Abstract
Regulated protein degradation in eukaryotes is performed by the 26S proteasome, which contains a 19-subunit regulatory particle (RP) that binds, processes, and translocates substrates to a 28-subunit hollow core particle (CP) where proteolysis occurs. In addition to its intrinsic subunits, myriad proteins interact with the proteasome transiently, including factors that assist and/or regulate its degradative activities. Efforts to identify proteasome-interacting components and/or to solve its structure have relied on over-expression of a tagged plasmid, establishing stable cell lines, or laborious purification protocols to isolate native proteasomes from cells. Here, we describe an engineered human cell line, derived from colon cancer HCT116 cells, with a biotin handle on the RP subunit hRpn1/PSMD2 (proteasome 26S subunit, non-ATPase 2) for purification of 26S proteasomes. A 75-residue sequence from Propionibacterium shermanii that is biotinylated in mammalian cells was added following a tobacco etch virus protease cut site at the C terminus of hRpn1. We tested and found that 26S proteasomes can be isolated from this modified HCT116 cell line by using a simple purification protocol. More specifically, biotinylated proteasomes were purified from the cell lysates by using neutravidin agarose resin and released from the resin following incubation with tobacco etch virus protease. The purified proteasomes had equivalent activity in degrading a model ubiquitinated substrate, namely ubiquitinated p53, compared to commercially available bovine proteasomes that were purified by fractionation. In conclusion, advantages of this approach to obtain 26S proteasomes over others is the simple purification protocol and that all cellular proteins, including the tagged hRpn1 subunit, remain at endogenous stoichiometry.
Keywords: proteasome, regulatory particle, CRISPR/Cas9, hRpn1, biotin
The 26S proteasome is a sophisticated protease that performs regulated protein degradation (1). It is formed by a regulatory particle (RP) that caps either end of a hollow core particle (CP), which contains three enzymatic activities (trypsin, chymotrypsin, and caspase-like) for substrate proteolysis (2). The RP contains a hexameric AAA+ ATPase ring composed of subunits Rpt1-Rpt6, the C termini of which dock into pockets formed at the subunit interface of the outer CP heptameric α-rings. Structures of the 26S proteasome have been solved by cryo-EM to reveal conformational switching driven by the ATPase ring as the proteasome engages and processes substrates, as reviewed in (3). Proteasome substrates are recognized by the RP through posttranslationally added ubiquitin chains, which bind RP receptors Rpn1, Rpn10, and Rpn13 (4). Small molecule binders have been developed for each of these proteasome substrate receptors (5, 6, 7, 8) and in the case of hRpn1, a macrocyclic peptide degrader molecule was developed that induced the proteasomal degradation of a model protein substate (9).
In addition to its intrinsic subunits, the proteasome, and in particular the substrate receptors, interact transiently with myriad proteins, including ubiquitination machinery (10, 11, 12, 13) or shuttle factors that can bind to both ubiquitin chains and the RP receptors (14, 15). The functional impact of these interactions are still being studied; for example, the Rad23 shuttle factor protein was recently shown to sequester proteasomes into cellular condensates in response to cell stress (16) and the E3 ligase E6AP/UBE3A found to bind to a C-terminal domain in the RP substrate receptor Rpn10 (13). Mass spectrometry studies have been paramount to identifying proteasome-interacting proteins (17, 18) and like single particle cryo-EM studies of proteasomes rely on methods of isolating such complexes with intact fidelity from cells.
The lentiviral transfection system was used to generate a cell line with stable expression of biotin-tagged hRpn11 from which samples of human proteasomes could be purified (19). Another approach to purify human proteasomes relies on its affinity for the ubiquitin-like domain of hHR23B, with elution by a competing polypeptide containing a ubiquitin-interacting motif (20). Since hHR23B binds to the same sites in the proteasome as ubiquitin (4), a limitation of this approach is not only the required displacement of endogenous hHR23B (18) but also displacement of ubiquitinated substrates. In addition, extraproteasomal binding partners compete with proteasomes for binding to hHR23B in this assay. More recently, gene editing was used to introduce a mNG2-StrepII tag onto the C-terminal end of CP subunit β4/PSMB2 for use as a purification handle (21). Disadvantages of these approaches include the altered cellular abundance of the expressed subunit, possible transfection efficiency issues, and for CP subunit targeting, heterogeneous CP mixtures. Native proteasomes have been purified from cells by using biochemical approaches, especially glycerol gradients (22, 23). Proteasomes purified biochemically from bovine red blood cells are available commercially (BMLPW9310; ENZO Life Sciences, Inc) and can be used to solve structures of proteasomes after addition of binding partners, for example (24). Recently, however, commercial proteasomes have been in short supply, and furthermore, commercial proteasome samples are restricted to red blood cells.
We sought to apply gene editing technology to engineer the human colon cancer cell line HCT116 with an affinity purification tag on an RP subunit for ready purification of 26S proteasomes. We chose hRpn1 as the subunit on which to knock-in a purification handle based on cryo-EM structures that indicated exposure of its C-terminal end by lack of intersubunit interactions with other proteasome components. By using our strategy, we developed a simple purification method to obtain purified samples of active 26S proteasome from the HCT116 cell line.
Results
Design and selection of HCT116 clones with a purification handle at the C terminus of proteasome subunit hRpn1
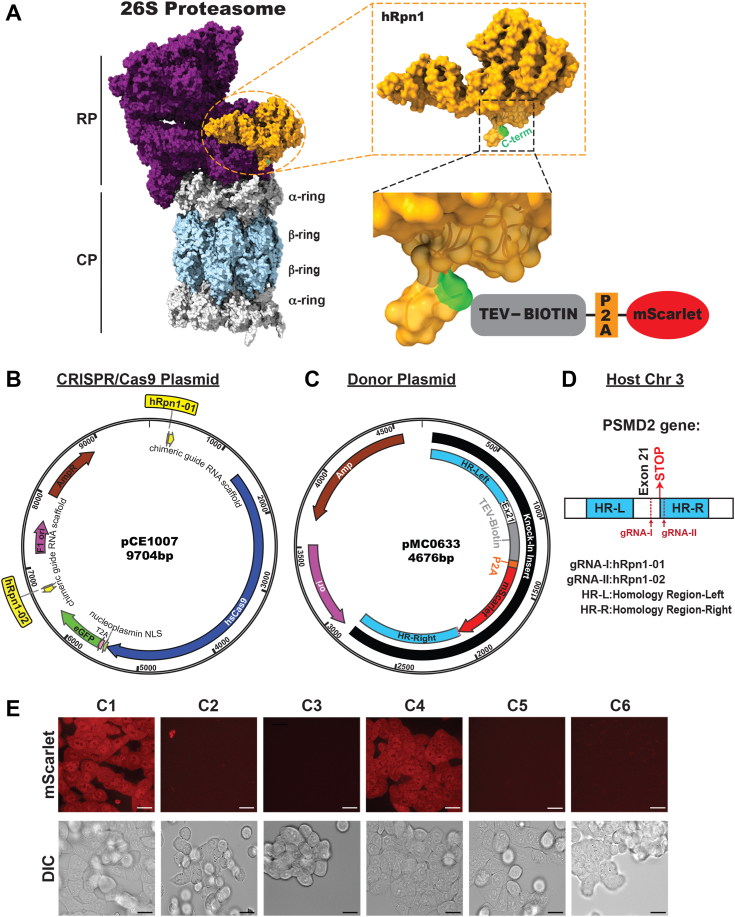
We used the cryo-EM structures of 26S proteasomes to search for RP subunits with available termini for tagging. We found both the N- and C-termini of the hRpn1 protein encoded by PSMD2 to be free from contacts with other subunits of the 26S proteasome, making it a good candidate for inserting an affinity tag. Rpn1 is an RP substrate receptor with a defined T1 site for ubiquitin chains (25) and the ubiquitin-like domain of shuttle factor Rad23 (15), T2 site for deubiquitinase Usp14/Ubp6 (25), and an additional region (spanning amino acids 214–355) near the T1 and T2 sites that has been demonstrated to also bind to ubiquitin chains in the isolated Rpn1 protein (26). Complete sequencing and characterization of the human PSMD2 complementary DNA revealed it to be expressed as three distinct isoforms (27); including isoform 1 representing the canonical variant encoding full length hRpn1 (908 residues) and isoform 2 and isoform 3 which are formed by alternative splicing variants at the N-terminal end. Isoform 2 yields replacement of isoform 1 residues 1 to 163 with four amino acids whereas isoform 3 yields deletion of isoform 1 residues 1 to 130. The C-terminal region of hRpn1 by contrast, encoded by exon 21, is identical in all three spliced variants. Hence, we chose the C terminus of hRpn1 as the location for affinity tag knock-in (Fig. 1A).
