Summary
Bacteria that assimilate synthetic nitroarene compounds represent unique evolutionary models, as their metabolic pathways are in the process of adaptation and optimization for the consumption of these toxic chemicals. We used Acidovorax sp. strain JS42, which is capable of growth on nitrobenzene and 2-nitrotoluene, in experiments to examine how a nitroarene degradation pathway evolves when its host strain is challenged with direct selective pressure to assimilate non-native substrates. Although the same enzyme that initiates the degradation of nitrobenzene and 2-nitrotoluene also oxidizes 4-nitrotoluene to 4-methylcatechol, which is a growth substrate for JS42, the strain is incapable of growth on 4-nitrotoluene. Using long-term laboratory evolution experiments, we obtained JS42 mutants that gained the ability grow on 4-nitrotoluene via a new degradation pathway. The underlying basis for this new activity resulted from the accumulation of specific mutations in the gene encoding the dioxygenase that catalyzes the initial oxidation of nitroarene substrates, but at positions distal to the active site and previously unknown to affect activity in this or related enzymes. We constructed additional mutant dioxygenases to identify the order of mutations that led to the improved enzymes. Biochemical analyses revealed a defined, step-wise pathway for the evolution of the improved dioxygenases.
Keywords: Nitrotoluene, dioxygenase, biodegradation, aromatic compound, pathway evolution
Introduction
Nitroaromatic compounds are an important class of industrial chemicals that is widely used in the synthesis of diverse products, including dyes, polymers, pesticides, and explosives (Booth, 2007). These chemicals are relatively rare in nature (Ju and Parales, 2010), and the majority of nitroaromatic compounds present in the environment have been introduced by human activities. However, bacteria have the ability to rapidly adapt to the presence of synthetic chemicals and take advantage of these potential sources of carbon and energy by developing new catabolic pathways (Cases and de Lorenzo, 2001; Copley, 2009; Johnson et al., 2002; Muller et al., 2003; van der Meer et al., 1992). Bacterial strains that use compounds such as nitrobenzene (Nishino and Spain, 1993; Nishino and Spain, 1995), mononitrotoluenes (Haigler and Spain, 1993; Haigler et al., 1994; Rhys-Williams et al., 1993; Spiess et al., 1998), and dinitrotoluenes (Nishino et al., 2000; Spanggord et al., 1991) as sole carbon, nitrogen, and energy sources have been isolated from contaminated soil and groundwater. Over the past 20 years, studies of the physiology, genetics, and biochemistry of nitroarene-degrading bacteria have not only revealed the mechanisms of catabolism of these compounds, but also provided clues to their evolutionary origin. Oxidative pathways for nitrobenzene, 2-nitrotoluene (2NT), 3-nitrotoluene (3NT), and 2,4- and 2,6-dinitrotoluene degradation all involve an initial oxidation of the aromatic ring that results in the formation of a catechol and release of nitrite (Haigler et al., 1994; Lessner et al., 2002; Nishino and Spain, 1995; Nishino et al., 2000; Spanggord et al., 1991). These pathways all appear to have evolved from a naphthalene degradation pathway similar to that in Ralstonia sp. strain U2 even though the strains were isolated from geographically distinct sites (Parales, 2000).
Acidovorax sp. strain JS42, which grows on 2NT and nitrobenzene, initiates degradation these compounds with the Rieske-type dioxygenase 2-nitrotoluenene 2,3-dioxygenase (2NTDO) (Haigler et al., 1994; Lessner et al., 2002). 2NTDO adds both atoms of molecular oxygen to the benzene ring, releasing nitrite and forming (3-methyl)catechol, which then enters a meta cleavage pathway (Haigler et al., 1994). Wild-type JS42, however, is unable to grow on 3- or 4-nitrotoluene (4NT) even though 2NTDO is able to oxidize these compounds to 3- or 4-methylcatechol (Parales et al., 1998a), both of which are growth substrates for the strain (Haigler et al., 1994). To directly observe how a degradation pathway changes as the host strain adapts to environmental conditions, we performed long-term laboratory-evolution experiments to select for evolved mutants of Acidovorax sp. JS42 that are able to grow on 4NT. Comparative analyses of the genes and proteins involved in nitrotoluene degradation in wild-type JS42 and the evolved strains led to identification of the genetic changes that allowed growth via a previously undescribed 4NT degradation pathway, and revealed the reason for the inability of the parental strain to grow on 4NT.
Results
Evolution of JS42 and initial characterization of 4NT+ strains.
Direct selection of 4NT-degrading variants of Acidovorax sp. JS42 on minimal medium plates with 4NT as the sole carbon source was unsuccessful. However, spontaneous mutants of JS42 that were able to utilize 4NT were obtained after prolonged selection in liquid culture, essentially using the classical enrichment culture technique. Isolates were then obtained on solid medium with 4NT as the sole carbon source, and three strains (designated JS42-KSJ9, JS42-KSJ10, and JS42-KSJ11; Table S1) were chosen for detailed characterization. Wild-type JS42 and the evolved strains were able to grow on nitrobenzene, 2NT, catechol, 3-methylcatechol, and 4-methylcatechol. The evolved strains had the added ability to grow on 4NT; none of the strains grew on 3NT.
2NTDO activity in the evolved Acidovorax strains.
Although 2NTDO is capable of oxidizing 4NT to 4-methylcatechol, the activity was 50-fold lower than that with 2NT (Parales et al., 1998a), so we hypothesized that increased oxidation of 4NT could have resulted in the ability to grow on 4NT. We first examined the induction of the genes encoding 2NTDO in order to test whether more enzyme was being produced due to increased gene expression. JS42 and the 4NT+ strains were grown in the presence of known inducers (nitrobenzene, 2NT, 3NT, 4NT, and salicylate (Lessner et al., 2003)), and expression was monitored as indicated by the specific activity of the enzyme with nitrobenzene. All four strains had comparable levels of basal and induced dioxygenase activity with the exception of slight increases (1.5 – 2-fold) in induction in the presence of nitrotoluenes (Fig. S1).
To determine if the substrate specificity of the dioxygenases in the 4NT+ strains had changed, salicylate-induced cultures were incubated with nitrobenzene, 2NT, 3NT, or 4NT as substrates, and the amount of nitrite released was measured. As expected (Parales et al., 1998a), wild-type 2NTDO in JS42 had high specific activity with nitrobenzene and 2NT, while activity with 3NT and 4NT was significantly lower (Fig. 1). In contrast, activity with 4NT was significantly higher in the 4NT+ strains compared to JS42 (Fig. 1), suggesting that the 4NT+ derivatives may have acquired mutations in the genes encoding 2NTDO.
Fig. 1.
