Abstract
Streptococcus mutans has been implicated as the major causative agent of human dental caries. S. mutans binds to saliva-coated tooth surfaces, and previous studies suggested that fimbriae may play a role in the initial bacterial adherence to salivary components. The objectives of this study were to establish the ability of an S. mutans fimbria preparation to bind to saliva-coated surfaces and determine the specific salivary components that facilitate binding with fimbriae. Enzyme-linked immunosorbent assay (ELISA) established that the S. mutans fimbria preparation bound to components of whole saliva. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot techniques were used to separate components of whole saliva and determine fimbria binding. SDS-PAGE separated 15 major protein bands from saliva samples, and Western blot analysis indicated significant binding of the S. mutans fimbria preparation to a 52-kDa salivary protein. The major fimbria-binding salivary protein was isolated by preparative electrophoresis. The ability of the S. mutans fimbria preparation to bind to the purified salivary protein was confirmed by Western blot analysis and ELISA. Incubation of the purified salivary protein with the S. mutans fimbria preparation significantly neutralized binding of the salivary protein-fimbria complex to saliva-coated surfaces. The salivary protein, whole saliva, and commercial amylase reacted similarly with antiamylase antibody in immunoblots. A purified 65-kDa fimbrial protein was demonstrated to bind to both saliva and amylase. These data indicated that the S. mutans fimbria preparation and a purified fimbrial protein bound to whole-saliva-coated surfaces and that amylase is the major salivary component involved in the binding.
The mechanism of Streptococcus mutans attachment to saliva-coated tooth surfaces has generated considerable interest, because blocking of attachment may lead to the prevention of dental caries. However, other than studies of salivary proline-rich polypeptides (PRP) (11, 12), little attention has been devoted to the specific salivary components responsible for the initial S. mutans adherence to saliva-coated tooth surfaces. S. mutans antigen I/II has been strongly implicated in the initial adherence to saliva-coated surfaces (13, 21). It is also well established that the later secondary attachment of S. mutans to tooth surfaces occurs with production of water-insoluble glucans by cell-associated glucosyltransferases (GTF) (21). Previously, members of our group characterized fimbrial surface components on S. mutans cells (7). Recently, Viscount et al. demonstrated Streptococcus parasanguinis fimA fimbrial gene homologs in S. mutans by hybridization (32). Because bacterial fimbriae play a significant role in the colonization of many pathogens, the function of S. mutans fimbriae may be to provide an additional mechanism for initial attachment to tooth surfaces. S. mutans strains from caries-active patients have significantly more fimbrial material on their surfaces than strains from caries-free subjects or a laboratory strain (27). In addition, our laboratory has generated data that indirectly suggest that S. mutans strains containing the most fimbriae may also induce the highest numbers of carious lesions (reference 27 and data not published).
Fimbriae have a particular tropism for certain tissues and, more specifically, carbohydrate moieties of glycoproteins associated with that tissue (2, 4, 19, 21). Many gram-negative bacterial fimbriae, including those from the oral microflora, have been well characterized (15). The interactions between Porphyromonas gingivalis recombinant fimbriae and individual salivary components have been examined (1). The greatest binding occurred with acidic PRP. It was also determined that statherin enhanced the binding of P. gingivalis fimbriae to hydroxyapatite (HA) beads. Understanding of the biology of gram-positive fimbriae is not as complete, and relatively little is known regarding gram-positive oral bacterial fimbriae. However, fimbriae from S. parasanguinis (3, 5, 6, 25), Streptococcus sanguinis (23), Streptococcus salivarius (33), and Actinomyces naeslundii (2) have been characterized. Studies conducted on S. parasanguinis FW213 (a member of the group of microorganisms formerly classified as S. sanguis [S. sanguinis]), which is one of the primary colonizers of dental plaque, have been extensive and demonstrated that attachment to saliva-coated HA is mediated by a 36-kDa adhesin protein, FimA. FimA is found on the fimbrial tips and is able to displace bound FW213 cells (5, 25). S. parasanguinis fimbriae are essential for the microorganism to attach, since wild-type fimbriated FW213 cells bind well to saliva-coated HA in an in vitro tooth model, whereas afimbriated FW213 mutants do not (6). Incubation of FimA with HA blocked the binding of 85% of whole cells added to saliva-coated HA (5, 6). The gene that encodes FimA has been cloned (25). Insertional and deletional FimA mutants produce fimbriae, suggesting that FimA is not the structural subunit. In addition, FimA mutants caused significantly less disease in an animal endocarditis model than did bacteria containing the fimbrial protein (3).
