Abstract
African American (AA) women in the United States have a 40% higher breast cancer mortality rate than Non-Hispanic White (NHW) women. The survival disparity is particularly striking among (estrogen receptor positive) ER+ breast cancer cases. The purpose of this study is to examine whether there are racial differences in metabolic pathways typically activated in patients with ER+ breast cancer. We collected pretreatment plasma from AA and NHW ER+ breast cancer cases (AA n = 48, NHW n = 54) and cancer-free controls (AA n = 100, NHW n = 48) to conduct an untargeted metabolomics analysis using gas chromatography mass spectrometry (GC–MS) to identify metabolites that may be altered in the different racial groups. Unpaired t-test combined with multiple feature selection and prediction models were employed to identify race-specific altered metabolic signatures. This was followed by the identification of altered metabolic pathways with a focus in AA patients with breast cancer. The clinical relevance of the identified pathways was further examined in PanCancer Atlas breast cancer data set from The Cancer Genome Atlas Program (TCGA). We identified differential metabolic signatures between NHW and AA patients. In AA patients, we observed decreased circulating levels of amino acids compared to healthy controls, while fatty acids were significantly higher in NHW patients. By mapping these metabolites to potential epigenetic regulatory mechanisms, this study identified significant associations with regulators of metabolism such as methionine adenosyltransferase 1A (MAT1A), DNA Methyltransferases and Histone methyltransferases for AA individuals, and Fatty acid Synthase (FASN) and Monoacylglycerol lipase (MGL) for NHW individuals. Specific gene Negative Elongation Factor Complex E (NELFE) with histone methyltransferase activity, was associated with poor survival exclusively for AA individuals. We employed a comprehensive and novel approach that integrates multiple machine learning and statistical methods, coupled with human functional pathway analyses. The metabolic profile of plasma samples identified may help elucidate underlying molecular drivers of disproportionately aggressive ER+ tumor biology in AA women. It may ultimately lead to the identification of novel therapeutic targets. To our knowledge, this is a novel finding that describes a link between metabolic alterations and epigenetic regulation in AA breast cancer and underscores the need for detailed investigations into the biological underpinnings of breast cancer health disparities.
Subject terms: Cancer metabolism, Endocrine cancer, Tumour biomarkers
Introduction
Breast cancer mortality rates in African American (AA) women are 40% higher than in Non-Hispanic White (NHW) women1, stemming from complex and multifaceted factors2. AA women experience earlier disease onset, exhibit biologically aggressive tumor phenotypes1–3, present with higher stages at diagnosis (4), experience increased likelihood of distant metastases4, and have lower survival rates post-diagnosis4–6. These disparities are particularly pronounced in individuals with estrogen receptor-positive (ER+) breast cancer1. While ER+ breast tumors typically respond favorably to endocrine therapy, leading to better outcomes, this improvement is not equally reflected in the overall survival rates of AA individuals. Thus, advancements in treatment that have significantly reduced breast cancer mortality overall, have not yielded equitable survival equally for all racial groups. Early studies suggested that the unfavorable outcomes among AA women was attributed to disparities in socioeconomic status, education and healthcare access7–11. Comprehensive meta-analysis at the population level confirmed that AA women continue to experience significantly higher breast cancer mortality rates compared with NHW women even after considering socioeconomic status12,13. Department of Defense studies furthered revealed that despite equal healthcare access and treatment protocols, AA women still faced poorer breast cancer outcomes than NHW women14,15. Additional investigations, adjusting for hormone receptor subtype, area-level factors, socioeconomic status and healthcare access consistently demonstrated significantly higher breast cancer mortality rates among AA women compared to NHW women16. Recent epidemiological findings from Chicago demonstrated that AA women with ER+ breast cancer faced a hazard of death that was 4–5 times higher than their white counterparts even after accounting for stage at diagnosis, tumor grade, and treatment1. Breast cancer mortality associated with ER+ cancers was higher in AA women treated in non-accredited hospitals, and from lower socioeconomic status neighborhoods17.
A deeper look into the biological mechanisms that affect tumor aggressiveness is crucial to understanding this disparity. The human metabolome reflects an individual metabolic state, which is influences by external factors like diet and environmental exposures. Usually, tumors exhibit increased demands for various nutrients in a frequently nutrient-poor environment to support sustained growth and proliferation. These alterations in demands and use of intracellular and extracellular metabolites are characteristic of cancer metabolic reprogramming, as it is well known that intermediate metabolites and products can act as signaling or epigenetic modulators18–21. Metabolomics profiling offers insights into cell function by directly influencing the phenotype of cancer cells22. In the context of breast cancer, this approach has successfully utilized to identified cancer risk factors23–25, prognostic indicators26–28, and early detection biomarkers26,29–31. Moreover, it has revealed disease-specific metabolic patterns and their associations with clinical outcomes32,33. Notably, metabolomics profiling has also demonstrated variations in metabolite levels across different racial groups34–37. These differences persists even after accounting for baseline characteristics such as lifestyle factors, health conditions, medications, and socioeconomic status.
Unfortunately, few studies have examined the role of circulating metabolites and the associated regulatory pathways that can help explain more aggressive tumor phenotypes and worsened disease outcomes in AA women. If racial differences exist in these associations, it may enhance understanding of the main biological and socioeconomic factors that drive racial disparities in ER+ breast cancer. Studying metabolic alterations in AA women, can provide insights into lifestyle, barriers to health access, dietary factors, and other contributors to the disparity in mortality in this racial group, which are often challenging to measure.
We previously used metabolomics analyses coupled with machine learning analysis to identify cancer-associated biomarkers38. Using a combination of multiple machine learning methods, statistics, and visualization coupled with human pathway analyses, our current study aims to identify differences in metabolic signatures and associated metabolic mechanisms perturbed in AA patients with ER+ breast cancer to help explain why they are more likely to die from the disease than their white counterparts are.
Methods
Study population
African American (AA) and Non-Hispanic White women (NHW) aged 20–79 years, with a new diagnosis of (American Joint Committee of Cancer) AJCC stage I–III, ER+ breast cancer were recruited from 3 hospitals in Chicago, IL between 2018 and 2019. This study was approved by the University of Illinois, Chicago Institutional Review Board (IRB protocol number 2017-1029). An informed consent was obtained from all subjects. All methods were performed in accordance with the relevant guidelines and regulations. Healthy AA and NHW women without breast symptoms or a personal history of breast cancer who presented for a screening mammogram at corresponding mammography centers were recruited as control subjects. Only control subjects whose screening mammogram was negative (BI-RADS category 1-2) were included in the analytic study population. Non-fasting whole blood was collected from AA and NHW breast cancer cases prior to breast surgery or any other breast cancer treatment. Control subjects donated a blood specimen at the time of study enrollment. Demographic and tumor pathologic characteristics are described in Table 1. Whole blood collected in serum separator tubes was allowed to clot for 40 min at room temperature, and plasma was separated by centrifugation within 1 h of collection (1680g for 10 min at room temperature in a horizontal rotor, tabletop centrifuge). Aliquots of plasma were immediately frozen at − 20 °C, and transferred to − 80 °C within 4 weeks. All samples were analyzed after a single freeze–thaw cycle.
Table 1.
