Abstract
Background
Intestinal flora has been proposed to mediate the occurrence of postmenopausal osteoporosis (PMO). However, the mechanism by which microbes and their metabolites interactively promote PMO remains unknown.
Methods
This study aimed to investigate changes in the intestinal flora and associated metabolites, and their role in PMO. 16S rRNA gene sequencing and metabolomics were performed to obtain postmenopausal women with osteopenia (lower bone mass, LBM), postmenopausal women with osteoporosis (OST), and healthy women as the control group.
Results
We identified taxa-specific and metabolite differences in the intestinal flora of the participants of this study. The pathogenic bacteria Klebsiella (0.59% and 0.71%, respectively) and Escherichia-Shigella (2.72% and 4.30%, respectively) were enriched in the LBM and OST groups (p < 0.05). Some short-chain fatty acid (SCFAs) producing bacteria, Lactobacillus, Akkermansia, Prevotella, Alistipes, and Butyricicoccus, were reduced in patients with LBM and OST compared to the control. Moreover, fecal metabolomic analyses suggested that the metabolites of indole-3-acetic acid and 7-ketodeoxycholic acid were altered in the LBM and OST groups compared to the control (p < 0.05). Enrichment analysis suggested that valine, leucine, and isoleucine biosynthesis; aromatic amino acid biosynthesis; and phenylalanine metabolism were significantly associated with the identified microbiota biomarkers and OST. Moreover, metabolite marker signatures distinguished patients in the OST from those in the control group with an area under the curve (AUC) of 0.978 and 1.00 in the negative and positive ion modes, respectively. Finally, we also found that the fecal level of interleukin-10 (IL-10) in the OST group was significantly lower than that in the control group and LBM group (p < 0.05), while tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6) were significantly higher in the OST group than that in the control group (p < 0.05).
Conclusions
This study provides robust evidence connecting the intestinal flora and fecal metabolomics with PMO. Integrated metabolite and microbiota analyses demonstrated that in addition to dysregulated bacteria, indole-3-acetic acid, 7-ketodeoxycholic acid, and other metabolites can be used for the distinguish of LBM and PMO.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-023-02927-5.
Keywords: Postmenopausal osteoporosis, Bone mineral density, Gut microbiota, Fecal metabolites, Biomarker
Introduction
Postmenopausal osteoporosis (PMO) was reported that increases the risk of fractures in postmenopausal women [1]. The most common complication associated with PMO is fragility fracture, which often occurs in non-traumatic or mildly traumatic conditions of the hip, femur, or spine, leading to pain, deformity, dysfunction [2]. Approximately 10% of the global human and more than 30% of PMO over the age of 50 suffer from osteoporosis [3]. Moreover, the first-year and second-year mortality rates for hip fractures are 17% and 12–20%, respectively [4]. Therefore, osteoporosis is a significant global public health, medical, and economic burden; however, the awareness of osteoporosis is low. Meanwhile, more studies are needed to allow the development of preventive strategies for osteoporosis in China.
Osteoporosis is diagnosed by bone imaging examination, ultrasound, biopsy, and metabolism biochemical index measurements [5]. However, it is difficult to perform large-scale screening and monitoring of osteoporosis. Early diagnosis and interventions to prevent PMO progression can also greatly reduce future healthcare costs, as most economic costs associated with PMO are incurred in advanced stages [6]. To date, several factors, including environmental factors, diet, lifestyle, hygiene, antibiotics, and probiotics, have been reported to contribute to the improvement of PMO [7]. However, at present, the available methods for the early prediction of PMO are limited and use only a few clinical parameters that may not reflect the heterogeneity and complexity of the disease. Thus, more convenient and non-invasive alternatives are required.
Alterations in the gut microbiota can drive the development of osteoporosis by regulating the immune system [8]. Various types of gut microbiota-targeted treatments can prevent the development of osteopenia and improve osteoporosis outcomes in humans [9]. For example, Li et al. reported that Lactobacillus rhamnosus GG can attenuate bone inflammation, inhibit bone loss, and reduce gut epithelial permeability in mice [10]. Bifidobacterium longum, Lactobacillus paracasei, and a mixture of Lactobacillus paracasei and Lactobacillus plantarum can decrease femoral bone loss and increase bone mineral density in rats [11]. A few studies have examined the role of intestinal flora in the occurrence of PMO; however, existing datas are inconsistent. He et al. reported that Klebsiella, Morganella, Escherichia/Shigella, Enterobacter, Citrobacter, Pseudomonas, Succinivibrio, and Desulfovibrio were enriched in women with PMO in Xiamen, China [12]. Ling et al. have observed that Actinobacillus, Blautia, Oscillospira, Bacteroides, and Phascolarctobacterium were positively associated with PMO in Guangzhou, China [13]. Mrinmoy et al. have suggested that Actinomycetes, Eggerthella, Clostridium X1Va, and Lactobacilli were more abundant in patients with PMO in Ireland [14]. To sum up, because of the differences in eating habits and climate conditions in each region, there was significant differences in the composition and structure of gut microbiota from different regions. Therefore, more studies should be conducted on the gut microbiota of patients with PMO from different regions to explain regional differences.
In addition, microbiota-associated metabolic pathways realted to the pathogenesis of osteoporosis. These metabolic pathways include enrichment pathways of lipopolysaccharide biosynthesis [15]; membrane transport, metabolism of tyrosine and tryptophan, valine, leucine, and isoleucine [13]; and metabolism of N-acetylmannosamine, deoxyadenosine, and adenosine [16]. However, the specific microbes and metabolites, as well as the mechanisms by which they interactively promote PMO, are still unclear. Thus, it is necessary to evaluate the mechanistic implications of the intestinal flora and their metabolites in PMO.
In the present study, we integrated the gut metabolomic and intestinal flora profiles of patients with postmenopausal osteopenia and osteoporosis in Xianyang, China, and compared them with those of healthy women. This study provides a valuable resource for understanding postmenopausal osteopenia and osteoporosis-specific microbiota/microbiome features and interactions and offers new insights into understanding postmenopausal osteopenia and osteoporosis.
Results
Analysis of clinical characteristics
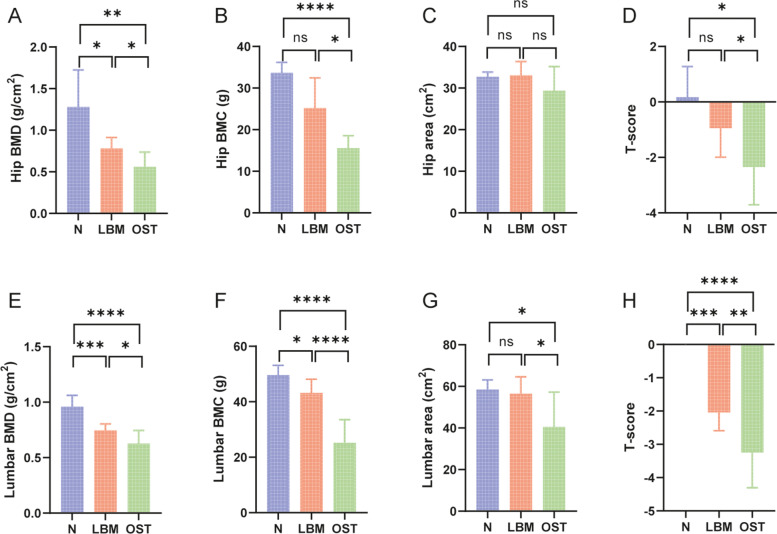
Included 26 individuals were analyzed, the patients were evenly divided into healthy (control, n = 6), osteopenia (lower bone mass, LBM, n = 10), and osteoporosis (OST, n = 10) groups according to the bone mineral density (BMD) index. The hip BMD, hip bone marrow concentrate (BMC), and T-score were significantly lower in the OST group than in the control group (p < 0.05). Hip BMD in the LBM group was lower than that in the control group (p < 0.05). Hip BMD, BMC, and T-score were higher in the LBM group than in the OST group (p < 0.05); however, there was no significant difference in the hip area among these three groups (p > 0.05). Moreover, the lumbar BMD, BMC, area, and T-score were significantly lower in the OST and LBM groups than in the control group (p < 0.05). However, lumbar BMD, BMC, area, and T-score were higher in the LBM group than in the OST group (p < 0.05), and there was no significant difference in the lumbar area between the LBM and control groups (p > 0.05, Fig. 1). Furthermore, no significant differences were observed in age, weight, BMI, or menopausal period among the three groups (Table 1). In addition, the results in this study suggested that the level of the anti-inflammatory factor interleukin-10 (IL-10) was significantly lower in the OST and LBM group than in the control groups (Fig. 2C) (p < 0.05). Moreover, the level of the inflammatory factor tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6) were significantly higher in the OST group than in the LBM and control group (Fig. 2A, B) (p < 0.05).
