Abstract

Nineteen biscoumarins were synthesized, well-characterized, and evaluated against α-glucosidases in vitro. Of these, six compounds (10, 12, 16, and 17–19) were newly synthesized and not previously reported in the chemical literature. The majority of the synthesized derivatives demonstrated significant inhibitory activity. A quantitative structure–activity relationship (QSAR) model was developed, revealing a strong correlation between the anti-α-glucosidase activity and selected molecular descriptors. Based on this model, two new compounds (18 and 19) were designed, which exhibited the strongest inhibition with IC50 values of 0.62 and 1.21 μM, respectively, when compared to the positive control (acarbose) with an IC50 value of 93.63 μM. Enzyme kinetic studies of compounds 18 and 19 revealed their competitive inhibition with Ki values of 3.93 and 1.80 μM, respectively. Computational studies demonstrated that compound 18 could be inserted into the original binding site (OBS) of α-glucosidase MAL12 and form multiple hydrophobic interactions with nearby amino acids, with the bromo group playing an essential role in enhancing the binding strength and stability at the OBS of the enzyme based on the quantum mechanical calculations using the fragment molecular orbital method. These findings provide valuable insights into the design of potent α-glucosidase inhibitors, which may have potential therapeutic applications in the treatment of diabetes and related diseases.
Introduction
Diabetes mellitus is one of the most common metabolic diseases in the world.1 The imbalance of glucose homeostasis from diabetes mellitus causes an increase in glucose levels in the blood or hyperglycemia.2 Type 1 diabetes is due to insulin deficiency, accounting for only 5–10% of the diabetic population, while type 2 diabetes, referred to as noninsulin-dependent diabetes, accounts for 90–95% of cases of diabetes.1 Diabetes mellitus can lead to serious complications in many parts of the body, such as stroke, blindness, heart attack, kidney failure, and amputation.3 Alarmingly, the worldwide prevalence of diabetes has been increasing sharply.2
α-Glucosidase plays a key role in the digestion of polysaccharides, oligosaccharides, and disaccharides to monosaccharides.4 α-Glucosidase inhibitors are oral antidiabetic drugs used to treat type 2 diabetes currently2 since they delay the absorption of sugars from the gut5 reducing the glucose uptake.6 Acarbose, miglitol, and voglibose are used as α-glucosidase inhibitors in the market.1 Acarbose is one of the most widely prescribed α-glucosidase inhibitors in diabetes.5 Nevertheless, using these α-glucosidase inhibitors might increase the risk of hepatotoxicity7 and cause gastrointestinal side effects such as flatulence and diarrhea,8 which is considered a limiting factor for treating diabetes. To discover better safety and efficacy of α-glucosidase inhibitors for drug development, scientists have been continuously further studying diverse compounds including the coumarin scaffold.
Coumarin compounds are oxygen-containing heterocycles with a typical benzopyrone framework that are essential in natural products and organic synthesis. Numerous studies have been proven about multiple potential activities of coumarins, including antiproliferative,9 anticancer,10,11 antihepatitis C virus (HCV),12 antihuman immunodeficiency virus (HIV),13 anti-Alzheimer,14 antimalarial,15,16 antibacterial,17 antifungal,18 antioxidant,19 anticonvulsant,20 anti-inflammatory,21 and enzyme inhibition.22 Coumarins were reported to inhibit several enzymes, i.e., cholinesterase, monoamine oxidase (A and B), aldehyde/aldose reductase, alkaline phosphatase, urease, carbonic anhydrase, lysine-specific demethylase, histone deacetylase, lipoxygenase, topoisomerase, tyrosinase, cyclooxygenase, and α-glucosidase.22 Many coumarins were investigated on α-glucosidase inhibitors such as substituted coumarins,23 hydroxycoumarin derivatives,24 sulfonamide coumarins,25 and biscoumarins (Figure 1).26−29 However, there are a few reported synthetic biscoumarin derivatives that exhibit potent α-glucosidase inhibition. In addition, biscoumarins displayed a wide range of biological versatility.30−41 This motivated us to design, synthesize biscoumarins, and evaluate biscoumarins for their inhibition effect and mechanism of action against α-glucosidase. In this research, we also performed quantitative structure–activity relationship (QSAR), molecular docking, molecular dynamics (MD) simulations, and fragment molecular orbital (FMO) method. The best active biscoumarin derivative could be further studied as a potential compound for the treatment of diabetes.
Figure 1.
α-Glucosidase inhibitors containing a biscoumarin skeleton.
Results and Discussion
Synthesis of Biscoumarins 1–16 and Their Inhibition against α-Glucosidase
To study QSAR of halogenated biscoumarins, 16 derivatives 1–16 (Scheme 1) were synthesized and tested against α-glucosidase. The inhibitory activity of these compounds at a concentration of 10 μM was screened, and their IC50 values were subsequently evaluated. The results are shown in Figure 2 and Table 1. In general, the synthesized derivatives showed better activity compared to the positive control (acarbose, IC50 = 93.63 μM) and starting material (4-hydroxycoumarin, IC50 > 200 μM). In addition, the substituents on the phenyl ring of the benzaldehydes affected the inhibition of α-glucosidase. The inhibition of compounds 2–16 was much higher than that of the unsubstituted biscoumarin 1.
Scheme 1. Synthesis of Biscoumarin Derivatives 1–16.
Figure 2.
α-Glucosidase inhibition of biscoumarin derivatives 1–16.
Table 1. In Vitro α-Glucosidase Inhibitory Activities of Biscoumarins 1–16.
| compound | IC50 (μM) | compound | IC50 (μM) | compound | IC50 (μM) |
|---|---|---|---|---|---|
| 1 | 30.77 ± 1.25 | 7 | 3.15 ± 0.31 | 13 | 3.80 ± 0.05 |
| 2 | 8.12 ± 0.59 | 8 | 1.90 ± 0.14 | 14 | 2.38 ± 0.24 |
| 3 | 6.06 ± 0.06 | 9 | 14.80 ± 0.32 | 15 | 15.23 ± 0.45 |
| 4 | 3.19 ± 0.41 | 10 | 3.41 ± 0.17 | 16 | 5.10 ± 0.18 |
| 5 | 12.31 ± 0.08 | 11 | 2.89 ± 0.25 | acarbose | 93.63 ± 0.49 |
| 6 | 2.95 ± 0.05 | 12 | 4.15 ± 0.06 |
Almost all biscoumarins containing halogen exhibited good to excellent inhibitory activity. Replacement of m-chloro with m-nitro led to reduced inhibitory activity, as observed in 3 and 9. Moreover, the introduction of a chloro group at different positions resulted in different inhibitory activities (2–4, 7, and 8). The activity was enhanced by placing the chloro substituent at the p-position.
Additionally, among chloro, fluoro, and bromo (4–6), the F group obviously led to lower inhibitory activity. The best result was obtained with p-bromo in biscoumarin 6 with an IC50 of 2.95 μM. It was worth noting that adding the bromo group at the m-position in compound 11 improved the inhibition to 2.89 μM compared to 5 bearing fluoro at the p-position (IC50 12.31 μM). The same effect could be seen when the inhibitory activities of 12 and 14, 15 and 16, or 9 and 10 (bromo at the p-position) were compared. When the bromo group was replaced by an electron-donating group like methoxy, the inhibitory activity decreased three times as in 12 and 15. Taken together, the above findings suggest that the existence of bromo was the most favorable to enhance the inhibitory activity against α-glucosidase.
QSAR Study
The α-glucosidase inhibitory activity and physical and chemical properties of 16 biscoumarin derivatives were analyzed using the Materials Studio program (MS)42 to establish a quantitative structure–activity relationship (QSAR) model. The model was developed using genetic algorithm (GA)-based multiple linear regression (MLR) techniques. The best obtained QSAR model as shown in eq 1 was used to predict the α-glucosidase inhibitory activity of biscoumarin derivatives with reasonable accuracy. The square correlation coefficient (R2) value was 0.835. Additionally, the cross-validated square correlation coefficient (Q2) with a value of 0.747 affirmed the robustness of this obtained model
| 1 |
According to the intuitive observations of the model, the high lipophilicity of the biscoumarin series (AlogP98) had a great positive effect on the inhibition of α-glucosidase. Combining the above discussions of 16 compounds that introducing more halogen groups was pivotal to enhance the inhibitory activities, this speculation was consistent with the previous consensus in the field of drug discovery that halogen substituents will increase the lipophilicity of molecules,43 which will help improve the penetration through the cell’s lipid membrane, especially when the halogen element became bulky and more polarized functional groups, leading to a corresponding increase in the London dispersion forces. Moreover, expanding the shadow area of the XY plane, which corresponded to the p-substituent of the R group, could bring about a moderate increase in the α-glucosidase suppression performance of biscoumarin derivatives. Thus, the bulky substitutions could slightly support the inhibitory activity of α-glucosidase, as found in a comparison between compounds 12 and 13.
