ABSTRACT
Powassan virus (POWV) is an emerging tick-borne flavivirus that causes neuroinvasive diseases, including encephalitis, meningitis, and paralysis. Similar to other neuroinvasive flaviviruses, such as West Nile virus (WNV) and Japanese encephalitis virus (JEV), POWV disease presentation is heterogeneous, and the factors influencing disease outcome are not fully understood. We used Collaborative Cross (CC) mice to assess the impact of host genetic factors on POWV pathogenesis. We infected a panel of Oas1b-null CC lines with POWV and observed a range of susceptibility, indicating that host factors other than the well-characterized flavivirus restriction factor Oas1b modulate POWV pathogenesis in CC mice. Among the Oas1b-null CC lines, we identified multiple highly susceptible lines (0% survival), including CC071 and CC015, and two resistant lines, CC045 and CC057 (>75% survival). The susceptibility phenotypes generally were concordant among neuroinvasive flaviviruses, although we did identify one line, CC006, that was specifically resistant to JEV, suggesting that both pan-flavivirus and virus-specific mechanisms contribute to susceptibility phenotypes in CC mice. We found that POWV replication was restricted in bone marrow-derived macrophages from CC045 and CC057 mice, suggesting that resistance could result from cell-intrinsic restriction of viral replication. Although serum viral loads at 2 days postinfection were equivalent between resistant and susceptible CC lines, clearance of POWV from the serum was significantly enhanced in CC045 mice. Furthermore, CC045 mice had significantly lower viral loads in the brain at 7 days postinfection than did CC071 mice, suggesting that reduced central nervous system (CNS) infection contributes to the resistant phenotype of CC045 mice.
IMPORTANCE Neuroinvasive flaviviruses, such as WNV, JEV, and POWV, are transmitted to humans by mosquitoes or ticks and can cause neurologic diseases, such as encephalitis, meningitis, and paralysis, and they can result in death or long-term sequelae. Although potentially severe, neuroinvasive disease is a rare outcome of flavivirus infection. The factors that determine whether someone develops severe disease after a flavivirus infection are not fully understood, but host genetic differences in polymorphic antiviral response genes likely contribute to the outcome of infection. We evaluated a panel of genetically diverse mice and identified lines with distinct outcomes following infection with POWV. We found that resistance to POWV pathogenesis corresponded to reduced viral replication in macrophages, more rapid clearance of virus in peripheral tissues, and reduced viral infection in the brain. These susceptible and resistant mouse lines will provide a system for investigating the pathogenic mechanisms of POWV and identifying polymorphic host genes that contribute to resistance.
KEYWORDS: Collaborative Cross mice, neuroinvasive flaviviruses, Powassan virus
INTRODUCTION
Neuroinvasive flaviviruses, such as West Nile virus (WNV), Japanese encephalitis virus (JEV), St. Louis encephalitis virus (SLEV), tick-borne encephalitis virus (TBEV), and Powassan virus (POWV), are transmitted to humans by mosquitoes or ticks and can spread from the circulation into the central nervous system (CNS) (1, 2). Flavivirus infections exhibit a heterogenous presentation, with approximately 80% of infections being asymptomatic and approximately 20% of infections presenting with febrile symptoms. A subset of individuals with symptomatic WNV, JEV, SLEV, TBEV, or POWV infection progress to severe neuroinvasive disease (e.g., encephalitis, meningitis, or paralysis), which can be fatal or lead to long-term cognitive and functional sequelae. Neurologic diseases can result from direct viral infection of neurons or the inflammatory response triggered by viral infection of the CNS. However, the factors influencing susceptibility to severe neuroinvasive disease remain incompletely understood.
POWV is an emerging tick-borne flavivirus within the tick-borne encephalitis serocomplex, and is transmitted by the same Ixodes ticks that transmit Lyme disease (3). POWV is the only tick-borne flavivirus found in North America. Like other tick-borne diseases, the incidence of POWV infection is increasing (4). POWV was first isolated from the brain of a young boy who died of encephalitis in 1958 in Powassan, Ontario, Canada (5). 40 years later, a virus sharing 94% amino acid identity with POWV was isolated from a deer tick (Ixodes scapularis) and was named deer tick virus (DTV) (6, 7). POWV circulates as two distinct, but serologically indistinguishable, genotypes: Lineage I containing the prototype POWV, and Lineage II, containing DTV (6–9). Infection with POWV can have devastating impacts, as approximately 10% of reported encephalitic cases of POWV are fatal, and over 50% of survivors experience long-term cognitive and functional sequelae (10).
Flavivirus infection in humans is characterized by significant variations in disease severity, suggesting that host genetic factors impact the probability and outcome of neuroinvasive disease (11–17). Host genes related to the antiviral immune response have been associated with the outcomes of flavivirus infections in humans (17). For example, polymorphisms in the dsRNA sensor OAS1 and the chemokine receptor CCR5 are associated with WNV and TBEV infection, symptomatic presentation, and neuroinvasive disease (11, 15, 18, 19). Flavivirus resistance is one of the earliest examples of a genetic determinant of pathogen susceptibility defined in mice. In the 1930s, resistance to flavivirus disease was shown to be inherited in mice (20), and in the 2000s, resistance was mapped to the 2′-5′ oligoadenylate synthetase 1b (Oas1b) gene (21, 22). The antiviral activity of Oas1b restricts all flaviviruses tested and appears to act exclusively against flaviviruses. Genetic resistance to tick-borne flavivirus disease was demonstrated in the 1930s via the selective breeding of mouse lines that were either resistant or susceptible to TBEV and louping ill virus (23, 24), and similar studies demonstrated differential susceptibility to the mosquito-borne flaviviruses SLEV and yellow fever virus (25–29).
The Collaborative Cross (CC) is a mouse genetic reference population of recombinant inbred lines. These lines were generated by crossing eight founder lines, representing three wild-derived and five classical laboratory mouse lines, and then independently inbreeding families deriving from these funnel breeding schemes (30, 31). The CC captures the genetic diversity of laboratory mice, which is roughly on par with the levels of common human genetic variation, in a reproducible manner, as each of the 63 lines has a known and fixed genome, thereby providing a valuable tool for mapping complex traits (30–34). As such, the CC enables the identification and study of polymorphic host genes underlying complex phenotypes, including the immune response to viral infection (30–36). Further, since each line is inbred, the CC can be used to facilitate the study of phenotypes that are diverse and dynamic through time (such as the response to infection) in a reproducible manner.
Common laboratory mouse lines (including the CC founder lines C57BL/6J, A/J, 129, NOD, and NZO) have truncated Oas1b alleles that lack 30% of the C-terminal sequence due to a premature stop codon, whereas wild-derived lines (including CC founder lines WSB, PWK, and CAST) each have unique full-length Oas1b alleles, meaning the CC lines carry either a full-length or truncated allele of Oas1b. Previous studies using F1 hybrids of CC mice to define genetic determinants of WNV pathogenesis found via genetic mapping that Oas1b had a major impact on WNV disease outcome (37, 38). However, the mechanism by which Oas1b restricts flavivirus infection remains unclear, as both full-length and truncated Oas1b proteins lack synthetase activity (39, 40). However, full-length Oas1b does inhibit Oas1a synthetase activity and reduces 2′-5′ linked oligoadenylate production (39).
In this study, we used CC mice to investigate the effect of host genetics on disease outcomes that follow neuroinvasive flavivirus infections. We found that a panel of Oas1bnull CC lines had a range of susceptibility phenotypes following POWV infections, and we used susceptible and resistant lines to investigate mechanisms of POWV pathogenesis. We found that resistance to POWV pathogenesis corresponded to reduced viral replication in macrophages, more rapid virus clearance in peripheral tissues, and reduced viral infection in the brain. These findings reveal diverse pathological outcomes of POWV infection in CC mice and suggest that rapid clearance of POWV in the periphery contributes to reduced neuroinvasion and resistance to lethality. These susceptible and resistant mouse lines will provide a system for investigating the pathogenic mechanisms of POWV and identifying polymorphic host genes that contribute to resistance.
RESULTS
Oas1b restricts the pathogenesis of neuroinvasive flaviviruses.
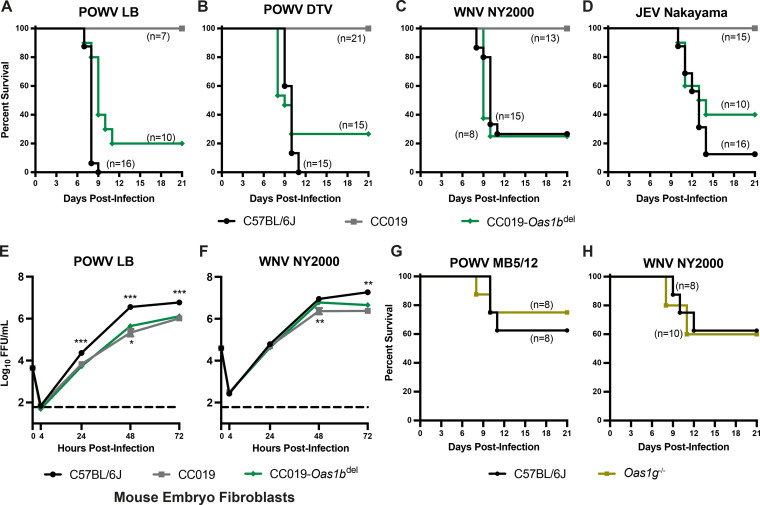
To assess the effect of Oas1b on neuroinvasive flavivirus pathogenesis in CC mice, we infected three strains of mice: wild-type C57BL/6J (nonfunctional Oas1b allele), CC019 (functional Oas1b allele derived from the WSB founder strain), and an Oas1bdel line on a CC019 background that was generated via CRISPR/Cas-9 gene editing (see Materials and Methods). We infected 5 to 6-week-old mice with 100 FFU of POWV (strains LB or DTV Spooner), WNV, or JEV, and we monitored survival for 21 days (Fig. 1A–D). As expected, CC019 mice were resistant to all three viruses, consistent with a strong effect of Oas1b on susceptibility to neuroinvasive flaviviruses. We found that CC019-Oas1bdel mice were susceptible to POWV LB, POWV DTV, WNV, and JEV (80%, 73%, 75%, and 60% lethality, respectively). To determine whether the genetic determinants of susceptibility in mice corresponded to differences in viral replication, we performed multistep growth curves in primary mouse embryo fibroblasts (MEFs). We generated MEFs from C57BL/6J, CC019, and CC019-Oas1bdel mice that were infected with POWV or WNV at a MOI of 0.01 and measured viral titers in the culture supernatant over 72 h (Fig. 1E and F). POWV replication was significantly higher in the MEFs that were derived from C57BL/6J mice, compared to CC019-Oas1bdel mice, starting at 24 hpi (Fig. 1E), with a maximum difference of 8-fold at 48 hpi and the WNV-infected MEFs only being significantly different at 72 hpi (4-fold) (Fig. 1F). POWV and WNV replication in CC019-Oas1bdel MEFs were only modestly increased compared to CC019 MEFs (2-fold and 3-fold, respectively), and this was observed only at 48 hpi. Taken together, these results suggest that non-Oas1b genetic factors are responsible for the differences in viral replication in MEFs.
