Abstract
Background
Breakthrough infections with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants are increasingly observed in vaccinated individuals. Immune responses towards SARS-CoV-2 variants, particularly Omicron-BA.5, are poorly understood. We investigated the humoral and cellular immune responses of hospitalized COVID-19 patients during Delta and Omicron infection waves.
Methods
The corresponding SARS-CoV-2 variant of the respective patients were identified by whole genome sequencing. Humoral immune responses were analyzed by ELISA and a cell culture-based neutralization assay against SARS-CoV-2 D614G isolate (wildtype), Alpha, Delta (AY.43) and Omicron (BA.1 and BA.5). Cellular immunity was evaluated with an IFN-γ ELISpot assay.
Results
On a cellular level, patients showed a minor IFN-γ response after stimulating PBMCs with mutated regions of SARS-CoV-2 variants. Neutralizing antibody titers against Omicron-BA.1 and especially BA.5 were strongly reduced. Double-vaccinated patients with Delta breakthrough infection showed a significantly increased neutralizing antibody response against Delta compared to double-vaccinated uninfected controls (median complete neutralization titer (NT100) 640 versus 80, p<0.05). Omicron-BA.1 infection increased neutralization titers against BA.1 in double-vaccinated patients (median NT100 of 160 in patients versus 20 in controls, p=0.07) and patients that received booster vaccination (median NT100 of 50 in patients versus 20 in controls, p=0.68). For boosted patients with BA.5 breakthrough infection, we found no enhancing effect on humoral immunity against SARS-CoV-2 variants.
Conclusion
Neutralizing antibody titers against Omicron-BA.1 and especially BA.5 were strongly reduced in SARS-CoV-2 breakthrough infections. Delta and Omicron-BA.1 but not Omicron-BA.5 infections boosted the humoral immunity in double-vaccinated patients and patients with booster vaccination. Despite BA.5 breakthrough infection, those patients may still be vulnerable for reinfections with BA.5 or other newly emerging variants of concern.
Keywords: SARS-CoV-2, breakthrough infections, Omicron, Delta, COVID-19
1. Introduction
Since the beginning of the coronavirus disease 2019 (COVID-19) pandemic, more than 700 million people worldwide have been infected with severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) and about seven million people have died as a result of COVID-19 (1). In an attempt to reduce the number of SARS-CoV-2 infections and severe COVID-19 cases, SARS-CoV-2 vaccines have been effectively deployed. The mRNA vaccines Comirnaty (BioNTech/Pfizer) and Spikevax (Moderna) have been administered most frequently in Germany, followed by Vaxzevria (AstraZeneca), Janssen (Johnson & Johnson) and Nuvaxovid (Novavax) (2). In particular, SARS-CoV-2 mRNA vaccines effectively protect against SARS-CoV-2 infection and severe COVID-19 (3, 4).
Throughout the COVID-19 pandemic, highly transmissible variants of concern (VOCs) have emerged, harboring multiple immune-escape mutations towards the available vaccines (5). By the end of 2021, the Omicron (B.1.1.529) variant displaced the Delta (B.1.617.2) variant as the leading VOC in Germany (6). Monoclonal antibodies as well as sera from vaccinated individuals are less effective in neutralizing Delta and Omicron compared to the D614G ancestral strain, with Omicron exhibiting the strongest immune evasiveness (7, 8). Despite the reduced neutralization capacity of vaccine-induced antibodies against these SARS-CoV-2 variants and the resulting increase of breakthrough infections among vaccinated individuals, most of the individuals with SARS-CoV-2 breakthrough infections were still protected against a lethal disease course (9–11). However, the humoral and cellular immune responses towards Omicron sub-variants BA.1 and in particular BA.5 are poorly understood.
In the present study, we assessed the humoral and cellular immune response in a group of patients hospitalized with SARS-CoV-2 breakthrough infection during Delta and Omicron infection waves. Our study sheds light on the extent of immune recall during breakthrough infection with Delta and Omicron-BA.1 and BA.5 in hospitalized patients and whether these infections provide a variant-specific immune boost or even cross-protective immunity.
2. Materials and methods
2.1. Study population
The study population consisted of 52 patients with a PCR-confirmed SARS-CoV-2 breakthrough infection hospitalized at the University Hospital Essen and a control group of 28 people without verified SARS-CoV-2 infection (Table 1). In total, 25 patients were infected with Delta, 15 with Omicron-BA.1 and 12 with Omicron-BA.5. The majority of Delta-infected patients were double vaccinated at the time of sample collection (88%). Patients with Omicron-BA.1 infection were predominantly double (53.3%) and triple (40%) vaccinated. All patients with Omicron-BA.5 infection was boosted, either with one booster dose (83.3%) or two booster doses (16.7%). Of the control group, 16 individuals were double vaccinated (57.1%), 10 were triple vaccinated (35.7%) and two were quadruple vaccinated (7.1%). Based on the definition of disease severity of COVID-19 by the World Health Organization (WHO), 42.2% of the patients had a non-severe course of COVID-19, 51.9% a severe course and 5.8% a non-severe course (12). Patient samples were collected from August 2021 to July 2022. Nasopharyngeal swabs and blood samples were collected to characterize the corresponding SARS-CoV-2 strain and the humoral and cellular immunity. Breakthrough infections were classified as Delta or Omicron based on sequencing information as well as information about infection waves from healthcare workers and patients at the University Hospital Essen (13).
Table 1.
Overview of study cohort. Data indicate median (interquartile range) or absolute numbers (percentage).