According to Figure 3, the QSAR results suggested that biscoumarins bearing 3,5-dibromo and 4-alkoxy groups may be potent compounds to study α-glucosidase inhibition further. Inspired by the result and aiming to investigate potent inhibitors, we synthesized three new compounds 17–19 according to the procedure in Scheme 1. Their inhibitory activity against α-glucosidase was determined. Interestingly, as expected, biscoumarin 17 enhanced the activity slightly compared to monobromo biscoumarin 13. 18 and 19 displayed excellent activity with IC50 values of 0.62 ± 0.01 and 1.21 ± 0.16 μM, respectively. The long-chain alkoxy substituent (–OC4H9, –OC8H17) at the p-position on the phenyl ring of benzaldehyde was highly preferred.
Figure 3.
(A) Experimental pIC50 versus predicted pIC50 from the QSAR model (compounds 1–16). (B) Compounds 17, 18, and 19 are newly designed candidate biscoumarin compounds.
Kinetic Study and Binding of Halogenated Biscoumarins to α-Glucosidase
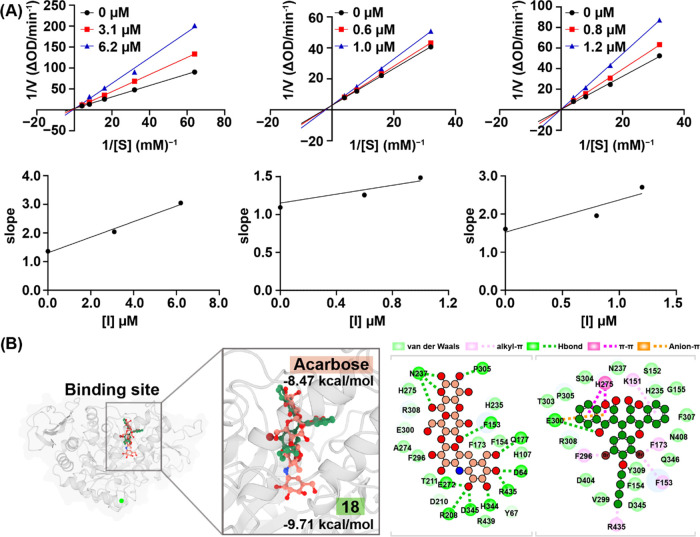
The enzyme kinetic study was performed to study the inhibition mode of 17–19 against α-glucosidase. The activity was determined at different concentrations of p-nitrophenyl-α-d-glucopyranoside (pNPG) in the presence or absence of 17–19 and analyzed using Lineweaver–Burk plots. As can be seen in Figure 4A, Km increased, while Vmax values were unaffected. Therefore, this study indicated that compounds 17–19 should be competitive inhibitors for α-glucosidase. The Ki values 4.81, 3.93, and 1.80 μM were calculated directly by the secondary replot of the Lineweaver–Burk plots against the different concentrations of 17, 18, and 19, respectively.
Figure 4.
(A) Kinetic study of 17–19. The Lineweaver–Burk plot for α-glucosidase inhibition and plots of slope versus concentration of 17–19 for determining the inhibition constant Ki. (B) Molecular docking analysis of acarbose (a native inhibitor) and 18 with α-glucosidase.
The binding affinity of acarbose and designed biscoumarins (17–19) was evaluated by docking them to the original binding site (OBS) of α-glucosidase MAL12. The docking results and inhibitory activity are presented in Table 2 and Figure 4B. 18 exhibited the most promising inhibitory potential by targeting the OBS of α-glucosidase MAL12 as a competitive inhibitor, based on the binding affinity score obtained from the molecular docking study. Computational studies provided insight into the molecular mechanism by which 18 inhibits the α-glucosidase activity. Compound 18 was found to be accessible to the OBS of the α-glucosidase, similar to acarbose, the native inhibitor, as shown in Figure 4B. The binding poses of these compounds were also similar, with the isomaltotriose group of acarbose aligned with one 4-hydroxycoumarin ring in biscoumarin 18. The residues N237, H275, E300, and P305 interacted with this moiety in both systems. Additionally, the core structure of acarbose and the 3,5-dibromo-4-butoxybenzene group of 18 revealed similar binding poses and common interacting residues, which were hydrophobic amino acids, such as F153, F154, F173, and F296. Although 18 did not show any interaction with the maltose-binding region, the double size of the 4-hydroxycoumarin ring could block the OBS entry pore by forming several hydrophobic interactions, such as π–π, alkyl–π, and anion–π, with nearby amino acids at the OBS.
Table 2. Molecular Docking Results and Inhibitory Activity of Acarbose and 17–19 against α-Glucosidase.
| binding affinity score (kcal/mol) | IC50 (μM) | %inhibition (10 μM) | |
|---|---|---|---|
| 17 | –9.00 | 3.31 ± 0.03 | 99.77 ± 0.49 |
| 18 | –9.71 | 0.62 ± 0.01 | 99.67 ± 0.42 |
| 19 | –8.90 | 1.21 ± 0.16 | 98.95 ± 0.26 |
| acarbose | –8.47 | 93.63 ± 0.49a | -b |
Data according to the literature.
-, not determined.
Dynamics and Binding Efficiency of Potent Compound
The binding of biscoumarin 18 to α-glucosidase at the OBS was found to be stable during the 500 ns MD simulation, and the resulting structural dynamics were compared with the apo form of the enzyme in Figure 5. The root-mean-square deviation (RMSD) value showed that the overall system had a lower fluctuation in the bound complex (2.00 ± 0.20 Å) relative to the apo form (2.20 ± 0.25 Å), as shown in Figure 5A. The normalized root-mean-square fluctuation (RMSF) was calculated for the free enzyme and 18-bound complex over the last 100 ns, revealing flexible and rigid domains in the protein (Figure 5B). Compared to the apo system, the residues 226–230, 270–275, and 300–360 with lower RMSF values were relatively stable in the 18-bound complex, indicating that the ligand restricts protein motion and binding site dynamics.44 The binding free energy of the 18/α-glucosidase complex of −21.74 ± 0.65 kcal/mol was estimated using the MM/GBSA method, showing a primary interaction toward α-glucosidase (−65.76 ± 0.32 kcal/mol) through van der Waals forces (Table S2 in the Supporting Information).
Figure 5.
Structural analysis of α-glucosidase by comparing (A) Cα-RMSD and (B) the normalized RMSF between apo (yellow) and holo (green) forms.
The presence of halogen atoms in a potent inhibitor prompted quantum mechanics calculations to investigate their influence on the binding interaction with the protein target.45−47 The FMO-RIMP2 method was used to calculate the total pair interaction energy (PIEDAtotal) between 18 and bound residues within 7 Å of the representative structure derived from RMSD clustering, revealing a robust binding interaction (−91.45 kcal/mol). The PIEDA was further decomposed into each interacting residue, as shown in Figure 6. The dispersion (EIJDI) and charge-transfer (EIJ) interactions between bromo substituents and nearby residues (F153, F154, F296, and Y309) were found to be crucial for enhancing the binding strength and stability to the OBS of α-glucosidase. Additionally, the electrostatic energy (EIJES) between bromine and Y309, combined with the hydrophobic interaction from H235, H275, G276, and S304 at the entry pore of the OBS, was also essential for 18 to maintain its binding in this site and reduce enzyme activity. Although the electrostatic interaction (EIJ), EIJDI, and EIJ in R308 displayed a good PIEDA value, it showed a moderate PIEDAtotal, which could be attributed to the presence of steric hindrance to 18 considered by charge exchange (EIJEX). Moreover, the repulsive effects caused by EIJ and EIJEX found in K151, T211, and N237 might affect ligand mobility. Overall, these findings provide insight into the role of brominated biscoumarin in α-glucosidase inhibition and highlight the importance of specific interactions with key residues in enhancing the binding affinity and stability of the inhibitor.