Binding of oral streptococci to specific salivary components such as amylase has been described. Several reports described the complex interactions between Streptococcus gordonii whole cells and human salivary amylase (28–30). In this regard, S. gordonii binding to amylase-coated HA was improved in the presence of maltotriose; however, S. sanguinis adhesion to amylase-coated HA was not enhanced by the presence of maltotriose (28). S. mutans cells have not been shown to bind to amylase.
It is clear that the fimbriae of certain oral bacteria have specific interactions with glycoproteins in the salivary pellicle that coats the tooth surface (26). The purpose of this study was to characterize the interactions between saliva and a preparation of S. mutans fimbriae. In Western blot analysis, a 52-kDa salivary protein displayed significant activity with the S. mutans fimbria preparation. We chose to isolate and identify the salivary protein and determine the characteristics of binding to the S. mutans fimbria preparation.
MATERIALS AND METHODS
Bacteria.
An S. mutans isolate from the saliva of a 7-year-old caries-active child (defined as having ≥5 unrestored surfaces) designated strain A32-2 was used in all experiments; it was maintained in 5% CO2 and 95% air at 37°C overnight in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.) and passaged a minimum number of times. This strain has previously been described to be heavily fimbriated (designated CS2 in reference 27).
Fimbrial preparation.
A modification (7, 27) of the technique of Morris and colleagues (23) for isolating fimbriae from S. sanguinis whole cells was used for the removal of S. mutans fimbriae. The procedure utilized alternating high- and low-speed centrifugations. S. mutans was grown in 9 liters of Todd-Hewitt broth for 18 h at 37°C in 5% CO2 and 95% air. Cells were pelleted and washed once gently at 16,274 × g at 4°C for 10 min in fimbria buffer (10 mM phosphate-buffered saline, 1 mM CaCl2, and 1 mM phenylmethylsulfonyl fluoride [pH 7.2]) and stored as a pellet at −20°C overnight. Phenylmethylsulfonyl fluoride was added to inhibit endogenous proteolytic digestion of fimbrial proteins, and CaCl2 was used to reduce fimbrial aggregation. Frozen cells were thawed and then suspended in fimbria buffer, and fimbriae were removed with a Waring blender by using two 1-min cycles at high speed. Following blending, the sample was centrifuged (16,274 × g, 4°C, 10 min) to remove intact cells and cell debris, and the fimbria preparation in the supernatant was isolated by ultracentrifugation (110,000 × g, 4°C, 2 h). The pellet containing the fimbria preparation was resuspended in fimbrial buffer and centrifuged (16,274 × g, 4°C, 10 min) to remove cellular debris and aggregated fimbriae, and the supernatant was divided into aliquots and stored at −80°C. The protein concentration was determined by using a micro-protein assay (Bio-Rad Laboratories, Hercules, Calif.).
Preparation of salivary components and the purified fimbrial protein.
Saliva was collected from seven healthy individuals (neither caries free [i.e., no decayed, missing, or filled surfaces] nor caries active [i.e., having ≥5 unrestored surfaces]) and stored at −20°C. Prior to use, the saliva samples were clarified by centrifugation (2,800 × g, 4°C, 10 min), and protein concentrations were determined. Saliva samples were diluted to 500 μg of protein/ml in physiological saline for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) or in 0.1 M carbonate-bicarbonate buffer (pH 9.6) for enzyme-linked immunosorbent assay (ELISA). In order to separate salivary protein fractions, preparative gel electrophoresis (Prep cell model 491; Bio-Rad) was utilized. The resolving and stacking gels were composed of 10 and 3% acrylamide (National Diagnostics, Atlanta, Ga.), respectively. A clarified whole-saliva sample (2 ml) was added to an equal volume of SDS-PAGE sample buffer, boiled for 7 min, and placed on a 6-cm column and subjected to 12 W of continuous power. The protein of interest, previously determined by immunoblotting of whole saliva to have a molecular mass of approximately 52 kDa, was eluted after approximately 8 h of electrophoresis. The proteins were collected and analyzed for molecular weight and purity by gel electrophoresis after staining with Coomassie brilliant blue. The fractions of interest were pooled and passed through an affinity column that removes SDS (Extracti-Gel; Pierce, Rockford, Ill.) and stored at −80°C. Purification of the immunodominant 65-kDa fimbrial protein identified earlier (7, 27) was also accomplished by preparative gel electrophoresis with an identical method. Rat antisera to the enriched A32-2 fimbrial preparation and to the 65-kDa fimbrial protein were obtained from eight animals, each immunized with 5 μg of protein/ml incorporated into the RIBI adjuvant system (RIBI ImmunoChem Research, Inc., Hamilton, Mont.). Rats were injected subcutaneously with 0.2 ml of fimbrial preparation in each of two sites, and 0.1 ml was injected intraperitoneally twice, 21 days apart; blood was collected 7 days after the last injection. The blood was allowed to clot, and serum was obtained and stored at −20°C until used.