Fig. 1.
Clinical information of the participants. A Hip BMD. B Hip BMC. C Hip area. D Hip T-score. E Lumbar BMD. F Lumbar BMC. G Lumbar area. H Lumbar T-score. *p < 0.05, **p < 0.01, *** p < 0.001, **** p < 0.0001, ns represent no significant difference between each two groups
Table 1.
Clinical information of the participants
| Participants, n = 26 | Control (n = 6) | LBM (n = 10) | OST (n = 10) | p-value |
|---|---|---|---|---|
| Basic characteristics | ||||
| Age (years) | 65 ± 2.9 | 66.9 ± 5.88 | 68.2 ± 5.9 | 0.5246 |
| Weight (kg) | 57.33 ± 5.75 | 62.4 ± 7.49 | 52.8 ± 13.93 | 0.1343 |
| BMI (kg/m2) | 21.37 ± 3.61 | 24.29 ± 2.54 | 22.09 ± 4.72 | 0.2660 |
| Menopausal period (years) | 50.33 ± 1.63 | 49.1 ± 4.65 | 49.4 ± 4.7 | 0.8481 |
Fig. 2.
Changes in systemic inflammation status. Fecal levels of (A) TNF-α, (B) IL-6 and (C) IL-10. *p < 0.05, **p < 0.01, *** p < 0.001, **** p < 0.0001
Characteristics of sample sequence and alpha and beta diversities of the gut microbiota
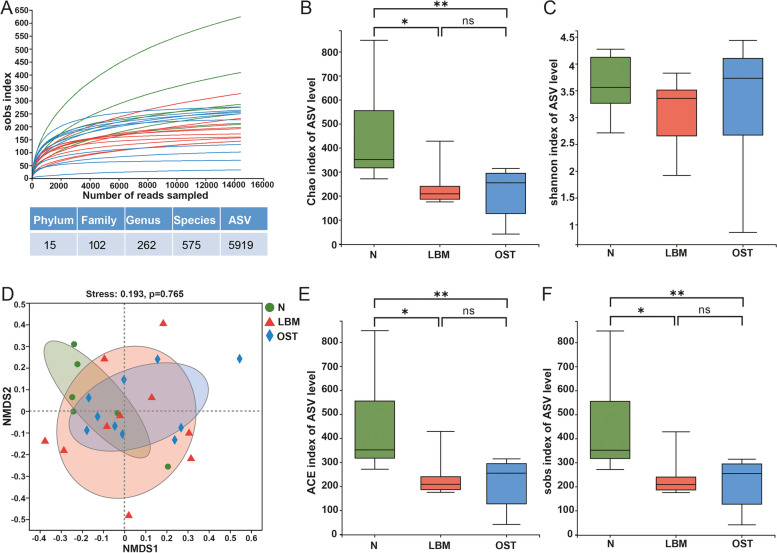
A total of 5,236,251 high-quality reads were obtained from 26 samples, with a mean of 201,394.27 ± 27,004.13 sequences per specimen. The sobs index of curves for each sample were near saturation ((Fig. 3A), indicating that the sequencing data were sufficiently robust, with considerably few new undetected species. Moreover, the bacterial community richness indicated by the Chao1, ACE, and Sobs indexes were significantly lower in the OST group than in the control and LBM groups, and was significantly higher in the control group than in the LBM group, whereas the indexes were not significantly different between the LBM and OST groups (Fig. 3B, E, F). Similarly, the community diversity estimated using the Shannon index was not significantly different among the three groups (Fig. 3C). Additionally, nonmetric multidimensional scaling (NMDS) analysis for beta diversity had a differed of the bacterial, nevertheless, after anosim analysis, there were no significant difference among the control, LBM, and OST groups (p > 0.05)( Fig. 3D).
Fig. 3.
Alpha-diversity and similarity of bacterial community among the group of LBM, OST and N. A Sobs curve for each sample, and number of species in phylum, family, genus, species and ASV. B Chao index of ASV level. C Shannon index of ASV level. D Nonmetric multidimensional scaling analysis (NMDS) at the ASV level. E ACE index of ASV level. F Sobs index of ASV level. LBM: Lower bone mass, OST: osteoporosis, N: Normal group
Taxonomic composition of bacterial community
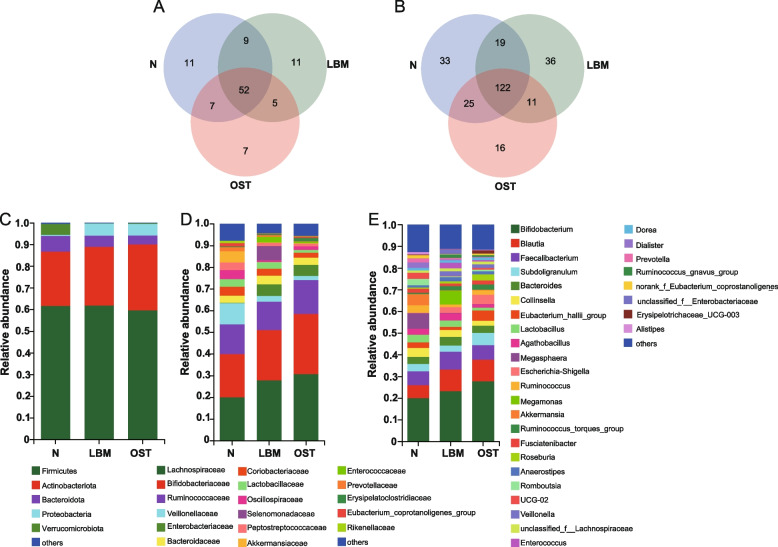
We assessed the gut microbiome structure of the control, LBM, and OST groups using 16S rRNA gene sequencing, which generated a total of 15 phyla, 102 families, 262 genera, and 575 species across all samples. As shown in Fig. 4A, 79, 77, and 71 families were obtained from the control, LBM, and OST groups, respectively, among which 52 families were common in all samples. The OST samples had the lowest number of unique families (seven families). In addition, as shown in Fig. 4B, 199, 188, and 174 genera were detected in samples from the control, LBM, and OST groups, respectively, among which only 122 were common to all 26 samples. The control, LBM, and OST samples had 33, 36, and 16 unique genera, respectively.
Fig. 4.
Composition of gut microbiota among the group of LBM, OST and N. A Venn diagrams of different groups in the bacterial biodiversity at the family level. B Venn diagrams of different groups in the bacterial biodiversity at the genus level. C Relative abundance of the bacteria community at the phylum level. D Relative abundance of the bacteria community at the family level. E Relative abundance of the bacteria community at the genus level. LBM: Lower bone mass, OST: osteoporosis, N: Normal group
At the phylum level (Fig. 4C), five major phyla, namely, Firmicutes, Actinobacteria, Bacteroidetes, Proteobacteria, and Verrucomicrobiota in all three sample groups. In most samples, Firmicutes and Actinobacteria were the two dominant phyla, with a total relative abundance accounting for 86.62%, 88.85%, and 90.00%, respectively. Moreover, Actinobacteria and Proteobacteria were enriched in the LBM (26.98% and 5.61%, respectively) and OST (30.39% and 5.40%, respectively) groups compared to the control group (25.01% and 0.004%, respectively), whereas Bacteroidetes and Verrucomicrobiota were depleted in the LBM (5.17% and 0.16%, respectively) and OST (4.13% and 0.30%, respectively) groups compared to the control group (7.36% and 5.15%, respectively).
