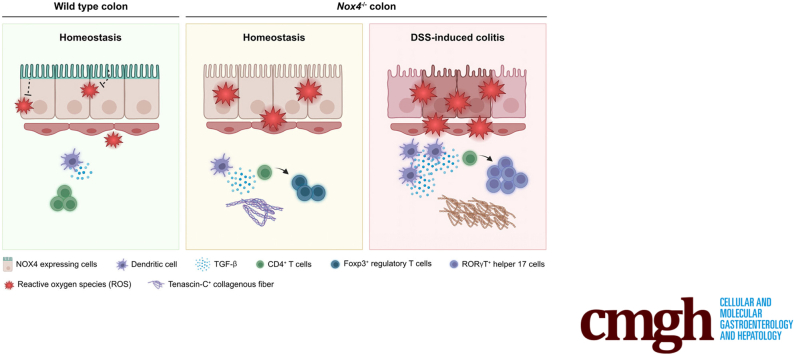
Abstract
Background & Aims
Fibrosis development in ulcerative colitis is associated directly with the severity of mucosal inflammation, which increases the risk of colorectal cancer. The transforming growth factor-β (TGF-β) signaling pathway is an important source of tissue fibrogenesis, which is stimulated directly by reactive oxygen species produced from nicotinamide adenine dinucleotide phosphate oxidases (NOX). Among members of the NOX family, NOX4 expression is up-regulated in patients with fibrostenotic Crohn's disease (CD) and in dextran sulfate sodium (DSS)-induced murine colitis. The aim of this study was to determine whether NOX4 plays a role in fibrogenesis during inflammation in the colon using a mouse model.
Methods
Acute and recovery models of colonic inflammation were performed by DSS administration to newly generated Nox4-/- mice. Pathologic analysis of colon tissues was performed, including detection of immune cells, proliferation, and fibrotic and inflammatory markers. RNA sequencing was performed to detect differentially expressed genes between Nox4-/- and wild-type mice in both the untreated and DSS-treated conditions, followed by functional enrichment analysis to explore the molecular mechanisms contributing to pathologic differences during DSS-induced colitis and after recovery.
Results
Nox4-/- mice showed increased endogenous TGF-β signaling in the colon, increased reactive oxygen species levels, intensive inflammation, and an increased fibrotic region after DSS treatment compared with wild-type mice. Bulk RNA sequencing confirmed involvement of canonical TGF-β signaling in fibrogenesis of the DSS-induced colitis model. Up-regulation of TGF-β signaling affects collagen activation and T-cell lineage commitment, increasing the susceptibility for inflammation.
Conclusions
Nox4 protects against injury and plays a crucial role in fibrogenesis in DSS-induced colitis through canonical TGF-β signaling regulation, highlighting a new treatment target.
Keywords: Fibrostenotic CD, T-Cell Lineage Commitment, RNA-Sequencing, Oxidative Stress
Graphical abstract
Ulcerative colitis (UC) is a common form of inflammatory bowel disease (IBD) resulting from long-term inflammatory damage and ulceration of the colonic mucosa. Continuous inflammation by commensal bacteria affects the intestinal epithelial barrier and is a major contributor to UC pathogenesis.1 Current therapies are ineffective for most patients with UC, and are associated with adverse events such as tissue function abnormalities, fibrostenosis, and malignant transformation.2 UC-associated carcinogenesis derives mainly from the inflammation-mediated production of excessive reactive oxygen species (ROS) in damaged tissues, and the consequent impairment of redox balance leads to oxidative stress after molecular damage (lipid, DNA, protein), along with genetic variation in the transforming colon cells, eventually causing colorectal cancer. Given the poor prognosis of UC-associated colorectal cancer, there is an urgent need to identify novel therapeutic targets.
Members of the nicotinamide adenine dinucleotide phosphate oxidase (NOX) family are mainly responsible for ROS production in the gastrointestinal tract, which play roles in pathogen clearance,3 maintenance of barrier function,4 and mucosal repair after injury.5 Specifically, NOX1 and dual oxidase (DUOX) 2 are expressed abundantly in the intestinal epithelium as the primary sources of ROS in the intestinal mucosa, and their mutation has been linked to the risk of adult IBD. NOX1 contributes to intestinal homeostasis by regulating mucus cell differentiation,6 migration, microbial defense, and mediation of mucus repair after injury.7,8 DUOX2 is activated by pathogen-derived uracil9 to produce DUOX-dependent ROS and governs gut microbe clearance10 caused by enteric infection.
Several recent studies have elucidated the roles of NOX4 in regulating inflammation-mediated pathophysiology in the colon. NOX4 produces H2O2, and is expressed predominately in vascular smooth muscle cells, endothelial cells, and fibroblasts in most tissues. NOX4 expression is up-regulated in drug-resistant patients with UC11 and fibrostenotic Crohn’s disease (CD),12 and in dextran sulfate sodium (DSS)-induced murine colitis models.13,14 Nox4 transcripts are detected at low levels in the normal colon,15 but increase to protect against DSS-induced16,17 and bacterial-induced17 murine colitis. The related mechanism likely involves NOX4-driven ROS production, which induces the phosphorylation of p65 to activate nuclear factor-κB signaling and the M1 phenotype of intestinal macrophages, leading to mucosal barrier injury to promote colitis progression.16 Although NOX4 has rarely been associated with the development of fibrogenic features in mouse colitis models,17 the main pathologic mechanism contributing to fibrotic disease in the lung,18 liver,19 kidney,20 and heart21 is attributed to ROS-induced canonical transforming growth factor-β (TGF-β) signaling, and redox signaling driven by NOX4 is closely related to canonical TGF-β signaling to cause a TGF-β–induced profibrotic response.22 However, the detailed molecular mechanism underlying fibrosis development in UC remains to be elucidated.
Based on this background, we hypothesized that NOX4 contributes to intestinal fibrotic injury in UC and is required for TGF-β signaling inducing fibrosis development. To test this hypothesis, we established NOX4-deficient mice (Nox4-/-) with DSS-induced colitis. RNA-sequencing (RNA-seq) analysis was performed to explore the effects of Nox4 on the response to intestinal damage. This study highlights the role of Nox4 in inducing inflammation in the colon, suggesting that exploiting the redox signal can be a double-edged sword and should be considered carefully for the effective treatment of UC.
Results
Nox4 Protects Against DSS-Induced Experimental Colitis
To study the role of Nox4 in fibrogenesis in DSS-induced colitis in mice, first we confirmed NOX4 expression in the tissues.
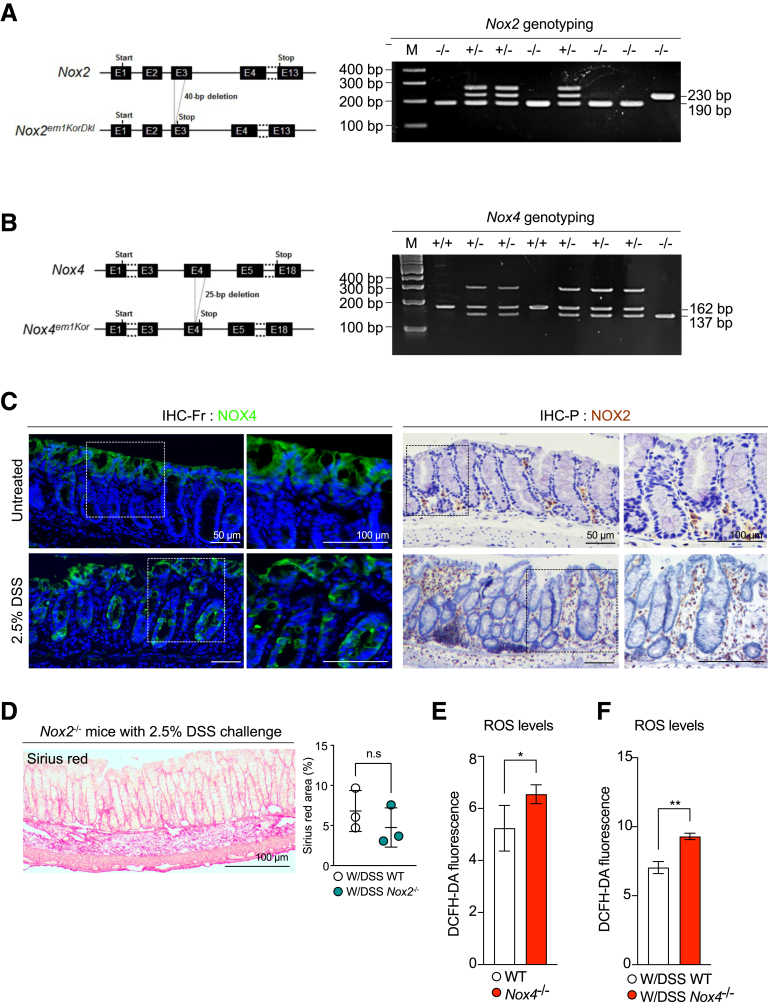
We performed immunohistochemistry in the normal and DSS-treated inflamed colon sections. In the normal colon, NOX4 was detected at the top of the crypt, and NOX4 proteins increased significantly in the injured mucosa (not only at the top, but also at the bottom, of the crypt) after 2.5% DSS-induced colitis (Figure 1C). Loss of Nox4 increased the production of ROS in the control (Figure 1E) and DSS-induced colitis model (Figure 1F), suggesting that Nox4 increase might be associated with disease progression.
Figure 1.
Generation of Nox4-/-mice. Schematic representation of (A) Nox2 and (B) Nox4 deletion using CRISPR/Cas9 technology. The resulting heterozygous and homozygous mice were determined by genotyping PCR using a specific Nox2 or Nox4 deletion site. (C) Immunofluorescence images for NOX4 on the untreated colon and 2.5% DSS-treated colon frozen sections (immunohistochemistry [IHC]-Fr) (left). IHC for NOX2 on the untreated colon and 2.5% DSS-treated colon paraffin-embedded sections (IHC-P) (right). (D) Sirius-red staining showing the fibrotic area in red. The graph represents the measurement of the fibrotic area using ImageJ (National Institutes of Health). Intracellular levels of ROS in the (E) untreated and (F) DSS-treated groups assessed using DCFH-DA staining followed by flow cytometry. Data are expressed as means ± SD. ∗P < .5 and ∗∗P < .01 compared with WT. bp, base pair; DCFH-DA, dichloro-dihydro-fluorescein diacetate.