Demographic and tumor pathologic characteristics.
| Cases | Controls | |||||
|---|---|---|---|---|---|---|
| AA | NHW | p value | AA | NHW | p value | |
| (%) | (%) | (%) | (%) | |||
| Number of participants | 49 | 54 | 0.121 | 105 | 45 | 0.1971 |
| Mean age, (years) | 60.5 | 56.8 | 55.5 | 58.1 | ||
| Missing (n) | 0 | 0 | 2 | 0 | ||
| Mean body mass index (kg) | 31.6 | 29.4 | 0.10a | 32.6 | 28.2 | 0.002a |
| Missing (n) | 0 | 0 | 2 | 1 | ||
| Tumor grade | ||||||
| Low | 15(33) | 13(24) | 0.89b | – | – | |
| Intermediate | 22(48) | 31(57) | ||||
| High | 9(20) | 10(19) | ||||
| Missing (n) | 3 | 0 | – | – | ||
| AJCC stage | ||||||
| 1 | 19(43) | 39(75) | 0.0023 | |||
| 2 | 19(43) | 12(23) | ||||
| 3 | 6(14) | 1(2) | ||||
| Missing (n) | 5 | 2 | ||||
AA African American, NHW non-Hispanic White, AJCC American Joint Committee on Cancer.
aStudent’s t-test.
bChi squared test for grade 1/2 vs. grade 3.
cFisher’s Exact test.
Metabolomics analysis
Sample preparation
To precipitate protein, 200 µL of thawed plasma was mixed with 1 mL isopropanol, acetonitrile, H2O 3:3:2 mix. The mixture was vortexed for 10 s and kept at − 20 °C for further processing. After spinning down, 600 µL of the supernatant were transferred to a separate Eppendorf tube and submitted to UIUC metabolomics core for a GC/MS whole metabolite profiling.
Mass spectrometry
A gas chromatography/mass spectrometry (GC/MS) based metabolic profiling was performed to detect and quantify the metabolites in plasma. Supernatants were collected by centrifugation (5 min at 15,000g), dried and derivatized with 50 μL of 40 mg/mL methoxyamine hydrochloride in pyridine (Sigma-Aldrich, MO, USA) for 60 min at 50 °C, then with 50 μL MSTFA + 1%TMCS (Thermo, MA, USA) at 70 °C for 120 min, and following 2-h incubation at room temperature. 50 μL of the internal standard (hentriacontanoic acid, 1 mg/mL) was added to each sample before derivatization. Metabolite profiles were acquired using a GC–MS system (Agilent Inc, CA, USA) consisting of an Agilent 7890 gas chromatograph, an Agilent 5975 MSD and an HP 7683B autosampler. Gas chromatography was performed on a ZB-5MS (60 m × 0.32 mm ID and 0.25 μm film thickness) capillary column (Phenomenex, CA, USA). The inlet and MS interface temperatures were 250 °C, and the ion source temperature was adjusted to 230 °C. An aliquot of 1 μL was injected with the split ratio of 10:1. The helium carrier gas was kept at a 2 mL/min constant flow rate. The temperature program was: 5-min isothermal heating at 70 °C, followed by an oven temperature increase of 5 °C/min to 310 °C and a final 10 min at 310 °C. The mass spectrometer was operated in positive electron impact mode (EI) at 69.9 eV ionization energy at m/z 30–800 scan range.
Spectra and data analysis
The spectra of all chromatogram peaks were evaluated using the AMDIS 2.71 (NIST, MD, USA) software using a custom-built database with 460 unique metabolites. All known artificial peaks were identified and removed before data mining. All data were normalized to the internal standard hentriacontanoic acid, 1 mg/mL in each chromatogram, and the cell’s dry weight (DW) to compare samples. The instrument variability was within the standard acceptance limit (5%).
Data processing
As part of data preprocessing, metabolites missing over 40% of data were removed. Remaining missing data values were completed using a trained data imputation algorithm using the available data. The imputation performance was tested one metabolite after another by removing one column and doing imputation on them. The imputation performance was evaluated for each specific metabolite considering R square metric. This process was repeated for each metabolite in the data set, removing and imputing them individually. Metabolites with R2 < 0.3, that indicated poor imputation performance were removed. To explore which metabolites (these are referred as features) are indicators of cancer and in each group, the data was sorted as African American (AA) data set and Non-Hispanic White (NHW) data set.
Statistical analyses
In order to compare the metabolite levels between cases and controls in each race group, we performed unpaired t-tests between cases and controls for each metabolic feature, separated by race groups. Differential metabolites were identified by adjusting the p values for multiple testing at a False discovery Rate (FDR) threshold of 0.05. A (*) indicates p value (p < 0.05) considered statistically significant. p value is reported as two-tailed p value, and standard error is included. All p values are corrected for multiple testing using FDR original method of Benjamini and Hochberg. R.4.02, and Graphpad Prism 9.2.0 software were used for statistical analyses (t-tests) (GraphPad Software Inc., La Jolla, CA, USA, www.graphpad.com).
Feature selection and prediction
To assess which features (metabolites) were associated with breast cancer in the cohort, we integrated a combination of machine learning feature selection and prediction algorithms. Algorithms used for feature selection included: Boruta test and Recursive Feature Elimination (RFE). Boruta algorithm identifies important features by creating shuffled shadow copies of all features and assigning threshold to each features. It selects the maximum threshold from the shadow features and applies it to the original features. Features that surpass this threshold are chosen. The algorithm iterates until all features are either confirmed or rejected as features of importance. Recursive Feature Elimination (RFE) in other hand begins with building a model on all predictors and calculating an importance score for each. The least important predictors are then removed, the model is rebuilt, and the importance scores are recalculated.
We explored the performance of each feature selection and classification performance method by comparing the AUC values obtained using each method combination. A complete diagram of the pipeline used is described in Fig. 1. Predictive approaches used included: Decision Tree, Random Forest, Logistic Regression, and Support Vector Machine (SVM). Boruta, caret, tidyverse, readxl, e1071, pROC, tree, randomForest R packages were used. Parameters used on each test are reported in Table 2.
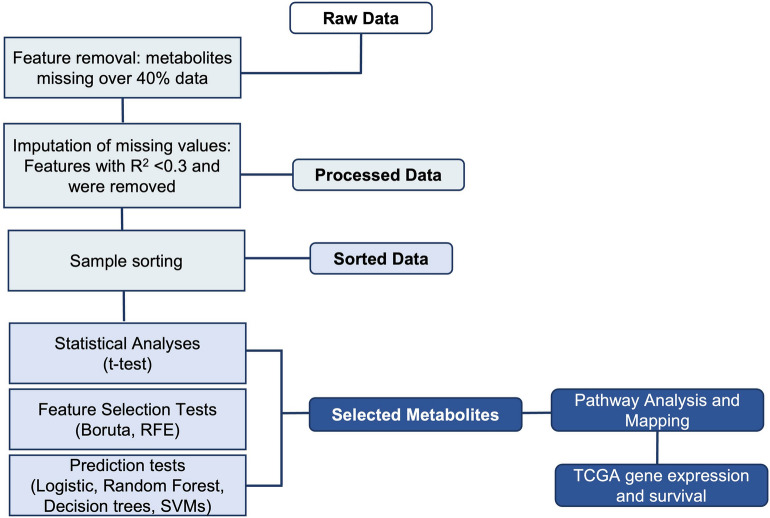
Figure 1.
The structure of the pipeline and different steps are shown. The method consist of data quality check data sorting by race, statistical analyses, feature selection and classification tests, pathway analysis and evaluation in gene expression and survival in TCGA cohort of selected genes associated to metabolites mapped.
Table 2.
Parameters used in each method.
| Parameters | |
|---|---|
| Feature selection algorithm | |
| Boruta | Default parameters |
| Recursive feature elimination | {functions: rfFuncs, method: repeatedcv, repeats: 5, verbose: FALSE} |
| Algorithm | |
| Random forests | {n_estimators: 100, criterion: gini, min_samples_split: 2, max_features: auto} |
| Decision trees | Default parameters, {criterion: gini, splitter: best, min_samples_split: 2} |
| Logistic regression | Default parameters, {family: binomial} |
| Support vector machine (SVM) | {kernel: linear, gamma: (depends on the data) (between 0.000001 and 0.1), cost: (depends on the data) (between 0.1 and 10)} |
Performance evaluation
Performance metric statistics were gathered, including Mathews Correlation Coefficient (MCC) and F1 score, as well as receiver operator characteristic and area under the curve (ROC, AUC) curves for each combination of feature selection and prediction method to visualize classification performance (true positive rate vs. false positive rate). We used the receiver operating characteristic curve (ROC) for each feature selection and prediction method as a quantitative performance metric. We further analyzed the performance of the combination methods by reporting F score statistics and MCC (Table 3). Once selected the best feature selection/prediction model combination, the top features of importance were identified. Their relative influence on each model (in %), in terms of Mean Decrease Gini were computed. These features were further explored for pathway analysis.