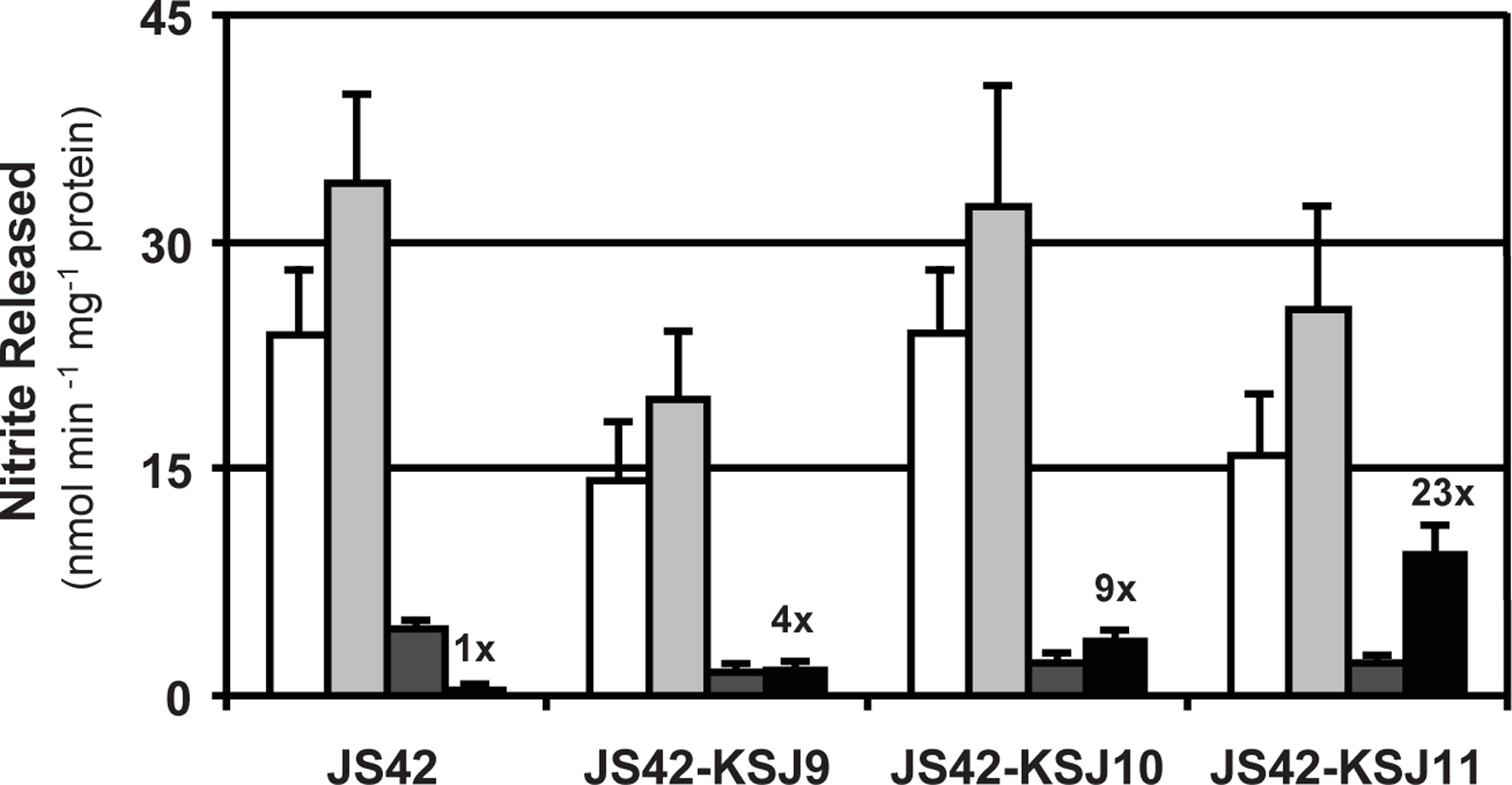
Nitroarene dioxygenase activity in Acidovorax sp. JS42 and 4NT+ evolved strains induced with salicylate. Substrates: nitrobenzene (white), 2NT (light grey), 3NT (dark grey), or 4NT (black) were provided at 1 mM and nitrite in culture supernatants was measured after 30 min. Fold increases in activity with 4NT are indicated. Error bars indicate standard deviations (N = 3).
Identification of mutations in 2NTDO.
To identify the changes responsible for the improved activity with 4NT in the evolved strains, the ntdAcAd genes encoding the 2NTDO oxygenase α and β subunits were amplified by PCR, cloned, and sequenced (see SI for details). A conserved missense mutation resulting in a M248I substitution was present in the gene encoding the catalytic (α) subunit (ntdAc) of the dioxygenase in all three 4NT+ strains. In addition, ntdAc in strains JS42-KSJ10 and JS42-KSJ11 each had a second independent missense mutation, resulting in S242N and L238V substitutions, respectively. Site-directed mutagenesis was used to create additional combinations of L238V, S242N, and M248I amino acid substitutions in 2NTDO (see SI for details) in order to assess the individual contributions of each substitution.
Substrate specificity of the mutant dioxygenases.
To evaluate the effects of the amino acid substitutions on substrate specificity and enzyme activity, whole-cell reactions were carried out in which recombinant E. coli strains expressing the dioxygenase variants were incubated with nitrobenzene, 2NT, 3NT, or 4NT as substrates. We first verified that dioxygenase levels were approximately equal by Western analysis with a polyclonal antiserum raised against NDO9816-4, which reacts with the α subunit of 2NTDO (Parales et al., 1998b). All proteins except the L238V S242N variant were present at levels comparable to wild-type 2NTDO (Fig. S2). Nitrite was measured at the end of six-hour biotransformation reactions and products were identified by GC-MS with standard compounds as controls. 2NTDO generated the characteristic ratios of methylcatechols and nitrobenzyl alcohols (Ju and Parales, 2006; Parales et al., 1998a) from the oxidation of 2NT, 3NT, and 4NT (Table 1). With the exception of the S242N and L238V S242N variants, which were inactive, the mutant enzymes generated significantly less or undetectable amounts of nitrobenzyl alcohols from 2NT, 3NT, and 4NT. In addition to these changes in regiospecificity, these enzymes produced significantly more nitrite from 4NT than 2NTDO (Fig. 2). In particular, the L238V M248I mutant showed a 50-fold increase in activity with 4NT. Some of the mutant enzymes also had slightly improved activity with other substrates compared to wild-type 2NTDO (Fig. 2), and some showed activity with 2,4-dinitrotoluene, which is not oxidized by wild-type 2NTDO (Table S2). However, the L238V S242N M248I triple mutant had reduced enzyme activity with substrates other than 4NT (Fig. 2).
TABLE 1.
Product formation from nitrotoluenes by wild-type and mutant dioxygenases expressed in E. coli.
| Enzymea | Source Strainb | Products (%) produced fromc: | ||||||
|---|---|---|---|---|---|---|---|---|
| 2NT | 3NT | 4NT | ||||||
|
| ||||||||
| 2NBA | 3MC | 3NBA | 3MC | 4MC | 4NBA | 4MC | ||
| 2NTDO | JS42 | 14 | 86 | 16 | 50 | 34 | 5 | 95 |
| L238V | n/a | --- | 100 | --- | 79 | 21 | --- | 100 |
| M248I | JS42-KSJ9 | 3 | 97 | --- | 78 | 22 | --- | 100 |
| L238V M248I |
JS42-KSJ11 | --- | 100 | --- | 73 | 27 | --- | 100 |
| S242N M248I |
JS42-KSJ10 | 2 | 98 | --- | 79 | 21 | --- | 100 |
| L238V S242N M248I |
n/a | --- | 100 | --- | 100 | --- | --- | 100 |
No products were detected with the S242N and L238V S242N variants.
n/a, not applicable; mutant enzymes were generated by site-directed mutagenesis.
N =3; standard deviations were < 3%; 3MC, 3-methylcatechol; 4MC, 4-methylcatechol; 2NBA, 2-nitrobenzyl alcohol; 3NBA, 3-nitrobenzyl alcohol; 4NBA, 4-nitrobenzyl alcohol.
, none detected.