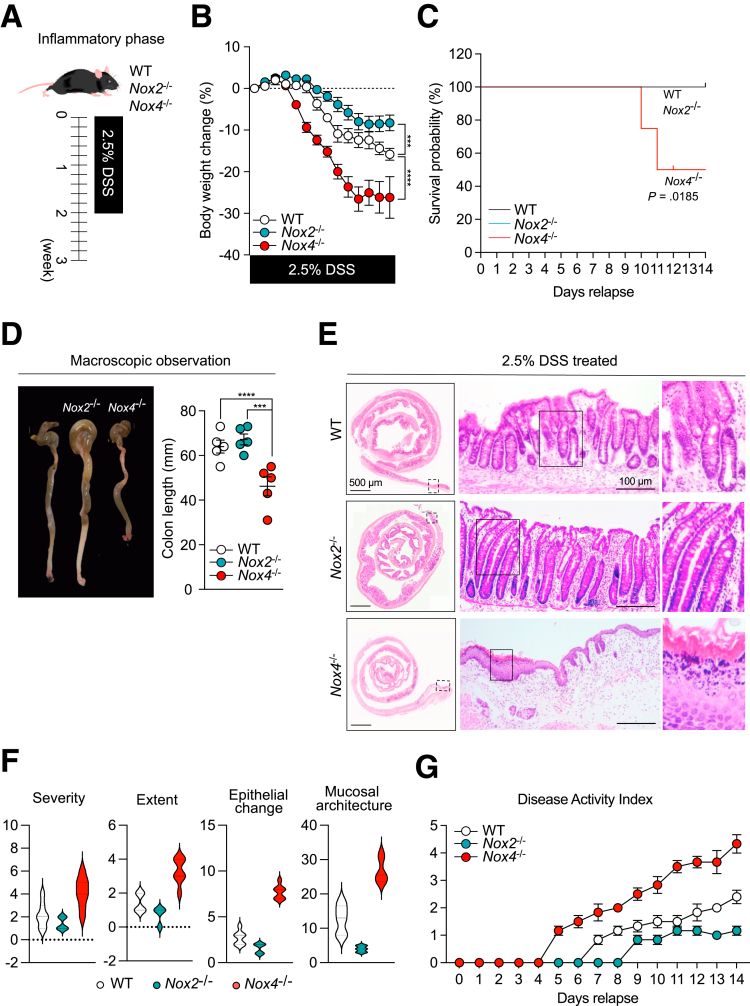
After inducing the inflammatory phase with DSS treatment (W/DSS) (Figure 2A), Nox4-/- mice showed severe inflammation with rapid body weight loss of approximately 20%–30% (Figure 2B) and decreased survival (Figure 2C) compared with that of wild-type (WT) mice. Macroscopic and histologic observations showed that W/DSS Nox4-/- mice had shorter colon lengths (Figure 2D) and more severe inflammatory regions in the distal colon, including infiltration of inflammatory cells into the lamina propria, transmural inflammation, and loss of the crypt and surface epithelium (Figure 2E and F). W/DSS Nox4-/- mice showed higher Disease Activity Index (DAI) scores from 1 to 14 days than the W/DSS WT group (Figure 2G). The W/DSS Nox4-/- colon showed 1.5-fold higher H2O2/ROS production than the W/DSS WT colon (Figure 1G). These results could suggest that Nox4 is involved in the regulation of ROS in the colonic issue, which contributes to the inflammatory response.
Figure 2.
Nox4 protects against DSS-induced experimental colitis. (A) Experimental scheme for the DSS-induced murine colitis model. (B) Body weight loss for WT and Nox4-/- mice with DSS-induced colitis was measured daily (n = 6). (C) Survival probability of WT or Nox4-/- mice with DSS-induced colitis was assessed until day 14 (n = 6). (D) Macroscopic observation of the colons from WT and Nox4-/- mice with DSS-induced colitis killed on day 14. Colon lengths were assessed after the mice were killed. (E) Representative H&E staining images of colon tissues from DSS-induced colitis WT and Nox4-/- mice. (F) Colitis scoring for pathologic assessment in DSS-induced colitis WT and Nox4-/- mice: severity, inflammation severity (0–3); extent, inflammation extent (0, none; 1, mucosa; 2, submucosa; and 3, transmural), and epithelial change. (G) Disease activity index comprising assessment of body weight loss, stool consistency, and rectal bleeding measured daily. Data are expressed as means ± SD. ∗∗∗P < .005, and ∗∗∗∗P < .001 compared with WT.
Nox2 was expressed mainly in the immune cells23 (Figure 1C). In line with the results of a previous study,24 W/DSS Nox2-/- mice did not develop severe colitis, and the body weight change decreased less than that found for W/DSS WT mice (Figure 2B). Moreover, W/DSS Nox2-/- mice showed high survival rates (Figure 2C), and there was no change in the colon length from that of WT mice (Figure 2D). Histologic examination of the DSS-treated W/DSS Nox2-/- colon revealed elongated crypts, immune cell infiltration, and goblet cell loss (Figure 1D), indicating an injured colon; however, the damage score was decreased significantly compared with the W/DSS WT and W/DSS Nox4-/- mice (Figure 2F and G). In line with these results, W/DSS Nox2-/- mice did not develop fibrotic regions in the colon after DSS administration (Figure 1D). Thus, Nox2 deficiency results in mild inflammation in DSS-induced colitis, whereas W/DSS Nox4-/- mice showed severe colonic damage upon DSS treatment; accordingly, we further investigated the characteristics of inflammation in Nox4-/- mice.
Nox4 Deficiency Increases Intestinal Fibrosis After DSS-Induced Colitis
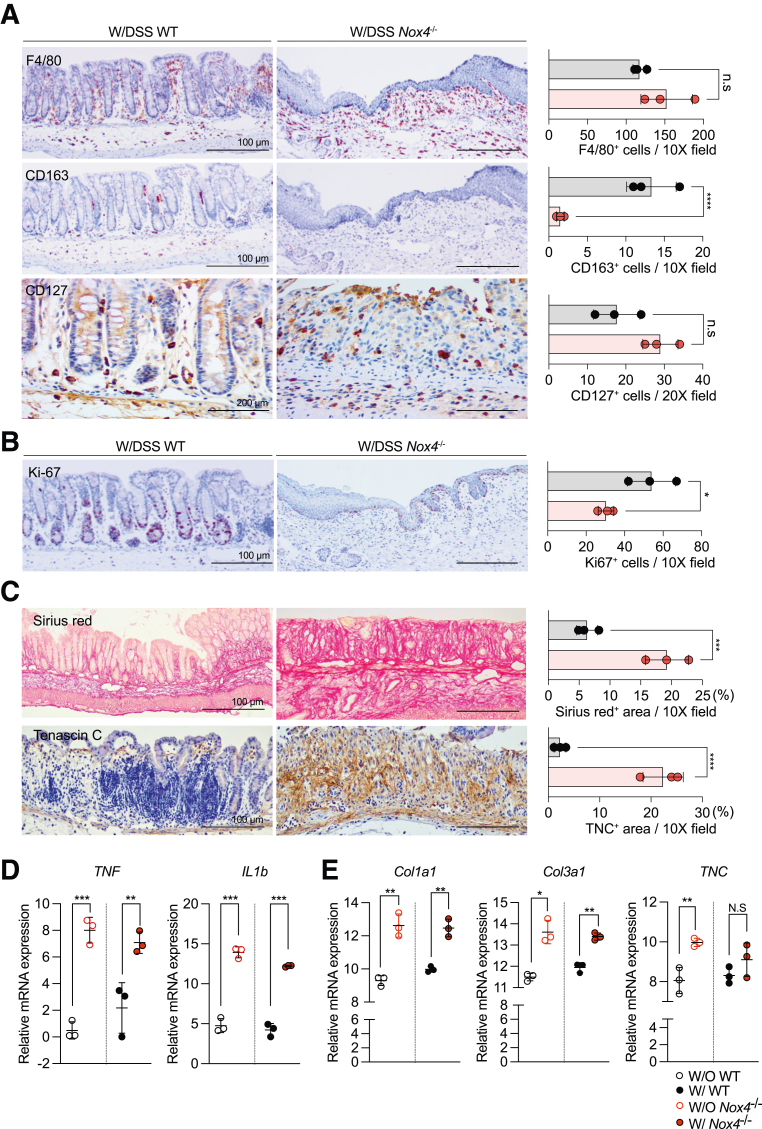
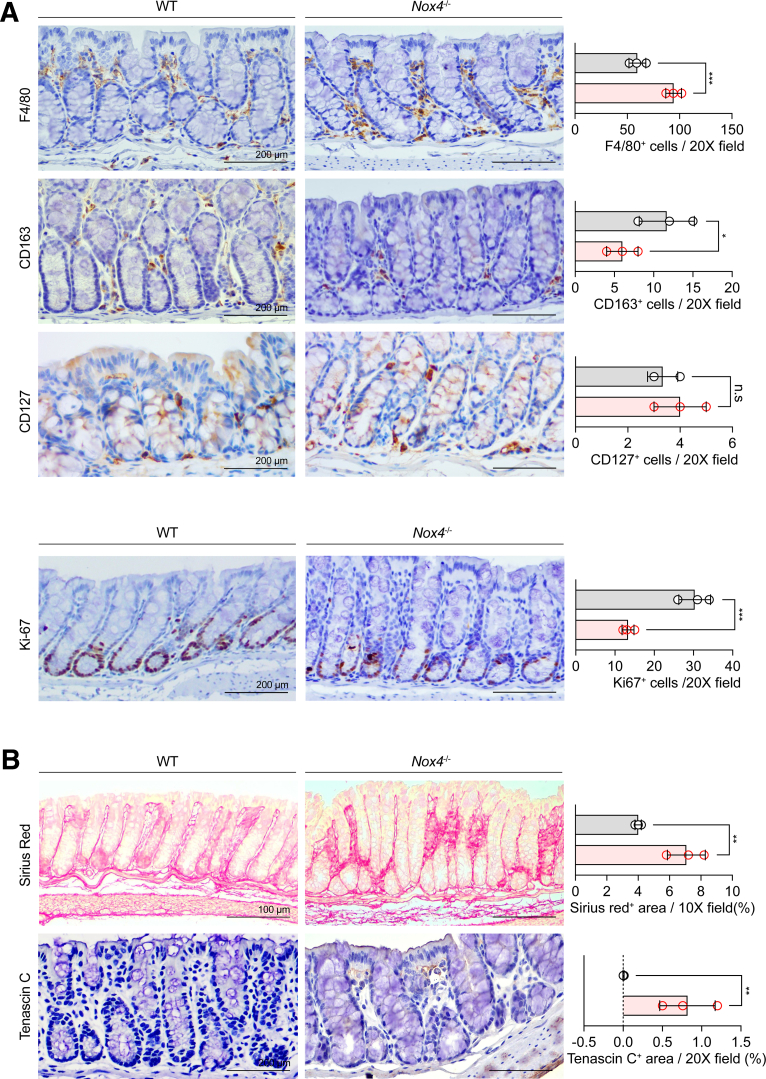
Immunohistochemistry showed that F4/80+ M1 macrophages were increased (Figure 3A, top), whereas CD163+ M2 macrophages were decreased significantly (Figure 3B, middle) in the Nox4-/- colon tissue compared with those of the WT colon. CD127+ lymphoid cells also were increased in DSS-induced colitis Nox4-/- mice (Figure 3A, bottom). Ki-67 immunostaining showed an increase in cell proliferation in the DSS-induced colitis WT colon compared with that of the DSS-induced colitis Nox4-/- mice (Figure 3B). This result indicated that Nox4-/- mice did not show a typical repair response to colitis-induced damage. Sirius red staining showed increased intestinal fibrosis (Figure 3C, top), which corresponded with increased expression of typical fibrosis markers such as Col1a1 and Col3a1 (Figure 3E) detected by reverse-transcription quantitative polymerase chain reaction (PCR). The specific diagnostic marker of collagenous colitis, Tenascin-C, was increased remarkably in the colitis-induced Nox4-/- colon at both the protein (Figure 3C, bottom) and messenger RNA (mRNA) (Figure 3E) levels compared with those in colitis-induced WT mice. Furthermore, the mRNA levels of the inflammatory cytokines tumor necrosis factor (Tnf) and interleukin 1β (Il1β) were increased significantly in colitis-induced Nox4-/- mice (Figure 3D) compared with those of WT mice at the same time points. Moreover, in the untreated condition (without DSS), Nox4-/- mice also showed an increase in the proteins of the F4/80+ macrophage and Ki-67+ proliferating cells (Figure 4A). Further, the mRNA levels of fibrosis-related markers and proinflammatory cytokines such as Tnf and Il1β were significantly increased in colitis induced Nox4-/- mice compared to those in WT mice at the same points. (Figure 3D and E). In addition, the colons of untreated Nox4-/- mice also had a greater fibrotic area, as detected by Tenascin-c immunostaining, compared with that of the WT colon (Figure 4B). These results suggest that the more severe inflammatory response to DSS induction in Nox4-/- mice is related to the activation of intestinal inflammation and fibrosis resulting from Nox4 deficiency.
