Abstract
Ornithine transcarbamylase (OTC) plays a significant role in the urea cycle, a metabolic pathway functioning in the liver to detoxify ammonia. OTC deficiency (OTCD) is the most prevalent urea cycle disorder. Here, we show that intravenously delivered human OTC (hOTC) mRNA by lipid nanoparticles (LNP) was an effective treatment for OTCD by restoring the urea cycle. We observed a homotrimer conformation of hOTC proteins produced by the mRNA-LNP in cells by cryo-electron microscopy. The immunohistochemistry revealed the mitochondria localization of produced hOTC proteins in hepatocytes in mice. In livers of mice intravenously injected with hOTC-mRNA/LNP at 1.0 mg/kg, the delivered hOTC mRNA levels steeply decreased with a half-life (t1/2) of 7.1 h, whereas the produced hOTC protein levels retained for 5 days and then declined with a t1/2 of 2.2 days. In OTCD model mice (high-protein diet-fed Otcspf-ash hemizygous males), a single dose of hOTC-mRNA/LNP at 3.0 mg/kg ameliorated hyperammonemia and weight loss with prolonged survival rate (22 days) compared with that of untreated mice (11 days). Weekly repeated doses at 0.3 and 1.0 mg/kg were well tolerated in wild-type mice and showed a dose-dependent amelioration of survival rate in OTCD mice, thus, showing the therapeutic potential of LNP-formulated hOTC mRNA for OTCD.
Keywords: MT: delivery strategies: ornithine transcarbamylase, messenger RNA therapy, lipid nanoparticles, homotrimer conformation, cryo-electron microscopy, mitochondria localization, Otcspf-ash mice
Graphical abstract
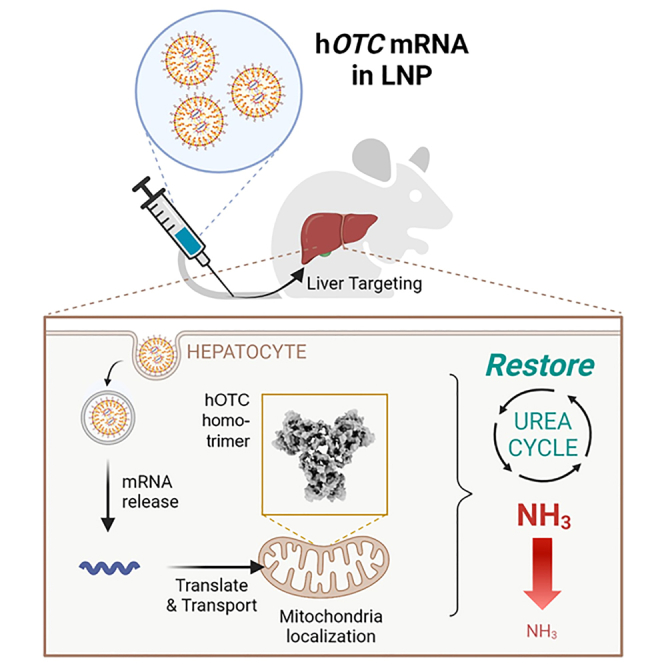
Yamazaki et al. used LNP to formulate hOTC-mRNA for delivery to hepatocytes to treat OTCD. hOTC proteins produced by hOTC-mRNA/LNP in cells shows the trimeric conformation and mitochondrial localization. Single and multiple doses of hOTC-mRNA/LNP demonstrated efficacy in an OTCD mouse model.
Introduction
The urea cycle is a pathway that converts toxic ammonia (NH3) into non-toxic urea, mainly in the liver. This pathway includes carbamoylphosphate synthetase 1 (CPS1), ornithine transcarbamylase (OTC), argininosuccinate synthetase, argininosuccinate lyase, and arginase 1 (ARG1).1 Urea cycle disorders (UCDs) are a group of diseases (e.g., N-acetylglutamate synthase deficiency, CPS1 deficiency, and argininemia) that have congenital abnormalities in the urea cycle and develop with symptoms such as hyperammonemia, neurological abnormalities, and coma, sometimes resulting in severe life-threatening conditions.1,2
OTC (EC 2.1.3.3) catalyzes carbamoyl phosphate and ornithine reaction to form citrulline and phosphate in the urea cycle (OMIM: 300461; https://omim.org/entry/300461). It is first synthesized as a precursor protein with a molecular mass of approximately 40,000 Da in the cytosol and then transported to the mitochondrial matrix, which is processed to the mature enzyme of approximately 36,000 Da. OTC becomes active after being formed into a homotrimer.3,4 OTC deficiency (OTCD) is the most common UCD (OMIM: 311250; https://omim.org/entry/311250).5,6 Although the prevalence of OTCD varies somewhat depending on literature, it has been reported as follows: 1/63,000,7 1/62,000–1/77,000,8 and 1/50,000–80,000 (Rare Disease Database: Ornithine Transcarbamylase Deficiency; https://rarediseases.org/rare-diseases/ornithine-transcarbamylase-deficiency/). At least 417 causative mutations have been reported for OTCD, including missense mutations, nonsense mutations, frameshifts, and splicing errors.4 OTCD patients generally have 5%–30% of normal OTC activity in the liver.9 As the hOTC locus is located within the band Xp11.4 of X chromosome (https://www.omim.org/entry/300461),10,11 OTCD shows a sex-linked inheritance: males are hemizygous and then develop the disease, while females are heterozygous carriers and asymptomatic in general, but 15%–20% exhibit symptoms during their lifetime (https://rarediseases.org/rare-diseases/ornithine-transcarbamylase-deficiency/),9 depending on the percentage of mosaicism of X chromosome inactivation in hepatocytes.12
Treatments are symptomatic, such as a low-protein diet, hemodiafiltration, and arginine and nitrogen scavengers (benzoate, phenylacetate, and phenylbutyrate).13 The mortality rate, however, remains high.7 Liver transplantation is performed in neonatal-onset and late-onset cases with repeated episodes of hyperammonemia.7 It has been reported that liver transplantation could correct hyperammonemia, but does not seem to significantly improve central nervous system injury predating transplantation.14 Therefore, OTC replacement by drugs is a direct and attractive therapy enabling early intervention. Gene therapy has been studied using adeno-associated virus (AAV) to produce OTC protein in the mitochondrial matrix in hepatocytes.15,16 However, issues include ineffectiveness because of pre-existing antibodies, generation of neutralizing antibodies against AAV,17,18 and long-term safety concerns caused by genome insertion.19
In contrast, mRNA-based drugs have the advantage of safety without the risk of genome insertion. Prieve et al.20 reported a hybrid mRNA technology (HMT) delivery system comprising an N-acetylgalactosamine (GalNAc)-targeted polymer micelle and cationic lipid-based nanoparticle that deliver mRNA to hepatocytes and induce endosomal escape. Their HMT system resulted in OTC synthesis in the mice’s liver and ameliorated OTCD model mice. However, in the case of the pharmacokinetics/pharmacodynamics (PK/PD) relationship by two independent entities in a single formulation, drug development of the HMT system would be extremely complicated.
In this study, we used our biodegradable ionizable-based lipid nanoparticles (LNP)21,22,23,24,25 to formulate engineered mRNA encoding hOTC in the LNP (hOTC-mRNA/LNP) for delivery to hepatocytes to treat OTCD. The three-dimensional structure of the hOTC protein produced in cells treated with hOTC-mRNA/LNP was determined, and the localization of the hOTC protein to mitochondria was demonstrated. In in vivo studies, we investigated changes in hOTC mRNA and hOTC protein levels in the livers of mice intravenously injected with hOTC-mRNA/LNP over time. Furthermore, we demonstrated the in vivo efficacy of hOTC-mRNA/LNP using OTCD model mice.
Results
Design of hOTC mRNA and LNP formulation
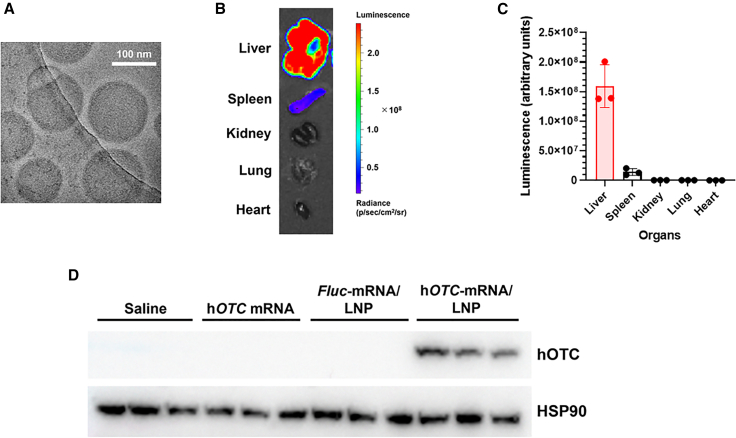
Our LNP formulation showed approximately 120 nm size with more than 90% mRNA encapsulation (Table S1). The representative micrographs acquired by cryo-transmission electron microscopy (cryo-EM) revealed that nanoparticles were homogeneous and had an electron-dense core (Figure 1A). First, we confirmed hOTC expression induction by codon-optimized hOTC mRNA in vitro. hOTC mRNA was transfected into Hep3B cells, and western blotting examined hOTC expression (Figure S1). Without the mRNA treatment, there was no band of hOTC, indicating that Hep3B cells have no endogenous hOTC expression. In the treatment, hOTC protein expression was induced, and its intensity was increased concentration dependently. Next, we evaluated the stability of mRNA/LNP in a medium with mouse plasma. No decomposition of mRNA was observed for 3 h (Figure S2), showing that LNP protects mRNA from RNase digestion in the presence of mouse plasma in vivo. To investigate the delivery of LNP to the liver, we injected LNP-formulated firefly luciferase (Fluc) mRNA (Fluc-mRNA/LNP) into mice intravenously and measured Fluc luminescence in several organs. The highest luminescence was observed in the liver, which was more than 10-fold higher than in the spleen. No luminescence was observed in the kidney, lung, or heart (Figures 1B and 1C).
Figure 1.
LNP characterization
(A) Representative cryo-electron microscopy image of LNP encapsulating hOTC mRNA. LNPs were homogeneous and had an electron-dense core. (B) Representative ex vivo luciferase imaging of organs of a mouse 24 h after intravenous injection of Fluc-mRNA/LNP at 1.0 mg/kg. (C) Quantified fluorescence of these organs. Values are expressed as means ± SD with individual values. n = 3. (D) Western blotting of hepatic hOTC proteins induced by hOTC-mRNA/LNP injection. Female BALB/c mice were intravenously injected with hOTC mRNA only, Fluc-mRNA/LNP, or hOTC-mRNA/LNP at 1.0 mg/kg, or saline. Liver specimens were sampled the next day and subjected to western blotting. Heat shock protein 90 (HSP90) proteins were used as an internal control. n = 3/group.
Finally, we tested hOTC-mRNA/LNPs. After intravenous injection of naked hOTC mRNA only, Fluc-mRNA/LNP, or hOTC-mRNA/LNP into mice, we performed western blotting for hepatic hOTC expression. hOTC-mRNA/LNP treatment induced hepatic hOTC protein expression, whereas there were no bands in the livers of mice treated with hOTC mRNA only or Fluc-mRNA/LNP (Figure 1D). hOTC-mRNA/LNP increased hepatic hOTC protein expression dose-dependently (Figures S3A and S3B).
