Abstract
Alternative polyadenylation (APA) plays important roles in modulating mRNA stability, translation, and subcellular localization, and contributes extensively to shaping eukaryotic transcriptome complexity and proteome diversity. Identification of poly(A) sites (pAs) on a genome-wide scale is a critical step toward understanding the underlying mechanism of APA-mediated gene regulation. A number of established computational tools have been proposed to predict pAs from diverse genomic data. Here we provided an exhaustive overview of computational approaches for predicting pAs from DNA sequences, bulk RNA sequencing (RNA-seq) data, and single-cell RNA sequencing (scRNA-seq) data. Particularly, we examined several representative tools using bulk RNA-seq and scRNA-seq data from peripheral blood mononuclear cells and put forward operable suggestions on how to assess the reliability of pAs predicted by different tools. We also proposed practical guidelines on choosing appropriate methods applicable to diverse scenarios. Moreover, we discussed in depth the challenges in improving the performance of pA prediction and benchmarking different methods. Additionally, we highlighted outstanding challenges and opportunities using new machine learning and integrative multi-omics techniques, and provided our perspective on how computational methodologies might evolve in the future for non-3′ untranslated region, tissue-specific, cross-species, and single-cell pA prediction.
Keywords: Polyadenylation, Predictive modeling, RNA-seq, scRNA-seq, Machine learning
Introduction
Precursor mRNA (pre-mRNA) polyadenylation is an essential two-step event in the post-transcriptional regulation of gene expression, which involves the cleavage of the pre-mRNA at the poly(A) site (pA) followed by the addition of an untemplated stretch of adenosines [1], [2]. The selective use of pAs of a single gene, termed alternative polyadenylation (APA), can generate a diversity of isoforms with different 3′ ends and/or encode distinct proteins [3], [4]. APA plays important roles in modulating mRNA stability, translation, and subcellular localization, which contributes extensively to shaping eukaryotic transcriptome complexity and proteome diversity. APA is a widespread regulatory mechanism in eukaryotes, which has been observed in more than 70% of mammalian and plant genes [5], [6], [7], [8], [9], [10], [11]. APA is highly tissue specific and dynamically modulated in various conditions, cell types, and/or states [2], [12]. Specific APA programs have been implicated in diverse biological processes and diseases, such as cell activation, proliferation, neurodegenerative disorders, and cancer [3], [4], [13], [14], [15], [16], [17], [18], [19], [20]. Given the functional significance of APA, identification and/or quantification of pAs on a genome-wide scale is crucial and may be the first step in understanding the underlying mechanism of APA-mediated gene regulation.
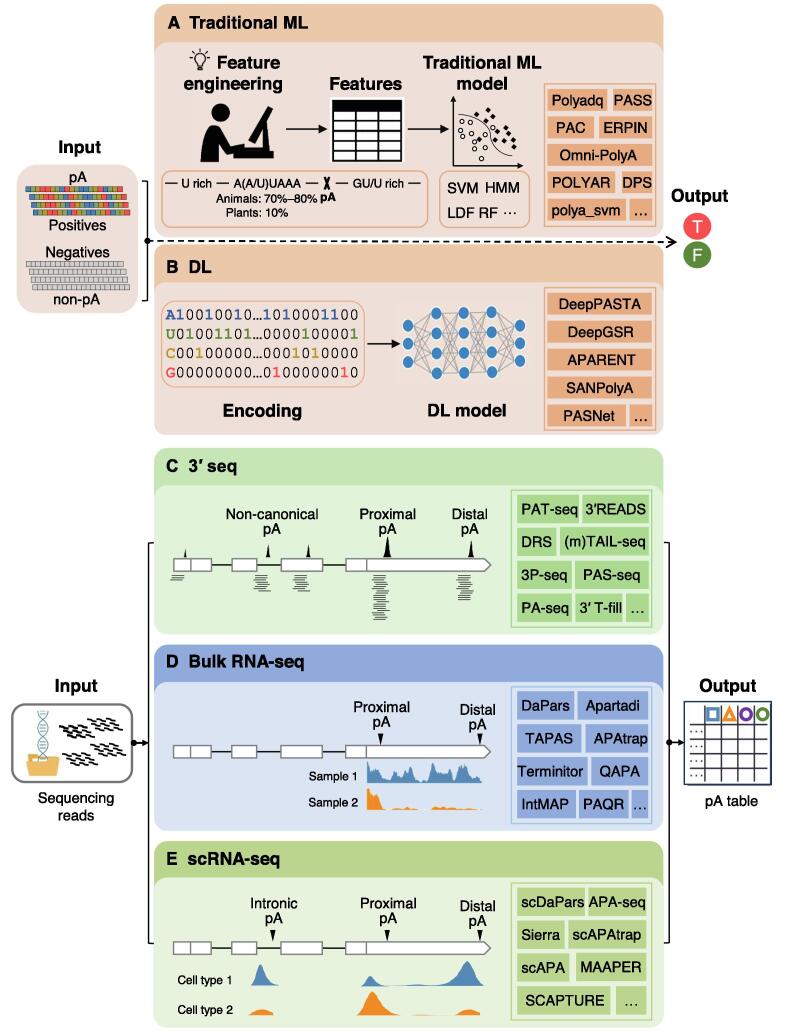
Early studies, dating back to the 1990s, predict pAs using conventional machine learning (ML) models like support vector machine (SVM) [21], [22], [23], [24], [25], which distinguish whether a nucleotide sequence contains a pA using a variety of hand-crafted features (Figure 1A). In recent years, deep learning (DL) models [26], [27], [28], [29] have been shown to provide better performance than traditional ML methods, owing to their great ability for direct and automatic feature extraction and high scalability with large amount of genomic data (Figure 1B). With the advance of next generation sequencing (NGS) technologies, experimental protocols have been designed to capture 3′ ends of mRNAs for direct profiling of genome-wide pAs (Figure 1C), such as DRS [10], [30], 3P-seq [7], [31], 3′READS [11], PAT-seq [32], TAIL-seq [33], [34], and several others (reviewed in [35], [36], [37]). Although these 3′ end sequencing (3′ seq) approaches are powerful and highly sensitive in detecting the precise locations of pAs, even for lowly expressed genes, they are too technically demanding and costly to be widely applied in genomic research. Alternatively, a myriad of computational tools [38], [17], [39], [40] have been developed for identifying and quantifying pAs by leveraging the explosively growing RNA sequencing (RNA-seq) data from diverse biological conditions, cell types, individuals, and organisms (Figure 1D). In recent years, the single-cell RNA sequencing (scRNA-seq) techniques, particularly those 3′ tag-based protocols such as cell expression by linear amplification sequencing (CEL-Seq) [41] and 10× Chromium [42], provide great potential to explore dynamics of APA usage during the process of cellular differentiation. Accordingly, a wide spectrum of tools have been proposed to profile APA from diverse scRNA-seq datasets at cell-type or even single-cell resolution [43], [44], [45] (Figure 1E).
Figure 1.
Schematic of computational approaches for predicting pAs from different kinds of sequencing data
A. Predicting pAs from DNA sequences based on traditional ML models. B. Predicting pAs from DNA sequences based on DL models. C. Identifying pAs from 3′ seq data. D. Predicting pAs from bulk RNA-seq data. E. Predicting pAs from scRNA-seq data. Some representative methods are listed in the text box on the right. pA, poly(A) site; ML, machine learning; T, true; F, false; DL, deep learning; 3′ seq, 3′ end sequencing; RNA-seq, RNA sequencing; scRNA-seq, single-cell RNA sequencing; HMM, hidden Markov model; SVM, support vector machine; LDF, linear discriminant function; RF, random forest.
The tsunami of genomic data especially bulk RNA-seq and scRNA-seq data and the emergence of ensemble DL methodologies have revolutionized computational methods for detecting pAs from diverse kinds of data. In the past decade, a few literature reviews have involved the computational tools for bioinformatic analysis of APA. In 2015, our group summarized computational tools for predicting pAs from DNA sequences and 3′ seq methods for mapping pAs [37]. Szkop and Nobeli [46] described experimental methods for probing 5′ untranslated regions (UTRs) and 3′ UTRs, and listed computational methods for discovering alternative transcription start sites (TSSs) and pAs from microarray and RNA-seq. Yeh et al. [47] reviewed experimental methods and technologies for studying APA, and briefly listed seven RNA-seq tools for analyzing APA dynamics in tabular form. Chen et al. [48] comprehensively reviewed 3′ seq methods for probing pAs, while their review did not cover the computational tools for APA analysis. Gruber and Zavolan [12] highlighted the importance of APA in health and disease, and briefly listed computational resources for studying APA in a table, including four pA databases, two databases of motifs of RNA-binding proteins (RBPs), eight RNA-seq tools for identifying and/or quantifying pAs, and three tools for APA analysis. Our group [49] benchmarked 11 tools for predicting pAs or dynamic APA events from RNA-seq data. Another benchmark study [50] benchmarked five tools for RNA-seq and compared their performance with 3′ seq, full-length isoform sequencing (Iso-Seq), and Pacific Biosciences (PacBio) single-molecule full-length RNA-seq method. Ye et al. [51] briefly summarized three computational methods for detecting APA dynamics from diverse single cell types. Zhang et al. [52] focused on the APA regulation in cancer, and briefly listed 14 computational tools for detecting APA. Kandhari et al. [53] highlighted the emerging role of APA as cancer biomarkers and provided an overview of existing relevant experimental and computational methods. However, these two reviews [52], [53] did not distinguish among the prediction of pAs, detection of APA dynamics, and analysis of APA. For example, APAlyzer [54] and movAPA [55] listed in these reviews are actually toolkits for analyzing APA rather than detecting APA dynamics or pAs, which are different from other tools they listed such as DaPars [17] or APAtrap [39]. Generally, although the aforementioned reviews have provided detailed overviews of the progress in the complex yet fruitful APA field, none of them has exhaustively summarized available tools for different kinds of data in this field, particularly the emerging DL-based methods and methods for scRNA-seq. Moreover, most reviews only briefly listed tools without delicate summary and sorting, which makes it difficult for the scientific community to decide desirable method for their data analysis. In this review, we described the principles of identifying pAs from different kinds of data and provided an extensive overview of available computational approaches. We cataloged these methods into different categories in terms of the underlying principles of the predictive models and the data they used, and summarized their performance and characteristics such as algorithms, features, and data used in the predictive model. Particularly, we examined several representative tools using bulk RNA-seq and scRNA-seq data from peripheral blood mononuclear cells (PBMCs) and put forward operable suggestions on how to assess the reliability of pAs predicted by different tools. We also described several notes on how to conduct objective benchmark analysis for these massive number of tools. Moreover, we proposed practical recommendations on choosing appropriate methods for different scenarios and discussed implications and future directions. Additionally, we highlighted outstanding challenges and opportunities using new ML and integrative multi-omics techniques. Lastly, we provided our perspective on how computational methodologies might evolve in the future for pA prediction, including non-3′ UTR, tissue-specific, cross-species, and single-cell pA prediction.
Computational approaches for pA prediction
Methods for predicting pAs from DNA sequences
The key trigger for cleavage and polyadenylation is the set of cis-regulatory elements surrounding a pA, including A(A/U)UAAA hexamer or variant thereof, the UGUA element, upstream and downstream U-rich elements, and downstream GU-rich elements [56]. Since poly(A) signals (PASs), the core AAUAAA and its variants, are in the vicinity of most mammalian pAs, the identification of the PASs is usually regarded as an alternative to determine the potential position of a pA. In this review, we refer to the task of predicting pAs or PASs as the “pA identification problem”. During the past few decades, a wide range of computational approaches have been proposed to predict pAs from DNA sequences using experimental and in silico mapping of 3′-end expressed sequence tags (ESTs) (Files S1 and S2).
