Summary
The FliG protein plays a pivotal role in switching the rotational direction of the flagellar motor between clockwise and counterclockwise. Although we previously showed that mutations in the Gly-Gly linker of FliG induce a defect in switching rotational direction, the detailed molecular mechanism was not elucidated. Here, we studied the structural changes in the FliG fragment containing the middle and C-terminal regions, named FliGMC, and the switch-defective FliGMC-G215A, using nuclear magnetic resonance (NMR) and molecular dynamics simulations. NMR analysis revealed multiple conformations of FliGMC, and the exchange process between these conformations was suppressed by the G215A residue substitution. Furthermore, changes in the intradomain orientation of FliG were induced by changes in hydrophobic interaction networks throughout FliG. Our finding applies to FliG in a ring complex in the flagellar basal body, and clarifies the switching mechanism of the flagellar motor.
Subject areas: Cell biology, Specialized functions of cells, Functional aspects of cell biology
Graphical abstract
Highlights
-
•
FliG protein determines the rotational direction of the flagellar motor
-
•
The structural changes in FliGMC and FliGMC-G215A were studied using NMR
-
•
G215A mutation suppressed the exchange process among multiple FliGMC conformations
-
•
Hydrophobic interaction networks induced intradomain orientation changes in FliG
Cell biology; Specialized functions of cells; Functional aspects of cell biology
Introduction
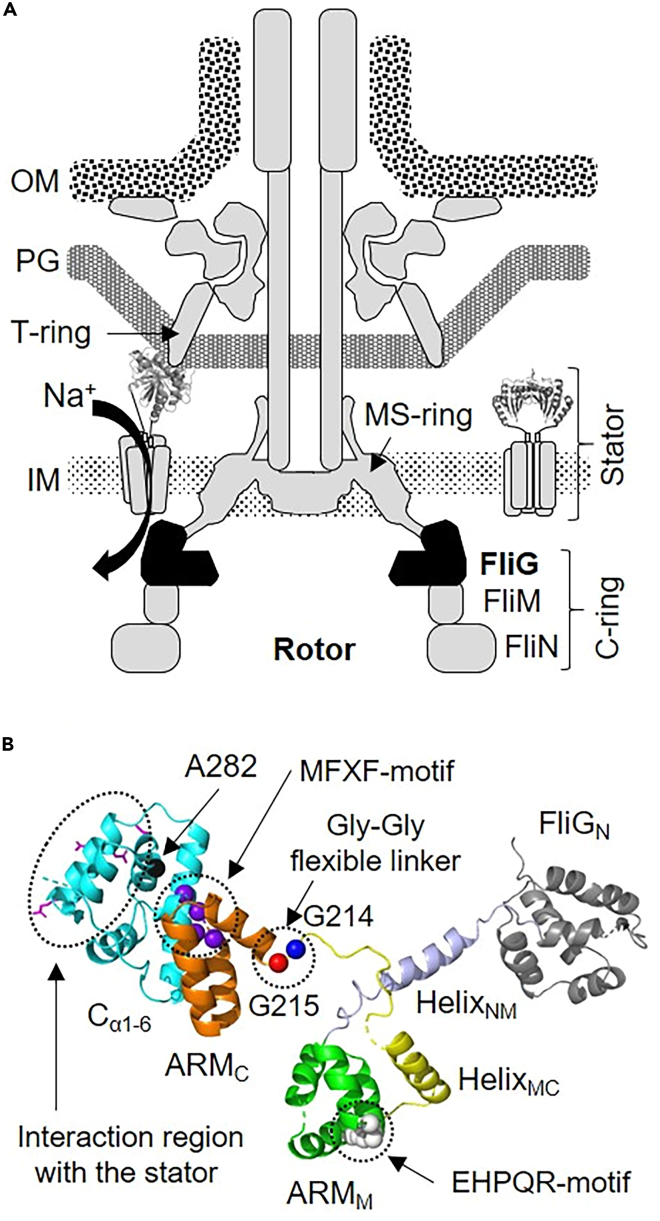
Most bacteria are highly sensitive to changes in the surrounding environment and exhibit regulated responsive movements, including efficient feeding and rapid avoidance of repellents. These movements are controlled by switching the rotational direction of the flagellar motor, which generates a rotational force in the flagellum. The flagellar motor consists of a stack of multiple ring structures from the outer cell membrane to the cytoplasm1 (Figure 1A). The stator complex, which is embedded in the cytoplasmic membrane, contains two membrane proteins, MotA/PomA and MotB/PomB. They act as an ion channel (generally for H+ or Na+) and mediate interactions with the C-ring of the motor.2,3,4 This interaction is crucial for generating motor torque using the electrochemical gradient developed across the cell membrane. To clarify the molecular mechanism underlying this interaction, functional and structural analyses of stator and C-ring components in various bacteria have been reported.5,6,7,8,9
Figure 1.
Polar flagellum model in V. alginolyticus
(A) Model of the rotary motor in the polar flagellum of Vibrio alginolyticus.
The C-ring is composed of three proteins; FliG, FliM and FliN. The inactive stator complex (stator in right side of the motor) diffuses in the inner membrane (IM). When the stator activates around the rotor (stator on the left side of the motor), it interacts with FliG to generate torque due to ion flex in the channel pore. Then, the C-terminal region of PomB (PomBC) binds to the curvature of the peptidoglycan layer (PG) of the T-ring. This binding allows the stator to remain around the rotor and undergo continued activation. OM represents the outer membrane.
(B) Structural model of V. alginolyticus FliG. FliG comprises three domains, N-terminal (FliGN), middle (FliGM), and C-terminal (FliGC). The latter two contain ARMM and helixMC and ARMC and Cα1-6, respectively. HelixNM connects FliGN with FliGM. The structure of FliGN, HelixNM contained N- and C-terminal loop, ARMM HelixMC contained N- and C-terminal loop, ARMC, and Cα1-6 are shown in gray, light blue, green, yellow, orange, and cyan, respectively, as a ribbon model. The atoms of white, blue, red, purple, and black balls show EHPQR motif, Gly-Gly flexible linker region (G214 and G215 residues), MFXF-motif and A282 residue. Side chains of highly conserved charged residues interacting with the stator are shown in magenta as a stick model.
The C-ring of the rotor complex is a three-layer structure consisting of FliG (facing the cell membrane), FliM (the middle part), and FliN (facing the cytoplasm)10 (Figure 1A). The C-ring plays a key role in switching the rotational direction between counterclockwise (CCW) and clockwise (CW). In Vibrio, CCW rotation drives the forward movement of bacteria, whereas CW rotation induces backward and flicking motions to change the swimming direction.11 Therefore, CCW-CW regulation of the flagellar motor is crucial for directing bacterial movement. The chemotaxis signaling pathway, which induces phosphorylation/dephosphorylation of the CheY protein, controls the switching of the rotational direction.12,13 Phosphorylated CheY (CheY-P) binds to FliM and FliN in the C-ring,14,15,16,17,18 and these interactions induce rotational switching. When CheY-P is dephosphorylated by CheZ, CheY dissociates from the C-ring, and motor rotation returns to CCW.16 As shown in Figure 1, FliM exists in the middle layer of the C-ring and connects FliG with FliN. The association/dissociation of CheY(-P) with the C-ring via FliM and/or FliN appears to induces conformational changes in FliG, FliM, and/or FliN; consequently, changes in the structure of the C-ring initiate rotational switching.
FliG has three structural domains: N-terminal (FliGN), middle (FliGM), and C-terminal (FliGC) (Figure 1B). Each domain has armadillo repeat motifs (ARM), denoted as ARMN, ARMM, and ARMC, which are involved in intramolecular and/or intermolecular interactions with FliG in the C-ring.19,20,21 Moreover, each domain interacts with other proteins. FliGN interacts with FliF, which is a component of the MS-ring, to tether the C-ring to the MS-ring.22,23,24 FliGM interacts with FliM via the EHPQR-motif.25,26,27 FliGC interacts with the cytoplasmic domain of MotA/PomA.9 These multiple protein-protein interaction networks mediated by FliG are important for the active formation and functioning of the flagellar motor machinery.19,28,29,30,31 FliG monomers can acquire multiple conformations due to the presence of flexible linkers: helixNM (between FliGN and FliGM), helixMC (between FliGM and FliGC), and the MFXF-motif (between ARMC and the C-terminal helical region of FliGC; Cα1-6).19,26,28,31,32,33 Therefore, it is believed that the drastic conformational change in FliG in the C-ring caused by its flexible linkers plays a crucial role in switching motor rotation between CCW and CW. Based on the biochemical and mutational analyses of FliG, we identified a FliG mutant from Vibrio alginolyticus with defective motor function.29,34,35 The wild-type (WT) flagellar motor rotates bidirectionally with a CCW:CW ratio of 7.0:3.0. However, flagellar motor rotation is strongly biased or locked in a single direction by residue substitutions at G214, G215, and A282. The mutation changing G214 to Ser (G214S) confers a CCW-biased (CCW:CW = 9.0:1.0) phenotype, whereas the G215A substitution confers a CW-locked phenotype. These Gly residues are located in the flexible helixMC region (V186-G215) in V. alginolyticus. The A282T substitution confers CW-biased flagellar motor rotation. A282 is located in the hydrophobic core region of Cα1-6 (F256-L351) and is close to the MFXF-motif (M253-F256). Previous 2D nuclear magnetic resonance (NMR), small angle X-ray scattering (SAXS) and molecular dynamics (MD) simulation studies demonstrated that the FliGM-FliGC fragment (FliGMC; K130-L351) of WT-FliG exists in an equilibrium state of multiple conformations.19,29,33 These conformational exchanges are suppressed in FliGMC (G214S), FliGMC (G215A), and FliGC (A282T). It is believed that the structural changes mediated by mutations affecting the FliG linker fix the rotor in the CCW or CW state. Although the structural change in FliG clearly play a crucial role in switching the rotational direction of the motor between CCW and CW, the detailed molecular mechanism of this allosteric conformational change in FliG remains unknown.
In general, proteins exist as ensembles with multiple structural states in the free energy landscape. Elucidation of the equilibrium process between these structures and the protein structure of the minor state with a high energy level provides important information for understanding protein function. Structural changes in proteins due to pressurization generally decrease their partial molar volume and expand the structural fluctuations seen at atmospheric pressure.36 The establishment of various pressure-variable experiments has elucidated the structures of protein folding intermediates and structural equilibrium processes. Recently, it was found that changes in the hydration state of proteins with pressurization are closely related to the structural dynamics of membrane proteins and protein-protein interactions.37
To study the molecular mechanism underlying switching of the flagellar motor, we performed structural analysis of FliGMC fragments of V. alginolyticus using NMR spectroscopy with variable pressure condition.