ELISA for binding of fimbriae and fimbrial protein to whole saliva and salivary components.
Whole saliva (undiluted and diluted 1:2 and 1:10), purified 52-kDa salivary protein (65.0 μg/ml), and human salivary α-amylase (10.0 μg/ml) (type IX A; Sigma Chemical Co., St. Louis, Mo.) were assayed to determine the abilities of fimbriae and the fimbrial protein to bind to salivary components. Polystyrene 96-well microtiter plates (Flow Laboratories, Inc., McLean, Va.) were coated (100 μl) with the salivary components or whole saliva (diluted in 0.1 M carbonate-bicarbonate buffer [pH 9.6]) and incubated for 3 h at 37°C or overnight at 4°C. The unbound salivary components were removed by washing the plates three times with normal saline containing 0.05% Tween 20 (Tween-saline). A solution of 1% bovine serum albumin (BSA) (Sigma) in carbonate-bicarbonate buffer was added (200 μl) to block any unbound sites, and the mixture was incubated for 1 h at 25°C. Following a wash step, 100 μl of the A32-2 fimbrial preparation (33.0 μg/ml of saline) and purified 65-kDa fimbrial protein (1 μg/ml) or Tween-saline (no-fimbria control) were added, and the mixture was incubated for 3 h at 37°C and washed three times. Rat antibody to the A32-2 fimbria preparation or rat antibody to the 65-kDa fimbrial protein (both diluted 1:4,000 in Tween-saline) was added (100 μl) and the mixture was incubated for 3 h at 37°C. After a wash step, goat antibody to rat immunoglobulin G (IgG) (Fc specific) conjugated to horseradish peroxidase (Sigma) was added (100 μl; 1:8,000 dilution) and the mixture was incubated for 3 h at 37°C. After a final wash step, the substrate (10 mg of orthophenylenediamine dihydrochloride and 14 μl of 30% H2O2 in 20 ml of 0.5 M citrate buffer [pH 5.0]) was added (100 μl), color development was monitored for 30 min, and the reaction was read at 490 nm with a microplate spectrophotometer (Molecular Devices Corp., Menlo Park, Calif.). In addition, a modification of the ELISA described above was used to determine the efficacy of the purified 52-kDa salivary protein in inhibiting the binding of S. mutans A32-2 fimbriae to a 1:10 dilution of whole saliva. Mixtures of the S. mutans fimbria preparation (33.0 μg/ml) and serially diluted 52-kDa salivary protein (0.5 to 65.0 μg/ml) were incubated for 30 min at 37°C and used in place of the untreated fimbria preparation. Controls included whole saliva and BSA.
Immunoblot analysis for binding of fimbriae to salivary components and amylase detection.
In order to determine which components in whole saliva bound S. mutans fimbriae, reducing SDS-PAGE was used (16). The resolving and stacking gels were composed of 10 and 3% acrylamide, respectively. Saliva samples (50-μl samples in saline) were boiled for 7 min and electrophoresed with a minigel electrophoresis apparatus (Mini-Protean II; Bio-Rad) for 60 min at 150 V. After electrophoresis, proteins separated on the gel were transferred to nitrocellulose paper (Bio-Rad) overnight at 4°C at a constant voltage of 30 V in a mini-transblot electrophoretic transfer cell (Bio-Rad) (31). The nitrocellulose paper was blocked in a solution of defatted milk (1% milk fat; Carnation instant milk; Carnation Company, Los Angeles, Calif.) diluted in washing buffer (0.9% NaCl containing 0.5% Tween-20) (WBT) for 2 h at 25°C. The nitrocellulose paper was washed with WBT three times for 10 min each, 2 ml of S. mutans fimbrial preparation (33 μg/ml) in WBT was added, and the paper was incubated for 1 h at 25°C. The membrane was washed to remove unbound protein and incubated with rat antibody to A32-2 fimbriae (diluted 1:500 in WBT) for 1 h at room temperature. Goat antibody to rat IgG (Fc specific)-alkaline phosphatase conjugate (1:1,000 in WBT; 100 μl) (Sigma) was added and the membrane was incubated for 1 h. Binding of the antibody was detected by addition of alkaline phosphatase substrate (p-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolylphosphate; Bio-Rad) dissolved in 100 mM Tris HCl (pH 9.5). In order to determine whether the 52-kDa salivary protein was amylase, the isolated salivary protein (65.0 μg/ml), commercial purified amylase (10.0 μg/ml), and undiluted whole saliva were electrophoresed by SDS-PAGE, transferred to nitrocellulose, and probed with rabbit anti-human α-amylase (Sigma) followed by alkaline phosphatase-labeled goat anti-rabbit IgG (Sigma) and a substrate, similar to the method described above.