At the family level (Fig. 4D), three major families. including Lachnospiraceae, Bifidobacteriaceae, and Ruminococcaceae, were identified in all samples. The relative abundances of Lachnospiraceae, Bifidobacteriaceae, and Ruminococcaceae in OST samples (30.75%, 27.70%, and 15.61%, respectively) were higher than those in LBM (27.80%, 23.18%, and 13.10%, respectively) and control samples (20.00%, 20.00%, and 13.72%, respectively). Moreover, the relative abundance of Enterobacteriaceae in the OST (5.02%) and LBM (5.39%) samples was higher than that in the control (0.21%) samples. However, control samples harbored other dominant families, including Veillonellaceae, Coriobacteriaceae, Lactobacillaceae, Oscillospiraceae, Peptostreptococcaceae, Eubacteriumcoprotanoligenesgroup, and Akkermansiaceae. Notably, other bacterial families, including Selenomonadaceae, Enterococcaceae, and Erysipelatoclostridiaceae, were also observed in LBM and OST samples but were not observed or were scarce in the control samples.
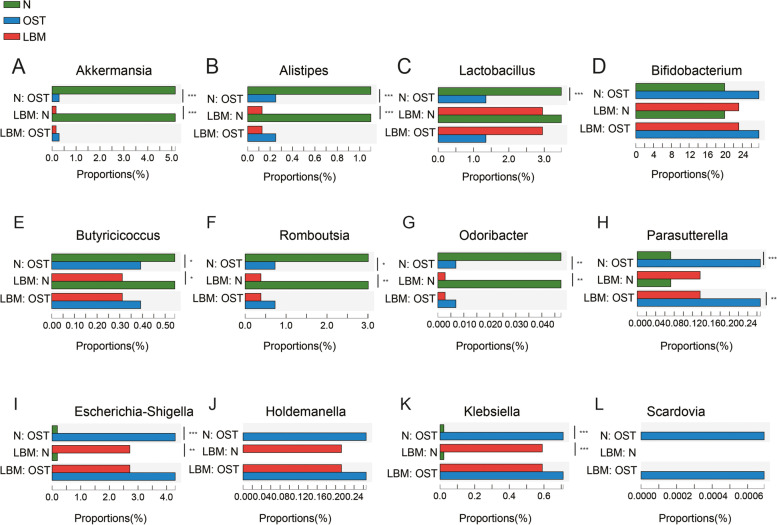
At the genus level (Figs. 4E and 5), Bifidobacterium, Blautia, Subdoligranulum, Ruminococcus_torques_group, Roseburia, Eubacterium hallii group, and Erysipelotrichaceae_UCG-003 were the most abundant in the OST and LBM groups. The relative abundance of Escherichia-Shigella was higher in the LBM and OST groups (2.72% and 4.30%, respectively) but was not observed or was scarce in the control (p < 0.05). Several bacterial species, including Lactobacillus, Megasphaera, Ruminococcus, Romboutsia, Akkermansia, Ruminococcaceae_UCG-02, Dialister, Prevotella, and Alistipes, were enriched in the control group compared to the LBM and OST groups (p < 0.05). Moreover, Megamonas, Anaerostipes, and Enterococcus were abundant in the LBM group whereas Erysipelotrichaceae_UCG-003 and Scardovia were enriched in the OST group. Furthermore, the relative abundances of Butyricicoccus and Odoribacter were higher in the control group than in the LBM and OST groups whereas those of Parasutterella, Holdemanella, and Klebsiella were higher in the LBM and OST groups (p < 0.05).
Fig. 5.
Difference analysis of microbial composition among the group of LBM, OST and N. AAkkermansia. BAlistipes. CLactobacillus. DBifidobacterium. EButyricicoccus. FRomboutsia. GOdoribacter. HParasutterella. IEscherichia-Shigella. JHoldemanella. KKlebsiella. LScardovia. LBM: Lower bone mass, OST: osteoporosis, N: Normal group
Fecal metabolite profiles
To assess whether the fecal metabolite profiles were associated with osteoporosis, we performed metabolic profiling of all stool samples. A total of 97 metabolites were quantified from stool samples using liquid chromatography-mass spectrometry (LC–MS). Partial least squares discriminant analysis (PLS-DA) (Fig. 6A-D) showed that there were differences in the gut metabolite profiles between the LBM and control group, OST and control group, after the fivefold cross validation, results shown that Q2 is all greater than 0.4, indicating the model is effective and a gut metabolite shift in the patients with osteoporosis.
Fig. 6.
Metabolomic data profiles and pathway enrichment analysis. A Partial least squares-discriminant analysis (PLS-DA) for LBM and N groups in negetive model. B Partial least squares-discriminant analysis (PLS-DA) for LBM and N groups in positive model. C Partial least squares-discriminant analysis (PLS-DA) for OST and N groups in negetive model. D Partial least squares-discriminant analysis (PLS-DA) for OST and N groups in positive model. E Enrichment analysis altered metabolites between LBM and N. F Enrichment analysis altered metabolites between OST and N. LBM: Lower bone mass, OST: osteoporosis, N: Normal group
The results revealed 15 significantly altered metabolites between the LBM and control groups; metformin, ornithine, stearic acid, amide C18, ursolic acid, 7-ketodeoxycholic acid, deanol, calcitriol, levalbuterol, hesperidin, and hypoxanthine were enriched in the LBM group, whereas dihydropteroate acid, diethyl phosphate, ofloxacin, and stearamide were depleted in the LBM group compared to the control group (Table 2). Moreover, 63 significantly altered metabolites were identified between the OST and control groups; metabolism of amino acids (L-proline, L-threonine, L-serine, L-phenylalanine, proline, and isoleucine) and organic acids (crotonic acid and stearic acid) was enriched in the OST group whereas that of (E)-ferulic acid, DL-tryptophan, L-( +)-leucine, L-tyrosine, L-theanine, creatine, and L-isoleucine was downregulated in the OST group compared to the control group (Table 3). Interestingly, the levels of 7-ketodeoxycholic acid (7-KDCA) and indole-3-acetic acid (IAA) were altered in the OST group compared to the control group, suggesting they have a potential impact on the development of osteoporosis.
Table 2.
Important features identified by volcano plot in LBM & N
| Compounds | FC | log2(FC) | pval | trends | |
|---|---|---|---|---|---|
| 1 | Dihydropteroic acid | 0.41853 | -1.2566 | 0.037848 | down |
| 2 | Metformin | 17.485 | 4.1281 | 0.0057087 | up |
| 3 | Diethyl phosphate | 0.41796 | -1.2586 | 0.014181 | down |
| 4 | Ornithine | 3.2711 | 1.7098 | 0.017052 | up |
| 5 | Stearic acid | 8.4884 | 3.0855 | 0.018507 | up |
| 6 | Ofloxacin | 0.4266 | -1.229 | 0.024241 | down |
| 7 | Stearamide | 0.48632 | -1.04 | 0.02806 | down |
| 8 | Amide C18 | 5.5551 | 2.4738 | 0.035269 | up |
| 9 | Ursolic acid | 5.1981 | 2.378 | 0.037459 | up |
| 10 | 7-ketodeoxycholic acid | 5.668 | 2.5028 | 0.038545 | up |
| 11 | Deanol | 5.801 | 2.5363 | 0.041853 | up |
| 12 | Calcitriol | 5.051 | 2.3366 | 0.043178 | up |
| 13 | Levalbuterol | 4.6834 | 2.2276 | 0.046517 | up |
| 14 | Hesperidin | 4.7183 | 2.2383 | 0.048639 | up |
| 15 | Hypoxanthine | 5.2503 | 2.3924 | 0.048701 | up |
Table 3.