Figure 1.
Overall strategy to generate an engineered cell line with a biotin handle on proteasome subunit hRpn1.A, surface view of a structural state of the human 26S proteasome (left, PDB: 5GJQ) illustrating hRpn1 within the RP (violet) in orange with its C-terminal residue Y908 highlighted in green. The CP α-ring and β-ring subunits are colored gray and light blue, respectively. Enlarged views of hRpn1 are provided to the right of a transparent rendering of the surface diagram with a ribbon diagram for the C-terminal region and in the lower image, including a representation of the knock-in tag containing the TEV-biotin segment (gray box), autocleavable P2A site (orange box), and mScarlet (red oval shaped box). B, CRISPR/Cas9 plasmid map with the two gRNAs (hRpn1-01 and hRpn1-02) represented in yellow. C, donor plasmid map with the knock-in tag consisting of homology region-left (HR-L, blue), exon 21 (E × 21), TEV-biotin (gray), the P2A site (orange), mScarlet (red), and homology region-right (HR-R, blue). D, representation of the region including and neighboring exon 21 of PSMD2 in chromosome 3, highlighting the respective homology regions (HR-L and HR-R) and location of the stop codon and gRNA-binding sites. E, confocal microscopy images captured for six clones (C1–C6) show fluorescent mScarlet signal (top panel) and differential interference contrast (DIC) reference images (bottom panel). The scale bar represents 12 μm. CP, core particle; gRNA, guide RNA; P2A, peptide 2a; PDB, Protein Data Bank; RP, regulatory particle; TEV, tobacco etch virus.
We used a 75-amino acid sequence from a Propionibacterium shermanii transcarboxylase that is biotinylated in mammalian cells (28) to generate a biotin tag; we selected biotin as the affinity tag based on its 10 fM affinity for neutravidin (29). Tobacco etch virus (TEV) cleavage site was included N terminal to biotin directly following hRpn1 for enzymatic separation of endogenous 26S proteasomes from neutravidin resin following pull-down (Fig. 1A). As a selection marker for cells with the engineered biotin tag incorporated, we included C terminally placed mScarlet with an intervening autocleavable peptide 2a (P2A) site for its removal from the hRpn1-biotin fusion protein. To insert biotin at the C terminus of hRpn1, CRISPR/Cas9 and donor plasmids were designed and transfected into HCT116 cells. The CRISPR/Cas9 plasmid contained two guide RNAs (gRNAs) (Fig. 1B, hRpn1-01 and hRpn1-02) that target distinct protospacer adjacent motifs (PAMs) within PSMD2 (Chr: 184,299,267–184,308,890). The gRNA hRpn1-01 (Chr:184,308,787–184,308,809) binds the PAM present in exon 21 whereas gRNA hRpn1-02 (Chr: 184,308,881–184,308,903) binds the PAM of the 3′ untranslated region (UTR) following exon 21 of PSMD2. In the donor plasmid, the TEV-biotin-P2A-mScarlet knock-in tag sequence is flanked by a total of 809 bp, 730 bp of sequence homologous to PSMD2, termed as homology region (HR)-left (Chr: 184,307,978–184,308,707) and the remaining 79 bp to exon 21, preceding the TEV-biotin sequence and 815 bp of homologous sequence, termed as HR-right (Chr: 184,308,888–184,309,702), following the mScarlet sequence (Fig. 1, C and D). The two HRs in the donor plasmid flanking the desired insert (TEV-biotin-P2A-mScarlet) are designed to replace the excised fragment by homology-directed repair (HDR).
For selection of the desired CRISPR/Cas9 engineered knock-in cells, fluorescence-activated cell sorting (FACS) was used to sort for a live, single-cell population (Fig. S1) that is positive for enhanced GFP (eGFP) (present in the CRISPR/Cas9-plasmid) and mScarlet (present in the donor plasmid). Unlike the transient expression of eGFP, mScarlet expression is permanent as it constitutes part of the desired knock-in tag and is integrated into the host genome. After propagating the FACS-sorted, single-cell population for 6 weeks, a total of six different single-cell populations (C1–C6) were assayed for mScarlet expression by confocal microscopy (Fig. 1E). C1 and C4 expressed mScarlet, suggesting insertion of the knock-in tag into either one or both alleles of these two clones.
Genotyping by PCR and immunoblotting identifies a clone that expresses biotin-tagged hRpn1
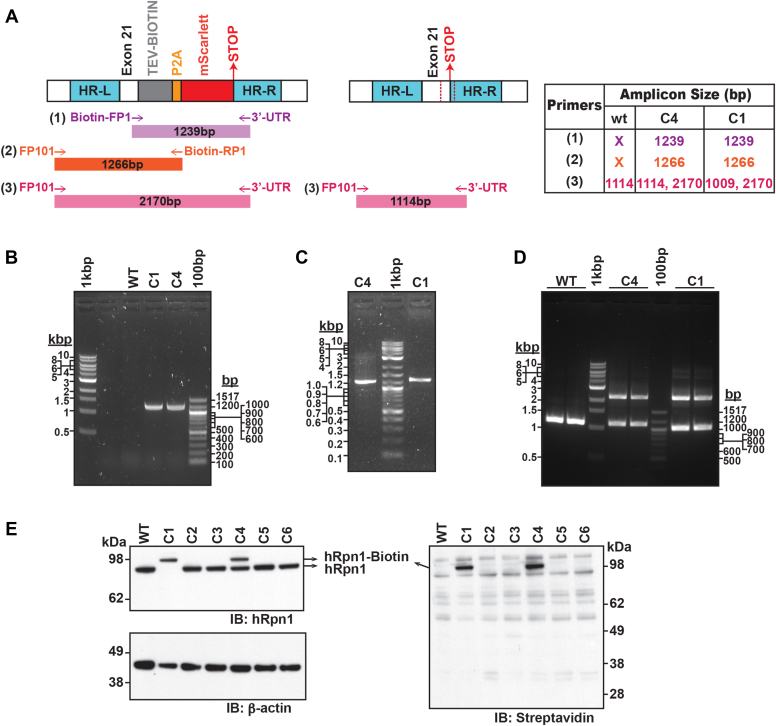
After the initial screening via confocal microscopy based on the presence of mScarlet, clones C1 and C4 were subjected to genotyping by PCR. Genomic DNA (gDNA) was isolated from these clones and primers designed for PCR reactions that span the PSMD2 and knock-in tag sequence (Fig. 2A). The PCR experiments were performed in parallel on the parental cell line as a control. Primers Biotin-FP1 and 3′-UTR-RP confirmed the presence of the knock-in tag in C1 and C4, as Biotin-FP1 recognizes the biotin region and 3′-UTR-RP binds the 3′-UTR of PSMD2. Clones with the knock-in tag integrated in either one or both alleles are expected to yield an amplicon of size 1239 bp, which was observed in C1 and C4 but not parental (WT) cells (Fig. 2B).
Figure 2.
