Abstract
The NADPH oxidase NOX4 has been proposed as necessary for the apoptosis induced by the Transforming Growth Factor-beta (TGF-β) in hepatocytes and hepatocellular carcinoma (HCC) cells. However, whether NOX4 is required for TGF-β-induced canonical (SMADs) or non-canonical signals is not fully understood yet, neither its potential involvement in other parallel actions induced by TGF-β. In this work we have used CRISPR Cas9 technology to stable attenuate NOX4 expression in HCC cells. Results have indicated that NOX4 is required for an efficient SMAD2/3 phosphorylation in response to TGF-β, whereas non-canonical signals, such as the phosphorylation of the Epidermal Growth Receptor or AKT, are higher in NOX4 silenced cells. TGF-β-mediated inhibition of cell proliferation and viability is attenuated in NOX4 silenced cells, correlating with decreased response in terms of apoptosis, and maintenance of high expression of MYC and CYCLIN D1. These results would indicate that NOX4 is required for all the tumor suppressor actions of TGF-β in HCC. However, analysis in human HCC tumors has revealed a worse prognosis for patients showing high expression of TGF-β1-related genes concomitant with high expression of NOX4. Deepening into other tumorigenic actions of TGF-β that may contribute to tumor progression, we found that NOX4 is also required for TGF-β-induced migratory effects. The Epithelial-Mesenchymal transition (EMT) program does not appear to be affected by attenuation of NOX4 levels. However, TGF-β-mediated regulation of cytoskeleton dynamics and focal adhesions require NOX4, which is necessary for TGF-β-induced increase in the chaperone Hsp27 and correct subcellular localization of Hic-5 within focal adhesions, as well for upregulation of the metalloprotease MMP9. All these results together point to NOX4 as a key element in the whole TGF-β signaling in HCC cells, revealing an unknown role for NOX4 as tumor promoter in HCC patients presenting activation of the TGF-β pathway.
Keywords: NADPH oxidase, NOX4, TGF-Beta, Hepatocellular carcinoma, HCC, Liver cancer
Graphical abstract
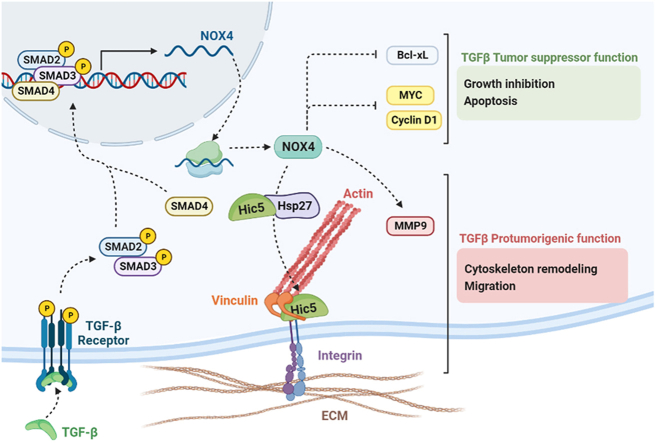
NOX4 mediates the Tumor suppressor actions of TGF-β by regulating Bcl-xL, C-Myc and Cyclin D1. However, NOX4 is also required for regulation of cytoskeleton dynamics and focal adhesions by TGF-β, correlating with changes in expression of Hsp27, Hic-5 and MMP9, which contribute to the migratory capacity of HCC cells. Altogether, NOX4 could participate in both suppressor and protumorigenic TGF-β actions in HCC cells.
Highlights
-
•
NOX4 mediates regulation of growth inhibition and apoptosis in response to TGF-β.
-
•
NOX4 is also required for TGF-β-induced cytoskeleton remodeling and cell migration.
-
•
HCC patients with high expression of NOX4 and TGF-β genes have worse prognosis.
-
•
Overall, NOX4 could participate in both suppressor and protumorigenic TGF-β actions.
Abbreviations
- ATPb
ATP synthase subnunit beta
- DAPI
4′, 6-diamidino-2-phenylindole
- DPI
diphenyleneiodonium
- EGFR
Epidermal Growth Factor Receptor
- EMT
Epithelial-Mesenchymal transition
- ER
endoplasmic reticulum
- FBS
fetal bovine serum
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- gRNA
short-guide RNA
- GSVA
gene set enrichment analysis
- H2DCFDA
2′,7′-dichlorodihydrofluorescein diacetate
- HBSS
Hank's balanced salt solution
- HCC
hepatocellular carcinoma
- HUB
Bellvitge University Hospital
- HVB
hepatitis virus B
- HVC
hepatitis virus C
- IF
immunofluorescence
- MMP9
matrix metallopeptidase 9
- NADPH
nicotinamide adenine dinucleotide phosphate
- NASH
non-acoholic steatohepatitis
- NOX
NADPH oxidase
- NT
Non-Tumor
- O2−
superoxide anion
- PBS
phosphate-buffered saline
- Q-VD-OPH
Quinoline-Val-Asp-Difluorophenoxymethylketone
- ROS
reactive oxygen species
- RT
room temperature
- RTCA
real-time cell analyzer
- SD
standard deviation
- SDS
sodium dodecyl sulfate
- siRNA
small interference RNA
- T
Tumor
- TCGA
The Cancer Genome Atlas
- TCGA-LIHC
the cancer genome atlas program - liver hepatocellular carcinoma
- TGF-β
Transforming Growth Factor-beta
- VSMC
vascular smooth muscle cells
- WB
western blot
1. Introduction
The Transforming Growth Factor-beta (TGF-β) pathway plays essential roles in liver, pancreas and gastrointestinal pathologies, particularly in chronic inflammatory processes and the transition from a noncancerous disease state to cancer [1]. The family of NADPH oxidases (NOXs) has emerged in recent years as targets of the TGF-β pathway mediating many of its effects on hepatocytes, stellate cells and macrophages [2]. In particular, up-regulation of NOX4 by TGF-β is required for its pro-apoptotic activity in hepatocytes and hepatocellular carcinoma (HCC) cells [3]. Different molecular mechanisms may impair TGF-β-induced up-regulation of NOX4, which offer an advantage to HCC cells to overcome its tumor-suppressor function [[4], [5], [6], [7]]. NOX4 also mediates TGF-β-induced senescence in well-differentiated liver tumor cells [8], which might also contribute to prevent tumorigenesis. For this reason, in the last years it has increased the interest on deepening into the role of NOX4 as a potential tumor suppressor in the liver. In vitro assays proved that stable knockdown of NOX4 expression in human liver tumor cells increased cell proliferation and conferred an advantage to human HCC cells in xenograft experiments in athymic mice [9]. NOX4 is also necessary to maintain parenchymal structures, increase cell-cell and cell-to-matrix adhesion, and impair actomyosin contractility and amoeboid invasion [10]. We have recently shown that loss of NOX4 in HCC tumor cells induces metabolic reprogramming in a Nrf2/MYC-dependent manner to promote HCC progression [11]. Interestingly, NOX4 expression has been proposed as a prognostic factor in patients of HCC after hepatectomy, low NOX4 expression reflecting shorter relapse-free and overall survival [12]. In the same line of evidence, increased NOX4 expression has been associated with genes that inhibit tumor progression in HCC patients [13]. However, no such significant trend regarding NOX4 predictive value in survival was seen in univariate analysis in another cohort of HCC patients after partial hepatectomy [14] and even it has been proposed that intranuclear localization of NOX4 significantly correlates with advanced pathological TNM stage and short overall survival [15]. Furthermore, it is very relevant to consider that in most of solid tumors (head and neck, esophagus, bladder, ovary, prostate, as well in melanoma) expression of NOX4 is high when compared to histologically uninvolved specimens from the same organs [16] and NOX4 is being considered as a promising therapeutic target of malignancy [17]. Considering all these discrepancies, much further work might be necessary to better understanding NOX4 role in the context of liver tumorigenesis.
TGF-β plays a dual role during liver carcinogenesis, behaving as a suppressor factor on the malignant cell at early stages, but contributing to later tumor progression once cells escape from its cytostatic effects [18]. During these last years, studies have been focused mainly on the role of NOX4, independently of TGF-β, in the biology and progression of liver tumors. However, TGF-β is the main inducer of NOX4 in the context of liver inflammation and fibrosis [19], previous stages of hepatocarcinogenesis, and in a relevant percentage of HCC tumors, levels of TGF-β are high [20]. Despite of this, the specific role of NOX4 in TGF-β-induced canonical (SMADs) or non-canonical signals is not fully understood yet, neither its potential involvement in other parallel pro-tumorigenic actions induced by TGF-β in HCC cells.
Here we have used CRISPR Cas9 technology to stable attenuate NOX4 expression in HCC cells, in order to explore the impact of NOX4 in all the different responses to TGF-β. We also have analyzed the relevance of the expression of NOX4 for patient prognosis in the context of high expression of TGF-β-related genes. Overall, results locate NOX4 as a key element in the anti- and pro-tumorigenic TGF-β effects in HCC cells and reveal an unknown role for NOX4 as tumor promoter in those HCC patients that would present activation of the TGF-β pathway.
2. Material and methods
2.1. Cell Culture
PLC/PRF/5 and Hep3B human HCC cells were obtained from the European Collection of Cell Cultures. All cell lines were submitted for authentication in 2014 before the generation of cells lacking NOX4 protein by CRISPR-Cas9 technology, and used in passages close to the date of authentication. Just before publication of this manuscript, authentication of the cells was corroborated successfully. PLC/PRF/5 cells were maintained in DMEM and Hep3B in MEM, supplemented with non-essential amino acids (Lonza, Basel, Switzerland), 10% foetal bovine serum (Sera Laboratories International Ltd, West Sussex, UK), Penicillin (100U/mL), Streptomycin (100 μg/mL), Amphotericin (2.5 μg/mL), and l-glutamine (2 mM). All of them were maintained in a humidified atmosphere of 37 °C, 5% CO2. Cell lines were never used in the laboratory for longer than 4 months after receipt or resuscitation. TGF-β (Sigma, #T70039) was used at 2 ng/mL.
To generate a pool of PLC/PRF/5 and Hep3B cells lacking NOX4 protein, we used the CRISPR-Cas9 system. Short-guide RNAs (gRNA) were designed to target the gene and then cloned into the pSpCas9(BB)-2A-puro vector (supplied by Addgene, Watertown, MA, USA), which encodes an RNA polymerase III promotor for the transcription of the guide, the Cas9 endonuclease, and a gene providing resistance to puromycin. Both cell lines were transfected overnight with Lipofectamine® LTX Reagent (Thermo Fisher Scientific, Waltham, Massachusetts, USA). An empty vector without gRNA was used as negative control. Puromycin was added for 24h at 2 μg/mL for selection. The selected cells were tested for gene deletion by endonuclease assay and checked for protein knockdown by immunoblot.