Important features identified by volcano plot in N & OST
| Compounds | FC | log2(FC) | p value | trends | |
|---|---|---|---|---|---|
| 1 | Phosphoarginine | 12.551 | 3.6498 | 8.7424e-06 | up |
| 2 | Xanthine | 8.5894 | 3.1026 | 0.00010962 | up |
| 3 | Aminolevulinic acid | 12.474 | 3.6409 | 0.001194 | up |
| 4 | o-Succinylbenzoate | 0.30746 | -1.7015 | 0.0019324 | down |
| 5 | 4-Nitrophenol | 7.6822 | 2.9415 | 0.0025816 | up |
| 6 | Dihydropteroic acid | 4.4263 | 2.1461 | 0.0051274 | up |
| 7 | 2-Methoxyestrone 3-sulfate | 3.2382 | 1.6952 | 0.0093508 | up |
| 8 | 2-(6’-methylthio)hexylmalic acid | 0.46843 | -1.0941 | 0.011958 | down |
| 9 | Zalcitabine | 0.41295 | -1.276 | 0.013035 | down |
| 10 | 3,4-Dihydroxyphenylglycol O-sulfate | 9.3213 | 3.2205 | 0.013586 | up |
| 11 | Norepinephrine sulfate | 0.22231 | -2.1693 | 0.029922 | down |
| 12 | fructosylglycine | 2.7231 | 1.4453 | 0.031641 | up |
| 13 | tetrathionic acid | 2.1023 | 1.0719 | 0.033801 | up |
| 14 | (E)-Ferulic acid | 2.0952 | 1.0671 | 0.048625 | up |
| 15 | L-Proline | 0.001889 | -9.0482 | 7.2329e-12 | down |
| 16 | Irbesartan | 0.00075929 | -10.363 | 3.7902e-11 | down |
| 17 | Phloionolic acid | 0.0028703 | -8.4446 | 1.3835e-10 | down |
| 18 | Enalapril | 0.0081559 | -6.9379 | 8.6221e-10 | down |
| 19 | L-Threonine | 0.0060897 | -7.3594 | 2.771e-09 | down |
| 20 | DL-Serine | 0.0042373 | -7.8826 | 5.8641e-09 | down |
| 21 | Amoxicillin | 0.011054 | -6.4993 | 4.104e-08 | down |
| 22 | urobilinogen | 19.09 | 4.2548 | 9.771e-08 | up |
| 23 | Pipecolic acid | 0.012253 | -6.3508 | 2.6359e-07 | down |
| 24 | N-Acetylhistamine | 0.020567 | -5.6035 | 8.5136e-07 | down |
| 25 | Crotonic acid | 0.030598 | -5.0304 | 5.1701e-06 | down |
| 26 | Aminolevulinic acid | 7.5296 | 2.9126 | 8.6311e-06 | up |
| 27 | Isoliquiritigenin | 0.072636 | -3.7832 | 9.9385e-06 | down |
| 28 | Cinnamic acid | 14.089 | 3.8165 | 1.3218e-05 | up |
| 29 | DL-Tryptophan | 16.092 | 4.0082 | 1.734e-05 | up |
| 30 | (-)-codeine | 7.5433 | 2.9152 | 1.7647e-05 | up |
| 31 | Ofoxacin | 17.909 | 4.1626 | 3.4116e-05 | up |
| 32 | 2,3,4,5-tetrahydrodipicolinic acid | 39.965 | 5.3207 | 4.278e-05 | up |
| 33 | D-Alanyl-D-alanine | 4.6462 | 2.2161 | 6.0808e-05 | up |
| 34 | Thymine | 14.686 | 3.8764 | 6.0812e-05 | up |
| 35 | g-Butyrobetaine | 5.2098 | 2.3812 | 6.7153e-05 | up |
| 36 | Daidzein | 12.957 | 3.6957 | 6.8347e-05 | up |
| 37 | Stearic acid | 0.1786 | -2.4852 | 7.4136e-05 | down |
| 38 | L-Phenylalanine | 0.1954 | -2.3555 | 9.463e-05 | down |
| 39 | Cholest-4-en-3-one | 15.548 | 3.9586 | 0.00023261 | up |
| 40 | L-( +)-Leucine | 38.539 | 5.2683 | 0.00023661 | up |
| 41 | L-Tyrosine | 2.9931 | 1.5817 | 0.00023666 | up |
| 42 | Styrene | 0.047016 | -4.4107 | 0.00030534 | down |
| 43 | L-Theanine | 56.025 | 5.808 | 0.00053619 | up |
| 44 | Acetylcholine | 2.6748 | 1.4194 | 0.00054292 | up |
| 45 | cholic acid | 5.7303 | 2.5186 | 0.00061697 | up |
| 46 | glutaral | 0.11453 | -3.1262 | 0.0007078 | down |
| 47 | Indole-3-acetic acid | 2.6714 | 1.4176 | 0.00097212 | up |
| 48 | 3-(3,4-dihydroxypheny]) propanoic acid | 2.6031 | 1.3803 | 0.0013811 | up |
| 49 | Chrysin | 161.83 | 7.3383 | 0.001399 | up |
| 50 | Tiglic acid | 2.6267 | 1.3932 | 0.0014194 | up |
| 51 | Creatine | 144.07 | 7.1706 | 0.0015611 | up |
| 52 | L-Isoleucine | 2.9603 | 1.5657 | 0.0016086 | up |
| 53 | Deanol | 0.30279 | -1.7236 | 0.0016419 | down |
| 54 | 7-ketodeoxycholic acid | 0.30624 | -1.7073 | 0.0017631 | down |
| 55 | Proline | 0.028507 | -5.1325 | 0.0018182 | down |
| 56 | Amide C18 | 0.32731 | -1.6113 | 0.0026659 | down |
| 57 | Hypoxanthine | 0.34182 | -1.5487 | 0.0028131 | down |
| 58 | Isoleucine | 4.2212 | 2.0777 | 0.0028639 | down |
| 59 | Ursolic acid | 0.35954 | -1.4758 | 0.0031063 | down |
| 60 | Berberine | 101.27 | 6.662 | 0.0034375 | up |
| 61 | Diethyl phosphate | 0.22342 | -2.1622 | 0.0036987 | down |
| 62 | Glycochenodeoxycholic acid | 2.5592 | 1.3557 | 0.0039483 | up |
| 63 | N(1)-acetylspermidine | 0.087937 | -3.5074 | 0.0041961 | down |
To further understanding the functions of these significantly changed metabolites, we conducted enrichment and pathway analyses (Fig. 6E–F). Several metabolite sets and pathways were enriched in the LBM group, including sulfur metabolism, purine metabolism, folate biosynthesis, porphyrin and chlorophyll metabolism, glycine, serine and threonine metabolism, tryptophan metabolism, and tyrosine metabolism, compared to the control group. The top four enriched sets and pathways in the control group compared with the OST group were aminoacyl-tRNA biosynthesis, valine, leucine, and isoleucine biosynthesis, arginine and proline metabolism, arginine biosynthesis, histidine metabolism, and tryptophan biosynthesis, indicating that they were significantly downregulated in the OST group. These results suggest that metabolic pathways but not individual metabolites are altered in osteoporosis.
Difference of metabolites between the group of osteopenia and osteoporosis
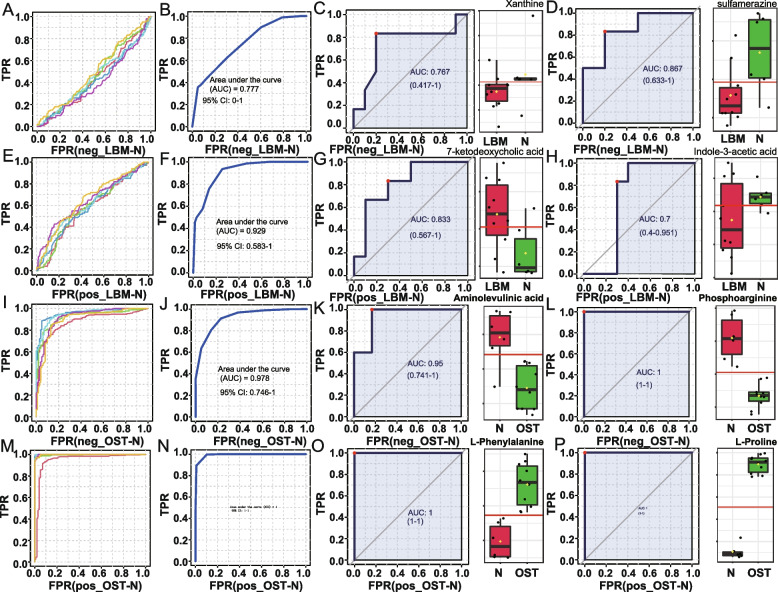
We further investigated potential difference metabolites between osteopenia and osteoporosis groups. We built a model for classifying the two groups based on the identified significantly altered metabolites. Our model selected six metabolites to classify and distinguish patients in the LBM from those in the control group in negative ion mode, with an area under the curve (AUC) of 0.777 (Fig. 7A, B) and found that the metabolites xanthine and sulfamerazine were significantly lower in the LBM group than in the control group (p < 0.05, Fig. 7C, D). The same six metabolites were identified in the LBM in positive ion mode with an AUC of 0.929 (Fig. 7E, F); however, the metabolites of 7-ketodeoxycholic acid and indole-3-acetic acid in the LBM group were significantly higher than those in the control group (p < 0.05) (Fig. 7G, H). To discriminate the OST from the control group, six metabolite markers were identified in the negative ion mode with an AUC of 0.978 (Fig. 7I, J), and aminolevulinic acid and phosphoarginine in were significantly lower in the OST group than in the control group (p < 0.05; Fig. 7K, L). Moreover, the distinction of the OST group from the control group in positive ion mode had an AUC of 1.0 (Fig. 7M, N), and the levels of L-phenylalanine and L-proline were higher in the OST group than in the control group (p < 0.05; Fig. 7O, P).