FIG 1.
Oas1b restricts the pathogenesis of diverse neuroinvasive flaviviruses. (A–D). Five to six-week-old male and female CC019, CC019-Oas1bdel, and C57BL/6J mice were infected with 100 FFU of POWV strain LB (A), POWV DTV strain Spooner (B), WNV strain NY2000 (C), or JEV strain Nakayama (D) via subcutaneous inoculation in the footpad. Lethality was monitored for 21 days. The data are combined from 4 to 5 experiments per virus. E and F. Mouse embryo fibroblasts (MEFs) were harvested from the indicated mouse lines for multistep growth curve analysis. MEFs were infected at a MOI of 0.01 with POWV strain LB (E) or WNV strain NY2000 (F). Supernatants were collected at 4, 24, 48, or 72 h postinfection and titered via focus-forming assay on Vero cells. The results shown are the mean ± SEM of two to three independent experiments that were performed in duplicate or triplicate. Asterisks represent statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001) via a two-way analysis of variance (ANOVA) compared to CC019-Oas1bdel. (G and H). 9 to 12-week-old C57BL/6J wild-type or Oas1g−/− male and female mice were infected with 100 FFU of POWV strain MB5/12 (G) or WNV strain NY2000 (H) via subcutaneous inoculation in the footpad. Lethality was monitored for 21 days.
Humans have four paralogous OAS genes: OAS1, OAS2, OAS3, and OASL. However, mice have single copies of Oas2 and Oas3, two copies of OasL, and eight copies of Oas1 (Oas1a-Oas1h) (40). To determine whether other Oas1 paralogs play a similar role to that of Oas1b in restricting flavivirus pathogenesis, we infected 9 to 12-week-old Oas1g−/− mice (C57BL/6N background) and wild-type mice (C57BL/6J background) with POWV strain MB5/12 or WNV and monitored lethality for 21 days. These older mice were more resistant to POWV and WNV, compared to the five to six week-old mice that were used in the previous experiments, but we found no significant difference in survival for the Oas1g−/− mice compared to the wild-type mice (Fig. 1G and H), suggesting that the function of Oas1b as a flavivirus restriction factor is not conserved among all Oas1 paralogs.
Oas1b-null Collaborative Cross lines exhibit a range of susceptibility to neuroinvasive flaviviruses.
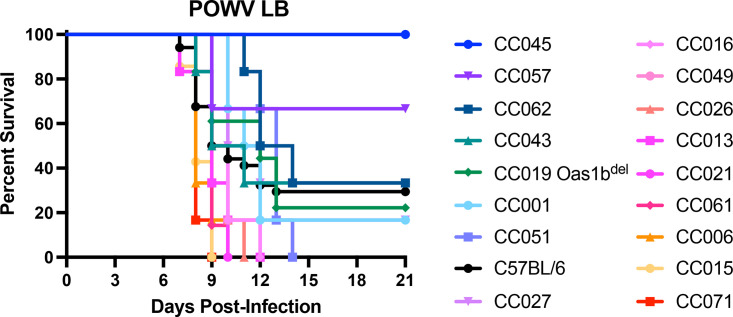
To investigate the role of host genetic factors outside the well-known flavivirus restriction factor Oas1b, we infected mice from a panel of 16 CC strains (9 to 12-week-old mice, all lines carrying Oas1bnull alleles), as well as CC019-Oas1bdel mice, with POWV strain LB and monitored lethality for 21 days (Fig. 2; Table 1). As expected, most lines were highly susceptible to POWV (100% lethality in 11 of 17 CC lines). However, we identified one resistant line (CC045, 0% lethality) and five lines with intermediate susceptibility (CC001, CC027, CC043, CC057, CC062; 50 to 83% lethality).
FIG 2.
Host factors influence POWV pathogenesis across Oas1b-null Collaborative Cross mouse lines. 9 to 12-week-old male and female mice were infected with 100 FFU of POWV (strain LB) via subcutaneous inoculation in the footpad. Lethality was monitored for 21 days. The lines are ordered by their mean time to death. N = 34 C67BL/6J mice, 18 CC019-Oas1bdel mice, and 6 to 7 mice for the other lines. The data are combined from 9 experiments.
TABLE 1.
Collaborative Cross mouse lines used in this studya
| CC line | Oas1b allele | POWV LB (Fig. 2) |
POWV MB5/12 (Fig. 3) |
||
|---|---|---|---|---|---|
| % Lethality | MTD | % Lethality | MTD | ||
| CC001 | Null | 83 | 12.7 | - | - |
| CC003 | WSB | - | - | 0 | NA |
| CC004 | PWK | - | - | 0 | NA |
| CC006 | Null | 100 | 8.8 | 56 | 15.9 |
| CC013 | Null | 100 | 9.3 | - | - |
| CC015 | Null | 100 | 8.3 | 100 | 9.3 |
| CC016 | Null | 100 | 10.3 | - | - |
| CC019-Oas1bdel | Null | 78 | 12.9 | 38 | 16.5 |
| CC021 | Null | 100 | 9.2 | - | - |
| CC026 | Null | 100 | 9.5 | - | - |
| CC027 | Null | 83 | 12.3 | - | - |
| CC030 | WSB | - | - | 0 | NA |
| CC043 | Null | 67 | 13.2 | - | - |
| CC045 | Null | 0 | NA | 22 | 18.4 |
| CC049 | Null | 100 | 10.2 | - | - |
| CC051 | Null | 100 | 12.7 | - | - |
| CC057 | Null | 33 | 17.0 | 0 | NA |
| CC061 | Null | 100 | 9.1 | - | - |
| CC062 | Null | 67 | 15.2 | - | - |
| CC071 | Null | 100 | 8.2 | 100 | 9.1 |
CC, Collaborative Cross; MTD, mean time to death (days); -, not done; NA, not applicable.
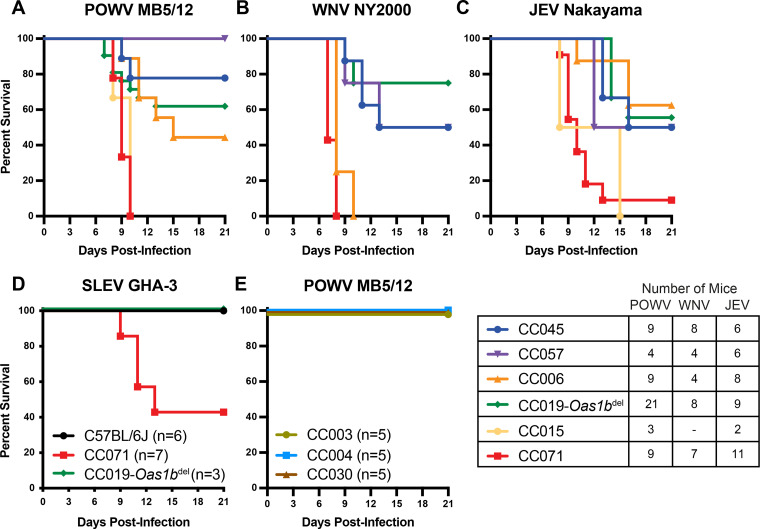
To validate the phenotypes observed with POWV strain LB, we infected a subset of the Oas1bnull CC lines with POWV strain MB5/12, WNV strain NY2000, JEV strain Nakayama, or St. Louis encephalitis virus (SLEV) strain GHA-3 and monitored lethality for 21 days (Fig. 3A–D). CC071 mice were highly susceptible to all viruses tested, with 100% lethality being observed after infection with POWV MB5/12 (Fig. 3A) or WNV (Fig. 3B) and 91% lethality being observed in JEV-infected mice (Fig. 3C). CC071 mice exhibited 57% lethality after SLEV infection, which was remarkable, as C57BL/6J and CC019-Oas1bdel mice exhibited no lethality after SLEV infection (Fig. 3D). Furthermore, CC045 mice, which were the most resistant CC line to POWV LB (0% lethality) (Fig. 2), also were relatively resistant to POWV MB5/12 (22% lethality) (Fig. 3A), WNV (50% lethality) (Fig. 3B), and JEV (50% lethality) (Fig. 3C). Thus, most susceptibility phenotypes were concordant among POWV, WNV, and JEV. However, some lines exhibited virus-specific susceptibility. CC006 mice were highly susceptible to WNV (100% lethality) (Fig. 3B) and intermediately susceptible to POWV MB5/12 (56% lethality) (Fig. 3A), yet this was the most resistant CC line to JEV (37% lethality) (Fig. 3C). This suggests that there are both pan-flavivirus and virus-specific mechanisms that control susceptibility to neuroinvasive flaviviruses. We also evaluated CC lines with functional Oas1b alleles (CC003, WSB allele; CC004, PWK allele; and CC030, WSB allele). As expected, these mice were resistant to POWV MB5/12 infection (0% lethality) (Fig. 3E), further supporting that functional Oas1b alleles derived from different CC founder lines restrict neuroinvasive flavivirus pathogenesis in CC mice.