| Characteristics | Patients with Delta breakthrough infection (N=25) | Patients with Omicron-BA.1 breakthrough infection (N=15) | Patients with Omicron-BA.5 breakthrough infection (N=12) | Uninfected controls (N=28) | p |
|---|---|---|---|---|---|
| Sex: | |||||
| Men (%) | 17 (68) | 10 (66.7) | 4 (33.3) | 14 (50) | n.s. |
| Women (%) | 8 (32) | 5 (33.3) | 8 (66.7) | 14 (50) | |
| Age: | |||||
| Total | 73 (51-82) | 60 (55-77) | 69 (62-79) | 53 (49-63) | C-BA.5 and C-D: p<0.01 |
| 2 doses of vaccine | 71 (49-83) | 58 (44-63) | N/A | 52 (48-64) | C-D: p=0.0457 |
| Booster vaccination | 71 (60-81) | 74 (55-82) | 69 (62-79) | 54 (50-61) | C-BA.1 and C-BA.5: p<0.01 |
| Vaccine: | |||||
| Comirnaty® (BioNTech/Pfizer) (%) | 23 (92) | 8 (53.3) | 6 (50) | 15 (53.6) | D-BA.1, D-BA.5 and C-D: p<0.01 |
| Spikevax® (Moderna) (%) | 1 (4) | 2 (13.3) | 0 | 12 (42.9) | C-BA.5 and C-D: p<0.01 |
| Janssen® (Johnson & Johnson) (%) | 1 (4) | 0 | 0 | 0 | n.s. |
| Combination (%) | 0 | 5 (33.3) | 5 (41.7) | 1 (3.6) | D-BA-1, D-BA-5 and C-BA.5: p<0.01; C-BA.1: p<0.05 |
| Unknown (%) | 0 | 0 | 1 (8.3) | 0 | n.s. |
| Vaccine doses: | |||||
| 1 (%) | 1 (4) | 1 (6.7) | 0 | 0 | n.s. |
| 2 (%) | 22 (88) | 8 (53.3) | 0 | 16 (57.1) | D-BA.1 and C-D: p<0.05; D-BA.5: p<0.0001; BA.1-BA.5: 0.01; C-BA.5: p<0.001 |
| 3 (%) | 2 (8) | 6 (40) | 10 (83.3) | 10 (35.7) | D-BA.1, BA.1-BA.5, C-D and C-BA.5: p<0.05; D-BA.5: p<0.0001 |
| 4 (%) | 0 | 0 | 2 (16.7) | 2 (7.1) | n.s. |
| Days since vaccination: | |||||
| Total | 149.5 (97-184.3) | 134.5 (66.25-192.5) | 184.5 (133-222.5) | 186 (45.75-199.5) | n.s. |
| Since 2nd vaccination | 160 (113-188) | 176 (90.5-229.3) | N/A | 54 (29-186) | D-C: p=0.0414 |
| Since booster | 96.5 (91-102) | 69 (46-140) | 184.5 (133-222.5) | 199 (192.3-208) | D-BA.5, BA.1-BA.5 and D-C: p<0.05; C-BA.1: p<0.01 |
| Unknown | 7 | 1 | 4 | 0 | |
Data indicate median (interquartile range) or absolute numbers (percentage).
Differences between groups for the categorical variables were analyzed by Fisher’s exact test and for the continuous variables by two-tailed Mann-Whitney U test. N/A, not applicable; D, Delta; C, uninfected control; n.s., not significant.
The study was approved by the local ethics committee and was performed in accordance with the ethical standards noted in the 1964 Declaration of Helsinki and its later amendments or comparable ethics standards (approval no. 20-9665-BO). Informed consent was obtained from all participants in the study.
2.2. Cells and viruses
A549-AT cells were cultivated in minimum essential media (MEM), supplemented with 10% (v/v) fetal calf serum (FCS), penicillin (100 IU/mL) and streptomycin (100 µg/mL) at 37 °C in an atmosphere of 5% CO2 (all Life Technologies Gibco, Darmstadt, Germany) (14). These cells overexpress both the carboxypeptidase angiotensin I converting enzyme 2 (ACE2) receptor and the cellular transmembrane protease serine 2 (TMPRSS2), allowing for high SARS-CoV-2 susceptibility and formation of cytopathic effects (CPEs).
Nasopharyngeal swabs from COVID-19 patients were used to isolate variants of SARS-CoV-2 (15, 16). In brief, the swab medium was incubated on A549-AT cells for several days until a profound CPE became apparent. Subsequently, supernatant was harvested, cleared from cell debris by centrifugation and stored at -80°. Viral titers were determined using A549-AT cells by a standard end-point dilution assay and calculated as 50% tissue culture infective dose (TCID50)/mL as previously described (17).
2.3. Sequencing and phylogenetic characterization
SARS-CoV-2 RNA of cell culture supernatants and nasopharyngeal swabs was purified using the QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany). SARS-CoV-2 whole genome libraries were obtained with the EasySeq™ SARS-CoV-2 Whole Genome NGS Sequencing kit (Nimagen, Nijmegen, Netherlands) after cDNA generation from 5.5 µl of viral RNA with the LunaScript RT SuperMix Kit (NEB). Pooled and normalized libraries were sequenced on an Illumina MiSeq instrument employing the V2 chemistry (300 cycles).
Data analysis was conducted by the opensource pipeline UnCoVar (18). Briefly, UnCoVar performs a series of QC steps, initially attempts de-novo assembly with reference guided scaffolding to achieve full genome reconstruction. Alternatively, the genome of recalcitrant samples is generated via incorporation of observed mutations to the Wuhan reference genome using variants called with Freebayes (19), Delly (20) and Varlociraptor (21). The workflow subsequently uses Pangolin (22) for genome lineage calling and Kallisto (23) for read based matching to 24 (25).
After obtaining whole genome sequences, sub-sequences were extracted according to the observed genomic features of the Wuhan reference genomes. For the selected features, e.g., the spike (S) protein coding region, as well as for the whole genome, sequences were aligned [mafft] and phylogenetic trees were calculated [iqtree] to obtain the evolutionary correlations between the samples.
2.4. SARS-CoV-2 S and NCP ELISA
IgG antibodies against subunit 1 of the SARS-CoV-2 S protein (S1; Wuhan-Hu-1 isolate) and IgG and IgM antibodies against the nucleocapsid protein (NCP) were measured from patient sera with an enzyme-linked immunosorbent assay (ELISA) (Euroimmun Medizinische Labordiagnostika, Lübeck, Germany). A ratio between the absorbance of the sample and calibrator of <0.8 was regarded as negative, ≥0.8 to <1.1 borderline, and ≥1.1 positive.
2.5. Neutralization Assay on A549-AT cells
The neutralization capacity of serum samples against a SARS-CoV-2 clinical isolate from September 2020 with the D614G mutation (wildtype) as well as the variants Alpha (B.1.1.7), Delta (AY.43) and Omicron (BA.1 and BA.5) was analyzed. Additionally, the neutralizing capacity of sera from ten patients was investigated (patient 1, 6, 9, 10, 24, 27, 33, 45, 48, 52) towards their equivalent clinical isolate that caused the SARS-CoV-2 breakthrough infection in comparison to wildtype isolate.
Neutralization assays were conducted as described previously (26). Briefly, two-fold serial dilutions of patient sera (1:20 to 1:2560) were pre-incubated with 100 TCID50/50 µL SARS-CoV-2 for one hour at 37 °C. These mixtures were added to A549-AT cells and incubated for three days at 37 °C and 5% CO2. Cell cultures were stained with 0.5% crystal violet (w/v) (Roth, Karlsruhe, Germany), solved in 20% (v/v) methanol (Merck, Darmstadt, Germany) and evaluated for CPEs by transmitted light microscopy. The highest serum dilution at which none of the triplicate cultures displayed CPEs was defined as the complete neutralization titer (NT100).