Validation by PCR and immunoblotting of a clone with a knock-in biotin tag at the hRpn1 C terminus.A, the binding site of each primer pairing in the PSMD2-edited (left) and PSMD2 parental (right) region is displayed including (1) Biotin-FP1 and 3′-UTR (purple) (2), FP101 and Biotin-RP1 (orange), and (3) FP101 and 3′-UTR (pink) with anticipated amplicon size. The measured amplicon size for each primer set is listed for parental, C1, and C4 in a table to the right. ‘X’ indicates not detected. B–D, representative agarose gel image showing the PCR amplicon generated from (B) primer set 1, (C) primer set 2, and (D) primer set 3, with inclusion of a 1-kilobase-pair (kbp) and 100 base-pair (bp) DNA ladder, as indicated. E, immunoblotting of the whole cell extract from C1 to C6 along with parental WT cells by anti-hRpn1 antibody (left) and HRP-conjugated streptavidin antibody (right). β-actin was also probed as a loading control. The bands corresponding to hRpn1 and hRpn1-biotin are labeled. HRP, horseradish peroxide, UTR, untranslated region.
The second set of primers, FP101 and Biotin-RP1, were designed to assay the integration of the knock-in tag at the desired chromosomal position. FP101 binds at the chromosomal region flanking the left HR of the tag whereas Biotin-RP1 binds in the biotin region of the knock-in tag (Fig. 2A). C1 and C4 resulted in an amplicon length of the expected size of 1266 bp (Fig. 2C), indicating the integration of the knock-in tag at the desired chromosomal location. This result was corroborated by sequencing for C1 (Fig. S2A).
The third and final PCR experiment was designed to distinguish heterozygous versus homozygous knock-in of the purification handle. For this purpose, a primer that binds to the PSMD2 intronic region (FP101) and 3′-UTR-RP were used (Fig. 2A). The expected amplicon size without any knock-in tag integration of 1114 bp was indicated for the parental cell line as expected, and as well as for C4 (Fig. 2D). These findings indicate that one of the C4 alleles lacks the knock-in tag. The expected amplicon size with the knock-in tag integrated is 2170 bp, and the corresponding PCR product was observed in both C4 and C1, but not in parental WT (Fig. 2D, upper bands). An additional lower molecular weight PCR product was also observed in C1 of ∼1000 bp that was smaller than the lower molecular weight species of C4 and WT cells (1114 bp). This smaller amplicon of C1 was sequenced by Taq-amplified cloning (Fig. S2B) and found to have undergone failed HDR repair following cleavage at the location specified by the two gRNAs. The identity of the amplicon from the parental cell line was also verified by sequencing (Fig. S2C). The genomic deletion in C1 could in principle result in a truncated protein, and we tested this possibility by using immunoblotting with anti-hRpn1 antibody as described below.
Whole cell extracts from all six clones (C1–C6) plus parental WT were immunoprobed for hRpn1 and biotin by using anti-hRpn1 and horseradish peroxide-conjugated streptavidin antibodies, respectively. The hRpn1 antibody recognizes the region surrounding K350, which is remote from the region targeted by the gRNAs. Immunoblotting with these antibodies indicated the presence of expressed hRpn1-TEV-biotin engineered protein in C1 with no altered/truncated hRpn1 protein (Fig. 2E). Therefore, the truncated PCR product observed in C1 does not appear to yield hRpn1 protein. C4 expressed biotin-tagged hRpn1, but also untagged hRpn1 protein (Fig. 2E), validating the heterozygous genotype indicated by the PCR experiments. Interestingly, the tagged protein is expressed at lower abundance compared to unmodified hRpn1. We quantified the abundance of tagged hRpn1 in C1 cells in comparison to the unaltered hRpn1 protein of WT cells. This measurement revealed a 16% decline in hRpn1 abundance, but without statistical significance (p-value = 0.073 for n = 3, Fig. S3). All other clones produced exclusively unmodified hRpn1.
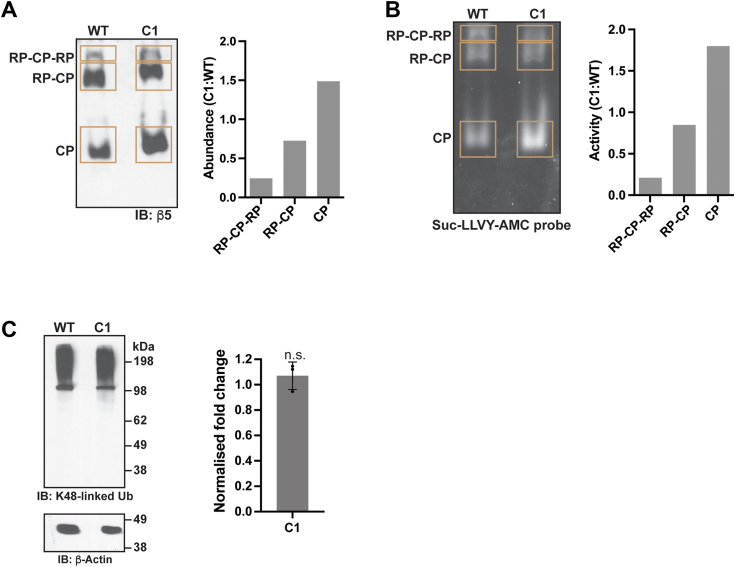
Assessment of proteasomes in HCT116 C1 cells
We next compared the abundance and activity of proteasomes in C1 and WT cells. We performed native gel electrophoresis on lysates from WT or C1 cells and immunoprobed for proteasomes by using antibodies against the CP subunit β5. CP, RP-CP, and RP-CP-RP were observed in both cell lines; however, the relative abundance of uncapped CP was higher and RP-capped CP lower in C1 cells compared to WT cells (Fig. 3A). To further examine this difference, we performed an in-gel peptidase activity assay by using Suc-LLVY-AMC. This experiment similarly indicated an increased relative signal for CP and decreased levels of RP-capped CP in C1 cells (Fig. 3B).
Figure 3.
Comparison of proteasome assembly and activity in C1 and WT cells.A, native gel electrophoresis of lysates from C1 or WT HCT116 cells immunoprobed for CP subunit β5. Whole cell extracts (∼30 μg) from WT or C1 cells were subjected to native gel electrophoresis and then incubated in solubilization buffer before transferring onto a PVDF membrane and immunoprobing with anti-β5 antibodies (left). Quantification of the C1:WT ratio of RP-CP-RP, RP-CP, and CP within the corresponding boxed regions (right). B, in-gel peptidase assay by using native gel electrophoresis with Suc-LLVY-AMC for lysates from WT and C1 cells. Equivalent quantities of WT and C1 cellular lysates were separated by native gel electrophoresis prior to staining with Suc-LLVY-AMC peptide (left). Quantification of the highlighted regions representing RP-CP-RP, RP-CP, and CP bands of C1 was performed by normalizing with the respective bands for WT and plotted as a bar graph (right). C, immunoprobing (left) and quantification (right, n = 3) of whole cell extracts from parental (WT) and C1 cells for K48-linked ubiquitin chains with β-actin as a loading control and used for normalization. n.s., not significant (p = 0.37, two-tailed paired t test). CP, core particle; PVDF, polyvinylidene fluoride; RP, regulatory particle.
To test whether there is a defect in the clearance of K48-linked ubiquitin chains in C1 cells, we immunoprobed lysates from WT or C1 cells in parallel for K48-linked ubiquitin chains. We did not detect any statistically significant difference however (Fig. 3C). Overall, these experiments indicate that C1 cells contain active RP-capped proteasomes, with no increase in K48-linked ubiquitinated substrates; however, the relative abundance of uncapped and RP-capped CP is altered. Future experiments are needed to define mechanistically how tagging the accessible C-terminal end of hRpn1 (Fig. 1A) alters CP and RP-CP abundance.