Figure 6.
Analysis of the binding pattern and interaction profile between 18 and α-glucosidase at the OBS using the FMO-RIMP2 method. The electrostatic (red), charge-transfer (yellow), dispersion (purple), and charge exchange (green) interactions of key residues with 18 are depicted, with the amino acids having PIEDAtotal values <−3 or >3 kcal/mol labeled.
Conclusions
In summary, biscoumarin derivatives, especially halogenated compounds, were successfully synthesized and their inhibitory activity toward α-glucosidase was evaluated. The result revealed that all synthesized derivatives exhibited prominent inhibitory activity with IC50 values of 0.62–30.77 μM. The enzyme kinetic study confirmed the competitive binding mode of the potential compound 18 with a Ki value of 3.93 μM. The computational study revealed that 18 could fit into the binding site of α-glucosidase MAL12 and create several hydrophobic interactions with the adjacent amino acids. The present findings suggest that biscoumarins containing 3,5-dibromo substituents and long-chain alkoxy groups at the p-position on the phenyl ring could be promising compounds for further study of α-glucosidase inhibitory activity.
Experimental Section
Chemistry
α-Glucosidase from Saccharomyces cerevisiae (EC 3.2.1.20) and pNPG were supplied by Sigma-Aldrich. All of the other commercially available reagents were used without further purification. 1H and 13C NMR spectra were recorded in CDCl3 or DMSO-d6 using a JEOL NMR 500 MHz spectrometer for 1H and 125 MHz for 13C. All solvents used in this research were distilled prior to use except those that were reagent grades. Thin-layer chromatography (TLC) was performed on aluminum sheets precoated with silica gel (Merck Kieselgel 60 PF254).
General Procedures
4-Hydroxycoumarin (2 equiv) was dissolved in ethanol, and benzaldehyde (1 equiv) was added. The mixture was refluxed until a precipitate occurred within approximately 24 h. After that, the reaction was cooled down. The precipitates were filtered and washed with ethanol to obtain pure products 1–19.
3,3′-(Phenylmethylene)bis(4-hydroxy-2H-chromen-2-one) 1
White solid; yield 70.3%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.57; 100.00% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.53 (s, 1H), 11.30 (s, 1H), 8.07 (d, J = 8.0 Hz, 1H), 8.00 (d, J = 7.5 Hz, 1H), 7.65–7.61 (m, 2H), 7.42–7.38 (m, 4H), 7.34–7.31 (m, 2H), 7.29–7.27 (m, 1H), 7.24–7.22 (m, 2H), and 6.11 (d, J = 1.0 Hz, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.4, 167.0, 165.9, 164.7, 152.7, 152.4, 135.3, 133.0 (2C), 128.8 (2C), 127.0, 126.6 (2C), 125.0 (2C), 124.5 (2C), 117.0, 116.8 (2C), 116.6, 105.8, 104.0, and 36.3.
3,3′-((2-Chlorophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 2
White solid; yield 72.0%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.48; 99.27% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.64 (s, 1H), 10.93 (s, 1H), 8.06 (s, 1H), 8.00 (s, 1H), 7.62 (m, 2H), 7.47–7.45 (m, 1H), 7.40–7.37 (m, 4H), 7.36–7.35 (m, 1H), 7.29–7.26 (m, 1H), 7.26–7.22 (m, 1H), 6.14 (s, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 168.9, 167.3, 165.3, 164.4, 152.6, 152.2, 133.6, 133.6, 133.0 (2C), 130.9, 129.4, 128.7, 126.9, 125.0 (2C), 124.5 (2C), 116.9, 116.7 (2C), 116.5, 105.8, 104.5, and 35.8.
3,3′-((3-Chlorophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 3
White solid; yield 75.0%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.54; 99.94% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.53 (s, 1H), 11.29 (s, 1H), 8.07 (d, J = 8.5 Hz, 1H), 8.01 (d, J = 8.0 Hz, 1H), 7.66–7.63 (m, 2H), 7.43–7.38 (m, 4H), 7.26 (m, 1H), 7.25 (d, 1H, J = 1.0 Hz), 7.19–7.18 (m, 1H), 7.13–7.10 (m, 1H), and 6.05 (s, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.3, 167.0, 166.2, 164.8, 152.7, 152.4, 137.7, 134.8, 133.2 (2C), 130.0, 127.3, 126.8, 125.1 (2C), 124.9, 124.6 (2C), 116.9, 116.9, 116.8, 116.5, 105.3, 103.6, and 36.1.
3,3′-((4-Chlorophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 4
White solid; yield 62.1%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.54; 99.41% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.54 (s, 1H), 11.32 (s, 1H), 8.06 (d, J = 8.0 Hz, 1H), 7.99 (d, J = 8.5 Hz, 1H), 7.65–7.62 (m, 2H), 7.42–7.38 (m, 4H), 7.30–7.29 (m, 1H), 7.28–7.27 (m, 1H), 7.16 (m, 1H), 7.14 (m, 1H), 6.04 (s, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.3, 167.0, 166.1, 164.7, 152.6, 152.4, 134.0, 133.2 (2C), 132.8, 128.9 (2C), 128.1 (2C), 125.1 (2C), 124.5 (2C), 116.9, 116.8, 116.8, 116.4, 105.4, 103.8, and 35.9.
3,3′-((4-Fluorophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 5
White solid; yield 65.7%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.51; 98.54% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.54 (s, 1H), 11.32 (s, 1H), 8.07 (1H, d, J = 7.0 Hz), 8.00 (d, J = 8.0 Hz, 1H), 7.65–7.62 (m, 2H), 7.42–7.37 (m, 4H), 7.20–7.17 (m, 2H), 7.03–6.99 (m, 2H), 6.05 (s, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.4, 167.0, 166.0, 164.8, 161.8 (d, J = 244.1 Hz, 1C), 152.6, 152.4, 133.1 (2C), 130.9 (d, J = 3.5 Hz, 1C), 128.2 (d, J = 7.8 Hz, 2C), 125.1 (2C), 124.5 (2C), 117.0, 116.8 (2C), 116.5, 115.6 (d, J = 21.4 Hz, 2C), 105.6, 104.0, and 35.8.
3,3′-((4-Bromophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 6
White solid; yield 65.0%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.51; 99.16% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.54 (s, 1H), 11.32 (s, 1H), 8.06 (d, J = 8.0 Hz, 1H), 7.99 (d, J = 8.0 Hz, 1H), 7.64 (m, 2H), 7.44–7.41 (m, 4H), 7.41–7.40 (m, 2H), 7.11–7.09 (m, 2H), and 6.01 (t, J = 1.5 Hz, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.4, 167.0, 166.2, 164.8, 152.6, 152.4, 134.5, 133.2 (2C), 131.8 (2C), 128.5 (2C), 125.1 (2C), 124.5 (2C), 120.9, 116.9, 116.8, 116.8, 116.4, 105.3, 103.7, and 36.0.
3,3′-((2,4-Dichlorophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 7
White solid; yield 65.5%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.46; 99.27% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.67 (s, 1H), 10.93 (s, 1H), 8.04 (s, 1H), 8.00 (s, 1H), 7.64–7.61 (m, 2H), 7.40–7.39 (m, 4H), 7.37–7.36 (m, 2H), 7.24 (dd, J = 8.5, 2.5 Hz, 1H), and 6.08 (d, J = 1.0 Hz, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 168.8, 167.4, 165.5, 164.7, 152.5, 152.3, 134.3, 133.8, 133.2 (2C), 132.4, 130.7, 130.3, 127.1, 125.1 (2C), 124.6 (2C), 116.8 (2C), 116.4 (2C), 105.4, 104.2, and 35.5.
