Abstract
Behavior genetics studies how genetic differences among people contribute to differences in their psychology and behavior. Here, I describe how the conclusions and methods of behavior genetics have evolved in the postgenomic era in which the human genome can be directly measured. First, I revisit the first law of behavioral genetics stating that everything is heritable, and I describe results from large-scale meta-analyses of twin data and new methods for estimating heritability using measured DNA. Second, I describe new methods in statistical genetics, including genome-wide association studies and polygenic score analyses. Third, I describe the next generation of work on gene × environment interaction, with a particular focus on how genetic influences vary across sociopolitical contexts and exogenous environments. Genomic technology has ushered in a golden age of new tools to address enduring questions about how genes and environments combine to create unique human lives.
Keywords: behavior genetics, twin studies, heritability, gene × environment interaction, genome-wide association study, polygenic scores
INTRODUCTION
“People vary in ability, energy, health, character, and other socially important traits, and there is good, though not absolutely conclusive, evidence that the variance of all these traits is in part genetically conditioned. Conditioned, mind you, not fixed or predestined.” This is how Theodosius Dobzhansky, the great evolutionary biologist, summarized the relationship between genetics and human behavior in an essay in Science in 1962 (Dobzhansky 1962, p. 112), and not much has happened in the intervening decades to prove him wrong. Genetics remains as essential to psychological science in the 2020s as it was in the 1960s, because all human psychological differences are conditioned (“conditioned, mind you, not fixed or predestined”) by one’s genes.
Yet, as in the 1960s, genetics remains controversial within psychological science, and within society in general, because it is widely misperceived as being a threat to cherished values about human equality and the potential to improve the human condition (Fox 2019, Panofsky 2014). As the bioethicist Erik Parens (2004, p. S31) described, “As long as creating an identity for ourselves entails specifying how we are different from others, a science of human differences will risk being appropriated to justify claims about why some enjoy more power than others.” Because of this risk, behavioral genetics has been characterized as “subversive science,” which can “shake the public’s faith in … democratic cornerstones,” including norms of human equality and personal responsibility (Fox 2019, p.153).
Reflecting the importance of behavioral genetics to psychology, many articles on this topic have appeared in the pages of the Annual Review of Psychology. And perhaps reflecting the perceived subversiveness of discussing genetic effects on behavior, most of the genetics papers published in this journal since 2010 have focused on the environmental moderation of genetic effects (e.g., Bakermans-Kranenburg & van IJzendoorn 2015, Halldorsdottir & Binder 2017, Manuck & McCaffery 2014, Sharma et al. 2016, Zhang & Meaney 2010). The emphasis of this article is different.
Here, I will review the major advances made in human behavioral genetics in the past decade, many of which have been made possible by the advent of cheap, noninvasive, genome-wide measurement of human DNA and by the formation of large, international scientific collaborations. On the one hand, these advances have ushered in a golden age of genetic research, with new discoveries and new tools arriving every day. On the other hand, these developments pose a serious challenge to the veracity of much of the published psychology literature, which has focused on methodologically flawed studies of candidate gene × environment interaction at the expense of more rigorous studies of genetic main effects. As a result, the field of psychology, which for much of the twentieth century was a leader in integrating genetics into the social sciences, now risks falling behind other disciplines in leveraging genetic technology to answer enduring questions about human development.
The article proceeds in three parts. First, I describe recent developments in the study of heritability: what we have learned from twin studies and about twin studies in the past decade. I further describe newer methods of estimating heritability from genomic data, which have raised new mysteries about the relationship between genotypes and phenotypes—the question of missing and hiding heritability. Second, I describe methodological advances in the study of measured DNA that have emerged in the postgenomic era, in particular, genome-wide association studies (GWASs). As we will see, results from GWASs have definitively undermined the validity of vast swathes of psychological research that claimed to find interactions between genetic variants and environmental context. But at the same time, GWASs have provided researchers with a new tool—polygenic scores—that can now be productively integrated into a wide variety of psychological data sets. Third, I consider the future of research on gene–environment interplay: What can the newer tools of molecular genetics, particularly when combined with the standard workhorses of classical behavioral genetics, reveal about environmental influences? This section considers, in particular, how genetic data can reveal the intergenerational transmission of social privilege, how genetic influences are refracted through the prisms of sociopolitical and historical context, and why the next generation of gene × environment interaction studies should focus largely on exogenous environments, such as interventions and policy reforms.
EVERYTHING IS HERITABLE
Heritability Is a Confusing Word but a Meaningful Concept
Twenty years ago, Eric Turkheimer summarized the already extensive body of twin and family research with his first law of behavior genetics: “All human behavioral traits are heritable” (Turkheimer 2000, p. 160). In other words, if one considers all the ways in which humans differ in their personality, cognition, emotion, behavior, and social relationships, at least some of the variation between people is due to the genetic differences between them.
The statement that “all human behavioral traits are heritable” seems straightforward enough. However, perhaps no concept in the social sciences has been the subject of as much confusion as heritability (Visscher et al. 2008). On the one hand, the heritabilities of psychological phenotypes like intelligence have been grossly misinterpreted to mean that those phenotypes are fixed and unchangeable through social policy or intervention (Goldberger 1979, Jensen 1969); that racialized disparities in a phenotype are due to genetic differences between races (Herrnstein & Murray 1996, Jencks & Philipps 1998); or that the phenotype is best understood at a biological level of analysis, i.e., at the level of the genome. These conclusions are all false, for reasons that have been extensively described previously (Goldberger 1979, Jencks & Philipps 1998, Turkheimer 1998). On the other hand, heritability is sometimes dismissed as a meaningless or trivial concept that gives no information about the causal role that genes play in human lives (Lerner 2006, Lewontin 1974, Manski 2011). This, too, is a mistake.
Instead, the concept of heritability is both more limited and more useful than is reflected in popular discussions about genetic research. The idea of apportioning observed phenotypic variance into a component due to genotypes was introduced by Ronald Fisher over a century ago (Fisher 1919), but even in the postgenomic era, “the importance of heritability remains central” (Visscher et al. 2008, p. 255). In agriculture and animal breeding, the heritability coefficient is an essential statistic, because it allows one to predict how the average phenotype in a population will change in response to natural or artificial selection. Indeed, heritability is one component of the breeder’s equation, R = h2S, where R is the change in the average phenotype across generations and S is the difference in the average phenotype between parents selected for breeding and the overall average in their generation. In human genetic research that aims to map specific genes that contribute to disease risk, the heritability coefficient is also an important statistic, as it is one factor determining the statistical power to detect significant genetic associations.
Among psychologists who are likely not interested in either selective breeding or gene mapping per se, heritability should nevertheless still be a quantity of interest. One rough analogy for the heritability coefficient is the Gini index, which is a metric for the level of economic inequality in a particular country at a particular time. Like heritability, the Gini can range from 0 to 1. A Gini of 0 means that everyone in the country has exactly equal shares of income; a Gini of 1 means that all of the income is held by a single person. Like heritability, the Gini coefficient is not static across time and place; it tells you about what is, not what could be. As the sources of inequality change with economic and political change, the Gini also changes, as does the heritability of a phenotype.
Yet, like heritability, the Gini is meaningful despite its dynamic nature. Knowing the extent to which wealth is concentrated in the hands of a few people tells us something useful about a particular society, even as one might imagine alternative political economies where wealth could be distributed differently. Similarly, knowing the extent to which inequality in life outcomes has been caused by genetic differences among people tells us something useful about a particular population, even as one might imagine alternative societies in which heritability could be different. Indeed, just as the Gini index provides an invaluable metric for examining how wealth inequality differs across time and place, so too does the heritability coefficient provide a metric for examining how the outcome of the genetic lottery differs across time and place. For instance, one paper compared the heritability of educational attainment in Estonia during and after a totalitarian Soviet regime (Rimfeld et al. 2018a). I consider more studies like this in the section titled The Future of Gene–Environment Interplay.
Estimating Heritability Using Twins
The first law of behavior genetics was a correct summary of the state of the behavioral genetics literature when it was written, and there has been nothing published in the intervening decades to demand serious revision. Rather, the pace of behavior studies has only continued to accelerate. A 2015 review of 50 years of twin studies analyzed data on 17,804 human traits from 2,748 publications, which collectively included over 14 million pairs of twins (Polderman et al. 2015).
The basic logic of the twin model is simple. Identical (monozygotic, or MZ) twins begin life as a single zygote and are (nearly) identical for their DNA sequence, except for postconception DNA mutations. Fraternal (dizygotic, or DZ) twins are conceived from two eggs and two sperm, and thus are no more genetically related than are nontwin full siblings. As both types of twins are raised together in the same home, the extent to which fraternal twins are more different in their phenotype (e.g., height, weight, intelligence, disease) than identical twins are is a metric of how much genetic differences between people cause differences in the phenotype. In technical terms, multiplying the difference between the MZ twin correlation and the DZ twin correlation by two yields an estimate of heritability (h2), i.e., the percentage of phenotypic variation within a population that is due to genetic differences.
Across all the traits studied by Polderman and colleagues (2015), the average heritability was 49%. This estimate is consistent with how textbooks and other sources tend to summarize the behavioral genetics literature, indicating that genes and environment make an equal contribution to human variation. This stylized fact, however, masks considerable heterogeneity across phenotypes (Figure 1).
Figure 1.
