Abstract

In this study, microwave-assisted Knoevenagel condensation was used to produce two novel series of derivatives (1–6) from benzylidenemalononitrile and ethyl 2-cyano-3-phenylacrylate. The synthesized compounds were characterized using Fourier transform infrared (FT-IR) and 1H NMR spectroscopies. The pharmacodynamics, toxicity profiles, and biological activities of the compounds were evaluated through an in silico study using prediction of activity spectra for substances (PASS) and Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) studies. According to the PASS prediction results, compounds 1–6 showed greater antineoplastic potency for breast cancer than other types of cancer. Molecular docking was employed to investigate the binding mode and interaction sites of the derivatives (1–6) with three human cancer targets (HER2, EGFR, and human FPPS), and the protein–ligand interactions of these derivatives were compared to those reference standards Tyrphostin 1 (AG9) and Tyrphostin 23 (A23). Compound 3 showed a stronger effect on two cell lines (HER2 and FPPS) than the reference drugs. A 20 ns molecular dynamics (MD) simulation was also conducted to examine the ligand’s behavior at the active binding site of the modeled protein, utilizing the lowest docking energy obtained from the molecular docking study. Enthalpies (ΔH), Gibbs free energies (ΔG), entropies (ΔS), and frontier molecular orbital parameters (highest occupied molecular orbital–lowest unoccupied molecular orbital (HOMO–LUMO) gap, hardness, and softness) were calculated to confirm the thermodynamic stability of all derivatives. The consistent results obtained from the in silico studies suggest that compound 3 has potential as a new anticancer and antiparasitic drug. Further research is required to validate its efficacy.
1. Introduction
Benzylidenemalononitrile (BMN) derivatives have been extensively studied for their biological properties, serving as both a target and transitional molecule in organic chemistry. The physiological activity of BMNs is noticeably impacted, in a significant way, by the position of the substituent on the ring structure. The 2-substituted position exhibits the highest activity, followed by a decrease in activity for the 3-substituted position, and the 4-substituent leads to a gradual reduction in activity until it disappears entirely.1−3 These derivatives have been utilized in various fields, including anticancer,4−6 antifungal,7,8 antibacterial,9−12 and anticorrosive applications.2,13−16 BMN derivatives have found widespread applications in biology, industry, agriculture, medicine, and chemical synthesis17,18 and have the potential to act as inhibitors of epidermal growth factor tyrosine kinase.16,19
The formation of BMN derivatives10,14,17,20−23via Knoevenagel condensation is based on the reaction of aldehydes with active methylene compounds, which contain a group (C=C).16 This type of reaction is considered a fundamental process in synthetic organic chemistry. The Knoevenagel condensation reaction has been facilitated by various catalysts, including ZnCl2,24 clays,25 silica gel,26 natural catalysts,27 ammonium acetate (NH4OAc)-basic alumina,28 microwave irradiation, and thermal heating conditions.29 NiCu@MWCNT has also been used as a catalyst for 10–180 min30 under mild conditions. Moreover, in some cases, the reaction was conducted without any solvent or catalyst.31 Microwave irradiation has several advantages when used in these reactions, including shorter reaction times, higher yields, improved selectivity, and a solvent-free approach that aligns with green principles.32 These benefits have resulted in a cost-effective and labor-affordable method for organic synthesis. Roskoski15 developed a new range of low-molecular-weight inhibitors for the protein tyrosine kinase of tyrphostins, which had the hydroxyl-cis-benzylidenemalonitrile group as a pharmacophore. These inhibitors proved to be effective in inhibiting EGF receptor kinase (HER1) and ErbB2/neu kinase (HER2). Bhuiyan et al.33 have presented an efficient method for synthesizing chalcones with high yields under solvent-free conditions using microwave irradiation.
There have been only a few computational studies9,34−36 conducted on the derivatives of benzylidenemalonitrile and ethyl 2-cyano-3-phenylacrylate. Mary et al.34 conducted a study on 1,3,5-triazine-2,4-diamine along with 1H-indole-2,3-dione (isatin), (2E)-1,3-diphenylprop-2-en-1-one (chalcone), and 10H-acridin-9-one (acridone). The study aimed to determine their molecular and electronic characteristics, energy gap, kinetic stability, and electrophilicity index of delocalized sites, as well as their relationship with biological activity. Molecular docking was conducted with different proteins to demonstrate that the products could serve as effective drugs for their corresponding activities. Therefore, the potential use of benzylidenemalononitrile and ethyl 2-cyano-3-phenylacrylate derivatives as bioactive molecules against human pathogenic microorganisms offers a vast scope of application in biological studies. However, further research is necessary to explore their full potential and develop them into safe and effective treatments for human use.
In this study, two novel series of benzylidenemalonitrile and ethyl 2-cyano-3-phenylacrylate derivatives (1–6) were synthesized using Knoevenagel condensation, and their chemical structures were determined by spectroscopic analysis. Scheme 1 and Tables S1–S6 and Figure S1 in the Supporting Information (SI) illustrate the synthesized compounds (Table 1). Notably, no computational studies have yet been carried out to predict the physicochemical, spectral, and biological characteristics of the newly synthesized derivatives (1–6). The main objective of this study is to conduct in silico analysis of the synthesized compounds, including molecular docking and molecular dynamics (MD) simulations. Moreover, the drug-likeness and pharmacokinetic properties of the selected compounds were evaluated using Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) prediction37 and SwissADME38 profiles. This study aims to understand better the binding affinities of the newly synthesized compounds (1–6), which could have significant implications in their biological applications. Furthermore, the thermophysical properties of these compounds have been calculated to determine their conformational stability. In addition, density functional theory (DFT) calculations were performed to determine the molecular structures, electronic density, NBO charges, frontier molecular orbitals, chemical hardness (η), and chemical potential (μ). These quantities are commonly used to explain reactivity and stability and complement the experimental study.
Scheme 1. Synthesis of Benzylidenemalononitrile and Ethyl 2-Cyano-3-phenylacrylate Derivatives (1–6).
Table 1. Structures of Benzylidenemalonitrile and Ethyl 2-Cyano-3-phenylacrylate Derivatives (1–6).
2. Materials and Methods
2.1. Materials and Reagents
All chemicals and reagents used in this study were of research grade (Merck, Germany) and were used without further purification. Solvents were purified using the standard distillation method before use. Throughout the experimental work, deionized water filtered through a Millipore filter was used. Melting points (mp) were determined using an electrothermal melting point apparatus, and the values were uncorrected. The progression of the reaction was monitored by thin-layer chromatography (TLC) on Kieselgel GF254 plates, using a mixture of n-hexane and ethyl acetate in a ratio of 3:1. The plates were then dried, and spots were detected using ultraviolet (UV) lamps. The solution’s ultraviolet–visible (UV–vis) spectra were recorded using an HP 8453 UV–vis scanning spectrophotometer within the 190–800 nm range. Fourier transform infrared (FT-IR) spectra were obtained using the KBr matrix and a FT-IR spectrophotometer (model 8900, Shimadzu, Japan) within the 4000–200 cm–1 range. 1H NMR (nuclear magnetic resonance) spectra were recorded using a Bruker DPX-400 (400 MHz NMR) spectrometer (Switzerland) with CDCl3 as a solvent and tetramethylsilane (TMS) as an internal standard. Chemical shifts were reported in δ unit (ppm), and J values were shown in Hz. The reactions were conducted using a commercially available LG microwave oven (MB-3947C, China) with a maximum power output of 800 W, operating at 2450 MHz.
2.2. Synthesis of Benzylidenemalononitrile Derivatives (1–3)
Benzylidenemalononitrile derivatives were synthesized using the microwave irradiation (MWI) technique with aromatic aldehyde, malononitrile, and ammonium acetate. The aldehyde (1.222 mmol) and malononitrile (1.222 mmol) were mixed in a porcelain dish, and 10 mg of ammonium acetate was added. The porcelain dish was then placed in the microwave oven and irradiated at 320 W for 20–50 s. The progress of the reaction was monitored by TLC (n-hexane/ethyl acetate, 3:1). The resulting crude solid was recrystallized using ethyl acetate and n-hexane to obtain pure crystals of the product. FT-IR and 1H NMR spectroscopies confirmed the formation of the product, and analytical data obtained from these techniques characterized the three benzylidenemalononitrile derivatives (1–3), as shown in Scheme 1 (Table 1 and Figures S2–S13 in the SI).
2.3. Synthesis of Ethyl 2-Cyano-3-phenylacrylate Derivatives (4–6)
The microwave (MW1) technique was employed to synthesize ethyl 2-cyano-3-phenylacrylate derivatives from aromatic aldehyde, ethyl cyanoacetate, and ammonium acetate (NH4OAc). The aldehyde (1.568 mmol) and ethyl cyanoacetate (1.568 mmol) were mixed in a porcelain dish, and 10 mg of ammonium acetate was added. The porcelain dish was then placed in the microwave oven and irradiated at 320 W for 50 s. The progress of the reaction was monitored by TLC using n-hexane/ethyl acetate (3:1) as the mobile phase. The resulting crude solid was recrystallized using ethyl acetate and n-hexane to obtain pure product crystals. FT-IR and 1H NMR spectroscopies were used to confirm the formation of the product and to get analytical data that characterized the three ethyl 2-cyano-3-phenylacrylate derivatives (4–6), as shown in Scheme 1 (Table 1 and Figures S14–S25 in the SI).
2.4. Computational Studies
Electronic structure calculations were carried out using Gaussian 16, Rev. C.01.39 The geometries and vibrational wave numbers of benzylidenemalononitrile and ethyl 2-cyano-3-phenylacrylate derivatives (1–6) were optimized using the B3LYP/6-31G(d,p) level of theory. The prior research40 that our group conducted consisted of exploring the deamination reactions of a variety of different nitrogenous bases using a variety of different computational methodologies. The results obtained from G3MP2, G3MP2B3, G4MP2, G3B3, and CBS-QB3 methods agreed with each other within a range of 10 kJ mol–1, while the B3LYP/6-31G(d,p) method had a deviation of up to 18 kJ mol–1. Thus, the B3LYP method can produce satisfactory outcomes while reducing computational time. GaussView 6 software was used to compute the energy distribution from the highest occupied molecular orbital (HOMO) to the lowest unoccupied molecular orbital (LUMO) and generate the molecular electrostatic potential (MEP) map. To ensure the absence of imaginary frequencies in the minima and the presence of only one imaginary frequency in the transition states, vibrational frequencies were calculated for all structures. The optimized structures can be seen in Tables S1–S11 and Figures S1–S44 in the Supporting Information (SI). The thermodynamic properties, including enthalpies (ΔH), Gibbs free energies (ΔG), and entropies (ΔS), were calculated at 298.15 K and reported in kJ mol–1.