3,3′-((3,4-Dichlorophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 8
White solid; yield 8.2%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.48; 99.20% purity by HPLC; 1H NMR (500 MHz, DMSO-d6) δH (ppm) 7.87 (dd, J = 8.0, 1.5 Hz, 2H), 7.58–7.55 (m, 2H), 7.45 (d, J = 8.0 Hz, 1H), 7.32 (d, J = 7.5 Hz, 2H), 7.30–7.27 (m, 3H), 7.14–7.11 (m, 1H), 6.27 (s, 1H); 13C NMR (125 MHz, DMSO-d6) δC (ppm) 166.5 (2C), 164.5 (2C), 152.5 (2C), 142.9, 131.8 (2C), 130.6, 130.1, 128.7, 127.9, 127.5, 124.1 (2C), 123.5 (2C), 118.7 (2C), 115.9 (2C), 103.3 (2C), and 35.9.
3,3′-((3-Nitrophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 9
White solid; yield 55.4%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.46; 96.11% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.58 (s, 1H), 11.38 (s, 1H), 8.17–8.13 (m, 1H), 8.08 (d, J = 8.0 Hz, 1H), 8.06 (m, 1H), 7.99 (d, J = 8.0 Hz, 1H), 7.69–7.65 (m, 2H), 7.59–7.56 (m, 1H), 7.51 (t, J = 8.0 Hz, 1H), 7.44 (m, 2H), 7.43–7.38 (m, 2H), 6.12 (d, J = 1.5 Hz, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.2, 167.1, 166.7, 165.0, 152.7, 152.4, 148.8, 138.1, 133.5 (2C), 132.9, 129.7, 125.3, 125.3, 124.6, 124.6, 122.2, 121.9, 116.9, 116.8, 116.8, 116.4, 104.7, 103.3, and 36.3.
3,3′-((4-Bromo-3-nitrophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 10
White solid; yield 22.3%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.43; 99.51% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.58 (s, 1H), 11.37 (s, 1H), 8.08 (d, J = 8.0 Hz, 1H), 7.99 (d, J = 8.0 Hz, 1H), 7.69–7.68 (m, 2H), 7.67–7.65 (m, 2H), 7.44–7.40 (m, 4H), 7.31–7.29 (m, 1H), 6.03 (t, J = 1.5 Hz, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.2, 167.0, 166.9, 165.0, 152.7, 152.5, 150.1, 137.4, 136.4, 133.6 (2C), 131.7, 125.4, 124.7 (2C), 124.2 (2C), 117.0, 116.9, 116.7, 116.3, 113.0, 104.3, 103.0, and 36.0; HRMS (ESI) calcd for C25H14BrNO8 [M + H]+: 535.9981, found 535.9982.
3,3′-((3-Bromo-4-fluorophenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 11
White solid; yield 42.8%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.48; 99.38% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.57 (s, 1H), 11.30 (s, 1H), 8.07 (d, J = 7.5 Hz, 1H), 8.00 (d, J = 8.0 Hz, 1H), 7.65 (m, 2H), 7.43–7.39 (m, 4H), 7.38–7.36 (m, 1H), 7.16–7.12 (m, 1H), 7.07 (t, J = 8.5 Hz, 1H), and 6.03 (t, J = 1.5 Hz, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.2, 167.3, 166.3, 164.9, 158.0 (d, J = 245.6 Hz, 1C), 152.7, 152.4, 133.3 (2C), 132.9 (d, J = 3.6 Hz, 1C), 131.7, 127.3 (d, J = 7.0 Hz, 1C), 125.2 (2C), 124.6 (2C), 116.9, 116.8, 116.6 (d, J = 22.2 Hz, 1C), 116.4 (2C), 109.5 (d, J = 21.4 Hz, 1C), 105.1, 103.6, and 35.6.
3,3′-((3-Bromo-4-hydroxyphenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 12
White solid; yield 38.9%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.34; 98.75% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.55 (s, 1H), 11.28 (s, 1H), 8.06 (d, J = 8.0 Hz, 1H), 8.00 (d, J = 7.5 Hz, 1H), 7.64 (m, 2H), 7.42–7.37 (m, 4H), 7.27 (m, 1H), 7.08–7.06 (m, 1H), 6.96 (d, J = 8.5 Hz, 1H), and 6.01 (t, J = 1.5 Hz, 1H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.3, 166.9, 166.1, 164.8, 152.7, 152.4, 151.3, 133.2 (2C), 130.1, 129.0, 127.6, 125.1 (2C), 124.6 (2C), 116.8, 116.8 (2C), 116.3 (2C), 110.7, 105.4, 103.8, and 35.4; HRMS (ESI) calcd for C25H15BrO7 [M + H]+: 507.0079, found 507.0074.
3,3′-((3-Bromo-4-methoxyphenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 13
White solid; yield 82.1%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.57; 98.89% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.55 (s, 1H), 11.28 (s, 1H), 8.06 (d, J = 8.0 Hz, 1H), 8.00 (d, J = 8.0 Hz, 1H), 7.64 (m, 2H), 7.42–7.38 (m, 4H), 7.35 (m, 1H), 7.11 (m, 1H), 6.84 (d, J = 7.5 Hz, 1H), 6.02 (t, J = 1.5 Hz, 1H), and 3.88 (s, 3H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.3, 166.9, 166.1, 164.8, 154.9, 152.7, 152.4, 133.1 (2C), 131.4, 128.9, 126.8, 125.1 (2C), 124.6 (2C), 117.0, 116.8 (2C), 116.5, 112.1, 111.9, 105.4, 103.9, 56.4, and 35.3.
3,3′-((3,5-Dibromo-4-hydroxyphenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 14
White solid; yield 68.3%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.43; 99.36% purity by HPLC; 1H NMR (500 MHz, DMSO-d6) δH (ppm) 7.87 (dd, J = 8.0, 2.0 Hz, 2H), 7.56 (m, 2H), 7.32 (dd, J = 8.0, 1.0 Hz, 2H), 7.29 (m, 2H), 7.20 (d, J = 1.0 Hz, 2H), and 6.20 (t, J = 1.0 Hz, 1H); 13C NMR (125 MHz, DMSO-d6) δC (ppm) 166.5 (2C), 164.4 (2C), 152.4 (2C), 148.4, 135.9, 131.7 (2C), 130.4 (2C), 124.1 (2C), 123.5 (2C), 118.7 (2C), 115.9 (2C), 111.8 (2C), 103.3 (2C), and 35.2.
3,3′-((4-Hydroxy-3-methoxyphenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 15
White solid; yield 80.2%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.37; 99.26% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.52 (s, 1H), 11.28 (s, 1H), 8.06 (brs, 1H), 8.02 (brs, 1H), 7.63 (m, 2H), 7.41–7.40 (m, 4H), 6.86 (d, J = 8.0 Hz, 1H), 6.73–6.71 (m, 1H), 6.68 (m, 1H), 6.06 (t, J = 1.0 Hz, 1H), and 3.75 (s, 3H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.4, 166.9, 165.8, 164.8, 152.6 (2C), 146.8, 144.7, 133.0, 127.0 (2C), 125.0 (2C), 124.5 (2C), 119.6, 116.8 (4C), 114.6, 109.6, 105.9, 104.3, 56.2, and 35.9.
3,3′-((3-Bromo-4-hydroxy-5-methoxyphenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 16
White solid; yield 83.4%; Rf (dichloromethane/methanol/acetic acid; 8:1:0.01) = 0.40; 99.85% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.56 (s, 1H), 11.26 (s, 1H), 8.06 (d, J = 7.5 Hz, 1H), 8.01 (d, J = 8.0 Hz, 1H), 7.64 (m, 2H), 7.42–7.40 (m, 4H), 6.92 (t, J = 1.5 Hz, 1H), 6.64 (d, J = 1.0 Hz, 1H), 6.03 (t, J = 1.0 Hz, 1H), and 3.76 (s, 3H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.4, 166.8, 166.1, 164.8, 152.4 (2C), 147.4, 142.2, 133.2, 128.1 (2C), 125.1, 124.5 (2C), 122.8 (2C), 116.8 (4C), 109.0, 108.6, 105.4, 103.8, 56.6, 35.7; DART-MS calcd for C26H17BrO8 [M + H]+: 537.0185, found 537.0244.
