Abstract
Insulin-like growth factor-1 receptor (IGF-1R) has been made an attractive anticancer target due to its overexpression in cancers. However, targeting it has often produced the disappointing results as the role played by cross talk with numerous downstream signalings. Here, we report a disobliging IGF-1R signaling which promotes growth of cancer through triggering the E3 ubiquitin ligase MEX3A-mediated degradation of RIG-I. The active β-arrestin-2 scaffolds this disobliging signaling to talk with MEX3A. In response to ligands, IGF-1Rβ activated the basal βarr2 into its active state by phosphorylating the interdomain domain on Tyr64 and Tyr250, opening the middle loop (Leu130‒Cys141) to the RING domain of MEX3A through the conformational changes of βarr2. The models of βarr2/IGF-1Rβ and βarr2/MEX3A could interpret the mechanism of the activated-IGF-1R in triggering degradation of RIG-I. The assay of the mutants βarr2Y64A and βarr2Y250A further confirmed the role of these two Tyr residues of the interlobe in mediating the talk between IGF-1Rβ and the RING domain of MEX3A. The truncated-βarr2 and the peptide ATQAIRIF, which mimicked the RING domain of MEX3A could prevent the formation of βarr2/IGF-1Rβ and βarr2/MEX3A complexes, thus blocking the IGF-1R-triggered RIG-I degradation. Degradation of RIG-I resulted in the suppression of the IFN-I-associated immune cells in the TME due to the blockade of the RIG-I-MAVS-IFN-I pathway. Poly(I:C) could reverse anti-PD-L1 insensitivity by recovery of RIG-I. In summary, we revealed a disobliging IGF-1R signaling by which IGF-1Rβ promoted cancer growth through triggering the MEX3A-mediated degradation of RIG-I.
KEY WORDS: IGF-1R, RIG-I, β-Arrestin-2 (βarr2), K48-linked ubiquitination, MEX3A, Anti-PD-L1
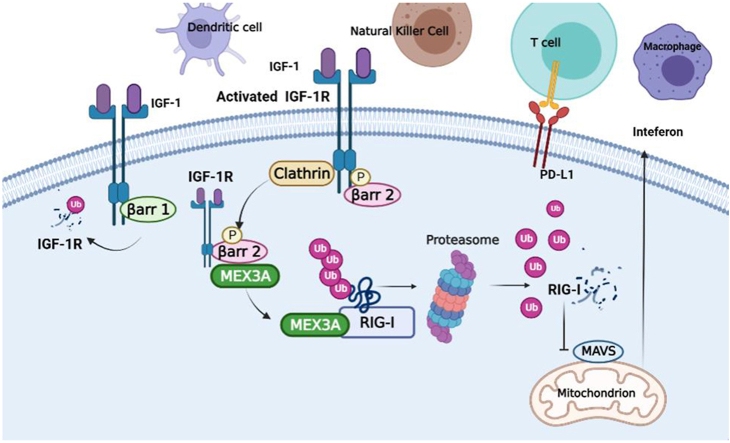
Graphical abstract
IGF-1R activation triggers the MEX3A-mediated degradation of RIG-I, leading to the suppression of the IFN-I-associated immune cells in TME due to the blockade of the RIG-I-MAVS-IFN-I pathway in colorectal cancer.
1. Introduction
Type 1 insulin-like growth factor receptor (IGF-1R), a receptor tyrosine kinase (RTK), composed of two extracellular α-subunits and two cytoplasmic β-subunits, has been known as the binary model1. Ligands binding to IGF-1R leads to activation of two main cascades: the insulin receptor substrate (IRS)-initiated phosphatidylinositol 3-kinase (PI3K)-AKT/mammalian target of rapamycin (mTOR) pathway, which predominantly leads to the metabolic outcomes, and the SHC-initiated Ras-mitogen-activated protein kinase (MAPK) pathway, which promotes the mitogenic outcomes. This receptor makes an attractive anticancer target due to its overexpression in cancers2, 3, 4. However, targeting it has often produced disappointing results as the role played by cross talk between IGF-1R and numerous downstream signalings, thus renders cancer cells resistant to anti-IGF-1R therapy5, 6, 7, 8. However, the mechanisms of these cross talks have not been understood completely, and so far, is still a baffling problem.
It is known that RTKs can be internalized by ligand binding via clathrin-or caveolin-mediated vesicles, following a process of the proteasome pathway to decide their fate9,10. The current studies are mainly concentrated on the following two modes11: one is postulated the internalization by a tyrosine-based motif. In response to ligands, IGF-1Rβ is targeted to the clathrin-coated membrane invagination to internalization. Another is a ubiquitin-based motif that “borrowed” the concept from the G-protein coupled receptor (GPCR) signal, which β-arrestins (βarr1/2) could mediate IGF-1Rβ to cease the IGF-1R signal through the ubiquitin-proteasome process. Actually, these “borrowed” concepts from GPCR signaling did not fully explain many pathological syndromes recently observed in tumorigenesis.
We have noted that although βarr1 and βarr2 share a high degree of the sequences and the structural homology, they actually are not functionally redundant12,13. They can act as signaling molecules in their own right and result in unique cellular, physiological, and pathophysiological consequences14. Accordingly, different β-arrestins isoforms might determine the differential fates of IGF-1R signal through their specificity of recognizing substrates. In this study, we identified a specific fate of IGF-1R signal determined by βarr2 in colonic cancer cells. In response to ligands, although IGF-1Rβ was bound with Clathrin but not followed the ubiquitin-proteasome process to cease IGF-1R signal but to interact with βarr2, leading to the E3 ligase MEX3A-mediated degradation of RIG-I. We proposed the βarr2/IGF-1Rβ and βarr2/MEX3A models to interpret the mechanism of the activated-IGF-1R in triggering MEX3A activity to degrade RIG-I. We designed the mutants of βarr2Y64A and βarr2Y250A to interpret the mechanism of the IGF-1Rβ-attached “phosphates” in phosphorylating Tyr64 and Tyr250 of the interdomain domain. Since the binding strength between two proteins is dependent on their interface size, we thus performed the assay of truncated-βarr2 to check the response of βarr2 to the activated-IGF-1R signaling, and the assay of the peptide to test the response of the active βarr2 to the RING domain of MEX3A. The middle loop (Leu130‒Cys141) of the active βarr2 was identified as the core region to adapt the structure of RING domain of MEX3A. It is the fully length RING domain of MEX3A but not the truncated-MEX3A that interacted with the middle loop of βarr2. The peptide that mimicked the contact surface A512‒F519 (sequencing ATQAIRIF) could block the interaction of the active βarr2 with the RING domain surface of MEX3A.
It is well known that RIG-I, an innate immunity sensor, plays an important role in the immunotherapy for cancers through activating the IFN-I-associated immune cells in tumor microenvironment (TME). Theoretically, RIG-I expression induces the release of type I interferons (IFNs). In the present study, RIG-I degradation resulted in the lower level of IFN-I in the TME due to the blockade of the RIG-I-MAVS-IFN pathway15, 16, 17. We thus tested the suppression of the IFN-I-associated immune cells in the TME. MC38 (a CRC line sensitive to PD-L1 ICI) and CT26 (a CRC line insensitive to PD-L1 ICI) were employed to evaluate the potential significance of these findings. MC38 cells exposed to IGF-1 become insensitive to anti-PD-L1 therapy due to the suppression of the IFN-I-associated immune cells. Poly(I:C) could reverse anti-PD-L1 insensitivity by recovery of RIG-I, suggesting the strategy for dealing with the IGF-1R-caused the “insensitive to PD-L1” tumors.
2. Methods
2.1. Cell lines and cell culture
Human colonic cancer cell lines SW620, RKO, normal colonic cells NCM460 and HEK293T cells were obtained from Cell Bank of China (Shanghai, China). All cells were maintained in DMEM (Gibco, Carlsbad, CA, USA) supplemented with 10% FBS (Procell Life Science &Technology Co., Ltd.) and incubated in a humidified incubator with 5% CO2 at 37 °C. All reagents were indicated in Supporting Information Table S1.
2.2. Construction of plasmids
Human βarr2 cDNA was subcloned in pEGFP-N1 (Sangon Biotech) with a Flag epitope tag. Human MEX3A cDNA were subcloned in pEGFP-N1 (Sangon Biotech) with a His tag. All constructs were confirmed by DNA sequencing.
2.3. Lentiviral infection
Lentivirus was used to establish individual stable cells. Empty vector was used as control for the shRNA-based knockdown. Virus was incubated with target cells for 24 h with 10 mg/mL polybrene, and the cells were allowed to recover for 24 h before selection. The infected cells were selected in 2 mg/mL puromycin until the uninfected control cells were dead.