The role of atomic charges in the deamination reactions was investigated by analyzing natural bonding orbitals (NBOs).41 The chemical potential (μ),42−45 hardness (η), and softness (S) of compounds 1–6 were calculated using the HOMO and LUMO energies. The formulas used for these calculations are given below
The global electrophilicity index, ω, for each of the compounds (1–6) was determined to assess their electrophilicity. The measurement was done following the method proposed by Parr et al.45 and calculated using the following expression
The electrophilicity indices were calculated using the values of μ and η. To obtain precise energy values and atomic charges, single-point-energy calculations and natural bond orbital (NBO) calculations were performed.
2.5. Evaluation of Physicochemical and Pharmacokinetic Properties
In this study, we utilized various free online tools such as AdmetSAR and SwissADME to predict the ADMET properties of benzylidenemalononitrile and ethyl 2-cyano-3-phenylacrylate derivatives (1–6).37,38 We also used Molinspiration46 to perform drug-likeness properties and ADME predictions for these compounds. The structures (1–6) were drawn using ChemBioDraw Ultra 14.0 and converted to a canonical simplified molecular input line entry system (SMILES) to collect MDL Molfile format. We predicted several physicochemical properties of these derivatives, including lipophilicity, water solubility, and pharmacokinetics parameters. Additionally, we evaluated the drug-likeness of these compounds as G-protein-coupled receptor (GPCR) ligands, ion channel modulators (ICMs), kinase inhibitors (KIs), nuclear receptor ligands (NRLs), protease inhibitors (PIs), and enzyme inhibitors (EIs). The results obtained from these in silico studies provide insights into the potential biological activity of these derivatives and their suitability as drug candidates.
2.6. Pharmacological Activities
In order to predict the probable pharmacological activities of the lead compounds (1–6), their sdf formats were submitted to the online tool prediction of activity spectra for substances (PASS).47,48 The tool was used to predict their antimicrobial spectra and toxicities.
2.7. Molecular Docking
2.7.1. Preparation of Target Protein
To identify the binding sites for the ligands, molecular docking software was utilized, and the required proteins were obtained from the RSCB Protein Data Bank.49 The human target proteins included the receptor tyrosine-protein kinase erbB2 (PDB ID: 7JXH), also known as HER2, a crystal structure of human FPPS (PDB ID: 4H5D) at a resolution of 2.02 Å, and an epidermal growth factor receptor (PDB ID: 4LRM).50 Validation parameters by X-ray diffraction were used to confirm the quality of the primary protein structure for all crystal structures. The protein molecules were optimized using Chimera v1.16.51 The optimization process involved the removal of water molecules and ligands, followed by energy minimization of the macromolecule for molecular docking.
2.7.2. Preparation of Ligands
The chemical structures of all of the synthesized compounds (1–6) were created in GaussView 6 software and then fully optimized in Gaussian 16 software at B3LYP/6-31G(d,p). The chemical structures of the selected ligands for docking were further subjected to energy minimization (EM) and converted to the PDBQT format using the OpenBabel plugin of PyRx 0.8 software, which is available at https://pyrx.sourceforge.io/.52
2.7.3. Docking of Protein–Ligand Complexes
In AutoDock Vina53 software, the ligand structures and target proteins were chosen for molecular docking. A grid box was selected to cover the complete protein at the center during the docking of the ligands. All of the proteins and ligands were stable throughout the docking experiment. The validation of all of the protein–ligand interactions was conducted through re-docking. In addition, Chimera v1.16,51 PyMol,54 and Discovery Studio visualizer were utilized for visualizing the binding modes of the receptor–ligand interactions.
2.7.4. Molecular Dynamics Simulation
Molecular dynamics (MD) simulations are computational techniques that track the movements of individual atoms within a system over time, providing information about their relative positions.55−57 In this study, MD simulations were performed using the GROMACS 2021.6 package58 and the AMBER99SB force field59 to investigate the docked complexes of compound 3 with PDB:7JXH and PDB: 4H5D. The topology parameters for the proteins in the system were generated using the Galaxy European Server.60 The complex was neutralized with added Na+ ions and placed in a triclinic box of SPC water molecules.61 To ensure the system’s stability, it was equilibrated through a position-restrained dynamics simulation (NVT) at 300 K for 3000 ps using the leapfrog algorithm.62 After equilibration, the full system underwent a production run for an additional 3000 ps at the same temperature and pressure conditions. MD simulations provide a highly precise means of investigating the behavior of the docked protein–ligand complexes, contributing to a better understanding of biological macromolecule structure and function. MD trajectories were generated using VMD, PyMol, and GROMACS tools, and the GROMACS utilities were used to determine RMSD, Rg, root-mean-square fluctuation (RMSF), and hydrogen bond analysis.
3. Results and Discussion
The present study focused on the investigation of two series of compounds: 4-hexyloxybenzylidenemalononitrile (1–3) and ethyl 2-cyano-3-(4-hexyloxy)phenylacrylate (4–6), as illustrated in Scheme 1. Detailed information of these compounds can be found in Table 1 and Figure S1 in the Supporting Information (SI).
3.1. Characterization of Benzylidenemalononitrile Derivatives (1–3)
The structure of 4-hexyloxybenzylidenemalononitrile (1) was confirmed by analyzing its 1H NMR spectrum (Figures S2–S10 in the SI). A singlet signal at δ 7.66 ppm was assigned to the vinylic proton, and two doublet signals at δ 7.92 and 7.01 ppm with a coupling constant (J) value of 8.80 Hz were assigned to the aromatic protons at H-2/H-6 and H-3/H-5 positions, respectively. Another two-proton triplet signal at δ 4.08 ppm with a J value of 6.80 Hz was attributed to the −OCH2– proton. Additionally, the spectrum displayed a two-proton multiplet signal at δ 1.82–1.35 ppm corresponding to the −CH2CH2CH2CH2– proton and three-proton triplet signals at δ 0.95 ppm corresponding to the −CH3 proton. The FT-IR spectrum of compound 1 (Figures S11–S13 in the SI) showed absorption bands at 3033 cm–1 for aromatic C–H, 2223 cm–1 for C≡N bond, 1605 cm–1 for C=C (alkene) bond, and 1583 cm–1 for aryl C=C bond. The 1H NMR spectra of compounds 2 and 3 were consistent with their structures, and almost identical FT-IR peaks were obtained for all three compounds.
3.1.1. 4-Hexyloxybenzylidenemalononitrile (1)
The synthesis of 4-hexyloxybenzylidenemalononitrile (1) yielded a chocolate-colored crystalline solid with a yield of 65%. Its molecular formula was determined to be C16H18N2O with a melting point range of 42–43 °C. Anal. Calcd for C16H18N2O: C, 75.56; H, 7.12; N, 11.01. Found: C, 75.21; H, 7.25; N, 10.87. FT-IR (KBr) νmax(cm–1): 3033 (str, C–H, Ar), 2223 (str, −CN), 1605 (str, C=C, alkene) and 1583 (str, C=C, Ar). 1H NMR (400 MHz, CDCl3) δ ppm: 7.92 (d, 2H, J = 8.8 Hz, Ar), 7.66 (s, 1H, −CH=C), 7.01 (2H, d, J = 8.8 Hz, Ar), 4.08 (t, 2H, J = 6.8 Hz, −OCH2−), 1.82 (m, 8H, (−CH2−)4), 0.95 (t, 3H, J = 2 Hz, −CH3).
3.1.2. 4-Octyloxybenzylidenemalononitrile (2)
Product (2), 4-octyloxybenzylidenemalononitrile, was obtained as a brown crystalline solid with a yield of 60%. Its molecular formula is C18H22N2O, with a melting point of 43–44 °C. Anal. Calcd for C18H22N2O: C, 76.56; H, 7.85; N, 9.92. Found: C, 76.31; H, 7.98; N, 9.88. FT-IR (KBr) νmax(cm–1): 3020 (str, C–H, Ar), 2224 (str, −CN), 1606 (str, C=C, alkene) and 1580 (str, C=C, Ar). 1H NMR (400 MHz, CDCl3)δ ppm: 7.92 (d, 2H, J = 8.8 Hz, Ar), 7.66 (s, 1H, −CH=C), 7.01 (2H, d, J = 8.8 Hz, Ar), 4.08 (t, 2H, J = 6.8 Hz, −OCH2−), 1.82 (m, 12H, (−CH2−)6), 0.91 (t, 3H, J = 2 Hz, −CH3).
3.1.3. 4-Decyloxybenzylidenemalonitrile (3)
Product (3), 4-decyloxybenzylidenemalononitrile, was obtained as a brown crystalline solid with a yield of 68%. Its molecular formula is C20H26N2O, and it has a melting point range of 49–51 °C. Anal. Calcd for C20H26N2O: C, 77.38; H, 8.44; N, 9.02. Found: C, 77.18; H, 8.51; N, 8.98. FT-IR (KBr) νmax(cm–1): 3030 (str, C–H, Ar), 2224 (str, −CN), 1620 (str, C=C, alkene), 1580 (str, C=C, Ar), 1186 (str, C–O–C). 1H NMR (400 MHz, CDCl3) δ ppm: 8.2 (s, 1H, −CH=C−), 7.97 (d, 2H, J = 8.4 Hz, Ar), 7.01 (d, 2H, J = 8 Hz, Ar), 4.06 (t, 2H, J = 6.4 Hz, OCH2), 1.85 m, 2H, (−OCH2CH2−), 1.49 (m, 14H, (−CH2−)7), 0.93 (t, 3H, J = 2 Hz, −CH3).
3.2. Characterization of Ethyl 2-Cyano-3-phenylacrylate Derivatives (4 to 6)
The structure of ethyl 2-cyano-3-(4-hexyloxy)phenylacrylate (4) was confirmed using the 1H NMR spectrum (Figures S14–S22 in the SI). A three-proton triplet signal at δ 0.93 ppm was identified as the −CH3 proton, while another three-proton triplet signal at δ 1.41 ppm indicated the presence of CH3CH2O– proton. A multiplet signal at δ 1.49 ppm was designated to the −CH2CH2CH2CH2– group proton, and a two-proton triplet signal at δ 4.06 ppm with a J value of 6.4 Hz was designated to the −OCH2CH2– proton. A two-proton quartet signal at δ 4.38 ppm indicated the presence of the −OCH2CH3 proton. Two two-proton doublet signals at δ 7.00 and 8.02 ppm with a J value of 8.4 Hz were designated to the aromatic proton H-3, H-5, and H-2, H-6 protons, respectively. A singlet signal at δ 8.2 ppm was designated to the vinylic proton. The FT-IR spectrum of compound 4 (Figures S23–S25 in the SI) showed an absorption stretching band at 3016 cm–1 due to the presence of an aromatic −C–H bond and a band at 2216 cm–1 due to the presence of C≡N bonds. Another band at 1722 cm–1 indicated the presence of an ester C=O bond. It should be noted that the 1H NMR and FT-IR spectra of compounds 5 and 6 were similar to those of compound 4.