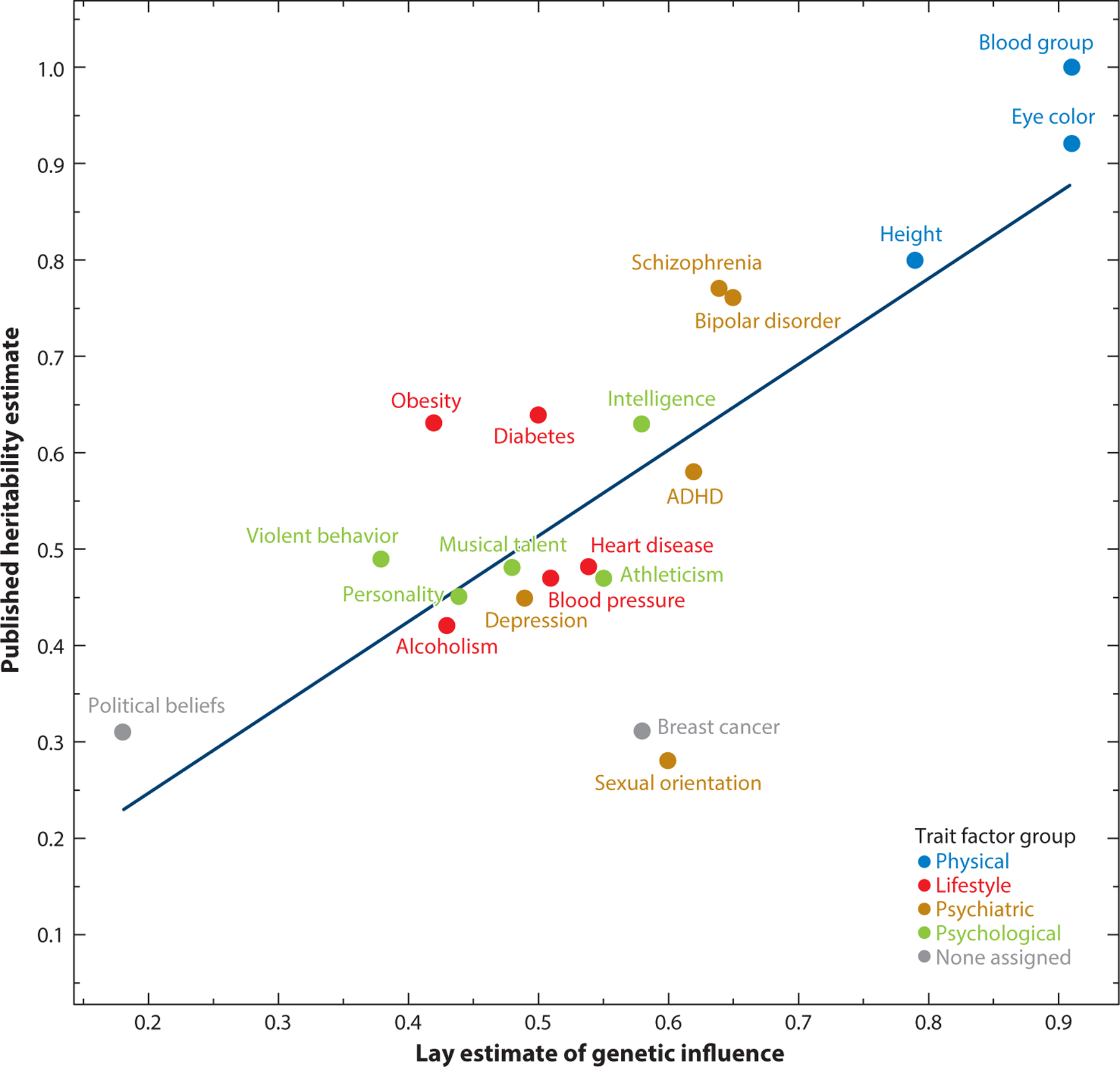
Heritabilities of human phenotypes and lay perceptions of heritability. The figure shows scatterplot and associated regression line of mean lay estimates of genetic influence for 20 human traits (excepting colorblindness) in a Mechanical Turk sample of 1,041 participants, along with published estimates of heritability for these traits from meta-analyses and large-scale twin studies. Data are converted from a 1-to-5 Likert scale to a 0-to-1 variance scale. The correspondence between lay estimates and published estimates is r = 0.77 (r2 = 0.59). Points are color-coded to their group membership according to the results of a four-factor solution of all responses. Abbreviation: ADHD, attention-deficit/hyperactivity disorder. Figure adapted with permission from Willoughby et al. (2019).
Generally, the lay public has fairly accurate intuitions about how heritabilities differ across phenotypes. When asked to estimate the heritability of phenotypes ranging from eye color to intelligence to political orientation, the average estimate given by convenience samples (e.g., Mechanical Turk participants) was correlated with consensus scientific estimates at r = 0.77 (Willoughby et al. 2019) (Figure 1). Mothers with more than one child, i.e., people who have had the opportunity to observe within-family differences as they emerge over development, are particularly accurate in their assessments.
There are, however, exceptions that prove the rule: The heritabilities of breast cancer and sexual orientation are consistently overestimated, perhaps because of public awareness around single-gene mutations (e.g., BRCA1/2) that cause breast cancer and of social narratives around sexual orientation being innate (i.e., the “born that way” argument).
While some phenotypes like breast cancer and sexual orientation are less heritable than popularly assumed, other psychological phenotypes have been shown in the past decade to be very strongly heritable, approaching the estimates seen for anthropometric traits. Perhaps most notably, autism spectrum disorders have been shown to be as heritable as height (∼90%) in both a meta-analysis of twin studies (Tick et al. 2016) and an analysis of large-scale population data including over 35,000 twin pairs and over 2 million sibling pairs (Sandin et al. 2017). This heritability is notably high not just because autism spectrum disorders are a psychological, rather than physiological, phenotype, but also because they onset during early childhood. Generally, heritabilities tend to be small in early childhood and increase with advancing age (Tucker-Drob et al. 2013). Another phenotype with nearly perfect heritability already in childhood is executive functioning, defined as a general ability to regulate attention (Engelhardt et al. 2015, Friedman et al. 2008).
Twin Studies Through the Lens of the Nonshared Environment
Very high heritabilities, as estimated from twin studies, imply that MZ (i.e., identical) twins are nearly perfectly concordant, that is, rMZ ≃ 1. This result can also be expressed in terms of what is often called the nonshared environment, or e2, which captures the extent to which MZ twins reared together in the same home are different from another other (1 − rMZ). The variable e2 captures the impact of environmental influences that are unique to each person instead of being shared by family members, as well as randomness and measurement error. Reviewing the twin literature from the perspective of e2 has the benefit of collapsing the specious nature-nurture debate, as the systematic effects of both genetics and family-level environments reduce e2.
Informed by philosophical perspectives on free will that emphasize whether or not a person could have done otherwise, Turkheimer (2011) proposed that e2 be known as the free-will coefficient. Whether one can glean from twin studies any information about a person’s metaphysical freedom to do otherwise is, to put it mildly, controversial. But even if we put aside metaphysical discussions about whether people really have free will, identical twins provide a glimpse into how constrained one’s life path is by one’s starting point in life, which encompasses both having a particular genotype and being born in a particular time and place. When e2 is low (<20%), people who begin life at the same starting point tend to show highly similar life outcomes, whereas a high e2 indicates that there is a large amount of “elbow room” (Dennett 2015) for the emergence of individuality. The method of estimating e2 in human twin studies parallels experimental research that uses isogenic animals, including fruit flies (Linneweber et al. 2020), mice (Freund et al. 2013), and fish (Bierbach et al. 2017), to study the emergence of individuality in behavior.
Not surprisingly, e2 is low for physical traits, like height and eye color, over which people are presumed to have little agency. But e2 is also low for many psychiatric diseases like schizophrenia, bipolar disorder, and autism spectrum disorders; for cognitive abilities like processing speed and executive functioning (Engelhardt et al. 2016); and for socially relevant outcomes like antisocial behavior (Krueger et al. 2002) and educational attainment (Branigan et al. 2013). At the same time, e2 is substantial for other traits and characteristics, like personality (Briley & Tucker-Drob 2017), income (Benjamin et al. 2012), and marital status ( Jerskey et al. 2010). You only have one life to live, but if you rewound the tape and started anew from the exact same genetic and environmental starting point, how differently could your life go? Overall, twin research suggests that, in your alternative life, you might not have gotten divorced, you might have made more money, you might be more extraverted or organized—but you are unlikely to be substantially different in your cognitive ability, education, or mental disease.
Heritability in the Post-Genomic Era
The Human Genome Project, which took over a decade to complete, sequenced a complete human genome at a cost of approximately $3 billion. Now, a few decades later, the cost of whole-genome sequencing has plummeted, now being less than $1,000 per person. And the cost of whole-genome sequencing, which measures every DNA letter in a person’s genome, still far exceeds the cost of genotyping a person on a genome-wide single-nucleotide polymorphism (SNP) array. Unlike whole-genome sequencing, a SNP array measures only a fraction of the genome, focusing on several million genetic variants that commonly vary between humans. SNP genotyping from noninvasive samples of human saliva is now available for less than $60 per person.
This rapid decrease in genotyping costs has revolutionized the commercial and research landscape. Direct-to-consumer genetics companies, such as 23andMe© or Ancestry.com, have now genotyped millions of people worldwide, providing them with information about health-relevant genetic variants and geographical ancestry. The explosion of direct-to-consumer genetic testing has raised critical questions about, for example, how to maintain one’s privacy when one is identifiable from genetic data given by one’s relatives, and what consumer safeguards are necessary when delivering potentially distressing information about one’s parentage, ancestry, or future health. At the same time, it has already proved a boon to genetic research by giving scientists access to genetic data on hundreds of thousands—or even millions—of people.
Enormous samples of genetic data have also been amassed by national biobanks, such as the UK Biobank and the Estonian Biobank. Thus far, the United States has lagged behind other countries in collecting and disseminating genetic data for the scientific community, but the launch by the National Institutes of Health of the All of Us research program, as well as the ongoing Million Veteran Program by the US Department of Veterans Affairs, has the potential to make it easier to conduct genetic research with American participants without relying on partnerships with corporations like 23andMe©. For now, the UK Biobank, which collected DNA from 500,000 participants in the United Kingdom, remains a primary source of data for large-scale genomic research, not only because of its size, but also because it has transparent and straightforward data access procedures and documentation.
Critics of the field of behavioral genetics have often predicted that the ability to measure the human genome directly would upend many of the assumptions and conclusions of twin and family studies (Charney 2012). “Another nine-inch nail for behavioral genetics!” was the title of one such prematurely triumphant paper (Lerner 2006). Instead, many of the core assumptions and conclusions of twin research have been validated using newer sources of genomic data.
