Objective
Cytochrome P450 1B1 (CYP1B1) genetic variants are relevant in the pathogenesis of breast cancer. Exploring the relationships between CYP1B1 functional variants and breast cancer could improve our understanding of breast cancer molecular pathophysiology.
Methods
This is a two-stage hospital-based case–control study of a Chinese Han population. Genotyping was performed to identify candidate gene variants. 3DSNP, ANNOVAR, and RegulomeDB were used to determine functional single nucleotide polymorphisms (SNPs). The relationship between candidate variants and breast cancer risk was evaluated through unconditional logistic regression analysis. The PancanQTL platform was used to perform cis and trans expression quantitative trait loci (eQTL) analysis of positive SNPs. The GSCA platform was then used to compare the gene expression levels of potential target genes between breast cancer tissue and normal tissue adjacent to the cancer.
Results
rs10175368-T acted as a protective factor against breast cancer based on an additive model [odds ratio (OR) = 0.722, 95% confidence interval (CI) = 0.613–0.850; P < 0.001], and was identified as a protective factor in the postmenopausal population (OR = 0.601; 95% CI, 0.474–0.764; P < 0.001). eQTL analysis and analysis of differential expression in carcinoma and paracancerous tissues revealed that the expression level of CYP1B1-AS1 was associated with rs10175368 and that CYP1B1-AS1 had significantly higher expression levels in breast cancer tissues than in paracancerous tissues.
Conclusion
We show, for the first time in a Chinese Han population, that the functional variant rs10175368 plays a protective role against breast cancer, especially in the postmenopausal population.
Keywords: breast cancer, case–control study, CYP1B1, genetic variant, susceptibility
Introduction
Breast cancer (Lukina et al., 2021) is currently the most prevalent malignancy in women and the second most common cancer worldwide (Sung et al., 2021). There were approximately 429 105 new cases of breast cancer and 124 002 fatalities among Chinese women in 2022 (Xia et al., 2022). Breast cancer incidence and mortality rates are continually rising, establishing it as a significant public health issue, affecting the physical and emotional well being of women worldwide (Park et al., 2021). Therefore, the identification of precise biomarkers of breast cancer susceptibility is crucial to improving disease prevention and outcome.
The pathophysiology and etiology of breast cancer are still not fully understood. Breast cancer, which is a complex disease, emerges due to a multitude of hereditary and environmental factors, as determined through extensive biochemical and population-based epidemiological research (Lichtenstein et al., 2000; Antoniou and Easton, 2006; Zhang et al., 2012; Jing et al., 2020; Barańska et al., 2022; Sarink et al., 2022). Estrogen is known to play a significant role in the development of breast cancer and it regulates various physiological processes, including cell growth, proliferation, development, and differentiation (Frasor et al., 2003; Kiyama and Wada-Kiyama, 2015). Further, higher estrogen levels may increase the risk of breast cancer (Ali and Coombes, 2000; Clemons and Goss, 2001; Platet et al., 2004). Cytochrome P450 1B1 (CYP1B1), a member of the second subfamily of cytochrome P450 family I, is a crucial metabolic enzyme in humans (Cavalieri and Rogan, 2006), and high expression levels of CYP1B1 were found in breast, ovarian, and uterine tissues (Saini et al., 2009; Min et al., 2022). As a critical factor in the initiation and development of hormone-related malignancies, CYP1B1 catalyzes the conversion of 17-estradiol to 4-hydroxylated estradiol (4-OH-E2), thus playing a central role in estrogen metabolism (Gajjar et al., 2012; Zhao et al., 2021). In breast cancer, 4-OH-E2 goes through a redox cycle that results in the production of oxides as well as chemically reactive estrogenic semiquinone and quinone intermediates (e.g. E2-3,4-semiquinone and E2-3,4-benzoquinone metabolites), which in turn react with purines in DNA to generate purine-free adducts and cause DNA damage (Cavalieri and Rogan, 2016; Li et al., 2017), thereby promoting estrogen-associated breast cancer (Park et al., 2012; An et al., 2019). Hanna et al. (2000) showed that genetic changes in CYP1B1 are linked to differential estrogen metabolism, which suggests that these may also contribute to individual variations in the risk of estrogen-driven breast cancer occurrence.
The association of genetic polymorphisms with breast cancer is of interest to researchers worldwide. Disease-causing mutations are implicated in most cancers (Apostolou and Fostira, 2013). Among the functional single nucleotide polymorphisms (SNPs) found in the CYP1B1 gene, those located in the promoter or exon regions are more likely to influence breast cancer development and progression than regular SNPs (Economopoulos and Sergentanis, 2010). Therefore, identifying functional SNPs is crucial for improving breast cancer prevention as well as our knowledge of its pathophysiology.
Research on genetic susceptibility to breast cancer has gradually transitioned from the genome-wide association study (GWAS) to the post-GWAS era (Fachal and Dunning, 2015). Scientists now utilize state-of-the-art bioinformatics tools to interpret and uncover the full range of susceptibility-associated SNPs. Population-based epidemiological studies are used for the identification of novel functional gene variants that are truly pathogenic. The use of bioinformatics tools is therefore imperative for the identification of functional CYP1B1 variants associated with breast cancer risk. Many of the currently identified breast cancer risk-associated SNPs need to be further explored and validated. In this study, we employed a bioinformatics approach to explore functional CYP1B1 gene variants. A two-stage case–control study was conducted to identify the variants most likely to influence breast cancer risk.