FIG 3.
Susceptibility phenotypes in Collaborative Cross mice are shared among diverse neuroinvasive flaviviruses. (A–D). 9 to 12-week-old male and female mice from Oas1b-null CC lines were infected with 100 FFU of POWV strain MB5/12 (A), WNV strain NY2000 (B), JEV strain Nakayama (C), or SLEV strain GHA-3 (D) via subcutaneous inoculation in the footpad. Lethality was monitored for 21 days. The data are combined from 4 to 8 experiments per virus. (E) 9 to 12-week-old male mice from Oas1b+/+ CC lines were infected with 100 FFU of POWV strain MB5/12. Lethality was monitored for 21 days. The data represent a single experiment.
The susceptibility of Collaborative Cross lines to neuroinvasive flavivirus pathogenesis does not correlate with early viremia levels.
To uncover the pathogenic mechanisms behind the differences in flavivirus susceptibility among Oas1bnull CC lines, we investigated whether resistance to lethality corresponded with decreased viremia. We measured viral loads in the serum via quantitative reverse transcription-PCR (qRT-PCR) in serum collected at 2 dpi from Oas1bnull CC lines (CC045, CC057, CC006, CC019-Oas1bdel, CC015, and CC071) that had been infected with POWV, WNV, or JEV (Fig. 4). Surprisingly, we found no concordance between viremia (Fig. 4A, C, and D) and susceptibility (Fig. 3A–C) among the Oas1bnull CC lines that were tested. Furthermore, the viral loads in the serum were similar between Oas1bnull CC mice (Fig. 4A) and Oas1b+/+ CC mice (Fig. 4B). Moreover, within CC lines, there was no difference in viremia between mice that survived (open symbols), compared to mice that succumbed to infection (closed symbols). These data suggest that controlling viremia at 2 dpi is not the mechanism of resistance to flavivirus pathogenesis in these CC lines.
FIG 4.
Serum viral loads at 2 dpi do not correlate with susceptibility to neuroinvasive flaviviruses. 9 to 12-week-old male and female mice were infected with 100 FFU of POWV strain MB5/12 (A and B), WNV strain NY2000 (C), or JEV strain Nakayama (D) via subcutaneous inoculation in the footpad. The mice were bled at 2 dpi, and viremia was assessed via qRT-PCR. Asterisks represent statistical significance (*, P < 0.05; ***, P < 0.001) via a one-way ANOVA, compared to CC045 (panels A, C, and D). The CC lines in panels A, C, and D are all Oas1b-null. The CC lines in panel B are Oas1b+/+. Open symbols denote surviving mice. The bar indicates the median.
Resistance to POWV pathogenesis in CC045 mice correlates with the rapid clearance of peripheral infection.
Although we observed no differences in viremia at 2 dpi between susceptible and resistant CC lines, we further investigated whether there were differences in the kinetics of clearance of POWV. To test this, we infected susceptible (CC071 and CC015) and resistant (CC057 and CC045) CC lines and performed serial measurements of viral loads in the serum at 2, 3, and 4 dpi to analyze the kinetics of viremia in individual mice (Fig. 5). Consistent with the results of our previous experiments, we found that CC071 mice had high viral loads at 2 dpi (mean of 6.5 log10 copies/mL), and these peaked at 3 dpi (mean of 7.5 log10 copies/mL) and remained high in 7 of 8 mice at 4 dpi (mean of 6.5 log10 copies/mL) (Fig. 5A). Similarly, CC015 mice had high viral loads at 2 dpi (mean of 8.0 log10 copies/mL), and these peaked at 3 dpi (mean of 8.5 log10 copies/mL) and remained high in all 8 mice at 4 dpi (mean of 7.0 log10 copies/mL) (Fig. 5B). In contrast, CC057 mice had variable viremia at 2 dpi (mean of 6.7 log10 copies/mL), and these remained high at 3 dpi (mean of 6.7 log10 copies/mL) and decreased by 4 dpi (mean of 6.1 log10 copies/mL) (Fig. 5C). Interestingly, whereas CC045 mice had relatively high viral loads in the serum at 2 dpi (mean of 5.7 log10 copies/mL), 6 of 8 mice had undetectable viremia at 3 dpi, and all mice had cleared by 4 dpi (Fig. 5D). These data suggest that although viremia at 2 dpi was not concordant with resistance (Fig. 4), CC045 mice cleared viremia more rapidly than did the susceptible mice (CC071 and CC015) and another resistant line, (CC057).
FIG 5.
CC045 mice clear viremia faster than do susceptible mice and another resistant CC line. 9 to 12-week-old male and female mice from susceptible (A and B) and resistant (C and D) CC lines were infected with 100 FFU of POWV (strain MB5/12) via subcutaneous inoculation in the footpad. The mice were serially bled at 2, 3, and 4 dpi, and viremia was assessed via qRT-PCR. All mice are Oas1b-null. Open symbols denote surviving mice.
Resistance to neuroinvasive flavivirus pathogenesis correlates with reduced replication in macrophages.
To evaluate whether resistance to POWV disease corresponds with cell-intrinsic restriction of viral replication, we generated bone marrow-derived macrophages (BMDM) from CC mice and performed multistep growth curves with POWV and WNV (Fig. 6). BMDMs from CC071, CC015, and CC019-Oas1bdel mice produced significantly higher viral titers of POWV, compared to BMDMs from CC045 mice (699-fold, 23-fold, and 8-fold higher at 72 hpi, respectively) (Fig. 6A), which is concordant with the increased susceptibility of these CC lines to POWV infection (Fig. 3A). Further, BMDMs from CC057 mice had only low levels of POWV replication, similar to CC045 mice.
FIG 6.
Susceptibility to flavivirus disease is concordant with restriction of viral replication in macrophages ex vivo. Bone marrow-derived macrophages (BMDM) were harvested from CC mice (CC045, CC057, CC019-Oas1bdel, CC015, and CC071) for multistep growth curve analysis. BMDMs were infected at a MOI of 0.01 with POWV strain MB5/12 (A) or WNV strain NY2000 (B). Supernatants were collected at 4, 24, 48, or 72 h postinfection and were titered via focus-forming assay on Vero cells. The results shown are the mean ± SEM of two to three independent experiments that were performed in duplicate or triplicate. Asterisks represent statistical significance (*, P < 0.05; ***, P < 0.001) via a two-way ANOVA, compared to CC045.
Similarly, BMDMs from CC015 mice produced significantly higher viral titers of WNV through 48 hpi, compared to BMDMs from CC045 mice (13-fold higher at 48 hpi) (Fig. 6B). Although viral titers from CC071 BMDMs were 5-fold and 4-fold higher than from CC045 BMDMs at 24 and 48 hpi, respectively, these differences were not statistically significant (Fig. 6B). BMDMs from CC019-Oas1bdel mice produced significantly lower viral titers of WNV, compared to BMDMs from CC045 mice (39-fold lower at 72 hpi) (Fig. 6B), which is in concordance with the enhanced resistance of CC019-Oas1bdel mice to WNV observed in vivo (75% survival) (Fig. 3B). Surprisingly, BMDMs from CC057 mice also supported reduced WNV replication, compared to BMDMs from CC045 mice (389-fold lower at 72 hpi) (Fig. 6B), despite equivalent resistance in vivo (Fig. 3B). Taken together, these data suggest that resistance to flavivirus disease in CC mice could result from the cell-intrinsic restriction of viral replication in macrophages, a key cellular target of flaviviruses in vivo. Although we observed similar trends between POWV and WNV replication in several CC lines, suggesting pan-flavivirus restriction mechanisms, we also observed notable differences, suggesting that there may be virus-specific mechanisms as well.
Resistance to POWV pathogenesis correlates with the rapid clearance of peripheral infection and lower CNS viral loads but can be overcome by bypassing the blood-brain barrier.
To further investigate the pathogenic mechanisms of POWV infection, we infected susceptible (CC071) and resistant (CC045) CC lines with POWV and measured the viral loads in the serum, spleen, brain, and spinal cord at 3 and 7 dpi. CC071 mice exhibited high viremia at 3 dpi (mean of 7.9 log10 copies/mL of serum), and POWV RNA was detected in the serum of all 8 of the CC071 mice that were harvested at 7 dpi (Fig. 7A). However, 2 of 5 CC045 mice had cleared POWV from the serum by 3 dpi, viremia was low in the remaining CC045 mice (maximum of 5.7 log10 copies/mL of serum), and POWV RNA was not detected in the serum of any of the 5 CC045 mice that were harvested at 7 dpi (Fig. 7A). These data in terminally bled mice are concordant with the viremia kinetics that were observed in individual CC045 and CC071 mice (Fig. 5A and D), further demonstrating the enhanced clearance of POWV in CC045 mice. Despite a 365-fold difference in the serum viral loads at 3 dpi between CC071 and CC045 mice, we found no significant difference in the spleen viral loads at 3 dpi (Fig. 7B). In contrast, by 7 dpi, all of the CC045 mice had cleared POWV from the spleen, whereas CC071 mice had viral loads in the spleen that were similar to those observed at 3 dpi (Fig. 7B), concordant with the sustained viremia in CC071 mice through 7 dpi (Fig. 7A).
FIG 7.
