Abstract
The growth physiology of Escherichia coli during colonization of the intestinal tract was studied with four animal models: the streptomycin-treated mouse carrying a reduced microflora, the monoassociated mouse with no other microflora than the introduced strain, the conventionalized streptomycin-treated mouse, and the conventionalized monoassociated mouse harboring a full microflora. A 23S rRNA fluorescent oligonucleotide probe was used for hybridization to whole E. coli cells fixed directly after being taken from the animals, and the respective growth rates of E. coli BJ4 in the four animal models were estimated by correlating the cellular concentrations of ribosomes with the growth rate of the strain. The growth rates thus estimated from the ribosomal content of E. coli BJ4 in vivo did not differ in the streptomycin-treated and the monoassociated mice. After conventionalization there was a slight decrease of the bacterial growth rates in both animal models.
Bacterial colonization of the mammalian gut plays two important roles. On one hand, it provides protection against pathogens and carries out digestive processes. On the other hand, it is a prerequisite of virulence for many pathogenic bacteria. The intestinal tract represents a complex ecosystem, harboring 400 to 500 different bacterial species. Densities of up to 1012 bacteria per g of contents have been reported (1, 5). This indigenous flora constitutes a highly competitive environment and a colonization barrier (3, 6, 8, 20, 24), which decreases the probability that ingested bacteria will be able to establish themselves in the intestinal ecosystem of a healthy host. It is important to understand how bacteria colonize the gut, how they find their optimal niche, and how they may be excreted as a consequence of the appearance of a new organism having more effective colonization factors, such as specific adhesion capacity, better utilization of nutrients, or faster growth (9).
In order to examine the influence of the microflora on the growth of a single species, four different animal models were investigated: germfree mice, streptomycin-treated mice, conventionalized germfree mice, and conventionalized streptomycin-treated mice. Germfree mice have no intestinal microflora and are excellent models for studies of single bacteria since there is no competition from other bacterial species, which makes it very easy for an incoming bacterium to establish itself, proliferate, and persist in the gastrointestinal tract at a high density. A streptomycin-treated mouse is a conventional mouse which is treated with streptomycin, after which most facultative aerobic, gram-negative rods, normally constituting only 0.1 to 1% of the flora, are removed (10). This is sufficient to remove the colonization barrier for incoming enteric bacteria, such as Escherichia coli, which will grow quickly to a level of 107 to 108 CFU/g of feces; this level is 10- to 50-fold lower than levels found in monoassociated animals but still 104-fold higher than those found in conventional animals (10, 19). Conventionalized mice received oral inoculation of a homogenate containing cecal contents from a conventional mouse.
In the present investigation (as in previous investigations [13, 18, 19]), we have monitored colonization and excretion of E. coli BJ4 in mouse intestines. E. coli BJ4 was originally isolated from a healthy Wistar rat at the Institute of Toxicology, National Food Agency, Copenhagen, Denmark. A streptomycin-resistant isolate which was found to be identical to its parental strain with respect to biochemical reactions and serology was used (12). The influence of the normal microflora on this bacterial species in a conventional gastrointestinal environment was investigated by addition of a complete mouse intestinal flora to monoassociated and inoculated, streptomycin-treated mice.
The strong correlation between cell growth rate and rRNA content (2, 11, 21) makes it possible to use quantitative in situ hybridization with fluorescent rRNA probes to estimate growth rates of single cells (16, 18, 19). For this estimation, an in vitro standard curve, based on suspended cultures of the organism under investigation growing at different rates in different media, is required. The cellular fluorescence of bacteria in samples from the respective animals was compared to that described by the standard curve, and generation times were estimated. Detailed protocols for such determinations have been presented previously (13, 17–19).
Eleven each of germfree and streptomycin-treated, albino, adult male mice of the NMRI/KI strain were used in the study. The mouse strain originates from the Institute fur Versuchstierzucht, Hanover, Germany, and has been inbred for 37 generations at the Department of Medical Microbial Ecology, Karolinska Institute, Stockholm, Sweden. The germfree mice were kept under standard conditions (22 ± 2°C; 50% ± 10% relative humidity; light-dark cycle, 12 h-24 h) as described previously (4). Eleven conventional mice were given drinking water containing 5 g of streptomycin sulfate per liter (starting 24 h before the colonization experiment), until the time of conventionalization (6 days after inoculation). Thereafter, they received tap water with no streptomycin added.
