Abstract
The pathogen Salmonella enterica encompasses a range of bacterial serovars that cause intestinal inflammation and systemic infections in humans. Mice are a widely used infection model due to their relative simplicity and versatility. Here, we provide standardized protocols for culturing the prolific zoonotic pathogen S. enterica serovar Typhimurium for intragastric inoculation of mice to model colitis or systemic dissemination, as well as techniques for direct extraintestinal infection. Furthermore, we present procedures for quantifying pathogen burden, and for characterizing the immune response by analyzing tissue pathology and inflammatory markers, and by isolating immune cells from intestinal tissues for analysis by flow cytometry.
INTRODUCTION
The genus Salmonella covers an array of Gram-negative facultative anaerobic bacteria that in general colonize a broad range of hosts. The species Salmonella enterica (S. enterica) is divided into six subspecies that in total represent over 2,500 serovars (Grimont, Weill, and Others 2007), with infection of humans and livestock being associated most commonly with serovars of subspecies enterica (S. enterica subsp. enterica). Based on disease presentation in humans, Salmonella serovars are often termed “typhoidal” or “non-typhoidal”. Non-typhoidal Salmonella (NTS) serovars (e.g., S. enterica subsp. enterica serovar Typhimurium; hereafter S. Typhimurium) are zoonotic generalists that cause inflammatory colitis during infection of otherwise healthy humans and represent the majority of serovars. In contrast, typhoidal serovars (e.g., S. Typhi) have evolved into human-restricted pathogens that typically present as a more severe extraintestinal disease known as typhoid or enteric fever (Gal-Mor, Boyle, and Grassl 2014; Hiyoshi et al. 2018).
Responsible for over 100 million human infections per year worldwide, S. enterica is a major concern for public health (Majowicz et al. 2010), and the emergence of multidrug resistance and invasive disease among NTS serovars is an increasingly significant problem (Ao et al. 2015; for Disease Control, Prevention, and Others 2019). Due to its well-characterized virulence systems (Sanchez-Garrido et al. 2021; Bao et al. 2020; LaRock, Chaudhary, and Miller 2015), relative ease of genetic manipulation, and availability of mutant strains and libraries (Geng et al. 2016; Porwollik et al. 2014), S. enterica has been widely studied as a model intestinal, extraintestinal, and intracellular bacterial pathogen, particularly using strains of the NTS serovar S. Typhimurium.
In humans, infections with NTS such as S. Typhimurium typically remain localized to the gut, where the pathogen triggers inflammatory diarrhea, a disease characterized by the recruitment of neutrophils (Hohmann 2001). Systemic NTS infection generally occurs in highly susceptible individuals (e.g., children and the elderly) or during coinfection with malaria or the human immunodeficiency virus (Hohmann 2001). However, unlike the typical NTS infection in humans, in commonly employed laboratory mouse strain backgrounds (e.g., C57BL/6 and BALB/c) S. Typhimurium causes a systemic infection with a limited intestinal inflammatory response, followed by rapid mortality (typically 3–10 days, depending on the infectious dose, the mouse strain, and the mouse microbiota). Thus, historically, S. Typhimurium infections of mice have been used to model aspects of the extraintestinal Salmonella disease caused by typhoidal serovars in humans, whereas alternative mammalian models (e.g., cow, monkey, pig, rabbit) have been used to investigate mechanisms of NTS-associated colitis in vivo (Rout et al. 1974; Everest et al. 1999; Santos et al. 2001; Tsolis et al. 2011).
More recently, researchers have developed models to investigate NTS colitis in mice. Some models use genetically resistant mouse lines that are more capable of controlling the extraintestinal infection of S. Typhimurium (Rivera-Chávez et al. 2016; Jacobson et al. 2018). Other models require the administration of antibiotics (e.g., streptomycin) to perturb the commensal microbiota and prime the exudative inflammatory responses that follow S. Typhimurium gut colonization (Palmer and Slauch 2017; Kaiser et al. 2012; Tsolis et al. 2011; Stecher et al. 2007). These advances have enabled researchers to employ “genetics-squared” approaches, wherein the host and the pathogen can each be genetically altered in order to probe the combined interactions of host immunity and pathogen virulence and metabolism on the outcome of infection (Persson and Vance 2007; Byndloss, Olsan, et al. 2017).
This article provides detailed protocols as examples of commonly used mouse infection models for studying Salmonella disease, primarily focused on using S. Typhimurium as a model pathogen. The protocols can be adapted to other S. enterica serovars. Basic Protocols 1 and 2 describe the steps for performing the streptomycin-pretreatment Salmonella colitis model in mice, including the preparation and administration of streptomycin (Basic Protocol 1), and the intragastric infection of mice with S. Typhimurium (Basic Protocol 2). Basic Protocol 3 provides procedures for a direct extraintestinal infection model of mice with S. Typhimurium by intraperitoneal injection. Support Protocol 1 describes the steps for preparing an inoculum of S. Typhimurium for use in these models, while Support Protocol 2 outlines the preparation of a mixed S. Typhimurium inoculum to study competitive fitness of the pathogen. Procedures for quantifying intestinal and tissue pathogen burden are explained in Basic Protocol 4. Support Protocol 3 outlines the assessment of infected tissue histopathology. Support Protocol 4 describes extraction of RNA from intestinal tissues and provides validated qPCR primers for markers and mediators of colitis. Support Protocol 5 describes the collection and preparation of intestinal content to measure markers of inflammation by ELISA, including myeloperoxidase and lipocalin-2. Finally, Support Protocol 6 provides methods for isolation of immune cells (including lymphocytes, infiltrating granulocytes, monocytes, and innate lymphoid cells) from infected intestinal tissues for immunophenotyping or ex vivo activity assays.
Strategic Planning:
Caution: Salmonella enterica strains are commonly regarded as Biosafety Level 2 (BSL-2) pathogens. All appropriate guidelines and regulations for the use and handling of pathogenic microorganisms should thus be followed in accordance with one’s local institution and government. Following from this, infected mice should be housed in a BSL-2 facility. Additionally, appropriate personal protective equipment (PPE) should be used when handling Salmonella cultures or infected animals/tissues.
Note 1: All protocols utilizing animal subjects require review and approval by an Institutional Animal Care and Use Committee (IACUC) or equivalent entity.
Note 2: General cleanliness, surface decontamination, materials sterilization, and aseptic technique are all critical elements to the preparation, performance, and conclusion of every experimental design and workflow. These help to ensure the safety of laboratory personnel and the surrounding community (by keeping infectious material contained), the integrity and reproducibility of results (by avoiding contamination), and that respect is shown for the privilege of utilizing animal subjects (by minimizing experimental repeats).
BASIC PROTOCOL 1: Murine colitis model utilizing oral streptomycin administration
Introduction:
Pre-treatment of mice with streptomycin inhibits microbiota-associated intestinal colonization resistance against S. Typhimurium, prompting a neutrophilic inflammatory response to the pathogen in the cecum and colon (the murine colitis model) (Barthel et al. 2003; Bohnhoff and Miller 1962). In this model, antibiotic administration is performed 24 hours prior to S. Typhimurium inoculation to allow for both reduced colonization resistance and clearance of most of the antibiotic from the gastrointestinal tract prior to the infection. Uninfected mice treated with streptomycin restore the microbiota by day 4 post-infection (Barthel et al. 2003; Stecher et al. 2007).
Materials
Streptomycin sulfate (EMD Millipore Corp)
Sterile water
Sterile 1.5 mL tubes
70% Ethanol
Sterile 1 mL syringes
0.22 μm syringe filter (Acrodisc; Pall Corporation)
Kimwipes
Reusable gavage needle/feeding tube (22-gauge, 1.5 inches), stored in 70% ethanol for sterility
Personal protective equipment (laboratory coat, gloves, and eye protection)
Procedure
- 24 hours prior to S. Typhimurium inoculation (i.e., Day −1), prepare a fresh solution of streptomycin sulfate in sterile water at 200 mg/mL, with enough volume to administer 0.1 mL (20mg) per mouse.
- This amount is used for 8-week-old mice of approximately 18–25 g body weight.
Filter sterilize the streptomycin solution into a sterile tube (e.g., autoclaved 1.5 mL microcentrifuge tubes), and keep the solution on ice until ready to administer.
- Immediately before administration to the mice, flush the gavage needle with 70% ethanol, then flush the needle with sterile water to remove any residual ethanol.
- Residual ethanol in the needle can yield difficult or inaccurate gavages.
Change the syringe on the gavage needle and draw up an adequate volume of the sterile streptomycin solution into the syringe.
Scruff the mouse firmly and insert the gavage needle down the esophagus into the stomach (i.e., intragastrically), withdrawing and re-inserting the needle if any resistance is encountered (an indicator you may be erroneously inserting down the trachea). Dispense 0.1 mL of the streptomycin solution and remove the needle.
Return the mouse to its cage and monitor for signs of respiratory distress, indicating accidental pulmonary administration, which may require immediate euthanasia.
Wipe down the gavage needle with 70% ethanol and a kimwipe between animals and repeat steps 3–6.
The next day, follow Basic Protocol 2 to inoculate the mice with S. Typhimurium, or vehicle for uninfected controls.
BASIC PROTOCOL 2: Intragastric infection with S. Typhimurium
This protocol provides directions for the intragastric inoculation of mice with S. Typhimurium. When performed in conjunction with streptomycin pre-treatment (Basic Protocol 1), S. Typhimurium oral infection should induce a severe inflammation of the cecum and the colon in mice beginning at 24 hours post-infection (Barthel et al. 2003). In susceptible mice (e.g., C57BL/6 mice; (Simon et al. 2011)), significant S. Typhimurium dissemination from the gut to systemic sites can be observed during infection over time, resulting in mortality after 4–5 days (Barthel et al. 2003), depending on the S. Typhimurium strain, on the vivarium, and on the gut microbiota. In the absence of streptomycin pre-treatment, susceptible mice will usually develop few intestinal signs of gastroenteritis and are considered a model for typhoid disease, given the substantial dissemination of Salmonella from the gut to systemic tissues, including the blood, liver, and spleen. However, in some mouse lines that display greater immunological control of systemic S. Typhimurium infection (“resistant” lines; e.g., CBA/J; (Simon et al. 2011)), S. Typhimurium infection without antibiotic pretreatment can generate significant intestinal inflammation by around 7–10 days of infection (Faber et al. 2017; Rivera-Chávez et al. 2016). For a more detailed classification of mouse lines displaying resistance and susceptibility to S. Typhimurium, please refer to Simon et al. (Simon et al. 2011).
Materials
Mice, typically 8–12 weeks old (sex-matched) and acclimated to their housing for ≥1 week
S. Typhimurium inoculum suspended in PBS or equivalent vehicle (See Support Protocol 1 for preparation)
70% ethanol
Sterile distilled water or PBS
Reusable gavage needle/feeding tube (22-gauge, 1.5 inches), stored in 70% ethanol for sterility
Sterile 1 mL syringe
Scale for weighing mice (accurate in the 10–50 g range)
Personal protective equipment (laboratory coat, gloves, and eye protection)
Procedure
- Prior to the infection, and at least daily after S. Typhimurium inoculation, mice should be weighed and the weights recorded. Mice can be identified by placing marker lines on tails or by ear tagging.
- S. Typhimurium infection can result in rapid morbidity and mortality, so regular assessment of behavior and weight loss is important to limit animal suffering. IACUC protocols commonly require euthanization after a maximum 20% reduction in body weight from the start of the experiment.
- Optional: The morning of the infection, remove the mouse chow to fast the mice 3–4 hours before intragastric inoculation.
- Pre-inoculation fasting can help limit discomfort associated with gavaging into a full stomach and may normalize differences in colonization resulting from acute variation in feeding behavior between the mice.
- Prepare the S. Typhimurium inoculum as specified in Support Protocols 1 or 2.
- Oral S. Typhimurium infection models usually use a dose of 5×107 to 1×109 total CFU per mouse, in a volume of 0.1 or 0.2 mL. A high inoculum helps to ensure more consistent infection dynamics between animals and experiments. Dose consistency (same CFU, same vehicle, same volume) between experiments is important.
Flush the gavage needle with 70% ethanol, then flush it again with sterile water or PBS to remove residual ethanol.
Agitate the prepared inoculum to mix evenly, draw up the inoculum into the syringe, then attach the gavage needle.
With the gavage needle pointing upwards, tap the syringe and depress the plunger to remove any air bubbles, then wipe down the outside of the gavage needle with 70% ethanol and a kimwipe to remove any external inoculum droplets.
Firmly scruff the mouse and move it to an upright position. Gently insert the gavage needle into its mouth, down its esophagus and into the stomach. If resistance is felt, do not continue to push the needle in. Instead, remove the gavage needle from the mouse and try inserting it again.
Once the needle is in place intragastrically, inject 0.1 mL of the inoculum into the stomach and gently withdraw the gavage needle. Repeat the procedure with additional mice.
Monitor mice for normal breathing patterns and behavior. If irregular breathing or respiratory distress is evident following inoculation, the inoculum may have been aspirated or the needle may have perforated the trachea, in which case the animal will need to be euthanized immediately.
- Clean and disinfect work surfaces and the gavage needle with 70% ethanol. If animals were fasted prior to infection, return food to the cage. Continue to monitor and weigh the animals at least 1–2 times daily to assess the course of the infection.
- Intragastric S. Typhimurium infection experiments in susceptible mouse lines pre-treated with streptomycin typically display peak pathology and need to be ended by 3–4 days post-infection. Resistant lines may take longer to see significant signs of morbidity, with some animals even apparently clearing the infection.
BASIC PROTOCOL 3: Intraperitoneal injection of S. Typhimurium for modeling extraintestinal infection
For some research questions it is desirable to determine whether an observed phenotype depends on the intestinal or extraintestinal stage of infection (e.g., whether higher levels of Salmonella systemically are due to increased extraintestinal growth/survival or are downstream of gut-associated factors). A simple model to investigate this question is performing an infection by intraperitoneal (i.p.) injection, allowing Salmonella to colonize systemic tissues while bypassing the selective pressures present in the gut. If the phenotype is absent during i.p. infection, it can be concluded that the mechanism driving the phenotype involves the intestinal stage of infection.
Materials
Mice, typically 6–8 weeks old (sex-matched) and acclimated to their space for ≥1 week
S. Typhimurium inoculum in sterile PBS or saline (refer to Support Protocol 1), generally prepared at a low concentration relative to oral infection (1×103 to 1×105 CFU/mL)
26-gauge PrecisionGlide needle (Becton Dickinson)
1 mL syringes (Becton Dickinson)
Scale for weighing mice (mice are typically 15–35 grams, depending on the strain and age)
Personal protective equipment (laboratory coat, gloves, and eye protection)
Procedure
- Weigh mice prior to inoculation of S. Typhimurium and at least daily during the infection.
- Generally, a starting weight of 18–20g is optimal for the i.p. infection model and for obtaining consistent, reproducible results. This is the typical weight of mice that are 6–8 weeks old.
- S. Typhimurium infection can result in rapid morbidity and mortality, so regular assessment of behavior and weight loss is important to limit animal suffering. Most IACUC protocols recommend euthanization at 20% reduction in body weight from the start of the experiment. Endpoints often occur 2–3 days following intraperitoneal S. Typhimurium inoculation.
- Prepare the S. Typhimurium inoculum as in Support Protocols 1 or 2.
- Typically, intraperitoneal S. Typhimurium infection models use a dose of 1×103 - 1×104 CFU per mouse, in a volume of 0.1 or 0.2 mL PBS or saline. This is sufficient to consistently infect systemic mouse tissues, while not causing the mice to rapidly succumb to septic shock. Dose consistency (same CFU, same vehicle, same volume) between experiments is important.