Methods based on traditional ML models
Earlier studies established traditional ML models to classify a sequence as containing a pA or not, using various algorithms (such as discriminant functions [21], [22], [57], hidden Markov model (HMM) [23], SVM [24], [58], Bayesian network [59], artificial neural networks, and random forests [60]) and combined classifiers [25], [61] (Figure 2; File S2). The ML frameworks of these methods are similar, except that different classification models were employed and/or diverse hand-crafted sequence features were compiled (File S1). As ML models rely heavily on manually designed features and the PASs of human/animals are considerably different from that of other species like plants or Saccharomyces cerevisiae (yeast) [37], [62], these ML-based methods can be divided into two categories according to the applicable species (File S1): (1) methods that are applicable to human or animals, including POLYAH [21], Polyadq [57], ERPIN [23], Poly(A) Signal Miner [63], polya_svm [24], PolyApred [58], POLYAR [22], Chang’s model [64], Dragon PolyA Spotter [60], Xie’s model [65], and Omni-PolyA [25]; and (2) methods that are applicable to other species, including the Graber’s method [66] for yeast, POLYA [67] for Caenorhabditis elegans, PASS [68], [69], PAC [59], and PASPA [70] for plants, and Wu’s model for Chlamydomonas reinhardtii [61]. These methods utilize diverse sequence features around pAs for pA prediction (File S1). The most commonly used features are position weight matrix for the poly(A) motifs, distance between motifs, and k-gram nucleotide acid patterns [21], [23], [24], [57], [58]. With the increase of the prior knowledge of DNA sequences, more carefully hand-crafted features were derived, such as Z-curve [59], RNA secondary structures [61], [64], physico-chemical, thermodynamic, and statistical characteristics [60], term frequency–inverse document frequency weight [61], and spectral latent features extracted by HMM [65]. Particularly, since the significance of PASs is different in pAs with different strengths, a few studies divided pAs into sub-groups based on the expression level [22] or pattern assembly [61], and then predicted pAs in each group. In terms of the availability and ease of use of tools, several tools were presented as website (Figure 2), which is particularly convenient for users with little program skill. However, since these tools were generally developed many years ago, the programming languages of many tools are outdated, such as Fortran or Perl, and many tools are no longer available or maintained.
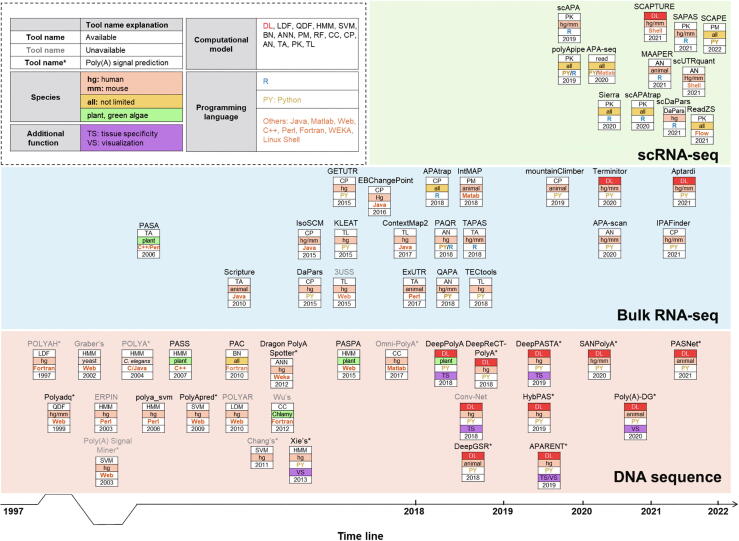
Figure 2.
Landscape of computational approaches for predicting pAsfrom DNA sequences, bulk RNA-seq, and scRNA-seq over time
QDF, quadratic discriminant function; BN, Bayesian network; ANN, artificial neural network; PM, probabilistic model; CC, combined classifier; CP, change point; AN, annotation-based method; TA, transcript assembly; PK, peak calling; TL, transcript assembly and read linking.
Methods based on DL models
Despite considerable progress has been made, the overall accuracy and generalizability of traditional ML-based methods remain moderate due to the limited experimentally verified pAs in the early years and the lack of prior domain knowledge to finely design and acquire useful features. In recent years, DL-based methods are emerging rapidly (Figure 2; File S2), which directly learn hidden features from input nucleotide sequences in a data-driven manner, without knowing any prior knowledge of sequence features. Most methods use convolution neural networks (CNNs), such as DeepPolyA [71], Conv-Net [72], DeeReCT-PolyA [26], DeepPASTA [28], DeepGSR [27], and APARENT [29]. Other DL techniques were also utilized, such as the recurrent neural network (RNN) employed in DeepPASTA [28], a hybrid model with four logistic regression models and eight neural networks used in HybPAS [73], and self-attention mechanisms used in SANPolyA [74] and PASNet [75]. All of these tools were implemented using DL frameworks in Python. In addition to pA prediction, several methods can be utilized for multiple tasks. For example, Conv-Net [72] is capable of inferring pA selection and predicting pathogenicity of polyadenylation variants. DeepPASTA [28] can be used for the prediction of the most dominant pA of a gene in a given tissue and the relative dominance of APA sites in a gene. DeepGSR [27] is able to predict genome-wide and cross-organism genomic signals such as translation initiation sites. APARENT [29] can also be utilized for the quantification of the impact of genetic variants on APA. Different from hand-picked features used in ML-based methods, one-hot encoding features without needing fine feature engineering are widely used in DL-based methods; however, DL-based models are generally of poor interpretability. To enhance the interpretability, several methods provide an additional function for visualization of signals. Xia et al. [26] showed the interpretability of their DeeReCT-PolyA model by transforming convolutional filters into sequence logos for the comparison between human and mouse. In APARENT [29], features learned across all network layers were visualized, which can reveal cis-regulatory elements known to recruit APA regulators and new sequence determinants of polyadenylation. In addition to performance improvement, DL-based methods have two significant advantages over ML-based methods, the higher generalizability for different species and the higher scalability with large amount of data. For example, DeeReCT-PolyA [26] is an interpretable and transferrable CNN model for recognition of 12 PAS variants, which enables transfer learning across datasets and species. APARENT [29] was trained using isoform expression data from more than three million synthetic APA reporters.
Methods for predicting pAs from bulk RNA-seq data
Methods that predict pAs only from DNA sequences conspicuously fail to consider in vivo expression. RNA-seq has become an indispensable approach for transcriptome profiling in diverse biological samples, and a number of methods have been proposed for identifying sample-specific pAs from RNA-seq data (File S3). Our group previously benchmarked 11 representative methods for predicting pAs and/or dynamic APA events from RNA-seq data [49]. Here we focus on prediction of pAs rather than dynamic APA events. We collected relevant methods summarized in our previous review [49] as well as newly emerging methods, and divided these methods into five categories according to their underlying strategies.
Methods that interrogate non-templated poly(A)-capped reads
RNA-seq data contain a small fraction (∼ 0.1%) of non-templated poly(A) tail-containing reads [hereinafter referred to as poly(A) reads] [46], which can be considered as direct evidence for polyadenylation. By interrogating poly(A) reads, an early study [76] identified ∼ 8000 novel pAs in Drosophila melanogaster from a total of 1.2 billion RNA-seq reads. Several other methods, such as KLEAT [77] and ContextMap 2 [78], not only employed direct evidence from poly(A) reads but also incorporated transcript assembly to identify pAs. These poly(A) read-based approaches have the advantage to determine the precise locations of pAs; however, it is still challenging to discover pAs of weakly expressed transcripts due to the decreased read coverage near the 3′ end and the low yield of poly(A) reads.
Methods based on transcript assembly
Another series of approaches identify pAs from inferred alternative 3′ UTRs by compiling transcript structures from RNA-seq data, including PASA [79], Scripture [80], 3USS [81], and ExUTR [82]. These transcriptome assembly-assisted methods deduce gene models first using transcriptome assembly tools, and then identify 3′ UTRs that are absent in the deduced gene models, which rely heavily on assembled gene structures. It is widely accepted that transcriptome assembly from RNA-seq data is a rather difficult and computationally demanding task, and it is more challenging to precisely determine 3′ UTRs, especially for lowly expressed genes, due to 3′ biases of read coverage inherent in RNA-seq. Therefore, the performance of these methods is inevitably hindered by potential limitations of existing transcriptome assembly tools.
Methods that rely on prior annotations of pAs
During the last decade, numerous experimental techniques have been developed to directly sequence 3′ ends of mRNAs, such as 3′ T-fill [83], 3′READS [11], TAIL-seq [33], [34], to name a few (Figure 1C). Accordingly, several pA databases built upon 3′ seq data of diverse species were continuously released, including PolyA_DB 3 [84], PolyAsite 2.0 [8], and PlantAPAdb [85]. These databases provide a large number high-confidence pAs, which can be used for establishing pA prediction models and evaluating pA prediction results. It is thus naturally to incorporate annotated pAs for predicting pAs from RNA-seq data. Several methods that rely on pre-defined pA annotations, including QAPA [38], PAQR [86], and APA-scan [87], were proposed for predicting pAs from RNA-seq data. For these methods, the quality of annotated pAs is particularly critical. Most studies establish a comprehensive compendium of well-annotated pAs by merging non-redundant annotations from diverse sources. By combining prior annotated pAs with RNA-seq data, the quality of predicted pAs can be greatly improved. However, currently available pA databases are far from complete and limited to only a few well-studied species, such as human, mouse, and Arabidopsis thaliana. Consequently, these tools are not capable of detecting novel pAs beyond existing poly(A) annotations.
Methods that infer pAs by detecting significant changes in RNA-seq read density
The majority of recent approaches predict pAs by modeling read density changes in terminal exons, including GETUTR [88], IsoSCM [89], DaPars/DaPars2 [17], [90], [91], EBChangePoint [92], APAtrap [39], TAPAS [40], moutainClimber [93], and IPAFinder [94]. According to our previous benchmark on 11 tools for RNA-seq [49], TAPAS generally obtained higher sensitivity than other tools across different datasets. Of note, unlike most methods that require at least two samples for change point detection, moutainClimber [93] is a de novo cumulative-sum-based approach, which runs on a single RNA-seq sample and simultaneously recognizes multiple TSSs or APA sites in a transcript. Using mountainClimber, Cass and Xiao analyzed 2342 genotype-tissue expression (GTEx) samples from 36 tissues of 215 individuals and found 75% of genes exhibited differential APA across tissues [93]. Different from most pA prediction tools focusing mainly on 3′ UTR, IPAFinder was specifically proposed for identifying intronic pAs from RNA-seq data [94]. Zhao et al. applied IPAFinder to pan-cancer datasets across six tumor types and discovered 490 recurrent dynamically changed intronic pAs [94]. Methods falling within this category rely on the detection of read density fluctuations which requires sufficient read coverage in terminal exons to detect APA sites. It is worth noting that data pre-processing (normalization or smoothing) is particularly important for reducing technical biases caused by non-biological variability [46]. Particularly, some methods, such as APAtrap and DaPars, re-define terminal exon boundaries based on RNA-seq read coverage before identifying pAs, which are capable of detecting pAs in previously unannotated regions.
Methods based on ML models
In recent years, some newly emerging methods employ traditional ML or DL model to identify pAs from RNA-seq, including TECtools [95], IntMAP [96], Terminitor [97], and Aptardi [98]. TECtools [95] first identifies terminal exons and transcript isoforms ending at known intronic pAs. Then a model was trained based on the aligned RNA-seq data for distinguishing terminal exons from internal exons and background regions, using diverse features reflecting differences in read coverage of these regions. TECtool can also be applied on scRNA-seq, which first pools reads of all cells to infer new transcripts and then quantify each transcript in individual cells. IntMAP [96] leverages one unified ML framework to combine the information from RNA-seq and 3′ seq to quantify different 3′ UTR isoforms using a global optimization strategy. Terminitor [97] is based on a deep neural network for three-label classification problem, which can determine whether an input sequence contains a pA with PAS, a site without PAS, or non-pA. Aptardi [98] is a multi-omics approach based on bidirectional long short-term memory (biLSTM) RNN, which predicts pAs by leveraging DNA sequences, RNA-seq data, and the predilection of transcriptome assemblers.
