Abstract
Soil health and root-associated microbiome are interconnected factors involved in plant health. The use of manure amendment on agricultural fields exerts a direct benefit on soil nutrient content and water retention, among others. However, little is known about the impact of manure amendment on the root-associated microbiome, particularly in woody species. In this study, we aimed to evaluate the effects of ovine manure on the microbial communities of the olive rhizosphere and root endosphere. Two adjacent orchards subjected to conventional (CM) and organic (OM) management were selected. We used metabarcoding sequencing to assess the bacterial and fungal communities. Our results point out a clear effect of manure amendment on the microbial community. Fungal richness and diversity were increased in the rhizosphere. The fungal biomass in the rhizosphere was more than doubled, ranging from 1.72 × 106 ± 1.62 × 105 (CM) to 4.54 × 106 ± 8.07 × 105 (OM) copies of the 18 S rRNA gene g-1 soil. Soil nutrient content was also enhanced in the OM orchard. Specifically, oxidable organic matter, total nitrogen, nitrate, phosphorous, potassium and sulfate concentrations were significantly increased in the OM orchard. Moreover, we predicted a higher abundance of bacteria in OM with metabolic functions involved in pollutant degradation and defence against pathogens. Lastly, microbial co-occurrence network showed more positive interactions, complexity and shorter geodesic distance in the OM orchard. According to our results, manure amendment on olive orchards represents a promising tool for positively modulating the microbial community in direct contact with the plant.
Keywords: Manure amendment, Microbial ecology, Microbial co-occurrence network, Oleae europaea, Rhizosphere, Root endosphere
Graphical Abstract
Highlights
-
●
Sheep manure increases nutrient levels in the soil of olive orchards.
-
●
Fungal community was more diverse in the root when sheep manure was applied.
-
●
Fungal biomass was increased in the rhizosphere of the organic managed trees.
-
●
Microbial co-occurrence networks were more complex in the organic-managed orchard.
1. Introduction
The cultivation of the olive tree (Olea europaea L. subsp. europaea var. europaea) has for millennia been a key element in the nutrition, economy and culture of many regions, particularly in countries located in the Mediterranean Basin [1]. Spain is the largest producer and exporter of olive oil, accounting for 36% of the worldwide production [2]. Olive oil is one of the main components of the Mediterranean diet, which has been declared as Intangible Cultural Heritage of Humanity (www.unesco.org). Moreover, consumption of olive oil is experiencing an outstanding expansion and increasing attention due to the known benefits for human’s health. For instance, it has been widely proved that the regular intake of extra virgin olive oil promotes a better lipid profile, successfully reducing thrombosis risk [3]. From an agronomic perspective, olive cultivation is less demanding in irrigation and nutrient levels than other trees, and its implementation in agroforestry can provide many ecosystem services (López-Escudero and Mercado-Blanco, 2011). All of this has led to an expansion of olive tree outside traditional cultivation areas, in areas of South Africa, China, Japan and Australia [1].
In the last decades, olive cultivation systems have faced important changes in order to increase yield and production, leading to the implementation of higher-density orchards [4]. Overall, most of the olive plantations are conventionally managed, involving the use of pesticides, herbicides and excessive tillage and clearing [5]. These agronomic practices have a broad impact on the environment, not only in terms of carbon released but also in terms of biodiversity [6]. They also affect the compaction and nutrient and water contents of the soil, factors that, among others, exert a direct effect on soil microbial communities as well as on those ones directly associated with trees [7], [8], [9], [10], [11]. In this sense, it is well-known that the interactions with microorganisms are essential for tree health, including those occurring in the rhizosphere and the root endosphere [7], [12], [13]. In fact, there is wide evidence of the role of root-associated microbiota in growth promotion, nutrient acquisition and protection against pathogens [7], [13], [14], [15].
Organic management of olive orchards is being increasingly implemented as a way to tackle the above-mentioned issues. For example, manure amendment constitutes a very complete source of nutrients, including nitrogen, phosphorous, potassium, micronutrients and organic matter [16]. Apart from being a rich nutritional supply, manure amendment promotes organic matter retention in the soil, thus enhancing water-holding capacity, porosity or aggregation, among others [17], [18]. This turns into a range of beneficial indirect effects such as preventing soil erosion, a matter of utmost importance in the Mediterranean region due to its intermittent heavy rainfall events and risk of desertification [19]. Some of these effects are exerted through modification of the soil microbiota. For instance, it has been shown that soil structure is enhanced by organic amendments through an increase in fungal abundance [20]. In addition, organic amendments can reduce the severity, incidence and even completely supress soilborne pathogens, being a source of microbial antagonists effective against phytopathogens [21]. In relation with this, it has also been shown that organic amendment application can increase the activity of soil microorganisms [22]. In summary, organic management in general, and manure inputs in particular, can generate important benefits to agricultural fields through a variety of interconnected mechanisms.
As mentioned above, nutrient levels directly affect microbial communities in soil. Therefore, manure amendment is known to modify soil as well as root microbiota. This has been widely studied for herbaceous plants but scarcely for woody species [23], [24], [25], [26], [27], [28]. Although several studies on the effect of manure amendment on olive tree yield are available[5], [29], [30], [31], [32], little is known about its influence on the olive root microbiome. Most of the studies conducted were based on culture-dependent analysis [33], [34], [35], focused on a specific group of bacteria [36], [37] or on other plant compartments, such as the xylem [38], [39].
Understanding to what extent the root microbial community of olive trees are shaped by manure inputs may contribute to a more complete picture of the impacts of organic management in such an important long-living, woody crop. Therefore, the aim of this study was to determine the effect of agricultural management (specifically, organic management vs. conventional management) on the root microbiome of olive trees. To achieve this target, the following objectives were pursued: assessment of the impact of sheep manure amendment in (i) shaping the belowground microbial (bacteria and fungi) community, (ii) altering the potential functions of the root bacterial community, and (iii) affecting microbial biomass in the olive trees rhizosphere, under field conditions. We test the hypothesis that sheep manure amendment has a positive impact on the belowground microbial communities associated with olive trees at different levels, such as diversity, metabolic functions, microbial biomass and co-occurrence network interactions.
2. Material and methods
2.1. Sample collection
Two adjacent olive orchards with different soil management (i.e., organic management [OM] and conventional management [CM)]) were surveyed. The CM orchard was subjected to traditional agronomic practices implemented in the region, including herbicide and pesticide treatments and mechanical plow. Specifically, herbicides were applied twice a year, on autumn and spring. The herbicides used were GLIFOVER (Nutesca, Jaén, Spain), Tragli Gold (BRANDT EUROPE, S.L., Sevilla, Spain), MINSK (KENOGARD, Barcelona, Spain) and PROTIBEL (PROBELTE, S.A.U, Murcia, Spain). Insecticides and fungicides (copper oxychloride, 25%) were used once a year. The specific products used as insecticides were RITMUS (PROBELTE, S.A.U, Murcia, Spain) and DAFENE PROGRESS (Bayer CropScience, S.L, Valencia, Spain). Chemical fertilization was applied to both leaves (AMISAN 80%, AMISAN REVITALIZADOR, ESTESAN 18–13–27 and ESTESAN 3.5–0–42–1MgO, all provided by Nutesca, S.L, Jaén, Spain) and soil (ammonium sulfate, 3.5 kg per olive tree and per year). In this orchard, the last application of herbicides was done 6 months before sampling. In contrast, the OM orchard did not receive any herbicide treatment. Sheep manure amendments (350 kg per hectare) were applied to the soil every two years for at least 7 years. Moreover, potassium chloride and Flecotec 4 SMART (ICL Specialty Fertilizers Iberia, Murcia, Spain), a certified ecological fertilizer that provides potassium, calcium, magnesium and sulfur, were applied to the soil as input of inorganic nutrients. Regarding irrigation, both olive orchards only received the natural rain regime. The sampling was carried out in the autumn of 2020 in the municipality of Lupión, Jaén province, Southern Spain (37˚ 59’ 56,2’’ N; 03˚ 35’ 16,3’’ W; 325 m above sea level). The owners were fully informed of our activities in advance and permission was granted. No further permissions were needed as no collection of genetic resources was intended. An image from the sampled fields can be seen in the supplementary figure 1 (Additional file 1: Fig. S1).
For each orchard, 8 trees of cultivar Picual were considered for analysis. From each tree, rhizosphere and endosphere samples were obtained. To assure representativeness, samples for each tree were collected from four different points (in two opposite zones, N = North side and S=South side). After removing the upper soil layer (less than 5 cm), the main roots of each plant were followed until finding young, cork-free roots at around 5–20 cm depth. For rhizosphere samples, soil firmly adhered to active roots was collected. Both soil and root samples were stored at 4 °C and processed in the next 48 h [40]. Root samples were washed and surfaced sterilized as previously described by Fernández-González et al. [41]. Once processed, samples were frozen in liquid nitrogen and stored at −80 °C. Aliquots of the last sterile rinse water were plated onto NA (Nutrient Agar) and LBA (Luria Bertani Agar) plates and incubated at 28 °C for 7 days to confirm the performance of the disinfection protocol. Additionally, three bulk soil samples of each orchard were obtained. In order to produce more representative samples of the bulk soil, samples were obtained on every tree from four different digs. After that, these samples were mixed and three bulk soil samples were obtained. These samples were analysed at the L.A.B Innovación Analítica (A Tentamus Company, Almería, Spain) to determine soil physicochemical properties and the presence of pesticides and biocides with a total of 516 compounds using standardized procedures implemented in this commercial service. The list of compounds is shown in the supplementary table 1 (Additional file 2: Table S1).
2.2. DNA extraction and quantitative PCR (qPCR)
For each replicate (tree), samples from the four digs were equally mixed. From this mixture, soil DNA was obtained using the Power Soil DNA Isolation kit (MoBio, Laboratories Inc., CA) following the manufacturer’s recommendations. Roots were ground in liquid nitrogen and 100 mg of this tissue was used for each replicate to extract DNA, using the Maxwell RSC (Rapid Sample Concentrator) with the ‘PureFood GMO and Authentication’ Kit (Promega Biotech Ibérica S.L, Madrid, Spain) according to the manufacturer’s instructions. For both compartments (rhizosphere and root endosphere), DNA yield and quality were checked both by electrophoresis in 0.8% (w/v) agarose gels stained with GelRed and visualization under UV light, and by using a Qubit 3.0 fluorometer (Life Technologies, Grand Island, NY).
Real-time PCR was used for absolute quantification of the copy number of the 16 S rRNA and 18 S rRNA genes present in the rhizosphere samples. 16 S rRNA quantification was performed as previously described by [42] in terms of primers used, qPCR conditions, calibration and DNA template concentration. For 18 S rRNA gene quantification, FR1 and FF390 primers were used [43]. The optimal conditions for amplification were 0.05 ng of template DNA and 56 °C of annealing temperature. Reaction conditions were: 95 °C for 30 s; 40 cycles consisting of 95 °C for 30 s, 56 °C for 30 s and 72 °C for 30 s. The melting curve was obtained at the end of each run by increasing the temperature from 62 °C to 95 °C. For calibration curves, the 18 S rRNA gene from template DNA of Rhizophagus irregularis (DAOM 197198) [44] was cloned into pGEM-T Easy Vector System (Promega) using primers Euk6–23 and REuk1755–1772 [45]. The cloning product was checked with NotI digestion and PCR with FR1 and FF390 primers followed by agarose gel electrophoresis.