Table 3.
Performance metrics.
| Race | Feature selection model | Prediction method | TPR (sensitivities) | FPR | MCC | F1 |
|---|---|---|---|---|---|---|
| AA | Boruta | Decision tree | 0.7 | 0.166667 | 0.526789 | 0.682927 |
| AA | Boruta | Random forest | 0.909091 | 0.215686 | 0.559715 | 0.625 |
| AA | Boruta | Logistic | 0.733333 | 0.212766 | 0.471053 | 0.611111 |
| AA | RFE | Random forest | 0.833333 | 0.22 | 0.512001 | 0.606061 |
| AA | RFE | SVM | 0.8 | 0.25 | 0.427428 | 0.516129 |
| AA | RFE | Decision tree | 0.368421 | 0.176471 | 0.213946 | 0.4375 |
| AA | Boruta | SVM | 0.857143 | 0.272727 | 0.390796 | 0.428571 |
| AA | RFE | Logistic | 0.75 | 0.277778 | 0.334493 | 0.413793 |
| NHW | Boruta | Random forest | 0.8 | 0.285714 | 0.515515 | 0.761905 |
| NHW | RFE | Logistic | 0.736842 | 0.285714 | 0.450564 | 0.717949 |
| NHW | Boruta | Logistic | 0.647059 | 0.277778 | 0.370494 | 0.666667 |
| NHW | Boruta | Decision tree | 0.631579 | 0.4375 | 0.194079 | 0.631579 |
| NHW | RFE | Decision tree | 0.631579 | 0.4375 | 0.194079 | 0.631579 |
| NHW | RFE | Random forest | 0.833333 | 0.22 | 0.512001 | 0.606061 |
| NHW | RFE | SVM | 0.714286 | 0.428571 | 0.280976 | 0.606061 |
| NHW | Boruta | SVM | 0.611111 | 0.470588 | 0.140984 | 0.594595 |
Comparison of the performance of various combinations of feature selection and classification methods.
Pathway analysis
To identify the most relevant pathways involved in each set (AA and NHW) a pathway analysis was performed using Metaboanalyst 4.0 Pathway Analysis Module. This module combines pathway enrichment analysis with pathway topology analysis. It uses high-quality KEGG metabolic pathways as the backend knowledge base and integrates many well-established (i.e., univariate analysis, over-representation analysis) methods, as well as novel algorithms and concepts (i.e., Global Test, GlobalAncova, network topology analysis) into pathway analysis. This analysis was limited to all metabolic features selected with the combination of Boruta test feature selection and Random Forest classification and that were consistent in the rest of the feature selection methods (Table 4A,B).
Table 4.
(A) Feature importance ranking with mean decrease Gini index calculated by the random forest (RF) algorithm for AA and (B) NHW.
| Feature selection method | Features selected | Classification method | Classification accuracy (%) |
|---|---|---|---|
| (A) | |||
| Boruta test—AA | 2-Oxoisocaproic acid, arginine, a-tocopherol, citric acid, histidine, maltose, methionine, n-acetylglutamic acid, o-phosphoethanolamine, oxalic acid | Random forest | 80.65 |
| Boruta test—AA | 2-Oxoisocaproic acid, arginine, a-tocopherol, citric acid, histidine, maltose, methionine, n-acetylglutamic acid, o-phosphoethanolamine, oxalic acid | Decision trees | 79.03 |
| Boruta test—AA | 2-Oxoisocaproic acid, arginine, a-tocopherol, citric acid, histidine, maltose, methionine, n-acetylglutamic acid, o-phosphoethanolamine, oxalic acid | Logistic regression | 77.42 |
| Boruta test—AA | 2-Oxoisocaproic acid, arginine, a-tocopherol, citric acid, histidine, maltose, methionine, n-acetylglutamic acid, o-phosphoethanolamine, oxalic acid | SVM | 74.19 |
| Recursive feature elimination—AA | Arginine, maltose, methionine, n-acetylglutamic acid, o-phosphoethanolamine | Random forest | 79.03 |
| Recursive feature elimination—AA | All features gave more accuracy than selecting some features | Decision trees | 66.04 |
| recursive feature elimination—AA | Arginine, maltose, methionine, n-acetylglutamic acid, o-Phosphoethanolamine | Logistic regression | 74.19 |
| Recursive feature elimination—AA | 2-Oxoisocaproic acid, arginine, histidine, maltose, methionine, n-acetylglutamic acid, o-phosphoethanolamine, oxalic acid | SVM | 75.81 |
| (B) | |||
| Boruta test—NHW | 3-Hydroxybutanoic acid, 9,12-octadecadienoic acid, 9-hexadecenoic acid, a-ketoglutaric acid, b-alanine, cholesterol, citric acid, lactamide, oxalic acid, palmitic acid, p-cresol, pyruvic acid, tetradecanoic acid | Logistic regression | 79.03 |
| Boruta test—NHW | 3-Hydroxybutanoic acid, 9-hexadecenoic acid, oxalic acid, palmitic acid, tetradecanoic acid | Random forest | 75.61 |
| Boruta test—NHW | 3-Hydroxybutanoic acid, 9-hexadecenoic acid, cholesterol, oxalic acid, palmitic acid, tetradecanoic acid | Decision trees | 60 |
| Boruta test—NHW | 3-Hydroxybutanoic acid, 9-hexadecenoic acid, cholesterol, oxalic acid, palmitic acid, tetradecanoic acid | SVM | 61.73 |
| Recursive feature elimination—NHW | 3-Hydroxybutanoic acid, 9-hexadecenoic acid, oxalic acid, palmitic acid, tetradecanoic acid | Logistic regression | 72.50 |
| Recursive feature elimination—NHW | 3-Hydroxybutanoic acid, 9-hexadecenoic acid, oxalic acid, palmitic acid, tetradecanoic acid | Random forest | 75.61 |
| Recursive feature elimination—NHW | 3-Hydroxybutanoic acid, 9-hexadecenoic acid, oxalic acid, palmitic acid, tetradecanoic acid | Decision trees | 60 |
| Recursive feature elimination—NHW | 3-Hydroxybutanoic acid, 9-hexadecenoic acid, oxalic acid, palmitic acid, tetradecanoic acid | SVM | 62.86 |
The top features selected are shown.
TCGA cohort mRNA and survival analyses
To gain molecular insights on the pathways altered in AA vs NHW individuals, the different abundances in AA vs NHW patients with breast cancer were mapped manually. Unpaired t-test was performed with Welch’s correction. To test the association of these metabolic genes with survival in AA breast cancer, we performed an analysis of the PanCancer atlas TCGA Cohort. The cohort was selected based on a cohort that contained information regarding self-reported race, and subtype. Cohort included AA cases (n = 82) and NHW cases (510) of ER+ breast cancer, that contained mRNA, protein expression and clinical outcomes.
Ethical approval
UIC IRB Protocl #2017-1029.
Results
Race-specific plasma metabolic signatures
Study population included 102 patients with breast cancer (AA, n = 48; NHW, n = 54) and 148 healthy women as controls (AA, n = 100; NHW, n = 48). Demographic and clinical characteristics are shown in Table 1. Following data processing with minimum observed values for each metabolite, 83 metabolites were included in the analysis. For statistical purposes, we report statistical differences in p value < 0.05 obtained by t-tests. (Table 5).