26S proteasomes are readily purified from HCT116 C1 cells
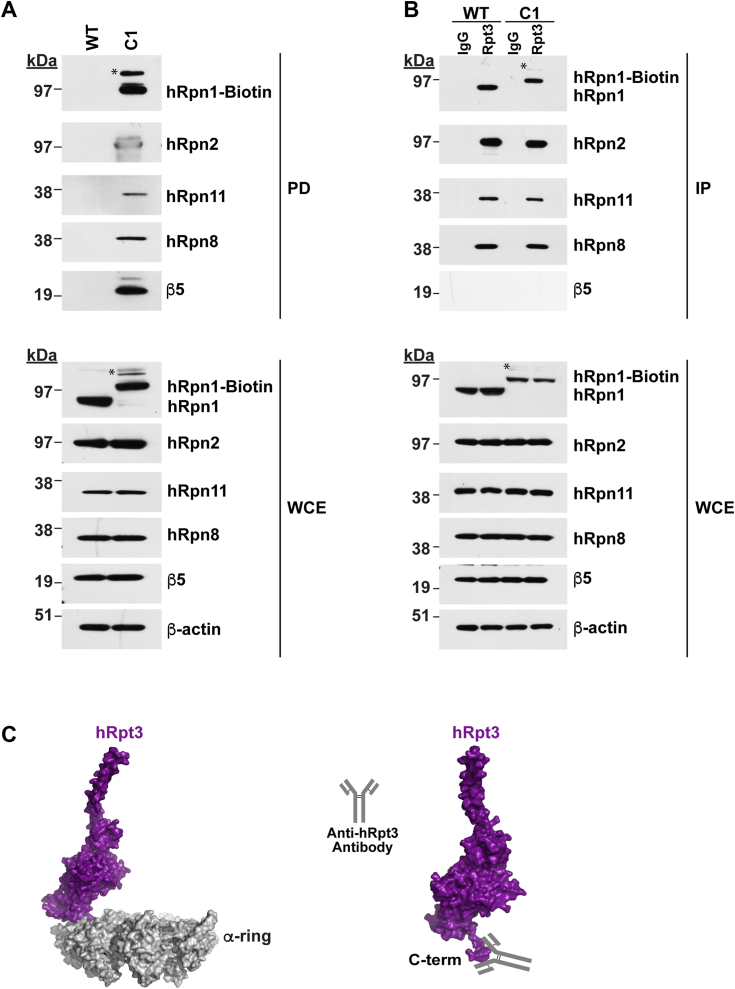
We next sought to determine whether the biotin handle can be used to purify RP-capped proteasomes from C1 cells. The whole cell extract from C1 or WT (as a negative control) was incubated with Streptavidin DynaBeads, which were then washed and immunoprobed for hRpn1. Immunoblotting confirmed the pull-down of biotin-tagged hRpn1 protein from C1 but not WT cells (Fig. 4A). To evaluate whether hRpn1-biotin was incorporated into the 26S proteasome, the membrane was also immunoprobed for RP components hRpn2, hRpn11, and hRpn8 and for CP subunit β5 (Fig. 4A). This experiment indicated pull-down of these proteasome subunits with hRpn1-biotin. For comparison, antibodies against the RP ATPase subunit hRpt3 were used to immunoprecipitate proteasomes from the whole cell extract of C1 or WT cells (Fig. 4B), as done previously (30). This experiment demonstrated hRpn1-biotin at the expected higher molecular weight compared to WT, but β5 was not co-immunoprecipitated with anti-Rpt3 antibodies. This lack of co-immunoprecipitation of β5 with hRpt3 is likely because this antibody recognizes the Rpt3 C-terminal region, which in the assembled 26S proteasome, is not accessible (Fig. 4C). Both by streptavidin (Fig. 4A) and by anti-Rpt3 immunoprecipitation (Fig. 4B), a higher molecular weight band above hRpn1-biotin is observed and is denoted by an asterisk symbol (∗) in Figure 4, A and B.
Figure 4.
26S proteasomes are isolated from C1 cells by using the biotin tag on hRpn1.A, pull-down of hRpn1-biotin from the C1 cell line by streptavidin magnetic beads. The experiment was performed in parallel with parental WT as a negative control. Proteins from the streptavidin-bound samples were eluted from the beads by SDS buffer and heating (pull-down, top) and, along with the whole cell extract (WCE, bottom), were immunoprobed for hRpn1, hRpn2, hRpn11, hRpn8, and β5. B, immunoprecipitation of 26S proteasomes from WT or C1 cells by hRpt3 or IgG (as a control) antibodies. The immunoprecipitants were dissolved in SDS buffer and heated and, along with WCE, immunoprobed for hRpn1, hRpn2, hRpn11, hRpn8, and β5. β-actin was used as a loading control. An asterisk symbol (∗) in (A) and (B) indicates a nonspecific higher molecular weight band above hRpn1-biotin. C, surface view of the 26S proteasome showing only hRpt3 (purple) with the antibody-binding site indicated (right) or also with the CP α-ring (gray, left) to illustrate the entrapment of the antibody-binding site within the CP α-ring. PDB 5GJQ was used to generate this figure. CP, core particle; PDB, Protein Data Bank.
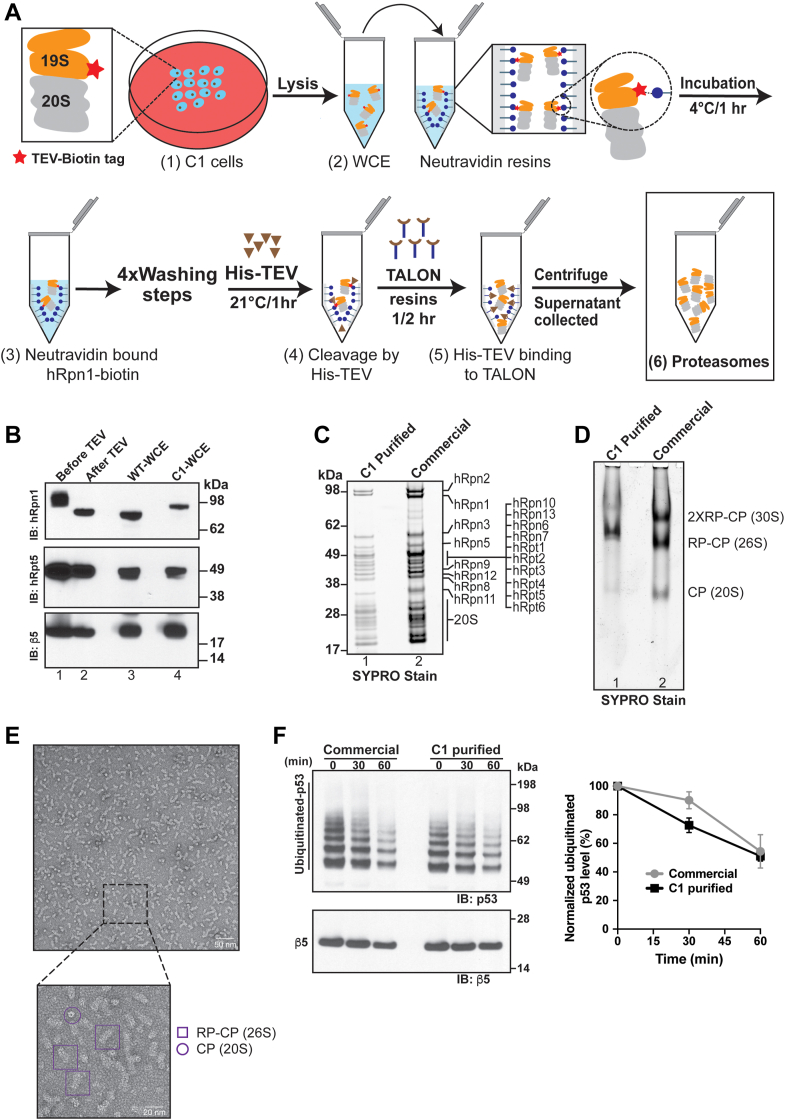
We next tried to purify 26S proteasomes from C1 cells (Fig. 5A). Ten dishes (150 × 25 mm) of C1 cells were seeded (step 1), which yielded approximately 1.1 g of dry pellet weight, and the whole cell lysates (step 2) from these dishes incubated with 500 μl of neutravidin resin for 1 h to bind hRpn1-biotin (step 3). Four wash cycles (20 column volumes per wash) of 10 min each were then performed, followed by 1 h incubation at 21 °C with His-tagged TEV protease (10 U/μl; 15–20 μl) to cleave hRpn1 from biotin (step 4). The sample was further incubated with TALON resin for 30 min (step 5) to remove His-tagged TEV protease from the elution and the TALON resin next removed by centrifugation. The supernatant containing 26S proteasomes (step 6) was concentrated to 0.5 mg/ml, flash frozen in liquid nitrogen, and then stored at −80 °C. The purification protocol was evaluated by immunoprobing for hRpn1, hRpt5, and β5 before (Fig. 5B, lane 1) and after TEV protease treatment (Fig. 5B, lane 2), with whole cell lysate from WT (Fig. 5B, lane 3) and C1 cells (Fig. 5B, lane 4) used for reference. All components of 26S proteasomes purified from C1 cells were visible by SDS-PAGE at the appropriate stoichiometry and comparable to commercial 26S proteasomes (Fig. 5C). In an effort to calculate the yield of our 26S proteasome preparation, we quantified the region spanning 17 to 100 kDa in the SDS-PAGE of purified proteasomes from C1 (Fig. 5C, lane 1), normalizing to the corresponding region of commercial proteasome (Fig. 5C, lane 2). This approach indicated a yield of 22.5 μg of 26S proteasomes purified from C1.