3.2.1. Ethyl 2-Cyano-3-(4-hexyloxy)phenylacrylate (4)
Ethyl 2-cyano-3-(4-hexyloxy)phenylacrylate (4) was obtained as a pure yellow crystalline solid in 62% yield by recrystallization from a mixture of ethyl acetate and n-hexane. Its molecular formula is C18H23NO3, and its melting point is 44–46 °C. Anal. Calcd for C18H23NO3: C,71.73; H, 7.69; N, 4.65. Found: C, 71.58; H, 7.75; N, 4.57. FT-IR (KBr) νmax(cm–1): 3016 (str, C–H, Ar), 2216 (str, −CN), 1722 (str, C=O), 1691 (str, C=C, alkene), 1589 (str, C=C, Ar), 1178 (str, C–O–C). 1H NMR (400 MHz, CDCl3) δ ppm: 8.2 (s, 1H, −CH=C−), 8.02 (d, 2H, J = 8.4 Hz, Ar), 7.00 (d, 2H, J = 8 Hz, Ar), 4.38 (q, 2H, J = 6.8 Hz, −OCH2−), 4.06 (t, 2H, J = 6.4 Hz, −OCH2−), 1.49 (m, 8H, (−CH2−)4), 1.41 (t, 3H, J = 2.1 Hz, −CH3), 0.93 (t, 3H, J = 2 Hz, −CH3).
3.2.2. Ethyl 2-Cyano-3-(4-octyloxy)phenylacrylate (5)
Product (5), ethyl 2-cyano-3-(4-octyloxy)phenylacrylate, was recrystallized from ethyl acetate and n-hexane to obtain a pure yellow crystalline solid in 75% yield, M.F. C20H27NO3, with a melting point of 42–44 °C. Anal. Calcd for C20H27NO3: C,72.92; H, 8.26; N, 4.25. Found: C, 72.85; H, 8.31; N, 4.19. FT-IR (KBr) νmax (cm–1): 3036 (str, C–H, Ar), 2216 (str, −CN), 1716 (str, C=O), 1692 (str, C=C, alkene), 1583 (str, C=C, Ar), 1180 (str, C–O–C).
1H NMR (400 MHz, CDCl3) δ ppm: 8.18 (s, 1H, −CH=C−), 8.02 (d, 2H, J = 9.2 Hz, Ar), 7.00 (d, 2H, J = 8.8 Hz, Ar), 4.38 (q, 2H, J = 7.2 Hz, −OCH2−), 4.06 (t, 2H, J = 6.8 Hz, −OCH2−), 1.84 (m, 2H, −OCH2CH2−), 1.49 (m, 10H, (−CH2−)5), 1.41 (t, 3H, J = 2.1 Hz, −CH3), 0.93 (t, 3H, J = 2 Hz, −CH3).
3.2.3. Ethyl 2-Cyano-3-(4-decyloxy)phenylacrylate (6)
Product (6) was recrystallized from ethyl acetate and n-hexane to obtain a pure yellow crystalline solid in 71% yield, M.F. C22H31NO3, with a melting point of 42–44 °C. Anal. Calcd for C22H31NO3: C, 73.92; H, 8.74; N, 3.92. Found: C, 73.86; H, 8.69; N, 3.84. FT-IR (KBr) νmax (cm–1): 3036 (str, C–H, Ar), 2216 (str, −CN), 1716 (str, C=O), 1692 (str, C=C, alkene), 1583 (str, C=C, Ar), 1180 (str, C–O–C). 1H NMR (400 MHz, CDCl3) δ ppm: 8.19 (s, 1H, −CH=C−), 8.02 (d, 2H, J = 9.2 Hz, Ar), 7.00 (d, 2H, J = 8.8 Hz, Ar), 4.38 (q, 2H, J = 7.2 Hz, −OCH2−), 4.06 (t, 2H, J = 6.8, −OCH2−), 1.85 (m, 2H, −OCH2CH2−), 1.49 (m, 14H, (−CH2−)7), 1.44 (t, 3H, J = 2.1 Hz, −CH3), 0.90 (t, 3H, J = 2 Hz, −CH3).
3.3. Evaluation of Thermodynamic Stability
Scheme 1 demonstrates that all of the reactions analyzed at B3LYP/6-31G(d,p) are both endothermic and endergonic, as confirmed by Table S7 and Figure S1 in the Supporting Information. The enthalpy difference between the reactants (4-hexyloxy-benzaldehyde + malonitrile) and the product (4-hexyloxybenzylidenemalonitrile (1) + H2O) indicates that this reaction is endothermic and endergonic, with a ΔH of 218.6 kJ mol–1 and a ΔG of 270.6 kJ mol–1 at B3LYP/6-31G(d,p). The formation of 3 + H2O and 4 + H2O is also endothermic and endergonic, with ΔH values of 113.6 and 10.5 kJ mol–1 and ΔG values of 171.6 and 21.1 kJ mol–1, respectively. The enthalpy differences between the reactions for the formation of three benzylidenemalonitrile derivatives (1, 2, and 3) by 1 + H2O and the other two reactions for the formation of 2 and 3 are 135.9 and 185.1 kJ mol–1, respectively, due to the increasing number of −CH2 groups. Similarly, the enthalpy differences between the reaction for forming 4 + H2O and the other two reactions for forming 5 and 6 are 103.1 and 15.7 kJ mol–1, respectively. The entropy (ΔS) difference between the separated reactants of all reactions at B3LYP/6-31G(d,p) ranges from −35.6 to −269.4 kJ mol–1 (Table S7 in the SI).
According to Table S7, an improved dipole moment enhances the polar nature of the molecule, promoting its binding properties, hydrogen bonding, and nonbonding interactions with the receptor protein.63 Compound 2 has a dipole moment of 7.30 Debye, and compound 3 has a dipole moment of 8.39 Debye, which are slightly higher than the dipole moment of 4-hexyloxybenzylidenemalonitrile (1) (5.92 Debye), resulting in their stronger binding affinity and interactions with the methylene (−CH2) residues of the receptor protein. Similarly, compound 5 has a dipole moment of 3.90 Debye, and compound 6 has a dipole moment of 3.72 Debye, which is slightly higher than that of ethyl 2-cyano-3-(4-hexyloxy)phenylacrylate (4) (3.15 Debye), resulting in better binding affinity and interactions with the methylene (−CH2) residues of the receptor protein (Table S7 in the SI).
3.4. Evaluation of In Silico Pharmacokinetic Properties
To assess the global reactivity and stability properties of synthesized compounds (1–6), their frontier orbital (HOMO and LUMO) analysis and HOMO–LUMO gap values were evaluated. Additionally, hardness (η), softness (S), chemical potential (μ), and electrophilicity index (ω) were calculated using B3LYP/6-31G(d,p) and are presented in Table 2 and Figure 1 (see Figures S26–S27 in the SI). The HOMO–LUMO gap plays a vital role in molecular chemical reactivity and is related to chemical hardness, softness, chemical potential, and electrophilic index.64 Compound (3) had the largest HOMO–LUMO gap value (3.881 eV), highest hardness (1.941 eV), and lowest softness (0.515 eV) among all compounds, likely due to its longer chain length than compounds 1 and 2. Compared to compound (3), reference compounds A23 and AG9 had lower stability and higher reactivity. The energy gap values decrease as follows: 3 (3.881 eV) > AG9 (3.838 eV) > 2 (3.821 eV) > 1 (3.822 eV) > A23 (3.648 eV). The electrophilicity index of compound (3) was 5.205 eV, indicating potential biological activity and reactive sites. Compound (6) had a lower electrophilicity index value of 4.369 eV than compounds 4 and 5, suggesting that it may also have biological activity. Molecular docking with an appropriate protein can further explore these potential activities.
Table 2. MO of the HOMO and LUMO of Chemical Hardness (η), Softness (S), Chemical Potential (μ), and Electrophilicity Index (ω) of All Compounds (1–6) (in eV) at 298.15 Ka.
| ligand | LUMO | HOMO | gap | η | S | μ | ω |
|---|---|---|---|---|---|---|---|
| 1 | –2.587 | –6.407 | 3.822 | 1.911 | 0.523 | –4.498 | 5.294 |
| 2 | –2.585 | –6.406 | 3.821 | 1.911 | 0.523 | –4.496 | 5.289 |
| 3 | –2.584 | –6.405 | 3.881 | 1.941 | 0.515 | –4.495 | 5.205 |
| 4 | –2.182 | –6.118 | 3.936 | 1.968 | 0.508 | –4.150 | 4.376 |
| 5 | –2.362 | –6.247 | 3.885 | 1.943 | 0.515 | –4.305 | 4.769 |
| 6 | –2.179 | –6.115 | 3.936 | 1.968 | 0.508 | –4.147 | 4.369 |
| A23 | –2.706 | –6.354 | 3.648 | 1.824 | 0.548 | –4.530 | 5.625 |
| AG9 | –2.642 | –6.480 | 3.838 | 1.919 | 0.521 | –4.561 | 5.420 |
Calculated by η = (ELUMO – EHOMO)/2; μ = (EHOMO + ELUMO)/2; S = 1/η; ω = μ2/2η.
Figure 1.

Molecular orbitals of isodensity surfaces (0.02 electrons Bohr–3 surface) (red = electron-rich, blue = electron-deficient) of HOMO and LUMO for the compounds 3 and 6.
Natural bond orbital (NBO)41 analysis was conducted to gain insights into the stability of molecules based on atomic bonds and charges. This analysis examined both intermolecular and intramolecular interactions and identified the most significant stabilization energies presented in Figures S28–S33 in the SI. The intramolecular charge transfer interactions were further investigated. Interestingly, changes in geometry observed in benzylidenemalononitrile derivatives (1–3) and ethyl 2-cyano-3-phenylacrylate derivatives (4–6) did not cause changes in the charges on several key atoms in the system. Figure 2 illustrates the molecular electrostatic potential (MEP) maps that show the most probable regions around the molecule for reactivity by depicting the net electrostatic effect of the total charge distribution of electrophilic and nucleophilic sites. Electrophilic sites, represented by blue regions, are electron acceptors, while nucleophilic sites, represented by red regions, are electron donors. Yellow regions indicate partial nucleophilic sites. The electrostatic potential is color-coded, with red, orange, and yellow regions showing the negative potential for electrophilic reactivity over electronegative atoms like oxygen and cyan and blue regions indicating the positive potential for nucleophilic reactivity over hydrogen atoms. The regions of zero potential are shown in green.
