Abstract
Omicron variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a health concern for both unvaccinated and vaccinated individuals against coronavirus disease 2019 (COVID-19). To date, the humoral immune response following vaccination and natural infection remains uncharacterized in children ages 17 years and younger. To address this concern, we performed clinical and immunological analyses of IgM and IgG antibody responses to SARS-CoV-2 Omicron BA.2.38 infection in 64 pediatric patients. COVID-19 symptom severity decreased with age in pediatric patients, from 70.8% (17/24) in patients 0–2 years of age to 50% (6/12) and 50% (14/28) in patients 3–5 years and 6–17 years of age, respectively. Furthermore, fewer patients experienced symptoms when vaccinated with the CoronaVac or BBIBP-CorV vaccine (50%, 13/26) than unvaccinated patients (71%, 22/31). Using a protein array, we found that the Omicron BA.2.38 infection induced antibody responses to other Omicron variants (Omicron BA.1-BA.5), which increased with vaccination. Notably, non-Omicron and Omicron variants showed distinct serotypes. Altogether, our results provide insight into the clinical and immunological characteristics of pediatric patients with COVID-19 Omicron BA.2.38 who have and have not been vaccinated against COVID-19. These data may help develop more effective diagnostic tests and vaccines in the future.
Keywords: COVID-19, SARS-CoV-2, Variant of concern, Pediatric patients, Antibody response, Proteomics
Research in Context
-
•
Characterization of IgM and IgG antibody responses to SARS-CoV-2 Omicron BA.2.38 infection in pediatric patients
-
•
The CoronaVac or BBIBP-CorV vaccine mays help prevent COVID-19 symptoms in pediatric patients
-
•
Different serotypes were identified for non-Omicron and Omicron variants, which could aid drug development efforts to fight COVID-19
1. Introduction
As of April 5, 2023, severe respiratory coronavirus 2 (SARS-CoV-2) has infected 762 million people and caused over 6 million deaths worldwide [1]. SARS-CoV-2 mutates, resulting in new variants of concern (VOCs) that decrease the efficacy of COVID-19 vaccines and lead to large-scale breakthrough infection [2,3]. Among Omicron lineages (Omicron BA.1-BA.5 and their sub-lineages), the BA.2 was detected in China for the first time in January 2022 [4,5]. Six months later, the Omicron variant sub-lineage BA.2.38, which originated from India [6] was identified in the Gansu province and induced a new pandemic wave of COVID-19 in China [7]. Compared to adults, the morbidity in pediatric patients is relatively low. Gudbjartsson et al. [8] found that children under 10 had a lower incidence of SARS-CoV-2 infection than adolescents or adults.
Compared to the wild-type, Omicron variants carries a large number of mutations that enable the variant escape from the immune protection by the vaccination that was developed based on the wild-type strain [[9], [10], [11], [12]]. Vaccine-resistant Omicron variants threaten the health of both unvaccinated and fully vaccinated individuals, including children. In a study of 465 pediatric patients (aged ≤14 years) who were admitted to the hospital, Li et al. [5] found that children vaccinated against Omicron BA.2 were still susceptible to infection by the variant. Moreover, IgM and IgG antibody expression were dependent on the number of vaccination doses. The immune response to SARS-CoV-2 variants following COVID-19 vaccination and the resulting clinical characteristics in children remains unclear.
To address this concern, we performed a retrospective cohort study of pediatric patients infected with Omicron BA.2.38 who were or were not previously vaccinated with an inactivated whole-virion SARS-CoV-2 vaccine (Sinovac-CoronaVac or BBIBP-CorV). More specifically, we used a protein microarray with amino acid resolution developed in our laboratory [13,14] to profile antibody production by the humoral immune response in serum. The binding epitopes of the antibodies to the SARS-CoV-2 Spike (S) protein of nine VOC variants (Alpha, D614G, Beta, Delta, Omicron BA.1, Omicron BA.2, Omicron BA.3, Omicron BA.4, Omicron BA.5) were characterized. We also analyzed the association of antibody antigenicity in the context of COVID-19 vaccination and symptoms.
2. Materials and methods
2.1. Study design and patients
This prospective cohort study was approved by the Ethics Committee of Guangdong Provincial Hospital of Chinese Medicine before initiation. Patients under 18 years of age infected with SARS-CoV-2 Omicron sub-variant BA.2.38 in the No.2 People's Hospital of Lanzhou from July 28, 2022 to August 5, 2022 were recruited for this study after obtaining informed consent from their guardians. The EC approval number of this experiment is ZF2022-246-01.
Patients were clinically confirmed to have COVID-19 according to the Protocol for Diagnosis and Treatment of Novel Coronavirus Pneumonia (9th edition), where the presence of SARS-CoV-2 was determined by RT-PCR. Asymptomatic infections were defined as no positive clinical signs or symptoms in patients with a positive nucleic acid test result for SARS-CoV-2. Participants with COVID-19 and having symptoms of the infection were classified as symptomatic cases. Patients vaccinated against COVID-19 were vaccinated with two doses of the inactivated whole-virion SARS-CoV-2 vaccines, including the Sinovac-CoronaVac vaccine (Sinovac Biotech Co.,Ltd, China) or BBIBP-CorV vaccine (Lanzhou Institute of Biological Products Co.,Ltd., China). All participants were uninfected prior to this study. Following a routine examination, serum samples were collected and then stored frozen at −80 °C until the broad antibody testing via a protein array. The total serum sample size was 64 patients, with one sample per patient (Fig. 1). Samples with incomplete data, particularly the antibody findings, were excluded. The experiments were executed according to the Declaration of Helsinki.
Fig. 1.
Characterization of pediatric patients with Omicron BA.2.38 infection. (a) Workflow of pediatric patient characterization in this study; (b) The time intervals between positive clinical nucleic acid test and blood collection in asymptomatic and symptomatic patients; (c) The proportion of asymptomatic and symptomatic pediatric patients classified based on age and vaccination status, respectively; (d) Clinical variables with a significant difference between asymptomatic (Asy) and symptomatic (Sym) patient groups. Statistical analysis was performed using the Mann-Whitney U test, and significant differences are defined as *p-value <0.05, **p-value <0.01, ***p-value <0.001 and ****p-value <0.0001.
