PCR experiment confirmed that Rnf151 and Rnf133 are downregulated in the Pmfbp1 knockout mouse testis at P21.
The chaperonin-containing TCP1 (CCT) complex is responsible for the ATP-dependent folding of client proteins [34]. CCT3 is an important member of the CCT family and participates in protein-folding reactions that include actin and tubulin to prevent protein aggregation in the crowded cellular environment [35]. Intriguingly, we observed that CCT3 may promote protein folding of PMFBP1 (Fig. 1d). There is already some evidence indicating that HDAC3 interacts with SMRT after activation of a cellular chaperone such as TRiC and regulates gene transcription and that CCT3 is a component of TRiC that is required for the assembly of HDAC3 into the SMRT complex [28, 36, 37]. HDAC1, HDAC2, and HDAC3 are class I HDACs that serve as the catalytic subunits of various transcriptional co-repressor complexes [38]. A prominent transcriptomic switch from late spermatocyte to RS is associated with a dynamic change in histone acetylation, which suggests that HDACs are involved in sperm formation in late meiosis and early RS stages via histone acetylation. However, a deficiency in HDAC3 catalytic activity does not cause infertility. Importantly, Hdac3 is the only Hdac with high expression levels during spermatogenesis, the only protein that shows a dynamic pattern of expression in germ cell development, and peaks at P21 in mice [29]. Our data show that Hdac3 was significantly downregulated in the Pmfbp1−/− mouse testis. We collected testis tissue at P21 for proteomic analysis, which indicates that Pmfbp1, Cct3, and Hdac3 could interact with each other in certain ways, and the assembly of the Cct3-Pmfbp1-Hdac3 complex would result in an abundance of Hdac3, which would impact a broad range of cellular processes during spermiogenesis in mice [29, 39–42]. However, we cannot rule out the possibility that other factors downstream of PMFBP1 affect expression of HDAC3 beyond the direct effect by PMFBP1-CCT3 on protein folding of HDAC3, thereby affecting the protein abundance of HDAC3. Hence, more experimental evidence is needed to confirm whether the spatiotemporal interaction of these three proteins regulates spermatogenesis.
The architecture of spermatozoa is remodeled to meet the requirement for fertilization [43]. During the maturation of spermatids from RS to the elongated sperm, unnecessary proteins, organelles, and bulk cytoplasm of the spermatid are discarded by extrusion of droplets from the cytoplasm, which, coupled with mitochondrial rearrangement around the flagella at the mid-piece of the tail, eventually leads to the formation of normal sperm [44]. Previous studies have identified many types of sterile sperm caused by inappropriate retention of cytoplasmic droplets and endoplasmic reticulum-associated degradation [32, 45]. The findings of proteomic analysis of sperm from the Pmfbp1 knockout mouse reflect the important role of PMFBP1 in sperm, namely, its function as a scaffold protein for the connection between HTCA and the nuclear membrane of sperm to maintain its head-tail structural integrity in the cytoplasm [7]. Furthermore, the sperm of Pmfbp1 knockout mice also exhibited cytoplasmic droplets outside the sperm flagella. The downregulated Rnf151 and Rnf133 genes in the Pmfbp1−/− mouse testis were confirmed to be the potential response factor in the Pmfbp1-Hdac3 signal axis when combined with previous reports on Hdac3 and Sox30 in mouse spermatogenesis [29–31]. These two E3 ubiquitin ligases of the RING family are testis-specific proteins that are expressed during spermatogenesis and are required for fecundity in mice [32, 46]. The Rnf133 knockout male exhibits aberrant head-neck morphology, and droplets of cytoplasm are retained in Rnf133 knockout spermatozoa, similar to the Pmfbp1 knockout [32]. Notably, a defect in the sperm head-tail junction does not affect fertilization or development of the embryo after intracytoplasmic sperm injection, which is an effective assisted reproductive technology strategy in patients with PMFBP1 mutations and Pmfbp1 knockout male mice, as previously described [3, 47]. Overall, our observations strongly suggest that RNF151 and RNF133 respond to the PMFBP1-HDAC3 axis during spermatogenesis. Despite the conservation and functional similarity of PMFBP1 that localizes within cells and related phenotypic variations between humans and mice, we cannot rule out the possibility that there are differences in the molecular mechanisms of spermatogenesis between mice and humans. Therefore, a straightforward strategy would be to generate gene-edited mice using gene editing technology such as CRISPR-Cas9 to replace the mouse Pmfbp1 gene with the human PMFBP1 gene in mice for functional investigation of human PMFBP1 in vivo.