2.6. ELISpot Assay for SARS-CoV-2 S and NCP
An IFN-γ enzyme-linked-immuno-spot (ELISpot) assay was conducted to evaluate the cell-mediated immune response to SARS-CoV-2, as described before (27, 28). Plates equipped with polyvinylidene difluoride (PVDF) membranes (MilliporeSigma™ MultiScreen™ HTS, Fisher Scientific, Schwerte, Germany) were activated with ethanol. Subsequently, plates were coated with 60 µL monoclonal antibodies against IFN-y (10 µg/mL of clone 1-D1K, Mabtech, Nacka, Sweden). After washing and blocking with 150 µL AIM-V® (Thermo Fisher Scientific, Grand Island, NY, USA) for 30 minutes at 37 °C, 250,000 peripheral blood mononuclear cells (PBMCs) in 150 µL of AIM-V® in the presence or absence of PepTivator® proteins (600 pmol/mL of each peptide, all Miltenyi Biotec, Bergisch Gladbach, Germany) were added. The NCP, S protein of Wuhan wildtype and selectively mutated regions of Alpha (B.1.1.7), Delta (AY.1) and Omicron (B.1.1.529) were incubated for 19 hours at 37 °C followed by washing. To detect captured IFN-y, 50 µL alkaline phosphatase conjugated monoclonal antibody against IFN-y (clone 7-B6-1, Mabtech) diluted 1:200 in PBS containing 0.5% bovine serum albumin (BSA) was incubated for one hour. Plates were washed again, and nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate was added. Spots were quantified using an ELISpot reader (AID Fluorospot, Autoimmun Diagnostika GmbH, Strassberg, Germany). Non-stimulated values were subtracted from stimulated values to obtain the SARS-CoV-2 specific spots. A spot increment of three was considered positive.
2.7. Statistical analyses
Statistical analyses and data visualization were conducted using GraphPad Prism 9.4.0 (San Diego, CA, USA) software. For continuous variables, the median and interquartile range were calculated. Significant differences were assessed using Kruskal-Wallis test with post-hoc Dunn’s test for multiple comparison, Mann-Whitney U test and Wilcoxon signed-rank test for analyses of more than two independent groups, two unpaired samples and two paired samples, respectively. Categorical variables were analyzed by Fisher’s exact test. Correlation coefficients were calculated using Spearman’s rank analysis. P-values <0.05 were considered significant.
3. Results
3.1. Sequencing and phylogenetic analysis of SARS-CoV-2 variants causing breakthrough infections
At the time of sample collection, all study participants had received at least one vaccine dose. Of the control group, 57.1% individuals were double-vaccinated, 35.7% were triple-vaccinated and 7.1% were quadruple-vaccinated. 88% of Delta-infected patients were double vaccinated. Of the patients with Omicron-BA.1 infection, 53.3% were double and 40% triple vaccinated. Omicron-BA.5 infected patients were all boosted with either one booster dose (83.3%) or two booster doses (16.7%).
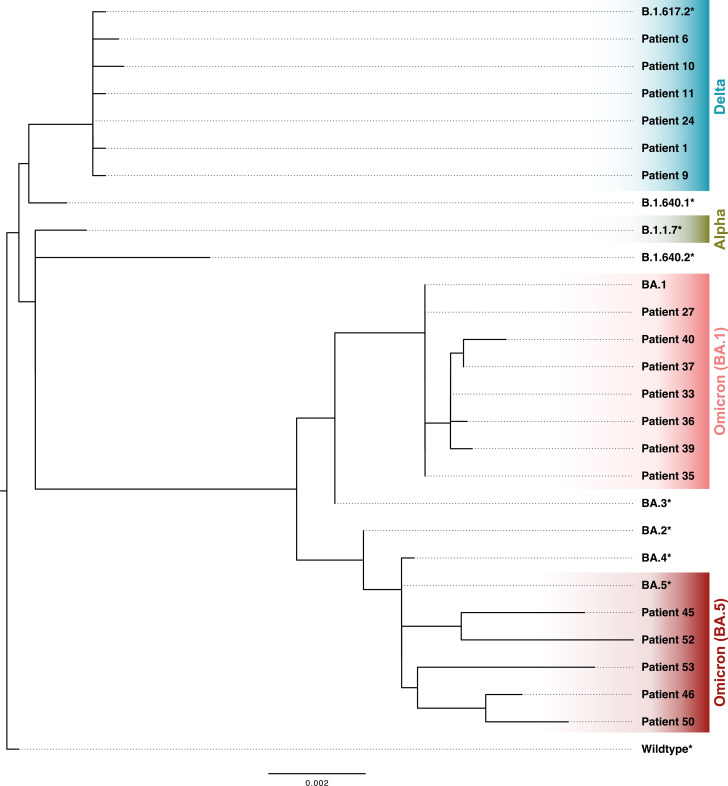
Clinical isolates of hospitalized patients with SARS-CoV-2 breakthrough infection were sequenced by whole genome sequencing (Figure 1). S region sequences were successfully assembled from 18 patients. These patients were infected with Delta (B.1.617.2) and Omicron sub-lineages BA.1 and BA.5 (Figure 2). The remaining patients were classified based on information about infection waves from healthcare workers and patients at the University Hospital Essen (13). The phylogenetic analysis highlights the continuous evolution of SARS-CoV-2, which poses a challenge for vaccine development.
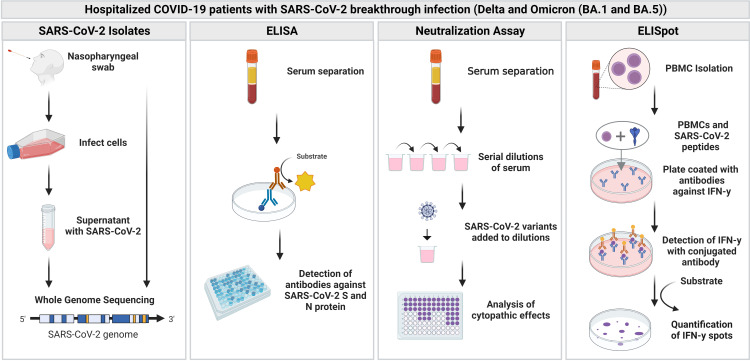
Figure 1.
Overview of the study. Blood samples and nasopharyngeal swabs were collected from hospitalized patients with SARS-CoV-2 breakthrough infection (Delta and Omicron). Blood samples were further analyzed with an enzyme-linked immunosorbent assay (ELISA), neutralization assay as well as enzyme-linked-immuno-spot (ELISpot) assay. Figure was created with BioRender.com.
Figure 2.
Phylogenetic tree of assembled SARS-CoV-2 spike (S) region sequences of clinical isolates of patients with SARS-CoV-2 breakthrough infection. * S region extracted from GISAID reference genomes (GISAID).