Statistical analysis.
The data were reduced by computing the means and standard errors of the means (SEM) of the absorbances of each sample, determined in triplicate. The data were analyzed by Student’s t test, and differences were defined as significant when P was ≤0.05.
RESULTS
Fimbria binding assays.
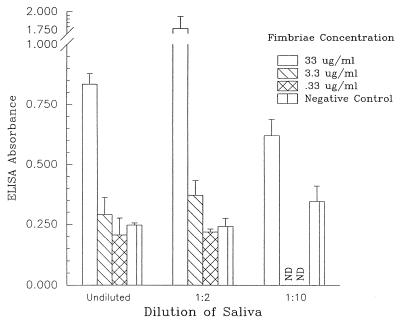
ELISA and immunoblotting were used to establish that the S. mutans fimbria preparation bound to saliva-coated surfaces. An ELISA was performed to determine if an S. mutans A32-2 fimbria preparation bound to human whole saliva. Fimbriae from S. mutans A32-2, a strain isolated from a caries-active subject, demonstrated significant binding compared with the corresponding Tween-saline control (i.e., with no fimbriae) (Fig. 1). The binding of fimbrial components to saliva was reduced when either the saliva or fimbriae were diluted. These data provided the first indication that S. mutans fimbriae had binding activity with saliva-coated surfaces. BSA-coated wells did not bind fimbriae (data not shown).
FIG. 1.
Binding of S. mutans fimbriae to whole-saliva-coated ELISA plates. The ability of S. mutans fimbriae (0.33 to 33.00 μg/ml) to bind to saliva (undiluted and diluted 1:2 and 1:10) was determined by ELISA. The negative controls were wells that did not contain fimbriae. The ELISA absorbances (means ± SEM) represent a relative measurement of binding between fimbriae and saliva. ND, not determined.
Immunoblot analysis of human whole saliva probed with S. mutans A32-2 fimbriae.
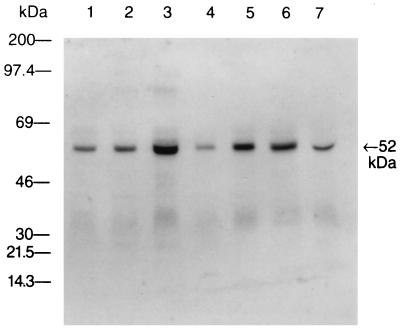
The binding of the S. mutans A32-2 fimbria preparation to separated salivary proteins was analyzed by immunoblotting. Human whole-saliva samples were collected from seven healthy subjects. Each saliva sample was electrophoresed, transferred to nitrocellulose paper, and probed with the S. mutans fimbria preparation. Fimbriae from the A32-2 strain bound strongly to a 52-kDa salivary protein in all seven saliva samples (Fig. 2). Controls with no fimbriae did not reveal any bands.
FIG. 2.
Representative immunoblot of whole saliva from seven different subjects. Blots were probed with fimbriae from S. mutans A32-2, followed by rat antibody to fimbriae of S. mutans A32-2 and alkaline phosphatase-labeled goat antibody to rat IgG. Whole-saliva samples from seven subjects (lanes 1 through 7) are shown. The arrow indicates the molecular mass of the major salivary component that bound fimbriae.
Isolation of a 52-kDa salivary protein with S. mutans fimbria-binding activity.
In order to better understand the interaction between the 52-kDa salivary protein and S. mutans fimbriae, the salivary protein was isolated by preparative gel electrophoresis. Following elution, the fractions were analyzed by gel electrophoresis, and fractions that contained only one band were identified (Fig. 3).
FIG. 3.