Cryo-EM analysis of hOTC proteins generated in hOTC-mRNA/LNP-treated Hep3B cells
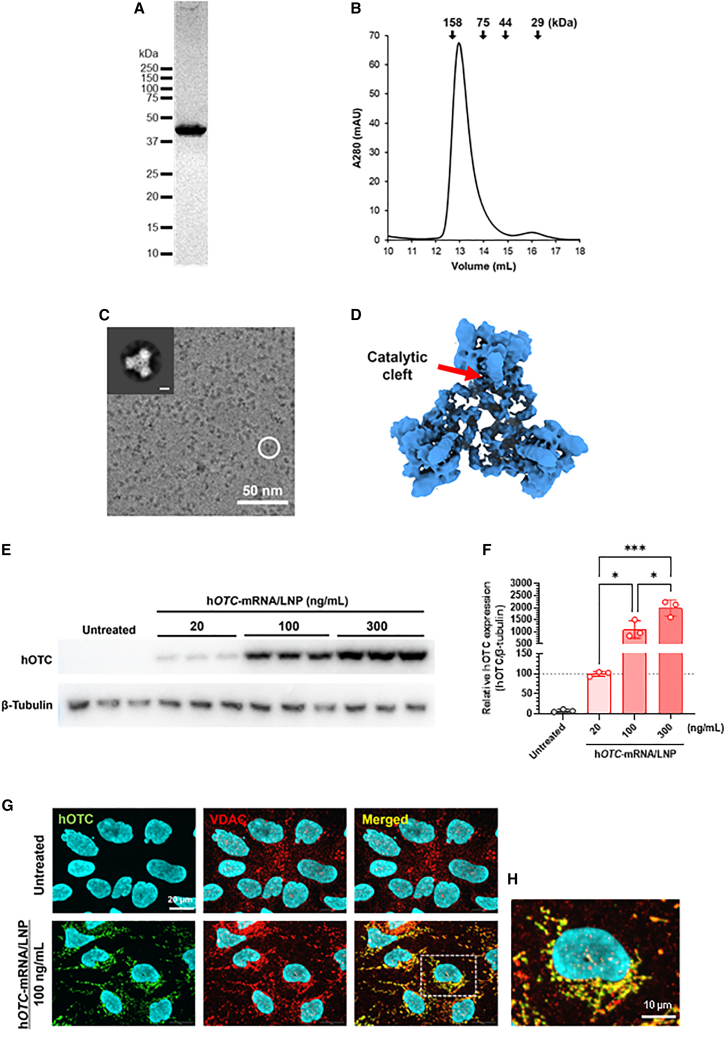
We purified the proteins and analyzed their structure to evaluate the conformational state of hOTC expressed in Hep3B cells using our hOTC-mRNA/LNP. The size-exclusion chromatogram of the FLAG-affinity purified hOTC proteins indicated their formation of the homotrimeric conformation (Figures 2A and 2B). Furthermore, the protein solution was flash frozen. The detailed molecular structure was explored by single-particle analysis using a 200-kV cryogenic microscope. Although the low resolution of the cryo-EM map hampered detailed analyses of the structures, it was revealed that hOTC-mRNA/LNP-derived hOTC proteins possessed the dish-like trimeric architecture harboring the catalytic clefts (Figures 2C and 2D), as previously reported crystal structures of heterologous expressed hOTC proteins,26,27,28 that is expected for the native OTC proteins.
Figure 2.
Function and structure of hOTC proteins expressed in Hep3B cells using hOTC-mRNA/LNP
(A) hOTC proteins were purified by M2 Anti-FLAG Agarose and size-exclusion chromatography and confirmed by SDS-PAGE. (B) The elution volume of the chromatogram indicates the proper trimeric folding of the purified hOTC. The arrows above the chromatogram are referred to the elution volumes of standard proteins. (C) A representative micrograph of purified hOTC proteins where the circle indicates a particle with the typical trimeric feature. The inset shows a 2D class average image of the hOTC particles (scale bar, 30 Å). (D) The overall cryo-EM structure of hOTC proteins. A catalytic cleft is indicated. (E) Western blotting analysis of hOTC proteins in Hep3B cells treated with hOTC-mRNA/LNP at 20, 100, or 300 ng/mL β-Tubulin proteins were used as an internal control. (F) Quantified hOTC protein expression (E) in Hep3B cells treated with hOTC-mRNA/LNP at 20, 100, or 300 ng/mL. β-Tubulin protein levels were used as an internal control. Data are relative to those at 20 ng/mL (%). Values are expressed as means ± SD with individual values (n = 3). The dotted line shows 100%. ∗p < 0.05 and ∗∗∗p < 0.001 by Tukey’s multiple comparisons tests. (G) Localization of hOTC proteins to mitochondria in Hep3B cells treated with hOTC-mRNA/LNP at 100 ng/mL. Voltage-dependent anion channel (VDAC) is an outer mitochondrial membrane marker. Nuclei were stained with Hoechst 33342. Scale bars, 20 μm. (H) Magnification of the square part in the merged image of cells treated with hOTC-mRNA/LNP. Scale bar, 10 μm.
In vitro expression of hOTC proteins by hOTC-mRNA/LNP
We examined whether hOTC-mRNA/LNP triggered hOTC protein expression in Hep3B cells and treated Hep3B cells with hOTC-mRNA/LNP at 20, 100, or 300 ng/mL for 24 h. The results showed that the mRNA/LNP treatment induced hOTC protein expression in a concentration-dependent manner (Figures 2E and 2F). Immunofluorescence analysis evidenced the mitochondrial localization of hOTC proteins (Figures 2G and 2H). hOTC immunofluorescence intensity was increased in a concentration-dependent manner (Figure S4).
Hepatic OTC activity in wild-type mice treated with hOTC-mRNA/LNP
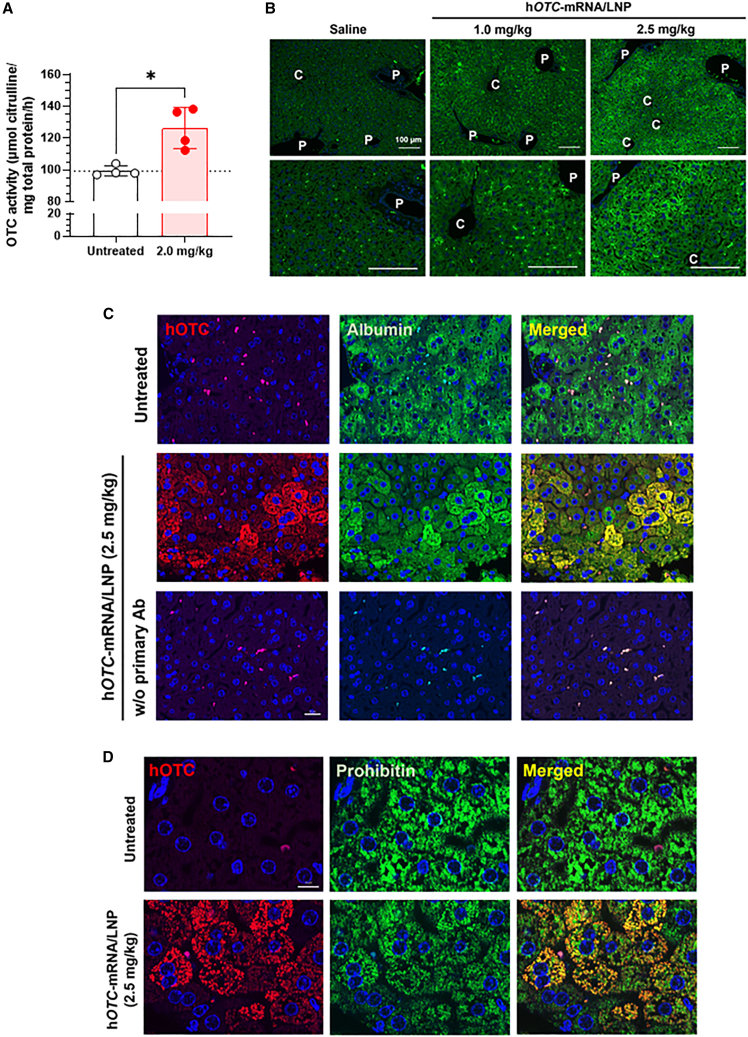
We compared hOTC protein expression and OTC enzymic activity in the liver of BALB/c mice 72 h after intravenous injection of hOTC-mRNA/LNP at 2.0 mg/kg. Western blotting analyzed the hepatic hOTC expression of untreated and treated mice (Figure S5A and S5B). Hepatic OTC activities are shown in Figure 3A. The hepatic OTC activity of hOTC-mRNA/LNP-treated mice was 127% relative to that of the untreated mice, showing that approximately 30% of the OTC activity was derived from exogenous hOTC mRNA. Figure S5C depicts the correlation between hepatic hOTC expression levels and hepatic OTC activity, indicating a positive correlation (r2 = 0.7931).
Figure 3.
In vivo hepatic expression of hOTC proteins in mice treated with hOTC-mRNA/LNP
(A) Hepatic OTC enzymic activity in female BALB/c mice intravenously injected with hOTC-mRNA/LNP at 2.0 mg/kg. Liver samples were collected three days after the injection. Values are expressed as means ± SD with individual values. ∗p < 0.05 by Mann-Whitney U test. The dotted line shows the mean of the untreated group. Untreated, n = 4; treated, n = 4. (B) Immunohistochemical distribution of hOTC proteins in the liver of BALB/c mice intravenously injected with hOTC-mRNA/LNP at 1.0 or 2.5 mg/kg, or saline. Livers were harvested 24 h after the injection. hOTC was stained using anti-hOTC mouse monoclonal antibody with nuclear staining with DAPI. C, central vein; P, portal vein. Scale bar, 100 μm. (C) Immunohistochemical staining of hOTC and albumin for liver specimens of hOTC-mRNA/LNP-treated (2.5 mg/kg) and untreated BALB/c mice. The bottom images are hOTC-mRNA/LNP-administered mouse liver samples processed without (w/o) the primary antibodies (Ab). Scale bar, 20 μm. (D) Immunohistochemical staining of hOTC proteins and prohibitin, a mitochondria maker, in the liver specimens of BALB/c mice intravenously injected with hOTC-mRNA/LNP at 2.5 mg/kg or untreated mice. Scale bar, 10 μm. Hepatic specimens were collected 24 h after the injection (C and D).
Immunohistochemical staining of hOTC proteins in livers of wild-type mice treated with hOTC-mRNA/LNP
Liver sections of mice were stained with hOTC-specific antibodies (Figure 3B). The samples were from mice subjected to intravenous injection of hOTC-mRNA/LNP (1.0 or 2.5 mg/kg) or saline. The overall degree of hOTC staining was increased in a dose-dependent manner. Interestingly, hOTC proteins were not uniformly stained in the liver, which was particularly noticeable at 2.5 mg/kg: high hOTC immunoreactivity was observed in periportal hepatocytes, whereas hepatocytes around central veins showed lower immunoreactivity. hOTC expression in hepatocytes was confirmed using immunohistochemistry (IHC) with albumin as a hepatocyte marker. Figure 3C shows that hOTC immunoreactivity is distributed in hepatocytes, which are albumin-positive cells. Moreover, we observed the localization of hOTC immunoreactivity to a mitochondria marker, prohibitin, and staining in the liver sections from mice treated with hOTC-mRNA/LNP (Figure 3D).