3.2. SARS-CoV-2 binding serum antibody levels
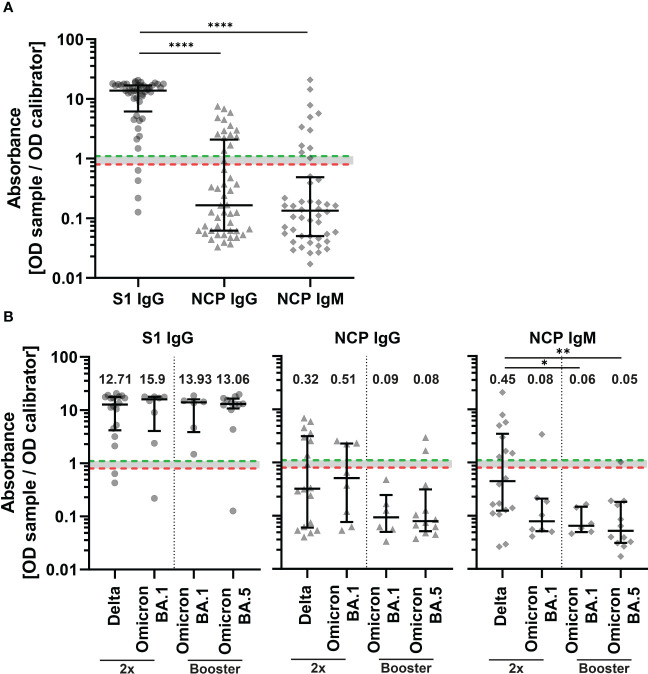
Sera of patients with SARS-CoV-2 breakthrough infection were tested for SARS-CoV-2 subunit 1 (S1) specific IgG antibodies and IgG and IgM antibodies against the nucleocapsid protein (NCP) by an enzyme-linked immunosorbent assay (ELISA) (Figure 1). Overall, 91.7% of samples were positive for S1 specific antibodies (Figure 3A). Next, we measured IgM and IgG antibody levels against NCP of SARS-CoV-2 to distinguish between the early and late humoral responses during infection. Antibody levels against the NCP were significantly lower compared to S1 (p<0.0001) (Figure 3A). In total, 20.8% of patient sera were positive for IgM antibodies and 29.2% for IgG antibodies. When dividing patients by breakthrough infection and number of vaccines, there were no significant differences in S1 and NCP IgG levels between groups (Figure 3B). However, patients with Delta breakthrough infection who received two vaccine doses had significantly higher levels of NCP IgM compared to patients with booster vaccination and Omicron BA.1 infection (p<0.05) as well as patients with booster vaccination and Omicron BA.5 infection (p<0.01).
Figure 3.
Binding serum antibody levels in COVID-19 patients with SARS-CoV-2 breakthrough infection. IgG antibodies against the subunit 1 of spike protein (S1) (Wuhan-Hu-1 isolate) and IgG and IgM antibodies against the nucleocapsid protein (NCP) of all patients (n=48) (A) and double vaccinated patients (2x) with Delta (n=18) and BA.1 (n=8) infection and patients with booster vaccination with BA.1 (n=6) and BA.5 (n=12) infection (B). Binding serum antibodies were measured with an enzyme-linked immunosorbent assay (ELISA). An absorbance of <0.8 was regarded as negative (red dotted line), ≥0.8 to <1.1 borderline, and ≥1.1 positive (green dotted line). Differences between groups were analyzed by Kruskal-Wallis test with post-hoc Dunn’s multiple comparisons test (* p<0.05; ** p<0.01, **** p<0.0001). Horizontal lines indicate median values, while error bars represent the interquartile range.
3.3. Neutralizing antibody titers in sera after Delta, BA.1 or BA.5 breakthrough infection
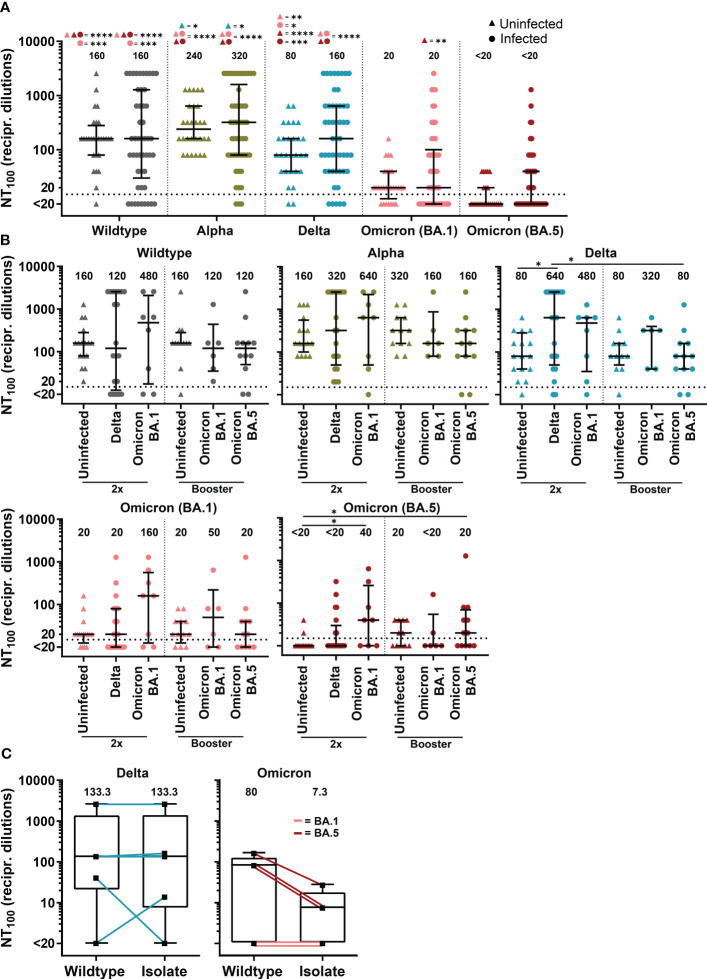
The humoral immunity of COVID-19 patients with SARS-CoV-2 breakthrough infections was further investigated using a cell culture-based neutralization assay. Serum samples from those patients were tested against a SARS-CoV-2 D614G wildtype clinical isolate and Alpha (B.1.1.7), Delta (AY.43) and the Omicron sub-lineages BA.1 and BA.5. Sera from COVID-19 patients as well as sera from non-infected but immunized individuals showed reduced complete neutralization titers (NT100) towards BA.1 and BA.5 compared to wildtype, Alpha and Delta (Figure 4A).
Figure 4.
Neutralizing antibody titers against SARS-CoV-2 variants of COVID-19 patients with SARS-CoV-2 breakthrough infection and uninfected, vaccinated controls. (A) Complete neutralization titer (NT100) against clinical isolate with D614G mutation (wildtype), Alpha (B.1.1.7), Delta (AY.43) and Omicron (BA.1 and BA.5) of patients with SARS-CoV-2 breakthrough infection (n=50) compared to vaccinated uninfected control (n=28). (B) NT100 against clinical isolate with D614G mutation (wildtype), Alpha (B.1.1.7), Delta (AY.43) and Omicron (BA.1 and BA.5) of double vaccinated patients (2x) with Delta (n=20) and BA.1 (n=8) infection and patients with booster vaccination with BA.1 (n=6) and BA.5 (n=12) infection compared to uninfected control with two vaccine doses (n=16) and booster vaccination (n=12). (A, B) Differences between groups were analyzed by Kruskal-Wallis test with post-hoc Dunn’s multiple comparisons test (* p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001). (C) NT100 of sera from patients with breakthrough infection with Delta and Omicron (BA.1 and BA.5) against their equivalent clinical isolate compared to wildtype. Differences between groups were analyzed by Wilcoxon signed-rank test. (A–C) Horizontal lines indicate median values, while error bars represent the interquartile range.