2.4. Cell transfection
HEK293T cells were seeded at a density of 6 × 105 cells in Glass Bottom Culture Dishes. After medium was replaced with Opti-MEM at approximately 60% confluence, cells were transfected with 5 μg of plasmid using Lipofectamine™ 3000 (Invitrogen) according to the manufacturer's instructions. Plasmids encoded either N-terminal Flag-tagged intact βarr2 or N-terminal His-tagged N-terminal truncated-MEX3A. siRNAs targeting IGF1R, MEX3A, βarr1, and βarr2 were synthesized by GenePharma. Information of siRNA, shRNA sequences, and the plasmids-based constructs was indicated in Supporting Information Table S2.
2.5. Western blotting, co-immunoprecipitation (Co-IP) and ubiquitination assays
Western blotting and Co-IP assays were routinely performed were routinely performed as described elsewhere. Ubiquitination assay was performed as described previously18. Cell lysates were sonicated in IP buffer with protease inhibitor cocktail and phosphorylate inhibitor for 10 min on ice. Supernatant was separated by 10% SDS-PAGE and then subjected to Western blotting with anti-polyubiquitin linkage-specific antibodies. Information of all antibodies was indicated in Supporting Information Table S3.
2.6. Immunofluorescence analysis, immunohistochemical (IHC) and multiplex immunohistochemistry (mIHC) assays
Immunofluorescence analysis, IHC and mIHC assays were routinely performed as described elsewhere. Information of all antibodies was indicated in Supporting Information Tables S4 and S5.
2.7. RT-qPCR assay
RT-qPCR assay was routinely performed as described elsewhere. Information of all primers was presented in Supporting Information Table S6.
2.8. Model construction
Crystal structures of basal βarr2 (PDB: 1G4R), active βarr2 (PDB: 6U1N), active IGF1Rβ (PDB: 1K3A), and inactive IGF1R (PDB: 1P4O) were all extracted from the Protein Data Bank (https://www.rcsb.org/). 3D structure of MEX3A generated by AlphaFold19 was downloaded from the Uniprot database, and only the RING domain (S462–S520) was retained. Protein–protein docking was performed using HDOCK (https://doi.org/10.1093/nar/gkx407), pyDock20 and ZDOCK21. Best model was chosen for energy minimization and further analysis.
2.9. Peptide synthesis
The peptide (ATQAIRIF and FITC-{beta-Ala}-ATQAIRIF-GRKKRRQRRRPQ-NH2) were synthesized by GenScript Biological Company (Nanjing, China). They were produced on an Apex 396 (Aapptec) automated peptide synthesizer using Rink amide AM LL resin (EMD Biosciences, 0.2 mmol/g resin), at 50 mmol scale. The peptide was purified by reversed-phase HPLC (Agilent) using a C18 column (Zorbax) and further quantified by amino acid analysis on a Beckman 6300 high-performance amino acid analyzer. Solvent was PBS containing 5% trehalose, pH 7.2.
2.10. Igf1r+/− mice colorectal cancer model
The protocol of mice experiments was approved by Animal Welfare Committee of Capital Medical University (AEEI‒2020–094). Igf1r+/− mice (both male and female, 6 weeks of age) were exposed to AOM/DSS for inducing colorectal cancer as described previously22. At 20 weeks of age, mice were sacrificed, and their colorectal cancers were analyzed.
2.11. Human colonic cancer cells xenografted in nude mice
Athymic nude mice (6–8 weeks and sex matched) were purchased from Charles River Laboratories (Beijing, China). SW620 cells and SW620shIGF1R cells were respectively injected into armpits. When tumors reached the desired volume, tumor tissues were cut into 1.0 mm thick pieces and inoculated to muscularis mucosa of junction of rectum and sigmoid colon. Mice were randomly divided into control group (n = 6, 0.2 mL NS by vena caudalis injection) and IGF-I-treated group (n = 6, 1 mg/kg of rhIGF-I in 0.2 mL NS, Genentech, San Francisco, USA). Four weeks later, mice were sacrificed and colonic tumors were analyzed.
2.12. Murine colon cancer models
MC38 colon cancer cell line and CT26 colon cancer cell line were obtained from China Cell Bank (Shanghai, China). To generate MC38 tumor, C57BL/6N mice (6–8 weeks and sex matched) were inoculated subcutaneously with 0.5 × 106 cells into flank. Mice were randomized into 4 groups when tumors reached the desired volume (Day 0), and treatment (n = 6, 0.2 mL by vena caudalis injection) was initiated on Day 0. Groups were as follows: vehicle control (NS 0.2 mL/per day); rhIGF-I (1 mg/kg/per day); Avelumab (anti-PDL-L1, 1 mg/kg per week); rhIGF-I (1 mg/kg) + Avelumab (1 mg/kg). To generate CT26 tumor, BALB/c mice (6–8 weeks and sex matched) were inoculated subcutaneously with 0.5 × 106 cells into flank. Mice were randomized into 7 groups when tumors reached the desired volume (Day 0), and treatment (n = 6, 0.2 mL by vena caudalis injection) was initiated on Day 0. Groups were as follows: vehicle (NS 0.2 mL/per day); rhIGF-I (1 mg/kg/per day); Avelumab (2 mg/kg per week); rhIGF-I (1 mg/kg) + Avelumab (2 mg/kg); rhIGF-I (1 mg/kg) + Poly (I:C) (Genentech, San Francisco) 10 μg/kg; rhIGF-I (1 mg/kg) + poly(I:C) (10 μg/kg) + Avelumab (2 mg/kg); Poly(I:C) (10 μg/kg) + Avelumab (2 mg/kg per week). Four weeks later, mice were sacrificed and their tumors were obtained for further analysis.
2.13. Statistical analysis
Data are presented as mean ± standard deviation (SD). Student's t test was used to evaluate differences between two groups and one-way ANOVA to analyze differences among different groups. Statistical significance was defined as P < 0.05. Analysis was performed using Graphpad Prism 8 software.
3. Results
3.1. Activation of IGF-1R promoted growth of colorectal cancers was statistically associated with the down regulation of RIG-I
Our previous study showed that activation of IGF-1R promoted the growth of colonic cancers through downregulation of tumor RIG-I. Inhibition of PI3K‒AKT pathway by LY294002 could not prevent the IGF-1R-induced down regulation of RIG-I, suggesting a disobliging IGF-1R signaling in promoting cancer growth23. However, the mechanism of this disobliging IGF-1R signaling in down regulating RIG-I has not been investigated as yet.
In the present study, the disobliging IGF-1R signaling in promoting cancer growth through downregulating RIG-I was further confirmed in Igf1r+/− mice and nude mice with human colonic cancer cells. Igf1r+/− mice exposed to AOM/DSS developed fewer and smaller colorectal tumors than their WT littermates (Fig. 1A-i). Western blotting assay showed a relatively higher level of IGF-1R and a consequently lower level of RIG-I in the tumor tissues than in the paired precancerous tissues in Igf1r+/− mice. Comparably, Igf1r+/− mice exhibited a higher level of RIG-I in tumor cells than that in WT mice (Fig. 1A-ii and Supporting Information Fig. S1A). The lower levels of IGF-1R were significantly associated with the higher levels of RIG-I in colorectal cancer cells of Igf1r+/− mice (Fig. 1A-iii), or, conversely, the higher levels of IGF-1R were significantly associated with the lower levels of RIG-I in WT mice (Fig. 1A-iv).
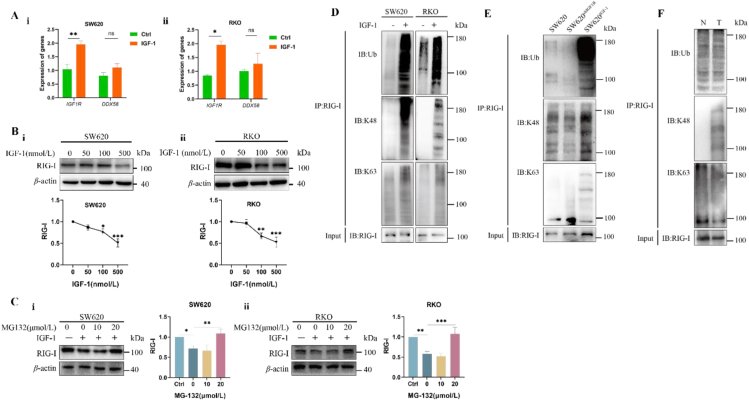
Figure 1.
Activation of IGF-1R promoted cancer growth through downregulation of RIG-I expression. (A) i) Exposed to AOM/DSS, Igf1r+/− mice developed fewer and the smaller-sized colorectal tumors than WT littermates. ii) Igf1r+/− mice had lower level of IGF-1R and higher level of RIG-I than WT littermates. iii) There was a correlation between the lower levels of IGF-1R and the higher levels of RIG-I in Igf1r+/− mice (n = 6). iv) The higher levels of IGF-1R were correlated with the lower levels of RIG-I in WT mice (n = 6). Data are presented as means ± SD; ∗∗P < 0.01, ∗∗∗P < 0.001. (B) i) As compared with WT SW620 xenografts, knockdown of IGF-1R retarded the IGF-1-stimulated growth in SW620shIGFR xenograft. ii) SW620 xenograft exposed to IGF-1 (SW620IGF-1) exhibited the higher IGF-1R level and the lower levels of RIG-I and IFN-I. SW620shIGFR xenograft had the lower level of IGF-1R and the higher levels of RIG-I and IFN-I. iii) There was a correlation between the lower levels of IGF-1R and the higher levels of RIG-I in SW620shIGFR xenograft (n = 6). iv) The higher levels of IGF-1R were correlated with the lower levels of RIG-I in SW620IGF-1 xenograft (n = 6). Data are presented as means ± SD; ∗P < 0.05. (C) i) Human colonic cancer tissues expressed the higher levels of IGF-1R and the lower levels of RIG-I (n = 20). ii) There was a correlation between the higher levels of IGF-1R and the lower levels of RIG-I (r = −0.5454). iii) A correlation between the lower levels of RIG-I and the lower levels of IFN-I (r = 0.7630). Data are presented as means ± SD; ∗∗P < 0.01 (n = 20). AOM = azoxymethane; DSS = dextran sodium sulfate.