Materials and methods
Selection of candidate single nucleotide polymorphisms
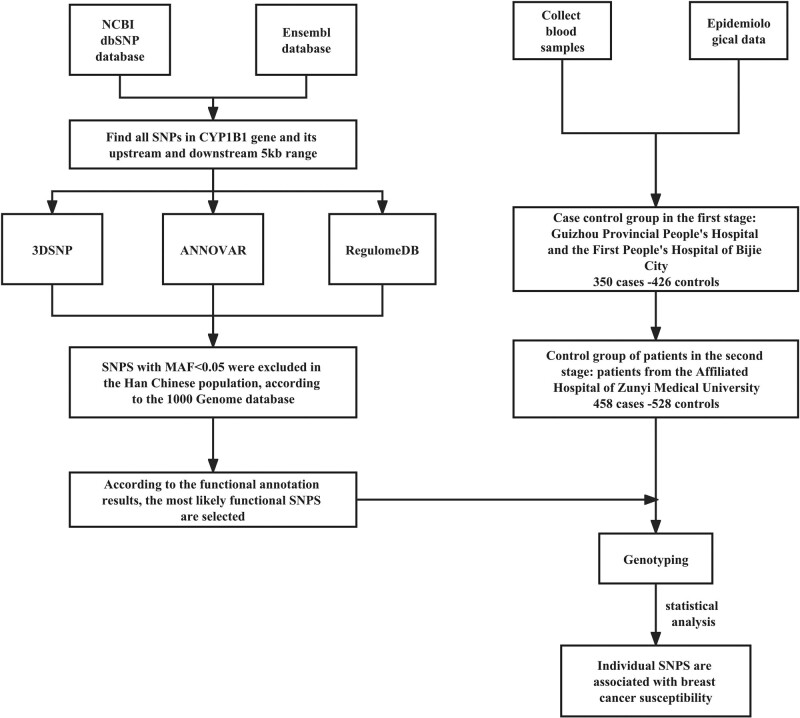
SNPs in the CYP1B1 gene and 5-kb flanking regions were obtained from the National Center for Biotechnology Information (NCBI) dbSNP database (https://www.ncbi.nlm.nih.gov/) and the Ensembl database (https://www.ensembl.org/index.html). SNPs with a minor allele frequency (MAF) < 0.05 in the Chinese Han population, as per the 1000 Genomes Project (http://www.1000genomes.org), were excluded. All SNPs were functionally annotated using 3DSNP (v2.0, https://omic.tech/3dsnpv2/) (Lu et al., 2017), ANNOVAR (Wang et al., 2010), and RegulomeDB (v2.1, https://regulomedb.org) (Boyle et al., 2012) (Fig. 1). SNPs were selected based on the functional annotation scores. The specific annotation scoring scheme is shown in Table 1.
Fig. 1.
Methodology roadmap.
Table 1.
Scoring schemes and related funtional notes for 3DSNP, ANNOVAR, and RegulomeDB
| Database | Functional categories | Score | Description | Note |
|---|---|---|---|---|
| 3DSNP | 3D interacting genes, Enhancer state, Promoter state, Transcription factor binding sites, sequence motifs altered, conservation | >0 | – | Functional categories with no record for a SNP are omitted and will not be measured in the total score. |
| ANNOVAR | Gene-based Annotation; Region-based Annotation; Filter-based Annotation | >0 | – | A higher annotation score indicates the stronger signal intensity observed. It indicates the higher probability that the SNP has the corresponding function. |
| RegulomeDB | Protein Binding, Motifs, Chromatin Structure, eQTLs, Histone Modifications, and Related Data | 1a | eQTL + TF binding + matched TF motif + matched DNase footprint + DNase peak | Likely to affect binding and linked to expression of a gene target |
| 1b | eQTL + TF binding + any motif + DNase footprint + DNase peak | |||
| 1c | eQTL + TF binding + matched TF motif + DNase peak | |||
| 1d | eQTL + TF binding + any motif + DNase peak | |||
| 1e | eQTL + TF binding + matched TF motif | |||
| 1f | eQTL + TF binding/DNase peak | |||
| 2a | TF binding + matched TF motif + matched DNase footprint + DNase peak | Likely to affect binding | ||
| 2b | TF binding + any motif + DNase footprint + DNase peak | |||
| 2c | TF binding + matched TF motif + DNase peak | |||
| 3a | TF binding + any motif + DNase peak | Less likely to affect binding | ||
| 3b | TF binding + matched TF motif | |||
| 4 | TF binding + DNase peak | Minimal binding evidence | ||
| 5 | TF binding or DNase peak | |||
| 6 | Motif hit |
eQTL, expression quantitative trait loci; SNP, single nucleotide polymorphism; TF, Transcription factor.