For both bacterial and fungal quantification, mixes were prepared using the TB Green Premix Ex Taq II (Tli RNase H Plus) (Takara Bio Europe SAS, Saint-Germain-en-Laye, France) and reactions were carried out with QuantStudio™ 3 Real-Time PCR System (Applied Biosystems™, Alcobendas, Spain). The qPCR reaction mixtures contained 5 μl of TB Green Premix, 0.4 μl of each primer (10 μM), 3.2 μl of H2O and 1 μl of DNA template 2 μl of H2O and 1 μl of DNA template. Results were analysed with QuantStudio Design & Analysis Software v1.5.2 (Applied Biosystems™, Alcobendas, Spain), and copy numbers were calculated per gram of soil.
2.3. Illumina sequencing and data processing
The DNA was sequenced with the Illumina MiSeq platform at the genomics service of the Institute of Parasitology and Biomedicine “López Neyra” (CSIC; Granada, Spain) following the recommended Illumina’s protocols. In the first run, a prokaryotic library was constructed amplifying the hyper-variable regions V3-V4 of the 16 S rRNA gene using the primer pair Pro341F (5’-CCTACGGGNBGCASCAG-3’) and Pro805R (5’-GACTACNVGGGTATCTAATCC-3’) according to [46]. These amplicons were tagged to be attached to PNA PCR clamps to reduce plastid and mitochondrial DNA amplification [47]. In the second run, a eukaryotic library was constructed amplifying the ITS2 region using the primer pair ITS4 (5’-TCCTCCGCTTATTGATATGC-3’) [48] and fITS7 (5’-GTGARTCATCGAATCTTTG-3’) [49]. Both runs were sequenced using a paired-end 2 × 300-bp (PE 300) strategy. Moreover, a ZymoBIOMICS Microbial Community Standard II (Log Distribution), ZYMO RESEARCH (https://www.zymoresearch.com/collections/zymobiomics-microbial-community-standards/products/zymobiomics-microbial-community-standard-ii-log-distribution), was added in triplicate in each run as metabarcoding sequencing control.
Raw reads were processed following our homemade tutorial, recently published on GitHub (https://nuriamw.github.io/micro4all/) and also available as an R package (Micro4all, see workflow scheme in Fig. 1). For inferring Amplicon Sequence Variants (ASVs), this tutorial makes use of DADA2. Various parameters were modified. In quality filtering, the function filterAndTrim was used and the parameter maxEE was set to 2 and 5 maximum expected errors for forward and reverse reads for both prokaryotic and eukaryotic datasets. Merging was done with default parameters. In prokaryotic dataset, reads smaller than 382 and larger than 444 nt were discarded. For eukaryotic dataset, only reads smaller than 100 nt were removed for further analyses. Finally, classification of bacterial and fungal ASVs was achieved using assignTaxonomy command against the Ribosomal Database Project II, training set v.18 [50] and the UNITE v.7.2 dynamic database [51], respectively.
Fig. 1.
Micro4all workflow. Pipeline summary for microbial community analysis. It allows users to easily analyze metabarcoding data from quality checking to ecological analyses. The pipeline includes functions from the R package Micro4all (https://nuriamw.github.io/micro4all/tutorial/package_workflow.html) to simplify analyses such as the calculation of a cut-off based on the MOCK community sequencing result, applying statistical tests to several diversity indices or performing ANCOM-BC between every possible comparison.
ASVs corresponding to host DNA were eliminated, i.e. those that were classified as mitochondria, chloroplast and unknown sequences. Moreover, eukaryotic ASVs not classified as fungi at the kingdom level were removed. Finally, MOCK community sequences were analysed to make the final trimming. According to these data, ASVs with relative abundance smaller than 0.002% of the total number of sequences were eliminated for each dataset.
2.4. α-diversity, β-diversity and differential abundance analysis
α-diversity indices (Observed Richness, Shannon, Inverse of Simpson and Evenness), rarefaction curves and β-diversity (determined by PCoA analysis based on Bray-Curtis, Unweighted Unifrac and Weighted Unifrac dissimilarities) were carried out as previously described by Fernández-González et al. [41]. Statistical tests included Kruskal-Wallis for α-diversity indices, and Permutational Multivariate Analysis of Variance (PERMANOVA) and Multivariate Homogeneity of Groups Dispersions (BETADISPER) for β-diversity analysis. Moreover, differences in taxonomical abundances were assessed with ANCOM-BC [52] tool, following our above-mentioned tutorial (https://nuriamw.github.io/micro4all/).
2.5. Metabolic functions prediction
To delve into the biogeochemical cycle functions of the bacteriome members, Functional Annotation of Prokaryotic Taxa (FAPROTAX) bioinformatic tool (v.1.2.4 release) was employed at the ASV level. FAPROTAX is a manually constructed database that maps prokaryotic taxa to establish metabolic or other ecologically relevant functions using the current literature on cultured strains. This tool was used following the developers' tutorial [53]. Briefly, it allows users to map their bacterial member's classification to functions. Afterwards, statistically significant differences between OM and CM for each metabolic function were determined with a Wilcoxon test with Benjamini-Hochberg FDR correction.
2.6. Microbial co-occurrence network construction
Four microbial (bacterial and fungal) networks were constructed (i.e., two orchards and two compartments). The online tool MENAP (ieg4.rccc.ou.edu/mena/main.cgi) was used following the developer’s instructions, including the new implementation to discern between direct and indirect interactions [54] as described in Gómez-Lama Cabanás et al. [55], with the only difference that the prevalence was set to 50% (as default) and correlation was calculated with Pearson correlation coefficient.
2.7. Prediction of fungal guilds
In order to obtain information about the functional characteristics of fungal taxa under study, FungalTraits database was used to predict the primary life style of fungi [56]. This was done at genera level (both in the rhizosphere and endosphere communities) using the database spreadsheet, according to the authors recommendations. Once the guilds were assigned to each genus, the guilds relative abundance was calculated. Statistically significant differences were assessed using a Wilcoxon test with Benjamini-Hochberg FDR correction.
3. Results
3.1. Effect of agricultural management on soil: physicochemical parameters and microbial biomass
3.1.1. Soil physicochemical parameters
The studied soils showed the same characteristics in terms of composition. Both OM and CM orchards are established on clay soils, with a percentage of clay ranging from 60.40% to 66.40% (Additional file 3: Table S2). Surprisingly, the pH was similar in both soils regardless of the management implemented. However, nutrient composition was clearly different. As expected, the OM soil was richer than the CM soil in terms of nutrient content. Indeed, oxidable organic matter, total nitrogen, assimilable K, nitrate, sulfate and bicarbonate were all statistically significantly (Student t test, p < 0.05) higher in OM soil. Assimilable P was also higher in this orchard, but no statistical test could be used as its concentration was below the detection limit for two out of three bulk soil samples analysed. Finally, soil conductivity was also statistically significantly (Student t test, p < 0.05) higher in OM soil. None of the 516 compounds (biocides and pesticides) measured were detectable in the samples, neither in OM nor in CM.
3.1.2. Rhizosphere microbial biomass
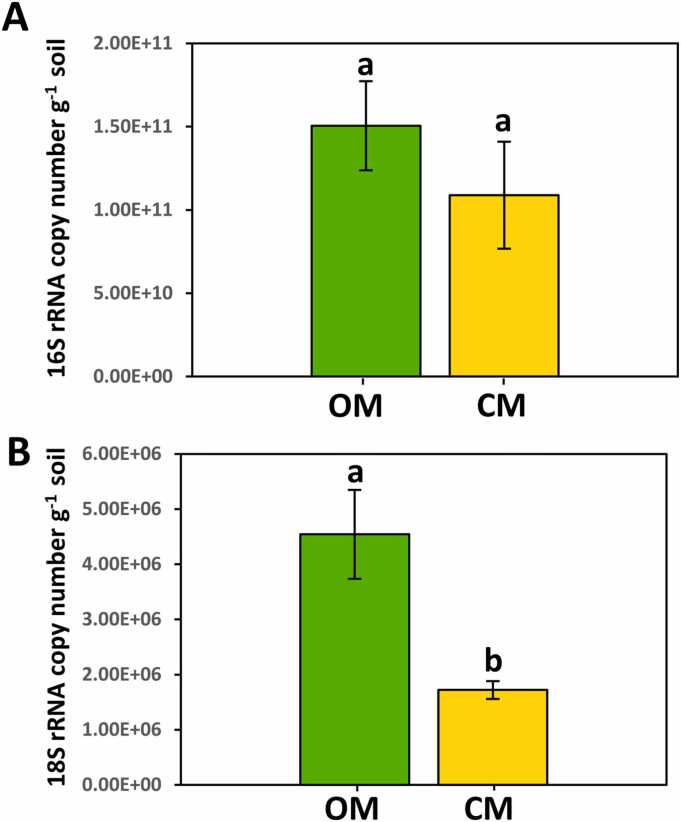
Bacterial and fungal biomasses present in the OM and CM rhizospheres were determined in terms of 16 S rRNA and 18 S rRNA genes copy number g-1 soil. Overall, organic management enhanced bacterial and fungal biomasses (Fig. 2). The 16 S rRNA gene copy number in the rhizosphere reached 1.50 × 1011 ± 2.68 × 1010 copies per gram of soil in the OM orchard, while in the CM orchard 1.09 × 1011 ± 3.21 × 1010 copies were scored (Fig. 2A). Nevertheless, differences observed were not statistically significant (Student’s t test; p = 0.38). In contrast, the fungal biomass showed a statistically significant (p = 0.01) increase in the OM orchard. In fact, OM samples reached 4.54 × 106 ± 8.07 × 105 copies of the 18 S rRNA gene per gram of soil while values for the CM soil remained at 1.72 × 106 ± 1.62 × 105 gene copies (Fig. 2B).
Fig. 2.
Bacterial and fungal biomass in the rhizosphere. Copy number g-1 soil of the 16 S rRNA (A) and 18 S rRNA genes (B) are presented for both studied conditions (OM and CM). The values significantly different at the 5% level (by a Student t test) are shown by different letters. OM: organic management; CM: conventional management.
3.2. General characteristics of sequenced datasets
The total number of raw reads obtained for bacterial and fungal datasets were 4,567,342 and 4,899,001, respectively. After quality filtering and ASVs inferring, 2,909,036 (bacterial) and 3,136,096 (fungal) sequences were retained. High-quality sequences represented at least 44,111 and at maximum 100,161 sequences per sample from the prokaryotic dataset; and 25,157 and 145,551 sequences from the fungal dataset. Finally, the mock community sequenced by triplicate for each library was analysed and ASVs with less than 0.002% of the bacterial and fungal reads were discarded. This yielded a final number of 3997 bacterial and 1650 fungal ASVs that were used for further analyses.