Human SW620 cells xenografted in nude mice responded sensitively to IGF-1, resulting in the larger-sized tumors. Conversely, knockdown of IGF-1R in SW620shIGF1R cells insensitively responded to IGF-1, leading to the smaller-sized tumors (Fig. 1B-i). The larger-sized SW620 xenograft demonstrated the higher levels of IGF-1R and the consequently lower levels of RIG-I (Fig. 1B-ii and Fig. 1B-iii, r = −0.8425). Conversely, tumor retardation of SW620shIGF1R xenograft was correlated with the lower IGF-1R level and the consequent higher level of RIG-I (r = −0.8996) (Fig. 1B-ii and Fig. 1B-iv). We compared the severity of the xenografts of WT SW620, SW620IGF-1 and SW620shIGFR under microscope, supporting the above results (Fig. S1B and S1C). Together, these results indicated that the activated-IGF-1R in promoting cancer growth was mainly associated with its role in downregulation of RIG-I. It is well known that RIG-I expression releases IFN-I through activating the RIG-I-MAVS-IFN-I pathway15, 16, 17. Due to the down regulation of RIG-I, we thus tested the lower level of IFN-I in SW620 xenograft and, conversely, the higher level of IFN-I in SW620shIGF1R xenograft (Fig. 1B-ii).
The role of the activated-IGF-1R in the regulating RIG-I was finally confirmed in human colorectal cancers. Higher IGF-1R levels, lower RIG-I levels, and the consequent lower levels of IFN-I were tested in colorectal cancers than in the paired paracancerous tissues (Fig. 1C-i). A correlation between the higher levels of IGF-1R and the lower levels of RIG-I was established (r = 0.5454) (Fig. 1C-ii); and correspondently, a correlation between the lower levels of RIG-I and the decreasing levels of IFN-I was also established in these colorectal cancers (Fig. 1C-iii, r = 0.7630, n = 20).
3.2. It is βarr2 but not βarr1 that mediated the transduction of the activated-IGF-1R in cancer cells
It is known that, in response to ligands, IGF-1R could internalize through binding with clathrin and β-arrestins, following the ubiquitin-proteasome process to cease IGF-1R signal7,8. In this study, IGF-1R was labeled by biotin for analyzing the trajectory of the activated-IGF-1R. Exposed to IGF-1 for 30 min, IGF-1Rβ was clearly internalized from cell membrane (Supporting Information Fig. S2A-i), and then bound with Clathrin in cytoplasm of RKO cells (Fig. S2A-ii). Similar results were also seen in SW620 cells (Fig. S2B-i and Fig. S2B-ii). The IGF-1Rβ/Clathrin complex was confirmed in SW620 xenografts injected with IGF-1 (rhIGF-1, 1 mg/kg) but not in SW620 xenografts injected with NS in nude mice (Fig. S2C). Further, Co-IP assay of subcellular proteins of SW620 cells determined a higher level of the IGF-1Rβ/Clathrin complex in membrane proteins than in cytoplasm proteins (Fig. 2A, first line). Meanwhile, the cytoplasmic protein had a higher level of the IGF-1Rβ/βarr2 complex than that of the membranous protein (Fig. 2A, third line). Interestingly, the IGF-1Rβ/Clathrin complex was gradually reduced (Fig. 2A, fifth line), and the consequent IGF-1Rβ/βarr2 complex was increased in the cytoplasm (Fig. 2A, seventh line). These results indicated a dynamic process of binding among IGF-1Rβ, Clathrin, and βarr2 in cancer cells. Supportive of this concept, SW620 cells exposed to the increasing levels of IGF-1 (0–1000 ng/mL) expressed an increasing βarr2 but not βarr1 (Fig. 2B). Normal colonic cells NCM460 expressed an appropriate level of IGF-1Rβ/βarr1 even under the higher level of IGF-1 (500 ng/mL), meanwhile a strong IGF-1Rβ/βarr2 complex was seen in the same concentration of IGF-1 (Fig. 2C). Comparably, a strong IGF-1Rβ/βarr2 complex but not an IGF-1Rβ/βarr1 complex was identified in SW620 cells under IGF-1 (Fig. 2D-i and Fig. 2D-ii), and SW620 xenograft grew in nude mice treated with IGF-1 (Fig. 2E-i and Fig. 2E-ii). Immunofluorescent analysis revealed the co-expression of IGF-1Rβ with βarr2 in cytoplasm of cancer cells. Pearson's correlation coefficient (PCC) analysis24 showed a higher colocalization in the IGF-1-treated RKO cells than control cells (0.71 vs. 0.52, P < 0.01) (Fig. S2D, above); and in the IGF-1-treated SW620 cells than control cells (0.70 vs. 0.39, P < 0.01) (Fig. S2D, below). Further, knockdown of βarr2 (Fig. 2F) or inhibition of βarr2 by Barbadin (Fig. 2G) both blocked the IGF-1-driven IGF-1Rβ/βarr2 complex in SW620 cells. Together, these results confirmed that it is βarr2 but not βarr1 that mediated the transduction of IGF-1Rβ in cancer cells.
Figure 2.
βarr2 mediated the transduction of IGF-1R signaling in cancer cells. (A) Exposed to IGF-1 for 30 min, IGF-1Rβ/Clathrin complex was decreased in cell membrane. Correspondingly, the IGF-1Rβ/βarr2 complex occurred in the cytoplasm of SW620 cells. (B) Upon IGF-1 (0–1000 ng/mL), βarr2 not βarr1 showed a dose-dependent increasing manner. Data are presented as means ± SD; n = 3. ∗∗P < 0.01 (one-way ANOVA). (C) Co-IP assay identified a weak IGF-1Rβ/βarr1 complex and a strong IGF-1Rβ/βarr2 complex under high dose of IGF-1 (500 ng/mL) in NCM460 cells. (D) Co-IP assay identified a strong IGF-1Rβ/βarr2 complex not IGF-1Rβ/βarr1 complex in the presence of IGF-1 (500 ng/mL) in SW620 cells. (E) SW620 xenograft was determined IGF-1Rβ/βarr2 complex not IGF-1Rβ/βarr1 complex in nude mice treated with IGF-1 (rhIGF-1, 1 mg/kg/day). (F) Knockdown of βarr2 failed the forming of IGF-1Rβ/βarr2 complex in SW620 cells. (G) Barbadin inhibited the forming of IGF-1Rβ/βarr2 complex in SW620 cells.
3.3. The K48-linked ubiquitination mediated the IGF-1R-induced RIG-I degradation
We investigated the mechanism of the activated-IGF-1R in the down regulation of RIG-I. Firstly, RT-qPCR analysis indicated that this regulation did not occur in the transcriptional levels of ddx58 and igf1r confirmed in both SW620 cells and RKO cells (Fig. 3A-i, ii). Then, cycloheximide (CHX) by 300 μg/mL, the inhibitor of protein synthesis, could completely inhibit the synthesis of RIG-I (Supporting Information Fig. S3A-i, ii) in cancer cells. Further, in the presence of IGF-1, the half-life of RIG-I was significantly shortened in cancer cells (Fig. S3A-iii). Meanwhile, under the increasing concentrations of IGF-1, these cancer cells still had the decreasing levels of RIG-I (Fig. 3B-i, ii). These results indicated that downregulation of RIG-I by IGF-1R was due to degradation in its protein level. Additionally, degradation of RIG-I could be reversed by proteasome inhibitor MG-132 (Fig. 3C-i, ii) but not autophagy inhibitor 3-methyladenine (3-MA) (Fig. S3B) and lysosome inhibitor NH4CL (Fig. S3C). These results suggested the proteasomal degradation of RIG-I in IGF-1-treated cancer cells. We finally identified that it is the K48- but not the K63-linked ubiquitination that mediated RIG-I degradation (Fig. 3D). SW620 xenografts but not SW620shIGF1R xenografts exhibited the K48-linked ubiquitination and proteasomal degradation of RIG-I in the IGF-1-treated nude mice (Fig. 3E). Importantly, human colonic cancers but not their precancerous tissues demonstrated the K48-linked ubiquitination and proteasomal degradation of RIG-I (Fig. 3F).