To a solution of 3,5-dibromo-4-hydroxybenzaldehyde (0.84 g, 3 mmol), methyl iodide or alkyl bromides (6.6 mmol) and anhydrous potassium carbonate (K2CO3, 0.828 g, 6 mmol) in DMF (10 mL) were added. The reaction mixture was stirred at 55 °C for 3 h, cooled to room temperature, and quenched with the addition of water (80 mL). The resulting white precipitate was filtered under vacuum, washed with water (40 mL), and dried to give the anisaldehyde intermediate (0.72 g, 2.44 mmol, 81.4%) as a white solid, Rf (n-hexane/ethyl acetate; 30:1) = 0.51.48 For the reaction with 1-bromobutane or 1-bromooctane, after the reaction was completed (analyzed by TLC), the crude product was extracted with EtOAc (3 × 20 mL), and the combined extracts were washed with water. The organic layer was dried over anhydrous Na2SO4 and evaporated to dryness. The synthesized 3,5-dibromo-4-alkoxybenzaldehydes yielded as a light yellow oil (50.0% yield); Rf (n-hexane/ethyl acetate; 30:1) = 0.51 was used to synthesize biscoumarins without further purification.
3,3′-((3,5-Dibromo-4-methoxyphenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 17
White solid; yield 21.0%; Rf (CH2Cl2/MeOH/AcOH; 8:1:0.01) = 0.51; 96.63% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.60 (s, 1H), 11.28 (s, 1H), 8.07 (d, J = 8.0 Hz, 1H), 8.02 (d, J = 8.0 Hz, 1H), 7.66 (m, 2H), 7.43–7.40 (m, 4H), 7.31 (d, J = 1.0 Hz, 2H), 6.00 (t, J = 1.0 Hz, 1H), and 3.89 (s, 3H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.2, 166.9, 166.4, 164.9, 153.1, 152.7, 152.4, 134.4, 133.4 (2C), 130.9 (2C), 125.2 (2C), 124.6 (2C), 118.5 (2C), 117.0 (2C), 116.8 (2C), 104.8, 103.3, 60.8, and 35.4; HRMS (ESI) calcd for C26H16Br2O7 [M + H]+: 598.9341, found 598.9283.
3,3′-((3,5-Dibromo-4-butoxyphenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 18
White solid; yield 54.0%; Rf (CH2Cl2/MeOH/AcOH; 8:1:0.01) = 0.54; 98.53% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.59 (s,1H), 11.27 (s, 1H), 8.06 (d, J = 7.5 Hz, 1H), 8.02 (d, J = 8.0 Hz, 1H), 7.65 (td, J = 7.5, 1.5 Hz, 2H), 7.43–7.40 (m, 4H), 7.31 (d, J = 1.5 Hz, 2H), 6.00 (t, J = 1.0 Hz, 1H), 4.01 (t, J = 6.5 Hz, 2H), 1.84 (quint, J = 6.5 Hz, 2H), 1.59–1.54 (m, 2H), and 1.00 (t, J = 7.5 Hz, 3H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.2, 166.9, 166.4, 164.9, 152.7, 152.5, 152.4, 134.0, 133.4 (2C), 130.8 (2C), 125.2 (2C), 124.6 (2C), 118.8 (2C), 116.9, 116.8 (2C), 116.4, 104.8, 103.3, 73.5, 35.4, 32.2, 19.3, and 14.1; HRMS (ESI) calcd for C29H22Br2O7 [M + H]+: 640.9811, found 640.9805.
3,3′-((3,5-Dibromo-4-(octyloxy)phenyl)methylene)bis(4-hydroxy-2H-chromen-2-one) 19
White solid; yield 50.6%; Rf (CH2Cl2/MeOH/AcOH; 8:1:0.01) = 0.63; 99.46% purity by HPLC; 1H NMR (500 MHz, CDCl3) δH (ppm) 11.59 (s, 1H), 11.27 (s, 1H), 8.07 (d, J = 8.0 Hz, 1H), 8.02 (d, J = 8.0 Hz, 1H), 7.65 (td, J = 7.5, 1.5 Hz, 2H), 7.43–7.38 (m, 4H), 7.30 (d, J = 1.5 Hz, 2H), 6.00 (t, J = 1.0 Hz, 1H), 4.00 (t, J = 6.5 Hz, 2H), 1.86 (quint, J = 6.5 Hz, 2H), 1.52 (quint, J = 6.5 Hz, 2H), 1.38–1.29 (m, 8H), and 0.88 (t, J = 6.5 Hz, 3H); 13C NMR (125 MHz, CDCl3) δC (ppm) 169.2, 166.9, 166.4, 164.9, 152.7, 152.5, 152.4, 134.0, 133.4 (2C), 130.8 (2C), 125.2 (2C), 124.6 (2C), 118.8 (2C), 116.9, 116.8 (2C), 116.4, 104.8, 103.3, 73.8, 35.4, 32.0, 30.2, 29.5, 29.4, 26.0, 22.8, and 14.2; DART-MS (ESI) calcd for C33H30Br2O7 [M + H]+: 697.0437, found 697.0509.
Inhibition of α-Glucosidase Activity In Vitro
The α-glucosidase inhibition of the synthesized compounds 1–19 was measured using a spectrophotometric method.49 Those compounds (4 mM) were dissolved in DMSO and diluted by 0.1 mM pH 6.9 phosphate buffer. The enzyme α-glucosidase from S. cerevisiae E.C. 3.2.1.20 (0.1 U/mL) and 1 mM pNPG as the substrate were dissolved in 0.1 mM pH 6.9 phosphate buffer. Then, 10 μL of a test compound or a positive control was added with 40 μL of the enzyme, and the mixture was then incubated at 37 °C for 10 min. Afterward, 50 μL of the substrate was added to the reaction mixture. The mixture was incubated further for 20 min at 37 °C, and then 1 M of Na2CO3 (100 μL) was added to stop the reaction. The activity was measured at 405 nm (Allsheng microplate reader, AMR100). All samples were analyzed in triplicate at different concentrations to obtain the IC50 values of each compound. The mean values and standard deviation were calculated. The percentage inhibition was calculated by the following equation
| 2 |
where % inhibition is the percentage of inhibition, Asample is the corrected absorbance of the synthesized compound under test [Asample(initial) – Ablank-sample], and Acontrol is the absorbance of the negative control.
QSAR Modeling
The three-dimensional (3D) structures of all biscoumarin derivatives were imported into the QSAR module of Material Studio (MS) software.42 In addition, 16 structural descriptors (1–16) generated via the model module in MS were selected as the independent variables. All molecular descriptors could be grouped into several categories: structural descriptors, thermodynamic descriptors, topological descriptors, E-state keys, fragment counts, shadow indices, spatial descriptors, and VAMP electrostatic descriptors. In addition, the α-glucosidase inhibitory experimental values in the unit of IC50 (μM) were transformed to be pIC50 (M) values, which act as the response data.
The statistics module in the MS program was used to establish the QSAR model to predict the inhibitory activity of α-glucosidase. First, the descriptor selection was performed via the genetic algorithm methods provided by the genetic function approximation50 (GFA) of the MS package. The 3000 population size, 200 maximum generations, and Friedman’s lack-of-fitness function51 were used for the GFA process. The regressions were carried out using two-five descriptors, and R2 and Q2 were monitored.
Molecular Docking, MD Simulation, and FMO Calculation
The 3D structure of α-glucosidase MAL12 was obtained from the alphafold2 database52,53 due to the lack of crystal structure available in the protein database. The 3D structure of acarbose, 17, 18, and 19 was constructed using GaussView 6.0.16 and optimized by Gaussian16 using the HF/6-31G+(d,p) basis set.54 Since the designed biscoumarins consist of halogen atoms, the complex structure was prepared by molecular docking using the AutoDock VinaXB program,55−57 which contains the halogen force field parameters. Acarbose and designed biscoumarins were docked to the original receptor binding site (OBS) with the grid dimensions of 16 × 16 × 16 Å3.58 The binding pattern of these compounds and their interaction were carried out using two-dimensional (2D) and 3D using Discovery Studio Visualizer Software59 and Chimera USCF.60 The MD trajectory was analyzed with regrading to the structural stability by RMSF and RMSD techniques using the Cpptraj module implemented in the AmberTools 21.61 The last 100 ns trajectories were used to calculate the binding free energy between 18 and α-glucosidase using the MM/GBSA method.62
The fragment molecular orbital method using the second-order Møller–Plesset perturbation theory with the resolution-of-identity approximation (FMO-RIMP2) at the B3LYP3/6-31G+(d,p) level of theory was applied for the representative model of 18/α-glucosidase complex extracted from the last 100 ns trajectories using RMSD clustering techniques.63,64 The pair interaction energy (PIE) can be additionally decayed into four terms of interaction: electrostatic (EIJES), exchange (EIJ), charge-transfer (EIJCT+mix), and dispersion (EIJ) contributions, which are called PIEDA (eq 3).65 These contributed energies can explain the essential halogen-containing compounds66
| 3 |
Acknowledgments
This work was financially supported by the ASEAN Scholarship.