Agitate the inoculum to mix evenly, then draw up the inoculum into the syringe (at least 100 μL per mouse to be infected) and attach the needle. With the needle pointing upwards, tap the syringe and depress the plunger to remove any air bubbles, then wipe down the outside of the needle with 70% ethanol and a kimwipe to remove any external inoculum droplets.
Firmly scruff the mouse and maneuver it underside-up, angling it away from you with the head pointed downward at an approximately 30° angle.
Insert the needle approximately 0.5 cm deep at an angle almost parallel to the body into the peritoneal cavity, aiming for either side between the nipples.
Inject the inoculum (0.1–0.2 mL) into the peritoneal cavity, then gently withdraw the needle. Confirm that the inoculum is not expelled out of the hole left by the needle, which would indicate it was not properly inserted into the peritoneal cavity. Repeat the injection in the additional mice.
Clean and disinfect the area with 70% ethanol, and dispose of the syringe and needle in a biohazardous sharps disposal container. Continue to monitor and weigh the animals at least daily to assess the course of the infection.
SUPPORT PROTOCOL 1: Preparation of Salmonella Typhimurium Inoculum
This protocol provides directions for the preparation of an inoculum of S. Typhimurium from a frozen glycerol stock, for infection in mice. This protocol requires 3 days.
Materials
Frozen stock of wild-type S. Typhimurium in 30% glycerol solution (e.g., strain ATCC 14028s or SL1344; or antibiotic-resistant derivatives, e.g., IR715), stored in a −80°C freezer
Sterile disposable plastic inoculation loops or a reusable metal inoculation loop
LB Agar or MacConkey Lactose Agar plates with appropriate selective antibiotic (50 μg/mL nalidixic acid for IR715; 100 μg/mL streptomycin for SL1344)
LB Broth (autoclaved to sterilize)
Sterile Phosphate-buffered saline (PBS)
15 mL sterile test tubes with loose-fitting caps
37°C incubator with apparatus for shaking or rolling tubes
Spectrophotometer for absorbance readings at OD600
Centrifuge capable of at least 3,200 × g
Personal protective equipment (laboratory coat, gloves, and eye protection)
Procedure
Put on appropriate personal protective equipment (e.g., lab coat, disposable gloves, safety glasses).
- To minimize contamination:
- Before beginning work and with flames off, clean work surfaces with 70% ethanol, fresh 10% bleach, or another approved/compatible disinfectant.
- Perform the following procedures near an active flame (within 2 feet) to maintain a ‘sterile field’ that will help to limit the chance of contaminating cultures with environmental microbes.
Obtain a frozen glycerol stock of S. Typhimurium from the −80°C freezer and place it in an insulated transport vessel containing dry ice, or one chilled to and capable of maintaining approximately −80°C.
- Gently scrape the top layer of the frozen glycerol stock with a sterile inoculation loop and run it across the top of a room-temperature LB agar plate containing selective antibiotics (see REAGENTS AND SOLUTIONS), using the quadrant or T-streak technique to generate isolated colonies.
- For more detailed instructions on streaking technique, refer to a recent Protocol by Bouladoux et al. (Bouladoux, Harrison, and Belkaid 2017).
- Warning: Work expeditiously, maintaining the stock tube on dry ice when not being handled. Freeze-thaw cycles of frozen bacterial stock culture kill a portion of the bacteria, so it is important to handle frozen stock vials briefly in order to limit thawing.
- Caution: Frozen stock vials are extremely cold. Consider double-gloving if you are unable to comfortably handle the frozen vial.
- Note: If the S. Typhimurium strain to be used lacks a common antibiotic resistance, it can be semi-selectively grown on MacConkey Lactose agar, where it forms white (non-lactose-fermenting) colonies.
- Incubate the agar plate upside-down (i.e., lid down, agar side up) in a 37°C incubator overnight (14–18 hours).
- Incubating agar plates upside-down prevents condensation build-up on the lid.
- When stored at 4°C, these plate cultures can be used for approximately 1 week for preparing overnight cultures for inocula, although they are best used fresh the next day.
The next day, aliquot approximately 5 mL of fresh LB broth in a sterile test tube.
- Use a sterile inoculation loop to collect a few colonies or confluent growth from the LB agar plate and transfer the loop to the LB broth. Gently rub the loop against the side of the tube in the broth to inoculate the culture, then remove the loop and loosely cap the tube.
- Culture derived from a few colonies or from confluent bacterial growth minimizes the chance of the entire inoculum being comprised of a stochastic mutant that exhibits attenuation. Multiple cultures derived from separate, isolated colonies is also acceptable, keeping in mind the caveat that a stochastic mutation could result in an altered phenotype.
- Incubate the culture at 37°C with aeration via shaking (~200 rpm) or rotating for 12–16 hours. As a control, also incubate a 5 mL culture of the LB broth alone, to check for contamination of the broth stock and supplies used.
- If antibiotic resistance is stably maintained by the strain and if good aseptic technique is used, there is no need to prepare the liquid culture in LB broth supplemented with selective antibiotics. Even if a strain is resistant to an antibiotic, growth with antibiotics can induce stress responses in the bacteria that could confound interpretation of results from experiments. That said, growth with antibiotics should be used if a strain harbors a selectable plasmid or unstable genetic element, to ensure that the element is stably maintained in the population.
- Note: Different liquid growth media (e.g., low salt LB) and/or static incubation (i.e., not aerated) are used in some protocols, depending on the growth conditions required for the experiment (e.g., favoring the expression of capsules or adhesins).
- Optional: Dilute the overnight culture 1:50 volume:volume in fresh LB broth (between 5–50 mL total volume, depending on the amount of inoculum required) and incubate this subculture for an additional 3–4 hours at 37°C with shaking.
- Subculturing helps restart logarithmic bacterial growth, which may be necessary to produce some infection results consistently such as invasion phenotypes early in the infection model (Lee and Falkow 1990; Ibarra et al. 2010).
- Use a spectrophotometer to measure the OD600 of the culture, and calculate the required volume necessary for the desired inoculum concentration (see Critical Parameters for more information on typical S. Typhimurium quantities used for inocula).
- In general, for wild-type S. Typhimurium, an OD600 of 1.0 corresponds with an estimated bacterial concentration of approximately 5×108 to 1×109 CFU/mL, although it is useful to empirically determine this for a given wild-type or mutant strain by performing a growth assay and correlating OD600 values with plated colony-forming units (CFU).
- Note: Spectrophotometer manufacturers specify a range (e.g., 0.1 to 1.5 absorbance units; AU) in which the instrument is most accurate. Overnight S. Typhimurium cultures are often in the range of 3–5 AU. As such, dilute cultures as necessary (e.g., 1:2 or 1:10) in fresh growth medium to reach the instrument’s prescribed measurement range, then factor the dilutions into your calculations.
Harvest the bacteria by centrifuging the culture for at least 5 minutes (3,200 x g, 4°C) to pellet the cells.
- Discard the supernatant and resuspend (e.g., vortex) the bacteria in 5–10 mL sterile PBS to wash the pellet at least once, and pellet the bacteria again by centrifugation.
- For intragastric infections, the inoculum may alternately be prepared in LB broth or sodium bicarbonate solutions, which may help buffer the acidic environments of the stomach and upper intestine and promote Salmonella implantation in the gut. Any of these vehicles are acceptable, but consistency across experiments is crucial.
Discard the supernatant and resuspend the bacterial pellet in the necessary volume of PBS calculated to produce an inoculum at the desired concentration for infection.
- Use the inoculum immediately (i.e., within 10 minutes) for infections, as described in Basic Protocol 2.
- Note: If the inoculum is not used immediately, it should be stored at 4°C or on ice, and should be used as soon as possible (within two hours).
Plate serial dilutions of the inoculum (typically 10−6, 10−7, 10−8, and 10−9 dilutions for an inoculum of 1×1010 CFU/mL), then count CFUs to confirm that the actual dose was close to the concentration estimated by OD600 approximation.
SUPPORT PROTOCOL 2: Preparation of Mixed S. Typhimurium Inoculum for Competitive Infection
This protocol is a variation of Support Protocol 1 providing directions for creating an inoculum made of two different S. Typhimurium strains (typically a wild-type and an isogenic mutant strain) that can be used for competitive fitness infection assays in vivo or in vitro. In a competitive infection experiment, the input ratio of the inoculum (Strain A:Strain B; i.e., Strain A’s CFU divided by Strain B’s CFU) is compared to the output ratio assessed at the end of the infection to determine the “Competitive Index” (CI), a measure of the relative fitness advantage of one strain over another in a given environment (Beuzón and Holden 2001). If there is no difference in fitness between strains, the average CI in a treatment group will be 1; if Strain A is more fit than Strain B, the average CI will be greater than 1, and if Strain B is more fit than Strain A, the average CI will be less than 1. This technique requires the two strains of interest to be discernible from each other when plated, typically by using different encoded antibiotic resistances (e.g., one strain is kanamycin-resistant and chloramphenicol-sensitive, whereas the other strain is chloramphenicol-resistant and kanamycin-sensitive; or, both strains are nalidixic acid resistant, but one strain is kanamycin-sensitive and the other is kanamycin-resistant).
Additional Materials to Support Protocol 2:
- Glycerol stocks of the two Salmonella strains, each with a different selectable resistance (Example 1), or two Salmonella strains that share one antibiotic resistance but differ in resistance to another antibiotic (Example 2; a common setup for comparisons of wild-type vs. mutant strains).
- Example 1: S. Typhimurium IR715 [Nalidixic acid-resistant, Streptomycin-sensitive; i.e., NalR StrepS] and S. Typhimurium SL1344 [NalS StrepR]
- Example 2: S. Typhimurium IR715 ΔiroN::KSAC [Nalidixic acid-resistant, Kanamycin-resistant; i.e., NalR KanR] and S. Typhimurium IR715 [NalR KanS]
- Important note: Be wary when using plasmids to “mark” strains with an antibiotic resistance for the purpose of quantification ex vivo from a host. Many commonly employed plasmids are not stably maintained within a population of cells without constant selective pressure for a bacterium to possess the plasmid (e.g., growth in an antibiotic-containing medium). As a result, the plasmid can begin to titrate out of the bacterial population; i.e., following cell division, daughter cells that do not receive a copy of an antibiotic resistance-mediating plasmid will still be able to replicate in the environment. Thus, plating on antibiotic-containing agar media selecting for the “marker” plasmid’s presence would undercount the strain of interest, as some percentage of the population will have lost the plasmid. As such, an observed infection phenotype could simply be measuring loss of a marker plasmid rather than the impact of a mutation of interest, and be further magnified the longer the experiment runs.
Procedure
- Follow Support Protocol 1 steps 1–13 for both strains, preparing each of the strains separately as their own inoculum.
- Both strain cultures should be prepared in liquid growth medium without antibiotics, as exposure to different antibiotics immediately prior to infection can influence the downstream competitive index.
Separately resuspend each strain to the same concentration in the desired inoculation vehicle (e.g., PBS), then mix the inocula 1:1 (vol:vol).
Follow Support Protocol 1, step 14.
- To more accurately quantify the concentration of each strain in the inoculum (the “input” ratio of Strain A:Strain B), serially dilute and plate a sample of the mixed inoculum on separate LB agar plates supplemented with appropriate selective antibiotics.
- Example 1: If Strain A is nalidixic acid-resistant (NalR) and Strain B is streptomycin-resistant (StrepR), then the mixed inoculum should be plated on both LB agar + nalidixic acid(LB+Nal) plates and on LB agar + streptomycin (LB+Strep) plates. The abundance of each strain is directly quantifiable from the colonies counted on the respective antibiotic-containing agar plates.
- Example 2: If both Strain A and Strain B are nalidixic acid-resistant (NalR), but Strain A is kanamycin-resistant (i.e., NalR KanR) and Strain B is kanamycin-sensitive (i.e., NalR KanS), then the mixed inoculum should be plated on both LB agar + nalidixic acid (LB+Nal) and on LB agar + kanamycin (LB+Kan). Strain A is directly quantifiable on LB+Kan agar, whereas the abundance of Strain B is ascertained by subtracting the LB+Kan agar quantification from the LB+Nal agar quantification. Of important note, this approach is valid only in the following scenarios: 1) Strain A (e.g., a mutant of Strain B) is less abundant than Strain B (e.g., the wild-type parent of Strain A); 2) Strain A and Strain B are roughly equal in abundance; 3) Strain A is at most a few-fold more abundant than Strain B (i.e., one loses the ability to easily quantify Strain B amidst Strain A as Strain A becomes relatively more abundant). In scenarios where Strain A is more abundant than Strain B, one may “pick and patch” several hundred colonies from each LB+Nal agar plate and transfer them to LB+Kan agar (test condition) and to LB agar (positive control) in order to quantify Strain B. Genetics permitting, in such a scenario one should consider reconstructing their strains for easier quantification.
- As the relationship between OD600 absorbance units and actual live bacterial cells can vary between strains and growth state, quantifying inoculum concentration by CFU levels of each strain provides a more accurate input ratio to assess the competitive index.
- Measuring infection outcomes: use the methods for quantifying S. Typhimurium burden in intestinal content and tissues described in Basic Protocol 4, while plating each sample’s dilutions on both selective agars to assess the output ratio of Strain A:Strain B in that sample. The CI is then calculated by dividing the output ratio by the input ratio.
- To statistically compare the CI between groups, logarithmically transform the values to properly weight for samples with inverted competitive outcomes (CI < 1).
BASIC PROTOCOL 4: Assessment of S. Typhimurium burden
This protocol describes methods for assessing the course of S. Typhimurium infection of mice, including both the intestinal and systemic arms of the infection.
Part 1: Quantification of fecal S. Typhimurium levels throughout infection
After orogastric infection, S. Typhimurium rapidly colonizes the mouse gut (terminal ileum, cecum, and colon). The increased levels of S. Typhimurium (especially in colitis models) in the distal gut are most readily measured over time by plating fecal samples. Feces are collected, processed and then spread onto selective agar media to assess the number of S. Typhimurium colony-forming units (CFUs). Colonization levels are more variable in mice that are not treated with antibiotics, and more consistent when mice are pretreated with streptomycin. This protocol provides instructions for the collection and preparation of murine fecal samples, followed by quantification of Salmonella abundance by counting CFUs.
Materials
Clean paper towels
500 mL clear glass beaker with a pouring lip (or similar hard container), disinfected with 70% ethanol
Forceps
Sterile microcentrifuge tubes (1.5 or 2.0 mL)
Milligram scale for weighing fecal samples in tubes
70% ethanol, for cleaning forceps and beaker between animals
Sterile PBS
Vortexer with a multi-tube holder attachment (such as the Horizontal-(24) Microtube Holder, Scientific Industries)
Micropipettes, 200 μL and 1000 μL
Sterile 200 μL and 1000 μL micropipette tips
Sterile wide-bore 200 μL micropipette tips
MacConkey or LB agar plates supplemented with appropriate antibiotics to select for S. Typhimurium growth
37°C Incubator
Personal protective equipment (laboratory coat, gloves, and eye protection)
Procedure
Prior to collecting intestinal content from the mice, prepare a labeled microcentrifuge tube for each infected mouse, then record the weight of each empty tube on a milligram scale.
- For tracking S. Typhimurium intestinal burden in live mice, fecal pellets can be collected daily. Place one mouse on a clean paper towel and cover it with an inverted, 70% ethanol-disinfected glass beaker. Mice typically produce fecal pellets within 10 minutes.