Figure 3.
The K48-linked ubiquitination mediated the IGF-1Rβ-induced RIG-I degradation. (A) RT-qPCR assay did not test a significant difference in ddx58 between IGF-1-treated cancer cells and vehicle-treated cancer cells. i) SW620 cells. ii) RKO cells. Data are presented as means ± SD; n = 3. ∗P < 0.05, ∗∗P < 0.01. (B) The decreasing levels of RIG-I were determined in cancer cells when they were exposed to the increasing concentrations of IGF-1 i) SW620 cells. ii) RKO cells. Data are presented as means ± SD; n = 3. ∗P < 0.05, ∗∗P < 0.01. ∗∗∗P < 0.001. (C) MG132 reversed the IGF-1R-induced degradation of RIG-I in i) SW620 cells and ii) RKO cells. Data are presented as means ± SD; n = 3. ∗P < 0.05, ∗∗P < 0.01. ∗∗∗P < 0.001. (D) Co-IP assay identified that K48- but not K63-linked ubiquitination mediated degradation of RIG-I in SW620 cells and RKO cells. (E) SW620 xenograft not SW620shIGF1R xenograft showed the K48-linked ubiquitination of RIG-I. (F) Human cancer tissues not paracancerous tissues showed the K48-linked ubiquitination of RIG-I.
3.4. The E3 ligase MEX3A was identified to be responsible for RIG-I degradation. However, IGF-1Rβ not directly interacted with MEX3A but induced βarr2 to interact with MEX3A
It is noted that E3 ligase can determine the substrate specificity in the process of proteasomal ubiquitination25. As RIG-I was recognized as a target of the K48-linked ubiquitination, we next searched for the E3 ligase isoform which was responsible for the proteasomal ubiquitination. Analysis of the total 667 E3 ligases which were collected in databases (https://hpcwebapps.cit.nih.gov/ESBL/Database/E3-ligases/ and https://hpcwebapps.cit.nih.gov/ESBL/Database/E3-ligases/RelatedProteins.html) identified all of E3 ligases which showed the difference between colonic cancers and normal colonic tissues (Supporting Information Fig. S4A). GEPIA (http://gepia.cancer-pku.cn/) showed the higher levels of MEX3A in colonic cancers than normal colonic tissues (Fig. S4B). RT-qPCR assay confirmed the relationship between MEX3A and DDX58 at their transcriptional level (Fig. S4C).
Analysis of TCGA indicated a positive correlation between mex3a and igf1r (Fig. 4A-i, r = 0.35, P = 2.8e-13), while a negative correlation between mex3a and ddx58 (Fig. 4A-ii, r = −0.16, P = 0.0013) in colonic cancers. These correlations among igf1r, mex3a and ddx58 were further confirmed in their protein levels. In cultured cells, level of MEX3A was upregulated as IGF-1R level was activated, while the level of RIG-I was consequently decreased as the levels of IGF-1R and MEX3A were increased in cancer cells exposed to IGF-1 (Fig. 4B). Knockdown of MEX3A failed the downregulation of RIG-I by the activated IGF-1R (Fig. 4C), indicating the essential role of MEX3A in the IGF-1R-induced degradation of RIG-I.
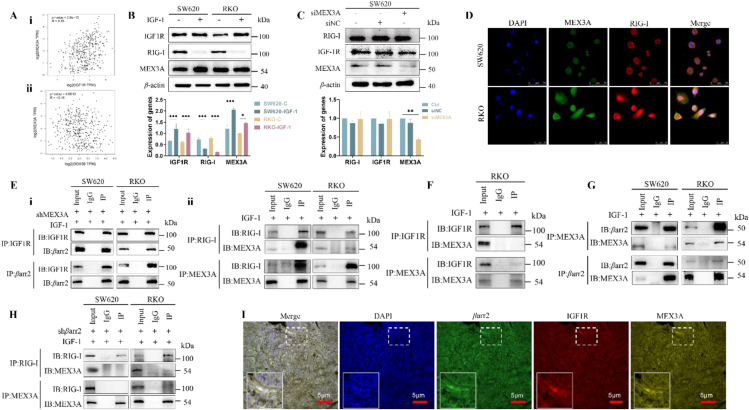
Figure 4.
MEX3A was identified to be responsible for RIG-I degradation. (A) i) MEX3A expression is positively correlated to the increase of IGF-1R (r = 0.35, P = 2.8e-13). ii) MEX3A expression is negatively correlated with RIG-I levels (r = −0.16, P = 0.0013). (B) Cancer cells showed a higher level of IGF-1R, a higher level of MEX3A, and a lower level of RIG-I. Data are presented as means ± SD. ∗P < 0.05, ∗∗∗P < 0.001. n = 3. (C) Knockdown of MEX3A failed the IGF-1Rβ-induced degradation of RIG-I. Data are presented as means ± SD; ∗∗P < 0.01. n = 3. (D) Activation of IGF-1R resulted in a co-expression of MEX3A with RIG-I. Scale bar, 50 μm. (E) Co-IP assay did not identify i) IGF-1Rβ‒MEX3A complex, while ii) MEX3A‒RIG-I complex was identified. (F) Silence of MEX3A did not affect the binding between IGF-1Rβ and βarr2 in cancer cells. (G) Co-IP assay identified βarr2‒MEX3A complex in the presence of IGF-1. (H) Silence of βarr2 failed the interaction of MEX3A with RIG-I in the presence of IGF-1. (I) mIHC analysis showed a co-expression of IGF-1Rβ, βarr2 and MEX3A in SW620 xenograft. Scale bar, 5 μm.
Interestingly, immunofluorescence analysis determined a co-staining of RIG-I with MEX3A in the IGF-1-treated cancer cells (Fig. 4D). However, Co-IP assay did not identify the IGF-1Rβ‒MEX3A complex (Fig. 4E-i) but MEX3A‒RIG-I complex (Fig. 4E-ii). Knockdown of MEX3A did not affect the interaction of IGF-1Rβ with βarr2 in SW620 and KRO cells (Fig. 4F). In the absence of IGF-1, only a weak MEX3A‒RIG-I complex was observed in cancer cells (Fig. S4D). Further, knockdown of MEX3A did not significantly affect the level of RIG-I in the absence of IGF-1; but RIG-I level was significantly increased in the presence of IGF-1 (Fig. S4E). Together, these results indicated that MEX3A was responsible for the IGF-1R-induced degradation of RIG-I in cancer cells.
It is well known that, as the scaffolds, β-arrestins are capable of conformation changes for adapting the structures of multiple downstream targets26, 27, 28, 29, 30, 31. Since MEX3A did not directly interact with IGF-1Rβ (Fig. 4E) but with βarr2 to form the MEX3A/βarr2 complex (Fig. 4G), we thus suggested that βarr2 might play the role of mediation between IGF-1Rβ and MEX3A during the process of RIG-I degradation. How does βarr2 scaffolds IGF-1Rβ to induce RIG-I degradation? Knockdown of βarr2 failed the interaction of MEX3A with RIG-I (Fig. 4H) in cancer cells (Fig. S4F), indicating that βarr2 was indispensable to the interaction between MEX3A and RIG-I. Multiplex IHC (mIHC) assay identified the colocalization of IGF-1Rβ, βarr2 and MEX3A in the IGF-1-treated SW620 xenograft (Fig. 4I). Accordingly, we proposed that, in response to IGF-1, IGF-1Rβ might firstly bind with βarr2 for activating βarr2, then the active βarr2 interact with MEX3A to form the βarr2/MEX3A complex through conformation changes of βarr2, thus leading to the promotion of MEX3A.
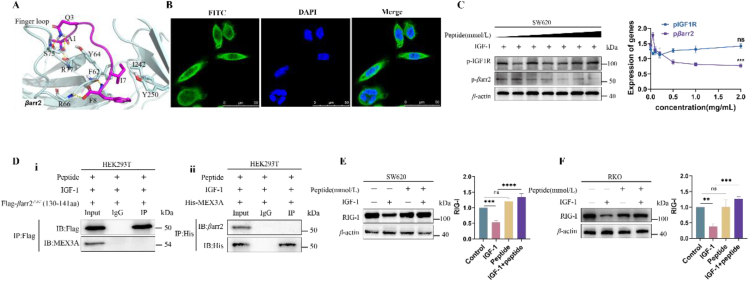
3.5. The IGF-1R-β-attached “phosphates” activated the basal βarr2 into its active state by phosphorylating Tyr64 and Tyr250 of the interdomain, opening the critical sequences Leu130‒Cys141 of the middle loop to the RING domain of MEX3A
We constructed the models of βarr2/IGF-1Rβ and βarr2/MEX3A for interpreting the above proposal. Naturally, βarr2 has a bi-lobed structure. It is stabilized by its C-terminal tail and interlobe loops, such as finger loop (G65‒R77) and middle loop (L130‒C141) (Fig. 5A)26, 27, 28, 29, 30, 31, 32. The phosphorylated IGF-1Rβ can interact with the interlobe loops of βarr2 (Fig. 5B). The IGF-1Rβ-attached “phosphates”28,29 could trigger the conformation changes of βarr2 by phosphorylating Tyr64 and Tyr250 of the interlobe. To confirm the role of the proposed residues of Tyr64 and Tyr250, the mutants of βarr2Y64A and βarr2Y250A were constructed and their response to the IGF-1Rβ-attached “phosphates” was analyzed in the IGF-1-treated cancer cells. As compared to control cells, the cells transfected with βarr2Y64A or βarr2Y250A were both expressed the lower levels of the phosphorylated βarr2 (pβarr2) in the presence of IGF-1 (Fig. 5C-i). Co-IP assay did not identify IGF-1R-βarr2 complex in these cells which were transfected with βarr2Y64A and βarr2Y250A (Fig. 5C-ii). Also, Co-IP assay did not identify βarr2-MEX3A complex in these cells which were transfected with βarr2Y64A and βarr2Y250A without IGF-1 (Fig. 5C-iii). Collectively, these results identified Tyr64 and Tyr250 as the key residues of βarr2 in response to the IGF-1Rβ-attached “phosphates” in the forming of the βarr2/IGF-1Rβ complex.