Figure 3.
Nox4 is involved in intestinal fibrosis and immune-mediated tissue regeneration. (A) Immunohistochemistry with antibodies against F4/80 (top) indicating M1 macrophages, CD164 (middle) indicating M2 macrophages, and CD127 (bottom) indicating lymphoid cells on DSS-treated WT and Nox4-/- mouse colon sections. (B) Immunohistochemistry with antibody against Ki-67 indicating proliferating cells. (C) Sirius red staining showing the fibrotic area in red (top). Immunolabeling with antibodies against Tenascin-C indicates fibrosis in the colon tissue (bottom). Each positive staining area was measured by ImageJ and shown as a bar graph. Grey, WT group (N = 3); red, Nox4-/- group (N = 3). (D) Reverse-transcription quantitative PCR results of proinflammatory-related genes TNF, and IL1b. (E) Reverse-transcription quantitative PCR results of fibrosis-related genes Col1a1, Col3a1, and TNC. Data are expressed as means ± SD. ∗P < .05, ∗∗P < .01, ∗∗∗P < .005, and ∗∗∗∗P < .001 compared with WT. Ki-67, marker of proliferation Ki(Kiel)-67.
Figure 4.
Immune cell infiltration in the WT and Nox4-/-mouse colon. (A) Immunohistochemistry with antibodies against F4/80 indicating M1 macrophages, CD164 indicating M2 macrophages, CD127 indicating lymphoid cells, and Ki-67 (bottom) indicating proliferating cells. (B) Sirius-red staining showing the fibrotic area in red (top). Immunolabeling with antibodies against Tenascin-C indicating fibrosis in the colon tissue (bottom). Each positive staining area and cell was measured by ImageJ and shown as a bar graph. The WT group (N = 3) is colored gray and the Nox4-/- group (N = 3) is colored red. Data are expressed as means ± SD. ∗P < .5, ∗∗P < .01, and ∗∗∗P < .005 compared with WT. Ki-67, marker of proliferation Ki(Kiel)-67.
Differential Gene Expression Under Nox4 Deficiency in the Distal Colon Tissue
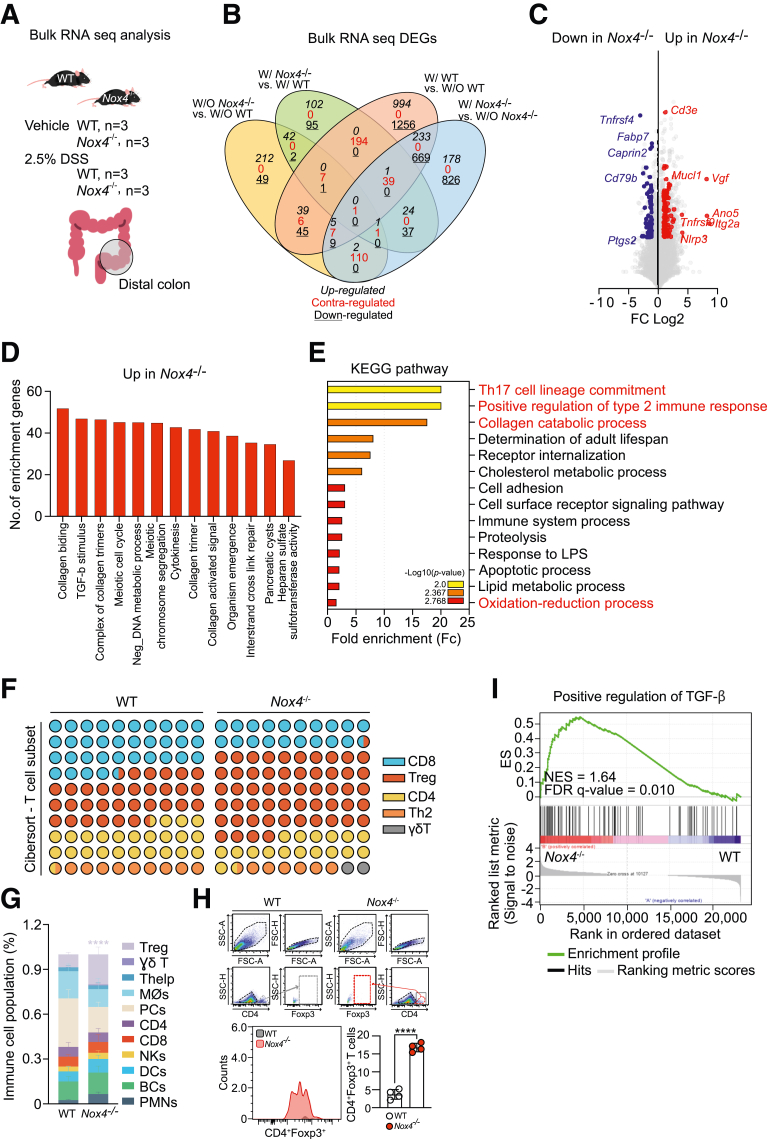
Bulk RNA-seq analysis was performed to identify the underlying mechanism contributing to the more severe inflammation and fibrosis in Nox4-/- mice (Figure 5A). A total of 5124 differentially expressed genes (DEGs) were identified among the 4 comparisons (W/O Nox4-/- vs W/O WT; W/Nox4-/- vs W/WT; W/WT vs W/O WT; and W/ Nox4-/- vs W/O Nox4-/-). Comparing the treated and untreated groups, 42 genes were co–up-regulated, and 2 genes were co–down-regulated (Figure 5B). The 42 co-regulated genes included T-cell–related genes (CD3e and Tnfrsf9),25 fibroblast activators (Itg2a26 and Vegf),27 inflammasome sensor (Nlrp3), and TGF-β target genes (Ano5 and Mucl1) with higher expression levels in Nox4-/- than in WT mice. For the untreated comparison, genes identified as colorectal cancer biomarkers, such as Tnfrsf4, Fabp47, CD79b, and Ptgs2,28 were down-regulated in Nox4-/- mice compared with those in WT mice (Figure 5C). These findings suggest that Nox4 deficiency suppresses TGF-β signaling, which acts as a tumor-suppressor factor in normal cells.29
Figure 5.
Loss of Nox4 leads to collagen synthesis and Treg lineage commitment. (A) Scheme for bulk RNA-seq analysis of freshly isolated distal colon tissues comparing transcriptomes from the vehicle group in WT (n = 3) and Nox4-/- (n = 3) mice or the 2.5% DSS-treated colitis group in WT (n = 3) and Nox4-/- (n = 3) mice. (B) Venn diagram analysis of the 5124 DEGs comparing the numbers of up-regulated, contraregulated, and down-regulated within each control and experimental group. (C) Volcano plot identifying genes significantly down-regulated (blue) and up-regulated (red) with a more than 2-fold express level difference in Nox4-/- compared with WT samples. (D) GO biological process analysis of up-regulated DEGs in Nox4-/- mice. (E) KEGG pathway enrichment analysis of Nox4-/- DEGs. The top 14 pathways are identified, and the colors indicate statistical significance. (F) T-cell subpopulation evaluated by CIBERSORT based on WT and Nox4-/- DEGs. (G) CIBERSORT analysis of WT and Nox4-/- colon tissue. The relative proportions of 11 different immune cell types are deconvoluted from the RNA-seq data using CIBERSORT. (H) Flow cytometry analysis of freshly isolated WT or Nox4-/- distal colon tissue showing the relative number of live cells gated on CD4+Foxp3+ T cells (top, histogram) and the number of CD4+Foxp3+ T cells (bar graph, bottom). (I) Gene set enrichment analysis based on WT or Nox4-/- DEGs. Positive regulation of the TGF-β gene set is enriched with genes up-regulated in the Nox4-/- mouse colon. BC, B cell; DC, Dendritic cell; FDR, False discovery rate; FSC-A, Forward scatter-area; FSC-H, Forward scatter-height; ES, Enrichement score; LPS, lipopolysaccharide; NES, Normalized enrichment socre; NK, Natural killer; PC, Plasma cell; PMN, Polymorphonuclear leukocyte; SSC-A, Side scatter-area; SSC-H, Side scatter-height.
Functional enrichment analysis of the DEGs, including Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment, was performed using Database for Annotation, Visualization, and Integrated Discovery tools to assess the biological functions increased in Nox4-/- compared with WT mice. The GO analysis indicated that most of the DEGs in Nox4-/- mice were enriched in collagen-associated responses, TGF-β stimulus, and cellular repair processes (Figure 5D). In addition, KEGG analysis showed high expression of in Nox4-/- DEGs related to the type 2 immune response, collagen process, and inflammatory response (immune system process, response to lipopolysaccharide, and redox process) (Figure 5E). Further investigation using Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) with 11 immune signature gene sets showed that the Nox4-/- colon had predominantly increased regulatory T cells (Tregs) and decreased plasma cells compared with the WT colon (Figure 5F and G). Flow cytometry of the colon tissue confirmed a 4-fold increase in CD4+Foxp3+ Tregs caused by loss of Nox4 (Figure 5H).