Fig. 2.
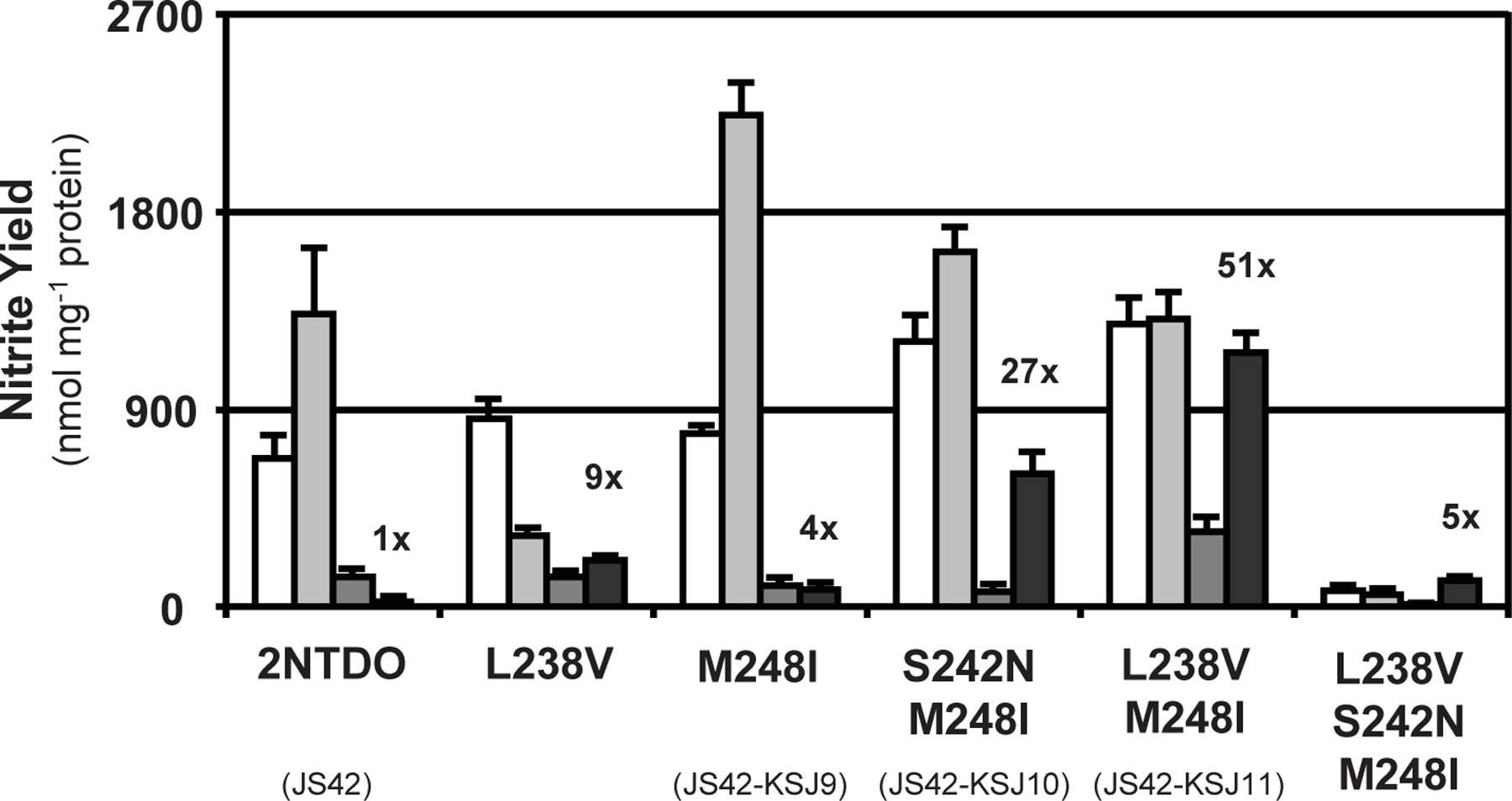
Nitrite produced by E. coli cultures expressing the 2NTDO variants in 6 h whole-cell reactions with nitrobenzene (white), 2NT (light grey), 3NT (dark grey), or 4NT (black) as substrates. The wild-type and evolved dioxygenases are labeled with the names of the parental strains; the other enzymes were generated by site-directed mutagenesis. The S242N and L238V S242N mutants did not produce detectable amounts of nitrite from any of the substrates. Fold increases in activity with 4NT are indicated. Error bars indicate standard deviations (N = 3).
Kinetics of nitroarene dioxygenases.
To gain further insight into the catalytic activities of the evolved dioxygenases relative to the parent enzyme, the kinetic coefficients of the substrates nitrobenzene, 2NT, and 4NT were measured in whole cells of E. coli expressing the relevant dioxygenase genes. As expected, 2NTDO had extremely low activity with 4NT, and did not exhibit Michaelis-Menten kinetics (Table 2). Nitrite was detected only when 0.1% (w/v) 4NT was present, a concentration in excess of the solubility of nitrotoluenes (~4 mM). In contrast, 4NT oxidation by the evolved enzymes was significantly improved. Both M248I and S242N M248I mutant dioxygenases had specific activities with 4NT that were comparable to wild-type 2NTDO with the native substrate 2NT. The L238V M248I double mutant from strain JS42-KSJ11 was the best 4NT-oxidizing enzyme, with the highest maximal activity (Vmax) and lowest Km for 4NT (Table 2).
TABLE 2.
Oxidation kinetics for nitrobenzene, 2NT, and 4NT determined from whole-cell reactions with E. coli expressing wild-type and mutant dioxygenases.
| Enzyme | Source | Kinetic coefficients from the oxidation of:a | |||||
|---|---|---|---|---|---|---|---|
| strain | NB | 2NT | 4NT | ||||
|
|
|||||||
| K m | Vmax | K m | Vmax | K m | Vmax | ||
| 2NTDO | JS42 | 317 ± 44 | 11 ± 1 | 188 ± 22 | 6 ± 1 | ---b | ---b |
| M248I | JS42-KSJ9 | 127 ± 24 | 7 ± 0 | 255 ± 47 | 14 ± 2 | 580 ± 66 | 7 ± 1 |
| S242N M248I |
JS42-KSJ10 | 232 ± 17 | 7 ± 0 | 239 ± 37 | 6 ± 1 | 637 ± 49 | 7 ± 0 |
| L238V M248I |
JS42-KSJ11 | 375 ± 89 | 13 ± 1 | 415 ± 97 | 22 ± 3 | 264 ± 19 | 14 ± 0 |
Km is reported as μM; Vmax is reported as maximal specific activity (nmoles min−1 mg−1 protein)
---, non-Michaelis-Menten behavior; Km and Vmax could not be determined. The maximal oxidation rate observed with saturating amounts of substrate was 0.75 ± 0.02 nmoles min−1 mg−1 protein.
N = 3; r2 > 0.95; standard deviations are reported.
The mutant dioxygenases are sufficient to allow growth on 4NT.