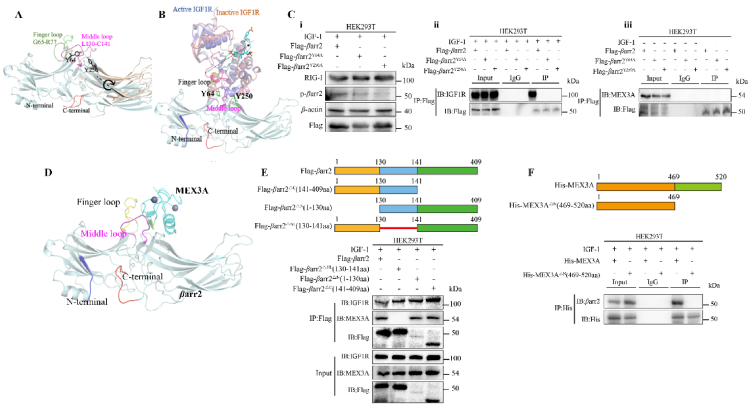
Figure 5.
The IGF-1R-β-attached “phosphates” activated basal βarr2 into its active state by phosphorylating Tyr64 and Tyr250 of interdomain, opening the sequences Leu130‒Cys141 of the middle loop to the RING domain of MEX3A. (A) Structures snapshot of βarr2 in basal state (light cyan) and active state (wheat). The key structural features of basal βarr2 were highlighted in bright colors, while in active state, structure was colored in dark. Arrow shows the rotation from basal state to active state. (B) The binding mode of IGR1R with active βarr2. Active IGF1R, inactive IGF1R, and βarr2 were colored blue, pink, and light cyan, respectively. The IGF-1Rβ-attached “phosphates” were shown in sticks. The arrow indicates long distance movement of the loop containing the phosphorated residues. (C) i) Either mutant βarr2Y64A or mutant βarr2Y250A did not respond to the IGF-1-driven IGF-1Rβ-attached “phosphates” in the cells transfected with βarr2Y64A or βarr2Y250A. ii) As compared to control cells, the cells transfected with βarr2Y64A or βarr2Y250A expressed a lower level of phosphorylated βarr2 (pβarr2). iii) Co-IP assay did not identify IGF-1R‒βarr2 complex in the cells transfected with βarr2Y64A and βarr2Y250A. Co-IP assay did not identify βarr2‒MEX3A complex in the cells transfected with βarr2Y64A and βarr2Y250A in the absence of IGF-1. Data are presented as means ± SD; n = 3. (D) Hypothetical binding pattern of the RING domain of MEX3A with active βarr2. (E) A schematic illustration of βarr2 with different domains including N-terminal amino acid (1–130 aa), middle region (130–141 aa), and C-terminal domain (141–409 aa). The truncated-βarr2 test identified core region of βarr2 in adapting the RING domain of MEX3A. HEK293T cells were transfected with Flag-βarr2ΔNL (130–141 aa), Flag-βarr2ΔN (1–130 aa), and Flag-βarr2ΔC (141–409 aa). Co-IP assay showed that Flag-βarr2ΔNL (130–141 aa) but not Flag-βarr2ΔN (1–130 aa) and Flag-βarr2ΔC (141–409 aa) interacted with the RING domain of MEX3A. (F) A schematic illustration of MEX3A with different domains including N-terminal amino acid (1–469 aa) and C-terminal RING domain (469–520 aa). Subclone of HEK293T cells expressed His-MEX3AΔN (469–520 aa) was constructed and the interaction of the truncated-MEX3A with βarr2 was analyzed. The fully length MEX3A not the truncated-MEX3A interacted with βarr2 in response to IGF-1.
We have understood that the binding strength between two proteins is mostly dependent on their interface size (number of residues at the interface)32. Theoretically, the middle loop (Leu130‒Cys141) of βarr2 could contact with the RING domain of MEX3A (Fig. 5D). Thus, the middle loop was considered as the core region in the mediation of the interaction between βarr2 with MEX3A. Certainly, we did not deny the roles of other two regions of βarr2 in mediating the interaction of IGF-1Rβ with MEX3A. To support this hypothesis, we performed the truncated-βarr2 test to identify the core region of βarr2 functioned the adapting to the structure of RING domain of MEX3A. To test these possibilities, we constructed three subclones of HEK293T cells expressed Flag-βarr2ΔNL (130–141 aa), Flag-βarr2ΔN (1–130 aa), and Flag-βarr2ΔC (141–409 aa), respectively. The interactions of IGF-1Rβ with these truncated-βarr2s were analyzed in the presence of IGF-1 (Fig. 5E, above). We identified that it is Flag-βarr2ΔNL (130–141 aa) but not Flag-βarr2ΔN (1–130 aa) and Flag-βarr2ΔC (141–409 aa) that interacted with IGF-1Rβ (Fig. 5E, below). These results confirmed that 130–141 aa is the core region of βarr2 by which βarr2 bound with the RING domain of MEX3A.
Further, the subclone of HEK293T cells expressed his-MEX3AΔN (469–520 aa) was constructed and the interaction of the truncated-MEX3A with βarr2 was analyzed (Fig. 5F, above). The full length of MEX3A interacted with βarr2, but the truncated-MEX3A lost this ability (Fig. 5F, below). These results interpreted the mechanism of the conformation changes of βarr2 in responding to the IGF-1Rβ-attached “phosphates” for adapting the RING domain of MEX3A.
The proposed βarr2/MEX3A model indicated that the major contact surface of MEX3A with βarr2 was around its sequences from A512 to F519 (sequencing ATQAIRIF). The linear peptide with the same sequence as ATQAIRIF could bind with βarr2 at the same binding site (Fig. 6A). Accordingly, we hypothesized that the peptide ATQAIRIF could block the interaction of active βarr2 with the RING domain of MEX3A by competing to bind to active βarr2. Supportive of this proposal, the peptide was designed and its active form FITC-{beta-Ala}-ATQAIRIF-GRKKRRQRRRPQ-NH2) was determined in SW620 cells (Fig. 6B). In the absence of the peptide, cancer cells demonstrated the higher level of the phosphorylated-βarr2 in response to IGF-1, while the levels of phosphorylated-βarr2 were gradually reduced in the presence of the peptide, although IGF-1Rβ was still accumulated in SW620 cells (Fig. 6C). This result indicated that the peptide only inhibited active βarr2 but not IGF-1Rβ. Further, the peptide strongly blocked the interaction of Flag-βarr2ΔN (130–141 aa) with RING domain of MEX3A (Fig. 6D-i), confirming the function of the core region of βarr2 in our proposed model, and the interaction between His-MEX3A and active βarr2 (Fig. 6D-ii), confirming the function of the peptide in blocking the binding of active βarr2 and the RING domain of MEX3A. Expectedly, the cancer cells treated with the peptide by 2 mmol/L resulted in the higher level of RIG-I as compared with cancer cells without the peptide in SW620 cells (Fig. 6E) and RKO cells (Fig. 6F). Together, we proved the rationality of the proposed βarr2/MEX3A model, in which the contact surface of the RING domain of MEX3A with active βarr2 might be around the sequences of A512‒F519, thus increasing MEX3A levels.
Figure 6.