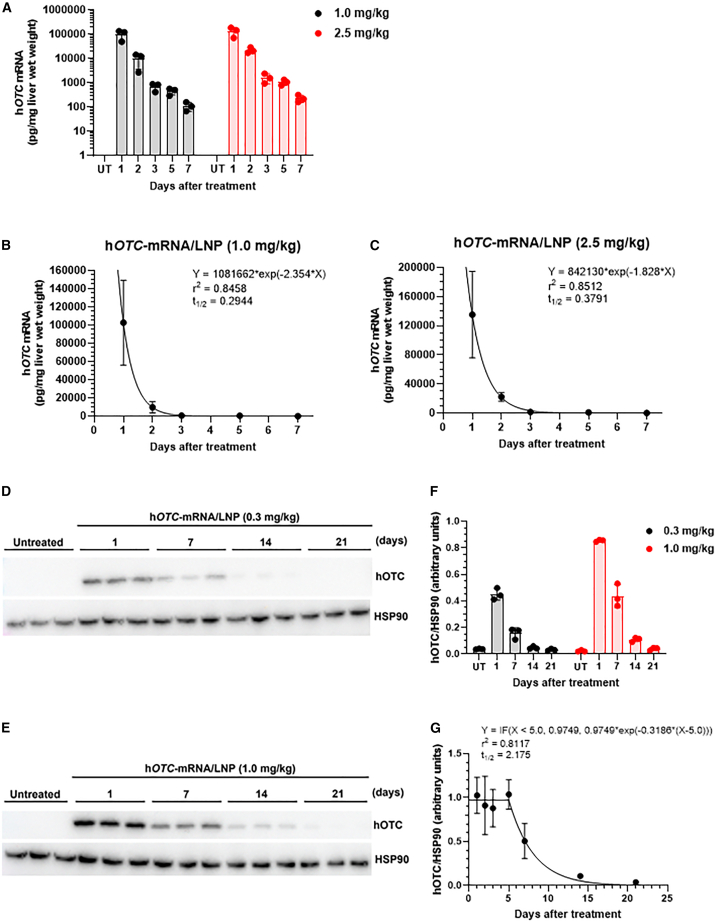
Time course changes of hepatic hOTC mRNA levels and hOTC protein expression in wild-type mice treated with hOTC-mRNA/LNP
First, we investigated chronological changes of hOTC mRNA delivered to the liver by LNP formulation on days 1–7. hOTC-mRNA/LNP at 1.0 or 2.5 mg/kg was intravenously injected into wild-type mice, and hepatic hOTC mRNA was determined up to 7 days after the administration. In both cases of doses, hOTC mRNA diminished sharply and exponentially (Figure 4A). These changes in the mRNA levels were fitted to one exponential phase decay to calculate the half-life (t1/2), which were 0.2944 days (7.066 h) and 0.3791 days (9.098 h) at 1.0 and 2.5 mg/kg, respectively (Figures 4B and 4C, respectively). Next, time course changes of hOTC protein expression were examined by western blotting analysis. In a short-term study, liver samples were harvested on days 1–7 after dosing of hOTC-mRNA/LNP at 1.0 or 2.5 mg/kg. In the case of the long-term study, they were collected on days 1–21 after dosing at 0.3 or 1.0 mg/kg. Western blots in the short-term study are shown in Figures S6A and S6C at 1.0 and 2.5 mg/kg, respectively. Their quantified data are indicated in Figures S6B and S6D, respectively. In the case of 1.0 mg/kg, hOTC protein levels were comparatively maintained on days 1–5, and the level significantly decreased on day 7 compared with that on day 1 (p < 0.05) (Figure S6C). Regarding 2.5 mg/kg, hOTC protein expression was kept weekly (Figure S6D). Western blotting of the long-term study is shown in Figures 4D and 4E at 0.3 and 1.0 mg/kg, respectively. Figure 4F summarizes quantified values in Figures 4D and 4E. Based on Figures 4F and S6B data at 1.0 mg/kg, we fitted changes of hOTC protein levels to plateau followed by a one-phase exponential decay (Figure 4G). hOTC expression was maintained for at least 5 days and subsequently decreased with a t1/2 of 2.175 days.
Figure 4.
PK/PD of hOTC-mRNA/LNP in wild-type mice
hOTC mRNA levels in the liver of female BALB/c mice intravenously injected with hOTC-mRNA/LNP at 1.0 or 2.5 mg/kg. Liver specimens were harvested 1, 2, 3, 5, and 7 days after the injection. Untreated (UT) mice were used as controls. (A) Time course changes of hOTC mRNA levels in the liver specimens. Values are expressed as means ± SD with individual values. n = 3 per each time. Fitting to one phase exponential equation in the case of 1.0 mg/kg (B) and 2.5 mg/kg (C). The equation is Y = (Y0 − Plateau)∗exp(−K∗X) + Plateau, where Y0 is the Y value when X is 0, Plateau is the Y value at infinite times, K is the rate constant, and the t1/2 is computed as ln(2)/K. Values are expressed as means ± SD. Time course changes of hOTC protein levels in the liver of mice intravenously injected with hOTC-mRNA/LNP at 0.3 or 1.0 mg/kg. Liver samples were collected 1, 7, 14, and 21 days after the treatment. n = 3 per time. Western blot of hepatic hOTC protein expression of mice treated with hOTC-mRNA/LNP at 0.3 mg/kg (D) and 1.0 mg/kg (E). HSP90 proteins were used as an internal control. (F) Quantified hOTC protein levels revealed by western blot in (D) and (E). Values are expressed as means ± SD with individual values. (G) Fitting to plateau followed by one-phase exponential decay, combined with data of (F) and Figure S6B at 1.0 mg/kg. The equation is Y = IF(X < X0, Y0, Plateau + (Y0 − Plateau)∗exp(−K∗(X-X0))), where X0 is the time at which the decay begins, Y0 is the average Y value up to time X0, Plateau is the Y value at infinite times, and K is the rate constant). In this case, Plateau was constrained to a constant value of zero. The t1/2 is computed as ln(2)/K. Values are expressed as means ± SD. n = 3 or 6 per time.
Effects of high-protein diet feeding on clinical signs and blood NH3 levels in wild-type and Otcspf-ash mice
Before starting in vivo efficacy evaluation of hOTC-mRNA/LNP using high-protein diet-fed Otcspf-ash mice as an OTCD model, we obtained primary data (changes of body weight and blood NH3 and survival) when feeding the mutant and wild-type mice a 40% high-protein diet prepared in this study. The high-protein diet did not affect body weight or blood NH3 concentrations in wild-type B6 mice during 7-day feeding (Figures S7A–S7C). However, body weight was lost immediately in Otcspf-ash mice (Figure S8A): the significant weight loss was already observed starting from day 1 when the mice were fed with a high-protein diet (p < 0.001), a large difference was observed from values fed on the normal diet (Figure S8B). From day 3, mutants began to die or were euthanized because of their moribund conditions, and only one mouse remained on day 5 (Figure S8C). Blood NH3 concentrations of high-protein diet-fed Otcspf-ash mice significantly increased on day 3 (Figure S8D). Figure S8E shows the correlation between blood NH3 concentrations and body weight changes on day 3 from day 0. A strong negative correlation was observed (r2 = 0.7616). This suggests that improving hyperammonemia leads to the ameliorating aggravation of general signs represented by weight loss.
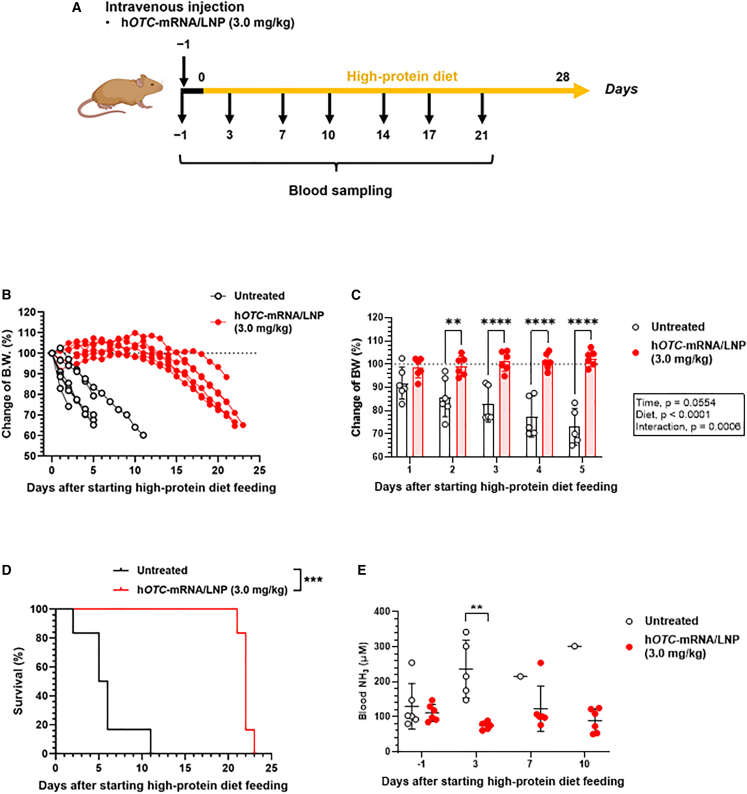
Single-dose efficacy of hOTC-mRNA/LNP in Otcspf-ash mice
Otcspf-ash mice were intravenously injected with hOTC-mRNA/LNP at 3.0 mg/kg on day 0, and the high-protein diet feeding started (Figure 5A). Similar to the initial investigation, the untreated control mice exhibited an initial body weight decrease of approximately 10% on the first day, followed by a rapid decrease in weight (Figure 5B). In contrast, the treated group maintained more than 90% body weight from day 0 until day 16. A significant decrease in body weight changes was seen from day 2 between the two groups (Figure 5C). Within the treated group, weight decreased significantly from day 19 compared with that of the time of pretreatment (day 0) (Figure S9A). The first dead mouse emerged on day 2, and on day 11 no survivors were found in the untreated control group, whereas, in the treated group, the first dead individual was observed on day 20, indicating that the treatment significantly improved the survival rate (p < 0.0001) (Figure 5D). Significant elevation of blood NH3 concentrations in the untreated group was detected on day 3 (p < 0.001) (Figure 5E). Figure S9B shows time course changes in blood NH3 concentrations of individual treated mice, indicating no significant increases in blood NH3 levels until day 14 compared with those on day 0.
Figure 5.