For instance, a critical assumption of the twin model is that identical twins are not treated more similarly by parents or by others in their environment, in ways that are relevant for a phenotype, simply because they are identical twins. One test of this equal-environments assumption is to consider twin pairs who have been misclassified by their parents (a not uncommon occurrence) and so are believed to be identical when they are, in fact, fraternal, or vice versa. Supporting the validity of the equal-environments assumption, the phenotypic similarity of twins tracks their actual genetic relationship, not their perceived zygosity (Conley et al. 2013).
Even more compellingly, genome-wide data can be used for so-called assumption-free methods that estimate heritability based on measured genetic relatedness (Visscher et al. 2006). One such method relies on the fact that sibling pairs randomly vary in the extent to which they inherit the same DNA segments from their parents (referred to as identity-by-descent sharing). The expectation is that siblings will share 50% of their segregating genetic variance on average, but this expectation is like the expectation that a coin will land on heads 50% of the time. In reality, any one pair of siblings can be more or less genetically similar, and variation across sibling pairs in their extent of identity-by-descent sharing can be leveraged to estimate heritability directly from DNA. Just as a twin model estimates heritability by testing whether dizygotic twins are more different in their phenotype than monozygotic twins, the assumption-free method for estimating heritability tests whether sibling pairs who have less identity-by-descent sharing are more different in their phenotypes than sibling pairs who have more. Other DNA-based methods for estimating heritability leverage small differences in the extent of measured genetic similarity found among pairs of people who would not typically be considered relatives (Yang et al. 2010): Are pairs of people who are more genetically similar also more phenotypically similar?
An early study using a DNA-based method estimated the heritability of height to be ∼80%, and it noted that this result was “consistent with results from independent twin and family studies but using an entirely separate source of information” (Visscher et al. 2006). However, although the results from DNA-based methods of estimating heritability scale with the estimates from twin and family studies, the former are typically smaller (Young et al. 2019). This discrepancy between heritability as estimated from classical twin and family studies and heritability as accounted for by measured DNA was labeled the missing heritability problem (Manolio et al. 2009). Recent work has suggested that some of the missing heritability is actually “hiding” in rare variants that are not typically measured and in the heterogeneity of genetic effects across populations (Tropf et al. 2017, Wainschtein et al. 2019, Young 2019). Whether missing or hiding, the continued gap between DNA-based estimates of heritability and estimates from twin/family studies means that the latter might still be overestimating heritability due to faulty assumptions. But it is no longer reasonable, contra some predictions, to expect that advances in human genomics will reveal that the heritability of psychological phenotypes is entirely illusory.
The Tacit Collusion to Ignore Heritability
The DNA revolution has confirmed what we have known for some time: Genetic differences between people are a nonignorable source of heterogeneity in psychological and behavioral traits, including cognition, achievement, personality, psychopathology, social relationships, health behaviors, fertility, and beyond. One corollary of this conclusion is that genetic differences potentially confound nearly all correlational studies of the relationships between individual psychological outcomes and people’s environmental contexts. This is because environments are not purely exogenously imposed on the individual. Rather, they are selected or actively crafted, either by the individual or by other people (such as parents) who are genetic relatives.
This is not a new observation. The problem of genetic confounding for social science, particularly for the nonexperimental research designs that are commonly employed in fields such as developmental and clinical psychology, has been repeatedly and cogently articulated for at least the past half-century. Yet, as Freese (2008, p. S19) noted,
Many quarters of social science still practice a kind of epistemological tacit collusion, in which genetic confounding potentially poses significant problems for inference but investigators do not address it in their own work or raise it in evaluating the work of others. Such practice implies wishful assumptions if our world is one in which “everything is heritable.”
Unfortunately, Freese’s (2008) assessment remains true more than a decade later. Perusing the contents of a single issue of a prominent journal in developmental psychology, for instance, one will find studies relating maternal depression and child intelligence, parental problem drinking and child sleep, maternal language and child executive function, child perceptions of caregivers and asthma-related immune function, family turbulence and child internalizing behaviors, parental structuring and child emotional eating, and maternal postpartum depression and child behavior problems. All of these studies use data from genetic relatives with only a cursory mention, at best, of the possibility that observed associations might be due to genetic inheritance. Whether one’s research goals are to identify targets for intervention or contribute to a basic understanding of developmental processes, the continued proliferation of research studies with, as Freese (2008, p. S19) describes it, an “incisive, significant, and easily explained flaw” represents an enormous waste of scientific resources.
THE ASSOCIATION STUDY IS DEAD; LONG LIVE THE ASSOCIATION STUDY
Genome-Wide Association Studies
GWASs typically measure hundreds of thousands or even millions of SNPs scattered throughout the entire genome. SNPs are genetic variants whereby people differ by a single DNA letter, or nucleotide. Because humans have two copies of every gene, a person might have 0, 1, or 2 copies of the alternative version of the SNP (i.e., the minor allele). The phenotype of interest—which could be as obviously biological as height or as socially complex as educational attainment or depression—is then used in a series of linear regression models, where it is regressed onto each SNP, one at a time, plus a set of standard covariates like age and sex. This exercise yields a set of summary statistics, or a set of beta weights representing the strength of the association between each SNP and the phenotype of interest. Because a GWAS conducts a large number of statistical tests, statistical significance is defined very strictly: A SNP is not typically considered a “hit” unless it is associated with the phenotype at p < 5 × 10−8 (what is called genome-wide significance).
Even if genome-wide significant association is detected, a SNP might not cause a phenotype. Rather, it could be correlated with the phenotype because it is structurally correlated with another (unmeasured) genetic variant that is the causal variant, or because it is correlated with an environmental factor that varies between human populations or subpopulations, a problem known as population stratification (Hamer & Sirota 2000). Researchers attempt to correct for population stratification by restricting their analyses to people who have similar genetic ancestry (e.g., people who are all of European ancestry). They then compute 100+ principal components that capture broad patterns of genomic similarity produced by human demographic history and include these as covariates in GWAS analyses. As a result, GWASs analyze genetic differences within groups of people that are fairly homogenous with regards to ancestry, and GWAS results are silent regarding the sources of differences between groups that differ in ancestry (e.g., between people with European versus African ancestry) (Martin et al. 2017). Despite these controls, follow-up work is necessary to confirm that any GWAS result is actually picking up on any causal signal. Within-sibling comparisons are particularly useful, as genotypes are randomized to siblings during meiosis (Young et al. 2019).
As genetic data have become more common (see the section titled Heritability in the Post-Genomic Era), GWAS research has accelerated rapidly. There have now been large-scale (N ≃ 30,000–1,500,000) GWASs conducted for a wide variety of phenotypes relevant for psychology, including psychiatric diseases, subjective well-being, personality traits, risk-taking behavior, substance use behaviors, educational attainment, cognitive ability, noncognitive skills, and body mass index (Buniello et al. 2019).
Overall, GWAS results have yielded two general lessons for psychology. First, traits of interest to psychologists are massively polygenic, meaning that they are associated with thousands upon thousands of genetic variants scattered throughout the genome, each of which has a tiny effect. This has been called the fourth law of behavior genetics (Chabris et al. 2015). Second, the aggregate predictive power of measured genetic variants, in some cases, rivals the predictive power of traditional social science variables, such as family socioeconomic status (SES) (Lee et al. 2018). In the subsequent sections, I consider the implications of each of these lessons for the future of psychological science.
Polygenicity and the Failed Candidate Gene Paradigm
Given what we now know about the effect size of individual genetic variants in relation to psychological and behavioral traits (R2 ≲ 4 0.1%), it is clear that psychological studies that interrogated single candidate genes, such as 5HTTLPR (a variant of the serotonin transporter gene), were massively underpowered statistically, and their results were false positives (Chabris et al. 2012). This methodological flaw—insufficient statistical power—is even more pronounced for studies of candidate gene × environment interaction (cG × E), such as studies of the interaction between 5HTTLPR and life stress on depression (Caspi et al. 2003). As early as 2011, analysts warned that most positive cG × E findings were false discoveries (Duncan & Keller 2011). In the next decade, subsequent review and editorial statements about candidate gene and cG × E findings warned that “it now seems likely that many of the published findings of the last decade are wrong or misleading and have not contributed to real advances in knowledge” (Hewitt 2012, p. 1), and that “the first decade of cG × E has produced few, if any, reliable results” (Duncan et al. 2014, p. 258). Most recently, an analysis using preregistered analysis plans and samples as large as ∼400,000 people concluded that there was “no support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples” (Border et al. 2019).
Damningly, Border et al.’s (2019) review found that psychology had continued to produce candidate gene studies at a steady rate from 2007 through 2016 (the last year analyzed), despite the fact that such studies did not meet editorial standards at genetics journals and had been rigorously criticized for fatal methodological flaws. Such studies were also prominently featured in the genetics-focused reviews published in these pages in the past decade. Even now, underpowered candidate gene studies continue to be regularly submitted to psychology journals. Clear editorial policies, such as the one enacted by the journal Behavior Genetics (Hewitt 2012), which required associations with single genetic variants to meet genome-wide significance levels, are necessary to prevent further proliferation of a discredited research paradigm. Clarity regarding the flaws of candidate gene research is additionally necessary, because studies on MAOA (the so-called warrior gene) and other candidate genes are still used in forensic settings (McSwiggan et al. 2017). Whether testimony that links propensity to violence to a single genetic variant can be excluded as scientifically unreliable is now being debated by the New Mexico Supreme Court [State v. Yepez (2018)].
Psychologists persisted in conducting candidate gene research long after the central flaws of the methodology had been incisively and convincingly described. What went wrong? The proliferation of cG × E research was doubtlessly influenced by more general factors that contributed to the replication crisis in psychology, such as investigator degrees of freedom and incentives against publishing null results. In addition, researchers might have been particularly eager to publish results that confirm their prior belief that genetic associations are ubiquitously moderated by variations in environmental context, because such results intuitively undermine ideas about genetic determinism and a nature-versus-nurture binary (Tabery 2014).