Figure 5.
hRpn1-biotin is an effective handle for purification of 26S proteasomes.A, schematic representation of the workflow for purification of 26S proteasomes (RP in orange, CP in gray) from C1 cells by hRpn1-biotin (red star). CRISPR/Cas9-engineered C1 cells were seeded in culture dishes (step 1) and the WCE post lysis (step 2) incubated with neutravidin resin for 1 h at 4 °C (step 3). After four washing steps, the mixture was treated with His-tagged TEV protease (step 4) for 1 h at 21 °C to release 26S proteasomes from the resin by cleaving hRpn1 from biotin. The eluant was incubated with TALON resin to remove His-tagged TEV protease (step 5) and the resin sedimented by centrifugation. Supernatant containing purified 26S proteasomes was flash frozen in liquid nitrogen and stored at −80 °C. B, immunoblot to evaluate the presence of 26S proteasomes by using hRpn1, hRpt5, and β5 antibodies before and after treatment with TEV protease. WCE from parental WT and C1 cells is also included. C, purified 26S proteasomes from the protocol described in (A) were evaluated by SDS-PAGE (4–12%) with SYPRO Ruby Fluorescent stain. Commercial proteasomes were included for comparison. D, purified and commercial 26S proteasomes were subjected to native-PAGE and evaluated with SYPRO Ruby Fluorescent stain. E, representative image of raw electron micrograph of negatively stained 26S proteasomes purified from C1 cells captured at 36k magnification, with a scale bar corresponding to 50 nm. The insert shows an expanded region captured at 92k magnification, with a 20 nm scale bar. Different orientations of 26S and CP proteasomes are highlighted with purple-colored boxes and circles, respectively. F, C1-purified proteasomes are active against ubiquitinated p53. Ubiquitinated p53 was incubated with C1 or commercial proteasomes for the indicated time points, after which the mixture was separated by SDS-PAGE and immunoblotted with anti-p53 or anti-β5 antibodies (left). Quantification of p53 normalized to β5 at the indicated time points following incubation with C1-purified (black squares) or commercial (gray circles) proteasomes plotting the mean value (n = 3) from three independent experiments with standard deviation (right). CP, core particle; RP, regulatory particle; TEV, tobacco etch virus; WCE, whole cell extract.
We further evaluated proteasomes purified from C1 cells by the protocol of Figure 5A by using native-PAGE with visualization by SYPRO Ruby fluorescent stain. This assay indicated the majority of proteasomes to be capped with one RP (Fig. 5D, lane 1). Commercial proteasomes from red blood cells indicated higher populations of 30S proteasomes with two RP subunits as well as of single CP without RP capping (Fig. 5D, lane two versus lane 1).
To further evaluate the integrity of 26S proteasomes purified from C1 cells, we used EM. Grids were made by staining the sample with uranyl formate and observed at 36k and 92k magnification on a L120C Talos microscope (Thermo Fisher Scientific). Various orientations of monodisperse 26S proteasome particles were observed, with a small population of uncapped CP (Fig. 5E). Overall, the electron micrographs indicate that the purified 26S proteasomes are intact and homogenous at the resolution detected.
We investigated the degradation activity of proteasomes purified from C1 cells as compared to commercial proteasomes by using ubiquitinated p53 as a substrate and β5 as a loading control. This experiment indicated equivalent clearance of ubiquitinated p53 (Fig. 5F).
Discussion
Functional pathways and activities in cells typically rely on machinery in the form of multisubunit complexes (large or small) that are modified and/or assemble dynamically in the native cellular environment. Reconstituting such complexes outside of the cellular environment is challenging, as is also the purification of proteins or their complexes from native cellular environments in the absence of an affinity tag handle. Here, we used CRISPR/Cas9 to engineer an affinity tag handle at the C-terminal end of proteasome subunit hRpn1 in the human colon cancer HCT116 cell line. This reagent complements previous efforts to obtain proteasomes from cells, including the generation of a stable HEK293 cell line with tagged proteasome subunit hRpn11, which was generated by using a lentiviral system (19, 31), and an engineered HEK293 cell line with an affinity tag on the CP subunit β4 (PSMB2). In contrast to our C1 cell line, the β4-tagging approach is not selective for RP-capped CP.
Our cell line incorporates an affinity tag at the C-terminal end of hRpn1 into one of the alleles and yields a truncation in the other allele that does not result in a translation product. The protocol that we established yielded a highly purified 26S proteasome complex. Interestingly, the commercial proteasome indicated two additional bands by SDS-PAGE that migrate between hRpn3 and hRpn1 (Fig. 5C) and also a greater abundance of uncapped CP and CP with two RP subunits (Fig. 5D). By contrast, proteasome purified from C1 cells with our protocol appear to be predominately RP-CP (Fig. 5, D and E).
Overall, this study provides a new approach for purification of the human 26S proteasomes from their native environment by using a simple protocol and without a need for overexpression. This approach can be applied to other cell lines to study the 26S proteasome in different cell types.
Experimental procedures
Design of CRISPR/Cas9 and donor plasmids
gRNAs were designed that target the final protein coding exon (exon 21) and 3′UTR by sgRNA Scorer 2.0 (Table 1) and tested for cutting activity in 293T cells as previously described (32, 33). The chromosomal location and gRNA-binding regions within PSMD2 are listed in Table 2. gRNAs for the knock-in tag were named hRpn1-01 and hRpn1-02, with corresponding oligonucleotides 3435 and 3437, respectively (Table 3), that were cloned into the pDG458 vector by golden gate ligation (34). pDG458 was a gift from Paul Thomas (Addgene plasmid # 100900).
Table 1.
Guide RNAs tested in 293T cells
| Candidate | Target site (PAM in bold) |
|---|---|
| 3432 | ACACTGGGGTTGTATGCGTCTGG |
| 3433 | TCCCCGTGGGCCAACAACACTGG |
| 3434 | GTTGTTGGCCCACGGGGAACGGG |
| 3435a | CAATTCTGCCCGTTCCCCGTGGG |
| 3436 | TCTTCCTGTTACCCCCATTCTGG |
| 3437a | TGATCTCTAAGTGACCACCAGGG |
Guide RNAs used for knock-in experiments.
Table 2.