POWV resistance is concordant with less frequent CNS invasion but can be overcome by bypassing the blood-brain barrier. 9 to 12-week-old male and female CC071 (susceptible) and CC045 (resistant) mice were infected with 100 FFU of POWV (strain MB5/12) via subcutaneous inoculation in the footpad. (A–C) At the indicated time points, the mice were perfused, and their tissues were harvested. (A) Mice were bled via cardiac puncture prior to perfusion, and viremia was assessed via qRT-PCR. (B and C) Spleen, brain, and spinal cord homogenates were titered via plaque assay on Vero cells. (D) 9 to 12-week-old male and female CC071 (n = 4) and CC045 (n = 6) mice were infected with 10 FFU of POWV (strain MB5/12) via intracranial inoculation. Lethality was monitored for 19 days. Asterisks represent statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001) via a two-way ANOVA, compared to CC045. The bars indicate the medians.
We also assessed viral loads in the CNS and detected no POWV in the brains of CC071 or CC045 mice at 3 dpi (data not shown), but, by 7 dpi, all CC071 mice had high viral loads in the brain (mean of 6.3 log10 PFU/mL) (Fig. 7C). Interestingly, whereas 4 of 6 CC045 mice had no detectable virus in the brain at 7 dpi, the remaining 2 mice had brain viral loads that were similar to those of CC071 mice (maximum of 6.3 log10 PFU/mL) (Fig. 7C). The observation that 33% of CC045 mice had detectable virus in the brain at 7 dpi (Fig. 7C) is concordant with our observation that CC045 mice had 22% lethality to POWV strain MB5/12 (Fig. 3A). We did not detect POWV in the spinal cords of CC071 mice at 3 dpi (data not shown), but, by 7 dpi, 2 of 9 CC071 mice had high viral loads in the spinal cord (maximum of 7.8 log10 PFU/mL) (Fig. 7D). These 2 mice also had the highest brain viral loads, which is consistent with a model in which POWV spreads to the spinal cord from the brain, rather than by separate stochastic breaches of the blood-brain barrier (BBB). We did not detect virus in the spinal cords of CC045 mice at 3 dpi (data not shown) or 7 dpi (Fig. 7C). To distinguish between reduced neuroinvasion and reduced neurovirulence as mechanisms for the resistant phenotype of CC045 mice, we infected CC071 and CC045 mice with POWV intracranially and monitored lethality. Both CC071 and CC045 mice were highly susceptible to intracranial infection (100% and 83% lethality, respectively) (Fig. 7D), demonstrating that the resistant phenotype of CC045 mice could be overcome by bypassing neuroinvasion. Taken together, these data support a model of POWV pathogenesis in which the rapid clearance of viremia reduces the likelihood of the virus breaching the BBB and accessing the CNS, where viral infection results in mortality.
DISCUSSION
In this study, we investigated the effect of host genetics on disease outcomes following neuroinvasive flavivirus infection. We found that a panel of Oas1bnull CC lines had a range of susceptibility phenotypes following POWV infection, indicating that polymorphic host genes other than Oas1b contribute to disease outcome after POWV infection. We identified Oas1bnull CC lines that are susceptible (100% lethality) or resistant (<25% lethality) to POWV and used these lines to investigate mechanisms of POWV pathogenesis. We observed reduced POWV and WNV replication in primary macrophages that were derived from resistant mice, suggesting that resistance to flavivirus disease in CC mice could result from the cell-intrinsic restriction of viral replication in macrophages. We found no differences in POWV viremia between susceptible and resistant CC mice at 2 dpi, but we did find that resistant mice cleared POWV from the periphery rapidly, whereas susceptible mice maintained high viremia throughout the infection. Furthermore, we observed significant differences in viral loads in the brains of susceptible and resistant CC mice, following POWV infection. These findings reveal diverse outcomes of POWV infection in CC mice and suggest that the rapid clearance of POWV in the periphery contributes to reduced neuroinvasion and protection from lethality.
Neurotropic viruses, including neuroinvasive flaviviruses, can cause disease by directly damaging infected neurons and stimulating pathogenic inflammatory responses (41–44). The pathogenic mechanisms of neuroinvasive flaviviruses have been studied extensively in mice, which recapitulate key features of human disease, such as neuronal infection, immune infiltration into the CNS, paralysis, encephalitis, and cognitive loss. Much of this work has focused on WNV, but more recent studies have investigated POWV pathogenesis in mice; these have studied POWV disease using laboratory mouse lines, such as C57BL/6J and BALB/c, as well as Peromyscus mice, which are thought to serve as reservoirs for Lineage II POWV (DTV) in nature (45–50). C57BL/6J mice are highly susceptible to POWV, which makes them useful for modeling severe human disease but makes them less useful for defining the mechanisms that allow most infected individuals to sustain mild or asymptomatic infections while a small subset of infected individuals develop severe neuroinvasive disease. Comparing POWV infection in resistant CC lines (e.g., CC045) to susceptible lines (e.g., CC071) has the potential to reveal which aspects of POWV infection and the resulting host immune response (e.g., viral loads in the periphery, persistent viremia, neuroinvasion, replication within the CNS, damage to CNS neurons, neuroinflammation, etc.) correlate with severe neurologic disease. CC mice can be useful models of relevant disease presentations that are not evident in conventional laboratory mouse lines, such as chronic WNV disease (51) or encephalitis caused by Rift Valley fever virus (35).
Taken together, our results support a model in which CC045 mice are resistant to severe POWV disease due to reduced viral replication in myeloid cells and rapid clearance of viremia, which thereby reduce the probability of neuroinvasion. This resistance is independent of a role for Oas1b in restricting flavivirus pathogenesis. This model suggests that the resistance mechanism of CC045 mice acts in peripheral tissues, not within the CNS. The clearance of arboviruses from the circulation occurs via a variety of mechanisms, including the binding of virions by antibodies, lectins, and scavenger receptors, and it is an important determinant of whether a host contributes to viral transmission to feeding arthropods (52). Future studies will determine whether the rapid clearance of viremia in resistant CC lines results solely from the reduced viral replication in myeloid cells or whether CC lines also vary in serum clearance activity.
Although the mechanisms by which flaviviruses cross the blood-brain barrier and invade the CNS remain incompletely understood (53), hematogenous neuroinvasion is likely somewhat stochastic, and prolonged high viremia (e.g., in CC071 mice) increases the probability of the virus crossing the BBB. We expect that once any POWV accesses the CNS, it encounters a highly permissive and sensitive environment, thereby resulting in uniform lethality. Accordingly, CC045 mice were susceptible to POWV when the BBB was bypassed via intracranial inoculation, and the approximately 22% lethality that we observed after subcutaneous inoculation is consistent with the detection of POWV in the brains of 2 of 6 CC045 mice at 7 dpi. The model that CC045 resistance results from the rapid clearance of viremia is somewhat at odds with the observation that susceptibility did not correlate with viremia at 2 dpi, either within or among CC lines. However, this model is supported by the distinct viremia kinetics between CC045 and CC071 mice, and it suggests that rapid clearance, but not peak viremia, is a key determinant of POWV susceptibility in CC mice. Future studies will characterize viral replication in cell types other than macrophages as well as compare CNS immune infiltrates and CNS pathology in CC045 mice versus CC071 mice.
In this study, we demonstrated that Oas1b restricts POWV pathogenesis, which was unsurprising, given that Oas1b has been shown to restrict all of the flaviviruses that have been tested to date. Despite observing marked differences in survival between CC019-Oas1bdel and CC019 mice, we found no significant difference in WNV or POWV replication in CC019-Oas1bdel MEFs, compared to CC019 MEFs, even though Oas1b has been shown to restrict flavivirus replication in MEFs that were derived from C3H.PRI-Flvr mice (54). This discordance could be due to distinct genetic features of CC019 mice and differences between MEFs and other cell types. We also demonstrated the ability to generate genetic knockouts on a CC background (CC019-Oas1bdel), which will enable the study of the function of single genes in the context of genetically diverse mouse models.
Host genes that are related to the antiviral immune response have been associated with the outcomes of flavivirus infections in humans (17). For example, a common polymorphism that ablates the expression of the chemokine receptor CCR5 (CCR5Δ32, which is best characterized by homozygotes being protected against HIV infection because CCR5 is the main coreceptor for HIV entry [55]) is associated with a higher risk of WNV and TBEV symptomatic presentation and neuroinvasive disease (11, 15, 18, 19). A protective role for CCR5 against WNV and TBEV disease is consistent with the results of studies showing that CCR5-deficient mice exhibit impaired trafficking of the CD8 T cells that are necessary to clear flavivirus CNS infections (56–59). Furthermore, polymorphisms in the dsRNA sensors OAS1, OAS2, and OAS3 are associated with WNV infection, TBEV infection, and neuroinvasive disease (11, 60, 61). The SNP rs10774671 of OAS1 corresponds to an A→G change in a splice acceptor site, where the G allele (protective) generates the p46 isoform of OAS1. The p46 isoform is prenylated, localizes to flavivirus replication complexes on ER membranes, and inhibits WNV replication, whereas the p42 isoform (resulting from the A allele) is not prenylated and lacks antiviral activity (62). Among the eight murine orthologs of OAS1, Oas1b and Oas1g both encode a C-terminal CaaX domain that is homologous to the prenylation site in the p46 isoform of human OAS1 (62). Whereas Oas1b plays a dominant role in restricting flavivirus pathogenesis in mice, we found no effect of Oas1g on survival after a WNV or POWV infection. This could indicate that Oas1g is not important for controlling flavivirus infections, that the antiviral effects of Oas1g are not strong enough to affect lethality, or that the antiviral effects of Oas1g are not evident on the Oas1bnull C57BL/6J genetic background.
Polymorphisms within additional antiviral response genes (e.g., CD209/DC-SIGN, TLR3, IL-10) are associated with TBEV infection (12–16). Similarly, polymorphisms within antiviral response genes, such as HERC5, IRF3, and MX1, are associated with WNV infection (11, 17, 63, 64). Other polymorphic host genes play a role in flavivirus infection, but their effects on human disease are less clear. TMEM41B is an ER-associated lipid scramblase that promotes the replication of a wide variety of flaviviruses (including POWV, TBEV, and WNV) (65). TMEM41B is polymorphic in humans, and TMEM41B alleles vary in their abilities to support flavivirus replication in cell cultures (65). However, the associations with the risk or outcome of a flavivirus infection remain to be demonstrated.