Subjects participating in the case–control study
The association between the candidate SNPs and breast cancer risk in a Chinese Han population was comprehensively assessed through a two-stage case–control study. The study subjects – all Han Chinese women from Guizhou and neighboring provinces within China – were unrelated. The exclusion criteria did not include age or histological subtypes. The first stage of the study was carried out at Guizhou Provincial People’s Hospital and The First People’s Hospital of Bijie city. The second stage of the study was carried out in the Affiliated Hospital of Zunyi Medical University. Subjects were patients with histopathologically confirmed breast cancer, newly diagnosed between June 2019 and June 2022, who had not yet received radiotherapy or chemotherapy. Patients with a history of metastasis to other organs or other cancer types as well as patients showing two or more malignancies at the same time were excluded. The control group included subjects who participated in health screening at the corresponding hospital during the same period. Control cases were frequently matched by age (±5 years). At the time of recruitment, each participant provided written informed consent before having their personal information and a peripheral venous blood sample (2 ml) taken. Subjects were defined as non-smokers if they had never smoked or smoked daily for less than a year. Otherwise, they were considered smokers. Subjects were defined as drinkers if they had consumed alcohol more than twice a week for at least a year, and as non-drinkers, if they had not.
Ethics committee approval for this study was granted by the Medical Ethics Committee of Zunyi Medical University (approval number: 2019-1-032). All of the subjects who volunteered to participate in the study provided written informed consent. Moreover, the study was conducted in accordance with the principles of the Declaration of Helsinki.
Genotyping
Genomic DNA was extracted from 2-ml peripheral venous blood samples collected from participants using the BloodZol kit (TransGen Biotech, China) according to the manufacturer’s recommended protocol. Patients were genotyped via TaqMan SNP genotyping analysis following PCR on a CFX96 real-time PCR detection system (CFX96; Bio-Rad, Hercules, California, USA). Genotyping was performed without knowledge of the case/control status of the subjects. Approximately 5% of the samples collected from breast cancer cases and controls were randomly selected for genotyping twice, and the results were 100% concordant. All methods were performed as per the approved guidelines.
Statistical analysis
Using Power v3.0 (available at https://dceg.cancer.gov/tools/design/power; García-Closas and Lubin, 1999), the statistical power to identify relationships between breast cancer risk and SNPs was determined. Differences in the distribution of the demographic characteristics between the case and control groups were tested using the t-test or chi-square test. A Hardy–Weinberg equilibrium test was performed to identify the gene frequencies in the control group using a goodness-of-fit chi-square test. After adjusting for smoking, alcohol consumption, menopausal status, and age, unconditional multivariate logistic regression analysis was used to calculate the odds ratios (ORs) and 95% confidence intervals (CIs) to estimate the association between genotype and breast cancer risk.
All P values were two-sided, and a P < 0.05 was considered statistically significant. All statistical analyses were performed using SPSS software (version 23.0; SPSS Inc., Chicago, Illinois, USA).
Expression quantitative trait loci analysis and analysis of differential expression in cancer and paracancerous tissues
We used the PancanQTL platform (http://gong_lab.hzau.edu.cn/PancanQTL/gwas) to conduct cis and trans expression quantitative trait loci (eQTL) analysis of genetic variants against the expression levels of genes in the Cancer Genome Atlas Program (TCGA) database to identify potential target genes that were statistically significantly different. The Gene Set Cancer Analysis (GSCA) platform (http://bioinfo.life.hust.edu.cn/GSCA/#/expression) was further used to compare the gene expression levels of potential target genes between breast cancer tissue and normal tissue adjacent to the cancer using RNA sequencing data in TCGA to determine whether the expression levels of the target genes were associated with breast cancer.
Results
Selection of candidate single nucleotide polymorphisms
A total of 2439 CYP1B1 SNPs were obtained from the NCBI dbSNP database and the Ensembl database. After 3DSNP, ANNOVAR, and RegulomeDB annotation, 312 SNPs were predicted to have biological function. Subsequently, SNPs with MAF < 0.05 in Southern Han Chinese individuals were excluded, and a total of 25 SNPs were obtained (Table 2). The inclusion criteria were a 3DSNP score > 0, RegulomeDB score > 4, and ANNOVAR score > 900 according to the scoring criteria in Table 1. Finally, the two SNPs most likely to have functional consequences, that is, rs10175368 and rs2551188, were selected (Table 3).
Table 2.