For small interference RNA (siRNA) transient silencing, cells were transfected by TransIT-Quest reagent (Mirus, Madison, WI, USA) at 50 nM, in complete media. After 8 h, cell plates were washed, and fresh media was added. siRNA sequences (5′-3′) used: Control: GUAAGACACGACUUAUCGC; CCND1 sequence #1: UGAACAAGCUCAAGUGGAA; CCND1 sequence #2: CCGAGGAGCUGCUGCAAAU. siRNAs were obtained from Sigma-Genosys (Suffold, UK). CCND1 #1 and #2 siRNA mix was used in all the experiments (ratio 1:1).
2.2. Human HCC tissues and ethics statement
Samples from non-tumor (NT) and tumor (T) tissues were obtained from patients during surgical procedures at the Bellvitge University Hospital (HUB). Samples derived from a cohort of 124 patients with different etiology came from liver explants at transplantation or resection and incorporated in the study, with most of them in histological grade 2–3, and few in grade 1 or 4 (Supplementary Table 1). Human tissues were collected with the required informed consent in written from each patient and the approval of the Institutional Review Board (Comité Ético de Investigación Clínica-CEIC, University Hospital of Bellvitge). Patients' written consent form and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki.
2.3. Analysis of NADPH oxidase activity
Cells were incubated with NADPH, and NADPH consumption was monitored by the decrease in absorbance at λ = 340 nm for 10 min. For analysis of specific NADPH oxidase activity, the rate of consumption of NADPH inhibited by diphenyleneiodonium (DPI; Sigma-Aldrich) was measured, by adding 10 μM DPI 30 min before the assays. Results were normalized to protein content and are expressed as picomoles per minute per μg of protein.
2.4. Measurement of intracellular reactive oxygen species (ROS)
To measure the intracellular content of ROS, the 2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA, Thermo Fisher, #C6827) kit was used. 24 h after seeding the cells in a 12-well plate, they were starved (≥4 h at 0% FBS). 1 h prior to the TGF-β activation (for 3 and 24 h), cells were treated with DPI. Then, a wash with PBS is done and incubated with 2.5 μM H2DCFDA in HBSS for 30 min at 37 °C in the dark. Finally, images were taken at the Olympus IX70 microscope.
MitoSOX™ Red reagent (#M36008, Invitrogen, UK) was used to determine mitochondrial O2− production. 24 h after seeding cells in 12-well plates, they were trypsinized and the pellet resuspended in a solution of 5 μM MitoSOX™ Red reagent in Hank's Balanced Salt Solution (HBSS, Gibco #14175-053) and incubated for 15 min. After washing with HBSS, the pellet was resuspended with 300 μL HBSS in the flow cytometry tubes. Flow cytometry was performed by using a BD FACS Canto II Cytometer and analyzed by BD FACSDiva ™ v9.0 Software (BD Biosciences, San Jose, CA, USA). BD FlowJo™ v10.8.1 Software (BD Biosciences, San Jose, CA, USA) was used for the generation of graphs. All was performed at the Scientific and Technological Centers of the University of Barcelona (CCiTUB).
2.5. Analysis of apoptosis by Annexin V/PI flow cytometry assay
Analysis was done following the specifications of the kit Annexin V-FITC apoptosis Staining (Abcam, Cambridge, UK, #ab14085) and the optimal concentrations were optimized before the experiments. Hep3B cells were seeded in a 6-well plate, incubated overnight and starved (≥4 h at 0% FBS) before treated with TGF-β for 48, 72 h. Some conditions were incubated with Q-VD-OPH 1 h before the treatment with TGF-β to inhibit the caspases. At the precise time, attached cells, after trypsinization, and those in culture medium were collected and added to a flow cytometry tube containing 1 mL of Annexin Binding Buffer (ABB). Tubes were centrifuged at 480g for 5 min and the supernatant discarded. Then, cells were resuspended in 100 μL of ABB with 6 μL of Annexin V-APC and 5 μL of Propidium Iodide (PI) and incubated for 15 min at room temperature in the dark. Finally, 100 μL of ABB are added, and the reading is done. Flow cytometry was performed by using a BD FACS Canto II Cytometer and analyzed by BD FACSDiva ™ v9.0 Software (BD Biosciences, San Jose, CA, USA). BD FlowJo™ v10.8.1 Software (BD Biosciences, San Jose, CA, USA) was used for the generation of graphs. All was performed at the Scientific and Technological Centers of the University of Barcelona (CCiTUB).
2.6. Analysis of the number of viable cells, migration and invasion
Number of viable cells was determined by crystal violet staining. Cells were seeded in 24-well plates. After treatment, culture medium was removed; cells were washed with PBS and stained with crystal violet (0.2% (w/v) in 2% ethanol solution) for 30 min. The staining solution was washed with distilled water and dissolved in 10% SDS on a shaker for 30 min. Absorbance was measured on a plate reader at 595 nm.
Cell migration was real-time examined through the xCELLigence System (Agilent, Santa Clara, CA, USA). 3 × 104 cells/well were seeded onto the top chamber of a CIM plate®, which was coated with a collagen IV solution (Sigma-Aldrich, St. Louis, MO, USA) and placed onto the RTCA station (Agilent, Santa Clara, CA, USA). Continuous values were represented as normalized cell index, which reflects a relative change in measured electrical impedance, and quantified as a slope (hours-1) of the first 16 h of cell migration.
For assays of invasion in collagen gels, 5 x105 cells/mL were resuspended in low viscosity media at 37°C-10%CO2 for 72 h to create the spheroids, with or without TGF-β treatment. Then, they were embedded in Pure Collagen Type I Bovine collagen solution (3 mg/mL) in DMEM media. They were incubated with or without the presence of TGF-β, continuing the treatment for 96 h. Phase contrast pictures were taken for 4 days at the Leica DMIRD in Scientific and Technological Centers of the University of Barcelona (CCiTUB).
2.7. Immunofluorescence and image acquisition
Cells were fixed with 4% paraformaldehyde and blocked using 1% BSA and 10% FBS in PBS during 1h at RT. Primary antibodies were diluted in 1% PBS-BSA (1:50 dilution, 1h, RT) (Supplementary Table 2). Samples were incubated with fluorescently-conjugated secondary antibodies (anti-mouse Alexa 488 or anti-rabbit Alexa 488, 1:200 dilution in 1% PBS-BSA, 1h, RT). At the end, samples were embedded using ProLong™ Gold antifade reagent with DAPI reagent (Invitrogen #P36935, Thermo Fisher Scientific, Massachusetts, USA). Cells were imagined in a Nikon Eclipse 80i vertical fluorescence microscope or a Carl Zeiss LSM880 Confocal Microscopy in Scientific and Technological Centers of the University of Barcelona (CCiTUB).
2.8. Western blot analysis
Western Blotting was carried out as previously described [10]. Cells were lysed in RIPA lysis buffer (supplemented with 1 mM PMSF, 5 μg/mL Leupeptin, 0.1 mM Na3VO4, 0.5 mM DTT, 20 mM β-Glycerol phosphate) for 1h at 4 °C. Protein was quantified with the Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Laboratories, Hercules, CA, USA). Polyacrylamide gels were prepared at 15%. Primary antibodies were diluted in 0.5% non-fat-dried milk in PBS with 0.05% Tween 20 (Supplementary Table 2). Peroxidase-conjugated secondary antibodies anti-mouse (NA931V) or anti-rabbit (NA934V) (GE Healthcare, 1/3000) were diluted in 0.5% non-fat-dried milk in PBS with 0.05% Tween 20.
2.9. Biochemical analysis of the localization of NOX4 protein
NOX4 subcellular localization was analyzed by several protein fractionation kits. The Subcellular Proten Fractionation Kit (Thermo Scientific, #78840) was used to isolate the whole cell in total, cytoplasm, membrane, soluble nucleus, and chromatin fractions. Additionally, the Endoplasmic Reticulum Isolation Kit (Sigma, #ER0100) was used to separate the organelle from the total fraction. Finally, the Mitochondria Isolation Kit (Sigma, #MITOISO2) was the kit used to divide the cytosol and the mitochondria.
2.10. Gene expression analysis
E.Z.N.A.® Total RNA Kit II (Omega bio-tek, Norcross, GA, USA) was used for total RNA isolation following manufacturer's instructions. Culture plates were washed with PBS and cells were scrapped with RLT lysis buffer containing 10 μL/mL of β-mercaptoethanol. 1 μg of total RNA isolated from each sample was reverse-transcribed with random primers for complementary DNA synthesis, using a High Capacity RNA to cDNA Master Mix Kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer's protocol. For the real time qPCR, expression levels were determined in duplicate in a LightCycler® 480 Real-time PCR system, using the LightCycler® 480 SYBR Green I Master (Roche Diagnostics GmbH, Mannheim, Germany). Gene expression obtained from in vitro data was normalized to housekeeping gene L32. Gene expression obtained from human HCC samples was normalized to the average of three different housekeeping genes: RPL32, HPRT1, TBP. See Supplementary Table 3 for primer sequences.
2.11. Data analysis
2.11.1. Statistical analyses
Differences between two groups were compared using parametric analysis (Student's t-test with Welch correction). In general, experiments were carried out at least 3 independent times. Data are represented as mean ± standard deviation (SD). Statistical calculation was developed using GraphPad Prism software (GraphPad for Science Inc., San Diego, CA, USA). Differences were considered statistically significant at p<0.05 (*), p<0.01 (**) and p<0.001 (***).
2.11.2. Specific analysis in human HCC tissue samples
expression of different genes in 124 HCC patients (HUB) were analyzed by RT-qPCR. The results were normalized to the average of three different housekeeping genes: RPL32, HPRT1, TBP. Correlation analysis between NOX4 and TGFB1 was analyzed by Pearson correlation criteria. p<0.05 was considered statistically significant. Survival curves were performed by Kaplan Meier analysis, and significance studied with log-rank test. All patients were classified among TGFB1 High/Low and NOX4 High/Low depending on the expression of each gene. All analysis were performed considering the tumoral region expression. All statistical calculation was done by GraphPad Prism software (GraphPad for Science Inc., San Diego, CA, USA).
2.11.3. TCGA database
The Cancer Genome Atlas (TCGA) gene expression profiles (log2TPM) from 327 hepatocellular carcinoma cases from TCGA Liver Hepatocellular Carcinoma (TCGA-LIHC) [21] were downloaded from TCGA2BED [22] FTP repository. Clinical data for these cases was downloaded from Genomics Data Commons data portal [23]. Samples were categorized as low stroma when the percentage of stromal cells was below <15%. Correlation was assessed using Pearson correlation tests and corrected for multiple testing. Gene expression levels were stratified using the median of each gene. For survival analyses, Kaplan-Meier curves were plotted, and statistical assessment was performed with Log-rank test. In the case of gene expression signatures (Supplementary Table 4), gene set variation analysis (GSVA) [24] was used to assess the relative activation of the signature in the samples. Differences in mean MMP9 or TGFB1I1 expression between NOX4 low and NOX4 high were tested using a Mann-Whitney U test. P-values were corrected for multiple testing with Bonferroni. All analyses were performed using R 4.0.4 [25].