Extensive studies using transgenic knockout mice have revealed the effects of various innate and adaptive immune genes on the pathogenesis of WNV and other flaviviruses (66), but investigating flavivirus pathogenesis in CC mice allows us to study complex traits and polymorphic alleles that better recapitulate the genetic diversity that is found in human populations (31, 34). A limitation of CC studies is that they can only reveal genetic factors that are polymorphic among the 8 CC founder lines (or private mutations that arose during the breeding of the CC lines). Future studies will use F2 crosses of resistant (e.g., CC045) and susceptible (e.g., CC071) CC lines and genetic mapping approaches to define the host genetic factors that contribute to the resistant phenotype of CC045 mice, analogous to previous studies that have used this approach with CC mice to map QTL and the underlying causal genes that contribute to the host control of the influenza A virus, SARS-CoV, SARS-CoV-2, and WNV (36, 38, 67–69) as well as similar approaches that were used for TBEV (70).
Previous studies have used CC mice to investigate the host genetic factors controlling WNV infection and pathogenesis (37, 38, 51, 71–73). These studies used F1 crosses of the CC parental lines, so it is not straightforward to compare the phenotypes that we identified in the CC parental lines. Further, our key lines of interest, CC045 and CC071, were not included in the earlier WNV studies. However, the QTL with the largest effect size that was identified in these studies mapped to Oas1b (37, 38). With this in mind, we designed our experiments to use only Oas1bnull CC lines, thereby allowing us to identify other polymorphic genes that were contributing to the disease outcomes. However, our design does not detect factors that are dependent upon or that synergize with Oas1b for their activities. Since we expected the Oas1bnull mice to be susceptible to neuroinvasive flaviviruses, the remarkable finding in our study was that CC045 mice were resistant to diverse neuroinvasive flaviviruses, even in the absence of Oas1b. We also identified other Oas1bnull CC lines (such as CC057) with more modest resistance phenotypes. Future studies will investigate whether the resistant phenotype of CC057 mice derives from the same mechanism as that of CC045 mice. Notably, CC057 mice were also relatively resistant to RVFV, exhibiting a delayed disease course that resulted in encephalitis rather than acute hepatitis (35). CC071 mice, which we identified as being highly susceptible to POWV, WNV, JEV, and SLEV, have previously been found to be highly susceptible to SARS-CoV-2 (74), RVFV (35), and rat hepacivirus (75), and they were highly susceptible to ZIKV when treated with an IFNAR1-blocking antibody (76). Taken together, this suggests that CC mice can reveal immune mechanisms that control the pathogenesis of diverse viruses.
MATERIALS AND METHODS
Cells and viruses.
Vero (African green monkey kidney epithelial) cells were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 5% heat-inactivated FBS at 37°C with 5% CO2. The POWV strains LB (Lineage I) and Spooner (DTV, Lineage II), WNV strain NY2000, JEV strain Nakayama, and SLEV strain GHA-3 were provided by Michael Diamond (Washington University, St. Louis). The POWV strain MB5/12 (DTV, Lineage II) was provided by Greg Ebel (Colorado State University). All of the viruses were handled under BSL3 containment. Virus stocks were grown in Vero cells and titered via focus-forming assay (FFA) (77). Duplicates of serial 10-fold dilutions of viruses in growth medium (DMEM containing 2% FBS and 20 mM HEPES) were applied to Vero cells in 96-well plates and were incubated at 37°C with 5% CO2. After 1 h, the cells were overlaid with 1% methylcellulose in Eagle’s minimum essential medium (MEM) containing 2% heat-inactivated fetal bovine serum (FBS). Following incubation for approximately 24 h (WNV), 24 to 36 h (JEV), or 48 h (POWV and SLEV), the plates were fixed with 2% paraformaldehyde for 2 h at room temperature. The fixed plates were incubated with 500 ng/mL flavivirus cross-reactive mouse MAb ZV13 (78) or E60 (79) for 2 h at room temperature or overnight at 4°C. After incubation at room temperature for 1 h with a 1:2,500 dilution of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Sigma), foci were detected via the addition of TrueBlue substrate (KPL). The foci were quantified using a CTL Immunospot instrument.
Mice.
All of the mouse procedures were performed under protocols that were approved by the Institutional Animal Care and Use Committee at the University of North Carolina at Chapel Hill. The CC mice were obtained from the UNC Systems Genetics Core Facility directly or as breeder pairs that were bred in-house. C57BL/6J mice were bred in-house. Oas1g−/− mice were obtained from Timothy Sheahan (UNC). All of the mouse work was performed under ABSL3 containment. 5-week-old or 9 to 12-week-old male and female mice were inoculated with a volume of 50 μL via a subcutaneous (footpad) route or with a volume of 20 μL via an intracranial route. For footpad inoculation, mice received 100 FFU of POWV strain LB, MB5/12, or Spooner (DTV), WNV strain NY2000, JEV strain Nakayama, or SLEV strain GHA-3, diluted in HBSS with Ca2+ and Mg2+, and supplemented with 1% heat-inactivated FBS. For intracranial inoculation, mice received 10 FFU of POWV MB5/12. The mice were monitored daily for disease signs for 21 days or until the time of tissue harvest. The mice were euthanized upon reaching humane endpoints, including the loss of ≥20% of the starting weight, nonresponsiveness, or severe neurologic disease signs (hunching, paralysis) that interfered with the ability to access food and water. Euthanized mice were scored as dead the following day. To evaluate viremia, blood was collected at 2 dpi via submandibular bleed with a 5 mm Goldenrod lancet or via cardiac puncture, prior to perfusion, in the tissue harvest experiments.
Generation of CC019-Oas1bdel mice.
CC019-Oas1bdel mice were generated through the UNC Mutant Mouse Resource and Research Center as part of a project to demonstrate the feasibility of performing CRISPR/Cas9 genome editing in CC mice. The CC019 line was chosen because it contains a functional Oas1b allele (derived from the WSB founder) and exhibited robust superovulation and in vitro fertilization (IVF) performance, with egg yields and embryo progression to the two-cell stage being comparable to those of C57BL/6J mice. Four CRISPR guide RNAs targeting the SNP position of the WSB Oas1b allele were designed and validated. Two guide RNAs showed full activity in vitro and were chosen for microinjection. A donor oligonucleotide was designed to introduce the susceptible SNP plus additional silent mutations to disrupt Cas9 binding and the cleavage of the introduced allele as well as to facilitate the genotyping of the resulting animals. Cas9 mRNA (20 or 40 ng/μL), 1 guide RNA (10 or 20 ng/μL), and donor oligonucleotide (20 or 50 ng/μL) were coinjected into the pronucleus of one-cell embryos that were produced via IVF. IVF/microinjection was performed for 3 days. The CC019 strain responded moderately to superovulation (average of 8.3 eggs/female). IVF was successful on each of the 3 days, yielding a total of 358 injectable embryos (6.2/female). The injection survival and progression to the two-cell stage in vitro were comparable to C57BL/6J mice. However, the production of live pups from injected embryos was low, with only 2 live pups being produced from 280 implanted embryos. Both pups had CRISPR-induced mutations at the Oas1b locus. CC019-Oas1bdel mice are homozygous for a 12 bp deletion in exon 4 of Oas1b, which generates an in-frame, 4 amino acid deletion. CC019-Oas1bdel mice were bred as knockout × knockout and exhibited similar breeding performance to that of the parental CC019 line. The CC019-Oas1bdel mice were genotyped by generating a PCR amplicon from tail snip DNA, using the forward primer CCACACACAACCACCAGGAACC and the reverse primer GGCTGTAGGACCTCATGTCAATCA and then sequencing with the forward primer TCTCATTGCCTTTCTCTTCTCAGTGTA. The wild-type sequence is GGGAGTATGGGAGTCCGAGTAACTAAATTCAACACAGCCCAGGGCTTCCGAACCGTCTTGGAACTGGTCACCAAGTACAAACAGCTTCGAATCTACTGGACAGTGTATTATGACTTTCGACATCAAGAGGTCTCTGAATACCTGCACCAA, and the Oas1bdel sequence is GGGAGTATGGGAGTCCGAGTAACTAAATTCAACACAGCCCAGGACTTGGAACTGGTCACCAAGTACAAACAGCTTCGAATCTACTGGACAGTGTATTATGACTTTCGACATCAAGAGGTCTCTGAATACCTGCACCAA.
Mouse embryo fibroblasts.
Mouse embryo fibroblasts (MEFs) were prepared from E15 embryos. Pregnant mice were euthanized, and the gravid uterus was isolated. The embryos were removed, placed in PBS, decapitated, and the gut and liver were removed. The embryos were then minced with scalpels, trypsinized (1 mL per embryo), pipetted up and down with a 10 mL serological pipette to break up any chunks, and incubated for 5 to 10 min at room temperature. The cells were resuspended in DMEM supplemented with nonessential amino acids, l-glutamine, Pen/Strep, and 10% heat-inactivated FBS, and they were then pelleted via centrifugation at 1,000 rpm for 5 min at 4°C. The supernatants were removed, and the cell pellets were resuspended in fresh medium and pelleted again via centrifugation at 1,000 rpm for 5 min at 4°C. The cell pellets were resuspended in 1 mL per embryo of fresh medium and were plated into culture flasks (1.5 embryos per T-150 flask) in 25 mL of fresh medium, and they were incubated at 37°C with 5% CO2. After 24 h, the medium was removed, the cells were washed with 1× PBS, and fresh medium was added. When the monolayers reached near-confluence, the MEFs were frozen down in DMEM supplemented with nonessential amino acids, l-glutamine, Pen/Strep, 30% heat-inactivated FBS, and 20% DMSO, and they were stored in liquid nitrogen. Thawed MEFs were seeded in 6-well plates at 2 × 105 cells per well in DMEM supplemented with nonessential amino acids, l-glutamine, Pen/Strep, and 10% heat-inactivated FBS. The MEFs were infected at a MOI of 0.01 with POWV strain LB or WNV strain NY2000. After 1 h, the inoculum was removed and replaced with fresh medium, and the plates were incubated at 37°C with 5% CO2. After 4, 24, 48, or 72 h, the supernatants were collected and titered via focus-forming assay on Vero cells.