In vivo efficacy of single dosing of hOTC-mRNA/LNP in OTCD model mice
We monitored body weight (BW), survival, and blood NH3 concentrations of male Otcspf-ash mice treated with hOTC-mRNA/LNP. Mice were administered an intravenous bolus of hOTC-mRNA/LNP at 3.0 mg/kg on day 0. After the injection, mice were provided a high-protein (40%/kcal) diet ad libitum. Untreated mice were used as controls. Blood was sampled from the cheek vein for NH3 determination on days −1, 3, 7, 10, 14, 17, and 21. The experimental scheme is shown in (A). (B) Time course changes of BW of individual mice (% of those on day 0). The dotted line indicates 100%. (C) Comparison of BW changes between the untreated and hOTC-mRNA/LNP-treated groups from days 1 to 5. Data were analyzed by two-way ANOVA (F4,78 = 9.684, p < 0.0001 for time; F1,78 = 341.5, p < 0.0001 for diet; F4,78 = 8.732, p < 0.0001 for interaction), followed by Šídák’s multiple comparisons test. ∗∗p < 0.01; ∗∗∗∗p < 0.0001. Values are expressed as means ± SD with individual values. (D) Kaplan-Meier survival curves of the untreated and hOTC-mRNA/LNP-treated groups. ∗∗∗ p < 0.001 by log-rank test. (E) Comparison of blood NH3 concentrations between the untreated and hOTC-mRNA/LNP-treated groups from days −1 to 10. ∗∗p < 0.01 by Mann-Whitney U test. n = 6 per group. Values are expressed as means ± SD with individual values.
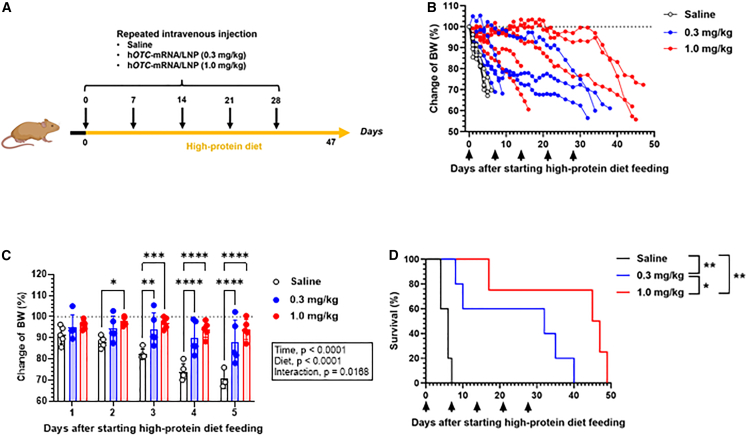
Repeated dose efficacy of hOTC-mRNA/LNP in Otcspf-ash mice
We evaluated the in vivo efficacy of repeated administration of hOTC-mRNA/LNP in high-protein diet-fed Otcspf-ash mice. Mice were intravenously injected with hOTC-mRNA/LNP (0.3 or 1.0 mg/kg) or saline on days 0, 7, 14, 21, and 28 after the high-protein diet feeding started from day 0 onward (Figure 6A). The saline-treated group exhibited rapid and drastic weight loss; all died by day 7 (Figure 6B). The body weight loss caused by the high-protein diet feeding was significantly attenuated by hOTC-mRNA/LNP administration (Figure 6C). Figure 6D depicts the survival curves of the three groups, indicating the following significant differences: p < 0.01 for the saline vs. hOTC-mRNA/LNP (0.3 mg/kg) groups; p < 0.01 for the saline vs. hOTC-mRNA/LNP (1.0 mg/kg) groups; and p < 0.05 for the hOTC-mRNA/LNP (0.3 mg/kg) vs. hOTC-mRNA/LNP (1.0 mg/kg) groups. Therefore, the survival rate was ameliorated in a dose-dependent fashion.
Figure 6.
In vivo efficacy of multiple dosing of hOTC-mRNA/LNP in OTCD model mice
We monitored body weight (BW) and survival of male Otcspf-ash mice, which were repeatedly treated with hOTC-mRNA/LNP. Mice were intravenously injected with hOTC-mRNA/LNP (0.3 or 1.0 mg/kg) or saline on days 0, 7, 14, 21, and 28. After the first injection on day 0, mice were provided a high-protein (40%/kcal) diet ad libitum. The experimental scheme is illustrated in (A). (B) Time course changes of BW of individual mice (% of those on day 0). The dotted line indicates 100%. (C) Comparison of BW changes among the three groups from days 1 to 5. Data were analyzed by two-way ANOVA (F4,58 = 9.939, p < 0.0001 for time; F2,58 = 47.74, p < 0.0001 for treatment; F8,58 = 2.601, p = 0.0168 for interaction), followed by Tukey’s multiple comparisons test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Values are expressed as means ± SD with individual values. (D) Kaplan-Meier survival curves of the three groups. ∗p < 0.05; ∗∗p < 0.01 by log-rank test. n = 5 per group.
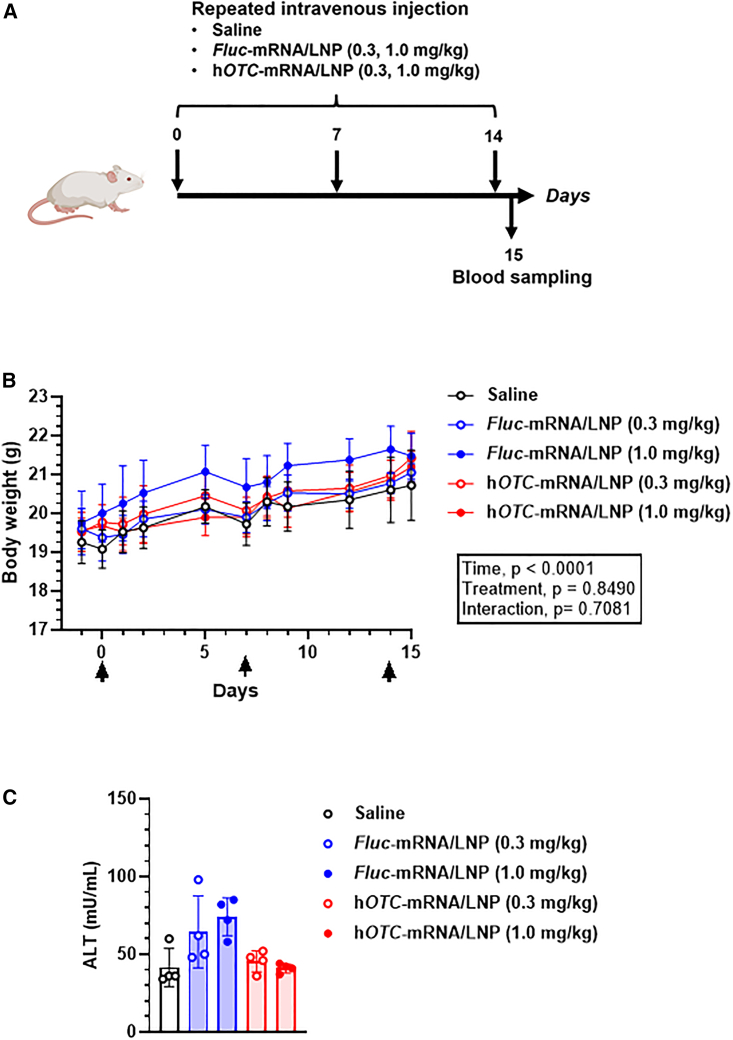
Tolerability of hOTC-mRNA/LNP in wild-type mice at multiple dosing
To test the tolerability of hOTC-mRNA/LNP under repeated administration, BALB/c mice were intravenously injected with Fluc-mRNA/LNP or hOTC-mRNA/LNP at 0.3 or 1.0 mg/kg or saline on days 0, 7, and 14. On day 15, plasma biochemistry was examined (Figure 7A). The groups had no significant differences in body weight changes (Figure 7B). Comprehensively, no remarkable effect of LNP-mRNA was observed on plasma biochemistry values (Table S2). Notably, no significant changes in hepatic function markers were observed, including ALT (Figure 7C). These results indicate that LNP-mRNA was well tolerated.
Figure 7.
Tolerability of repeated doses of hOTC-mRNA/LNP in wild-type mice
Female BALB/c mice were injected intravenously with hOTC-mRNA/LNP or Fluc-mRNA/LNP at 0.3 or 1.0 mg/kg, or saline on days 0, 7, and 14, and blood was collected by cardiac puncture from fed mice on day 15. The experimental scheme is shown in (A). (B) Body weight changes from days −1 to 15. Mice were weighed on days −1, 0, 1, 2, 5, 7, 8, 9, 12, 14, and 15 under fed conditions. On days, 0, 7, and 14, mice were weighed before the injection. Data were analyzed by two-way ANOVA (F3.599,53.99 = 55.46, p < 0.0001 for time; F4,15 = 0.3356, p = 0.8490 for treatment; F40,150 = 0.8583, p = 0.7081 for interaction), followed by Dunnett’s multiple comparisons test. Values show means ± SD. (C) Plasma alanine aminotransferase (ALT) levels. Data were analyzed by Kruskal-Wallis test, followed by Dunn’s multiple comparisons test. There was no significant difference between any groups. Data show means ± SD with individual values. n = 4 per group.
Discussion
OTCD is a rare X-linked genetic disorder characterized by a complete or partial lack of OTC (https://omim.org/entry/311250). OTC is an enzyme in the urea cycle that eliminates excess nitrogen. The lack of OTC results in excess accumulation of nitrogen in the form of NH3 in the blood (hyperammonemia), which affects the central nervous system. OTCD becomes evident at any age; however, the most severe form occurs in the first few days of life. Complications from OTCD may include developmental delay and intellectual disability. Enzyme replacement therapy for mitochondrial disorders is challenging because of the need for organelle-specific delivery,29 and this difficulty may be true for OTCD, because OTC is a mitochondrial matrix enzyme. Preclinical studies using AAV have been reported in Otcspf-ash15,30 and Otc knock-out mice.31 However, AAV gene therapy has issues with neutralizing antibodies17,18 and genome insertion.19 Under these circumstances, mRNA-based therapy may be an alternative approach. Truong et al.32 reported LNP-targeted mRNA therapy for arginase deficiency (argininemia; OMIM: 207800; https://www.omim.org/entry/207800), one of the UCDs. Therefore, OTCD is also considered a good target for mRNA LNP therapy.
Prieve et al.20 used the following components of LNP: (2,3-dioleoyloxy-propyl)-trimethylammonium chloride (DOTAP; cationic lipid), cholesteryl hemisuccinate, cholesterol, and polyethylene glycol (PEG) 2,000 lipid. Because DOTAP-based LNP has a weak endosomal escape function, they separately prepared a polymer with an endosomal escape function and combined it with mRNA-LNP. LNPs reach the hepatocytes via apolipoprotein E-low-density lipoprotein receptor interaction and the polymer conjugated with GalNAc promoted hepatic delivery and endosomal escape. However, drug development of the mixture of polymer micelle and mRNA/LNP should be highly complicated. From the perspective of safety, drug metabolism, and PK, individual toxicity studies on the two components may be mandatory because their PK differ. Manufacturing and quality control should also be complicated, specifically for a polymer that requires strict control of the percentage of functionalization and molecular weight. In addition, the two agents must be reconstituted when used because the mixed formulation is unstable. In contrast, our LNP formulation is a simple, single agent without an extra polymer and reconstitution step. Our biodegradable ionizable lipid-based LNP can induce efficient endosomal escape of mRNA and be simply used after thawing.
OTC proteins are synthesized on cytosolic free ribosomes33 as a precursor34 with the NH2-terminal leader sequence of 32 amino acid residues.35 The precursor proteins are imported into the mitochondrial matrix. During or immediately after the import, the NH2-terminal sequence is removed by a protease in the mitochondrial matrix.36,37 The resultant mature OTC proteins are assembled into the enzymatically active trimer. Therefore, for hOTC-mRNA/LNP to exhibit in vivo efficacy, (1) mRNA is delivered to hepatocytes by our LNP formulation, (2) hOTC proteins translated from hOTC mRNA are transported to mitochondria, and (3) hOTC proteins form the enzymatically active trimeric structure in the mitochondrial matrix. We observed liver-specific biodistribution of Fluc-mRNA/LNP, the trimeric conformation of hOTC proteins by cryo-EM, and the localization of hOTC proteins to mitochondria by immunocytochemistry and IHC in Hep3B cells and mouse hepatocytes, respectively, treated with hOTC-mRNA/LNP. Accordingly, these findings satisfy the above conditions.