To determine if the mutant dioxygenases were sufficient to confer the ability to grow on 4NT, genes encoding the wild-type and evolved dioxygenases were introduced into JS42Ac (Table S1), a mutant strain that does not make a functional 2NTDO and cannot grow on nitrobenzene or nitrotoluenes (Ju et al., 2009). Complementation of JS42Ac with pKSJ44 (carrying wild-type 2NTDO) restored growth on 2NT, but did not confer the ability to grow on 4NT (Table 3), indicating that the increased copy number of the wild-type ntdAaAbAcAd genes did not increase activity to a level sufficient for growth. However, introduction of plasmids pKSJ93 (M248I), pKSJ94 (S242N M248I), and pKSJ95 (L238V M248I) each allowed growth on 2NT and 4NT (Table 3). In addition, strains carrying the enzyme with the highest activity and lowest Km values (L238V M248I; Fig. 2; Table 2) had the fastest doubling times on 4NT (Table 3).
Table 3.
Growth of Acidovorax strains in liquid culture
| Strain | Dioxygenase Present | Doubling Timea (h) |
Cell Yieldb (μg protein ml−1) | ||
|---|---|---|---|---|---|
|
| |||||
| 2NT | 4NT | 2NT | 4NT | ||
| JS42 | 2NTDO | 4.3 | --- c | 113 | --- |
| JS42-KSJ9 | M248I | 5.2 | 4.5 | 126 | 64 |
| JS42-KSJ10 | S242N M248I |
4.0 | 4.3 | 131 | 63 |
| JS42-KSJ11 | L238V M248I |
5.1 | 3.1 | 126 | 63 |
| JS42Ac (pBBR1MCS5) | none | --- | --- | --- | --- |
| JS42Ac (pKSJ44) | 2NTDO | 4.5 | --- | 113 | --- |
| JS42Ac (pKSJ93) | M248I | 6.0 | 6.8 | 130 | 64 |
| JS42Ac (pKSJ94) | S242N M248I |
4.5 | 6.6 | 132 | 64 |
| JS42Ac (pKSJ95) | L238V M248I |
4.0 | 5.5 | 128 | 61 |
n = 3; standard deviations were 7% or less
n = 3; standard deviations were 5% or less
---, no growth observed
Growth yields and contributions of the meta cleavage pathway
Wild-type JS42 and the complemented 2NTDO mutant JS42Ac(pKSJ44) reached average cells yields of 113 μg protein ml−1 when grown with 2 mM 2NT, while the 4NT+ strains and JS42Ac complemented with the mutant dioxygenases yielded 10–15% more protein (Table 3). This increased cell yield may be attributable to the shift in regiospecificity of 2NT oxidation by the 2NTDO variants (Table 1), which reduced or eliminated the formation of the dead-end metabolite 2-nitrobenzyl alcohol. Although the 4NT+ strains and the complemented JS42Ac strains grew on 4NT, the yields were approximately half of those on 2NT (Table 3), suggesting that only half of the carbon from 4NT was utilized for growth.
Studies of the meta cleavage pathway in Pseudomonas putida mt-2 have demonstrated that all seven carbons from 3-methylcatechol can be assimilated, but due to a decarboxylation that occurs in the oxalocrotonate branch of the pathway, only 6 of the 7 carbons from 4-methylcatechol are utilized (Fig. S3); propionaldehyde and pyruvate are the products of 4-methylcatechol degradation (Murray et al., 1972). Analysis of the JS42 genome (GenBank accession NC_008782) indicated that the strain contains all of the necessary genes to encode both branches of the meta-cleavage pathway (Fig. S3; Table S3). When crude cell extracts of JS42-KSJ11 were incubated with 3 mM 4-methylcatechol, 37 ± 6 mg of pyruvate and 42 ± 11 mg of propionaldehyde were produced after 2.5 hours (see SI for details), demonstrating that enzymes of the meta cleavage pathway are functional and the expected products were generated in approximately equal amounts. However, we found that propionaldehyde accumulated in the headspace of the 4NT+ cultures grown on 4NT (see SI), suggesting that the acylating aldehyde dehydrogenase (ADA) that catalyzes the conversion of acetaldehyde and propionaldehyde (produced from 3- and 4-methylcatechol metabolism, respectively) into acetyl- and propionyl-CoA might not be functional with propionaldehyde. However, assays of extracts of an E. coli strain expressing cloned ctdQ (encoding ADA; Fig. S3) indicated that the enzyme was equally active with acetaldehyde and propionaldehyde (see SI). As expected, propionaldehyde was not produced when cells were grown on nitrobenzene or 2NT, nor was it detected in cultures of P. putida mt-2 growing on p-toluate, which is degraded through 4-methylcatechol (see SI).
We attempted to engineer pathways for propionaldehyde utilization by introducing additional copies of the genes encoding the 4-hydroxy-2-ketovalerate aldolase (HOA) and ADA enzymes from JS42 or Pseudomonas sp. strain CF600 (Shingler et al. 1993) and the genes encoding the conversion of propionyl-CoA to propionate (which is a good growth substrate for JS42) from Salmonella enterica LT2 (Liu and Bobik et al. 2007). These efforts were only partially successful at increasing cell yields during growth on 4NT (see SI and Table S4).
Discussion
The evolved strains contain a new pathway for the degradation of 4NT that has not been reported to date (Fig. 3), as all previously described 4NT degradation pathways involve reduction of the nitro-group and release of ammonia (Fig. 3), rather than oxidation of the aromatic ring and release of nitrite (Haigler and Spain, 1993; Spiess et al., 1998). In this pathway, 4NT is converted to the substrate for ring cleavage in a single step, similar to pathways for 2NT and nitrobenzene degradation (Haigler et al., 1994; Lessner et al., 2003; Mulla et al., 2010).
Fig. 3.