In contrast, in the absence of environmental moderation, genetic research on human psychology, particularly on socially valued traits like intelligence and self-control, is widely seen as subversive, because it is perceived as posing a challenge to “ideals of responsibility and equality so central to our democratic system” (Fox 2019, p. 164). Consistent with Fox’s theoretical analysis, scientists who report genetic main effects on intelligence are perceived as having less egalitarian values—e.g., to be less likely to endorse views like “People and social groups should be treated equally, independently of ability,” and more likely to endorse views such as “Some people … should be treated as superior” (Hannikainen 2019). The cG × E paradigm allowed researchers who appreciated the relevance of genetics for human psychology to escape the perceived danger of producing subversive science, as the method appeared to yield bounteous evidence for interaction effects in small, easily acquired samples. Older, more rigorous behavioral genetic methods, in contrast, required large samples, special populations (such as twins), and yielded statistically significant interaction effects much less consistently (e.g., Tucker-Drob & Bates 2016).
In summary, large-scale GWASs provided definitive evidence that the past two decades of psychological research on candidate gene associations and cG × E interaction effects yielded few real scientific insights. As psychology begins to move away from these discredited paradigms to more rigorous approaches (such as the polygenic score analyses described in the next section), it will be important to guard against letting the field’s enthusiasm for the idea of interaction supersede its commitment to methodological rigor.
Polygenic Scores Are Social Science Variables
The extremely small effect sizes of associations with individual genetic variants means that the types of samples that psychologists typically work with (with N ranging from tens to thousands) are not useful for discovering new genetic variants that are relevant for social and behavioral phenotypes or for following up the effects of individual variants. However, the effects of genetic variants scattered throughout the genome can be aggregated together in the form of a polygenic score. Briefly, polygenic scoring takes the summary statistics from a GWAS and uses them as a type of scoring key for genetic data from a new (independent) sample of people. A person’s allele count (0, 1, or 2) at each SNP is multiplied by that SNP’s beta weight from the GWAS, and then the weighted counts are summed across the genome. The resulting polygenic score is a single number for each person that reflects an estimate, based entirely on DNA information, of their likelihood to display the target phenotype.
Because of differences between human populations in genetic ancestry, polygenic scores created using GWASs conducted in one population are generally less predictive of phenotypes in other populations (Duncan et al. 2019). Moreover, average polygenic scores cannot be compared across different ancestral groups (Martin et al. 2017). Currently, the vast majority of GWASs are conducted with European-ancestry populations, with large amounts of data coming from just a few countries, such as the United States and Iceland (Martin et al. 2019). Criticisms of the Eurocentrism of genetic research mirror criticisms of psychology’s focus on so-called WEIRD (Western, educated, industrialized, rich, and democratic) populations (Henrich et al. 2010).
Within European-ancestry samples, however, some polygenic scores now rival the predictive power of traditional variables used in social science research. This was powerfully demonstrated by a GWAS of educational attainment (defined as years of schooling) conducted among 1.1 million people. In an independent sample of European-ancestry participants from the National Longitudinal Study of Adolescent Health (Add Health), a polygenic score of education-associated genetic variants was as strongly associated with educational attainment as family income (Lee et al. 2018). Recognizing that polygenic scores allow researchers an opportunity to integrate genetic measures in modestly sized samples, many commonly used social science data sets—like Add Health, the Wisconsin Longitudinal Study, the Health and Retirement Survey, and the Panel Study of Income Dynamics—have moved toward releasing polygenic score information to researchers (Okbay et al. 2019).
Unfortunately, the burgeoning predictive power of polygenic scores has led some high-profile academics to oversell them, referring to polygenic scores as fortune-tellers (Plomin 2018) or as measures of inborn or innate traits (Murray 2020). These statements reinforce the tendency to interpret genetic associations as evidence for genetic determinism or genetic essentialism (Dar-Nimrod & Heine 2011) and are not supported by the evidence. First, the magnitudes of the correlations with even the best polygenic scores, while mirroring the effect sizes typical in social and behavioral science, are far from the level of prediction accuracy necessary for valid individual prediction (Morris et al. 2019). Non-heritable variation in the phenotype, sampling error in the original GWAS used to create the polygenic score (Dudbridge 2013), and differences in sample composition between the original GWAS sample and the polygenic score sample (Mostafavi et al. 2020) attenuate the predictive power of the polygenic score.
Second, associations with polygenic scores—like the SNP associations detected in GWAS—can be tapping uncorrected population stratification; that is, they could be driven by environmental differences that systematically covary with genetics. The effect size of polygenic scores obtained from sibling fixed-effects models (i.e., from within-family designs) is a better measure of the association free from population stratification, as genes are randomized within sibling pairs (Trejo & Domingue 2019). Often, for behavioral and social phenotypes like education or childbearing, the within-family effect size is about half the between-family effect size (Lee et al. 2018, Mostafavi et al. 2020).
Third, even the causal effects of genetic variation might depend on transactions with environmental processes (Tucker-Drob et al. 2013). For instance, genetically influenced early traits in children might elicit greater cognitive stimulation from caregivers, which further facilitates the development of intelligence (Tucker-Drob & Harden 2012). Such “outside the skin” processes for genotype–phenotype relationships are expected to be the norm for psychological development. Other research has found that the polygenic score of a focal person is associated with their phenotype because it is indirectly measuring the genotype of the focal person’s parent, who is providing environments relevant for the phenotype. One clever demonstration of this was a study showing that polygenic scores are less predictive of a person’s educational attainment if they are adopted than if they are raised by their biological parents (Cheesman et al. 2019). In the case of adoptees who are not genetically related to their parents, the polygenic score is not revealing anything about the environment provided by those parents.
In this way, polygenic scores are similar to most variables used in social science research. Consider, for example, a researcher who reports a correlation between a child outcome and family SES, or who includes SES simply as a covariate in a study of another focal variable. Correlations with SES are not evidence for a deterministic process: Children who grow up in poverty are not destined to experience a particular outcome, there is considerable variability in life trajectories across the SES spectrum, and family SES is expected to function differently across societies and historical times. SES is itself a composite variable, typically calculated by combining information about parental education, income, and occupational status, each of which captures an array of subordinate processes. Correlations with SES or any of its components reflect a combination, in largely unknown proportions, of casual processes and confounding by other variables (including genotype) that themselves systematically vary with SES. Controlling for SES is more difficult than it might originally appear, as the role of measurement error must be taken into account (Westfall & Yarkoni 2016). Like SES, a polygenic score is a risk factor that is measured with error, is probabilistically correlated with developmental outcomes, and operates through largely unspecified mechanisms that are socially and historically contingent.
There are, of course, some differences between polygenic scores and other social science variables that make them interesting for research purposes. DNA sequence is fixed at conception; thus, polygenic scores do not change over the course of an individual’s lifespan. As such, they are immune from reciprocal causation. This makes them useful for studying recursive processes, such as peer influence or academic achievement. For instance, one study examined the peer metagenomic context to study peer influences on smoking (Sotoudeh et al. 2019). One person’s behavior might change their peers’ behavior, but it cannot change their peers’ DNA. Consequently, relationships between peer DNA and one person’s smoking can offer new insights about peer influence. Another study used polygenic scores to examine students’ trajectories through the high school math curriculum. Unlike measures of math ability or interest, which are dynamic and could be affected by the difficulty of one’s previous math classes, polygenic scores are inert and can thus be used as molecular tracers to see how students with fixed characteristics progress in their STEM education (Harden et al. 2019).
The immutability of polygenic scores also makes it possible for researchers to add genetic information long after the conclusion of the study. For example, people who participated in a study of early childhood intervention could be genotyped as adults, thus adding a new source of information about their childhood that is not tainted by recollection biases or errors (see Rietveld et al. 2013, supplement, for a discussion of this idea). Moreover, a person’s genetic information can be used to create multiple polygenic scores—as many as there are available GWASs—and these polygenic scores can be updated as new genetic insights become available, without new contact with the participants.
Summary
The past two decades of genetic research in psychology have witnessed the explosion of candidate gene studies and the (later, slower, but ultimately more enduring) ascension of GWASs. The latter approach, which has emphasized international collaboration and enormous sample sizes, has finally yielded replicable insights about which genetic variants are associated with social and behavioral phenotypes. As the predictive power of polygenic scores begins to rival traditional social science variables, genetic insights can now be leveraged by psychological scientists in modestly sized samples. Thus, the field is now finally able to address some of the questions about gene–environment interplay that animated the candidate gene era, particularly when the newer methods that rely on measured genotypes are combined with the familiar workhorse of behavioral genetics—the family design. It is to the future of studying gene–environment interplay that I turn in the final section.
THE FUTURE OF GENE–ENVIRONMENT INTERPLAY
As I described at the beginning of this article, it was already clear in the early 1960s that genetics conditioned the variation we observe in people’s “ability, energy, health, character, and other socially important traits” (Dobzhansky 1962, p. 112). But the scientists working in the 1960s could have scarcely imagined the tools that we can now bring to bear on studying the genetics of behavior. The ability to sequence human genomes cheaply, noninvasively, and at scale has ushered in a golden age of genetics research.
Since the earliest days of genetics, there has been enduring interest in superseding a nature-versus-nurture binary to understand what has come to be known as gene–environment interplay (Rutter & Silberg 2002, Rutter et al. 2006). The term gene–environment interplay captures a variety of ways in which genetic and environmental effects can be dependent on each other, including epigenetics, gene–environment correlation, and gene × environment interaction. Even more simply, research designs that include genetic information can be used to study environmental main effects on behavior. Such insights are not “interplay” as usually conceptualized, but they emphasize a more general point: that genetic research is not useful solely for understanding biological effects.