3. Results
3.1. Generation of cell models where expression of NOX4 is stably silenced, to analyze the different responses to TGF-β in HCC cells
We and others previously demonstrated that cells that respond to TGF-β in terms of tumor suppressor functions undergo inhibition of proliferation and/or apoptosis, which is related to SMADs phosphorylation (the canonical TGF-β signals) [20]. However, many HCC cells simultaneously respond to it transactivating survival/proliferative signals (non-canonical TGF-β signals) that protect them from apoptosis, undergoing Epithelial-Mesenchymal Transition (EMT), which increases their capacity to migrate and invade [18]. Previous experience from our group had demonstrated that the PLC/PRF/5 cell line offers a good tool to analyze the response to TGF-β in terms of growth inhibition, whereas the Hep3B cells offer an excellent model to analyze the response in terms of apoptosis and EMT. We decided to use CRISPR Cas9 technology to silence NOX4 in these cells. Experimental procedure is detailed in Materials and Methods section and presented in Supplementary Fig. 1. The efficiency of the silencing was demonstrated by analyzing NOX4 protein levels (Supplementary Fig. 2A) and NADPH oxidase activity (Supplementary Fig. 2B). Response to TGF-β was specifically impaired in terms of NOX4 up-regulation, whereas increase in the expression of other TGF-β target genes, SMAD7 and SERPINE1, was not affected (Supplementary Figs. 2C–D). Analysis of intracellular ROS production, by using H2DCFDA, revealed increase in CRISPR Control cells after TGF-β treatment, an effect that was clearly attenuated in CRISPR NOX4 cells (Supplementary Fig. 2E). TGF-β did not affect the levels of mitochondrial ROS, analyzed with MitoSOX™, neither in Control nor in NOX4 silenced cells (Supplementary Fig. 2F). Under the experimental conditions used in this study, we could not observe compensation by other members of the NOX family neither at basal levels nor after treatment with TGF-β in NOX4 silenced cells (results not shown).
3.2. Silencing NOX4 affects the response to TGF-β in terms of both canonical and non-canonical signals
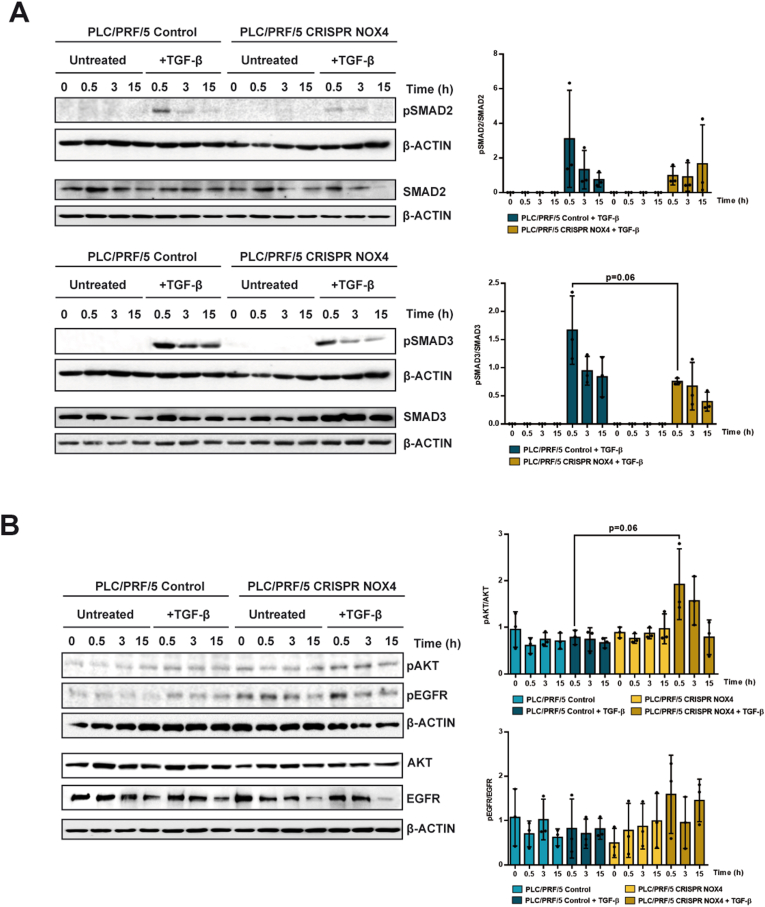
We started by analyzing how silencing NOX4 affects SMADs phosphorylation (Fig. 1A). We observed a tendency to attenuation in the response to TGF-β in terms of SMAD2 or SMAD3 phosphorylation in NOX4 silenced PLC/PRF/5 cells. When quantified as ratio phospho-SMADs versus total SMADs, CRISPR NOX4 cells showed an almost significant decrease in SMAD3 phosphorylation. However, SMAD-dependent response to TGF-β does not appear to be strongly affected, according to the observed SMAD7 and SERPIN1 up-regulation previously mentioned (Supplementary Fig. 2D) and response in the expression of other genes that we will comment later.
Fig. 1.
Role of NOX4 on TGF-β-mediated canonical and non-canonical signals. Analysis made in PLC/PRF/5 (Control and CRISPR NOX4) cells, untreated or TGF-β-treated. A) Phospho-SMAD2/3 and total SMAD2/3; B) Phospho-AKT, phospho-EGFR and their corresponding total protein levels. In both cases, Western blot, β-Actin was used as loading control. Left: Representative experiments. Right: quantification of the Phospho/Total ratio in each case, after densitometric analysis of the levels taking into account the specific loading control [(Phospho)/(phospho loading)]/[(Total)/(total loading)]. Data are Mean ± SD (n = 3).
Attenuation of the response to TGF-β in terms of tumor suppressor functions in HCC cells is mainly mediated by transactivation of the Epidermal Growth Factor Receptor (EGFR) pathway [6], which activates the phosphatidylinositol-3-kinase/AKT pathway. A crosstalk between EGFR/AKT and NOX4 has been demonstrated, AKT impairing up-regulation of NOX4 by TGF-β [3]. Here we observed that a crosstalk in the opposite way also exits, since silencing NOX4 provokes an increase in EGFR and AKT phosphorylation both in untreated cells and in TGF-β-treated cells (Fig. 1B). Levels of AKT did not show differences in any of the conditions, whereas the total level of EGFR decreased during the culture regardless TGF-β is or not present, in both CRISPR Control and CRISPR NOX4. The levels of EGFR were high in the CRISPR NOX4 cells, which contributes to the higher levels of phospho-EGFR, even at basal levels, observed in CRISPR NOX4 cells, as it is observed when the ratio phospho-EGFR/total EGFR is analyzed.
These results evidence for the first time that stable silencing of NOX4 in HCC cells may attenuate the TGF-β-induced intracellular canonical signals (SMADs) and situate the cells in a context of activation of other signals (EGFR/AKT) that could contribute to decrease the response to TGF-β in terms of growth inhibition and apoptosis.
3.3. Silencing NOX4 attenuates TGF-β response in terms of growth inhibition and apoptosis in HCC cells
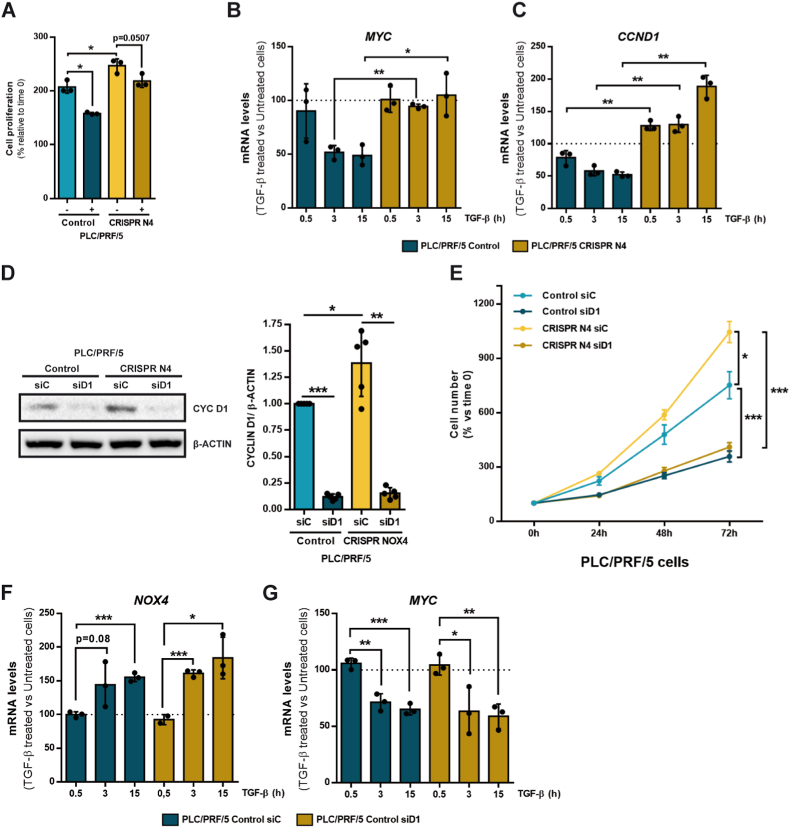
We previously published that stable knockdown of NOX4 expression in human liver tumor cells increased cell proliferation, which correlated with a higher percentage of cells in S/G2/M phases of the cell cycle and increase in MYC mRNA and c-Myc protein levels [10,11]. We observed here that after silencing NOX4 by CRISPR Cas9, expression of MYC was much higher both at the mRNA and protein levels in PLC/PRF/5 and Hep3B cells (Supplementary Fig. 3). Since repression of the mitogen-induced MYC early expression was described as the main mechanism regulated by TGF-β to inhibit proliferation [26], also in hepatocytes [27], we wondered whether silencing NOX4 might affect the response to TGF-β in terms of cell proliferation in HCC cells. Fig. 2A shows that inhibition of proliferation by TGF-β is not so clearly observed in CRISPR NOX4 PLC/PRF/5 cells. Analysis of MYC mRNA levels revealed that the downregulation induced by TGF-β in CRISPR Control cells, was impaired in CRISPR NOX4 PLC/PRF/5 cells (Fig. 2B). Furthermore, in CRISPR Control cells, TGF-β mediated down-regulation of Cyclin D1 (CCND1), one of the most relevant cell-cycle-related genes, regulated by c-Myc [28], that belongs to the highly conserved Cyclin family that functions as regulators of Cyclin dependent kinases (CDK) and cell cycle G1/S transition. Interestingly, this effect was inhibited in CRISPR NOX4 cells (Fig. 2C). In parallel to the results observed for c-Myc, basal expression of Cyclin D1 is much higher both at the mRNA and protein levels in CRISPR NOX4 PLC/PRF/5 and Hep3B cells (Supplementary Fig. 3) and the expression of CCND1 even increased at longer times of TGF-β treatment (Fig. 2C). In order to analyze the relevance of Cyclin D1 in HCC cell proliferation, we transiently knocked down it with specific siRNA in PLC/PRF/5 cells (Fig. 2D) and analyzed cell number along the culture. Cyclin D1 is crucial for PLC/PRF/5 cells to proliferate and its silencing strongly inhibited growth in both control and CRISPR NOX4 cells (Fig. 2E), minimizing their differences in cell proliferation. Silencing Cyclin D1 did not affect the up-regulation of NOX4 or the downregulation of MYC by TGF-β in PLC/PRF/5 cells (Fig. 2F-G), which indicates that it acts downstream of these signals regulating cell cycle. Considering the essential role played by Cyclin D1 in the proliferation of HCC cells, its overexpression in NOX4 silenced cells, and the lack of down-regulation by TGF-β, may impair TGF-β-induced growth inhibition.