Candidate genetic loci screened by bioinformatics analysis
| SNP ID | Ref/Alt | 3DSNP | ANNOVAR | RegulomeDB | MAFa |
|---|---|---|---|---|---|
| rs10175368 | C/T | 4.38 | 999 | 2c | 0.2048 |
| rs2551188 | C/T | 82.31 | 934 | 3b | 0.2238 |
| rs162332 | A/G | 14.05 | 897 | 3a | 0.0667 |
| s149148 | G/A | 10.58 | 892 | 3a | 0.0762 |
| rs162331 | T/G | 55.11 | 890 | 4 | 0.0897 |
| rs4646430 | C/G | 9.36 | 889 | 4 | 0.0967 |
| rs162328 | G/A | 12.99 | 887 | 3a | 0.0619 |
| rs458704 | G/A | 3.89 | 829 | 5 | 0.1002 |
| rs10175472 | C/T | 4.54 | 666 | 4 | 0.1049 |
| rs13386936 | T/A | 9.55 | 599 | 3a | 0.0570 |
| rs9341266 | G/A | 3.89 | 579 | 5 | 0.0609 |
| rs2855655 | T/C | 158.12 | 529 | 2b | 0.0667 |
| rs3057354 | T/TAA | 54.81 | 389 | 3a | 0.0714 |
| rs9341254 | GA/G | 14.35 | 379 | 2b | 0.2133 |
| rs162556 | G/A | 8.25 | 320 | 3a | 0.2381 |
| rs151096 | C/A | 2.48 | 317 | 1f | 0.0619 |
| rs10175338 | G/T | 7.30 | 313 | 4 | 0.1657 |
| rs162552 | C/A | 93.81 | 287 | 1f | 0.0680 |
| rs4670814 | G/C | 35.55 | 234 | 4 | 0.1379 |
| rs162327 | G/A | 5.71 | 206 | 5 | 0.1557 |
| rs10201389 | A/T | 2.60 | 129 | 3a | 0.1786 |
| rs10178134 | G/A | 12.13 | 111 | 2b | 0.2048 |
| rs162329 | T/C | 19.96 | 107 | 1f | 0.0762 |
| rs1056827 | C/A | 67.59 | 102 | 4 | 0.1796 |
| rs186661 | G/A | 1.71 | 100 | 7 | 0.1141 |
MAF, minor allele frequency; SNP, single nucleotide polymorphism.
MAF was downloaded from the data for Southern Han Chinese individuals in the 1000 Genomes Project.
Table 3.
Details of the two single nucleotide polymorphisms identified as most likely to have functional consequences
| Chr | SNP position | SNP ID | Ref/Alt | MAFa | Annotationb |
|---|---|---|---|---|---|
| chr2 | 38307860 | rs10175368 | C/T | 0.2048 | Influencing transcription factor binding |
| chr2 | 38302793 | rs2551188 | C/T | 0.2238 | Affecting transcription |
MAF, minor allele frequency; SNP, single nucleotide polymorphism.
MAF was downloaded from the data for Southern Han Chinese individuals in the 1000 Genomes Project.
Prediction results based on 3DSNP, ANNOVAR, and RegulomeDB.
Participant characteristics
The characteristics of the participants in the two-stage case–control study are presented in Table 4. In stage 1, the cases and controls were well matched based on age and menopausal status, with P values of 0.152 and 0.47, respectively. Similar distributions of age and menopausal status between the two groups were also observed in stage 2 (P > 0.05).
Table 4.
Demographic characteristics of the participants in the two-stage case–control study
| Variables | Stage 1 | Stage 2 | Combined | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Case (350) No (%) | Control (426) No (%) | P value | Case (458) No (%) | Control (528) No (%) | P value | Case (808) No (%) | Control (954) No (%) | P value | |
| Age (mean ± SD) | 49.78 ± 9.69 | 49.68 ± 10.44 | 0.152a | 50.58 ± 10.53 | 49.65 ± 10.59 | 0.156a | 50.24 ± 10.19 | 49.70 ± 10.26 | 0.265a |
| Menopausal status | 0.476b | 0.347b | 0.434b | ||||||
| Premenopausal | 186 (53.14) | 230 (53.99) | 0.476a | 230 (50.22) | 281 (53.22) | 0.737a | 416 (51.49) | 509 (53.35) | 0.449a |
| Postmenopausal | 164 (46.86) | 196 (46.01) | 0.525a | 228 (49.78) | 247 (46.78) | 0.183a | 392 (48.51) | 445 (46.65) | 0.680a |
| Estrogen receptor | |||||||||
| Negative | 149 (42.57) | 195 (42.58) | 344 (42.57) | ||||||
| Positive | 178 (50.86) | 241 (52.62) | 419 (51.86) | ||||||
P value was calculated via t-test.
P value was calculated via χ2test.
A total of 808 cases of breast cancer and 954 healthy controls were analyzed in the combined study. The mean age was 50.24 years (±10.19) in the combined case group and 49.70 years (±10.26) in the combined control group. There was no statistical difference in the distribution of age (P = 0.265) and menopausal status (P = 0.434) between the combined cases and controls. In total, 416 (51.49%) and 392 (48.51%) patients with breast cancer were of premenopausal and postmenopausal status, respectively. In total, 419 (51.86%), 344 (42.57%), and 45 (5.57%) patients with breast cancer were estrogen receptor-positive, estrogen receptor-negative, and estrogen receptor unknown, respectively.
Association between candidate single nucleotide polymorphisms and breast cancer risk
Results from the first stage revealed a significant association between rs10175368 and breast cancer development. After adjusting for smoking, drinking, menopausal status, and age, individuals with the CT and TT genotypes were shown to have a lower risk of breast cancer than those carrying the CC genotype, with ORs of 0.607 (95% CI, 0.480–0.831; P = 0.008) and 0.338 (95% CI, 0.202–0.681; P = 0.001), respectively. Additive model results also indicated that the T allele at this locus was a protective factor against breast cancer, with an OR of 0.628 (95% CI, 0.532–0.736; P < 0.001). No statistical association was found between rs2551188 and breast cancer risk. An OR of 0.860 (95% CI, 0.718–1.052; P = 0.123) was obtained via the additive model. The recessive and dominant models were not statistically significant (Table 5).
Table 5.