We prepared a stereo-array isotope labeling (SAIL) FliGMC, which can specifically provide the δ1 methyl signal of isoleucine residues and the δ aromatic CH signal of phenylalanine residues with high sensitivity.38,39,40,41,42,43 The 1H-13C 2D NMR spectrum of FliGMC showed two conformational states in several Ile and Phe residues. In the CW-locked mutant FliGMC-G215A, chemical shift change and exchange broadening were observed in the proximal region of the Gly-Gly linker (G214-G215) as well as in the distal region containing the EHPQR-motif (E144-I148 and R179), helixMC (V186-G215), ARMC (L216-V255), the MFXF-motif (M253-F256) and Cα1-6 (F256-F349). MD simulations showed that the relative orientation between FliGM and FliGC changed in FliGMC-G215A. Furthermore, contact between FliGMC and water molecules changed remarkably. FliGMC NMR spectra under high-pressure conditions were comparable to those of the FliGMC-G215A under ambient pressure. These results suggest that the G215A substitution causes changes in the hydrophobic interaction network within FliGMC that modulate the relative orientation of FliGM and FliGC. Considering the changes observed in the relative orientation of FliGM and FliGC C-ring structure,44 we propose that this mechanism is the primary factor accounting for determining the rotational direction of the flagellar motor.
Results
Role of the Gly-Gly linker in switching rotational direction
Single amino acid substitutions at Gly214-Gly215 (i.e., in the Gly-Gly linker) dramatically switch the rotational direction of the flagellar motor. The FliG (G214S) mutation in V. alginolyticus causes CCW-biased motility. In contrast, FliG (G215A) mutation causes CW-locked motility.35 These phenomena were first shown by analysis of mutations in E. coli FliG.45 The Gly-Gly linker is conserved in various species, suggesting that the structural properties of this region are closely related to the mechanism for changing the direction of flagellar rotation. The amino acid substitutions in the Gly-Gly linker may induce steric hindrance between FliGM and FliGC,29 but the details of the molecular mechanism remain to be discovered.
To elucidate the structural properties of the Gly-Gly linker region, we produced various FliG variants (G214X and/or G215X) in V. alginolyticus and conducted motility assays (Figure S1). By observing the swimming of V. alginolyticus, we calculated the CCW:CW ratio and frequency of directional switching. The CCW: CW ratio of cells expressing WT-FliG was 7.0:3.0 and switching occurred 1.1 times per second. The G214S and G215A substitutions resulted in CCW-biased (CCW: CW ratio of 9.9: 0.1 and switching 0.08 times per second) and CW-locked rotation, respectively, as previously reported.35 The CCW: CW ratio of the G214A motor was 9.9: 0.1, and switching occurred 0.01 times per second (Figure S1). Cells expressing G214C or G214P FliG showed CCW-locked rotation like cells expressing G214S FliG. In contrast, cells expressing G215P or G215S FliG had the same CW-locked phenotype as cells expressing G215A FliG. Cells expressing a G214S/G215A doubly substituted FliG showed a CW-locked rotation. Thus, the mutation that generates G215A is dominant over the mutation that generates G214S. Because Gly residue causes the least steric hindrance, the WT protein can more easily assume multiple conformations, including a hairpin turn structure. The limitation of the backbone dihedral angles in the Gly-Gly linker may contribute to maintaining the CW state.
Using 1H-13C 2D NMR to observe the Ile δ1 methyl and Phe δ aromatic ring CH signals of FliGMC
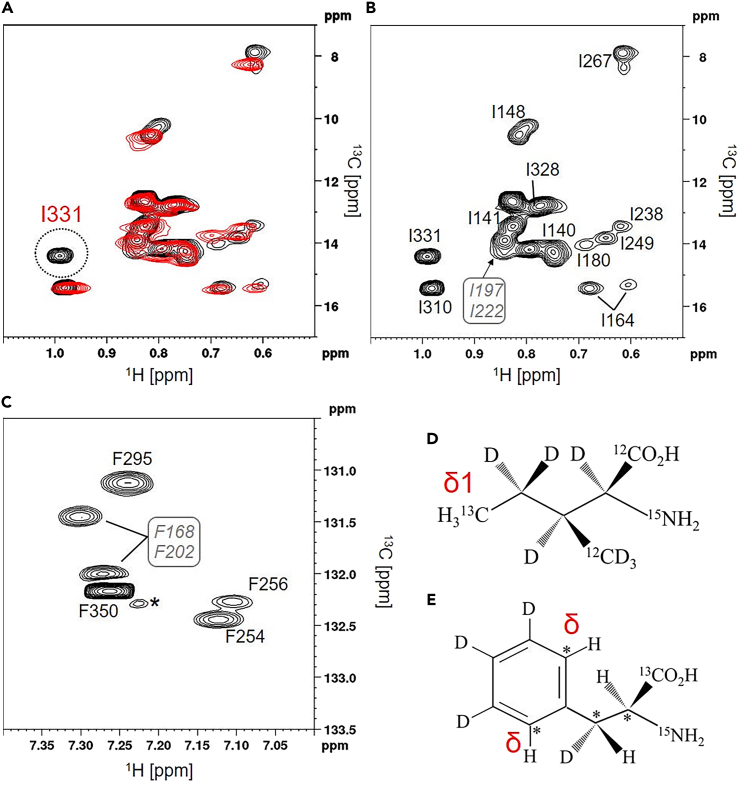
The motility patterns of cells expressing mutant FliG suggested that changes in the flexibility of the Gly-Gly linker may affect the relative orientation of FliGM relative to FliGC. To examine the structural differences between FliGMC and FliGMC -G215A in detail, 2D NMR measurements were performed. We prepared FliGMC that was isotope labeled at the δ1 methyl of Ile and the aromatic δ-CH of Phe (SAIL-Phe) and measured the 1H-13C 2D HMQC and 2D aromatic TROSY spectra.43 These experiments yield highly sensitive NMR signals, even for proteins with >100 kDa molecular weight.43,46,47 We succeeded in observing the Ile δ1 methyl signals and Phe δ-CH signals from 27.8 kDa FliGMC and its G215A variant (Figure 2). For sequence-specific signal assignments for the Ile δ1 methyl group, we compared the singles from proteins with single-residue substitution. Because FliGMC contains 16 Ile residues, including one at the N-terminal Factor Xa cleavage site, we prepared 15 mutants–I140L, I141L, I148L, I151L, I164L, I180L, I197L, I213L, I222L, I238L, I249L, I267L, I310L, I328L, and I331L– that were selectively labeled with δ1-13CH3 Ile. To assign the Ile residue in the Factor Xa cleavage site, the I140L substitution fragment was treated with Factor Xa and investigated by NMR spectroscopy. Figure 2A shows the overlaid 1H-13C HMQC spectra for FliGMC (black) and its I331L mutant (red). The result showed that the missing signal in the mutant was assigned to the I331 δ1 methyl. In the analyses of mutant FliGMC (Figure S2), several Ile δ1 signals could not be unambiguously assigned to a specific Ile residue. We also performed nuclear Overhauser effect spectroscopy (NOESY) experiments with FliGMC and FliGMC-G215A to obtain intra- and inter-residue NOEs, which helped achieve unambiguous signal assignments (Figure S3). The end result was that δ1 methyl signals could be assigned for all of the Ile residues in FliGMC except I151 and I213 (Figure 2B).
Figure 2.
2D 1H-13C NMR spectra for Ile δ1 methyl and Phe δ-CH of Vibrio FliGMC fragment
The spectra were measured using an 800 MHz spectrometer at 288 K equipped with a cryogenic probe.
(A) Overlaid HMQC spectra for FliGMC (black) and its I331L mutant (red). The I331 signal was not detected in the mutant spectra.
(B) Sequence-specific signal assignment for Ile δ1 methyl. The assignment of each of the 16 Ile residues is shown in Figure S2. For I151 and I213 residues, an unambiguous signal could not be assigned.
(C) Sequence-specific signal assignment for Phe δ-CH mapped on 1H-13C TROSY-HSQC spectrum. The six Phe residues were assigned using 13C edited NOESY measurement (Figures S3B–S3D). ∗ indicates impurity.
(D and E) Structure of the isotope labeled at δ1 methyl of isoleucine and SAIL Phenylalanine are shown, respectively. ∗ indicates 13C.
The systematic analysis of mutant FliGMC fragments (Figure S2) revealed simultaneous changes in chemical shift in response to Ile-to-Leu substitutions. Comparing 1H-13C HMQC between FliGMC and its I249L variant showed concomitant chemical shift changes for the δ1 methyl signals of I222 and I238 (Figure S4A). Because these three Ile residues are located in the ARMC domain, the three Ile δ1 methyl groups are located in proximity to each other and may form a hydrophobic cluster (Figure S4C). As discussed later, these local core structures may provide useful information regarding the structural rearrangement of FliGMC.
FliGMC contains six Phe residues: F168, F202, F254, F256, F295, and F350. Notably, the F254 and F256 residues are present in the MFXF-motif. The Phe δ-CH signal of FliGMC was assigned using intra-residue NOE connectivity, based on previously assigned amide signals33 (Figures 2C and S3). Owing to the lack of signal assignment for amide protons in the FliGM region, we could not unambiguously assign the signal for F168 and F202. The signal intensity of the F256 δCH was significantly lower than those of the others, suggesting the presence of a conformational exchange state in F256. A previous NMR study also indicated broadening of the amide signal of F256 due to the existence of an exchange process.33
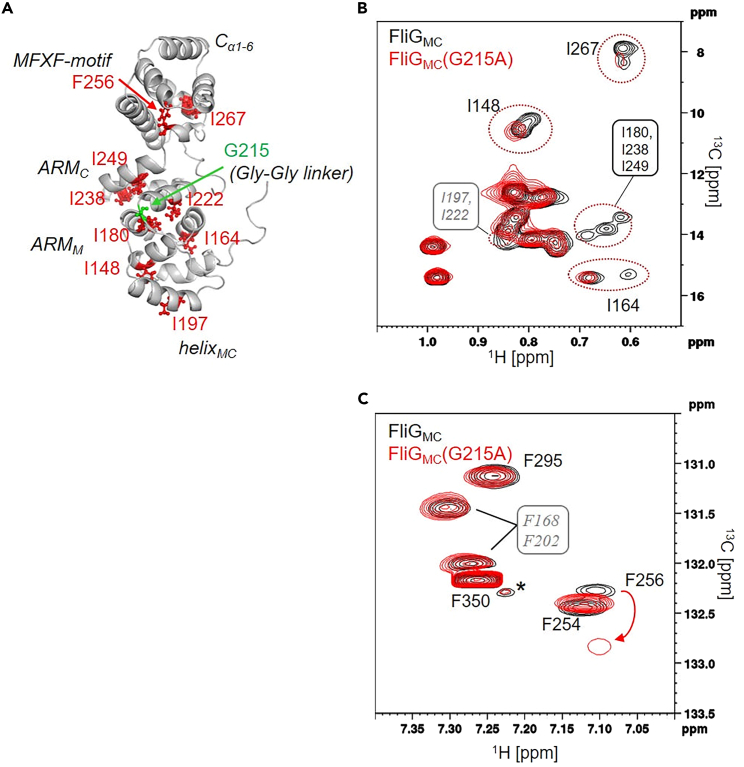
After sequence-specific signal assignment, we also measured the 1H-13C 2D NMR spectra for the Ile δ1 methyl and Phe δ-CH signals of FliGMC-G215A. The G215A substitution caused structural changes not only in the Gly-Gly linker but also in the entire FliGMC (Figure 3A). In the 1H-13C HMQC spectrum of FliGMC-G215A (Figure 3B), the δ1 methyl signal from I180, I197, I222, I238, and I249 disappeared. Moreover, the δ1 methyl signal of I148, I164, and I267 gave two peaks in the 1H-13C HMQC spectrum of FliGMC, but these signals appeared as a single peak in FliGMC-G215A (Figure 3B). These Ile residues were distributed in ARMM (I148, I164, I180), helixMC (I197), ARMC (I222, I238, I249) and Cα1-6 (I267) (Figure 3A). A comparison of the aromatic CH TROSY spectra of FliGMC and its G215A variant showed line broadening and chemical shift changes at F168 (in ARMM), F202 (in helixMC), and F254 and F256 (in the MFXF motif), respectively (Figure 3C). In general, hydrophobic amino acid residues, such as Ile and Phe, form a network through hydrophobic interactions and maintain the overall structure of proteins. Therefore, the G215A substitution probably change the hydrophobic interaction network and influences intradomain orientation.
