Significance
Bacterial lytic polysaccharide monooxygenases (LPMOs) are understudied in disease contexts. We show the trimodular Pseudomonas aeruginosa LPMO CbpD is a virulence determinant in pneumonia, and mice immunized with CbpD generate robust antibody responses that promote opsonophagocytosis and protect against lethal infection. Our findings can impact research on this broadly distributed enzyme class, their roles in bacterial pathogenesis, and their potential targeting by antivirulence therapies and vaccines.
Keywords: plytic polysaccharide monooxygenase, vaccine antigen, pneumonia, mouse models, crystallography
Abstract
Pseudomonas aeruginosa (PA) CbpD belongs to the lytic polysaccharide monooxygenases (LPMOs), a family of enzymes that cleave chitin or related polysaccharides. Here, we demonstrate a virulence role of CbpD in PA pneumonia linked to impairment of host complement function and opsonophagocytic clearance. Following intratracheal challenge, a PA ΔCbpD mutant was more easily cleared and produced less mortality than the wild-type parent strain. The x-ray crystal structure of the CbpD LPMO domain was solved to subatomic resolution (0.75Å) and its two additional domains modeled by small-angle X-ray scattering and Alphafold2 machine-learning algorithms, allowing structure-based immune epitope mapping. Immunization of naive mice with recombinant CbpD generated high IgG antibody titers that promoted human neutrophil opsonophagocytic killing, neutralized enzymatic activity, and protected against lethal PA pneumonia and sepsis. IgG antibodies generated against full-length CbpD or its noncatalytic M2+CBM73 domains were opsonic and protective, even in previously PA-exposed mice, while antibodies targeting the AA10 domain were not. Preexisting antibodies in PA-colonized cystic fibrosis patients primarily target the CbpD AA10 catalytic domain. Further exploration of LPMO family proteins, present across many clinically important and antibiotic-resistant human pathogens, may yield novel and effective vaccine antigens.
Pseudomonas aeruginosa (PA) is an often highly multidrug-resistant bacterial pathogen that poses a serious threat to human health, particularly in immunocompromised and/or hospitalized patients. Chronic PA respiratory infections are seen in patients with cystic fibrosis (CF), bronchiectasis, and chronic obstructive pulmonary disease(1–3), where tracheobronchial colonization is linked to deterioration of pulmonary function, poor clinical outcome, and increased mortality (4–7). PA is also a leading agent of potentially fatal ventilator-associated pneumonia in intensive care units (8, 9) and of dangerous bloodstream infections and sepsis in patients with cancer, neutropenia, burns, and/or disruption of the microbiota from prior broad-spectrum antibiotic therapy (10–13). As PA strains have proven capable of deploying high-level intrinsic and acquired resistance mechanisms to counter all major classes of antibiotics in clinical use (14, 15), the development of vaccine-induced immunity against PA is considered an essential, albeit challenging pillar of future prevention strategies (16, 17).
Antigens possessing one or more of the following characteristics including strong immunogenicity, surface exposure, high abundance, and/or a role in bacterial virulence can serve in the foundation of an effective vaccine (18). Due to its medical importance, PA has been extensively studied in terms of its molecular epidemiology (19), fundamental biochemical composition (20, 21), gene regulation (22), and host–pathogen interactions (23, 24). A number of conserved PA surface molecules that proved promising in animal vaccination models later entered Phase I–III human clinical trials, including lipopolysaccharide(25–28), flagellins (29), alginate (30), and outer membrane proteins OprF-OprI (31, 32). Despite these many investigations, no licensed vaccines are available against PA, and the search for an optimal antigen or formulation of antigens continues.
Recently, we reported that chitin-binding protein D (CbpD), a highly conserved protein across PA clinical isolates (33, 34), contributed strongly to systemic virulence and immune evasion by interference with the terminal complement cascade (35). CbpD belongs to a class of ancient microbial copper-dependent redox enzymes, the lytic polysaccharide monooxygenases (LPMOs) (35), commonly known to degrade recalcitrant polysaccharides such as chitin [β-1,4(GlcNAc)n] and cellulose [β-1,4(Glc)n] through oxidative cleavage of glycosidic bonds (36–39). In PA, CbpD is secreted by a type II secretion system (40, 41), regulated by quorum-sensing control (42, 43), and induced in the presence of human serum and respiratory mucus (35, 44). Abundant CbpD was reported in the secretome of a transmissible CF-associated PA strain under conditions mimicking CF lung physiology (45) and cbpD gene transcription was strongly induced by human respiratory tract mucus in a mucoid PA isolate from a CF patient (44).
Here, we show that the inducible expression and protein abundance of CbpD in the respiratory tract context is corroborated by an essential virulence role of the protein in PA in a murine pneumonia model. Combining structural biology and immunological studies, we demonstrate the potential of CbpD and its various domains as a protective vaccine antigen against PA lung infection. As similar LPMOs are present in many important human bacterial pathogens, their role(s) in virulence and immunogenicity merit further in-depth exploration.
Results
Loss of CbpD Reduces PA Virulence and Modifies Host Responses in Murine Lung Infection.
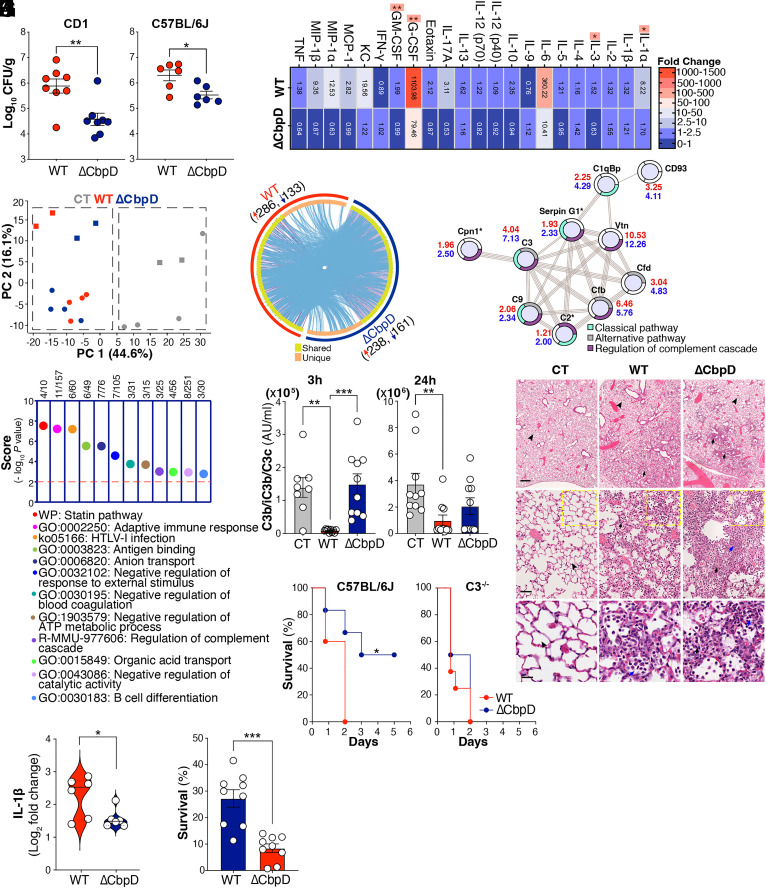
An isogenic CbpD knockout mutant (ΔCbpD) of the well-characterized virulent wild-type (WT) PA clinical isolate PA14 (46) was kindly provided by Deborah Hogan (Dartmouth) and confirmed biochemically (immunoblotting and proteomics) and by whole genome sequencing in our earlier work (35). There we reported a preliminary finding of significantly reduced mortality of the ΔCbpD mutant compared to the WT strain following intratracheal (IT) challenge of outbred CD-1 mice (35). Here, we show that complementation of the mutant with the cbpD gene expressed on a plasmid vector restored the virulence phenotype in the IT challenge model, with 100% mortality by 48 h while 50% of mutant mice survived through 96 h (SI Appendix, Fig. S1A). By 24 h post-IT challenge of both outbred CD-1 or inbred C57BL/6 mice, bacterial CFU recovered from the lungs were significantly lower than those infected with the WT parent strain (Fig. 1A). Reduced bacterial burdens in the lungs of ΔCbpD-infected mice were accompanied by significantly lower levels of inflammatory serum cytokines and chemokines at the same 24 h time point, including IL-1α, IL-3, G-CSF, and GM-CSF (FDR-adjusted P or q < 0.05) (Fig. 1B).
Fig. 1.