3.3. Sheep manure amendment differently affects bacterial and fungal communities
Diversity and richness (α-diversity), expressed as Observed Richness, Shannon and Inverse of Simpson indices, showed statistically significant differences only in root (rhizosphere and endosphere) fungal community, according to the Wilcoxon test (Fig. 3). Moreover, differences in α-diversity can be explained because of a variation in observed richness, as the evenness index comparison was not statistically significant. In contrast, the root bacterial α-diversity showed no differences either in the rhizosphere or in the endosphere (Figs. 3A and 3B).
Fig. 3.
Bacterial (A, B) and Fungal (C, D) α-diversity indices of each treatment and compartment (A and C, rhizosphere; B and D, endosphere). For every panel, it is shown five summary statistics (the median, two hinges and two whiskers) and outlying points. Asterisks show statistically significant differences according to a Wilcoxon test test (p value < 0,05). OM: organic management, CM: conventional management.
Interestingly, community structure (β-diversity) differed in both microhabitats (rhizosphere and root endosphere) and both kingdoms (Bacteria and Fungi). Specifically, a PERMANOVA test showed statistically significant differences between orchards (OM versus CM) for all analyses (Table 1). In order to elucidate variations in dispersion between treatments, a BETADISPER test was carried out and variance differences were not statistically significant, except for the fungal community rhizosphere. However, the graphical representation showed a sharp separation between OM and CM fungal communities in this compartment (Additional file 4: Fig. S2). Therefore, we can certainly say that in all cases the manure amendment had an effect on the belowground microbial community structure of the studied olive trees.
Table 1.
p values of beta diversity indices.
| PERMANOVA |
R2 |
BETADISPERSION |
||||
|---|---|---|---|---|---|---|
| Compartment | Bacteria | Fungi | Bacteria | Fungi | Bacteria | Fungi |
| Rhizosphere | 0.0011 | 0.0012 | 0.57 | 0.22 | 0.89 | 0.02 |
| Endosphere | 0.0012 | 0.0012 | 0.18 | 0.15 | 0.31 | 0.658 |
Numbers in boldface and italics show statistically significant p values (p < 0.05) according to PERMANOVA and BETADISPER tests between the OM and CM studied olive orchards. OM: organic management, CM: conventional management. 1: with Weighted Unifrac dissimilarities, 2: with Bray-Curtis dissimilarities.
3.4. Influence of soil management on the taxonomical profile of the olive rhizosphere
Bacterial taxonomical profile (phylum level) in the rhizosphere was dominated by Acidobacteria, Proteobacteria, Actinobacteria, Gemmatimonadetes, Bacteroidetes and Candidate division WPS-1 (Fig. 4A). These phyla accounted for at least 80.91% of the sequences. Among phyla that represented at least 0.9% of the total sequences, Acidobacteria, Gemmatimonadetes and Candidate division WPS-1 were statistically significantly (ANCOM-BC test, p < 0.05) more abundant in CM, while Bacteroidetes, Chloroflexi, Firmicutes and Nitrospirae were prevalent in OM (Table S2).
Fig. 4.
Bacterial taxonomic profiles of the rhizosphere and endosphere. Bacterial taxonomic profile at phylum (A, C) and genus level (B, D) for the rhizosphere (above panel) and endosphere (below panel) compartments in the two studied orchards. Only main taxa are displayed. Bold letters show statistically significant differences (p-value < 0.05) according to ANCOM-BC test. OM: organic management; CM: conventional management.
At genus level the community was dominated by members of genera Gp6, Sphingomonas, Ohtaekwangia, Gp4 and Rubrobacter (Fig. 4B). Among the genera with statistically significant differences, the 20 most abundant were analysed in detail. In this sense, Gp6, Sphingomonas, Gp4, Rubrobacter, Roseisolibacter, Gp7, Blastococcus, Stenotrophobacter, Arenimicrobium, Gemmatirosa, Flavitalea, Flavisolibacter, Ramlibacter and Actinomarinicola were more relatively abundant in the CM orchard, while Ohtaekwangia, Agromyces, Opitutus, Pseudoartrobacter, Nitrospira and Nocardioides were more relatively abundant in the rhizosphere of OM trees (Table S3).
Regarding the fungal community phyla Ascomycota, Glomeromycota and Basidiomycota were the most abundant, accounting for at least 88.52% of the sequences (Fig. 5A). Moreover, Basidiomycota and Ascomycota showed statistically significantly (ANCOM-BC test, p < 0.05) higher relative abundances in the CM orchard, while Glomeromycota was more relatively abundant in the rhizosphere of OM trees (Additional file 5: Table S4).
Fig. 5.
Fungal taxonomic profiles of the rhizosphere and endosphere. Fungal taxonomic profile at phylum (A, C) and genus level (B, D) for the rhizosphere (above panel) and endosphere (below panel) compartments in the two studied orchards. Only main taxa are displayed. Bold letters show statistically significant differences (p-value < 0.05) according to ANCOM-BC test. OM: organic management; CM: conventional management.
Fungal taxonomical profiles at genus level varied greatly between management conditions (Fig. 5B). Nevertheless, Solicoccozyma was the most abundant genus in both orchards, representing 9.86% (OM) and 9.60% (CM) of the sequences. Other relevant genera in terms of relative abundance were Mortierella (7.85%) and Lophotricus (5.78%) for the OM orchard and Tricharina (5.92%) and Coniosporium (4.59%) for the CM orchard. Among the genera with statistically significant differences, the 20 most abundant were Coniosporium, Paratricharina, Kamienskia, Montagnula and Bradymyces (more abundant in the CM orchard), and Mortierella, Lophotricus, Preussia, Chrysosporium, Wallemia, Heydenia, Phyloctochytrium, Ajellomyces, Plectopsphaerella, Scopulariopsis, Coniochaeta, Volutella, Stagnosporopsis, Diaporthe and Monodictys (more abundant in the OM orchard) (Additional file 5: Table S5).
3.5. Two main taxa in the endosphere taxonomical profiles: Actinophytocola and Ascomycota
Intriguingly, bacterial and fungal root endosphere communities were mostly represented by members of Actinobacteria and Ascomycota phyla, respectively (Fig. 4C and Fig. 5C). Indeed, Actinobacteria accounted for 70.72% of relative abundance in OM and 70.96% in CM while Ascomycota comprised 66.57% in OM and 72.63% in CM of total sequences. In contrast with the rhizosphere, the root endosphere of trees grown under the two management conditions here examined showed similarities at phylum level. For instance, statistically significant (ANCOM-BC test, p < 0.05) differences were only found for minor phyla (i.e. Gemmatimonadetes for the bacterial dataset, representing less than a 0.9% of total sequences) (Additional file 5: Table S6). However, for the fungal dataset, statistically significant (ANCOM-BC test, p < 0.05) differences were found for two of the most abundant phyla, i.e., Ascomycota and Glomeromycota, both with higher dominance in the CM orchard (Additional file 5: Table S8).
At genus level, and concerning the bacterial dataset, Actinophytocola, Kibdelosporangium, Streptomyces and Pseudonocardia (phylum Actinobacteria) accounted for at least 55.41% of the sequences. Moreover, Actinophytocola alone represented 22.51% (OM) and 21.79% (CM) of the bacterial root endosphere community. The most abundant fungal genus was Diaporthe (phylum Ascomycota), which alone made up 13.8 2% (OM) and 3.63% (CM) of the total sequences (Fig. 4D and Fig. 5D).
It should be pointed out that organic management affected mostly minor genera (relative abundance <0.9%) in the bacterial root endosphere community (Additional file 5: Table S7). This was not the case for the fungal dataset, where differences affected 6 genera, Preussia, Mycenella and Cladophialophora being more abundant in the OM orchard and Aureobasidium, Rhizophagus and Alternaria in the CM grove (Table S8).
3.6. Fungal guilds
For both analysed compartments, only guilds with a relative abundance higher than 0.7% were considered. In the rhizosphere, the most abundant guilds were soil saprotroph, plant pathogen, dung saprotroph and arbuscular mycorrhizal. Interestingly enough, primary lifestyles related to organic matter degradation were significantly increased in OM, such as dung saprotrophs, unspecified saprotrophs and litter saprotrophs. Animal parasites were significantly more abundant in OM (Additional file 6: Fig. S3, panel A).
In the endosphere, the most abundant guilds corresponded to plant pathogens, litter saprotrophs, soil saprotrophs, dung saprotrophs and arbuscular mycorrhizal. Following the rhizosphere results, dung saprotrophs were significantly more abundant in OM. Moreover, arbuscular mycorrhizal was significantly enhanced in CM (Additional file 6: Fig. S3, panel B).
3.7. Inferred metabolic functions
FAPROTAX results retrieved function assignment for 37.0% and 87.0% of the total ASVs in the rhizosphere and root endosphere, respectively. As expected, the predominant functions were related to the carbon (C) cycle, with “chemoheterotrophy” and “aerobic chemoheterotrophy” as the two most abundant functions (Table 2). In the communities of both compartments, functions related to degradation of recalcitrant compounds (i.e., “aromatic compound degradation”, “aromatic hydrocarbon degradation”, “hydrocarbon degradation”, “aliphatic non-methane hydrocarbon degradation”) were statistically significantly (Wilcoxon test, p < 0.05) more represented in OM than in CM. These functions were mainly related to members of the genera Rhodococcus and Nocardioides, as well as members of the Methylocystaceae family.
Table 2.
Potential metabolic functions predicted by FAPROTAX. Only functions with an abundance greater than 0.1% are presented. Main taxa contribution shows the 10 most abundant taxa. OM: organic management, CM: conventional management.