Association of the candidate single nucleotide polymorphisms with breast cancer risk in stage 1
| Variables | Case (350) | Control (426) | OR (95% CI); P valuea |
|---|---|---|---|
| No (%) | No (%) | ||
| rs10175368 | 345 (98.57) | 425 (99.77) | |
| CC | 226 (64.57) | 243 (57.04) | 1.00 |
| CT | 109 (31.14) | 156 (36.62) | 0.607 (0.480–0.831); 0.008 |
| TT | 10 (2.86) | 26 (6.10) | 0.338 (0.202–0.681); 0.001 |
| Recessive | 0.413 (0.216–0.854); 0.006 | ||
| Dominant | 0.526 (0.414–0.745); <0.001 | ||
| Additive | 0.628 (0.532–0.736);<0.001 | ||
| rs2551188 | 340 (97.14) | 425 (99.77) | |
| CC | 215 (61.43) | 252 (59.15) | 1.00 |
| CT | 111 (31.71) | 150 (35.21) | 0.857 (0.674–1.109); 0.235 |
| TT | 14 (4.00) | 23 (5.40) | 0.728 (0.413–1.240); 0.218 |
| Recessive | 0.719 (0.436–1.296); 0.321 | ||
| Dominant | 0.843 (0.636–1.086); 0.158 | ||
| Additive | 0.860 (0.718–1.052); 0.123 |
CI, confidence interval; OR, odds ratio.
ORs and 95% CIs were calculated using unconditional logistic regression after adjusting for smoking, drinking, menopausal status, and age.
The significance of the values in bold emphasise that the result is statistically significant.
Association between candidate single nucleotide polymorphisms and breast cancer risk
The genotype distribution of rs10175368 and rs2551188 in the second stage and the combined stage and their association with the risk of breast cancer are shown in Table 6. At the combined stage, after adjusting for smoking, drinking, menopausal status, and age, the individuals with the CT and TT genotypes were shown to have a lower risk of breast cancer than those carrying the CC genotype, with ORs of 0.756 (95% CI, 0.618–0.925; P = 0.007) and 0.456 (95% CI, 0.283–0.735; P = 0.001), respectively. The additive model results also indicated that the T allele at this locus was a protective factor against breast cancer, with an OR of 0.722 (95% CI, 0.613–0.850; P < 0.001). The distribution of the control rs10175368 genotype was consistent with the Hardy–Weinberg equilibrium (P = 0.924), and the MAF was 0.245, which is close to that obtained in the 1000 Genomes Project. For rs10175368, the statistical power required to obtain an OR of 1.5 for our sample size was 0.967. No statistical association was found between rs2551188 and breast cancer risk. The recessive, dominant, and additive models were not statistically significant.
Table 6.
Association of the candidate single nucleotide polymorphisms with breast cancer risk in stage 2 and combined stages
| Variables | Stage 2 | Combined | ||||
|---|---|---|---|---|---|---|
| Case (458) | Control (528) | OR (95% CI); Pa | Case (808) | Control (954) | OR (95% CI); P valuea | |
| No (%) | No (%) | No (%) | No (%) | |||
| rs10175368 | 458 (100) | 525 (99.43) | 803 (99.38) | 950 (99.58) | ||
| CC | 299 (65.28) | 301 (57.01) | 1.00 | 525 (64.98) | 544 (57.26) | 1.00 |
| CT | 143 (31.22) | 191 (36.17) | 0.817 (0.365–0.932); 0.008 | 252 (31.19) | 347 (36.53) | 0.756 (0.618–0.925); 0.007 |
| TT | 16 (3.49) | 33 (6.25) | 0.516 (0.234–0.695); 0.002 | 26 (3.22) | 59 (6.21) | 0.456 (0.283–0.735); 0.001 |
| Recessive | 0.593 (0.315–0.887); 0.007 | 0.524 (0.329–0.839); 0.007 | ||||
| Dominant | 0.836 (0.515–0.945); 0.001 | 0.708 (0.583–0.860); <0.001 | ||||
| Additive | 0.738 (0.562–0.886);0.001 | 0.722 (0.613–0.850); <0.001 | ||||
| rs2551188 | 440 (96.07) | 523 (99.05) | 780 (96.53) | 948 (99.37) | ||
| CC | 287 (62.67) | 320 (60.61) | 1.00 | 493 (61.01) | 571 (59.85) | 1.00 |
| CT | 132 (28.82) | 183 (34.66) | 0.795(0.604–1.048);0.103 | 252 (31.19) | 330 (34.59) | 0.879(0.717–1.070);0.603 |
| TT | 21 (4.59) | 20 (3.79) | 1.191(0.631–2.245);0.589 | 35(4.33) | 46(4.82) | 0.886(0.561–1.398);0.215 |
| Recessive | 1.017(0.687–2.410);0.431 | 0.927(0.591–1.455);0.742 | ||||
| Dominant | 0.834(0.640–1.086);0.177 | 0.880(0.723–1.070);0.199 | ||||
| Additive | 0.907(0.726–1.133);0.390 | 0.905(0.768–1.066);0.234 | ||||
CI, confidence interval; OR, odds ratio.
ORs and 95% CIs were calculated using unconditional logistic regression after adjusting for smoking, drinking, menopausal status, and age.