2.2. Data collection
Clinical information, including demographic data, vaccination, symptoms, signs, and laboratory results during hospitalization, was collected by reviewing patient records using an electronic medical record system. Laboratory records included complete blood counts, acute phase reactants (C-reactive protein, procalcitonin, serum amyloid, and interleukin-6), cycle threshold of SARS-CoV-2 (N-gene, ORF1ab), parameters of liver and kidney function (alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, gamma-glutamyltransferase, creatinine, urea), cardiac enzymes, and coagulation profile. To minimize high doses of radiation as much as possible, only some of the children received chest computerized tomography (CT) scans.
2.3. Analysis of broad antibody responses using a SARS-CoV-2 Spike protein variant array
The protein microarray displaying the Spike VOCs (Alpha, D614G, Beta, Delta, Omicron BA.1, Omicron BA.2, Omicron BA.3, Omicron BA.4, Omicron BA.5) (Sino Biological, Beijing, China) was prepared by ProteomicsEra Medical Co. (Beijing, China) as previously described [13]. The SARS-CoV-2 N (N) protein (Sino Biological, Beijing, China) and human IgG and IgM proteins (Jackson Immuno Research, Human IgG: cat no. 009-000-003; Human IgM: cat no. 009-000-012) served as positive controls while 1x phosphate-buffered saline (PBS) buffer served as the negative control. After printing, the microarray was manually examined, vacuum sealed, and stored at 4 °C until use.
For the experiment with serum samples, the microarray was blocked with 5% (w/v) milk in 1 × PBS with 0.05% (v/v) Tween-20 (PBST) for 10 min at 25 °C. After washing three times with PBST, the microarray was incubated with serum (1:100) and a mixture of antibodies [Cy™3 AffiniPure Donkey Anti-Human IgG (H + L) (4 μg/mL) (Jackson Immuno Research, cat no. 709-165-149) and Alexa Fluor® 647 AffiniPure Goat Anti-Human IgM (4 μg/mL) (Jackson Immuno Research, cat no. 109-605-043)] for 30 min. The resulting array was scanned with a GenePix 4300A microarray scanner (Molecular Devices, LLC, San Jose, CA, USA). The median fluorescent signal intensity (MFI) was extracted using GenePix Pro7 software (Molecular Devices, USA). The average signal across triplicate spots was calculated and then normalized using the signal-to-noise ratio (SNR), which was the average signal divided by the 25th percentile of all negative control spots as the background.
To compare the antibody responses to different SARS-CoV-2 variants, the fluorescent signal intensity of each VOC was further normalized by dividing the signal intensity by the average signal of all variant proteins. The expression of VOCs on the microarray was determined using an anti-his-tag mouse monoclonal antibody (0.1 μg/mL) (CWBio, cat no. CW0286).
Yv is the normalized signal of a variant protein.
Fv is the normalization factor, which is the normalized fluorescent signal of a variant protein divided by the average fluorescent signal of all variants.
2.4. Data analysis
Continuous variables with a normal distribution were expressed as a mean and standard deviation (SD), while continuous variables with a non-normal distribution were expressed as a median and interquartile range (IQR). The distributions were compared using the Student's t-test for parametric data and the Mann-Whitney U test for non-parametric data. Categorical variables were described as percentages. Categorical variables were compared using the χ2 test or, if the categorical variables had expected frequencies of less than five, Fisher's exact test. A two-sided p-value less than 0.05 was considered significant. All statistical analyses were done using R Version 4.1.1 statistical software.
3. Results
3.1. Demographic characteristics of pediatric patients with Omicron BA.2.38 infection
Sixty-four (64) children infected with Omicron BA.2.38 were split into three groups: 0–2 years (24 patients), 3–5 years (12 patients), and 6–17 years (28 patients) (Table 1) [15]. The data analysis workflow is shown in Fig. 1a, with the employed samples shown in Fig. 1b.
Table 1.
Basic characteristics of pediatric patients infected with SARS-CoV-2 Omicron BA.2.38
| COVID-19 Symptoms |
Total | p-value | ||||
|---|---|---|---|---|---|---|
| Asymptomatic | Symptomatic | |||||
| Age (years) | 0–2 | 7 (25.9) | 17 (45.9) | 24 | 2.669 | 0.255 |
| 3–5 | 6 (22.2) | 6 (16.2) | 12 | |||
| 6–17 | 14 (51.9) | 14 (37.8) | 28 | |||
| Sex | male | 13 (48.1) | 18 (48.6) | 31 | 0.002 | 1.000 |
| female | 14 (51.9) | 19 (51.4) | 33 | |||
| Vaccination | No | 9 (40.9) | 22 (62.9) | 31 | – | 0.172 |
| Yes | 13 (59.1) | 13 (37.1) | 26 | |||
* Continuous variables with a normal distribution were expressed as a mean and standard deviation (SD), while continuous variables with a non-normal distribution were expressed as a median and interquartile range (IQR). The distributions were compared using the Student's t-test for parametric data and the Mann-Whitney U test for non-parametric data. Categorical variables were compared using the χ2 test or, if the categorical variables had expected frequencies of less than five, Fisher's exact test. A two-sided p-value of less than 0.05 was considered significant. All statistical analyses were done using R Version 4.1.1 statistical software.
COVID-19 symptoms ranged from asymptomatic [42.2% (27/64)] to symptomatic [57.8% (37/64)] (Fig. 1c, Table 1). The percentage of asymptomatic patients increased with age, from 29.2% (7/24, 0–2 years) to 50% (6/12, 3–5 years; 14/28, 6–17 years).
To better understand the influence of vaccination on the immune response's ability to protect against natural SARS-CoV-2 infection, the percentage of asymptomatic and symptomatic patients who were vaccinated was compared. The average percentage of asymptomatic patients was 29% (9/31) in the unvaccinated group, whereas the average percentage of symptomatic patients was 71% (22/31) (Fig. 1c, Table 1). The average percentage of symptomatic patients dropped to 50% (13/26) in the vaccinated group, indicating that vaccination may potentially help protect against COVID-19 progression following natural infection. No statistically significant differences were identified by age group due to the limited size of patients identified in this study.