Methods for predicting pAs from scRNA-seq data
scRNA-seq is a powerful high-throughput technique for interrogating transcriptome of individual cells and measuring cell-to-cell variability in transcription [99]. Particularly, several 3′ tag-based scRNA-seq methods enriching for mRNA 3′ ends via poly(A) priming, such as CEL-Seq [41], Drop-seq [100], and 10× Chromium [42], provide great potential to dissect APA at the single-cell resolution. However, the extremely high dropout rate and cell-to-cell variability inherent in scRNA-seq make it difficult to directly apply bulk RNA-seq methods to scRNA-seq data. During the last few years, a wide range of computational approaches specifically designed for pA identification from scRNA-seq data have emerged (Figure 2; File S4). We divided these methods into three categories according to their underlying strategies.
Methods based on peak calling
The peak calling strategy is widely used by most methods for pA identification from scRNA-seq data, including scAPA [101], polyApipe (https://github.com/MonashBioinformaticsPlatform/polyApipe), Sierra [43], scAPAtrap [44], SAPAS [102], and SCAPE [103]. The underlying principle of these methods is that aligned reads from 3′ tag-based scRNA-seq accumulate to form peaks at genomic intervals upstream of pAs [101]. In scAPA [101], a set of non-overlapping 3′ UTRs is first defined from the genome annotation and then peaks within 3′ UTRs are identified using an existing peak calling tool. As adjacent pAs may situate in a single peak, the Gaussian finite mixture model was implemented in scAPA to split large peaks into smaller ones. polyApipe is a pipeline for identifying pAs from 10× Chromium scRNA-seq data, which defines peaks of poly(A)-containing reads. Sierra [43] employs the splice-aware peak calling based on Gaussian curve fitting to determine potential peaks with pAs and then the peaks are annotated and quantified in individual cells. Our group proposed scAPAtrap [44] for identifying and quantifying pAs in individual cells from 3′ tag-based scRNA-seq. scAPAtrap incorporates a genome-wide sensitive peak calling strategy and poly(A) read anchoring, which can accurate locate pAs without using prior genome annotation, even for those with very low read coverage. Yang et al. proposed SAPAS for identifying pAs from poly(A)-containing reads and quantifying pAs in peak regions determined by a parametric clustering algorithm [102]. They further applied SAPAS to the scRNA-seq data of GABAergic neurons and detected cell type-specific APA events and cell-to-cell modality of APA for different GABAergic neuron types. Very recently, Zhou et al. proposed the SCAPE method based on a probabilistic mixture model for identification and quantification of pAs in single cells by utilizing insert size information [103]. The parametric modeling of peaks in most tools based on peak calling such as scAPA and Sierra may cause biases and reduce statistical power in detecting APA events. Alternatively, ReadZS [104], an annotation-free statistical approach, was proposed to characterize read distributions that bypasses parametric peak calling and identifies differential APA usages at single-cell resolution among ≥ 2 cell types. ReadZS can not only detect pAs in normal peak shape, but also identify distributional shifts that are not.
Methods that rely on prior annotations of pAs
In contrast to the peak calling-based methods used for de novo pA identification, a few approaches identify pAs base on prior pA annotations, including MAAPER [105], SCAPTURE [106], and scUTRquant [107]. Li et al. developed MAAPER [105] for predicting pAs from both bulk RNA-seq and scRNA-seq data, which incorporates annotated pAs in PolyA_DB 3 [84] and pools single cells of the same type to mimic pseudo-bulk samples. MAAPER also provides a likelihood-based statistical framework for analyzing APA changes and can identify common and distinct APA events in cell groups from different individuals. The group of MAAPER later developed SCAPTURE [106] which embedded a DL model DeepPASS for evaluating called peaks from scRNA-seq. The DL model was trained by sequence shifting, using annotated pAs from PolyA_DB 3, PolyA-seq, PolyASite 2.0, and GENCODE v39. The authors used SCAPTURE to profile APA dynamics between COVID-19 patients and healthy individuals, and found the preference of proximal pA usage in numerous immune response-associated genes upon SARS-CoV-2 infection. Fansler et al. developed scUTRquant [107] for measuring 3′ UTR isoform expression from scRNA-seq data, which relies on a cleavage site atlas established from GENCODE annotation and a mouse Microwell-seq dataset of 400,000 single cells [108].
Other methods for predicting pAs from scRNA-seq data
Additionally, some other methods do not use the peak calling strategy, including APA-seq [109] and scDaPars [45]. Levin et al. [109] designed the APA-seq approach to detect and quantify pAs from CEL-Seq, which interrogates the gene identity and poly(A) information in the paired read 1 and read 2. Although APA-seq is in principle applicable to other 3′ tag-based scRNA-seq methods, it may not be universally applied in practice in that only sample barcodes rather than the whole 3′ end sequence of the transcript are retained in read 1 of many public scRNA-seq data [44]. Unlike most tools that are only applicable to 3′ tag-based scRNA-seq, scDaPars [45] that was proposed by the group of DaPars [17] can identify and quantify APA events from either 3′ tag (e.g., 10× Chromium) or full-length (e.g., Smart-seq2) scRNA-seq. In the scDaPars pipeline, DaPars, a tool for identifying APA events from bulk RNA-seq, was first adopted to calculate raw relative APA usage in individual cells, and then a regression model was utilized to impute missing values in the sparse single-cell APA usage matrix. By applying scDaPars to cancer and human endoderm differentiation data, Gao et al. revealed cell type-specific APA regulation and detected novel cell subpopulations that were not found in conventional gene expression analysis.
Methods for APA analysis rather than pA prediction
In addition to the task of pA prediction (hereinafter termed task 1), there are additional tasks related to the bioinformatic analysis of APA, mainly including the prediction of tissue-specific pAs (task 2), prediction of dominant pAs (task 3), prediction of APA site switching (task 4), and other kinds of APA analysis (task 5). Although most tools described in this review are developed for task 1, several tools are capable of performing multiple tasks. For example, DeepPASTA [28] is able to perform tasks 1–3; Conv-Net [72] can perform tasks 1 and 3. In this review, we focus only on tools that are applicable to task 1. Of note, NGS-based techniques specially designed for probing pAs, generally known as 3′ seq, such as DRS [10], [30], 3P-Seq [7], [31], and 3′READS [11], are experimental methods rather than computational methods for identifying pAs. Genome-wide pAs generated from 3′ seq are highly confident and are usually regarded as the true reference (i.e., prior information) for building models or evaluating computational methods. These 3′ seq methods are beyond the scope of this review, while have been reviewed in several other reviews [12], [46], [48], [53]. In addition, we have briefly summarized tools or resources designed for APA analysis rather than pA prediction in File S5. Tools such as DeeReCT-APA [110], polyA code [111], and TSAPA [112] are not targeted at task 1 but for tasks 2 and 3. Among the five tasks, detection of APA site switching (task 4) is usually a routine step involved in the analysis of RNA-seq or scRNA-seq data. APA site switching reflects the differential usage of APA sites between samples, which does not necessarily need the prediction of pAs (task 1) as a prerequisite. Of note, there are other commonly used phrases similar to “APA site switching” mentioned in this review, such as differential APA site usage [8], [17], 3′ UTR shortening/lengthening [44], [101], and APA dynamics [17], [44], [98], [113]. Some approaches for RNA-seq, such as PHMM [114], ChangePoint [115], MISO [116], and roar [117], directly discover APA site switching by detecting sudden change of read density at terminal exons without identifying APA sites. Recently, several tools have been developed for scRNA-seq, such as SCUREL [118], scMAPA [119], and scDAPA [120]. For example, our group developed scDAPA [120] for characterizing differential usages of APA in different cell types using 10× Chromium data, and found that APA plays an important role in acute myeloid leukemia [113]. Additionally, some toolkits have been developed for routine analyses of APA (e.g., annotation and visualization, task 5) using annotated pAs and/or RNA-seq, such as APAlyzer [54] and movAPA [55], while they are not capable of predicting pAs. These diverse tools provide a wide range of complementary resources and opportunities to address the more complex but fruitful field of APA.
Discussion
Performance of pA prediction models
At present, there are only a few benchmark studies that systematically evaluate the performance of different tools. Previously, our group benchmarked 11 tools for RNA-seq [49] and found that the sensitivity of some methods varied greatly among different species. For instance, QAPA [38] performs the second best on human data, while it performs the worst on mouse data. APAtrap [39] is the top performer for Arabidopsis data, while TAPAS [40] performs the best on human or mouse data. Recently, Shah et al. [50] benchmarked five tools for RNA-seq against 3′ seq, Iso-Seq, and a full-length RNA-seq method and found that pAs from 3′ seq and Iso-Seq are more reliable than pAs predicted from RNA-seq. They suggested that incorporating the RNA-seq prediction tool QAPA [38] with pA annotations derived from 3′ seq or Iso-Seq can reliably quantify APA dynamics across conditions.
The performance of different tools described in the respective studies is summarized in Files S1–S4. Generally, for predicting pAs from DNA sequences, DL-based models significantly outperform ML-based methods and are more suitable for large-scale analysis, owning to the good ability of automatic feature extraction and scalability for big data analysis (File S2). For example, DeepPASTA [28] has an area under the curve (AUC) score over 93% in predicting pAs on a DNA sequence dataset, which performs much better than ML-based tools like PolyAR [22] or Dragon PolyA Spotter [60]. APARENT [29], based on deep neural network, was trained on over three million synthetic APA reporter genes, which overcomes inherent size limitations of traditional biological datasets. In contrast, traditional ML-based methods like POLYAR [22] and Omni-PolyA [25] require a considerable amount of prior knowledge and are unable to cope with the rapidly growing data. In terms of the model generalizability, methods for RNA-seq or scRNA-seq are generally applicable to different species if the reference genome and the genome annotation are available. In contrast, the cross-species applicability of methods for DNA sequences is more complex. Models applied to human are normally applicable to other mammals like mouse due to similar PASs among mammals [56]. However, although most models can be in principle trained using data from a different species, users need collect training data from the other species which are not always available, and most models use hand-crafted features that may not be generalized well across species. Recent techniques like DL and transfer learning greatly enhance the generalizability of models. Cross-species experiments have been performed for evaluating the generalizability of some tools, such as DeepGSR [27] and Poly(A)-DG [121], and for these tools single model trained over one species can be generalized well to datasets of other species without retraining. We need to point out that, the evaluation results in a single study may be biased and should be treated with caution, because different datasets and performance indicators were used for the performance evaluation in different studies (Files S1–S4). In the following section of “Conclusion and prospects”, we also put forward several notes on how to conduct more objective benchmarking in order to make a fairer comparison of different tools.
How reliable are the obtained results?