Representative dually stained (Coomassie brilliant blue and silver) SDS-PAGE gel containing purified salivary protein. Purified salivary protein was collected by preparative gel electrophoresis and analyzed by SDS-PAGE. The arrow on the right indicates the molecular mass of the isolated salivary component.
ELISA for binding of S. mutans fimbriae and purified 65-kDa fimbrial protein to isolated salivary protein, amylase, and whole saliva.
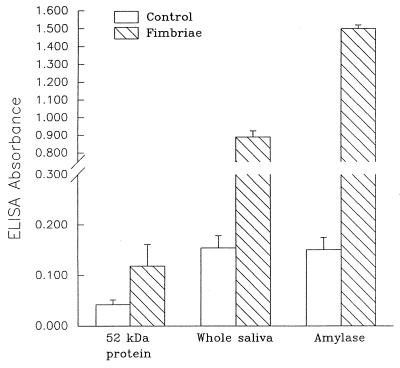
In order to ascertain that both the salivary protein and amylase have fimbria-binding characteristics, an ELISA was employed to measure binding. Amylase was chosen because its molecular mass is near 52 kDa and because several oral streptococci have demonstrated the ability to bind to amylase (26–28). In this assay, amylase (10.0 μg/ml) had significantly greater fimbria-binding activity than the no-fimbria Tween-saline control (Fig. 4). Amylase also had an absorbance significantly greater than that of diluted whole saliva (0.5 μg/ml). The isolated salivary protein (65.0 μg/ml) had a lower absorbance than either amylase or whole saliva, but the absorbance was significantly higher than that of the no-fimbria control. Purified 65-kDa fimbrial protein bound similarly to amylase (optical density at 490 nm [OD], 0.250 ± 0.026 [mean ± SEM]) as to a 1:2 dilution of saliva (OD, 0.260 ± 0.030) but not to a Tween-saline negative control (OD, 0.070 ± 0.012).
FIG. 4.
S. mutans A32-2 fimbria binding to salivary proteins. ELISA plate wells were coated with the purified salivary protein (65.0 μg/ml), whole saliva (diluted 1:10), and amylase (10.0 μg/ml). After blocking with 1% BSA, the S. mutans A32-2 fimbria preparation (33.0 μg/ml) was incubated with the various salivary proteins. The controls did not include fimbriae. The ELISA absorbances (means ± SEM) represent a relative measurement of binding between fimbriae and salivary components.
Inhibition of binding of S. mutans fimbriae to whole-saliva-coated surfaces.
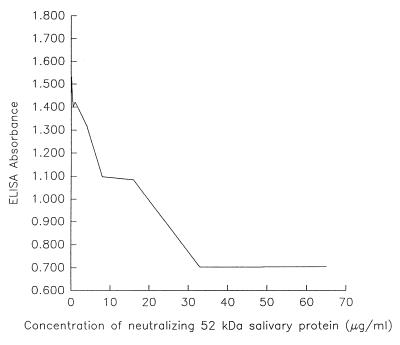
In binding assays, an important feature is the ability to inhibit the interaction. The ability to inhibit binding suggests that the interaction is specific. In this system, the purified salivary protein was incubated with the fimbria preparation from S. mutans A32-2. Following incubation with the salivary protein, the mixture was added to whole saliva. The data indicated an inverse relationship between the concentration of the salivary protein and the extent of binding of the S. mutans fimbria preparation to whole saliva (Fig. 5). Whole saliva and BSA controls yielded complete and no inhibition, respectively.
FIG. 5.
Neutralization of S. mutans fimbria binding to saliva-coated surfaces by a purified salivary protein. The S. mutans A32-2 fimbria preparation (33.0 μg/ml) was incubated with various concentrations (0.5 to 65.0 μg/ml) of the purified salivary protein and assayed for binding to saliva.
Immunoblot analysis of the purified salivary protein probed with anti-human α-amylase antibody.
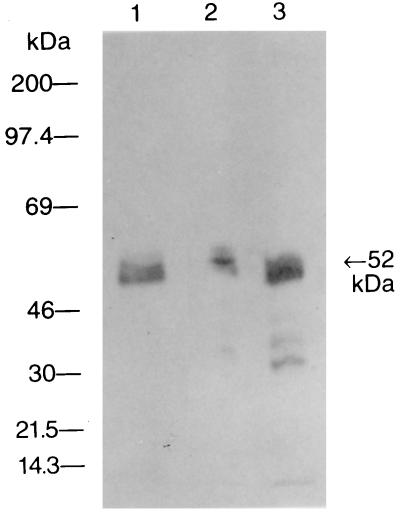
The purified salivary protein, human amylase, and whole saliva were assayed for reactivity with rabbit antibody to human α-amylase. The results indicated that all three salivary preparations contained components that were recognized by the antiamylase antibody (Fig. 6).