Figure 2.
Maps of electrostatic potential (0.02 electrons Bohr–3) (red = electron-rich, blue = electron-deficient) for 4-hexyloxybenzylidenemalononitrile (1–3) and ethyl 2-cyano-3-(4-hexyloxy)phenylacrylate (4–6) derivatives.
3.5. Predicted Biological Activities Using PASS
The chemical structures (1–6) were analyzed using multilevel neighborhoods of atoms (MNA)65 creative descriptors in PASS (prediction of activity spectra for substances) online to predict thousands of potential biological activities, including pharmacological effects, mechanisms of action, mutagenicity, carcinogenicity, teratogenicity, and embryotoxicity. The biological activity of a compound depends on several factors, such as its structure, physicochemical properties, the biological entity it interacts with (species, gender, age, etc.), and mode of treatment (dose, route of administration, etc.). PASS predicts two probabilities for each compound based on its MNA descriptors: probable activity (Pa) and probable inactivity (Pi). Pa and Pi values range from 0.000 to 1.000, indicating the likelihood of a compound being active or inactive, respectively. In general, Pa + Pi ≠ 1. PASS predictions are usually interpreted flexibly: (i) when Pa > 0.7, the chance of finding the activity experimentally is high; (ii) if 0.5 < Pa < 0.7, the chance of finding the activity experimentally is lower, but the compound is likely dissimilar to known pharmaceutical agents; and (iii) if Pa < 0.5, the chance of finding the activity experimentally is lower.
Table 3 provides evidence that the benzylidenemalononitrile derivatives (1–3) were more effective as antineoplastic (breast cancer) agents with a Pa value of 0.512, compared to their activity as cancer-associated disorder treatment agents with a Pa value of 0.322. In contrast, the ethyl 2-cyano-3-phenylacrylate derivatives (4–6) showed a lower Pa value of 0.266 for cancer-associated disorder treatment but a similar Pa value of 0.512 for antineoplastic (breast cancer) activity, indicating that they may be less potent than the benzylidenemalononitrile derivatives. These findings are consistent with the structural similarity between these compounds and the reference compound AG9.
Table 3. Predicted Biological Activity of All Compounds (1–6) Using PASS Softwarea,b,c.
| cancer
disorder treatment |
antineoplastic
(breast cancer) |
|||
|---|---|---|---|---|
| ligand | Pa | Pi | Pa | Pi |
| 1 | 0.266 | 0.149 | 0.512 | 0.018 |
| 2 | 0.266 | 0.149 | 0.512 | 0.018 |
| 3 | 0.266 | 0.149 | 0.512 | 0.018 |
| 4 | – | – | – | – |
| 5 | – | – | – | – |
| 6 | – | – | – | – |
| A23 | 0.337 | 0.052 | 0.720 | 0.005 |
| AG9 | 0.259 | 0.158 | 0.716 | 0.005 |
Pa: probability “to be active”.
Pi: probability “to be inactive”.
AG9: Tyrphostin 1; A23: Tyrphostin 23.
The results shown in Table 3 demonstrate the potential of the benzylidenemalononitrile and ethyl 2-cyano-3-phenylacrylate derivatives as therapeutic agents for cancer-associated disorders. However, further validation is crucial to confirm the safety and efficacy of these compounds through in vitro and in vivo experiments, as well as in silico studies. The latter can provide valuable insights into the molecular mechanisms of these compounds and guide further experimental work.
3.6. Evaluation of Physicochemical Properties of Synthesized Organic Compounds
The oral bioavailability of benzylidenemalononitrile derivatives (1–3) and ethyl 2-cyano-3-phenylacrylate derivatives (4–6) was assessed by analyzing their physicochemical properties using Lipinski and Veber rules.66,67 Lipinski’s rules require a compound to meet at least three of the following conditions for oral administration: molecular weight below 500, log P below 5, no more than 5 hydrogen bond donor (HBD) atoms, and no more than 10 hydrogen bond acceptor (HBA) atoms. Veber rules add that nrotb must be less than 10 and TPSA must be less than 140 Å2. Lipinski’s rule of five, M log P, log S, X log P3, and sp3 hybridization were used to calculate drug-likeness values. All six compounds satisfied the Lipinski and Veber rules with a molecular weight less than 500 g/mol, M log P < 4.15, and log S (ESOL) values less than 6,68 and lipophilicity, X log P3 between −0.7 and +5.0,69 and fraction of carbons in the sp3 hybridization not less than 0.25 were considered for drug-likeliness calculated using Lipinski’s rule of five.66,70 SwissADME was used to assess its physicochemical properties, and Table 4 presents the results.
Table 4. In Silico Prediction of Physicochemical Parameters for the Benzylidenemalononitrile and Ethyl 2-Cyano-3-phenylacrylate Derivatives (1–6)a.
| ligand | MW | log P | HBD | HBA | nRB | TPSA |
|---|---|---|---|---|---|---|
| Lipinsli* | ≤500 | ≤5 | ≤5 | ≤10 | ||
| Veber** | ≤10 | |||||
| 1 | 254.33 | 2.31 | 0 | 3 | 7 | ≤140 |
| 2 | 282.38 | 2.79 | 0 | 3 | 9 | 59.3 |
| 3 | 310.43 | 3.25 | 0 | 3 | 9 | 59.3 |
| 4 | 301.38 | 2.75 | 0 | 4 | 10 | 56.8 |
| 5 | 329.43 | 3.21 | 0 | 4 | 10 | 56.8 |
| 6 | 357.49 | 3.65 | 0 | 4 | 10 | 56.8 |
| AG9 | 184.19 | 0.98 | 0 | 3 | 2 | 59.8 |
| A23 | 186.17 | 0.11 | 2 | 4 | 1 | 56.8 |
*Lipinski reference values; **Veber reference values; MW, molecular weight; log P, lipophilicity (O/W); HBD, number of hydrogen bond donors; HBA, number of hydrogen bond acceptors; nRB, number of rotatable bonds; TPSA, topological polar surface area (Å2).
The study further investigated the compounds by applying different computational filters, such as Lipinski, Ghose, Veber, Egan, and Muegge rules, as shown in Table S8 in the SI. Results revealed that compounds 2, 3, and 6 had one Veber violation, while compounds 2–6 had one Muegge violation. However, none of the six compounds violated Lipinski or Ghose rules (Tables 4 and S8 in the SI). A drug-like score of 1 indicates that the compound has potential as a therapeutic agent,71 and all six compounds satisfied the drug-likeness evaluation in this study. Furthermore, the bioavailability score for each compound met the drug-likeness test, suggesting high bioactivity and a promising outcome in developing new drugs.
Table 5 displays the bioactivity scores of all six compounds (1–6) based on the rule of five classifications into ion channel modulators (ICMs), kinase inhibitors (KIs), nuclear receptor ligands (NRLs), protease inhibitors (PIs), and enzyme inhibitors (EIs). According to Paramashivam et al.,72 a score greater than 0.00 denotes high activity, while scores between 0.00 and −0.5 denote moderate activity, and scores less than −0.5 indicate inactivity. The bioactivity data obtained from this study showed that the benzylidenemalononitrile derivatives (1, 2, and 3) and ethyl 2-cyano-3-phenylacrylate derivatives (4, 5, and 6) demonstrated moderate activity (Table 5 and Figures S34–S41 in the Supporting Information).
Table 5. Prediction of Drug-Likeness Properties of Benzylidenemalononitrile and Ethyl 2-Cyano-3-phenylacrylate Derivatives (1–6) via Molinspiration Softwarea.
| ligand | GPCR | ICM | KI | NRL | PI | EI |
|---|---|---|---|---|---|---|
| 1 | –0.32 | –0.35 | –0.36 | –0.15 | –0.43 | –0.22 |
| 2 | –0.21 | –0.32 | –0.25 | –0.05 | –0.30 | –0.19 |
| 3 | –0.15 | –0.29 | –0.19 | –0.00 | –0.21 | –0.17 |
| 4 | –0.37 | –0.36 | –0.53 | –0.06 | –0.35 | –0.19 |
| 5 | –0.32 | –0.33 | –0.47 | –0.03 | –0.28 | –0.17 |
| 6 | –0.29 | –0.31 | –0.43 | –0.03 | –0.24 | –0.16 |
| AG9 | –0.94 | –0.65 | –0.85 | –0.88 | –1.12 | –0.66 |
| A23 | –0.81 | –0.47 | –0.74 | –0.66 | –1.03 | –0.48 |
Physicochemical parameters of drug-likeness property data for benzylidenemalononitrile derivatives (1–6) defined in Figures S34–S40 in the SI.
3.7. Assessment of ADMET Properties of Compounds 1–6
Evaluating pharmacokinetic properties such as absorption, distribution, metabolism, excretion, and toxicity is crucial for developing new drugs to save time and cost. To evaluate the ADMET properties of the active compounds (1–6), three free access websites, including PreAdmet, admetSAR, and SwissADME software, were used to calculate various ADMET-associated properties, such as water solubility, oral bioavailability, blood–brain barrier penetration, and toxicity. A total of seven ADMET parameters, including human intestinal absorption (HIA), blood–brain barrier (BBB), plasma protein binding (PPB), CYP3A4, CYP2C19, lead-likeness, and synthetic accessibility (SA) core, were tested for compounds (1–6) as listed in Table 6.
Table 6. Calculation of Selected ADMET Parameters for Derivatives of Benzylidenemalononitrile and Ethyl 2-Cyano-3-phenylacrylatea,b,c,d,e.
| ligand | bHIA | cBBB | bPPB | cCYP3A4 inhibition | cCYP2C19 inhibition | dsynthetic accessibility score | ehERG_pIC50 |
|---|---|---|---|---|---|---|---|
| 1 | 98.27 | +(0.96) | 96.93 | +(0.5477) | +(0.5194) | 2.47 | 4.94 |
| 2 | 98.36 | +(0.96) | 99.83 | +(0.5477) | +(0.5194) | 2.70 | 5.25 |
| 3 | 98.25 | +(0.96) | 100 | +(0.5477) | +(0.5194) | 2.92 | 5.51 |
| 4 | 98.43 | +(0.94) | 93.24 | –(0.7273) | –(0.5071) | 3.02 | 4.97 |
| 5 | 98.40 | +(0.94) | 94.83 | –(0.7273) | –(0.5071) | 3.26 | 5.26 |
| 6 | 98.21 | +(0.94) | 96.17 | –(0.7273) | –(0.5071) | 3.48 | 5.50 |
| AG9 | 98.80 | +(0.95) | 85.62 | +(0.7822) | –(0.9025) | 1.84 | 3.82 |
| A23 | 97.75 | –(0.32) | 83.19 | +(0.8192) | –(0.9080) | 1.99 | 4.12 |
HIA: Human Intestinal absorption (%); BBB: blood–brain barrier penetration; PPB: plasma protein binding; CYP3A4: cytochrome P450 3A4; CYP2C19: cytochrome P4502C19; hERG: human ether-a-go-go-related gene, hERG inhibition potential (pIC50), the potential risk for inhibitors ranges 5.5–6.