Our immunohistochemical study confirmed hOTC protein expression of wild-type mice 24 h after intravenous injection of hOTC-mRNA/LNP. In particular, high expression was observed in hepatocytes around the portal veins. NH3 detoxication in livers via the urea cycle and glutamine synthesis is strictly compartmentalized, occurring in periportal hepatocytes and pericentral hepatocytes, respectively.38 The mRNA expression of genes involved in ureagenesis, including OTC in the liver lobule, is zoned, highest in periportal hepatocytes and declining toward pericentral hepatocytes.38 In ureagenesis, OTC is activated by peroxisome proliferator-activated receptor gamma coactivator 1-alpha, which is highly expressed in periportal hepatocytes.39 Considering the OTC zonation, the high expression of hOTC revealed by IHC may be very favorable for OTCD treatment. Prieve et al.20 reported no difference in the distribution of hOTC protein expression between the central vein and portal regions of Otcspf-ash mice dosed with hOTC-mRNA/HMT. The cause of this difference is unknown; however, it may be due to the difference in genotypes of Otc locus (Otcspf-ash vs. Otc wild-type) and sampling timing.
In general, mRNA levels in mouse livers delivered by LNP disappears quickly, within 2–3 days after administration (e.g., human methylmalonyl-CoA mutase40; human glucose 6-phosphatase α (G6PC)41; and human ARG1).32 However, the duration of protein expression in the liver induced by LNP-formulated mRNA varies depending on the protein. Truong et al.32 showed that luciferase protein in the liver was undetectable 72 days after luc-mRNA/LNP. Sabnis et al.42 reported that hEPO induced by a bolus of hEPO-mRNA/LNP almost disappeared from the blood after 48 h in rats and cynomolgus monkeys. In contrast, an example shows a long t1/2: severe acute respiratory syndrome coronavirus 2 neutralizing antibody had a t1/2 of 15.38 days in mice treated with a bolus of LNP-formulated antibody mRNA because of the aid of Fc portion.43 Compared with the rapid decline and a short t1/2 of hOTC mRNA (0.2944 days [7.066 h] for 1.0 mg/kg and 0.3791 days [9.098 h] for 2.5 mg/kg), hOTC protein was retained relatively long. hOTC expression was maintained for 5 days and subsequently reduced with a t1/2 of 2.2 days from day 5 at 1.0 mg/kg; hOTC protein was sustained for at least 7 days at 2.5 mg/kg. The relatively long duration of hOTC expression can explain the results of the single-dose experiment; significant weight loss was observed from day 19, and the first death of mice was observed from day 22, whereas all untreated mice died on day 11. A t1/2 of hepatic OTC protein was reported to be comparatively long in rats (t1/2 = 8 days).44 In addition, Prieve et al.20 reported that induced hOTC protein was maintained until 10 days after a bolus of hOTC-mRNA/HMT. These findings are in accord with our results. To further extend the duration of hOTC protein expression, enhancing mRNA stability and translation, as well as decreasing mRNA degradation are attractive strategies through nucleoside modification.45 Beside mRNA modification, amino acid modification of protein is also plausible. For example, introducing the triple mutation into the Fc portion improves the t1/2 and activity of monoclonal antibodies.46 Amino acid substitution to serine to cysteine at position 298 showed improvement in expression levels and activity of human G6PC.41 Regarding intracellular proteins, OTC activity was improved by mutation insertion based on deep latent variable modeling.47
Several companies are developing LNP-formulated hOTC-mRNA drugs. Among the companies, Arcturus Therapeutics leads clinical studies with ARCT-810 (LUNAR-OTC) in phase 2 studies (ClinicalTrial.gov: NCT05526066; https://clinicaltrials.gov/ct2/show/NCT05526066). Arcturus Therapeutics reported PK/PD studies of single dosing of LUNAR-OTC in Otcspf-ash mice.48 In addition, they demonstrated a positive correlation between hepatic OTC concentrations and hepatic OTC enzymic activities, as well as a positive correlation between hepatic OTC concentrations and plasma OTC enzymic activities. In particular, the latter is an important finding that indicates the possibility of using plasma OTC activity as a surrogate marker in clinical trials.48 Our present study revealed critical aspects (e.g., hOTC protein conformation produced by hOTC-mRNA/LNP, cytological and histological hOTC protein expression, and efficacy of single-dosing and repeated administration) that were not described by Yu et al.48 These findings further demonstrate the potential usefulness of an LNP-mRNA approach for the therapy of OTCD.
In summary, we found the trimeric conformation of hOTC proteins in cells treated with hOTC-mRNA/LNP and their localization to mitochondria. Intravenous injection of hOTC-mRNA/LNP into mice induced hepatic hOTC protein expression and increased activity. Using the OTCD model mice, efficacy was observed with its single and continuous administration. No severe changes in biochemical values were detected with continuous administration. However, this study has key limitations. First, the in vivo experiments of our study used mice. The amount and duration of hOTC protein expression induced by hOTC-mRNA/LNP must be examined in larger animals, such as non-human primates. Second, we used Otcspf-ash mice, whose OTCD symptoms are induced only when fed a high-protein diet. Authentic models are essential for the precise prediction of clinical efficacy in humans. Third, we used adult Otcspf-ash mice for the efficacy evaluation of hOTC-mRNA/LNP. Patients with the neonatal-onset form have a high risk of neurological damage and death. Subsequently, we have to develop new in vivo models to solve this issue. Nevertheless, our data provide a strong rationale for using an mRNA-based enzyme replacement approach to OTCD and demonstrate the potential usefulness of our LNP-mRNA therapeutics.
Materials and methods
mRNA synthesis and LNP formulation
The mRNAs encoding Fluc and hOTC and hOTC labeled with 3× FLAG at the C-terminus were synthesized by in vitro transcription using a linearized DNA template. The codon optimization was performed toward the open reading frame of Fluc and hOTC (NM_000531.6) (hOTC).49 In the designed mRNA, uridine was completely replaced by N1-methylpseudouridine, with the Cap 1 structure (#N-7113, TriLink BioTechnologies, San Diego, CA), the 5′ and 3′ untranslated regions and a poly(A) tail of around 120 nucleotides. As previously described, the nucleoside-modified mRNAs were purified and formulated into LNPs for intravenous delivery.25 The obtained LNPs in 8% sucrose/20 mM Tris buffer (pH 7.5) were used immediately or cryopreserved at −80°C before in vivo study.
LNP characterization
The particle sizes and polydispersity index were determined by dynamic light scattering using a Zeta Sizer (Malvern Panalytical, Malvern, Worcestershire, UK). As previously described, total mRNA concentrations in LNP were determined using a Quant-iT RiboGreen RNA assay kit (ThermoFisher Scientific, Waltham, MA).50 Encapsulation efficiency (EE, %) was calculated as follows: EE (%) = (1 − free mRNA concentration/total mRNA concentration) × 100. The physical properties of mRNA-encapsulating LNPs are summarized in Table S1.
To confirm delivery to the liver by LNP formulation, we examined the biodistribution of Fluc luminescence after treatment of LNP-formulated Fluc mRNA (Fluc-mRNA/LNP). Female BALB/c mice were injected intravenously with Fluc-mRNA/LNP at 1.0 mg/kg. The liver, spleen, kidney, lung, and heart were harvested 24 h after the injection, and their Fluc luminescence was determined by an IVIS imaging system (PerkinElmer, Waltham, MA).
Cells
Hep3B cells were obtained from the American Type Culture Collection (Cat# HB-8064; Gaithersburg, MD). Cells were cultured in E-MEM (FUJIFILM Wako, Osaka, Japan) containing 10% fetal bovine serum (Gibco, Grand Island, NY) and 1× penicillin-streptomycin (FUJIFILM Wako) at 37°C in a humidified atmosphere of 5% CO2.
In vitro hOTC protein expression by transfection of hOTC mRNA in Hep3B cells
In vitro, hOTC protein expression by codon-optimized hOTC mRNA was confirmed in Hep3B cells using Lipofectamine MessengerMAX mRNA Transfection Reagent (ThermoFisher Scientific). On day 0, 5.0 × 104 cells were plated in 24-well plates, and hOTC mRNA (0, 20, 100, or 500 ng/mL) was added the next day. After 24 h, cells were harvested for western blotting.
Protein production and purification for cryo-electron microscopy
Hep3B cells were transiently transfected with 200 ng/mL hOTC-mRNA/LNP, and protein expression was induced for 1–2 days. Harvested cells were resuspended in lysis buffer (25 mM HEPES-NaOH [pH 8.0], 200 mM NaCl, and 10% [v/v] glycerol supplemented with cOmplete EDTA-free Protease Inhibitor Cocktail [Roche, Basel, Switzerland]) and were disrupted by sonication on ice. After centrifugation at 10,000×g for 30 min at 4°C, the supernatant was mixed with M2 Anti-FLAG Agarose (Sigma-Aldrich, St. Louis, MO) and gently stirred at 4°C for 2 h. The resin was subsequently loaded onto an Econo-Pac column (Bio-Rad, Hercules, CA) and washed with purification buffer (50 mM HEPES-NaOH [pH 7.5] and 200 mM NaCl with cOmplete). We eluted 3× FLAG-tagged hOTC proteins with a purification buffer containing 400 μg/mL 3× FLAG peptides (Sigma-Aldrich). The eluate was concentrated by Amicon Ultra 10-kDa MWCO (Millipore, Burlington, MA) and further purified by size-exclusion chromatography using Superdex 200 Increase 10/300 column (Cytiva, Tokyo, Japan) pre-equilibrated with the purification buffer. The peak fractions were concentrated with Amicon Ultra 30-kDa MWCO, flash frozen in liquid nitrogen, and stored at −80°C until use.
PEGylation51 was introduced to the purified sample (Figures S10A–S10C) to improve the quality of vitrified hOTC specimens on cryo-EM grids. hOTC solution was diluted to 0.7 mg/mL in a purification buffer and mixed with 2 mM methyl-PEG8-NHS ester (Tokyo Chemical Industry, Tokyo, Japan). After incubation on ice for 1 h, the reaction was quenched by adding 1/10 volume of 1 M Tris-HCl (pH 8.0). Then the PEGylated hOTC was isolated by Superdex 200 Increase 10/300 column pre-equilibrated with the purification buffer and concentrated with Amicon Ultra 30-kDa MWCO to 0.8 mg/mL.