FIG. 6.
Representative antiamylase immunoblot. Purified salivary protein (65.0 μg/ml), undiluted whole saliva, and human α-amylase (10.0 μg/ml) were probed with rabbit antibody to human α-amylase. Human whole saliva (lane 1), human α-amylase (lane 2), and purified salivary protein (lane 3) are shown. The arrow on the right indicates the molecular mass of the isolated salivary component.
DISCUSSION
It is generally accepted that pathogenic bacteria must first attach to a host surface to cause infection. The structures that provide attachment are referred to as adhesins. It is of great importance to characterize not only the bacterial adhesin but also the host ligand. Understanding the mechanism of attachment may aid in prevention of the disease. Several investigators have examined salivary components as potential receptors for S. mutans and other oral bacteria (4, 11, 13, 14, 17, 18, 24). Gibbons et al. (10–12) documented that PRP attach with great affinity to HA and S. mutans whole cells attach to PRP-coated HA beads. The majority of research has focused on the attachment of S. mutans whole cells to saliva-coated surfaces. Our laboratory was interested in determining the ability of the fimbrial preparation to bind to saliva.
Our data provided evidence of binding between the fimbrial preparation and whole saliva. S. mutans A32-2 fimbriae demonstrated a significant increase in binding as indicated by ELISA absorbance, depending on saliva and fimbria concentrations, compared to the no-fimbria control. After determining that a component of saliva bound to S. mutans fimbriae, we separated saliva by electrophoresis and identified the component(s) which bound fimbriae. Immunoblots of S. mutans A32-2 fimbriae demonstrated significant activity with a salivary component at about 52 kDa. Perhaps the best-characterized salivary protein with this molecular mass is amylase. Amylase is a receptor for several oral streptococci (28–30) and has a reported molecular mass of approximately 55 kDa.
In order to determine if the 52-kDa protein was amylase, whole saliva was subjected to preparative gel electrophoresis to separate the salivary protein from other salivary components. Isolation of the salivary protein was successful; however, the separation technique, which used SDS and boiling, denatured the protein and inactivated amylase enzymatic activity (data not shown). Thus, confirmation required utilization of antibodies specific for human α-amylase to detect specific epitopes within the molecule. The purified salivary protein had epitopes that were recognized by antibody to human salivary α-amylase. These data suggest that S. mutans may bind to amylase-coated surfaces. These data are contradictory to published reports that other oral streptococci, such as S. gordonii but not S. mutans, bind salivary amylase (28). There are several possible explanations for this finding. The first explanation is that most investigators have analyzed S. mutans whole-cell, but not fimbria, binding activity with salivary components (12, 28). The second reason may be that different systems of measurement provide different results (20, 30). The most commonly utilized techniques for binding determinations are systems that include radionuclides incorporated into whole cells or the receptor. Generally, HA beads have served as the surface for binding amylase or whole saliva. In our studies, although amylase binding to a nitrocellulose membrane or an ELISA plate may not be representative of dental plaque, the binding surfaces used may expose a binding site that is not exposed by attachment to HA. The A32-2 strain was isolated from a caries-active subject, so the fimbriae may have increased the pathogenic potential of this strain by acquiring amylase-binding capabilities. In this regard, Mintz and Fives-Taylor (22) reported that strain variants of Actinobacillus actinomycetemcomitans demonstrated differences in adhesion to a human oral carcinoma cell line, suggesting that such alterations may occur in the oral cavity. However, our earlier data indicated that the amylase-binding 65-kDa fimbrial protein was present in various levels in isolates from both caries-active and caries-free subjects and in a laboratory strain (27), suggesting that amylase-binding activity resides in many strains of S. mutans.
An important characteristic of binding is the ability to inhibit the interaction. We were able to inhibit the binding of S. mutans fimbriae to whole saliva with competitive inhibition by incubating the isolated salivary protein with the S. mutans A32-2 fimbria preparation. The highest concentration of the purified salivary protein (65.0 μg/ml) caused more than a twofold decrease in ELISA absorbance compared to the negative control (i.e., with no salivary protein added).