3.2. Clinical assessment of pediatric patients with Omicron BA.2.38 infection
The common clinical manifestations are fever, rhinorrhoea, cough, cough-up phlegm and headache. Upon hospital admission, all pediatric patients were examined systematically in terms of viral infection, blood count, blood biochemistry, coagulation function, and liver and renal function (Table 2) according to the COVID-19 Treatment Guidelines (9th edition) from the National Health Commission of China. The antibody expression to SARS-CoV-2 ORF1ab and N proteins did not differ between asymptomatic and symptomatic patient groups (Table 2).
Table 2.
Clinical testing of pediatric patients with SARS-CoV-2 Omicron BA.2.38 infection.
| Index | Classification | n | M(Q) | Mann-Whitney U test | p-value |
|---|---|---|---|---|---|
| ALT first timea | Asymptomatic | 26 | 14.0 (11, 23) | 332 | 0.052 |
| Symptomatic | 36 | 19.0 (14, 28) | |||
| AST first time | Asymptomatic | 26 | 25.0 (21, 34) | 301 | 0.017 |
| Symptomatic | 36 | 34.0 (26, 43) | |||
| CRP first time | Asymptomatic | 23 | 0.0 (0, 1) | 184 | 0.004 |
| Symptomatic | 30 | 3.0 (1, 9) | |||
| SARS-CoV-2 N-gene first time | Asymptomatic | 10 | 27.0 (22.2, 31.5) | 135 | 0.32 |
| Symptomatic | 22 | 25.0 (20.2, 28.0) | |||
| SARS-CoV-2 ORF1ab first time | Asymptomatic | 10 | 26.0 (22.0, 30.5) | 134 | 0.33 |
| Symptomatic | 22 | 24.0 (19.2, 27.8) | |||
| D2 dimer first time | Asymptomatic | 25 | 0.3 (0.22, 0.39) | 322 | 0.12 |
| Symptomatic | 34 | 0.35 (0.28, 0.52) | |||
| WBC first time | Asymptomatic | 26 | 6.5 (5.40, 7.80) | 454 | >0.99 |
| Symptomatic | 35 | 7.5 (4.80, 9.70) | |||
| γ-Glutamyl transferase first time | Asymptomatic | 24 | 14.0 (13, 16) | 240 | 0.32 |
| Symptomatic | 24 | 15.0 (12, 21) | |||
| Neutrophils first time | Asymptomatic | 26 | 2.95 (2.20, 3.98) | 438 | 0.81 |
| Symptomatic | 35 | 2.9 (2.30, 3.95) | |||
| Lactate dehydrogenase first time | Asymptomatic | 26 | 229.0 (204, 308) | 380 | 0.27 |
| Symptomatic | 35 | 263.0 (217, 319) | |||
| Urea nitrogen first time | Asymptomatic | 26 | 4.9 (3.90, 5.45) | 487 | 0.65 |
| Symptomatic | 35 | 4.4 (3.55, 5.50) | |||
| Leukomonocyte first time | Asymptomatic | 26 | 2.5 (2.00, 4.35) | 489 | 0.62 |
| Symptomatic | 35 | 2.5 (1.35, 5.20) | |||
| Interleukin 6 first time | Asymptomatic | 22 | 4.1 (2.23, 4.66) | 207 | 0.034 |
| Symptomatic | 29 | 5.42 (3.01, 7.87) | |||
| Alkaline phosphatase first time | Asymptomatic | 24 | 224.0 (197, 267) | 272 | 0.75 |
| Symptomatic | 24 | 231.0 (198, 267) | |||
| Fibrinogen first time | Asymptomatic | 25 | 2.09 (1.88, 2.31) | 302 | 0.061 |
| Symptomatic | 34 | 2.28 (1.99, 2.75) | |||
| Creatinine first time | Asymptomatic | 26 | 38.0 (32, 42) | 478 | 0.74 |
| Symptomatic | 35 | 36.0 (29, 49) | |||
| Creatine kinase isoenzyme MB first time | Asymptomatic | 26 | 18.0 (14, 22) | 384 | 0.3 |
| Symptomatic | 35 | 20.0 (16, 30) | |||
| Creatine kinase first time | Asymptomatic | 26 | 74.0 (57, 108) | 428 | 0.7 |
| Symptomatic | 35 | 89.0 (55, 111) | |||
| Platelet first time | Asymptomatic | 26 | 270.0 (230, 335) | 492 | 0.6 |
| Symptomatic | 35 | 260.0 (197, 312) | |||
| Serum amyloid A measure first time | Asymptomatic | 19 | 18.0 (1, 27) | 233 | 0.22 |
| Symptomatic | 31 | 24.0 (16, 29) | |||
| Hemoglobin first time | Asymptomatic | 26 | 127.0 (122, 134) | 489 | 0.62 |
| Symptomatic | 35 | 127.0 (120, 132) | |||
| Procalcitonin first time | Asymptomatic | 22 | 0.08 (0.07, 0.10) | 221 | 0.064 |
| Symptomatic | 29 | 0.1 (0.08, 0.18) |
First laboratory test results after admission.
We found that the level of aspartate aminotransferase (AST), a marker of liver damage, was significantly lower in vaccinated patients than in unvaccinated patients regardless of symptoms (Mann-Whitney U test, p < 0.05) (Fig. 1d). Two proteins related to inflammation, C-reactive protein (CRP) and interleukin-6 (IL-6), were also measured. While vaccination did not affect CRP and IL-6 protein expression, their protein levels were significantly higher in symptomatic patients than in asymptomatic patients (Fig. 1d). These results suggest that CRP and IL-6 are potential biomarkers of asymptomatic to the symptomatic progression of COVID-19 in pediatric patients.