Fig. 2.
Role of NOX4 in the response to TGF-β in terms of cell proliferation. Analysis made in PLC/PRF/5 (Control and CRISPR NOX4) cells. A) Cell viability assay analyzed by crystal violet staining after 72h of TGF-β treatment, normalized to initial time (time 0h). Data represents mean ± SD of triplicates from one representative experiment. B) Relative MYC and C)CCND1 mRNA expression analyzed by RT-qPCR, normalized to housekeeping gene L32, after TGF-β treatment at 0, 0.5, 3 and 15 h. Represented as percentage of TGF-β treated versus untreated cells in each time. D) Cyclin D1 protein levels analyzed by Western blot after transiently transfecting either with control or Cyclin D1 specific siRNA sequences. β-Actin was used as loading control. Representative experiment (left) and densitometric analysis of Cyclin D1 levels relative to β-Actin (right). E) Cell proliferation assay analyzed by crystal violet staining after transiently transfecting with either control or Cyclin D1 specific siRNA sequences, normalized to initial time (time 0h). F) Relative NOX4 and G)MYC mRNA expression analyzed by RT-qPCR, normalized to housekeeping gene L32, in Control CRISPR PLC/PRF/5 cells transiently transfecting with either control or Cyclin D1 specific siRNA sequences: effect of TGF-β treatment at 0, 0.5, 3 and 15 h. Data are represented as percentage of TGF-β treated versus untreated cells in each time. In all the panels, data are Mean ± SD (n = 3). *p<0.05 **p<0.01 ***p<0.001. (siC: Control siRNA; siD1: Cyclin D1 siRNA). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
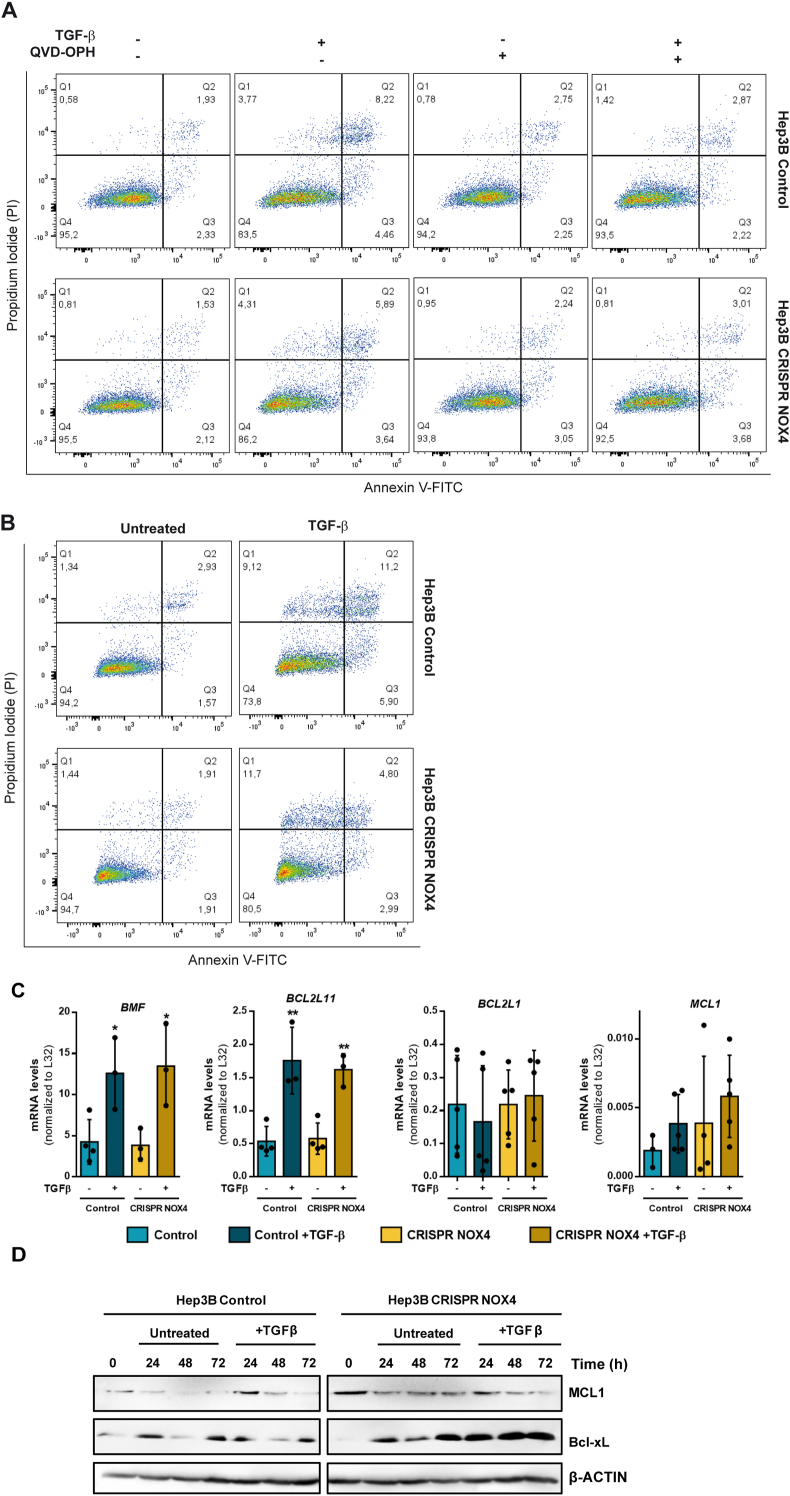
With the aim of deepening into the impact of NOX4 in TGF-β-induced apoptosis, we moved to the Hep3B cells. Treating these cells with TGF-β induced appearance of apoptotic (fragmented)/picnotic nuclei, as a hallmark of caspase-3 activation. This response was significantly attenuated in CRISPR NOX4 cells (Supplementary Fig. 4). The analysis of apoptosis by Annexin V/propidium iodide (PI) staining, using Q-VD-OPH as a caspase-3 inhibitor, revealed an increase in the cells accumulating in Q3 (Annexin+, PI-, early apoptosis) and Q4 (Annexin+, PI+, late apoptosis) in CRISPR Control cells after 48 h of treatment with TGF-β, an effect that was not observed in the presence of Q-VD-OPH (Fig. 3A). In the case of the Hep3B CRISPR NOX4, the percentage of Annexin + cells (in Q3 or Q4) was clearly lower when analyzed at 48 or 72 h (Fig. 3A-B).
Fig. 3.
Role of NOX4 on TGF-β-induced apoptosis. Analysis made in Hep3B (Control and CRISPR NOX4) cells. A-B) Representative flow cytometry plots using Annexin V-FITC/PI staining for analysis of apoptosis in Hep3B (Control and CRISPR NOX4) treated with TGF-β for 48h (A) or 72h (B). In A, co-treatment with Q-VD-OPH, supporting the role of caspases in the process. C) Proapoptotic (BMF and BCL2L11) and antiapoptotic (BCL2L1 and MCL1) mRNA expression analyzed by RT-qPCR, normalized to housekeeping gene L32, after TGF-β treatment at 48 h. D) MCL1 and Bcl-xL protein levels analyzed by western blot after TGF-β treatment at 24, 48 and 72 h. β-Actin was used as loading control. In A and C data are Mean ± SD (n = 3–6). *p<0.05 **p<0.01, ***p<0.001. In B and D, a representative experiment is shown.
Looking for the molecular mechanisms, we analyzed TGF-β-induced up-regulation of BMF and BCL2L11 (BIM), proapoptotic Bcl-2 family members previously identified as induced by TGF-β and required for its pro-apoptotic effects [29,30]. Expression of both genes increased in response to TGF-β, but no differences were observed in CRISPR NOX4 cells (Fig. 3C). Then, we moved to anti-apoptotic members of the family widely expressed in HCC cells: BCL2L1 (Bcl-xL) and MCL-1. None of these genes suffered significant changes in TGF-β treated cells, with great heterogeneity in the mRNA levels in the different experiments (Fig. 3C). However, western blot analysis revealed a clear increase in Bcl-xL levels, but not in MCL1, in CRISPR NOX4 cells: along the culture in untreated cells, and particularly at any time after treatment with TGF-β (Fig. 3D). These results together indicate that stable silencing of NOX4 inhibits TGF-β-induced apoptosis, but it is not related to transcriptional regulation of pro-apoptotic genes, but post-transcriptional regulation of anti-apoptotic genes, particularly Bcl-xL.