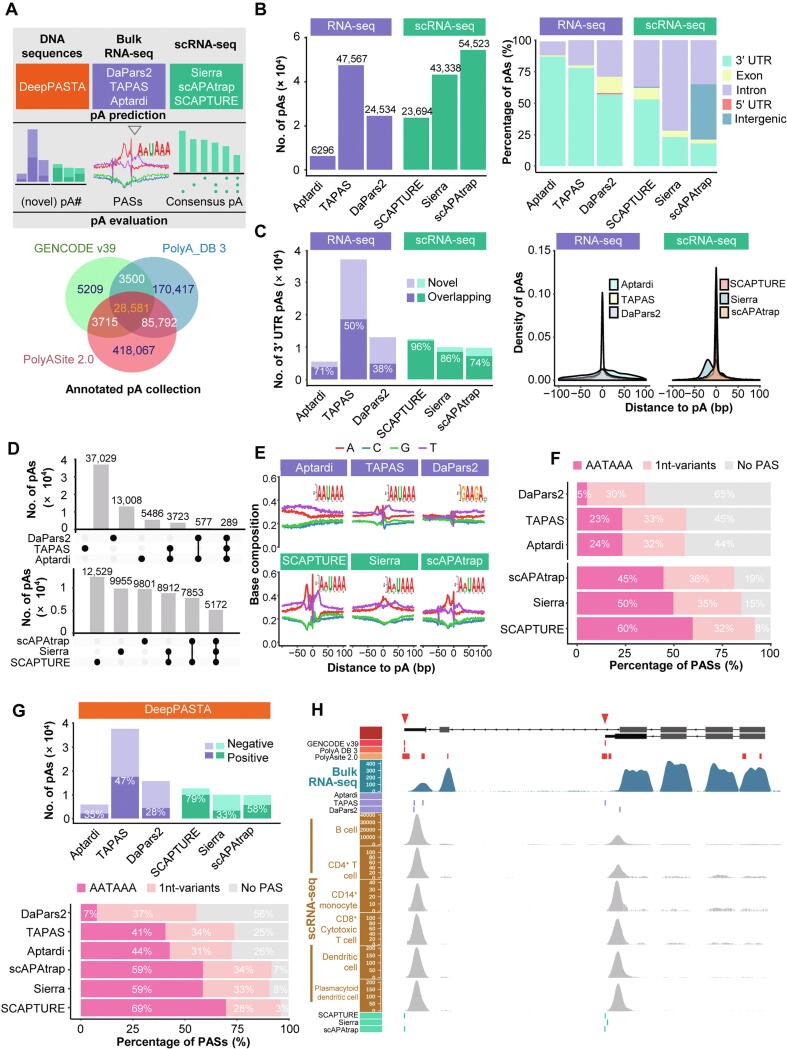
Currently, there is no benchmark evaluation of tools for DNA sequences or scRNA-seq data. Here we attempted to make a preliminary examination of the reliability of results obtained from different pA prediction tools, using a matched bulk RNA-seq and 10× Chromium scRNA-seq data of human PBMCs (File S6). We chose representative tools from each category, including DaPars2 [90], TAPAS [40], and Aptardi [98] for bulk RNA-seq data, Sierra [43], scAPAtrap [44], and SCAPTURE [106] for scRNA-seq data, and DeepPASTA [28] for DNA sequences (Figure 3A, top). We collected a total of 676,424 non-redundant pAs from GENCODE v39, PolyASite 2.0, and PolyA_DB 3, which were compiled from 3′ seq and can be used as the true reference (Figure 3A, bottom). The number of pAs predicted by different tools, even those under the same category, varied greatly (Figure 3B, left). For example, the numbers of pAs predicted from bulk RNA-seq by TAPAS and DaPars2 were nearly 8 times and 4 times that of Aptardi. The numbers of pAs predicted from scRNA-seq by Sierra and scAPAtrap were about twice that of SCAPTURE. Of note, scAPAtrap can predict pAs for the whole genome including intergenic regions, and all the three tools predict a large number of pAs in introns (Figure 3B, right). If only 3′ UTR regions are considered, the numbers of pAs predicted by the three scRNA-seq tools were much closer (Figure 3C, left). As most tools only identify pAs in 3′ UTR, here we used 3′ UTR pAs for subsequent evaluation. Next, we assessed the authenticity of the predicted pAs by checking whether they are supported by annotated pAs in the true reference. The overlap of pAs predicted from RNA-seq with annotated pAs is much lower than that of scRNA-seq (Figure 3C, left). Particularly, the overlap rate between pAs predicted by SCAPTURE and annotated pAs was as high as 96%, which may be because that the DL model embedded in SCAPTURE was trained with annotated pAs. The positions of pAs predicted by TAPAS, SCAPTURE, and scAPAtrap were much more precise than those by other tools (Figure 3C, right). Further, we examined the consistency of the results predicted by different tools. Generally, the consistency among different tools was very low (Figure 3D). For bulk RNA-seq data, only 289 pAs were identified by all the three tools, whereas the vast majority of pAs were identified exclusively by a single tool (Figure 3D, top). In contrast, the consistency of pAs predicted from scRNA-seq data by different tools was relatively higher (Figure 3D, bottom). In addition, we assessed the reliability of predicted pAs by investigating sequence features. The single nucleotide profiles around pAs predicted by TAPAS, SCAPTURE, and scAPAtrap resembled the general profile [49] (Figure 3E), which is also consistent with the fact that they determine more precise locations for pAs (Figure 3C, right). The percentage of AATAAA around pAs predicted from scRNA-seq was much higher than that from bulk RNA-seq (Figure 3F), indicating that predicted pAs from scRNA-seq tend to be more reliable and more accurate than those from bulk RNA-seq. Next, we used the pA prediction tool for DNA sequences, DeepPASTA, to examine how many pAs identified from bulk RNA-seq and scRNA-seq data were predicted as true solely based on the sequence characteristics. We extracted the upstream and downstream sequences of pAs predicted by RNA-seq tools as the input for DeepPASTA. The proportion of pAs obtained by different tools to be predicted as true by DeepPASTA was not high and varied greatly, ranging from 28% to 79% (Figure 3G, top), indicating again the low overlap of pAs predicted by different tools. Considering only positive pAs by DeepPASTA, the percentage of AATAAA and 1-nt variants of different tools increased slightly (Figure 3G, bottom vs. Figure 3F), reflecting that positive pAs confirmed by DeepPASTA are relatively more reliable than negative ones. Finally, we examined predicted pAs of the immunoglobulin M heavy chain (IGMM) gene, which was reported to be expressed as a secreted form using the proximal pA and the membrane-bound form using the distal one [122]. The proximal pA of IGHM has been recently found preferentially used in B cells and plasma cells of COVID-19 samples [106]. SCAPTURE and scAPAtrap predicted the precise locations of both proximal and distal pAs from scRNA-seq data, while Sierra only predicted the proximal one (Figure 3H). TAPAS predicted three pAs from bulk RNA-seq data, of which two perfectly matched the reference pAs in PolyASite 2.0. In contrast, Aptardi failed to predict any pA for this gene and DaPars2 predicted two pAs yet not verified by reference pAs.
Figure 3.
Comparison of representative tools for predicting pAs frommatched bulk RNA-seq and scRNA-seq data of human PBMCs
A. Schematic of the benchmark (top) and the collection of reference pAs from GENCODE v39, PolyASite 2.0, and PolyA_DB 3 (bottom). B. Number of pAs obtained by different tools (left) and distribution of pAs in different genomic regions (right). C. Overlap of 3′ UTR pAs predicted by different tools with reference pAs (left) and distribution of distance from predicted 3′ UTR pAs to reference pAs (right). D. Overlap of 3′ UTR pAs predicted by different tools from bulk RNA-seq data (top) and scRNA-seq data (bottom). E. Single nucleotide profile around 3′ UTR pAs predicted by different tools. For each tool, the sequence logo of the most dominant motif around the pA identified by DREME was also shown. F. Percentage of AATAAA and 1-nt variants around pAs predicted by different tools. G. The number of pAs obtained by different tools to be predicted as positive or negative by DeepPASTA (top) and the percentage of AATAAA and 1-nt variants around positive pAs (bottom). The upstream and downstream sequences of pAs predicted by each RNA-seq tool were extracted as the input for DeepPASTA. H. Predicted 3′ UTR pAs by different tools for the IGHM gene. Tracks from top to the bottom are gene model, reference pAs from three databases, read coverage from bulk RNA-seq, predicted pAs from bulk RNA-seq data, read coverage for each cell type of scRNA-seq, and predicted pAs from scRNA-seq data. The red triangles on the chromosome strip highlight the two representative pAs of IGHM. PBMC, peripheral blood mononuclear cell; PAS, poly(A) signal; UTR, untranslated region; IGHM, immunoglobulin M heavy chain.
Although this preliminary benchmark is far from objective or exhaustive to reflect the advantages and disadvantages of different tools, it reveals several potential issues when using the results obtained by different pA prediction methods. First, although a considerable number of pAs are identified by most tools, the overall prediction accuracy and sensitivity of these tools are low (Figure 3C). Our previous comparative study [49] on tools for bulk RNA-seq have also revealed that a considerable number of predicted pAs are not annotated in 3′ seq, and the overall prediction accuracy of these tools, even the best one, TAPAS, is not high (40%–60% for human/mouse data). It is still challenging to determine whether a pA not present in prior annotations is false or novel. We anticipate that at least part of predicted pAs that are not overlapping with annotated ones may potentially be true due to that the current pA annotations are still far from complete. Second, the number of pAs identified by different tools, either for bulk RNA-seq or scRNA-seq, varies greatly, and the consensus of results obtained by different tools is limited (Figure 3D). This is also similar to the observation in our previous benchmark that each tool predicts an independent set of pAs and the overlap of results from different tools is extremely low (< 7% for human/mouse data) [49]. Third, as some tools incorporate additional information to predict pAs, e.g., prior pAs used by SCAPTURE and poly(A) reads used by scAPAtrap, the resolution of pAs predicted by different tools varies greatly (Figure 3C and E). Fourth, 21%–72% of the predicted pAs by different tools are not recognized as true pAs based on their sequence features (Figure 3G). Fifth, although scRNA-seq data suffers from extremely high level of noise and sparsity, prediction results from scRNA-seq seem to be more reliable and consistent than those from bulk RNA-seq (Figure 3C, D, and F). However, this is not unexpected because it may be less challenging to computationally predict pAs from the 3′ tag-based scRNA-seq data than the full-length-based bulk RNA-seq data. Still, further benchmark study with more complete prior annotations, diverse datasets, and performance indicators is needed in order to assess the results obtained from different tools more fairly and objectively.
Here we try to give some operable suggestions on how to obtain high-confidence pAs. The most straightforward way may be making a consensus set of pAs that are predicted by multiple tools; however, this may result in a relatively small number of pAs due to the limited overlap between different tools. Another way is to obtain the intersection of predicted sites and real sites, using annotated pAs that are manually curated and available in several databases such as PolyASite 2.0 and PolyA_DB 3. However, it should be noted that these annotated data sources are compiled from limited biological samples and species; they are far from complete to cover all real sites especially tissue-specific ones. Similar to our benchmark analysis on bulk RNA-seq and scRNA-seq PBMCs (Figure 3), users can also use data from another omics from similar biological samples, if available, to predict pAs for mutual verification. In addition, since many sequence motifs, e.g., AAUAAA and its variants, have been reported to have a positional preference relative to the pA, it is naturally to examine sequence patterns surrounding each predicted pA to get pAs with explicit PASs. This is particularly useful for assessing the authenticity of pAs from animals because AAUAAA and its 1-nt variants appeared in > 90% of animal pAs [8]. In contrast, AAUAAA only accounts for < 10% of pAs in plants, and therefore it is not practical to validate plant pAs through sequence features. Moreover, the general single nucleoside compositions surrounding pAs in different species have been clearly reported, so we can inspect the base composition around predicted pAs. Of note, this way is applicable to evaluation of the overall quality of the pAs, while it cannot be used to assess the reliability of a single pA. The movAPA package [55] can be used for most of the aforementioned quality assessments.
Practical guidelines for choosing appropriate methods
Based on the summary of different methods (Files S1–S4), we attempted to choose representative tools from each category and propose a set of practical guidelines for users (Table 1). As methods in different categories use different kinds of data as the input, the choice of the method first depends on the users’ own data. For bulk RNA-seq data, the choice of the method should be mainly driven by the availability of pA annotations. For scRNA-seq data, the choice of the method mainly depends on the protocol of the scRNA-seq (e.g., 3′ tag or full-length) and the availability of pA annotations. For methods predicting pAs from DNA sequences, the choice of the method should be primarily driven by the algorithm used, DL or traditional ML. Particularly, for cross-species pA prediction from DNA sequences, users should pay extra attention to whether they need to retrain the model for individual species, which may require users to have certain programming ability. Additionally, several tools are in the form of web servers, providing a portable platform for predicting pAs from DNA sequences for researchers with limited programming ability. Several other factors also affect the choice of methods, such as the availability of the tool or code, the popularity, the ease of use, the clarity of documentation, and the scale of the data. When predicting pAs on a dataset of interest, it is important to further consider two points. First, it is critical that the obtained pAs and/or the downstream results (e.g., differential APA events) are confirmed by multiple pA prediction methods. This is to ensure that the prediction is not biased due to predefined parameter settings or the specific algorithm used in the method. The merit of using different methods is also demonstrated by the benchmark results in previous studies [49], [50] and in this study (Figure 3), which show substantial complementarity between different methods. Second, even if prior pA annotations are available, it can be also beneficial to try out methods that do not rely on prior annotations. When predicted pAs, even a small portion, are confirmed using such a different method, it provides users with additional evidence.
Table 1.