CbpD virulence role in a murine pneumonia model. (A) Female CD-1 and C57BL/6J mice were inoculated IT with 1×106 CFU PA WT or ΔCbpD per mouse. Bacterial loads in the lung (CFU/g) were enumerated 24 h postinfection. Data are plotted as the mean ± SEM, representing 8 (CD-1) and 6 (C57BL/6J) mice per group, and were analyzed by the two-tailed t test (CD-1: P = 0.0030; C57BL/6J: P = 0.0143). (B) Categorical heatmap of cytokines, chemokines, and growth factors in the serum of CD-1 mouse 24 h post IT infection (as described in A). The samples were examined using the Bio-Plex ProTM mouse cytokine assay. Data depict mean fold-change of the cytokine values in WT- or ΔCbpD-infected relative to mock-infected mice (10 mice/group). The data were analyzed by the multiple unpaired t test and the FDR-adjusted P value (or q value) was determined using the two-stage step-up method of Benjamini, Krieger, and Yekutieli. Significant differences between WT and ΔCbpD are indicated by asterisks (*). IL-1α: q = 0.0181, IL-3: q = 0.0181, G-CSF: q = 0.0052, GM-CSF: q = 0.0054. (C) Principal component analysis (PCA) of identified proteins showing segregation of lung proteomes into infected (WT and ΔCbpD) and uninfected groups; quantified proteins are plotted in two-dimensional principal component space by PC1= 44.6% and PC2= 16.1%. Each circle represents one mouse/biological replicate. One mouse sample per treatment was utilized as a technical replicate for each TMT group and depicted using square (n = 5 mice/group) (D) Dimension and overlap of differentially expressed proteins (π score ≥ 2) in the lung proteome of WT (red arc)- and ΔCbpD (dark blue arc)-infected relative to mock-infected mice. The dark orange arc and purple lines reflect the regulated proteins shared in both datasets; light orange color represents regulated proteins unique to WT and ΔCbpD-infected vs. control mice; blue lines indicate the ontology term overlap among the significantly regulated proteins; plot generated with Metascape. (E) Dot plot of enrichment score (−log10 P value ≥ 2) showing pathways and cellular processes enriched in the lung proteome (Dataset S4) of ΔCbpD-infected mice. Enrichment analysis was performed using the list of up-regulated proteins belonging to the unique category by Metascape; the P value was calculated based on cumulative hypergeometric distribution. (F) STRING network analysis showing connection of regulated proteins (π score ≥ 2) associated with complement activation in the lung proteome of WT- and/or ΔCbpD-infected mice vs. control (Datasets S2–S4). The π score values are depicted next to the nodes in red (WT) and blue (ΔCbpD). Regulated proteins unique to ΔCbpD-infected versus control mice are marked with asterisks. Proteins without any interaction partners within the network are omitted from the graphics. GO:0006958: Complement activation, classical pathway (cyan), GO:0006957: Complement activation, alternative pathway (gray), MMU:977606: Regulation of complement cascade (purple). (G) Quantification of soluble complement factor C3b in EDTA-treated murine blood 3 h and 24 h post IT infection with WT or ΔCbpD (as described in A); mock (PBS)-infected mice included as control (CT). Results are given in complement arbitrary units (AU) per mL; data are plotted as mean ± SEM, representing 10 CD-1 mice/group. Data were analyzed by two-way ANOVA (Tukey’s multiple comparisons). (Left) (3 h) CT vs WT: P = 0.0025, CT vs. ΔCbpD: P = 0.9893, WT vs. ΔCbpD: P = 0.0009; (Right) (24 h) CT vs. WT: P = 0.0075, CT vs. ΔCbpD: P = 0.1162, WT vs. ΔCbpD: P = 0.3876. (H) Wild-type C57BL/6J (n = 5 to 6 mice/group) and C3 knock-out (C3−/−) mice (n = 8 mice/group) were inoculated IT with 3×106 CFU PA WT or ΔCbpD per mouse. Survival is represented by Kaplan–Meier survival curves and was analyzed with the log-rank (Mantel–Cox) test. (Left): P = 0.0445, (Right): P = 0.3464. (I) Representative hematoxylin and eosin–stained sections of spleen tissues from infected (WT or ΔCbpD) and uninfected mice (n = 5 mice/group) were collected 24 h post IT infection (as described in A), analyzed by light microscopy. Arrowheads indicate normal alveolar structure characterized by patent airspaces lined by a thin border of alveolar epithelium with no inflammatory cells. Arrows show areas of the lung with an infiltration of neutrophils (black) and macrophages (blue) in the alveoli and the interstitium leading to loss of the pulmonary surface. (The scale bar represents 500, 50, and 20 µm in the Upper, Middle, and Lower, respectively.) (J) Mean fold change values of IL-1β in the lung proteome of ΔCbpD and WT infected compared to mock-infected (control) mice (as described in A); data are plotted as the mean (log2 fold change) ± SEM, representing 5 CD-1 mice/group (one mouse sample/treatment was utilized as technical replicate in each TMT group) and analyzed by the two-tailed t test. P = 0.004. (K) Survival of PA WT and ΔCbpD upon incubation with freshly isolated human neutrophils. Bacterial survival was calculated relative to inoculum in percentage (%). Data are plotted as the mean ± SEM, representing three experiments performed in triplicate, and analyzed by the two-tailed t test (P = 0.0001). Each data point represents an individual mouse, and the significant differences are marked with asterisks (*): *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
High-resolution tandem mass tag (TMT)–based quantitative proteomics was used to study the effect of CbpD deletion on lung responses to IT PA infection at the 24 h experimental endpoint; 3,944 proteins were quantified (Datasets S1 and S2). Principal component analysis (Fig. 1C) and similarity clustering (SI Appendix, Fig. S2A) segregated the lung proteomes into PA-infected (WT and ΔCbpD) and control (mock-infected) clusters. Strong ontology term overlap was identified among the regulated proteins (π score ≥ 2) in WT-infected or ΔCbpD-infected vs. mock-infected mice, particularly among proteins belonging to the shared proteome (n = 285 proteins, Fig. 1D, Dataset S3). However, functional exploration of differentially regulated markers unique to the WT-infected (Dataset S4, n = 134) or ΔCbpD-infected lung (Dataset S4, n = 114) identified more distinct strain-dependent immune responses (Fig. 1E and SI Appendix, Fig. S2B). The lung proteome data suggested effects on the complement system (Fig. 1E) or neutrophil responses (SI Appendix, Fig. S2C), which we therefore pursued through additional experimentation.
CbpD and Complement Function in PA Lung Infection.
Among the 12 different immune-related categories that were differentially abundant in the unique proteome of ΔCbpD-infected vs. WT-infected mice (Fig. 1E) were elements of the complement cascade (Serpin G1, C2, and Cpn1, Fig. 1F); proteins were further highly interconnected (Fig. 1F) to other regulated complement factors in the shared lung proteome (Dataset S3). Since CbpD reduced the deposition of lytic membrane attack complexes (MAC) on the bacterial surface (35), we explored serum complement cascade function both early (3 h) and later (24 h) post-IT PA infection in WT or ΔCbpD strains vs. mock-infected controls. Plasma C3 activation (reviewed in refs. 47 and 48) as measured by C3b/iC3b/C3c (C3b-assay) was significantly reduced (below basal level) at both 3 h and 24 h time points in WT PA-infected mice compared to either mock- or ΔCbpD mutant-infected mice (P < 0.05, Fig. 1G). Correspondingly, total C3 protein ranged lower in the lung proteome of WT-infected vs. ΔCbpD-infected mice at the 24 h experimental endpoint (SI Appendix, Fig. S1B). We found that CbpD promotes PA resistance to complement-mediated killing, as the WT PA strain showed higher survival than the ΔCbpD in normal human serum, but the two strains survived equally in serum that was heat-inactivated or depleted of key complement proteins (reviewed in refs. 47 and 48) C3 (49) (ΔC3) or C3 and C5 (50) (ΔC3/C5) (SI Appendix, Fig. S1 C and D). CbpD-mediated interference with C3-dependent complement activity contributed to PA pathogenicity in vivo, since the virulence defect of the ΔCbpD mutant observed upon IT challenge of WT C57/BL6 mice was fully rescued in C3 knockout (C3−/−) mice (Fig. 1H).
CbpD and Neutrophil Function in PA Lung Infection.
Strong infiltration of neutrophils was observed on histopathological examination of lungs of mice infected with either the WT or ΔCbpD mutant PA strains (Fig. 1I and SI Appendix, Fig. S1E), and the importance of neutrophils in protection against mortality in the murine pneumonia model was confirmed by depletion of the cells with an anti-Ly6G antibody (SI Appendix, Fig. S1 H and I). Functional exploration of the top regulated proteins (Dataset S3, π score ≥ 10 at least in one group) in the shared lung proteome following infection with both PA strains revealed a strong association with leukocyte migration, neutrophil infiltration, and inflammation (SI Appendix, Fig. S2C). Among these, IL-1β was localized in the central part of the STRING network analysis and significantly reduced in the lung of ΔCbpD-infected compared to WT-infected mice (Fig. 1J), fitting the overall pattern of diminished proinflammatory serum cytokines in the mutant-infected animals (Fig. 1B). As heavy lung neutrophil infiltration occurred in both groups, we hypothesized that the reduced bacterial loads seen in ΔCbpD-infected animals (Fig. 1A) might reflect increased susceptibility to neutrophil opsonophagocytic killing. Indeed, while opsonized WT and ΔCbpD mutant bacteria were killed with efficiency (SI Appendix, Fig. S1G) by murine alveolar macrophages (SI Appendix, Fig. S1F), ΔCbpD mutants were more rapidly killed by freshly isolated human neutrophils (P < 0.05, Fig. 1K).
Considerations for Vaccine Design—CbpD Structural Analysis.
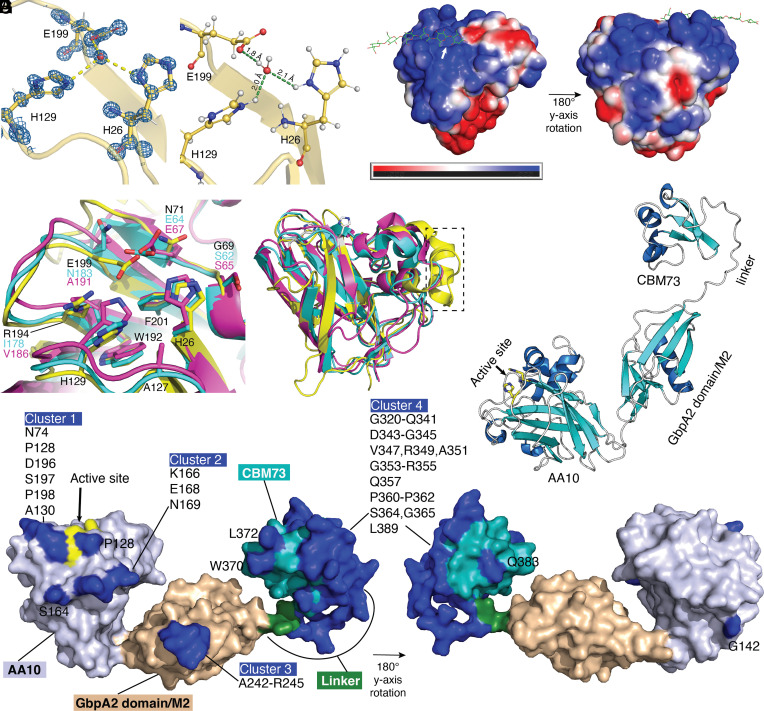
The strong conservation and high prevalence of CbpD across PA clinical isolates (33–35), coupled with experimental animal studies indicating a virulence role in systemic (35) and lung infection (Fig. 1 A and H and SI Appendix, Fig. S1A), suggests that the protein or its subdomain(s) could represent attractive vaccine antigens. Since CbpD catalytic activity is important for PA virulence (35), we determined the X-ray crystal structure of the CbpD catalytic domain (AA10) to subatomic resolution (0.75Å; PDB ID: 8C5N)—the highest-resolution LPMO structure to date, followed by 0.95Å structures with PDB ID: 4A02 (51) and 5O2X (51, 52) (Fig. 2A and SI Appendix, Table S1). The protein crystallized in space group P21, with a single chain in the asymmetric unit, and the model was refined to final Rwork/Rfree values of 14.0%/15.2%. The structure closely resembles other AA10 LPMOs, such as those from Tectaria macrodonta (PDB code: 6IF7 (53); all-atom r.m.s.d.: 0.7Å) or Vibrio cholerae (PDB code: 2XWX) (54); all-atom r.m.s.d.: 1.1Å). All LPMOs have a pyramidal shape formed by a core β-sandwich fold with protruding loops, often hosting clusters of α-helices, with an N-terminal histidine, which is a part of a characteristic histidine brace catalytic motif. The anomalous difference map showed no peak between His26 and His129 corresponding to the catalytically essential copper ion; instead, a water molecule was modeled ~1Å away from the putative copper position, additionally coordinated by a glutamate residue (Glu199), which may potentially be involved in proton delivery to the cosubstrate (55).