Figure 3.
Comparison of NMR spectra between the wild-type and G215A-mutated FliGMC
(A) Mapping of Ile and Phe residues exhibiting chemical shift perturbations and signal broadening upon G215A mutation (red ball-and-stick and red letters). The mutated site (G215) is shown with green. The position of each domain in FliGMC is shown in black italic letters.
(B) 1H-13C HMQC Spectra of Ile δ1 methyl region and (C) 1H-13C TROSY HSQC spectra of Phe δ-CH region overlaid for FliGMC (black) and its G215A mutant (red). The peaks whose signal intensity and chemical shift changed with the G215A mutation are highlighted by dotted circles and arrows in spectra (B, C). The spectra were measured using an 800 MHz spectrometer at 288 K equipped with a cryogenic probe.
The pressure dependence of structural changes in FliGMC
The results of the motility assays and the NMR experiment described above suggested that the G215 replacement caused a structural change throughout FliGMC that placed it in a form that leads to CW rotation. In particular, WT FliGMC showed multiple NMR signals in some residues that converged into a single signal in G215A FliGMC. This phenomenon is consistent with previous MD simulations and SAXS analysis.19,29,33 It suggests that the chemical exchange process in FliGMC controls the direction of flagellar rotation. The relaxation dispersion and variable temperature NMR experiments can provide information that is useful for elucidating the chemical exchange properties of proteins. However, it was difficult to perform these experiments because FliGMC undergoes degradation and precipitation with temperature change. As an alternative, we considered variable pressure experiments.
A previous motility assay in E. coli cells lacking the cheY gene (i.e., CCW-locked cells) demonstrated that the number of flagellar motors rotating in the CW direction increased at 288 K and 60 MPa.48 In addition, CW rotation increased in a sigmoidal manner with increasing pressure, consistent with the increasing concentration of CheY-P.48 Therefore, the flagellar motor machinery, including FliG, may exhibit structural changes similar to those induced by the binding of CheY-P upon pressurization, leading to CW rotation. To gain insight into the structural change in FliGMC during the change from the CCW state to the CW state, we measured 2D NMR spectra of FliGMC after pressure perturbation.
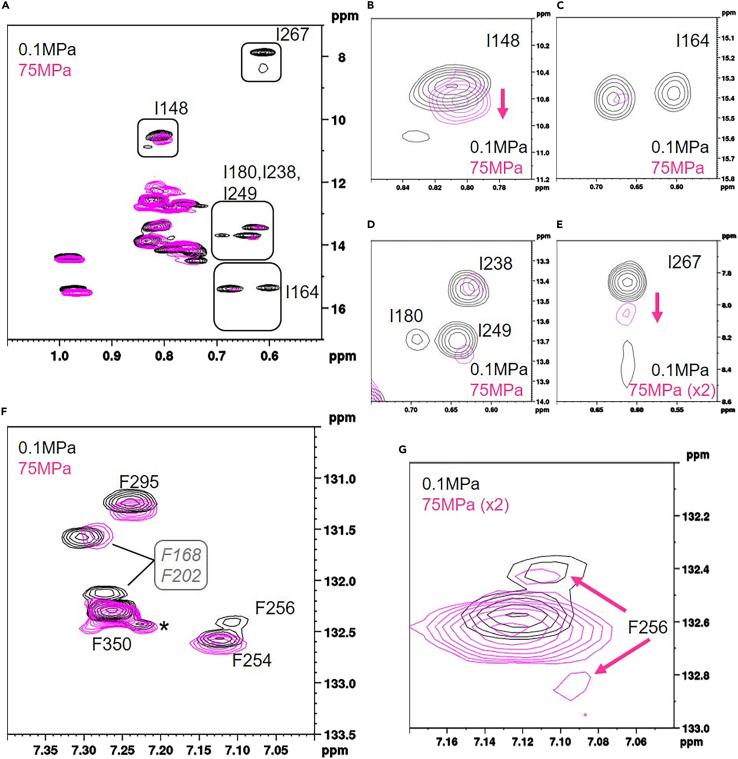
The 2D NMR spectra of FliGMC at ambient pressure and high pressure (75 MPa; FliGMC-HP) were compared. In the 1H-13C HMQC spectrum, the signal intensity of the Ile δ1 was reduced at high pressure, particularly for I180, I238, I249, and I267 (Figure 4). Although the δ1 of I148, I164, and I267 at 0.1 MPa produced a double signal, at 75 MPa the δ1 of these residues gave a single peak (Figure 4). The δ-CH signal of Phe residue also changed under high-pressure conditions. Marked changes in chemical shift and a reduction in intensity were observed for F168, F202, F256, and F295. Notably, the δCH signal of F256 at 75 MPa appeared as a double peak (Figure 4G), and the lower peak was close to the peak of F256 δCH in FliGMC-G215A (Figure 3B). Thus, FliGMC-HP seemed to exhibit an intermediate state between CCW/CW and CW-biased/locked states.
Figure 4.
Changes in 1H-13C signals of Ile and Phe residues in FliGMC subjected to pressure
(A–E) Overlaid HMQC spectra of FliGMC at 0.1 MPa (black) and 75 MPa (magenta). (B), (C), (D), and (E) highlight the signals from I148; I164; I180, I238, and I249; and I267 residues, respectively, that exhibited a significant change in the chemical shift.
(F and G) Overlaid aromatic CH TROSY HSQC spectra of FliGMC at 0.1 MPa (black) and 75 MPa (magenta). (G) The signal from F256 residue is highlighted. The spectral display threshold for 75 MPa in (E) and (G) is doubled.
In general, structural changes in a protein are reversible under a pressure of <500 MPa. In this pressure range, changes in the hydration state of the protein and structural changes that decreases the partial molar volume are exhibited.48,49,50,51,52,53 Our results thus suggest that the hydration state of FliGMC changes under high pressure, leading to the formation of a compact structure corresponding to a partial unfolded state. The Ile and Phe residues altered by pressurization led to characteristic changes in FliGMC-G215A as well, suggesting that the signal changes exhibited by these residues reveal the process which converts FliGMC into its CW conformation.
MD simulation of FliGMC under high pressure
To clarify the structural properties of FliGMC in the CW state, we performed MD simulations under three conditions: (1) FliGMC corresponding to a mixed CCW and CW state; (2) FliGMC-G215A, corresponding to a CW-locked state; and (3) FliGMC-HP, corresponding to a CW-biased state. The initial structures of FliGMC and FliGMC-G215A for these simulations were obtained using homology modeling (see STAR Methods). The distribution of the FliGMC structures based on simulations performed at each condition was evaluated (Figure S5, Videos S1, S2, and S3). During the simulation, for each structural domain, namely ARMM, helixMC, ARMC, and Cα1-6, the root-mean-square deviations (RMSD) from the initial conformation were at most 4 Å apart, thus the native conformations and secondary structure were approximately retained under each condition (Figure S5, Videos S1, S2, and S3). However, the RMSD of full-length proteins varied significantly from the initial structures, indicating that the orientations of the connections among the three domains were highly flexible, likely because of the flexibility of the random coil regions of helixMC and the MFXF motif. Such domain linker regions may play a pivotal role in the structural rearrangement FliGMC undergoes during conversion between the CCW and CW states. A similar change in the relative orientation of FliGM and FliGC was observed in the CW-state C-ring structure determined by cryo-electron tomography (Cryo-ET) analysis.44
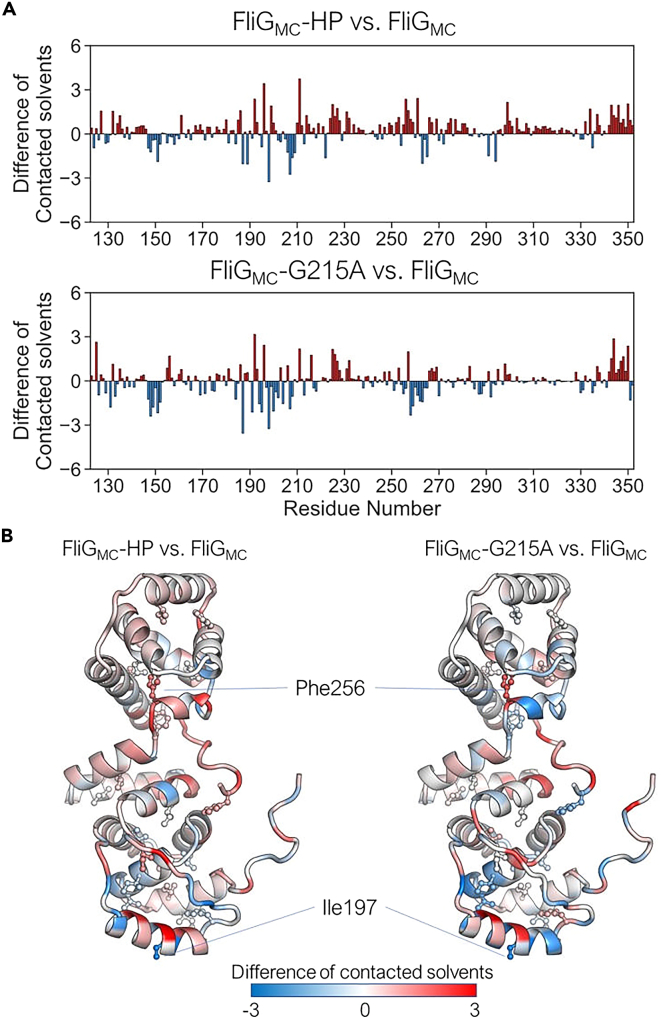
To gain structural insight into the G215A mutation, we determined the water accessibility of FliGMC during simulations (Figures 5 and S6). Overall, the number of water molecules in contact with atoms in FliGMC increased significantly in FliGMC-G215A and FliGMC-HP. Moreover, for each residue, the increase in accessibility was comparable for FliGMC-G215A and FliGMC-HP (Pearson’s correlation coefficient: 0.56; p-value <0.001, Figure S6). The change in accessibility was largest for helixMC and the MFXF motif. The number of water molecules in contact with I197 in helixMC decreased for both FliGMC-G215A and FliGMC-HP but increased for both at F256 in the MFXF motif (Figure 5, Table S1). Consistent with the NMR results, the G215A mutation and pressurization of FliGMC induce similar conformational changes by altering intramolecular hydrophobic interactions and protein–solvent interactions (Figures 3, 4 and S7). These changes are associated with the preference of FliGMC-G215A and FliGMC-HP for the CW conformation.