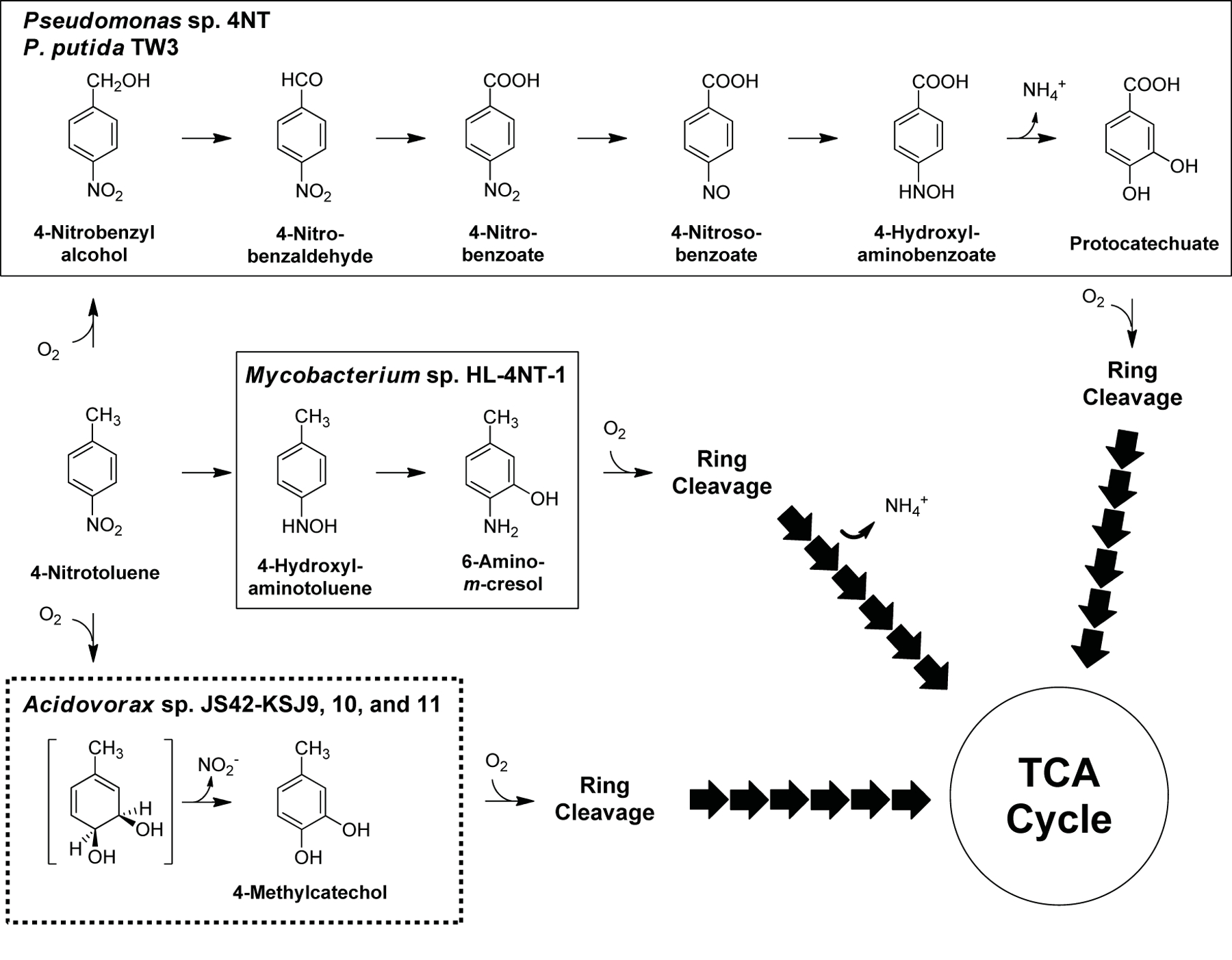
Comparison of 4NT degradation pathways in Pseudomonas sp. strain 4NT (Haigler and Spain, 1993), P. putida TW3 (Hughes and Williams, 2001; James and Williams, 1998; James et al., 2000; Rhys-Williams et al., 1993), Mycobacterium sp. strain HL4-NT-1 (Spiess et al., 1998) and evolved variants of Acidovorax sp. strain JS42.
Interestingly, only three missense mutations were detected in the mutant forms of 2NTDO that allowed growth on 4NT. Analysis of the substrate specificities and oxidation activities of the seven 2NTDO enzymes containing all of the possible combinations of the L238V, S242N, and M248I mutations (Table 1; Fig. 2) suggests that the dioxygenase mutations present in the 4NT+ strains arose by a defined sequence of changes (Fig. 4). The mutation resulting in the M248I substitution was most likely the first to arise, followed independently by the L238V M248I and S242N M248I double mutants. Alternatively, a mutation resulting in the L238V substitution could have been followed by the M248I change; our limited sampling may explain why this variant was not detected among the 4NT+ strains. Given that the S242N mutation is only beneficial in the context of the M248I substitution, and in all other combinations it dramatically reduced or eliminated activity with 4NT, it is unlikely that the S242N M248I variant arose from an initial change at this position. Likewise, there would be no selective advantage of the L238V or the L238V M248I variants to acquire S242N, or the S242N M248I dioxygenase to acquire L238V (Fig. 4). The successful enrichment of spontaneous mutants of Acidovorax sp. JS42 that are able to grow on 4NT may have been facilitated by the natural mutagenicity of 4NT (Padda et al., 2003), although this was not specifically investigated.
Fig. 4.
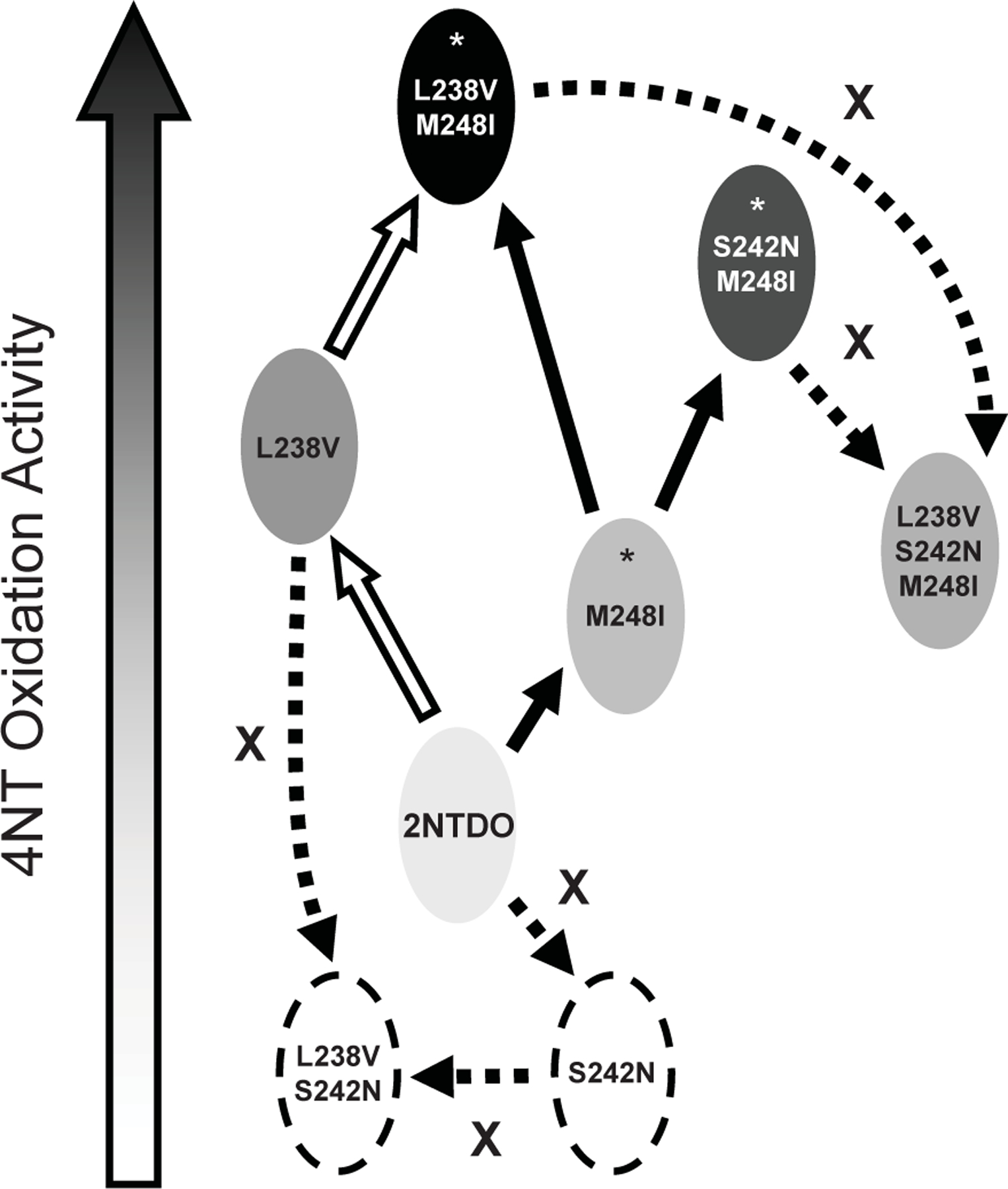
Proposed pathway for the evolution of 2NTDO to variant enzymes with improved 4NT oxidation activity. Dioxygenases with higher relative activities with 4NT are indicated with darker shading; those with no detectable activity are shown with dashed-borders. Solid black arrows indicate steps that lead to enzymes with improved activity (enzymes identified in this study indicated by asterisks), and solid white arrows indicate another possible route to enzymes with improved activity. Dashed arrows indicate routes to enzymes with decreased activity.