The germfree mice were orally dosed with a 0.1-ml aliquot of a stationary-phase E. coli BJ4 suspension containing 5 × 106 CFU/ml. The streptomycin-treated mice were given 0.1 ml containing 100-fold more cells to ensure colonization. Before oral dosing, aliquots of homogenized fecal samples (negative controls) were fixed for hybridization (13, 18, 19). Six days after inoculation, the mice (germfree and streptomycin treated) were conventionalized per os with 100 μl of a homogenate of cecal contents isolated from a conventional mouse, diluted 10-fold in 0.9% NaCl. The homogenate contained 1.3 × 106 E. coli cells/ml. No streptomycin-resistant colonies were detected in the homogenate. Two mice of each animal model were sacrificed at 1, 3, and 5 h and on days 7 and 10. On day 15, one mouse of each model was sacrificed. From each of the sacrificed mice, samples of feces (excluding the 1-, 3-, and 5-h samplings, since the bacteria reach the cecum faster than they do the feces), cecal contents, and cecal mucus were taken for CFU determination and fixation. The cecal samplings were carried out as described previously (13, 19).
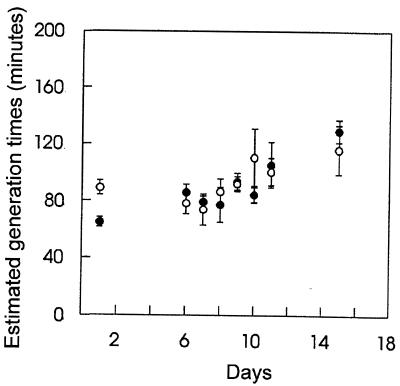
When the ribosomal probe EC 1531, which is specific for E. coli 23S rRNA (18, 19), was used for quantification of the fluorescence in E. coli BJ4 in fecal samples from the streptomycin-treated and the monoassociated mice, it was estimated that the generation time was constant from days 1 to 6 in both animal models, varying between 70 and 89 min (Fig. 1). This relatively fast growth of E. coli BJ4 in the two animal models is consistent with previous findings for streptomycin-treated mice (19). After inoculation with a normal cecal flora on day 6, the estimated generation times of the E. coli cells increased to 130 min for the streptomycin-treated and 116 min for the monoassociated mice on day 15. Statistical analysis revealed that the increasing trend was significant, although weak. The similarity of the estimated growth rates of E. coli BJ4 in monoassociated mice and streptomycin-treated mice indicates that the large number of bacteria in the streptomycin-treated mice had no influence on the growth of the E. coli BJ4 strain.
FIG. 1.
Estimated generation times (means ± standard errors of the means) of E. coli BJ4 in feces from streptomycin-treated (closed circles) and the monoassociated (open circles) mice, as determined by hybridization of fixed fecal samples. Animals were orally inoculated with E. coli BJ4 on day 0. On day 6, they were conventionalized by oral inoculation of a homogenate containing cecal contents from a conventional mouse.
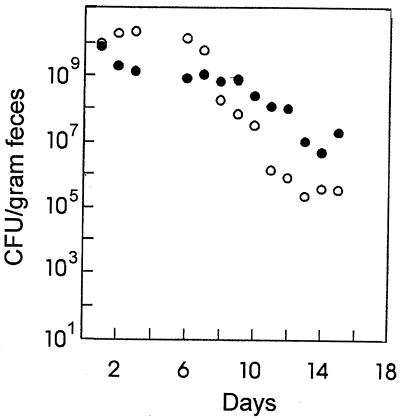
The population sizes of E. coli BJ4 had reached steady states of 1.2 × 1010 CFU/g of feces in the monoassociated mice and 7.1 × 108 CFU/g in the streptomycin-treated mice by day 6 (Fig. 2). At day 6 after inoculation, the mice were conventionalized with a normal cecal flora, leading to a fast reduction of E. coli BJ4 levels in both animal models. The decrease in BJ4 levels in the monoassociated mice was particularly high; the number of CFU per gram was reduced from 1.2 × 1010 to a new stable level of 2.1 × 105 in 8 days (Fig. 2), a drop of 4 orders of magnitude, consistent with the studies by Freter et al. (9). However, in the same time period, in conventionalized streptomycin-treated mice (Fig. 2) the BJ4 level dropped less than 100-fold to a level of 107 CFU/g.
FIG. 2.
Observed concentration of E. coli BJ4 cells before and after conventionalization of streptomycin-treated (closed circles) and monoassociated (open circles) mice. Mice were conventionalized on day 6 by oral inoculation of a homogenate containing cecal contents from a conventional mouse.
The prevalent theory as to how bacteria colonize the mammalian gut is that all species can coexist as long as each member of the flora is able to utilize one or a few limiting nutrients better than all the others and that the rate of bacterial growth during the colonization process is greater than the washout rate from the intestine (8, 9). It is also possible for a species which does not compete well for limiting nutrients to avoid washout and to colonize if it is able to adhere to the intestinal wall (8, 9).