Fig. 2.
Structural analysis of CbpD. (A) Electron density and protonation state of the LPMO domain. σA-weighted 2mFo-DFc map contoured at 1.0 σ showing the histidine brace motif. (B) Protonation state of the histidine side chains of the active site at pH 7.3, calculated by H++ (56) to be fully protonated. (C) APBS-calculated surface potential for the CbpD LPMO domain, based on the protonated model generated by H++ for pH 7.3. The cellulose substrate (green) from the Lentinus similis LPMO9A (PDB ID: 5ACI) (57) was used to mark the putative substrate–binding region, and the active site is indicated by a white arrow. (D) Comparison of the active sites of CbpD (yellow; PDB ID: 8C5N, this work), GbpA from Vibrio cholerae (purple; PDB ID: 2XWX) and EfCBM33 from Enterococcus faecalis (cyan; PDB ID: 4ALE). Amino acids are labeled and color-coded, unless identical in all three proteins (in which case they are labeled by the residue number of CbpD, in black). (E) Superimposition of the three structures from panel (D) in cartoon representation. A dashed rectangle indicates the additional α-helix and loop that extend the putative substrate–binding site of CbpD. (F) Ab initio model of full-length CbpD, generated using Alphafold2 (UniProt entry Q02I11 from the Alphafold database). The three domains of CbpD and the linker region connecting the second domain [referred to as (‘GbpA2/M2’ domain) and third domain (‘CBM73’ domain) are labeled. An arrow points to the active site of the AA10 domain, where the two histidine residues are shown in stick representation. (G) Surface representation of the CbpD structure predicted with Alphafold2, showing the AA10 LPMO domain (light purple; active site histidines colored yellow and indicated by arrow], the GbpA2/M2 domain (wheat), the linker connecting domains 2 and 3 (forest green), and the CBM73 domain (teal). The potential discontinuous structural epitopes predicted using Discotope 2.0 (58) are colored blue and labeled.
The CbpD LPMO domain protonation state was evaluated using the H++ server (56). Side chains of the histidine brace motif were fully protonated under neutral (pH 7.3; and acidic conditions (pH 5.0, 6.2, 6.7) conditions (Fig. 2B). Reduced pH during PA respiratory tract infection could theoretically impair activity of epithelial-derived antimicrobial peptides (59) or enzymatic performance of CbpD. At pH 7.3, the adaptive Poisson–Boltzmann Solver (APBS)-calculated surface potential (60) of the catalytic site, and the CbpD enzyme surface as a whole, is dominated by positive charges (Fig. 2C). Compared with the structures of virulence-related LPMOs, such as GbpA from Vibrio cholerae and EfCBM33 form Enterococcus faecalis, the CbpD active site has several unique features (Fig. 2D), as also observed recently by Dade et al (61). At the position where both GbpA and EfCBM33 carry nonpolar amino acids (lle and Val), CbpD possesses an arginine residue (R194). The presumed catalytically important residue E199 in CbpD aligns with N183 and A191 in GbpA and EfCBM33, respectively. However, these two proteins have a glutamate residue (E64 and E67) spatially overlapping with the E199 side chain and likely play a similar role in catalysis. Finally, at a position where both GbpA and EfCBM33 exhibit a conserved serine residue, CbpD carries a glycine (G69). This makes CbpD more similar to the LPMO CelS2 (also known as ScAA10C) from Streptomyces coelicolor (39), which unlike CbpD oxidizes cellulose rather than chitin. CbpD also appears to have an extension of the putative substrate–binding surface, constituted by an α-helix and a loop that is not present in GbpA or EfCBM33 (Fig. 2E, boxed). Finally, AlphaFold2 (62) was used to predict the full-length structure of CbpD, which is a three-domain protein. The top-ranked model (Fig. 2F) shows an elongated arrangement of the three domains, which is supported by the previously reported small-angle X-ray scattering (SAXS) data of the protein (35). A recent low-resolution (3Å) structure of the CbpD first and second domains (61) shows an interaction between the two domains. A similar interaction is seen for the Vibrio cholerae LPMO, GbpA (54), indicating that multidomain LPMOs may reorganize their domains upon substrate binding.
Considerations for Vaccine Design—CbpD In Silico B Cell Epitope Prediction.
To gauge potential immunogenicity of CbpD, the Discotope 2.0 server (58) was used to predict continuous and discontinuous B-cell structural epitopes. Predicted epitopes were present in all three CbpD domains but were most abundant on the CBM73 domain (almost covering the complete domain) and the linker connecting the two domains (M2/CBM73) (Fig. 2G). In silico analysis revealed discontinuous epitopes that may be represented by four distinct clusters of amino acids: 1) N74, P128, A130, D196, S197, P198, and with P128 in close vicinity; 2) K166, E168, N169, and with S164 in close vicinity; 3) A242-R245 and 4) G320-Q341, D343-G345, V347, R349, A351, G353-R355, Q357, P360-P362, S364, G365, L389, and with L372 and W370 in close vicinity.
Immunization with CbpD Provides Protection against Lethal PA Pneumonia and Sepsis.
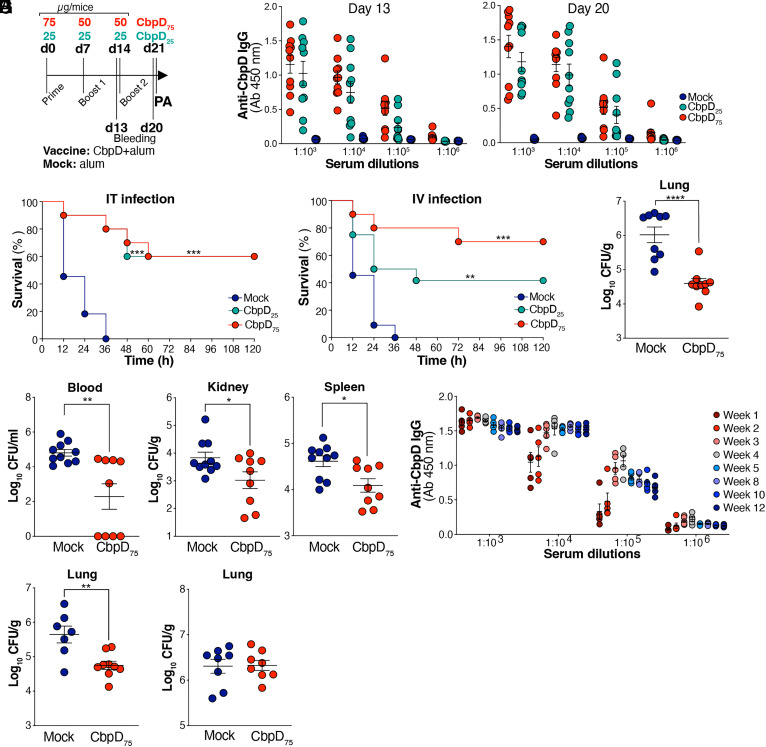
We applied the IT challenge model of lethal PA pneumonia (Fig. 1 A and H) to test the vaccine efficacy of CbpD adjuvanted with alum administered at two different dosing regimens (Fig. 3A): 75 µg prime (day 0) + 50 µg booster doses on days 7 and 14 (high dose); or 25 µg prime (day 0) + 25 µg booster doses on days 7 and 14 (low-dose). Blood was collected on days 13 and 20 postprime for antibody titer determination prior to experimental pneumonia (IT) or sepsis (intravenous, IV) challenge at day 21 postprime with a lethal dose of WT PA (Fig. 3A). CbpD immunization with either the high- or low-dose regimen induced high levels of anti-CbpD immunoglobulin G (IgG) measured by ELISA on days 13 and 20 postprime (Fig. 3B), which occurred without evidence of generalized inflammatory cytokine perturbations in the day 20 postprime sera comparing CbpD-immunized to mock-immunized mice (SI Appendix, Fig. S3A). Both CbpD vaccine dosing regimens provided significant protection against lethal infectious challenges with PA. In the pneumonia model, all mock-immunized control mice died within 36 h of IT infection, whereas 60% of animals in the low- or high-dose CbpD vaccine groups survived (Fig. 3C). Likewise in the sepsis model, all mock-immunized control mice died within 36 h of IV infection, whereas the low-dose CbpD vaccination series led to 40% survival and the high-dose regimen of CbpD vaccination series yielded 70% survival (Fig. 3D). Given the superiority of the higher dose protocol in the systemic challenge, additional experiments were performed with high-dose (75 µg prime + 50 µg boosters) to assess bacterial clearance and cytokine responses. Here, we observed significantly reduced bacterial load in CbpD-immunized vs. control mice in the lung 24 h after IT challenge (Fig. 3E) and in the blood, kidney, and spleen 24 h after IV challenge (Fig. 3F). A simultaneous assessment by multiplexed immunoassay of 23 cytokines and chemokines in homogenates of lung tissue (post-IT challenge) or splenic tissue (post-IV challenge) showed no significant differences between CbpD-immunized and mock-immunized mice (SI Appendix, Fig. S3B), indicating that IgG-mediated capture of CbpD does not impair the acute cytokine response. Stability of anti-CbpD IgG titers was confirmed until 9 wk after the second booster in the high-dose vaccination regimen (Fig. 3G and SI Appendix, Fig. S3C). Additional intratracheal challenge experiments were performed to evaluate the vaccine efficacy of high-dose CbpD regimen against a hypervirulent clinical PA strain, PA103UCSD, isolated from a CF patient and shown to harbor cbpD in its genome (SI Appendix, Fig. S3D). The bacterial load in the lung of CbpD-immunized mice was significantly lower than that of mock-immunized mice 24 h post IT challenges (Fig. 3H). Furthermore, 24 h after IT challenge, the bacterial load of the isogenic PA ΔCbpD mutant was unaffected by the high-dose vaccination protocol demonstrating the protective specificity to CbpD (Fig. 3I).