Nox4 Deficiency Activates the Canonical TGF-β Signaling Pathway
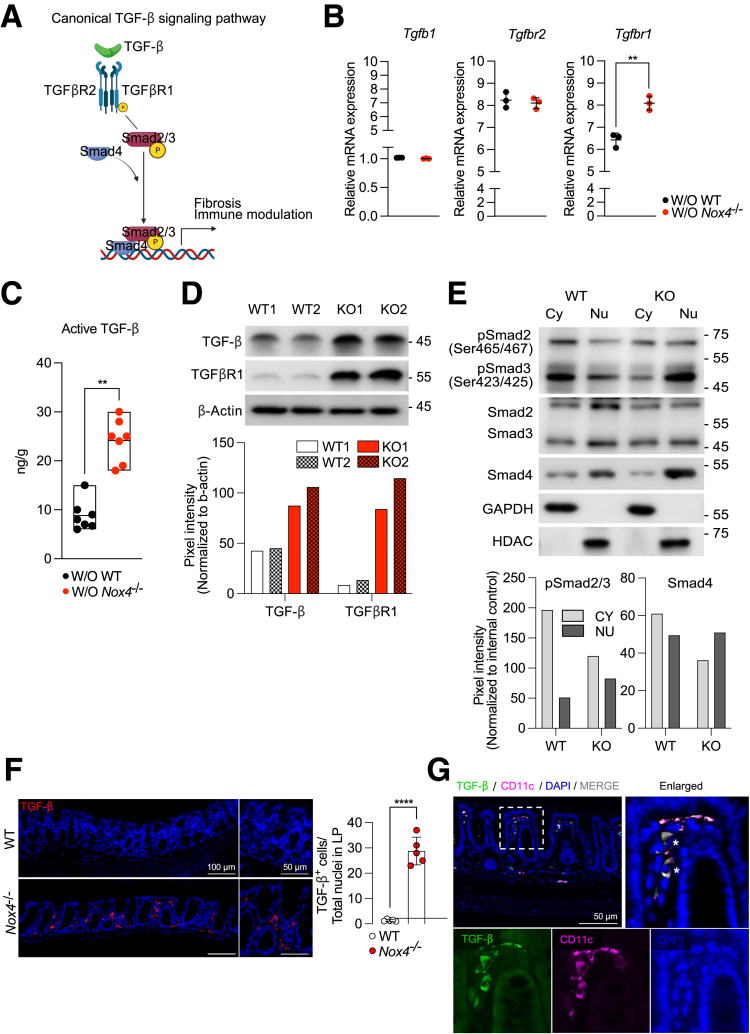
Gene set enrichment analysis confirmed up-regulation of the TGF-β signaling stimulatory gene set in Nox4-/- DEGs compared with WT DEGs (Figure 5I). Given recent studies highlighting a role of Nox4 in regulating the canonical TGF-β signaling pathway in lung fibrosis30 and pancreatic cancer,31 we further focused on the role of Nox4 in canonical TGF-β signaling in the murine colon. Activation of canonical TGF-β signaling involves binding of TGF-β to TGFβR2, which then phosphorylates TGFβR1 to subsequently induce the phosphorylation of Smad2/3 (pSmad2/3) and Smad4, ultimately promoting the translocation of pSmad2/3 and Smad4 to the nucleus (Figure 6A). We observed higher mRNA expression of Tgfbr1 in the Nox4-/- colon tissue than in the nontreated WT colon, whereas there were no differences in the mRNA levels of Tgfbr2 and Tgfb1 between the 2 groups (Figure 6B). An enzyme-linked immunosorbent assay (ELISA) showed that an active form of TGF-β was increased in the Nox4-/- colon compared with the WT colon (Figure 6C). Consistently, Western blot showed that TGF-β and TGFβR1 protein levels were increased in the Nox4-/- colon tissue lysate compared with those of the WT group (Figure 6D). To validate the TGFβR1-induced phosphorylation of Smad2/3, total Smad2/3, and Smad4, the cytosol–nuclear fraction of each colon tissue lysate was evaluated, showing increased translocation of cytosolic pSmad2/3 and Smad4 to the nucleus of the Nox4-/- colon compared with the WT colon (Figure 6E). TGF-β+ cells were expressed in the lamina propria rather than in the epithelium in the Nox4-/- colon, along with a significantly higher number of TGF-β+ cells (Figure 6F). Immunofluorescence staining further showed co-expression of CD11c+ dendritic cells in TGF-β+ cells in the colon (Figure 6G). Overall, these results strongly suggested that Nox4 depletion leads to canonical TGF-β activation in the colon, thereby inducing fibrotic gene expression and Treg commitment.
Figure 6.
Nox4 suppresses activation of canonical TGF-β signaling in vivo. (A) Scheme representing the canonical TGF-β signaling pathway. (B) Reverse-transcription quantitative PCR analysis of the expression of indicated TGF-β regulatory genes in the WT or Nox4-/- mouse colon tissue; these mice differ from those used for RNA-seq (n = 3). Black dots represent untreated WT mice, and red dots represent untreated Nox4-/- mice. (C) Active TGF-β level in WT (black) or Nox4-/- (red) colon tissue lysates measured by enzyme-linked immunosorbent assay. (D) Immunoblotting of TGF-β and TGFβR1 protein levels in WT or Nox4-/- colon tissue extracts; β-actin was used as an internal control. The band size was quantified by ImageJ. (E) Immunoblotting of pSmad2/3, Smad2/3, and Smad4 protein levels in cytosolic and nucleic fraction extracts from WT and Nox4-/- colon tissues. Smad2/3 was used as the pSmad2/3 control proteins, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and HDAC were used as the cytosol and nucleus control proteins, respectively. The band size was quantified by ImageJ. Representative immunofluorescence images of WT or Nox4-/- colon sections stained with (F) TGF-β (red) and 4′,6-diamidino-2-phenylindole (DAPI) (blue), or (G) TGF-β (green), CD11c (violet), and DAPI. Data are expressed as means ± SD. ∗∗P < .01, and ∗∗∗∗P < .001 compared with WT. Cy, Cytosel; KO, Knock-out; LP, Lamina propria; Nu, Nucleus; P, phosporelated.
Nox4 Deficiency Exacerbates Experimental Colitis via Inducing the TGF-β Pathway to Promote Intestinal Inflammation
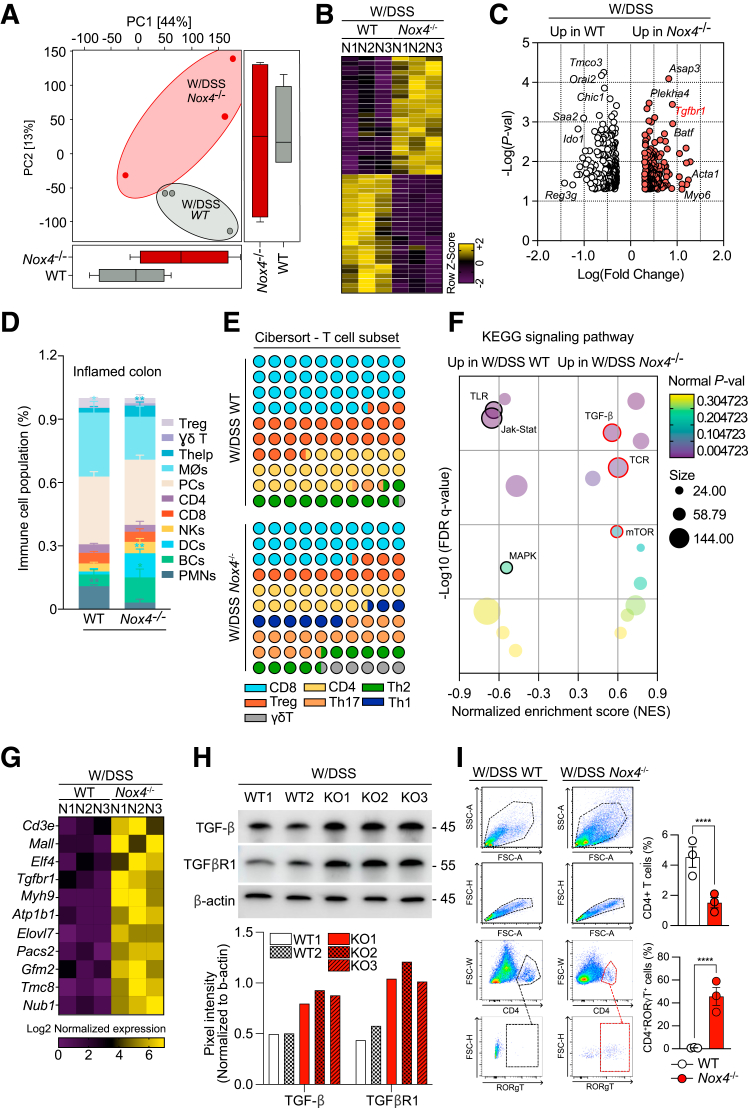
Based on these results, we further analyzed the bulk RNA-seq data of DSS-treated WT and Nox4-/- mice to determine whether the TGF-β signal continuously affects intestinal inflammation during DSS-induced colitis. Principal component analysis showed that the W/DSS Nox4-/- transcripts were clearly discriminated from those of W/DSS WT mice (Figure 7A). A total of 427 DEGs (P < .05; fold change, >2) were identified between the 2 groups, with 247 up-regulated genes and 179 down-regulated genes in W/DSS Nox4-/- mice compared with W/DSS WT mice (Figure 7B). Notably, in the W/DSS Nox4-/- mice, the expression levels of TGF-β–related genes such as Asap3 and Tgfbr1, which are positive regulators of the TGF-β pathway; inflammation-mediated Th17 cell differentiation genes such as Plekha414 and Batf32,33; and fibrosis-related genes such as Acta1 and Myo6, which promote the synthesis of collagen,34 were increased. However, in the W/DSS WT mice, UC-related genes such as Chic1, Ido1, and Reg3g,35,36 and immune infiltrate–related genes such as Tmco3,37 Orai2,38 and Saa2,39 were up-regulated (Figure 7C). Using CIBERSORT, we confirmed that the proportion of Th17 cells was increased significantly in the W/DSS Nox4-/- colon compared with that of the W/DSS WT colon (Figure 7D and E).
Figure 7.
TGF-β signaling and its downstream molecules are up-regulated in DSS-induced colitis Nox4-/-mice. (A) Local Fisher discriminant analysis plot for principal component analysis. Two-dimensional scores are readily distinguished between W/DSS WT (green) and W/DSS Nox4-/- (red) samples. The shared areas indicate 95% CIs. (B) Total colonic cells sorted from W/DSS WT or W/DSS Nox4-/- were subject to RNA-seq. The heatmap of RNA-seq data showing up-regulated (purple) and down-regulated (yellow) genes. (C) Volcano plot identifying significantly up-regulated genes in W/DSS WT (white) and W/DSS Nox4-/- (red). CIBERSORT analysis of WT and Nox4-/- colon tissues. The relative proportions of (D) 11 different immune cell types and (E) 7 T-cell lineage cell types are deconvoluted from the RNA-seq data using CIBERSORT. (F) Bubble plot showing significantly enriched KEGG pathways in the W/DSS WT and W/DSS Nox4-/- DEGs. The bubble colors indicate P values, and the bubble size represents the expression level. (G) Heatmap of the expression of TGF-β–related genes from W/DSS Nox4-/- DEGs. (H) Immunoblotting of TGF-β and TGFβR1 protein levels from W/DSS WT or W/DSS Nox4-/- distal colon tissue extracts. β-actin was used as an internal control. The band size was quantified by ImageJ. (I) Flow cytometric plots showing the expression of RORγT on CD4+ T cells from W/DSS WT and W/DSS Nox4-/- colon tissues. Data are expressed as means ± SD. ∗∗∗∗P < .001 compared with W/DSS WT. JAK-STAT, Janus kinase–signal transducer and activator of transcription pathway; MAPK, mitogen-activated protein kinase; mTOR, Mammalian target of rapamycin; NK, natural killer cell; p-val, P value; TCR, T cell receptor.