Glossary
Abbreviations
- OBS
original binding site
- HCV
hepatitis C virus
- HIV
human immunodeficiency virus
- QSAR
quantitative structure–activity relationship
- MS
Material Studio program
- GA
genetic algorithm
- MLR
multiple linear regression
- MD
molecular dynamics
- RMSF
root-mean-square fluctuation
- RMSD
root-mean-square deviation
- pNPG
p-nitrophenyl-α-d-glucopyranoside
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c02868.
1H and 13C NMR spectra, HPLC analysis of synthesized compounds 1–19, HRMS of 10, 12, 17, 18, DART-MS of 16 and 19 (Figures S1–S63), SMILES (Table S1), and MM/GBSA binding free energy (ΔGbindMM/GBSA) for 18 binding to α-glucosidase at the OBS site (Table S2) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Ghani U. Re-exploring promising α-glucosidase inhibitors for potential development into oral anti-diabetic drugs: Finding needle in the haystack. Eur. J. Med. Chem. 2015, 103, 133–162. 10.1016/j.ejmech.2015.08.043. [DOI] [PubMed] [Google Scholar]
- Pan Y.; Liu T.; Wang X.; Sun J. Research progress of coumarins and their derivatives in the treatment of diabetes. J. Enzyme Inhib. Med. Chem. 2022, 37, 616–628. 10.1080/14756366.2021.2024526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove A. Seeking sweet relief for diabetes. Nat. Biotechnol. 2002, 20, 977–981. 10.1038/nbt1002-977. [DOI] [PubMed] [Google Scholar]
- Tan M.-H. α-Glucosidase inhibitors in the treatment of diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 1997, 4, 48–55. 10.1097/00060793-199702000-00007. [DOI] [Google Scholar]
- Laar F. Alpha-glucosidase inhibitors in the early treatment of type 2 diabetes. Vasc. Health Risk. Manag. 2008, 4, 1189–1195. 10.2147/VHRM.S3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob G. S. Glycosylation inhibitors in biology and medicine. Curr. Opin. Struct. Biol. 1995, 5, 605–611. 10.1016/0959-440X(95)80051-4. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Chen Q.; Li L.; Kwong J. S.; Jia P.; Zhao P.; Wang W.; Zhou X.; Zhang M.; Sun X. Alpha-glucosidase inhibitors and hepatotoxicity in type 2 diabetes: a systematic review and meta-analysis. Sci. Rep. 2016, 6, 32649 10.1038/srep32649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krentz A. J.; Bailey C. J. Oral antidiabetic agents. Drugs 2005, 65, 385–411. 10.2165/00003495-200565030-00005. [DOI] [PubMed] [Google Scholar]
- Ueberschaar N.; Xu Z.; Scherlach K.; Metsä-Ketelä M.; Bretschneider T.; Dahse H.-M.; Görls H.; Hertweck C. Synthetic remodeling of the chartreusin pathway to tune antiproliferative and antibacterial activities. J. Am. Chem. Soc. 2013, 135, 17408–17416. 10.1021/ja4080024. [DOI] [PubMed] [Google Scholar]
- Amin K. M.; Abou-Seri S. M.; Awadallah F. M.; Eissa A. A.; Hassan G. S.; Abdulla M. M. Synthesis and anticancer activity of some 8-substituted-7-methoxy-2H-chromen-2-one derivatives toward hepatocellular carcinoma HepG2 cells. Eur. J. Med. Chem. 2015, 90, 221–231. 10.1016/j.ejmech.2014.11.027. [DOI] [PubMed] [Google Scholar]
- Liu M.-M.; Chen X.-Y.; Huang Y.-Q.; Feng P.; Guo Y.-L.; Yang G.; Chen Y. Hybrids of phenylsulfonylfuroxan and coumarin as potent antitumor agents. J. Med. Chem. 2014, 57, 9343–9356. 10.1021/jm500613m. [DOI] [PubMed] [Google Scholar]
- Tsay S.-C.; Hwu J. R.; Singha R.; Huang W.-C.; Chang Y. H.; Hsu M.-H.; Shieh F.-k.; Lin C.-C.; Hwang K. C.; Horng J.-C.; et al. Coumarins hinged directly on benzimidazoles and their ribofuranosides to inhibit hepatitis C virus. Eur. J. Med. Chem. 2013, 63, 290–298. 10.1016/j.ejmech.2013.02.008. [DOI] [PubMed] [Google Scholar]
- Kamiyama H.; Kubo Y.; Sato H.; Yamamoto N.; Fukuda T.; Ishibashi F.; Iwao M. Synthesis, structure–activity relationships, and mechanism of action of anti-HIV-1 lamellarin α-20-sulfate analogues. Bioorg. Med. Chem. 2011, 19, 7541–7550. 10.1016/j.bmc.2011.10.030. [DOI] [PubMed] [Google Scholar]
- Shaik J. B.; Palaka B. K.; Penumala M.; Kotapati K. V.; Devineni S. R.; Eadlapalli S.; Darla M. M.; Ampasala D. R.; Vadde R.; Amooru G. D. Synthesis, pharmacological assessment, molecular modeling and in silico studies of fused tricyclic coumarin derivatives as a new family of multifunctional anti-Alzheimer agents. Eur. J. Med. Chem. 2016, 107, 219–232. 10.1016/j.ejmech.2015.10.046. [DOI] [PubMed] [Google Scholar]
- Sashidhara K. V.; Kumar A.; Dodda R. P.; Krishna N. N.; Agarwal P.; Srivastava K.; Puri S. Coumarin–trioxane hybrids: Synthesis and evaluation as a new class of antimalarial scaffolds. Bioorg. Med. Chem. Lett. 2012, 22, 3926–3930. 10.1016/j.bmcl.2012.04.100. [DOI] [PubMed] [Google Scholar]
- Pingaew R.; Saekee A.; Mandi P.; Nantasenamat C.; Prachayasittikul S.; Ruchirawat S.; Prachayasittikul V. Synthesis, biological evaluation and molecular docking of novel chalcone-coumarin hybrids as anticancer and antimalarial agents. Eur. J. Med. Chem. 2014, 85, 65–76. 10.1016/j.ejmech.2014.07.087. [DOI] [PubMed] [Google Scholar]
- Hu Y.-Q.; Xu Z.; Zhang S.; Wu X.; Ding J.-W.; Lv Z.-S.; Feng L.-S. Recent developments of coumarin-containing derivatives and their anti-tubercular activity. Eur. J. Med. Chem. 2017, 136, 122–130. 10.1016/j.ejmech.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Puttaraju K. B.; Shivashankar K.; Mahendra M.; Rasal V. P.; Vivek P. N. V.; Rai K.; Chanu M. B. Microwave assisted synthesis of dihydrobenzo[4,5]imidazo[1,2-α] pyrimidin-4-ones; synthesis, in vitro antimicrobial and anticancer activities of novel coumarin substituted dihydrobenzo[4,5]imidazo [1,2-α]pyrimidin-4-ones. Eur. J. Med. Chem. 2013, 69, 316–322. 10.1016/j.ejmech.2013.07.015. [DOI] [PubMed] [Google Scholar]
- Nagamallu R.; Srinivasan B.; Ningappa M. B.; Kariyappa A. K. Synthesis of novel coumarin appended bis(formylpyrazole) derivatives: Studies on their antimicrobial and antioxidant activities. Bioorg. Med. Chem. Lett. 2016, 26, 690–694. 10.1016/j.bmcl.2015.11.038. [DOI] [PubMed] [Google Scholar]