- To increase collection efficiency, additional mice may be placed under separate beakers.
- To limit the potential for cross-contamination of Salmonella between animals or samples, use a fresh, clean paper towel for each mouse and wipe down beakers and forceps with 70% ethanol between mice, then let dry by evaporation before use.
- Collection will usually take less time in the early light cycle. Late in the infection, mice may start to eat less, and the time to produce a fecal pellet can go up considerably (30 minutes or more). Additionally, very morbid mice may not defecate.
- A glass beaker with a pouring lip is an ideal container: glass, because the weight will lessen the chance of the beaker moving or tipping over; a pouring lip, because it allows air exchange, and can be positioned over the mouse tail when setting down the beaker, allowing one to cover the mouse while holding the tail and not pinching it.
- Depending on the size of the mouse, an additional weight on top of the beaker may be necessary to prevent the beaker from moving or being knocked over.
Use 70% ethanol-disinfected forceps to collect 1–3 pellets from each mouse (approximately 25–100 mg fecal content) into the empty tubes, close firmly, then immediately place the tubes on ice.
Weigh the closed, feces-containing tubes on a milligram scale, then record the weight. Subtract each tube’s empty weight to determine the weight of the feces.
- Add sterile PBS to the fecal pellet samples at a 9:1 ratio (e.g., 900μL PBS to 100mg feces).
- For faster sample dilution, all samples can instead be suspended in the same volume of PBS (e.g., 1 mL) and then be weight-corrected mathematically, although this method has the drawback of producing a variable limit of detection for each animal when correcting for fecal weight.
- Homogenize the pellets into a fecal slurry by vortexing at top speed for 5–10 minutes.
- If any fecal pellets remain intact after homogenization, a sterile micropipette tip can be used to manually break the pellet up by mashing it against the tube, then vortex-homogenize for an additional 5 minutes.
- Perform 10-fold serial dilutions of the fecal slurry in PBS (e.g., 100μL slurry in 900μL PBS), vortexing each dilution before proceeding to the next.
- The first two 10-fold serial dilutions require the use of wide-bore micropipette tips to prevent clogging. The remaining dilutions can be done with standard micropipette tips.
- The number of 10-fold dilutions to perform is based on the predicted CFU at a given timepoint. Typically, early infection feces (e.g., 24 hrs) will have a countable number of wild-type S. Typhimurium CFUs at the 10−1, 10−2, or 10−3 dilutions, while later timepoints (e.g., 96 hrs) and throughout the streptomycin-colitis model will likely require plating dilutions out to 10−6 or further.
- Plate the serial dilutions using wide-bore tips on LB agar with selective antibiotics. Incubate the plates inverted, overnight (14–16 hours), at 37°C.
- Dilutions can either be spread-plated as 100–200 μL samples on individual agar plates, or to save agar plates, up to 6 dilutions can be plated as multiple, discrete 25–50 μL drops on a single plate. Note that drop-plating may require shorter incubation times (e.g., 12–14 hours) to ensure that discrete colonies are obtained.
- Store the undiluted fecal slurry samples at 4°C to limit growth, in case additional dilutions are required or replating is needed.
- After incubation, record the number of colonies (“colony forming units”, CFUs) that form from each dilution exhibiting separated, distinct, countable colonies.
- For accurate counts, colonies from spread-plated samples should be counted from plates that have between 30 and 300 discrete colonies. For drop or line plating, it will be difficult to discern more than 200 colonies per dilution due to the reduced surface area
- The fecal load (CFU/mg) of S. Typhimurium in the sample can be calculated as: (CFUs on the least-diluted countable sample) divided by (mg/mL fecal slurry * dilution * volume plated). For example: 108 CFU obtained from plating 0.1 mL of a 10−3 dilution of 54 mg/mL fecal slurry = 108 CFU / (54mg/mL * 0.1 mL * 10−3 dilution) = 2×104 CFU/mg).
Part 2: Quantification of S. Typhimurium loads at the termination of infection
In addition to intestinal content, intestinal tissue from different sections (terminal ileum, colon, cecum) may be collected and homogenized to quantify S. Typhimurium levels in the gut. S. Typhimurium also colonizes Peyer’s patches, intestinal lymphoid tissues that are found mostly throughout the small intestine (see Figure 1). Because wild-type mice are incapable of stopping S. Typhimurium from spreading beyond the intestine, measuring the level of S. Typhimurium burden in extraintestinal sites can help assess the relative severity of the infection in a mouse. Commonly collected tissues that display substantial S. Typhimurium extraintestinal colonization following infection by oral gavage include the mesenteric lymph nodes, spleen, liver, and blood. For intraperitoneal infection, blood, spleen and liver are typically collected.
Figure 1. Representative gross pathology in the streptomycin-colitis model of S. Typhimurium infection.
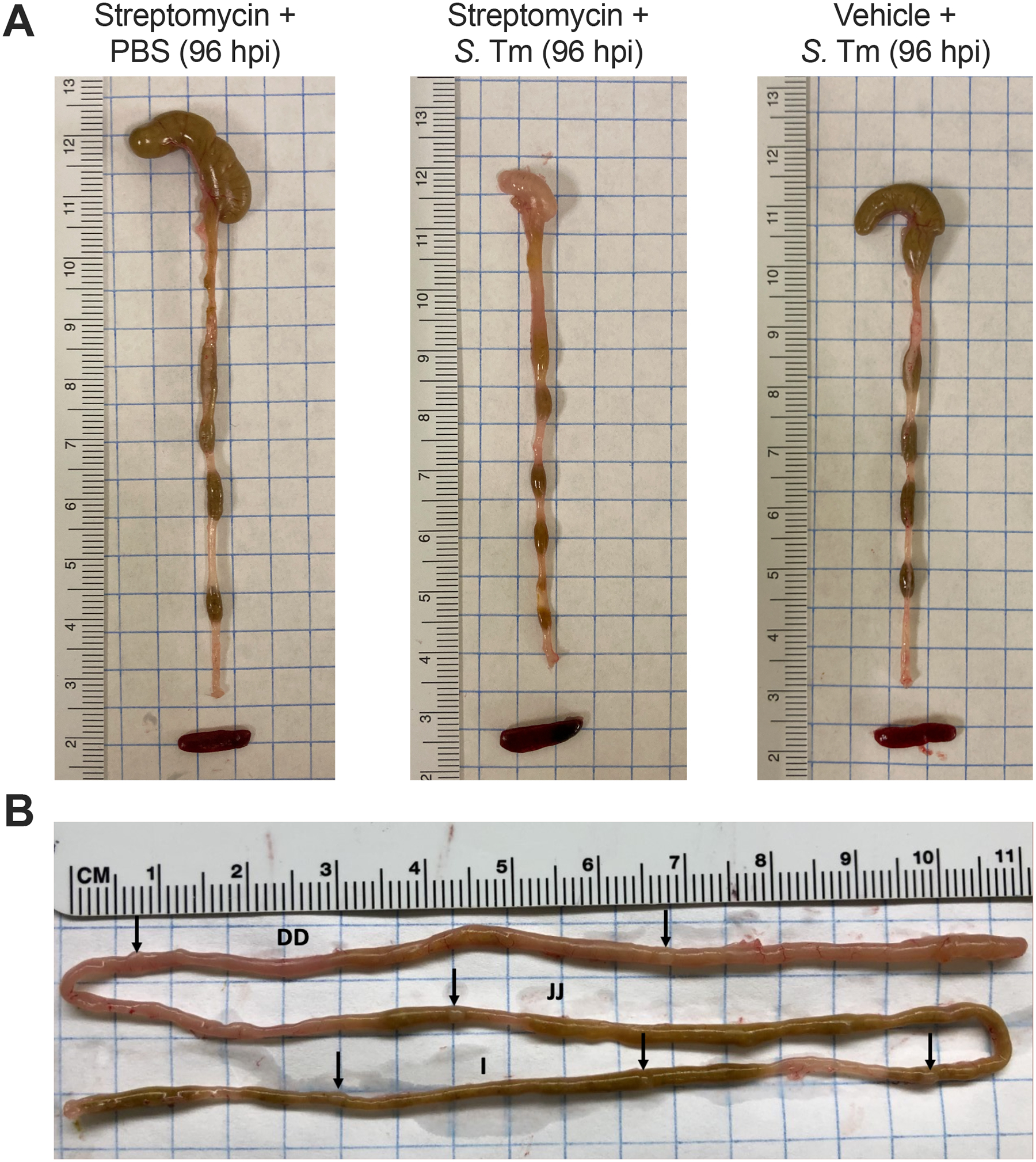
(A) Gross pathology of the cecum, colon, and spleen collected from C57BL6/J mice pretreated with 20 mg of oral streptomycin and mock-infected with PBS, or inoculated intragastrically with 1×109 CFU of S. Typhimurium IR715 (S. Tm) for 4 days following pretreatment gavage with streptomycin or vehicle (sterile water). The small, thick cecum devoid of content is a hallmark of the inflammatory response triggered by S. Typhimurium in mice pretreated with oral streptomycin. (B) Representative photograph of small intestine on day 4 of Salmonella infection. Black arrows indicate the Peyer’s patches, which are easily identified by their protruding appearance on the anti-mesenteric site. I, ileum; JJ, jejunum; DD, duodenum.
Materials
Sterile dissection tools for mouse necropsy (forceps, scissors)
Sterile 15 or 50 mL conical tubes (Corning)
Sterile microcentrifuge tubes (1.5 or 2.0 mL)
26-gauge PrecisionGlide needles (Becton Dickinson)
1 mL syringe (Becton Dickinson)
0.5 M EDTA (sterile, for anticoagulant)
Sterile PBS
70% ethanol, for disinfecting equipment between samples
Tissue homogenizer/disperser (such as the IKA T25 digital ULTRA-TURRAX), fixed on a lab stand support, inside a biosafety cabinet
MacConkey or LB agar plates supplemented with appropriate antibiotics to select for S. Typhimurium growth
Micropipettes (200 μL and 1000 μL)
Sterile 200 μL micropipette tips
Milligram scale for weighing samples in tubes
37°C Incubator
Personal protective equipment (laboratory coat, gloves, and eye protection)
Procedure
- Preparations
- Intestinal content can be collected from the small intestine, cecum, and colon at necropsy. Label and weigh microcentrifuge tubes as described in Part 1.
- For collecting intestinal and extraintestinal tissues, label and weigh 15 mL or 50 mL sterile polypropylene tubes containing 1–5 mL of sterile PBS.
- For collecting blood, prepare 26-gauge needles and 1mL syringes by drawing up and expelling 1 mL of sterile 0.5 M EDTA, leaving behind a residual amount as an anticoagulant. Prepare 1.5 mL sterile microcentrifuge tubes by adding 50 μL of 0.5 M EDTA to the bottom and weighing the tubes prior to blood collection.
Humanely euthanize the mouse by CO2 asphyxiation, followed by cervical dislocation, or perform a similar method of euthanasia as approved in your IACUC protocol.
Disinfect the fur and outer skin by wetting with 70% ethanol.
In general: Use disinfected scissors and forceps to open the mouse and collect relevant tissues intended for measurement in pre-weighed/labeled sterile tubes. Store the tissue-containing tubes on ice until they can be processed. Disinfect tools with 70% ethanol between collecting different tissues to minimize the potential of cross-contamination between samples.
Blood: Locate the heart in the thoracic cavity and perform a cardiac puncture in either ventricle with the EDTA-treated needle. Pull the plunger back slowly to draw up blood, and gently relocate the needle without withdrawing it from the heart to relieve pressure. Once an adequate volume of blood (200–600 μL) has been collected, remove the needle and expel the blood from the syringe into the pre-weighed sterile 1.5 mL microcentrifuge tube containing 50μL EDTA.
Spleen: Excise the spleen by blunt dissection from the left side of the body cavity. Part or all of the spleen can be collected into a prepared tube.
Mesenteric Lymph Nodes: Locate the mesenteric lymph nodes, which look like a white translucent tube in the fatty mass adjacent to the intestines (they will often appear larger/more prominent in S. Typhimurium-infected animals). Dissect out the nodes and collect them into a prepared tube.
- Intestinal content/tissues: Gently move the intestines to locate the distal end of the colon. Cut the colon as distally as possible, and use forceps to pull and straighten-out the intestines onto a clean paper towel, being careful not to tear the organs. The colon, cecum, and small intestine may be separated by cutting where they join at the cecum. Make sure to orient the tissue before cutting it, as the colon and small intestine can be hard to distinguish once cut. If the mice are pretreated with streptomycin before infection with S. Typhimurium wild-type, observe that the cecum is small, thick, whitish in color, and devoid of content (Figure 1A). Luminal content from each section, if present, may be squeezed out and collected into prepared tubes. If desired, samples of the terminal ileum, cecum, and colon tissue may also be collected into separate tubes with PBS for homogenization.
- Important: The cecum can be sectioned with scissors during necropsy for different downstream analyses (CFUs, RNA, histopathology, etc.). If performing histopathological analysis of the cecum, be sure to only minimally handle the cecum while collecting the cecal tip (distal third-to-half of cecum) for this analysis.
- Peyer’s Patches: The Peyer’s patches can be located at various points along the small intestine, appearing as white, raised, oval spots, and will often appear larger/more prominent in S. Typhimurium infected animals (Figure 1B). They can be identified more easily if the intestinal content is present. Peyer’s patches may be cut off the small intestine most readily using fine-tip curved dissecting scissors. For CFU assessment, the 3–4 most distal Peyer’s patches in the ileum from each animal may be pooled into a single prepared tube.
- Due to their small size, it is often easier to collect a consistent number (e.g., 3) of Peyer’s patches from each subject and then normalize CFU counts as an average per Peyer’s patch, rather than per milligram tissue.
- Liver: Mice have four liver lobes, which may be found immediately below the lungs, attached to the diaphragm. Cut a section of liver tissue (e.g., ⅓ to ½ of a lobe) and collect it into a prepared tube.
- Avoid rupturing the gallbladder (the small, yellow-to-black colored sack in the middle of the median liver lobe), which may contain high loads of Salmonella and can contaminate liver tissue samples if it is damaged.
Infected mouse carcasses should be properly disposed of as biohazardous waste.
To process the EDTA-treated blood for CFU enumeration, weigh the tubes to ascertain the total volume of blood collected (subtract the weight of the tube before collection; blood density is ~ 1g/mL), then perform 10-fold serial dilutions in sterile PBS and plate the dilutions on appropriate selective agar media.
Weigh the tubes with collected tissues and intestinal contents to determine the weight of the collected material (subtract the weight of the tube before collection).
- To process tissue for CFU enumeration, use a homogenizer to thoroughly disrupt the tissues in their tubes with the PBS. This typically takes 3–10 seconds per sample tube.
- Clean and disinfect the homogenizer’s dispersing element between samples by running it through two washes of 70% ethanol (two separate 50 mL conical tubes containing enough volume to sufficiently cover the disperser) and one wash of sterile PBS. Use tweezers (disinfected with 70% ethanol after each usage) to remove any residual tissue that remains stuck in the homogenizer. Process one type of tissue at a time, and change the washing media between tissues, and additionally as needed.
To process intestinal contents (small intestine, colon, and cecum) for CFU enumeration, follow the same method described for quantifying S. Typhimurium CFUs in the feces.
To enumerate CFUs of processed samples, perform 10-fold serial dilutions and then plate them as described in this protocol for quantifying S. Typhimurium CFUs in the feces.