Fig. 3.
CbpD-immunized mice are protected against PA infectious challenge. (A) The timeline depicts the immunization of CD-1 mice with CbpD plus alum (CbpD) or alum alone (Mock), bleeding and PA WT challenge. (B) Total serum anti-CbpD IgG levels following CbpD immunization with 0 µg/mouse (mock, alum), 25 µg/mouse (CbpD25) or 75 µg/mouse (CbpD75). The serum was collected on days 13 and 20 postprime (as described in A). The data are plotted as the mean ± SEM, representing 10 mice per group. (C) CbpD25 (n = 10), CbpD75 (n = 10), and mock (alum)-immunized (n = 11) CD-1 mice were inoculated IT with 3×106 CFU/mouse PA WT at day 21 of the vaccination schedule (as described in A). Survival is represented by Kaplan–Meier survival curves and was analyzed by the log-rank (Mantel–Cox) test. CbpD25 vs. Mock: P = 0.0001, CbpD75 vs. Mock: P = 0.001. (D) CbpD25- (n = 12 mice), CbpD75- (n = 10 mice) and mock (alum)-immunized (n = 11 mice) CD-1 mice were inoculated IV with 2 ×107 CFU/mouse PA WT at day 21 postprime. Survival is represented by Kaplan–Meier survival curves and was analyzed by the log-rank (Mantel–Cox) test. CbpD25 vs. Mock: P = 0.0091, CbpD75 vs. Mock: P = 0.0002. (E) CbpD75- (n = 9) and mock (alum)-immunized (n = 9) CD-1 mice were IT infected with 1×106 PA WT per mice at day 21 postprime. The bacterial burden in the lung (CFU/g) was enumerated 24 h postinfection. The data are plotted as the mean ± SEM, representing 9 to 10 mice per group, and were analyzed by the unpaired t test. P < 0.0001. (F) CbpD75- (n = 9) and mock (alum)-immunized (n = 10) CD-1 mice were enumerated 4 h post IV infection with 2 ×107 CFU WT PA per mice at day 21 postprime. Bacterial load in blood (CFU/mL) and tissues (CFU/g) was enumerated 24 h postinfection. The data are plotted as the mean ± SEM, representing 9 to 10 mice per group, and were analyzed by the unpaired t test. Blood: P = 0.0029; spleen: P = 0.0112; kidney: P = 0.0348. (G) Dynamics of changes in the titers of CbpD-IgG in CD-1 murine sera in the period from 1 to 12 wk postprime immunization. The timeline of the immunization is described in A. CbpD-IgG titers in the mock-immunized group are shown in SI Appendix, Fig. S3C. The data are plotted as the mean ± SEM, representing five mice per group. (H) CbpD75- (n = 9) and mock (alum)-immunized (n = 7) CD-1 mice were enumerated 24 h post IT infection with 0.5 ×106 PA103UCSD per mice at day 21 postprime. Bacterial load in the lung (CFU/g) was enumerated 24 h postinfection. The data are plotted as the mean ± SEM, representing seven to nine mice per group, and were analyzed by the unpaired t test. P = 0.0032. (I) CbpD75- (n = 8) and mock (alum)-immunized (n = 8) CD-1 mice were enumerated 24 h post IT infection with 2 ×106 CFU ΔCbpD per mice at day 21-post prime, respectively. Bacterial load in the lung (CFU/g) was enumerated 24 h postinfection. The data are plotted as the mean ± SEM, representing eight mice per group, and were analyzed by the unpaired t test. Each data point represents an individual mouse, and the significant differences are marked with asterisks (*): *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Anti-CbpD Postvaccine Serum Promotes Neutrophil Opsonophagocytic Killing of PA Independent of Antibody Binding to the Bacterial Surface.
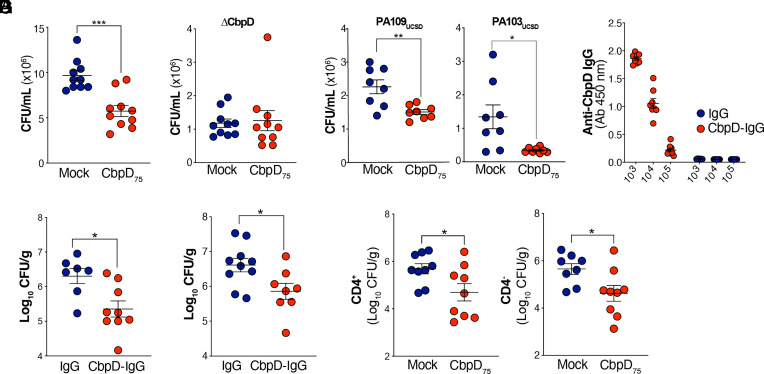
Given that CbpD is a secreted enzyme, CbpD-IgG binding to the WT PA14 surface was unaffected by sera from CbpD-immunized mice (SI Appendix, Fig. S3G). However, sera of mice fully immunized with the high-dose CbpD regimen (collected day 20 postprime) significantly boosted human neutrophil opsonophagocytic killing of WT PA compared to normal preimmune serum (Fig. 4A). Confirming antigen specificity of the antibody response to CbpD, the postimmune serum did not increase neutrophil opsonophagocytic killing of the isogenic PA ΔCbpD (Fig. 4B). High-dose CbpD postimmune sera also improved the opsonophagocytic killing of two clinical PA strains (PA103UCSD and PA109UCSD) isolated from CF patients, and (Fig. 4C) that was confirmed to harbor cbpD in their genome (SI Appendix, Fig. S3D).
Fig. 4.
Opsonophagocytic and protective activities of anti-CbpD IgG. (A) Opsonophagocytic killing of WT PA by freshly purified human neutrophils at MOI = 15 in the presence of 5% immunized murine sera (collected at day 20 postprime). The assay lasted 30 min. Data are plotted as the mean ± SEM, representing 10 mice per group, and were analyzed by the unpaired t test. P = 0.0002. (B and C) Opsonophagocytic killing of (B) ΔCbpD (n = 10 mice), (C) PA109UCSD (n = 8 mice), and PA103UCSD (n = 8 mice) at MOI = 5 by freshly isolated human neutrophils in the presence of 5% fully immunized murine sera (collected at day 20 postprime). The assay lasted 30 min. The data are plotted as mean ± SEM, representing 8 to 10 mice per group, and analyzed by the unpaired t test. PA109UCSD P = 0.0036, PA103UCSD P = 0.0140. (D and E) Anti-CbpD IgG (70 µg/mouse, n = 8 mice) or mouse IgG (70 µg/mouse, n = 7 mice) was IV transferred to naive CD-1 mice. The recipient mice were IT challenged with 1 × 106 CFU/mouse WT PA 16 h after adoptive transfer. (D) The serum was collected 16 h post adoptive transfer for anti-CbpD IgG titer analysis. (E) The recipient mice were IT challenged with 1 × 106 WT PA 16 h post adoptive transfer. Lungs were collected 24 h postinfection for enumeration. Data are plotted as the mean ± SEM of two experiments, representing seven to nine mice per group. (F) Two doses of antibodies including 200 µg/mouse (16 h prior infection) and 100 µg/mouse (30 min postinfection) µg/mouse anti-CbpD IgG (n = 8 mice) or mouse IgG (n = 10 mice) were IV transferred to naive CD-1 mice. The recipient mice were IT challenged with 2.5 × 106 CFU/mouse WT PA. The lung was collected 24 h postinfection for enumeration. The data are plotted as the mean ± SEM of two experiments, representing 8 to 10 mice per group. (G) CbpD-immunized (CbpD75) and mock-immunized (alum) CD-1 mice were treated IV with anti-CD4 or isotype control (n = 9 mice/group) 48 h and 24 h prior PA challenge. Next, 24 h post CD4 cells depletion, mice were inoculated IT with 1×106 CFU/mouse PA WT. The bacterial load in the lung (CFU/g) was enumerated 24 h postinfection. Representative flow plots are presented in SI Appendix, Fig. S4A. The data are plotted as the mean ± SEM, representing nine mice/group and were analyzed with the unpaired t test. Anti-CD4: P = 0.0311, Isotype control: P = 0.0246. Each data point represents an individual mouse, and the significant differences are marked with asterisks (*): *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Transfer of Anti-CbpD Postvaccine Serum Provides Passive Immunity to Naïve Animals.