Measurement of viremia.
Blood was collected via submandibular bleed or terminal cardiac puncture in serum separator tubes (BD). Serum was separated via centrifugation at 8,000 rpm for 4 min and stored at −80°C until RNA isolation. RNA was extracted using a Viral RNA Mini Kit (Qiagen). Viral RNA levels were determined via TaqMan one-step qRT-PCR on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad), using standard cycling conditions. Viremia is expressed on a log10 scale as copies per mL, based on a standard curve that was produced using serial 10-fold dilutions of a DNA plasmid that contained a 400 bp gBlock (Integrated DNA Technologies) encoding a portion of the viral envelope (E) protein sequence. All of the primers and probes were purchased from Integrated DNA Technologies. The primers that were used to detect POWV MB5/12 were as follows: forward, GAAGCTGAAAGGCACAACTTAC; reverse, CACCTCCATGACCACTGTATC; and probe, AAGAGTTCCTGTGGACAGTGGTCA. The primers that were used to detect WNV NY2000 were: forward, TCAGCGATCTCTCCACCAAAG; reverse, GGGTCAGCACGTTTGTCATTG; and probe, TGCCCGACCATGGGAGAAGCTC. The primers that were used to detect JEV Nakayama were: forward, CAGCGTGGAGAAACAGAGAA; reverse, TGTGACCCAAGAGCAACAA; and probe, CATGGAATTTGAAGAGGCGCACGC.
Bone marrow-derived macrophages.
Bone marrow-derived macrophages (BMDM) were generated from CC mice. The mice were euthanized, and their femurs and tibias were isolated from their hind limbs. Bone marrow was flushed out with 10 mL DMEM delivered via syringe with a 25G ½ inch needle. The bone marrow was pooled and pipetted up and down using a 5 mL serological pipette to break up large chunks. The cells were pelleted via centrifugation at 1,500 rpm for 5 min at 4°C. Supernatants were removed, and the cell pellets were resuspended in ACK Red Blood Cell Lysis Buffer that contained 150 mM NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA (pH 7.3). Then, they were incubated for 2 to 3 min. The cells were resuspended in DMEM containing 10% heat-inactivated FBS and were then pelleted via centrifugation at 1,500 rpm for 5 min at 4°C. The cell pellets were resuspended in DMEM containing 10% heat-inactivated FBS and counted. 12-well non-TC treated plates were seeded with 1.5 × 105 cells/well in 1 mL of DMEM containing l-glutamine, NaPyr, Pen/Strep, 10% heat-inactivated FBS, and 40 ng/mL mouse M-CSF (BioLegend 576406), and they were incubated for 7 days at 37°C with 5% CO2. BMDMs were infected at a MOI of 0.01 with POWV strain MB5/12 or WNV strain NY2000 in DMEM containing l-glutamine, NaPyr, Pen/Strep, 10% heat-inactivated FBS, and 20 ng/mL mouse M-CSF. After 1 h, the inoculum was removed and replaced with fresh medium, and the plates were incubated at 37°C with 5% CO2. After 4, 24, 48, or 72 h, the supernatants were collected and titered via focus-forming assay on Vero cells.
Tissue titers.
9 to 12-week-old male and female mice were infected with 100 FFU of POWV strain MB5/12. At 3 or 7 dpi, the mice were bled via cardiac puncture and perfused with 20 mL of PBS, and their tissues were harvested. Spleens, brains, and spinal cords were collected into 2 mL screwcap tubes that contained 1 mL (spleens and brains) or 0.5 mL (spinal cords) of DMEM supplemented with 2% heat-inactivated FBS and homogenizer beads. The tissues were stored at −80°C until processing. Whole tissues were thawed and homogenized using a MagNA Lyser (Roche) that was set to 6,000 for 1 min. The homogenates were titered via plaque assay on Vero cells. Serial 10-fold dilutions of the tissue homogenates were applied to Vero cells in 6-well plates and were incubated at 37°C with 5% CO2. After 1 h, the cells were overlaid with 1% methylcellulose in MEM containing 2% heat-inactivated FBS. Following incubation for 6 days, the plates were fixed with 2% paraformaldehyde overnight at room temperature. The fixed plates were stained with 1% crystal violet in 20% ethanol and washed with tap water. The plaques were counted manually.
Data analysis.
The data were analyzed using the GraphPad Prism software package. The growth curves were analyzed via a two-way analysis of variance (ANOVA) to assess the impact of time and the CC line on viral replication, compared to CC019-Oas1bdel (Fig. 1E and F) or CC045 (Fig. 6) mice. Viremia was compared to the CC045 mice via a one-way ANOVA (Fig. 4). The tissue titers were analyzed via a two-way ANOVA to assess the impact of time and the CC line on viral loads, compared to the CC045 (Fig. 7) mice. A P value of <0.05 was considered to be indicative of a statistically significant result.
ACKNOWLEDGMENTS
This work was supported by R21 AI145377 (H.M.L.), R01 AI170625 (H.M.L.), and U19 AI100625 (M.T.H. and M.T.F.), by start-up funds from the UNC Lineberger Comprehensive Cancer Center and Department of Microbiology & Immunology, and by Systems Genetics Pilot Projects from the UNC School of Medicine. B.A.J. was supported by F32 AI161786. K.E.N. was supported by T32 AI007419. We appreciate the support of the Systems Genetics Core Facility and the Mutant Mouse Research and Resource Center. We also acknowledge Dale Cowley and the UNC Animal Models Core Facility for generating the CC019-Oas1bdel mice.
Contributor Information
Helen M. Lazear, Email: helen.lazear@med.unc.edu.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Gould EA, Solomon T. 2008. Pathogenic flaviviruses. Lancet 371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- 2.Solomon T. 2004. Flavivirus encephalitis. N Engl J Med 351:370–378. doi: 10.1056/NEJMra030476. [DOI] [PubMed] [Google Scholar]
- 3.Hermance ME, Thangamani S. 2017. Powassan virus: an emerging arbovirus of public health concern in North America. Vector Borne Zoonotic Dis 17:453–462. doi: 10.1089/vbz.2017.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paules CI, Marston HD, Bloom ME, Fauci AS. 2018. Tickborne diseases - confronting a growing threat. N Engl J Med 379:701–703. doi: 10.1056/NEJMp1807870. [DOI] [PubMed] [Google Scholar]
- 5.McLean DM, Donohue WL. 1959. Powassan virus: isolation of virus from a fatal case of encephalitis. Can Med Assoc J 80:708–711. [PMC free article] [PubMed] [Google Scholar]
- 6.Beasley DW, Suderman MT, Holbrook MR, Barrett AD. 2001. Nucleotide sequencing and serological evidence that the recently recognized deer tick virus is a genotype of Powassan virus. Virus Res 79:81–89. doi: 10.1016/s0168-1702(01)00330-6. [DOI] [PubMed] [Google Scholar]
- 7.Kuno G, Artsob H, Karabatsos N, Tsuchiya KR, Chang GJ. 2001. Genomic sequencing of deer tick virus and phylogeny of Powassan-related viruses of North America. Am J Trop Med Hyg 65:671–676. doi: 10.4269/ajtmh.2001.65.671. [DOI] [PubMed] [Google Scholar]
- 8.Vogels CBF, Brackney DE, Dupuis AP, Robich RM, Fauver JR, Brito AF, Williams SC, Anderson JF, Lubelczyk CB, Lange RE, Prusinski MA, Kramer LD, Gangloff-Kaufmann JL, Goodman LB, Baele G, Smith RP, Armstrong PM, Ciota AT, Dellicour S, Grubaugh ND. 2023. Phylogeographic reconstruction of the emergence and spread of Powassan virus in the northeastern United States. Proc Natl Acad Sci USA 120:e2218012120. doi: 10.1073/pnas.2218012120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebel GD, Spielman A, Telford SR. 2001. Phylogeny of North American Powassan virus. J Gen Virol 82:1657–1665. doi: 10.1099/0022-1317-82-7-1657. [DOI] [PubMed] [Google Scholar]
- 10.Ebel GD. 2010. Update on Powassan virus: emergence of a North American tick-borne flavivirus. Annu Rev Entomol 55:95–110. doi: 10.1146/annurev-ento-112408-085446. [DOI] [PubMed] [Google Scholar]
- 11.Bigham AW, Buckingham KJ, Husain S, Emond MJ, Bofferding KM, Gildersleeve H, Rutherford A, Astakhova NM, Perelygin AA, Busch MP, Murray KO, Sejvar JJ, Green S, Kriesel J, Brinton MA, Bamshad M. 2011. Host genetic risk factors for West Nile virus infection and disease progression. PLoS One 6:e24745. doi: 10.1371/journal.pone.0024745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barkhash AV, Babenko VN, Voevoda MI, Romaschenko AG. 2016. Association of IL28B and IL10 gene polymorphism with predisposition to tick-borne encephalitis in a Russian population. Ticks Tick Borne Dis 7:808–812. doi: 10.1016/j.ttbdis.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 13.Barkhash AV, Perelygin AA, Babenko VN, Brinton MA, Voevoda MI. 2012. Single nucleotide polymorphism in the promoter region of the CD209 gene is associated with human predisposition to severe forms of tick-borne encephalitis. Antiviral Res 93:64–68. doi: 10.1016/j.antiviral.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Barkhash AV, Voevoda MI, Romaschenko AG. 2013. Association of single nucleotide polymorphism rs3775291 in the coding region of the TLR3 gene with predisposition to tick-borne encephalitis in a Russian population. Antiviral Res 99:136–138. doi: 10.1016/j.antiviral.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Kindberg E, Mickiene A, Ax C, Akerlind B, Vene S, Lindquist L, Lundkvist A, Svensson L. 2008. A deletion in the chemokine receptor 5 (CCR5) gene is associated with tickborne encephalitis. J Infect Dis 197:266–269. doi: 10.1086/524709. [DOI] [PubMed] [Google Scholar]