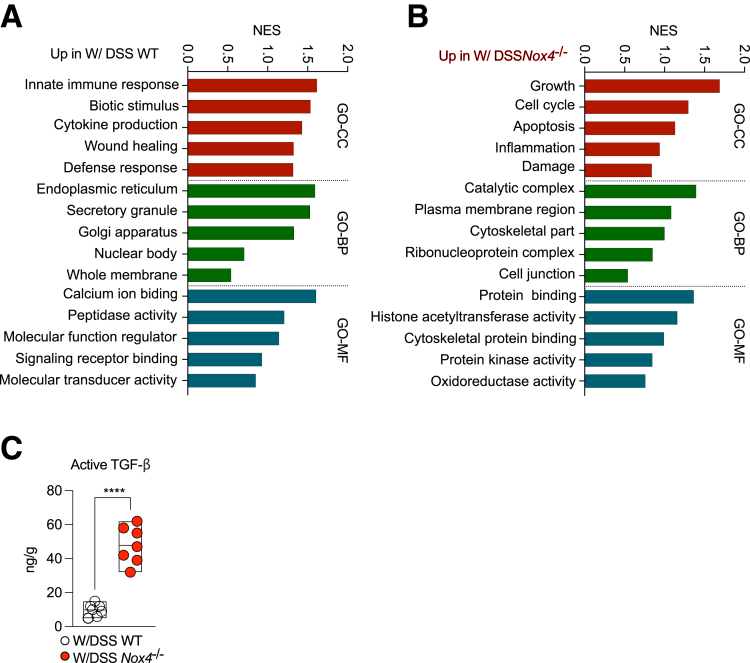
GO analysis of the W/DSS WT or W/DSS Nox4-/- DEGs showed enrichment of up-regulated W/DSS WT DEGs in the cellular component (GO-CC) related to colitis-mediated functions such as innate immune response, biotic stimulus, and cytokine production, and tissue repair programs such as wound healing and defense response (Figure 8A). Similarly, up-regulated W/DSS WT DEGs were enriched in biological processes (GO-BP) of endoplasmic reticulum stress associated with intestinal inflammation, such as innate immune response and cytokine production, and tissue recovery–related processes.40 The DEGs were enriched in molecular function GO terms (GO-MF) included in the IBD pathophysiology-related gene set,41 such as calcium ion binding, peptidase activity, and signal transduction–related functions (Figure 8A). However, the W/DSS Nox4-/- DEGs showed a different functional pattern. For GO-CC, the DEGs were enriched in tissue damage gene sets such as apoptosis, inflammation, and damage. Furthermore, the top-enriched gene sets in biological processes were identified as tissue fibrosis–related gene sets,42 such as the catalytic complex, cytoskeletal part, and ribonucleoprotein complex. For GO-MF, highly enriched DEGs were verified as UC-associated oncogenic gene sets such as protein binding, protein kinase activity, histone acetyltransferase activity, and oxidoreductase activity (Figure 8B). KEGG pathway analysis indicated that the DEGs in W/DSS WT were enriched in the Toll-like receptor pathway, Janus kinase–signal transducer and activator of transcription pathway, and mitogen-activated protein kinase pathway, whereas DEGs in W/DSS Nox4-/- were enriched in the TGF-β, T-cell receptor, and mammalian target of rapamycin pathways (Figure 7F). The heatmap confirmed that the expression levels of TGF-β target genes were increased significantly in DEGs of the W/DSS Nox4-/- group compared with those of the W/DSS group (Figure 7G).
Figure 8.
Related physiologic features in the WT and Nox4-/-mouse colon. GO-term analysis of the up-regulated gene sets in (A) W/DSS WT DEGs and (B) W/DSS Nox4-/- DEGs. (C) Active TGF-β levels in W/DSS WT or W/DSS Nox4-/- colon tissue lysates determined by enzyme-linked immunosorbent assay. Data are expressed as means ± SD. ∗∗∗∗P < .001 compared with W/DSS WT.
These findings suggest that TGF-β signaling and its related fibrogenic and T-cell signals are increased consistently in the colons lacking Nox4, contributing to the maintenance of more severe inflammation. In line with these results from RNA-seq analyses, Western blot showed that TGF-β and TGFβR1 protein levels were increased in the W/DSS Nox4-/- colon lysate compared with those of the W/DSS WT colon lysate (Figure 7H). ELISA confirmed that the active TGF-β level was increased by 2-fold in the W/DSS Nox4-/- colon compared with the W/DSS WT colon (Figure 8C).
Flow cytometry analysis showed an increase in total CD4+ T cells in the W/DSS WT colon; however, approximately 59% of the CD4+ cells expressed RORγT+, a marker of Th17 cells, in the W/DSS Nox4-/- colon, which was greater than that of the W/DSS WT colon (Figure 7I). Together, these findings confirmed an association of DSS-induced inflammation in the Nox4-/- colon with Th17 cell lineage commitment.
Nox4 is Required for Intestinal Recovery After DSS-Induced Experimental Acute Colitis
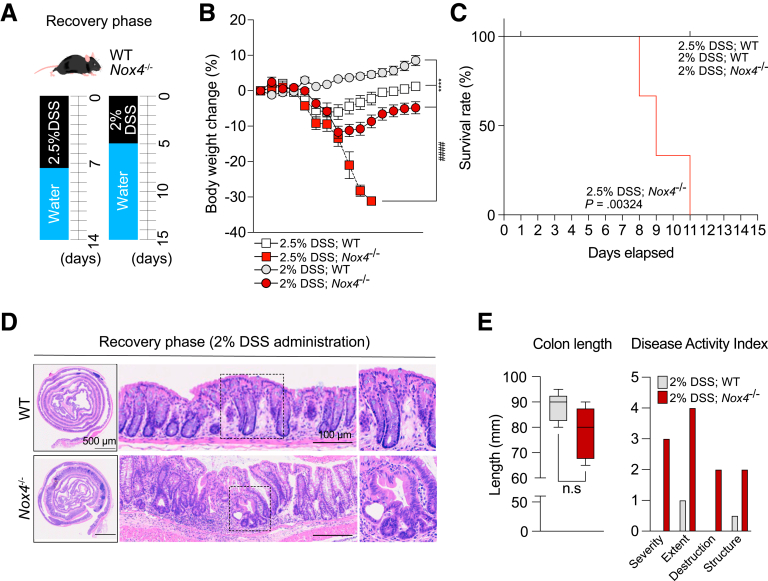
Finally, we investigated whether Nox4 is required for recovery of intestinal inflammation (Figure 9A, left). Body weight began to improve in W/DSS WT mice on day 9 (2 days after DSS treatment), whereas the body weight of W/DSS Nox4-/- mice continuously reduced after 2.5% DSS administration and more rapid weight reduction was observed in the recovery phase. None of the W/DSS Nox4-/- mice that experienced more than 30% weight loss survived (Figure 9B, 2.5% DSS; Nox4-/- group); hence, the overall survival probability was diminished significantly in W/DSS Nox4-/- mice (Figure 9C). We further performed experiments using 2% DSS administration to enter the recovery phase (Figure 9A, right). After 2% DSS administration, the 2% DSS; Nox4-/- group showed a significant reduction in weight loss compared with the 2% DSS; WT group (Figure 9B), although there was no significant difference in survival rate (Figure 9C).
Figure 9.
Loss of Nox4 disrupts tissue repair from DSS-induced colitis. (A) Experimental scheme for the recovery phase from DSS-induced murine colitis. (B) Body weight change for WT and Nox4-/- mice with DSS-induced colitis and recovery phase measured daily (n = 5). (C) Survival probability of the recovery phase after DSS-induced colitis in WT and Nox4-/- mice until day 15 (n = 5 per group). (D) Representative H&E staining images of colon tissues with the recovery phase after DSS-induced colitis in WT and Nox4-/- mice. (E) Quantification of colon length and DAI score. DAI: severity, inflammatory severity (0–3); extent, inflammation extent (0, none; 1, mucosa; 2, submucosa; and 3, transmural), and epithelial change. Data are expressed as means ± SD. ∗∗∗∗P < .001 compared with 2% DSS; WT vs. 2% DSS; Nox4-/-; ####P < .001 compared with 2.5% DSS; WT vs. 2.5% DSS; Nox4-/-.
During the recovery phase, 2% DSS; WT mice displayed epithelial repair, mucosal healing, and only moderate inflammation on day 15. Conversely, the 2% DSS; Nox4-/- mice still showed crypt atrophy, extended goblet cells, and severe immune cell infiltration (Figure 9D). In addition, colon length did not differ between the 2% DSS; Nox4-/- group compared with the 2% DSS; WT group (Figure 9E, left), and the DAI showed that the 2% DSS; Nox4-/- group had more severe symptoms that the WT group (Figure 9E, right).
Collectively, these results indicate that Nox4 is crucial in the recovery process, and its absence can slow down the recovery rate.
Discussion
This study provides evidence that lack of Nox4 leads to activation of TGF-β signaling in the normal colon, followed by a shift to a moderate fibrogenic phenotype and Treg recruitment as a type 2 immune response. Consequently, Nox4-/- mice suffered from more intense mucosal damage, an increased immune response, and more severe fibrosis from DSS-induced colitis than WT mice, resulting in recovery failure leading to death.