SUPPORT PROTOCOL 3: Preservation and assessment of pathology of S. Typhimurium infected-tissues
Non-typhoidal Salmonella colitis is characterized by the influx of inflammatory monocytes and neutrophils into the lamina propria and lumen of the gut, primarily in the cecum and colon (Faber et al. 2017; Rivera-Chávez et al. 2016; Barthel et al. 2003). In the absence of streptomycin treatment, a slowly-developing mononuclear infiltrate can be observed, primarily in the small intestine (Santos et al. 2001). Here, we describe procedures for preservation of S. Typhimurium-infected intestinal tissues for histopathology, and provide a sample rubric for quantifying inflammation in the intestine based on scoring hematoxylin & eosin (H&E) stained tissues. This scoring is most effectively performed by a trained professional (medical or veterinary pathologist), and the samples should be blinded to limit potential bias in the analysis.
Materials
S. Typhimurium-infected mice (see Basic Protocol 3)
Dissecting instruments (see Basic Protocol 4)
Dissection cutting board
20 mL syringe and 21-gauge needle (BD)
70% ethanol
Sterile PBS
Sterile petri dishes
Biopsy foam pad (Simport)
10% (v/v) neutral-buffered formalin (Fisher Scientific)
Tissue-processing histology cassettes (Sakura Finetek)
Histology pencil (VWR International or lead pencil)
Personal protective equipment (laboratory coat, gloves, and eye protection)
Paraffin-embedding of formalin-fixed tissues, sectioning of paraffin-embedded tissues, and hematoxylin/eosin (H&E) staining of sections is performed according to standard histological procedures.
Prepare required material prior to the necropsy. Assign each sample a unique ID and label the histology cassette using a histology pencil.
At critical time points after infection (3–4 days post-S. Typhimurium inoculation for streptomycin-treated C57BL/6 mice; 4–8 days post-inoculation without streptomycin in susceptible mice, or 7–10 days post-inoculation in resistant mice) or upon reaching a preset criterion to terminate the experiment (e.g., weight loss or morbidity), humanely euthanize mice using an approved method such as CO2 asphyxiation followed by cervical dislocation.
- Thoroughly wet the fur with 70% ethanol and open the abdomen to dissect out the entire large intestine (cecum and colon). Separate the cecum from the colon using scissors and place them in a Petri dish containing 10% neutral-buffered formalin.
- Formalin is a carcinogen and irritates the eyes, skin and respiratory tract. Work under a fume hood and wear disposable gloves and eye protection. Because tissue such as the intestine undergoes autolysis immediately post-mortem, rapid transfer of tissue into fixative is critical for optimal morphological preservation.
While tissue is in the fixative, organs that are needed for other purposes (e.g., CFU enumeration) can be collected and handled accordingly.
If parts of the cecum are needed for RNA or protein analysis, the cecal tip is usually sufficient for histology scoring. If the entire organ is available for histology preservation, cut open the cecum and gently flush the tissue with PBS utilizing a syringe to remove fecal content/pus, and then carefully pat dry on a clean paper towel. Place the cecum luminal side up onto a biopsy foam pad and transfer it into a labeled histology cassette placed in the Petri dish with formalin.
- Cut 1 cm portions of the proximal, middle, and distal part of the colon and gently push out luminal content using forceps. Tissue pieces can be transferred either to the same histology cassette or to separate ones.
- Distinct anatomy of colon and cecum can be distinguished by a trained pathologist.
- Fix samples in 10% neutral-buffered formalin for at least 24–48 h to allow for proper preservation of morphology. After fixation, transfer cassettes to a container with 70% ethanol. Collect used fixatives containing formalin in a waste container for appropriate disposal.
- Formalin fixation for at least 24h is sufficient to kill Salmonella. Histology cassettes may be stored long-term submerged in 70% ethanol, but need to be kept in a flammable storage cabinet.
- Over-fixation in formalin (>48 h) should be avoided, as it can negatively impact tissue sectioning and analysis.
Standard histological processing including paraffin-embedding, sectioning, and staining can be performed in-house or samples can be submitted to a histology lab.
Analyze H&E-stained sections by light microscopy or scan sections with a digital pathology scanner (e.g., Nanozoomer by Hamamatsu) to determine the extent of intestinal inflammation according to the semi-quantitative scoring scheme described in Table 1. Of note, slightly different scoring systems have been reported in the literature (Barthel et al. 2003).
Table 1.
Histopathological parameters for assessment of cecal or colonic inflammation. Each sample is investigated for the described parameters and multiplied by the factor of the respective extent. The total score is a sum score of each parameter multiplied by the extent. Abbreviations: lamina propria, LP; submucosa, SM.
| A. Mononuclear inflammation | Description | Area affected (extent) | Total (max. score) |
|---|---|---|---|
|
0
1 2 3 |
Absent (normal sparse lymphocytic infiltrate) Mild (diffuse increase in LP) Moderate (LP with increased basal immune aggregates displacing crypts) Severe (LP with submucosal infiltrate) |
0 – 4 | (12) |
| B. Neutrophilic inflammation | |||
|
0
1 2 3 |
None Mild (LP only) Moderate (LP and cryptitis/crypt abscesses) Sheet-like or submucosal infiltration |
0 – 4 | (9) |
| C. Submucosal edema/width | |||
|
0
1 2 3 |
No pathological changes Mild (less than 50% of the diameter of the intestinal wall) Moderate (SM accounts for 50–80% of the intestinal wall diameter) Severe (SM accounts for >80% of the diameter of the intestinal wall) |
0 – 4 | (12) |
| D. Epithelial injury | |||
|
0
1 2 3 |
None Mild (crypt dropout, epithelial desquamation) Moderate (focal ulceration) Severe (multifocal or extensive ulceration) |
0 – 4 | (9) |
| Total (sum score) | 0–42 |
|
0 1 2 3 4 |
None < 10 % of section < 10–25 % of section < 25–50 % of section > 50% of section |
SUPPORT PROTOCOL 4: Measurement of inflammatory marker expression in intestinal tissues by qPCR
This protocol provides recommended methods for the collection and preparation of cecal tissue mRNA to measure S. Typhimurium-associated changes in gene expression, and provides a sample set of validated qPCR primers of known inflammatory markers relevant to S. Typhimurium infection. Increased expression of these factors in infected tissues is a hallmark of the host response to S. Typhimurium infection, and can be measured by qPCR of the tissue mRNA.
Materials
Cryovials (2.0 mL, Thermo Fisher)
Liquid nitrogen and an appropriate transport vessel
Mortars and pestles (cleaned, then treated with RNaseZAP™ [Sigma] between uses)
Sterile 50 mL conical tubes (Corning)
Kimwipes
TRI Reagent (Sigma-Aldrich)
Ultrapure RNase-free water
Isopropanol
Chloroform or 1-bromo-3-chloropropane
70% ethanol
75% ethanol
SuperScript VILO cDNA Synthesis Kit (Invitrogen)
SYBR Green Master Mix (Applied BioSystems)
Personal protective equipment (laboratory coat, gloves, cryoprotective gloves, face shield, and eye protection)
Procedures
Extracting Tissue RNA
- At necropsy (described in detail in Basic Protocol 4), carefully remove the cecal contents and collect a portion of cecal tissue (approximately 1 cm2) into an empty cryotube and immediately snap-freeze the tissue in liquid nitrogen.
- For consistency, the same portion of the cecum should be collected from all mice. Frozen tissues may be stored in a −80°C freezer for later mRNA extraction.
Prepare a benchtop by first disinfecting with 70% ethanol, then treating surfaces with RNaseZAP™ to eliminate environmental RNase contamination.
Aliquot 1 ml of TRI Reagent in a 50 ml conical tube per sample in a fume hood.
- Clean a mortar and pestle set for each sample with RNaseZAP™. Thoroughly dry with a Kimwipe. Cool down each mortar and pestle set with liquid nitrogen, until the liquid no longer evaporates.
- Always handle liquid nitrogen with appropriate PPE, including a face shield and cryoprotective gloves.
- Add the tissue sample to the mortar and make a very fine powder by working tissue with the pestle, adding liquid nitrogen as needed to keep everything as cold as possible.
- Be careful not to lose the tissue sample. It can be helpful to cover the mortar with a clean paper towel and a gloved hand while working it with the pestle.
- Condense the powdered tissue by adding liquid nitrogen and tilting the mortar as it evaporates. Gently transfer the powder into a 50 mL conical tube containing the previously aliquoted TRI Reagent, then vortex to mix.
- Conical tubes with the powdered tissue can be kept on ice until all of the samples have been processed to this step.
- Store each homogenate for 5 min at room temperature and transfer to RNAse-free 1.5 mL tubes in the fume hood. Add 200 μL chloroform (or 100 μL 1-bromo-3-chloropropane, BCP) to each tube and vortex to thoroughly mix. Let the tubes stand at room temperature for 5–10 minutes, then centrifuge at 12,000 × g for 10 minutes at 4°C to separate the upper clear aqueous phase containing RNA from the middle interphase (DNA) and red organic phase (protein).
- Interphase and organic phase may be saved and stored at −20°C.
Carefully remove the aqueous phase (~500 μL) to a clean, RNAse-free microcentrifuge tube and add an equal volume (500 μL) of isopropanol to precipitate nucleic acids. Store samples at −20°C for 30 minutes, or overnight.
Centrifuge samples at 12,000 × g for 8 min at 4°C and discard the supernatant. Wash the RNA pellet by adding 1 mL of 75% ethanol (made with ultrapure, DEPC-treated water) and invert to mix.
Centrifuge samples at 12,000 × g for 8 min at 4°C. Decant the ethanol and air-dry the pellet with tubes inverted on a clean surface for 3–5 min or longer.
Resuspend the pellet in ultrapure, RNase-free water (30–100 μL, depending on pellet size), recording how much water was used for each sample. Incubate for 10 min at 50–55°C to dissolve RNA thoroughly.
- Quantify the RNA concentration by NanoDrop or Qubit.
- RNA tubes can be stored at −80°C for later analysis.
- Use a reverse transcription kit (such as the Invitrogen SuperScript VILO cDNA Synthesis Kit) to prepare cDNA from 1 μg of total RNA for each sample.
- DNase treatment should be performed prior to cDNA synthesis. DNase treatment is included with the SuperScript VILO cDNA Synthesis Kit
Quantifying Expression by real-time quantitative PCR (qPCR)
Reactions for qPCR can be set up for each gene of interest and a “housekeeping” gene such as Actb (β-actin) for normalization, using SYBR Green Master Mix (Applied Biosystems) with approximately 35 ng cDNA and 500 nM mRNA-targeted primers per 11 μL reaction. Table 2 lists validated sets of mouse transcript-targeted primers (preferably intron-spanning) for a sample panel of genes expected to display increased expression during S. Typhimurium colitis.
Table 2.
Primer sets for S. Typhimurium colitis-associated transcripts
| Mouse Target Gene | Primer Sequence (5’ to 3’) | Source | |
|---|---|---|---|
| Il1 (IL-1β) | Forward: | 5’-CTCTCCAGCCCAAGCTTCCTTGTGC-3’ | (Perez-Lopez et al. 2019) |
| Reverse: | 5’-GCTCTCATCAGGACAGCCCAGGT-3’ | ||
| Il12a (IL-12a) | Forward: | 5’-GGAAGCACGGCAGCAGAATA-3’ | (Perez-Lopez et al. 2019) |
| Reverse: | 5’-AACTTGAGGGGAGAATGAGGAATGG-3’ | ||
| Il17a (IL-17A) | Forward: | 5’-GCTCCAGAAGGCCCTCAGA-3’ | (Perez-Lopez et al. 2019; Godinez et al. 2009) |
| Reverse: | 5’-AGCTTTCCCTCCGCATTGA-3’ | ||
| Il22 (IL-22) | Forward: | 5’-GGCCAGCCTTGCAGATAACA-3’ | (Perez-Lopez et al. 2019; Godinez et al. 2009) |
| Reverse: | 5’-GCTGATGTGACAGGAGCTGA-3’ | ||
| Ifng (IFN-γ) | Forward: | 5’-TCAAGTGGCATAGATGTGGAAGAA-3’ | (Godinez et al. 2009; Deriu et al. 2013) |
| Reverse: | 5’-TGGCTCTGCAGGATTTTCATG-3’ | ||
| Tnf (TNF-α) | Forward: | 5’-CATCTTCTCAAAATTCGAGTGACAA-3’ | (Deriu et al. 2013) |
| Reverse: | 5’-TGGGAGTAGACAAGGTACAACCC-3’ | ||
| Cxcl1 (CXCL1) | Forward: | 5’-TGCACCCAAACCGAAGTCAT-3’ | (Perez-Lopez et al. 2019; Godinez et al. 2009) |
| Reverse: | 5’-TTGTCAGAAGCCAGCGTTCAC-3’ | ||
| Lcn2 (LCN2) | Forward: | 5’-ACATTTGTTCCAAGCTCCAGGGC-3’ | (Deriu et al. 2013) |
| Reverse: | 5’-CATGGCGAACTGGTTGTAGTCCG-3’ | ||
| Actb (β-actin, Housekeeping) | Forward: | 5’-GGCTGTATTCCCCTCCATCG-3’ | (Perez-Lopez et al. 2019) |
| Reverse: | 5’-CCAGTTGGTAACAATGCCATGT-3’ | ||
SUPPORT PROTOCOL 5: Preparation of intestinal content for inflammatory marker quantification by ELISA
Many inflammatory mediators and host-derived byproducts of the response to Salmonella in the gut are soluble and can be directly detected by ELISA on the supernatant from intestinal content samples or from homogenized tissue samples. This protocol provides directions for acquiring intestinal content samples suitable for quantification of inflammatory markers by ELISA, such as the antimicrobial peptide lipocalin-2 and myeloperoxidase (a product of neutrophil activity).
Materials
Sterile 1.5 mL microcentrifuge tubes
Sterile PBS
Protease inhibitor cocktail, such as cOmplete™ Protease Inhibitor Cocktail Tablets (Roche)
Microcentrifuge
Milligram scale for weighing samples in tubes
ELISA kits of interest
Procedure
Prior to necropsy, label and weigh sterile microfuge tubes filled with 1 mL of PBS or an appropriate reagent diluent (usually 1% BSA in PBS) supplemented with protease inhibitor cocktail.
Collect the cecal or other intestinal content into the labeled tubes. Weigh the tubes after collection to determine the intestinal content weight. Tubes can be stored on ice until processing.
Homogenize the intestinal content by vortexing as directed in Basic Protocol 4.
- Centrifuge the samples at 10,000 × g for 5–10 minutes and transfer the supernatant (containing soluble inflammatory markers) to new labeled and sterile microcentrifuge tubes.
- Samples may be stored at −80°C for later quantification.
Perform the ELISA per the manufacturer’s instructions, diluting the samples as necessary to be measured in the linear zone of the provided standard.
Correct concentration measurements to the weight of content collected for comparisons between samples.
Recommended ELISA Kits and additional reagents
Mouse Myeloperoxidase Duoset ELISA (R&D Systems)
Mouse Lipocalin-2/NGAL Duoset ELISA (R&D Systems)
96-well NUNC MaxiSorp Plates, flat bottom (ThermoFisher)
Plate reader with a tunable monochromator or absorbance filters at 450 nm and 540 or 570 nm
SUPPORT PROTOCOL 6: Immune cell isolation from Salmonella-infected intestinal tissues
S. Typhimurium colitis is associated with the influx and activation of several cell types at the intestinal mucosa, including lymphocytes, macrophages, neutrophils, and innate lymphoid cells (ILCs). This protocol provides directions for the isolation of intraepithelial lymphocytes (IELs) and lamina propria lymphocytes (LPLs) along with associated myeloid cells and ILCs from Salmonella-infected intestinal tissues for use in analysis by downstream procedures, including immunophenotyping by flow cytometry or ex vivo activation assays. All steps in this protocol should be completed on the same day as the tissues are collected.