To date, substitutions at positions corresponding to 238, 242, and 248 have not been shown to affect substrate specificity or enzyme activity in other nitroarene or naphthalene dioxygenases (Ju and Parales, 2006; Keenan et al., 2004; Keenan et al., 2005; Lee et al., 2005; Leungsakul et al., 2005; Parales et al., 2000a; Parales et al., 2000b). To gain structural insight into the 4NT-evolved variants of 2NTDO, a homology model of the α-subunit of 2NTDO was generated based on the crystal structure of NBDO. The residues at positions 238, 242, and 248 are located in one region, adjacent to but not directly within the active site of the enzyme (Fig. S4). Although the exact mechanism by which these three residues affect enzyme activity is unclear, their positions outside the catalytic center suggest that activity may be modulated by indirect effects such as through subtle changes in the topology of the protein (Morley and Kazlauskas, 2005).
In long-term evolution experiments, phenotypic changes in fitness reflect underlying genetic mutations (Elena and Lenski, 2003; Finkel, 2006). The only change required for growth on 4NT resulted from mutations that improved the activity of 2NTDO with 4NT. This result indicates that the primary bottleneck in 4NT degradation was due to inadequate metabolic flux from the low activity of 2NTDO with 4NT. It is of interest to note, however, that the doubling times of the complemented JS42Ac strains were longer than those of the evolved strains on 4NT (Table 3). It is unlikely that the longer doubling times were due to increased metabolic burden of maintaining the plasmids, as JS42Ac complemented in trans with wild-type 2NTDO had a similar doubling time as the wild-type strain on 2NT (Table 3). These results suggest that additional beneficial mutations may have occurred in the evolved strains. However, other mutations have not yet been identified.
Both the evolved and complemented strains had growth yields that suggested only half of the total available carbon from 4NT was utilized (Table 3). For reasons that we do not understand, propionaldehyde is not efficiently metabolized by JS42 or its derivatives. Propionaldehyde and pyruvate are also generated by Pseudomonas putida CF600 during growth on 3,4-dimethylphenol, and the acylating acetaldehyde dehydrogenase (DmpF) in this strain is capable of acting on both acetaldehyde and propionaldehyde as substrates (Powlowski et al., 1993). Although JS42 grows well on propionate, it is apparently lacking one or more enzymes for the efficient conversion of propionaldehyde to propionate, and our attempts to improve growth and substrate utilization by introducing some of these enzymes were only partially successful (SI; Table S3).
This study demonstrates the power of direct selection of mutant strains in response to the presence of alternative carbon sources to generate enzymes and pathways with new or improved activities. In addition, this strategy mimics the selection process occurring in natural ecosystems in response to the introduction of new chemicals. Cells containing mutations that confer the largest improvements in activity will out-compete others in the population, while cells harboring no mutations, deleterious mutations, or mutations that only minimally improve enzyme activity are at a growth disadvantage, and are eliminated from the population during multiple rounds of selection in liquid culture. Using this approach, specific amino acid positions or regions of the enzyme important for activity at non-obvious locations can be identified; such positions are likely to be missed when using in vitro protein engineering methods. This strategy also bypasses the need for separate optimization of engineered proteins after introduction into the host strain of choice, since the enzyme of interest is already an integral part of the degradation pathway. In this case, JS42 was able to take advantage of the promiscuity of the dioxygenase and develop the ability to grow on multiple nitroaromatic compounds. From a biodegradation perspective, these newly evolved strains are valuable because of their ability to completely degrade three different nitroarene compounds. It is conceivable that the range of nitroaromatic compounds that serve as growth substrates for JS42 could be further extended using this approach.
Long-term evolutionary adaptation to a single environmental condition can lead to the formation of specialists, which exhibit tradeoffs in relative fitness in different environments (Bull et al., 2000; Cooper and Lenski, 2000; Elena and Lenski, 2003). Interestingly, our strains did not lose the ability to grow with nitrobenzene or 2NT, which may reflect the absence of negative trade-offs that would otherwise constrict specificity towards 4NT. Future studies are currently underway to characterize the evolution of JS42 for growth on other nitroaromatic compounds, identify co-occurring mutations, and the biochemical analysis of the mutant dioxygenases.
Experimental procedures
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table S1. Plasmids for construction of mutations and for complementation were prepared as described in SI Materials and Methods. Minimal-salts broth (MSB) (Stanier et al., 1966) containing Balch’s vitamins (Gerhardt et al., 1994) (minus thiamine and p-aminobenzoate) and supplemented with the appropriate carbon source, inducer, and/or antibiotic(s) was used for growth of Acidovorax strains. E. coli strains were grown on LB (1% tryptone, 0.5% yeast extract, 0.5% NaCl) unless otherwise indicated, and tryptone-yeast extract medium (TY; 1% tryptone, 0.5% yeast extract) was used for conjugative matings. MSB was solidified with 1.8 % Noble agar for plates, while LB and TY were solidified with 1.6% (w/v) Bacto agar. Ampicillin was used at 200 μg ml−1 for plasmid maintenance in E. coli, and kanamycin and tetracycline were used at 50 μg ml−1 and 20 μg ml−1 for both E. coli and Acidovorax strains. Gentamicin was used at 7.5 μg ml−1 for E. coli and 15 μg ml−1 for Acidovorax strains.
Chemicals.
With the following exceptions, chemicals were obtained commercially and were of this highest purity available. 4-Methyl-5-nitrocatechol, 4-methyl-3-nitrocatechol, 3-methyl-4-nitrocatechol, 3-amino-4-methyl-5-nitrocatechol, and 3-amino-6-methyl-5-nitrocatechol were generously provided by Jim C. Spain (Georgia Institute of Technology). (+)-(1R, 2S)-cis-1,2-Dihydroxy-1,2-dihydronaphthalene was prepared as previously described (Jeffrey et al., 1975) using Pseudomonas sp. strain 9816/11 (Klecka and Gibson, 1979).
Evolution strategy.
Succinate-grown Acidovorax sp. JS42 was diluted in MSB, and 50 μl volumes were plated onto MSB agar with 4NT crystals placed in the cover of the Petri dishes as the sole carbon source. Plates were sealed with Parafilm, and incubated at 28° C. The remaining culture was harvested by centrifugation, resuspended in 100 ml fresh MSB and incubated with 4NT crystals (final concentration, 100 μg ml−1) at 28° C in with shaking (225 rpm). After approximately 3 weeks, turbidity was observed and the culture was transferred into fresh medium. After three transfers, the culture was diluted and plated onto MSB plates with crystals of 4NT in the covers and incubated at 28° C. After 2 weeks, individual colonies were randomly picked, streaked onto new plates with 4NT, and incubated. These strains were repeatedly transferred on MSB plates with 4NT every seven to ten days for six months.
Nitroarene dioxygenase activity assays in JS42.