3.3. Humoral responses to SARS-CoV-2 variants in pediatric patients with and without vaccination
The humoral response to the Spike protein of different SARS-CoV-2 VOCs (Alpha, D614G, Beta, Delta, Omicron BA.1-BA.5) was detected using a protein array that was fabricated as previously described [13,14] (Supplementary Fig. S1). The correlations (r) of antibody detection within an array and between different arrays resulted were 0.9902 and 0.9464, respectively (Supplementary Fig. S2). To determine the potential influence of the sampling time on the antibody results [16], we compared the difference in sampling time between unvaccinated and vaccinated groups as well as between asymptomatic and symptomatic groups. No statistical differences were found, indicating that the array data obtained in this work was not associated with when the clinical samples were obtained (Supplementary Table S1).
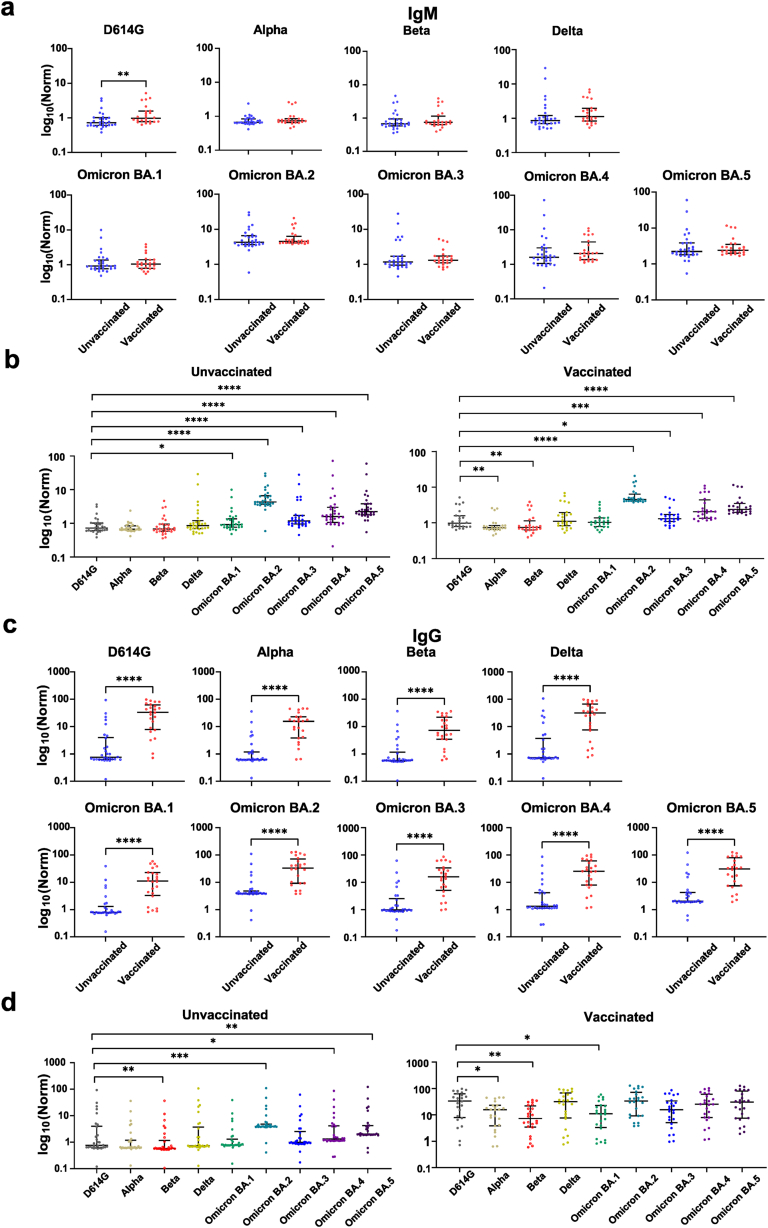
The production of IgM antibodies by the humoral immune response was first analyzed with the protein array. No difference between unvaccinated and vaccinated groups to eight variants (Alpha, Beta, Delta, Omicron BA.1-BA.5) was observed. However, IgM levels to the D614G variant were upregulated in vaccinated patients compared to unvaccinated patients (p < 0.05) (Fig. 2a). For both the vaccinated and unvaccinated groups, IgM antibodies showed the highest responses to Omicron BA.2, followed by BA.5, BA.4, BA.3, BA.1, Delta, D614G, Alpha, and Beta in the vaccinated group (Fig. 2b).
Fig. 2.
Humoral responses to SARS-CoV-2 variants in pediatric patients with and without vaccination. (a, c) IgM and IgG antibody responses to SARS-CoV-2 variants in unvaccinated and vaccinated pediatric patients, respectively. (b, d) Differential expression of IgM and IgG antibodies to the non-Omicron and Omicron variants, respectively. Statistical analysis was performed using the Mann-Whitney U test, and significant differences are defined as *p-value <0.05, **p-value <0.01, ***p-value <0.001 and ****p-value <0.0001.
Next, IgG levels were measured. Compared to the IgM data, IgG antibody levels to all VOCs were significantly higher in vaccinated patients than in unvaccinated patients (Fig. 2c). Notably, IgG antibody responses in vaccinated patients were similar to IgM antibody responses, with antibody expression to Omicron BA.2 being the highest, followed by BA.5, BA.4, BA.3, BA.1, Delta, D614G, Alpha, and Beta (Fig. 2d). Vaccinated patients also had high IgG antibody responses to most variants, with no significant differences between Omicron and non-Omicron variants (Fig. 2d). It is possible that the high IgG antibody response was the result of vaccination and BA 2.38 infection (Fig. 2c).