Double-vaccinated patients with Delta breakthrough infection displayed a significantly increased neutralizing antibody response against Delta compared to double-vaccinated uninfected controls (median NT100 640 versus 80, p<0.05, Figure 4B). In double-vaccinated patients, infection with Omicron sub-lineage BA.1 boosted immunity against BA.1 just above statistical significance (median NT100 of 160 in patients versus 20 in controls, p=0.07) as well as against BA.5 (median NT100 of 40 in patients versus <20 in controls, p<0.05) (Figure 4B). A higher median NT100 against BA.1 was also observed for boosted Omicron-BA.1 infected patient compared to boosted controls (median NT100 of 50 versus 20, p=0.68). Interestingly, results suggest cross-reactive immunity for patients with Omicron-BA.1 infection against Delta, as double-vaccinated had a 6-fold (median NT100 of 480 versus 80, p=0.24) and boosted a 4-fold (median NT100 of 320 versus 80, p=0.44) higher NT100 than control. For individuals with BA.5 infection, we observed no immune boost against BA.5 or other variants.
Next, we investigated neutralization capacity of patient sera against the SARS-CoV-2 clinical isolate from these respective patients compared to wildtype. In total, ten different SARS-CoV-2 isolates from patients infected with sub-lineages of Delta and Omicron could be propagated in cell culture to investigate the respective neutralizing antibody titers. Patients infected with Delta showed similar neutralization efficacy against their isolate compared to wildtype (Figure 4C). In contrast, we found reduced neutralization capacity against isolates from Omicron-infected patients in comparison to wildtype (median NT100 of 7.3 versus 80, p=0.25).
In summary, we found that Delta infections exhibit a strong immune boosting effect against the Delta variant. Patients infected with BA.1 showed an increased neutralizing antibody response against both tested Omicron variants. Compared to Delta and BA.1, BA.5 was the least immunogenic variant, as BA.5 infections did not boost immunity against BA.5 or other variants.
3.4. Cellular immunity in patients with SARS-CoV-2 breakthrough infection
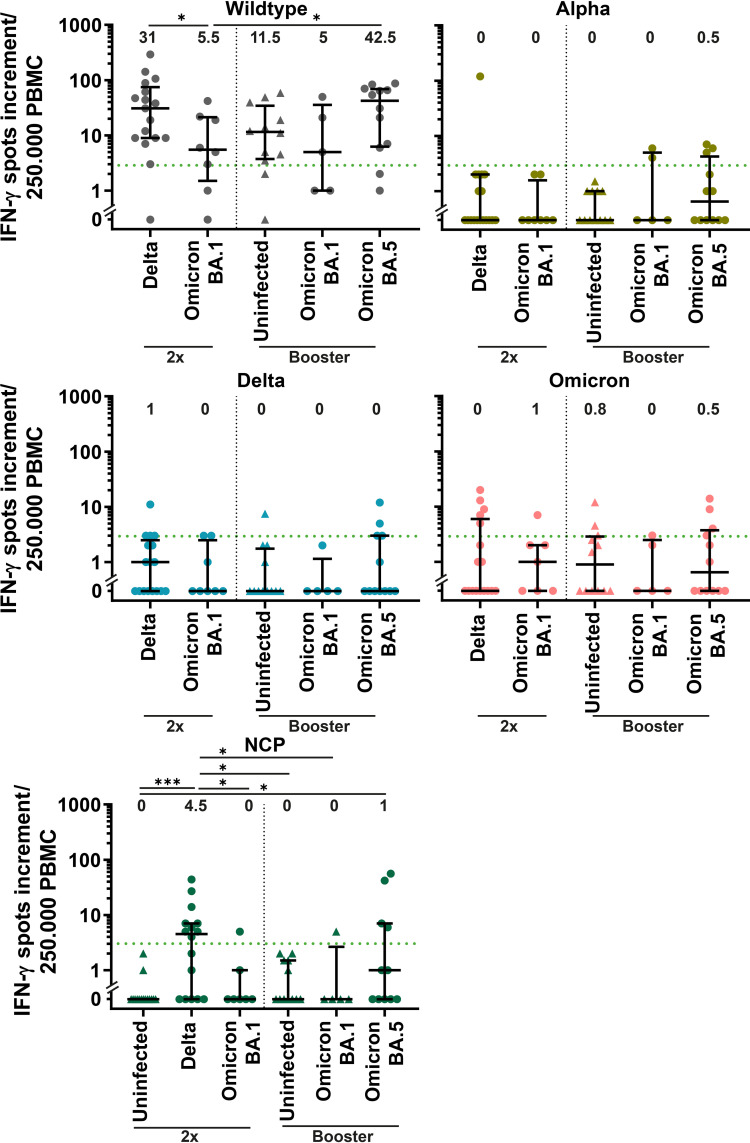
Cellular immunity against SARS-CoV-2 was measured using an IFN-γ enzyme-linked-immuno-spot (ELISpot) assay. We stimulated peripheral blood mononuclear cells (PBMCs) with the NCP, spike (S) protein of Wuhan wildtype and with selectively mutated regions of Alpha (B.1.1.7), Delta (AY.1) and Omicron (B.1.1.529). An IFN-γ-spots increment of three was considered positive.
Double-vaccinated patients with Delta infection showed the highest positivity in response to NCP stimulation, followed by boosted BA.5-infected patients (56.2% and 33.3%, respectively, Figure 5). As expected, infection-naïve participants did not show a positive NCP response. Among dually vaccinated patients, the IFN-γ spots increment was significantly higher for Delta-infected patients than for patients infected with BA.1 (31 versus 5.5, p<0.05) after stimulation with wildtype S protein. A significantly higher response to S wildtype was also observed for BA.5-infected patients compared to double-vaccinated BA.1-infected patients (42.5 versus 5.5, p<0.05). All groups showed a median IFN-γ spots increment below positivity to mutated regions of SARS-CoV-2 variants (Figure 5).
Figure 5.
Cellular response against SARS-CoV-2 variants in COVID-19 patients with SARS-CoV-2 breakthrough infection. Cellular immunity was assessed by an IFN-γ enzyme-linked-immuno-spot (ELISpot) assay using peripheral blood mononuclear cells (PBMCs) and is displayed for double vaccinated patients (2x) with Delta (n=18) and BA.1 (n=8) infection and patients with booster vaccination with BA.1 (n=5) and BA.5 (n=12) infection compared to uninfected control with two vaccine doses (n=15) and booster vaccination (n=12). PBMCs were stimulated with S protein of Wuhan wildtype, nucleocapsid protein (NCP) and selectively mutated regions Alpha (B.1.1.7), Delta (AY.1) and Omicron (B.1.1.529). A spot increment of three was considered positive (green dotted line). Differences between groups were analyzed by Kruskal-Wallis test with post-hoc Dunn’s multiple comparisons test (* p<0.05; *** p<0.001). Horizontal lines indicate median values, while error bars represent the interquartile range.