- Amin K. M.; Rahman D. E. A.; Al-Eryani Y. A. Synthesis and preliminary evaluation of some substituted coumarins as anticonvulsant agents. Bioorg. Med. Chem. Lett. 2008, 16, 5377–5388. 10.1016/j.bmc.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Aggarwal R.; Kumar S.; Kaushik P.; Kaushik D.; Gupta G. K. Synthesis and pharmacological evaluation of some novel 2-(5-hydroxy-5-trifluoromethyl-4,5-dihydropyrazol-1-yl)4-(coumarin-3-yl)thiazoles. Eur. J. Med. Chem. 2013, 62, 508–514. 10.1016/j.ejmech.2012.11.046. [DOI] [PubMed] [Google Scholar]
- Ibrar A.; Shehzadi S. A.; Saeed F.; Khan I. Developing hybrid molecule therapeutics for diverse enzyme inhibitory action: Active role of coumarin-based structural leads in drug discovery. Bioorg. Med. Chem. 2018, 26, 3731–3762. 10.1016/j.bmc.2018.05.042. [DOI] [PubMed] [Google Scholar]
- Raju B. C.; Tiwari A. K.; Kumar J. A.; Ali A. Z.; Agawane S. B.; Saidachary G.; Madhusudana K. α-Glucosidase inhibitory antihyperglycemic activity of substituted chromenone derivatives. Bioorg. Med. Chem. 2010, 18, 358–365. 10.1016/j.bmc.2009.10.047. [DOI] [PubMed] [Google Scholar]
- Shen Q.; Shao J.; Peng Q.; Zhang W.; Ma L.; Chan A. S.; Gu L. Hydroxycoumarin derivatives: Novel and potent α-glucosidase inhibitors. J. Med. Chem. 2010, 53, 8252–8259. 10.1021/jm100757r. [DOI] [PubMed] [Google Scholar]
- Wang S.; Yan J.; Wang X.; Yang Z.; Lin F.; Zhang T. Synthesis and evaluation of the α-glucosidase inhibitory activity of 3-[4-(phenylsulfonamido)benzoyl]-2H-1-benzopyran-2-one derivatives. Eur. J. Med. Chem. 2010, 45, 1250–1255. 10.1016/j.ejmech.2009.12.031. [DOI] [PubMed] [Google Scholar]
- Khan K. M.; Rahim F.; Wadood A.; Kosar N.; Taha M.; Lalani S.; Khan A.; Fakhri M. I.; Junaid M.; Rehman W.; et al. Synthesis and molecular docking studies of potent α-glucosidase inhibitors based on biscoumarin skeleton. Eur. J. Med. Chem. 2014, 81, 245–252. 10.1016/j.ejmech.2014.05.010. [DOI] [PubMed] [Google Scholar]
- Zawawi N. K. N. A.; Taha M.; Ahmat N.; Ismail N. H.; Wadood A.; Rahim F.; Rehman A. U. Synthesis, in vitro evaluation and molecular docking studies of biscoumarin thiourea as a new inhibitor of α-glucosidases. Bioorg. Chem. 2015, 63, 36–44. 10.1016/j.bioorg.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Asgari M. S.; Mohammadi-Khanaposhtani M.; Kiani M.; Ranjbar P. R.; Zabihi E.; Pourbagher R.; Rahimi R.; Faramarzi M. A.; Biglar M.; Larijani B.; et al. Biscoumarin-1, 2, 3-triazole hybrids as novel anti-diabetic agents: Design, synthesis, in vitro α-glucosidase inhibition, kinetic, and docking studies. Bioorg. Chem. 2019, 92, 103206 10.1016/j.bioorg.2019.103206. [DOI] [PubMed] [Google Scholar]
- Mohammadi-Khanaposhtani M.; Yahyavi H.; Barzegaric E.; Imanparast S.; Faramarzi M. A.; et al. New biscoumarin derivatives as potent α-glucosidase inhibitors: Synthesis, biological evaluation, kinetic analysis, and docking study. Polycyclic Aromat. Compd. 2020, 40, 915–926. 10.1080/10406638.2018.1509359. [DOI] [Google Scholar]
- Zhao H.; Neamati N.; Hong H.; Mazumder A.; Wang S.; Sunder S.; Milne G. W.; Pommier Y.; Burke T. R. Coumarin-based inhibitors of HIV integrase. J. Med. Chem. 1997, 40, 242–249. 10.1021/jm960450v. [DOI] [PubMed] [Google Scholar]
- Khan K. M.; Iqbal S.; Lodhi M. A.; Maharvi G. M.; Choudhary M. I.; Perveen S.; et al. Biscoumarin: new class of urease inhibitors; economical synthesis and activity. Bioorg. Med. Chem. 2004, 12, 1963–1968. 10.1016/j.bmc.2004.01.010. [DOI] [PubMed] [Google Scholar]
- Lodhi M. A.; Nawaz S. A.; Iqbal S.; Khan K. M.; Rode B. M.; Choudhary M. I.; et al. 3D-QSAR CoMFA studies on bis-coumarine analogues as urease inhibitors: A strategic design in anti-urease agents. Bioorg. Med. Chem. 2008, 16, 3456–3461. 10.1016/j.bmc.2005.09.048. [DOI] [PubMed] [Google Scholar]
- Arif Lodhi M.; Wadood A.; Iqbal S.; Khan K. M.; Choudhary M. I.; et al. Three-dimensional quantitative structure–activity relationship (CoMSIA) analysis of bis-coumerine analogues as urease inhibitors. Med. Chem. Res. 2013, 22, 498–504. 10.1007/s00044-012-9999-8. [DOI] [Google Scholar]
- Lodhi M. A.; Shams S.; Choudhary M. I.; Lodhi A.; Ul-Haq Z.; Jalil S.; Nawaz S. A.; Khan K. M.; Iqbal S.; Rahman A.-u. Structural basis of binding and rationale for the potent urease inhibitory activity of biscoumarins. Biomed Res. Int. 2014, 2014, 935039 10.1155/2014/935039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisal M.; Saeed A.; Shahzad D.; Fattah T. A.; Lal B.; Channar P. A.; Mahar J.; Saeed S.; Mahesar P. A.; Larik F. A. Enzyme inhibitory activities an insight into the structure–Activity relationship of biscoumarin derivatives. Eur. J. Med. Chem. 2017, 141, 386–403. 10.1016/j.ejmech.2017.10.009. [DOI] [PubMed] [Google Scholar]
- Alomari M.; Taha M.; Imran S.; Jamil W.; Selvaraj M.; Uddin N.; Rahim F. Design, synthesis, in vitro evaluation, molecular docking and ADME properties studies of hybrid bis-coumarin with thiadiazole as a new inhibitor of Urease. Bioorg. Chem. 2019, 92, 103235 10.1016/j.bioorg.2019.103235. [DOI] [PubMed] [Google Scholar]
- Salar U.; Nizamani A.; Arshad F.; Khan K. M.; Fakhri M. I.; Perveen S.; Ahmed N.; Choudhary M. I. Bis-coumarins; non-cytotoxic selective urease inhibitors and antiglycation agents. Bioorg. Chem. 2019, 91, 103170 10.1016/j.bioorg.2019.103170. [DOI] [PubMed] [Google Scholar]
- Mukhtar F.; Stieglitz K.; Ali S.; Ejaz A.; Choudhary M. I.; Fakhri M. I.; Salar U.; Khan K. M. Coumarin and biscoumarin inhibit in vitro obesity model. Adv. Biol. Chem. 2016, 6, 71593 10.4236/abc.2016.65014. [DOI] [Google Scholar]
- Rahim F.; Ullah H.; Fakhri M. I.; Salar U.; Perveen S.; Khan K. M.; Choudhary M. I. Anti-Leishmanial Activities of Synthetic Biscoumarins. J. Chem. Soc. Pak. 2017, 39, 79–82. [Google Scholar]
- Mayank; Kumar D.; Kaur N.; Giri R.; Singh N. A biscoumarin scaffold as an efficient anti-Zika virus lead with NS3-helicase inhibitory potential: in vitro and in silico investigations. New J. Chem. 2020, 44, 1872–1880. 10.1039/C9NJ05225A. [DOI] [Google Scholar]
- Ullah H.; Fahad K. Synthetic Biscoumarin Analogs: Their PC3 Cell Line and Antioxidant Inhibitory Potentials. J. Ongoing Chem. Res. 2020, 5, 1–6. [Google Scholar]
- Materials Studio Modeling;release 7.0; Accelrys Software Inc., 2013.