3.4. Broad humoral responses to the SARS-CoV-2 variants in asymptomatic and symptomatic pediatric patients
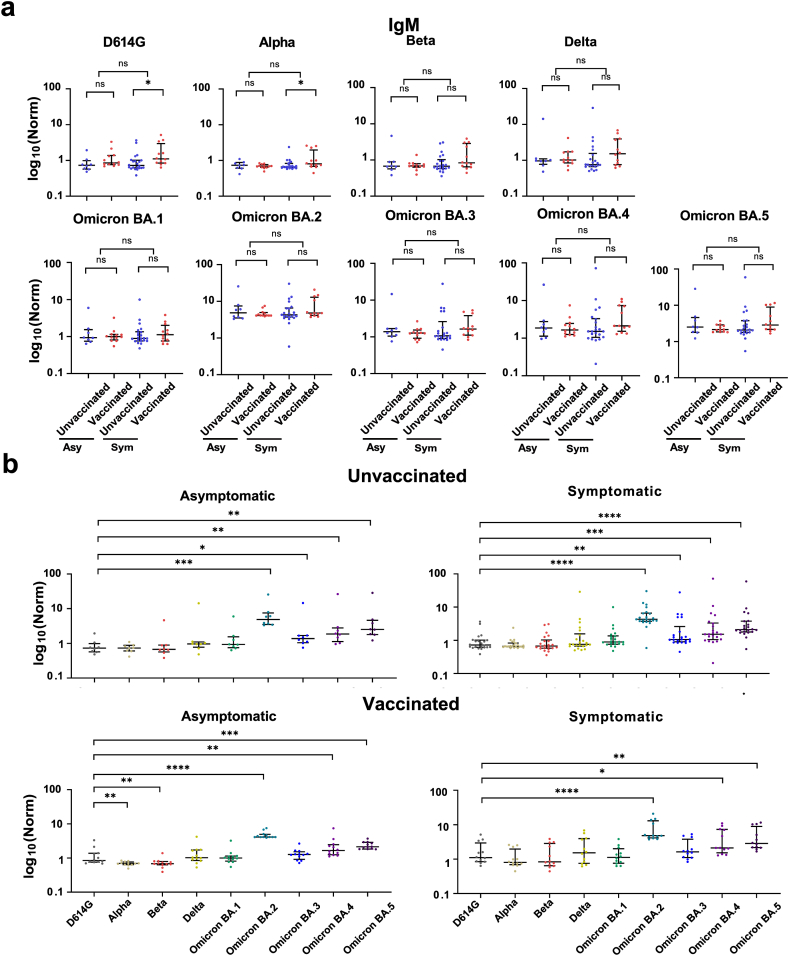
The broad humoral responses to the SARS-CoV-2 variants were analyzed. No significant difference in IgM levels was found between the asymptomatic and symptomatic patient groups (Fig. 3). In symptomatic patients, higher IgM binding to the D614G and Alpha variants was observed in vaccinated patients than in unvaccinated patients (Fig. 3a). Patients in both asymptomatic and symptomatic groups had high IgM levels to the Omicron BA.2 variant, followed by the BA.5, BA.4, BA.3, BA.1, Delta, D614G, Beta, and Alpha (Fig. 3b).
Fig. 3.
IgM antibody responses to the SARS-CoV-2 variants in asymptomatic and symptomatic pediatric patients. (a) IgM antibody responses to SARS-CoV-2 variants in asymptomatic (Asy) and symptomatic (Sym) pediatric patients with and without vaccination. (b) Differential expression of IgM antibodies to the non-Omicron and Omicron variants, respectively. Statistical analysis was performed using the Mann-Whitney U test, and significant differences are defined as *p-value <0.05, **p-value <0.01, ***p-value <0.001 and ****p-value <0.0001.
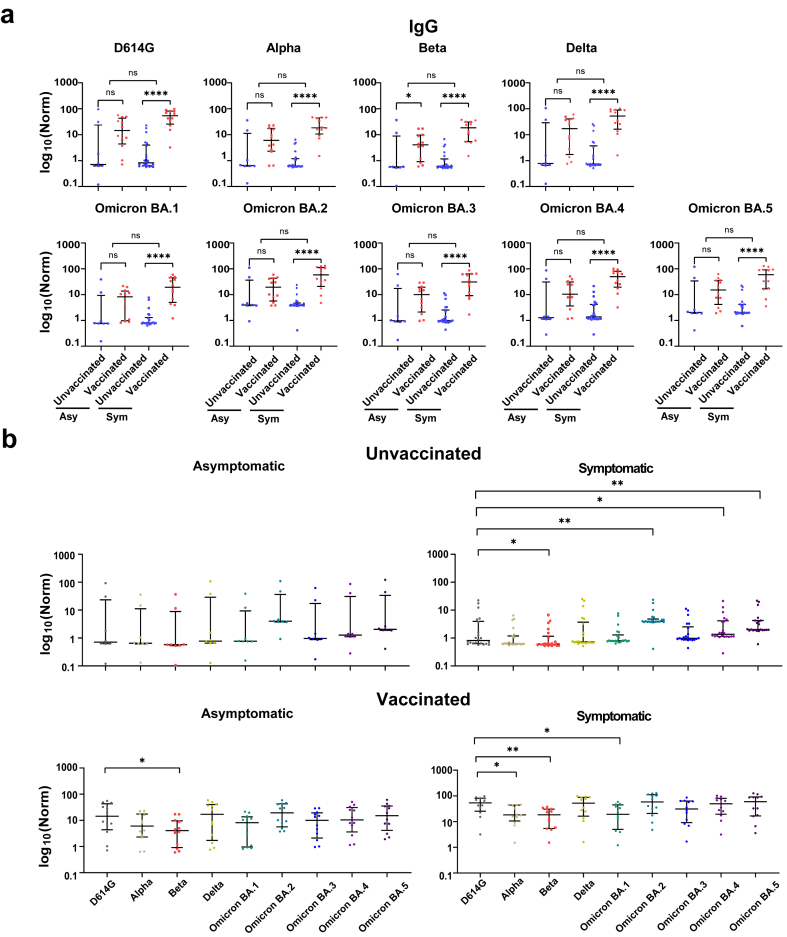
Like IgM, IgG levels to SARS-CoV-2 variants did not change between asymptomatic and symptomatic groups. However, vaccinated patients did have higher IgG levels than unvaccinated patients for the Beta variant, and this difference was more pronounced in the symptomatic group (p < 0.0001) than in the asymptomatic group (p < 0.05) (Fig. 4a).
Fig. 4.
IgG antibody responses to the SARS-CoV-2 variants in asymptomatic and symptomatic pediatric patients. (a) IgG antibody responses to SARS-CoV-2 variants in asymptomatic (Asy) and symptomatic (Sym) pediatric patients with and without vaccination. (b) Differential expression of IgM antibodies to the non-Omicron and Omicron variants, respectively. Statistical analysis was performed using the Mann-Whitney U test, and significant differences are defined as *p-value <0.05, **p-value <0.01, ***p-value <0.001 and ****p-value <0.0001.