Other studies in this laboratory have demonstrated that protective mucosal immune responses to the fimbriae are able to reduce S. mutans colonization and caries in experimental animals following intranasal immunization with a fimbria-cholera toxin conjugate (8). Furthermore, antibodies to the fimbriae or the 65-kDa fimbrial protein inhibited caries formation and S. mutans colonization in an in vitro caries model study (9). The 65-kDa fimbrial protein that binds amylase was present in fimbrial preparations from all S. mutans strains examined which did not react with specific antibodies to either antigen I/II or GTF (27), suggesting that all S. mutans strains carry an amylase-binding fimbrial protein distinct from either antigen I/II or GTF.
Future studies are planned to investigate the genomic strain variations between various S. mutans isolates carrying different levels of fimbriae by utilizing pulsed-field gel electrophoresis and restriction fragment length polymorphism. We have raised antibody specific for fimbrial proteins, so the screening of cDNA libraries may allow the detection of the gene(s) of interest. Once that is accomplished, the gene can be cloned and a pure polypeptide can be analyzed for binding activity with amylase. Nevertheless, it is clear that a surface fimbrial component of S. mutans A32-2 has binding reactivity primarily with salivary amylase.
ACKNOWLEDGMENT
We are grateful to Margherita Fontana for helpful discussions and critical comments on the manuscript.
REFERENCES
- 1.Amano A, Sojar H T, Lee J-Y, Sharma A, Levine M J, Genco R J. Salivary receptors for recombinant fimbrillin of Porphyromonas gingivalis. Infect Immun. 1994;62:3372–3380. doi: 10.1128/iai.62.8.3372-3380.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babu J P, Dabbous M K, Abraham S N. Isolation and characterization of a 180-kilodalton salivary glycoprotein which mediates the attachment of Actinomyces naeslundii to human buccal epithelial cells. J Periodontal Res. 1991;26:97–106. doi: 10.1111/j.1600-0765.1991.tb01632.x. [DOI] [PubMed] [Google Scholar]
- 3.Burnette-Curley D, Wells V, Viscount H, Munro C L, Fenno J C, Fives-Taylor P, Macrina F L. FimA, a major virulence factor associated with Streptococcus parasanguis endocarditis. Infect Immun. 1995;63:4669–4674. doi: 10.1128/iai.63.12.4669-4674.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowley P J, Brady L J, Piacentini D A, Bleiweis A S. Identification of a salivary agglutinin-binding domain within cell surface adhesin P1 of Streptococcus mutans. Infect Immun. 1993;61:1547–1552. doi: 10.1128/iai.61.4.1547-1552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fachon-Kalweit S, Elder B L, Fives-Taylor P. Antibodies that bind to fimbriae block adhesion of Streptococcus sanguis to saliva-coated hydroxyapatite. Infect Immun. 1985;48:617–624. doi: 10.1128/iai.48.3.617-624.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fives-Taylor P M, Thompson D W. Surface properties of Streptococcus sanguis FW213 mutants nonadherent to saliva-coated hydroxyapatite. Infect Immun. 1985;47:752–759. doi: 10.1128/iai.47.3.752-759.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fontana M, Gfell L E, Gregory R L. Characterization of preparations enriched for Streptococcus mutans fimbriae: salivary immunoglobulin A antibodies in caries-free and caries-active subjects. Clin Diagn Lab Immunol. 1995;2:719–725. doi: 10.1128/cdli.2.6.719-725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontana M, Dunipace A J, Stookey G K, Gregory R L. Intranasal immunization against dental caries with a Streptococcus mutans-enriched fimbrial preparation. Clin Diagn Lab Immunol. 1999;6:405–409. doi: 10.1128/cdli.6.3.405-409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fontana, M., A. J. Dunipace, G. K. Stookey, and R. L. Gregory. 1998. Unpublished data.