Materials
Necropsy tools (forceps, scissors)
Sterile PBS
Sterile 50 mL conical tubes (Corning)
Sterile petri dishes
Wide-bore needles (18 gauge or larger)
3 mL or 10 mL syringes (Becton Dickinson)
1 M DTT
0.5 M EDTA
Shaking Medium (recipe in REAGENTS AND SOLUTIONS)
Harvest Medium (recipe in REAGENTS AND SOLUTIONS)
Digest Medium (recipe in REAGENTS AND SOLUTIONS)
40, 70, and 100 μm strainers (Corning)
Sterile serological pipettes (5, 10, 25mL)
Tabletop shaker
37°C incubator
Vortex
Light microscope and hemocytometer, or an automated cell counter
Procedure
Remove the desired intestinal tissue (small intestine, cecum, and/or colon) from the mouse and place the tissues in ice-cold sterile PBS in a sterile petri dish.
- Using a wide-bore needle, flush the luminal contents out of the intestine with ice-cold sterile PBS.
- Once flushed, individual tissues can be kept in PBS on ice until all tissues are collected.
- Carefully remove fatty tissue adherent to the intestine.
- This fatty tissue may contain portions of mesenteric lymph nodes, which are effectively a contaminant that will skew results.
For small intestinal tissue, optionally remove the Peyer’s patches.
- Use scissors to open the intestine longitudinally, then dice it into fragments of approximately 5 mm length, then transfer the tissue into a 50 mL conical tube containing 10 mL Shaking Medium at room temperature.
- All media should contain an antibiotic/antimycotic cocktail when processing tissues from Salmonella-infected mice.
Add 5 μL of 1 M DTT and 40 μL of 0.5 M EDTA to each 50 mL conical tube. Shake tubes vertically on a tabletop shaker (250–300 rpm) for 10 minutes at room temperature. Add another 5 μL of 1 M DTT and shake for another 10 minutes.
Vortex the tubes for 4 minutes at maximum speed (e.g., Setting 10 on a Vortex Genie II instrument) to dislodge intraepithelial cells.
Let the tissue fragments settle, then use a serological pipette to collect the supernatant containing intraepithelial cells (including IELs), and dispense it through a 100 μm cell strainer into a new sterile 50 mL conical tube (“IEL Fraction Tube”), which should be stored on ice.
- To increase the yield of intraepithelial cells, add 10 mL fresh Harvest Medium to the tube containing the remaining tissue fragments and vortex them for 2 min to release additional cells. The resulting supernatant can be collected and pooled with the initial supernatant by straining through the 100 μm cell strainer into the IEL Fraction Tube.
- This step can be repeated 1–2 more times to maximize collection of intraepithelial cells.
- If cells in the intraepithelial cell fraction are to be analyzed separately from the lamina propria cells, the pooled supernatant cells can be strained once more through a 70 μm strainer, centrifuged for 5 min at 350 × g, 4°C, then resuspended in the desired media for analysis.
To isolate lamina propria cells (including LPLs, neutrophils, and other immune cells of potential interest), add 5 mL pre-warmed (to 37°C) Digest Medium to the tissue fragments. Shake the tissue in a 37°C incubator for 60 minutes.
- Decant and strain the digest consecutively through 100 μm, 70 μm, then 40 μm cell strainers. At each filtration step, mash the digested tissue fragments on the filter with the plunger of a 1 mL syringe (changing the plunger between samples) and flush the strainers with 5 mL IMDM.
- To obtain a general sample of intestinal immune cells without discriminating between intraepithelial and lamina propria-associated populations, strained intraepithelial cell preparations can be mixed back into the lamina propria cell digests at this stage.
- Optional: For a cleaner cell preparation, intraepithelial and lamina propria cells can be centrifuged through a Percoll gradient.
Pellet the filtered cells by centrifugation for 5 min at 350 × g, 4°C. Decant the supernatant and resuspend the cells in sterile PBS or harvest medium. Count the viable cells using a hemocytometer or automated cell counter
- Centrifuge the cells at 350 × g, 4°C, for 5 min, decant the supernatant and resuspend the cells in sterile PBS or desired media at an appropriate cell/mL concentration. Cells should be stored on ice and used as soon as possible (no more than a few hours later).
- For general immunophenotyping by flow cytometry, intestinal immune cells can be processed and stained using the same markers described by Bouladoux and colleagues for Citrobacter rodentium infection models (Bouladoux, Harrison, and Belkaid 2017).
REAGENTS AND SOLUTIONS
Antibiotics for Selective Media
Typical concentrations used for selection of S. Typhimurium 14028s and its derivatives:
Carbenicillin, 100 mg/L
Chloramphenicol, 30 mg/L
Kanamycin, 100 mg/L
Nalidixic acid, 50 mg/L
Streptomycin, 100 mg/L
Important note: Antibiotic concentrations may differ between wild-type strain backgrounds (e.g., S. Typhimurium ATCC 14028s vs. SL1344), or even within a given strain background depending on the location of an antibiotic resistance cassette within the genome. It is necessary to first validate the minimum inhibitory concentrations for your wild-type strain and mutants, and that the concentrations used for selection are sufficient but not excessive (e.g., a kanamycin-resistance cassette may be underexpressed when located in a particular region of the genome, resulting in a lower level of resistance to kanamycin for that particular mutant strain).
LB Broth (Miller)
10 g/L tryptone
5 g/L yeast extract
10 g/L NaCl
Dissolve in ultrapure water and autoclave. Store at room temperature.
LB Agar (Miller)
10 g/L tryptone
5 g/L yeast extract
10 g/L NaCl
15 g/L agar
Antibiotics (as needed, diluted at functional concentrations for selection)
Disperse in ultrapure water with a stir bar (leave in container) and autoclave. Use stir bar on a low speed (to avoid generating bubbles) after autoclaving to disperse the settled agar layer, and let cool to ~55°C (until ‘hand hot’; i.e., when one can just handle the flask base without autoclave gloves). Add desired antibiotics to working concentrations and stir for one minute (adding antibiotics at too hot of a temperature can inactivate them; adding at too cool of a temperature can locally solidify the medium and prevent uniform antibiotic dispersal). Aliquot into labeled (e.g., color-coded markings), sterile, disposable petri dishes (~25–30 mL per plate) and let cool at room temperature to solidify overnight. Invert plate stacks the next day and let dry an additional 2–3 days (depending laboratory humidity) at room temperature. Store plate stacks in closed plastic sleeves with the lids facing down and store at 4°C. 1 L should make approximately 30–35 plates.
MacConkey Lactose Agar
40 g/L MacConkey agar base
10 g/L lactose
Lactose is necessary for differentiating Lac- colonies (like Salmonella) from Lac+ colonies (including most commensal E. coli) on the media. Some premixed MacConkey agar media includes lactose; check the ingredient list to see if it is already included)
Follow instructions for preparing LB Agar (Miller).
Phosphate-buffered saline (PBS)
8 g NaCl
0.2g KCl
1.44g Na2HPO4 (anhydrous)
0.25g KH2PO4 (anhydrous)
Dissolve the reagents into 900 mL of MilliQ H2O. Adjust the pH to 7.4 with HCl, then add H2O to 1 L total. Aliquot the solution and autoclave to sterilize. PBS may be stored at room temperature.
Salmonella glycerol stocks
- 60% glycerol in ultrapure H2O, sterile-filtered (0.22 μm) or autoclaved
- Measure glycerol by weight (density = 1.26g/mL)
S. Typhimurium culture (grown in LB broth overnight, OD600 absorbance at ~1–4)
1.5mL or 2.0mL screw-cap cryovials, labelled with appropriate strain/stock identification
Add 500 μL of sterile 60% glycerol and 500 μL of S. Typhimurium culture in a cryovial for a final stock concentration of 30% glycerol, close the cap tightly, then vortex to mix. Store glycerol stock at −80°C. NEVER thaw frozen stocks. An aliquot of S. Typhimurium can be collected by gently scraping the top of the frozen stock with a sterile inoculating loop, then streaking onto LB or MacConkey agar plates as described in Support Protocol 1.
Shaking Medium (500 mL)
445 mL Hank’s Buffered Saline Solution (Gibco)
50 mL FBS (10% Final)
5 mL 100x Antibiotic/Antimycotic (Gibco)
Harvest Medium (500 mL)
395 mL IMDM (Cat. 21980, Gibco)
100 mL FBS (20% Final)
5 mL 100x Antibiotic/Antimycotic (Gibco)
Digest Medium (100 mL)
89 mL IMDM (Cat. 21980, Gibco)
10 mL FBS (20% Final)
1 mL 100x Antibiotic/Antimycotic (Gibco)
33 mg Collagenase VII (Sigma, Catalog #C2139)
1.75 mg DNase I (Sigma, Catalog #DN25)
For better enzyme activity, prepare the collagenase and DNase as fresh 100x stocks the day of isolation and add 50 μL per 5 mL medium.
COMMENTARY
Background Information
To infect a host and cause illness, both non-typhoidal and typhoidal Salmonella rely on a core set of virulence factors that includes two distinct Type-3 secretion systems (T3SS-1 and T3SS-2) and their related effector proteins. These virulence factors enable Salmonella to interact directly with mammalian host cells and shape a host response that benefits pathogen growth and transmission (Byndloss, Rivera-Chávez, et al. 2017). Studies using secretion system mutants have shown that T3SS-1, encoded by the Salmonella pathogenicity island 1 (SPI-1) genetic locus, is necessary for invasion of the intestinal epithelium (LaRock, Chaudhary, and Miller 2015). T3SS-2, encoded by SPI-2, promotes Salmonella intracellular survival inside host cells, particularly phagocytes (Jennings, Thurston, and Holden 2017).
Infection with NTS triggers an inflammatory response characterized by the secretion of proinflammatory cytokines and chemokines that attract immune cells, particularly neutrophils, to the gut. This process helps restrict the pathogen to the gut and limit its dissemination to systemic sites (Perez-Lopez et al. 2016). In contrast, typhoidal serovars such as S. Typhi have acquired virulence mechanisms to avoid detection by the innate immune system and persist in the host for weeks to years, with some hosts becoming chronic carriers of the pathogen. The capacity to evade immunity has been linked to functional stealth adaptations of S. Typhi, including the loss of fepE-regulated very-long O-antigen chains and the acquisition of a polysaccharide capsule (the Vi antigen) encoded by the viaB locus, which helps to mask LPS and limits complement deposition and phagocytosis of the pathogen (Wilson et al. 2011; Wangdi et al. 2014; Crawford et al. 2013; Wilson et al. 2008). Further adaptations by S. Typhi include the altered structure and decreased expression of pathogen-associated molecules, such as flagellin, that otherwise allow for rapid immune recognition of non-typhoidal serovars via Toll-like receptors (Atif et al. 2014; Winter et al. 2014).
The generation of relevant and appropriate animal models to study the pathogenesis of different Salmonella serovars concerning human disease has been a long journey, nicely summarized by Tsolis et al. (Tsolis et al. 2011). Typhoidal Salmonella serovars are human-adapted and neither colonize nor cause disease in conventional mice. Thus, experimental research with typhoidal Salmonella serovars such as S. Typhi has been primarily limited to in vitro assays. More recently, humanized mice have been used to investigate S. Typhi virulence mechanisms in vivo (Song et al. 2010; Libby et al. 2010), but these models present challenges and are not widely available. Of note, although a Tlr11−/− mouse model of S. Typhi infection was proposed (Mathur et al. 2012), the model was subsequently disproven by multiple leaders in the field (Song et al. 2016). Meanwhile, oral infection of commonly used mouse lines (such as C57BL/6 and BALB/c) with non-typhoidal serovars such as S. Typhimurium yields a typhoid-like disease with minimal colitis and significant extraintestinal pathogen spread, resulting in rapid mortality (Santos et al. 2001). S. Typhimurium infection of susceptible mice has been used to model human typhoid disease and contributed to identifying virulence factors important for pathogenesis, including those mentioned above. Nevertheless, an important limitation of this model is that NTS serovars, including S. Typhimurium, lack many important features of S. Typhi’s genome (Nuccio and Bäumler 2014) and do not cause enteric fever in humans, instead causing inflammatory diarrhea.
To interrogate the virulence and immunological bases of NTS colitis, much in vivo research prior to the 2000s employed bovine and rabbit intestinal loop infection models, which display substantial inflammation and neutrophil influx to the infected intestine compared to conventional mice (Everest et al. 1999; Santos et al. 2001). In 2003, Barthel and colleagues revived an infection model originally described in a series of studies by Marjorie Bonhoff and C. Philip Miller 40 years prior (Bohnhoff, Drake, and Miller 1954; Bohnhoff and Miller 1962), utilizing mice pretreated with streptomycin to transiently disrupt colonization resistance prior to S. Typhimurium oral infection (Barthel et al. 2003). Compared to S. Typhimurium oral infection without pretreatment, streptomycin administration prior to infection resulted in efficient colonization of the pathogen to high levels in the intestinal lumen (>109 CFU/g in the cecum and colon) and pronounced cecal inflammation, including edema, epithelial degradation, and neutrophil infiltration (Barthel et al. 2003). Similar to the calf model (Santos et al. 2001), intestinal inflammation was found to depend on the T3SS virulence factors, and the model has been adopted as the gold standard for studying the host-microbe molecular interactions involved in Salmonella colitis (Hapfelmeier et al. 2005; Kaiser et al. 2012). This model is independent of the wild-type strain and mouse lineage, so it can be adapted to interrogate the interplay between Salmonella virulence factors and mammalian genetics on colitis.
In streptomycin-pretreated mice, S. Typhimurium colitis is accompanied by a neutrophil-dominated inflammatory response akin to that observed in humans (Hohmann 2001). This inflammatory response results from T3SS-dependent and PAMP/TLR-mediated transcriptional activation of proinflammatory cytokines produced by macrophages (TNF-α, IL-1β) and of Type-1 (IFN-γ, IL-12, IL-18) and Type-17 (IL-23, IL-17, IL-22) cytokines, which collectively promote antibacterial functions to limit the infection. IL-23 produced by dendritic and mononuclear cells in the gut in response to S. Typhimurium infection triggers IL-17 and IL-22 production by TH17 cells, γδ T cells, and innate lymphoid cells (ILCs). IL-17 and IL-22 stimulate intestinal epithelial cells to rapidly amplify the production of antimicrobial proteins (REG3γ, lipocalin-2, and calprotectin) and chemokines. Upregulated chemokines include CXCL1, CXCL2, and CXCL5, which recruit neutrophils to the gut (Perez-Lopez et al. 2016). Even though aspects of these inflammatory responses benefit the host and promote control of S. Typhimurium infection, numerous studies have demonstrated that the pathogen exploits host immunity to colonize the host and thrive (Behnsen et al. 2015).