In the streptomycin-treated mouse, E. coli BJ4 should occupy its normal narrow nutrient niche (22, 23) and, once established, be able to compete well with incoming enterics. In the germfree mouse, however, E. coli BJ4 should be free to utilize any nutrient, including many for which it would compete very poorly against normal intestinal residents. Moreover, the flora in the streptomycin-treated mouse should make it more difficult for incoming bacteria to grow and colonize, whereas in the germfree mouse no such barrier exists. Therefore, if the colonization theory is correct, it would be expected that E. coli BJ4 in the streptomycin-treated mouse model would be more resistant to administration of the cecal flora of a conventional mouse than BJ4 would be in the germfree mouse model. This is, in fact, observed in the present experiments (Fig. 2). Since the estimated growth rates of BJ4 in the two models decreased after conventionalization but were essentially identical to each other at all times (Fig. 1), the more rapid loss of BJ4 in the conventionalized germfree mice was independent of growth rate.
The data presented here show that E. coli grows in the mouse gut at rates that are quite similar whether the animal is monoassociated (germfree) or streptomycin treated (clearing the major groups of gram-negative bacteria); i.e., in both models doubling times of about 75 min were observed (Fig. 2). The data also show that in both the streptomycin-treated mouse model and the monoassociated mouse model, the estimated doubling times of E. coli BJ4 increased to between 116 and 130 min by nine days after conventionalization (Fig. 1). In streptomycin-treated mice, E. coli grows well in the intestinal mucus layer, which covers the epithelial cells, but not in the luminal contents in vivo and in vitro (14, 25). Moreover, nonmotile and nonchemotactic E. coli mutants, when parents and mutants are fed simultaneously to streptomycin-treated mice, colonize just as well as their parents (15). Since the intestinal population stays constant, it seems likely that E. coli must grow in the mucus layer at a rate similar to that with which it is sloughed into the luminal contents. If it is assumed that there is no specific adhesion to any permanent surfaces in the intestine (18), the rate of release of the mucus should determine the growth rate of the bacteria. If so, the present experiments suggest that conventionalization leads to a decrease in the rate of mucus sloughing. Alternatively, it is possible that E. coli can grow slower than the rate of mucus sloughing and still maintain a stable population if a large proportion of bacteria is continuously pulled back into the mucus layer as water is resorbed to form fecal pellets. In any event, the decrease in the bacterial growth rate is in fact quite small, and the actual biological impact of such a decrease remains unclear.
The different densities of E. coli in the different mouse models are most likely caused by differences in nutrient availabilities. The spectrum of potential nutrients for one species depends strictly on the competition from the other species present. Finally, growth may be limited to specific parts of the intestinal system in some but not all of the different animal models; for example, it has been found that in germfree mice E. coli grows very well in the intestinal contents, whereas, as stated above, in streptomycin-treated (and most likely also conventional) mice, growth in the contents is severely inhibited (7, 14).
In conclusion, several factors appear to be important in defining the growth conditions for bacteria in an ecosystem like the mouse large intestine, factors such as nutrient availability, flow rates, adhesion to permanent sites, presence of inhibitors, competition from coresidents, etc. The in situ methods useful for characterization of subpopulations of bacteria in complex environments are important tools in building up a better understanding of the significance of these parameters.
Acknowledgments
We thank Anna-Carin Persson and Johannes Bergstedt, Karolinska Institute, for excellent assistance with the animals.
We acknowledge the financial support provided by NorFa (Nordic Academy for Advanced Study), Oslo, Norway.