Experiments were undertaken to determine whether anti-CbpD antibodies (humoral response) were the key factor explaining protection against PA challenge in immunized mice. High-titer anti-CbpD IgG was purified from mice immunized with the high-dose CbpD protocol and used in adoptive transfer to naive mice, with an equivalent dose of normal purified mouse IgG serving as a control (Fig. 4D). When these antibodies were provided to naive mice (single 70 µg IV dose) followed by IT challenge with a sublethal inoculum of WT PA, mice that received the anti-CbpD postimmune IgG had a slightly lower bacterial CFU recovered from the lung (Fig. 4E) at a statistically significant level. A similar result was obtained using a two-dose passive transfer protocol (200 µg + 100 µg anti-CbpD vs. control IgG) followed by higher-dose IT PA challenge and reduced bacterial CFU in the lung observed with postvaccine IgG transfer (Fig. 4F). We confirmed no overall difference in lung inflammatory cytokine responses in post-CbpD vaccine IgG-treated mice vs. control IgG-treated mice, excluding a nonspecific mechanism of protection (SI Appendix, Fig. S3E). Finally, we performed an experiment in which two groups of mice underwent the high-dose CbpD vs. mock immunization protocol but were depleted of CD4+ T cells using a monoclonal antibody (SI Appendix, Fig. S4A) prior to WT PA pneumonia challenge via examination of splenocytes. Though we did not specifically evaluate the efficiency of this treatment on depletion of tissue-resident memory C4 T cells in the lung, the CbpD-immunized mice showed reduced lung CFU compared to mock-immunized mice (Fig. 4G).
Repeated Prior PA Infection Does Not Impair CbpD Vaccine Efficacy.
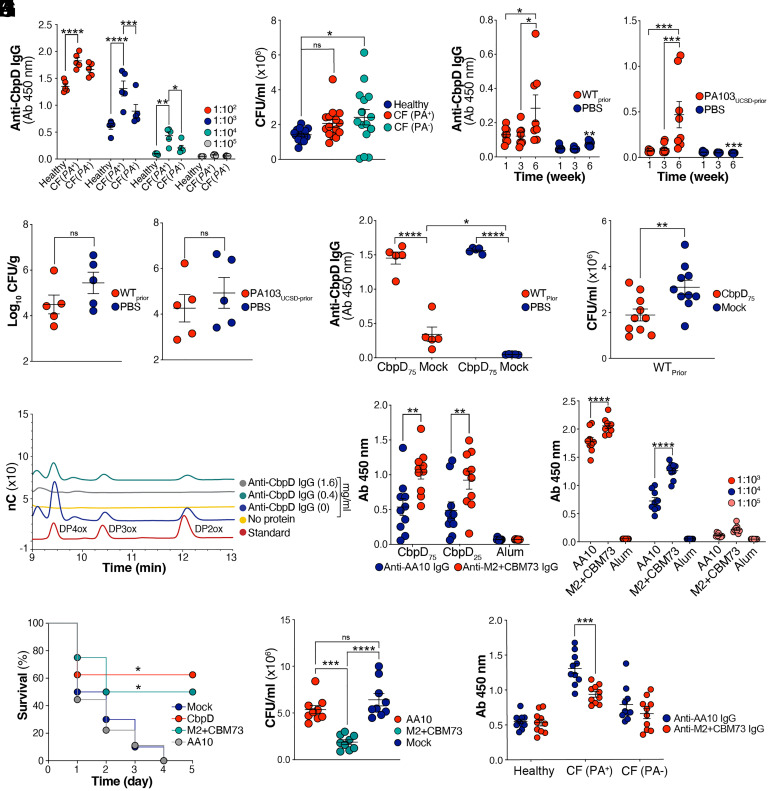
Human serum IgG recognizing CbpD was identified in healthy individuals (SI Appendix, Fig. S3F and Fig. 5A) and in CF patients colonized (PA+) or noncolonized (PA-) with PA at the time of blood collection (Fig. 5A). While the level of anti-CbpD IgG was significantly higher in CF(PA+) individuals compared to the two other groups, the serum failed to improve human neutrophil opsonophagocytic killing of WT PA (Fig. 5B). This observation raised a concern of whether preexisting IgG directed against CbpD in chronically infected individuals might present a form of “original antigenic sin” (63) that interferes with effective anti-CbpD vaccine responses. To model chronic host exposure to PA and to determine whether preexisting antibodies could revoke protection conferred by CbpD immunization, 4-wk-old CD-1 mice were intratracheally infected with sublethal WT PA or PA103UCSD (3 to 5 × 105 CFU/mouse) vs. PBS control at weekly intervals for a total of six exposures. A gradual rise of anti-CbpD IgG titers in murine serum with substantial mouse-to-mouse heterogeneity was identified upon repeated IT infection with either PA strain compared to the mock-infected group (Fig. 5C). Next, we investigated the sensitization of the chronically exposed mice to a subsequent IT PA challenge. Regardless of the examined PA strain, the assessment of the lung bacterial loads revealed variation among individuals in the chronically exposed and nonexposed groups, with a general tendency to lower counts in the preexposed mice. (Fig. 5D). Nevertheless, prior PA infection did not interfere with the induction of robust anti-CbpD IgG titers in mice by the high-dose CbpD vaccination protocol (Fig. 5E) nor suppress the functional efficacy of the post-CbpD immune serum in boosting human neutrophil opsonophagocytic killing of WT PA (Fig. 5F).
Fig. 5.
Immunization with CbpD elicits neutralizing antibodies. (A) Total plasma anti-CbpD IgG levels in healthy human individuals and cystic fibrosis (CF) patients who were colonized (PA+) or noncolonized (PA-) with PA at the time of blood collection. Data are plotted as the mean ± SEM, representing 5 individuals per group. Data were analyzed by two-way ANOVA (Dunnett’s multiple comparisons test). 102: CF (PA+) vs. Healthy: P = <0.0001; 103: CF (PA+) vs. Healthy: P = <0.0001, CF (PA+) vs. CF (PA-) A: P = 0.0001; 104: CF (PA+) vs. Healthy: P = 0.0017, CF (PA+) vs. CF (PA-): P = 0.0360. (B) Opsonophagocytic killing of WT PA at MOI = 7 by freshly isolated human neutrophils in the presence of 5% human plasma obtained from healthy individuals or CF patients (as described in A). The assay lasted 30 min. Data are plotted as the mean ± SEM, representing five individuals per group that were analyzed in triplicate and examined by the unpaired t test. Healthy vs. CF (PA-): P = 0.0319. (C) CD-1 mice were IT challenged with PA once per week for 6 wk (6×). Serum anti-CbpD IgG levels were measured (dilution: 1:103) at week 1, 3, and 6 postinfection at week 0 with WT (Left, WTPrior) or PA103UCSD (Right, PA103UCSD-Prior). Mock-infected mice (PBS) served as control. The data are plotted as the mean ± SEM, representing 8 mice per group. Data were analyzed by two-way ANOVA (Tukey’s multiple comparisons test). WT: week (1 vs. 6): P = 0.0240; week (3 vs. 6): P = 0.0157; PA103UCSD: week (1 vs. 6): P = 0.0004, week (3 vs. 6): P = 0.0009. (D) The lung bacterial burden in the mice that were recurrently exposed to WT PA (WTPrior) or PA103UCSD-Prior and PBS (Mock-infected mice) as described in C. The mice were mock-immunized (alum) (as described in Fig. 3A) and rechallenged IT with a sublethal dose of 1 ×106 WT PA or 0.5 ×106 PA103UCSD. Bacterial load was enumerated 24 h postinfection. The data are plotted as the mean ± SEM, representing 5 individuals per group. Data were analyzed by the two-tailed paired t test. (E) Total anti-CbpD IgG in sera (dilution: 1:103) from immunized (CbpD75)- and mock-immunized mice that were recurrently exposed to WT PA (as described in C). The data are plotted as the mean ± SEM, representing five mice per group. Data were analyzed by two-way ANOVA (Tukey’s multiple comparisons test). WTPrior P = < 0.0001, PA103UCSD-Prior P = <0.0001. (F) Opsonophagocytic killing by freshly isolated human neutrophils in the presence of 5% immunized (CbpD75)- and mock-immunized mice that were recurrently exposed to WT PA (as described in C). The assay lasted 30 min at MOI = 5. Data are plotted as the mean ± SEM of two experiments, representing five mice per group. Data were analyzed by the unpaired t test. P = 0.0075. (G) HPLC analysis of reaction products emerging from a mixture of copper-saturated recombinant CbpD, β-chitin, ascorbate as reducing agent (in all samples), and mouse anti-CbpD IgG, in buffer (20 mM Tris-HCl pH 7.0), for 3 h at 37 °C under shaking. The degrees of polymerization (DP) of chitooligosaccharide aldonic acids in a standard sample are depicted. The reactions without 1 mM ascorbate, which resulted in generation of no oxidized product is shown in SI Appendix, Fig. S3H. (H) Total serum anti-AA10 IgG and anti-M2+CBM73 IgG levels (dilution: 1:103) following CbpD immunization with 0 µg/mouse regimen (mock, alum), 25 µg/mouse regimen (CbpD25), and 75 µg/mouse regiment (CbpD75) as described in Fig. 3A. Serum was collected on day 20 postprime and recombinant AA10 and M2+CBM73 were immobilized on an ELISA plate. Data are plotted as the mean ± SEM, representing 10 mice per group, and analyzed by two-way ANOVA (Tukey’s multiple comparisons test). CbpD75 P = 0.0018; CbpD25 P = 0.0078. (I) Total serum anti-AA10 IgG and anti-M2+CBM73 IgG level following immunization with truncated variants of CbpD (25 µg/mouse regimen) as described in Fig. 3A. The serum was collected on day 20 postprime and recombinant CbpD were immobilized on an ELISA plate. The data are plotted as the mean ± SEM, representing nine mice per group. Data were analyzed by two-way ANOVA (Tukey’s multiple comparisons test). 1:103 P = < 0.0001; 1:104 P = < 0.0001. (J) CbpD- (n = 8 mice, 25 µg/mouse regimen), AA10- (n = 9 mice, 25 µg/mouse regimen), M2+CBM73 (n = 8 mice, 25 µg/mouse regimen), and mock (alum)-immunized (n =10 mice) were inoculated IT with 2 ×106 CFU/mouse PA WT. Survival is represented by Kaplan–Meier survival curves and analyzed by the log-rank (Mantel–Cox) test. CbpD vs. Mock: P = 0.0130, M2+CBM73 vs. Mock: P = 0.0360. (K) Opsonophagocytic killing of WT PA by freshly purified human neutrophils at MOI = 12 in the presence of 5% immunized mouse sera (collected at day 20 postprime). The assay lasted 30 min. Data are plotted as the mean ± SEM, representing nine mice per group, and were analyzed by two-way ANOVA (Tukey’s multiple comparisons test). AA10 vs. M2+CBM73: P = 0.0008; Mock vs. M2+CBM73: P = < 0.0001. (L) Total plasma anti-AA10 and M2+CBM73 IgG levels in healthy human individuals and cystic fibrosis (CF) patients (dilution: 1:102) who were colonized (PA+) or noncolonized (PA-) with PA at the time of blood collection. Recombinant AA10 and M2+CBM73 were immobilized on an ELISA plate. The data are plotted as the mean ± SEM of two independent ELISA, representing five individuals per group. Data were analyzed by two-way ANOVA (Šídák’s multiple comparisons test). AA10 vs. M2+CBM73 CF (PA+): P = 0.0002. Each data point represents an individual mouse/human, and the significant differences are marked with asterisks (*): *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
Antibodies Elicited by CbpD Immunization Block LPMO Activity of the Enzyme.