The peptide mimicked the contact surface of MEX3A and blocked the interaction of active βarr2 with the RING domain of MEX3A. (A) Hypothetical binding modes indicated that the contact surface of MEX3A is around A512‒F519 (pink color). (B) The peptide (FITC-{beta-Ala}-ATQAIRIF-GRKKRRQRRRPQ-NH2) was absorbed into cancer cells. (C) Colonic cancer cells treated by the peptide exhibited decreasing levels of pβarr2. (D) Co-IP assay proved that the peptide blocked the interaction between Flag-βarr2▵N (130–141 aa) and the RING domain of MEX3A. (E) SW620 cells exposed to the peptide resulted in an increase of RIG-I. (F) RKO cells exposed to the peptide resulted in the increase of RIG-I. Data are presented as means ± SD; n = 3. ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
3.6. The IGF-1Rβ-triggered degradation of RIG-I resulted in the suppression of IFN-I-associated immune cells in the TME due to the blockade of the RIG-I-MAVS-IFN-I pathway
It is well known that tumor intrinsic RIG-I plays an essential role in sensitizing tumor cells to immune checkpoint inhibitors (ICIs) through activating the RIG-I/MAVS/IFN-I pathway15,33, 34, 35. However, the levels of RIG-I and IFN-I are suppressed in the majority of cancers36. These cancers are considered as the “cold tumors” which are characterized by suppression of the IFN-I-associated immune cells15,37, 38, 39. MC38 murine colon tumor cells, a CRC line sensitive to PD-L1 ICIs, expressed the lower level of IGF-1R and the higher level of RIG-I, MAVS and IFN-I. Comparably, CT26 murine colon tumor cells, a CRC line insensitive to PD-L1 ICIs, expressed the higher level of IGF-1R and the lower level of RIG-I, MAVS and IFN-I (Supporting Information Fig. S5). Thus, CT26 cell line and MC38 cell line were used to evaluate the potential significance of our finding.
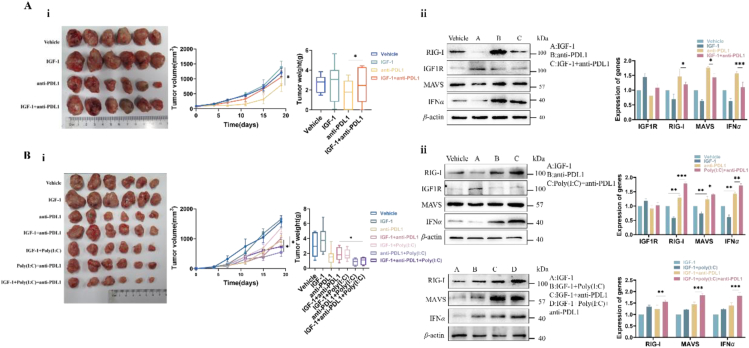
Injection of rhIGF-1 (1 mg/kg/day) promoted the growth of MC38 cells in mice. Anti-PD-L1 (1 mg/kg/week) strongly inhibited MC38 cells, while they became insensitive to anti-PD-L1 in the presence of IGF-1 (Fig. 7A-i, 64.2% vs. 19.6%). Western blotting analysis determined the lower levels of RIG-I and MAVS and the consequent lower level of IFN-I in the IGF-1-treated MC38 tumors than those in control group. Anti-PD-L1 reversed the IGF-1-induced lower levels of RIG-I, MAVS and IFN-I in the anti-PD-L1-treated MC38 tumor cells (Fig. 7A-ii). Expectedly, the suppression of these IFN-I-associated immune cells was reversed by anti-PD-L1. The anti-PD-L1-treated MC38 demonstrated the higher levels of natural killer cells (marked by CD11b), dendritic cells (marked by CD68), total macrophages (marked by F4/80), and CD8+ T cells; and the lower level of M2 macrophage (marked by CD206) than MC38 tumors without anti-PD-L1 (Supporting Information Fig. S6A and Fig. S7A and B).
Figure 7.
Degradation of RIG-I worsened the TME by suppressing the IFN-associated immune cells, leading to anti-PD-L1 insensitivity in murine colon cancer models. (A) i) MC38 colonic cancer cells were injected in C57BL/6N mice and then were treated with IGF-1, anti-PD-L1 (1 mg/kg), and IGF-1 plus anti-PD-L1, respectively. MC38 tumor models were sensitive to anti-PD-L1; while this sensitivity of anti-PD-L1 became blunt in the presence of IGF-1. ii) Western blotting analysis showed the lower levels of RIG-I, MAVS and IFN-I in the IGF-1-treated MC38 tumors. Anti-PD-L1 reversed the suppression of RIG-I, MAVS and IFN-I in the anti-PD-L1-treated MC38 tumor models; while the activity of anti-PD-L1 was weakened by IGF-1 in MC38 tumor models simultaneously exposed to anti-PD-L1 plus IGF-1. (B) i) CT26 colonic cancer cells were injected in BALB/c mice and then were treated with IGF-1, anti-PD-L1 (1 mg/kg), IGF-1 plus anti-PD-L1, IGF-1 plus poly(I:C), anti-PD-L1 plus poly(I:C), and IGF-1 plus anti-PD-L1 and poly(I:C), respectively. Anti-PD-L1 weakly inhibited growth of CT26 tumors; this weak activity of anti-PD-L1 was further blunt in the presence of IGF-1. Poly(I:C) promoted the sensitivity of CT26 tumors to anti-PD-L1. ii) CT26 tumors exhibited the lower levels of RIG-I, MAVS and IFN-I, and their levels were further decreased in the presence of IGF-1. Anti-PD-L1 weakly increased levels of RIG-I, MAVS and IFN-I in the anti-PD-L1-treated CT26 tumor models. Poly(I:C) reversed the lower levels of RIG-I, MAVS and IFN-I. Data are presented as means ± SD; ∗P < 0.05; ∗∗P < 0.01, ∗∗∗P < 0.001 (n = 6).
The anti-PD-L1 insensitivity could be reversed through the recovery of RIG-I by poly(I:C)40. As shown in Fig. 7B-i, CT26 tumor models demonstrated the resistance to anti-PD-L1 (inhibition rate only 36.5% by 2 mg/kg/week, line 3), and this inhibition was further reduced in the presence of IGF-1. As compared to CT26 tumor model treated with anti-PD-1 in the presence of IGF-1 (Fig. 7B-i, line 4), the effect of anti-PD-L1 on the IGF-1-treated CT26 group was significantly increased by 58.8% by delivering poly(I:C) (Fig. 7B-i, line 7). Accordingly, we proposed that this achievement might be due to the recovery of RIG-I by poly(I:C). Supportive of this proposal, the levels of RIG-I, MAVS and IFN-I were suppressed in CT26 tumor models in the presence of IGF-1. Anti-PD-L1 significantly increased the levels of RIG-I, MAVS and IFN-I, and their levels were further increased in the presence of poly(I:C). Particularly, the higher levels of RIG-I were determined in the poly(I:C)-treated CT26 group than in the CT26 group without poly(I:C) (Fig. 7B-ii). These results confirmed that the achievement in the poly(I:C)-treated CT26 tumor model was associated with the recovery of RIG-I. Due to the IGF-1R-induced suppression of the RIG-I-MAVS-IFN-I pathway, the levels of the IFN-I-associated immune cells including natural killer cells, dendritic cells, total macrophages and CD8+ T cells were significantly reduced in the IGF-1-treated CT26 tumors. Expectedly, higher levels of immune cells were determined in the poly(I:C)-treated CT26 than in the CT26 without poly(I:C) treatment (Supporting Information Fig. S8 and Fig. S9).
4. Discussion
RIG-I plays a crucial role in the development of cancer and cancer therapy. RIG-I can activate innate immune cells in TME41,42. Mechanically, RIG-I can promote the polymerization of MAVS on mitochondria, and MAVS further recruits its downstream adaptors, activating the transcription factors IRF3 and nuclear factor κB (NF-κB), which trigger IFNs and pro-inflammatory cytokines to activate numerous innate immune cells and CD8+ T cells43, 44, 45. Therapeutic targeting RIG-I could sensitize “cold tumors” to ICIs through increasing the infiltration of NK cells, CD8+ T cells and numerous other immune cells in the TME46,47. Thus, RIG-I agonists are used to treat cancers48. However, the effects of RIG-I therapy can be influenced by numerous factors. Previous studies identified a negative correlation between RIG-I and MEX3A in tumorigenesis47,49. MEX3A, an RNA-binding protein (RBP), post-transcriptionally regulates a number of biological processes, including tumor immunologically relevant ones, thereby, leading to the immune evasion of tumor cells. Indeed, overexpression of MEX3A degraded RIG-I in the TME, which was significantly associated with a poor survival in patients with liver cancer, colorectal cancer, and glioblastoma50,51. In the present study, activation of IGF-1R led to the degradation of RIG-I through triggering the βarr2-mediated MEX3A activity, resulting in the suppression of the IFNs-related immune cells in the TME. These cancer cells became resistant to anti-PD-L1. From this point of view, IGF-1/IGF-1R signaling might be the root cause of “cold tumors”. As expected, poly(I:C) enhanced anti-PD-L1 activity through recovery of RIG-I. Therefore, delivery of RIG-I agonists could be a workable strategy for dealing with the IGF-1/IGF-1R-caused “cold tumors”.