Recent research has focused on how intestinal inflammation benefits NTS and related pathogens/pathobionts, such as E. coli, by creating new nutrient and respiratory metabolic niches in the gut that promote the transient expansion of Enterobacteriaceae (Winter and Bäumler 2014). Other models, like the high-fat diet model and a model with a low-complexity microbiota, have been shown to promote intestinal inflammation and enhance S. Typhimurium colonization (Stecher et al. 2005, 2010; Wotzka et al. 2019). One major drawback of the streptomycin model is the ablation of microbiota-mediated colonization resistance, which is an important factor in understanding NTS infection dynamics (Rogers, Tsolis, and Bäumler 2021). Using fecal microbiota transfer to germ-free mice, comparative studies of the microbiota found in mice from different vendors have begun elucidating critical factors in microbiota-derived colonization resistance (Velazquez et al. 2019; Stecher 2021). Additionally, genetically resistant CBA/J mice infected with S. Typhimurium have been found to eventually develop cecal inflammation without antibiotic treatment (Faber et al. 2017; Rivera-Chávez et al. 2016), even though the inflammation levels are typically lower and more variable. These mice have been used, for example, to determine whether phenotypes observed after streptomycin pretreatment are also present in mice with intact microbiota. Systemic infection still occurs in such resistant mice, but the host controls S. Typhimurium replication at systemic sites, and infection studies can be continued for two weeks or more. Long-term infection models using mice from the resistant (Nramp+/+) 129Sv-lineage have also been developed to study S. Typhimurium dynamics at extraintestinal sites such as the mesenteric lymph nodes for up to a year (Monack, Bouley, and Falkow 2004). These mice allow for the study of the underlying genetics and mechanisms by which Salmonella promotes granuloma formation during infection and how the microbiota impacts transmission by chronic carriers, both aspects observed in human typhoid fever (Monack, Bouley, and Falkow 2004; Lawley et al. 2006, 2008; Gopinath et al. 2013; Nagy et al. 2013).
There has been an interest in using mouse Salmonella infections to model and probe the potential immune deficiencies associated with NTS bacteremia, which is responsible for most of the “all-cause” Salmonella-associated mortality (Ao et al. 2015). Based on epidemiological evidence, underlying risk factors promoting NTS bacteremia include compromised immunity resulting from HIV infection in adults, nutritional deficiencies, and co-infection with malaria in very young children (Ao et al. 2015; Feasey et al. 2012; Bronzan et al. 2007). The latter risk factors have been modeled in diet studies and parasite co-infection models in mice, which may help explain potential mechanisms boosting extraintestinal Salmonella (Stull-Lane et al. 2020; Mottram et al. 2016; Roux et al. 2010). For a detailed protocol on murine malaria models, which may be combined with the S. Typhimurium infection models presented here to study co-infection, please refer to Huang et al. (Huang, Pearman, and Kim 2015).
Critical Parameters
When planning S. Typhimurium infection experiments, choosing the most appropriate model for the relevant research question is important. All animal experiments require Institutional Animal Care and Use Committee review and approval, to ensure that animal welfare is always given consideration.
Mouse considerations and sources of model variability
As with any mouse infection model, mice should be age, strain, and sex-matched between experimental groups to ensure robust and replicable data. Typically, young adult mice between 8 and 12 weeks of age are used for infections. Genetically susceptible mice will typically succumb to infection with S. Typhimurium in 3–5 days with streptomycin pretreatment or 5–10 days without streptomycin pretreatment. Resistant mouse lines can usually control S. Typhimurium for longer (10 days or more) and may even clear the infection. Mouse resistance and susceptibility to S. Typhimurium are multifactorial, but in general are largely impacted by allelic variation (point mutation) in the phagosomal transition metal transporter gene Slc11a1 (also known as Nramp1) of susceptible lines such as C57BL/6 and BALB/c (Monack, Bouley, and Falkow 2004; Vidal et al. 1996). Resistant mice (i.e., encode functional SLC11A1; particularly mouse strain 129SvJ) have been used as models of chronic Salmonella carriage, a phenotype observed in human S. Typhi cases (Gunn et al. 2014; Ruby et al. 2012; Stecher et al. 2006). Resistant mice (particularly CBA/J) have also proved to be useful models for studying gut microbiota-associated phenotypes during colitis that can be hard to track following administration of streptomycin or other antibiotics (Faber et al. 2017; Rivera-Chávez et al. 2016). One caveat of using 129SvJ or CBA/J mice is the general lack of readily available transgenic and knockout strains, which are mostly available in more commonly used susceptible backgrounds (e.g., C57BL/6). Because many knockout mouse lines were generated as a chimera of 129S2 and C57BL/6, knock-out mice should be genotyped for the Slc11a1 allele. Moreover, knockouts of loci (e.g., Il17a, Cxcr2) in relatively close proximity to the Slc11a1 gene should also be genotyped for Slc11a1 status before use, as wild-type and knockout littermates may harbor different alleles.
The same strain of mice from different vendors can harbor vastly different microbiota compositions that may influence the colonization and competition of S. Typhimurium in the gut. For example, the specific pathogen-free C57BL/6J mice from the Jackson Laboratory typically lack common commensal Enterobacteriaceae (such as E. coli), which are present in many other vendors’ mice and can compete with Salmonella in the gut for nutrients (Velazquez et al. 2019; Sassone-Corsi et al. 2016). While co-housing different mouse strains (e.g., wild-type and knockout) prior to infection experiments can help normalize their microbiota, this is not always effective. As such, breeding littermate controls is the preferred option for maintaining consistent microbiota between groups. Characterizing microbiota-associated infection findings will often require fecal microbiota transfer to germ-free animals. Diet composition also influences the microbiota, inflammation, and Salmonella pathogenesis (Desai et al. 2016; Daniel et al. 2014; Nickerson et al. 2014; Kreuzer and Hardt 2020). For example, a high-fat diet stimulates bile acid production, which affects the gut microbiota but not Salmonella (Wotzka et al. 2019). It is best to work with a veterinary dietitian to select an appropriate control diet when testing the impact of food components.
The timing of infection can also be important due to circadian fluctuations in feeding behaviors and inflammatory mediators, so infection times should remain consistent between experiments (Bellet et al. 2013).
Salmonella considerations
For each experiment, following the same procedure to generate the inoculum is necessary for reproducibility of results. Usually, a dose of 5×107 - 1×109 CFU of S. Typhimurium per mouse is used for oral infection inocula. Lower concentrations are necessary for intraperitoneal infection, typically around 1×103 CFU per mouse. While some protocols may call for direct inoculation of liquid cultures from the glycerol stock, even small amounts of glycerol can alter Salmonella gene expression in the inoculum, which may impact experimental outcomes. Instead, inoculating cultures from colonies or confluent growth on a plate, as this protocol directs, promotes both active Salmonella growth and more consistently reproducible infections with the resulting inoculum (Kragh et al. 2018). We have also found that growing overnight cultures in LB with antibiotics for selection can influence stress responses in the bacteria, potentially altering disease phenotypes and competitive fitness. As such, antibiotics should in general only be added to cultures used for inocula when necessary to maintain selection for an unstable genetic locus or plasmid. In both intragastric and intraperitoneal infection models, uninfected control mice should be mock-infected with the appropriate vehicle (LB or PBS) to control for the impact of the inoculum and for stress responses induced by the procedure.
Additionally, it is unfortunately common in some research fields to utilize a Salmonella infection model while not being aware of the specific serovar or substrain used for experiments, despite that being critical to understanding the relevance of the findings. In particular, many immunological studies utilize an S. Typhimurium aroA mutant, which is a metabolically deficient strain often used as a live-attenuated vaccine vector in mice (Felgner et al. 2016; Hoiseth and Stocker 1981). Infection studies with this strain are not equivalent to fully virulent wild-type strains such as SL1344 and ATCC 14028s (including its IR715 derivative) (Everest et al. 1999), and findings should always be conveyed in that context. Even between wild-type strains, genomic variations such as the presence or absence of a single gene (e.g., the phage-encoded effector gene sopE; present in SL1344, absent in ATCC 14028s) have been shown to impact colitis severity (Hapfelmeier et al. 2004; Lopez et al. 2012), which can impede the comparison of infection phenotypes between model strains. Of note, although initial experiments in the streptomycin pretreatment model were performed using an S. Typhimurium strain with relatively high resistance to streptomycin (SL1344), strains more sensitive to streptomycin (such as ATCC 14028s and its derivative IR715) have also been shown to colonize to high levels and induce colitis in the model. Nevertheless, when the streptomycin pretreatment model is used for competitive infections, the two competing strains must have similar sensitivity/resistance to the antibiotic, to limit the potential of the antibiotic itself directly influencing the competitive index. One option to normalize resistance between strains with dissimilar resistances is to transform them with a plasmid conferring streptomycin resistance (e.g., pHP45Ω).
The intragastric gavage technique described here is the most commonly employed method for delivering an S. Typhimurium inoculum to the gut. However, some alternate oral inoculation techniques have been reported to generate equivalent colonization results in mice; for example, foodborne inoculation by providing mice small amounts of feed coated with a defined quantity of S. Typhimurium was shown to successfully implant the infection (Nilsson, Kari, and Steele-Mortimer 2019), and better mimics the typical food-associated route of infection. Non-invasive techniques like this also help to limit animal distress and reduce the odds of inoculum misdelivery during inoculation, although they have the downside of the inoculum being less precise and less consistent from animal to animal.
Anticipated Results and Analysis
The overall outcomes of the S. Typhimurium infection models will be influenced by many variables, including but not limited to: mouse strain genetics, gut microbiota composition, S. Typhimurium strain(s) used, how the inoculum was prepared, the route of infection, and the timing of the infection. Maintaining consistency in these factors helps generate reproducible results between experiments. Wild-type S. Typhimurium colonization tends to follow log-normal distributions in mouse infection studies, so raw CFU/g values should be logarithmically transformed prior to comparing S. Typhimurium loads between infected mouse groups by parametric statistical tests. Similarly, competitive indices should usually be log-transformed for comparisons to properly weight ratios less than 1 (i.e., more mutant strain recovered than wildtype). For an intragastric infection of 1×109 CFU S. Typhimurium 14028s in Salmonella-susceptible C57BL/6J mice (no streptomycin pretreatment), one would expect to detect colonization of 103 to 106 CFU/g of feces 1 day after infection, and anywhere from 105 to 109 CFU/g feces or cecal content by 4 days post-infection (Velazquez et al. 2019). In Salmonella-resistant CBA/J mice, in the absence of streptomycin pretreatment, one would expect 103 to 106 CFU/g feces early in the infection (1–3 days) with a transient increase up to 106 to 1010 CFU/g after 5–7 days (Rivera-Chávez et al. 2016). The same intragastric infection in streptomycin pre-treated susceptible mice should result in substantially higher fecal S. Typhimurium loads at 1 day post-infection (≥107 CFU/g), while inflammation-associated phenotypes will usually develop within 3–4 days. S. Typhimurium levels in systemic tissues such as the liver and spleen can vary throughout the infection and are dependent on mouse lines, but are generally held to <109 CFU/g tissue in susceptible mice. Resistant mice exhibit low-level S. Typhimurium colonization in the spleen, as they can control the pathogen’s replication at this site. Planning to continue an S. Typhimurium infection past 4 days in susceptible mice is risky, as it will result in significant mortality and a consequent loss of important experimental samples.
On a gross pathology level, inflamed colons are generally shorter than healthy colons and can appear hyperemic, and the cecum typically shrinks during infection (Figure 1). Exemplary H&E-stained sections of streptomycin-pretreated uninfected and S. Typhimurium-infected cecal tissues are provided in Figure 2. Histological inflammation can be quite severe, as shown in Figure 2, whereas no signs of inflammation are observed in preserved tissue from uninfected animals.
Figure 2. Representative H&E-stained ceca from uninfected mice or 96 h after intestinal S. Typhimurium infection.
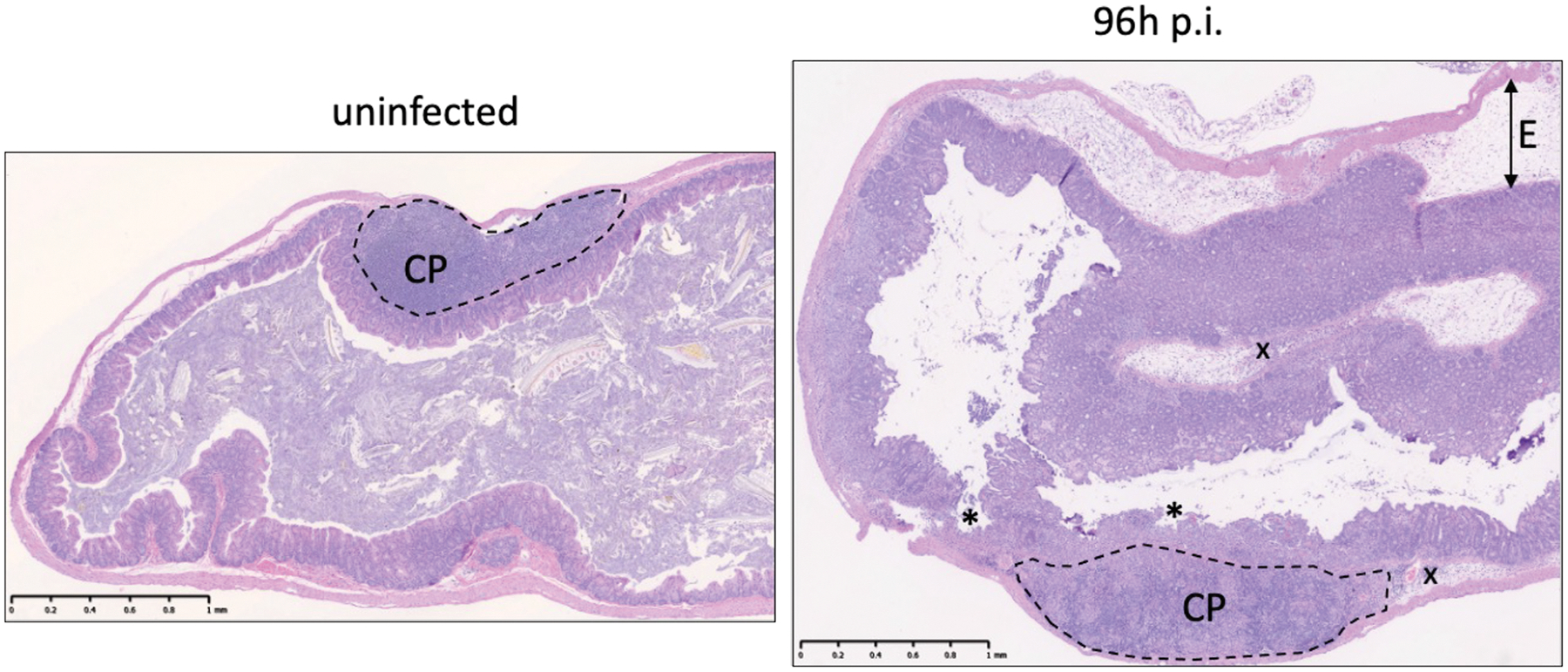
The dashed line indicates the cecal patch (CP), which shouldn’t be confused with tissue inflammation. Asterisks (*) indicate severe ulceration of the epithelial lining. The arrow indicates severe submucosal edema (E). The x indicates submucosal infiltration of immune cells. 60 x magnification, scale bar 1mm.
In colitis models, S. Typhimurium can induce 10-fold to 1,000-fold changes in the expression of colitis-associated genes (see Table 2) relative to uninfected controls by day 3–4 of infection (Godinez et al. 2008). Changes in gene expression can be measured by qPCR of cecal RNA (ΔΔCt method, normalized to mouse Actb levels) (Godinez et al. 2008; Perez-Lopez et al. 2019; Deriu et al. 2013). In intestinal content and tissue of streptomycin-pretreated mice, protein levels of inflammatory markers (e.g., myeloperoxidase, lipocalin-2, calprotectin) will typically be higher (2 to 1,000-fold) in samples from S. Typhimurium-infected mice relative to healthy control mice at the peak of infection. A general increase of T cell numbers and T cell cytokine production (particularly IL-22, IL-17, IFNγ) can be observed in the gut beginning at 24–48h after S. Typhimurium infection (Godinez et al. 2008, 2009; Raffatellu et al. 2008; Perez-Lopez et al. 2019). These cells amplify the inflammatory response during S. Typhimurium colitis, including the recruitment of neutrophils. Together with lymphocyte and neutrophil isolation, analysis of changes in gene expression and histopathology provide a comprehensive understanding of the immune response during NTS colitis.