| Metabolic functions rhizosphere | OM | CM | Main taxa contribution |
|---|---|---|---|
| C cycle | |||
| Chemoheterotrophy | 16.28* | 14.50 | Methylocystaceae unclassified, Sphingomonas, Rubrobacter, Blastococcus, Agromyces, Solirubrobacter, Opitutus, Lysobacter, Streptomyces, Nocardioides |
| Aerobic chemoheterotrophy | 14.44 | 13.99 | Erythrobacteraceae unclassified, Sphingomonas, Rubrobacter, Blastococcus, Agromyces, Solirubrobacter, Streptomyces, Nocardioides, Microvirga, Mesorhizobium |
| Fermentation | 1.04* | 0.28 | Opitutus, Cellulosimicrobium, Lautropia, Isptericola, Enterococcus, Kluyvera, Geothrix, Cellulomonas, Olsenella, Streptococcus |
| Aromatic compound degradation | 0.58* | 0.17 | Rhodococcus, Nocardioides |
| Hydrocarbon degradation | 0.15* | 0.01 | Rhodococcus, Methylocystaceae unclassified |
| Phototrophy | 0.20* | 0.15 | Blastochloris, Rhodopseudomonas, Rhodomicrobium, Craurococcus, Ectothiorhodospiraceae unclassified |
| Photoautotrophy | 0.18* | 0.09 | Ectothiorhodospiraceae unclassified |
| Anoxygenic photoautotrophy | 0.18* | 0.09 | Ectothiorhodospiraceae unclassified |
| Aromatic hydrocarbon degradation | 0.11* | 0.00 | Rhodococcus |
| Aliphatic non-methane hydrocarbon degradation | 0.11* | 0.00 | Rhodococcus |
| Methylotrophy | 0.13* | 0.03 | Methanomassiliicoccus, Methylobacterium, Methylocystaceae unclassified, Paracoccus, Methylobacillus, Methylophilus, Methylotenera |
| C and N cycles | |||
| Chitinolysis | 0.60* | 0.19 | Lysobacter |
| Ureolysis | 0.46* | 0.17 | Afipia, Methylobacterium, Mesorhizobium, Roseomonas, Azospirillum, Massilia, Methylophilus |
| N cycle | |||
| Nitrate reduction | 3.45 | 3.17 | Rubrobacter, Nitrobacter, Ensifer, Paracoccus, Craurococcus, Achromobacter, Klebsiella, Stenotrophomonas, Kluyvera, Opitutus |
| Nitrification | 1.02* | 0.28 | Nitrososphaera, Nitrospira, Nitrobacter, Nitrosospira |
| Aerobic nitrite oxidation | 0.74* | 0.18 | Nitrospira, Nitrobacter |
| Aerobic ammonia oxidation | 0.28* | 0.09 | Nitrososphaera, Nitrosospira |
| Nitrogen fixation | 0.22 | 0.30 | Bradyrhizobium, Azospirillum |
| S cycle | |||
| Anoxygenic photoautotrophy S oxidizing | 0.18* | 0.05 | Ectothiorhodospiraceae unclassified |
| Others | |||
| Manganese oxidation | 0.11* | 0.24 | Geodermatophilus |
| Not assigned | 57.93 | 64.24 |
Several metabolic functions related to the nitrogen (N) cycle was significant increased statistically (Wilcoxon test, p < 0.05) in the OM grove in both compartments. In the OM rhizosphere, nitrification was favored, as functions related to this process were enhanced, i.e., “nitrification”, “aerobic ammonia oxidation” and “aerobic nitrite oxidation”. In this sense, ureolysis was also enhanced in the OM rhizosphere. Nitrification involves the transformation of ammonia into nitrate, making nitrogen more easily assimilated by the plant. Along with this process, denitrification was enhanced in the OM endosphere. This was shown by the increase in abundance of metabolic functions such as “nitrate reduction”, “nitrate respiration” and “nitrogen respiration”. Lastly, other beneficial metabolic functions were enhanced in OM rhizosphere, as is the case of chitinolysis.
3.8. Opposite effect of soil management on co-occurrence networks in both compartments
Microbial networks were separately constructed and analysed for each orchard (OM and CM) and each compartment (rhizosphere and root endosphere). Overall, agronomic management produced notable differences in the network topologies (Table 3). Moreover, the impact of each management was different depending on the compartment. Indeed, an opposite pattern was observed in the endosphere compared to the rhizosphere, except for the average path distance (GD), which was shorter in the OM networks for both compartments. Specifically, the OM rhizosphere network was more complex than that of the CM, with higher number of links, average degree (avgK) and average clustering coefficient (avgCC) (Table 3). Moreover, the modularity was lower along with a fewer number of modules. Finally, the percentage of positive edges was higher in the OM than in the CM rhizosphere network (Additional file 7: Fig. S4). The opposite pattern was observed in the root endosphere network; that is, the OM network was less complex (according to the number of links, avgK and avgCC), while the modularity was significantly higher. Moreover, the percentage of positive interactions was lower than that observed for the CM endosphere network (Additional file 8: Fig. S5).
Table 3.
The major topological properties of OM and CM co-occurrence networks. Statistically significant p values (p < 0.05) between orchards (OM and CM) are shown in boldface with asterisks. OM: organic management; CM: conventional management. The brackets in Modularity values mean the number of modules in that network. OM: organic management, CM: conventional management.
| Community | No. of original OTUs | Similarity thershold (St) | Total nodes | Totallinks | R2 of power-law | Average degree (avgK) | Average clustering coefficient (avgCC) | Average path distance (GD) | Modularity (nº of modules) | Percentage of positive edges (PEP) |
|---|---|---|---|---|---|---|---|---|---|---|
| OM rhizosphere | 1268 | 0.92 | 871 | 2047 | 0.80 | 4.70 | 0.10* | 7.51* | 0.82(74)* | 55.30% |
| CM rhizosphere | 1333 | 0.93 | 835 | 1887 | 0.83 | 4.50 | 0.08* | 7.81* | 0.90(89)* | 33.70% |
| OM endosphere | 226 | 0.85 | 173 | 239 | 0.74 | 2.76 | 0.04* | 6.79* | 0.77(21)* | 31.80% |
| CM endospohere | 322 | 0.87 | 257 | 705 | 0.74 | 5.49 | 0.17* | 7.08* | 0.59(25)* | 64.40% |
3.9. DATA AVAILABILITY
The sequencing dataset generated during the current study were deposited and are publicly available at the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) repository under the BioProject accession number PRJNA925330.
4. Discussion
Organic amendments have a strong impact on soil properties (e.g. enhanced nutrient levels and moisture), thereby influencing the microbial community in soil, rhizosphere and root endosphere [22], [57], [58]. In this study, we aimed to evaluate the effect of two different agronomic practices, namely organic management and conventional management, on the root microbiome of olive trees. The two studied orchards were adjacent, thus making environmental and pedological conditions similar and reducing variability in this regard. Indeed, both orchards did not show statistically significant differences regarding clay, sand and silt content. However, manure input drastically changed the soil’s nutrient composition, as shown by the observed increases in oxidable organic matter, nitrates, total nitrogen and phosphorous. Although sulfate and potassium concentrations were also increased, this could be due to the amendment with an inorganic ecological fertilizer implemented in the OM orchard. Interestingly enough, levels of nitrates and total nitrogen were higher in this grove, despite the fact that in the CM orchard ammonium sulfate was applied to the soil. Due to changes in the nutrient content, a shift in the microbial community could be expected. In fact, we observed an increment in microbial diversity and a change in community structure in both root compartments studied. Nevertheless, the effect on the α-diversity was only observed for the fungal community, while diversity and richness of the bacteriome remained mostly unchanged. This agrees with previous studies dealing with herbaceous plants under different managements [26], [27], [28], [59], [60]. In fact, a stronger effect on fungal diversity can be related to differences in soil organic matter content [61], in agreement with our results.
Although soil microbial biomass has been studied in olive orchards [62], little is known about the effect of organic amendment on the microbial biomass of the olive rhizosphere. This would be of interest because of the potential implications with plant nutrition. Our results showed that microbial biomass increased upon organic amendment, although statistically significant differences were only found for the fungal community. Several studies demonstrated increases in microbial biomass associated with organic amendments, both in long-term and short-term settings [63], [64], [65], [66], [67]. In our study, bacterial and fungal biomasses were scored separately. By using this approach, we found that sheep manure amendment had a larger impact on the fungal biomass, which can be related to the above-mentioned results on community diversity and carbon content of the soil. Olive trees could thus benefit from this type of amendments by increasing soil nutrients availability, microbial diversity and biomass [68]. These results, while supporting previous studies performed in herbaceous plants [26], [27], [28], [59], set a precedent in woody plants, in which the effect of manure amendment on the belowground microbial communities has not been yet studied.
Differences observed in microbial community structure and microbial biomass correlated with distinctive taxonomical profiles. Indeed, specific phyla and genera enhanced or diminished their relative abundances depending on the soil management implemented in the orchard. Concerning the bacterial community, Acidobacteria were considerably more abundant in the rhizosphere of olive trees subjected to conventional management. Acidobacteria are regarded as slow-growth oligotrophs [69], thus being enhanced in poor soils. Although the CM orchard received a regime of conventional fertilizers, our analysis showed a higher content of nutrients in the OM orchard, hence likely diminishing the presence of this phylum. Likewise, Gemmatimonadetes was also found to be more abundant in the CM rhizosphere. This phylum has been detected in a broad range of soils but its presence seems to be related to water retention, being more abundant in dry soils [70]. Since manure amendments can increase water content in the soil by improving soil structure, their use can diminish members of this phylum. In contrast, Firmicutes and Bacteroidetes members were more abundant in the OM orchard. Both phyla are usually present in faecal matter, hence their abundance can be related to manure application [69], [71]. Furthermore, they could be affected by the higher carbon content found in OM. In fact, Firmicutes members are regarded as copiotrophic bacteria [69]. Concerning Bacteroidetes, they can secrete enzymes to degrade organic compounds present in the soil [72]. Moreover, several studies pointed out their sensitivity to soil management [73], [74], [75]. It is likely that the higher proportion of organic matter found in the OM orchard promoted the growth of Bacteroidetes members. Differences found for other phyla upon manure amendment are more difficult to explain. This was the case of Chloroflexi; while this phylum has not been widely studied and characterized, its abundance has been earlier correlated with nutrient-poor soils [75], [76], [77]. Intriguingly, our results showed that the rhizosphere of olive trees subjected to organic management was enriched in Chloroflexi. Nonetheless, a positive correlation between Chloroflexi and the content of NO3- in soil has been reported in the rhizosphere of wheat plants subjected to increasing N fertilization [78]. This outcome was also observed in our study, since the OM orchard had a significantly higher amount of nitrates. The contrasting nutrient content observed between OM and CM soils could also explain the differences found for the phylum Nitrospirae, particularly the genus Nitrospira, which was significantly more abundant in the OM orchard. Nitrospira spp. have recently gained attention because of their ability to perform complete ammonia oxidation (comammox), thus playing an important role in the N cycle [79], [80], [81]. Niche variability within the Nitrospira group has been recently shown. Although the relationship between nutrient levels and the growth of members of each clade is not fully understood, it seems clear that organic fertilization through changing nitrogen levels can affect their growth [81], [82], [83]. Moreover, the increase in relative abundance in Nitrospira members is related to an increase in nitrification, as shown by FAPROTAX results (see Table 2).
The relative abundances of genera Rubrobacter and Sphingomonas also differed between the OM and CM orchards. Members of these genera are able to colonize harsh environments and have been reported to metabolize pesticides and pollutants [84], [85], [86], [87], [88]. In fact, they were significantly more abundant in the rhizosphere of olives trees cultivated in the CM grove, where herbicides and insecticides were continuously applied. Moreover, the degradation of aromatic compounds was enhanced in the OM rhizosphere, according to FAPROTAX results. This was related to genera such as Nocardioides and Rhodococcus, which were significantly more abundant in this orchard. In fact, it has been shown that the addition of organic fertilizers such as manure to the soil improves the degradation of contaminants, likely because of an increase in diversity, nutrients levels and better soil texture [42], [89], [90]. The FAPROTAX analysis also highlighted chitinolysis as one of the metabolic functions enhanced in the rhizosphere of olive trees grown in the OM orchard. It has been shown that chitinolysis is involved in pathogenic fungi and nematodes defense [91]. Ureolysis was also significantly increased in the OM rhizosphere. Manure amendments are known to provide urea to the soil, that can constitute a source of nitrogen for plants due to the presence of ureolytic microorganisms [92], [93]. Overall, our results suggest that sheep manure favor the presence of bacteria involved in beneficial processes such as improved nutrient cycling, degradation of recalcitrant pollutants and defense against soil borne pathogens.