The IgG expression of different SARS-CoV-2 variants was compared across patient groups based on vaccination and symptom status (Fig. 4b). For the vaccinated, asymptomatic group, there was a minor but significant difference (p < 0.05) in IgG levels between D614G and Beta. In the vaccinated and symptomatic group, IgG levels to D614G were higher than Alpha, Beta, and Omicron BA.1.
3.5. Proteomic analysis identifies different serotypes between non-Omicron and Omicron variants in pediatric patients
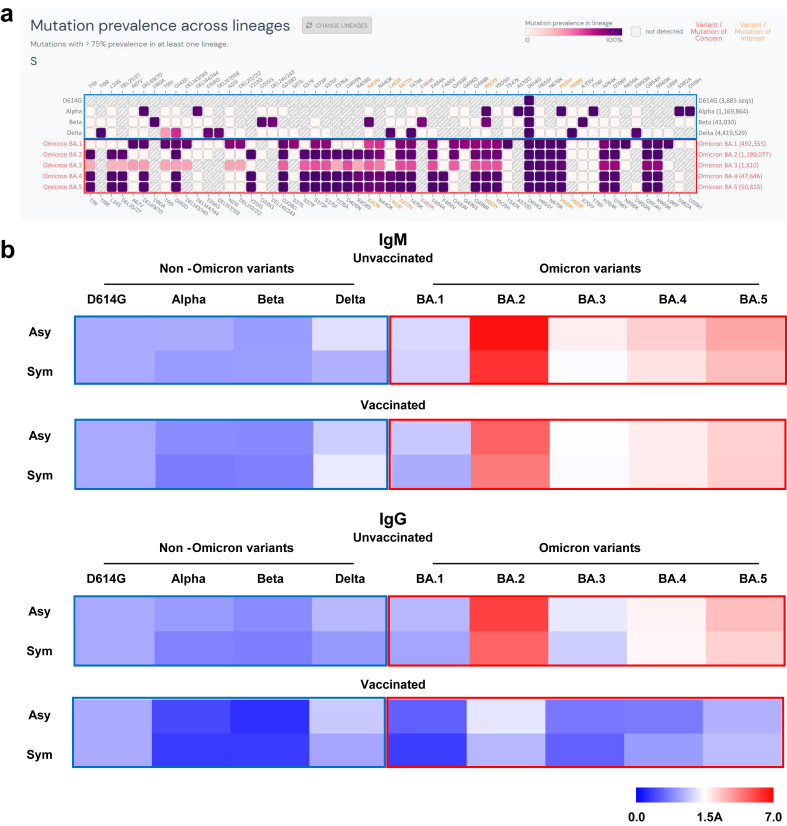
Accumulated evidence in previous studies has indicated that antibodies to Alpha and D614G assist the immune response in resisting infection with SARS-CoV-2 Omicron variants. In a recent review, Simon-Loriere and Schwartz [17] proposed that the humoral response to Omicron and non-Omicron variants should be classified into different serotypes based on their sequences (Fig. 5a, Supplementary Fig. S3) as well as results from previous studies. However, these previous studies used adult patients. The humoral response of pediatric patients to Omicron (BA.1-BA.5) and non-Omicron variants has not been documented to date.
Fig. 5.
Different serotypes between Omicron and non-Omicron variants were identified in pediatric patients. (a) Sequence homology analysis of the SARS-CoV-2 Spike protein in non-Omicron and Omicron variants. (b) Heatmap illustration of different serotypes between non-Omicron and Omicron variants in pediatric patients.
Here, the relative changes of antibodies to SARS-CoV-2 variants in pediatric patients were measured using D614G as the reference (Fig. 5b). D614G was selected as the reference because it was the first SARS-CoV-2 variant carrying a point mutation in the Spike protein that rapidly surpassed the Alpha strain in prevalence [18]. Independent of vaccination, IgM antibodies that were produced in response to infection with Omicron BA2.38 targeted more Omicron variant epitopes than non-Omicron variants. Similar results were obtained with IgG antibodies in patients who were not vaccinated. In the vaccinated group, little difference was observed between IgG Omicron BA.1-BA.5 and D614G antibody expression. These data suggest that IgM antibodies could act as candidate biomarkers to distinguish Omicron BA2.38 infection from vaccination. Furthermore, IgG antibody production initially stimulated by vaccination to non-Omicron variants might be enhanced by Omicron BA2.38 infection (Fig. 2, Fig. 4).
4. Discussion
The SARS-CoV-2 Omicron BA.2.38 variant recently received attention following a surge of COVID-19 infections in China in 2022. However, little evidence has been collected regarding the clinical and immunological characteristics of pediatric patients (<18 years of age) to SARS-CoV-2 variants. In this work, we analyzed the clinical characteristics and IgM and IgG humoral responses in pediatric patients infected with Omicron BA.2.38 to different SARS-CoV-2 variants using array-based proteomics technology.
First, the number of children with SARS-CoV-2 Omicron BA.2.38 presenting with asymptomatic infection increased with age (Fig. 1c, Table 1), with 29.2% (7/24) of patients 0–2 years of age and 50% (20/40) of patients 3–17 years of age being asymptomatic. One possible explanation for these results is that young children (2 years of age and younger) may not be able to clear the virus quickly due to their underdeveloped immunity. Indeed, higher viral loads have been measured in young children (<6 months in age) than in children older than 6 months [19].