Cryo-EM
One droplet of 3 μL of mRNA/LNP solution was applied to a glow-discharged holey carbon grid (Cu R1.2/1.3, 300 mesh, #M2955C-1–300; Quantifoil Micro Tools, Jena, Germany). The grid was blotted for 3 s with a blot force of 15 and flash frozen in liquid ethane using a Vitrobot Mark IV (ThermoFisher Scientific) at 18°C and 100% humidity. Data were collected on a Talos Arctica electron microscope (ThermoFisher Scientific) equipped with a Falcon 4 direct electron detector (ThermoFisher Scientific) at 200 kV. Images were acquired at a defocusing of −1 μm with a nominal magnification of ×73,000 at a pixel size of 1.35 Å. The images were processed and analyzed with ImageJ 1.52p Fiji software.52
The hOTC grid and data acquisition were prepared the same way as mRNA/LNP. Images were acquired at a range of defocus from −1.0 to −2.5 μm with a nominal magnification of ×120,000, corresponding with a pixel size of 0.841 Å at the electron exposure of 11.00 e−Å−2 per second and a total exposure time of 4.55 s, resulting in an accumulated exposure of 50 e−Å−2. A total of 4,206 movies were collected.
Cryo-EM image processing of 3× FLAG-tagged hOTC proteins
Image processing was performed using RELION-4.0.53 Movies were aligned and dose weighted by RELION’s implementation of MotionCor2.54 Contrast transfer function parameters were estimated by CTFFIND4.1.55 Particles were first picked by the Laplacian-of-Gaussian function and used for two-dimensional (2D) classification to make templates. The subsequent template-based autopicking found 5,698,069 particles, then extracted with a pixel size of 3.364 Å. The particles were subjected to multiple rounds of 2D and three-dimensional (3D) classifications with an initial 3D reference model that was generated from an hOTC crystal structure (PDB: 1C9Y) using UCSF ChimeraX 1.4.56 hOTC particles exhibited a strong preferred orientation, whereas finer angular sampling (3.7°) slightly decreased alignment bias. After choosing classes with visible secondary structural elements and the ternary feature, the selected subset of 87,434 particles was re-extracted with a pixel size of 0.841 Å. The 3D refinement was carried out with C3 symmetry in RELION and SIDESPLITTER v.1.3.57 The resulting 3D model and particles were subjected to iterative CTF refinement and Bayesian polishing.58 The final 3D refinement and post-processing yielded a map with a resolution of 4.3 Å, estimated by the Fourier shell correlation criterion at 0.143. The processing workflow is summarized in Figure S10D. Figures were generated using ChimeraX 1.4. Because of the low resolution of the map, an atomic model of the OTC proteins has not been built. The map has been deposited to the Electron Microscopy DataBank under accession code EMD-35535.
Mice
Male “sparse fur-abnormal skin and hair” hemizygous (Otcspf-ash) mice 55 of B6EiC3Sn a/A-Otcspf-ash/J (Strain# 001811) were obtained from The Jackson Laboratory (Bar Harbor, ME). OTC activity of Otcspf-ash mice is 5%–10%.59 Male C57BL/6J (B6) and female BALB/cAnNCrlCrlj (BALB/c) mice were obtained from The Jackson Laboratory Japan (Yokohama, Kanagawa, Japan). Animals were maintained under conventional conditions of controlled temperature, humidity, and lighting (23 ± 3°C, 55 ± 15%, and a 12-h light/dark cycle with lights on at 07:00). Animal care and experimental procedures were performed in an animal facility accredited by the Health Science Center for Accreditation of Laboratory Animal Care and Use of the Japan Health Sciences Foundation. All protocols were approved by the Institutional Animal Care and Use Committee of Eisai Co., Ltd. (Tokyo, Japan) and carried out in accordance with the Animal Experimentation Regulations.
Hepatic hOTC protein expression induced by hOTC-mRNA/LNP
To demonstrate specific hOTC expression in the liver by LNP-formulated hOTC mRNA, female BALB/c mice were intravenously injected with hOTC mRNA only, Fluc-mRNA/LNP, or hOTC-mRNA/LNP at 1.0 mg/kg, or saline. Liver tissues were collected 1 day after the injection. Then, we examined the concentration dependency of hepatic hOTC expression by hOTC-mRNA/LNP treatment. Livers were collected after intravenous bolus dosing at 0.3 or 1.0 mg/kg to BALB/c mice.
Time course changes of hOTC mRNA amounts and hOTC expression
Female BALB/c mice were intravenously injected with hOTC-mRNA/LNP at 1 or 2.5 mg/kg. Liver tissues (left lateral lobes) were collected on days 1, 2, 3, 4, and 7 to investigate short-term expression and on days 1, 7, 14, and 21 for long-term expression.
Measurement of OTC enzymic activity
We administered hOTC-mRNA/LNP at 2.0 mg/kg to female BALB/c mice intravenously, and liver tissues were harvested after 72 h (untreated, n = 4; treated, n = 4). The liver tissues (left lateral lobes) were used for western blotting and activity determination. OTC enzymic activity was determined basically according to the method of Pastra-Landis et al.60 Briefly, liver tissues were homogenized in mitochondrial lysis buffer: 0.5% Triton X-100, 10 mM HEPES sodium salt, 0.5 mM DTT, 2 mM EDTA, and 1× protease inhibitor cocktail (pH 7.4) (Sigma-Aldrich). We added 2 μg total liver protein extract to 700 μL of the reaction mixture (5 mM L-ornithine, 25 mM carbamyl phosphate, and 50 mM Tris-acetate [pH 8.3]) and incubated at 37°C for 30 min. The reaction was stopped by adding 250 μL of 3:1 phosphoric/sulfuric acid solution, followed by adding 50 μL of 3% 2,3-butanedione monoxime, and followed by incubation at 95°C for 15 min in the dark. Citrulline production was determined by measuring the absorbance at 490 nm with a CRALIOstar microplate reader (BMG LABTECH, Ortenberg, Germany). Results were expressed as μmol citrulline mg−1 total protein h−1. The materials used in this study were purchased as follows: L-ornithine, L-citruline, phosphoric acid, sulfuric acid, and acetic acid from Fujifilm Wako (Osaka, Japan); carbamyl phosphate from Sigma-Aldrich; and 2,3-butanedione monoxime from Nacalai (Kyoto, Japan).
Western blotting analysis
hOTC protein expression in Hep3B cell lysates or liver specimens was measured by standard western blotting. Total protein concentrations were determined by Pierce 660 nm Protein Assay Kit (ThermoFisher Scientific). Samples were separated by 10% SDS-PAGE gel and transferred to nitrocellulose membranes by Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad). Membranes were incubated with mouse anti-hOTC monoclonal antibody (Cat# TA802590, OriGene, Rockville, MD) at 1:20,000, mouse anti-β-tubulin monoclonal antibody (Cat# sc-53140, Santa Cruz Biotechnology, Dallas, TX) at 1:5,000, and mouse anti-HSP90AB1 (HSP90) monoclonal antibody (Cat# TA500494, OriGene) at 1:2,000. The signals were visualized by chemiluminescence using LuminataForte HRP substrate (Merck Millipore) with a FUSION (Vilber, Paris, France). The densitometry of each band was quantified by BIO-1D Software (Vilber).
RT-qPCR
Total RNA from livers was isolated using a Maxwell RSC simply RNA Tissue Kit (Promega, Madison, WI). Briefly, a portion of the liver (left lateral lobes) was weighed and stored in RNAlater Stabilization Solution (ThermoFisher Scientific). The tissues were homogenized in Homogenization Solution with a TissueLyser II (Qiagen, Hilden, Germany) for 4 min at 30 Hz and centrifuged at 17,000×g for 5 min to deplete tissue debris. Subsequently, the supernatants were transferred to cartridges and further processed according to the kit protocol. RNA was quantified with QIAxpert (Qiagen), and 800 ng of total RNA was converted into cDNA using SuperScript IV VILO Master Mix (ThermoFisher Scientific). hOTC mRNA was quantified using TaqMan probe assays (5′-ACCGGCGAGGAGATCAAGTAC-3′ as a forward primer, 5′-TGCGCTTCTCGAAGATCATG-3′ as a reverse primer; ThermoFisher Scientific) on ViiA 7 (ThermoFisher Scientific). The thermal cycling conditions were: 50°C for 2 min, followed by an initial denaturation step at 95°C for 20 s, 40 cycles at 95°C for 1 s, and 60°C for 20 s. To generate the standard curve, pure hOTC mRNA was reverse-transcribed and the cDNA was serially diluted. The amounts of hOTC mRNA in livers (pg) were normalized to liver wet weights (g). In addition, the following primers were used: Mm00493267_m1 for Otc; Mm99999915_g1 for Gapdh; and Mm04394036_g1 for Actb.
Immunocytochemistry
Hep3B cells were fixed in cold methanol (FUJIFILM Wako) for 15 min. The cells were washed next with D-PBS(−) (FUJIFILM Wako) at room temperature (approximately 25°C) three times and incubated in 1% BSA/PBST for 30 min at room temperature. Cells were incubated overnight at 4°C with primary antibodies diluted in 1% BSA/PBS with 0.1% Tween 20 (PBST). Subsequently, they were incubated for 1 h at room temperature with secondary antibodies and Hoechst 33342 (0.2 μg/mL; Sigma-Aldrich) diluted in 1% BSA/PBST. The cells were washed again with PBS three times. The primary and secondary antibodies are listed in Table S3. Image acquisition and analyses were performed with a CellVoyager CV7000 High-throughput Cytological Discovery System (Yokogawa) and a CellPathFinder image analysis software (Yokogawa).
IHC
To examine hOTC protein expression histologically, we stained hOTC in the liver of mice treated with hOTC-mRNA/LNP immunohistochemically. Twenty-four hours after intravenous injection of hOTC-mRNA/LNP (1.0 or 2.5 mg/kg) or saline, BALB/c mice were euthanized by exsanguination, and livers (left lateral lobes) were collected. The liver tissues were fixed in a 4% paraformaldehyde phosphate buffer solution (PFA; FUJIFILM Wako, Osaka, Japan). For double staining of hOTC and albumin (a hepatocyte marker) or prohibitin (a mitochondria marker), mice were intravenously injected with hOTC-mRNA/LNP at 2.5 mg/kg. After 24 h, liver specimens were collected after being perfused with saline (Otsuka Pharmaceutical, Tokyo, Japan) containing heparin (10 U/mL; AY Pharmaceuticals, Tokyo, Japan), and fixed with 4% PFA. Sections of the paraffin-embedded tissues were processed for immunohistochemical staining in a Full Automated IHC & ISH System, BOND RX (Leica Microsystems, Wetzlar, Germany). The antibodies and reagents used are summarized in Table S3. Images were captured with an All-in-One Fluorescence Microscope, BZ-X800 (KEYENCE, Osaka, Japan) and analyzed with a BZ-X Analyzer (KEYENCE).
Diets
MF (Oriental Yeast, Tokyo, Japan) was used as the standard regular diet. Oriental Yeast manufactured a high-protein diet (40%/kcal). Table S4 shows the results of the main ingredient analysis of these diets.
Effects of a high-protein diet on hyperammonemia and survival of Otcspf-ash and wild-type mice
First, as a preliminary study, we observed the progress of the disease in the OTCD model of Otcspf-ash mice when fed the high-protein diet prepared this time, compared with wild-type mice. Male Otcspf-ash and male wild-type (B6 mice were fed on MF [n = 9] or a 40% high-protein diet [n = 10]). We weighed mice every day and observed them twice daily (08:00–10:00 and 15:00–17:00) after their conditions began to deteriorate. Mice were euthanized if they were moribund: the moribund state was defined as a state in which the following observation predicted death based on symptoms, such as decreased activity, abnormal breathing, hypothermia, anemic symptoms, and persistent convulsions (the conditions for euthanasia were the same for the hOTC-mRNA/LNP efficacy tests described below). Blood samples for NH3 determination were collected from the cheek vein at 08:00–11:00 on days −1, 3, and 7 (the starting day of the feeding was day 0). In the case of wild-type B6 mice, blood sampling was conducted on days 0, 3, and 7 in the feeding. Blood NH3 was determined using a CicaLiquid NH3 (Kanto Chemical Co., Ltd., Tokyo, Japan) and a 7180 Clinical Analyzer (Hitachi High-Technologies, Tokyo, Japan).