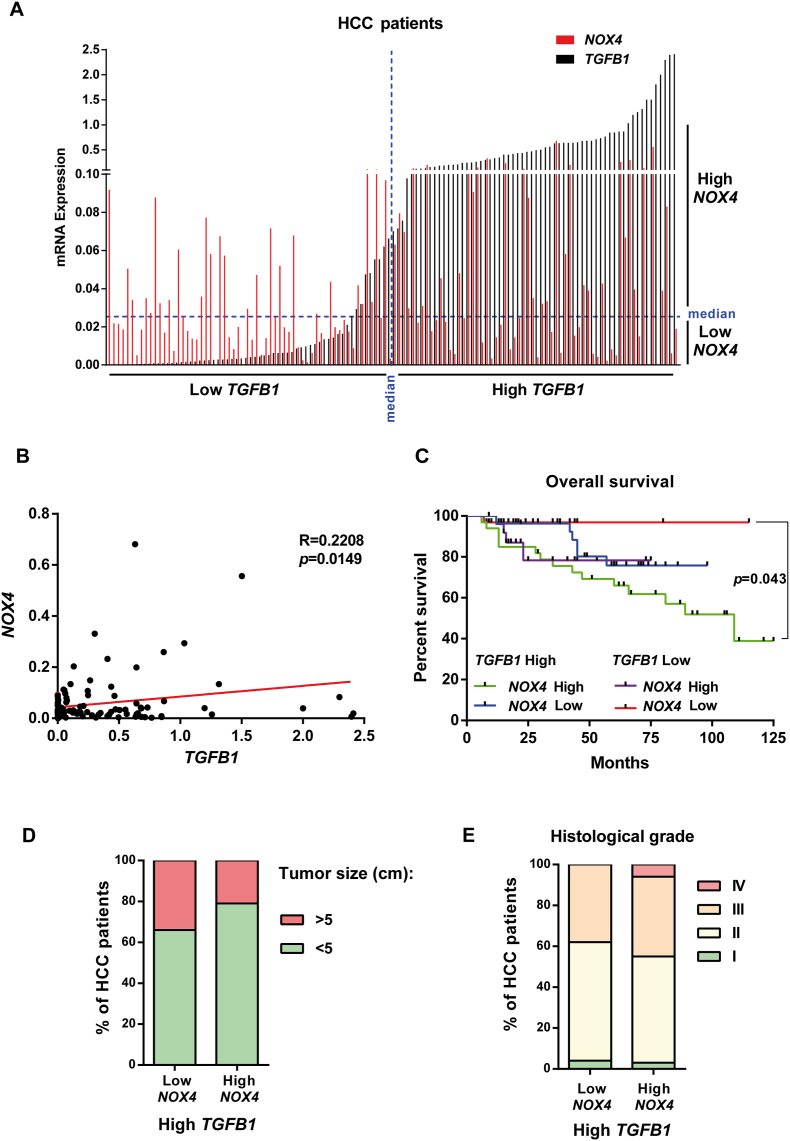
3.4. Concomitant high expression of TGF-β-related genes and NOX4 in HCC patients is related to worse overall survival
Considering the essential role played by NOX4 in mediating TGF-β-induced tumor suppressor actions, we decided to explore the expression of TGFB1 and NOX4 in a cohort of 124 patients, whose tumors were collected during surgical procedures, at transplantation or resection, in the Bellvitge University Hospital (HUB) (Fig. 4A). NOX4 and TGFB1 expression presented a positive correlation that was statistically significant (Fig. 4B). We defined “high” or “low” expression depending on whether the data were above or below the median, respectively (Fig. 4A). Contrary to our initial hypothesis, patients showing high NOX4 and high TGFB1 expression presented the worse prognosis in term of overall survival (Fig. 4C). Looking in the TCGA public data bases, it could also be observed a tendency to a worse overall survival in the NOX4high versus the NOX4low in patients presenting high expression of TGF-β ligands, particularly in the case of TGFB2 (Supplementary Fig. 5A, Left) or high expression of the receptor TGFBR1 (Supplementary Fig. 5B, Left). These differences were never observed in patients with low expression of TGF-β ligands or receptors (Supplementary Figs. 5A–B, Right). Furthermore, TCGA data also reflected a positive correlation between NOX4 and TGFB1, TGFB2, TGFB3, TGFBR1 and TGFBR2 expression that was statistically significant (Supplementary Fig. 6A). Using a stablished TGF-β gene signature that reflects hallmarks of cancer (TGF-β-signaling Hallmarks of Cancer, see Supplementary Table 4 in Material and Methods Section for details), we found a positive correlation between NOX4 expression and this gene signature (Supplementary Fig. 6B), which was observed even when selected only the samples with a low stroma, indicating a role for NOX4 expression in the tumor cells. Interestingly, Kaplan-Meier curve for overall survival probability for NOX4low versus NOX4high patients when “TGF-β-signaling Hallmarks of Cancer” gene signature is high revealed significant differences, lower overall survival in the NOX4high patients (Supplementary Fig. 6C).
Fig. 4.
Impact of the expression of NOX4 and TGFB1 on different parameters in HCC patients. HCC patients n = 124 from HUB. A) Distribution of the patients according to the expression of TGFB1 and NOX4. B) Pearson correlation analysis between NOX4 and TGFB1 gene expression in 120 patients for which we had data of survival: TGFB1 High/NOX4 High (green: 33 patients); TGFB1 High/NOX4 Low (blue: 27 patients); TGFB1 Low/NOX4 High (purple, 28 patients); TGFB1 High/NOX4 High (red, 32 patients). C) Kaplan-Meier curve for overall survival percentage when TGFB1 and NOX4 expression are high/low. D) Percentage of HCC patients that have a tumor size bigger or smaller than 5 cm when TGFB1 expression is high, depending on their NOX4 expression. E) Percentage of HCC patients in each histological grade (I-IV) when TGFB1 expression is high, depending on their NOX4 expression. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Interestingly, in our cohort of NOX4low/TGFB1high HUB-patients, the percentage of tumors with a size above 5 cm was higher, which may correlate with the previous presented results indicating that NOX4 mediates TGF-β-induced growth inhibition (Fig. 4D). However, histologically these tumors presented a more advanced stage (Fig. 4E). Since the cohort comes from surgical procedures, grade IV is infrequent, but all the patients with grade IV were in the NOX4high/TGFB1high group. We also observed lower percentage of relapse-free survival patients in the NOX4high/TGFB1high cohort when compared to the NOX4low/TGFB1high (Supplementary Fig. 7A). Looking at etiology, we could not observe any difference related to virus infection, alcohol or NASH between the NOX4high/TGFB1high and the NOX4low/TGFB1high groups (Supplementary Fig. 7B). The NOX4high/TGFB1high patients presented a higher percentage of liver with mild fibrosis, but no differences were observed in terms of cirrhosis (Supplementary Fig. 7C).
Altogether, these results would indicate that contrary to expectations regarding the strong role of TGF-β/NOX4 in mediating inhibition of growth and induction of apoptosis in liver tumor cells, the activation of both TGF-β and NOX4 pathways would reflect higher tumor progression.
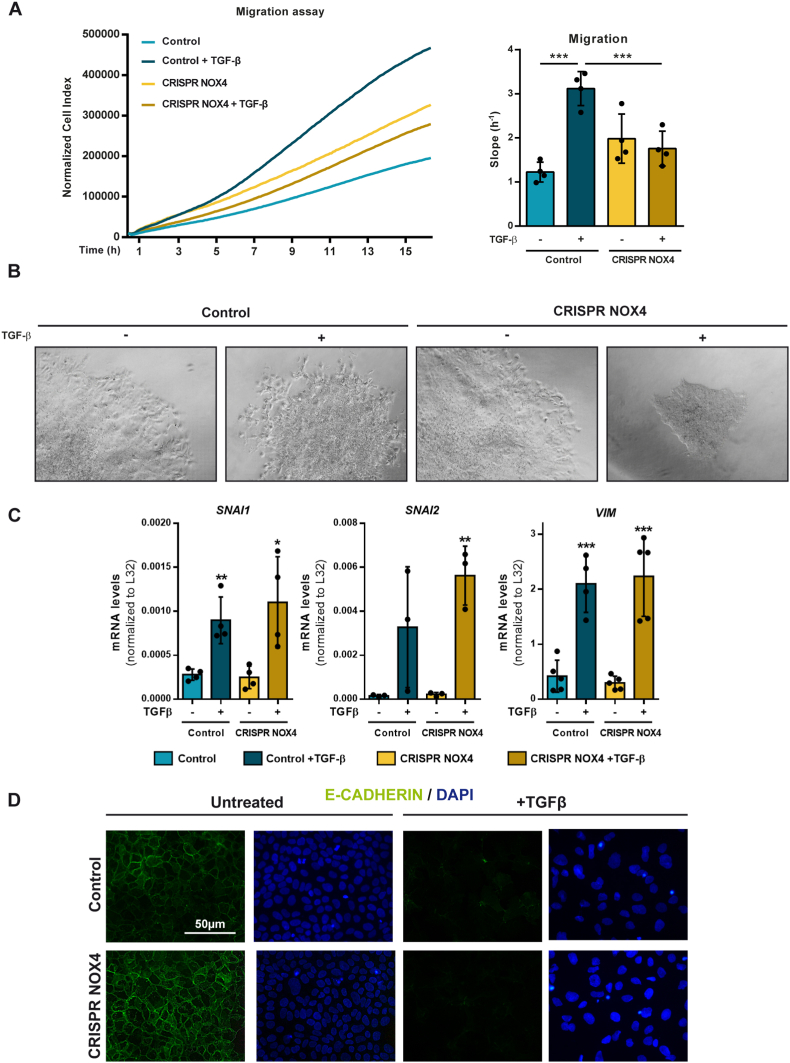
3.5. NOX4 mediates TGF-β effects on cytoskeleton remodeling and migration in HCC cells
Results observed in patients led us to take into consideration other parallel effects of TGF-β in HCC cells that may contribute to tumor progression. Indeed, we analyzed the role of NOX4 on TGF-β-mediated migration in the Hep3B cells (Fig. 5A). Continuous monitoring of cell migration by the xCelligence system clearly revealed a significant effect of TGF-β on the migratory capacity of control cells. However, this TGF-β-induced migratory effect was not observed in CRISPR NOX4 cells. Next, we performed invasive growth experiments in 3D where spheroids, previously formed with the HCC cells, were later immersed in collagen gels to monitor the capacity of the cells to expand and invade the matrix. Control Hep3B cells in the presence of TGF-β showed spreading of the cells, which invade the collagen. However, this effect could not be observed in TGF-β-treated CRISPR NOX4 cells (Fig. 5B).
Fig. 5.