Fig. 7.
Metabolite markers for pairwise discriminations of OST, LBM and N groups. A Receiver operating characteristic (ROC) analysis for the 6 metabolite markers discriminating LBM from N in negetive model. B Area under the curve (AUC) applying the 6 LBM vs N metabolite markers to discriminate LBM from N in negetive model. C AUC value of metabolite of Xanthine discriminating LBM from N in negetive model. D AUC value of metabolite of Sulfamerazine discriminating LBM from N in negetive model. E ROC analysis for the 6 metabolite markers discriminating LBM from N in positive model. F AUC applying the 6 LBM vs N metabolite markers to discriminate LBM from N in positive model. G AUC value of metabolite of 7-ketodeoxycholic acid discriminating LBM from N in positive model. H AUC value of metabolite of indole-3-acetic acid discriminating LBM from N in positive model. I ROC analysis for the 6 metabolite markers discriminating OST from N in negetive model. J AUC applying the 6 OST vs N metabolite markers to discriminate OST from N in negetive model. K AUC value of metabolite of Aminolevulinic discriminating OST from N in negetive model. L AUC value of metabolite of Phosphoarginine discriminating OST from N in negetive model. M ROC analysis for the 6 metabolite markers discriminating OST from N in positive model. N AUC applying the 6 OST vs N metabolite markers to discriminate OST from N in positive model. O)AUC value of metabolite of L-Phenylalanine discriminating OST from N in positive model. P AUC value of metabolite of L-proline discriminating OST from N in positive model. LBM: Lower bone mass, OST: osteoporosis, N: Normal group
Relationship among the different bacteria, different metabolites, and clinical profiles
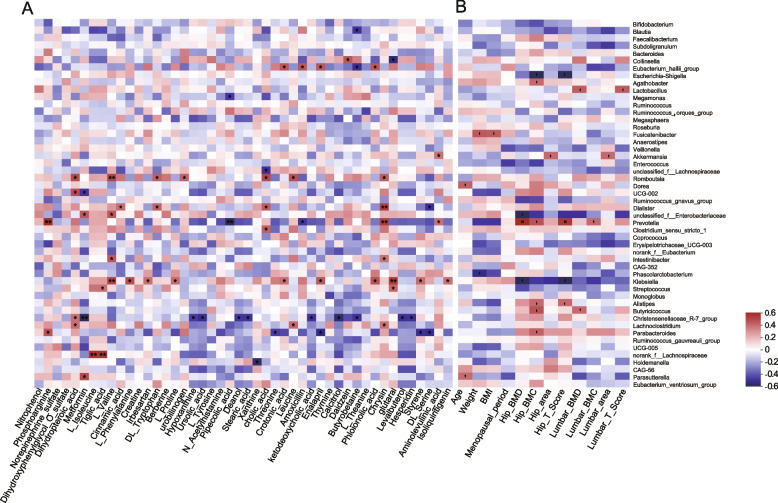
We investigated the significant associations among different bacteria, metabolites, and clinical profiles and found that LBM and OST groups enriched with Klebsiella were positively correlated with L_Valine, L_Proline, Crotonicacid, Phloionolicacid, Glutara, styrene, isoliquiritigenin, and L_ phenylalanine. Eubacteriumhalliigroup was positively associated with phloionolic and crotonic acids. Romboutsia was positively associated with chrysin, leucine, cholic acid, tryptophan, valine, and dihydropteroic acid levels. Prevotella was positively correlated with aminolevulinic acid, chrysin, and phosphoarginine levels but negatively associated with amoxicillin and pipecolic acid levels. LBM and OST groups with decreased Christensenellaceae_R-7_group were negatively correlated with the levels of hesperidin, levalbutero, butyrobetaine, calcitrio, 7ɑ-ketodeoxycholicacid, stearic acid, deanol, ursolic acid, and hypoxanthine. In addition, we found that the decrease in Parabacteroides in the LBM and OST groups was negatively correlated with the levels of threonine, enalapril, styrene, and DL_Serine, but was positively correlated with phosphoarginine (Fig. 8A). In addition, the LBM and OST groups enriched with Klebsiella and Escherichia-Shigella were negatively correlated with hip BMD, BMC, and T scores. Conversely, LBM and OST groups with a decreased abundance of Lactobacillus, Akkermansia, Prevotella, Alistipes, and Butyricicoccus, were positively correlated with hip BMD, hip BMC, hip area, hip T-score, lumbar BMD, lumbar BMC, lumbar area, and lumbar T-score (Fig. 8B).
Fig. 8.
Correlational analyses. A Heatmap of the Spearman’s correlation coefficients between gut metabolites and microbiota. B Heatmap of the Spearman’s correlation coefficients between clinical parameters and microbiota. *p < 0.05, **p < 0.01, *** p < 0.001
Discussion
Previous studies have revealed that intestinal flora and their metabolites play an essential in PMO [17]. The intestinal flora affects the development and function of the host immune system by releasing metabolites and immune cells (including T and B cells), stimulating the immune system at the intestinal mucosal barrier, releasing proinflammatory or anti-inflammatory mediators and cytokines, and regulating bone metabolism through blood circulation [18]. In this study, we analyzed and compared the intestinal flora and metabolites of patients with PMO, patients with postmenopausal osteopenia, and healthy individuals. It was obeserved that key intestinal flora and metabolites are disrupted during the development of PMO. Moreover, integrated microbiome and metabolomic analyses showed that interactions between osteoporosis-associated bacteria and metabolites were altered during the progress of PMO. Importantly, we demonstrated the intestinal flora and metabolites for the non-invasive diagnosis of PMO.
The richness and diversity of the gut microbiota are significantly altered in patients with osteoporosis [19]. He et al. have suggested that patients with PMO had lower bacterial richness and diversity than healthy controls [12]. Consistent with this study, our findings suggested decreased bacterial community richness in the PMO group. Moreover, Ling et al. have shown that the gut microbiome of patients with osteoporosis had an increased abundance of Actinobacillus, Blautia, Oscillospira, Bacteroides, and Phascolarctobacterium, and decreased abundance of Veillonellaceae, Collinsella, and Ruminococcaceae [13]. In our study, we identified a few PMO-related bacterial biomarkers. We observed that the abundance of pathogenic bacteria Escherichia-Shigella [20, 21] and Klebsiella [22], which are significantly correlated with systemic inflammatory cytokines, was significantly more increased in the PMO than in the control group. Moreover, Ling et al. have suggested that Blautia is abundant in patients with osteoporosis group [13], which is consistent with our findings. McGinty et al. reported that Bifidobacterium spp. could elevate bone density through increasing the absorption of minerals[23]. However, the abundance of Bifidobacterium was enriched in the osteoporosis group in our study. This conflicting result may be attributed to geographical differences. It has been reported that human living in different altitudes and climates may be responsible for the different microbiota compositions [24]. Xianyang, Shaanxi Province, in northwest China, has a continental monsoon climate, with distinct cold, hot, dry, and wet seasons. Meanwhile, as is well known, the diet of Shaanxi people is mainly based on pasta, which may have caused the unique intestinal microbiota of Shaanxi people, thus, this regional factor may also be the reason for obtaining inconsistent results.
In addition, Lactobacillus [25], Romboutsia [26], Akkermansia [27], Butyricicoccus [28], Ruminococcaceae_UCG-02, and Alistipes [29], which are bacteria involved in the production of short chain fatty acid, were depleted in the LBM and OST groups. Therefore, the enrichment of Escherichia-Shigella and Klebsiella and the decrease in Lactobacillus, Romboutsia, Akkermansia, Butyricicoccus, Ruminococcaceae_UCG-02, and Alistipes may predict the development of PMO. Collectively, these results provide insight into the association of intestinal flora with PMO in humans.