In summary, we have shown that dynamic assembly of the CCT3-PMFBP1-HDAC3 complex by regulation of target gene expression is essential for spermatogenesis in humans and mice, and a steady-state imbalance would result in a phenotype of defective sperm. The findings of this study further highlight the important role of Pmfbp1 during spermatogenesis in mice.
Supplementary Information
(PDF 1158 kb)
(PDF 158 kb)
Acknowledgements
We thank Yin H. et al., Bai S. et al., and Zhang D. et al. for the transcriptomic data and ChIP-seq data (numbers GSE153065 and GSE113073) and the National Omics Data Encyclopedia (accession number OEP000012). The ChIP-seq data in this study were generated from Sox30−/− RS using the Hdac3 antibody and wild-type mouse RS using the Sox30 antibody and downloaded from the Gene Expression Omnibus (GSE153065) database. We thank Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
Author Contribution
F. Wang, F. Zhu, and H. Zhang designed the research. W. Xu, Y. Li, Z. Yao, and K. Wang performed the research. S. Kong, Y. Wang, and M. Xiang analyzed data. H. Zhang and W. Xu wrote the paper. F. Wang and F. Zhu revised the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (81972641 to F.W.; 82071701 to F.Z.), the Scientific Research Foundation of the Institute for Translational Medicine of Anhui Province (2021zhyx-C25), the Basic and Clinical Cooperative Research Promotion Program of Anhui Medical University (2022xkjT015), and the Natural Science Research Project for Anhui Universities (KJ2021A0242).
Data Availability
All data are available in the main text or the supporting information materials.
Declaration
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Weilong Xu, Zhoujuan Yao, Yunzhi Li, and Ke Wang contributed equally to this work.
Contributor Information
Fuxi Zhu, Email: fxzhu@ahmu.edu.cn.
Fengsong Wang, Email: fengsongw@ahmu.edu.cn.
Hui Zhang, Email: zhanghui520627@hotmail.com.
References
- 1.Ni FD, Hao SL, Yang WX. Multiple signaling pathways in Sertoli cells: recent findings in spermatogenesis. Cell Death Dis. 2019;10:541. doi: 10.1038/s41419-019-1782-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharma S, Hanukoglu A, Hanukoglu I. Localization of epithelial sodium channel (ENaC) and CFTR in the germinal epithelium of the testis, Sertoli cells, and spermatozoa. J Mol Histol. 2018;49:195–208. doi: 10.1007/s10735-018-9759-2. [DOI] [PubMed] [Google Scholar]
- 3.Zhu F, Liu C, Wang F, Yang X, Zhang J, Wu H, Zhang Z, He X, Zhang Z, Zhou P, et al. Mutations in PMFBP1 cause acephalic spermatozoa syndrome. Am J Hum Genet. 2018;103:188–199. doi: 10.1016/j.ajhg.2018.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mazaheri Moghaddam M, Mazaheri Moghaddam M, Hamzeiy H, Baghbanzadeh A, Pashazadeh F, Sakhinia E. Genetic basis of acephalic spermatozoa syndrome, and intracytoplasmic sperm injection outcomes in infertile men: a systematic scoping review. J Assist Reprod Genet. 2021;38:573–586. doi: 10.1007/s10815-020-02008-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Liu C, Wu B, Li L, Li W, Yuan L. The missing linker between SUN5 and PMFBP1 in sperm head-tail coupling apparatus. Nat Commun. 2021;12:4926. doi: 10.1038/s41467-021-25227-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie H, Tang Y, Zhang X, Tan Y, Qin W. Novel mutations of PMFBP1 in a man with acephalic spermatozoa defects. Mol Genet Genomic Med. 2022;10:e2020. doi: 10.1002/mgg3.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sha YW, Wang X, Xu X, Ding L, Liu WS, Li P, Su ZY, Chen J, Mei LB, Zheng LK, et al. Biallelic mutations in PMFBP1 cause acephalic spermatozoa. Clin Genet. 2019;95:277–286. doi: 10.1111/cge.13461. [DOI] [PubMed] [Google Scholar]
- 8.Deng TQ, Xie YL, Pu JB, Xuan J, Li XM. Compound heterozygous mutations in PMFBP1 cause acephalic spermatozoa syndrome: a case report. World J Clin Cases. 2022;10:12761–12767. doi: 10.12998/wjcc.v10.i34.12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang D, Xu J, Chen K, Liu Y, Yang X, Tang L, Luo X, Liu Z, Li M, Walters JR, et al. BmPMFBP1 regulates the development of eupyrene sperm in the silkworm. Bombyx mori. PLoS Genet. 2022;18:e1010131. doi: 10.1371/journal.pgen.1010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu G, Xing X, Zhang H, Zhu W, Lin G, Lu G, Li W. Patients with acephalic spermatozoa syndrome linked to novel TSGA10/PMFBP1 variants have favorable pregnancy outcomes from intracytoplasmic sperm injection. Clin Genet. 2021;100:334–339. doi: 10.1111/cge.14007. [DOI] [PubMed] [Google Scholar]