3.5. Correlation between SARS-CoV-2 ELISA IgG antibody levels and neutralizing antibody titers as well as cellular IFN-γ response
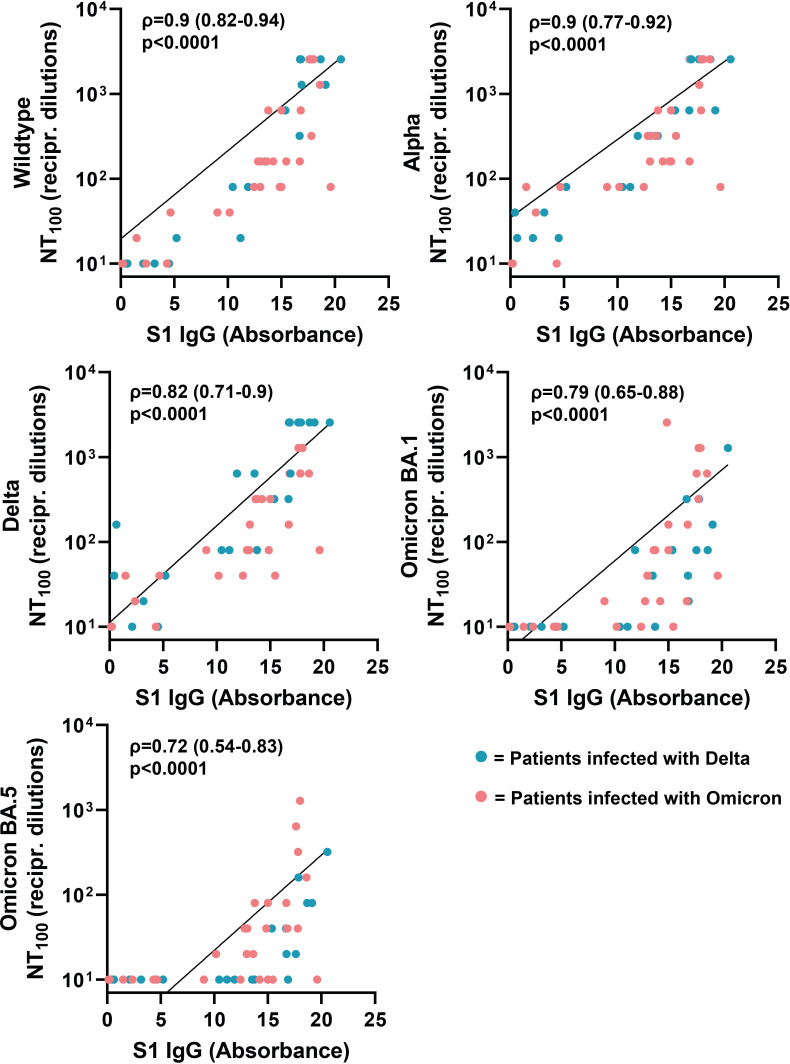
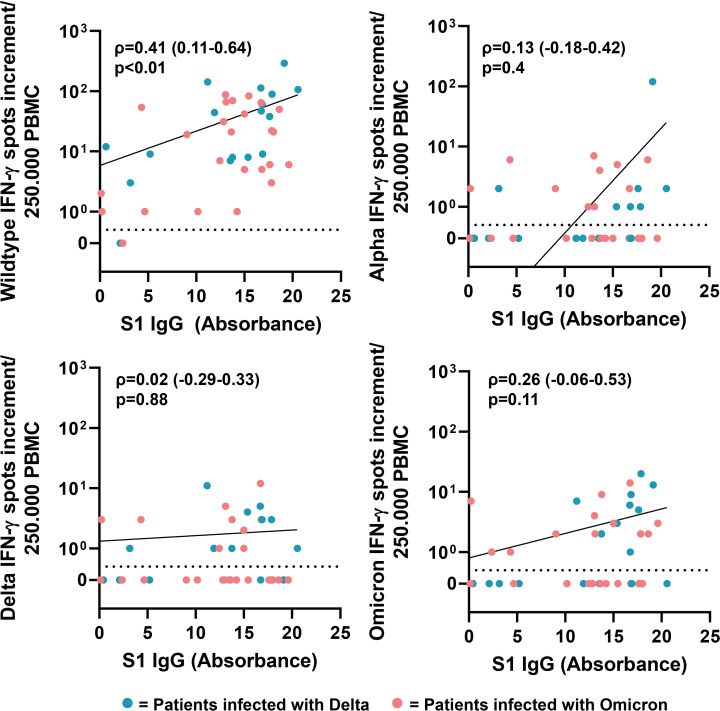
Next, we analyzed if there is a correlation between neutralizing antibody titers of the respective sera against SARS-CoV-2 wildtype, Alpha, Delta and Omicron (BA.1 and BA.5) and SARS-CoV-2 ELISA IgG antibody levels against S1 (Wuhan-Hu-1 isolate). The neutralizing antibody titers correlated positively with ELISA IgG antibody levels (Figure 6). The highest correlation was observed for neutralizing antibody titers against wildtype and Alpha (Spearman’s ρ=0.9, respectively). Compared to wildtype and Alpha, we observed a lower correlation for Delta, Omicron-BA-1 and Omicron-BA.5, with Spearman’s rank coefficients of 0.82, 0.79 and 0.72, respectively. Next, we analyzed the correlation between IgG antibodies against S1 and cellular IFN-γ production in response to stimulation with SARS-CoV-2 variants. The results only revealed a correlation between SARS-CoV-2 ELISA IgG antibody levels and the cellular immune response against wildtype (ρ=0.41), but not SARS-CoV-2 variants (Figure 7).
Figure 6.
Correlation between serum antibody levels and neutralizing antibody titers against SARS-CoV-2 variants. Correlation coefficients (ρ) and p-values were calculated using Spearman’s rank analysis.
Figure 7.
Correlation between serum antibody levels and cellular IFN-γ response after stimulation with SARS-CoV-2 variants. Correlation coefficients (ρ) and p-values were calculated using Spearman’s rank analysis.
4. Discussion
Throughout the COVID-19 pandemic, several SARS-CoV-2 variants with immune-escape mutations have emerged, leading to an increase of SARS-CoV-2 breakthrough infections (5). In the present study, we report on the humoral and cellular immunity in response to Delta and Omicron (BA.1 and BA.5) infection in a group of vaccinated patients with SARS-CoV-2 breakthrough infections. We compared the results to vaccinated uninfected controls, to assess the additive effect of the infection on immunity.