To interpret the mechanism of MEX3A in degrading RIG-I, we underlined the role of βarr2 in mediating IGF-1Rβ to trigger MEX3A activity. Recently, the roles of β-arr isoforms are acknowledged to be relevant for their physiological or pathological processes52. Thus, β-arr isoforms might play different roles in deciding the IGF-1Rβ′s fates. β-arrs function as a major hub for controlling the IGF-1R's functions and the fates of IGF-1Rβ. Further, β-arrs appear to be so pleiotropic with one conformation favoring one specific function but dampening another that defining an “active” β-arrestin becomes difficult. How does IGF-1R identify its partnerships β-arr isoforms? In our study, normal cells expressed an appropriate level of βarr1 and a lower level of βarr2 in their resting state. We determined IGF-1Rβ/βarr1 complex under the lower levels of IGF-1, but IGF-1Rβ/βarr2 complex only under the high levels of IGF-1. Comparably, cancer cells expressed a relatively strong IGF-1Rβ/βarr2 complex but not IGF-1Rβ/βarr1 complex. Thus, IGF-1Rβ might select it is the partnerships of β-arr isoforms according to the pathological situation of IGF-1R signaling. We concluded that it was βarr2 but not βarr1 that mediated the activated-IGF-1R in cancer cells. The forming of IGF-1Rβ/βarr2 complex might be indispensable in the decision of IGF-1Rβ′s fate. Thus, the IGF-1Rβ/βarr2 complex might play an important role in tumorigenesis.
Since the K48-linked ubiquitination was identified to mediate the IGF-1Rβ-triggered degradation of RIG-I, we searched for the E3 ligase isoform which was responsible for this process. MEX3A was identified from a total 667 E3 ligases. Interestingly, IGF-1Rβ did not directly interact with MEX3A but induced βarr2 to interact with MEX3A. How does βarr2 mediate IGF-1Rβ to stimulate MEX3A?
It is well known that β-arrestins are capable of adapting multiple conformations through which β-arrestins could identify their specific interacting domains28, 29, 30, 31. We established the models of βarr2/IGF-1Rβ and βarr2/MEX3A for interpreting the mechanism of IGF-1Rβ in triggering degradation of RIG-I. We underlined the functions of βarr2 in mediating IGF-1Rβ through the conformational changes of βarr2. Theoretically, βarr2 could undergo the conformational changes once it captures the phosphorylated receptor, which displaces C-tail27, 28, 29. The mechanism of βarr activation has been proved to be conserved29,31. Our proposed models suggested that, in response to ligands, the IGF-1Rβ-attached “phosphates” could activate basal βarr2 into its active state by phosphorylating βarr2 on Tyr64 and Tyr250 of the interlobe. The electron repulsive force between the phosphorate groups of Tyr64 and Tyr250 could break the interlobe hydrogen-bond interactions, which make middle loop and finger loop more outward. After dephosphorylation, IGF-1Rβ underwent a conformational change and was released from active βarr2 for the steric clashes between the unphosphorylated IGF-1Rβ and βarr2. In the active βarr, the breakage of central polar core and other interlobe hydrogen-bond networks leads to a rotation of two lobes27, 28, 29, 30, 31. Similarly, the diminished interlobe interaction unlocks the two lobes and thus increases the flexibility between the two lobes. The extending of finger loop together with middle loop and the C-terminal lobe form a hydrophobic groove which fits the RING domain surface of MEX3A, thus promoting MEX3A activity.
To confirm Tyr64 and Tyr250 as the key residues of the interlobe in the response to the IGF-1Rβ-attached “phosphates”, we constructed mutant of βarr2Y64A and mutant of βarr2Y250A for analyzing their responses to the IGF-1-driven IGF-1Rβ in cancer cells. As compared to control cells, the cells transfected with βarr2Y64A or βarr2Y250A both expressed the lower levels of phosphorylated βarr2 (pβarr2) in the presence of IGF-1. Co-IP assay did not identify the IGF-1R‒βarr2 complex in these cells. Thus, we confirmed that Tyr64 and Tyr250 were the key residues in the interlobe of βarr2 through which the active βarr2 responded to the IGF-1Rβ-attached “phosphates”, leading to the forming βarr2/IGF-1Rβ complex.
The binding strength between two proteins was mostly dependent on their interface size32. Theoretically, middle loop (Leu130‒Cys141) of βarr2 could contact RING domain of MEX3A. Thus, middle loop was considered as the core region in the mediation of interaction between βarr2 with MEX3A. Certainly, we did not deny the roles of other two regions of βarr2 in mediating the interaction of IGF-1Rβ with MEX3A. To support this hypothesis, we performed the truncated-βarr2 test to identify the core region of βarr2 functioned the adapting to the RING domain of MEX3A. To test these possibilities, we constructed three subclones expressed Flag-βarr2ΔNL (130–141 aa), Flag-βarr2ΔN (1–130 aa), and Flag-βarr2ΔC (141–409 aa). The interactions of IGF-1Rβ with these truncated-βarr2s were analyzed, respectively, in the presence of IGF-1. We identified that it is Flag-βarr2ΔNL (130–141 aa) but not Flag-βarr2ΔN (1–130 aa) and Flag-βarr2ΔC (141–409 aa) that interacted with IGF-1Rβ. These results confirmed that the 130–141 aa is the core region of active βarr2 which functioned in the forming βarr2‒MEX3A complex. Further, subclone of HEK293T cells expressed his-MEX3AΔN(469–520 aa) was constructed and the interaction of truncated-MEX3A with active βarr2 was analyzed. We identified that the fully length of MEX3A interacted with βarr2. The truncated-MEX3A lost this ability. These results could interpret the mechanism of the conformation changes of βarr2 in the response to the IGF-1Rβ-attached “phosphates” for adapting the RING domain of MEX3A.
Further, the βarr2/MEX3A model showed that the major contact surface of MEX3A with βarr2 were around A512‒F519 in the RING domain of MEX3A. Theoretically, the linear peptide with the same sequences of ATQAIRIF could bind with active βarr2 in the same binding site. We hypothesized that the peptide ATQAIRIF could block the interaction of the active βarr2 with the RING domain of MEX3A through competing to binding to 130–141 aa of active βarr2. In the presence of IGF-1, the peptide effectively blocked the interaction of Flag-βarr2ΔN (130–141 aa) with the RING domain of MEX3A, confirming the function of core region of βarr2 in our proposed model, and the interaction between His-MEX3A and active βarr2, confirming the function of the peptide in blocking the interaction of active βarr2 and the RING domain of MEX3A.
5. Conclusions
Activation of IGF-1R promotes cancer growth through triggering the MEX3A-mediated degradation of RIG-I. IGF-1Rβ activated basal βarr2 into its active state by phosphorylating the interdomain domain on Tyr64 and Tyr250, opening the middle loop (Leu130‒Cys141) to adapt the RING domain of MEX3A, leading to the promotion of MEX3A. Degradation of RIG-I resulted in the suppression of the IFN-I-associated immune cells in TME due to the blockade of the RIG-I-MAVS-IFN-I pathway. Poly(I:C) could reverse anti-PD-L1 insensitivity by recovery of RIG-I.
Acknowledgments
This work was supported by Beijing Natural Science Foundation (7222253, China) and National Natural Science Foundation of China (81973350/82173841). This work also supported by Beijing Natural Science Foundation (7212149, China).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2023.04.001.
Contributor Information
Shuxiang Cui, Email: cuisx@ccmu.edu.cn.
Xianjun Qu, Email: qxj@ccmu.edu.cn.
Author contributions
Xianjun Qu and Shuxiang Cui conceived the projects. Qiaobo Xie, Yanan Li, Xinfeng Wu, Keqin Li, Xiaohui Liu, Rui Xu and Wenmin Yuan performed experiments. Yanyan Chu constructed the molecular binding model. Qiaobo Xie and Xianjun Qu wrote the manuscript, which was edited by all authors.