Interestingly, the bacterial community in the root endosphere was dominated by Actinobacteria representatives, in agreement with our previous study [94]. Moreover, Actinophytocola was the most abundant genus in the olive root endosphere. This emphasizes the key role that this genus seems to play in the inner of the olive roots, and that different agronomic managements do not affect its high prevalence within this plant organ. Actinobacteria are well-known for the production of secondary metabolites that play important roles in tree health, i.e., pathogen defense. Particularly, several isolates of Actinophytocola have already exhibited potential for antimicrobial activities [95]. Moreover, we have previously reported the correlation of Actinophytocola abundance with the expression of genes related with pathogen defense in the olive tree root [96]. We emphasize here that further research is needed to isolate and characterize the relation between Actinophytocola and olive tree health.
Concerning the fungal community, the taxonomical composition was dominated by Ascomycota members in both compartments. Moreover, this phylum was significantly more abundant in the CM orchard, both in the root endosphere and in the rhizosphere. This agrees with previous studies focused on endophytic communities of diverse plant species, including olive [94], [97]. Some studies have reported high abundance of Ascomycota members in soils with higher nitrogen content[98], [99], [100]. However, our results showed the opposite since the CM orchard clearly had a lower nutrient content, including N. Furthermore, differences in Glomeromytoca relative abundance were also found between conditions and in both compartments. In fact, in the root endosphere, Glomeromycota was more abundant in the CM orchard. Moreover, FungalTraits results showed a significantly higher abundance of arbuscular mycorrhiza in CM orchard. It is known that olive trees form symbiotic associations with arbuscular mycorrhizal fungi which are increasingly implemented during the propagation stage in nurseries to improve the establishment of the plants in fields [101], [102]. In this sense, the reduction of Glomeromycota members, specifically Rhizophagus, in the OM orchard can be due to the higher amount of phosphorus in this soil, as this element is known to inhibit the establishment of symbiosis with Glomeromycota representatives [103], [104]. At the genus level, the composition of the fungal community confirmed our previous studies showing high abundance of Solicoccozyma, Mortierella and Coniosporium in the rhizosphere [40]. In the root endosphere, Diaporthe was identified as the most abundant genera in both orchards. This genus includes endophytes earlier described in herbaceous and woody plants, particularly in olive trees [105], [106]. Interestingly, members of this genus are reported to produce secondary metabolites such as antimicrobial compounds [105], [107], [108]. Other genera showing differences in their abundances between conditions are worth mentioning. For instance, Preussia was significantly more abundant in the manure-amended orchard, both in the root endosphere and in the rhizosphere. Preussia species have mainly been described as coprophilous endophytes, and their abundance has been shown to increase under manure amendment fields. Moreover, its members can produce secondary metabolites, such as antimicrobial compounds, conferring them the ability to outcompete pathogens [109], [110], [111]. Other genera were only found as endophytes, such as Cladophialophora members. Representatives of this genus belong to the so-called dark septate endophytic fungi which have been described as plant-growth promoters. Moreover, some of them have been reported as effective biocontrol agents in several crops [112], [113]. Nonetheless, the CM orchard also showed high abundance of diverse genera with antifungal capacities. For example, Aerobasidium and Alternaria which have been reported as olive tree endophytes, particularly in the phyllosphere [114], [115], [116], [117]. Furthermore, Aerobasidium has been effectively used for the control of olive anthracnose [115], [117].
Finally, co-occurrence networks were constructed for both treatments and compartments. Previous studies have shown an increase in the complexity of soil microbial networks subjected to manure application [118], [119], [120], [121]. Related to the root microbiome, correlation between network complexity and agricultural intensification, with organic farming showing a higher complexity of root-associated fungi, has been reported in wheat [122]. Wang et al. (2022) studied the effect of manure application on the wheat rhizosphere microbial community (bacteria and fungi), finding a higher complexity associated with manure application [123]. This has also been proved for woody plants, as in poplar (Populus) trees [124]. Manure amendment can affect microbial networks in two ways [99]. On the one hand, manure provides the soil with a variety of nutrient sources, thus increasing soil niches and microbial interactions. On the other hand, manure contains recalcitrant organic matter and its degradation would require cooperation among microorganisms. According to our study, OM increased the complexity of the networks in the rhizosphere of olive trees as well as the percentage of positive interactions, likely related with a higher level of nutrients. Interestingly, several studies also reported a decrease in the average geodesic distance (GD) upon manure application[118], [124]. In our study, this trend was observed for both the rhizosphere and the root endosphere. A lower GD means that interactions in the network are more efficiently spread [125]. Thus, a microbial community with a smaller GD may have higher stability in the presence of perturbations[126], [127], [128]. In this sense, our results provide more evidence of a positive effect of manure amendment on microbial communities, thereby improving tree health. Indeed, it has been proposed that complex networks might help the plant to face pathogen attacks and environmental stresses [41], [119], [120], [126], [129]. Conversely, a decrease in network complexity was found in the root endosphere compartment of trees grown in the OM orchard. Little is known about the effect of agricultural practices in the endosphere microbial community, particularly with regard to co-occurrence networks. Thus, we would like to emphasize that further investigation would be needed to understand the effect of agricultural management on root endosphere microbiome.
5. Conclusions
Overall, our results showed an influence of sheep manure application on soil properties and on the belowground microbial communities. This influence was evidenced by several results. Firstly, an increased nutrient content of the soil was observed. In addition, a higher fungal diversity was found in the OM orchard, as well as a higher biomass in the rhizosphere. As for the bacterial dataset, potential metabolic functions of interest involved in pollutant degradation and defence against pathogens were shown. Finally, microbial co-occurrence network showed a more favorable scenario, with more positive interactions, complexity and shorter GD. Based on our results, we conclude that the application of sheep manure to olive orchards has a potential beneficial impact on soil properties and on the belowground microbial communities.
Funding
This research was funded by grants AGL2016-75729-C2-1-R from the Spanish Ministerio de Economía y Competitividad and PID2019-106283RB-I00 from Spanish Ministerio de Ciencia e Innovación/Agencia Estatal de Investigación, both co-financed by the European Regional Development Fund (ERDF). These funding sources had no involvement in this work preparation.
CRediT authorship contribution statement
Nuria M. Wentzien: Conceptualization, Methodology, Software, Formal analysis, Investigation, Writing – original draft; Antonio J. Fernández-González: Software, Formal analysis; Pablo J. Villadas: Formal analysis, Investigation, Resources Antonio Valverde-Corredor: Resources, Investigation; Jesús Mercado-Blanco: Conceptualization, Writing – review & editing, Supervision, Funding acquisition; Manuel Fernández-López: Conceptualization, Methodology, Resources Writing – review & editing, Supervision, Project administration, Funding acquisition. All authors read and approved the submitted version.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
We thank Antonio Estévez and Rocío Estévez (Nutesca, S.L.) for their help to identify and select olive orchards, as well as María Dolores Molina-Sánchez for the help with qPCR.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.07.015.
Appendix A. Supplementary material
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
Supplementary material.
.
References
- 1.López-Escudero F.J., Mercado-Blanco J. Verticillium wilt of olive: a case study to implement an integrated strategy to control a soil-borne pathogen. Plant Soil. 2011;344:1–50. doi: 10.1007/s11104-010-0629-2. [DOI] [Google Scholar]
- 2.FAO. FAOSTAT, production statistics. 2019. 〈http://faostat.fao.org〉 (accessed September 29, 2022).
- 3.Pérez-Jiménez F., Ruano J., Perez-Martinez P., Lopez-Segura F., Lopez-Miranda J. The influence of olive oil on human health: Not a question of fat alone. Mol Nutr Food Res. 2007;51:1199–1208. doi: 10.1002/mnfr.200600273. [DOI] [PubMed] [Google Scholar]
- 4.Montes-Osuna N., Mercado-Blanco J. Verticillium wilt of olive and its control: what did we learn during the last decade. Plants. 2020;9:1–31. doi: 10.3390/plants9060735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zipori I., Erel R., Yermiyahu U., Ben‐gal A., Dag A. Sustainable management of olive orchard nutrition: a review. Agriculture. 2020;10:11. doi: 10.3390/agriculture10010011. [DOI] [Google Scholar]
- 6.Gabel V.M., Meier M.S., Köpke U., Stolze M. The challenges of including impacts on biodiversity in agricultural life cycle assessments. J Environ Manag. 2016;181:249–260. doi: 10.1016/j.jenvman.2016.06.030. [DOI] [PubMed] [Google Scholar]
- 7.Berg G., Grube M., Schloter M., Smalla K. Unraveling the plant microbiome: looking back and future perspectives. Front Microbiol. 2014:5. doi: 10.3389/fmicb.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang Y., Wu L., Clark I.M., Xue K., Yang Y., van Nostrand J.D., et al. Over 150 years of long-term fertilization alters spatial scaling of microbial biodiversity. MBio. 2015:6. doi: 10.1128/mBio.00240-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bevivino A., Paganin P., Bacci G., Florio A., Pellicer M.S., Papaleo M.C., et al. Soil bacterial community response to differences in agricultural management along with seasonal changes in a mediterranean region. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobsen C.S., Hjelmsø M.H. Agricultural soils, pesticides and microbial diversity. Curr Opin Biotechnol. 2014;27:15–20. doi: 10.1016/j.copbio.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Lo C.C. Effect of pesticides on soil microbial community. J Environ Sci Health B. 2010;45:348–359. doi: 10.1080/03601231003799804. [DOI] [PubMed] [Google Scholar]