The values are using ADME.
The values are using admetSAR.
The values are using swissADME.
The values are using ADMET 9.5.
The ADMET properties of the compounds (1–6) were evaluated using seven parameters, including HIA, BBB, PPB, CYP3A4, CYP2C19, lead-likeness, and SA core. HIA is crucial in drug discovery, with absorption levels classified as high (70–100%), medium (20–70%), and low (0–20%). The compounds (1–6), along with Tyrphostin 1 (AG9) and Tyrphostin 23 (A23) references, were found to have high HIA, indicating their potential as oral drug candidates. BBB penetration is also important for drugs that affect the CNS, with high BBB penetration (>2) being beneficial. The compounds (1, 2, and 3) showed moderate BBB penetration values (+0.96), and compounds (4, 5, and 6) showed similar results (+0.94). These findings align with the reference compound AG9, which showed BBB penetration of +0.95.
Plasma protein binding (PPB) can affect the duration of a drug in the body, with a higher protein-bound percentage resulting in a slower release of the drug. A binding of >90% is considered high, while that of <90% is considered weak.73 Based on computational calculations, the benzylidenemalonitrile derivative (3) showed a PPB of 100, and the ethyl 2-cyano-3-phenylacrylate derivative (6) showed a PPB of 96.17, indicating a high binding to plasma proteins due to the increasing methylene (−CH2) residues of the receptor protein. In silico calculations suggest that CYP3A4 and CYP2C19, which are cytochrome P450 enzymes, are responsible for drug metabolism, and inhibiting these proteins can increase the plasma levels and toxicity of these compounds (as shown in Table 6).
A new method has been developed to calculate the synthetic accessibility (SA)74 score of drug-like molecules (1–6). The SA score ranges from 1 (very easy) to 10 (very difficult), and the benzylidenemalononitrile derivative (3) showed a score of 2.92. In contrast, the ethyl 2-cyano-3-phenylacrylate derivative (6) scored 3.48, indicating that these compounds are relatively easy to synthesize. The SA score is helpful for virtual screening and predicting the identified molecules’ hERG activity (pIC50) (as shown in Table 6). For example, 4-hexyloxybenzylidenemalononitrile (1) showed a higher hERG_pIC50 value of 4.94 compared to 4-methoxybenzylidenemalononitrile (Tyrphostin 1, AG9) due to the increasing methylene (−CH2) residues. Compound 3 had a higher value than compounds 1 and 2, with a reference value of the potential risk for inhibitors ranging from 5.5 to 6.74
3.8. Computational Drug Discovery and Design Using In Silico Molecular Docking
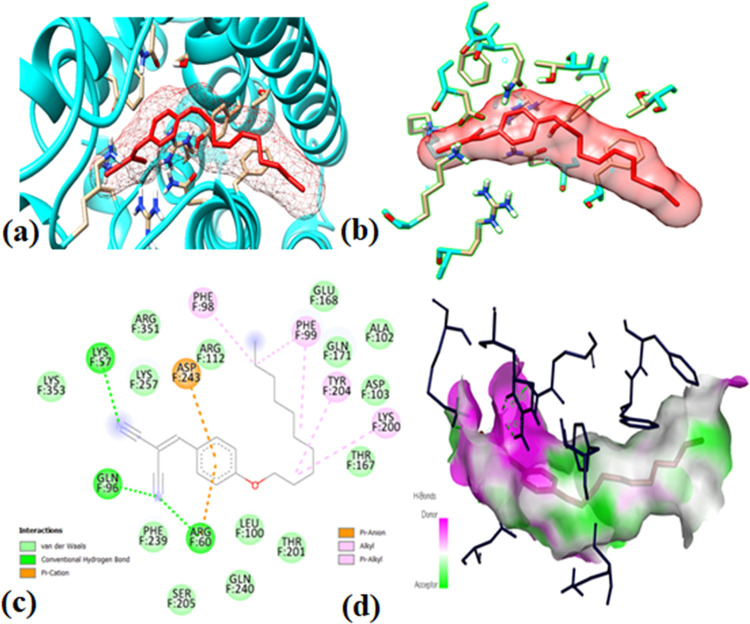
The use of molecular docking is a crucial computational approach in drug discovery. The synthesized compounds (1–6) and reference compounds AG9 and A23 were subjected to molecular docking studies against the targets HER2 (7JXH), human FPPS (4H5D), and EGFR (4LRM), and the results are presented in Table 4. Compounds 3 and 4, namely, 4-decyloxybenzylidenemalonitrile and 2-cyano-3-(4-hexyloxy)phenylacrylate, respectively, exhibited high binding affinity against human FPPS (−8.0 kcal mol–1) and HER2 (−7.9 kcal mol–1). Compound 3 also demonstrated binding affinity of −7.4 kcal mol–1 against HER2. The docking values obtained for all compounds were superior regarding anticancer activity compared to the reference drugs AG9 (−6.4 kcal mol–1) and A23 (−6.5 kcal mol–1), as displayed in Table 7. Additionally, compounds 3 and 4 exhibited robust binding abilities, followed by compounds 1, 2, 5, and 6 (refer to Table 7 and Figure 3 in the SI).
Table 7. Results of Molecular Docking Simulation for the Selected Compounds Targeting Three Proteins.
Figure 3.
Molecular docking results: (a) ligand binding in protein pocket; (b) hydrogen bonding; (c) ligand–protein interaction in two-dimensional (2D) diagram; and (d) hydrogen bonding in the solid state for compound 3 in HER2.
The binding site interactions of the docked protein and ligand and their derivatives (1–6) are defined in Tables S9–S11 in the supplementary information. The synthesized organic compounds (1–6) were found to have lower binding affinity for the epidermal growth factor receptor as compared to human FPPS and HER2, as shown in Table 7. The chemical bonds between the ligand and protein at the active site were analyzed to understand how the ligand affects the disease-causing pathogens. The bond length and residue numbers for each type of bond, including the ligand in protein pocket, hydrogen bonding, ligand–protein interaction for 2D diagram, and hydrogen bonding in solid for compound 3 in human FPPS, are shown in Figure 3. The ligand–protein interaction through the amino acid residues for compound 3 in FPPS is illustrated in Figure 3, where approx. 10 different bonds were observed. Seven of them were hydrophobic, which included Pi-Alkyl on the residues PHE F:98, PHE F:99, PHE F:99, TYR F:204, LYS F:200, and Pi-Anion on the residues ASP F:243, ARG F:60 with various bond lengths. The other three types of bonds observed were conventional hydrogen bonds at residues ARG F:60, GLN F:96, and LYS F:27.
3.9. Molecular Dynamics Simulation Using In Silico Methods
To understand the stability and interactions of the protein–ligand complex with HER2 for compound 3 (Figures 4–7) as well as human FPPS (see Figures S42–S44 in the SI), as well as HER2, the results of the MD simulations were analyzed. The analysis included various metrics, such as the RMSD values for the backbone and ligand and the Lennard-Jones short-range and potential energies. The RMSD analysis of the protein–ligand complex showed that both the protein and ligand were stable in the binding site, with RMSD values ranging from 0.2 to 0.35 nm (Figure 4). The whole protein system remained stable throughout the trajectory. These findings indicate that the docked complex could maintain its structure despite the thermal and dynamic motions of the atoms within the system. Overall, the MD simulations provide evidence that the protein–ligand complex is stable and that the ligand binding to the protein is likely strong and persistent.
Figure 4.
RMSD evolution for (a) protein–ligand complex, (b) protein (HER2), and (c) ligand (3) during 20 ns MD simulation.
Figure 7.
Molecular dynamics simulation and trajectory analysis of the ligand (3) with human FPPS: (a) LJ analysis, (b) total energy analysis, (c) Coulomb (SR) analysis, and (d) hydrogen bond stabilization.
The degree of structural flexibility of a protein is often captured by a metric called root-mean-square fluctuation (RMSF), which measures localized variations in the protein’s conformation. In this study, the RMSF analysis revealed that the N- and C-termini of the protein tend to have greater mobility compared to other rigid segments, such as the loop regions of α helices and β strands within its secondary structures (shown in Figure 5). The RMSF plot of the protein–ligand complex also showed a similar trend, with the RMSF values of both the protein and the ligand complex measured at 0.1 nanometers.
Figure 5.
RMSF evolution for (a) protein–ligand complex, (b) protein (HER2), and (c) ligand (3) during 20 ns MD simulation.
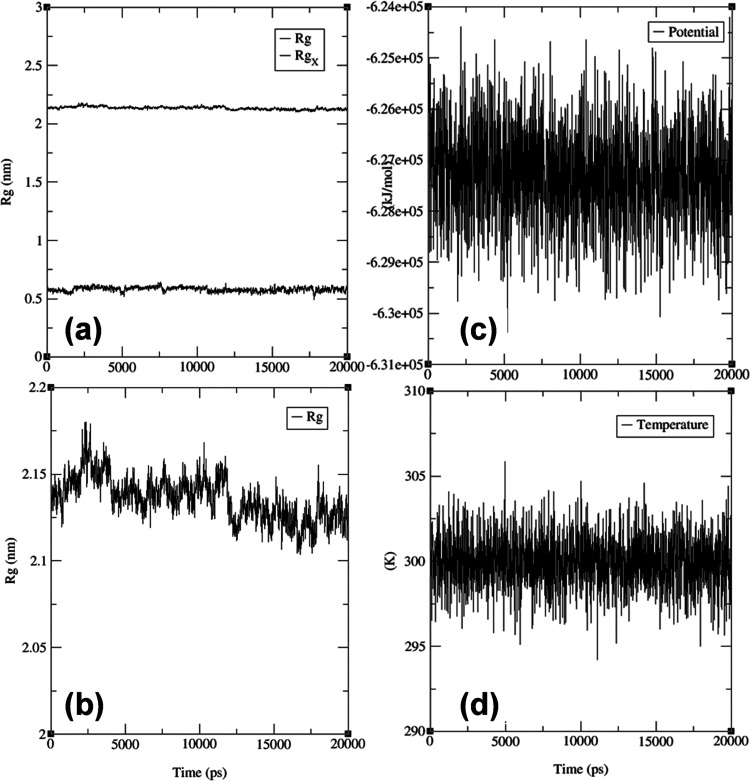
The radius of gyration (Rg) is a parameter that characterizes the size and shape of a macromolecule, such as a protein–ligand complex. In this study, the Rg value of the protein–ligand complex was determined to be in the range of 2.12–2.15 nm (Figure 6a,b). This finding suggests that including the ligand did not significantly change the protein’s overall size or shape. This result is consistent with the idea that the ligand has bound to the protein in a way that did not cause significant disruptions to its tertiary structure. Moreover, a similar Rg value between the protein and the protein–ligand complex may imply that the binding has not induced significant structural changes within the protein. It is essential to note that while Rg values can provide insights into a protein’s overall size and shape, they may not reveal the exact structural changes that have occurred at the molecular level. Further experiments, such as X-ray crystallography or NMR spectroscopy, may be necessary to fully understand the molecular interactions between the protein and the ligand.