Sera from CbpD-immunized mice did not enhance IgG binding to WT PA surface (SI Appendix, Fig. S3G), consistent with the understanding that the enzyme is a secreted protein. To assess the neutralizing characteristics of CbpD IgG, we evaluated the enzymatic activity of CbpD on a model substrate, β-chitin, in the presence or absence of the anti-CbpD IgG (Fig. 5G) purified from the serum of immunized mice with a higher dose of CbpD (Fig. 3A). Copper-saturated and ascorbate-reduced full-length CbpD was fully active toward β-chitin in the absence of anti-CbpD IgG, leading to the release of chitooligosaccharide aldonic acids with a degree of polymerization ranging from four to six (Fig. 5G). Incubation of the recombinant CbpD with a lower dose (0.4 mg/mL) anti-CbpD antibodies reduced activity against the chitin substrate, while a higher dose (1.6 mg/mL) abolished activity almost completely (Fig. 5G). No oxidized products were observed in the non-ascorbate-driven control reactions (SI Appendix, Fig. S3H).
The Noncatalytic Domains of CbpD Are Important for Protective Immunogenicity.
The trimodular CbpD structure consists of an N-terminal catalytic (AA10) domain followed by a module with an unknown function (M2) and a C-terminal CBM domain (CBM73)(35). Antibodies elicited by CbpD immunization at the low- and high-dose series (Fig. 3A) bound more broadly to M2+CBM73 domains than to the AA10 domain (Fig. 5H). Immunization with truncated variants of CbpD, either the AA10 domain or the M2+CBM73 domains, showed a significantly higher level of antibody response to the latter (Fig. 5I). While mice vaccinated with the CbpD AA10 domain were not protected (full mortality within 96 h post IT infection), 50% of those immunized with the CbpD M2+CBM73 domains survived the lethal challenge (Fig. 5J). Additionally, sera of mice fully immunized with the M2+CBM73 regimen (collected at day 20 postprime), but not those immunized with AA10, significantly increased human neutrophil opsonophagocytic killing of WT PA compared to mock-immunized serum (Fig. 5K). Of note, preexisting antibodies in CF patients with PA respiratory tract colonization (Fig. 5L and SI Appendix, Fig. S3I) were primarily directed against the AA10 domain, which may in part explain their failure to improve the opsonophagocytic killing of WT PA (Fig. 5B) or impede the ongoing susceptibility to PA pneumonia in this patient population.
Discussion
Here, we provide evidence that the LPMO CbpD contributes to PA virulence in the lower respiratory tract, as shown by reduced mortality and increased clearance of an isogenic PA ΔCbpD mutant in a murine pneumonia model (Fig. 1). While a comparable inflammatory cytokine response and robust neutrophil recruitment occurred in the lungs of both WT PA- and ΔCbpD-infected mice, the reduced bacterial burden seen with ΔCbpD challenge mirrored enhanced opsonophagocytic killing of the isogenic mutant by human neutrophils (Fig. 1). C3b is a pivotal opsonin for PA (64) and increased levels of C5 convertase assembly and deposition of C3b on the ΔCbpD surface were demonstrated in our previous study (35). Increased soluble C3b, as measured by a neoepitope expressed in C3b, iC3b, and C3c, thus reflecting specific activation of C3 in the plasma of intratracheally challenged CbpD-infected mice (Fig. 1) and vulnerability of the isogenic mutant to complement-mediated killing (SI Appendix, Fig. S1) could also contribute to accelerated lung bacterial clearance. Neutrophils (65, 66) and the complement system (67, 68) are crucial immune defenses against PA, and neutropenic (69) and complement-deficient mice (70–72) suffer more severe PA infections. In contrast to normal mice, we found comparable mortality upon ΔCbpD- and WT PA IT challenge of C3−/−(Fig. 1) or neutropenic mice (SI Appendix, Fig. S1), indicating that complement activation and neutrophil functions are involved in CbpD-mediated virulence functions.
Both cell-mediated immunity and opsonizing antibodies have been linked to protective immunological responses against PA (26, 73–77). Immunization with CbpD elicited protective antibodies that promoted neutrophil opsonophagocytic killing, independent of their binding to the bacterial surface, and exhibited neutralizing characteristics (Figs. 3–5). The encoding cbpD gene is highly conserved and prevalent across multiple PA clinical isolates (33–35), an important characteristic of a vaccine candidate. Immunization of naive mice with recombinant CbpD provided antibody-mediated protection against murine acute pneumonia and systemic infections (Fig. 3). Given CbpD is a secreted virulence factor (40, 41), enhanced opsonophagocytic killing of PA (Fig. 4) is likely related to the potency of anti-CbpD IgG in immunized murine sera to block enzyme activity (Fig. 5) and mitigate CbpD-mediated immune subversion (Fig. 3) to the advantage of the host. In contrast to the biomass-degradation potential of LPMOs (reviewed in refs. 78 and 79), their role in bacterial pathogenesis (reviewed in refs. 80 and 81) and protective antigenicity are important but understudied aspects of LPMO biology. Among other LPMO family members, passive immunization of mice with rabbit antisera raised against Vibrio cholerae GbpA reduced intestinal colonization with the pathogen (82). The results of our in silico discontinuous epitopes analysis (Fig. 2) corroborate our observation that CbpD-immunized mice developed significant amounts of protective antibodies against the M2+CBM73 domains and that immunization of CD-1 mice with these two domains provided nearly comparable protection to full-length CbpD in IT-challenged mice and promoted opsonophagocytic mediated killing of the bacterium (Fig. 5).
Previously, a recombinant-based vaccine candidate (IC43) containing recombinant OprF and OprI, two conserved and prevalent PA outer membrane proteins, progressed to clinical trials (31, 83 –85). Although IC43 elicited a significant level of immunogenicity, it expressed no therapeutic advantage over the sham immunized group in terms of overall mortality and invasive PA infections in severely ill ICU patients, who were on mechanical ventilation (31). Recently, prior Staphylococcus aureus exposure was shown to render several protective recombinant-based vaccine candidates against this bacterium, such as IsdB, MntC, and FhuD2, ineffective in vivo (86). Importantly, despite offering substantial protection against invasive S. aureus infection during preclinical animal evaluation, these vaccines underperformed in human clinical trials (87, 88). In contrast, we observed immunization with CbpD-induced robust IgG titers with high levels of opsonophagocytic killing even in mice that were recurrently exposed to PA (Fig. 5). Protective immunogenicity is not evenly distributed through the trimodular architecture of CbpD. Our studies suggest that prior exposure to PA in healthy individuals or chronic colonization with the bacterium in CF patients may lead to a propensity for the development of antibodies against the nonprotective AA10 domain of CbpD. Immunization with recombinant CbpD in naive mice generates high titers of antibodies against the protective noncatalytic domains (M2+CBM73), and these domains combined function effectively as an antigen for vaccine protection (Fig. 5). This could offer a theoretical advantage for immunization since the AA10 domain carries a potentially toxic copper ion in its holo form or may act as a copper scavenger in its apo-form.
Chitin or chitosan polymers can be converted into oligosaccharides (COS) that contain β-(1,4)-linked d-glucosamine monomers, N-acetyl-glucosamine monomers, or a combination of these. Due to the absence of such biopolymers in the mammalian host, they are able to evoke host humoral and cellular immunological responses (reviewed in ref. 89), and many variables including the degree of polymerization and chain length may alter their immunostimulatory properties (reviewed in ref. 89). Despite the low likelihood of such molecules arising from CbpD activity in the mammalian host, it cannot be ruled out that products arising from CbpD activity could have properties similar to COS and thereby provide an immunopotentiation activity. The study of such effects would be of great interest once the CbpD substrate is defined. PA lung infections are a major concern in vulnerable populations, and we show the involvement of the conserved LPMO CbpD in exacerbation of PA pneumonia through interference with complement and neutrophil-mediated killing. Our immunization studies provide evidence on protective characteristics of CbpD as a potential PA vaccine candidate and suggest that further exploration of the functional immunogenicity provided by the LPMO family of proteins, represented across many clinically important and antibiotic-resistant human pathogens, is warranted.
Materials and Methods
Materials and methods, describing in detail reagents, bacterial strains, constructs, cells, and serum collection; protein production and purification; murine model of immunization; murine model of intratracheal (IT) PA infection; lung histopathology analysis; murine model of systemic PA infection; isolation and infection of murine alveolar macrophages (AMs); fluorescence microscopy analysis of AMs; serum killing assay; neutrophil killing assay; CbpD-specific antibody response in human and murine sera; cytokine profiling in murine sera or organ homogenates; complement analysis (C3b-assay); in vivo depletion of neutrophils; in vivo depletion of CD4 cells; adoptive transfer of anti-CbpD IgG in vivo; CbpD-IgG neutralization assay; flow cytometric analysis of CbpD antibody binding to PA cell surface; analysis of lung proteome; X-ray crystallography of CbpD catalytic domain; modeling the protonation state of CbpD catalytic domain (AA10); epitope prediction; and statistical analysis are provided in SI Appendix.