- 12.Berg G., Köberl M., Rybakova D., Müller H., Grosch R., Smalla K. Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol Ecol. 2017;93:50. doi: 10.1093/femsec/fix050. [DOI] [PubMed] [Google Scholar]
- 13.Compant S., Clément C., Sessitsch A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role colonization mechanisms involved and prospects for utilization. Soil Biol Biochem. 2010;42:669–678. doi: 10.1016/j.soilbio.2009.11.024. [DOI] [Google Scholar]
- 14.Reinhold-Hurek B., Hurek T. Living inside plants: bacterial endophytes. Curr Opin Plant Biol. 2011;14:435–443. doi: 10.1016/j.pbi.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 15.Mercado-Blanco J., Abrantes I., Caracciolo A.B., Bevivino A., Ciancio A., Grenni P., et al. Belowground microbiota and the health of tree crops. Front Microbiol. 2018;9:1006. doi: 10.3389/fmicb.2018.01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dadrasnia A., de Bona Muñoz I., Yáñez E.H., Lamkaddam I.U., Mora M., Ponsá S., et al. Sustainable nutrient recovery from animal manure: a review of current best practice technology and the potential for freeze concentration. J Clean Prod. 2021;315 doi: 10.1016/j.jclepro.2021.128106. [DOI] [Google Scholar]
- 17.Celik I., Ortas I., Kilic S. Effects of compost, mycorrhiza, manure and fertilizer on some physical properties of a Chromoxerert soil. Soil Tillage Res. 2004;78:59–67. doi: 10.1016/j.still.2004.02.012. [DOI] [Google Scholar]
- 18.Tejada M., Gonzalez J.L., García-Martínez A.M., Parrado J. Application of a green manure and green manure composted with beet vinasse on soil restoration: effects on soil properties. Bioresour Technol. 2008;99:4949–4957. doi: 10.1016/j.biortech.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 19.Celik I., Gunal H., Budak M., Akpinar C. Effects of long-term organic and mineral fertilizers on bulk density and penetration resistance in semi-arid Mediterranean soil conditions. Geoderma. 2010;160:236–243. doi: 10.1016/j.geoderma.2010.09.028. [DOI] [Google Scholar]
- 20.Lucas S.T., D’Angelo E.M., Williams M.A. Improving soil structure by promoting fungal abundance with organic soil amendments. Appl Soil Ecol. 2014;75:13–23. doi: 10.1016/j.apsoil.2013.10.002. [DOI] [Google Scholar]
- 21.Vida C., Vicente A., Cazorla F.M. The role of organic amendments to soil for crop protection: induction of suppression of soilborne pathogens. Ann Appl Biol. 2020;176:1–15. doi: 10.1111/aab.12555. [DOI] [Google Scholar]
- 22.Chernov T.I., Semenov M. v. Management of soil microbial communities: opportunities and prospects (a Review) Eurasia Soil Sci. 2021;54:1888–1902. doi: 10.1134/S1064229321120024. [DOI] [Google Scholar]
- 23.Sharaf H., Thompson A.A., Williams M.A., Peck G.M. Compost applications increase bacterial community diversity in the apple rhizosphere. Soil Sci Soc Am J. 2021;85:1105–1121. doi: 10.1002/saj2.20251. [DOI] [Google Scholar]
- 24.Sun J., Zhang Q., Zhou J., Wei Q. Pyrosequencing technology reveals the impact of different manure doses on the bacterial community in apple rhizosphere soil. Appl Soil Ecol. 2014;78:28–36. doi: 10.1016/j.apsoil.2014.02.004. [DOI] [Google Scholar]
- 25.Qiao C., Penton C.R., Xiong W., Liu C., Wang R., Liu Z., et al. Reshaping the rhizosphere microbiome by bio-organic amendment to enhance crop yield in a maize-cabbage rotation system. Appl Soil Ecol. 2019;142:136–146. doi: 10.1016/j.apsoil.2019.04.014. [DOI] [Google Scholar]
- 26.Windisch S., Sommermann L., Babin D., Chowdhury S.P., Grosch R., Moradtalab N., et al. Impact of long-term organic and mineral fertilization on rhizosphere metabolites, root–microbial interactions and plant health of lettuce. Front Microbiol. 2021;11:3157. doi: 10.3389/fmicb.2020.597745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H., Luo N., Ji C., Li J., Zhang L., Xiao L., et al. Liquid organic fertilizer amendment alters rhizosphere microbial community structure and co-occurrence patterns and improves sunflower yield under salinity-alkalinity stress. Micro Ecol. 2021:1–16. doi: 10.1007/s00248-021-01870-0. [DOI] [PubMed] [Google Scholar]
- 28.Wu X., Hu H., Li S., Zhao J., Li J., Zhang G., et al. Chemical fertilizer reduction with organic material amendments alters co-occurrence network patterns of bacterium-fungus-nematode communities under the wheat–maize rotation regime. Plant Soil. 2022;473:605–623. doi: 10.1007/s11104-022-05314-7. [DOI] [Google Scholar]
- 29.Atmaca S., Demiral S., Ülger S. Effects of chicken manure application on olive (Olea europaea) growth. Agrotech Ind Crops. 2021;1:61–70. doi: 10.22126/atic.2021.6514.1013. [DOI] [Google Scholar]
- 30.Chatzistathis T., Papadakis I.E., Papaioannou A., Chatzissavvidis C., Giannakoula A. Comparative study effects between manure application and a controlled-release fertilizer on the growth, nutrient uptake, photosystem II activity and photosynthetic rate of Olea europaea L. (cv. ‘Koroneiki’) Sci Hortic. 2020;264 doi: 10.1016/j.scienta.2020.109176. [DOI] [Google Scholar]
- 31.Hadawi I., Safari M., Arji I. Effects of different organic manures and chemical fertilizers on yield and yield component of Olive (Olea europaea L.,) cv Zard In Kermanshah Province 2. Agrotech Ind Crops. 2021;1:61–70. doi: 10.22126/ATIC.2021.6514.1013. [DOI] [Google Scholar]
- 32.Pekcan A., Turan T., Sevim Çolakoğlu H. Effects of organomineral, mineral and farmyard manures on the yield and quality of olive trees. J Appl Sci Res. 2009;3:1152–1155. [Google Scholar]
- 33.Ahmad Yassin Majjami, Abdullah Saad Al-Modaihsh, Fahad Nasser Al-Barakah, Mohamed Hamza El-Saeid, Samir Gamil Al-Solimani, Fahad Mohammed Alghabari. Comparing microbial community heterogeneity in soil under organic and conventional agricultural system cultivated with olive and peach trees in three fields differ with age at Al-Jouf Saudi Arabia. World J Adv Res Rev. 2020;7:283–290. doi: 10.30574/wjarr.2020.7.3.0350. [DOI] [Google Scholar]
- 34.Hassan A., Haggag L.F., Hassan H., Abdelhafez A.A., Hassan A.M. Effect of mineral and bio-fertilization on yield and fruit quality of Manzanillo Olive trees. Artic Int J ChemTech Res. 2015;8:63–73. [Google Scholar]
- 35.Montes-Borrego M., Navas-Cortés J.A., Landa B.B. Linking microbial functional diversity of olive rhizosphere soil to management systems in commercial orchards in southern Spain. Agric Ecosyst Environ. 2013;181:169–178. doi: 10.1016/j.agee.2013.09.021. [DOI] [Google Scholar]
- 36.Caliz J., Montes-Borrego M., Triadó-Margarit X., Metsis M., Landa B.B., Casamayor E.O. Influence of Edaphic, Climatic, and agronomic factors on the composition and abundance of nitrifying microorganisms in the rhizosphere of commercial olive crops. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sofo A., Ricciuti P., Fausto C., Mininni A.N., Crecchio C., Scagliola M., et al. The metabolic and genetic diversity of soil bacterial communities depends on the soil management system and C/N dynamics: the case of sustainable and conventional olive groves. Appl Soil Ecol. 2019;137:21–28. doi: 10.1016/j.apsoil.2018.12.022. [DOI] [Google Scholar]
- 38.Llimós M., Segarra G., Sancho-Adamson M., Trillas M.I., Romanyà J. Impact of olive saplings and organic amendments on soil microbial communities and effects of mineral fertilization. Front Microbiol. 2021;12:1248. doi: 10.3389/fmicb.2021.653027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fausto C., Mininni A.N., Sofo A., Crecchio C., Scagliola M., Dichio B., et al. Olive orchard microbiome: characterisation of bacterial communities in soil-plant compartments and their comparison between sustainable and conventional soil management systems. Plant Ecol Divers. 2018;11:597–610. doi: 10.1080/17550874.2019.1596172. [DOI] [Google Scholar]
- 40.Fernández-González A.J., Wentzien N.M., Villadas P.J., Valverde-Corredor A., Lasa A. v, Gómez-Lama Cabanás C., et al. Comparative study of neighboring Holm oak and olive trees-belowground microbial communities subjected to different soil management. PLoS One. 2020;15 doi: 10.1371/journal.pone.0236796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fernández-González A.J., Cardoni M., Gómez-Lama Cabanás C., Valverde-Corredor A., Villadas P.J., Fernández-López M., et al. Linking belowground microbial network changes to different tolerance level towards Verticillium wilt of olive. Microbiome. 2020;8:11. doi: 10.1186/s40168-020-0787-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medina R., Fernández-González A.J., García-Rodríguez F.M., Villadas P.J., Rosso J.A., Fernández-López M., et al. Exploring the effect of composting technologies on the recovery of hydrocarbon contaminated soil post chemical oxidative treatment. Appl Soil Ecol. 2020:150. doi: 10.1016/J.APSOIL.2019.103459. [DOI] [Google Scholar]
- 43.Chemidlin Prévost-Bouré N., Christen R., Dequiedt S., Mougel C., Lelièvre M., Jolivet C., et al. Validation and application of a PCR Primer Set to quantify fungal communities in the soil environment by real-time quantitative PCR. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herrera-Medina M.J., Steinkellner S., Vierheilig H., Ocampo Bote J.A., García Garrido J.M. Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 2007;175:554–564. doi: 10.1111/j.1469-8137.2007.02107.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y., Tian R.M., Gao Z.M., Bougouffa S., Qian P.Y. Optimal Eukaryotic 18S and Universal 16S/18S Ribosomal RNA primers and their application in a study of symbiosis. PLoS One. 2014;9 doi: 10.1371/JOURNAL.PONE.0090053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi S., Tomita J., Nishioka K., Hisada T., Nishijima M. Development of a Prokaryotic Universal Primer for simultaneous analysis of bacteria and archaea using next-generation sequencing. PLoS One. 2014;9 doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundberg D.S., Yourstone S., Mieczkowski P., Jones C.D., Dangl J.L. Practical innovations for high-throughput amplicon sequencing. Nat Methods. 2013;10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 48.White T.J., BTLSTJ . 315-322. - References - Scientific Research Publishing. Elsevier,; 1999. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols, a Guide to Methods and Applications; pp. 315–322. [Google Scholar]
- 49.Ihrmark K., Bödeker I.T.M., Cruz-Martinez K., Friberg H., Kubartova A., Schenck J., et al. New primers to amplify the fungal ITS2 region - evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–677. doi: 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- 50.Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.UNITE Community. UNITE Community (2019): UNITE general FASTA release for Fungi n.d.;10. https://doi.org/15156/BIO/786343.