Time Considerations
Mice should be allowed to acclimate to a new room at least one week before infection. Preparation of S. Typhimurium cultures for inoculation will take 3 days in total, although the active portions involving streaking of glycerol stocks onto agar plates and inoculating the LB broth cultures should only take minutes. Streptomycin preparation will take 10–20 minutes, and should be used as soon as possible. Scruffing restraint and gavage or intraperitoneal infection of mice can take 1–5 minutes per mouse, depending on the experience level of the operator. After inoculation, S. Typhimurium infection will typically result in mortality within 1 week in susceptible mice, and the animals should be monitored and weighed at least daily during that time period.
Collection of fecal samples from live mice that are infected typically takes 5 to 15 minutes per mouse, although this may take 30 minutes or longer when colitis is more severe. Humanely euthanizing animals takes approximately 5 minutes, and dissection and tissue removal will take 10–20 minutes per mouse. Blood collection should be performed as soon as possible after euthanasia to limit coagulation in the mouse. Weighing sample tubes, homogenization, preparing serial dilutions and plating of fecal or tissue slurries can take 20 minutes per mouse, but processing multiple samples in parallel will greatly speed the per-mouse time (i.e., processing samples from 10 mice would take approximately 1 hour total). Plated dilutions should be incubated at 37°C for 14–18 hours to generate colony forming units (CFUs) of appropriate size for enumeration, whereas counting CFUs can take 1–2 minutes per plate.
Organs recovered for histopathological assessment during necropsy should be expeditiously moved into formalin to start fixation in order to limit degradation of tissue structures. Histological cassettes should be fixed in formalin for 24–48 hours, then moved into 70% ethanol for long term storage. Analysis and scoring of H&E-stained tissue sections can take 5–10 minutes per section.
Tissues sampled for RNA extraction should be snap frozen in liquid nitrogen immediately at necropsy, then stored at −80°C for later processing. Mortar-and-pestle grinding of tissues can take 5 minutes per sample. Phenol-chloroform RNA extraction from tissue homogenates takes about 2 hours for 4–10 samples. Synthesizing cDNA takes 1–2 hours, depending on the number of RNA samples needed to be diluted. Preparing SYBR Green reaction mixes and samples in a 96- or 384-well plate for qPCR takes 1–2 hours; the qPCR reaction takes 1.5–2 hours.
ELISAs that do not come with pre-coated plates will need to be set up to incubate with capture antibodies the evening before performing the assay. These assays will typically take 5–7 hours, including incubation steps.
Media for immune cell isolation from intestinal tissues can be made prior to necropsy, while isolation steps should be performed as soon as possible (within a few hours) following collection of tissues in harvest medium for good quality cell preparations. The isolation takes 3–4 hours overall for tissues from 6–10 mice. Cells should be used for immunophenotyping or other ex vivo stimulation assays the same day as collection.
Acknowledgments
G.T.W. was funded by the National Institutes of Health (NIH) grant T32AI007036. R.R.G. was funded by a fellowship from the Crohn’s & Colitis Foundation, award number 649744. M.R. was funded by the NIH grants AI126277, AI145325, AI154644, AI114625, by the Chiba University-University of California-San Diego (UCSD) Center for Mucosal Immunology, Allergy, and Vaccines, and by the UCSD Department of Pediatrics. M.R. also holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. The authors declare no conflict of interest.
References
- Ao Trong T., Feasey Nicholas A., Gordon Melita A., Keddy Karen H., Angulo Frederick J., and Crump John A.. 2015. “Global Burden of Invasive Nontyphoidal Salmonella Disease, 2010(1).” Emerging Infectious Diseases 21 (6). 10.3201/eid2106.140999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atif Shaikh M., Winter Sebastian E., Winter Maria G., McSorley Stephen J., and Bäumler Andreas J.. 2014. “Salmonella Enterica Serovar Typhi Impairs CD4 T Cell Responses by Reducing Antigen Availability.” Infection and Immunity 82 (6): 2247–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Hongxia, Wang Shuang, Zhao Jian-Hua, and Liu Shu-Lin. 2020. “Salmonella Secretion Systems: Differential Roles in Pathogen-Host Interactions.” Microbiological Research 241 (December): 126591. [DOI] [PubMed] [Google Scholar]
- Barthel Manja, Hapfelmeier Siegfried, Quintanilla-Martínez Leticia, Kremer Marcus, Rohde Manfred, Hogardt Michael, Pfeffer Klaus, Rüssmann Holger, and Hardt Wolf-Dietrich. 2003. “Pretreatment of Mice with Streptomycin Provides a Salmonella Enterica Serovar Typhimurium Colitis Model That Allows Analysis of Both Pathogen and Host.” Infection and Immunity 71 (5): 2839–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnsen Judith, Perez-Lopez Araceli, Nuccio Sean-Paul, and Raffatellu Manuela. 2015. “Exploiting Host Immunity: The Salmonella Paradigm.” Trends in Immunology 36 (2): 112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet Marina M., Deriu Elisa, Liu Janet Z., Grimaldi Benedetto, Blaschitz Christoph, Zeller Michael, Edwards Robert A., et al. 2013. “Circadian Clock Regulates the Host Response to Salmonella.” Proceedings of the National Academy of Sciences of the United States of America 110 (24): 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuzón CR, and Holden DW. 2001. “Use of Mixed Infections with Salmonella Strains to Study Virulence Genes and Their Interactions in Vivo.” Microbes and Infection / Institut Pasteur 3 (14–15): 1345–52. [DOI] [PubMed] [Google Scholar]
- Bohnhoff M, Drake BL, and Miller CP. 1954. “Effect of Streptomycin on Susceptibility of Intestinal Tract to Experimental Salmonella Infection.” Proceedings of the Society for Experimental Biology and Medicine. Society for Experimental Biology and Medicine 86 (1): 132–37. [DOI] [PubMed] [Google Scholar]
- Bohnhoff M, and Miller CP. 1962. “Enhanced Susceptibility to Salmonella Infection in Streptomycin-Treated Mice.” The Journal of Infectious Diseases 111 (September): 117–27. [DOI] [PubMed] [Google Scholar]
- Bouladoux Nicolas, Harrison Oliver J., and Belkaid Yasmine. 2017. “The Mouse Model of Infection with Citrobacter Rodentium.” Current Protocols in Immunology / Edited by John E. Coligan … [et Al.] 119 (November): 19.15.1–19.15.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronzan Rachel N., Taylor Terrie E., Mwenechanya James, Tembo Madalitso, Kayira Kondwani, Bwanaisa Lloyd, Njobvu Alfred, et al. 2007. “Bacteremia in Malawian Children with Severe Malaria: Prevalence, Etiology, HIV Coinfection, and Outcome.” The Journal of Infectious Diseases 195 (6): 895–904. [DOI] [PubMed] [Google Scholar]
- Byndloss Mariana X., Olsan Erin E., Rivera-Chávez Fabian, Tiffany Connor R., Cevallos Stephanie A., Lokken Kristen L., Torres Teresa P., et al. 2017. “Microbiota-Activated PPAR-γ Signaling Inhibits Dysbiotic Enterobacteriaceae Expansion.” Science 357 (6351): 570–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byndloss Mariana X., Rivera-Chávez Fabian, Tsolis Renée M., and Bäumler Andreas J.. 2017. “How Bacterial Pathogens Use Type III and Type IV Secretion Systems to Facilitate Their Transmission.” Current Opinion in Microbiology 35 (February): 1–7. [DOI] [PubMed] [Google Scholar]
- Crawford Robert W., Wangdi Tamding, Spees Alanna M., Xavier Mariana N., Tsolis Renée M., and Bäumler Andreas J.. 2013. “Loss of Very-Long O-Antigen Chains Optimizes Capsule-Mediated Immune Evasion by Salmonella Enterica Serovar Typhi.” mBio 4 (4). 10.1128/mBio.00232-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel Hannelore, Gholami Amin Moghaddas, Berry David, Desmarchelier Charles, Hahne Hannes, Loh Gunnar, Mondot Stanislas, et al. 2014. “High-Fat Diet Alters Gut Microbiota Physiology in Mice.” The ISME Journal 8 (2): 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriu Elisa, Liu Janet Z., Pezeshki Milad, Edwards Robert A., Ochoa Roxanna J., Contreras Heidi, Libby Stephen J., Fang Ferric C., and Raffatellu Manuela. 2013. “Probiotic Bacteria Reduce Salmonella Typhimurium Intestinal Colonization by Competing for Iron.” Cell Host & Microbe 14 (1): 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai Mahesh S., Seekatz Anna M., Koropatkin Nicole M., Kamada Nobuhiko, Hickey Christina A., Wolter Mathis, Pudlo Nicholas A., et al. 2016. “A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility.” Cell 167 (5): 1339–53.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disease Control, Centers for, Prevention, and Others. 2019. Antibiotic Resistance Threats in the United States, 2019. US Department of Health and Human Services, Centres for Disease Control and …. [Google Scholar]
- Everest P, Ketley J, Hardy S, Douce G, Khan S, Shea J, Holden D, Maskell D, and Dougan G. 1999. “Evaluation of Salmonella Typhimurium Mutants in a Model of Experimental Gastroenteritis.” Infection and Immunity 67 (6): 2815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber Franziska, Thiennimitr Parameth, Spiga Luisella, Byndloss Mariana X., Litvak Yael, Lawhon Sara, Andrews-Polymenis Helene L., Winter Sebastian E., and Bäumler Andreas J.. 2017. “Respiration of Microbiota-Derived 1,2-Propanediol Drives Salmonella Expansion during Colitis.” PLoS Pathogens 13 (1): e1006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey Nicholas A., Dougan Gordon, Kingsley Robert A., Heyderman Robert S., and Gordon Melita A.. 2012. “Invasive Non-Typhoidal Salmonella Disease: An Emerging and Neglected Tropical Disease in Africa.” The Lancet 379 (9835): 2489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner Sebastian, Frahm Michael, Kocijancic Dino, Rohde Manfred, Eckweiler Denitsa, Bielecka Agata, Bueno Emilio, et al. 2016. “aroA-Deficient Salmonella Enterica Serovar Typhimurium Is More Than a Metabolically Attenuated Mutant.” mBio 7 (5). 10.1128/mBio.01220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mor Ohad, Boyle Erin C., and Grassl Guntram A.. 2014. “Same Species, Different Diseases: How and Why Typhoidal and Non-Typhoidal Salmonella Enterica Serovars Differ.” Frontiers in Microbiology 5 (August): 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Shizhong, Tian Qin, An Shuming, Pan Zhiming, Chen Xiang, and Jiao Xinan. 2016. “High-Efficiency, Two-Step Scarless-Markerless Genome Genetic Modification in Salmonella Enterica.” Current Microbiology 72 (6): 700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinez Ivan, Haneda Takeshi, Raffatellu Manuela, George Michael D., Paixão Tatiane A., Rolán Hortensia G., Santos Renato L., Dandekar Satya, Tsolis Renée M., and Bäumler Andreas J.. 2008. “T Cells Help to Amplify Inflammatory Responses Induced by Salmonella Enterica Serotype Typhimurium in the Intestinal Mucosa.” Infection and Immunity 76 (5): 2008–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinez Ivan, Raffatellu Manuela, Chu Hiutung, Paixão Tatiane A., Haneda Takeshi, Santos Renato L., Bevins Charles L., Tsolis Renée M., and Bäumler Andreas J.. 2009. “Interleukin-23 Orchestrates Mucosal Responses to Salmonella Enterica Serotype Typhimurium in the Intestine.” Infection and Immunity 77 (1): 387–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath Smita, Hotson Andrew, Johns Jennifer, Nolan Garry, and Monack Denise. 2013. “The Systemic Immune State of Super-Shedder Mice Is Characterized by a Unique Neutrophil-Dependent Blunting of TH1 Responses.” PLoS Pathogens 9 (6): e1003408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont Patrick A. D., Weill François-Xavier, and Others. 2007. “Antigenic Formulae of the Salmonella Serovars.” WHO Collaborating Centre for Reference and Research on Salmonella 9: 1–166. [Google Scholar]
- Gunn John S., Marshall Joanna M., Baker Stephen, Dongol Sabina, Charles Richelle C., and Ryan Edward T.. 2014. “Salmonella Chronic Carriage: Epidemiology, Diagnosis, and Gallbladder Persistence.” Trends in Microbiology 22 (11): 648–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier Siegfried, Ehrbar Kristin, Stecher Bärbel, Barthel Manja, Kremer Marcus, and Hardt Wolf-Dietrich. 2004. “Role of the Salmonella Pathogenicity Island 1 Effector Proteins SipA, SopB, SopE, and SopE2 in Salmonella Enterica Subspecies 1 Serovar Typhimurium Colitis in Streptomycin-Pretreated Mice.” Infection and Immunity 72 (2): 795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier Siegfried, Stecher Bärbel, Barthel Manja, Kremer Marcus, Müller Andreas J., Heikenwalder Mathias, Stallmach Thomas, et al. 2005. “The Salmonella Pathogenicity Island (SPI)-2 and SPI-1 Type III Secretion Systems Allow Salmonella Serovar Typhimurium to Trigger Colitis via MyD88-Dependent and MyD88-Independent Mechanisms.” Journal of Immunology 174 (3): 1675–85. [DOI] [PubMed] [Google Scholar]
- Hiyoshi Hirotaka, Tiffany Connor R., Bronner Denise N., and Bäumler Andreas J.. 2018. “Typhoidal Salmonella Serovars: Ecological Opportunity and the Evolution of a New Pathovar.” FEMS Microbiology Reviews 42 (4): 527–41. [DOI] [PubMed] [Google Scholar]
- Hohmann EL 2001. “Nontyphoidal Salmonellosis.” Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 32 (2): 263–69. [DOI] [PubMed] [Google Scholar]
- Hoiseth SK, and Stocker BA. 1981. “Aromatic-Dependent Salmonella Typhimurium Are Non-Virulent and Effective as Live Vaccines.” Nature 291 (5812): 238–39. [DOI] [PubMed] [Google Scholar]
- Huang Brian W., Pearman Emily, and Kim Charles C.. 2015. “Mouse Models of Uncomplicated and Fatal Malaria.” Bio-Protocol 5 (13). 10.21769/bioprotoc.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra J. Antonio, Knodler Leigh A., Sturdevant Daniel E., Virtaneva Kimmo, Carmody Aaron B., Fischer Elizabeth R., Porcella Stephen F., and Steele-Mortimer Olivia. 2010. “Induction of Salmonella Pathogenicity Island 1 under Different Growth Conditions Can Affect Salmonella-Host Cell Interactions in Vitro.” Microbiology 156 (Pt 4): 1120–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson Amanda, Lam Lilian, Rajendram Manohary, Tamburini Fiona, Honeycutt Jared, Pham Trung, Van Treuren Will, et al. 2018. “A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection.” Cell Host & Microbe 24 (2): 296–307.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings Elliott, Thurston Teresa L. M., and Holden David W.. 2017. “Salmonella SPI-2 Type III Secretion System Effectors: Molecular Mechanisms And Physiological Consequences.” Cell Host & Microbe 22 (2): 217–31. [DOI] [PubMed] [Google Scholar]