Although a previous study reported that Nox4 does not play a role in fibrosis in the murine colitis model, an increase in Tgfbr1 expression was detected with 2.5% DSS-induced colitis in Nox4-/- mice.17 This result was observed after 6 days of administration of 2.5% DSS and 9 days after recovery with water for only 3 days. In our experiment, 2.5% DSS was administered for 14 days, which induced more severe damage in the colon. This is in line with a previous study showing differences in the severity of tissue damage depending on the DSS concentration and duration of administration in the establishment of the DSS-induced murine colitis model.43
Although Nox4 is considered to be closely related to tissue fibrosis, its specific role in fibrosis progression remains controversial. Nox4-induced ROS promotes cell apoptosis and mesenchymal cell differentiation and activation, leading to tissue fibrosis.18,19,30,44,45 Nox4 was reported to play a critical role in the murine lung, leading to alveolar cell death and subsequent bleomycin-induced fibrosis by myofibroblast activation,18,30 and to attenuate Carbon tetrachloride (CCl4)-induced liver injury, hepatocyte apoptosis, and liver fibrosis.45 Conversely, another study showed that Nox4 contributed to protecting against renal fibrosis in murine kidneys with chronic renal injury by inhibiting tubular cell apoptosis and oxidative stress.20 Specifically, Nox4 reduced Hif1a expression and antioxidant molecules in the Kelch-like ECH-associated protein 1 (Keap1) – Nuclear factor (erythroid-derived 2) (Nrf) pathway, thereby inhibiting ROS production, suggesting that Nox4 may positively regulate antioxidant activity in the kidney.20
Similarly, our results showed that loss of Nox4 caused increased oxidation-reduction signaling with a consequent increase of ROS levels in the mouse colon tissue. The increase in ROS caused by deletion of Nox4 in the murine colon is similar to the previously reported role of Nox4 in the kidney, which inhibits ROS and regulates downstream signaling. These results correlated with the finding that Nox4 suppressed ROS production in the colon and inhibited TGF-β signaling and its downstream fibrosis-related and type 2 immunity–related signals. Based on the functional enrichment analysis results of DEGs identified in the Nox4-/- colon, collagen-associated signaling was up-regulated, consistent with changes in collagen protein level, and Treg cell lineage activation was up-regulated along with positive regulation of type 2 immunity. These data suggest that increased ROS stimulate the TGF-β–mediated immune response, such as type 2 immunity for host defense, and also activated immune suppression by Treg cells for immune tolerance.46
The TGF-β signaling pathway is broadly associated with tissue physiology, including collagen synthesis47 and Treg cell lineage commitment.48 Moreover, the loss of Nox4 altered the subsets of T cells stimulated by consistently activated TGF-β in the normal and inflammatory colons. TGF-β signaling can directly induce the differentiation of Foxp3+ Tregs and RORγT+ Th17 cells from naïve CD4+ T cells.49,50 Under normal conditions (ie, without DSS treatment), Nox4-/- mice showed significantly increased infiltration of Foxp3+ Treg cells. Tregs are involved in immune suppression, especially against commensal bacteria in the intestine. Infiltration of Foxp3+ Tregs along with increased ROS in the Nox4-/- colon triggered TGF-β activation, proinflammatory cytokine (Tnf and Il1β) expression, and F4/80+ macrophage infiltration. Therefore, the increase in Tregs is considered to be owing not only to TGF-β activation, but also represents a defense mechanism to regulate the inflammatory immune response in the Nox4-/- mouse colon.
After induction of DSS-induced inflammation, the Treg population decreased and RORγT+ Th17 cells expanded. This result shows that Nox4 regulates the Treg/Th17 balance during colitis; however, further studies are needed to determine the underlying mechanism. Recent studies have shown that Th17 cells are more abundant in patients with active IBD and play a causative role in IBD-associated colorectal cancer.51, 52, 53 Colorectal cancer typically shows a compact infiltrate of proinflammatory cytokine-producing cells. Among the tumor-infiltrating immune cells, T cells are switched into functional populations that secrete a large number of Th17-driven cytokines (interleukin [IL]17A, IL12, and IL22), tumor necrosis factor-α, and IL6. This population then promotes colon tumorigenesis and survival via signal transducer and activator of transcription 3/nuclear factor-κB activation.54 Although we did not identify the role of Nox4 in colorectal cancer, Helfinger et al15 reported that Nox4 can recognize DNA damage and suppress the oxidation of Protein kinase B (AKT), thereby playing a protective role in the DSS/Azoxymethane (AOM) murine colorectal cancer model. Although they did not describe the immune response of DSS/AOM-induced colorectal cancer in Nox4-/- mice, our findings suggest that Th17 cells and related cytokines may enhance the survival and growth of colorectal cancer in Nox4-/- mice.
Increased oxidative stress has long been implicated in the development of murine colitis. The NOX family has been suggested as a major contributor to intestinal inflammation progression.7,55 Nox1 and Duox2 are expressed in the intestinal epithelium and are the primary sources of ROS in the gastrointestinal tract; a deficit of these genes is related to increased disease severity of colitis. Recently, several studies have shown that deletion of Nox4 induces more severe colitis16,17 and increases the risk of colorectal cancer.15 Conditional deletion of Nox4 in fibroblasts and enterocytes was reported to lead to a higher tumor burden, especially from endothelial cells, and fibroblast-specific deletion of Nox4 resulted in more rapidly developed tumors than the epithelial cell–specific deletion of Nox4 mice. We found that Nox4 is expressed in a nonepithelial cell population, suggesting that Nox4 may determine the severity of colonic damage by regulating ROS-driven TGF-β signaling in the colonic epithelial cells.
In conclusion, we elucidated that genetic deletion of Nox4 leads to the progression of more severe fibrotic colitis and subsequent recovery failure. Loss of Nox4 increased oxidative stress in the colon tissues and subsequently activated canonical TGF-β signaling permanently. Furthermore, TGF-β–mediated T-cell lineage commitment (Treg and Th17 cells) and fibrosis-related signal influenced colitis progression. Our findings on the role of Nox4 in murine colitis highlight that regulation of the redox signal can be a valuable target for the successful treatment of patients with IBD.
Materials and Methods
Mice and Model Establishment
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC 2017-0258). Eight-week-old C57BL6/J male mice were used for in vivo experiments. Animals were housed in a specific pathogen-free facility under a 12-hour light-dark cycle and fed PicoLab Rodent Diet 20 (LabDiet, St. Louis, MO).
Nox4-null mice (Nox4-/-) were established using clustered regularly interspaced short palindromic repeats and CRIPR-assocated protein 9 (CRISPR/Cas9) technology with the deletion of exon 4 (Figure 1A). In addition, to test the influence of immune cells expressing Nox family members on fibrotic features in DSS colitis, we also generated Nox2-null (Nox2-/-) mice (Figure 1B).
Previous studies have reported that Nox4-/- mice are susceptible to damage by a DSS regimen.16,17 In this study, WT or Nox4-/- 8-week-old male mice were administered 2.5% DSS (MP Biochemicals, Santa Ana, CA) in drinking water for 14 days to induce fibrotic colitis; fresh DSS solution was prepared every 2 days.24 For the recovery phase, mice were administered 2% (w/v) DSS in the drinking water for 5 days, followed by a recovery period of 10 days with autoclaved tap water. Control mice received only autoclaved tap water. Daily weight changes and the DAI of mice were recorded.56 DSS-induced colitis activity was scored according to body weight loss (0–4), stool frequency (0–3), and rectal bleeding (0–3).
Pathologic Analysis
Mice were killed and immediately perfused with ice-cold PBS in the left ventricle. The colon tissue was fixed with ice-cold 4% paraformaldehyde overnight at 4°C. The colon tissue was processed into a Swiss roll, then dehydrated, paraffin-embedded, and sectioned (5 μm). The sections were deparaffinized with xylene 3 times for 20 minutes each, absolute EtOH 3 times for 10 minutes each, 90% EtOH twice for 10 minutes each, and 75% EtOH for 10 minutes, and then stained with H&E. The sectioned tissue slides were dehydrated and mounted with a Shandon synthetic mount solution (Thermo Scientific).
Immunohistochemistry
During an inflammatory reaction in the colon, immune cell infiltration is characterized by an increase in proinflammatory cytokines in the colitis tissue.57 To assess the immunologic phenotype in DSS-induced WT and Nox4-/- mice, we performed immunohistochemistry of immune cell markers in the colon sections. Paraffin-embedded mouse colon sections were heated at 60°C for 1 hour and cooled to room temperature (22°C∼25°C). Deparaffinization and tissue rehydration were performed in xylene, graded ethanol, and distilled water, as described earlier. Antigen retrieval was performed using Target Retrieval Solution (Dako) at high pressure for 15 minutes, and cooled down at 4°C for 1 hour. Sections were incubated with 3% H2O2 for 30 minutes to block endogenous peroxidase, followed by a reaction with Protein Block, Serum Free reagent (Dako) at room temperature for 2 hours. The sections were incubated with primary antibodies overnight at 4°C (Table 1). The sections were washed using PBS and incubated with Envision+ system horseradish peroxidase–labeled polymer anti-rabbit or mouse secondary antibody (Dako) at room temperature for 20 minutes. Slides were developed with a liquid 3,3′-diaminobenzidine tetra hydrochloride+ substrate chromogen system (Dako), counterstained with Mayer’s hematoxylin (Dako) solution, dehydrated, and mounted. For immunofluorescence staining, primary antibody incubation was performed as described earlier, followed by incubation with cyanine-3 (Cy3)-labeled goat anti-mouse IgG (Jackson ImmunoResearch) or Alexa488-labeled donkey anti-rabbit IgG (Invitrogen) at room temperature for 1 hour. Slides were washed using PBS, counterstained with 4,6-diamino-2-phenylindole (Sigma), and mounted using ProLong Gold Antifade Reagent (Invitrogen).
Table 2.
Primer Sequences Used in This Study
| Genes | Sequence | Accession number |
|---|---|---|
| TNF | F: GGTGCCTATGTCTCAGCCTCTT R: GCCATAGAACTGATGAGAGGGAG |
NM_013693 |
| Il1b | F: TGGACCTTCCAGGATGAGGACA R: GTTCATCTCGGAGCCTGTAGTG |
NM_008361 |
| Col1a1 | F: CCTCAGGGTATTGCTGGACAAC R: CAGAAGGACCTTGTTTGCCAGG |
NM_007742 |
| Col3a1 | F: GACCAAAAGGTGATGCTGGACAG R: CAAGACCTCGTGCTCCAGTTAG |
NM_009930 |
| Tnc | F: GAGACCTGACACGGAGTATGAG R: CTCCAAGGTGATGCTGTTGTCTG |
NM_011607 |
| Tgfb1 | F: TGATACGCCTGAGTGGCTGTCT R: CACAAGAGCAGTGAGCGCTGAA |
NM_011577 |
| Tgfbr1 | F: TGCTCCAAACCACAGAGTAGGC R: CCCAGAACACTAAGCCCATTGC |
NM_009370 |
| Tgfbr2 | F: CCTACTCTGTCTGTGGATGACC R: GACATCCGTCTGCTTGAACGAC |
NM_009371 |
F, forward; R, reverse.
Table 1.
Antibodies Used in This Study
| Antibody | Dilution | Vendor | Catalog |
|---|---|---|---|
| NOX4 | 1:100 | LsBio | LS-C313066 |
| Cytochrome B245 | 1:1000 | Bio-Rad | MCA4685 |
| F4/80 | 1:500 | Cell Signaling | 70076 |
| CD163 | 1:1000 | Novus | NB110-59935 |
| CD127 | 1:1000 | eBioscience | 14-1271-82 |
| Ki-67 | 1:1000 | Abcam | ab16667 |
| TGF-β | 1:100 | Genetex | GTX21279 |
| TGFβR1 | 1:100 | R&D Systems | MAB5871 |
| pSmad2/3 | 1:1000 | Cell Signaling | 8828 |
| Smad2/3 | 1:1000 | Cell Signaling | 8685 |
| Smad4 | 1:1000 | Cell Signaling | 46535 |
| CD11c | 1:300 | Cell Signaling | 97585 |
| β-actin | 1:5000 | Abcam | ab8227 |
| GAPDH | 1:5000 | Abcam | ab8245 |
| HDAC1 | 1:1000 | Abcam | ab280198 |
| PE anti-mouse CD4 | 1:100 | BioLegend | 100408 |
| Pacific blue anti-Foxp3 | 1:100 | BioLegend | 126409 |
| Alexa 488 anti-RORγT | 1:100 | R&D Systems | IC9125G-025 |
| Cy3 AffiniPure donkey anti-mouse | 1:500 | Jackson Immunoresearch | 715-165-151 |
| Donkey anti-mouse IgG Alexa 488 | 1:500 | Invitrogen | A-21202 |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HDAC, Histone deacetylase; PE, Phycoerythrin.