In the following sections, I consider three streams of research on gene–environment interplay, broadly defined: (a) research that uses genetic information to reveal the ways that social privilege is transmitted across generations; (b) research that considers how genetic influences vary as a function of sociopolitical or historical context, the life course, and other social structures; and (c) research that uses exogenous environments (e.g., interventions, policy reforms, and natural experiments) to understand the mechanisms of genetic effects and heterogeneity in response to environmental change. Throughout, I focus on examples from genetic research on cognitive ability and educational attainment but suggest how similar designs could be used for the study of other psychological phenotypes.
Studying the Genetic Lottery Reveals the Importance of the Social Lottery
The importance of genetic influences is often misinterpreted to mean that family-level environments, such as school contexts, neighborhood conditions, family SES, and parenting behaviors, are unimportant. One prominent behavioral geneticist, for instance, summarized twin studies as demonstrating that parents and schools “matter, but don’t make a difference” (Plomin 2018, p. 82). This is false. Twin and adoption studies do indeed pose a challenge to a naïve environmentalism that presumes that all correlations between parents and children are due to the effects of the former on the latter. Resemblance among family members for some psychological phenotypes is, indeed, primarily due to the fact that they are genetically related (Polderman et al. 2015). But twin studies (in conjunction with molecular genetic studies, which are discussed in more detail below) provide some of the strongest evidence that families reproduce their social privilege across generations through environmental mechanisms, not just genetic ones. When it comes to whether one is poor or rich, educated or uneducated, the family-level environment in which the person was raised does certainly make a difference.
Evidence from twin and adoption studies.
The importance of shared environmental influences is most clearly apparent for educational attainment, defined as the number of years of schooling that one completes. People with more education make more money, are more likely to be employed in prestigious occupations, are more likely to be married and to avoid nonmarital childbearing, have higher subjective well-being, and live longer (Case & Deaton 2017, Deaton 2013). A meta-analysis of 15 twin studies spanning 10 countries found that 36% of the variance in educational attainment, on average, was due to environmental factors shared by children raised in the same home, i.e., the family-level environment (Branigan et al. 2013). In one-third of the included studies, the proportion of variance attributable to the shared environment exceeded the estimated heritability; that is, for many cohorts of people, the social lottery remains even more important than the genetic lottery in shaping how far they go in school.
Shared environmental influences on educational outcomes are also evident at earlier points in the educational trajectory. Whereas more basic cognitive abilities like executive functioning and processing speed are nearly perfectly heritable even in childhood (Engelhardt et al. 2015), academic skills in reading and mathematics, which are the direct targets of instruction, show substantial shared environmental variance, even when all participants are drawn from schools in a single geographical area (Engelhardt et al. 2019, Rimfeld et al. 2018b). In Germany, shared environmental influences are minimal for cognitive test performance but are substantial in relation to whether a student is tracked into gymnasium, i.e., the college-preparatory academic track that permits matriculation to university (Schulz et al. 2017). In societies that reproduce privilege across generations, the family environment in which one is raised makes a difference in how far one goes in school.
In contrast to what is seen for educational attainment, most studies find a minimal effect of shared environmental factors on cognitive abilities, particularly when measured in adulthood. It has been suggested, however, that this near-zero main effect of the family-level environment masks the heterogeneity of the effects of the shared environment across the SES spectrum. An early paper by Turkheimer et al. (2003) analyzed data from a sample of twins with an unusual overrepresentation of children in poverty and found substantial effects of the shared environment on cognitive ability at age 7. Subsequent research on the genotype × SES interaction effect yielded mixed results, with several studies finding null effects or even effects in the opposite direction. However, a meta-analysis of this literature (Tucker-Drob & Bates 2016) found evidence of a significant interaction effect (albeit with a smaller effect size than estimated by Turkheimer and colleagues, an example of the winner’s curse), particularly in the United States.
The importance of the shared environment for cognitive ability has also been demonstrated using adoption studies. In particular, population-wide data from Sweden allowed researchers to estimate the impact of the family environment using a unique sample of male-male sibling pairs where one brother was adopted while the other brother was raised by his biological parents (Kendler et al. 2015). The IQ score of the adopted brother was, on average, ∼4 points higher, an increase that varied with the education level of the adopting parents.
Evidence from polygenic score analyses.
In addition to evidence from classical twin studies and adoption studies, the advent of polygenic scores has provided even more evidence that families reproduce their social privilege through environmental mechanisms. One particularly noteworthy study analyzed a large sample of trios (a focal person and both of their parents) from Iceland, using a design that allows researchers to examine the genetic variants that children inherit from their parents as well as the genetic variants that they do not inherit (Koellinger & Harden 2018, Kong et al. 2018). (Recall that humans are diploid organisms with two copies of every gene, only one of which is randomly transmitted to any child.) If the parental genes that a child does not inherit are nevertheless associated with the child’s phenotype, this association must be due to environmental transmission from parent to child, as genetic inheritance has been ruled out by design. This is exactly what was observed for educational attainment: The nontransmitted parental genotypes were associated with the child’s educational attainment.
Other research has compared how children with similar polygenic scores fare differently in life as a function of their social background. A notable study by Belsky and colleagues (2018), which pooled data from five samples that spanned several countries and birth cohorts, examined how polygenic scores created from a GWAS of educational attainment predicted intergenerational social mobility, i.e., whether a person increased or decreased in social class relative to their parents. They found that higher polygenic scores predicted greater social mobility, even when comparing within sibling pairs. As sibling genetic differences are random, the within-family analysis is compelling evidence that genetic differences between people are causally related to social mobility. At the same time, children with high polygenic scores who were raised in low-SES families still ended up worse off, as adults, than children with low polygenic scores who were raised in high-SES families.
A complementary analysis of overlapping data (from the Health and Retirement Survey) from an independent group of investigators found additional evidence for an interaction between polygenic score and child SES in predicting rates of college graduation, with stronger genetic associations evident among high-SES families (Papageorge & Thom 2020). [The direction of this interaction is consistent with twin studies of G × SES interactions (Tucker-Drob & Bates 2016).] A college degree, in turn, was a strong predictor of labor market earnings at all levels of the polygenic score.
An interaction between polygenic score and social privilege was also evident in a study that we conducted on the relationship between educational attainment polygenic scores and progress through the high school math curriculum (Harden et al. 2019). On average, higher polygenic scores were associated with tracking to more advanced math classes at the transition to high school and with greater persistence in math across all four years of high school, even when controlling for family SES and math course grades in the previous year. However, students at socioeconomically advantaged schools (those primarily serving families with higher levels of parental education) were buffered from dropping out of math even if they had relatively low polygenic scores. Together, these studies reveal environmentally rooted inequities in the extent to which genetic differences between people are translated into human capital and socioeconomic attainments.
Summary and future directions.
In summary, recent behavioral genetic studies on educational attainment and social mobility illustrate that genetic research can actually provide strong evidence for the power of social privilege. As mentioned in the Introduction, a recurrent fear about behavioral genetic studies is that they will be used to naturalize and justify social inequalities (Columbia Law Sch. 2017, Parens 2004), but recent studies have, in fact, spotlighted how social privilege is replicated across generations via environmental mechanisms.
Genetic Influences in Sociopolitical and Historical Context
The biologist Richard Lewontin was one of the twentieth century’s most vehement critics of behavioral genetics. In a still widely cited paper, Lewontin (1974) took aim at twin studies and, more generally, at linear regression models, path analysis, and the analysis of variance. He criticized the results of twin models, because they depend on the particular distribution of genes and environments in the population being studied, for having a “historical (i.e., spatiotemporal) limitation”; the “spatiotemporally local analysis of variance” was described as “useless,” in contrast to the “global analysis” of “functional relations,” i.e., mechanisms of gene action (Lewontin 1974, pp. 403, 410).
Lewontin was correct, of course, that behavioral genetic methods are local analyses that make statements about specific populations of people living in a specific time and place; but the biologist’s trash has turned out to be the psychologist’s and sociologist’s treasure. Nearly a half-century after Lewontin’s critique, it is now clear that analyzing genetic associations with human psychological outcomes in spatiotemporally local samples has yielded interesting insights about how such associations differ across historical and sociopolitical context—and how they do not.
One major theme that has emerged from historical and cross-national comparisons of twin data is that heritabilities are generally higher at times and places that provide large amounts of social opportunity. Four recent papers, in particular, support this idea. First, data from the World Bank on national differences in intergenerational social mobility, defined by the parent-child correlation in years of schooling, were matched to results from twin studies of educational attainment (Engzell & Tropf 2019). In countries with lower social mobility (such as Italy and the United States), shared environmental variance in educational attainment was higher (and heritability was lower) than in countries with higher social mobility (such as Denmark). Second, an earlier paper compared just two twin samples and similarly concluded that “family background buys an education in Minnesota but not Sweden” ( Johnson et al. 2010).
Third, within the United States, the association between polygenic score and educational attainment differed by birth cohort and gender. In an older birth cohort (born 1939–1940), the genetic association was stronger for men than for women. However, as structural constraints on women’s access to education diminished over the course of the twentieth century, the gender difference in the predictive power of the polygenic score similarly diminished; that is, social opportunity led to stronger genetic associations.
Fourth, analyses of data from Estonia show a similar pattern, albeit over a more dramatic social transition (Rimfeld et al. 2018a). The heritability of educational attainment, as estimated based on measured DNA rather than twin data, was compared for Estonians who came of age during and after the Soviet regime. In comparison to earlier cohorts who were raised in a totalitarian government that provided little social opportunity, later cohorts of Estonians showed higher heritability of educational attainment.
Together, these results reveal that more open societies—e.g., those with intergenerational social mobility, gender equality in education, and nontotalitarian government—are typically associated with stronger effects of genetics and weaker effects of the family social background. Whether or not inequalities tied to genetics are more palatable than inequalities tied to family social class is a different question. As political philosopher John Rawls stated, “once we are troubled by the influence of either social contingencies or natural chance on the determination of distributive shares, we are bound on reflection to be bothered by the influence of the other. From a moral standpoint the two seem equally arbitrary” [Rawls 1999 (1971), pp. 64–65].