Of note, the neutralizing antibody titers against Omicron sub-variants BA.1 and especially BA.5 were strongly reduced when compared to Alpha, Delta or wildtype. These findings are consistent with recently published data using pseudovirus-neutralization assays, showing a substantial immune escape of BA.5 sub-variant against antibodies of vaccinated individuals or individuals infected with BA.1 or BA.2 (29–31).
In line with recent studies, we showed that Delta and Omicron-BA.1 infections lead to a strain-specific boost of neutralizing immunity (32, 33). Previous data indicated that Delta breakthrough infections increase Delta specific neutralization titers to levels comparable to wildtype neutralization (32). In our study, Delta infection markedly increased neutralizing antibody titers against Delta in double-vaccinated patients, even with a 5.3-fold higher neutralizing antibody titer against Delta compared to wildtype. Omicron-BA.1 breakthrough infection enhanced the neutralizing antibody titer against BA.1 and Delta (33). Notably, the neutralizing antibody titer of sera from uninfected controls was 8-fold reduced against BA.1 when compared to wildtype. In contrast, in double-vaccinated patients with BA.1 infection the ratio between BA.1 and wildtype neutralizing antibody titers reduced to 3 and in boosted patients to 2.4.
Additionally, our study provides insight into the immunity in BA.5 breakthrough infections. We found no evidence of a boosting effect on humoral immunity for this sub-variant, which could increase the likelihood of reinfections in people who have recovered from BA.5 infection. Our results suggest that BA.5 sub-variant is capable not only of bypassing humoral immunity boosted by SARS-CoV-2 infection, but also leads to a weak enhancement of humoral immunity itself. In contrast to our data, recent data indicated an enhanced neutralization against BA.5 following BA.5 infection in triple-vaccinated individuals (34). In the study by Wang et al. (34), serum samples were collected from already recovered patients at a mean of 32 days after infection, whereas in our study the sera were collected during the acute phase at hospitalization.
Interestingly, we found a weak IFN-γ response after stimulating PBMCs with selectively mutated regions of SARS-CoV-2 variants. One reason could be that the participants were still early in the infection and a measurable T-cell immunity against the mutated regions had not yet developed. Overall, all patient groups had a high positivity after cellular stimulation with S protein of Wuhan wildtype, regardless of vaccination status and variant responsible for breakthrough infection. PBMCs of patients with Omicron-BA.5 breakthrough infection showed the strongest IFN-γ response against Wuhan wildtype, followed by patients with Delta infection.
One limitation of this study are differences between cohorts regarding to demographic characteristics. For instance, among the BA.1-infected patients, 87.5% were under 70 years of age in the dually vaccinated group compared to 33.3% of boosted patients. That might be an explanation for the weaker humoral immune enhancement through BA-1 infection we observed in the group with booster vaccination. For instance, data has shown a reduced antibody neutralization response for elderly above 70 years after vaccination or infection (35, 36). Furthermore, the uninfected control group received a higher percentage of Spikevax (Moderna) vaccines than the patient groups, which could have influenced the results. However, studies found a similar high neutralization potential for individuals vaccinated with Spikevax (Moderna), Comirnaty (BioNTech/Pfizer) and a combination of vaccines (37).
In conclusion, we found strongly reduced neutralizing antibody titers against Omicron sub-variants BA.1 and BA.5. Furthermore, humoral immunity was boosted through Delta and Omicron-BA.1 infections in hospitalized double-vaccinated patients and patients with booster vaccination. This finding does not apply to BA.5 infections, in which we found no enhancing effect on humoral immunity. Despite BA.5 breakthrough infection, those patients may still be vulnerable for reinfections with BA.5 or other newly emerging SARS-CoV-2 variants. Further studies are needed to investigate the humoral and cellular immune response after breakthrough infection with BA.5 and its role in protecting from subsequent breakthrough infections.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJEB59607 (ENA; https://www.ebi.ac.uk/ena/browser/view/PRJEB59607).
Ethics statement
The studies involving human participants were reviewed and approved by Ethik-Kommission der Medizinischen Fakultät der Universität Duisburg-Essen. The patients/participants provided their written informed consent to participate in this study.
Author contributions
MB, LB, MA, MO, LT, LS, IK, AT, JG, PB, SC and MW performed the experiments. MB, LB, MA and MO were involved in sample collection. MB, AT, SD, UD, OW, FM and ML analyzed the data. AK, AS, HR and MB planned the study. AK and MB wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding Statement
This study was supported by the Stiftung Universitätsmedizin Essen (awarded to A. Krawczyk) and the Rudolf Ackermann Foundation (awarded to O. Witzke).
Acknowledgments
The work was in part supported by the cluster project ENABLE, the Innovation Center TheraNova, and the LOEWE Priority Program CoroPan funded by the Hessian Ministry for Science and the Arts (HMWK).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1. World Health Organization . WHO coronavirus (COVID-19) dashboard (2023). Available at: https://covid19.who.int/ (Accessed 6 June 2023).
- 2. Federal Ministry Of Health . Impfdashboard (2022). Available at: https://impfdashboard.de/ (Accessed 15 September 2022).
- 3. Chung H, He S, Nasreen S, Sundaram ME, Buchan SA, Wilson SE, et al. Effectiveness of BNT162b2 and mRNA-1273 covid-19 vaccines against symptomatic SARS-CoV-2 infection and severe covid-19 outcomes in Ontario, Canada: test negative design study. BMJ (2021) 374:n1943. doi: 10.2139/ssrn.3845993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ssentongo P, Ssentongo AE, Voleti N, Groff D, Sun A, Ba DM, et al. SARS-CoV-2 vaccine effectiveness against infection, symptomatic and severe COVID-19: a systematic review and meta-analysis. BMC Infect Dis (2022) 22:439. doi: 10.1186/s12879-022-07418-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dyson L, Hill EM, Moore S, Curran-Sebastian J, Tildesley MJ, Lythgoe KA, et al. Possible future waves of SARS-CoV-2 infection generated by variants of concern with a range of characteristics. Nat Commun (2021) 12:5730. doi: 10.1038/s41467-021-25915-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Robert Koch Institut . Besorgniserregende SARS-CoV-2-Virusvarianten (VOC). Available at: https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Virusvariante.html (Accessed 15 September 2022).