- 10.Gibbons R J, Hay D I, Schlesinger D H. Delineation of a segment of adsorbed salivary acidic proline-rich proteins which promotes adhesion of Streptococcus gordonii to apatitic surfaces. Infect Immun. 1991;59:2948–2954. doi: 10.1128/iai.59.9.2948-2954.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbons R J, Hay D I. Adsorbed salivary acidic proline-rich proteins contribute to the adhesion of Streptococcus mutans JBP to apatitic surfaces. J Dent Res. 1989;68:1303–1307. doi: 10.1177/00220345890680090201. [DOI] [PubMed] [Google Scholar]
- 12.Gibbons R J, Cohen L, Hay D I. Strains of Streptococcus mutans and Streptococcus sobrinus attach to different pellicle receptors. Infect Immun. 1986;52:555–561. doi: 10.1128/iai.52.2.555-561.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajishengallis G, Koga T, Russell M W. Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res. 1994;73:1493–1502. doi: 10.1177/00220345940730090301. [DOI] [PubMed] [Google Scholar]
- 14.Handley P S. Structure, composition and functions of surface structures on oral bacteria. Biofouling. 1990;2:239–264. [Google Scholar]
- 15.Klemm P. Fimbrial adhesins of Escherichia coli. Rev Infect Dis. 1985;7:321–340. doi: 10.1093/clinids/7.3.321. [DOI] [PubMed] [Google Scholar]
- 16.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Lamont R J, Demuth D R, Davis C A, Malamud D, Rosan B. Salivary-agglutinin-mediated adherence of Streptococcus mutans to early plaque bacteria. Infect Immun. 1991;59:3446–3450. doi: 10.1128/iai.59.10.3446-3450.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S F, Progulske-Fox A, Erdos G W, Piacentini D A, Ayakawa G Y, Crowley P J, Bleiweis A S. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II) Infect Immun. 1989;57:3306–3313. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine M J, Herzberg M C, Levine M S, Ellison S A, Stinson M W, Li H C, Van Dyke T. Specificity of salivary-bacterial interactions: role of terminal sialic acid residues in the interaction of salivary glycoproteins with Streptococcus sanguis and Streptococcus mutans. Infect Immun. 1978;19:107–115. doi: 10.1128/iai.19.1.107-115.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ligtenberg A J M, Walgreen-Weterings E, Veerman E C I, de Soet J J, de Graaff J, Amerongen A V N. Influence of saliva on aggregation and adherence of Streptococcus gordonii HG 222. Infect Immun. 1992;60:3878–3884. doi: 10.1128/iai.60.9.3878-3884.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loesche W J. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mintz K P, Fives-Taylor P M. Adhesion of Actinobacillus actinomycetemcomitans to a human oral cell line. Infect Immun. 1994;62:3672–3678. doi: 10.1128/iai.62.9.3672-3678.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris E J, Ganeshkumar N, Song M, McBride B C. Identification and preliminary characterization of a Streptococcus sanguis fibrillar glycoprotein. J Bacteriol. 1987;169:164–171. doi: 10.1128/jb.169.1.164-171.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogier J A, Klein J P, Sommer P, Frank R M. Identification and preliminary characterization of saliva-interacting surface antigens of Streptococcus mutans by immunoblotting, ligand blotting, and immunoprecipitation. Infect Immun. 1984;45:107–112. doi: 10.1128/iai.45.1.107-112.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oligino L, Fives-Taylor P. Overexpression and purification of a fimbria-associated adhesin of Streptococcus parasanguis. Infect Immun. 1993;61:1016–1022. doi: 10.1128/iai.61.3.1016-1022.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orstavik D, Kraus F W. The acquired pellicle: enzyme and antibody activities. Scand J Dent Res. 1974;82:202–205. doi: 10.1111/j.1600-0722.1974.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 27.Perrone M, Gfell L E, Fontana M, Gregory R L. Antigenic characterization of fimbria preparations from Streptococcus mutans isolates from caries-free and caries-susceptible subjects. Clin Diagn Lab Immunol. 1997;4:291–296. doi: 10.1128/cdli.4.3.291-296.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scannapieco F A, Torres G I, Levine M J. Salivary amylase promotes adhesion of oral streptococci to hydroxyapatite. J Dent Res. 1995;74:1360–1366. doi: 10.1177/00220345950740070701. [DOI] [PubMed] [Google Scholar]
- 29.Scannapieco F A, Haraszthy G G, Cho M-I, Levine M J. Characterization of an amylase-binding component of Streptococcus gordonii G9B. Infect Immun. 1992;60:4726–4733. doi: 10.1128/iai.60.11.4726-4733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scannapieco F A, Bergey E J, Reddy M S, Levine M J. Characterization of salivary α-amylase binding to Streptococcus sanguis. Infect Immun. 1989;57:2853–2863. doi: 10.1128/iai.57.9.2853-2863.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viscount H B, Munro C L, Burnette-Curley D, Peterson D L, Macrina F L. Immunization with FimA protects against Streptococcus parasanguis endocarditis in rats. Infect Immun. 1997;65:994–1002. doi: 10.1128/iai.65.3.994-1002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weerkamp A H, Jacobs T. Cell wall-associated protein antigens of Streptococcus salivarius: purification, properties, and function in adherence. Infect Immun. 1982;38:233–242. doi: 10.1128/iai.38.1.233-242.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]