Chromosomal locations of different regions of PSMD2 as per GRCh38/Hg38
| No. | Regions | Chromosomal location |
|---|---|---|
| 1. | PSMD2 | 184,299,267–184,308,890 |
| 2. | PSMD2-Exon21 | 184,308,708–184,308,890 |
| 3. | PSMD2-3′UTR | 184,308,891–184,309,050 |
| 4. | 3435: hRpn1-01 | 184,308,787–184,308,809 |
| 5. | 3437: hRpn1-02 | 184,308,881–184,308,903 |
| 6. | HR-left | 184,307,978–184,308,707 |
| 7. | HR-right | 184,308,888–184,309,702 |
Table 3.
Oligos for guide RNAs
| Guide | Forward oligo | Reverse oligo |
|---|---|---|
| 3435 | CACCGCAATTCTGCCCGTTCCCCGT | AAACACGGGGAACGGGCAGAATGC |
| 3437 | ACCGTGATCTCTAAGTGACCACCAGT | TAAAACTGGTGGTCACTTAGAGATCA |
A donor construct for HDR-based knock-in was generated by using synthesized DNA (Twist Bioscience) of ∼800 bp of homology sequence 5′ and 3′ of the point of insertion and complementary DNA sequence encoding TEV-biotin-P2A-mScarlet. This DNA sequence was then cloned into the pRC0082 vector (Addgene plasmid # 195320) by using two sequential isothermal assembly cloning reactions (35). The Cas9/dual sgRNA and HDR donor plasmids were each verified by Sanger sequencing with primers hU6-FP, DG1-FP, DG1-RP, donor-FP, and donor-RP (Table 4). To facilitate identification of successful knock-in, eGFP in the CRISPR/Cas9 plasmid and mScarlet in the donor plasmid were used as fluorogenic selection markers with cell sorting by FACS.
Table 4.
Details of the primers used in the study
| Primer name | Primer sequence | Tm (°C) |
|---|---|---|
| hU6-FP | 5′– GAG GGC CTA TTT CCC ATG ATT CC –3′ | 57.5 |
| DG1-FP | 5′– GGT TTC GCC ACC TCT GAC TTG –3′ | 58.4 |
| DG2-RP | 5′– TGC ATC GCA TTG TCT GAG TAG G –3′ | 57.2 |
| Donor-FP | 5′– ACC TGA CGT CGC TAG CTG TAC –3′ | 58.7 |
| Donor-RP | 5′– TTA CGG TTC CTG GCC TTT TG –3′ | 55.5 |
| Biotin-FP1 | 5′– CCC CAA TTA TGA CAG G –3′ | 49.8 |
| 3′-UTR-RP | 5′– GAC CTT GAT AAA AGT CTT T –3′ | 52.6 |
| FP101 | 5′– GAC CGG CAG CTT ATG AG –3′ | 52.9 |
| Biotin-RP1 | 5′– GGC GCT ACT AAC TTC AGC C –3′ | 55.8 |
Transfection into HCT116 cells
1.5 × 105 HCT116 cells (P3) purchased from American Tissue Culture Collection (CCL-247) were reverse transfected in a 6-well plate (3506; Costar) with 2 μg of Cas9 plasmid and 3 μg of donor (hRpn1-TEV-Biotin-P2A-mScarlet) plasmid by using Lipofectamine 3000 (L3000015; Thermo Fisher Scientific, Inc) with Opti-MEM reduced serum medium (31985070; Life Technologies) according to the manufacturer’s instructions. Transfected cells were incubated at 37 °C with 5% CO2 humidity for 72 h and then single sorted by FACS.
FACS selection for transfected HCT116 cells
Seventy-two hours after transfection, cells were detached by trypsin (25200056; Life Technologies) and counted with a countess automated cell counter (Invitrogen). Ten thousand cells were resuspended in PBS (1X; pH 7.4; 10010-023; Thermo Fisher Scientific, Inc) containing 20% fetal bovine serum (Atlanta Biologicals) and treated with DNA-staining DAPI (D3571; Thermo Fisher Scientific, Inc) prior to sorting by FACS with a BD FACSymphony S6 Cell Sorter (BD Sciences). Forward scatter (FSC)-area and side scatter (SSC)-area settings were used to gate for intact cells and exclude debris. For singlet isolation, the samples were gated by SSC-height and SSC-width. To further discriminate against doublet events, the cells were subsequently gated using FSC-height and FSC-width parameters. Following debris and doublet exclusion, live singlets were gated using 4′,6-diamidino-2-phenylindole in the BV421-A channel and FSC-area parameters were used to exclude dead cells. The identified live singlet population was then analyzed and sorted for double positive eGFP- and mScarlet-expressing cells. Single cells were sorted into 96-well plates (3596; Costar) prefilled with 100 μl of media using FITC-A and PE-Texas Red-A settings. The selected cells were then incubated at 37 °C in a humidified environment of 5% CO2 for a minimal of 6 weeks, changing media every 2 weeks.
Confocal microscopy
After 6 weeks of incubation post FACS-sorting, six clones from the 96-well plate exhibited cell growth. These clones were trypsinized and expanded into a T25 flask (Corning; 430639) and incubated at 37 °C with 5% CO2 humidity for an additional 2 weeks of growth. To validate the integration of the biotin tag into PSMD2, live cells expressing mScarlet were visualized by confocal microscopy. Imaging was performed by an Eclipse Ti inverted microscope (Nikon Inc) equipped with a Yokogawa spinning disc (Yokogawa Electric Corporation), back-illuminated EMCCD camera (Andor, DU888), and a 2x relay lens placed before the spinning disc. A 60x, NA 1.45, Plan Apo objective and a 561 nm laser (Agilent Technology MCL-400) was used to acquire fluorescence (mScarlet) images in parallel with differential interference contrast images for reference.
Genotyping
Isolation of gDNA
All six clones (C1–C6) were subcultured in a 6-well plate. Once the wells were 70 to 80% confluent, the cells were harvested and the gDNA isolated for each clone using a DNeasy kit (69504, Qiagen).
PCR
PCR genotyping was used to confirm integration of the knock-in tag at the genomic level. Different sets of primers (Integrated DNA Technologies, Inc) were designed against different regions of PSMD2 as well as the knock-in tag (Table 4). All PCR was performed using the KAPA-Hifi enzyme mixture (KK2601; Roche) with the basic composition of the PCR master mix and the PCR cycling protocol provided in Tables 5 and 6, respectively. All PCR products were observed by 0.8 to 1% agarose gels made with 1X Tris-acetate-EDTA buffer.
Table 5.
Preparation of PCR mixture
| Components | Reaction | Final concentration |
|---|---|---|
| PCR-grade water | Add to 50 μl | N.A |
| 2X KAPA HiFi HotStart Ready Mix | 25 μl | 1X |
| Forward primer (10 μM) | 1.5 μl | 0.3 μM |
| Reverse primer (10 μM) | 1.5 μl | 0.3 μM |
| Template gDNA | 50 ng | 1 ng/μl |
Table 6.
PCR cycling protocol
| No. | Steps | Temperature (°C) | Time | Cycle |
|---|---|---|---|---|
| 1 | Initial denaturation | 98 | 3 min | 1 |
| 2 | Denaturation | 98 | 30 s | 33 |
| 3 | Annealing | 53–60 | 30 s | |
| 4 | Extension | 72 | 15–60 s/kb | |
| 5 | Final extension | 72 | 1 min/kb | 1 |
TA cloning and Sanger sequencing
The amplicon of clone C1 and parental WT DNA was gel extracted using a gel elution kit (GFX PCR DNA and Gel Band Purification Kit from GE HealthCare). Postextraction, the amplicons were processed to add 3′ A overhangs by Taq polymerase and dATP. This reaction was incubated for 15 min at 72 °C. The TOPO-cloning reaction (Table 7) was performed by using the PCR amplicons following addition of the 3′ overhangs by a TOPO kit (Thermo Fisher Scientific, Inc). The TA cloning reaction mixture was incubated for 5 min at room temperature before proceeding to transformation. All TA clones for the different PCR amplicons were transformed into TOPO-One Shot competent cells and plated to isolate individual colonies. Three to five colonies were selected for plasmid DNA isolation and further screened by Sanger sequencing with T3 and T7 primers flanking either side of the PCR amplicon within the TA vector.