Figure 5.
Comparison of the number of water molecules interacting with the amino acid residues between FliGMC proteins in the simulations
(A) Residue-wise difference in the number of contacting water molecules between FliGMC and FliGMC-HP (top) and between FliGMC and FliGMC-G215A (bottom). The residues in red (positive values) and blue (negative values) depict the increased number of interacting water molecules in FliGMC-HP or FliGMC-G215A compared with FliGMC and in FliGMC compared with the other conditions, respectively.
(B) Mapping of the difference in the number of contacting water molecules between FliGMC and FliGMC-HP (left) and between FliGMC and FliGMC-G215A (right) on the initial structure. The side-chain atoms of Phe and Ile residues are shown using the ball-and-stick model.
Discussion
Correlation between the conformation of FliGMC and the rotational direction of the flagellar motor
In this study, we analyzed structural changes in the conformation of FliGMC and its G215A variant from V. alginolyticus. The G215A residue substitution generates a CW-locked rotational phenotype in the flagellar motor (Figure S1). In the 1H-13C HMQC spectrum of WT FliGMC, the δ1 methyl signal from I148, I164, and I267 showed broadened and double signals. However, these signals appeared as a single peak in FliGMC-G215A (Figure 3). These I148 and I164 residues are located close to the EHPQR motif in ARMM, and I267 is positioned close to the Cα1-6 domain.
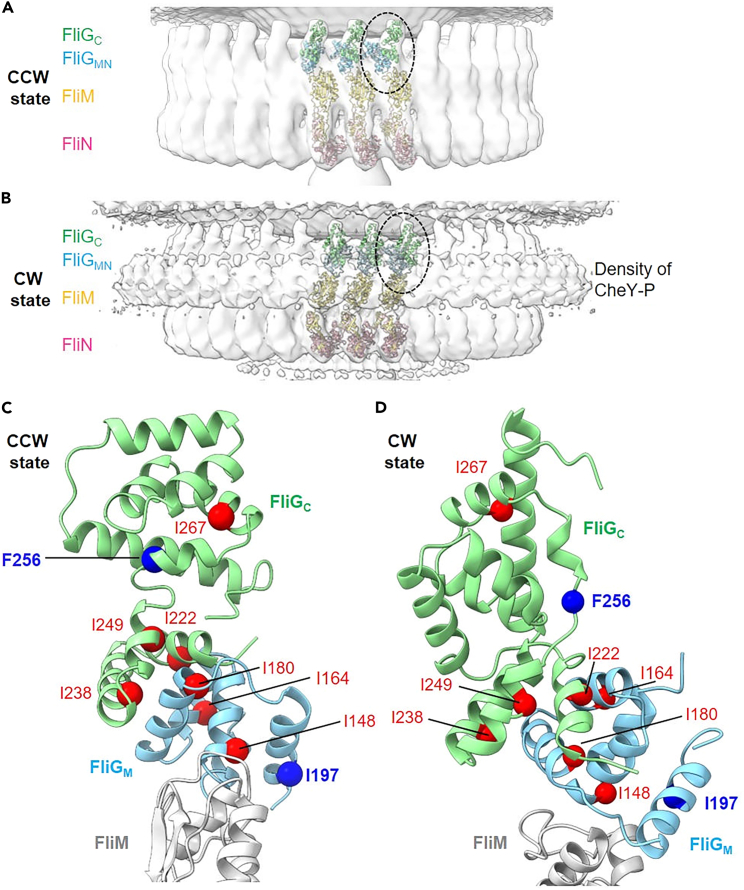
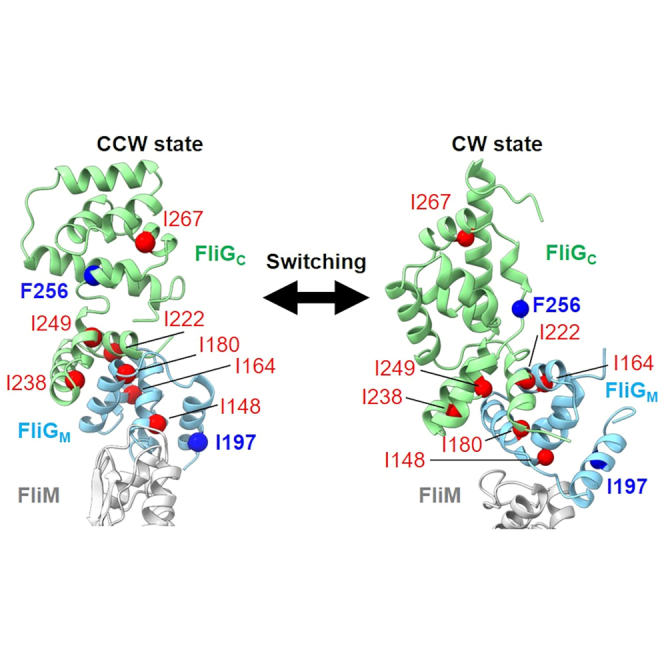
Recently, Cryo-ET analysis revealed high-resolution images of flagellar motor complexes constructed by FliG (G214S), which corresponds to the CCW state of the motor complex, and FliG (G215A), which corresponds to the CW state.44 Figure 6 shows the model structures of FliG (G214S) and FliG (G215A) derived from Cryo-ET mapping. The EHPQR motif and the charged residue cluster are believed to undergo marked structural changes during the transition between the CCW and CW states of the motor. Both domains contribute to the interaction between FliG, FliM and the stator complex. The broadening and doubling of the δ1 methyl signals of Ile residues in WT FliGMC correspond to a mixture of CCW and CW conformations. The existence of multiple conformations in such protein–protein interaction sites is advantageous for quickly switching the direction of motor rotation.
Figure 6.
Structural models of the conformational change in the C-ring during switching between CCW and CW states
(A–D) The structural models show CCW (A) and CW (B) states based on the C-ring volumes of the G214S and G215A mutation; the model was generated based on Cryo-ET analysis.44 FliGC, FliGM FliM and FliN, are shown in light green, light blue, yellow, pink, respectively, as a ribbon model. Side view of FliGC, FliGM and FliM from the dotted circle in (A) and (B) are shown in (C) and (D), respectively. The Ile/Phe residues whose NMR signals changed with the G215A mutation or applying pressure to FliGMC were mapped in the model structure of each FliG by ball representation (red). Among those residues, I197 and F256, which showed particularly large changes, are indicated by blue balls.
The G215A substitution blocks conformational changes in FliG
The G215A substitution in causes structural changes throughout FliGMC, from the N-terminal ARMM domain to the C-terminal Cα1-6 cluster of charged residues. HelixMC connects ARMM and the Gly-Gly linker. In the model structure, helixMC lies close to ARMM, and this interface contains many hydrophobic residues (Figure S4C). The MD simulation results indicated that G215A significantly decreased the accessibility of water molecules to helixMC (Figure 5). A systematic analysis of the FliGMC mutant revealed that the δ1 methyl signal of I148, located close to the EHPQR motif, caused a secondary chemical shift change upon Ile to Leu substitution in various regions (Figure S2). Furthermore, previous mutational analysis indicated that mutations targeting the EHPQR motif of FliG induce severe defects in the switching of the direction of flagellar motor rotation, causing a CW-biased phenotype.29 These results suggest that structural changes in ARMM are transmitted to the Gly-Gly linker through changes in hydrophobic interactions with the flexible helixMC region.
A hydrophobic interaction network between ARMC and the MFXF-motif may be involved in transmitting structural changes in the Gly-Gly linker to the Cα1-6 domain. The δ1 signals of I222, I238, and I249 present in ARMC disappeared in FliGMC-G215A (Figure 3A). Because these residues are located close to each other, a secondary chemical shift change associated with Ile-to-Leu substitution was observed at these residues (Figures 3 and S2). Furthermore, inter-residue methyl–methyl NOE signals were observed among these residues (Figure S3A). Therefore, the G215A substitution may alter hydrophobic interactions in ARMC.
Previous studies have suggested that the MFXF motif regulates the relative orientation between ARMC and Cα1-6.24,25,26 F256 in the MFXF motif may interact with the A282 residue in Cα1-6. In the MD simulations for the A282T variant of FliG, which induced a CW-biased rotation of the flagellar motor, interactions between the MFXF-motif and Cα1-6 were restricted by the steric hindrance between F256 and T282.33 In the present study, a marked chemical shift change was observed in the F256 δCH and I267 δ1 methyl signals in presence of the G215A substitution (Figures 3 and S7). Phe residues have a bulky aromatic ring in their side chain that can interact strongly with various hydrophobic amino acid residues. Therefore, the chemical shift changes and/or line-width broadening of aromatic ring NMR signals often reflect structural changes in the entire protein. Recent studies have reported that the “large amplitude slow breathing motion” of proteins is associated with the aromatic ring flipping, which induces structural changes that can affect biological functions.54,55,56 In Cα1-6 clusters of charged residues are involved in the interaction with MotA/PomA in the stator unit. The intradomain rearrangements in FliGMC mediated by hydrophobic residues may affect the interaction between FliG and MotA/PomA, thereby regulating switching in the direction of motor rotation.
Previous studies have demonstrated that the FliG Gly-Gly linker is the central node of the dynamic network controlling motor rotation switching, but the detailed mechanism was poorly understood. In this study, using SAIL NMR methods, variable pressure NMR experiments and MD simulations, we show that single residue mutations in the glycine linker induce large scale changes in the coupled domain cores through extensive hydrophobic interaction networks. The Gly-Gly and Gly-Ser linkers are ubiquitous in a variety of multidomain proteins, including signal proteins and engineered fusion proteins, and their flexibility allows them to control interdomain interactions.57,58 The mechanism characterized in this study may be applicable as a strategy for switching between functional conformations in other multi domain proteins.