Table 5.
Significant features in AA and NHW according to statistical analysis: t-test.
| p value | |
|---|---|
| Significant features in AA | |
| Arginine | 0.0024 |
| Methionine | 0.005 |
| Mimosine | 0.012 |
| 2-Oxoisocaproic acid | 0.0132 |
| Citric acid | 0.0307 |
| Threonine | 0.0378 |
| Tyrosine | 0.0389 |
| Maltose | 0.0393 |
| Serine | 0.0512 |
| Proline | 0.077 |
| Tryptophan | 0.078 |
| Oxalic acid | 0.0821 |
| Significant features in NHW | |
| Palmitic acid | 0.0001 |
| 9-Hexadecenoic acid | 0.0001 |
| 3-Hydroxybutanoic acid | 0.0003 |
| Tetradecanoic acid | 0.0005 |
| 9,12-Octadecadienoic acid | 0.0012 |
| P-Cresol | 0.0013 |
| Citric acid | 0.0015 |
| Pyruvic acid | 0.003 |
| Tryptophan | 0.005 |
| Oxalic acid | 0.0068 |
| Nicotinic acid | 0.0177 |
| Cysteine | 0.0224 |
| Lactic acid | 0.0288 |
| Tyrosine | 0.0578 |
| Glyceric acid | 0.0799 |
| Methionine | 0.0874 |
| Cholesterol | 0.0962 |
Aminoacids are good predictors of ER+ breast cancer in African American patients, while fatty acids are good predictors in Non-Hispanic White patients
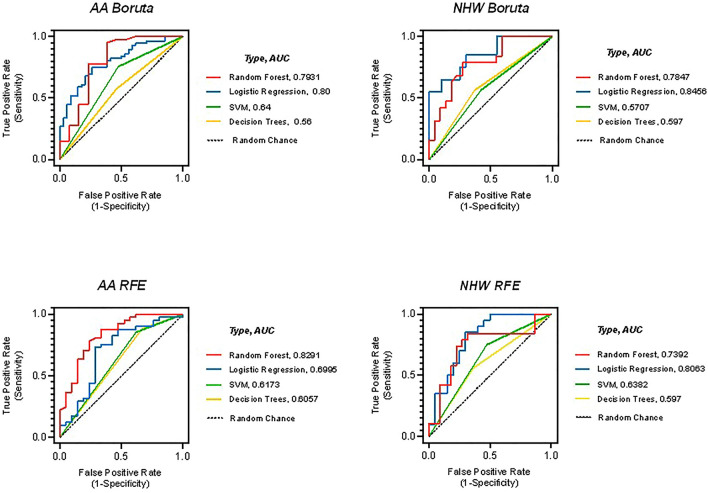
We compared feature selection and prediction combinations instead of relying on only one machine learning method as previously described39. Boruta/Random Forest combination was the most accurate, or best combination of feature selection/prediction model for AA (AUC = 0.79, MCC = 0.56) and NHW (AUC = 0.78, MCC = 0.52), according to the Matthews correlation coefficient (MCC) (Fig. 2).
Figure 2.
Feature selection and selection prediction method combinations identified top features that discriminate between cases and controls in AA and NHW.
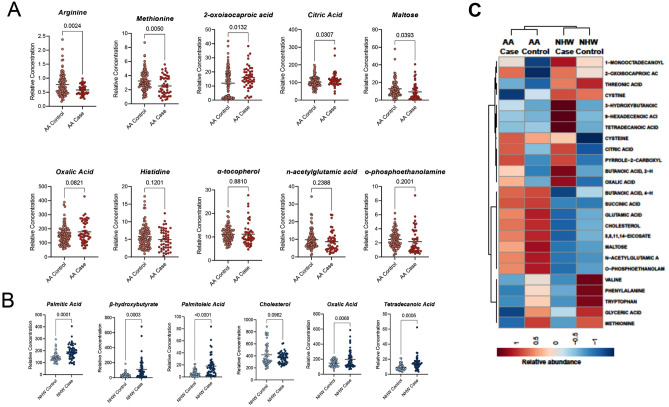
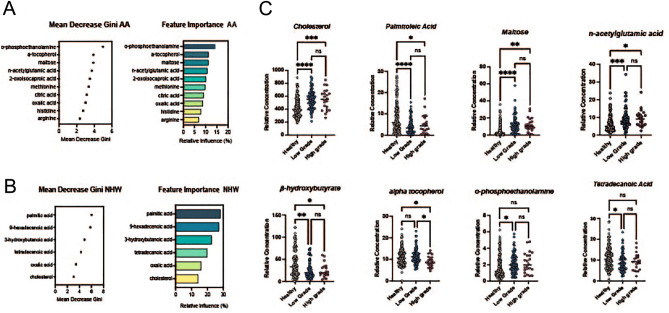
This optimal method identified 16 features that discriminate between cases and controls in both races. In AA individuals, a total of 10 features contributed to the signature to distinguish between case and controls and 9 of these features were exclusive for AA patients. These metabolites were: alpha ketoisocaproic acid, arginine, alpha tocopherol, citric acid, histidine, maltose, methionine, n-acetylglutamic acid, o-phosphoethanolamine, and oxalic acid. In AA individuals with breast cancer, we observed higher plasma levels of alpha ketoisocaproic acid a ketoacid, product of branched-chain aminoacid metabolism, alpha tocopherol oxalic acid, and citric acid, when compared to the healthy control group. In contrast, we observed lower arginine, histidine, maltose, methionine, n-acetyl glutamic acid, and o-phosphoethanolamine levels (Fig. 3A). In NHW individuals, a total of six features were identified as part of a signature that discriminates between case and controls: β-hydroxybutyrate, cholesterol, oxalic acid, palmitic acid, palmitoleic acid, and tetra decanoic acid. In NHW, we observed higher levels of all six metabolites in cases when compared with race-matched controls, and five of these features were exclusive for NHW patients (Fig. 3B,C). Overall, each of these differentially abundant features contributed to the metabolic signature significantly (Fig. 4A,B).
Figure 3.
(A) Boxplot of the relative concentration of 10 differential metabolites in AA individuals divided by cancer cases and healthy controls. (B) Boxplot of the relative concentration of six differential metabolites in NHW individuals divided by cancer cases and healthy controls. The relative contribution of each feature towards the accuracy of predicting cancer cases is reported as mean Gini (A,B). The percentage of importance or relative influence that each feature contributes to the model are shown in the figure. (C) Heatmap showing metabolite levels in AA cases vs controls and NHW cases vs controls.
Figure 4.
(A) Feature importance ranking with Mean Decrease Gini index calculated by the Random Forest (RF) algorithm for AA. The top features selected are shown. (B) Feature importance ranking with Mean Decrease Gini index calculated by the Random Forest (RF) algorithm for NHW The top features selected are shown. (C) Boxplot of relative concentration of 8 differential metabolites in cohort divided by healthy controls, low tumor grade and high tumor grade.
In order to evaluate the association of differential metabolite levels and tumor grade, a total of 148 breast cancer samples (with known tumor grade 1, 2, 3), were compared with 102 healthy controls. We grouped grades I and II breast cancer patients as “low grade” and grade III as “high grade” similar to previous studies40–42. We evaluated differential levels of all metabolites (n = 15) that were good predictors of breast cancer in Fig. 4A,B. Eight metabolites were found to be significantly changed among healthy controls and two grades of breast cancer (Fig. 4C).