Recommended tools for predicting pAs from DNA sequences, bulk RNA-seq, and scRNA-seq
| Category | Tool | Year | Description | Refs. |
|---|---|---|---|---|
| Web servers for predicting pAs from DNA sequences | Dragon PolyA Spotter | 2012 | A web server for predicting 12 poly(A) motifs from human DNA sequences, using an artificial neural network and a random forest | [60] |
| PolyApred | 2009 | An SVM-based web server for predicting 13 poly(A) motifs in human, using sequence features of different types of nucleotide frequencies and binary pattern | [58] | |
| Polyadq | 1999 | An early web server based on two quadratic discriminant functions for predicting AAUAAA/AUUAAA signals, using features encoded by position weight matrix | [57] | |
| DL-based tools for predicting pAs from DNA sequences | PASNet | 2021 | A hybrid DL framework for identifying 16 poly(A) motifs in different species, which integrates gated convolutional highway networks with self-attention mechanisms | [75] |
| SANPolyA | 2020 | A self-attention DL model for predicting 18 poly(A) motifs in human and mouse | [74] | |
| HybPAS | 2019 | A hybrid model for predicting 12 poly(A) motifs in human, using eight neural networks and four logistic regression models | [73] | |
| APARENT | 2019 | The model trained on isoform expression data from more than three million synthetic APA reporters | [29] | |
| DeepPASTA | 2019 | A model based on CNN and RNN for predicting pAs from both sequence and RNA secondary structure | [28] | |
| DeeReCT-PolyA | 2018 | A transferrable CNN model for recognition of 12 poly(A) motifs, which enables transfer learning across datasets and species | [26] | |
| DeepGSR | 2018 | An approach based on CNN and one-hot features to predict genome-wide and cross-organism genomic signals and regions | [27] | |
| DeepPolyA | 2018 | A model for predicting pAs in Arabidopsis with one-hot encoding features | [71] | |
| Traditional ML-based tools for predicting pAs from DNA sequences | PASS | 2007 | A GHMM-based model for predicting pAs in plants | [68] |
| polya_svm | 2006 | An SVM-based tool for predicting pAs using position-specific scoring matrices to score 15 cis-regulatory elements | [24] | |
| Methods for bulk RNA-seq that rely on prior annotations of pAs | QAPA | 2018 | It compiles an expanded compendium of known pA annotations for identifying and quantifying pAs, which was suggested by Shah et al. [50] to be used in combination with pAs derived from 3′ seq or Iso-Seq | [38] |
| PAQR | 2018 | It uses read coverage to segment 3′ UTRs at annotated pAs | [86] | |
| Methods for bulk RNA-seq that based on detecting changes in read density | moutainClimber | 2019 | It runs on a single RNA-seq sample and can recognize multiple TSSs or pAs | [93] |
| APAtrap | 2018 | It can detect all pAs along the 3′ UTR and can be used to improve 3′ end annotations | [39] | |
| TAPAS | 2018 | It adopts a method originally used for time-series data to detect change points, which was suggested to have overall high performance in several benchmark studies [49], [50] | [40] | |
| DaPars, DaPars2 | 2014, 2018 | DaPars is probably the first and the most widely used tool for bulk RNA-seq and DaPars2 is its updated version | [17], [90], [91] | |
| Methods for bulk RNA-seq that based on ML models | Aptardi | 2021 | A multi-omics DL-based approach for predicting pAs by leveraging DNA sequences, RNA-seq, and the predilection of transcriptome assemblers; however, its sensitivity may be low according to our preliminary test (Figure 3) | [98] |
| Terminitor | 2020 | A DL-based model for three-label classification problem, which determines a poly(A) cleavage site, a non-polyadenylated cleavage site, or non-cleavage site | [97] | |
| TECtool | 2018 | It is based on transcriptome assembly and prior pA annotations, and can predict novel terminal exons | [95] | |
| Methods for predicting pAs from scRNA-seq | scDaPars | 2021 | It is applicable to both full-length and 3′ tag scRNA-seq, which uses DaPars to infer pAs and may be slow for large-scale scRNA-seq | [45] |
| MAAPER | 2021 | An annotation-assisted method for both bulk RNA-seq and 3′ tag scRNA-seq data, which incorporates prior pAs in the PolyA_DB for identifying pAs in 3′ UTRs and introns | [105] | |
| SCAPTURE | 2021 | An annotation-assisted pipeline that implements a DL model to evaluate called peaks from 3′ tag scRNA-seq, using prior pAs from four databases for model training | [106] | |
| scUTRquant | 2021 | An annotation-assisted method that incorporates pA atlas established from a mouse full-length Microwell-seq dataset of 400,000 single cells [108] for filtering pAs predicted from 3′ tag scRNA-seq | [107] | |
| SCAPE | 2022 | A peak calling-based method based on a probabilistic mixture model for identification and quantification of pAs in 3′ tag scRNA-seq by utilizing insert size information | [103] | |
| ReadZS | 2021 | A statistical approach to characterize read distributions that bypasses parametric peak calling and identifies pAs from 3′ tag scRNA-seq | [104] | |
| scAPAtrap | 2020 | A peak calling-based method that incorporates poly(A) reads for genome-wide pA prediction from 3′ tag scRNA-seq | [44] | |
| Sierra | 2020 | A splice-aware peak calling-based method that can identify pAs in 3′ UTRs and introns from 3′ tag scRNA-seq | [43] |
Note: Tools are chosen based on criteria such as availability, function, ease of use, and popularity. pA, poly(A) site; ML, machine learning; SVM, support vector machine; GHMM, generalized hidden Markov model; CNN, convolution neural network; RNN, recurrent neural network; scRNA-seq, single-cell RNA sequencing; 3′ seq, 3′ end sequencing; Iso-Seq, isoform-sequencing; RNA-seq, RNA sequencing; DL, deep learning; UTR, untranslated region; TSS, transcription start site.
Conclusion and prospects
Challenges in improving the performance of pA prediction
The field of pA prediction is progressing rapidly, primarily in the aspects of using DL models and predicting pAs at the single-cell resolution. However, the overall accuracy, sensitivity, and specificity of currently available methods remain moderate (Figure 3). The coming flood of extensive sequencing data, especially multi-omics and single-cell data, will provide new opportunities but also demand new computational methods to exploit this new information. Potential challenges of improving the prediction performance include but are not limited to: paucity of annotated pAs covering diverse tissues and species; mis-assemblies caused by the low complexity of 3′ UTR sequences; mis-alignment of short reads or incomplete sequence coverage near 3′ ends; difficulty in capturing pAs in low-expression genes; poor knowledge on primary, secondary, or higher structure information of PASs, particularly in plants; gaps in our knowledge on understanding APA regulators in different omics layers; limited success in integrating the quantitative features from multiple omics layers; lack of transferrable intelligent methods for cross-species prediction; lack of interpretability in models based on deep neural networks; hurdle in constructing negative datasets due to the prevalence of unconventional pAs in coding sequences (CDSs) and introns; difficulty in identifying multiple pAs anywhere in a transcript; and lack of effective algorithms to deal with the extremely high isoform-level dropout rate and noise inherent in scRNA-seq. Furthermore, higher standards for software quality assurance and documentation would help improve the ease of use of these tools and facilitate their application in the broader community. Finally, new algorithms should be designed to cope with ever-increasing amount of different kinds of data, especially the explosion in single-cell data with multi-omics features.
Notes on benchmarking different methods for predicting pAs
Till now, there are few reports on the exhaustive evaluation of computational tools for predicting pAs. Previously, our group benchmarked 11 representative tools for predicting pAs and/or dynamic APA events from RNA-seq [49]. Lately, Shah et al. [50] evaluated five tools for RNA-seq against 3′ seq, Iso-Seq, and a full-length RNA-seq method in identifying pAs and quantifying pA usage. However, there is no study to provide an exhaustive evaluation of existing tools for pA prediction from different kinds of data, particularly those tools for scRNA-seq. Here we attempt to give some notes on benchmarking analysis in this field. First, the real pA dataset is very critical for performance evaluation; however, the reference datasets used in different studies are quite different. Therefore, it is imperative to compile reliable reference datasets with uniform standards. In particular, RNA-seq or scRNA-seq data are sample-specific, so the reference pA dataset from matched samples should also be considered. Moreover, due to the paucity of real pA datasets at the single-cell level, possible deviations need to be considered when using real pA data from bulk data for evaluation. For example, pAs exclusively recognized in single cells may be authentic pAs from rare transcripts or rare cells, even though they may not be present in the bulk pA reference. Second, most tools were evaluated using data only in mammals (mainly human and mouse), and therefore the scalability of these tools in different species, especially their applicability to plants, needs to be further evaluated. Third, almost all published prediction tools provide their own benchmark pipelines using different datasets, which potentially favors their prediction efficiency. These benchmark protocols might be credible, but may lack objectivity, simplicity, and effectiveness. We have sorted out the data used for performance evaluation in the respective study of each tool in detail (Files S1–S4), which can facilitate researchers to compile more diverse and standard data for objective benchmark in the future. So far, the most widely used datasets for evaluating pA tools for DNA sequences are the PASS dataset [68], [69] of plant species, the ERPIN dataset [23] of human, and the DeepGSR dataset [27] of animal species; datasets for bulk RNA-seq are the MAQC dataset [123] and the HEK293 dataset [124]; and datasets for scRNA-seq are the 10× human PBMC data and the Tabula Muris atlas [125] (Files S1–S4). Moreover, genomic data could be small sample data and large-scale data; it is also necessary to evaluate the performance of different tools under different sizes of data. Fourth, the output format varies among different tools. For example, most tools for DNA sequences generate binary output or probabilities between 0 and 1; some tools for bulk RNA-seq or scRNA-seq output potential regions of pA instead of exact pA position. Therefore, how to unify the output of different tools for objective evaluation needs to be carefully considered. Fifth, compared with the benchmark of tools for DNA sequence data, the benchmark for scRNA-seq tools is much less uniform (Files S1–S4). Almost all studies examined the consensus between the identified pAs and annotated pAs, while there is still no commonly used objective evaluation strategies with diverse indicators. Therefore, it is necessary to use a variety of performance indicators (e.g., sensitivity, specificity, and precision) that are complementary in nature for comprehensive performance evaluation, particularly for the evaluation of the emerging scRNA-seq tools. At the same time, it is also important to simply present an overall ranking of different tools. The last but not least, many tools have parameters that can be adjusted; however, only the default parameters are normally used for evaluation. Therefore, some strategies (e.g., grid search) should be proposed to evaluate the impact of different parameters of a method.
Predicting pAs in non-3′ UTRs
With the advance of 3′ seq, more and more unconventional pAs located in non-3′ UTRs like introns and CDSs were discovered [3], [48], [126]. These non-3′ UTR pAs may generate mRNA isoforms encoding distinct proteins or resulting in the creation of premature stop codons. Intronic polyadenylation has been found associated with cancer through the inactivation of tumor-suppressor genes [94], [127]. The differential use of intronic pAs is a potential indicator for the differential expression of pre-spliced mRNA transcripts, which contributes to detecting newly transcribed genes and ultimately helps estimate the rate and direction of cell differentiation [128]. Till now, almost all computational tools focus on pA prediction in 3′ UTRs. Many tools, particularly those for DNA sequences, usually consider random sequences from introns as negative datasets for model training, which would cause some real intronic pAs to be mistakenly regarded as negative instances. Therefore, even for the pA prediction in 3′ UTRs, it is necessary to consider the prevalence of unconventional pAs when constructing the negative dataset. Lately, some tools for bulk RNA-seq or scRNA-seq have found a considerable number of pAs in introns. By applying IPAFinder [94] on pan-cancer data from bulk RNA-seq, 490 recurrent dynamically changed intronic pAs were found. Sierra [43] utilized a splice-aware strategy and identified a considerable number of intronic peaks from scRNA-seq; however, the majority of these peaks may be internal priming artifacts as they are proximal to A-rich regions. SCAPTURE [106] also found > 16,000 candidate intronic pAs from 10× PBMC samples, while < 20% pAs overlapped with known intronic sites and a large number of false positives were present in lowly expressed genes. Therefore, further careful inspection or filtering is critical to obtain true non-3′ UTR pAs or new intelligent algorithms are demanded to effectively call non-3′ UTR pAs.