Table 7.
TA cloning reaction mixture
| Reagent | Volume |
|---|---|
| Fresh PCR-amplicons | 3 μl |
| Salt solution | 1 μl |
| Water | Add to 6 μl |
| TOPO vector | 1 μl |
| Final volume | 6 μl |
Immunoblotting
Whole cell lysates from the six clones (C1–C6) and parental cell line (WT) were prepared using Triton lysis buffer (1% Triton X-100, 50 mM Tris–HCl [pH 7.5], and 150 mM NaCl) supplemented with protease inhibitor cocktail (25765800; Roche) and PMSF (195381; MP Biomedicals). Protein concentration was determined by BCA Protein Assay (23225; Thermo-Pierce) according to the manufacturer’s instructions. A total of 10 to 20 μg of protein was loaded onto 4 to 12% Bis-Tris gels (NP0322; Thermo Fisher Scientific, Inc) and wet transferred onto polyvinylidene fluoride (PVDF) (0.45 μm) membranes. Immunoblotting was performed by probing with antibodies against streptavidin (N100; Thermo Fisher Scientific, Inc), hRpn1 (25430, Cell Signaling Technology, Inc), K48-linked ubiquitin chains (140601; Abcam, Plc.), or β-actin (3700; Cell Signaling Technology, Inc) in 5% bovine serum albumin (5217; Tocris Biosciences) or 5% skim milk (LP0031B; Thermo Fisher Scientific, Inc) made in Tris-buffered saline with 0.1% Tween 20, followed by washing, and then an hour of incubation with secondary antibodies. The PVDF membranes were developed on HyBlot CL autoradiography films using Pierce ECL chemiluminescent substrates (32106; Thermo Fisher Scientific, Inc) for horseradish peroxide-conjugated secondary antibodies.
The whole cell extracts of WT and C1 cells were separated on a 3.5% native-PAGE gel that was run for 4 h at 100 V. The gel was then incubated in solubilization buffer (2% (w/v) SDS, 66 nM Na2CO3, 1.5% (v/v) β-mercaptoethanol) for 30 min at room temperature, before transferring onto a PVDF membrane and immunoprobed with anti-β5 antibodies (BML-PW8895-0100; Enzo Lifesciences, Inc). Quantification and analyses were done by ImageJ (https://imagej.nih.gov/ij/index.html) and GraphPad Prism 9.4.1 (https://www.graphpad.com), respectively.
In-gel peptidase activity assay
The whole cell extract of WT or C1 cells was separated by 3.5% native-PAGE and the gel then incubated with 50 μM of Suc-LLVY-AMC peptide (S-280; R&D Systems, Inc) in 100 mM Tris (pH 7.5), 5 mM ATP, 5 mM MgCl2, and 1 mM DTT for 1 h at 37 °C. Fluorescence was measured by using a Gel Doc EZ imager (Bio-Rad Laboratories, Inc).
Immunoprecipitation
For Rpt3 immunoprecipitation, lysates (0.5 mg total protein) were incubated overnight at 4 °C with anti-hRpt3 antibodies followed by 3-h incubation with Dynabead protein G (10004; Thermo Fisher Scientific). For pull-down with streptavidin, lysates (0.5 mg total protein) were incubated at 4 °C for 2 h with the Dynabeads MyOne Streptavidin T1 (65601; Thermo Fisher Scientific, Inc). The beads were washed at least 4 times with Triton lysis buffer supplemented with protease inhibitor cocktail (25765800; Roche), PMSF (195381; MP Biomedicals), and 1mM ATP, and the bound proteins resolved in 2X SDS with 10% DTT and visualized by SDS-PAGE. Immunoblotting was done by overnight incubation with primary antibodies in 5% skim milk–Tris-buffered saline with 0.1% Tween 20, followed by washing and 1 h incubation with secondary antibodies. Blots were detected by addition of Pierce ECL chemiluminescent substrates (32106; Thermo Fisher Scientific, Inc).
Purification of 26S proteasomes
Cells from ten 150 × 25 mm confluent dishes were harvested and lysed with lysis buffer (50 mM Tris–HCl [pH 7.5], 100 mM NaCl, 1% Triton X-100, 10% glycerol, 5 mM ATP, and 5 mM MgCl2, supplemented with protease inhibitor cocktail) using a Dounce homogenizer and incubated on ice for 15 min. The lysed sample was centrifuged at 16,000g for 15 min in a prechilled centrifuge at 4 °C. The supernatant was incubated for 1 h at 4 °C with NeutrAvidin Agarose resin (29202; Thermo Fisher Scientific, Inc) that was preequilibrated with lysis buffer. The resins were next washed with wash buffer (50 mM Tris–HCl [pH 7.5], 100 mM NaCl, 10% glycerol, 5 mM ATP, and 5 mM MgCl2) and then incubated for 1 h at 21 °C with TEV-digestion buffer (50 mM Tris–HCl [pH 7.5], 100 mM NaCl, 10% glycerol, 5 mM ATP, 5 mM MgCl2, and 1 mM DTT) containing His-tagged TEV protease (12575015; Thermo Fisher Scientific, Inc). To remove TEV enzyme, TALON resin, preequilibrated with TEV-digestion buffer, was added to the mixture. The unbound mixture containing cleaved 26S proteasome was collected and flash frozen in liquid nitrogen before storing at −80 °C. To test for purity and fidelity, an aliquot of purified 26S proteasome was subjected to SDS-PAGE analyses with a 4 to 12% Bis-Tris gel as well as native-PAGE (36) with a 3.5% acrylamide gel and SYPRO Ruby fluorescent staining.
Negative staining electron microscopy
For negative staining EM, a standard thickness carbon coated grid with a 200-mesh copper support (CF200-CU; Electron Microscopy Sciences) was used. The grids were glow discharged using a standard protocol before loading 4 μl of sample. Freshly prepared 0.75% uranyl formate solution was used to stain the sample on the grid before capturing EM images with a Talos L120C TEM (Thermo Fisher Scientific, Inc) microscope.
Activity assay with ubiquitinated p53
Fifty nanomolar of ubiquitinated p53 (50 nM, SBB-US0012; South Bay Bio, LLC) was incubated with 25 nM of proteasomes from C1 cells or commercially obtained (A1101; UBPBio, LLC) in 50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, and 5 mM ATP at 37 °C. The mixture was left for 0, 30, or 60 min and the reaction terminated by adding SDS loading buffer. The proteins were separated by SDS-PAGE and p53 status immunoprobed by anti-p53 antibodies (1101; Abcam, PLC), with β5 immunoblotting by anti-β5 antibodies used as a loading control. Three independent experiments were performed, and quantification and analyses done by using ImageJ and GraphPad Prism 9.4.1, respectively.
Data availability
All data are available in the main text and reagents distributed upon request. Request for materials used in this study can be addressed to Kylie Walters (kylie.walters@nih.gov).
Supporting information
The article contains the supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank the CCR Frederick Flow Core, NCI, NIH, Frederick, MD for providing the cell sorting services and Dr Tapan Kanai for technical support with the electron microscopy.
Author contributions
H. N., V. O.-A., B. I., C. N. E., C. S., J. L., and R. C. methodology; H. N., V. O.-A., B. I., C. N. E., C. S., J. L., and R. C. investigation; H. N. and V. O.-A. validation; H. N. and V. O.-A. writing–original draft; V. O.-A. and K. J. W. writing–review and editing; K. J. W. conceptualization; K. J. W. supervision.