REFERENCES
- 1.Borriello S P. Microbial flora of the gastrointestinal tract. In: Hill M J, editor. Microbial metabolism in the digestive tract. Boca Raton, Fla: CRC Press, Inc.; 1986. pp. 2–16. [Google Scholar]
- 2.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- 3.Dubos R, Schadler R W, Costello R. Composition, alterations, and effects of the intestinal flora. Fed Proc. 1963;22:1322–1329. [PubMed] [Google Scholar]
- 4.El-Bayoumy K, Hecht S S, Hoffmann D. Comparative tumor initiating activity on mouse skin of 6-nitrobenzol[a]pyrene, 6-nitrochrysene, 3-nitroperylene, 1-nitropyrene and their parent hydrocarbons. Cancer Lett. 1982;16:333–337. doi: 10.1016/0304-3835(82)90015-5. [DOI] [PubMed] [Google Scholar]
- 5.Finegold S M, Sutter V L, Mathisen G E. Normal indigenous intestinal flora. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press; 1983. pp. 3–31. [Google Scholar]
- 6.Freter R. Experimental enteric Shigella and Vibrio infections in mice and guinea pigs. J Exp Med. 1956;104:411–418. doi: 10.1084/jem.104.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freter R. Possible effects of foreign DNA on pathogenic potential and intestinal proliferation of Escherichia coli. J Infect Dis. 1978;137:624–629. doi: 10.1093/infdis/137.5.624. [DOI] [PubMed] [Google Scholar]
- 8.Freter R. Mechanisms that control the microflora in the large intestine. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press; 1983. pp. 33–54. [Google Scholar]
- 9.Freter R, Brickner H, Fekete J, Vickerman M M, Carey K E. Survival and implantation of Escherichia coli in the intestinal tract. Infect Immun. 1983;39:686–703. doi: 10.1128/iai.39.2.686-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hentges D J. Role of the intestinal microflora in host defense against infection. In: Hentges D J, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press; 1983. pp. 311–331. [Google Scholar]
- 11.Kjeldgaard N O, Kurland C G. The distribution of soluble and ribosomal RNA as a function of growth rate. J Mol Biol. 1963;6:341–348. [Google Scholar]
- 12.Krogfelt K A, Poulsen L K, Molin S. Identification of coccoid Escherichia coli BJ4 cells in the large intestine of streptomycin-treated mice. Infect Immun. 1993;61:5029–5034. doi: 10.1128/iai.61.12.5029-5034.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Licht T R, Krogfelt K A, Cohen P S, Poulsen L K, Urbance J, Molin S. Role of lipopolysaccharide in colonization of the mouse intestine by Salmonella typhimurium studied by in situ hybridization. Infect Immun. 1996;64:3811–3817. doi: 10.1128/iai.64.9.3811-3817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Licht T R, Tolker-Nielsen T, Holmstrøm K, Krogfelt K A, Molin S. Inhibition of Escherichia coli precursor-16S processing by mouse intestinal contents. Environ Microbiol. 1999;1:23–32. doi: 10.1046/j.1462-2920.1999.00001.x. [DOI] [PubMed] [Google Scholar]
- 15.McCormick B A, Laux D C, Cohen P S. Neither motility nor chemotaxis plays a role in the ability of Escherichia coli F-18 to colonize the streptomycin-treated mouse large intestine. Infect Immun. 1990;58:2957–2961. doi: 10.1128/iai.58.9.2957-2961.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Møller S, Kristensen C S, Poulsen L K, Carstensen J M, Molin S. Bacterial growth on surfaces: automated image analysis for quantification of growth rate-related parameters. Appl Environ Microbiol. 1995;61:741–748. doi: 10.1128/aem.61.2.741-748.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poulsen L K, Ballard G, Stahl D A. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl Environ Microbiol. 1993;59:1354–1360. doi: 10.1128/aem.59.5.1354-1360.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poulsen L K, Lan F, Kristensen C S, Hobolth P, Molin S, Krogfelt K A. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect Immun. 1994;62:5191–5194. doi: 10.1128/iai.62.11.5191-5194.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poulsen L K, Licht T R, Rang C, Krogfelt K A, Molin S. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J Bacteriol. 1995;177:5840–5845. doi: 10.1128/jb.177.20.5840-5845.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salminen S, Isolauri E, Onnela T. Gut flora in normal and disordered states. Chemotherapy (Basel) 1995;41(Suppl. 1):5–15. doi: 10.1159/000239391. [DOI] [PubMed] [Google Scholar]
- 21.Schaechter M, Maaloe O, Kjeldgard N O. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. J Gen Microbiol. 1958;19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 22.Sweeney N J, Klemm P, McCormick B A, Moller-Nielsen E, Utley M, Schembri M A, Laux D C, Cohen P S. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect Immun. 1996;64:3497–3503. doi: 10.1128/iai.64.9.3497-3503.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sweeney N J, Laux D C, Cohen P S. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect Immun. 1996;64:3504–3511. doi: 10.1128/iai.64.9.3504-3511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van der Waaij D, Berghuis-de-Vries J M, Lekkerkerk-van der Wees J E C. Colonization resistance of the digestive tract in conventional and antibiotic treated mice. J Hyg Camb. 1971;69:405–411. doi: 10.1017/s0022172400021653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wadolkowski E A, Laux D C, Cohen P S. Colonization of the streptomycin-treated mouse large intestine by a human fecal Escherichia coli strain: role of growth in mucus. Infect Immun. 1988;56:1030–1035. doi: 10.1128/iai.56.5.1030-1035.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]