Predicting tissue-specific pAs
APA plays a significant role in tissue-specific regulation of gene expression [2], [12]. Profiling APA dynamics or differential APA usages under different physiological or pathological conditions has become a routine analysis in most APA studies. Computational prediction of tissue-specific pAs may be an alternative yet cost-effective solution for analyzing tissue specificity of APA. The pA prediction problem described in this review is essentially a binary classification problem, which aims to distinguish between nucleotide sequences or genomic regions that contain a pA and those do not. Studies are currently in progress to solve the problem of pA quantification, which aims to predict the strength or dominance of a given pA across tissues. Weng et al. [111] and Hafez et al. [129] predicted whether a given pA is tissue-specific or not, whereas they did not tackle the question of alternative choice of APA sites. One way to study tissue specificity of pAs is to explore the differential usage of APA sites in a gene (e.g., proximal and distal pAs). Several tools, such as Conv-Net [72], have been proposed to predict the strength of APA sites. Leung et al. [72] predicted relative dominance of pAs within 3′ UTR in human tissues solely based on nucleotide sequences using a DL model. However, these methods only make predictions based on sequence features, while fail to consider sample specificity and in vivo expression. In contrast, many tools for bulk RNA-seq or scRNA-seq can be used for pairwise comparisons between two samples, while they are not very suitable for profiling APA across multiple tissues. Ever-larger bulk RNA-seq or scRNA-seq data comprising of growing number of samples from diverse tissues are increasingly available, which places new demands on developing new methods to efficiently tackle the question of tissue specificity of APA.
Cross-species prediction of pAs
Traditional ML methods, such as those based on SVM, can hardly adapt to different species, because they use hand-crafted features learnt for a specific species. Although many DL-based tools have been proposed to improve the performance of pA prediction, most tools still need species-specific real pA collection for model training. Consequently, these tools may suffer from high risks of overfitting and are not applicable to species without any prior pA annotations. Therefore, it is a promising direction to design new transferrable algorithms for cross-species pA prediction or to improve the generalizability of existing tools, which allows a well-trained model from a species with rich annotations to be transferred to data from a different species without retraining or prior knowledge. Annotation-assisted methods, compared to methods without using prior annotations, generally ensure higher data quality and achieve better performance; however, their application is limited to data from specific species or biological conditions. Collection of more extensive pA annotations from different sources would definitely contribute to predicting novel sites and increasing the coverage of pAs in diverse cell types, biological conditions, and species. Therefore, an alternative solution for predicting pAs in poorly annotated species could be building an elegant model for well-annotated species and then transferring the model to a different but related species, even without an established pA collection. An initial attempt has been made by some existing methods like Poly(A)-DG [121], which extracts shared features from multiple species and can be generalized to the target species without fine-tuning. However, Poly(A)-DG was only tested between four animals. Till now, tools applicable to plants are still limited. It is widely accepted that the sequence conservation in PASs in plants is very low, where the most dominant AAUAAA only appears in less than 10% of pAs [56]. Our group recently developed a tool called QuantifyPoly(A) [62] to profile genome-wide polyadenylation choices, which found plant pAs generally exhibit higher micro-heterogeneity than animal ones, and UGUA, UAAA, and/or AAUAAA are used in a species-dependent manner. Still, more efforts are needed to explore additional motifs and/or higher-order structures associated with plant polyadenylation, and more intelligent algorithms are demanded in order to better predict pAs in multiple species.
Predicting pAs by integrating multi-omics data
pAs can be derived from different kinds of data. For example, 3′ seq has the unique advantage of acquiring high-quality pAs transcriptome-wide, which contributes to a larger compendium of authentic pAs. Third-generation sequencing technologies, such as PacBio sequencing, are powerful in profiling full-length transcriptome, which could provide a more accurate transcriptome annotation. Widely conducted bulk RNA-seq data can be used for capturing and quantifying pAs of low-abundance transcripts, and the rapid growing scRNA-seq data support the identification of relatively rare transcripts in single cells. In addition to the genome or transcriptome layers, APA modulation has been found associated with other layers of gene regulation, such as nucleosome positioning, transcription rate, DNA methylation, and RBPs [2], [130], [131], [132]. By integrating multi-omics data, weak signals from one layer can be amplified or noises be reduced to avoid false negative predictions by referring to the complementary information from additional layers. For instance, potential pAs identified from RNA-seq without well-recognized PASs could be eliminated if there is no evidence in 3′ seq or full-length RNA-seq data. Initial attempts have been made for APA analysis using multi-omics data. scUTRquant [107] incorporates a cleavage site atlas established from a mouse full-length Microwell-seq dataset of 400,000 single cells [108] for filtering high-confidence pAs predicted from 3′ tag scRNA-seq. Leung et al. [72] predicted strength of pAs using nucleotide sequences, considering features from additional layers like nucleosome positioning and RBP motifs. IntMAP [96] is a unified ML-based framework, which can fine-tune the contributions of RNA-seq and 3′ seq data by tailoring the parameter λ. Currently, DL models have been widely used in predicting pAs from DNA sequences. However, in many cases, DL models fail to make accurate prediction, while patterns of RNA-seq coverage provide clear evidence of polyadenylation, and vice versa [110]. Accordingly, several DL-based tools have integrated bulk RNA-seq or scRNA-seq with DNA sequences for pA prediction, such as SCAPTURE [106] and Aptardi [98]. It is promising yet challenging to formulate one unified computational framework, especially leveraging the strength of intelligent algorithms, to integrate the quantitative information from multiple omics layers, e.g., genomic DNA, transcriptome data, methylation data, and chromatin accessibility data, to identify and quantify pAs genome-wide.
Predicting pAs at the single-cell level
With the rapid development of scRNA-seq technology, different tools continue to emerge for pA identification in single cells. Currently, most methods, like scAPA [101], Sierra [43], and MAAPER [105], construct pseudo-bulk RNA-seq data by pooling reads from cells of the same cell cluster (or cell type) to address the high dropout rate and variability inherent in scRNA-seq. Although many of these tools, like scAPA or Sierra, can still quantify the expression of a pA in each cell by counting reads within a poly(A) region, single cell-based quantification may have a high noise level and missing values due to biological and technical variances [105]. As such, APA usage is characterized at the cell-cluster resolution rather than the single-cell resolution, which somewhat contradicts the ultimate goal of single-cell sequencing. Moreover, cell clusters or cell types in these studies were inferred by the conventional gene–cell expression profile, consequently, the APA analysis is limited to predefined cell types and the result may be affected by different cell type annotations. Alternatively, scDaPars [45] quantifies single-cell APA usage based on the model for bulk RNA-seq introduced in DaPars [17], and then recovers missing APA usage by leveraging APA information of the same gene in similar cells. Another limitation of most tools for scRNA-seq is that they are only applicable to 3′ tag-based scRNA-seq like 10× Chromium or CEL-Seq. Till now, only scDaPars can be applied to both 3′ tag and full-length scRNA-seq, e.g., Smart-seq2. However, although scDaPars was reported to be able to quantify APA usage in individual cells independent of gene expression, pAs were actually predicted from the bulk RNA-seq tool DaPars that was not specifically designed for scRNA-seq. Moreover, it is challenging to identify and verify low-expression pAs in highly sparse scRNA-seq, particularly those in rare cells. In addition, the tsunami of complex scRNA-seq datasets with various tissue sources, batch effects, and library sizes also have brought huge computational and analytical challenges. Therefore, more efforts are needed to develop new methods to address inherent issues in scRNA-seq for establishing a more comprehensive landscape of pAs at the single-gene and single-cell resolutions.
Competing interests
The authors have declared no competing interests.
CRediT authorship contribution statement
Wenbin Ye: Investigation, Data curation, Visualization, Writing – original draft, Writing – review & editing. Qiwei Lian: Data curation, Writing – review & editing. Congting Ye: Investigation, Writing – review & editing. Xiaohui Wu: Conceptualization, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition. All authors have read and approved the final manuscript.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 61871463 to XW) and the Natural Science Foundation of Fujian Province of China (Grant No. 2020J01047 to CY).
Handled by Leng Han
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2022.09.005.
Supplementary material
The following are the Supplementary material to this article:
List of methods for predicting pAs or PASs from DNA sequences based on traditional machine learning models
List of methods for predicting pAs or PASs from DNA sequences based on deep learning models
List of methods for predicting pAs from RNA-seq
List of methods for predicting pAs from scRNA-seq
List of tools or resources for APA analysis rather than pA prediction
Predicting pAs from bulk RNA-seqand scRNA-seq
References
- 1.Wu X., Bartel D.P. Widespread influence of 3′-end structures on mammalian mRNA processing and stability. Cell. 2017;169:905–917. doi: 10.1016/j.cell.2017.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian B., Manley J.L. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18:18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Giammartino D.C., Nishida K., Manley J.L. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian B., Manley J.L. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X., Liu M., Downie B., Liang C., Ji G., Li Q.Q., et al. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci U S A. 2011;108:12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lianoglou S., Garg V., Yang J.L., Leslie C.S., Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013;27:2380–2396. doi: 10.1101/gad.229328.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ulitsky I., Shkumatava A., Jan C.H., Subtelny A.O., Koppstein D., Bell G.W., et al. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber A.J., Schmidt R., Gruber A.R., Martin G., Ghosh S., Belmadani M., et al. A comprehensive analysis of 3′ end sequencing data sets reveals novel polyadenylation signals and the repressive role of heterogeneous ribonucleoprotein C on cleavage and polyadenylation. Genome Res. 2016;26:1145–1159. doi: 10.1101/gr.202432.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derti A., Garrett-Engele P., MacIsaac K.D., Stevens R.C., Sriram S., Chen R., et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozsolak F., Kapranov P., Foissac S., Kim S.W., Fishilevich E., Monaghan A.P., et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoque M., Ji Z., Zheng D.H., Luo W.T., Li W.C., You B., et al. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gruber A.J., Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet. 2019;20:599–614. doi: 10.1038/s41576-019-0145-z. [DOI] [PubMed] [Google Scholar]