Second, we found that symptomatic patients were more common in unvaccinated patients (71%, 22/31) than in vaccinated patients (50%, 13/26). The results indicate that vaccination with the inactivated whole-virion SARS-CoV-2 vaccine (Sinovac-CoronaVac or BBIBP-CorV) induces the production of antibodies that recognize different Omicron and non-Omicron variants (Fig. 2, Fig. 4), which may protect BA.2.38 infected patients from disease progression. This finding agrees with a previous study of adults in which two doses of the CoronaVac or BBIBP-CorV vaccine prevented symptomatic COVID-19 [20]. In addition, we found that the IgM levels to the D614G were upregulated in vaccinated patients (Fig. 2a). The reason might be due to the homology of protein sequences between D614G and Omicron variants [21,22]. In Fig. 3a, it can be observed that the level of IgM antibodies was higher in symptomatic patients than symptomatic patients as well as in vaccinated patients than unvaccinated patients. The results can be explained by the strengthening of immunity through vaccination and following infection [23,24]. Like IgM, IgG levels to SARS-CoV-2 variants did not change between asymptomatic and symptomatic groups. However, vaccinated patients did have higher IgG levels than unvaccinated patients for the Beta variant, and this difference was more pronounced in the symptomatic group (p < 0.0001) than in the asymptomatic group (p < 0.05) (Fig. 4a). The reason might be caused by the higher expression of IgG antibodies in symptomatic patients compared to asymptomatic patients [25]. However, the mechanism is unknown, which remains to be investigated in future.
Third, we collected data characterizing the binding of antibodies produced by unvaccinated patients following Omicron BA.2.38 infection to the prevalent VOC variants (Alpha, Beta, Delta, Omicron BA.1, BA.2, BA.3, BA.4, BA.5). The antibodies cross-reacted with the other Omicron variants (Fig. 2, Fig. 3, Fig. 4), but had low cross-reactivity to non-Omicron variants in some patients (Fig. 5b). The results support the perspective raised by Simon-Loriere E. and Schwartz O [17]. and suggest that the effectiveness of antibody and neutralization antibody tests, which were developed based on the early variants, is reduced with the newly evolved Omicron variants. Therefore, it is necessary to develop new tests for different variants. One option for testing is a protein array like the one used here, which can analyze SARS-CoV-2 variants simultaneously [[26], [27], [28]].
There are several limitations to this study. First, the number of clinical samples from pediatric patients was limited due to the low availability. Second, we did not analyze the antibodies’ ability to neutralize infection with SARS-CoV-2 VOCs. However, numerous studies have demonstrated that there is a correlation between antibodies to the S protein and neutralizing capability [29]. In addition, only the CoronaVac or BBIBP-CorV vaccine was studied in this work.
This study provides clinical and broad immunological characteristics of pediatric patients with Omicron BA.2.38 infection who have and have not been vaccinated with the CoronaVac or BBIBP-CorV vaccine. The data underscore the potential value of vaccination in reducing COVID-19 symptoms and may help to develop more effective diagnostic tests and vaccines against COVID-19.
Author contribution statement
Xiaobo Yu: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Yu Liu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Liunuobei Zhao: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Li Wang: Performed the experiments; Wrote the paper.
Yuxia Li; Longde Wang; Bo Yu: Performed the experiments.
Di Hu: Contributed reagents, materials, analysis tools or data.
Heng Weng: Analyzed and interpreted the data.
Jianwen Guo; Jinghua Yang; Jing Yang: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Beijing Municipal Natural Science Foundation (M21003 and M23010), National Key R&D Program of China (2022YFE0210400, 2021YFA1301604, 2020YFE0202200), State Key Laboratory of Proteomics (SKLP-O202007), Guangdong Province Science and Technology Planning Project (2020B1111100006), Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine. (No: ZYYCXTD-C-202204). We thank Dr. Brianne Petritis for her critical review and editing of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e18093.
Contributor Information
Jianwen Guo, Email: jianwen_guo@qq.com.
Jinghua Yang, Email: doumiaomama@126.com.
Jing Yang, Email: yangjing54@hotmail.com.
Xiaobo Yu, Email: xiaobo.yu@hotmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A., Gayed S., Jahun A., Visualization Johnson Rob 82 SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592(7853):277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng V.C.C., Ip J.D., Chu A.W.H., Tam A.R., Chan W.M., Abdullah S.M.U., Chan B.P.C., Wong S.C., Kwan M.Y.W., Chua G.T., Ip P., Chan J.M.C., Lam B.H.S., To W.K., Chuang V.W.M., Yuen K.Y., Hung I.F.N., To K.K.W. Rapid spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) omicron subvariant BA.2 in a single-source community outbreak. Clin. Infect. Dis. 2022;75:e44–e49. doi: 10.1093/cid/ciac203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li X., Wu L., Qu Y., Cao M., Feng J., Huang H., Liu Y., Lu H., Liu Q., Liu Y. Clinical characteristics and vaccine effectiveness against SARS-CoV-2 Omicron subvariant BA.2 in the children. Signal Transduct. Targeted Ther. 2022;7:203. doi: 10.1038/s41392-022-01023-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh P., Sharma K., Shaw D., Bhargava A., Negi S.S. Mutational characterization of Omicron SARS-CoV-2 lineages circulating in Chhattisgarh, a central state of India. Front. Med. 2022;9 doi: 10.3389/fmed.2022.1082846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao Y., Song W., Wang L., Liu P., Yue C., Jian F., Yu Y., Yisimayi A., Wang P., Wang Y., Zhu Q., Deng J., Fu W., Yu L., Zhang N., Wang J., Xiao T., An R., Wang J., Liu L., Yang S., Niu X., Gu Q., Shao F., Hao X., Jin R., Wang Y., Xie X.S., Wang X. vol. 2. 2022. (Characterizations of Enhanced Infectivity and Antibody Evasion of Omicron BA). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson D.F., Helgason A., Jonsson H., Magnusson O.T., Melsted P., Norddahl G.L.