Key metabolic pathways activated in ER+ breast cancer patients differ by race
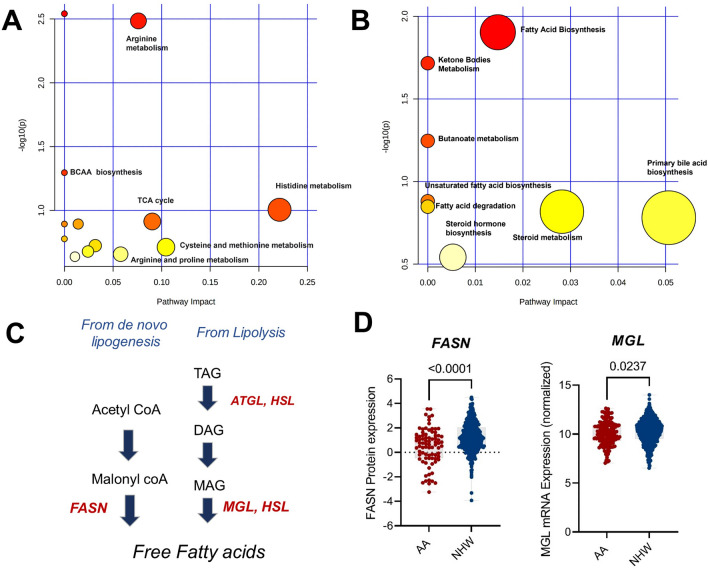
Pathway-based analysis was performed to explore metabolic pathway differences according to race in breast cancer cases. For AA individuals, metabolites that are associated with aminoacyl-tRNA biosynthesis, arginine metabolism (p < 0.05), branched amino acid metabolism (p = 0.05), and histidine metabolism (p = 0.09) were differentially abundant in plasma from individuals with breast cancer (Fig. 5A). Other pathways including, TCA cycle, beta-alanine metabolism, sphingolipid metabolism, alanine, aspartate, and glutamate metabolism glyoxylate and dicarboxylate metabolism, cysteine and methionine metabolism, glycerophospholipid metabolism, arginine and proline metabolism, were not statistically significant, but were identified in the global pathway analysis.
Figure 5.
(A) Bubble plot showing metabolic pathways related to important features in African American individuals. The node color is based on its p value and the node radius is determined based on their pathway impact values. Pathway analysis performed with MetaboAnalyst 5.0. (B) Bubble plot showing Metabolic pathways related to important features in Non-Hispanic white individuals. The node color is based on its p value and the node radius is determined based on their pathway impact values. Pathway analysis performed with MetaboAnalyst 5.0. (C) Schematic of hypothesis in NHW patients, (D) protein levels of fatty acid synthase (FASN) and monoacylglycerol lipase (MGL) in NHW and AA patients with ER+ breast cancer.
In NHW individuals, three significant pathways were identified including fatty acid metabolism (p < 0.05), ketone body metabolism (p < 0.05), and butanoate metabolism (p = 0.05). Pathways like unsaturated fatty acid synthesis, fatty acid elongation and degradation, steroid metabolism, primary bile acid synthesis and steroid hormone synthesis were not statistically significant but were identified in the global pathway analysis (Fig. 5B). One potential explanation for this is that there is a higher production of free fatty acids coming from de novo lipogenesis or from triacylglycerol lipolysis. Therefore, we analyzed mRNA levels of key enzymes responsible for the generation of free fatty acids in tumors from AA vs NHW patients with ER+ breast cancer in TCGA. Expression of fatty acid synthase and Monoglycerol lipase was higher in tumors from NHW patients compared with AA patients (Fig. 5C,D).
Metabolic pathway analysis suggests a role for epigenetic regulation differences that result in worse survival in African-American breast cancer patients
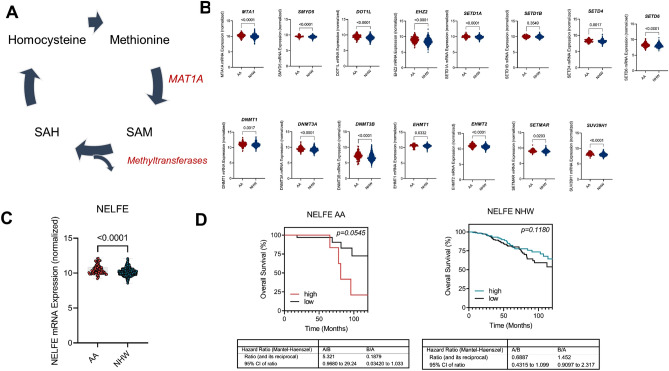
In AA women with breast cancer, we observed lower methionine (Met) levels, which led us to hypothesize that there is a trend of higher demand for Met consumption and metabolism. One potential explanation for the utilization of Met is that it is being used as substrate for methylation processes. Therefore, we analyzed samples from Pan Cancer Atlas cohort (TCGA) for mRNA levels of MAT1A (methionine adenosyl transferase 1A) and methyltransferases (DNA and Histone) in AA (n = 183) compared with NHW individuals (n = 757). MAT1A is responsible for catalyzing the reaction from methionine to SAM. SAM then is used as a substrate for methyltransferases. We hypothesized and found that AA individuals have higher mRNA levels of methyl transferases and MAT1A. We identified 200 genes with histone methylation activity, and 71 genes with DNA methylation activity that were differentially expressed between AA and NHW individuals. Out of the 271 genes studied, 49 had increased expression in AA individuals, 15 of these were statistically significant (Fig. 6A). To obtain additional insights, we further investigated the associations of these genes with survival in AA and NHW breast cancer. A Kaplan Meier survival analysis of NELFE showed that high expression was associated with poor survival in AA women, but it was not associated with survival among NHW patients (Fig. 6B–D) Together these findings suggest that this gene may play role in the biology of breast cancer in AA individuals.
Figure 6.
(A) Schematics of methionine metabolism to generate SAM. (B) Boxplot of mRNA levels of different DNA methyltransferases and histone methyl transferases. (C) Boxplots of mRNA levels of NELFE. (D) Kaplan Meier Curve sorted by low and high levels of NELFE in AA and NHW individuals.
Discussion
In this study, we established blood metabolite profiles of 102 patients with breast cancer and 148 healthy controls and we observed that metabolic profiles differed by race. We found that levels of amino acids were significantly lower in AA patients with breast cancer than healthy controls. One possible explanation might be the high demand for amino acids in breast tumor metabolism. Tumor cells require amino acids as alternative fuels, and as precursors of biosynthetic materials, including those for DNA synthesis, for building new blood vessels and supporting their rapid growth and proliferation. The alterations in these metabolic pathways cause differential activation of downstream signaling that further translate to altered oncogenic regulation. Breast cancer is associated with marked metabolic shifts. However, these shifts in metabolism within tumors can vary between stages, subtypes, and race25,43,44 Most differences involve variation in amino acid, fatty acids and TCA intermediates. Previous studies have shown that pathways associated with energy metabolism: glycolysis, amino acid metabolism and TCA cycle, are dominant in AA women with ER+ tumors, which potentially indicates the aggressiveness of their tumors44,45.
Additionally, changes in metabolite levels affect gene and protein expression by altering substrate availability of epigenetic modifiers that mark to the DNA, RNA, and histones, ultimately dictating cancer progression and outcome46–48. Previous studies19–21, 49–51 and studies from our lab52–55 showed how systemic or cellular metabolic changes impact epigenetic landscape, which is important for ERα activity and response to clinical drugs. Amino acids provide metabolic intermediates for epigenetic regulation. For example, carbon units from methionine cycle serve as a methyl donor for DNA and histone methyltransferases (DNMTs and HMTs). HMTs catalyze the transfer of methyl groups to lysine and arginine residues in histone proteins. Depending on the type of residue being modified, histone methylation regulates activation or repression of gene expression. In AA women, poverty levels correlate with hypermethylation of cancer-associated pathways including glucocorticoid receptor, p53, estrogen dependent breast cancer signaling, and cell proliferation56. Therefore, hypermethylation is a possible biological mechanism that can explain worse outcomes in AA women with breast cancer conferred by living in low socioeconomical status (SES) neighborhoods. We found that the methionine levels are lower in plasma samples from AA women with breast cancer. Methionine is a methyl group donor for methylation and represents the major contributor for epigenetic regulation. Both DNMTs and HMTs use S-adenosylmethionine (SAM) as a methyl donor, which is generated in the methionine cycle by the action of methionine adenolsyltransferase (MAT). Increased uptake of methionine leads to an excess of SAM, resulting in hypermethylation and aberrant histone methylation.