Role of NOX4 on TGF-β-induced cell migration and Epithelial-Mesenchymal Transition. Analysis made in Hep3B (Control and CRISPR NOX4) cells. A) Cell migration after TGF-β treatment, real-time monitored using xCELLigence system. Results expressed as Normalized Cell Index (left) or slope (h-1) (right) of the first 16 h. B) Representative images of spheroids from cells either untreated or treated with TGF-β, embedded in a collagen I matrix for 96h. C)SNAI1, SNAI2 and VIM mRNA expression analyzed by RT-qPCR, normalized to housekeeping gene L32, after 48h TGF-β treatment. D) Immunofluorescence of E-Cadherin (green) and DAPI (blue) for nuclei staining after 48h TGF-β treatment. In A-Right and C data are Mean ± SD (n = 3–4). *p<0.05 **p<0.01, ***p<0.001. In B and D, representative images are shown. Scale bar, 50 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
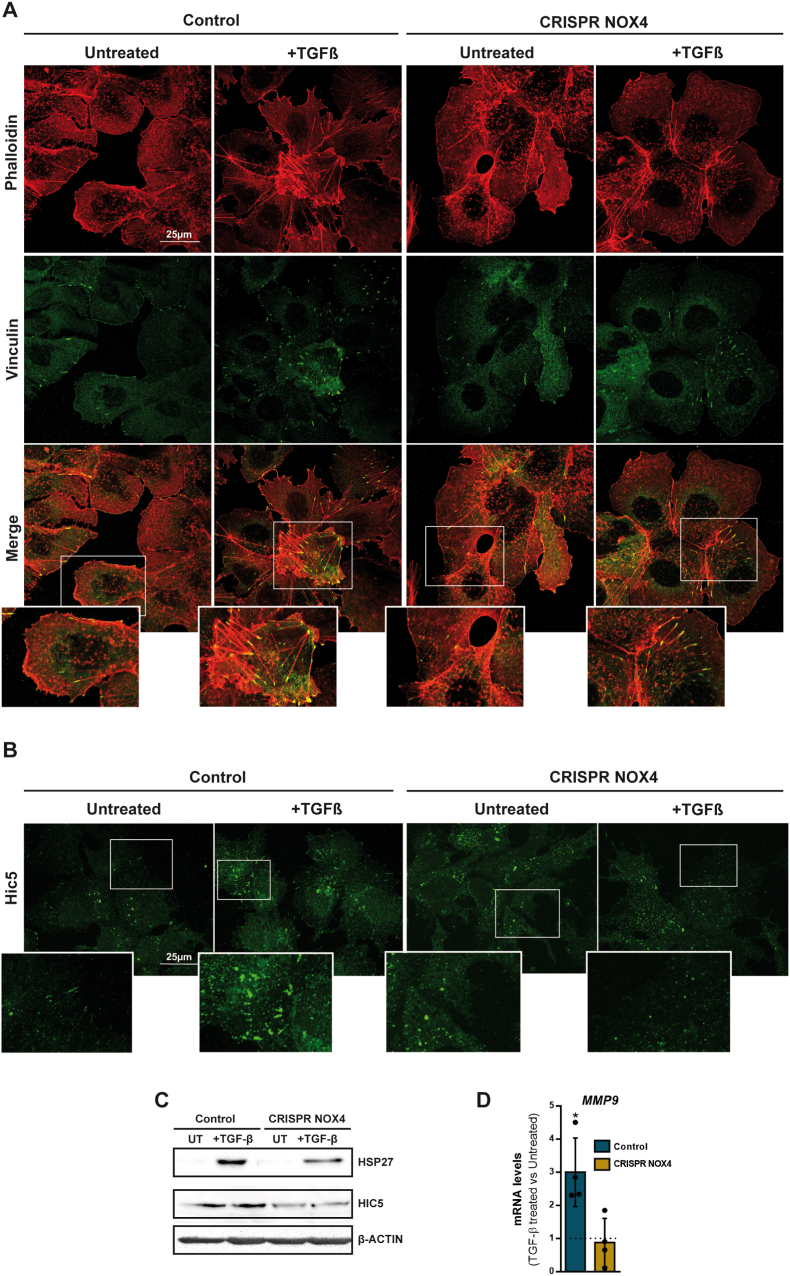
Hep3B cells respond to TGF-β undergoing EMT. Indeed, we next evaluated the potential role of NOX4 in upregulating SNAIL1 (SNAI1), SNAIL2 (SNAI2) or Vimentin (VIM), but we observed that NOX4 is not required for the upregulation of these EMT-related genes (Fig. 5C). In correlation with this, E-Cadherin, which is the main target of SNAIL that down-regulates it, was lost regardless NOX4 is expressed or not (Fig. 5D). But, additionally to the regulation of the EMT-transcriptional program, the increase in cell migration by TGF-β requires cytoskeleton remodeling and changes in focal adhesions [31]. Analysis of F-Actin localization and structure indicated clear differences in the CRISPR NOX4 cells, regardless they were treated or not with TGF-β (Fig. 6A). In control cells, F-Actin is organized pericellularly, whereas in the CRISPR NOX4 cells, it is not so well organized. TGF-β induced a complete disorganization of F-Actin in the control cells, with appearance of some stress fibers. In CRISPR NOX4 cells changes were not so evident, since these cells already presented disorganized F-Actin. Moreover, more importantly, in TGF-β-treated control cells changes in F-Actin were accompanied by notable increase in focal adhesions, analyzed by immunostaining of Vinculin, an effect that was attenuated in CRISPR NOX4 cells (Fig. 6A). Indeed, NOX4 is required for an efficient TGF-β regulation of cytoskeleton remodeling and increase in focal adhesions.
Fig. 6.
Role of NOX4 on TGF-β-induced cytoskeleton remodeling. Analysis made in Hep3B (Control and CRISPR NOX4) cells. A) Immunofluorescence of Phalloidin (red) and Vinculin (green) after 48h TGF-β treatment. B) Immunofluorescence of Hic-5 (green) after 48h TGF-β treatment. C) Hsp27 and Hic-5 protein levels analyzed by western blot after 48 h of TGF-β treatment. β-Actin was used as loading control D)MMP9 mRNA expression analyzed by RT-qPCR, normalized to housekeeping gene L32, after 48h TGF-β treatment and represented as fold induction (TGF-β-treated versus untreated cells). In A and B, representative images are shown. Scale bar, 25 μm (A). Scale bar, 25 μm (B). In D, data are Mean ± SD (n = 4–6). *p<0.05. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
It was previously suggested in VSMCs that NOX4 is a focal adhesion resident protein [32]. Here, immunocytochemical analysis of NOX4 and focal adhesions in Control Hep3B cells, analyzed by Vinculin staining, revealed co-localization in only some few points. In fact, NOX4 was mainly located in intracellular vesicles (Supplementary Fig. 8A) in both untreated and TGF-β-treated cells. Analysis by western blot after separation of different fractions in untreated cells revealed the majority of the NOX4 protein in the cytosolic fraction (Supplementary Fig. 8B), correlating with high levels in the endoplasmic reticulum fraction (Supplementary Fig. 8C). We cannot exclude that a fraction of NOX4 may be in the nucleus (Supplementary Fig. 8A) or in the mitochondria (Supplementary Fig. 8D).
Looking for alternative molecular mechanisms that would explain the participation of NOX4 in regulating TGF-β-induced focal adhesions, we first analyzed the level of expression of RAC/RHO family genes, which are upregulated by TGF-β, but we could not find significant differences between Control and CRISPR NOX4 cells (Supplementary Fig. 9). However, immunostaining of Hic-5, which was proposed to mediate TGF-β-induced adhesion in Vascular Smooth Muscle cells (VSMC) [33], showed significant differences between Control and CRISPR NOX4 cells (Fig. 6B). In Control cells, in the presence of TGF-β, Hic-5 levels increased and localized specifically in areas resembling focal adhesions. In CRISPR NOX4 cells, Hic-5 levels were lower, and no change was observed after treatment with TGF-β. The correct subcellular localization of Hic-5 within focal adhesions requires the chaperone protein Hsp27, also a target of NOX4-mediated gene transcription [33]. The analysis of Hsp27 protein levels revealed an increase in TGF-β-treated CRISPR Control cells that was attenuated in CRISPR NOX4 cells (Fig. 6C). These results suggest that TGF-β-mediated regulation of cytoskeleton dynamics and focal adhesions require NOX4, which is necessary for TGF-β-induced increase in the chaperone Hsp27 and Hic-5, and correct subcellular localization of Hic-5 within focal adhesions. Finally, worthy to mention that the expression of the metalloprotease MMP9 was upregulated by TGF-β in Control but not in CRISPR NOX4 cells (Fig. 6D). Looking at TCGA data, analysis of MMP9 expression in the cohort of TGFB1high patients revealed significant higher levels in NOX4high versus NOX4low patients, a difference that could not be observed in the cohort of TGFB1low patients (Supplementary Fig. 10A), which reinforces the role of NOX4 in regulating MMP9 in the context of TGF-β signaling. The analysis of TGFB1l1 (Hic-5) also revealed that the maximum level of expression was found in TGFB1highNOX4high patients. Again, results were similar when low stroma samples were selected (Supplementary Fig. 10B), indicating that differences are mainly due to the tumor cells.
Altogether, these results indicate that NOX4 is required for TGF-β induction of migration in HCC cells, through the regulation of cytoskeleton dynamics and focal adhesions, correlating with changes in expression of Hsp27, Hic-5 and MMP9.
4. Discussion
Hepatocellular carcinoma (HCC) is one of the deadliest malignancies worldwide because of its high recurrence rate, high metastatic potential, and resistance to drugs [34]. Elucidation of the mechanisms underlying malignancy in HCC is needed to improve diagnosis, therapy, and prognosis. Results here point to a crucial role for NOX4 axis in regulating both suppressor and pro-tumorigenic signals induced by TGF-β in HCC cells. On one side, NOX4 is required for an efficient SMADs phosphorylation and full inhibition of cell proliferation by TGF-β. Although the specific targets of the NOX4-mediated ROS are unclear, we may speculate that inactivation of protein phosphatases could be a probable possibility. In this sense, it has been proposed that protein phosphatase 2A (PP2A), physically interact with SMAD2/3, thereby promoting their dephosphorylation [35] and absence of Nox4 facilitates nuclear PP2A translocation in murine models of carcinogen-induced solid tumors [36]. However, although we could expect that the attenuated SMAD pathway would be related to a less efficient regulation of TGF-β target genes, we observed a correct response to TGF-β in CRISPR NOX4 cells in upregulating SMAD7, SERPINE1, BMF, BCL2L11, SNAI1, or VIM. In terms of proliferation, probably the most relevant aspect is that the absence of NOX4 provoked increase in the expression of MYC and Cyclin D1, being TGF-β unable of properly down-regulate the expression of these genes. We here show how Cyclin D1 is required for HCC cell proliferation and previous results strongly demonstrated that its overexpression impairs TGF-β-induced growth inhibition [37]. Indeed, the capacity of TGF-β to inhibit proliferation in CRISPR NOX4 cells may be significantly attenuated due to the high levels of Cyclin D1. Furthermore, NOX4 appears to be required for an efficient TGF-β-induced apoptosis. Although these results could be expected, the previous experiments were performed by transiently knock-down of NOX4 [3]. Results here reinforces the crucial role of NOX4 in TGF-β-induced apoptosis, demonstrating that it does not exist a compensatory mechanism when NOX4 is stably silenced for long time, as may occur in patients that present NOX4 deletions. TGF-β induces pro-apoptotic genes (such as BMF or BCL2L11) in primary cultures of hepatocytes, but also increases the levels of anti-apoptotic genes, such as Bcl-xL, the balance among them decides cell fate [30,38]. Inhibition of survival signals, such as the EGFR or the MAPK/ERKs pathways, abrogates the increase in the expression of the anti-apoptotic genes, in particular Bcl-xL, and significantly enhances cell death [5,30,38,39]. Results here shown demonstrate a role for NOX4 not through transcriptional regulation of pro-apoptotic genes, but through post-transcriptional regulation of anti-apoptotic proteins, particularly Bcl-xL, which appears also increased along the culture even in the absence of TGF-β, probably related to the higher activation of the EGFR/AKT pathway observed in these cells. Previous studies had identified Bcl-xL as a critical apoptosis antagonist in hepatocytes [40]. Indeed, in the absence of NOX4, apoptosis may be prevented by the high levels of Bcl-xL.
Considering this essential role of NOX4 in TGF-β-induced tumor suppressor functions, it should have been expected that in HCC patients presenting high expression levels of TGF-β ligands and/or receptors, high level of NOX4 expression may define a better prognosis. Contrary to these expectations, analysis in a cohort of patient from the HUB in our campus, as well as in silico data from TCGA public data, revealed that high expression of NOX4 defines a worse overall survival in patients presenting high expression of TGF-β-related genes or a TGF-β cancer-related gene signature. A more detailed analysis in the HUB cohort correlated this NOX4high/TGFB1high group with a more aggressive phenotype. This stratification of patients has facilitated a better comprehension of the role of NOX4 in HCC and could explain the controversial data in the literature, with some publications defending a better prognosis [12,13], not any relevance [14], or even worse prognosis for NOX4high patients [15].