- 16.Mickienė A, Pakalnienė J, Nordgren J, Carlsson B, Hagbom M, Svensson L, Lindquist L. 2014. Polymorphisms in chemokine receptor 5 and Toll-like receptor 3 genes are risk factors for clinical tick-borne encephalitis in the Lithuanian population. PLoS One 9:e106798. doi: 10.1371/journal.pone.0106798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cahill ME, Conley S, DeWan AT, Montgomery RR. 2018. Identification of genetic variants associated with dengue or West Nile virus disease: a systematic review and meta-analysis. BMC Infect Dis 18:282. doi: 10.1186/s12879-018-3186-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glass WG, McDermott DH, Lim JK, Lekhong S, Yu SF, Frank WA, Pape J, Cheshier RC, Murphy PM. 2006. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J Exp Med 203:35–40. doi: 10.1084/jem.20051970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim JK, McDermott DH, Lisco A, Foster GA, Krysztof D, Follmann D, Stramer SL, Murphy PM. 2010. CCR5 deficiency is a risk factor for early clinical manifestations of West Nile virus infection but not for viral transmission. J Infect Dis 201:178–185. doi: 10.1086/649426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster LT. 1937. Inheritance of resistance of mice to enteric bacterial and neurotropic virus infections. J Exp Med 65:261–286. doi: 10.1084/jem.65.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perelygin AA, Scherbik SV, Zhulin IB, Stockman BM, Li Y, Brinton MA. 2002. Positional cloning of the murine flavivirus resistance gene. Proc Natl Acad Sci USA 99:9322–9327. doi: 10.1073/pnas.142287799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mashimo T, Lucas M, Simon-Chazottes D, Frenkiel MP, Montagutelli X, Ceccaldi PE, Deubel V, Guenet JL, Despres P. 2002. A nonsense mutation in the gene encoding 2'-5'-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc Natl Acad Sci USA 99:11311–11316. doi: 10.1073/pnas.172195399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Webster LT. 1933. Inherited and acquired factors in resistance to iInfection: I. Development of resistant and susceptible lines of mice through selective breeding. J Exp Med 57:793–817. doi: 10.1084/jem.57.5.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casals J, Schneider HA. 1943. Natural resistance and susceptibility to Russian spring-summer encephalitis in mice. Proceedings of the Society for Experimental Biology and Medicine 54:201–202. doi: 10.3181/00379727-54-14367. [DOI] [Google Scholar]
- 25.Webster LT, Clow AD. 1936. Experimental encephalitis (St. Louis Type) in mice with high inborn resistance: aAchronic subclinical infection. J Exp Med 63:827–845. doi: 10.1084/jem.63.6.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Webster LT, Johnson MS. 1941. Comparative virulence of St. Louis encephalitis virus cultured with brain tissue from innately susceptible and innately resistant mice. J Exp Med 74:489–494. doi: 10.1084/jem.74.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sawyer WA, Lloyd W. 1931. The use of mice in tests of immunity against yellow fever. J Exp Med 54:533–555. doi: 10.1084/jem.54.4.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch CJ, Hughes TP. 1936. The inheritance of susceptibility to yellow fever encephalitis in mice. Genetics 21:104–112. doi: 10.1093/genetics/21.2.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brinton MA, Perelygin AA. 2003. Genetic resistance to flaviviruses. Adv Virus Res 60:43–85. doi: 10.1016/S0065-3527(03)60002-3. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Cross C. 2012. The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics 190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noll KE, Ferris MT, Heise MT. 2019. The Collaborative Cross: a systems genetics resource for studying host-pathogen interactions. Cell Host Microbe 25:484–498. doi: 10.1016/j.chom.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Srivastava A, Morgan AP, Najarian ML, Sarsani VK, Sigmon JS, Shorter JR, Kashfeen A, McMullan RC, Williams LH, Giusti-Rodriguez P, Ferris MT, Sullivan P, Hock P, Miller DR, Bell TA, McMillan L, Churchill GA, de Villena FP. 2017. Genomes of the mouse Collaborative Cross. Genetics 206:537–556. doi: 10.1534/genetics.116.198838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graham JB, Swarts JL, Mooney M, Choonoo G, Jeng S, Miller DR, Ferris MT, McWeeney S, Lund JM. 2017. Extensive homeostatic T cell phenotypic variation within the Collaborative Cross. Cell Rep 21:2313–2325. doi: 10.1016/j.celrep.2017.10.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leist SR, Baric RS. 2018. Giving the genes a shuffle: using natural variation to understand host genetic contributions to viral infections. Trends Genet 34:777–789. doi: 10.1016/j.tig.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartwright HN, Barbeau DJ, Doyle JD, Klein E, Heise MT, Ferris MT, McElroy AK. 2022. Genetic diversity of collaborative cross mice enables identification of novel Rift Valley fever virus encephalitis model. PLoS Pathog 18:e1010649. doi: 10.1371/journal.ppat.1010649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schafer A, Leist SR, Gralinski LE, Martinez DR, Winkler ES, Okuda K, Hawkins PE, Gully KL, Graham RL, Scobey DT, Bell TA, Hock P, Shaw GD, Loome JF, Madden EA, Anderson E, Baxter VK, Taft-Benz SA, Zweigart MR, May SR, Dong S, Clark M, Miller DR, Lynch RM, Heise MT, Tisch R, Boucher RC, Pardo Manuel de Villena F, Montgomery SA, Diamond MS, Ferris MT, Baric RS. 2022. A multitrait locus regulates Sarbecovirus pathogenesis. mBio 13:e0145422. doi: 10.1128/mbio.01454-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graham JB, Thomas S, Swarts J, McMillan AA, Ferris MT, Suthar MS, Treuting PM, Ireton R, Gale M, Jr, Lund JM. 2015. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. mBio 6:e00493-15–e00415. doi: 10.1128/mBio.00493-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Green R, Wilkins C, Thomas S, Sekine A, Hendrick DM, Voss K, Ireton RC, Mooney M, Go JT, Choonoo G, Jeng S, de Villena FP, Ferris MT, McWeeney S, Gale M, Jr. 2017. Oas1b-dependent immune transcriptional profiles of West Nile virus infection in the Collaborative Cross. G3 (Bethesda) 7:1665–1682. doi: 10.1534/g3.117.041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elbahesh H, Jha BK, Silverman RH, Scherbik SV, Brinton MA. 2011. The Flvr-encoded murine oligoadenylate synthetase 1b (Oas1b) suppresses 2-5A synthesis in intact cells. Virology 409:262–270. doi: 10.1016/j.virol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kakuta S, Shibata S, Iwakura Y. 2002. Genomic structure of the mouse 2',5'-oligoadenylate synthetase gene family. J Interferon Cytokine Res 22:981–993. doi: 10.1089/10799900260286696. [DOI] [PubMed] [Google Scholar]
- 41.Shives KD, Tyler KL, Beckham JD. 2017. Molecular mechanisms of neuroinflammation and injury during acute viral encephalitis. J Neuroimmunol 308:102–111. doi: 10.1016/j.jneuroim.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milora KA, Rall GF. 2019. Interferon control of neurotropic viral infections. Trends Immunol 40:842–856. doi: 10.1016/j.it.2019.07.005. [DOI] [PubMed] [Google Scholar]
- 43.Klein RS, Hunter CA. 2017. Protective and pathological immunity during central nervous system infections. Immunity 46:891–909. doi: 10.1016/j.immuni.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klein RS, Garber C, Howard N. 2017. Infectious immunity in the central nervous system and brain function. Nat Immunol 18:132–141. doi: 10.1038/ni.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone ET, Hassert M, Geerling E, Wagner C, Brien JD, Ebel GD, Hirsch AJ, German C, Smith JL, Pinto AK. 2022. Balanced T and B cell responses are required for immune protection against Powassan virus in virus-like particle vaccination. Cell Rep 38:110388. doi: 10.1016/j.celrep.2022.110388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scroggs SLP, Offerdahl DK, Stewart PE, Shaia C, Griffin AJ, Bloom ME. 2023. Of murines and humans: modeling persistent Powassan disease in C57BL/6 mice. mBio 14:e0360622. doi: 10.1128/mbio.03606-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Santos RI, Hermance ME, Reynolds ES, Thangamani S. 2021. Salivary gland extract from the deer tick, Ixodes scapularis, facilitates neuroinvasion by Powassan virus in BALB/c mice. Sci Rep 11:20873. doi: 10.1038/s41598-021-00021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mlera L, Meade-White K, Saturday G, Scott D, Bloom ME. 2017. Modeling Powassan virus infection in Peromyscus leucopus, a natural host. PLoS Negl Trop Dis 11:e0005346. doi: 10.1371/journal.pntd.0005346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hermance ME, Thangamani S. 2015. Tick saliva enhances Powassan virus transmission to the host, influencing its dissemination and the course of disease. J Virol 89:7852–7860. doi: 10.1128/JVI.01056-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pesko KN, Torres-Perez F, Hjelle BL, Ebel GD. 2010. Molecular epidemiology of Powassan virus in North America. J Gen Virol 91:2698–2705. doi: 10.1099/vir.0.024232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Graham JB, Swarts JL, Wilkins C, Thomas S, Green R, Sekine A, Voss KM, Ireton RC, Mooney M, Choonoo G, Miller DR, Treuting PM, Pardo Manuel de Villena F, Ferris MT, McWeeney S, Gale M, Jr, Lund JM. 2016. A mouse model of chronic West Nile virus disease. PLoS Pathog 12:e1005996. doi: 10.1371/journal.ppat.1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ander SE, Li FS, Carpentier KS, Morrison TE. 2022. Innate immune surveillance of the circulation: a review on the removal of circulating virions from the bloodstream. PLoS Pathog 18:e1010474. doi: 10.1371/journal.ppat.1010474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cain MD, Salimi H, Diamond MS, Klein RS. 2019. Mechanisms of pathogen invasion into the central nervous system. Neuron 103:771–783. doi: 10.1016/j.neuron.2019.07.015. [DOI] [PubMed] [Google Scholar]