Conflicts of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Holzenberger M., et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 2.Luey B.C., May F.E. Insulin-like growth factors are essential to prevent anoikis in oestrogen-responsive breast cancer cells: importance of the type I IGF receptor and PI3-kinase/Akt pathway. Mol Cancer. 2016;15:8. doi: 10.1186/s12943-015-0482-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mao K., et al. Late-life targeting of the IGF-1 receptor improves healthspan and lifespan in female mice. Nat Commun. 2018;9:2394. doi: 10.1038/s41467-018-04805-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H.X., Sharon E. IGF-1R as an anti-cancer target-trials and tribulations. Chin J Cancer. 2013;32:242–252. doi: 10.5732/cjc.012.10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gombos A., Metzger-Filho O., Dal Lago L., Awada-Hussein A. Clinical development of insulin-like growth factor receptor-1 (IGF-1R) inhibitors: at the crossroad?. Invest New Drugs. 2012;30:2433–2442. doi: 10.1007/s10637-012-9811-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C., Zhang Z., Tang H., Jiang Z., You L., Liao Y. Crosstalk between IGF-1R and other tumor promoting pathways. Curr Pharm Des. 2014;20:2912–2921. doi: 10.2174/13816128113199990596. [DOI] [PubMed] [Google Scholar]
- 7.Stefani C., et al. Growth factors, PI3K/AKT/mTOR and MAPK signaling pathways in colorectal cancer pathogenesis: where are we now?. Int J Mol Sci. 2021;22 doi: 10.3390/ijms221910260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.King H., Aleksic T., Haluska P., Macaulay V.M. Can we unlock the potential of IGF-1R inhibition in cancer therapy?. Cancer Treat Rev. 2014;40:1096–1105. doi: 10.1016/j.ctrv.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen M.K., Hung M.C. Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases. FEBS J. 2015;282:3693–3721. doi: 10.1111/febs.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delos S.R., Garay C., Antonescu C.N. Charming neighborhoods on the cell surface: plasma membrane microdomains regulate receptor tyrosine kinase signaling. Cell Signal. 2015;27:1963–1976. doi: 10.1016/j.cellsig.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Girnita L., Worrall C., Takahashi S., Seregard S., Girnita A. Something old, something new and something borrowed: emerging paradigm of insulin-like growth factor type 1 receptor (IGF-1R) signaling regulation. Cell Mol Life Sci. 2014;71:2403–2427. doi: 10.1007/s00018-013-1514-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith J.S., Rajagopal S. The β-arrestins: multifunctional regulators of protein-coupled receptors. J Biol Chem. 2016;291:8969–8977. doi: 10.1074/jbc.R115.713313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aydin Y., Coin I. Biochemical insights into structure and function of arrestins. FEBS J. 2021;288:2529–2549. doi: 10.1111/febs.15811. [DOI] [PubMed] [Google Scholar]
- 14.Wess J. The two β-arrestins regulate distinct metabolic processes: studies with novel mutant mouse models. Int J Mol Sci. 2022;23:495. doi: 10.3390/ijms23010495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang B.J., et al. Sensitizing immune unresponsive colorectal cancers to immune checkpoint inhibitors through MAVS overexpression. J Immunother Cancer. 2022;10 doi: 10.1136/jitc-2021-003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snider D.L., Park M., Murphy K.A., Beachboard D.C., Horner S.M. Signaling from the RNA sensor RIG-I is regulated by ufmylation. Proc Natl Acad Sci U S A. 2022;119 doi: 10.1073/pnas.2119531119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y., et al. Goose MAVS functions in RIG-I-mediated IFN-β signaling activation. Dev Comp Immunol. 2019;93:58–65. doi: 10.1016/j.dci.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Liu B., et al. Sustained ER stress promotes hyperglycemia by increasing glucagon action through the deubiquitinating enzyme USP14. Proc Natl Acad Sci U S A. 2019;116:21732–21738. doi: 10.1073/pnas.1907288116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jumper J., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiménez-García B., Pons C., Fernández-Recio J. PyDockWEB: a web server for rigid-body protein-protein docking using electrostatics and desolvation scoring. Bioinformatics. 2013;29:1698–1699. doi: 10.1093/bioinformatics/btt262. [DOI] [PubMed] [Google Scholar]
- 21.Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., Weng Z. ZDOCK server: interactive docking prediction of protein‒protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y.H., et al. SphK2 confers 5-fluorouracil resistance to colorectal cancer via upregulating H3K56ac-mediated DPD expression. Oncogene. 2020;39:5214–5227. doi: 10.1038/s41388-020-1352-y. [DOI] [PubMed] [Google Scholar]
- 23.Wang S.Q., Yang X.Y., Yu X.F., Cui S.X., Qu X.J. Knockdown of IGF-1R triggers viral RNA sensor MDA5- and RIG-I-mediated mitochondrial apoptosis in colonic cancer cells. Mol Ther Nucleic Acids. 2019;16:105–117. doi: 10.1016/j.omtn.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunn K.W., Kamocka M.M., Mcdonald J.H. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moududee S.A., et al. Structural and functional characterization of hMEX-3C ring finger domain as an E3 ubiquitin ligase. Protein Sci. 2018;27:1661–1669. doi: 10.1002/pro.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gurevich V.V., Gurevich E.V., Uversky V.N. Arrestins: structural disorder creates rich functionality. Protein Cell. 2018;9:986–1003. doi: 10.1007/s13238-017-0501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurevich E.V., Gurevich V.V. Arrestins: ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han M., Gurevich V.V., Vishnivetskiy S.A., Sigler P.B., Schubert C. Crystal structure of beta-arrestin at 1.9 a: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 29.Kim Y.J., Hofmann K.P., Ernst O.P., Scheerer P., Choe H.W., Sommer M.E. Crystal structure of pre-activated arrestin p44. Nature. 2013;497:142–146. doi: 10.1038/nature12133. [DOI] [PubMed] [Google Scholar]
- 30.Qu C., et al. Scaffolding mechanism of arrestin-2 in the cRaf/MEK1/ERK signaling cascade. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2026491118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheerer P., Sommer M.E. Structural mechanism of arrestin activation. Curr Opin Struct Biol. 2017;45:160–169. doi: 10.1016/j.sbi.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 32.Nilofer C., Sukhwal A., Mohanapriya A., Kangueane P. Protein‒protein interfaces are vdW dominant with selective H-bonds and (or) electrostatics towards broad functional specificity. Bioinformation. 2017;13:164–173. doi: 10.6026/97320630013164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam K.C., et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell. 2021;184:5338–5356. doi: 10.1016/j.cell.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brägelmann J., et al. MAPK-pathway inhibition mediates inflammatory reprogramming and sensitizes tumors to targeted activation of innate immunity sensor RIG-I. Nat Commun. 2021;12:5505. doi: 10.1038/s41467-021-25728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heidegger S., et al. RIG-I activation is critical for responsiveness to checkpoint blockade. Sci Immunol. 2019;4 doi: 10.1126/sciimmunol.aau8943. [DOI] [PubMed] [Google Scholar]
- 36.Miar A., et al. Hypoxia induces transcriptional and translational downregulation of the type I IFN pathway in multiple cancer cell types. Cancer Res. 2020;80:5245–5256. doi: 10.1158/0008-5472.CAN-19-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pomponio R., et al. An integrative approach of digital image analysis and transcriptome profiling to explore potential predictive biomarkers for TGFβ blockade therapy. Acta Pharm Sin B. 2022;12:3594–3601. doi: 10.1016/j.apsb.2022.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancel J.C., Crozat K., Dalod M., Mattiuz R. Are conventional type 1 dendritic cells critical for protective antitumor immunity and how?. Front Immunol. 2019;10:9. doi: 10.3389/fimmu.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruzicka M., et al. RIG-I-based immunotherapy enhances survival in preclinical AML models and sensitizes AML cells to checkpoint blockade. Leukemia. 2020;34:1017–1026. doi: 10.1038/s41375-019-0639-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du X., et al. Cytosolic delivery of the immunological adjuvant poly I:C and cytotoxic drug crystals via a carrier-free strategy significantly amplifies immune response. Acta Pharm Sin B. 2021;11:3272–3285. doi: 10.1016/j.apsb.2021.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X.Y., Guo H.Z., Zhu J. Tumor suppressor activity of RIG-I. Mol Cell Oncol. 2014;1 doi: 10.4161/23723548.2014.968016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z., Han C., Fu Y.X. Targeting innate sensing in the tumor microenvironment to improve immunotherapy. Cell Mol Immunol. 2020;17:13–26. doi: 10.1038/s41423-019-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thoresen D., Wang W., Galls D., Guo R., Xu L., Pyle A.M. The molecular mechanism of RIG-I activation and signaling. Immunol Rev. 2021;304:154–168. doi: 10.1111/imr.13022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belgnaoui S.M., Paz S., Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 45.Duewell P., et al. RIG-I-like helicases induce immunogenic cell death of pancreatic cancer cells and sensitize tumors toward killing by CD8(+) T cells. Cell Death Differ. 2014;21:1825–1837. doi: 10.1038/cdd.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Such L., et al. Targeting the innate immunoreceptor RIG-I overcomes melanoma-intrinsic resistance to T cell immunotherapy. J Clin Invest. 2020;130:4266–4281. doi: 10.1172/JCI131572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang Y., et al. Exploiting RIG-I-like receptor pathway for cancer immunotherapy. J Hematol Oncol. 2023;16:8. doi: 10.1186/s13045-023-01405-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rameshbabu S., Labadie B.W., Argulian A., Patnaik A. Targeting innate immunity in cancer therapy. Vaccines (Basel) 2021;9:138. doi: 10.3390/vaccines9020138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Z., et al. Grass carp MEX3A promotes ubiquitination and degradation of RIG-I to inhibit innate immune response. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.909315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bufalieri F., et al. The RNA-binding ubiquitin ligase MEX3A affects glioblastoma tumorigenesis by inducing ubiquitylation and degradation of RIG-I. Cancers (Basel) 2020;12:321. doi: 10.3390/cancers12020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bufalieri F., Basili F., Marcotullio L.D., Infante P. Harnessing the activation of RIG-I like receptors to inhibit glioblastoma tumorigenesis. Front Mol Neurosci. 2021;14 doi: 10.3389/fnmol.2021.710171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Defea K.A. Beta-arrestins as regulators of signal termination and transduction: how do they determine what to scaffold?. Cell Signal. 2011;23:621–629. doi: 10.1016/j.cellsig.2010.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.