…Stefansson K. Spread of SARS-CoV-2 in the Icelandic population. N. Engl. J. Med. 2020;382(24):2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noor R., Shareen S., Billah M. COVID-19 vaccines: their effectiveness against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its emerging variants. Bull. Natl. Res. Cent. 2022;46(1):96. doi: 10.1186/s42269-022-00787-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shrestha L.B., Foster C., Rawlinson W., Tedla N., Bull R.A. Evolution of the SARS‐CoV‐2 omicron variants BA. 1 to BA. 5: implications for immune escape and transmission. Rev. Med. Virol. 2022;32(5) doi: 10.1002/rmv.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flemming A. Omicron, the great escape artist. Nat. Rev. Immunol. 2022;22(2) doi: 10.1038/s41577-022-00676-6. 75-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J.…Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602(7898):671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 13.Wang H., Wu X., Zhang X., Hou X., Liang T., Wang D., Teng F., Dai J., Duan H., Guo S., Li Y., Yu X. SARS-CoV-2 proteome microarray for mapping COVID-19 antibody interactions at amino acid resolution. ACS Cent. Sci. 2020;6:2238–2249. doi: 10.1021/acscentsci.0c00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X., Zheng M., Liang T., Zhou H., Wang H., Zhang J., Ren J., Peng H., Li S., Bian H., Wei C., Yin S., He C., Han Y., Li M., Hou X., Zhang J., Xie L., Lv J., Kan B., Wang Y., Yu X. medRxiv; 2021. Inhibitor Screening of Spike Variants Reveals the Heterogeneity of Neutralizing Antibodies to COVID-19 Infection and Vaccination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams K., Thomson D., Seto I., Contopoulos-Ioannidis D.G., Ioannidis J.P., Curtis S., Constantin E., Batmanabane G., Hartling L., Klassen T., Sta R.C.H.G. Standard 6: age groups for pediatric trials. Pediatrics. 2012;129(Suppl 3):S153–S160. doi: 10.1542/peds.2012-0055I. [DOI] [PubMed] [Google Scholar]
- 16.Cheng L., Zhang X., Chen Y., Wang D., Zhang D., Yan S., Wang H., Xiao M., Liang T., Li H., Xu M., Hou X., Dai J., Wu X., Li M., Lu M., Wu D., Tian R., Zhao J., Zhang Y., Cao W., Wang J., Yan X., Zhou X., Liu Z., Xu Y., He F., Li Y., Yu X., Zhang S. Dynamic landscape mapping of humoral immunity to SARS-CoV-2 identifies non-structural protein antibodies associated with the survival of critical COVID-19 patients. Signal Transduct. Targeted Ther. 2021;6:304. doi: 10.1038/s41392-021-00718-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon-Loriere E., Schwartz O. Towards SARS-CoV-2 serotypes? Nat. Rev. Microbiol. 2022;20:187–188. doi: 10.1038/s41579-022-00708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O'Toole A., Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Consortium C.-U., Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Evaluating the effects of SARS-CoV-2 Spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75. doi: 10.1016/j.cell.2020.11.020. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochoa V., Diaz F.E., Ramirez E., Fentini M.C., Carobene M., Geffner J., Arruvito L., Remes Lenicov F., Group I.C.-S. Infants younger than 6 Months infected with SARS-CoV-2 show the highest respiratory viral loads. J. Infect. Dis. 2022;225:392–395. doi: 10.1093/infdis/jiab577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin L., Li Z., Zhang X., Li J., Zhu F. CoronaVac: a review of efficacy, safety, and immunogenicity of the inactivated vaccine against SARS-CoV-2. Hum. Vaccines Immunother. 2022 doi: 10.1080/21645515.2022.2096970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng S.M., Mok C.K.P., Chan K.C., Ng S.S., Lam B.H., Luk L.L., Peiris M. SARS-CoV-2 Omicron variant BA. 2 neutralisation in sera of people with Comirnaty or CoronaVac vaccination, infection or breakthrough infection, Hong Kong, 2020 to 2022. Euro Surveill. 2022;27(18) doi: 10.2807/1560-7917.ES.2022.27.18.2200178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavot V., Berry C., Kishko M., Anosova N.G., Li L., Tibbitts T.…Lecouturier V. Beta variant COVID-19 protein booster vaccine elicits durable cross-neutralization against SARS-CoV-2 variants in non-human primates. Nat. Commun. 2023;14(1):1309. doi: 10.1038/s41467-023-36908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmed T.I., Rishi S., Irshad S., Aggarwal J., Happa K., Mansoor S. Inactivated vaccine Covaxin/BBV152: a systematic review. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.863162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLean G., Kamil J., Lee B., Moore P., Schulz T.F., Muik A., Pather S. The impact of evolving SARS-CoV-2 Mutations and variants on COVID-19 vaccines. mBio. 2022;13(2) doi: 10.1128/mbio.02979-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dingemans J., van der Veer B.M., Gorgels K.M., Hackert V., Hensels A.Y., den Heijer C.D., van Alphen L.B. Investigating SARS-CoV-2 breakthrough infections per variant and vaccine type. medRxiv. 2021 doi: 10.3389/fmicb.2022.1027271. 2021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang H., Li Y., Zhang H., Wang W., Men D., Yang X., Qi H., Zhou J., Tao S. Global profiling of SARS-CoV-2 specific IgG/IgM responses of convalescents using a proteome microarray. medRxiv. 2020 doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., Zheng M., Wang H., Zhou H., Liang T., Zhang J., Ren J., Peng H., Li S., Bian H., Wei C., Yin S., He C., Han Y., Li M., Hou X., Zhang J., Xie L., Lv J., Kan B., Wang Y., Yu X. Inhibitor screening using microarray identifies the high capacity of neutralizing antibodies to Spike variants in SARS-CoV-2 infection and vaccination. Theranostics. 2022;12:2519–2534. doi: 10.7150/thno.67038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Becker M., Strengert M., Junker D., Kaiser P.D., Kerrinnes T., Traenkle B., Dinter H., Haring J., Ghozzi S., Zeck A., Weise F., Peter A., Horber S., Fink S., Ruoff F., Dulovic A., Bakchoul T., Baillot A., Lohse S., Cornberg M., Illig T., Gottlieb J., Smola S., Karch A., Berger K., Rammensee H.G., Schenke-Layland K., Nelde A., Marklin M., Heitmann J.S., Walz J.S., Templin M., Joos T.O., Rothbauer U., Krause G., Schneiderhan-Marra N. Exploring beyond clinical routine SARS-CoV-2 serology using MultiCoV-Ab to evaluate endemic coronavirus cross-reactivity. Nat. Commun. 2021;12:1152. doi: 10.1038/s41467-021-20973-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J., Teng F., Zhang X., Wang H., Liang T., Guo S., Yu X. Down-regulation of SARS-CoV-2 neutralizing antibodies in vaccinated smokers. MedComm. 2022;3:e166. doi: 10.1002/mco2.166. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supplementary material/referenced in article.