Figure 6.
Molecular dynamics simulation and trajectory analysis of the ligand (3) with HER2: Radius of gyration (Rg) for the ligand (3) with the protein–ligand complex (RgX), (b) Rg for the whole protein, (c) temperature analysis, and (d) potential analysis.
The simulation demonstrated that the temperature of the system remained steady during the 20,000 ps run, fluctuating between 298.5 and 303.5 Kelvin, as indicated by Figure 6c,d. This suggests that the protein–ligand complex did not significantly affect the thermal energy of the system. However, the potential energy of the system showed some fluctuations, with values ranging from −6.29 × 105 to −6.25 × 105 kJ mol–1 (Figure 6c,d), suggesting changes in the electrostatic interactions between the atoms. Meanwhile, the Lennard-Jones short-range energy (Figure 7a), which characterizes the van der Waals forces between atoms, remained consistent between 76,000 and 79,000, indicating that the attractive and repulsive forces between the atoms remained stable throughout the simulation.
During molecular dynamics simulations, the total energy of a system is a crucial measure for assessing the stability and interactions of a protein–ligand complex. In this study, the total energy of compound 3 was computed and found to range from −5.02 × 105 to −4.98 × 105 (Figure 7b). The consistently negative values and limited range of the total energy suggest that the complex was stable throughout the simulation and did not undergo significant fluctuations in energy. Moreover, analyzing the magnitude and distribution of the energy values can offer insights into the specific protein–ligand interactions that occurred.
The Coulomb (SR) energy values obtained from the MD simulation, which varied from −8.07 × 105 to −8.04 × 105 (Figure 7c), indicate that the electrostatic interactions between the protein and ligand are energetically favorable and stable throughout the simulation. The consistent and negative values suggest that the protein–ligand complex did not experience significant changes in electrostatic interactions. By analyzing the magnitude and distribution of the Coulomb (SR) energy, researchers can gain insights into the specific interactions between the protein and the ligand. Further studies, such as free energy calculations, may be necessary to gain a comprehensive understanding of the energetics of the protein–ligand interaction.
The MD simulation results highlight the significance of hydrogen-bonding interactions in determining the stability of protein–ligand binding. The interactions between the ligand and the protein through hydrogen bonds are illustrated in Figure 7d. The simulation demonstrated that the compound in the active site of the protein formed an average of 0.98 ± 0.23 hydrogen bonds, ranging from zero to two hydrogen bonds throughout the 20 ns simulation. The relatively high mean number of hydrogen bonds suggests that the ligand–protein complex is likely to be stable, as the hydrogen-bonding interactions contribute to the thermodynamic stability of the complex. However, it is essential to note that additional studies, such as thermodynamic analyses, are necessary to gain a complete understanding of the protein–ligand interactions.
4. Conclusions
In this study, six derivatives (1–6) of benzylidenemalononitrile and ethyl 2-cyano-3-phenylacrylate were confirmed by FT-IR and 1H NMR spectroscopies. A computational study was performed on these compounds against three cancer-related proteins and one parasite target to determine their potential as anticancer and antiparasitic medicines. Various computational methods were utilized, including DFT calculations, molecular docking calculations, binding energy calculations, thermodynamic properties analysis, HOMO and LUMO investigation, drug-likeness analyses, ADMET property evaluation, and MD simulations. Compound 3 showed the highest binding affinity as an anticancer target, which suggests that it could be an effective therapeutic molecule. In comparison to the reference drugs, compound 3 also displayed strong effects on HER2 and FPPS cell lines, and all of the compounds satisfied numerous drug-likeness requirements. All of the compounds exhibited moderate-to-low acute oral toxicity, demonstrating that they are safe for oral administration. However, additional experimental validation is necessary to validate the potential of these chemicals as anticancer and antiparasite medications. Such results are expected in the near future.
Acknowledgments
This study received financial support from the Ministry of Science and Technology (MOST) under SL 534 PHY’s for 2020–2021 in Bangladesh. The authors wish to acknowledge the Princess Nourah bint Abdulrahman University Researchers Supporting Project (reference number PNURSP2023R29) in Riyadh, Saudi Arabia, for their assistance. The authors are also grateful to the Deanship of Scientific Research, King Saud University, for funding the research through the Vice Deanship of Scientific Research Chairs, Research Chair for Tribology, Surface, and Interface Sciences. Finally, the authors thank Professor Raymond A. Poirier and the Digital Research Alliance of Canada (alliancecan.ca) for providing the computational resources in this study.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c01123.
Experimental section, FT-IR and NMR; in silico section; DFT output files; drug-likeness; PASS; SwissADME; ADMET; HOMO–LUMO, NBO, docking, and MD simulation studies (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Lindsay C. D.; Green C.; Bird M.; Jones J. T. A.; Riches J. R.; McKee K. K.; Sandford M. S.; Wakefield D. A.; Timperley C. M. Potency of Irritation by Benzylidenemalononitriles in Humans Correlates with TRPA1 Ion Channel Activation. R. Soc. Open Sci. 2015, 2, 140160 10.1098/rsos.140160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpaev K.; Ermolenko M.; Cresteil T.; Drapier J. C. Benzylidenemalononitrile Compounds as Activators of Cell Resistance to Oxidative Stress and Modulators of Multiple Signaling Pathways. A Structure–Activity Relationship Study. Biochem. Pharmacol. 2011, 82, 535–547. 10.1016/j.bcp.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Güller P.; Dağalan Z.; Güller U.; Çalışır U.; Nişancı B.. Enzymes Inhibition Profiles and Antibacterial Activities of Benzylidenemalononitrile Derivatives J. Mol. Struct., 1239130498 10.1016/j.molstruc.2021.130498. [DOI] [Google Scholar]
- Viel C.; Doré J. C. [New Synthetic Cytotoxic and Antitumoral Agents Derived from Aristolochic Acid, an Antitumoral Nitrophenanthrenic Acid Extracted from Aristolochiaceae]. Farmaco, Ed. Sci. 1972, 27, 257–312. [PubMed] [Google Scholar]
- Hu Q.; Shi X.-L.; Chen Y.; Han X.; Duan P.; Zhang W. Revisiting the Knoevenagel Condensations: A Universal and Flexible Bis-Ammoniated Fiber Catalyst for the Mild Synthesis of α,β-Unsaturated Compounds. J. Ind. Eng. Chem. 2017, 54, 75–81. 10.1016/j.jiec.2017.05.020. [DOI] [Google Scholar]
- Maltsev S. S.; Mironov M. A.; Bakulev V. A. Synthesis of Cyclopentene Derivatives by the Cyclooligomerization of Isocyanides with Substituted Benzylidenemalononitriles. Mendeleev Commun. 2006, 16, 201–202. 10.1070/mc2006v016n04abeh002354. [DOI] [Google Scholar]
- Sidhu A.; Sharma J. R.; Rai M. Chemoselective Reaction of Malononitrile with Imine-Ones and Antifungal Potential of Products. Indian Journal of Chemistry 2010, 49B, 247–250. [Google Scholar]
- Alwarappan S.; Boyapalle S.; Kumar A.; Li C.-Z.; Mohapatra S. Comparative Study of Single-, Few-, and Multilayered Graphene toward Enzyme Conjugation and Electrochemical Response. J. Phys. Chem. C 2012, 116, 6556–6559. 10.1021/jp211201b. [DOI] [Google Scholar]
- Khan S. A.; Asiri A. M.; Rahman R. M.; Elroby S. A.; Aqlan F. M. S.; Wani M. Y.; Sharma K. Multistep Synthesis of Fluorine-Substituted Pyrazolopyrimidine Derivatives with Higher Antibacterial Efficacy Based on in Vitro Molecular Docking and Density Functional Theory. J. Heterocycl. Chem. 2017, 54, 3099–3107. 10.1002/jhet.2923. [DOI] [Google Scholar]
- Bhuiyan M. M. H.; Rahman K. M. M.; Alam M. A.; Mahmud M. M. Microwave Assisted Knoevenagel Condensation: Synthesis and Antimicrobial Activities of Some A-Cyanoacrylates. Pak. J. Sci. Ind. Res., Ser. A 2013, 56, 131–137. 10.52763/PJSIR.PHYS.SCI.56.3.2013.131.137. [DOI] [Google Scholar]