The metabolic profile may help identify biomarkers to predict diseases including osteoporosis [13]. In this study, several amino acids, including L-proline, L-threonine, L-serine, L-phenylalanine, and isoleucine, were abundant in the OST group compared to the control group, whereas DL-tryptophan, L-( +)-leucine, and L-tyrosine were depleted in the OST group. Ling et al. have suggested that amino acid metabolism could be a target for intervention in osteoporosis [13], supporting our findings in this study. Interestingly, IAA (downregulated) and 7-KDCA (upregulated) were simultaneously altered in the LBM and OST groups compared with the control group. IAA, has been reported to be produced by the flora from tryptophan metabolism and can alleviate inflammation related to alcoholic liver disease [30] and obesity [31] by directly or indirectly regulating the balance between proinflammatory and anti-inflammatory cytokines (TGF-β, TNF-α, IL-10, and IL-22). Additionally, 7-KDCA is a bile acid of microbial origin, which is associated with advanced stages of fibrosis and non-alcoholic fatty liver disease [32]. Tom et al. [33] reported that patients with chronic liver disease are more likely to develop osteoporosis owing to abnormal vitamin D metabolism, calcium malabsorption, and other factors. Therefore, our results suggest that the simultaneous reduction in IAA and increase in 7-KDCA can directly or indirectly, promote the development of PMO, suggesting that they are potential biomarkers for PMO.
In this study, the pathway enrichment analysis showed that the biosynthesis of aminoacylt-RNA, valine, leucine, and isoleucine, and aromatic amino acid (phenylalanine, tyrosine, and tryptophan), as well as phenylalanine metabolism, were downregulated in the PMO group compared to the healthy control. However, a previous study has suggested that the upregulation of aminoacyl-tRNA biosynthesis pathway is associated with prostate cancer cell development [34]; therefore, this needs to be further studied. Dietary proteins have also been reported to increase calcium absorption [19]. Branched-chain amino acids, including valine, leucine, and isoleucine, were downregulated in the PMO, which is consistent with the study by Ling et al., reporting that patients with osteoporosis had decreased concentrations of serum valine and leucine [13]. Additionally, Isley et al. reported that aromatic amino acid (AAA) intake induces an increase in serum IGF-1 levels, which promotes bone production [19]. In our study, we observed that AAAs, including tyrosine, phenylalanine, and tryptophan, were downregulated in the PMO group. In summary, our results suggest that patients with osteoporosis should consume more proteins, especially food rich in branched-chain and aromatic amino acids.
We further explored the relationships between clinical factors, metabolites, and gut microbiota. Notably, Klebsiella and Escherichia-Shigella were negatively correlated with hip BMD, hip BMC, and hip T scores. Klebsiella has been reported to cause bone and joint infections that are associated with serious morbidity and mortality [19]. Escherichia-Shigella was reported to produce propionate ester and other metabolites, thereby inducing the expression of chromatin Acid hydroxylase (Tph) 1 in intestinal chromaffin cells, which increases peripheral 5-HT levels in germ-free mice, promotes osteoclast generation, reduces osteoblast proliferation, and inhibits bone growth [19, 35, 36]. Therefore, targeting Klebsiella and Escherichia-Shigella in the intestines might delay the progression of osteoporosis. In addition, we found that short-chain fatty acid (SCFA)-producing bacteria, including Lactobacillus [37], Akkermansia [27], Prevotella [38], Alistipes [39], and Butyricicoccus [28], were positively correlated with hip BMD, hip BMC, hip area, hip T-score, lumbar BMD, lumbar BMC, lumbar area, and lumbar T-score. We also found that Romboutsia [26], an SCFA producer, was positively associated with the levels of chrysin, leucine, cholic acid, DL_ tryptophan, L_ valine, and dihydropteroic acid. All above these results suggest a significant interaction between clinical factors, metabolites, and gut microbiota, which might affect the progess of osteoporosis.
This study had some limitations. In this study, all patients were recruited from Xianyang, Shaanxi Province, a small modern city in the mainland area of northwest China. Because the gut microbiota and its metabolites are significantly influenced by geographical, climatic, and dietary habit factors, our findings need validation in other regions. Other limitations are the limited sample size, the cross-sectional design of the study, and the lack of a comprehensive mechanistic analysis and validation cohorts, it is necessary to expand the sample size in the follow-up studies. In the future, the potential identified fecal metabolites and key strains that were associated with PMP need to be validated in vitro or in vivo studies using metagenomic sequencing.
Conclusion
In summary, this study described the disordered profiles of intestinal bacteria and fecal metabolomes in menopausal patients with osteopenia or osteoporosis. We identified key strains and metabolite differences in the intestinal flora of the participants of this study. We found that the pathogenic bacteria Klebsiella and Escherichia-Shigella were enriched in patients with LBM and PMO. Some SCFAs producers, including Lactobacillus, Akkermansia, Prevotella, Alistipes, and Butyricicoccus, were reduced in patients with LBM and PMO. Moreover, the metabolites of IAA and 7-KDCA were altered in patients with LBM and PMO. The pathways of aminoacyl-tRNA biosynthesis, valine, leucine, and isoleucine biosynthesis, aromatic amino acid (phenylalanine, tyrosine, and tryptophan) biosynthesis, and phenylalanine metabolism were downregulated in patients with PMO. Additionally, the relationship between these parameters and the bone parameters that can effect osteoporosis is discussed. These findings provide deeper understanding the relationship between gut microbiota, metabolites, and PMO.
Materials and methods
Patients and specimen collection
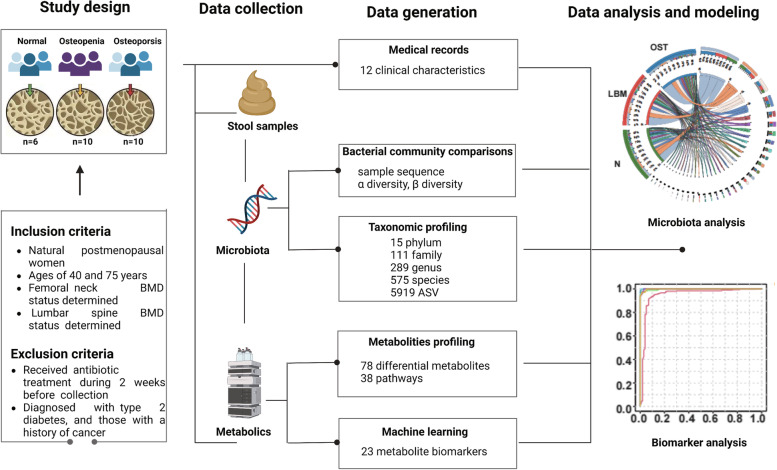
All participants were from Shaanxi Province and were admitted to the Affiliated Hospital of Shaanxi University of Traditional Chinese Medicine (Xianyang, China) from October 2021 to December 2021. This trial was registered in the Chinese Clinical Trial Registry (SZFYIEC-PJ-KY-202130). A total of 26 women were recruited for the study based on the inclusion and exclusion criteria (listed below): 10 postmenopausal patients with lower bone mass (LBM: T-score between -1 and -2.5), 10 postmenopausal patients with OST, with T-score less than -2.5, and 6 healthy patients as the control group (Fig. 9). The inclusion criteria and exclusion criteria for the patients were Table 4.
Fig. 9.
Overview of the prospective study design. A graphical representation summarizing the study design, data collection, and the methodologies of data generation and analysis
Table 4.
The inclusion criteria and exclusion criteria for the patients
| Inclusion criteria |
| (1) had completed bone mineral density examination and clinical data |
| (2) naturally postmenopausal women |
| (3) had no previous systemic diseases |
| Exclusion criteria |
| (1) secondary osteoporosis |
| (2) incomplete case data or patients and their families who were unable to complete the questionnaire |
| (3) under the age of 50 years |
| (4) use of antibiotics, probiotics, probiotics, or co-organisms within two months before collection of stool samples |
| (5) patients suffering from severe malnutrition, infections, drug or alcohol abuse |
| (6) patients with digestive system diseases |
| (7) patients with complicated with serious systemic diseases, tumors, or other serious primary diseases |
| (8) patients with previous lumbar and hip surgery history |
Enzyme-linked immunosorbent assay
Sample pretreatment: 50-100 g fecal was collected, and added with the same volume of PBS. The supernatant was centrifuged at 2000 g and stored at -70. The TNF-α, IL-6 and IL-10 were determined by ELISA kits.