TGF-β Activity
The level of active TGF-β was measured using a TGFβR1 kinase ELISA system according to the manufacturer’s instructions (Promega, Madison, WI). To measure active TGFβR1, freshly isolated colon lysates were activated with 1 N HCl.
RNA-seq
Total RNA was isolated using TRIzol reagent (Invitrogen). RNA quality was assessed by an Agilent 2100 bioanalyzer using the RNA 6000 Nano Chip (Agilent Technologies, Amstelveen, The Netherlands), and RNA quantification was performed on an ND-2000 spectrophotometer (Thermo, Inc).
According to the manufacturer's instructions, library construction was performed for control and test RNAs using the QuantSeq 3ʹ mRNA-Seq Library Prep Kit (Lexogen, Inc). In brief, each 500 ng total RNA sample was hybridized with an oligo-dT primer containing an Illumina-compatible sequence at its 5′ end, and reverse-transcription was performed. After degradation of the RNA template, second-strand synthesis was initiated by a random primer containing an Illumina-compatible linker sequence at its 5′ end. The double-stranded library was purified using magnetic beads to remove all reaction components. The library was amplified to add the complete adapter sequences required for cluster generation. The library was purified from PCR components. High-throughput sequencing was performed as single-end 75-bp sequencing using the NextSeq 500 platform (Illumina, Inc).
QuantSeq 3ʹ mRNA-Seq reads were aligned using Bowtie2.58 Bowtie2 indices were either generated from the genome assembly sequence or the representative transcript sequences for aligning to the genome and transcriptome. The alignment file was used for assembling transcripts, estimating their abundances, and detecting DEGs between untreated WT (W/O WT) and Nox4-/- (W/O Nox4-/-) distal colon samples and W/DSS WT and W/DSS Nox4-/- distal colon samples based on counts from unique and multiple alignments using coverage in bedtools.59 The read count data were processed based on the quantile normalization method using the EdgeR package of R software (version 3.6.1) with Bioconductor (http://www.bioconductor.org).60 The raw count data were first preprocessed using quality-control checks and lowly expressed genes were removed. We then normalized the data using the trimmed mean of M values method. We defined DEGs using linear models in DESeq2 v1.22.1 and applied significance criteria based on Benjamini–Hochberg false-discovery rate–adjusted P values less than .05.
Three-dimentional principal component analysis was performed using Python (version 3.6 or higher) (Python software foundation) with the following libraries installed: NumPy (for generating sample data) (Numpy community), Pandas (for loading the data set) (Pandas development team), Scikit-learn (for applying principal component analysis) (Scikit-learn community), and Matplotlib (for visualizing data) (Matplotlib development team). To explore the functional processes and pathways, we performed functional enrichment analysis for each module's eigengenes separately. We used the Database for Annotation, Visualization, and Integrated Discovery online tool version 6.8 to extract significant GO functional terms, including biological processes, molecular function, and cellular components, as well as KEGG pathways. The results of the KEGG pathway were visualized using Prism 9 (GraphPad software), and Enrichment Map (Enrichemnt map developers) was sorted based on enrichment score and false discovery rate values. To ensure the accuracy of the analysis, we performed gene set permutations 100 times. We selected significant items based on a P value threshold of less than .05. Gene set enrichment analysis for comparisons of 2 sample groups (Nox4-/- vs WT) was performed in Gene set enrichment analysis v4.3.2 (Broad Institute, Cambridge, MA) using updated guidelines. To identify the cell types present and estimate their relative subsets, we used the CIBERSORT tool available at http://cibersort.stanford.edu. We used the RNA expression profiles of each sample to characterize the infiltration of 22 immune cell types and obtained an abundance ratio matrix of 15 immune cell types based on a significance threshold of P < .05.
Reverse-Transcription Quantitative PCR
Freshly isolated colon tissues were immersed in RNAlater stabilization solution (Invitrogen; Thermo Fisher Scientific, Inc) and incubated overnight at 4°C. Total RNA extraction was performed with TRIzol reagent (Invitrogen; Thermo Fisher Scientific) following the manufacturer’s instructions. Contaminating genomic DNA was removed with Recombinant DNase1 (Takara Bio, Inc, Shiga, Japan), and the RNA was quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific). One milligram template RNA was reverse-transcribed using the ImProm-II reverse transcription System (Promega).
Western Blot
Freshly isolated colon tissues were immersed in RIPA solution (Sigma Aldrich). Colon lysates were loaded on 8%–15% polyacrylamide gels and transferred to polyvinylidene fluoride membranes (Millipore). Specific primary antibodies were used to detect the expression of proteins (Table 1). After incubation with horseradish peroxidase–conjugated secondary antibodies, membranes were developed using Signal West Femto Maximum Sensitivity Substrate (Thermo Scientific) with the LAS chemiluminescent imaging system (Amersham).
Flow Cytometry
To detect CD4+ and Foxp3+ Tregs or RORγT + Th17 cells in the colon, single cells were isolated from the colon sections, as described previously.61 The cells were blocked with fragment crystallizable (Fc) Block (BD Biosciences) in fluorescence-activated cell sorter (FACS) buffer (0.5% fetal bovine serum, 10 mmol/L EDTA, and 0.05% NaN3 in PBS) for 30 minutes at 4°C, followed by washing with FACS buffer. The cells were incubated with phycoerythrin-conjugated anti-mouse CD4 (BioLegend) and Pacific blue–conjugated anti-mouse Foxp3 (BioLegend) or Alexa 488-conjugated anti-mouse RORγT (R&D Systems) for 1 hour at 4°C. After washing with FACS buffer, the cells were sorted after gating for the live CD4+ and Foxp3+ cell fractions on a FACS Aria II cell sorter (BD Biosciences) for quantification.
Statistical Analysis
Data are presented as the means ± SD. Significant differences were evaluated by Student t test, Mann–Whitney U tests, or 1-way analysis of variance as appropriate using Prism9 (GraphPad Software). P < .05 was considered statistically significant.
Acknowledgments
CRediT Authorship Contributions
Yura Lee (Investigation: Lead; Software: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead)
Sung-Hee Kim, PhD (Conceptualization: Supporting; Data curation: Supporting; Funding acquisition: Supporting; Project administration: Supporting; Writing – original draft: Supporting)
Haengdueng Jeong (Data curation: Supporting; Investigation: Supporting; Software: Supporting; Visualization: Supporting)
Kwang H. Kim (Formal analysis: Supporting; Investigation: Supporting)
Donghun Jeon (Formal analysis: Supporting; Methodology: Supporting; Software: Supporting)
Yejin Cho (Formal analysis: Supporting; Methodology: Supporting; Visualization: Supporting)
Daekee Lee (Methodology: Supporting; Resources: Lead)
Ki Taek Nam, DVM, PhD (Conceptualization: Lead; Data curation: Lead; Funding acquisition: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This research was supported by the Brain Korea 21 Project for Medical Science at Yonsei University (K.T.N.), the Bio and Medical Technology Development Program of the National Research Foundation of Korea grant 2022R1A2C3007850 (K.T.N.), National Research Foundation of Korea grant 2022M3A9F3016364 (K.T.N.), the Korean Mouse Phenotyping Project grant 2016M3A9D5A01952416) (K.T.N.), and National Research Foundation of Korea grant 2020R1I1A1A01052033 (S.-H.K.).
Data Availability The raw data have been submitted and can be accessed at the Korean Nucleotide Archive using accession number PRJKA230579.