The Next Generation of Gene × Environment Studies: Improving Causal Inferences About Environmental Effects
In contrast to the macro-environmental contexts that I reviewed in the previous section, most previous G × E studies have examined more micro-level environments that vary within a population, such as urbanicity, family environment, peer context, school privilege, etc. However, variation in these environments is typically not exogenous to an individual’s genotype, complicating attempts to estimate the causal effects of specific environments in observational data. Estimating G × E in the presence of gene–environment correlation is challenging and can lead to false positive results (Keller 2014, van der Sluis et al. 2012, Van Hulle et al. 2013). More generally, the problem of wresting causal inferences from correlations among variables is still a problem even when the variable is entered as part of an interaction term (Fletcher & Conley 2013, Keller 2014).
To overcome this problem, future psychological research on G × E should seek to surmount its reliance on endogenous environmental variation and instead integrate experimental and econometric methods that allow for more rigorous inferences about causality. That is, researchers should focus on natural experiments and quasi-experimental designs, such as instrumental variables, differences-in-differences, or regression continuity designs, which leverage quasi-random variation in environmental exposure (Schmitz & Conley 2017). Alternatively, they should integrate genetic data into fully randomized experiments, such as randomized controlled trials of psychological interventions (Burt et al. 2019). Psychologists have been interested in integrating genetics into intervention studies before (Bakermans-Kranenburg & van IJzendoorn 2015), but previous attempts at such an integration were stymied by insufficient statistical power and a reliance on the flawed candidate gene paradigm. It is imperative for the next generation of gene × intervention (G × I) research to improve its measurement of the genome, whether by using polygenic scores or by using twin/family designs. Here, I will describe three examples of G × E research that focus on exogenous environmental exposures and high-quality measures of the genome and that showcase the promise of this approach.
In the first example, researchers used the Vietnam draft lottery to examine an interaction between genetic risk and military service on smoking behavior and health (Schmitz & Conley 2016). Notably, the environment variable (E) that was entered into the analysis was not military service itself, which is nonrandomly associated with socioeconomic background and other potentially omitted variables; rather, researchers used an instrumental variable, date of birth, that governed eligibility for the military draft. Among draft-eligible men, a polygenic score created from an independent GWAS of smoking initiation predicted ever being a smoker, being a heavy smoker, and having smoking-related health problems such as cancer. This genetic association was not evident among draft-ineligible men, suggesting that some aspect of exposure to military service activated a genetic risk for smoking. Interestingly, a significant main effect of draft eligibility on smoking was not detected, illustrating how modest or null average treatment effects might mask substantial gains for particularly at-risk populations (Bakermans-Kranenburg & van IJzendoorn 2015, Kent et al. 2010, Yeager et al. 2019).
In the second example, researchers examined the effects of a policy reform in the United Kingdom that increased the years of compulsory schooling (Barcellos et al. 2018). Because the reform applied only to students who were born after a certain date, there was quasi-random assignment to the reform, which could be exploited to estimate its causal effect. On average, the educational reform reduced body size. However, this salutary treatment effect was particularly strong for people at high genetic risk for obesity, as measured by a polygenic score created from a GWAS of body mass index.
As the policy reform preferentially benefitted people at high risk for obesity, it effectively narrowed genetically associated health disparities. This outcome is not a foregone conclusion: Policy reforms could, instead, operate equally across genotypes or could even exacerbate pre-existing genetically associated disparities (e.g., produce a Matthew effect) (Merton 1968). Closing gaps between subpopulations to achieve greater equality of outcome is frequently a goal of intervention and policy (Ceci & Papierno 2005), but in the absence of genetically informed analyses such as those conducted by Barcellos and colleagues (2018), whether gaps between people at high versus low genetic risk are closed or widened is typically unknown by policy evaluators.
A third example of the next generation of G × E research examined interactions between polygenic scores created from a GWAS of alcohol dependence and a randomized intervention in adolescence, the Family Check-Up (Kuo et al. 2019). The Family Check-Up is designed to improve parents’ management of their children’s problem behavior by improving the accuracy of parental appraisals and providing training in skills such as limit setting and supervision (Dishion & Kavanagh 2003). Among people who had been randomized to the treatment condition of the Family Check-Up as adolescents (15 years prior to follow-up), polygenic scores were no longer associated with rates of alcohol dependence.
This G × I result unites two previously separate lines of research that had independently converged on similar environmental mechanisms. Specifically, twin research has found that the genetic influences on externalizing behaviors are weaker in conditions of higher social control, such as in families with high parental monitoring, whereas genetic influences are higher in conditions that provide greater social opportunity for norm-violating behavior, such as involvement with deviant peer groups (Barr & Dick 2019; Cooke et al. 2015; Dick et al. 2001, 2007; Harden et al. 2008; Mann et al. 2016; Slutske et al. 2019). At the same time, previous research on the Family Check-Up identified parental monitoring and involvement with deviant peers as mechanisms of treatment effects on problem behavior (Kuo et al. 2019). Together, these converging lines of inquiry suggest that parental monitoring is an environmental switch that can disrupt the pathway between genotype and alcohol dependence phenotype. Future research could mine similar points of convergence between existing interventions and the behavioral genetic literature.
CONCLUSION
The study of how genes contribute to individual differences in human psychology will probably always be an object of fascination and fear. The field of behavioral genetics connects some of our most cherished aspects of human identity and our most prized accomplishments to an accident of birth that preceded our conscious awareness. This fascination and fear fuel continuing controversies over statistical parameters like heritability and over the legitimacy of conducting behavioral genetic research at all. In the past few decades, there have been incredible technological advances that have made it possible to measure the human genome directly. This genomic technology has not, as some people feared, vindicated a biodeterministic view of human development. As described in this article, genetic studies have provided some of the strongest evidence for the continued importance of the social environment for the human life course. At the same time, neither has genomic technology invalidated the central methods and conclusions of human behavioral genetics. Instead, the future of behavior genetics is both more nuanced and more scientifically interesting than the picture painted by its ardent champions or its vociferous critics. Never before have behavior geneticists had such a wide array of powerful tools; never before have the methodological and theoretical challenges of connecting DNA to human thoughts, feelings, behaviors, and identities been more apparent.
Heritability:
the proportion of population variation in a phenotype that is due to genotypic differences
Genotype:
the unique genetic makeup of an organism
Phenotype:
the observable characteristics of an organism (e.g., height)
Genome-wide association study (GWAS):
a method for testing the correlations between a phenotype and a large number of measured genetic differences, most commonly SNPs
Polygenic score:
a single number that aggregates information from an individual’s genotype to estimate the individual’s likelihood of showing a particular phenotype
Gene–environment interplay:
an umbrella term that captures various statistical and biological concepts about the joint action of genes and environments on the development of phenotypes
Nonshared environment (e2):
the extent to which identical twins raised in the same home differ in their phenotype
Genotyping:
measuring the unique genetic makeup of an organism by measuring some or all of its DNA sequences
Single-nucleotide polymorphism (SNP):
a type of genetic difference whereby individuals differ in a single DNA nucleotide
Zygosity:
the genetic relationship between twins; monozygotic twins are genetically identical, whereas dizygotic twins have the same genetic relationship as nontwin full siblings
Missing heritability:
the gap between the heritability of a phenotype estimated from a twin study and the variation in a phenotype accounted for by measured genotypes
Nucleotide:
part of the DNA molecule; a DNA sequence is made up of 4 types of nucleotides, which are abbreviated G, C, T, and A
Allele:
one of two or more versions of a gene; the rarer version of a gene is the minor allele
Summary statistics:
list of genetic variants and association statistics resulting from a GWAS that reflect the strength of those variants’ association with a target phenotype
Population stratification:
systematic differences in allele frequencies between groups of people; uncontrolled population stratification can confound GWASs, leading to spurious results
Genetic ancestry:
patterns of genetic similarity and dissimilarity in human populations that are the result of human demographic history
Polygenic:
influenced by many genetic variants
Candidate gene:
a gene that is studied in relation to a phenotype based on an a priori hypothesis about the gene’s biological function
Genetic determinism:
the flawed idea that a person’s behavior and life outcomes are conclusively determined by their DNA
SUMMARY POINTS.
Genetic differences between people matter for every aspect of their thinking, feeling, and behavior. Psychological characteristics and behaviors are typically influenced by very many genes.
We can now measure the human genome cheaply and easily. Results from genomic research have validated many of the assumptions of traditional behavioral genetics, and family studies are more important than ever.
Studies on candidate gene × environment interactions, as well as studies that correlate aspects of child development with aspects of environments provided by biological relatives, continue to be popular within psychology. These studies are often methodologically flawed, are unlikely to yield true insights, and can waste valuable scientific resources.
Advances in genotyping technology, open science practices, massive sample sizes, and large-scale international collaborations have finally begun to yield replicable knowledge about specific genes associated with human psychology and behavior. This knowledge can most readily be put to use by psychological researchers in the form of polygenic scores.
Genetic research has provided strong evidence that families reproduce their social privilege across generations via environmental mechanisms. It has also shown that the effects of the natural lottery of genetic inheritance play out differently across historical eras, sociopolitical contexts, and other social structures.
Future psychological research on gene × environment interaction should prioritize experimental and econometric methods that allow for stronger inferences about the causality of environments. This approach has the potential to answer critical questions about who is being served by existing interventions and how genetic differences are translated into complex human outcomes.
Instead of shunning genetics out of fear that it will subvert cherished values about human equality, or embracing a socially dangerous and scientifically flawed biodeterminism, psychologists have a new opportunity to use the tools of the postgenomic age to improve human health and well-being.