DNA extraction and 16S rRNA gene Sequencing
Total DNA from the fecal samples was extracted using the QIAamp® DNA Stool Mini kit (QIAGEN, Hilden, Germany), according to the manufacturer’s protocol. The DNA quality control and library construction according to the manufacturer’s instructions. Paired-end sequencing was carried out using the Illumina MiSeq (PE300) sequencing platform. The detailed information is described in our previous study [12].
Sequencing data analysis
After de-multiplexing, the obtained sequences were merged in FLASH (v1.2.11) [40] and quality-filtered using fastp (v0.19.6) [41]. The high-quality sequences were then denoised using the DADA2 [42] plugin in the Qiime2 (v2020.2) pipeline [43]. DADA2–denoised sequences are amplicon sequence variants (ASVs). Finally, sequencing data were analyzed using the free online Majorbio Cloud Platform (www.majorbio.com).
Untargeted metabolomics
Fecal metabolites were extracted from fecal samples as described previously, and metabolites were analyzed using UHPLC system (1290; Agilent Technologies, Santa Clara, CA, USA). The detailed information is described in our previous study [12].
Metabolite data analysis
The mass spectrum data were processed using Compounds Discovered 3.1 software (Thermo Fisher Scientific) for noise reduction, peak alignment, and identification. Differential metabolites were identified using the SIMCA-P v14.1 software (Umetrics AB, Umea, Sweden). PLS-DA analysis method was used to distinguished the metabolites composition of patients with LBM and OST from healthy controls.
Statistical analysis
Some data are shown as mean ± standard deviation (SD), and some data are shown as mean with quartile. Statistical significance with one-way analysis of variance (ANOVA) followed by Duncan’s multiple comparison test was set at p < 0.05. Microbiological analysis was performed using the free online Majorbio Cloud Platform (www.majorbio.com). The metabolite analysis was performed using the free online platform MetaboAnalyst5.0 (www.metaboanalyst.ca/).
Supplementary Information
Additional file 1: Supplemental Table 1. Result from pathwayanalysis between LBM & N. SupplementalTable 2. Result frompathway analysis between OST & N. Fig. S1. Changes in the composition ofkey metabolites among OST, LBM and N groups. LBM: Lower bone mass, OST:osteoporosis, N: Normal group.
Acknowledgements
The authors are grateful to the Editor and anonymous reviewers for their suggestions to improve the manuscript.
Abbreviations
- PMO
Postmenopausal osteoporosis
- LBM
Lower bone mass
- OST
Postmenopausal women with osteoporosis
- SCFAs
Short-chain fatty acid
- AUC
Area under the curve
- IL-10
Interleukin-10
- TNF-α
Tumor necrosis factor-α
- IL-6
Interleukin-6
- BMD
Bone mineral density
- BMC
Bone marrow concentrate
- NMDS
Nonmetric multidimensional scaling
- LC–MS
Liquid chromatography-mass spectrometry
- 7-KDCA
7-Ketodeoxycholic acid
- IAA
Indole-3-acetic acid
- AAA
Aromatic amino acid
- SD
Standard deviation
- ANOVA
One-way analysis of variance
- SRA
Sequence Read Archive
Authors’ contributions
S/b ZL, YH and LY designed and prepared the manuscript. ZL, TL, and BD recruited and supervised experimental procedures. PY and WK performed statistical analysis. ZL, TL, and BG performed the amplifiers sequencing and metabolomics analysis. TL, BG, and BD are responsible for the full reviews, verifying that all data, figures, materials. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Key R & D project of Shaanxi Provincial Department of science and Technology (2021sf419), Graduate innovation project of school level scientific research project of Shaanxi University of traditional Chinese medicine (2021cx12), Postdoctoral fund of Guangdong Provincial People's Hospital (BY012022017), Guangdong Province Basic and Applied Basic Research Fund Project (2022A1515110447). The funding bodies played no role in the design of the study and collection, analysis, interpretation of data, and in writing the manuscript.
Availability of data and materials
The detail data and materials available please see https://www.ncbi.nlm.nih.gov/sra/PRJNA916764. The amplicon sequencing data are available in National Center for Biotechnology Information with accession numbers of Sequence Read Archive (SRA) submission: PRJNA916764.
Declarations
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of the Affiliated Hospital of Shannxi University of Traditional Chinese Medicine (reference number: SZFYIEC-PJ-KY-202130, signed on 31/05/2021. All participants provided written informed consent. This work was carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for Experiments in Humans.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhuang Liang, Yuqi Hao and Lei Yang contributed equally to this work.
Contributor Information
Tingting Liang, Email: 1320199507@qq.com.
Bing Gu, Email: gubing@gdph.org.cn.
Bo Dong, Email: dahe_78@163.com.
References
- 1.Watts N, Bilezikian J, Camacho P, Greenspan S, Harris S, Hodgson S, et al. American association of clinical endocrinologists medical guidelines for clinical practice for the diagnosis and treatment of menopause. Endocr Pract. 2011;17:1–25. doi: 10.4158/ep.16.s3.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu X, Jia X, Mo L, Liu C, Zheng L, Yuan Q, et al. Intestinal microbiota: a potential target for the treatment of postmenopausal osteoporosis. Bone Res. 2017;5:177–194. doi: 10.1038/boneres.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cipriani C, Pepe J, Bertoldo F, Bianchi G, Cantatore FP, Corrado A, et al. The epidemiology of osteoporosis in Italian postmenopausal women according to the National Bone Health Alliance (NBHA) diagnostic criteria: a multicenter cohort study. J Endocrinol Invest. 2018;41:431–438. doi: 10.1007/s40618-017-0761-4. [DOI] [PubMed] [Google Scholar]
- 4.Forsén L, Søgaard AJ, Meyer HE, Edna TH, Kopjar B. Survival after hip fracture: short- and long-term excess mortality according to age and gender. Osteoporosis Int. 1999;10:73–78. doi: 10.1007/s001980050197. [DOI] [PubMed] [Google Scholar]
- 5.Morris RM, Lang Y, Martín-Fernández M, Pozo JM, Frangi AF, Mark WJ. High-spatial-resolution bone densitometry with dual-energy X-ray absorptiometric region-free analysis. Radiology. 2015;275(1):310. doi: 10.1148/radiol.15154008. [DOI] [PubMed] [Google Scholar]
- 6.Tucci JR. Importance of early diagnosis and treatment of osteoporosis to prevent fractures. Am J Manag Care. 2006;12(7):3. [PubMed] [Google Scholar]
- 7.Zarei A, Morovat A, Javaid K, Brown CP. Vitamin D receptor expression dose-dependent activation in human bone tissue and resorbing osteoclasts. Bone Res. 2016;4(1):1–10. [DOI] [PMC free article] [PubMed]
- 8.Tyagi AM, Yu M, Darby TM, Vaccaro C, Li JY, Owens JA, et al. The microbial metabolite butyrate stimulates bone formation via t regulatory cell-mediated regulation of WNT10B expression. Immunity. 2018;49:1116–1131. doi: 10.1016/j.immuni.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preidis GA, Saulnier DM, Blutt SE, Mistretta T-A, Riehle KP, Major AM, et al. Probiotics stimulate enterocyte migration and microbial diversity in the neonatal mouse intestine. FASEB J. 2012;26:1960–1969. doi: 10.1096/fj.10-177980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J-Y, Chassaing B, Tyagi AM, Vaccaro C, Luo T, Adams J, et al. Sex steroid deficiency–associated bone loss is microbiota dependent and prevented by probiotics. J Clin Invest. 2016;126:2049–2063. doi: 10.1172/JCI86062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parvaneh K, Ebrahimi M, Sabran M R, Karimi G, Hwei A N M, Abdul-Majeed S, et al. Probiotics increase bone mass density and upregulate genes in rats with bone loss resulting from ovariectomy. Biomed Res Int. 2015;2015:897639. [DOI] [PMC free article] [PubMed]