- 54.Scherbik SV, Paranjape JM, Stockman BM, Silverman RH, Brinton MA. 2006. RNase L plays a role in the antiviral response to West Nile virus. J Virol 80:2987–2999. doi: 10.1128/JVI.80.6.2987-2999.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lopalco L. 2010. CCR5: from natural resistance to a new anti-HIV strategy. Viruses 2:574–600. doi: 10.3390/v2020574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glass WG, Lim JK, Cholera R, Pletnev AG, Gao JL, Murphy PM. 2005. Chemokine receptor CCR5 promotes leukocyte trafficking to the brain and survival in West Nile virus infection. J Exp Med 202:1087–1098. doi: 10.1084/jem.20042530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larena M, Regner M, Lobigs M. 2012. The chemokine receptor CCR5, a therapeutic target for HIV/AIDS antagonists, is critical for recovery in a mouse model of Japanese encephalitis. PLoS One 7:e44834. doi: 10.1371/journal.pone.0044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Durrant DM, Daniels BP, Pasieka T, Dorsey D, Klein RS. 2015. CCR5 limits cortical viral loads during West Nile virus infection of the central nervous system. J Neuroinflammation 12:233. doi: 10.1186/s12974-015-0447-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Michlmayr D, Bardina SV, Rodriguez CA, Pletnev AG, Lim JK. 2016. Dual function of Ccr5 during Langat virus encephalitis: reduction in neutrophil-mediated central nervous system inflammation and increase in T cell-mediated viral clearance. J Immunol 196:4622–4631. doi: 10.4049/jimmunol.1502452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim JK, Lisco A, McDermott DH, Huynh L, Ward JM, Johnson B, Johnson H, Pape J, Foster GA, Krysztof D, Follmann D, Stramer SL, Margolis LB, Murphy PM. 2009. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog 5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barkhash AV, Perelygin AA, Babenko VN, Myasnikova NG, Pilipenko PI, Romaschenko AG, Voevoda MI, Brinton MA. 2010. Variability in the 2'-5'-oligoadenylate synthetase gene cluster is associated with human predisposition to tick-borne encephalitis virus-induced disease. J Infect Dis 202:1813–1818. doi: 10.1086/657418. [DOI] [PubMed] [Google Scholar]
- 62.Soveg FW, Schwerk J, Gokhale NS, Cerosaletti K, Smith JR, Pairo-Castineira E, Kell AM, Forero A, Zaver SA, Esser-Nobis K, Roby JA, Hsiang TY, Ozarkar S, Clingan JM, McAnarney ET, Stone AE, Malhotra U, Speake C, Perez J, Balu C, Allenspach EJ, Hyde JL, Menachery VD, Sarkar SN, Woodward JJ, Stetson DB, Baillie JK, Buckner JH, Gale M, Jr, Savan R. 2021. Endomembrane targeting of human OAS1 p46 augments antiviral activity. Elife 10. doi: 10.7554/eLife.71047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Danial-Farran N, Eghbaria S, Schwartz N, Kra-Oz Z, Bisharat N. 2015. Genetic variants associated with susceptibility of Ashkenazi Jews to West Nile virus infection. Epidemiol Infect 143:857–863. doi: 10.1017/S0950268814001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long D, Deng X, Singh P, Loeb M, Lauring AS, Seielstad M. 2016. Identification of genetic variants associated with susceptibility to West Nile virus neuroinvasive disease. Genes Immun 17:298–304. doi: 10.1038/gene.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoffmann HH, Schneider WM, Rozen-Gagnon K, Miles LA, Schuster F, Razooky B, Jacobson E, Wu X, Yi S, Rudin CM, MacDonald MR, McMullan LK, Poirier JT, Rice CM. 2021. TMEM41B is a pan-Flavivirus host factor. Cell 184:133–148. doi: 10.1016/j.cell.2020.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manet C, Roth C, Tawfik A, Cantaert T, Sakuntabhai A, Montagutelli X. 2018. Host genetic control of mosquito-borne Flavivirus infections. Mamm Genome 29:384–407. doi: 10.1007/s00335-018-9775-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferris MT, Aylor DL, Bottomly D, Whitmore AC, Aicher LD, Bell TA, Bradel-Tretheway B, Bryan JT, Buus RJ, Gralinski LE, Haagmans BL, McMillan L, Miller DR, Rosenzweig E, Valdar W, Wang J, Churchill GA, Threadgill DW, McWeeney SK, Katze MG, Pardo-Manuel de Villena F, Baric RS, Heise MT. 2013. Modeling host genetic regulation of influenza pathogenesis in the Collaborative Cross. PLoS Pathog 9:e1003196. doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gralinski LE, Ferris MT, Aylor DL, Whitmore AC, Green R, Frieman MB, Deming D, Menachery VD, Miller DR, Buus RJ, Bell TA, Churchill GA, Threadgill DW, Katze MG, McMillan L, Valdar W, Heise MT, Pardo-Manuel de Villena F, Baric RS. 2015. Genome wide identification of SARS-CoV susceptibility loci using the Collaborative Cross. PLoS Genet 11:e1005504. doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maurizio PL, Ferris MT, Keele GR, Miller DR, Shaw GD, Whitmore AC, West A, Morrison CR, Noll KE, Plante KS, Cockrell AS, Threadgill DW, Pardo-Manuel de Villena F, Baric RS, Heise MT, Valdar W. 2018. Bayesian diallel analysis reveals Mx1-dependent and Mx1-independent effects on response to influenza avirus in mice. G3 (Bethesda) 8:427–445. doi: 10.1534/g3.117.300438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palus M, Sohrabi Y, Broman KW, Strnad H, Šíma M, Růžek D, Volkova V, Slapničková M, Vojtíšková J, Mrázková L, Salát J, Lipoldová M. 2018. A novel locus on mouse chromosome 7 that influences survival after infection with tick-borne encephalitis virus. BMC Neurosci 19:39. doi: 10.1186/s12868-018-0438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Green R, Wilkins C, Thomas S, Sekine A, Ireton RC, Ferris MT, Hendrick DM, Voss K, de Villena FP, Baric R, Heise M, Gale M, Jr. 2016. Identifying protective host gene expression signatures within the spleen during West Nile virus infection in the collaborative cross model. Genom Data 10:114–117. doi: 10.1016/j.gdata.2016.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Green R, Wilkins C, Thomas S, Sekine A, Ireton RC, Ferris MT, Hendrick DM, Voss K, Pardo-Manuel de Villena F, Baric RS, Heise MT, Gale M, Jr. 2016. Transcriptional profiles of WNV neurovirulence in a genetically diverse Collaborative Cross population. Genom Data 10:137–140. doi: 10.1016/j.gdata.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Graham JB, Swarts JL, Menachery VD, Gralinski LE, Schafer A, Plante KS, Morrison CR, Voss KM, Green R, Choonoo G, Jeng S, Miller DR, Mooney MA, McWeeney SK, Ferris MT, Pardo-Manuel de Villena F, Gale M, Heise MT, Baric RS, Lund JM. 2020. Immune predictors of mortality after ribonucleic acid virus infection. J Infect Dis 221:882–889. doi: 10.1093/infdis/jiz531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montagutelli X, Prot M, Jouvion G, Levillayer L, Conquet L, Reyes-Gomez E, Donati F, Albert M, van der Werf S, Jaubert J, Simon-Lorière E. 2021. A mouse-adapted SARS-CoV-2 strain replicating in standard laboratory mice. bioRxiv. doi: 10.1101/2021.07.10.451880:2021.07.10.451880. [DOI]
- 75.Brown AJ, Won JJ, Wolfisberg R, Fahnøe U, Catanzaro N, West A, Moreira FR, Batista MN, Ferris MT, Linnertz CL, Leist SR, Nguyen C, GDl C, Midkiff BR, Xia Y, Montgomery SA, Billerbeck E, Bukh J, Scheel TKH, Rice CM, Sheahan TP. 2023. Host genetic variation guides hepacivirus clearance, chronicity, and liver fibrosis in mice. bioRxiv. doi: 10.1101/2023.03.18.533278:2023.03.18.533278. [DOI] [PMC free article] [PubMed]
- 76.Manet C, Simon-Loriere E, Jouvion G, Hardy D, Prot M, Conquet L, Flamand M, Panthier JJ, Sakuntabhai A, Montagutelli X. 2020. Genetic diversity of Collaborative Cross mice controls viral replication, clinical severity, and brain pathology induced by Zika virus infection, independently of Oas1b. J Virol 94. doi: 10.1128/JVI.01034-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brien JD, Lazear HM, Diamond MS. 2013. Propagation, quantification, detection, and storage of West Nile virus. Curr Protoc Microbiol 31:15D.3.1–15D.3.18. [DOI] [PubMed] [Google Scholar]
- 78.Zhao H, Fernandez E, Dowd KA, Speer SD, Platt DJ, Gorman MJ, Govero J, Nelson CA, Pierson TC, Diamond MS, Fremont DH. 2016. Structural basis of Zika virus-specific antibody protection. Cell 166:1016–1027. doi: 10.1016/j.cell.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi BE, Olshevsky U, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol 80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]