Expression of the genes encoding 2NTDO in Acidovorax strains was compared by measuring nitroarene dioxygenase activity after growth in the presence of different inducer compounds. Strains were grown in MSB containing 10 mM succinate, with or without 100 μM salicylate, nitrobenzene, 2NT, 3NT, or 4NT provided as inducers. Cultures were incubated at 28° C and harvested by centrifugation when an OD660 between 0.7 and 0.8 was reached. Cells were resuspended in 40 mM phosphate buffer (pH 7.3) to an OD660 of 0.5 and a saturating amount of nitrobenzene (~ 3 μl/ ml) was added to cultures, which were incubated at 30° C, 200 RPM for 30 min. Nitrite was measured as previously described (Gerhardt et al., 1994; Ju and Parales, 2006). Protein concentrations were determined by the method of Bradford (Bradford, 1976) with bovine serum albumin as the standard after resuspending cell pellets in 100 mM NaOH and boiling for 10 minutes (Ju and Parales, 2006). The activity of the nitroarene dioxygenases in JS42 and the 4NT+ derivatives with different substrates was compared after growing strains in MSB with 10 mM succinate and 100 μM salicylate as described above. Cells were harvested and incubated as described above after addition of 1 mM (final concentration) nitrobenzene, 2NT, 3NT, or 4NT. Nitrite was measured after 30 minutes.
Product formation in E. coli.
Whole-cell biotransformation reactions with E. coli DH5α expressing recombinant dioxygenase enzymes from plasmids pDTG850, pKSJ90, pKSJ92, or pKSJ93 (Table S1) with 0.1% nitrobenzene, 2NT, 3NT, 4NT, 2,4-dinitrotoluene, 2,6- dinitrotoluene, and naphthalene were carried out as previously described (Ju and Parales, 2006). After six hours, nitrite in the supernatant of each culture was analyzed (Gerhardt et al., 1994; Ju and Parales, 2006), before extracting with ethyl acetate (Ju and Parales, 2006). To identify the products, the extracts were concentrated and analyzed by gas chromatography-mass spectrometry (GC-MS) as previously described (Ju and Parales, 2006; Resnick et al., 1994).
Oxidation kinetics.
E. coli cultures expressing recombinant dioxygenases were grown and harvested as described previously (Ju and Parales, 2006), and nitrobenzene, 2NT and 4NT were provided from methanol stock solutions to final concentrations between 25 μM and 4 mM. Cultures were incubated in a shaking water bath at 30° C and 300 RPM, and 500 μl volumes were periodically withdrawn at 5-min intervals and immediately frozen on dry ice. For analysis, frozen samples were rapidly thawed in a 65° C water bath, clarified by centrifugation, and analyzed for nitrite and protein as described (Ju and Parales, 2006). Kinetic coefficients (apparent Michaelis constant [Km], and maximum observed oxidation rate, [Vmax]) for nitrobenzene, 2NT, and 4NT were derived by non-linear regression of the initial oxidation rates using the Michaelis-Menten model (Michaelis and Menten, 1913) in Grapher 2.0.
Growth experiments and detection of products.
Acidovorax strains were tested for growth on nitrobenzene, 2NT, 3NT, 4NT, catechol, 3-methylcatechol, 4-methylcatechol, pyruvate, propionaldehyde, and propionate on MSB plates. Catechols, propionate, and pyruvate were incorporated into the medium at final concentrations of 3, 5, and 20 mM, respectively. Nitrobenzene, 2NT, 3NT, 4NT and propionaldehyde were supplied as vapor. Plates were incubated at 28° C. To test growth in liquid media, strains were inoculated into MSB containing 2 mM 2NT or 4NT, and sealed with butyl stoppers. Tubes were incubated at 28° C on a roller drum at 75 RPM, and culture turbidity was monitored. Propionaldehyde production was examined by sampling the atmosphere of the cultures during exponential growth with a gas-tight syringe and analyzing 100 μl of headspace by GC-MS (Ju and Parales, 2006). Crude-cell extracts of Acidovorax sp. JS42-KSJ11 were prepared as described in SI Experimental Procedures, and pyruvate and propionaldehyde formed from 4-methylcatechol were detected by TLC following derivatization with 2,4-dinitrophenylhydrazone as previously described (Dagley and Gibson, 1965), and quantified as described in SI. Cell yields were determined as a function of total protein content, and measured as described above.
SDS-PAGE and Western blot analysis.
Cell pellets were resuspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample loading buffer, boiled for 10 minutes, and resolved by 12% SDS-PAGE (Maniatis et al., 1982). Gels were subjected to Western analysis (Ausubel et al., 1993) using a polyclonal antibody against the oxygenase component of naphthalene dioxygenase from Pseudomonas sp. strain 9816-4 (Parales et al., 1998b), with crude cell extracts from DH5α (pDTG850) expressing 2NTDO as a positive control. Antigens were visualized using alkaline-phosphatase-conjugated goat anti-mouse immunoglobin G (Invitrogen, Carlsbad, Calif.).
Supplementary Material
Acknowledgements
This work was supported by the National Science Foundation (MCB 02627248 and MCB 1022362). K.-S.J. was supported by an NIH Traineeship in Molecular and Cellular Biology (NIH TM32 GM070377) and a fellowship from the University of California Toxic Substances and Teaching Program (http://tsrtp.ucdavis.edu). We thank Victoria Shingler for providing cloned dmp genes, John Roth and Joseph Penrod for Salmonella strains and helpful discussions, Juan Parales for carrying out RT-PCR experiments, and David Gibson for critical reading of the manuscript and for suggesting additional experiments.