The significance of the values in bold emphasise that the result is statistically significant.
Association of rs10175368 with breast cancer risk based on stratified analysis
Stratified analysis was carried out according to the menopausal status. No association was observed in the premenopausal population (Table 7); however, among postmenopausal women, the risk of breast cancer was shown to be lower in individuals with CT and TT genotypes than in those carrying the CC genotype (OR = 0.625, 95% CI, 0.463–0.844, P = 0.002; OR = 0.327, 95% CI, 0.165–0.645, P = 0.001; Table 5). Additive model results also indicated that the T allele at this site was a protective factor against breast cancer in postmenopausal women (OR = 0.601, 95% CI, 0.474–0.764; P < 0.001).
Table 7.
Stratified analysis of rs10175368 and menopausal status for breast cancer risk
| rs10175368 | Cases No (%) | Control No (%) | OR (95% CI); P valuea |
|---|---|---|---|
| Premenopausal | 413 | 508 | |
| CC | 254 (61.50) | 294 (57.88) | 1.00 |
| CT | 145 (35.11) | 189 (37.20) | 0.888 (0.675–1.167); 0.394 |
| TT | 14 (3.39) | 25 (4.92) | 0.645 (0.328–1.647); 0.203 |
| Recessive | 0.725 (0.377–1.394); 0.335 | ||
| Dominant | 0.850 (0.652–1.109); 0.231 | ||
| Additive | 0.854 (0.681–1.072); 0.173 | ||
| Postmenopausal | 390 | 442 | |
| CC | 271 (69.48) | 250 (56.56) | 1.00 |
| CT | 107 (27.44) | 158 (35.75) | 0.625 (0.463–0.844); 0.002 |
| TT | 12 (3.08) | 34 (7.69) | 0.327 (0.165–0.645); 0.001 |
| Recessive | 0.383 (0.195–0.750); 0.005 | ||
| Dominant | 0.573 (0.430–0.763); <0.001 | ||
| Additive | 0.601 (0.474–0.764); <0.001 |
CI, confidence interval; OR, odds ratio.
ORs and 95% CIs were calculated via unconditional logistic regression after adjusting for smoking, drinking, and age.
The significance of the values in bold emphasise that the result is statistically significant.
Association of rs10175368 with breast cancer risk based on stratified analysis
Stratified analysis was carried out based on the expression of the estrogen receptor. The results showed that rs10175368 was significantly associated with breast cancer in the different estrogen receptor subgroups and that a lower risk of breast cancer was strongly connected to the T allele (Table 8).
Table 8.
Estrogen receptor stratification analysis of rs10175368 and breast cancer risk in a Chinese population
| rs10175368 | Cases (763) No (%) | Control (954) No (%) | OR (95% CI); P valuea |
|---|---|---|---|
| Estrogen receptor + | 417 | 950 | |
| CC | 272 (65.23) | 544 (57.26) | 1.00 |
| CT | 132 (31.65) | 347 (36.53) | 0.764 (0.597–0.980); 0.034 |
| TT | 13 (3.12) | 59 (6.21) | 0.441 (0.238–0.819); 0.010 |
| Recessive | 0.525 (0.289–0.951); 0.034 | ||
| Dominant | 0.710 (0.559–0.902); 0.005 | ||
| Additive | 0.724 (0.591–0.887); 0.002 | ||
| Estrogen receptor − | 341 | 950 | |
| CC | 226 (66.28) | 544 (57.26) | 1.00 |
| CT | 103 (30.20) | 347 (36.53) | 0.717 (0.548–0.938); 0.015 |
| TT | 12 (3.52) | 59 (6.21) | 0.485 (0.256–0.920); 0.027 |
| Recessive | 0.545 (0.289–1.027); 0.060 | ||
| Dominant | 0.683 (0.527–0.884); 0.004 | ||
| Additive | 0.709 (0.569–0.882); 0.002 |
CI, confidence interval; OR, odds ratio.
ORs and 95% CIs were calculated using unconditional logistic regression after adjusting for smoking, drinking, menopausal status, and age.
The significance of the values in bold emphasise that the result is statistically significant.