Altered interactions between the C-ring and the stator complex associated with structural changes in FliG
Single particle analysis using cryo-electron microscopy (cryo-EM) determined the 3D structures of MotA-MotB stator complexes at near atomic resolution.59,60 These studies revealed that the transmembrane helices of the MotB dimer insert into the center hole of a ring complex formed by five MotA monomers. Based on these structures, a working hypothesis was proposed that stated that the rotational motion of the stator complex driven by the ion flux acted as the driving force for C-ring rotation.59,60,61 Cryo-ET analysis has produced high resolution images of flagellar motor complexes formed with FliG (G214S) (the CCW state) and FliG (G215A) (the CW state).44 Comparing the structural models for the CCW and CW states revealed that the molecular architecture of FliG in the C-ring was changed drastically by these single amino acid substitutions (Figure 6). In the CW-state motor, FliG(G215A) forms a compact structure via intramolecular interactions between FliGM and FliGC.28,62 In contrast, the elongated structure of FliG observed in previous crystal structural analysis19,26 fits well into the Cryo-ET map of the C-ring in the CCW-state FliG(G214S) motor. The region showing a large structural change between the CCW and CW-forms of FliG corresponded to the region containing the Ile/Phe residues that showed structural changes in the NMR and MD simulation analyses (Figure 6). This correspondence suggests that changes in the hydrophobic interaction network of FliG play an important role in switching the rotational direction of flagellar motor.
Besides the C-ring model from Vibrio, the C-ring structures from Salmonella,63 determined by Cryo-EM single particle analysis using purified flagellar motors, and from Borrelia,64 using Cryo-ET analysis, have been reported. In the latter two models, stoichiometry of the C-ring components, the diameter of the C-ring in both the CW and CCW states varied and is not consistent with the C-ring model for Vibrio. However, our findings are consistent with the idea that the diameter of the FliG region in the C-ring changes substantially between the CCW and CW states. However, the mechanism whereby FliG cooperatively changes its conformation in the C-ring due to association and dissociation of CheY with FliM and FliN is still unclear.
Our NMR analysis and MD simulations show that applying pressure to FliGMC yields structural changes similar to those caused by the G215A substitution (Figures 3, 4 and S7). This structural change may be related to changes in the diameter of C-rings between the CW and CCW states. Both the G215A substitution and the application of pressure to FliGMC affect the relative orientation between the EHPQR-motif in ARMM and Cα1-6 via a hydrophobic interaction network. These regions are involved in interactions with FliM and the stator complex, respectively. These findings are consistent with the torque generation model with stator rotation powering the C-ring rotation and the conformation of FliG determining the rotational direction. The findings from the present study suggest that an interdomain rearrangement of FliG through the hydrophobic network alters the interaction between FliG within the C-ring and the stator complex to induce switching in the rotational direction of the flagellar motor.
Conclusion
In this study, we performed SAIL-NMR analysis combined with variable pressure experiment, and MD simulations for FliGMC and FliGMC-G215A to elucidate the molecular mechanism of the change in the rotational direction of the flagellar motor. The G215A substitution in FliGMC alters the hydrophobic interaction networks mediated by Ile and Phe residues and results in the formation of a more compact structure. Since polyglycine linkers are scattered in various proteins, the mechanism of FliG regulation by Gly-Gly linker characterized in this study could be used for switching between functional conformations in other proteins.
Limitations of the study
A limitation of our study is that detailed 3D structures of FliGMC have not been provided. Solution NMR methods cannot obtain NMR signals from all amino acid residues for highly molecular weight proteins due to several limitations. In this study, we observed the NMR signal for only the Ile and Phe residues of FliGMC. Therefore, the structural dynamics of the other hydrophobic amino acid residues could not be observed, and the details of the hydrophobic interaction network of FliGMC remain to be elucidated.
In addition, FliG is known to interact with other proteins to form a C-ring structure in vivo. Since this study focused on the FliGMC monomer, we believe that we have captured part of the molecular mechanism for changing the direction of flagellar motor.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Vibrio alginolyticus VIO5, see Table S2 | Okunishi et al.65 | N/A |
| Vibrio alginolyticus NMB198, see Table S2 | Yorimitsu et al., 200366 | N/A |
| Escherichia coli DH5α, See Table S2 | Grant et al., 199067 | N/A |
| Escherichia coli BL21(DE3), See Table S2 | Novagen | Cat#69450-3 |
| Escherichia coli S17-1, See Table S2 | Simon et al., 198368 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Deuterium oxide | Wako | Cat#040-18831 |
| Ammonium 15N chloride | ISOTEC | Cat#299251-10G |
| TALON Metal Affinity Resin | Clontech Laboratories, Inc. | Cat#635501 |
| Factor Xa | New England Biolabs | Cat#P8010S |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Roche Life Science | Cat#11873580001 |
| sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) | Cambridge Isotope Laboratories | Cat#DLM-32-1 |
| α-ketobutyric acid sodium salt (Methyl-13C; 3,3-2H2) | Cambridge Isotope Laboratories | Cat#CDLM-7318-0.5 |
| [δ-13C; α, β, γ12, γ13, γ2, γ2, γ2-2H7; 15N] isoleucine | SAIL Technologies | Cat#I-004 |
| [0, α, β, δ1, δ2-13C5; β2, ε1, ε2, ζ-2H4; 15N] phenylalanine | SAIL Technologies | Cat#F-013a |
| Critical commercial assays | ||
| QuikChange Site-directed Mutagenesis kit | Stratagene | Cat#200516 |
| Deposited data | ||
| Structure of FliG-FliM from thermotoga maritima | Vartanian et al.62 | PDB: 4FHR |
| Cryo-ET volume of Vibrio alginolyticus clockwise locked rotor | Carroll et al.44 | EMDB: EMD-21819 |
| Cryo-ET volume of Vibrio alginolyticus counter-clockwise locked rotor | Carroll et al.44 | EMDB: EMD-21837 |
| Oligonucleotides | ||
| Primer: To generate I140L mutation: CAAGTGGCGAGCTTGATTGTTAACGAACAC |
This study | N/A |
| Primer: To generate I140L mutation: GTGTTCGTTAACAATCAAGCTCGCCACTTG |
This study | N/A |
| Primer: To generate I141L mutation: CAAGTGGCGAGCATCTTGGTTAACGAACACC |
This study | N/A |
| Primer: To generate I141L mutation: GGTGTTCGTTAACCAAGATGCTCGCCACTTG |
This study | N/A |
| Primer: To generate I148L mutation: GAACACCCGCAGTTGCAAACCATCGTATTG |
This study | N/A |
| Primer: To generate I148L mutation: CAATACGATGGTTTGCAACTGCGGGTGTTC |
This study | N/A |
| Primer: To generate I151L mutation: CCGCAGATCCAAACCTTGGTATTGTCTTATTTAG |
This study | N/A |
| Primer: To generate I151L mutation: CTAAATAAGACAATACCAAGGTTTGGATCTGCGG |
This study | N/A |
| Primer: To generate I164L mutation: CCAATCCGCGGAGTTGTTGTCTCAGTTCC |
This study | N/A |
| Primer: To generate I164L mutation: GGAACTGAGACAACAACTCCGCGGATTGG |
This study | N/A |
| Primer: To generate I180L mutation: CCTAATGATGCGTTTGGCCAACCTAGAAG |
This study | N/A |
| Primer: To generate I180L mutation: CTTCTAGGTTGGCCAAACGCATCATTAGG |
This study | N/A |
| Primer: To generate I197L mutation: CAGAGCTGAACGAATTGATGGAGAAACAGTTC |
This study | N/A |
| Primer: To generate I197L mutation: GAACTGTTTCTCCATCAATTCGTTCAGCTCTG |
This study | N/A |
| Primer: To generate I213L mutation: CAAGCAGCCAAGTTAGGCGGCCTGAAAG |
This study | N/A |
| Primer: To generate I213L mutation: CTTTCAGGCCGCCTAACTTGGCTGCTTG |
This study | N/A |
| Primer: To generate G214A mutation: CAGCCAAGATTGCGGGCCTGAAAGCGGCA |
This study | N/A |
| Primer: To generate G214A mutation: TGCCGCTTTCAGGCCCGCAATCTTGGCTG |
This study | N/A |
| Primer: To generate G214S mutation: CAGCCAAGATTAGCGGCCTGAAAGCGG |
This study | N/A |
| Primer: To generate G214S mutation: CCGCTTTCAGGCCGCTAATCTTGGCTG |
This study | N/A |
| Primer: To generate G214C mutation: CAGCCAAGATTTGTGGCCTGAAAGCGGCA |
This study | N/A |
| Primer: To generate G214C mutation: TGCCGCTTTCAGGCCACAAATCTTGGCTG |
This study | N/A |
| Primer: To generate G214P mutation: CAGCCAAGATTCCAGGCCTGAAAGCGGCA |
This study | N/A |
| Primer: To generate G214P mutation: TGCCGCTTTCAGGCCTGGAATCTTGGCTG |
This study | N/A |
| Primer: To generate G215A mutation: CAAGATTGGCGCACTGAAAGCGGCAG |
This study | N/A |
| Primer: To generate G215A mutation: CTGCCGCTTTCAGTGCGCCAATCTTG |
This study | N/A |
| Primer: To generate G215S mutation: CAGCCAAGATTGGCTCTCTGAAAGCGGCA |
This study | N/A |
| Primer: To generate G215S mutation: TGCCGCTTTCAGAGAGCCAATCTTGGCTG |
This study | N/A |
| Primer: To generate G215P mutation: CAGCCAAGATTGGCCCACTGAAAGCGGCA |
This study | N/A |
| Primer: To generate G215P mutation: TGCCGCTTTCAGTGGGCCAATCTTGGCTG |
This study | N/A |
| Primer: To generate G214S/G215A mutation: CAGCCAAGATTAGCGCACTGAAAGCGGCA |
This study | N/A |
| Primer: To generate G214S/G215A mutation: TGCCGCTTTCAGTGCGCTAATCTTGGCTG |
This study | N/A |
| Primer: To generate I222L mutation: CTGAAAGCGGCAGCGGAGCTGATGA ACTATCTAGACAAC |
This study | N/A |
| Primer: To generate I222L mutation: GTTGTCTAGATAGTTCATCAGCTCCG CTGCCGCTTTCAG |