- 52.Lin H., Peddada S.Das. Analysis of compositions of microbiomes with bias correction. Nat Commun. 2020;11:1–11. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Louca S., Parfrey L.W., Doebeli M. Decoupling function and taxonomy in the global ocean microbiome. Science. 2016;353:1272–1277. doi: 10.1126/science.aaf4507. [DOI] [PubMed] [Google Scholar]
- 54.Xiao N., Zhou A., Kempher M.L., Zhou B.Y., Shi Z.J., Yuan M., et al. Disentangling direct from indirect relationships in association networks. Proc Natl Acad Sci USA. 2022:119. doi: 10.1073/PNAS.2109995119/-/DCSUPPLEMENTAL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cabanás C.G.L., Fernández-González A.J., Cardoni M., Valverde-Corredor A., López-Cepero J., Fernández-López M., et al. The banana root endophytome: differences between mother plants and suckers and evaluation of selected bacteria to control fusarium oxysporum f.sp. cubense. J Fungi. 2021:7. doi: 10.3390/jof7030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Põlme S., Abarenkov K., Henrik Nilsson R., Lindahl B.D., Clemmensen K.E., Kauserud H., et al. FungalTraits: a user-friendly traits database of fungi and fungus-like stramenopiles. Fungal Divers. 2020;105:1–16. doi: 10.1007/S13225-020-00466-2/FIGURES/3. [DOI] [Google Scholar]
- 57.French E., Kaplan I., Iyer-Pascuzzi A., Nakatsu C.H., Enders L. Emerging strategies for precision microbiome management in diverse agroecosystems. Nat Plants. 2021;7:256–267. doi: 10.1038/s41477-020-00830-9. [DOI] [PubMed] [Google Scholar]
- 58.Lucas S.T., D’Angelo E.M., Williams M.A. Improving soil structure by promoting fungal abundance with organic soil amendments. Appl Soil Ecol. 2014;75:13–23. doi: 10.1016/j.apsoil.2013.10.002. [DOI] [Google Scholar]
- 59.Qiao C., Penton C.R., Xiong W., Liu C., Wang R., Liu Z., et al. Reshaping the rhizosphere microbiome by bio-organic amendment to enhance crop yield in a maize-cabbage rotation system. Appl Soil Ecol. 2019;142:136–146. doi: 10.1016/j.apsoil.2019.04.014. [DOI] [Google Scholar]
- 60.Laconi A., Mughini-Gras L., Tolosi R., Grilli G., Trocino A., Carraro L., et al. Microbial community composition and antimicrobial resistance in agricultural soils fertilized with livestock manure from conventional farming in Northern Italy. Sci Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.143404. [DOI] [PubMed] [Google Scholar]
- 61.Sun R., Dsouza M., Gilbert J.A., Guo X., Wang D., Guo Z., et al. Fungal community composition in soils subjected to long-term chemical fertilization is most influenced by the type of organic matter. Environ Microbiol. 2016;18:5137–5150. doi: 10.1111/1462-2920.13512. [DOI] [PubMed] [Google Scholar]
- 62.Moreno B., Garcia-Rodriguez S., Cañizares R., Castro J., Benítez E. Rainfed olive farming in south-eastern Spain: long-term effect of soil management on biological indicators of soil quality. Agric Ecosyst Environ. 2009;131:333–339. doi: 10.1016/j.agee.2009.02.011. [DOI] [Google Scholar]
- 63.Luan H., Gao W., Huang S., Tang J., Li M., Zhang H., et al. Substitution of manure for chemical fertilizer affects soil microbial community diversity, structure and function in greenhouse vegetable production systems. PLoS One. 2020:15. doi: 10.1371/journal.pone.0214041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi R., Li J., Lin Z., Li Z., Li Y., Yang X., et al. Temperature effects on soil organic carbon, soil labile organic carbon fractions, and soil enzyme activities under long-term fertilization regimes. Appl Soil Ecol. 2016;102:36–45. doi: 10.1016/j.apsoil.2016.02.004. [DOI] [Google Scholar]
- 65.Li J., Cooper J.M., Lin Z., Li Y., Yang X., Zhao B. Soil microbial community structure and function are significantly affected by long-term organic and mineral fertilization regimes in the North China Plain. Appl Soil Ecol. 2015;96:75–87. doi: 10.1016/j.apsoil.2015.07.001. [DOI] [Google Scholar]
- 66.Zhang Q.C., Shamsi I.H., Xu D.T., Wang G.H., Lin X.Y., Jilani G., et al. Chemical fertilizer and organic manure inputs in soil exhibit a vice versa pattern of microbial community structure. Appl Soil Ecol. 2012;57:1–8. doi: 10.1016/j.apsoil.2012.02.012. [DOI] [Google Scholar]
- 67.Das S., Jeong S.T., Das S., Kim P.J. Composted cattle manure increases microbial activity and soil fertility more than composted swine manure in a submerged rice paddy. Front Microbiol. 2017;8:1702. doi: 10.3389/fmicb.2017.01702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pii Y., Mimmo T., Tomasi N., Terzano R., Cesco S., Crecchio C. Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. a review. Biol Fertil Soils. 2015;51:403–415. doi: 10.1007/S00374-015-0996-1/TABLES/1. [DOI] [Google Scholar]
- 69.Francioli D., Schulz E., Lentendu G., Wubet T., Buscot F., Reitz T. Mineral vs. organic amendments: Microbial community structure, activity and abundance of agriculturally relevant microbes are driven by long-term fertilization strategies. Front Microbiol. 2016;7:1446. doi: 10.3389/fmicb.2016.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DeBruyn J.M., Nixon L.T., Fawaz M.N., Johnson A.M., Radosevich M. Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl Environ Microbiol. 2011;77:6295–6300. doi: 10.1128/AEM.05005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang S., Sun L., Wang Y., Fan K., Xu Q., Li Y., et al. Cow manure application effectively regulates the soil bacterial community in tea plantation. BMC Microbiol. 2020;20:190. doi: 10.1186/s12866-020-01871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larsbrink J., McKee L.S. vol. 110. Academic Press Inc,; 2020. Bacteroidetes bacteria in the soil: Glycan acquisition, enzyme secretion, and gliding motility; pp. 63–98. (Adv Appl Microbiol). [DOI] [PubMed] [Google Scholar]
- 73.Yang Y., Wang N., Guo X., Zhang Y., Ye B. Comparative analysis of bacterial community structure in the rhizosphere of maize by high-throughput pyrosequencing. PLoS One. 2017;12 doi: 10.1371/journal.pone.0178425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang S., Sun L., Wang Y., Fan K., Xu Q., Li Y., et al. Cow manure application effectively regulates the soil bacterial community in tea plantation. BMC Microbiol. 2020;20:190. doi: 10.1186/s12866-020-01871-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolińska A., Kuźniar A., Zielenkiewicz U., Izak D., Szafranek-Nakonieczna A., Banach A., et al. Bacteroidetes as a sensitive biological indicator of agricultural soil usage revealed by a culture-independent approach. Appl Soil Ecol. 2017;119:128–137. doi: 10.1016/j.apsoil.2017.06.009. [DOI] [Google Scholar]
- 76.Lee S.A., Kim J.M., Kim Y., Joa J.H., Kang S.S., Ahn J.H., et al. Different types of agricultural land use drive distinct soil bacterial communities. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-74193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trivedi P., Delgado-Baquerizo M., Anderson I.C., Singh B.K. Response of soil properties and microbial communities to agriculture: Implications for primary productivity and soil health indicators. Front Plant Sci. 2016;7:990. doi: 10.3389/fpls.2016.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma G., Kang J., Wang J., Chen Y., Lu H., Wang L., et al. Bacterial community structure and predicted function in wheat Soil From the North China Plain are closely linked with soil and plant characteristics after seven years of irrigation and nitrogen application. Front Microbiol. 2020;11:506. doi: 10.3389/fmicb.2020.00506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu J., Zhao Y., Yao X., Wang J., Zheng P., Xi C., et al. Dominance of comammox Nitrospira in soil nitrification. Sci Total Environ. 2021;780 doi: 10.1016/j.scitotenv.2021.146558. [DOI] [PubMed] [Google Scholar]
- 80.Daims H., Lebedeva E. v, Pjevac P., Han P., Herbold C., Albertsen M., et al. Complete nitrification by Nitrospira bacteria. Nature. 2015;528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li C., Hu H.W., Chen Q.L., Yan Z.Z., Thi Nguyen B.A., Chen D., et al. Niche specialization of comammox Nitrospira clade A in terrestrial ecosystems. Soil Biol Biochem. 2021;156 doi: 10.1016/j.soilbio.2021.108231. [DOI] [Google Scholar]
- 82.Han S., Zeng L., Luo X., Xiong X., Wen S., Wang B., et al. Shifts in nitrobacter- and nitrospira-like nitrite-oxidizing bacterial communities under long-term fertilization practices. Soil Biol Biochem. 2018;124:118–125. doi: 10.1016/j.soilbio.2018.05.033. [DOI] [Google Scholar]
- 83.He Z.Y., Sun A., Jiao X.Y., Ge A.H., Hu H.W., Jin S., et al. Fertilization has a greater effect than rhizosphere on community structures of comammox nitrospira in an alkaline agricultural soil. Appl Soil Ecol. 2022;175 doi: 10.1016/j.apsoil.2022.104456. [DOI] [Google Scholar]
- 84.Holmes A.J., Bowyer J., Holley M.P., O’Donoghue M., Montgomery M., Gillings M.R. Diverse, yet-to-be-cultured members of the rubrobacter subdivision of the actinobacteria are widespread in Australian arid soils. FEMS Microbiol Ecol. 2000;33:111–120. doi: 10.1111/j.1574-6941.2000.tb00733.x. [DOI] [PubMed] [Google Scholar]
- 85.Moffett B.F., Nicholson F.A., Uwakwe N.C., Chambers B.J., Harris J.A., Hill T.C.J. Zinc contamination decreases the bacterial diversity of agricultural soil. FEMS Microbiol Ecol. 2003;43:13–19. doi: 10.1111/j.1574-6941.2003.tb01041.x. [DOI] [PubMed] [Google Scholar]
- 86.Li X., Bond P.L., van Nostrand J.D., Zhou J., Huang L. From lithotroph- to organotroph-dominant: directional shift of microbial community in sulphidic tailings during phytostabilization. Sci Rep. 2015;5:1–12. doi: 10.1038/srep12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sørensen S.R., Ronen Z., Aamand J. Isolation from agricultural soil and characterization of a sphingomonas sp. able to mineralize the phenylurea herbicide isoproturon. Appl Environ Microbiol. 2001;67:5403–5409. doi: 10.1128/AEM.67.12.5403-5409.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leys NMEJ Ryngaert A., Bastiaens L., Top EM Verstraete W., Springael D. Occurrence and phylogenetic diversity of sphingomonas strains in soils contaminated with polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 2004;70:1944–1955. doi: 10.1128/AEM.70.4.1944-1955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu M., Guo X., Wu J., Chen K. Effect of compost amendment and bioaugmentation on PAH degradation and microbial community shifting in petroleum-contaminated soil. Chemosphere. 2020:256. doi: 10.1016/j.chemosphere.2020.126998. [DOI] [PubMed] [Google Scholar]
- 90.Haritash A.K., Kaushik C.P. Biodegradation aspects of Polycyclic Aromatic Hydrocarbons (PAHs): a review. J Hazard Mater. 2009;169:1–15. doi: 10.1016/j.jhazmat.2009.03.137. [DOI] [PubMed] [Google Scholar]