- 7. Shrestha LB, Tedla N, Bull RA. Broadly-neutralizing antibodies against emerging SARS-CoV-2 variants. Front Immunol (2021) 12. doi: 10.3389/fimmu.2021.752003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Planas D, Saunders N, Maes P, Guivel-Benhassine F, Planchais C, Buchrieser J, et al. Considerable escape of SARS-CoV-2 omicron to antibody neutralization. Nature (2022) 602:671–5. doi: 10.1038/s41586-021-04389-z [DOI] [PubMed] [Google Scholar]
- 9. Collie S, Champion J, Moultrie H, Bekker L-G, Gray G. Effectiveness of BNT162b2 vaccine against omicron variant in south Africa. New Engl J Med (2021) 386:494–6. doi: 10.1056/NEJMc2119270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang P, Hasan MR, Chemaitelly H, Yassine HM, Benslimane FM, Al Khatib HA, et al. BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 delta variant in Qatar. Nat Med (2021) 27:2136–43. doi: 10.1038/s41591-021-01583-4 [DOI] [PubMed] [Google Scholar]
- 11. Tseng HF, Ackerson BK, Luo Y, Sy LS, Talarico CA, Tian Y, et al. Effectiveness of mRNA-1273 against SARS-CoV-2 omicron and delta variants. Nat Med (2022) 28:1063–71. doi: 10.1038/s41591-022-01753-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. World Health Organization . Therapeutics and COVID-19 - living guideline (2022). Available at: https://apps.who.int/iris/bitstream/handle/10665/362843/WHO-2019-nCoV-therapeutics-2022.5-eng.pdf (Accessed 6 June 2023). [PubMed]
- 13. Möhlendick B, Čiučiulkaitė I, Elsner C, Anastasiou OE, Trilling M, Wagner B, et al. Individuals with weaker antibody responses after booster immunization are prone to omicron breakthrough infections. Front Immunol (2022) 13:907343. doi: 10.3389/fimmu.2022.907343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Widera M, Wilhelm A, Toptan T, Raffel JM, Kowarz E, Roesmann F, et al. Generation of a sleeping beauty transposon-based cellular system for rapid and sensitive screening for compounds and cellular factors limiting SARS-CoV-2 replication. Front Microbiol (2021) 12:701198–8. doi: 10.3389/fmicb.2021.701198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heilingloh CS, Aufderhorst UW, Schipper L, Dittmer U, Witzke O, Yang D, et al. Susceptibility of SARS-CoV-2 to UV irradiation. Am J Infect Control (2020) 48:1273–5. doi: 10.1016/j.ajic.2020.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bormann M, Alt M, Schipper L, Van De Sand L, Otte M, Meister TL, et al. Disinfection of SARS-CoV-2 contaminated surfaces of personal items with UVC-LED disinfection boxes. Viruses (2021) 13:1–7. doi: 10.3390/v13040598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krah DL. A simplified multiwell plate assay for the measurement of hepatitis a virus infectivity. Biologicals (1991) 19:223–7. doi: 10.1016/1045-1056(91)90039-M [DOI] [PubMed] [Google Scholar]
- 18. Uncovar . Available at: https://github.com/IKIM-Essen/uncovar (Accessed 8 September 2022).
- 19. Erik G, Marth G. Haplotype-based variant detection from short-read sequencing. (2012). doi: 10.48550/arXiv.1207.3907 [DOI] [Google Scholar]
- 20. Rausch T, Zichner T, Schlattl A, Stütz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics (2012) 28:i333–9. doi: 10.1093/bioinformatics/bts378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Köster J, Dijkstra LJ, Marschall T, Schönhuth A. Varlociraptor: enhancing sensitivity and controlling false discovery rate in somatic indel discovery. Genome Biol (2020) 21:98. doi: 10.1186/s13059-020-01993-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O’toole Á., Scher E, Underwood A, Jackson B, Hill V, Mccrone JT, et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol (2021) 7:veab064. doi: 10.1093/ve/veab064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol (2016) 34:525–7. doi: 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 24. GISAID . Available at: https://gisaid.org/hcov19-variants/ (Accessed 13 October 2022).
- 25. Khare S, Gurry C, Freitas L, Schultz MB, Bach G, Diallo A, et al. GISAID's role in pandemic response. China CDC Wkly (2021) 3:1049–51. doi: 10.46234/ccdcw2021.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lindemann M, Lenz V, Knop D, Klump H, Alt M, Aufderhorst UW, et al. Convalescent plasma treatment of critically ill intensive care COVID-19 patients. Transfusion (2021) 61:1394–403. doi: 10.1111/trf.16392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwarzkopf S, Krawczyk A, Knop D, Klump H, Heinold A, Heinemann FM, et al. Cellular immunity in COVID-19 convalescents with PCR-confirmed infection but with undetectable SARS-CoV-2–specific IgG. Emerg Infect Dis (2021) 27:122–9. doi: 10.3201/eid2701.203772 [DOI] [PubMed] [Google Scholar]
- 28. Wünsch K, Anastasiou OE, Alt M, Brochhagen L, Cherneha M, Thümmler L, et al. COVID-19 in elderly, immunocompromised or diabetic patients-from immune monitoring to clinical management in the hospital. Viruses (2022) 14:1–17. doi: 10.3390/v14040746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cao Y, Yisimayi A, Jian F, Song W, Xiao T, Wang L, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by omicron infection. Nature (2022) 608:593–602. doi: 10.1038/s41586-022-04980-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hachmann NP, Miller J, Collier AY, Ventura JD, Yu J, Rowe M, et al. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N Engl J Med (2022) 387:86–8. doi: 10.1056/NEJMc2206576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tuekprakhon A, Nutalai R, Dijokaite-Guraliuc A, Zhou D, Ginn HM, Selvaraj M, et al. Antibody escape of SARS-CoV-2 omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell (2022) 185:2422–2433.e13. doi: 10.1016/j.cell.2022.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Servellita V, Syed AM, Morris MK, Brazer N, Saldhi P, Garcia-Knight M, et al. Neutralizing immunity in vaccine breakthrough infections from the SARS-CoV-2 omicron and delta variants. Cell (2022) 185:1539–1548.e5. doi: 10.1016/j.cell.2022.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quandt J, Muik A, Salisch N, Lui BG, Lutz S, Krüger K, et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory b cell formation against conserved epitopes. Sci Immunol (2022) 7:eabq2427. doi: 10.1126/sciimmunol.abq2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Q, Bowen A, Valdez R, Gherasim C, Gordon A, Liu L, et al. Antibody response to omicron BA.4–BA.5 bivalent booster. New Engl J Med (2023) 388:567–9. doi: 10.1056/NEJMc2213907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Doria-Rose N, Suthar MS, Makowski M, O'connell S, Mcdermott AB, Flach B, et al. Antibody persistence through 6 months after the second dose of mRNA-1273 vaccine for covid-19. N Engl J Med (2021) 384:2259–61. doi: 10.1056/NEJMc2103916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Konik M, Lindemann M, Zettler M, Meller L, Dolff S, Rebmann V, et al. Long-term SARS-CoV-2 specific immunity is affected by the severity of initial COVID-19 and patient age. J Clin Med (2021) 10:1–17. doi: 10.3390/jcm10194606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adjobimey T, Meyer J, Sollberg L, Bawolt M, Berens C, Kovačević P, et al. Comparison of IgA, IgG, and neutralizing antibody responses following immunization with moderna, BioNTech, AstraZeneca, Sputnik-V, Johnson and Johnson, and sinopharm's COVID-19 vaccines. Front Immunol (2022) 13:917905. doi: 10.3389/fimmu.2022.917905 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: PRJEB59607 (ENA; https://www.ebi.ac.uk/ena/browser/view/PRJEB59607).