The reason for the potential role of NOX4 as pro-tumorigenic in the context of overactivation of the TGF-β pathway can be explained by the results demonstrating that NOX4 is required for TGF-β-induced migration of HCC cells. Although nothing was known in the context of HCC, previous results in other solid tumors, such as breast, lung or pancreas, suggested that NOX4 is required for an efficient TGF-β-mediated EMT and increase in cell migration [[41], [42], [43], [44]]. Here we propose the molecular mechanism that may explain these effects. We found that NOX4 is not required for regulation of the EMT-related transcriptional program, but for TGF-β-induced regulation of cytoskeleton dynamics and focal adhesions. Although poorly described in the context of tumorigenesis, a role for NOX4 in regulating cytoskeleton dynamics and focal adhesions was elegantly demonstrated in VSMC [32,[45], [46], [47], [48]]. Particularly, it was proposed that TGF-β, by a NOX4-dependent mechanism, increases Hic-5 expression and localization in focal adhesions, which is essential for the TGF-β-mediated increase in focal adhesions number, adhesive forces and migration [33]. In the context of HCC, our results indicate that NOX4 is also required to regulate the levels of Hic-5 and its intracellular localization. Regarding the potential molecular mechanism, as previously proposed in VSMCs [33], NOX4 is required for TGF-β-induced increase in the chaperone Hsp27, a target of NOX4-mediated gene transcription that is required for correct subcellular localization of Hic-5 within focal adhesions [33]. Additionally, we also observed that NOX4 is essential for TGF-β-mediated regulation of MMP9 expression. Interestingly, it has been proposed that NOX4 and associated reactive oxygen species (ROS) regulate MMP9 expression both at promoter activation and mRNA stability levels [49,50]. Poorly studied in the context of the TGF-β actions in cancer, MMP9 has been described as an essential regulator of proteins involved in actin polymerization and cell migration during TGF-β-induced EMT in epithelial cells [51,52]. In vitro results are reinforced by data in patients, indicating the highest expression of MMP9 or TGFB1l1 in TGFB1highNOX4high patients.
In conclusion, data here presented open new insights about the role of NOX4 in the context of overactivation of the TGF-β pathway in HCC (see graphical abstract). NOX4 mediates the tumor suppressor actions of TGF-β by regulating Bcl-xL, C-Myc and Cyclin D1. However, NOX4 is also required for regulation of cytoskeleton dynamics and focal adhesions by TGF-β, correlating with changes in expression of Hsp27, Hic-5 and MMP9, which contribute to the migratory capacity of HCC cells. Altogether, NOX4 could participate in both suppressor and protumorigenic TGF-β actions in HCC cells.
Funding
This work was supported by: 1) Agencia Estatal de Investigación (AEI), Ministry of Science and Innovation, Spain, cofounded by FEDER funds/Development Fund—a way to build Europe to I.F. [grants numbers RTI2018-094079-B-100, RED2018-102576-T, RTC2019-007125-1, PID2021-122551OB-I00] and the FPI fellowship to RE-S [grant number PRE2019-089144]; 2) Asociación Española contra el Cáncer (AECC) to I.F. [grant number PRYGN211279FABR]; 3) Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR, Generalitat de Catalunya) to I.F. [grant number 2021SGR00029]. The CIBEREHD, National Biomedical Research Institute on Liver and Gastrointestinal Diseases, is funded by the Instituto de Salud Carlos III, Spain. We thank CERCA Programme/Generalitat de Catalunya for institutional support.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Isabel Fabregat reports financial support was provided by Agencia Estatal de Investigación, Ministry of Science and Innovation, Spain. Rut Espinosa-Sotelo reports financial support was provided by Agencia Estatal de Investigación, Ministry of Science and Innovation, Spain. Isabel Fabregat reports financial support was provided by Asociación Española contra el Cáncer (AECC), Spain. Isabel Fabregat reports financial support was provided by Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR, Generalitat de Catalunya). Isabel Fabregat reports financial support was provided by CIBER, National Biomedical Research Institute, Instituto de Salud Carlos III, Spain.
Acknowledgements
We acknowledge the technical support of Benjamin Torrejón, Beatriz Barroso and Esther Castaño, from the Scientific and Technological Centers of the University of Barcelona (CCiTUB), for support in confocal microscopy and flow cytometry. We thank Dr. Susan M.E. Smith for reading of the manuscript and helpful comments.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102818.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The work here did not generate massive data. But we used public data related to gene expression in HCC patients, as specified in the Materials and Methods Section and in the figures.
References
- 1.Gough N.R., Xiang X., Mishra L. TGF-Β signaling in liver, pancreas, and gastrointestinal diseases and cancer. Gastroenterology. 1 de agosto de. 2021;161(2):434–452.e15. doi: 10.1053/j.gastro.2021.04.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herranz-Itúrbide M., Peñuelas-Haro I., Espinosa-Sotelo R., Bertran E., Fabregat I. The TGF-β/NADPH oxidases axis in the regulation of liver cell biology in health and disease. Cells. 1 de septiembre de. 2021;10(9) doi: 10.3390/cells10092312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmona-Cuenca I., Roncero C., Sancho P., Caja L., Fausto N., Fernández M., et al. Upregulation of the NADPH oxidase NOX4 by TGF-beta in hepatocytes is required for its pro-apoptotic activity. Journal of Hepatology. diciembre de. 2008;49(6):965–976. doi: 10.1016/j.jhep.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Yu J.H., Zhu B.M., Riedlinger G., Kang K., Hennighausen L. The liver-specific tumor suppressor STAT5 controls expression of the reactive oxygen species-generating enzyme NOX4 and the proapoptotic proteins PUMA and BIM in mice. Hepatology. diciembre de. 2012;56(6):2375–2386. doi: 10.1002/hep.25900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caja L., Sancho P., Bertran E., Iglesias-Serret D., Gil J., Fabregat I. Overactivation of the MEK/ERK pathway in liver tumor cells confers resistance to TGF-β-induced cell death through impairing up-regulation of the NADPH oxidase NOX4. Cancer Research. 1 de octubre de. 2009;69(19):7595–7602. doi: 10.1158/0008-5472.CAN-09-1482. [DOI] [PubMed] [Google Scholar]
- 6.Caja L., Sancho P., Bertran E., Fabregat I. Dissecting the effect of targeting the epidermal growth factor receptor on TGF-β-induced-apoptosis in human hepatocellular carcinoma cells. Journal of Hepatology. agosto de. 2011;55(2):351–358. doi: 10.1016/j.jhep.2010.10.041. [DOI] [PubMed] [Google Scholar]
- 7.Caballero-Díaz D., Bertran E., Peñuelas-Haro I., Moreno-Càceres J., Malfettone A., López-Luque J., et al. Clathrin switches transforming growth factor-β role to pro-tumorigenic in liver cancer. Journal of Hepatology. 1 de enero de. 2020;72(1):125–134. doi: 10.1016/j.jhep.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Senturk S., Mumcuoglu M., Gursoy-Yuzugullu O., Cingoz B., Akcali K.C., Ozturk M. Transforming growth factor-beta induces senescence in hepatocellular carcinoma cells and inhibits tumor growth. Hepatology. septiembre de. 2010;52(3):966–974. doi: 10.1002/hep.23769. [DOI] [PubMed] [Google Scholar]
- 9.Crosas-Molist E., Bertran E., Sancho P., López-Luque J., Fernando J., Sánchez A., et al. The NADPH oxidase NOX4 inhibits hepatocyte proliferation and liver cancer progression. Free Radic. Biol. Med. 2014;69:338–347. doi: 10.1016/j.freeradbiomed.2014.01.040. [DOI] [PubMed] [Google Scholar]
- 10.Crosas-Molist E., Bertran E., Rodriguez-Hernandez I., Herraiz C., Cantelli G., Fabra A., et al. The NADPH oxidase NOX4 represses epithelial to amoeboid transition and efficient tumor dissemination. Oncogene. 25 de mayo de. 2017;36(21):3002–3014. doi: 10.1038/onc.2016.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.I. Peñuelas-Haro, R. Espinosa-Sotelo, E. Crosas-Molist, M. Herranz-Itúrbide, D. Caballero-Díaz, A. Alay, et al., The NADPH oxidase NOX4 regulates redox and metabolic homeostasis preventing hepatocellular carcinoma progression, Hepatology (2022). Online ahead of print. [DOI] [PMC free article] [PubMed]
- 12.Ha S.Y., Paik Y.H., Yang J.W., Lee M.J., Bae H., Park C.K. NADPH Oxidase 1 and NADPH oxidase 4 have opposite prognostic effects for patients with hepatocellular carcinoma after hepatectomy. Gut and Liver. 1 de septiembre de. 2016;10(5):826–835. doi: 10.5009/gnl15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eun H.S., Cho S.Y., Joo J.S., Kang S.H., Moon H.S., Lee E.S., et al. Gene expression of NOX family members and their clinical significance in hepatocellular carcinoma. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-11280-3. 1 de diciembre de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu C., Qiu J., Huang P., Zou R., Hong J., Li B., et al. NADPH oxidase DUOX1 and DUOX2 but not NOX4 are independet predictors in hepatocellular carcinoma after hepatectomy. Tumor Biology. 1 de julio de. 2011;32(6):1173–1182. doi: 10.1007/s13277-011-0220-3. [DOI] [PubMed] [Google Scholar]
- 15.Eun H., Chun K., Song I., Oh C., Seong I., Yeo M., et al. High nuclear NADPH oxidase 4 expression levels are correlated with cancer development and poor prognosis in hepatocellular carcinoma. Pathology. 1 de febrero de. 2019;51(6):579–585. doi: 10.1016/j.pathol.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Meitzler J.L., Makhlouf H.R., Antony S., Wu Y., Butcher D., Jiang G., et al. Decoding NADPH oxidase 4 expression in human tumors. Redox Biol. 2017;13:182–195. doi: 10.1016/j.redox.2017.05.016. 1 de octubre de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong S., Wang S., Shao M. NADPH oxidase 4: a potential therapeutic target of malignancy. Frontiers in Cell and Developmental Biology. 11 de mayo de. 2022:10. doi: 10.3389/fcell.2022.884412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez-Sanchez E., Vaquero J., Férnandez-Barrena M.G., Lasarte J.J., Avila M.A., Sarobe P., et al. The TGF-β pathway: a pharmacological target in hepatocellular carcinoma? Cancers. 1 de julio de. 2021;13(13) doi: 10.3390/cancers13133248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang F., Liu G.S., Dusting G.J., Chan E.C. NADPH oxidase-dependent redox signaling in TGF-β-mediated fibrotic responses. Redox Biol. 2014;2(1):267–272. doi: 10.1016/j.redox.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaidi S., Gough N.R., Mishra L. En: Advances in Cancer Research. Academic Press Inc.; 2022. Mechanisms and clinical significance of TGF-β in hepatocellular cancer progression; pp. 227–248. [DOI] [PubMed] [Google Scholar]