ACKNOWLEDGMENTS
The author was supported by an Early Career Research Fellowship from the Jacobs Foundation.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Bakermans-Kranenburg MJ, van IJzendoorn MH. 2015. The hidden efficacy of interventions: gene × environment experiments from a differential susceptibility perspective. Annu. Rev. Psychol 66:381–409 [DOI] [PubMed] [Google Scholar]
- Barcellos SH, Carvalho LS, Turley P. 2018. Education can reduce health differences related to genetic risk of obesity. PNAS 115(42):E9765–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr PB, Dick DM. 2019. The genetics of externalizing problems. Curr. Top. Behav. Neurosci In press. 10.1007/7854_2019_120 [DOI] [PubMed]
- Belsky DW, Domingue BW, Wedow R, Arseneault L, Boardman JD, et al. 2018. Genetic analysis of social-class mobility in five longitudinal studies. PNAS 115(31):E7275–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DJ, Cesarini D, Chabris CF, Glaeser EL, Laibson DI, et al. 2012. The promises and pitfalls of genoeconomics. Annu. Rev. Econ 4:627–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierbach D, Laskowski KL, Wolf M. 2017. Behavioural individuality in clonal fish arises despite near-identical rearing conditions. Nat. Commun 8:15361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Border R, Johnson EC, Evans LM, Smolen A, Berley N, et al. 2019. No support for historical candidate gene or candidate gene-by-interaction hypotheses for major depression across multiple large samples. Am. J. Psychiatry 176(5):376–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branigan AR, McCallum KJ, Freese J. 2013. Variation in the heritability of educational attainment: an international meta-analysis. Soc. Forces 92(1):109–40 [Google Scholar]
- Briley DA, Tucker-Drob EM. 2017. Comparing the developmental genetics of cognition and personality over the life span. J. Personal 85(1):51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, et al. 2019. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 47(D1):D1005–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt SA, Plaisance KS, Hambrick DZ. 2019. Understanding “what could be”: a call for “experimental behavioral genetics.” Behav. Genet 49(2):235–43 [DOI] [PubMed] [Google Scholar]
- Case A, Deaton A. 2017. Mortality and morbidity in the 21st century. Brook. Pap. Econ. Act 2017(1):397–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, et al. 2003. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science 301(5631):386–89 [DOI] [PubMed] [Google Scholar]
- Ceci SJ, Papierno PB. 2005. The rhetoric and reality of gap closing: when the “have-nots” gain but the “haves” gain even more. Am. Psychol 60(2):149–60 [DOI] [PubMed] [Google Scholar]
- Chabris CF, Hebert BM, Benjamin DJ, Beauchamp J, Cesarini D, et al. 2012. Most reported genetic associations with general intelligence are probably false positives. Psychol. Sci 23(11):1314–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabris CF, Lee JJ, Cesarini D, Benjamin DJ, Laibson DI. 2015. The fourth law of behavior genetics. Curr. Dir. Psychol. Sci 24(4):304–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charney E 2012. Behavior genetics and postgenomics. Behav. Brain Sci 35(5):331–58 [DOI] [PubMed] [Google Scholar]
- Cheesman R, Hunjan A, Coleman JRI, Ahmadzadeh Y, Plomin R, et al. 2019. Comparison of adopted and non-adopted individuals reveals gene–environment interplay for education in the UK Biobank. bioRxiv 707695 10.1101/707695 [DOI] [PMC free article] [PubMed]
- Columbia Law Sch. 2017. Faculty statement on Charles Murray Lecture. Columbia Law School, March 20. https://web.law.columbia.edu/open-university-project/academic-freedom/faculty-murray-statement
- Conley D, Rauscher E, Dawes C, Magnusson PKE, Siegal ML. 2013. Heritability and the equal environments assumption: evidence from multiple samples of misclassified twins. Behav. Genet 43(5):415–26 [DOI] [PubMed] [Google Scholar]
- Cooke ME, Meyers JL, Latvala A, Korhonen T, Rose RJ, et al. 2015. Gene–environment interaction effects of peer deviance, parental knowledge and stressful life events on adolescent alcohol use. Twin Res. Hum. Genet 18(5):507–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar-Nimrod I, Heine SJ. 2011. Genetic essentialism: on the deceptive determinism of DNA. Psychol. Bull 137(5):800–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton A 2013. The Great Escape: Health, Wealth, and the Origins of Inequality Princeton, NJ: Princeton Univ. Press [Google Scholar]
- Dennett DC. 2015. Elbow Room: The Varieties of Free Will Worth Wanting Cambridge, MA: MIT Press. 2nd ed. [Google Scholar]
- Dick DM, Rose R, Viken R, Kaprio J, Koskenvuo M. 2001. Exploring gene–environment interactions: socioregional moderation of alcohol use. J. Abnorm. Psychol 110(4):625–32 [DOI] [PubMed] [Google Scholar]
- Dick DM, Viken R, Purcell S, Kaprio J, Pulkkinen L, Rose RJ. 2007. Parental monitoring moderates the importance of genetic and environmental influences on adolescent smoking. J. Abnorm. Psychol 116(1):213–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dishion TJ, Kavanagh K. 2003. Intervening in Adolescent Problem Behavior: A Family-Centered Approach New York: Guilford [Google Scholar]
- Dobzhansky T 1962. Genetics and equality: equality of opportunity makes the genetic diversity among men meaningful. Science 137(3524):112–15 [DOI] [PubMed] [Google Scholar]
- Dudbridge F 2013. Power and predictive accuracy of polygenic risk scores. PLOS Genet 9(3):e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. 2011. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am. J. Psychiatry 168(10):1041–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Pollastri AR, Smoller JW. 2014. Mind the gap: why many geneticists and psychological scientists have discrepant views about gene–environment interaction (G×E) research. Am. Psychol 69(3):249–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Shen H, Gelaye B, Meijsen J, Ressler K, et al. 2019. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun 10:3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt LE, Briley DA, Mann FD, Harden KP, Tucker-Drob EM. 2015. Genes unite executive functions in childhood. Psychol. Sci 26(8):1151–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt LE, Church JA, Harden KP, Tucker-Drob EM. 2019. Accounting for the shared environment in cognitive abilities and academic achievement with measured socioecological contexts. Dev. Sci 22(1):e12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt LE, Mann FD, Briley DA, Church JA, Harden KP, Tucker-Drob EM. 2016. Strong genetic overlap between executive functions and intelligence. J. Exp. Psychol. Gen 145(9):1141–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engzell P, Tropf FC. 2019. Heritability of education rises with intergenerational mobility. PNAS 116(51):25386–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. 1919. The correlation between relatives on the supposition of Mendelian inheritance. Earth Environ. Sci. Trans. R. Soc. Edinb 52(2):399–433 [Google Scholar]
- Fletcher JM, Conley D. 2013. The challenge of causal inference in gene–environment interaction research: leveraging research designs from the social sciences. Am. J. Public Health 103(Suppl. 1):S42–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D 2019. Subversive science. Penn State Law Rev 124(1):153–91 [Google Scholar]
- Freese J 2008. Genetics and the social science explanation of individual outcomes. Am. J. Sociol 114(Suppl. 1):S1–35 [DOI] [PubMed] [Google Scholar]
- Freund J, Brandmaier AM, Lewejohann L, Kirste I, Kritzler M, et al. 2013. Emergence of individuality in genetically identical mice. Science 340(6133):756–59 [DOI] [PubMed] [Google Scholar]
- Friedman NP, Miyake A, Young SE, DeFries JC, Corley RP, Hewitt JK. 2008. Individual differences in executive functions are almost entirely genetic in origin. J. Exp. Psychol. Gen 137(2):201–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberger AS. 1979. Heritability. Economica 46(184):327–47 [Google Scholar]
- Halldorsdottir T, Binder EB. 2017. Gene × environment interactions: from molecular mechanisms to behavior. Annu. Rev. Psychol 68:215–41 [DOI] [PubMed] [Google Scholar]
- Hamer D, Sirota L. 2000. Beware the chopsticks gene. Mol. Psychiatry 5(1):11–13 [DOI] [PubMed] [Google Scholar]
- Hannikainen IR. 2019. Ideology between the lines: lay inferences about scientists’ values and motives. Soc. Psychol. Personal. Sci 10(6):832–41 [Google Scholar]
- Harden KP, Domingue BW, Belsky DW, Boardman JD, Crosnoe R, et al. 2019. Genetic associations with mathematics tracking and persistence in secondary school. bioRxiv 598532 10.1101/598532 [DOI] [PMC free article] [PubMed]
- Harden KP, Hill JE, Turkheimer E, Emery RE. 2008. Gene–environment correlation and interaction in peer effects on adolescent alcohol and tobacco use. Behav. Genet 38(4):339–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich J, Heine SJ, Norenzayan A. 2010. The weirdest people in the world? Behav. Brain Sci 33(2–3):61–83 [DOI] [PubMed] [Google Scholar]
- Herrnstein RJ, Murray C. 1996. The Bell Curve: Intelligence and Class Structure in American Life New York: Free Press [Google Scholar]
- Hewitt JK. 2012. Editorial policy on candidate gene association and candidate gene-by-environment interaction studies of complex traits. Behav. Genet 42(1):1–2 [DOI] [PubMed] [Google Scholar]
- Jencks C, Philipps M. 1998. The black-white test score gap: why it persists and what can be done. Brookings, March 1
- Jensen A 1969. How much can we boost IQ and scholastic achievement? Harv. Educ. Rev 39(1):1–123 [Google Scholar]
- Jerskey BA, Panizzon MS, Jacobson KC, Neale MC, Grant MD, et al. 2010. Marriage and divorce: a genetic perspective. Personal. Individ. Differ 49(5):473–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W, Deary IJ, Silventoinen K, Tynelius P, Rasmussen F. 2010. Family background buys an education in Minnesota but not in Sweden. Psychol. Sci 21(9):1266–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MC. 2014. Gene × environment interaction studies have not properly controlled for potential confounders: the problem and the (simple) solution. Biol. Psychiatry 75(1):18–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Turkheimer E, Ohlsson H, Sundquist J, Sundquist K. 2015. Family environment and the malleability of cognitive ability: a Swedish national home-reared and adopted-away cosibling control study. PNAS 112(15):4612–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent DM, Rothwell PM, Ioannidis JP, Altman DG, Hayward RA. 2010. Assessing and reporting heterogeneity in treatment effects in clinical trials: a proposal. Trials 11(1):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koellinger PD, Harden KP. 2018. Using nature to understand nurture. Science 359(6374):386–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, et al. 2018. The nature of nurture: effects of parental genotypes. Science 359(6374):424–28 [DOI] [PubMed] [Google Scholar]
- Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M. 2002. Etiologic connections among substance dependence, antisocial behavior and personality: modeling the externalizing spectrum. J. Abnorm. Psychol. 111(3):411–24 [PubMed] [Google Scholar]
- Kuo SI-C, Salvatore JE, Aliev F, Ha T, Dishion TJ, Dick DM. 2019. The Family Check-Up intervention moderates polygenic influences on long-term alcohol outcomes: results from a randomized intervention trial. Prev. Sci. 20(7):975–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, et al. 2018. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet 50(8):1112–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner RM. 2006. Another nine-inch nail for behavioral genetics! Hum. Dev 49(6):336–42 [Google Scholar]
- Lewontin RC. 1974. Annotation: the analysis of variance and the analysis of causes. Am. J. Hum. Genet 26(3):400–11 [PMC free article] [PubMed] [Google Scholar]
- Linneweber GA, Andriatsilavo M, Dutta SB, Bengochea M, Hellbruegge L, et al. 2020. A neurodevelopmental origin of behavioral individuality in the Drosophila visual system. Science 367(6482):1112–19 [DOI] [PubMed] [Google Scholar]
- Mann FD, Patterson MW, Grotzinger AD, Kretsch N, Tackett JL, et al. 2016. Sensation seeking, peer deviance, and genetic influences on adolescent delinquency: evidence for person-environment correlation and interaction. J. Abnorm. Psychol 125(5):679–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, et al. 2009. Finding the missing heritability of complex diseases. Nature 461(7265):747–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manski CF. 2011. Genes, eyeglasses, and social policy. J. Econ. Perspect 25(4):83–94 [Google Scholar]
- Manuck SB, McCaffery JM. 2014. Gene–environment interaction. Annu. Rev. Psychol 65:41–70 [DOI] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, et al. 2017. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet 100(4):635–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. 2019. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet 51(4):584–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSwiggan S, Elger B, Appelbaum PS. 2017. The forensic use of behavioral genetics in criminal proceedings: case of the MAOA-L genotype. Int. J. Law Psychiatry 50:17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merton RK. 1968. The Matthew effect in science: The reward and communication systems of science are considered. Science 159(3810):56–63 [PubMed] [Google Scholar]
- Morris TT, Davies NM, Smith GD. 2019. Can education be personalised using pupils’ genetic data? bioRxiv 645218 10.1101/645218 [DOI] [PMC free article] [PubMed]
- Mostafavi H, Harpak A, Agarwal I, Conley D, Pritchard JK, Przeworski M. 2020. Variable prediction accuracy of polygenic scores within an ancestry group. eLife 9:e48376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C 2020. Human Diversity: The Biology of Gender, Race, and Class New York: Twelve [Google Scholar]
- Okbay A, Becker J, Benjamin D, Burik CA, Cesarini D, Turley P. 2019. A repository of polygenic scores. Behav. Genet 49(6):507 (Abstr.) [Google Scholar]
- Panofsky A 2014. Misbehaving Science Chicago: Univ. Chicago Press [Google Scholar]
- Papageorge NW, Thom K. 2020. Genes, education, and labor market outcomes: evidence from the Health and Retirement Study. J. Eur. Econ. Assoc 18:1351–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parens E 2004. Genetic differences and human identities: on why talking about behavioral genetics is important and difficult. Hastings Cent. Rep 34(1):S4–35 [PubMed] [Google Scholar]
- Plomin R 2018. Blueprint: How DNA Makes Us Who We Are Cambridge, MA: MIT Press [Google Scholar]
- Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, et al. 2015. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet 47(7):702–9 [DOI] [PubMed] [Google Scholar]
- Rawls J 1999 (1971). A Theory of Justice Cambridge, MA: Harvard Univ. Press. Rev. ed. [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, et al. 2013. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340(6139):1467–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimfeld K, Krapohl E, Trzaskowski M, Coleman JRI, Selzam S, et al. 2018a. Genetic influence on social outcomes during and after the Soviet era in Estonia. Nat. Hum. Behav 2(4):269–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimfeld K, Malanchini M, Krapohl E, Hannigan LJ, Dale PS, Plomin R. 2018b. The stability of educational achievement across school years is largely explained by genetic factors. npj Sci. Learn 3(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter M, Moffitt TE, Caspi A. 2006. Gene–environment interplay and psychopathology: multiple varieties but real effects. J. Child Psychol. Psychiatry 47(3–4):226–61 [DOI] [PubMed] [Google Scholar]
- Rutter M, Silberg J. 2002. Gene–environment interplay in relation to emotional and behavioral disturbance. Annu. Rev. Psychol 53:463–90 [DOI] [PubMed] [Google Scholar]
- Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A. 2017. The heritability of autism spectrum disorder. JAMA 318(12):1182–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz L, Conley D. 2016. The long-term consequences of Vietnam-era conscription and genotype on smoking behavior and health. Behav. Genet 46:43–58 [DOI] [PubMed] [Google Scholar]
- Schmitz L, Conley D. 2017. Modeling gene–environment interactions with quasi-natural experiments. J. Personal 85(1):10–21 [DOI] [PubMed] [Google Scholar]
- Schulz W, Schunck R, Diewald M, Johnson W. 2017. Pathways of intergenerational transmission of advantages during adolescence: social background, cognitive ability, and educational attainment. J. Youth Adolesc 46(10):2194–214 [DOI] [PubMed] [Google Scholar]
- Sharma S, Powers A, Bradley B, Ressler KJ. 2016. Gene × environment determinants of stress- and anxiety-related disorders. Annu. Rev. Psychol 67:239–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutske WS, Deutsch AR, Piasecki TM. 2019. Neighborhood density of alcohol outlets moderates genetic and environmental influences on alcohol problems. Addiction 114(5):815–22 [DOI] [PubMed] [Google Scholar]
- Sotoudeh R, Harris KM, Conley D. 2019. Effects of the peer metagenomic environment on smoking behavior. PNAS 116(33):16302–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- State v. Yepez, A-1-CA-35330 (N.M. Ct. App. 2018)
- Tabery J 2014. Beyond Versus: The Struggle to Understand the Interaction of Nature and Nurture Cambridge, MA: MIT Press [Google Scholar]
- Tick B, Bolton P, Happé F, Rutter M, Rijsdijk F. 2016. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J. Child Psychol. Psychiatry 57(5):585–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trejo S, Domingue BW. 2019. Genetic nature or genetic nurture? Quantifying bias in analyses using polygenic scores. bioRxiv 524850 10.1101/524850 [DOI]
- Tropf FC, Lee SH, Verweij RM, Stulp G, van der Most PJ, et al. 2017. Hidden heritability due to heterogeneity across seven populations. Nat. Hum. Behav 1(10):757–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker-Drob EM, Bates TC. 2016. Large cross-national differences in gene × socioeconomic status interaction on intelligence. Psychol. Sci 27(2):138–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker-Drob EM, Briley DA, Harden KP. 2013. Genetic and environmental influences on cognition across development and context. Curr. Dir. Psychol. Sci 22(5):349–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker-Drob EM, Harden KP. 2012. Early childhood cognitive development and parental cognitive stimulation: evidence for reciprocal gene–environment transactions. Dev. Sci 15(2):250–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkheimer E 1998. Heritability and biological explanation. Psychol. Rev 105(4):782–91 [DOI] [PubMed] [Google Scholar]
- Turkheimer E 2000. Three laws of behavior genetics and what they mean. Curr. Dir. Psychol. Sci 9(5):160–64 [Google Scholar]
- Turkheimer E 2011. Genetics and human agency: comment on Dar-Nimrod and Heine 2011. Psychol. Bull 137(5):825–28 [DOI] [PubMed] [Google Scholar]
- Turkheimer E, Haley A, Waldron M, D’Onofrio B, Gottesman II. 2003. Socioeconomic status modifies heritability of IQ in young children. Psychol. Sci 14(6):623–28 [DOI] [PubMed] [Google Scholar]
- van der Sluis S, Posthuma D, Dolan CV. 2012. A note on false positives and power in G × E modelling of twin data. Behav. Genet 42(1):170–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hulle CA, Lahey BB, Rathouz PJ. 2013. Operating characteristics of alternative statistical methods for detecting gene-by-measured environment interaction in the presence of gene-environment correlation in twin and sibling studies. Behav. Genet 43(1):71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Hill WG, Wray NR. 2008. Heritability in the genomics era—concepts and misconceptions. Nat. Rev. Genet 9(4):255–66 [DOI] [PubMed] [Google Scholar]
- Visscher PM, Medland SE, Ferreira MAR, Morley KI, Zhu G, et al. 2006. Assumption-free estimation of heritability from genome-wide identity-by-descent sharing between full siblings. PLOS Genet 2(3):e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainschtein P, Jain DP, Yengo L, Zheng Z, TOPMed Anthropometry Work. Group, et al. 2019. Recovery of trait heritability from whole genome sequence data. bioRxiv 588020 10.1101/588020 [DOI]
- Westfall J, Yarkoni T. 2016. Statistically controlling for confounding constructs is harder than you think. PLOS ONE 11(3):e0152719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby EA, Love AC, McGue M, Iacono WG, Quigley J, Lee JJ. 2019. Free will, determinism, and intuitive judgments about the heritability of behavior. Behav. Genet 49(2):136–53 [DOI] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, et al. 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet 42(7):565–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager DS, Hanselman P, Walton GM, Murray JS, Crosnoe R, et al. 2019. A national experiment reveals where a growth mindset improves achievement. Nature 573(7774):364–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AI. 2019. Solving the missing heritability problem. PLOS Genet 15(6):e1008222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AI, Benonisdottir S, Przeworski M, Kong A. 2019. Deconstructing the sources of genotype-phenotype associations in humans. Science 365(6460):1396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T-Y, Meaney MJ. 2010. Epigenetics and the environmental regulation of the genome and its function. Annu. Rev. Psychol 61:439–66 [DOI] [PubMed] [Google Scholar]