- Wilcken R.; Zimmermann M. O.; Lange A.; Joerger A. C.; Boeckler F. M. Principles and applications of halogen bonding in medicinal chemistry and chemical biology. J. Med. Chem. 2013, 56, 1363–1388. 10.1021/jm3012068. [DOI] [PubMed] [Google Scholar]
- Sakulkeo O.; Wattanapiromsakul C.; Pitakbut T.; Dej-adisai S. Alpha-glucosidase inhibition and molecular docking of isolated compounds from traditional Thai medicinal plant, Neuropeltis racemosa Wall. Molecules 2022, 27, 639. 10.3390/molecules27030639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F.-Y.; MacKerell A. D. Jr Do halogen–hydrogen bond donor interactions dominate the favorable contribution of halogens to ligand–protein binding?. J. Phys. Chem. B 2017, 121, 6813–6821. 10.1021/acs.jpcb.7b04198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzec E.; Świtalska M.; Winiewska-Szajewska M.; Wójcik J.; Wietrzyk J.; Maciejewska A. M.; Poznański J.; Mieczkowski A. The halogenation of natural flavonoids, baicalein and chrysin, enhances their affinity to human protein kinase CK2. IUBMB Life 2020, 72, 1250–1261. 10.1002/iub.2298. [DOI] [PubMed] [Google Scholar]
- Deetanya P.; Hengphasatporn K.; Wilasluck P.; Shigeta Y.; Rungrotmongkol T.; Wangkanont K. Interaction of 8-anilinonaphthalene-1-sulfonate with SARS-CoV-2 main protease and its application as a fluorescent probe for inhibitor identification. Comput. Struct. Biotechnol. J. 2021, 19, 3364–3371. 10.1016/j.csbj.2021.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M.; Yamamoto A.; Inuzuka T.; Sengoku T.; Yoda H. Synthesis of lipophilic bisanthracene fluorophores: versatile building blocks toward the synthesis of new light-harvesting dendrimers. Tetrahedron 2011, 67, 9484–9490. 10.1016/j.tet.2011.10.021. [DOI] [Google Scholar]
- Ramadhan R.; Phuwapraisirisan P. New arylalkanones from Horsfieldia macrobotrys, effective antidiabetic agents concomitantly inhibiting α-glucosidase and free radicals. Bioorg. Med. Chem. Lett. 2015, 25, 4529–4533. 10.1016/j.bmcl.2015.08.069. [DOI] [PubMed] [Google Scholar]
- Wu W.; Zhang C.; Lin W.; Chen Q.; Guo X.; Qian Y.; Zhang L. Quantitative structure-property relationship (QSPR) modeling of drug-loaded polymeric micelles via genetic function approximation. PLoS One 2015, 10, e0119575 10.1371/journal.pone.0119575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. H. Multivariate adaptive regression splines. Ann. Stat. 1991, 19, 1–67. 10.1214/aos/1176347963. [DOI] [Google Scholar]
- Jumper J.; Evans R.; Pritzel A.; Green T.; Figurnov M.; Ronneberger O.; Tunyasuvunakool K.; Bates R.; Žídek A.; Potapenko A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M.; Anyango S.; Deshpande M.; Nair S.; Natassia C.; Yordanova G.; Yuan D.; Stroe O.; Wood G.; Laydon A.; et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch M.; Trucks G.; Schlegel H.; Scuseria G.; Robb M.; Cheeseman J.; Scalmani G.; Barone V.; Petersson G.; Nakatsuji H.. Gaussian 16; Gaussian, Inc.: Wallingford, CT, 2016.
- Koebel M. R.; Schmadeke G.; Posner R. G.; Sirimulla S. AutoDock VinaXB: implementation of XBSF, new empirical halogen bond scoring function, into AutoDock Vina. J. Cheminf. 2016, 8, 27 10.1186/s13321-016-0139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengphasatporn K.; Kaewmalai B.; Jansongsaeng S.; Badavath V. N.; Saelee T.; Chokmahasarn T.; Khotavivattana T.; Shigeta Y.; Rungrotmongkol T.; Boonyasuppayakorn S. Alkyne-tagged apigenin, a chemical tool to navigate potential targets of flavonoid anti-dengue leads. Molecules 2021, 26, 6967. 10.3390/molecules26226967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengphasatporn K.; Wilasluck P.; Deetanya P.; Wangkanont K.; Chavasiri W.; Visitchanakun P.; Leelahavanichkul A.; Paunrat W.; Boonyasuppayakorn S.; Rungrotmongkol T.; et al. Halogenated baicalein as a promising antiviral agent toward SARS-CoV-2 main protease. J. Chem. Inf. Model. 2022, 62, 1498–1509. 10.1021/acs.jcim.1c01304. [DOI] [PubMed] [Google Scholar]
- Zafar M.; Khan H.; Rauf A.; Khan A.; Lodhi M. A. In silico study of alkaloids as α-glucosidase inhibitors: Hope for the discovery of effective lead compounds. Front. Endocrinol. 2016, 7, 153 10.3389/fendo.2016.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BIOVIA . Dassault Systèmes, Discovery Studio Visualizer; Dassault Systèmes: San Diego, 2021. [Google Scholar]
- Pettersen E. F.; Goddard T. D.; Huang C. C.; Couch G. S.; Greenblatt D. M.; Meng E. C.; Ferrin T. E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Case D. A.; Belfon K.; Ben-Shalom I. Y.; Brozell S. R.; Cerutti D. S.; Cheatham T. E. III; Cruzeiro V. W. D.; Darden T. A.; Duke R. E.; Giambasu G.; Gilson M. K.; Gohlke H.; Goetz A. W.; Harris R.; Izadi S.; Izmailov S. A.; Kasavajhala K.; Kovalenko A.; Krasny R.; Kurtzman T.; Lee T. S.; LeGrand S.; Li P.; Lin C.; Liu J.; Luchko T.; Luo R.; Man V.; Merz K. M.; Miao Y.; Mikhailovskii O.; Monard G.; Nguyen H.; Onufriev A.; Pan F.; Pantano S.; Qi R.; Roe D. R.; Roitberg A.; Sagui C.; Schott-Verdugo S.; Shen J.; Simmerling C. L.; Skrynnikov N. R.; Smith J.; Swails J.; Walker R. C.; Wang J.; Wilson L.; Wolf R. M.; Wu X.; Xiong Y.; Xue Y.; York D. M.; Kollman P. A.. AMBER 2020; University of California: San Francisco, 2020.
- Mishra S. K.; Koča J. Assessing the performance of MM/PBSA, MM/GBSA, and QM–MM/GBSA approaches on protein/carbohydrate complexes: effect of implicit solvent models, QM methods, and entropic contributions. J. Phys. Chem. B 2018, 122, 8113–8121. 10.1021/acs.jpcb.8b03655. [DOI] [PubMed] [Google Scholar]
- Pham B. Q.; Gordon M. S. Development of the FMO/RI-MP2 fully analytic gradient using a hybrid-distributed/shared memory programming model. J. Chem. Theory Comput. 2020, 16, 1039–1054. 10.1021/acs.jctc.9b01082. [DOI] [PubMed] [Google Scholar]
- Nutho B.; Mahalapbutr P.; Hengphasatporn K.; Pattaranggoon N. C.; Simanon N.; Shigeta Y.; Hannongbua S.; Rungrotmongkol T. Why are lopinavir and ritonavir effective against the newly emerged coronavirus 2019? Atomistic insights into the inhibitory mechanisms. Biochemistry 2020, 59, 1769–1779. 10.1021/acs.biochem.0c00160. [DOI] [PubMed] [Google Scholar]
- Fedorov D. G.; Kitaura K. Pair interaction energy decomposition analysis. J. Comput. Chem. 2007, 28, 222–237. 10.1002/jcc.20496. [DOI] [PubMed] [Google Scholar]
- Hengphasatporn K.; Garon A.; Wolschann P.; Langer T.; Yasuteru S.; Huynh T. N.; Chavasiri W.; Saelee T.; Boonyasuppayakorn S.; Rungrotmongkol T. Multiple virtual screening strategies for the discovery of novel compounds active against dengue virus: A hit identification study. Sci. Pharm. 2020, 88, 2 10.3390/scipharm88010002. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.