Expression quantitative trait loci analysis of rs10175368 and differential expression of target genes in cancer and paracancerous tissues
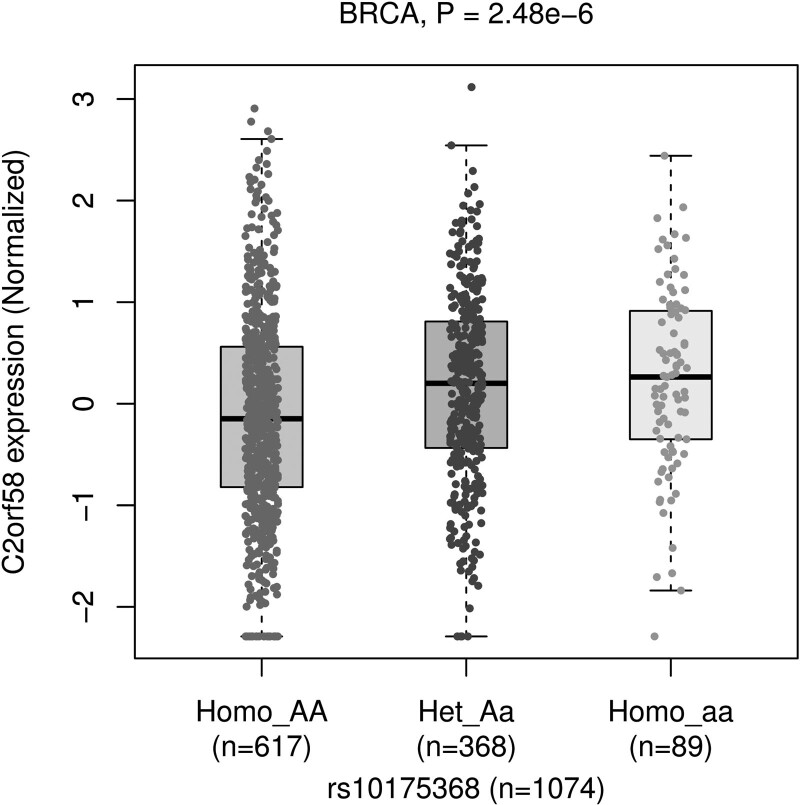
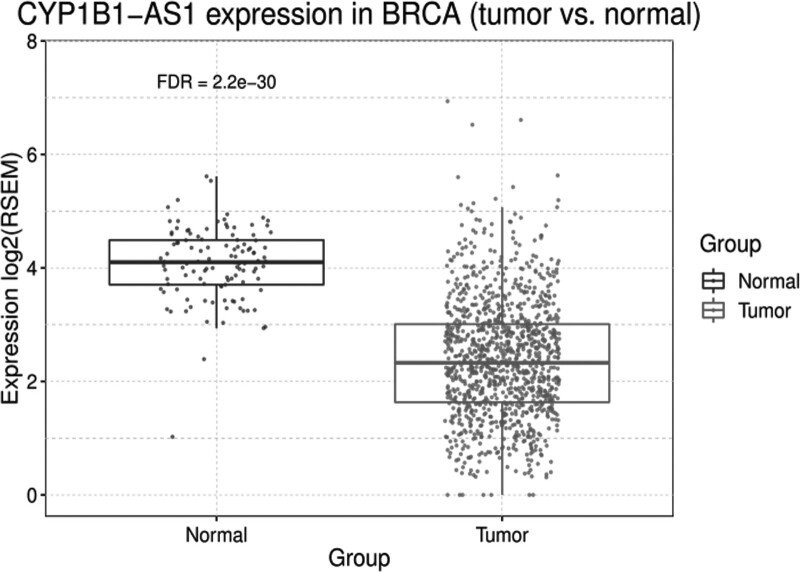
We used the PancanQTL platform to perform cis and trans eQTL analysis of genetic variants against the expression levels of genes in the TCGA database to identify potential target genes that were statistically significantly different, finding that the expression level of CYP1B1-AS1 (also known as C2orf58) was associated with rs10175368 (P = 2.8 × 10−6) and that CYP1B1-AS1 had a higher expression level in individuals with wild genotype AA (Fig. 2). We used the GSCA platform to investigate the difference in the expression of CYP1B1-AS1 between breast cancer tissues and paracancerous tissues. The results showed that CYP1B1-AS1 expression was significantly higher in breast cancer tissues than in paracancerous normal tissues (P = 2.2 × 10−30) (Fig. 3).
Fig. 2.
eQTL analysis of rs10175368 in BRCA. eQTL, expression quantitative trait loci.
Fig. 3.
CYP1B1-AS1 expression in BRCA (tumor vs. normal). CYP1B1, cytochrome P450 1B1.
Discussion
In the current study, we screened variations in the CYP1B1 gene and its 5-kb flanking regions to identify functional variants by integrating data from 3DSNP, ANNOVAR, and RegulomeDB. A two-stage hospital-based case–control study was conducted in a Chinese Han population to examine the potential association between CYP1B1 functional variants and breast cancer risk. To our knowledge, our findings are the first to show that the rs10175368 mutant T allele plays a protective role against breast cancer in the Chinese population, especially in postmenopausal women.
Breast cancer develops as a result of a multitude of environmental, genetic, and lifestyle factors (Sun et al., 2017). This is exemplified by the observation that only a small number of individuals subjected to a given mutagenic exposure develop breast cancer (Zheng et al., 2000; Hidaka et al., 2016). That is, the individual genetic makeup is a major determinant of carcinogenesis. Patients with breast, lung, kidney, and prostate cancer have elevated levels of CYP1B1, the main phase I drug-metabolizing enzyme (Muskhelishvili et al., 2001; Anttila et al., 2011; Hollis et al., 2022). Genetic polymorphisms in the CYP1B1 gene are responsible for inter-individual and inter-ethnic differences in disease susceptibility (Manikandan and Nagini, 2018). Previously reported immunohistochemical analysis results reveal that the minor rs10175368 genotype is linked to higher CYP1B1 expression, with the mobility shift and higher luciferase activity at rs10175368 demonstrating greater nuclear factor binding at the minor allele (Kato et al., 2018). Thus, the functional SNP rs10175368 influences CYP1B1 expression. CYP1B1 gene polymorphisms have been shown to alter its catalytic characteristics as a 17-estradiol metabolic enzyme, eventually leading to an increase or decrease in enzyme activity (Agundez, 2004). Owing to its role in enzyme metabolism, CYP1B1 influences the development of estrogen-dependent tumors (Crooke et al., 2006). Studies have suggested that heterozygous or homozygous variants of CYP1B1 may lead to significantly compromised enzyme function, resulting in lower levels of 4-OH-E2 and lower corresponding adduct production, thereby reducing breast cancer risk (Qiu et al., 2018; Zhao et al., 2021).