Aberrant hypermethylation in DNA can occur in promoter and enhancer regions of cancer related genes, such as tumor suppressor genes, resulting in loss of expression57. These abnormalities are involved in oncogenesis and tumor progression. Many cancer suppressor genes are silenced by DNA methylation in tissues58. A study in 2019 reported that residing in residential areas with high poverty may impact DNA methylation patterns. The authors reported that in AA women, poverty levels affected hypermethylation of important pathways including glucocorticoid receptor, p53, estrogen dependent breast cancer signaling, cell proliferation (BCL2, JUN, ESR1, ESR2, CYP19A1)56. Therefore, hypermethylation is a candidate biological mechanism that may contribute to worse outcomes in AA women with breast cancer because of a higher likelihood of residing in low SES neighborhoods.
Besides its function as methyl donor, SAM is involved in polyamine synthesis. Polyamines are essential for cell growth, and enzymes that catalyze this synthesis are often overexpressed in cancer. Studies have shown that polyamines can alter gene expression by modulating global chromatin structure and ultimately affecting cell proliferation59,60. Aminoacyl tRNAs (aa-tRNA) biosynthesis associated metabolites were enriched in AA individuals with breast cancer. The production of aminoacyl-tRNA is directly associated with the protein synthesis process. The main role of aa-tRNAs is to deliver the amino acid to the ribosome for incorporation into the polypeptide chain that is being produced during translation.
Moreover, we observed that the level of fatty acids in NHW patients with breast cancer was higher compared with healthy controls or AA individuals. These results might suggest higher rates of de novo lipogenesis or triacylglycerol lipolysis in the tumor. Highly proliferative cells use citrate produced in TCA cycle to generate fatty acids in the cytosol. The principal fatty acid synthesis enzyme, fatty acid synthase (FASN), is often upregulated in tumors61. Moreover, FASN has been detected in cell lysate and supernatant of breast cancer cells derived from NHW patients62. The regulation of de novo lipogenesis occurs mainly at the transcriptional level through the activation of sterol regulatory element binding proteins (SREBP). Its active N terminal fragment translocate to the nucleus and induces the transcription of gene that contain sterol regulatory elements (SRE) such as lipogenic enzymes (fatty acid synthase) FAS, ACLY (ATP citrate lyase) and ACC (acetyl coA carboxylase). In addition to its function in de novo lipogenesis, FASN is part of crosstalk between PI3K-AKT, MAPK-ERK2 pathways and their interaction with estradiol-ERα. The overexpression of lipogenic enzymes mentioned above, have been observed early in tumorigenesis. In ER+ cancer models, both genetic and pharmacological inhibition of FASN, hypersensitizes ERα to estrogen dependent transactivation, inducing estrogen receptor element (ERE) transcriptional activity and MAPK-ERK signaling63. Our results are consistent with previously published metabolomics studies that show fatty acids levels are higher in plasma from patients with breast cancer. In addition, our group previously reported that higher levels of free fatty acids in plasma were an indicator of high breast cancer risk, and result in mTOR and MAPK signaling and ERα recruitment to chromatin to increase transcriptional activity of factors that regulate cancer cell metabolism53. Consistently, other studies also reported a role for free fatty acids in other cancer types including lung64 childhood tumors65, and colon cancer66. Recent studies using biological deuterium fractionation and discrimination points out diet as the main source of increased fatty acid pool in plasma, which is delivered to cells via circulation. Thus, fatty acids act as the intermediate proton carrying carbon source for mitochondrial respiration. Further, generation of ketones using these deuterium-depleted fatty acids might explain benefit of ketogenic diets. Food insecurity and inequality in food quality and availability resulting in metabolic inefficiency might contribute to differential fatty acid profiles in AA vs. NHW women and breast cancer disparities.
One weakness of our study is the data imbalance. When data imbalance occurs, the number of samples in one class is larger than number of samples in the other class, and we can encounter an overoptimistic estimation of the classifier ability on the class with majority number of samples, such as our case (for AA individuals we have 100 healthy controls vs 48 cancer cases). To overcome this, we use the MCC score since produces a high score only if the prediction obtained good results in all the four confusion matrix categories (true positives, false negatives, true negatives, and false positives, Table 3). MCC score is a more reliable statistical rate and performance metric for binary classification when dealing with imbalanced data in binary data as it has been reported previously67,68.
In conclusion, our metabolic and bioinformatics analyses provided valuable information regarding metabolic alterations associated with cancer within each race, and provide insights on potential metabolic vulnerabilities that should be considered for depth studies of tumor biology that interrogate the role of candidate oncometabolites in breast cancer disparities, which may inform novel therapeutic development and biologically-informed epidemiologic studies to provide greater insight into biological mechanisms underlying racial disparities in breast cancer survival.
Acknowledgements
The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov.tcga. We kindly acknowledge the expertise of personnel in the UIUC Roy J. Carver Biotechnology Center. Special thanks to Dr. Alex Ulanov and Dr. Lucas Li for providing training in Metabolomics analyses. We thank Elonia Martin, Anita Fareeduddin, Archana Bargaje and Carlos Garcia for their assistance with sample collection at Mercy Hospital.
Abbreviations
- AA
African American
- NHW
Non-Hispanic White
- ER
Estrogen receptor
- GC–MS
Gas chromatography–mass spectrometry
- TCGA
The Cancer Genome Atlas
- MAT1A
Methionine Adenosyl transferase
- FASN
Fatty acid synthase
- MGL
Monoacyl glycerol lypase
- NELFE
Negative elongation factor
- SES
Socioeconomical status
- AJCC
American Joint Committee on Cancer
- FDR
False discovery rate
- AUC
Area under the curve
- MCC
Matthews correlation coefficient
- TCA
Tricarboxylic acid cycle
- DNMTS
DNA methyl transferases
- HMTS
Histone methyl transferases
- SAM
S-adenosyl methionine
- AATRNA
Aminoacyl-t-RNA
Author contributions
Z.M.E., J.F. and K.F.H. designed the study. A.S.C. and Z.M.E. wrote the main manuscript text, and A.S.C. prepared the figures. All authors reviewed the manuscript. O.C.D., H.P., L.B., A.Z., J.B., G.R., L.S., L.R.W., D.T., E.L.H., N.P., S.M.F., S.K., J.K., W.G. and S.H. contributed to sample procurement. A.S.C. performed the metabolomics analysis (sample preparation, running the GC/MS, peak identification and quantification, preliminary analysis). D.M. performed the machine learning analysis of the data.
Funding
This work was supported by Grants U54CA203000; U54CA2022995; U54CA2022997 from the National Institutes of Health/National Cancer Institute (to KFH, JF and ZME), the University of Illinois, Office of the Vice Chancellor for Research, Future Interdisciplinary Research Endeavors Grant from college of ACES, National Center for Supercomputing Application Faculty Fellows Program (to ZME), and National Institute of Food and Agriculture, U.S. Department of Agriculture, award ILLU-698-924 and ILLU-698-331 (to ZME). Research reported in this publication was supported by the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health under Award Number T32EB019944 (To ASC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data availability
The datasets generated and analyzed during the current study will be available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors jointly supervised this work: Jonna Frasor, Kent F. Hoskins and Zeynep Madak-Erdogan.