- Panicker R. K. G.; Krishnapillai S. Synthesis of on Resin Poly(Propylene Imine) Dendrimer and Its Use as Organocatalyst. Tetrahedron Lett. 2014, 55, 2352–2354. 10.1016/j.tetlet.2014.02.084. [DOI] [Google Scholar]
- Almáši M.; Zeleňák V.; Opanasenko M.; Čejka J. A Novel Nickel Metal–Organic Framework with Fluorite-like Structure: Gas Adsorption Properties and Catalytic Activity in Knoevenagel Condensation. Dalton Trans. 2014, 43, 3730 10.1039/c3dt52698d. [DOI] [PubMed] [Google Scholar]
- Fouda A. S.; El-Ewady Y. A.; Abo-El-Enien O. M.; Agizah F. A. Cinnamoylmalononitriles as Corrosion Inhibitors for Mild Steel in Hydrochloric Acid Solution. Anti-Corros. Methods Mater. 2008, 55, 317–323. 10.1108/00035590810913105. [DOI] [Google Scholar]
- Sheikhhosseini E.; Soltaninejad S. Design and Efficient Synthesis of Novel Biological Benzylidenemalononitrile Derivatives Containing Ethylene Ether Spacers. Iran. J. Sci. Technol., Trans. A: Sci. 2019, 43, 111–117. 10.1007/s40995-017-0376-9. [DOI] [Google Scholar]
- Roskoski R. The Role of Small Molecule Kit Protein-Tyrosine Kinase Inhibitors in the Treatment of Neoplastic Disorders. Pharmacol. Res. 2018, 133, 35–52. 10.1016/j.phrs.2018.04.020. [DOI] [PubMed] [Google Scholar]
- Levitzki A.; Mishani E. Tyrphostins and Other Tyrosine Kinase Inhibitors. Annu. Rev. Biochem. 2006, 75, 93–109. 10.1146/annurev.biochem.75.103004.142657. [DOI] [PubMed] [Google Scholar]
- a Doni E.; Murphy J. A. Reductive Decyanation of Malononitriles and Cyanoacetates Using Photoactivated Neutral Organic Super-Electron-Donors. Org. Chem. Front. 2014, 1, 1072–1076. 10.1039/C4QO00202D. [DOI] [Google Scholar]; b Fatiadi A. J. New Applications of Malononitrile in Organic Chemistry - Part I. Synthesis 1978, 1978, 165–204. 10.1055/s-1978-24703. [DOI] [Google Scholar]
- Freeman F. Properties and Reactions of Ylidenemalononitriles. Chem. Rev. 1980, 80, 329–350. 10.1021/cr60326a004. [DOI] [Google Scholar]
- Hoffknecht P.; Tufman A.; Wehler T.; Pelzer T.; Wiewrodt R.; Schütz M.; Serke M.; Stöhlmacher-Williams J.; Märten A.; Maria Huber R.; Dickgreber N. J. Afatinib Compassionate Use Consortium (ACUC). Efficacy of the Irreversible ErbB Family Blocker Afatinib in Epidermal Growth Factor Receptor (EGFR) Tyrosine Kinase Inhibitor (TKI)-Pretreated Non-Small-Cell Lung Cancer Patients with Brain Metastases or Leptomeningeal Disease. J. Thorac. Oncol. 2015, 10, 156–163. 10.1097/JTO.0000000000000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y. New Synthetic Methodologies for Carbon–Carbon Double Bond Formation. Acc. Chem. Res. 1998, 31, 584–592. 10.1021/ar970060i. [DOI] [Google Scholar]
- Yi X.-C.; Huang M.-X.; Qi Y.; Gao E.-Q. Synthesis, Structure, Luminescence and Catalytic Properties of Cadmium(II) Coordination Polymers with 9H-Carbazole-2,7-Dicarboxylic Acid. Dalton Trans. 2014, 43, 3691–3697. 10.1039/C3DT53109K. [DOI] [PubMed] [Google Scholar]
- Ammar H. B.; Chtourou M.; Frikha M. H.; Trabelsi M. Green Condensation Reaction of Aromatic Aldehydes with Active Methylene Compounds Catalyzed by Anion-Exchange Resin under Ultrasound Irradiation. Ultrason. Sonochem. 2015, 22, 559–564. 10.1016/j.ultsonch.2014.07.018. [DOI] [PubMed] [Google Scholar]
- a Lolak N.; Kuyuldar E.; Burhan H.; Goksu H.; Akocak S.; Sen F. Composites of Palladium–Nickel Alloy Nanoparticles and Graphene Oxide for the Knoevenagel Condensation of Aldehydes with Malononitrile. ACS Omega 2019, 4, 6848–6853. 10.1021/acsomega.9b00485. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Appaturi J. N.; Ratti R.; Phoon B. L.; Batagarawa S. M.; Din I. U.; Selvaraj M.; Ramalingam R. J. Correction: A Review of the Recent Progress on Heterogeneous Catalysts for Knoevenagel Condensation. Dalton Trans. 2021, 50, 5370. 10.1039/D1DT90055B. [DOI] [PubMed] [Google Scholar]; c Li X.; Lin B.; Li H.; Yu Q.; Ge Y.; Jin X.; Liu X.; Zhou Y.; Xiao J. Carbon Doped Hexagonal BN as a Highly Efficient Metal-Free Base Catalyst for Knoevenagel Condensation Reaction. Appl. Catal., B 2018, 239, 254–259. 10.1016/j.apcatb.2018.08.021. [DOI] [Google Scholar]
- Rao P. S.; Venkataratnam R. V. Zinc Chloride as a New Catalyst for Knoevenagel Condensation. Tetrahedron Lett. 1991, 32, 5821–5822. 10.1016/s0040-4039(00)93564-0. [DOI] [Google Scholar]
- Chakrabarty M.; Mukherji A.; Arima S.; Harigaya Y.; Pilet G. Expeditious Reaction of Ninhydrin with Active Methylene Compounds on Montmorillonite K10 Clay. Monatsh. Chem. Chem. Mon. 2008, 140, 189–197. 10.1007/s00706-008-0066-6. [DOI] [Google Scholar]
- de la Cruz P.; Díez-Barra E.; Loupy A.; Langa F. Silica Gel Catalysed Knoevenagel Condensation in Dry Media under Microwave Irradiation. Tetrahedron Lett. 1996, 37, 1113–1116. 10.1016/0040-4039(95)02318-6. [DOI] [Google Scholar]
- Patil S. S.; Jadhav S. D.; Deshmukh M. B. Eco-Friendly and Economic Method for Knoevenagel Condensation by Employing Natural Catalyst. Indian Journal of Chemistry 2013, 52B, 1172–1175. [Google Scholar]
- Balalaie S.; Nemati N. Ammonium Acetate-Basic Alumina Catalyzed Knoevenagel Condensation under Microwave Irradiation under Solvent-Free Condition. Synth. Commun. 2000, 30, 869–875. 10.1080/00397910008087099. [DOI] [Google Scholar]
- Bigi F.; Conforti M. L.; Maggi R.; Piccinno A.; Sartori G. Clean Synthesis in Water: Uncatalysed Preparation of Ylidenemalononitriles. Green Chem. 2000, 2, 101–103. 10.1039/b001246g. [DOI] [Google Scholar]
- Zengin N.; Burhan H.; Şavk A.; Göksu H.; Şen F. Synthesis of Benzylidenemalononitrile by Knoevenagel Condensation through Monodisperse Carbon Nanotube-Based NiCu Nanohybrids. Sci. Rep. 2020, 10, 12758 10.1038/s41598-020-69764-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.-W.; Cheng B. Solvent-Free and Aqueous Knoevenagel Condensation of Aromatic Ketones with Malononitrile. Arkivoc 2004, 2004, 4–8. 10.3998/ark.5550190.0005.902. [DOI] [Google Scholar]
- Varma R. S. Solvent-Free Organic Syntheses. Using Supported Reagents and Microwave Irradiation. Green Chem. 1999, 1, 43–55. 10.1039/A808223E. [DOI] [Google Scholar]
- Bhuiyan M.; Hossain M.; Mahmud M.; Al-Amin M.. Microwave-assisted Efficient Synthesis of Chalcones as Probes for Antimicrobial Activities. https://www.semanticscholar.org/paper/Microwave-assisted-Efficient-Synthesis-of-Chalcones-Bhuiyan-Hossain/22ec2d1c5e4b93f865e6a71b14a9fe1ab025df55. (accessed 27 January, 2023).
- Mary Y. S.; Mary Y. S.; Ciltaş A. C. Biological Perspective of a Triazine Derivative with Isatin/Chalcone/Acridone: DFT and Docking Investigations. Struct. Chem. 2021, 32, 19–26. 10.1007/s11224-020-01609-6. [DOI] [Google Scholar]
- Matin M. M.; Uzzaman M.; Chowdhury S. A.; Bhuiyan M. M. H. In Vitro Antimicrobial, Physicochemical, Pharmacokinetics and Molecular Docking Studies of Benzoyl Uridine Esters against SARS-CoV-2 Main Protease. J. Biomol. Struct. Dyn. 2022, 40, 3668–3680. 10.1080/07391102.2020.1850358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matin M. M.; Iqbal M. Z. Methyl 4-O-(2-Chlorobenzoyl)-α-L-Rhamnopyranosides: Synthesis, Characterization, and Thermodynamic Studies. Orbital: Electron. J. Chem. 2021, 13, 19–27. 10.17807/orbital.v13i1.1532. [DOI] [Google Scholar]
- van de Waterbeemd H.; Gifford E. ADMET in Silico Modelling: Towards Prediction Paradise?. Nat. Rev. Drug Discovery 2003, 2, 192–204. 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- Daina A.; Michielin O.; Zoete V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M. J.; Trucks G. W.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Mennucci B.; Petersson G. A.; Nakatsuji H.; Caricato M.; Li X.; Hratchian H. P.; Izmaylov A. F.; Bloino J.; Zheng G.; Sonnenberg J. L.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T. Jr.; Montgomery J. A.; Peralta J. E.; Ogliaro F.; Bearpark M.; Heyd J. J.; Brothers E.; Kudin K. N.; Staroverov V. N.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Rega N.; Millam J. M.; Klene M.; Knox J. E.; Cross J. B.; Bakken V.; Adamo C.; Jaramillo J.; Gomperts R.; Stratmann R. E.; Yazyev O.; Austin A. J.; Cammi R.; Pomelli C.; Ochterski J. W.; Martin R. L.; Morokuma K.; Zakrzewski V. G.; Voth G. A.; Salvador P.; Dannenberg J. J.; Dapprich S.; Daniels A. D.; Farkas Ö.; Foresman J. B.; Ortiz J. V.; Cioslowski J.; Fox D. J.. Gaussian 16, Revision C.01; Gaussian, Inc.: Wallingford CT, 2016.