This study | N/A |
| Primer: To generate I238L mutation: GGTTTGTTGATGGAGCAGCTGCGC GATCAAGACGAAGAC |
This study | N/A |
| Primer: To generate I238L mutation: GTCTTCGTCTTGATCGCGCAGCTG CTCCATCAACAAACC |
This study | N/A |
| Primer: To generate I249L mutation: GAAGACATGGCGACGCAACTGCA AGACTTGATGTTTGTC |
This study | N/A |
| Primer: To generate I249L mutation: GACAAACATCAAGTCTTGCAGTT GCGTCGCCATGTCTTC |
This study | N/A |
| Primer: To generate I267L mutation: GAAGTGGACGATCAAGGTCTGCA GAAATTGCTGCGTGAT |
This study | N/A |
| Primer: To generate I267L mutation: ATCACGCAGCAATTTCTGCAGACC TTGATCGTCCACTTC |
This study | N/A |
| Primer: To generate I310L mutation: GAGATGATGCGTGATGACCTGGA AGCGATGCCGCCAGTT |
This study | N/A |
| Primer: To generate I310L mutation: AACTGGCGGCATCGCTTCCAGGT CATCACGCATCATCTC |
This study | N/A |
| Primer: To generate I328L mutation: GAAGCGGCACAGAAAGAACTGC TAGCGATCGCTCGTCGC |
This study | N/A |
| Primer: To generate I328L mutation: GCGACGAGCGATCGCTAGCAGT TCTTTCTGTGCCGCTTC |
This study | N/A |
| Primer: To generate I331L mutation: CAGAAAGAAATCCTAGCGCTGGC TCGTCGCATGGCCGAT |
This study | N/A |
| Primer: To generate I331L mutation: ATCGGCCATGCGACGAGCCAGC GCTAGGATTTCTTTCTG |
This study | N/A |
| Primer: To generate F254 mutation: CAAGACTTGATGTACGTCTTCGA AAACTTAG |
This study | N/A |
| Primer: To generate F254 mutation: CTAAGTTTTCGAAGACGTACAT CAAGTCTTG |
This study | N/A |
| Primer: To generate F256 mutation: GACTTGATGTTTGTCTACGAAAA CTTAGTCGAA |
This study | N/A |
| Primer: To generate F256 mutation: TTCGACTAAGTTTTCGTAGACA AACATCAAGTC |
This study | N/A |
| Recombinant DNA | ||
| pMMB206, see Table S2 | Morales et al., 199169 | N/A |
| pNT1, see Table S2 | Takekawa et al.8 | N/A |
| pColdI, see Table S2 | Takara | Cat#3361 |
| pColdI-FliGMC, see Table S2 | Onoue et al., 201670 | N/A |
| Software and algorithms | ||
| TopSpin version 3.6.2 | Bruker BioSpin | https://www.bruker.com/service/supportupgrades/software-downloads/nmr.html |
| UCSF Chimera X Version 1.5 | UCSF | https://www.cgl.ucsf.edu/chimerax/download.html |
| UCSF Chimera, Microsoft Windows 64-bit, Ver 1.16 | UCSF | https://www.cgl.ucsf.edu/chimera/download.html |
| GROMACS version 2016.5 | Pronk et al.71 | http://www.gromacs.org |
| Pymol, version 2.5.0 | Schrödinger LLC | https://pymol.org/2/ |
| MODELLER version 9.6 | Marti-Renom et al. 200072 | https://salilab.org/modeller/ |
| NMRFAM-SPARKY | Lee et al.73 | https://nmrfam.wisc.edu/nmrfam-sparky-distribution/ |
| Other | ||
| Xtreme-60 Syringe pump system | Daedalus Innovations | Cat#Xtreme-60 |
| 5 mm high pressure zirconia tube | Daedalus Innovations | https://daedalusinnovations.com/high-pressure-nmr/ |
| Superdex™ 200 Increase 10/300 GL column | Citiva | Cat#28990944 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yohei Miyanoiri (y-miyanoiri.protein@osaka-u.ac.jp).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
For motility, growth and biochemical assays, Vibrio alginolyticus NMB198 containing appropriate plasmids were cultured in VC broth or VPG broth at 30°C as described in the Methods details. The strain was transformed with the pNT1 plasmids carrying mutations. The FliGMC proteins used for structural were manipulated in Escherichia coli DH5a and expressed in Escherichia coli BL21 (DE3). The strain was transformed with the pColdI-FliGMC plasmids carrying mutations. The protein source organism for this study is Vibrio alginolyticus.
Method details
Bacterial strains, media and growth conditions
The bacterial strains and plasmids used are listed in Table S2. The growth condition and mutagenesis are detailed in SI Appendix. V. alginolyticus cells were cultured at 30°C in VC medium (0.5% [w/v] polypeptone or hi-polypeptone, 0.5% [w/v] yeast extract, 3% [w/v] NaCl, 0.4% [w/v] K2HPO4, and 0.2% [w/v] glucose) or in VPG medium (1% [w/v] polypeptone or hi-polypeptone, 3% [w/v] NaCl, 0.4% [w/v] K2HPO4, and 0.5% [w/v] glycerol). During culture of cells harboring pMMB206 or pNT1 plasmid, chloramphenicol was added at a final concentration of 2.5 μg/mL. E. coli cells were cultured at 37°C in LB medium (1% [w/v] bactotrypeptone, 0.5% [w/v] yeast extract, 0.5% [w/v] NaCl). During culture of cells harboring pColdI-FliGMC, or pMMB206 and pNT1 plasmid, ampicillin, or chloramphenicol was added to a final concentration of 100 μg/mL or 25 μg/mL, respectively.
Mutagenesis
To introduce mutations in the fliG gene cloned into the plasmids pNT1 and pColdI-FliGMC, site-directed mutagenesis was performed using the QuikChange method, following the manufacturer’s instructions (Stratagene). The core facility of Nagoya University or Eurofins genomic was used to confirm all the constructs via DNA sequencing. Transformation of V. alginolyticus with plasmids pMMB206 and pNT1 was performed using conjugational transfer from E. coli S17-1, as described previously.65 Aliquots (20 mL each) of fresh overnight cultures of E. coli S17-1 cells carrying plasmid pMMB206 and pNT1 (donor) and V. alginolyticus NMB198 cells were mixed on a VC–1.5% agar plate and incubated at 30°C overnight. Cells were scraped from the plate and suspended in 300 μL of VC medium. To select transconjugants, the suspension was plated on a VC–1.5% agar plate supplemented with 2.5 μg/mL of chloramphenicol and incubated at 30°C overnight.
Flagellar rotation analysis of FliG mutants
Rotational direction and switching events were observed as described previously.35 Briefly, cells were grown overnight in VC medium at 30°C, diluted 1/100 in VPG medium, and incubated for 4 h at 30°C. The cells were washed twice in buffer V (50 mM Tris-HCl (pH 7.5), 5 mM MgCl2, and 300 mM NaCl). The cell suspension was diluted 1:1 with fresh buffer V. Flagellar rotational direction and switching events in swimming cells were measured for 10 s. The rotational direction was determined from the position of the flagellum and the direction of cell swimming. The flagellum pushes the cell body during CCW rotation and pulls it during CW rotation. Measurements were performed at least six times.
Preparation of isotope labeled FliGMC fragment
FliGMC proteins were prepared following previously described methods29 with slight modifications. Briefly, E. coli BL21 (DE3) cells were transformed with pColdI-FliGMC, which encodes the FliGMC protein fused with translation enhancing element, 6 x His-tag and Factor Xa cleavage site at its N-terminus.
The E. coli cells were cultured at 37°C in M9-D2O medium.29,39 At the start of the culture, 3 mg/L of [0, α, β1, δ1, δ2-13C5; β2, ε1, ε2, ζ-2H4; 15N] phenylalanine (SAIL Phe) (Figure 2E) and 5 mg/L [δ-13C; α, β, γ12, γ13, γ2, γ2, γ2-2H7; 15N] isoleucine (δ1-Ile) (Figure 2D) were added. When the cell OD660nm was 0.38-0.50, cells were incubated on ice for 30 min. Subsequently, 12 mg/L of SAIL Phe and 15 mg/L of δ1-Ile were added, and culture was resumed at 15°C. After 30–60 min culture, isopropyl β-D-thiogalactopyranoside was added to a final concentration of 0.5 mM to induce overexpression and grown for 1 day at 15°C. The cells were harvested by centrifugation and stored at −80°C to break the cell membrane. For sequence-specific signal assignment, 15 Ile residue mutants (I140L, I141L, I148L, I151L, I164L, I180L, I197L, I213L, I222L, I238L, I249L, I267L, I310L, I328L, and I331L) were prepared. For isotope labeling of Ile residues in FliGMC, α-ketobutyric acid sodium salt (Methyl-13C; 3,3-2H2) (CIL, Andover, MA) was used as the isoleucine precursor in following samples; WT, I140L, I151L, I164L, I180L, I197L, I213L, G215A, I222L, I238L, I310L, and I328L. In this case, 70 mg/L α-ketobutyric acid was used instead of δ1-Ile during culture. The procedure for mutant culture and purification was identical to that for the WT.
The frozen cells were suspended with T7.0-N150 buffer (50 mM Tris-HCl, pH 7.0, and 150 mM NaCl) and sonicated using a sonicator (Branson) set on duty cycle 50% and power 5 with proteinase inhibitor, complete EDTA free (Roche Life Science). Unbroken cells were removed by low-speed centrifugation. The samples were ultra-centrifuged at 118,000 × g for 30 min. The resultant supernatants were mixed with 5 mg of Talon Metal Affinity Resin (Clontech Laboratories, Inc.) and incubated at least 10 min at room temperature in a polypropylene column by batch method. After eluting the supernatant in the column, 15 mL (3 fractions of the volume) of T7.0-N150 buffer was added to wash the column. To further wash the column, 5 mL (1 fraction volume) of I30 buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, and 30 mM imidazole) was added. To elute His-tag protein from the resin, 20 mL (4 fraction volume) of I120 buffer (50 mM Tris-HCl, pH 7.0, 150 mM NaCl, and 120 mM imidazole) was added and collected by 1 mL fractions. The His-tag affinity-purified proteins were concentrated to 1 mL using 10 K Amicon device (Millipore). The samples were subjected to size exclusion chromatography using Superdex 200 Increase 10/300 column (Cytiva) in T7.0-N150 buffer with the flow rate at 0.75 mL per min. The peak fractions were collected, and the concentration of samples was measured using Nanodrop (Millipore).
Additionally, to identify the NMR signal of the Ile residue in the N-terminus-fused Tag sequence, which was derived from the pCold I vector, the purified FliGMC (I140L) was digested with Factor Xa. Briefly, 3 μg Factor Xa (New England Biolabs) was added to 2 mL FliGMC (I140L) protein solution (0.2 mM) and incubated at 4°C for 2 days.