REFERENCES
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, and Struhl K (1993) Current protocols in molecular biology New York: John Wiley & Sons, Inc. [Google Scholar]
- Booth G (2007) Nitro Compounds, Aromatic. In Ullmann’s Encyclopedia of Industrial Chemistry New York: John Wiley & Sons. [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Badgett MR, and Wichman HA (2000) Big-benefit mutations in a bacteriophage inhibited with heat. Mol Biol Evol 17: 942–950. [DOI] [PubMed] [Google Scholar]
- Cases I, and de Lorenzo V ( 2001) The black cat/white cat principle of signal integration in bacterial promoters. EMBO J 20: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, and Lenski RE (2000) The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407: 736–739. [DOI] [PubMed] [Google Scholar]
- Copley SD (2009) Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat. Chem. Biol 5: 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagley S, and Gibson DT (1965) The bacterial degradation of catechol. Biochem. J 95: 466–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, and Lenski RE (2003) Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet 4: 457–469. [DOI] [PubMed] [Google Scholar]
- Finkel SE (2006) Long-term survival during stationary phase: evolution and the GASP phenotype. Nat. Rev. Microbiol 4: 113–120. [DOI] [PubMed] [Google Scholar]
- Gerhardt P, Murray RGE, Wood WA, and Krieg NR, (eds) (1994) Methods for general and molecular bacteriology Washington, D.C.: American Society for Microbiology. [Google Scholar]
- Haigler BE, and Spain JC (1993) Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl. Environ. Microbiol 59: 2239–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigler BE, Wallace WH, and Spain JC (1994) Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl. Environ. Microbiol 60: 3466–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, and Williams PA (2001) Cloning and characterization of the pnb genes, encoding enzymes for 4-nitrobenzoate catabolism in Pseudomonas putida TW3. J. Bacteriol 183: 1225–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James KD, and Williams PA (1998) ntn genes determine the early steps in the divergent catabolism of 4-nitrotoluene and toluene in Pseudomonas sp. strain TW3. J. Bacteriol 180: 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James KD, Hughes MA, and Williams PA (2000) Cloning and expression of ntnD, encoding a novel NAD(P)+-independent 4-nitrobenzyl alcohol dehydrogenase from Pseudomonas sp. strain TW3. J. Bacteriol 182: 3136–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey AM, Yeh HJC, Jerina DM, Patel TR, Davey JF, and Gibson DT (1975) Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry 14: 575–583. [DOI] [PubMed] [Google Scholar]
- Johnson GR, Jain RK, and Spain JC (2002) Origins of the 2,4-dinitrotoluene pathway. J. Bacteriol 184: 4219–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju K-S, and Parales RE (2006) Control of substrate specificity by active site residues in nitrobenzene 1,2-dioxygenase. Appl. Environ. Microbiol 72: 1817–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju K-S, Parales JV, and Parales RE (2009) Reconstructing the evolutionary history of nitrotoluene detection in the transcriptional regulator NtdR. Mol. Microbiol 74: 826–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju K-S, and Parales RE (2010) Nitroaromatic compounds: From synthesis to biodegradation. Microbiol. Mol. Biol. Rev 74: 250–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan BG, Leungsakul T, Smets BF, and Wood TK (2004) Saturation mutagenesis of Burkholderia cepacia R34 2,4-dinitrotoluene dioxygenase at DntAc valine 350 for synthesizing nitrohydroquinone, methylhydroquinone, and methoxyhydroquinone. Appl. Environ. Microbiol 70: 3222–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan BG, Leungsakul T, Smets BF, Mori MA, Henderson DE, and Wood TK (2005) Protein engineering of the archetypal nitroarene dioxygenase of Ralstonia sp. strain U2 for activity on aminonitrotoluenes and dinitrotoluenes through alpha-subunit residues leucine 225, phenylalanine 350, and glycine 407. J. Bacteriol 187: 3302–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecka GM, and Gibson DT (1979) Metabolism of dibenzo[1,4]dioxan by a Pseudomonas species. Biochem. J 180: 639–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-S, Parales JV, Friemann R, and Parales RE (2005) Active site residues controlling substrate specificity in 2-nitrotoluene dioxygenase from Acidovorax sp. strain JS42. J. Ind. Microbiol. Biotechnol 32: 465–473. [DOI] [PubMed] [Google Scholar]
- Lessner DJ, Johnson GR, Parales RE, Spain JC, and Gibson DT (2002) Molecular characterization and substrate specificity of nitrobenzene dioxygenase from Comamonas sp. strain JS765. Appl. Environ. Microbiol 68: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessner DJ, Parales RE, Narayan S, and Gibson DT (2003) Expression of nitroarene dioxygenase genes in Comamonas sp. strain JS765 and Acidovorax sp. strain JS42 is induced by multiple aromatic compounds. J. Bacteriol 185: 3895–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leungsakul T, Keenan BG, Yin H, Smets BF, and Wood TK (2005) Saturation mutagenesis of 2,4-DNT dioxygenase of Burkholderia sp. strain DNT for enhanced dinitrotoluene degradation. Biotechnol. Bioeng 92: 416–420. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, and Sambrook J (1982) Molecular cloning: a laboratory manual Cold Spring Harbor, New York: Cold Spring Harbor Laboratory. [Google Scholar]
- Michaelis L, and Menten ML (1913) Die kinetik der invertinwirkung. Biochem. Z 49: 333–369. [Google Scholar]
- Morley KL, and Kazlauskas RJ (2005) Improving enzyme properties: when are closer mutations better? Trends Biotechnol 23: 231–237. [DOI] [PubMed] [Google Scholar]
- Mulla SI, Hoskeri RS, Shouche YS, and Ninnekar HZ (2010) Biodegradation of 2-nitrotoluene by Micrococcus sp. strain SMN-1. Biodegradation In press [DOI] [PubMed] [Google Scholar]
- Muller TA, Werlen C, Spain JC, and Van Der Meer JR (2003) Evolution of a chlorobenzene degradative pathway among bacteria in a contaminated groundwater mediated by a genomic island in Ralstonia. Environ. Microbiol 5: 163–173. [DOI] [PubMed] [Google Scholar]
- Murray K, Duggleby CJ, Sala-Trepat JM, and Williams PA (1972) The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur. J. Biochem 28: 301–310. [DOI] [PubMed] [Google Scholar]
- Nishino SF, and Spain JC (1993) Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl. Environ. Microbiol 59: 2520–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino SF, and Spain JC (1995) Oxidative pathway for the biodegradation of nitrobenzene by Comamonas sp. strain JS765. Appl. Environ. Microbiol 61: 2308–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino SF, Paoli GC, and Spain JC (2000) Aerobic degradation of dinitrotoluenes and the pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol 66: 2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padda RS, Wang C, Hughes JB, Kutty R, and Bennett GN (2003) Mutagenicity of nitroaromatic degradation compounds. Environ. Toxicol. Chem 22: 2293–2297. [DOI] [PubMed] [Google Scholar]
- Parales JV, Parales RE, Resnick SM, and Gibson DT (1998a) Enzyme specificity of 2-nitrotoluene 2,3-dioxygenase from Pseudomonas sp. strain JS42 is determined by the C-terminal region of the a subunit of the oxygenase component. J. Bacteriol 180: 1194–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales RE, Emig MD, Lynch NA, and Gibson DT (1998b) Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J. Bacteriol 180: 2337–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales RE (2000) Molecular biology of nitroarene degradation. In Biodegradation of nitroaromatic compounds and explosives Spain JC, Hughes JB and Knackmuss H-J (eds). Boca Raton, FL.: CRC Press, pp. 63–89. [Google Scholar]
- Parales RE, Lee K, Resnick SM, Jiang H, Lessner DJ, and Gibson DT (2000a) Substrate specificity of naphthalene dioxygenase: effect of specific amino acids at the active site of the enzyme. J. Bacteriol 182: 1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales RE, Resnick SM, Yu CL, Boyd DR, Sharma ND, and Gibson DT (2000b) Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: control by phenylalanine 352 in the a subunit. J. Bacteriol 182: 5495–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powlowski J, Sahlman L, and Shingler V (1993) Purification and properties of the physically associated meta-cleavage pathway enzymes 4-hydroxy-2-ketovalerate aldolase and aldehyde dehydrogenase (acylating) from Pseudomonas sp. strain CF600. J. Bacteriol 175: 377–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick SM, Torok DS, Lee K, Brand JM, and Gibson DT (1994) Regiospecific and stereoselective hydroxylation of 1-indanone and 2-indanone by naphthalene dioxygenase and toluene dioxygenase. Appl. Environ. Microbiol 60: 3323–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhys-Williams W, Taylor SC, and Williams PA (1993a) A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas. J. Gen. Microbiol 139: 1967–1972. [DOI] [PubMed] [Google Scholar]
- Spanggord RJ, Spain JC, Nishino SF, and Mortelmans KE (1991) Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl. Environ. Microbiol 57: 3200–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess T, Desiere F, Fischer P, Spain JC, Knackmuss HJ, and Lenke H (1998) A new 4-nitrotoluene degradation pathway in a Mycobacterium strain. Appl. Environ. Microbiol 64: 446–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier RY, Palleroni NJ, and Doudoroff M (1966) The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol 43: 159–271. [DOI] [PubMed] [Google Scholar]
- van der Meer JR, de Vos WM, Harayama S, and Zehnder AJB (1992) Molecular mechanisms of genetic adaptation to xenobiotic compounds. Microbiol. Rev 56: 677–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