Ethics Declarations.
Animal experiments followed all ethical regulations for animal research and were carried out in accordance with the rules and regulations of the Institutional Animal Care and Use Committee, which was approved by the UC San Diego IRB protocol S00227M. Blood was collected from several healthy volunteers (male and female) and cystic fibrosis (CF) patients in accordance with the Helsinki Declaration’s ethical principles. The two blood collection processes were approved by the UC San Diego Human Research Protection Program under IRB protocol numbers 131,002 and 160,078, respectively. All blood donors signed a written informed consent form.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
This work was supported by grants from the NIH (HD090259 to V.N. and 1KL2TR001444 to A.M.), Norwegian Research Council (NRC grant nos. 326272 to G.V.-K. and 272201 to U.K.). We are grateful for services provided by the Proteomics Core Facility at UiT-The Arctic University of Norway. We acknowledge the use of the University of Oslo (UiO) Structural Biology core facilities, part of the Norwegian Macromolecular Crystallography Consortiumand funding by the Norwegian INFRASTRUKTUR-program (project no. 245828) and UiO (core facility funds). MS-based proteomic analyses were performed by UiT Proteomics and Metabolomics Core Facility (PRiME), a member of the National Network of Advanced Proteomics Infrastructure, funded by the Norwegian INFRASTRUKTUR-program (project number: 295910). We thank Thomas Schneider for valuable comments regarding SHELXL refinement, Oskar Aurelius for assistance in X-ray diffraction experiments at the BioMAX beamline at MAX IV in Lund, Sweden, and the technical assistance of Prof. Alison Coady, Dr. Alexandria Hoffman, and Samira Dahesh.
Author contributions
F.A., C.-M.T., G.V.-K., and V.N. designed research; F.A., C.-M.T., G.C., R.H.Z., E.B., O.G., H.V.S., A.K., A.M., E.C., J.-A.B., J.A.L., D.T., S.D., P.K.T.E., and U.K. performed research; D.J.C., T.E.M., and G.Y.L. contributed new reagents/analytic tools; F.A., C.-M.T., G.C., E.B., H.V.S., E.C., J.-A.B., B.C., T.E.M., U.K., G.V.-K., and V.N. analyzed data; and F.A., G.C., U.K. and V.N. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Fatemeh Askarian, Email: faaskarian@health.ucsd.edu.
Victor Nizet, Email: vnizet@health.ucsd.edu.
Data, Materials, and Software Availability
Mass spectrometry proteomics data are deposited to the ProteomeXchange Consortium (90) via the PRIDE (91) partner repository with dataset identifier PXD039298. The crystal structure of CbpD is provided by publicly available protein structures with PDB identifiers 8C5N. All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Folkesson A., et al. , Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: An evolutionary perspective. Nat. Rev. Microbiol. 10, 841–851 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Rossi E., et al. , Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat. Rev. Microbiol. 19, 331–342 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Cystic Fibrosis Foundation, “Cystic fibrosis foundation patient registry 2020 annual data report” (Cystic Fibrosis Foundation Patient Registry, Bethesda, MD, 2021).
- 4.Almagro P., et al. , Pseudomonas aeruginosa and mortality after hospital admission for chronic obstructive pulmonary disease. Respiration 84, 36–43 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Finch S., McDonnell M. J., Abo-Leyah H., Aliberti S., Chalmers J. D., A comprehensive analysis of the impact of Pseudomonas aeruginosa colonization on prognosis in adult bronchiectasis. Ann. Am. Thorac. Soc. 12, 1602–1611 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Jacobs D. M., et al. , Impact of Pseudomonas aeruginosa isolation on mortality and outcomes in an outpatient chronic obstructive pulmonary disease cohort. Open Forum. Infect. Dis. 7, ofz546 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenfeld M., et al. , Diagnostic accuracy of oropharyngeal cultures in infants and young children with cystic fibrosis. Pediatr. Pulmonol. 28, 321–328 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez-Aguirregabiria M., Asensio-Martín M. J., Nanwani-Nanwani K. L., Recurrent ventilator-associated pneumonia caused by “difficult to treat” resistance Pseudomonas aeruginosa. Rev. Esp. Quimioter. 35, 117–119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albin O. R., Kaye K. S., McCreary E. K., Pogue J. M., Less is more? Antibiotic treatment duration in Pseudomonas aeruginosa ventilator-associated pneumonia. Clin. Infect. Dis. 76, 745–749 (2023). [DOI] [PubMed] [Google Scholar]
- 10.Herrera S., Bodro M., Soriano A., Predictors of multidrug resistant Pseudomonas aeruginosa involvement in bloodstream infections. Curr. Opin. Infect. Dis. 34, 686–692 (2021). [DOI] [PubMed] [Google Scholar]
- 11.Shupp J. W., et al. , Epidemiology of bloodstream infections in burn-injured patients: A review of the national burn repository. J. Burn Care Res. 31, 521–528 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Trecarichi E. M., Tumbarello M., Antimicrobial-resistant Gram-negative bacteria in febrile neutropenic patients with cancer: Current epidemiology and clinical impact. Curr. Opin. Infect. Dis. 27, 200–210 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Rolston K. V. I., Infections in cancer patients with solid tumors: A review. Infect. Dis. Ther. 6, 69–83 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pang Z., Raudonis R., Glick B. R., Lin T.-J., Cheng Z., Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 37, 177–192 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Botelho J., Grosso F., Peixe L., Antibiotic resistance in Pseudomonas aeruginosa—Mechanisms, epidemiology and evolution. Drug Resist. Updat. 44, 100640 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Killough M., Rodgers A. M., Ingram R. J., Pseudomonas aeruginosa: Recent advances in vaccine development. Vaccines (Basel) 10, 1100 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sousa S. A., Seixas A. M. M., Marques J. M. M., Leitão J. H., Immunization and immunotherapy approaches against Pseudomonas aeruginosa and Burkholderia cepacia complex infections. Vaccines (Basel) 9, 670 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunham R. C., Plummer F. A., Stephens R. S., Bacterial antigenic variation, host immune response, and pathogen-host coevolution. Infect. Immun. 61, 2273–2276 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sawa T., et al. , Molecular epidemiology of clinically high-risk Pseudomonas aeruginosa strains: Practical overview. Microbiol. Immunol. 64, 331–344 (2020). [DOI] [PubMed] [Google Scholar]
- 20.Wilkinson S. G., Composition and structure of lipopolysaccharides from Pseudomonas aeruginosa. Rev. Infect. Dis. 5, S941–S949 (1983). [DOI] [PubMed] [Google Scholar]
- 21.Magnowska Z., Hartmann I., Jänsch L., Jahn D., Membrane proteomics of Pseudomonas aeruginosa. Methods Mol. Biol. 1149, 213–224 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Pletzer D., et al. , The stringent stress response controls proteases and global regulators under optimal growth conditions in Pseudomonas aeruginosa. mSystems 5, e00495-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sainz-Mejías M., Jurado-Martín I., McClean S., Understanding Pseudomonas aeruginosa-Host interactions: The ongoing quest for an efficacious vaccine. Cells 9, 2617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurado-Martín I., Sainz-Mejías M., McClean S., Pseudomonas aeruginosa: An audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci. 22, 3128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young L. S., Meyer R. D., Armstrong D., Pseudomonas aeruginosa vaccine in cancer patients. Ann. Intern. Med. 79, 518–527 (1973). [DOI] [PubMed] [Google Scholar]
- 26.Langford D. T., Hiller J., Prospective, controlled study of a polyvalent pseudomonas vaccine in cystic fibrosis–Three year results. Arch. Dis. Child. 59, 1131–1134 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Döring G., Prevention of Pseudomonas aeruginosa infection in cystic fibrosis patients. Int. J. Med. Microbiol. 300, 573–577 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Alexander J. W., Fisher M. W., Vaccination for Pseudomonas aeruginosa. Am. J. Surg. 120, 512 (1970). [DOI] [PubMed] [Google Scholar]
- 29.Döring G., Meisner C., Stern M., Flagella vaccine trial study group, A double-blind randomized placebo-controlled phase III study of a Pseudomonas aeruginosa flagella vaccine in cystic fibrosis patients. Proc. Natl. Acad. Sci. U.S.A. 104, 11020–11025 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pier G. B., et al. , Human immune response to Pseudomonas aeruginosa mucoid exopolysaccharide (alginate) vaccine. Infect. Immun. 62, 3972–3979 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adlbrecht C., et al. , Efficacy, immunogenicity, and safety of IC43 recombinant Pseudomonas aeruginosa vaccine in mechanically ventilated intensive care patients-a randomized clinical trial. Crit. Care 24, 74 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rello J., et al. , A randomized placebo-controlled phase II study of a Pseudomonas vaccine in ventilated ICU patients. Crit. Care 21, 22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Folders J., Tommassen J., van Loon L. C., Bitter W., Identification of a chitin-binding protein secreted by Pseudomonas aeruginosa. J. Bacteriol. 182, 1257–1263 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salunkhe P., et al. , A cystic fibrosis epidemic strain of Pseudomonas aeruginosa displays enhanced virulence and antimicrobial resistance. J. Bacteriol. 187, 4908–4920 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Askarian F., et al. , The lytic polysaccharide monooxygenase CbpD promotes Pseudomonas aeruginosa virulence in systemic infection. Nat. Commun. 12, 1230 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaaje-Kolstad G., et al. , An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Quinlan R. J., et al. , Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proc. Natl. Acad. Sci. U.S.A. 108, 15079–15084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips C. M., Beeson W. T., Cate J. H., Marletta M. A., Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 6, 1399–1406 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Forsberg Z., et al. , Structural and functional characterization of a conserved pair of bacterial cellulose-oxidizing lytic polysaccharide monooxygenases. Proc. Natl. Acad. Sci. U.S.A. 111, 8446–8451 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Douzi B., et al. , Unraveling the self-assembly of the XcpQ secretin periplasmic domain provides new molecular insights into type II secretion system secreton architecture and dynamics. mBio 8, e01185-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michel-Souzy S., et al. , Direct interactions between the secreted effector and the T2SS components GspL and GspM reveal a new effector-sensing step during type 2 secretion. J. Biol. Chem. 293, 19441–19450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arevalo-Ferro C., et al. , Identification of quorum-sensing regulated proteins in the opportunistic pathogen Pseudomonas aeruginosa by proteomics. Environ. Microbiol. 