NMR spectroscopy
NMR measurements were performed using Avance III 950, 900 and Avance III HD 800, 600 spectrometers equipped with a cryogenic probe (Bruker Biospin) at 288K.
We used the fraction containing isotope-labeled FliGMC fragments (obtained by the size-exclusion chromatography) with the highest peak at A280. The concentrations of the samples were 0.1–0.3 mM. NMR sample buffer contained 50 mM Tris-HCl (pH 7.0), 150 mM NaCl, 0.01% (w/v) sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS), and 5% (w/v) D2O.
In 2D 1H-13C HMQC experiments to observe methyl signals, the data size and spectral width were 128 (t1) x 2048 (t2) and 4020 Hz (ω1, 13C) x 8015 Hz (ω2, 1H), respectively. The carrier frequencies of 1H and 13C were 4.7 and 10 ppm, respectively.
In 2D 1H-13C TROSY HSQC experiments for observing aromatic signals, the data size and spectral width were 128 (t1) x 2048 (t2) and 2010 Hz (ω1, 13C) x 12820 Hz (ω2, 1H), respectively. The carrier frequencies of 1H and 13C were 4.7 and 130 ppm.
For sequence-specific assignments of the Ile methyl signals, we performed 13C-edited NOESY. In the 3D NOESY-HMQC experiments, the data size and spectral width were 256 (t1) x 24 (t2) x 2048 (t3) and 12820 Hz (ω1, 1H) × 2012 Hz (ω2, 13C) × 12820 Hz (ω3, 1H), respectively. The carrier frequencies of 1H and 13C were 4.7 and 10 ppm, respectively. The NOE mixing time was set to 200 ms. To assign the aromatic CH signal in Phe residues, the 13C-edited NOESY-HSQC experiment was performed with a NOE mixing time of 300 ms. The data size and spectral width were 160 (t1) x 16 (t2) x 2048 (t3) and 11160 Hz (ω1, 1H) × 1610 Hz (ω2, 13C) × 11160 Hz (ω3, 1H), respectively. The carrier frequencies of 1H and 13C were 4.7 and 128 ppm, respectively.
In high-pressure NMR experiments, a 5 mm high pressure zirconia tube and an Xtreme-60 Syringe pump system were used (Daedalus Innovations).
All NMR data were processed using the TopSpin software (Bruker Biospin) and NMRFAM-SPARKY.73
MD simulations
The model structure of FliGMC was built using MODELLER version 9.6 with the crystal structure of FliG obtained from Thermotoga maritima (PDB code: 4FHR) used as a template. For the model structure of FliGMC (G215A), a point mutation (G215A) was introduced using the mutagenesis wizard of the PyMOL package (Schrödinger LLC). These models were used for the initial structures in the MD simulations. MD simulations were performed using GROMACS version 2016.5.71 The topology was generated using standard amino acid protonation states at pH 7.0. The force field of AMBER99SB-ILDN and the TIP3P water model were used for the simulation. The starting structure was placed in a cubic box with 1.0 nm space around the solute, and the box was filled with water molecules. In total, there were 89,425 atoms for FliGMC (WT) and 89,431 atoms for FliGMC (G215A) in the systems. Energy minimization was performed using the steepest descent method. Subsequently, the system was equilibrated for 1 ns at 300 K under the NVT (constant number of particles, volume, and temperature) and NPT (constant number of particles, pressure, and temperature) conditions. After equilibration, all-atom production simulations were performed at 300 K under NPT condition at a pressure of 0.1 or 75 MPa for 1 μs without restraints.
Quantification and statistical analysis
In Figures 5 and S6, the statistical analysis of the difference in the number of contact solvent or relative solvent accessibility for each residue in the MD trajectories between conditions; FliGMC 0.1 MPa vs. 75 MPa, and FliGMC 0.1 MPa vs. FliGMC (G215A) were performed with Welch’s t-test. The computation was performed using stats module in the SciPy package.74 In Figure S6, the statistical significance of the Pearson’s correlation coefficient was also evaluated using the same module.
Acknowledgments
This research was supported by Grants-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan (24117004 and 23247024 to MH, and JP18K19293 to S.K.), and Program for leading Graduate Schools of Japan, Science for the Promotion of Science (17J11237 and 20J00329 to T.N.). Work by T.N. was supported in part by the Integrative Graduate Education and Research program of Nagoya University. This work was performed in part using the NMR spectrometers with the ultra-high magnetic fields under the Collaborative Research Program of Institute for Protein Research, Osaka University, NMRCR-17-05. This research was partially supported by Research Support Project for Life Science and Drug Discovery (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED.
Author contributions
T.N., A.H., S.K., T.S., M.K., M.H., and Y.M. designed the study; T.N., A.H., and Y.M. performed the research; T.N., A.H., and Y.M. analyzed the data; T.N., A.H., S.K., T.S., M.H., and Y.M. wrote the paper.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: July 11, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107320.
Contributor Information
Michio Homma, Email: g44416a@cc.nagoya-u.ac.jp.
Yohei Miyanoiri, Email: y-miyanoiri.protein@osaka-u.ac.jp.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Terashima H., Kojima S., Homma M. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 2008;270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 2.Kojima S. Dynamism and regulation of the stator, the energy conversion complex of the bacterial flagellar motor. Curr. Opin. Microbiol. 2015;28:66–71. doi: 10.1016/j.mib.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Blair D.F., Berg H.C. Restoration of torque in defective flagellar motors. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 4.Blair D.F., Berg H.C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhou J., Lloyd S.A., Blair D.F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA. 1998;95:6436–6441. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lloyd S.A., Blair D.F. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 1997;266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 7.Yakushi T., Yang J., Fukuoka H., Homma M., Blair D.F. Roles of charged residues of rotor and stator in flagellar rotation: comparative study using H+-driven and Na+-driven motors in Escherichia coli. J. Bacteriol. 2006;188:1466–1472. doi: 10.1128/JB.188.4.1466-1472.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takekawa N., Kojima S., Homma M. Contribution of many charged residues at the stator-rotor interface of the Na+-driven flagellar motor to torque generation in Vibrio alginolyticus. J. Bacteriol. 2014;196:1377–1385. doi: 10.1128/JB.01392-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Terashima H., Kojima S., Homma M. Site-directed crosslinking identifies the stator-rotor interaction surfaces in a hybrid bacterial flagellar motor. J. Bacteriol. 2021;203:e00016–e00021. doi: 10.1128/JB.00016-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francis N.R., Sosinsky G.E., Thomas D., DeRosier D.J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 11.Xie L., Altindal T., Chattopadhyay S., Wu X.L. Bacterial flagellum as a propeller and as a rudder for efficient chemotaxis. Proc. Natl. Acad. Sci. USA. 2011;108:2246–2251. doi: 10.1073/pnas.1011953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barak R., Eisenbach M. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry. 1992;31:1821–1826. doi: 10.1021/bi00121a034. [DOI] [PubMed] [Google Scholar]
- 13.Cluzel P., Surette M., Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 14.Welch M., Oosawa K., Aizawa S., Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bren A., Eisenbach M. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J. Mol. Biol. 1998;278:507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 16.McEvoy M.M., Bren A., Eisenbach M., Dahlquist F.W. Identification of the binding interfaces on CheY for two of its targets, the phosphatase CheZ and the flagellar switch protein fliM. J. Mol. Biol. 1999;289:1423–1433. doi: 10.1006/jmbi.1999.2830. [DOI] [PubMed] [Google Scholar]
- 17.Dyer C.M., Vartanian A.S., Zhou H., Dahlquist F.W. A molecular mechanism of bacterial flagellar motor switching. J. Mol. Biol. 2009;388:71–84. doi: 10.1016/j.jmb.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarkar M.K., Paul K., Blair D. Chemotaxis signaling protein CheY binds to the rotor protein FliN to control the direction of flagellar rotation in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2010;107:9370–9375. doi: 10.1073/pnas.1000935107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee L.K., Ginsburg M.A., Crovace C., Donohoe M., Stock D. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature. 2010;466:996–1000. doi: 10.1038/nature09300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baker M.A.B., Hynson R.M.G., Ganuelas L.A., Mohammadi N.S., Liew C.W., Rey A.A., Duff A.P., Whitten A.E., Jeffries C.M., Delalez N.J., et al. Domain-swap polymerization drives the self-assembly of the bacterial flagellar motor. Nat. Struct. Mol. Biol. 2016;23:197–203. doi: 10.1038/nsmb.3172. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita M., Namba K., Minamino T. Effect of a clockwise-locked deletion in FliG on the FliG ring structure of the bacterial flagellar motor. Gene Cell. 2018;23:241–247. doi: 10.1111/gtc.12565. [DOI] [PubMed] [Google Scholar]
- 22.Ogawa R., Abe-Yoshizumi R., Kishi T., Homma M., Kojima S. Interaction of the C-terminal tail of FliF with FliG from the Na+-driven flagellar motor of Vibrio alginolyticus. J. Bacteriol. 2015;197:63–72. doi: 10.1128/JB.02271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lynch M.J., Levenson R., Kim E.A., Sircar R., Blair D.F., Dahlquist F.W., Crane B.R. Co-folding of a FliF-FliG split domain forms the basis of the MS:C Ring interface within the bacterial flagellar motor. Structure. 2017;25:317–328. doi: 10.1016/j.str.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue C., Lam K.H., Zhang H., Sun K., Lee S.H., Chen X., Au S.W.N. Crystal structure of the FliF-FliG complex from Helicobacter pylori yields insight into the assembly of the motor MS-C ring in the bacterial flagellum. J. Biol. Chem. 2018;293:2066–2078. doi: 10.1074/jbc.M117.797936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lam K.H., Lam W.W.L., Wong J.Y.K., Chan L.C., Kotaka M., Ling T.K.W., Jin D.Y., Ottemann K.M., Au S.W.N. Structural basis of FliG-FliM interaction in Helicobacter pylori. Mol. Microbiol. 2013;88:798–812. doi: 10.1111/mmi.12222. [DOI] [PubMed] [Google Scholar]