- 13.Oktaba K., Zhang W., Lotz T.S., Jun D.J., Lemke S.B., Ng S.P., et al. ELAV links paused Pol II to alternative polyadenylation in the Drosophila nervous system. Mol Cell. 2015;57:341–348. doi: 10.1016/j.molcel.2014.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blazie S.M., Babb C., Wilky H., Rawls A., Park J.G., Mangone M. Comparative RNA-seq analysis reveals pervasive tissue-specific alternative polyadenylation in Caenorhabditis elegans intestine and muscles. BMC Biol. 2015;13:4. doi: 10.1186/s12915-015-0116-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkovits B.D., Mayr C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batra R., Manchanda M., Swanson M.S. Global insights into alternative polyadenylation regulation. RNA Biol. 2015;12:597–602. doi: 10.1080/15476286.2015.1040974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia Z., Donehower L.A., Cooper T.A., Neilson J.R., Wheeler D.A., Wagner E.J., et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han T., Kim J.K. Driving glioblastoma growth by alternative polyadenylation. Cell Res. 2014;24:1023–1024. doi: 10.1038/cr.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta I., Clauder-Munster S., Klaus B., Jarvelin A.I., Aiyar R.S., Benes V., et al. Alternative polyadenylation diversifies post-transcriptional regulation by selective RNA-protein interactions. Mol Syst Biol. 2014;10:719. doi: 10.1002/msb.135068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruber A.R., Martin G., Muller P., Schmidt A., Gruber A.J., Gumienny R., et al. Global 3′ UTR shortening has a limited effect on protein abundance in proliferating T cells. Nat Commun. 2014;5:5465. doi: 10.1038/ncomms6465. [DOI] [PubMed] [Google Scholar]
- 21.Salamov A.A., Solovyev V.V. Recognition of 3′-processing sites of human mRNA precursors. Comput Appl Biosci. 1997;13:23–28. doi: 10.1093/bioinformatics/13.1.23. [DOI] [PubMed] [Google Scholar]
- 22.Akhtar M.N., Bukhari S.A., Fazal Z., Qamar R., Shahmuradov I.A. POLYAR, a new computer program for prediction of poly(A) sites in human sequences. BMC Genomics. 2010;11:646. doi: 10.1186/1471-2164-11-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legendre M., Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng Y., Miura R.M., Tian B. Prediction of mRNA polyadenylation sites by support vector machine. Bioinformatics. 2006;22:2320–2325. doi: 10.1093/bioinformatics/btl394. [DOI] [PubMed] [Google Scholar]
- 25.Magana-Mora A., Kalkatawi M., Bajic V.B. Omni-PolyA: a method and tool for accurate recognition of poly(A) signals in human genomic DNA. BMC Genomics. 2017;18:620. doi: 10.1186/s12864-017-4033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia Z., Li Y., Zhang B., Li Z., Hu Y., Chen W., et al. DeeReCT-PolyA: a robust and generic deep learning method for PAS identification. Bioinformatics. 2019;35:2371–2379. doi: 10.1093/bioinformatics/bty991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalkatawi M., Magana-Mora A., Jankovic B., Bajic V.B. DeepGSR: an optimized deep-learning structure for the recognition of genomic signals and regions. Bioinformatics. 2019;35:1125–1132. doi: 10.1093/bioinformatics/bty752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arefeen A., Xiao X., Jiang T. DeepPASTA: deep neural network based polyadenylation site analysis. Bioinformatics. 2019;35:4577–4585. doi: 10.1093/bioinformatics/btz283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogard N., Linder J., Rosenberg A.B., Seelig G. A deep neural network for predicting and engineering alternative polyadenylation. Cell. 2019;178:91–106. doi: 10.1016/j.cell.2019.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherstnev A., Duc C., Cole C., Zacharaki V., Hornyik C., Ozsolak F., et al. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol. 2012;19:845–852. doi: 10.1038/nsmb.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jan C.H., Friedman R.C., Ruby J.G., Bartel D.P. Formation, regulation and evolution of Caenorhabditis elegans 3′ UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrison P.F., Powell D.R., Clancy J.L., Preiss T., Boag P.R., Traven A., et al. PAT-seq: a method to study the integration of 3′-UTR dynamics with gene expression in the eukaryotic transcriptome. RNA. 2015;21:1502–1510. doi: 10.1261/rna.048355.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J.E., Yi H., Kim Y., Chang H., Kim V.N. Regulation of poly(A) tail and translation during the somatic cell cycle. Mol Cell. 2016;62:462–471. doi: 10.1016/j.molcel.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Chang H., Lim J., Ha M., Kim V.N. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell. 2014;53:1044–1052. doi: 10.1016/j.molcel.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y. Alternative polyadenylation: new insights from global analyses. RNA. 2012;18:2105–2117. doi: 10.1261/rna.035899.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elkon R., Ugalde A.P., Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- 37.Ji G., Guan J., Zeng Y., Li Q.Q., Wu X. Genome-wide identification and predictive modeling of polyadenylation sites in eukaryotes. Brief Bioinform. 2015;16:304–313. doi: 10.1093/bib/bbu011. [DOI] [PubMed] [Google Scholar]
- 38.Ha K.C.H., Blencowe B.J., Morris Q. QAPA: a new method for the systematic analysis of alternative polyadenylation from RNA-seq data. Genome Biol. 2018;19:45. doi: 10.1186/s13059-018-1414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye C., Long Y., Ji G., Li Q.Q., Wu X. APAtrap: identification and quantification of alternative polyadenylation sites from RNA-seq data. Bioinformatics. 2018;34:1841–1849. doi: 10.1093/bioinformatics/bty029. [DOI] [PubMed] [Google Scholar]
- 40.Arefeen A., Liu J., Xiao X., Jiang T. TAPAS: tool for alternative polyadenylation site analysis. Bioinformatics. 2018;34:2521–2529. doi: 10.1093/bioinformatics/bty110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hashimshony T., Wagner F., Sher N., Yanai I. CEL-Seq: single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patrick R., Humphreys D.T., Janbandhu V., Oshlack A., Ho J.W.K., Harvey R.P., et al. Sierra: discovery of differential transcript usage from polyA-captured single-cell RNA-seq data. Genome Biol. 2020;21:167. doi: 10.1186/s13059-020-02071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu X., Liu T., Ye C., Ye W., Ji G. scAPAtrap: identification and quantification of alternative polyadenylation sites from single-cell RNA-seq data. Brief Bioinform. 2021;22:bbaa273. doi: 10.1093/bib/bbaa273. [DOI] [PubMed] [Google Scholar]
- 45.Gao Y., Li L., Amos C.I., Li W. Analysis of alternative polyadenylation from single-cell RNA-seq using scDaPars reveals cell subpopulations invisible to gene expression. Genome Res. 2021;31:1856–1866. doi: 10.1101/gr.271346.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szkop K.J., Nobeli I. Untranslated parts of genes interpreted: making heads or tails of high-throughput transcriptomic data via computational methods. Computational methods to discover and quantify isoforms with alternative untranslated regions. Bioessays. 2017;39:1700090. doi: 10.1002/bies.201700090. [DOI] [PubMed] [Google Scholar]
- 47.Yeh H.S., Zhang W., Yong J. Analyses of alternative polyadenylation: from old school biochemistry to high-throughput technologies. BMB Rep. 2017;50:201–207. doi: 10.5483/BMBRep.2017.50.4.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen W., Jia Q., Song Y., Fu H., Wei G., Ni T. Alternative polyadenylation: methods, findings, and impacts. Genomics Proteomics Bioinformatics. 2017;15:287–300. doi: 10.1016/j.gpb.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen M., Ji G., Fu H., Lin Q., Ye C., Ye W., et al. A survey on identification and quantification of alternative polyadenylation sites from RNA-seq data. Brief Bioinform. 2020;21:1261–1276. doi: 10.1093/bib/bbz068. [DOI] [PubMed] [Google Scholar]
- 50.Shah A., Mittleman B.E., Gilad Y., Li Y.I. Benchmarking sequencing methods and tools that facilitate the study of alternative polyadenylation. Genome Biol. 2021;22:291. doi: 10.1186/s13059-021-02502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ye C., Lin J., Li Q.Q. Discovery of alternative polyadenylation dynamics from single cell types. Comput Struct Biotechnol J. 2020;18:1012–1019. doi: 10.1016/j.csbj.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y., Liu L., Qiu Q., Zhou Q., Ding J., Lu Y., et al. Alternative polyadenylation: methods, mechanism, function, and role in cancer. J Exp Clin Cancer Res. 2021;40:51. doi: 10.1186/s13046-021-01852-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kandhari N., Kraupner-Taylor C.A., Harrison P.F., Powell D.R., Beilharz T.H. The detection and bioinformatic analysis of alternative 3′ UTR isoforms as potential cancer biomarkers. Int J Mol Sci. 2021;22:5322. doi: 10.3390/ijms22105322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang R., Tian B. APAlyzer: a bioinformatic package for analysis of alternative polyadenylation isoforms. Bioinformatics. 2020;36:3907–3909. doi: 10.1093/bioinformatics/btaa266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye W., Liu T., Fu H., Ye C., Ji G., Wu X. movAPA: modeling and visualization of dynamics of alternative polyadenylation across biological samples. Bioinformatics. 2021;37:2470–2472. doi: 10.1093/bioinformatics/btaa997. [DOI] [PubMed] [Google Scholar]
- 56.Tian B., Graber J.H. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA. 2012;3:385–396. doi: 10.1002/wrna.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tabaska J.E., Zhang M.Q. Detection of polyadenylation signals in human DNA sequences. Gene. 1999;231:77–86. doi: 10.1016/s0378-1119(99)00104-3. [DOI] [PubMed] [Google Scholar]
- 58.Ahmed F., Kumar M., Raghava G.P.S. Prediction of polyadenylation signals in human DNA sequences using nucleotide frequencies. In Silico Biol. 2009;9:135–148. [PubMed] [Google Scholar]
- 59.Ji G., Wu X., Shen Y., Huang J., Li Q.Q. A classification-based prediction model of messenger RNA polyadenylation sites. J Theor Biol. 2010;265:287–296. doi: 10.1016/j.jtbi.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 60.Kalkatawi M., Rangkuti F., Schramm M., Jankovic B.R., Kamau A., Chowdhary R., et al. Dragon PolyA Spotter: predictor of poly(A) motifs within human genomic DNA sequences. Bioinformatics. 2012;28:127–129. doi: 10.1093/bioinformatics/btr602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu X., Ji G., Zeng Y. In silico prediction of mRNA poly(A) sites in Chlamydomonas reinhardtii. Mol Genet Genomics. 2012;287:895–907. doi: 10.1007/s00438-012-0725-5. [DOI] [PubMed] [Google Scholar]
- 62.Ye C., Zhao D., Ye W., Wu X., Ji G., Li Q.Q., et al. QuantifyPoly(A): reshaping alternative polyadenylation landscapes of eukaryotes with weighted density peak clustering. Brief Bioinform. 2021;22:bbab268. doi: 10.1093/bib/bbab268. [DOI] [PubMed] [Google Scholar]
- 63.Liu H., Han H., Li J., Wong L. An in-silico method for prediction of polyadenylation signals in human sequences. Genome Inform. 2003;14:84–93. [PubMed] [Google Scholar]
- 64.Chang T.H., Wu L.C., Chen Y.T., Huang H.D., Liu B.J., Cheng K.F., et al. Characterization and prediction of mRNA polyadenylation sites in human genes. Med Biol Eng Comput. 2011;49:463–472. doi: 10.1007/s11517-011-0732-4. [DOI] [PubMed] [Google Scholar]
- 65.Xie B., Jankovic B.R., Bajic V.B., Song L., Gao X. Poly(A) motif prediction using spectral latent features from human DNA sequences. Bioinformatics. 2013;29:i316–i325. doi: 10.1093/bioinformatics/btt218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graber J.H., McAllister G.D., Smith T.F. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 2002;30:1851–1858. doi: 10.1093/nar/30.8.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hajarnavis A., Korf I., Durbin R. A probabilistic model of 3′ end formation in Caenorhabditis elegans. Nucleic Acids Res. 2004;32:3392–3399. doi: 10.1093/nar/gkh656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ji G., Zheng J., Shen Y., Wu X., Jiang R., Lin Y., et al. Predictive modeling of plant messenger RNA polyadenylation sites. BMC Bioinformatics. 2007;8:43. doi: 10.1186/1471-2105-8-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen Y., Ji G., Haas B.J., Wu X., Zheng J., Reese G.J., et al. Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation. Nucleic Acids Res. 2008;36:3150–3161. doi: 10.1093/nar/gkn158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ji G., Li L., Li Q.Q., Wu X., Fu J., Chen G., et al. PASPA: a web server for mRNA poly(A) site predictions in plants and algae. Bioinformatics. 2015;31:1671–1673. doi: 10.1093/bioinformatics/btv004. [DOI] [PubMed] [Google Scholar]
- 71.Gao X., Zhang J., Wei Z., Hakonarson H. DeepPolyA: a convolutional neural network approach for polyadenylation site prediction. IEEE Access. 2018;6:24340–24349. [Google Scholar]
- 72.Leung M.K.K., Delong A., Frey B.J. Inference of the human polyadenylation code. Bioinformatics. 2018;34:2889–2898. doi: 10.1093/bioinformatics/bty211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albalawi F., Chahid A., Guo X., Albaradei S., Magana-Mora A., Jankovic B.R., et al. Hybrid model for efficient prediction of poly(A) signals in human genomic DNA. Methods. 2019;166:31–39. doi: 10.1016/j.ymeth.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 74.Yu H., Dai Z. SANPolyA: a deep learning method for identifying poly(A) signals. Bioinformatics. 2020;36:2393–2400. doi: 10.1093/bioinformatics/btz970. [DOI] [PubMed] [Google Scholar]
- 75.Guo Y., Zhou D., Li W., Cao J., Nie R., Xiong L., et al. Identifying polyadenylation signals with biological embedding via self-attentive gated convolutional highway networks. Appl Soft Comput. 2021;103:107133. [Google Scholar]