References
- 1.Rauscher GH, et al. Racial disparity in survival from estrogen and progesterone receptor-positive breast cancer: Implications for reducing breast cancer mortality disparities. Breast Cancer Res. Treat. 2017;163:321–330. doi: 10.1007/s10549-017-4166-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams DR, Mohammed SA, Shields AE. Understanding and effectively addressing breast cancer in African American women: Unpacking the social context. Cancer. 2016;122:2138–2149. doi: 10.1002/cncr.29935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guth AA, et al. The relationship of race, oncotype DX, and Ki67 in a population highly screened for breast cancer. Breast J. 2017;23:177–181. doi: 10.1111/tbj.12781. [DOI] [PubMed] [Google Scholar]
- 4.Iqbal J, Ginsburg O, Rochon PA, Sun P, Narod SA. Differences in breast cancer stage at diagnosis and cancer-specific survival by race and ethnicity in the United States. JAMA. 2015;313:165–173. doi: 10.1001/jama.2014.17322. [DOI] [PubMed] [Google Scholar]
- 5.Ooi SL, Martinez ME, Li CI. Disparities in breast cancer characteristics and outcomes by race/ethnicity. Breast Cancer Res. Treat. 2011;127:729–738. doi: 10.1007/s10549-010-1191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howlader N, et al. Overview of breast cancer collaborative stage data items–their definitions, quality, usage, and clinical implications: A review of SEER data for 2004–2010. Cancer. 2014;120(Suppl 23):3771–3780. doi: 10.1002/cncr.29059. [DOI] [PubMed] [Google Scholar]
- 7.Fedewa SA, Ward EM, Stewart AK, Edge SB. Delays in adjuvant chemotherapy treatment among patients with breast cancer are more likely in African American and Hispanic populations: A national cohort study 2004–2006. J. Clin. Oncol. 2010;28:4135–4141. doi: 10.1200/jco.2009.27.2427. [DOI] [PubMed] [Google Scholar]
- 8.Fedewa SA, et al. Race and ethnicity are associated with delays in breast cancer treatment (2003–2006) J. Health Care Poor Underserved. 2011;22:128–141. doi: 10.1353/hpu.2011.0006. [DOI] [PubMed] [Google Scholar]
- 9.Freedman RA, et al. The association of race/ethnicity, insurance status, and socioeconomic factors with breast cancer care. Cancer. 2011;117:180–189. doi: 10.1002/cncr.25542. [DOI] [PubMed] [Google Scholar]
- 10.Gullate M. The influence of spirituality and religiosity on breast cancer screening delay in African American women: Application of the Theory of Reasoned Action and Planned Behavior (TRA/TPB) Abnf. J. 2006;17:89–94. [PubMed] [Google Scholar]
- 11.Wang J, John EM, Horn-Ross PL, Ingles SA. Dietary fat, cooking fat, and breast cancer risk in a multiethnic population. Nutr. Cancer. 2008;60:492–504. doi: 10.1080/01635580801956485. [DOI] [PubMed] [Google Scholar]
- 12.Newman LA, et al. African-American ethnicity, socioeconomic status, and breast cancer survival: A meta-analysis of 14 studies involving over 10,000 African-American and 40,000 White American patients with carcinoma of the breast. Cancer. 2002;94:2844–2854. doi: 10.1002/cncr.10575. [DOI] [PubMed] [Google Scholar]
- 13.Newman LA, et al. Meta-analysis of survival in African American and white American patients with breast cancer: Ethnicity compared with socioeconomic status. J. Clin. Oncol. 2006;24:1342–1349. doi: 10.1200/jco.2005.03.3472. [DOI] [PubMed] [Google Scholar]
- 14.Wojcik BE, Spinks MK, Optenberg SA. Breast carcinoma survival analysis for African American and white women in an equal-access health care system. Cancer. 1998;82:1310–1318. doi: 10.1002/(sici)1097-0142(19980401)82:7<1310::aid-cncr14>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 15.Jatoi I, Becher H, Leake CR. Widening disparity in survival between white and African-American patients with breast carcinoma treated in the US Department of Defense Healthcare system. Cancer. 2003;98:894–899. doi: 10.1002/cncr.11604. [DOI] [PubMed] [Google Scholar]
- 16.Akinyemiju T, Moore JX, Ojesina AI, Waterbor JW, Altekruse SF. Racial disparities in individual breast cancer outcomes by hormone-receptor subtype, area-level socio-economic status and healthcare resources. Breast Cancer Res. Treat. 2016;157:575–586. doi: 10.1007/s10549-016-3840-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.John EM, et al. Racial/ethnic disparities in survival after breast cancer diagnosis by estrogen and progesterone receptor status: A pooled analysis. Cancer Epidemiol. Biomark. Prev. 2021;30:351–363. doi: 10.1158/1055-9965.epi-20-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YP, Lei QY. Metabolite sensing and signaling in cell metabolism. Signal Transduct. Target Ther. 2018;3:30. doi: 10.1038/s41392-018-0024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faubert B, Solmonson A, DeBerardinis RJ. Metabolic reprogramming and cancer progression. Science. 2020 doi: 10.1126/science.aaw5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pascual G, Domínguez D, Benitah SA. The contributions of cancer cell metabolism to metastasis. Dis. Model Mech. 2018 doi: 10.1242/dmm.032920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boukouris AE, Zervopoulos SD, Michelakis ED. Metabolic enzymes moonlighting in the nucleus: Metabolic regulation of gene transcription. Trends Biochem. Sci. 2016;41:712–730. doi: 10.1016/j.tibs.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Spratlin, J. L., Serkova Nj Fau-Eckhardt, S. G. & Eckhardt, S. G. Clinical applications of metabolomics in oncology: A review. [DOI] [PMC free article] [PubMed]
- 23.Moore SC, et al. A metabolomics analysis of body mass index and postmenopausal breast cancer risk. J. Natl. Cancer Inst. 2018;110:588–597. doi: 10.1093/jnci/djx244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Playdon MC, et al. Nutritional metabolomics and breast cancer risk in a prospective study. Am. J. Clin. Nutr. 2017;106:637–649. doi: 10.3945/ajcn.116.150912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen J, Yan L, Liu S, Ambrosone CB, Zhao H. Plasma metabolomic profiles in breast cancer patients and healthy controls: By race and tumor receptor subtypes. Transl. Oncol. 2013;6:757–765. doi: 10.1593/tlo.13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hart CD, et al. Serum metabolomic profiles identify ER-positive early breast cancer patients at increased risk of disease recurrence in a multicenter population. Clin. Cancer Res. 2017;23:1422–1431. doi: 10.1158/1078-0432.ccr-16-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenori L, et al. Serum metabolomic profiles evaluated after surgery may identify patients with oestrogen receptor negative early breast cancer at increased risk of disease recurrence. Results from a retrospective study. Mol. Oncol. 2015;9:128–139. doi: 10.1016/j.molonc.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oakman C, et al. Identification of a serum-detectable metabolomic fingerprint potentially correlated with the presence of micrometastatic disease in early breast cancer patients at varying risks of disease relapse by traditional prognostic methods. Ann. Oncol. 2011;22:1295–1301. doi: 10.1093/annonc/mdq606. [DOI] [PubMed] [Google Scholar]