- 11.Lu M, Kong S, Xiang M, Wang Y, Zhang J, Duan Z, Zha X, Wang F, Cao Y, Zhu F. A novel homozygous missense mutation of PMFBP1 causes acephalic spermatozoa syndrome. J Assist Reprod Genet. 2021;38:949–955. doi: 10.1007/s10815-021-02075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wörheide MA, Krumsiek J, Kastenmüller G, Arnold M. Multi-omics integration in biomedical research-a metabolomics-centric review. Anal Chim Acta. 2021;1141:144–162. doi: 10.1016/j.aca.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan J, Risacher SL, Shen L, Saykin AJ. Network approaches to systems biology analysis of complex disease: integrative methods for multi-omics data. Brief Bioinform. 2018;19:1370–1381. doi: 10.1093/bib/bbx066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W, Murugan S, Lindberg J, Chellappa V, Shen X, Pawitan Y, Vu TN. Fusion gene detection using whole-exome sequencing data in cancer patients. Front Genet. 2022;13:820493. doi: 10.3389/fgene.2022.820493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Göös H, Kinnunen M, Salokas K, Tan Z, Liu X, Yadav L, Zhang Q, Wei GH, Varjosalo M. Human transcription factor protein interaction networks. Nat Commun. 2022;13:766. doi: 10.1038/s41467-022-28341-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Rezelj VV, Guo JZ, Swaney DL, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doncheva NT, Morris JH, Gorodkin J, Jensen LJ. Cytoscape StringApp: network analysis and visualization of proteomics data. J Proteome Res. 2019;18:623–632. doi: 10.1021/acs.jproteome.8b00702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res. 2010;38(Database issue):D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–d612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, Kolas N, O'Donnell L, Leung G, McAdam R, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47:D529–d541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Knethen A, Tzieply N, Jennewein C, Brüne B. Casein-kinase-II-dependent phosphorylation of PPARgamma provokes CRM1-mediated shuttling of PPARgamma from the nucleus to the cytosol. J Cell Sci. 2010;123:192–201. doi: 10.1242/jcs.055475. [DOI] [PubMed] [Google Scholar]
- 23.Mlecnik B, Galon J, Bindea G. Automated exploration of gene ontology term and pathway networks with ClueGO-REST. Bioinformatics. 2019;35:3864–3866. doi: 10.1093/bioinformatics/btz163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otasek D, Morris JH, Bouças J, Pico AR, Demchak B. Cytoscape Automation: empowering workflow-based network analysis. Genome Biol. 2019;20(1):185. doi: 10.1186/s13059-019-1758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freund A, Zhong FL, Venteicher AS, Meng Z, Veenstra TD, Frydman J, Artandi SE. Proteostatic control of telomerase function through TRiC-mediated folding of TCAB1. Cell. 2014;159:1389–1403. doi: 10.1016/j.cell.2014.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seo S, Baye LM, Schulz NP, Beck JS, Zhang Q, Slusarski DC, Sheffield VC. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci U S A. 2010;107:1488–1493. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guenther MG, Yu J, Kao GD, Yen TJ, Lazar MA. Assembly of the SMRT-histone deacetylase 3 repression complex requires the TCP-1 ring complex. Genes Dev. 2002;16:3130–3135. doi: 10.1101/gad.1037502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams SR, Aldred MA, Der Kaloustian VM, Halal F, Gowans G, McLeod DR, Zondag S, Toriello HV, Magenis RE, Elsea SH. Haploinsufficiency of HDAC4 causes brachydactyly mental retardation syndrome, with brachydactyly type E, developmental delays, and behavioral problems. Am J Hum Genet. 2010;87:219–228. doi: 10.1016/j.ajhg.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin H, Kang Z, Zhang Y, Gong Y, Liu M, Xue Y, He W, Wang Y, Zhang S, Xu Q, et al. HDAC3 controls male fertility through enzyme-independent transcriptional regulation at the meiotic exit of spermatogenesis. Nucleic Acids Res. 2021;49:5106–5123. doi: 10.1093/nar/gkab313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bai S, Fu K, Yin H, Cui Y, Yue Q, Li W, Cheng L, Tan H, Liu X, Guo Y, et al. Sox30 initiates transcription of haploid genes during late meiosis and spermiogenesis in mouse testes. Development. 2018;145 [DOI] [PMC free article] [PubMed]