- 76.Pickrell J.K., Marioni J.C., Pai A.A., Degner J.F., Engelhardt B.E., Nkadori E., et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Birol I., Raymond A., Chiu R., Nip K.M., Jackman S.D., Kreitzman M., et al. Kleat: cleavage site analysis of transcriptomes. Pac Symp Biocomput. 2015:347–358. [PMC free article] [PubMed] [Google Scholar]
- 78.Bonfert T., Friedel C.C. Prediction of poly(A) sites by poly(A) read mapping. PLoS One. 2017;12:e0170914. doi: 10.1371/journal.pone.0170914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Campbell M.A., Haas B.J., Hamilton J.P., Mount S.M., Buell C.R. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7:327. doi: 10.1186/1471-2164-7-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guttman M., Garber M., Levin J.Z., Donaghey J., Robinson J., Adiconis X., et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Le Pera L., Mazzapioda M., Tramontano A. 3USS: a web server for detecting alternative 3′ UTRs from RNA-seq experiments. Bioinformatics. 2015;31:1845–1847. doi: 10.1093/bioinformatics/btv035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huang Z., Teeling E.C. ExUTR: a novel pipeline for large-scale prediction of 3′-UTR sequences from NGS data. BMC Genomics. 2017;18:847. doi: 10.1186/s12864-017-4241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilkening S., Pelechano V., Jarvelin A.I., Tekkedil M.M., Anders S., Benes V., et al. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res. 2013;41:e65. doi: 10.1093/nar/gks1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang R., Nambiar R., Zheng D., Tian B. PolyA_DB 3 catalogs cleavage and polyadenylation sites identified by deep sequencing in multiple genomes. Nucleic Acids Res. 2018;46:D315–D319. doi: 10.1093/nar/gkx1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhu S., Ye W., Ye L., Fu H., Ye C., Xiao X., et al. PlantAPAdb: a comprehensive database for alternative polyadenylation sites in plants. Plant Physiol. 2020;182:228–242. doi: 10.1104/pp.19.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gruber A.J., Schmidt R., Ghosh S., Martin G., Gruber A.R., van Nimwegen E., et al. Discovery of physiological and cancer-related regulators of 3′ UTR processing with KAPAC. Genome Biol. 2018;19:44. doi: 10.1186/s13059-018-1415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fahmi N.A., Ahmed K.T., Chang J.W., Nassereddeen H., Fan D., Yong J., et al. APA-Scan: detection and visualization of 3′-UTR alternative polyadenylation with RNA-seq and 3′-end-seq data. BMC Bioinformatics. 2022;23:396. doi: 10.1186/s12859-022-04939-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim M., You B.H., Nam J.W. Global estimation of the 3′ untranslated region landscape using RNA sequencing. Methods. 2015;83:111–117. doi: 10.1016/j.ymeth.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 89.Shenker S., Miura P., Sanfilippo P., Lai E.C. IsoSCM: improved and alternative 3′ UTR annotation using multiple change-point inference. RNA. 2015;21:14–27. doi: 10.1261/rna.046037.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li L., Huang K.L., Gao Y., Cui Y., Wang G., Elrod N.D., et al. An atlas of alternative polyadenylation quantitative trait loci contributing to complex trait and disease heritability. Nat Genet. 2021;53:994–1005. doi: 10.1038/s41588-021-00864-5. [DOI] [PubMed] [Google Scholar]
- 91.Feng X., Li L., Wagner E.J., Li W. TC3A: the cancer 3′ UTR atlas. Nucleic Acids Res. 2018;46:D1027–D1030. doi: 10.1093/nar/gkx892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang J., Wei Z. An empirical Bayes change-point model for identifying 3′ and 5′ alternative splicing by next-generation RNA sequencing. Bioinformatics. 2016;32:1823–1831. doi: 10.1093/bioinformatics/btw060. [DOI] [PubMed] [Google Scholar]
- 93.Cass A.A., Xiao X. mountainClimber identifies alternative transcription start and polyadenylation sites in RNA-seq. Cell Syst. 2019;9:393–400. doi: 10.1016/j.cels.2019.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Z., Xu Q., Wei R., Wang W., Ding D., Yang Y., et al. Cancer-associated dynamics and potential regulators of intronic polyadenylation revealed by IPAFinder using standard RNA-seq data. Genome Res. 2021;31:2095–2106. doi: 10.1101/gr.271627.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gruber A.J., Gypas F., Riba A., Schmidt R., Zavolan M. Terminal exon characterization with TECtool reveals an abundance of cell-specific isoforms. Nat Methods. 2018;15:832–836. doi: 10.1038/s41592-018-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang J.W., Zhang W., Yeh H.S., Park M., Yao C., Shi Y., et al. An integrative model for alternative polyadenylation, IntMAP, delineates mTOR-modulated endoplasmic reticulum stress response. Nucleic Acids Res. 2018;46:5996–6008. doi: 10.1093/nar/gky340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang C., Li C., Nip K.M., Warren R.L., Birol I. Terminitor: cleavage site prediction using deep learning models. bioRxiv. 2020:710699. [Google Scholar]
- 98.Lusk R., Stene E., Banaei-Kashani F., Tabakoff B., Kechris K., Saba L.M. Aptardi predicts polyadenylation sites in sample-specific transcriptomes using high-throughput RNA sequencing and DNA sequence. Nat Commun. 2021;12:1652. doi: 10.1038/s41467-021-21894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., et al. Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017;65:631–643. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 100.Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shulman E.D., Elkon R. Cell-type-specific analysis of alternative polyadenylation using single-cell transcriptomics data. Nucleic Acids Res. 2019;47:10027–10039. doi: 10.1093/nar/gkz781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang Y., Paul A., Bach T.N., Huang Z.J., Zhang M.Q. Single-cell alternative polyadenylation analysis delineates GABAergic neuron types. BMC Biol. 2021;19:144. doi: 10.1186/s12915-021-01076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhou R., Xiao X., He P., Zhao Y., Xu M., Zheng X., et al. SCAPE: a mixture model revealing single-cell polyadenylation diversity and cellular dynamics during cell differentiation and reprogramming. Nucleic Acids Res. 2022;50:e66. doi: 10.1093/nar/gkac167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Meyer E., Chaung K., Dehghannasiri R., Salzman J. ReadZS detects cell type-specific and developmentally regulated RNA processing programs in single-cell RNA-seq. Genome Biol. 2022;23:226. doi: 10.1186/s13059-022-02795-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li W.V., Zheng D., Wang R., Tian B. MAAPER: model-based analysis of alternative polyadenylation using 3′ end-linked reads. Genome Biol. 2021;22:222. doi: 10.1186/s13059-021-02429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li G.W., Nan F., Yuan G.H., Liu C.X., Liu X., Chen L.L., et al. SCAPTURE: a deep learning-embedded pipeline that captures polyadenylation information from 3′ tag-based RNA-seq of single cells. Genome Biol. 2021;22:221. doi: 10.1186/s13059-021-02437-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fansler M.M., Zhen G., Mayr C. Quantification of alternative 3′ UTR isoforms from single cell RNA-seq data with scUTRquant. bioRxiv. 2021:469635. [Google Scholar]
- 108.Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., et al. Mapping the mouse cell atlas by Microwell-seq. Cell. 2018;172:1091–1107. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 109.Levin M., Zalts H., Mostov N., Hashimshony T., Yanai I. Gene expression dynamics are a proxy for selective pressures on alternatively polyadenylated isoforms. Nucleic Acids Res. 2020;48:5926–5938. doi: 10.1093/nar/gkaa359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Z., Li Y., Zhang B., Li Y., Long Y., Zhou J., et al. DeeReCT-APA: prediction of alternative polyadenylation site usage through deep learning. Genomics Proteomics Bioinformatics. 2022;20:483–495. doi: 10.1016/j.gpb.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Weng L., Li Y., Xie X., Shi Y. Poly(A) code analyses reveal key determinants for tissue-specific mRNA alternative polyadenylation. RNA. 2016;22:813–821. doi: 10.1261/rna.055681.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ji G., Chen M., Ye W., Zhu S., Ye C., Su Y., et al. TSAPA: identification of tissue-specific alternative polyadenylation sites in plants. Bioinformatics. 2018;34:2123–2125. doi: 10.1093/bioinformatics/bty044. [DOI] [PubMed] [Google Scholar]
- 113.Ye C., Zhou Q., Hong Y., Li Q.Q. Role of alternative polyadenylation dynamics in acute myeloid leukaemia at single-cell resolution. RNA Biol. 2019;16:785–797. doi: 10.1080/15476286.2019.1586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu J., Bushel P.R. Dynamic expression of 3′ UTRs revealed by Poisson hidden Markov modeling of RNA-Seq: implications in gene expression profiling. Gene. 2013;527:616–623. doi: 10.1016/j.gene.2013.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang W., Wei Z., Li H. A change-point model for identifying 3′ UTR switching by next-generation RNA sequencing. Bioinformatics. 2014;30:2162–2170. doi: 10.1093/bioinformatics/btu189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Grassi E., Mariella E., Lembo A., Molineris I., Provero P. Roar: detecting alternative polyadenylation with standard mRNA sequencing libraries. BMC Bioinformatics. 2016;17:423. doi: 10.1186/s12859-016-1254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Burri D., Zavolan M. Shortening of 3′ UTRs in most cell types composing tumor tissues implicates alternative polyadenylation in protein metabolism. RNA. 2021;27:1459–1470. doi: 10.1261/rna.078886.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bai Y., Qin Y., Fan Z., Morrison R.M., Nam K., Zarour H.M., et al. scMAPA: identification of cell-type-specific alternative polyadenylation in complex tissues. Gigascience. 2022;11:giac033. doi: 10.1093/gigascience/giac033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ye C., Zhou Q., Wu X., Yu C., Ji G., Saban D.R., et al. scDAPA: detection and visualization of dynamic alternative polyadenylation from single cell RNA-seq data. Bioinformatics. 2020;36:1262–1264. doi: 10.1093/bioinformatics/btz701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zheng Y., Wang H., Zhang Y., Gao X., Xing E.P., Xu M. Poly(A)-DG: a deep-learning-based domain generalization method to identify cross-species poly(A) signal without prior knowledge from target species. PLoS Comput Biol. 2020;16:e1008297. doi: 10.1371/journal.pcbi.1008297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Singh I., Lee S.H., Sperling A.S., Samur M.K., Tai Y.T., Fulciniti M., et al. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nat Commun. 2018;9:1716. doi: 10.1038/s41467-018-04112-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bullard J.H., Purdom E., Hansen K.D., Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N6-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schaum N., Karkanias J., Neff N.F., May A.P., Quake S.R., Wyss-Coray T., et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.de Lorenzo L., Sorenson R., Bailey-Serres J., Hunt A.G. Noncanonical alternative polyadenylation contributes to gene regulation in response to hypoxia. Plant Cell. 2017;29:1262–1277. doi: 10.1105/tpc.16.00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee S.H., Singh I., Tisdale S., Abdel-Wahab O., Leslie C.S., Mayr C. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature. 2018;561:127–131. doi: 10.1038/s41586-018-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.La Manno G., Soldatov R., Zeisel A., Braun E., Hochgerner H., Petukhov V., et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hafez D., Ni T., Mukherjee S., Zhu J., Ohler U. Genome-wide identification and predictive modeling of tissue-specific alternative polyadenylation. Bioinformatics. 2013;29:i108–i116. doi: 10.1093/bioinformatics/btt233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Neve J., Patel R., Wang Z., Louey A., Furger A.M. Cleavage and polyadenylation: ending the message expands gene regulation. RNA Biol. 2017;14:865–890. doi: 10.1080/15476286.2017.1306171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mayr C. Regulation by 3′-untranslated regions. Annu Rev Genet. 2017;51:171–194. doi: 10.1146/annurev-genet-120116-024704. [DOI] [PubMed] [Google Scholar]
- 132.MacDonald C.C. Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond (2018 update) Wiley Interdiscip Rev RNA. 2019;10:e1526. doi: 10.1002/wrna.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of methods for predicting pAs or PASs from DNA sequences based on traditional machine learning models
List of methods for predicting pAs or PASs from DNA sequences based on deep learning models
List of methods for predicting pAs from RNA-seq
List of methods for predicting pAs from scRNA-seq
List of tools or resources for APA analysis rather than pA prediction
Predicting pAs from bulk RNA-seqand scRNA-seq