- 12.He J, Xu S, Zhang B, Xiao C, Chen Z, Si F, et al. Gut microbiota and metabolite alterations associated with reduced bone mineral density or bone metabolic indexes in postmenopausal osteoporosis. Aging. 2020;12(9):8583–8604. doi: 10.18632/aging.103168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling CW, Miao Z, Xiao ML, Zhou H, Jiang Z, Fu Y, et al. The association of gut microbiota with osteoporosis is mediated by amino acid metabolism: multiomics in a large cohort. J Clin Endocr Metab. 2021;106(10):e3852–e3864. doi: 10.1210/clinem/dgab492. [DOI] [PubMed] [Google Scholar]
- 14.Mrinmoy D, Owen C, Keohane DM, Cormac EM, Helena N, Michelle N, et al. Gut microbiota alterations associated with reduced bone mineral density in older adults. Rheumatology. 2019;12:12. doi: 10.1093/rheumatology/kez302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Dai Z, Tao W. Gut microbiota composition and bone mineral loss-epidemiologic evidence from individuals in Wuhan China. Osteoporosis Int. 2019;30(5):1003–1013. doi: 10.1007/s00198-019-04855-5. [DOI] [PubMed] [Google Scholar]
- 16.Xu Z, Xie Z, Sun J, Huang S, Chen Y, Li C, et al. Gut microbiome reveals specific dysbiosis in primary osteoporosis. Front Cell Infect Mi. 2020;10:160. doi: 10.3389/fcimb.2020.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sjogren K, Engdahl C, Henning P, Lerner UH, Tremaroli V, Lagerquist MK, et al. The gut microbiota regulates bone mass in mice. Bone. 2012;50:S91–S92. doi: 10.1016/j.bone.2012.02.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang D Wu, Dong YS, Zhong C. Mutual interaction of microbiota and host immunity during health and diseases. Biophys Rep. 2021;7:326–340. doi: 10.52601/bpr.2021.200045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Wang Y, Gao W, Wang B, Zhao H, Zeng Y, et al. Diversity analysis of gut microbiota in osteoporosis and osteopenia patients. PeerJ. 2017;5:e3450. doi: 10.7717/peerj.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Bai M, Ning X, Qin Y, Wang Y, Yu Z, et al. Expansion of Escherichia-Shigella in gut is associated with the onset and response to immunosuppressive therapy of iga nephropathy. J Am Soc Nephrol. 2022;20:189. doi: 10.1681/ASN.2022020189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radhakrishnan ST, Alexander JL, Mullish BH, Gallagher Kate I, Powell N, Hicks LC, et al. Systematic review: the association between the gut microbiota and medical therapies in inflammatory bowel disease. Aliment Pharm Ther. 2022;55:26–48. doi: 10.1111/apt.16656. [DOI] [PubMed] [Google Scholar]
- 22.Sequeira RP, McDonald JAK, Marchesi JR, Clarke TB. Commensal bacteroidetes protect against klebsiella pneumoniae colonization and transmission through IL-36 signalling. Nat Microbiol. 2020;5:304–313. doi: 10.1038/s41564-019-0640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGinty T, Mallon PWG. Fractures and the gut microbiome. Current Opinion in HIV and AIDS. 2018;13(1):28. doi: 10.1097/COH.0000000000000425. [DOI] [PubMed] [Google Scholar]
- 24.Gaulke CA, Sharpton TJ. The influence of ethnicity and geography on human gut microbiome composition. Nat Med. 2018;24:1495–1496. doi: 10.1038/s41591-018-0210-8. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, D'Aigle J, Atadja L, Quaicoe V, Venna V R. Gut Microbiota-Derived Short-chain fatty acids promote post-stroke recovery in aged mice. Circ Res. 2020;127(4):453–465. [DOI] [PMC free article] [PubMed]
- 26.Yang X, Chang T, Yuan Q, Wei W, Wang P, Song X, et al. Changes in the composition of gut and vaginal microbiota in patients with postmenopausal osteoporosis. Front Immunol. 2022;13:930244. doi: 10.3389/fimmu.2022.930244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Z, Jiang Z, Zhang Z, Liu T, Fan Y, Liu T, Peng N. Bacillus coagulans in combination with chitooligosaccharides regulates gut microbiota and ameliorates the dss-induced colitis in mice. Microbiol Spectr. 2022;10:e00641–e722. doi: 10.1128/spectrum.00641-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sh HI, Chang Y, Gao Y, Wang X, Chen X, Wang Y, et al. Dietary fucoidan of Acaudina molpadioides alters gut microbiota and mitigates intestinal mucosal injury induced by cyclophosphamide. Food Funct. 2017;8:3383–3393. doi: 10.1039/C7FO00932A. [DOI] [PubMed] [Google Scholar]
- 29.Huang Anni, Cai Rujian, Wang Qun, et al. Dynamic change of gut microbiota during porcine epidemic diarrhea virus infection in suckling piglets. Front Microbiol. 2019;10:322. doi: 10.3389/fmicb.2019.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duan Hendrikx, Yanhan Wang, Alexander Jee-Hwan, et al. Bacteria engineered to produce IL-22 in intestine induce expression of REG3G to reduce ethanol-induced liver disease in mice. Gut. 2019;68(8):1504–1515. doi: 10.1136/gutjnl-2018-317232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su X, Zhang M, Qi H, Gao Y, Yang Y, Yun H, et al. Gut microbiota–derived metabolite 3-idoleacetic acid together with LPS induces IL-35+ B cell generation. Microbiome. 2022;10:13. doi: 10.1186/s40168-021-01205-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smirnova E, Muthiah MD, Narayan N, Siddiqui MS, Puri P, Luketic VA, et al. Metabolic reprogramming of the intestinal microbiome with functional bile acid changes underlie the development of NAFLD. Hepatology. 2022;76(6):1811–1824. doi: 10.1002/hep.32568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tom B, Compston JE, Ninkovic M, Love SA, Alexander GJ. High prevalence of osteoporosis in patients with chronic liver disease prior to liver transplantation. Calcified Tissue Int. 2001;69(6):321. doi: 10.1007/s00223-001-2028-4. [DOI] [PubMed] [Google Scholar]
- 34.Adaikkalam V. Proteomic interrogation of androgen action in prostate cancer cells reveals roles of aminoacyl tRNA synthetases. PLoS One. 2009;4:e7075. doi: 10.1371/journal.pone.0007075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roshchina VV. New trends and perspectives in the evolution of neurotransmitters in microbial, plant, and animal cells. Adv Exp Med Biol. 2015;874:25–77. doi: 10.1007/978-3-319-20215-0_2. [DOI] [PubMed] [Google Scholar]
- 36.Spohn SN, Mawe GM. Non-conventional features of peripheral serotonin signalling - the gut and beyond. Nat Rev Gastro Hepat. 2017;14:412–420. [DOI] [PMC free article] [PubMed]
- 37.Chen LH, Chang SS, Chang HY, Wu CH, Pan CH, Chang CC, et al. Probiotic supplementation attenuates age-related sarcopenia via the gut–muscle axis in SAMP8 mice. J Cachexia Sarcopeni. 2022;13:515–531. doi: 10.1002/jcsm.12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu E, Yang H, Liu X, Ren M, Shen L, Wentao L, et al. Study on bacterial community structure and short chain fatty acid content in different parts of intestines of yorkshire pigs. Chin J Anim Nutr. 2019;31(10):4509–4518. [Google Scholar]
- 39.Wang G, Song J, Huang Y, Li X, Wang H, Zhang Y, et al. Lactobacillus plantarum SHY130 isolated from yak yogurt attenuates hyperglycemia in C57BL/6J mice by regulating the enteroinsular axis. Food Funct. 2022;13(2):675–687. doi: 10.1039/D1FO02387J. [DOI] [PubMed] [Google Scholar]
- 40.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen S, Zhou Y, Chen Y, Gu J. fastp : an ultra-fast all-in-one FASTQ preprocessor. Cold Spring Harbor Laboratory. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolyen E, Rideout JR. Reproducible, interactive, scalable and extensible microbiome data science using QIIME. Nat Biotechnol. 2019;37:852. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental Table 1. Result from pathwayanalysis between LBM & N. SupplementalTable 2. Result frompathway analysis between OST & N. Fig. S1. Changes in the composition ofkey metabolites among OST, LBM and N groups. LBM: Lower bone mass, OST:osteoporosis, N: Normal group.
Data Availability Statement
The detail data and materials available please see https://www.ncbi.nlm.nih.gov/sra/PRJNA916764. The amplicon sequencing data are available in National Center for Biotechnology Information with accession numbers of Sequence Read Archive (SRA) submission: PRJNA916764.