- 26.Brown P.N., Hill C.P., Blair D.F. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 2002;21:3225–3234. doi: 10.1093/emboj/cdf332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown P.N., Terrazas M., Paul K., Blair D.F. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 2007;189:305–312. doi: 10.1128/JB.01281-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minamino T., Imada K., Kinoshita M., Nakamura S., Morimoto Y.V., Namba K. Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishikino T., Hijikata A., Miyanoiri Y., Onoue Y., Kojima S., Shirai T., Homma M. Rotational direction of flagellar motor from the conformation of FliG middle domain in marine Vibrio. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-35902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pandini A., Kleinjung J., Rasool S., Khan S. Coevolved mutations reveal distinct architectures for two core proteins in the bacterial flagellar motor. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandini A., Morcos F., Khan S. The gearbox of the bacterial flagellar motor switch. Structure. 2016;24:1209–1220. doi: 10.1016/j.str.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam K.H., Ip W.S., Lam Y.W., Chan S.O., Ling T.K.W., Au S.W.N. Multiple conformations of the FliG C-terminal domain provide insight into flagellar motor switching. Structure. 2012;20:315–325. doi: 10.1016/j.str.2011.11.020. [DOI] [PubMed] [Google Scholar]
- 33.Miyanoiri Y., Hijikata A., Nishino Y., Gohara M., Onoue Y., Kojima S., Kojima C., Shirai T., Kainosho M., Homma M. Structural and functional analysis of the C-Terminal region of FliG, an essential motor component of Vibrio Na+-driven flagella. Structure. 2017;25:1540–1548.e3. doi: 10.1016/j.str.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Kojima S., Nonoyama N., Takekawa N., Fukuoka H., Homma M. Mutations targeting the C-terminal domain of FliG can disrupt motor assembly in the Na(+)-driven flagella of Vibrio alginolyticus. J. Mol. Biol. 2011;414:62–74. doi: 10.1016/j.jmb.2011.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Nishikino T., Zhu S., Takekawa N., Kojima S., Onoue Y., Homma M. Serine suppresses the motor function of a periplasmic PomB mutation in the Vibrio flagella stator. Gene Cell. 2016;21:505–516. doi: 10.1111/gtc.12357. [DOI] [PubMed] [Google Scholar]
- 36.Hirata F., Akasaka K. Structural fluctuation of proteins induced by thermodynamic perturbation. J. Chem. Phys. 2015;142 doi: 10.1063/1.4906071. [DOI] [PubMed] [Google Scholar]
- 37.Hata H., Nishihara Y., Nishiyama M., Sowa Y., Kawagishi I., Kitao A. High pressure inhibits signaling protein binding to the flagellar motor and bacterial chemotaxis through enhanced hyfration. Sci. Rep. 2020;10:2351. doi: 10.1038/s41598-020-59172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gardner K.H., Kay L.E. The use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 39.Goto N.K., Gardner K.H., Mueller G.A., Willis R.C., Kay L.E. A robust and cost-effective method for the production of Val, Leu, Ile (delta 1) methyl-protonated 15N-13C-2H-labeled proteins. J. Biomol. NMR. 1999;13:369–374. doi: 10.1023/a:1008393201236. [DOI] [PubMed] [Google Scholar]
- 40.Kainosho M., Torizawa T., Iwashita Y., Terauchi T., Mei Ono A., Güntert P. Optimal isotope labelling for NMR protein structure determinations. Nature. 2006;440:52–57. doi: 10.1038/nature04525. [DOI] [PubMed] [Google Scholar]
- 41.Kainosho M., Miyanoiri Y., Terauchi T., Takeda M. Perspective: next generation isotope-aided methods for protein NMR spectroscopy. J. Biomol. NMR. 2018;71:119–127. doi: 10.1007/s10858-018-0198-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyanoiri Y., Takeda M., Kainosho M. Stereo-array isotope labeling method for studying protein structure and dynamics. Adv. Exp. Med. Biol. 2012;992:83–93. doi: 10.1007/978-94-007-4954-2_5. [DOI] [PubMed] [Google Scholar]
- 43.Miyanoiri Y., Takeda M., Terauchi T., Kainosho M. Recent developments in isotope-aided NMR methods for supramolecular protein complexes -SAIL aromatic TROSY. Biochim. Biophys. Acta Gen. Subj. 2020;1864 doi: 10.1016/j.bbagen.2019.129439. [DOI] [PubMed] [Google Scholar]
- 44.Carroll B.L., Nishikino T., Guo W., Zhu S., Kojima S., Homma M., Liu J. The flagellar motor of Vibrio alginolyticus undergoes major structural remodeling during rotational switching. Elife. 2020;9 doi: 10.7554/eLife.61446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Way S.M., Millas S.G., Lee A.H., Manson M.D. Rusty, jammed, and well-oiled hinges: muations affecting the interdomain region of FliG, a rotor element of the Escherichia coli flagellar motor. J Bactriol. 2004;186:3173–3181. doi: 10.1128/JB.186.10.3173-3181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruschak A.M., Kay L.E. Proteasome allostery as a population shift between interchanging conformers. Proc. Natl. Acad. Sci. USA. 2012;109:E3454–E3462. doi: 10.1073/pnas.1213640109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kerfah R., Hamelin O., Boisbouvier J., Marion D. CH3-specific NMR assignment of alanine, isoleucine, leucine and valine methyl groups in high molecular weight proteins using a single sample. J. Biomol. NMR. 2015;63:389–402. doi: 10.1007/s10858-015-9998-4. [DOI] [PubMed] [Google Scholar]
- 48.Nishiyama M., Sowa Y., Kimura Y., Homma M., Ishijima A., Terazima M. High hydrostatic pressure induces counterclockwise to clockwise reversals of the Escherichia coli flagellar motor. J. Bacteriol. 2013;195:1809–1814. doi: 10.1128/JB.02139-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kamatari Y.O., Kitahara R., Yamada H., Yokoyama S., Akasaka K. High-pressure NMR spectroscopy for characterizing folding intermediates and denatured states of proteins. Methods. 2004;34:133–143. doi: 10.1016/j.ymeth.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Roche J., Caro J.A., Norberto D.R., Barthe P., Roumestand C., Schlessman J.L., Garcia A.E., García-Moreno B.E., Royer C.A. Cavities determine the pressure unfolding of proteins. Proc. Natl. Acad. Sci. USA. 2012;109:6945–6950. doi: 10.1073/pnas.1200915109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins M.D., Hummer G., Quillin M.L., Matthews B.W., Gruner S.M. Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc. Natl. Acad. Sci. USA. 2005;102:16668–16671. doi: 10.1073/pnas.0508224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xue M., Wakamoto T., Kejlberg C., Yoshimura Y., Nielsen T.A., Risør M.W., Sanggaard K.W., Kitahara R., Mulder F.A.A. How internal cavities destabilize a protein. Proc. Natl. Acad. Sci. USA. 2019;116:21031–21036. doi: 10.1073/pnas.1911181116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abiko L.A., Grahl A., Grzesiek S. High pressure shifts the β1-adrenergic receptor to the active conformation in the absence of G protein. J. Am. Chem. Soc. 2019;141:16663–16670. doi: 10.1021/jacs.9b06042. [DOI] [PubMed] [Google Scholar]
- 54.Mariño Pérez L., Ielasi F.S., Bessa L.M., Maurin D., Kragelj J., Blackledge M., Salvi N., Bouvignies G., Palencia A., Jensen M.R. Visualizing protein breathing motions associated with aromatic ring flipping. Nature. 2022;602:695–700. doi: 10.1038/s41586-022-04417-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang C.J., Takeda M., Terauchi T., Jee J., Kainosho M. Differential large-amplitude breathing motions in the interface of FKBP12-drug complexes. Biochemistry. 2015;54:6983–6995. doi: 10.1021/acs.biochem.5b00820. [DOI] [PubMed] [Google Scholar]
- 56.Tomita A., Sato T., Ichiyanagi K., Nozawa S., Ichikawa H., Chollet M., Kawai F., Park S.Y., Tsuduki T., Yamato T., et al. Visualizing breathing motion of internal cavities in concert with ligand migration in myoglobin. Proc. Natl. Acad. Sci. USA. 2009;106:2612–2616. doi: 10.1073/pnas.0807774106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshida M., Aizawa T., Nakamura T., Shitara K., Hayakawa Y., Matsubara K., Miura K., Kouno T., Clark K.D., Strand M.R., et al. The Gly-Gly linker region of the insect cytokine growth-blocking peptide is essential for activity. J. Biol. Chem. 2004;279:51331–51337. doi: 10.1074/jbc.M409382200. [DOI] [PubMed] [Google Scholar]
- 58.van Rosmalen M., Krom M., Merkx M. Tuning the flexibility of Glycine-Serine linkers to allow rational design of multidomain proteins. Biochemistry. 2017;56:6565–6574. doi: 10.1021/acs.biochem.7b00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deme J.C., Johnson S., Vickery O., Aron A., Monkhouse H., Griffiths T., James R.H., Berks B.C., Coulton J.W., Stansfeld P.J., Lea S.M. Structures of the stator complex that drives rotation of the bacterial flagellum. Nat. Microbiol. 2020;5:1553–1564. doi: 10.1038/s41564-020-0788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santiveri M., Roa-Eguiara A., Kühne C., Wadhwa N., Hu H., Berg H.C., Erhardt M., Taylor N.M.I. Structure and function of stator units of the bacterial flagellar motor. Cell. 2020;183:244–257.e16. doi: 10.1016/j.cell.2020.08.016. [DOI] [PubMed] [Google Scholar]
- 61.Chang Y., Moon K.H., Zhao X., Norris S.J., Motaleb M.A., Liu J. Structural insights into flagellar stator-rotor interactions. Elife. 2019;8 doi: 10.7554/eLife.48979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vartanian A.S., Paz A., Fortgang E.A., Abramson J., Dahlquist F.W. Structure of flagellar motor proteins in complex allows for insights into motor structure and switching. J. Biol. Chem. 2012;287:35779–35783. doi: 10.1074/jbc.C112.378380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakai T., Miyata T., Terahara N., Mori K., Inoue Y., Morimoto Y.V., Kato T., Namba K., Minamino T. Novel insights into conformational rearrangements of the bacterial flagellar switch complex. mBio. 2019;10:e00799-19. doi: 10.1128/mBio.00079-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang Y., Zhang K., Carroll B.L., Zhao X., Charon N.W., Norris S.J., Motaleb M.A., Li C., Liu J. Molecular mechanism for rotational switching of the bacterial flagellar motor. Nat. Struct. Mol. Biol. 2020;27:1041–1047. doi: 10.1038/s41594-020-0497-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okunishi I., Kawagishi I., Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yorimitsu T., Mimaki A., Yakushi T., Homma M. The conserved charged residues of the C-terminal region of FliG, a rotor component of the Na+-driven flagellar motor. J. Mol. Biol. 2003;334:567–583. doi: 10.1016/j.jmb.2003.09.052. [DOI] [PubMed] [Google Scholar]
- 67.Grant S.G., Jessee J., Bloom F.R., Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simon R., Priefer U.B., Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio-Technology. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 69.Morales V.M., Bäckman A., Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 70.Onoue Y., Abe-Yoshizumi R., Gohara M., Nishino Y., Kobayashi S., Asami Y., Homma M. Domain-based biophysical characterization of the structural and thermal stability of FliG, an essential rotor component of the Na+-driven flagellar motor. Biophys. Physicobiol. 2016;13:227–233. doi: 10.2142/biophysico.13.0_227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pronk S., Páll S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D., et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., Sali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 73.Lee W., Tonelli M., Markley J.L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.