Effects of a single dose of hOTC-mRNA/LNP on clinical signs and blood NH3 concentrations of Otcspf-ash mice fed the high-protein diet
Setting the start day of the high-protein diet feeding to day 0, Otcspf-ash mice were intravenously injected with hOTC-mRNA/LNP at 3.0 mg/kg or saline on day −1. Blood was sampled from the cheek vein on days −1, 3, 7, 10, 14, 17, and 21 (Figure 5A). Mice were weighed from days 0 to 28 daily.
Effects of multiple doses of hOTC-mRNA/LNP on clinical signs of Otcspf-ash mice fed the high-protein diet
Assuming day 0 as the start day of high-protein diet feeding, Otcspf-ash mice were intravenously injected with hOTC-mRNA/LNP (0.3 or 1.0 mg/kg) or saline on days 0, 7, 14, 21, and 28 (Figure 6A). Body weights were measured every day from day 0 to day 24 and every 2 days until day 47.
Tolerability for multiple dosing of hOTC-mRNA/LNP
Female BALB/c mice were injected intravenously with hOTC-mRNA/LNP at 0.3 or 1.0 mg/kg on days 0, 7, and 14 (Figure 7A). On day 15, blood was collected by cardiac puncture under isoflurane anesthesia for plasma biochemical examinations (n = 4/group). As a control mRNA formulation, Fluc-mRNA was used at the same doses. After separating plasma from collected blood, we determined plasma biochemistry using a 7180 Clinical Analyzer (Hitachi High-Technologies, Tokyo, Japan).
mRNA stability
The stability of mRNA was tested in the presence of mouse plasma. hOTC mRNA and hOTC-mRNA/LNP were incubated in the absence or presence of 50% mouse plasma at 37°C for 3 h. The samples were purified by RNeasy mini kit (Qiagen) and electrophoresed on a 1% agarose gel (E-Gel precast agarose cassette, ThermoFisher Scientific), together with those immediately mixed with 50% mouse plasma. Fresh mouse plasma of female BALB/c mice was prepared from heparinized blood collected by cardiac puncture under isoflurane anesthesia.
Statistics
Graphic drawing and statistical analysis were performed using GraphPad Prism 9.5.1 (GraphPad Software, San Diego, CA). Results were expressed as means ± SD or median. The following tests were performed as indicated in the figure and table legends: two-tailed unpaired Student t test, Mann-Whitney U test, log rank test, two-way ANOVA, one-way repeated measures ANOVA, two-way repeated measures ANOVA, Tukey’s multiple comparisons test, Dunn’s multiple comparisons test, and Šídák’s multiple comparisons test. A p value of less than 0.05 was considered statistically significant.
Data for hOTC mRNA levels and hOTC protein levels over time were fit using one phase exponential decay (Y = (Y0 − Plateau)∗exp(−K∗X) + Plateau, where Y0 is the Y value when X is 0, Plateau is the Y value at infinite times, and K is the rate constant) or plateau followed by one-phase exponential decay (Y = IF(X < X0, Y0, Plateau + (Y0 − Plateau)∗exp(−K∗(X-X0))), where X0 is the time at which the decay begins, Y0 is the average Y value up to time X0, Plateau is the Y value at infinite times, and K is the rate constant). In this study, Plateau was constrained to a constant value of zero. The t1/2 is computed as ln(2)/K.
Acknowledgments
The authors would like to thank Professor Kandai Nozu, Professor Kazumoto Iijima, Professor Masanori Miyanishi, Assistant Professor Tomoko Horinouchi, and Assistant Professor Ryosuke Bo of Kobe University Graduate School of Medicine (Kobe, Japan) for the helpful discussion on OTCD. We are grateful to the members of the cryo-EM facility in KEK (Ibaraki, Japan) for cryo-EM data collection, which was supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research [BINDS]) from AMED under Grant Number JP22ama121001. We also thank Kenji Nakano and Ayami Amano of Sunplanet Co., Ltd. for immunohistochemical preparation, and Nao Komatsu of WORLD INTEC Co., Ltd. for subculturing Hep3B cells used for hOTC expression. Part of the ToC Figure, and Figures 5A, 6A, as well as 7A were created using BioRender.com. We would like to thank Editage (www.editage.com) for their English language editing.
Author contributions
K.Y., K. Kubara, K. Kondo, and Y.S. conceived and designed the experiments. K.Y., K. Kondo, and Y.S. wrote the manuscript. K.Y., K. Kubara, S.I., K. Kondo, Y.S., T.M., and K.M. performed experiments. M.I. and K.T. supervised the study.
Declaration of interests
The authors declare no conflict of interest.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.06.023.
Supplemental information
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.
References
- 1.Mitchell S., Ellingson C., Coyne T., Hall L., Neill M., Christian N., Higham C., Dobrowolski S.F., Tuchman M., Summar M., Urea Cycle Disorder Consortium Genetic variation in the urea cycle: a model resource for investigating key candidate genes for common diseases. Hum. Mutat. 2009;30:56–60. doi: 10.1002/humu.20813. [DOI] [PubMed] [Google Scholar]
- 2.Burton B.K. Urea cycle disorders. Clin. Liver Dis. 2000;4:815–830.vi. doi: 10.1016/s1089-3261(05)70143-4. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda I. Hyperammonemia in pediatric clinics: a review of ornithine transcarbamylase deficiency (OTCD) based on our case studies. JMAJ. 2004;47:160–165. [Google Scholar]
- 4.Caldovic L., Abdikarim I., Narain S., Tuchman M., Morizono H. Genotype-phenotype correlations in ornithine transcarbamylase deficiency: a mutation update. J. Genet. Genomics. 2015;42:181–194. doi: 10.1016/j.jgg.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCullough B.A., Yudkoff M., Batshaw M.L., Wilson J.M., Raper S.E., Tuchman M. Genotype spectrum of ornithine transcarbamylase deficiency: correlation with the clinical and biochemical phenotype. Am. J. Med. Genet. 2000;93:313–319. doi: 10.1002/1096-8628(20000814)93:4<313::aid-ajmg11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 6.Arranz J.A., Riudor E., Marco-Marín C., Rubio V. Estimation of the total number of disease-causing mutations in ornithine transcarbamylase (OTC) deficiency. Value of the OTC structure in predicting a mutation pathogenic potential. J. Inherit. Metab. Dis. 2007;30:217–226. doi: 10.1007/s10545-007-0429-x. [DOI] [PubMed] [Google Scholar]
- 7.Batshaw M.L., Tuchman M., Summar M., Seminara J., Members of the Urea Cycle Disorders Consortium A longitudinal study of urea cycle disorders. Mol. Genet. Metabol. 2014;113:127–130. doi: 10.1016/j.ymgme.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wasim M., Awan F.R., Khan H.N., Tawab A., Iqbal M., Ayesha H. Aminoacidopathies: prevalence, etiology, screening, and treatment options. Biochem. Genet. 2018;56:7–21. doi: 10.1007/s10528-017-9825-6. [DOI] [PubMed] [Google Scholar]
- 9.Nagamani S.C.S., Lichter-Konecki U. In: Swaiman's Pediatric Neurology, 6th Edition Principles and Practice. Swaiman K.F., Ashwal S., Ferriero D.M., Schor N.F., Finkel R.S., Gropman A.L., Pearl P.L., Shevell M., editors. Elsevier; Amsterdam, Netherlands: 2017. Inborn errors of urea synthesis; pp. 298–304. [Google Scholar]
- 10.Deardorff M.A., Gaddipati H., Kaplan P., Sanchez-Lara P.A., Sondheimer N., Spinner N.B., Hakonarson H., Ficicioglu C., Ganesh J., Markello T., et al. Complex management of a patient with a contiguous Xp11.4 gene deletion involving ornithine transcarbamylase: a role for detailed molecular analysis in complex presentations of classical diseases. Mol. Genet. Metabol. 2008;94:498–502. doi: 10.1016/j.ymgme.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono M., Tsuda J., Mouri Y., Arai J., Arinami T., Noguchi E. Contiguous Xp11.4 Gene deletion leading to ornithine transcarbamylase deficiency detected by high-density single-nucleotide array. Clin. Pediatr. Endocrinol. 2010;19:25–30. doi: 10.1297/cpe.19.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ricciuti F.C., Gelehrter T.D., Rosenberg L.E. X-chromosome inactivation in human liver: confirmation of X-linkage of ornithine transcarbamylase. Am. J. Hum. Genet. 1976;28:332–338. [PMC free article] [PubMed] [Google Scholar]
- 13.De Las Heras J., Aldámiz-Echevarría L., Martínez-Chantar M.L., Delgado T.C. An update on the use of benzoate, phenylacetate and phenylbutyrate ammonia scavengers for interrogating and modifying liver nitrogen metabolism and its implications in urea cycle disorders and liver disease. Expet Opin. Drug Metabol. Toxicol. 2017;13:439–448. doi: 10.1080/17425255.2017.1262843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitington P.F., Alonso E.M., Boyle J.T., Molleston J.P., Rosenthal P., Emond J.C., Millis J.M. Liver transplantation for the treatment of urea cycle disorders. J. Inherit. Metab. Dis. 1998;21(Suppl. 1):112–118. doi: 10.1023/a:1005317909946. [DOI] [PubMed] [Google Scholar]
- 15.De Sabbata G., Boisgerault F., Guarnaccia C., Iaconcig A., Bortolussi G., Collaud F., Ronzitti G., Sola M.S., Vidal P., Rouillon J., et al. Long-term correction of ornithine transcarbamylase deficiency in Spf-Ash mice with a translationally optimized AAV vector. Mol. Ther. Methods Clin. Dev. 2021;20:169–180. doi: 10.1016/j.omtm.2020.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baruteau J., Cunningham S.C., Yilmaz B.S., Perocheau D.P., Eaglestone S., Burke D., Thrasher A.J., Waddington S.N., Lisowski L., Alexander I.E., Gissen P. Safety and efficacy of an engineered hepatotropic AAV gene therapy for ornithine transcarbamylase deficiency in cynomolgus monkeys. Mol. Ther. Methods Clin. Dev. 2021;23:135–146. doi: 10.1016/j.omtm.2021.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calcedo R., Morizono H., Wang L., McCarter R., He J., Jones D., Batshaw M.L., Wilson J.M. Adeno-associated virus antibody profiles in newborns, children, and adolescents. Clin. Vaccine Immunol. 2011;18:1586–1588. doi: 10.1128/CVI.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selot R.S., Hareendran S., Jayandharan G.R. Developing immunologically inert adeno-associated virus (AAV) vectors for gene therapy: possibilities and limitations. Curr. Pharmaceut. Biotechnol. 2014;14:1072–1082. doi: 10.2174/1389201015666140327141710. [DOI] [PubMed] [Google Scholar]