- 29.Asiago VM, et al. Early detection of recurrent breast cancer using metabolite profiling. Cancer Res. 2010;70:8309–8318. doi: 10.1158/0008-5472.can-10-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang AH, Sun H, Qiu S, Wang XJ. Metabolomics in noninvasive breast cancer. Clin. Chim. Acta. 2013;424:3–7. doi: 10.1016/j.cca.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Hadi NI, et al. Serum metabolomic profiles for breast cancer diagnosis, grading and staging by gas chromatography–mass spectrometry. Sci. Rep. 2017;7:1715. doi: 10.1038/s41598-017-01924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra P, et al. ADHFE1 is a breast cancer oncogene and induces metabolic reprogramming. J. Clin. Invest. 2018;128:323–340. doi: 10.1172/jci93815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terunuma A, et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J. Clin. Invest. 2014;124:398–412. doi: 10.1172/jci71180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patel MJ, et al. Race and sex differences in small-molecule metabolites and metabolic hormones in overweight and obese adults. OMICS. 2013;17:627–635. doi: 10.1089/omi.2013.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walejko JM, et al. Gut microbiota and serum metabolite differences in African Americans and White Americans with high blood pressure. Int. J. Cardiol. 2018;271:336–339. doi: 10.1016/j.ijcard.2018.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu J, et al. Differences in metabolomic profiles between black and white women and risk of coronary heart disease: An observational study of women from four US cohorts. Circ. Res. 2022;131:601–615. doi: 10.1161/circresaha.121.320134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vantaku V, et al. Large-scale profiling of serum metabolites in African American and European American patients with bladder cancer reveals metabolic pathways associated with patient survival. Cancer. 2019;125:921–932. doi: 10.1002/cncr.31890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oktay K, et al. A computational statistics approach to evaluate blood biomarkers for breast cancer risk stratification. Horm. Cancer. 2020;11:17–33. doi: 10.1007/s12672-019-00372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith BP, et al. Identification of early liver toxicity gene biomarkers using comparative supervised machine learning. Sci. Rep. 2020;10:19128. doi: 10.1038/s41598-020-76129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bathen TF, et al. MR-determined metabolic phenotype of breast cancer in prediction of lymphatic spread, grade, and hormone status. Breast Cancer Res. Treat. 2007;104:181–189. doi: 10.1007/s10549-006-9400-z. [DOI] [PubMed] [Google Scholar]
- 41.Hilvo M, et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 2011;71:3236–3245. doi: 10.1158/0008-5472.can-10-3894. [DOI] [PubMed] [Google Scholar]
- 42.Lean C, et al. Determination of grade and receptor status from the primary breast lesion by magnetic resonance spectroscopy. Technol. Cancer Res. Treat. 2004;3:551–556. doi: 10.1177/153303460400300604. [DOI] [PubMed] [Google Scholar]
- 43.Kanaan YM, et al. Metabolic profile of triple-negative breast cancer in African-American women reveals potential biomarkers of aggressive disease. Cancer Genom. Proteom. 2014;11:279–294. [PubMed] [Google Scholar]
- 44.Tayyari F, et al. Metabolic profiles of triple-negative and luminal A breast cancer subtypes in African-American identify key metabolic differences. Oncotarget. 2018;9:11677–11690. doi: 10.18632/oncotarget.24433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H, et al. Breast cancer risk in relation to plasma metabolites among Hispanic and African American women. Breast Cancer Res. Treat. 2019;176:687–696. doi: 10.1007/s10549-019-05165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu W, et al. One-carbon metabolism supports S-adenosylmethionine and histone methylation to drive inflammatory macrophages. Mol. Cell. 2019;75:1147–1160.e1145. doi: 10.1016/j.molcel.2019.06.039. [DOI] [PubMed] [Google Scholar]
- 47.Sulkowski PL, et al. Oncometabolites suppress DNA repair by disrupting local chromatin signalling. Nature. 2020;582:586–591. doi: 10.1038/s41586-020-2363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huo M, Zhang J, Huang W, Wang Y. Interplay among metabolism, epigenetic modifications, and gene expression in cancer. Front. Cell Dev. Biol. 2021;9:793428. doi: 10.3389/fcell.2021.793428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garcia-Martinez L, Zhang Y, Nakata Y, Chan HL, Morey L. Epigenetic mechanisms in breast cancer therapy and resistance. Nat. Commun. 2021;12:1786. doi: 10.1038/s41467-021-22024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong CC, Qian Y, Yu J. Interplay between epigenetics and metabolism in oncogenesis: Mechanisms and therapeutic approaches. Oncogene. 2017;36:3359–3374. doi: 10.1038/onc.2016.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Achinger-Kawecka J, et al. Epigenetic reprogramming at estrogen-receptor binding sites alters 3D chromatin landscape in endocrine-resistant breast cancer. Nat. Commun. 2020;11:320. doi: 10.1038/s41467-019-14098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cotul EK, et al. Combined targeting of estrogen receptor alpha and Exportin 1 in metastatic breast cancers. Cancers (Basel) 2020 doi: 10.3390/cancers12092397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madak-Erdogan Z, et al. Free fatty acids rewire cancer metabolism in obesity-associated breast cancer via estrogen receptor and mTOR signaling. Cancer Res. 2019;79:2494–2510. doi: 10.1158/0008-5472.can-18-2849. [DOI] [PubMed] [Google Scholar]
- 54.Zuo Q, et al. Targeting metabolic adaptations in the breast cancer-liver metastatic niche using dietary approaches to improve endocrine therapy efficacy. Mol. Cancer Res. 2022;20:923–937. doi: 10.1158/1541-7786.MCR-21-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mogol AN, et al. NAD+ metabolism generates a metabolic vulnerability in endocrine-resistant metastatic breast tumors in females. Endocrinology. 2023 doi: 10.1210/endocr/bqad073. [DOI] [PubMed] [Google Scholar]
- 56.Hsu PC, et al. County poverty levels influence genome-wide DNA methylation profiles in African American and European American women. Transl. Cancer Res. 2019;8:683–692. doi: 10.21037/tcr.2019.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 58.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 59.Stjernborg L, Heby O, Mamont P, Persson L. Polyamine-mediated regulation of S-adenosylmethionine decarboxylase expression in mammalian cells. Studies using 5'-([(Z)-4-amino-2-butenyl]methylamino)-5'-deoxyadenosine, a suicide inhibitor of the enzyme. Eur. J. Biochem. 1993;214:671–676. doi: 10.1111/j.1432-1033.1993.tb17967.x. [DOI] [PubMed] [Google Scholar]
- 60.Hobbs CA, Paul BA, Gilmour SK. Deregulation of polyamine biosynthesis alters intrinsic histone acetyltransferase and deacetylase activities in murine skin and tumors. Cancer Res. 2002;62:67–74. [PubMed] [Google Scholar]
- 61.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, et al. Fatty acid synthase (FAS) expression in human breast cancer cell culture supernatants and in breast cancer patients. Cancer Lett. 2001;167:99–104. doi: 10.1016/s0304-3835(01)00464-5. [DOI] [PubMed] [Google Scholar]
- 63.Menendez JA, et al. Fatty acid synthase confers tamoxifen resistance to ER+/HER2+ breast cancer. Cancers (Basel) 2021 doi: 10.3390/cancers13051132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hao D, et al. Temporal characterization of serum metabolite signatures in lung cancer patients undergoing treatment. Metabolomics. 2016;12:58. doi: 10.1007/s11306-016-0961-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gros SJ, Holland-Cunz SG, Supuran CT, Braissant O. Personalized treatment response assessment for rare childhood tumors using microcalorimetry-exemplified by use of carbonic anhydrase IX and aquaporin 1 inhibitors. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20204984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arima K, et al. Metabolic profiling of formalin-fixed paraffin-embedded tissues discriminates normal colon from colorectal cancer. Mol. Cancer Res. 2020;18:883–890. doi: 10.1158/1541-7786.mcr-19-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chicco D, Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genom. 2020;21:6. doi: 10.1186/s12864-019-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boughorbel S, Jarray F, El-Anbari M. Optimal classifier for imbalanced data using Matthews correlation coefficient metric. PLoS One. 2017;12:e0177678. doi: 10.1371/journal.pone.0177678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the current study will be available from the corresponding author on reasonable request.