- 91.Kielak A.M., Cretoiu M.S., Semenov A. v, Sørensen S.J., van Elsas J.D. Bacterial chitinolytic communities respond to chitin and pH alteration in soil. Appl Environ Microbiol. 2013;79:263–272. doi: 10.1128/AEM.02546-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhao R., Liu J., Xu N., He T., Meng J., Liu Z. Urea hydrolysis in different farmland soils as affected by long-term biochar application. Front Environ Sci. 2022;10:1304. doi: 10.3389/fenvs.2022.950482. [DOI] [Google Scholar]
- 93.Harder Nielsen T., Bonde T.A., Sørensen J. Significance of microbial urea turnover in N cycling of three Danish agricultural soils. FEMS Microbiol Ecol. 1998;25:147–157. doi: 10.1111/j.1574-6941.1998.tb00468.x. [DOI] [Google Scholar]
- 94.Fernández-González A.J., Villadas P.J., Gómez-Lama Cabanás C., Valverde-Corredor A., Belaj A., Mercado-Blanco J., et al. Defining the root endosphere and rhizosphere microbiomes from the World Olive Germplasm Collection. Sci Rep. 2019;9:1–13. doi: 10.1038/s41598-019-56977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bouznada K., Bouras N., Schumann P., Spröer C., Sabaou N., Klenk H.P. Actinophytocola algeriensis sp. Nov., an actinobacterium isolated from saharan soil. Int J Syst Evol Microbiol. 2016;66:2760–2765. doi: 10.1099/ijsem.0.001136. [DOI] [PubMed] [Google Scholar]
- 96.Fernández-González A.J., Ramírez-Tejero J.A., Nevado-Berzosa M.P., Luque F., Fernández-López M., Mercado-Blanco J. Coupling the endophytic microbiome with the host transcriptome in olive roots. Comput Struct Biotechnol J. 2021;19:4777–4789. doi: 10.1016/j.csbj.2021.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hardoim P.R., van Overbeek L.S., Berg G., Pirttilä A.M., Compant S., Campisano A., et al. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320. doi: 10.1128/mmbr.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Weber C.F., Vilgalys R., Kuske C.R. Changes in fungal community composition in response to elevated atmospheric CO2 and nitrogen fertilization varies with soil horizon. Front Microbiol. 2013;4:78. doi: 10.3389/fmicb.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ye G., Banerjee S., He J.Z., Fan J., Wang Z., Wei X., et al. Manure application increases microbiome complexity in soil aggregate fractions: results of an 18-year field experiment. Agric Ecosyst Environ. 2021;307 doi: 10.1016/j.agee.2020.107249. [DOI] [Google Scholar]
- 100.Zhou J., Jiang X., Zhou B., Zhao B., Ma M., Guan D., et al. Thirty four years of nitrogen fertilization decreases fungal diversity and alters fungal community composition in black soil in northeast China. Soil Biol Biochem. 2016;95:135–143. doi: 10.1016/j.soilbio.2015.12.012. [DOI] [Google Scholar]
- 101.Montes-Borrego M., Metsis M., Landa B.B. Arbuscular mycorhizal fungi associated with the olive crop across the Andalusian landscape: factors driving community differentiation. PLoS One. 2014:9. doi: 10.1371/journal.pone.0096397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Calvente R., Cano C., Ferrol N., Azcón-Aguilar C., Barea J.M. Analysing natural diversity of arbuscular mycorrhizal fungi in olive tree (Olea europaea L.) plantations and assessment of the effectiveness of native fungal isolates as inoculants for commercial cultivars of olive plantlets. Appl Soil Ecol. 2004;26:11–19. doi: 10.1016/j.apsoil.2003.10.009. [DOI] [Google Scholar]
- 103.Nouri E., Surve R., Bapaume L., Stumpe M., Chen M., Zhang Y., et al. Phosphate suppression of arbuscular mycorrhizal symbiosis involves gibberellic acid signaling. Plant Cell Physiol. 2021;62:959–970. doi: 10.1093/pcp/pcab063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Balzergue C., Chabaud M., Barker D.G., Bécard G., Rochange S.F. High phosphate reduces host ability to develop arbuscular mycorrhizal symbiosis without affecting root calcium spiking responses to the fungus. Front Plant Sci. 2013:4. doi: 10.3389/fpls.2013.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gomes R.R., Glienke C., Videira S.I.R., Lombard L., Groenewald J.Z., Crous P.W. Diaporthe: a genus of endophytic, saprobic and plant pathogenic fungi. Pers: Mol Phylogeny Evol Fungi. 2013;31:1–41. doi: 10.3767/003158513x666844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Martins F., Pereira J.A., Bota P., Bento A., Baptista P. Fungal endophyte communities in above- and belowground olive tree organs and the effect of season and geographic location on their structures. Fungal Ecol. 2016;20:193–201. doi: 10.1016/j.funeco.2016.01.005. [DOI] [Google Scholar]
- 107.Xu T.-C., Lu Y.-H., Wang J.-F., Song Z.-Q., Hou Y.-G., Liu S.-S., et al. Bioactive secondary metabolites of the genus diaporthe and anamorph phomopsis from terrestrial and marine habitats and endophytes: 2010–2019. Microorganisms. 2021;9:217. doi: 10.3390/microorganisms9020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li G., Kusari S., Kusari P., Kayser O., Spiteller M. Endophytic Diaporthe sp. LG23 produces a potent antibacterial tetracyclic triterpenoid. J Nat Prod. 2015;78:2128–2132. doi: 10.1021/acs.jnatprod.5b00170. [DOI] [PubMed] [Google Scholar]
- 109.Hartmann M., Frey B., Mayer J., Mäder P., Widmer F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2015;9:1177–1194. doi: 10.1038/ismej.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mapperson R.R., Kotiw M., Davis R.A., Dearnaley J.D.W. The diversity and antimicrobial activity of preussia sp. endophytes isolated from australian dry rainforests. Curr Microbiol. 2014;68:30–37. doi: 10.1007/s00284-013-0415-5. [DOI] [PubMed] [Google Scholar]
- 111.Semenov M. v, Krasnov G.S., Semenov V.M., van Bruggen A. Mineral and organic fertilizers distinctly affect fungal communities in the crop rhizosphere. J Fungi. 2022;8:251. doi: 10.3390/jof8030251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Harsonowati W., Marian M., Surono, Narisawa K. The effectiveness of a Dark Septate Endophytic Fungus, Cladophialophora chaetospira SK51, to Mitigate Strawberry Fusarium Wilt Disease and With Growth Promotion Activities. Front Microbiol. 2020;11:585. doi: 10.3389/fmicb.2020.00585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thambugala K.M., Daranagama D.A., Phillips A.J.L., Kannangara S.D., Promputtha I. Fungi vs. fungi in biocontrol: an overview of fungal antagonists applied against fungal plant pathogens. Front Cell Infect Microbiol. 2020:10. doi: 10.3389/fcimb.2020.604923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nicoletti R., di Vaio C., Cirillo C. Endophytic fungi of olive tree. Microorganisms. 2020;8:1321. doi: 10.3390/microorganisms8091321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Costa D., Fernandes T., Martins F., Pereira J.A., Tavares R.M., Santos P.M., et al. Illuminating Olea europaea L. endophyte fungal community. Microbiol Res. 2021;245 doi: 10.1016/j.micres.2020.126693. [DOI] [PubMed] [Google Scholar]
- 116.Materatski P., Varanda C., Carvalho T., Dias A.B., Campos M.D., Rei F., et al. Spatial and temporal variation of fungal endophytic richness and diversity associated to the phyllosphere of olive cultivars. Fungal Biol. 2019;123:66–76. doi: 10.1016/j.funbio.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 117.Martins F., Pereira J.A., Baptista P. Springer International Publishing,; 2019. Olive Anthracnose and Its Management by Fungal Endophytes: An Overview. Plant Microbe Interface; pp. 253–269. [DOI] [Google Scholar]
- 118.Ye G., Banerjee S., He J.Z., Fan J., Wang Z., Wei X., et al. Manure application increases microbiome complexity in soil aggregate fractions: results of an 18-year field experiment. Agric Ecosyst Environ. 2021;307 doi: 10.1016/j.agee.2020.107249. [DOI] [Google Scholar]
- 119.Ling N., Zhu C., Xue C., Chen H., Duan Y., Peng C., et al. Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol Biochem. 2016;99:137–149. doi: 10.1016/j.soilbio.2016.05.005. [DOI] [Google Scholar]
- 120.Liu Z., Guo Q., Feng Z., Liu Z., Li H., Sun Y., et al. Long-term organic fertilization improves the productivity of kiwifruit (Actinidia chinensis Planch.) through increasing rhizosphere microbial diversity and network complexity. Appl Soil Ecol. 2020;147 doi: 10.1016/j.apsoil.2019.103426. [DOI] [Google Scholar]
- 121.Shi W., Zhao H.Y., Chen Y., Wang J.S., Han B., Li C.P., et al. Organic manure rather than phosphorus fertilization primarily determined asymbiotic nitrogen fixation rate and the stability of diazotrophic community in an upland red soil. Agric Ecosyst Environ. 2021;319 doi: 10.1016/j.agee.2021.107535. [DOI] [Google Scholar]
- 122.Banerjee S., Walder F., Büchi L., Meyer M., Held A.Y., Gattinger A., et al. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019;13:1722–1736. doi: 10.1038/s41396-019-0383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang P., Wang X., Nie J., Wang Y., Zang H., Peixoto L., et al. Manure application increases soil bacterial and fungal network complexity and alters Keystone Taxa. J Soil Sci Plant Nutr. 2022;22:607–618. doi: 10.1007/s42729-021-00673-z. [DOI] [Google Scholar]
- 124.Wang G., Chen J., Zhu Y. Distinct bacterial community compositions in the Populus rhizosphere under three types of organic matter input across different soil types. Plant Soil. 2022;470:51–63. doi: 10.1007/s11104-021-04859-3. [DOI] [Google Scholar]
- 125.Strang A., Haynes O., Cahill N.D., Narayan D.A. Generalized relationships between characteristic path length, efficiency, clustering coefficients, and density. Soc Netw Anal Min. 2018;8:1–6. doi: 10.1007/s13278-018-0492-3. [DOI] [Google Scholar]
- 126.Tao J., Meng D., Qin C., Liu X., Liang Y., Xiao Y., et al. Integrated network analysis reveals the importance of microbial interactions for maize growth. Appl Microbiol Biotechnol. 2018;102:3805–3818. doi: 10.1007/s00253-018-8837-4. [DOI] [PubMed] [Google Scholar]
- 127.Qu Z., Zhao H., Zhang H., Wang Q., Yao Y., Cheng J., et al. Bio-priming with a hypovirulent phytopathogenic fungus enhances the connection and strength of microbial interaction network in rapeseed. NPJ Biofilms Micro. 2020;6:1–13. doi: 10.1038/s41522-020-00157-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gómez-Lama Cabanás C., Wentzien N.M., Zorrilla-Fontanesi Y., Valverde-Corredor A., Fernández-González A.J., Fernández-López M., et al. Impacts of the biocontrol strain pseudomonas simiae PICF7 on the Banana Holobiont: alteration of root microbial co-occurrence networks and effect on host defense responses. Front Microbiol. 2022:13. doi: 10.3389/fmicb.2022.809126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wagg C., Schlaeppi K., Banerjee S., Kuramae E.E., van der Heijden M.G.A. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat Commun. 2019;10:1–10. doi: 10.1038/s41467-019-12798-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Supplementary material.
Data Availability Statement
The sequencing dataset generated during the current study were deposited and are publicly available at the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) repository under the BioProject accession number PRJNA925330.