- Kaiser Patrick, Diard Médéric, Stecher Bärbel, and Hardt Wolf-Dietrich. 2012. “The Streptomycin Mouse Model for Salmonella Diarrhea: Functional Analysis of the Microbiota, the Pathogen’s Virulence Factors, and the Host’s Mucosal Immune Response.” Immunological Reviews 245 (1): 56–83. [DOI] [PubMed] [Google Scholar]
- Kragh Kasper Nørskov, Alhede Maria, Rybtke Morten, Stavnsberg Camilla, Jensen Peter Ø., Tolker-Nielsen Tim, Whiteley Marvin, and Bjarnsholt Thomas. 2018. “The Inoculation Method Could Impact the Outcome of Microbiological Experiments.” Applied and Environmental Microbiology 84 (5). 10.1128/AEM.02264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer Markus, and Hardt Wolf-Dietrich. 2020. “How Food Affects Colonization Resistance Against Enteropathogenic Bacteria.” Annual Review of Microbiology 74 (September): 787–813. [DOI] [PubMed] [Google Scholar]
- LaRock Doris L., Chaudhary Anu, and Miller Samuel I.. 2015. “Salmonellae Interactions with Host Processes.” Nature Reviews. Microbiology 13 (4): 191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley Trevor D., Bouley Donna M., Hoy Yana E., Gerke Christine, Relman David A., and Monack Denise M.. 2008. “Host Transmission of Salmonella Enterica Serovar Typhimurium Is Controlled by Virulence Factors and Indigenous Intestinal Microbiota.” Infection and Immunity 76 (1): 403–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley Trevor D., Chan Kaman, Thompson Lucinda J., Kim Charles C., Govoni Gregory R., and Monack Denise M.. 2006. “Genome-Wide Screen for Salmonella Genes Required for Long-Term Systemic Infection of the Mouse.” PLoS Pathogens 2 (2): e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CA, and Falkow S. 1990. “The Ability of Salmonella to Enter Mammalian Cells Is Affected by Bacterial Growth State.” Proceedings of the National Academy of Sciences of the United States of America 87 (11): 4304–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby Stephen J., Brehm Michael A., Greiner Dale L., Shultz Leonard D., McClelland Michael, Smith Kelly D., Cookson Brad T., et al. 2010. “Humanized Nonobese Diabetic-Scid IL2rgammanull Mice Are Susceptible to Lethal Salmonella Typhi Infection.” Proceedings of the National Academy of Sciences of the United States of America 107 (35): 15589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Christopher A., Winter Sebastian E., Rivera-Chávez Fabian, Xavier Mariana N., Poon Victor, Nuccio Sean-Paul, Tsolis Renée M., and Bäumler Andreas J.. 2012. “Phage-Mediated Acquisition of a Type III Secreted Effector Protein Boosts Growth of Salmonella by Nitrate Respiration.” mBio 3 (3). 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majowicz Shannon E., Musto Jennie, Scallan Elaine, Angulo Frederick J., Kirk Martyn, O’Brien Sarah J., Jones Timothy F., Fazil Aamir, Hoekstra Robert M., and International Collaboration on Enteric Disease “Burden of Illness” Studies. 2010. “The Global Burden of Nontyphoidal Salmonella Gastroenteritis.” Clinical Infectious Diseases: An Official Publication of the Infectious Diseases Society of America 50 (6): 882–89. [DOI] [PubMed] [Google Scholar]
- Mathur Ramkumar, Oh Hyunju, Zhang Dekai, Park Sung-Gyoo, Seo Jin, Koblansky Alicia, Hayden Matthew S., and Ghosh Sankar. 2012. “A Mouse Model of Salmonella Typhi Infection.” Cell 151 (3): 590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack Denise M., Bouley Donna M., and Falkow Stanley. 2004. “Salmonella Typhimurium Persists within Macrophages in the Mesenteric Lymph Nodes of Chronically Infected Nramp1+/+ Mice and Can Be Reactivated by IFNgamma Neutralization.” The Journal of Experimental Medicine 199 (2): 231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottram Lynda, Speak Anneliese O., Selek Reza M., Cambridge Emma L., McIntyre Zoe, Kane Leanne, Mukhopadhyay Subhankar, et al. 2016. “Infection Susceptibility in Gastric Intrinsic Factor (Vitamin B12)-Defective Mice Is Subject to Maternal Influences.” mBio 7 (3). 10.1128/mBio.00830-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Toni A., Moreland Sarah M., Andrews-Polymenis Helene, and Detweiler Corrella S.. 2013. “The Ferric Enterobactin Transporter Fep Is Required for Persistent Salmonella Enterica Serovar Typhimurium Infection.” Infection and Immunity 81 (11): 4063–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson Kourtney P., Homer Craig R., Kessler Sean P., Dixon Laura J., Kabi Amrita, Gordon Ilyssa O., Johnson Erin E., de la Motte Carol A., and McDonald Christine. 2014. “The Dietary Polysaccharide Maltodextrin Promotes Salmonella Survival and Mucosal Colonization in Mice.” PloS One 9 (7): e101789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson Olof R., Kari Laszlo, and Steele-Mortimer Olivia. 2019. “Foodborne Infection of Mice with Salmonella Typhimurium.” PloS One 14 (8): e0215190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio Sean-Paul, and Bäumler Andreas J.. 2014. “Comparative Analysis of Salmonella Genomes Identifies a Metabolic Network for Escalating Growth in the Inflamed Gut.” mBio 5 (2): e00929–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer Alexander D., and Slauch James M.. 2017. “Mechanisms of Salmonella Pathogenesis in Animal Models.” Human and Ecological Risk Assessment: HERA 23 (8): 1877–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Lopez Araceli, Behnsen Judith, Nuccio Sean-Paul, and Raffatellu Manuela. 2016. “Mucosal Immunity to Pathogenic Intestinal Bacteria.” Nature Reviews. Immunology 16 (3): 135–48. [DOI] [PubMed] [Google Scholar]
- Perez-Lopez Araceli, Nuccio Sean-Paul, Ushach Irina, Edwards Robert A., Pahu Rachna, Silva Steven, Zlotnik Albert, and Raffatellu Manuela. 2019. “CRTAM Shapes the Gut Microbiota and Enhances the Severity of Infection.” Journal of Immunology 203 (2): 532–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson Jenny, and Vance Russell E.. 2007. “Genetics-Squared: Combining Host and Pathogen Genetics in the Analysis of Innate Immunity and Bacterial Virulence.” Immunogenetics 59 (10): 761–78. [DOI] [PubMed] [Google Scholar]
- Porwollik Steffen, Santiviago Carlos A., Cheng Pui, Long Fred, Desai Prerak, Fredlund Jennifer, Srikumar Shabarinath, et al. 2014. “Defined Single-Gene and Multi-Gene Deletion Mutant Collections in Salmonella Enterica Sv Typhimurium.” PloS One 9 (7): e99820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu Manuela, Santos Renato L., Verhoeven David E., George Michael D., Wilson R. Paul, Winter Sebastian E., Godinez Ivan, et al. 2008. “Simian Immunodeficiency Virus-Induced Mucosal Interleukin-17 Deficiency Promotes Salmonella Dissemination from the Gut.” Nature Medicine 14 (4): 421–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chávez Fabian, Zhang Lillian F., Faber Franziska, Lopez Christopher A., Byndloss Mariana X., Olsan Erin E., Xu Gege, et al. 2016. “Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella.” Cell Host & Microbe 19 (4): 443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers Andrew W. L., Tsolis Renée M., and Bäumler Andreas J.. 2021. “Salmonella versus the Microbiome.” Microbiology and Molecular Biology Reviews: MMBR 85 (1). 10.1128/MMBR.00027-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout WR, Formal SB, Dammin GJ, and Giannella RA. 1974. “Pathophysiology of Salmonella Diarrhea in the Rhesus Monkey: Intestinal Transport, Morphological and Bacteriological Studies.” Gastroenterology 67 (1): 59–70. [PubMed] [Google Scholar]
- Roux Christelle M., Butler Brian P., Chau Jennifer Y., Paixao Tatiane A., Cheung Kong Wai, Santos Renato L., Luckhart Shirley, and Tsolis Renée M.. 2010. “Both Hemolytic Anemia and Malaria Parasite-Specific Factors Increase Susceptibility to Nontyphoidal Salmonella Enterica Serovar Typhimurium Infection in Mice.” Infection and Immunity 78 (4): 1520–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby Thomas, McLaughlin Laura, Gopinath Smita, and Monack Denise. 2012. “Salmonella’s Long-Term Relationship with Its Host.” FEMS Microbiology Reviews 36 (3): 600–615. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garrido Julia, Ruano-Gallego David, Choudhary Jyoti S., and Frankel Gad. 2021. “The Type III Secretion System Effector Network Hypothesis.” Trends in Microbiology, November. 10.1016/j.tim.2021.10.007. [DOI] [PubMed] [Google Scholar]
- Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, and Bäumler AJ. 2001. “Animal Models of Salmonella Infections: Enteritis versus Typhoid Fever.” Microbes and Infection / Institut Pasteur 3 (14–15): 1335–44. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi Martina, Nuccio Sean-Paul, Liu Henry, Hernandez Dulcemaria, Vu Christine T., Takahashi Amy A., Edwards Robert A., and Raffatellu Manuela. 2016. “Microcins Mediate Competition among Enterobacteriaceae in the Inflamed Gut.” Nature 540 (7632): 280–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon Raphael, Tennant Sharon M., Galen James E., and Levine Myron M.. 2011. “Mouse Models to Assess the Efficacy of Non-Typhoidal Salmonella Vaccines: Revisiting the Role of Host Innate Susceptibility and Routes of Challenge.” Vaccine 29 (32): 5094–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Jeongmin, Wilhelm Cara L., Wangdi Tamding, Maira-Litran Tomas, Lee Seung-Joo, Raetz Megan, Sturge Carolyn R., et al. 2016. “Absence of TLR11 in Mice Does Not Confer Susceptibility to Salmonella Typhi.” Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Jeongmin, Willinger Tim, Rongvaux Anthony, Eynon Elizabeth E., Stevens Sean, Manz Markus G., Flavell Richard A., and Galán Jorge E.. 2010. “A Mouse Model for the Human Pathogen Salmonella Typhi.” Cell Host & Microbe 8 (4): 369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher Bärbel. 2021. “Establishing Causality in Salmonella-Microbiota-Host Interaction: The Use of Gnotobiotic Mouse Models and Synthetic Microbial Communities.” International Journal of Medical Microbiology: IJMM 311 (3): 151484. [DOI] [PubMed] [Google Scholar]
- Stecher Bärbel, Chaffron Samuel, Käppeli Rina, Hapfelmeier Siegfried, Freedrich Susanne, Weber Thomas C., Kirundi Jorum, et al. 2010. “Like Will to like: Abundances of Closely Related Species Can Predict Susceptibility to Intestinal Colonization by Pathogenic and Commensal Bacteria.” PLoS Pathogens 6 (1): e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher Bärbel, Macpherson Andrew J., Hapfelmeier Siegfried, Kremer Marcus, Stallmach Thomas, and Hardt Wolf-Dietrich. 2005. “Comparison of Salmonella Enterica Serovar Typhimurium Colitis in Germfree Mice and Mice Pretreated with Streptomycin.” Infection and Immunity 73 (6): 3228–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher Bärbel, Paesold Günther, Barthel Manja, Kremer Marcus, Jantsch Jonathan, Stallmach Thomas, Heikenwalder Mathias, and Hardt Wolf-Dietrich. 2006. “Chronic Salmonella Enterica Serovar Typhimurium-Induced Colitis and Cholangitis in Streptomycin-Pretreated Nramp1+/+ Mice.” Infection and Immunity 74 (9): 5047–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher Bärbel, Robbiani Riccardo, Walker Alan W., Westendorf Astrid M., Barthel Manja, Kremer Marcus, Chaffron Samuel, et al. 2007. “Salmonella Enterica Serovar Typhimurium Exploits Inflammation to Compete with the Intestinal Microbiota.” PLoS Biology. 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stull-Lane Annica R., Lokken-Toyli Kristen L., Diaz-Ochoa Vladimir E., Walker Gregory T., Cevallos Stephanie A., Winter Andromeda L. N., Del Hoyo Muñoz Ariel, et al. 2020. “Vitamin A Supplementation Boosts Control of Antibiotic-Resistant Salmonella Infection in Malnourished Mice.” PLoS Neglected Tropical Diseases 14 (10): e0008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis Renée M., Xavier Mariana N., Santos Renato L., and Bäumler Andreas J.. 2011. “How to Become a Top Model: Impact of Animal Experimentation on Human Salmonella Disease Research.” Infection and Immunity 79 (5): 1806–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez Eric M., Nguyen Henry, Heasley Keaton T., Saechao Cheng H., Gil Lindsey M., Rogers Andrew W. L., Miller Brittany M., et al. 2019. “Endogenous Enterobacteriaceae Underlie Variation in Susceptibility to Salmonella Infection.” Nature Microbiology 4 (6): 1057–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal SM, Pinner E, Lepage P, Gauthier S, and Gros P. 1996. “Natural Resistance to Intracellular Infections: Nramp1 Encodes a Membrane Phosphoglycoprotein Absent in Macrophages from Susceptible (Nramp1 D169) Mouse Strains.” Journal of Immunology 157 (8): 3559–68. [PubMed] [Google Scholar]
- Wangdi Tamding, Lee Cheng-Yuk, Spees Alanna M., Yu Chenzhou, Kingsbury Dawn D., Winter Sebastian E., Hastey Christine J., Wilson R. Paul, Heinrich Volkmar, and Bäumler Andreas J.. 2014. “The Vi Capsular Polysaccharide Enables Salmonella Enterica Serovar Typhi to Evade Microbe-Guided Neutrophil Chemotaxis.” PLoS Pathogens 10 (8): e1004306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. Paul, Raffatellu Manuela, Chessa Daniela, Winter Sebastian E., Tükel Cagla, and Bäumler Andreas J.. 2008. “The Vi-Capsule Prevents Toll-like Receptor 4 Recognition of Salmonella.” Cellular Microbiology 10 (4): 876–90. [DOI] [PubMed] [Google Scholar]
- Wilson R. Paul, Winter Sebastian E., Spees Alanna M., Winter Maria G., Nishimori Jessalyn H., Sanchez Jesus F., Nuccio Sean-Paul, Crawford Robert W., Tükel Çagla, and Bäumler Andreas J.. 2011. “The Vi Capsular Polysaccharide Prevents Complement Receptor 3-Mediated Clearance of Salmonella Enterica Serotype Typhi.” Infection and Immunity 79 (2): 830–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter Sebastian E., and Bäumler Andreas J.. 2014. “Dysbiosis in the Inflamed Intestine: Chance Favors the Prepared Microbe.” Gut Microbes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter Sebastian E., Winter Maria G., Poon Victor, Keestra A. Marijke, Sterzenbach Torsten, Faber Franziska, Costa Luciana F., et al. 2014. “Salmonella Enterica Serovar Typhi Conceals the Invasion-Associated Type Three Secretion System from the Innate Immune System by Gene Regulation.” PLoS Pathogens 10 (7): e1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotzka Sandra Y., Kreuzer Markus, Maier Lisa, Arnoldini Markus, Nguyen Bidong D., Brachmann Alexander O., Berthold Dorothée L., et al. 2019. “Escherichia Coli Limits Salmonella Typhimurium Infections after Diet Shifts and Fat-Mediated Microbiota Perturbation in Mice.” Nature Microbiology 4 (12): 2164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]