- a Uddin K. M.; Alrawashdeh A. I.; Henry D. J.; Warburton P. L.; Poirier R. A. Hydrolytic Deamination Reactions of Amidine and Nucleobase Derivatives. Int. J. Quantum Chem. 2019, 120, e26059 10.1002/qua.26059. [DOI] [Google Scholar]; b Uddin K. M.; Henry D. J.; Alrawashdeh A. I.; Warburton P. L.; Poirier R. A. Mechanism for the deamination of ammeline, guanine, and their analogues. Struct. Chem. 2017, 28, 1467–1477. 10.1007/s11224-017-0941-z. [DOI] [Google Scholar]; c Uddin K. M.; Almatarneh M. H.; Shaw D. M.; Poirier R. A. Mechanistic Study of the Deamination Reaction of Guanine: A Computational Study. J. Phys. Chem. A 2011, 115, 2065–2076. 10.1021/jp1120806. [DOI] [PubMed] [Google Scholar]; d Uddin K. M.; Poirier R. A. Computational Study of the Deamination of 8-Oxoguanine. J. Phys. Chem. B 2011, 115, 9151–9159. 10.1021/jp202098k. [DOI] [PubMed] [Google Scholar]; e Uddin K. M.; Flinn C. G.; Poirier R. A.; Warburton P. L. Comparative computational investigation of the reaction mechanism for the hydrolytic deamination of cytosine, cytosine butane dimer and 5,6-saturated cytosine analogues. Comput. Theor. Chem. 2014, 1027, 91–102. 10.1016/j.comptc.2013.10.027. [DOI] [Google Scholar]; f Alrawashdeh A. I.; Almatarneh M. H.; Poirier R. A. Computational study on the deamination reaction of adenine with OH–/nH2O (n = 0, 1, 2, 3) and 3H2O. Can. J. Chem 2013, 91, 518–526. 10.1139/cjc-2012-0416. [DOI] [Google Scholar]; g Almatarneh M. H.; Flinn C. G.; Poirier R. A.; Sokalski W. A. Computational Study of the Deamination Reaction of Cytosine with H2O and OH-. J. Phys. Chem. A 2006, 110, 8227–8234. 10.1021/jp062300u. [DOI] [PubMed] [Google Scholar]; h Uddin K. M.; Hosen M. A.; Khan M. F.; Ozeki Y.; Kawsar S. M. A. Investigation of Structural, Physicochemical, Pharmacokinetics, PASS Prediction, and Molecular Docking Analysis of Methyl 6-O-Myristoyl-α-D -Glucopyranoside Derivatives against SARS-CoV-2. Philipp. J. Sci. 2022, 151, 2215–2231. 10.56899/151.6A.13. [DOI] [Google Scholar]
- a Pearson R. G. Absolute Electronegativity and Absolute Hardness of Lewis Acids and Bases. J. Am. Chem. Soc. 1985, 107, 6801–6806. 10.1021/ja00310a009. [DOI] [Google Scholar]; b Das R.; Vigneresse J.-L.; Chattaraj P. K. Redox and Lewis Acid–Base Activities through an Electronegativity-Hardness Landscape Diagram. J. Mol. Model. 2013, 19, 4857–4864. 10.1007/s00894-013-1986-6. [DOI] [PubMed] [Google Scholar]
- a Chamizo J. A.; Morgado J.; Sosa P. Organometallic Aromaticity. Organometallics 1993, 12, 5005–5007. 10.1021/om00036a047. [DOI] [Google Scholar]; b Zhu J. Open Questions on Aromaticity in Organometallics. Commun. Chem. 2020, 3, 161 10.1038/s42004-020-00419-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasstone S.; Laidler K. J.; Eyring H.. The Theory of Rate Processes: The Kinetics of Chemical Reactions, Viscosity, Diffusion and Electrochemical Phenomena; McGraw-Hill Book Company, Incorporated, 1941. [Google Scholar]
- Alberty R. A. The Foundations of Chemical Kinetics (Benson, Sidney W.). J. Chem. Educ. 1960, 37, 660. 10.1021/ed037p660.1. [DOI] [Google Scholar]
- a Parr R. G.; Szentpály Lv.; Liu S. Electrophilicity Index. J. Am. Chem. Soc. 1999, 121, 1922–1924. 10.1021/ja983494x. [DOI] [Google Scholar]; b Pal R.; Chattaraj P. K. Electrophilicity Index Revisited. J. Comput. Chem. 2023, 44, 278–297. 10.1002/jcc.26886. [DOI] [PubMed] [Google Scholar]
- Srivastava A. K.; Singh D.; Roy B. K. Structural Interactions of Curcumin Biotransformed Molecules with the N-Terminal Residues of Cytotoxic-Associated Gene a Protein Provide Insights into Suppression of Oncogenic Activities. Interdiscip. Sci.: Comput. Life Sci. 2017, 9, 116–129. 10.1007/s12539-016-0142-2. [DOI] [PubMed] [Google Scholar]
- Lagunin A.; Stepanchikova A.; Filimonov D.; Poroikov V. PASS: Prediction of Activity Spectra for Biologically Active Substances. Bioinformatics 2000, 16, 747–748. 10.1093/bioinformatics/16.8.747. [DOI] [PubMed] [Google Scholar]
- Poroikov V. V.; Filimonov D. A.; Ihlenfeldt W.-D.; Gloriozova T. A.; Lagunin A. A.; Borodina Y. V.; Stepanchikova A. V.; Nicklaus M. C. PASS Biological Activity Spectrum Predictions in the Enhanced Open NCI Database Browser. J. Chem. Inf. Comput. Sci. 2003, 43, 228–236. 10.1021/ci020048r. [DOI] [PubMed] [Google Scholar]
- Berman H. M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J.; Jang J.; Beyett T. S.; Eum Y.; Haikala H. M.; Verano A.; Lin M.; Hatcher J. M.; Kwiatkowski N. P.; Eser P. Ö.; Poitras M. J.; Wang S.; Xu M.; Gokhale P. C.; Cameron M. D.; Eck M. J.; Gray N. S.; Jänne P. A. A Novel HER2-Selective Kinase Inhibitor Is Effective in HER2 Mutant and Amplified Non–Small Cell Lung Cancer. Cancer Res. 2022, 82, 1633–1645. 10.1158/0008-5472.can-21-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T. D.; Huang C. C.; Ferrin T. E. Visualizing Density Maps with UCSF Chimera. J. Struct. Biol. 2007, 157, 281–287. 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Dallakyan S.; Olson A. J.. Small-Molecule Library Screening by Docking with PyRx. In Methods in Molecular Biology; Springer, 2014; pp 243–250. [DOI] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L.The PyMOL Molecular Graphics System; De-Lano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Hansson T.; Oostenbrink C.; van Gunsteren W. Molecular Dynamics Simulations. Curr. Opin. Struct. Biol. 2002, 12, 190–196. 10.1016/s0959-440x(02)00308-1. [DOI] [PubMed] [Google Scholar]
- Allen M. P. Introduction to molecular dynamics simulation. Comput. Soft Matter: Synth. Polym. Proteins 2004, 23, 1–28. [Google Scholar]
- Karplus M.; McCammon J. A. Molecular Dynamics Simulations of Biomolecules. Nat. Struct. Biol. 2002, 9, 646–652. 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D.; Lindahl E.; Hess B.; Groenhof G.; Mark A. E.; Berendsen H. J. C. GROMACS: Fast, Flexible, and Free. J. Comput. Chem. 2005, 26, 1701–1718. 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Showalter S. A.; Brüschweiler R. Validation of Molecular Dynamics Simulations of Biomolecules Using NMR Spin Relaxation as Benchmarks: Application to the AMBER99SB Force Field. J. Chem. Theory Comput. 2007, 3, 961–975. 10.1021/ct7000045. [DOI] [PubMed] [Google Scholar]
- Bray S. A.; Lucas X.; Kumar A.; Grüning B. A. The ChemicalToolbox: Reproducible, User-Friendly Cheminformatics Analysis on the Galaxy Platform. J. Cheminf. 2020, 12, 40 10.1186/s13321-020-00442-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presti D.; Pedone A.; Mancini G.; Duce C.; Tiné M. R.; Barone V. Insights into Structural and Dynamical Features of Water at Halloysite Interfaces Probed by DFT and Classical Molecular Dynamics Simulations. Phys. Chem. Chem. Phys. 2016, 18, 2164–2174. 10.1039/C5CP05920H. [DOI] [PubMed] [Google Scholar]
- Cuendet M. A.; van Gunsteren W. F. On the Calculation of Velocity-Dependent Properties in Molecular Dynamics Simulations Using the Leapfrog Integration Algorithm. J. Chem. Phys. 2007, 127, 184102 10.1063/1.2779878. [DOI] [PubMed] [Google Scholar]
- Lien E. J.; Guo Z.-R.; Li R.-L.; Su C.-T. Use of Dipole Moment as a Parameter in Drug–Receptor Interaction and Quantitative Structure–Activity Relationship Studies. J. Pharm. Sci. 1982, 71, 641–655. 10.1002/jps.2600710611. [DOI] [PubMed] [Google Scholar]
- Cohen N.; Benson S. W. Estimation of Heats of Formation of Organic Compounds by Additivity Methods. Chem. Rev. 1993, 93, 2419–2438. 10.1021/cr00023a005. [DOI] [Google Scholar]
- Filimonov D.; Poroikov V.; Borodina Y.; Gloriozova T. Chemical Similarity Assessment through Multilevel Neighborhoods of Atoms: Definition and Comparison with the Other Descriptors. J. Chem. Inf. Comput. Sci. 1999, 39, 666–670. 10.1021/ci980335o. [DOI] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Delivery Rev. 1997, 23, 3–25. 10.1016/s0169-409x(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Veber D. F.; Johnson S. R.; Cheng H.-Y.; Smith B. R.; Ward K. W.; Kopple K. D. Molecular Properties That Influence the Oral Bioavailability of Drug Candidates. J. Med. Chem. 2002, 45, 2615–2623. 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Delaney J. S. ESOL: Estimating Aqueous Solubility Directly from Molecular Structure. J. Chem. Inf. Comput. Sci. 2004, 44, 1000–1005. 10.1021/ci034243x. [DOI] [PubMed] [Google Scholar]
- Cheng T.; Zhao Y.; Li X.; Lin F.; Xu Y.; Zhang X.; Li Y.; Wang R.; Lai L. Computation of Octanol–Water Partition Coefficients by Guiding an Additive Model with Knowledge. J. Chem. Inf. Model. 2007, 47, 2140–2148. 10.1021/ci700257y. [DOI] [PubMed] [Google Scholar]
- Fukunishi Y.; Nakamura H. Definition of Drug-Likeness for Compound Affinity. J. Chem. Inf. Model. 2011, 51, 1012–1016. 10.1021/ci200035q. [DOI] [PubMed] [Google Scholar]
- Ertl P.; Schuffenhauer A. Estimation of Synthetic Accessibility Score of Drug-like Molecules Based on Molecular Complexity and Fragment Contributions. J. Cheminf. 2009, 1, 8 10.1186/1758-2946-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramashivam S. K.; Elayaperumal K.; Natarajan B.; Ramamoorthy M.; Balasubramanian S.; Dhiraviam K. In Silico Pharmacokinetic and Molecular Docking Studies of Small Molecules Derived from Indigofera Aspalathoides Vahl Targeting Receptor Tyrosine Kinases. Bioinformation 2015, 11, 73–84. 10.6026/97320630011073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung A. B.; Bhattacharjee A.; Ali M. A. Exploring the Physicochemical Profile and the Binding Patterns of Selected Novel Anticancer Himalayan Plant Derived Active Compounds with Macromolecular Targets. Inf. Med. Unlocked 2016, 5, 1–14. 10.1016/j.imu.2016.09.004. [DOI] [Google Scholar]
- Shadrack D. M.; Ndesendo V. M. K.. Molecular Docking and ADMET Study of Emodin Derivatives as Anticancer Inhibitors of NAT 2, COX 2 and TOP 1 Enzymes Comput. Mol. Biosci., 71–18. 10.4236/cmb.2017.71001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.