Funding and additional information
This research was supported by the Intramural Research Program through the Center for Cancer Research, National Cancer Institute, National Institutes of Health (1 ZIA BC011490 and 1 ZIA BC011627 to K. J. W.) and in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261201500003I. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by George DeMartino
Supporting information
References
- 1.Finley D., Chen X., Walters K.J. Gates, channels, and switches: elements of the proteasome machine trends. Biochem. Sci. 2016;41:77–93. doi: 10.1016/j.tibs.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehlinger A., Walters K.J. Structural insights into proteasome activation by the 19S regulatory particle. Biochemistry. 2013;52:3618–3628. doi: 10.1021/bi400417a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X., Htet Z.M., López-Alfonzo E., Martin A., Walters K.J. Proteasome interaction with ubiquitinated substrates: from mechanisms to therapies. FEBS J. 2021;288:5231–5251. doi: 10.1111/febs.15638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osei-Amponsa V., Walters K.J. Proteasome substrate receptors and their therapeutic potential Trends. Biochem. Sci. 2022;47:950–964. doi: 10.1016/j.tibs.2022.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anchoori R.K., Karanam B., Peng S., Wang J.W., Jiang R., Tanno T., et al. A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer Cancer. Cell. 2013;24:791–805. doi: 10.1016/j.ccr.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trader D.J., Simanski S., Kodadek T. A reversible and highly selective inhibitor of the proteasomal ubiquitin receptor rpn13 is toxic to multiple myeloma cells. J. Am. Chem. Soc. 2015;137:6312–6319. doi: 10.1021/jacs.5b02069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu X., Sabbasani V.R., Osei-Amponsa V., Evans C.N., King J.C., Tarasov S.G., et al. Structure-guided bifunctional molecules hit a DEUBAD-lacking hRpn13 species upregulated in multiple myeloma. Nat. Commun. 2021;12:7318. doi: 10.1038/s41467-021-27570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du T., Song Y., Ray A., Wan X., Yao Y., Samur M.K., et al. Ubiquitin receptor PSMD4/Rpn10 is a novel therapeutic target in multiple myeloma. Blood. 2023;141:2599–2614. doi: 10.1182/blood.2022017897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bashore C., Prakash S., Johnson M.C., Conrad R.J., Kekessie I.A., Scales S.J., et al. Targeted degradation via direct 26S proteasome recruitment. Nat. Chem. Biol. 2023;19:55–63. doi: 10.1038/s41589-022-01218-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leggett D.S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R.T., et al. Multiple associated proteins regulate proteasome structure and function. Mol. Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 11.Martínez-Noël G., Galligan J.T., Sowa M.E., Arndt V., Overton T.M., Harper J.W., et al. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol. Cell. Biol. 2012;32:3095–3106. doi: 10.1128/MCB.00201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu B.W., Kovary K.M., Guillaume J., Chen L.C., Teruel M.N., Wandless T.J. The E3 ubiquitin ligase UBE3C enhances proteasome processivity by ubiquitinating partially proteolyzed substrates. J. Biol. Chem. 2013;288:34575–34587. doi: 10.1074/jbc.M113.499350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buel G.R., Chen X., Chari R., O'Neill M.J., Ebelle D.L., Jenkins C., et al. Structure of E3 ligase E6AP with a proteasome-binding site provided by substrate receptor hRpn10. Nat. Commun. 2020;11:1291. doi: 10.1038/s41467-020-15073-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Ebelle D.L., Wright B.J., Sridharan V., Hooper E., Walters K.J. Structure of hRpn10 bound to UBQLN2 UBL illustrates basis for complementarity between shuttle factors and substrates at the proteasome. J. Mol. Biol. 2019;431:939–955. doi: 10.1016/j.jmb.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X., Randles L., Shi K., Tarasov S.G., Aihara H., Walters K.J. Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 reveal distinct binding mechanisms between substrate receptors and shuttle factors of the proteasome. Structure. 2016;24:1257–1270. doi: 10.1016/j.str.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yasuda S., Tsuchiya H., Kaiho A., Guo Q., Ikeuchi K., Endo A., et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature. 2020;578:296–300. doi: 10.1038/s41586-020-1982-9. [DOI] [PubMed] [Google Scholar]
- 17.Wang X., Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol. Cell. Proteomics. 2008;7:46–57. doi: 10.1074/mcp.M700261-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Yu C., Yang Y., Wang X., Guan S., Fang L., Liu F., et al. Characterization of dynamic UbR-proteasome subcomplexes by in vivo cross-linking (X) assisted bimolecular tandem affinity purification (XBAP) and label-free quantitation. Mol. Cell. Proteomics. 2016;15:2279–2292. doi: 10.1074/mcp.M116.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Chen C.F., Baker P.R., Chen P.L., Kaiser P., Huang L. Mass spectrometric characterization of the affinity-purified human 26S proteasome complex. Biochemistry. 2007;46:3553–3565. doi: 10.1021/bi061994u. [DOI] [PubMed] [Google Scholar]
- 20.Besche H.C., Haas W., Gygi S.P., Goldberg A.L. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry. 2009;48:2538–2549. doi: 10.1021/bi802198q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J., Makhija S., Zhou C., Zhang H., Wang Y., Muralidharan M., et al. Structural insights into the human PA28-20S proteasome enabled by efficient tagging and purification of endogenous proteins. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2207200119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim Y.C., DeMartino G.N. C termini of proteasomal ATPases play nonequivalent roles in cellular assembly of mammalian 26 S proteasome. J. Biol. Chem. 2011;286:26652–26666. doi: 10.1074/jbc.M111.246793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koulich E., Li X., DeMartino G.N. Relative structural and functional roles of multiple deubiquitylating proteins associated with mammalian 26S proteasome. Mol. Biol. Cell. 2008;19:1072–1082. doi: 10.1091/mbc.E07-10-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen X., Dorris Z., Shi D., Huang R.K., Khant H., Fox T., et al. Cryo-EM reveals unanchored M1-ubiquitin chain binding at hRpn11 of the 26S proteasome. Structure. 2020;28:1206–1217. doi: 10.1016/j.str.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi Y., Chen X., Elsasser S., Stocks B.B., Tian G., Lee B.H., et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016 doi: 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boughton A.J., Liu L., Lavy T., Kleifeld O., Fushman D. A novel recognition site for polyubiquitin and ubiquitin-like signals in an unexpected region of proteasomal subunit Rpn1. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ota T., Suzuki Y., Nishikawa T., Otsuki T., Sugiyama T., Irie R., et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 28.Cronan J.E., Jr. Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J. Biol. Chem. 1990;265:10327–10333. [PubMed] [Google Scholar]
- 29.Green N.M. Avidin. 3. The nature of the biotin-binding site. Biochem. J. 1963;89:599–609. doi: 10.1042/bj0890599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osei-Amponsa V., Sridharan V., Tandon M., Evans C.N., Klarmann K., Cheng K.T., et al. Impact of losing hRpn13 Pru or UCHL5 on proteasome clearance of ubiquitinated proteins and RA190 cytotoxicity. Mol. Cell. Biol. 2020;40 doi: 10.1128/MCB.00122-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., et al. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol. Cell. Proteomics. 2006;5:737–748. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Chari R., Yeo N.C., Chavez A., Church G.M. sgRNA Scorer 2.0: a species-independent model to predict CRISPR/Cas9 activity. ACS Synth. Biol. 2017;6:902–904. doi: 10.1021/acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gooden A.A., Evans C.N., Sheets T.P., Clapp M.E., Chari R. dbGuide: a database of functionally validated guide RNAs for genome editing in human and mouse cells. Nucleic Acids Res. 2021;49:D871–D876. doi: 10.1093/nar/gkaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adikusuma F., Pfitzner C., Thomas P.Q. Versatile single-step-assembly CRISPR/Cas9 vectors for dual gRNA expression. PLoS One. 2017;12 doi: 10.1371/journal.pone.0187236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 36.Roelofs J., Suppahia A., Waite K.A., Park S. Native gel approaches in studying proteasome assembly and chaperones methods. Mol. Biol. 2018;1844:237–260. doi: 10.1007/978-1-4939-8706-1_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text and reagents distributed upon request. Request for materials used in this study can be addressed to Kylie Walters (kylie.walters@nih.gov).