- 19.Dalwadi D.A., Calabria A., Tiyaboonchai A., Posey J., Naugler W.E., Montini E., Grompe M. AAV integration in human hepatocytes. Mol. Ther. 2021;29:2898–2909. doi: 10.1016/j.ymthe.2021.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prieve M.G., Harvie P., Monahan S.D., Roy D., Li A.G., Blevins T.L., Paschal A.E., Waldheim M., Bell E.C., Galperin A., et al. Targeted mRNA therapy for ornithine transcarbamylase deficiency. Mol. Ther. 2018;26:801–813. doi: 10.1016/j.ymthe.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki Y., Hyodo K., Suzuki T., Tanaka Y., Kikuchi H., Ishihara H. Biodegradable lipid nanoparticles induce a prolonged RNA interference-mediated protein knockdown and show rapid hepatic clearance in mice and nonhuman primates. Int. J. Pharm. 2017;519:34–43. doi: 10.1016/j.ijpharm.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Kawase W., Kurotaki D., Suzuki Y., Ishihara H., Ban T., Sato G.R., Ichikawa J., Yanai H., Taniguchi T., Tsukahara K., Tamura T. Irf5 siRNA-loaded biodegradable lipid nanoparticles ameliorate concanavalin A-induced liver injury. Mol. Ther. Nucleic Acids. 2021;25:708–715. doi: 10.1016/j.omtn.2021.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishigooka H., Katsumata H., Saiga K., Tokita D., Motoi S., Matsui C., Suzuki Y., Tomimatsu A., Nakatani T., Kuboi Y., et al. Novel complement C5 small-interfering RNA lipid nanoparticle prolongs graft survival in a hypersensitized rat kidney transplant model. Transplantation. 2022;106:2338–2347. doi: 10.1097/TP.0000000000004207. [DOI] [PubMed] [Google Scholar]
- 24.Kuboi Y., Suzuki Y., Motoi S., Matsui C., Toritsuka N., Nakatani T., Tahara K., Takahashi Y., Ida Y., Tomimatsu A., et al. Identification of potent siRNA targeting complement C5 and its robust activity in pre-clinical models of myasthenia gravis and collagen-induced arthritis. Mol. Ther. Nucleic Acids. 2023;31:339–351. doi: 10.1016/j.omtn.2023.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki Y., Miyazaki T., Muto H., Kubara K., Mukai Y., Watari R., Sato S., Kondo K., Tsukumo S.I., Yasutomo K., et al. Design and lyophilization of lipid nanoparticles for mRNA vaccine and its robust immune response in mice and nonhuman primates. Mol. Ther. Nucleic Acids. 2022;30:226–240. doi: 10.1016/j.omtn.2022.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi D., Morizono H., Ha Y., Aoyagi M., Tuchman M., Allewell N.M. 1.85-Å resolution crystal structure of human ornithine transcarbamoylase complexed with N-phosphonacetyl-L-ornithine. Catalytic mechanism and correlation with inherited deficiency. J. Biol. Chem. 1998;273:34247–34254. doi: 10.1074/jbc.273.51.34247. [DOI] [PubMed] [Google Scholar]
- 27.Shi D., Morizono H., Aoyagi M., Tuchman M., Allewell N.M. Crystal structure of human ornithine transcarbamylase complexed with carbamoyl phosphate and L-norvaline at 1.9 Å resolution. Proteins. 2000;39:271–277. [PubMed] [Google Scholar]
- 28.Shi D., Morizono H., Yu X., Tong L., Allewell N.M., Tuchman M. Human ornithine transcarbamylase: crystallographic insights into substrate recognition and conformational changes. Biochem. J. 2001;354:501–509. doi: 10.1042/0264-6021:3540501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darvish-Damavandi M., Ho H.K., Kang T.S. Towards the development of an enzyme replacement therapy for the metabolic disorder propionic acidemia. Mol. Genet. Metab. Rep. 2016;8:51–60. doi: 10.1016/j.ymgmr.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Cai Z., Stothard P., Moore S., Goebel R., Wang L., Lin G., Yu H., Batshaw M.L., Wilson J.M. Sustained correction of OTC deficiency in Spfash mice using optimized self-complementary AAV2/8 vectors. Gene Ther. 2012;5:404–410. doi: 10.1038/gt.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L., Bell P., Morizono H., He Z., Pumbo E., Yu H., White J., Batshaw M.L., Wilson J.M. AAV gene therapy corrects OTC deficiency and prevents liver fibrosis in aged OTC-knock out heterozygous mice. Mol. Genet. Metabol. 2017;120:299–305. doi: 10.1016/j.ymgme.2017.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Truong B., Allegri G., Liu X.B., Burke K.E., Zhu X., Cederbaum S.D., Häberle J., Martini P.G.V., Lipshutz G.S. Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase deficiency. Proc. Natl. Acad. Sci. USA. 2019;116:21150–21159. doi: 10.1073/pnas.1906182116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morita T., Mori M., Tatibana M., Cohen P.P. Site of synthesis and intracellular transport of the precursor of mitochondrial ornithine carbamoyltransferase. Biochem. Biophys. Res. Commun. 1981;99:623–629. doi: 10.1016/0006-291x(81)91790-3. [DOI] [PubMed] [Google Scholar]
- 34.Mori M., Miura S., Tatibana M., Cohen P.P. Processing of a putative precursor of rat liver ornithine transcarbamylase, a mitochondrial matrix enzyme. J. Biochem. 1980;88:1829–1836. doi: 10.1093/oxfordjournals.jbchem.a133158. [DOI] [PubMed] [Google Scholar]
- 35.McIntyre P., Graf L., Mercer J.F., Wake S.A., Hudson P., Hoogenraad N. The primary structure of the imported mitochondrial protein, ornithine transcarbamylase from rat liver: mRNA levels during ontogeny. DNA. 1985;4:147–156. doi: 10.1089/dna.1985.4.147. [DOI] [PubMed] [Google Scholar]
- 36.Miura S., Mori M., Amaya Y., Tatibana M. A mitochondrial protease that cleaves the precursor of ornithine carbamoyltransferase. Purification and properties. Eur. J. Biochem. 1982;122:641–647. [PubMed] [Google Scholar]
- 37.Conboy J.G., Fenton W.A., Rosenberg L.E. Processing of pre-ornithine transcarbamylase requires a zinc-dependent protease localized to the mitochondrial matrix. Biochem. Biophys. Res. Commun. 1982;105:1–7. doi: 10.1016/s0006-291x(82)80002-8. [DOI] [PubMed] [Google Scholar]
- 38.Dingemanse M.A., de Jonge W.J., de Boer P.A., Mori M., Lamers W.H., Moorman A.F. Development of the ornithine cycle in rat liver: zonation of a metabolic pathway. Hepatology. 1996;24:407–411. doi: 10.1002/hep.510240219. [DOI] [PubMed] [Google Scholar]
- 39.Li L., Zhang P., Bao Z., Wang T., Liu S., Huang F. PGC-1α promotes ureagenesis in mouse periportal hepatocytes through SIRT3 and SIRT5 in response to glucagon. Sci. Rep. 2016;6:24156. doi: 10.1038/srep24156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.An D., Schneller J.L., Frassetto A., Liang S., Zhu X., Park J.S., Theisen M., Hong S.J., Zhou J., Rajendran R., et al. Systemic messenger RNA therapy as a treatment for methylmalonic acidemia. Cell Rep. 2017;21:3548–3558. doi: 10.1016/j.celrep.2017.11.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao J., Choi M., Guadagnin E., Soty M., Silva M., Verzieux V., Weisser E., Markel A., Zhuo J., Liang S., et al. mRNA therapy restores euglycemia and prevents liver tumors in murine model of glycogen storage disease. Nat. Commun. 2021;12:3090. doi: 10.1038/s41467-021-23318-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deng Y.Q., Zhang N.N., Zhang Y.F., Zhong X., Xu S., Qiu H.Y., Wang T.C., Zhao H., Zhou C., Zu S.L., et al. Lipid nanoparticle-encapsulated mRNA antibody provides long-term protection against SARS-CoV-2 in mice and hamsters. Cell Res. 2022;32:375–382. doi: 10.1038/s41422-022-00630-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grisolía S., Hernández-Yago J., Knecht E. Regulation of mitochondrial protein concentration: a plausible model which may permit assessing protein turnover. Curr. Top. Cell. Regul. 1985;27:387–396. doi: 10.1016/b978-0-12-152827-0.50040-2. [DOI] [PubMed] [Google Scholar]
- 45.Granados-Riveron J.T., Aquino-Jarquin G. Engineering of the current nucleoside-modified mRNA-LNP vaccines against SARS-CoV-2. Biomed. Pharmacother. 2021;142:111953. doi: 10.1016/j.biopha.2021.111953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dall'Acqua W.F., Kiener P.A., Wu H. Properties of human IgG1s engineered for enhanced binding to the neonatal Fc receptor (FcRn) J. Biol. Chem. 2006;281:23514–23524. doi: 10.1074/jbc.M604292200. [DOI] [PubMed] [Google Scholar]
- 47.Giessel A., Dousis A., Ravichandran K., Smith K., Sur S., McFadyen I., Zheng W., Licht S. Therapeutic enzyme engineering using a generative neural network. Sci. Rep. 2022;12:1536. doi: 10.1038/s41598-022-05195-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu H., Brewer E., Shields M., Crowder M., Sacchetti C., Soontornniyomkij B., Dou D., Clemente B., Sablad M., Chivukula P., et al. Restoring ornithine transcarbamylase (OTC) activity in an OTC-deficient mouse model using LUNAR-OTC mRNA. Clin. Transl. Discov. 2022;2:e33. [Google Scholar]
- 49.Gustafsson C., Govindarajan S., Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Walsh C., Ou K., Belliveau N.M., Leaver T.J., Wild A.W., Huft J., Lin P.J., Chen S., Leung A.K., Lee J.B., et al. Microfluidic-based manufacture of siRNA-lipid nanoparticles for therapeutic applications. Methods Mol. Biol. 2014;1141:109–120. doi: 10.1007/978-1-4939-0363-4_6. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Z., Shigematsu H., Shimizu T., Ohto U. Improving particle quality in cryo-EM analysis using a PEGylation method. Structure. 2021;29:1192–1199.e4. doi: 10.1016/j.str.2021.05.004. [DOI] [PubMed] [Google Scholar]
- 52.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kimanius D., Dong L., Sharov G., Nakane T., Scheres S.H.W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 2021;478:4169–4185. doi: 10.1042/BCJ20210708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rohou A., Grigorieff N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramlaul K., Palmer C.M., Nakane T., Aylett C.H.S. Mitigating local over-fitting during single particle reconstruction with SIDESPLITTER. J. Struct. Biol. 2020;211:107545. doi: 10.1016/j.jsb.2020.107545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zivanov J., Nakane T., Scheres S.H.W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hodges P.E., Rosenberg L.E. The spfash mouse: a missense mutation in the ornithine transcarbamylase gene also causes aberrant mRNA splicing. Proc. Natl. Acad. Sci. USA. 1989;86:4142–4146. doi: 10.1073/pnas.86.11.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pastra-Landis S.C., Foote J., Kantrowitz E.R. An improved colorimetric assay for aspartate and ornithine transcarbamylases. Anal. Biochem. 1981;118:358–363. doi: 10.1016/0003-2697(81)90594-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary material.