- 21.Ally A., Balasundaram M., Carlsen R., Chuah E., Clarke A., Dhalla N., et al. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 15 de junio de. 2017;169(7):1327–1341.e23. doi: 10.1016/j.cell.2017.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cumbo F., Fiscon G., Ceri S., Masseroli M., Weitschek E. TCGA2BED: extracting, extending, integrating, and querying the cancer genome atlas. BMC Bioinformatics. 3 de enero de. 2017;18(1) doi: 10.1186/s12859-016-1419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossman R.L., Heath A.P., Ferretti V., Varmus H.E., Lowy D.R., Kibbe W.A., et al. Toward a shared vision for cancer genomic data. N. Engl. J. Med. 22 de septiembre de 2016;375(12):1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hänzelmann S., Castelo R., Guinney J. Vol. 14. BMC Bioinformatics; 2013. p. 7.http://www.biomedcentral.com/1471-2105/14/7http://www.bioconductor.org.Background (GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data [Internet]). Disponible en: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celià-Terrassa T., Liu D.D., Choudhury A., Hang X., Wei Y., Zamalloa J., et al. Normal and cancerous mammary stem cells evade interferon-induced constraint through the miR-199a–LCOR axis. Nat Cell Biol. 1 de junio de. 2017;19(6):711–723. doi: 10.1038/ncb3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zentella A., Weis F.M.B., Ralph D.A., Laiho M., Massaguel J. Vol. 11. MOLECULAR AND CELLULAR BIOLOGY; 1991. pp. 4952–4958.https://journals.asm.org/journal/mcb (Early Gene Responses to Transforming Growth Factor-,B in Cells Lacking Growth-Suppressive RB Function [Internet]). Disponible en: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez A., Alvarez A., Benito M., Fabregat I. Transforming growth factor beta modulates growth and differentiation of fetal hepatocytes in primary culture. Journal of Cell Physiology. 1 de marzo de. 1995;165(2):398–405. doi: 10.1002/jcp.1041650221. [DOI] [PubMed] [Google Scholar]
- 28.Mateyak M.K., Obaya A.J., Sedivy J.M. Vol. 19. MOLECULAR AND CELLULAR BIOLOGY; 1999. pp. 4672–4683.https://journals.asm.org/journal/mcb (c-Myc Regulates Cyclin D-Cdk4 And-Cdk6 Activity but Affects Cell Cycle Progression at Multiple Independent Points [Internet]). Disponible en: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramjaun A.R., Tomlinson S., Eddaoudi A., Downward J. Upregulation of two BH3-only proteins, Bmf and Bim, during TGFβ-induced apoptosis. Oncogene. 15 de febrero de. 2007;26(7):970–981. doi: 10.1038/sj.onc.1209852. [DOI] [PubMed] [Google Scholar]
- 30.Caja L., Bertran E., Campbell J., Fausto N., Fabregat I. The transforming growth factor-beta (TGF-β) mediates acquisition of a mesenchymal stem cell-like phenotype in human liver cells. Journal of Cellular Physiology. mayo de. 2011;226(5):1214–1223. doi: 10.1002/jcp.22439. [DOI] [PubMed] [Google Scholar]
- 31.Ungefroren H., Witte D., Lehnert H. The role of small GTPases of the rho/rac family in TGF-B-induced EMT and cell motility in cancer. Dev Dyn [Internet] 2018 doi: 10.1002/dvdy.24505. https://anatomypubs.onlinelibrary.wiley.com/doi/10.1002/dvdy.24505 Disponible en: [DOI] [PubMed] [Google Scholar]
- 32.Hilenski L.L., Clempus R.E., Quinn M.T., Lambeth J.D., Griendling K.K. Distinct subcellular localizations of Nox1 and Nox4 in vascular smooth muscle cells. Arteriosclerosis, thrombosis, and vascular biology. abril de. 2004;24(4):677–683. doi: 10.1161/01.ATV.0000112024.13727.2c. [DOI] [PubMed] [Google Scholar]
- 33.Fernandez I., Martin-Garrido A., Zhou D.W., Clempus R.E., Seidel-Rogol B., Valdivia A., et al. Hic-5 mediates TGFβ-induced adhesion in vascular smooth muscle cells by a Nox4-dependent mechanism. Arterioscler. Thromb. Vasc. Biol. 27 de mayo de 2015;35(5):1198–1206. doi: 10.1161/ATVBAHA.114.305185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruix J., da Fonseca L.G., Reig M. Insights into the success and failure of systemic therapy for hepatocellular carcinoma. Nature Reviews Gastroenterology and Hepatology. 1 de octubre de. 2019;16(10):617–630. doi: 10.1038/s41575-019-0179-x. [DOI] [PubMed] [Google Scholar]
- 35.Rizvi F., Siddiqui R., DeFranco A., Homar P., Emelyanova L., Holmuhamedov E., et al. Simvastatin reduces TGF-β1-induced SMAD2/3-dependent human ventricular fibroblasts differentiation: role of protein phosphatase activation. Int. J. Cardiol. 2018;270:228–236. doi: 10.1016/j.ijcard.2018.06.061. noviembre de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helfinger V., Freiherr von Gall F., Henke N., Kunze M M., Schmid T., Rezende F., et al. Genetic deletion of Nox4 enhances cancerogen-induced formation of solid tumors. Proc. Natl. Acad. Sci. USA. 2021;118(11) doi: 10.1073/pnas.2020152118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jong H.S., Lee H.S., Kim T.Y., Im Y.H., Park J.W., Kim N.K., et al. Attenuation of transforming growth factor β-induced growth inhibition in human hepatocellular carcinoma cell lines by cyclin D1 overexpression. Biochem. Biophys. Res. Commun. 29 de marzo de 2002;292(2):383–389. doi: 10.1006/bbrc.2002.6666. [DOI] [PubMed] [Google Scholar]
- 38.Meyer C., Liu Y., Kaul A., Peipe I., Dooley S. Caveolin-1 abrogates TGF-β mediated hepatocyte apoptosis. Cell Death Dis. 2013;4 doi: 10.1038/cddis.2012.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nass S.J., Li M., Amundadottir L.T., Furth P.A., Dickson R.B. Role for bcl-xL in the regulation of apoptosis by EGF and TGF beta1 in c-myc overexpressing mammary epithelial cells. Biochem. Biophys. Res. Commun. 1996;227:248–256. doi: 10.1006/bbrc.1996.1497. [DOI] [PubMed] [Google Scholar]
- 40.Takehara T., Tatsumi T., Suzuki T., Rucker E.B., Hennighausen L., Jinushi M., et al. Hepatocyte-specific disruption of Bcl-xL leads to continuous hepatocyte apoptosis and liver fibrotic responses. Gastroenterology. 2004;127(4):1189–1197. doi: 10.1053/j.gastro.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 41.Tobar N., Guerrero J., Smith P.C., Martínez J. NOX4-dependent ROS production by stromal mammary cells modulates epithelial MCF-7 cell migration. Br. J. Cancer. 28 de septiembre de 2010;103(7):1040–1047. doi: 10.1038/sj.bjc.6605847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boudreau H.E., Casterline B.W., Rada B., Korzeniowska A., Leto T.L. Nox4 involvement in TGF-beta and SMAD3-driven induction of the epithelial-to-mesenchymal transition and migration of breast epithelial cells. Free Radic. Biol. Med. 2012;53(7):1489–1499. doi: 10.1016/j.freeradbiomed.2012.06.016. 1 de octubre de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiraga R., Kato M., Miyagawa S., Kamata T. Nox4-derived ROS signaling contributes to TGF-beta -induced Epithelial-mesenchymal transition in pancreatic cancer cells. Anticancer Res. 2013;10:4431–4439. [PubMed] [Google Scholar]
- 44.Ma M., Shi F., Zhai R., Wang H., Li K., Xu C., et al. TGF-B promote epithelial-mesenchymal transition via NF-KB/NOX4/ROS signal pathway in lung cancer cells. Mol. Biol. Rep. 2021;48(3):2365–2375. doi: 10.1007/s11033-021-06268-2. 1 de diciembre de. [DOI] [PubMed] [Google Scholar]
- 45.Clempus R.E., Sorescu D., Dikalova A.E., Pounkova L., Jo P., Sorescu G.P., et al. Nox4 is required for maintenance of the differentiated vascular smooth muscle cell phenotype. Arteriosclerosis, Thrombosis, and Vascular Biology. enero de. 2007;27(1):42–48. doi: 10.1161/01.ATV.0000251500.94478.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyle A.N., Deshpande N.N., Taniyama Y., Seidel-Rogol B., Pounkova L., Du P., et al. Poldip2, a novel regulator of Nox4 and cytoskeletal integrity in vascular smooth muscle cells. Circ. Res. 31 de julio de 2009;105(3):249–259. doi: 10.1161/CIRCRESAHA.109.193722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiménez-Altayó F., Meirelles T., Crosas-Molist E., Sorolla M.A., del Blanco D.G., López-Luque J., et al. Redox stress in Marfan syndrome: dissecting the role of the NADPH oxidase NOX4 in aortic aneurysm. Free Radic. Biol. Med. 2018;118:44–58. doi: 10.1016/j.freeradbiomed.2018.02.023. 1 de abril de. [DOI] [PubMed] [Google Scholar]
- 48.Datla S.R., Mcgrail D.J., Vukelic S., Huff L.P., Lyle A.N., Pounkova L., et al. Poldip2 controls vascular smooth muscle cell migration by regulating focal adhesion turnover and force polarization. Am. J. Physiol. Heart Circ. Physiol. 2014;307:945–957. doi: 10.1152/ajpheart.00918.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Z.M., Tseng H.Y., Tsai H.W., Su F.C., Huang H.S. Transforming growth factor β-interacting factor-induced malignant progression of hepatocellular carcinoma cells depends on superoxide production from Nox4. Free Radic. Biol. Med. 2015;84:54–64. doi: 10.1016/j.freeradbiomed.2015.03.028. 1 de julio de. [DOI] [PubMed] [Google Scholar]
- 50.Doppler W., Jansen-Dürr P. Regulation of mitochondrial ROS production by HIC-5: a common feature of oncogene-induced senescence and tumor invasiveness? FEBS Journal. 1 de febrero de. 2019;286(3):456–458. doi: 10.1111/febs.14746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santibáñez J.F., Kocić J., Fabra A., Cano A., Quintanilla M. Rac1 modulates TGF-β1-mediated epithelial cell plasticity and MMP9 production in transformed keratinocytes. FEBS (Fed. Eur. Biochem. Soc.) Lett. 2010;584(11):2305–2310. doi: 10.1016/j.febslet.2010.03.042. junio de. [DOI] [PubMed] [Google Scholar]
- 52.Liu Z.Z., Taiyab A., West-Mays J.A. MMP9 differentially regulates proteins involved in actin polymerization and cell migration during TGF-β-induced EMTin the Lens. International Journal of Molecular Sciences. 1 de noviembre de. 2021;(21):22. doi: 10.3390/ijms222111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The work here did not generate massive data. But we used public data related to gene expression in HCC patients, as specified in the Materials and Methods Section and in the figures.