- 32.Zhang D, Xie D, Lin X, Ma L, Chen J, Zhang D, Wang Y, Duo S, Feng Y, Zheng C, et al. The transcription factor SOX30 is a key regulator of mouse spermiogenesis. Development. 2018;145 [DOI] [PubMed]
- 33.Nozawa K, Fujihara Y, Devlin DJ, Deras RE, Kent K, Larina IV, Umezu K, Yu Z, Sutton CM, Ye Q, et al. The testis-specific E3 ubiquitin ligase RNF133 is required for fecundity in mice. BMC Biol. 2022;20:161. doi: 10.1186/s12915-022-01368-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Zhang L, Li W, Huang Q, Yuan S, Li Y, Liu J, Zhang S, Pin G, Song S, et al. The sperm-associated antigen 6 interactome and its role in spermatogenesis. Reproduction. 2019;158:181–197. doi: 10.1530/REP-18-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger J, Berger S, Li M, Jacoby AS, Arner A, Bavi N, Stewart AG, Currie PD. In vivo function of the chaperonin TRiC in α-actin folding during sarcomere assembly. Cell Rep. 2018;22:313–322. doi: 10.1016/j.celrep.2017.12.069. [DOI] [PubMed] [Google Scholar]
- 36.Liu W, Lu Y, Yan X, Lu Q, Sun Y, Wan X, Li Y, Zhao J, Li Y, Jiang G. Current understanding on the role of CCT3 in cancer research. Front Oncol. 2022;12:961733. doi: 10.3389/fonc.2022.961733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Millán-Zambrano G, Rodríguez-Gil A, Peñate X, de Miguel-Jiménez L, Morillo-Huesca M, Krogan N, Chávez S. The prefoldin complex regulates chromatin dynamics during transcription elongation. PLoS Genet. 2013;9:e1003776. doi: 10.1371/journal.pgen.1003776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Banks CAS, Miah S, Adams MK, Eubanks CG, Thornton JL, Florens L, Washburn MP. Differential HDAC1/2 network analysis reveals a role for prefoldin/CCT in HDAC1/2 complex assembly. Sci Rep. 2018;8:13712. doi: 10.1038/s41598-018-32009-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei W, Liu X, Chen J, Gao S, Lu L, Zhang H, Ding G, Wang Z, Chen Z, Shi T, et al. Class I histone deacetylases are major histone decrotonylases: evidence for critical and broad function of histone crotonylation in transcription. Cell Res. 2017;27:898–915. doi: 10.1038/cr.2017.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chini CC, Escande C, Nin V, Chini EN. HDAC3 is negatively regulated by the nuclear protein DBC1. J Biol Chem. 2010;285:40830–40837. doi: 10.1074/jbc.M110.153270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emmett MJ, Lim HW, Jager J, Richter HJ, Adlanmerini M, Peed LC, Briggs ER, Steger DJ, Ma T, Sims CA, et al. Histone deacetylase 3 prepares brown adipose tissue for acute thermogenic challenge. Nature. 2017;546:544–548. doi: 10.1038/nature22819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, Zumbo P, Kirouac K, Bhaskara S, Polo JM, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013;4:578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zakrzewski P, Lenartowski R, Rędowicz MJ, Miller KG, Lenartowska M. Expression and localization of myosin VI in developing mouse spermatids. Histochem Cell Biol. 2017;148:445–462. doi: 10.1007/s00418-017-1579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fayomi AP, Orwig KE. Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res. 2018;29:207–214. doi: 10.1016/j.scr.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nozawa K, Zhang Q, Miyata H, Devlin DJ, Yu Z, Oura S, Koyano T, Matsuyama M, Ikawa M, Matzuk MM. Knockout of serine-rich single-pass membrane protein 1 (Ssmem1) causes globozoospermia and sterility in male mice†. Biol Reprod. 2020;103:244–253. doi: 10.1093/biolre/ioaa040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nian H, Fan C, Liao S, Shi Y, Zhang K, Liu Y, Han C. RNF151, a testis-specific RING finger protein, interacts with dysbindin. Arch Biochem Biophys. 2007;465:157–163. doi: 10.1016/j.abb.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Fang J, Zhang J, Zhu F, Yang X, Cui Y, Liu J. Patients with acephalic spermatozoa syndrome linked to SUN5 mutations have a favorable pregnancy outcome from ICSI. Hum Reprod. 2018;33:372–377. doi: 10.1093/humrep/dex382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 1158 kb)
(PDF 158 kb)
Data Availability Statement
All data are available in the main text or the supporting information materials.