5, 1350–1369 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Schuster M., Lostroh C. P., Ogi T., Greenberg E. P., Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: A transcriptome analysis. J. Bacteriol. 185, 2066–2079 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cattoir V., et al. , Transcriptional response of mucoid Pseudomonas aeruginosa to human respiratory mucus. mBio 3, e00410–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scott N. E., Hare N. J., White M. Y., Manos J., Cordwell S. J., Secretome of transmissible Pseudomonas aeruginosa AES-1R grown in a cystic fibrosis lung-like environment. J. Proteome Res. 12, 5357–5369 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Rahme L. G., et al. , Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Reis E. S., Mastellos D. C., Hajishengallis G., Lambris J. D., New insights into the immune functions of complement. Nat. Rev. Immunol. 19, 503–516 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heesterbeek D. A. C., Angelier M. L., Harrison R. A., Rooijakkers S. H. M., Complement and bacterial infections: From molecular mechanisms to therapeutic applications. J. Innate Immun. 10, 455–464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssen B. J. C., et al. , Structures of complement component C3 provide insights into the function and evolution of immunity. Nature 437, 505–511 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Fredslund F., et al. , Structure of and influence of a tick complement inhibitor on human complement component 5. Nat. Immunol. 9, 753–760 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Vaaje-Kolstad G., et al. , Characterization of the chitinolytic machinery of Enterococcus faecalis V583 and high-resolution structure of its oxidative CBM33 enzyme. J. Mol. Biol. 416, 239–254 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Forsberg Z., et al. , Structural determinants of bacterial lytic polysaccharide monooxygenase functionality. J. Biol. Chem. 293, 1397–1412 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yadav S. K., Archana R., Singh P. K., Singh P. G., Vasudev, Insecticidal fern protein Tma12 is possibly a lytic polysaccharide monooxygenase. Planta 249, 1987–1996 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Wong E., et al. , The Vibrio cholerae colonization factor GbpA possesses a modular structure that governs binding to different host surfaces. PLoS Pathog. 8, e1002373 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caldararu O., Oksanen E., Ryde U., Hedegård E. D., Mechanism of hydrogen peroxide formation by lytic polysaccharide monooxygenase. Chem. Sci. 10, 576–586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anandakrishnan R., Aguilar B., Onufriev A. V., H++ 3.0: Automating pK prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulations Nucleic Acids Res. 40, W537–W541 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frandsen K. E. H., et al. , The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases. Nat. Chem. Biol. 12, 298–303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kringelum J. V., Lundegaard C., Lund O., Nielsen M., Reliable B cell epitope predictions: Impacts of method development and improved benchmarking. PLoS Comput. Biol. 8, e1002829 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres I. M., Demirdjian S., Vargas J., Goodale B. C., Berwin B., Acidosis increases the susceptibility of respiratory epithelial cells to—Induced cytotoxicity. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L126–L137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jurrus E., et al. , Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27, 112–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dade C. M., et al. , The crystal structure of CbpD clarifies substrate-specificity motifs in chitin-active lytic polysaccharide monooxygenases. Acta Crystallogr. D Struct. Biol. 78, 1064–1078 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Francis T., On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 104, 572–578 (1960). [Google Scholar]
- 64.Mishra M., et al. , Pseudomonas aeruginosa Psl polysaccharide reduces neutrophil phagocytosis and the oxidative response by limiting complement-mediated opsonization. Cell. Microbiol. 14, 95–106 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gudiol C., et al. , Clinical predictive model of multidrug resistance in neutropenic cancer patients with bloodstream infection due to Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 64, e02494-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carratalà J., Rosón B., Fernández-Sevilla A., Alcaide F., Gudiol F., Bacteremic pneumonia in neutropenic patients with cancer: Causes, empirical antibiotic therapy, and outcome. Arch. Intern. Med. 158, 868–872 (1998). [DOI] [PubMed] [Google Scholar]
- 67.Kawakami T., et al. , Severe infection of Pseudomonas aeruginosa during Eculizumab therapy for paroxysmal nocturnal hemoglobinuria. Intern. Med. 57, 127–130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Webb B. J., et al. , Recurrent infection with Pseudomonas aeruginosa during eculizumab therapy in an allogeneic hematopoietic stem cell transplant recipient. Transpl. Infect. Dis. 18, 312–314 (2016). [DOI] [PubMed] [Google Scholar]
- 69.Koh A. Y., Priebe G. P., Ray C., Van Rooijen N., Pier G. B., Inescapable need for neutrophils as mediators of cellular innate immunity to acute Pseudomonas aeruginosa pneumonia. Infect. Immun. 77, 5300–5310 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Younger J. G., et al. , Murine complement interactions with Pseudomonas aeruginosa and their consequences during pneumonia. Am. J. Respir. Cell Mol. Biol. 29, 432–438 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mueller-Ortiz S. L., Drouin S. M., Wetsel R. A., The alternative activation pathway and complement component C3 are critical for a protective immune response against Pseudomonas aeruginosa in a murine model of Pneumonia. Infect. Immun. 72, 2899–2906 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gross G. N., Rehm S. R., Pierce A. K., The effect of complement depletion on lung clearance of bacteria. J. Clin. Invest. 62, 373–378 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buret A., Dunkley M. L., Pang G., Clancy R. L., Cripps A. W., Pulmonary immunity to pseudomonas aeruginosa in intestinally immunized rats roles of alveolar macrophages, tumor necrosis factor alpha, and interleukin-1 alpha. Infect. Immun. 62, 5335–5343 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kooguchi K., et al. , Role of alveolar macrophages in initiation and regulation of inflammation in Pseudomonas aeruginosa pneumonia. Infect. Immun. 66, 3164–3169 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pennington J. E., Kuchmy D., Mechanism for pulmonary protection by lipopolysaccharide Pseudomonas vaccine. J. Infect. Dis. 142, 191–198 (1980). [DOI] [PubMed] [Google Scholar]
- 76.Schaad U. B., et al. , Safety and immunogenicity of Pseudomonas aeruginosa conjugate A vaccine in cystic fibrosis. Lancet 338, 1236–1237 (1991). [DOI] [PubMed] [Google Scholar]
- 77.Worgall S., et al. , Protection against pulmonary infection with Pseudomonas aeruginosa following immunization with P. aeruginosa-pulsed dendritic cells. Infect. Immun. 69, 4521–4527 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chylenski P., et al. , Lytic polysaccharide monooxygenases in enzymatic processing of lignocellulosic biomass. ACS Catal. 9, 4970–4991 (2019). [Google Scholar]
- 79.Moon M., et al. , Lytic polysaccharide monooxygenase (LPMO)-derived saccharification of lignocellulosic biomass. Bioresour. Technol. 359, 127501 (2022). [DOI] [PubMed] [Google Scholar]
- 80.Vandhana T. M., et al. , On the expansion of biological functions of lytic polysaccharide monooxygenases. New Phytol. 233, 2380–2396 (2022). [DOI] [PubMed] [Google Scholar]
- 81.Agostoni M., Hangasky J. A., Marletta M. A., Physiological and molecular understanding of bacterial polysaccharide monooxygenases. Microbiol. Mol. Biol. Rev. 81, e00015-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kirn T. J., Jude B. A., Taylor R. K., A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438, 863–866 (2005). [DOI] [PubMed] [Google Scholar]
- 83.Larbig M., et al. , Safety and immunogenicity of an intranasal Pseudomonas aeruginosa hybrid outer membrane protein F-I vaccine in human volunteers. Vaccine 19, 2291–2297 (2001). [DOI] [PubMed] [Google Scholar]
- 84.Göcke K., et al. , Mucosal vaccination with a recombinant OprF-I vaccine of Pseudomonas aeruginosa in healthy volunteers: Comparison of a systemic vs. a mucosal booster schedule. FEMS Immunol. Med. Microbiol. 37, 167–171 (2003). [DOI] [PubMed] [Google Scholar]
- 85.Baumann U., Göcke K., Gewecke B., Freihorst J., von Specht B. U., Assessment of pulmonary antibodies with induced sputum and bronchoalveolar lavage induced by nasal vaccination against Pseudomonas aeruginosa: a clinical phase I/II study. Respir. Res. 8, 57 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsai C.-M., et al. , Non-protective immune imprint underlies failure of Staphylococcus aureus IsdB vaccine. Cell Host Microbe 30, 1163–1172.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Redi D., Raffaelli C. S., Rossetti B., De Luca A., Montagnani F., Staphylococcus aureus vaccine preclinical and clinical development: Current state of the art. New Microbiol. 41, 208–213 (2018). [PubMed] [Google Scholar]
- 88.Fowler V. G. Jr., Proctor R. A., Where does a Staphylococcus aureus vaccine stand? Clin. Microbiol. Infect. 20, 66–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang Y., et al. , A review of the immune activity of chitooligosaccharides. Food Sci. Technol. 43, 97822 (2023), 10.1590/fst.97822. [DOI] [Google Scholar]
- 90.Deutsch E. W., et al. , The ProteomeXchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 48, D1145–D1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Perez-Riverol Y., et al. , The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
Mass spectrometry proteomics data are deposited to the ProteomeXchange Consortium (90) via the PRIDE (91) partner repository with dataset identifier PXD039298. The crystal structure of CbpD is provided by publicly available protein structures with PDB identifiers 8C5N. All study data are included in the article and/or supporting information.