References
- 1.Choi Y.Y., Lee J.K., Kim H.S., et al. Medications and the risk of colorectal cancer in patients with inflammatory bowel diseases: use of the landmark method. Yonsei Med J. 2021;62:997–1004. doi: 10.3349/ymj.2021.62.11.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mak J.W.Y., Ng S.C. Epidemiology of fibrostenosing inflammatory bowel disease. J Dig Dis. 2020;21:332–335. doi: 10.1111/1751-2980.12853. [DOI] [PubMed] [Google Scholar]
- 3.Pircalabioru G., Aviello G., Kubica M., et al. Defensive mutualism rescues NADPH oxidase inactivation in gut infection. Cell Host Microbe. 2016;19:651–663. doi: 10.1016/j.chom.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Ahrne S., Hagslatt M.L. Effect of lactobacilli on paracellular permeability in the gut. Nutrients. 2011;3:104–117. doi: 10.3390/nu3010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy S., Khanna S., Nallu K., et al. Dermal wound healing is subject to redox control. Mol Ther. 2006;13:211–220. doi: 10.1016/j.ymthe.2005.07.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coant N., Ben Mkaddem S., Pedruzzi E., et al. NADPH oxidase 1 modulates WNT and NOTCH1 signaling to control the fate of proliferative progenitor cells in the colon. Mol Cell Biol. 2010;30:2636–2650. doi: 10.1128/MCB.01194-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato M., Marumo M., Nakayama J., et al. The ROS-generating oxidase Nox1 is required for epithelial restitution following colitis. Exp Anim. 2016;65:197–205. doi: 10.1538/expanim.15-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aviello G., Knaus U.G. NADPH oxidases and ROS signaling in the gastrointestinal tract. Mucosal Immunol. 2018;11:1011–1023. doi: 10.1038/s41385-018-0021-8. [DOI] [PubMed] [Google Scholar]
- 9.Lee K.A., Cho K.C., Kim B., et al. Inflammation-modulated metabolic reprogramming is required for DUOX-dependent gut immunity in Drosophila. Cell Host Microbe. 2018;23:338–352 e5. doi: 10.1016/j.chom.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Kim S.H., Lee W.J. Role of DUOX in gut inflammation: lessons from Drosophila model of gut-microbiota interactions. Front Cell Infect Microbiol. 2014;3:116. doi: 10.3389/fcimb.2013.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X., Jiang L., Han W., et al. Artificial neural network analysis-based immune-related signatures of primary non-response to infliximab in patients with ulcerative colitis. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.742080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadler T., Bhasin J.M., Xu Y., et al. Genome-wide analysis of DNA methylation and gene expression defines molecular characteristics of Crohn's disease-associated fibrosis. Clin Epigenetics. 2016;8:30. doi: 10.1186/s13148-016-0193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan L.L., Ren Z., Liu Y., et al. A novel danshensu derivative ameliorates experimental colitis by modulating NADPH oxidase 4-dependent NLRP3 inflammasome activation. J Cell Mol Med. 2020;24:12955–12969. doi: 10.1111/jcmm.15890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong S., Chen M., Dai F., et al. 5-Hydroxytryptamine (5-HT)-exacerbated DSS-induced colitis is associated with elevated NADPH oxidase expression in the colon. J Cell Biochem. 2019;120:9230–9242. doi: 10.1002/jcb.28198. [DOI] [PubMed] [Google Scholar]
- 15.Helfinger V., Freiherr von Gall F., Henke N., et al. Genetic deletion of Nox4 enhances cancerogen-induced formation of solid tumors. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2020152118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han C., Sheng Y., Wang J., et al. NOX4 promotes mucosal barrier injury in inflammatory bowel disease by mediating macrophages M1 polarization through ROS. Int Immunopharmacol. 2022;104 doi: 10.1016/j.intimp.2021.108361. [DOI] [PubMed] [Google Scholar]
- 17.Stenke E., Aviello G., Singh A., et al. NADPH oxidase 4 is protective and not fibrogenic in intestinal inflammation. Redox Biol. 2020;37 doi: 10.1016/j.redox.2020.101752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hecker L., Vittal R., Jones T., et al. NADPH oxidase-4 mediates myofibroblast activation and fibrogenic responses to lung injury. Nat Med. 2009;15:1077–1081. doi: 10.1038/nm.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Q., Li C., Yang C.F., et al. Methyl ferulic acid attenuates liver fibrosis and hepatic stellate cell activation through the TGF-beta1/Smad and NOX4/ROS pathways. Chem Biol Interact. 2019;299:131–139. doi: 10.1016/j.cbi.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Nlandu Khodo S., Dizin E., Sossauer G., et al. NADPH-oxidase 4 protects against kidney fibrosis during chronic renal injury. J Am Soc Nephrol. 2012;23:1967–1976. doi: 10.1681/ASN.2012040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cucoranu I., Clempus R., Dikalova A., et al. NAD(P)H oxidase 4 mediates transforming growth factor-beta1-induced differentiation of cardiac fibroblasts into myofibroblasts. Circ Res. 2005;97:900–907. doi: 10.1161/01.RES.0000187457.24338.3D. [DOI] [PubMed] [Google Scholar]
- 22.Jiang F., Liu G.S., Dusting G.J., et al. NADPH oxidase-dependent redox signaling in TGF-beta-mediated fibrotic responses. Redox Biol. 2014;2:267–272. doi: 10.1016/j.redox.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sareila O., Kelkka T., Pizzolla A., et al. NOX2 complex-derived ROS as immune regulators. Antioxid Redox Signal. 2011;15:2197–2208. doi: 10.1089/ars.2010.3635. [DOI] [PubMed] [Google Scholar]
- 24.Bao S., Carr E.D., Xu Y.H., et al. Gp91(phox) contributes to the development of experimental inflammatory bowel disease. Immunol Cell Biol. 2011;89:853–860. doi: 10.1038/icb.2011.4. [DOI] [PubMed] [Google Scholar]
- 25.Vinay D.S., Kwon B.S. 4-1BB (CD137), an inducible costimulatory receptor, as a specific target for cancer therapy. BMB Rep. 2014;47:122–129. doi: 10.5483/BMBRep.2014.47.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adorno-Cruz V., Liu H. Regulation and functions of integrin alpha2 in cell adhesion and disease. Genes Dis. 2019;6:16–24. doi: 10.1016/j.gendis.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito T.K., Ishii G., Chiba H., et al. The VEGF angiogenic switch of fibroblasts is regulated by MMP-7 from cancer cells. Oncogene. 2007;26:7194–7203. doi: 10.1038/sj.onc.1210535. [DOI] [PubMed] [Google Scholar]
- 28.Susmi T.F., Rahman A., Khan M.M.R., et al. Prognostic and clinicopathological insights of phosphodiesterase 9A gene as novel biomarker in human colorectal cancer. BMC Cancer. 2021;21:577. doi: 10.1186/s12885-021-08332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seoane J., Gomis R.R. TGF-beta family signaling in tumor suppression and cancer progression. Cold Spring Harb Perspect Biol. 2017;9:a022277. doi: 10.1101/cshperspect.a022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carnesecchi S., Deffert C., Donati Y., et al. A key role for NOX4 in epithelial cell death during development of lung fibrosis. Antioxid Redox Signal. 2011;15:607–619. doi: 10.1089/ars.2010.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hiraga R., Kato M., Miyagawa S., et al. Nox4-derived ROS signaling contributes to TGF-beta-induced epithelial-mesenchymal transition in pancreatic cancer cells. Anticancer Res. 2013;33:4431–4438. [PubMed] [Google Scholar]
- 32.Park S.H., Rhee J., Kim S.K., et al. BATF regulates collagen-induced arthritis by regulating T helper cell differentiation. Arthritis Res Ther. 2018;20:161. doi: 10.1186/s13075-018-1658-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galbavy W., Lu Y., Kaczocha M., et al. Transcriptomic evidence of a para-inflammatory state in the middle aged lumbar spinal cord. Immun Ageing. 2017;14:9. doi: 10.1186/s12979-017-0091-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roma-Rodrigues C., Fernandes A.R. Genetics of hypertrophic cardiomyopathy: advances and pitfalls in molecular diagnosis and therapy. Appl Clin Genet. 2014;7:195–208. doi: 10.2147/TACG.S49126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng C., Hua J., Tan J., et al. Identification of differentially expressed genes, associated functional terms pathways, and candidate diagnostic biomarkers in inflammatory bowel diseases by bioinformatics analysis. Exp Ther Med. 2019;18:278–288. doi: 10.3892/etm.2019.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saeterstad S., Ostvik A.E., Royset E.S., et al. Profound gene expression changes in the epithelial monolayer of active ulcerative colitis and Crohn's disease. PLoS One. 2022;17 doi: 10.1371/journal.pone.0265189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Oh W.K., Zhu J. Disease-specific classification using deconvoluted whole blood gene expression. Sci Rep. 2016;6 doi: 10.1038/srep32976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grimes D., Johnson R., Pashos M., et al. ORAI1 and ORAI2 modulate murine neutrophil calcium signaling, cellular activation, and host defense. Proc Natl Acad Sci U S A. 2020;117:24403–24414. doi: 10.1073/pnas.2008032117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye Z., Bayron Poueymiroy D., Aguilera J.J., et al. Inflammation protein SAA2.2 spontaneously forms marginally stable amyloid fibrils at physiological temperature. Biochemistry. 2011;50:9184–9191. doi: 10.1021/bi200856v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eugene S.P., Reddy V.S., Trinath J. Endoplasmic reticulum stress and intestinal inflammation: a perilous union. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.543022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carroll I.M., Maharshak N. Enteric bacterial proteases in inflammatory bowel disease- pathophysiology and clinical implications. World J Gastroenterol. 2013;19:7531–7543. doi: 10.3748/wjg.v19.i43.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y., Xiong L.L., Zhang P., et al. Microarray expression profiles of genes in lung tissues of rats subjected to focal cerebral ischemia-induced lung injury following bone marrow-derived mesenchymal stem cell transplantation. Int J Mol Med. 2017;39:57–70. doi: 10.3892/ijmm.2016.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J.J., Shajib M.S., Manocha M.M., et al. Investigating intestinal inflammation in DSS-induced model of IBD. J Vis Exp. 2012;60:3678. doi: 10.3791/3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z.M., Xu S.Y., Feng Y.Z., et al. The role of NOX4 in pulmonary diseases. J Cell Physiol. 2021;236:1628–1637. doi: 10.1002/jcp.30005. [DOI] [PubMed] [Google Scholar]
- 45.Jiang J.X., Chen X., Serizawa N., et al. Liver fibrosis and hepatocyte apoptosis are attenuated by GKT137831, a novel NOX4/NOX1 inhibitor in vivo. Free Radic Biol Med. 2012;53:289–296. doi: 10.1016/j.freeradbiomed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chapoval S., Dasgupta P., Dorsey N.J., et al. Regulation of the T helper cell type 2 (Th2)/T regulatory cell (Treg) balance by IL-4 and STAT6. J Leukoc Biol. 2010;87:1011–1018. doi: 10.1189/jlb.1209772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yun S.M., Kim S.H., Kim E.H. The molecular mechanism of transforming growth factor-beta signaling for intestinal fibrosis: a mini-review. Front Pharmacol. 2019;10:162. doi: 10.3389/fphar.2019.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang S., Gang X., Yang S., et al. The alterations in and the role of the Th17/Treg balance in metabolic diseases. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.678355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominguez-Villar M., Hafler D.A. Regulatory T cells in autoimmune disease. Nat Immunol. 2018;19:665–673. doi: 10.1038/s41590-018-0120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Veldhoen M., Hocking R.J., Atkins C.J., et al. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Amicarella F., Muraro M.G., Hirt C., et al. Dual role of tumour-infiltrating T helper 17 cells in human colorectal cancer. Gut. 2017;66:692–704. doi: 10.1136/gutjnl-2015-310016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kempski J., Brockmann L., Gagliani N., et al. TH17 cell and epithelial cell crosstalk during inflammatory bowel disease and carcinogenesis. Front Immunol. 2017;8:1373. doi: 10.3389/fimmu.2017.01373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez L.G., Kempski J., McGee H.M., et al. TGF-beta signaling in Th17 cells promotes IL-22 production and colitis-associated colon cancer. Nat Commun. 2020;11:2608. doi: 10.1038/s41467-020-16363-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.De Simone V., Franze E., Ronchetti G., et al. Th17-type cytokines, IL-6 and TNF-alpha synergistically activate STAT3 and NF-kB to promote colorectal cancer cell growth. Oncogene. 2015;34:3493–3503. doi: 10.1038/onc.2014.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burgueno J.F., Fritsch J., Gonzalez E.E., et al. Epithelial TLR4 signaling activates DUOX2 to induce microbiota-driven tumorigenesis. Gastroenterology. 2021;160:797–808 e6. doi: 10.1053/j.gastro.2020.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wirtz S., Popp V., Kindermann M., et al. Chemically induced mouse models of acute and chronic intestinal inflammation. Nat Protoc. 2017;12:1295–1309. doi: 10.1038/nprot.2017.044. [DOI] [PubMed] [Google Scholar]
- 57.Xue G., Hua L., Zhou N., et al. Characteristics of immune cell infiltration and associated diagnostic biomarkers in ulcerative colitis: results from bioinformatics analysis. Bioengineered. 2021;12:252–265. doi: 10.1080/21655979.2020.1863016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gentleman R.C., Carey V.J., Bates D.M., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Graves C.L., Harden S.W., LaPato M., et al. A method for high purity intestinal epithelial cell culture from adult human and murine tissues for the investigation of innate immune function. J Immunol Methods. 2014;414:20–31. doi: 10.1016/j.jim.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]