Our present findings indicate that the minor T allele of rs10175368 protects against breast cancer (OR = 0.722, 95% CI, 0.613–0.850). Predictions based on the RegulomeDB database showed that rs10175368 affects transcription factor binding. Even though the precise mechanism underlying the protective effect is unclear, we hypothesized that the minor T allele of rs10175368 may lead to a reduction in CYP1B1 expression. Lower CYP1B1 enzyme levels then cause a reduction in DNA adduct formation via 4-OH-E2 as well as via estrogenic semiquinone and quinone intermediates. The lessening of DNA damage reduces the risk of breast cancer.
Meanwhile, we found that the minor T allele of rs10175368 acts as a protective factor against breast cancer in postmenopausal women. Estrogen exposure causes phenotypic changes and chromosomal abnormalities in human non-malignant breast epithelial cells, thus driving tumorigenesis (Fernandez et al., 2006). Through the effects of downstream metabolites, estrogen exposure may indirectly damage DNA, causing chromosomal instability and subsequently cancer (Li et al., 2004; Blackburn et al., 2015). Changes in progesterone and estrogen levels have been linked to genetic variants of CYP1B1 (García-Closas et al., 2002; Schilling et al., 2007). Moreover, estrogen levels vary among women with different menopausal statuses, with the levels being higher in premenopausal women (Zheng et al., 2000). Thus, we hypothesize that the protective effect of the rs10175368 T allele against breast cancer in postmenopausal women is associated with reduced estrogen levels.
rs10175368 was not found to be correlated with breast cancer risk among Caucasian and Jordanian women (Huang et al., 2009; Al-Eitan et al., 2019), suggesting that inter-ethnic genetic variation influences the effects of certain cytochrome P450 variants (McGraw and Waller, 2012; Polimanti et al., 2012). Furthermore, the present findings highlight the existence of genetic heterogeneity across populations in relation to breast cancer risk. Hence, future research should address racial differences in the relationship between rs10175368 and breast cancer risk in larger, more diverse populations.
Herein, cis and trans eQTL analysis of the expression levels of rs10175368 against the expression of target genes in the TCGA database using the PancanQTL platform revealed that the expression level of CYP1B1-AS1 was associated with rs10175368. Furthermore, we found that CYP1B1-AS1 had significantly higher expression levels in breast cancer tissues than in normal tissue adjacent to the cancer. In recent years, the long-stranded non-coding RNA CYP1B1-AS1 is gaining attention as an important factor in the development of tumors, such as breast cancer and glioblastoma (Molaei Ramshe et al., 2021; Ye et al., 2021). A study noted that CYP1B1 was identified as a regulatory target of CYP1B1-AS1 (Ye et al., 2021). We, therefore, speculate that the polymorphism rs10175368 may indirectly affect CYP1B1 gene expression by affecting the target gene CYP1B1-AS1; however, this hypothesis still needs to be tested by evaluating relevant clinical samples.
The present study had certain limitations. Selection bias may exist due to the hospital-based case–control study design. It is also critical to carry out comprehensive molecular biology experiments to clarify the precise mechanism underlying the protective effect of the rs10175368 variant.
Conclusion
Our present findings suggest that the minor T allele in rs10175368 protects against breast cancer development among Chinese Han women. Given that a multitude of factors influences breast cancer risk, future studies should more comprehensively evaluate the significance of this variant. That is, researchers should consider the importance of gene–gene as well as gene–environment interactions. A more comprehensive assessment of breast cancer susceptibility and etiology can be achieved through bioinformatics analysis using public databases, with the ultimate goal of improving breast cancer prevention, early detection, and treatments, to altogether reduce the disease burden.
Acknowledgements
The authors wish to thank all the study participants, research staff, and students who participated in this study, especially the blood sample donors. J.L., L.Z., and Y.Z. contributed to study design, data analysis and interpretation, and drafting of the manuscript. J.L., L.Z., and X.C. contributed to study design and blood sample acquisition. Y.L., Y.D., and J.F. contributed to blood sample acquisition. J.L., L.Z., X.C., M.T., and C.Y. contributed to DNA preparation and data acquisition. J.L., L.Z., and X.C. contributed to DNA preparation and genotyping. J.L., M.T., and Y.Z. contributed to study design. X.W. and Y.Z. contributed to conceptualization and study design, data acquisition, critical revision of the manuscript for important intellectual content, study supervision, and obtained funding. All data generated and analyzed during this study are included in this manuscript. Ethics committee approval for this study was granted from Zunyi Medical University (approval number: 2019-1-032). All study subjects who volunteered to participate provided written informed consent. The study was conducted in accordance with the principles of the Declaration of Helsinki. This work was financially supported by the National Natural Science Foundation of China (Grant No. 31960209) to X.W., Guizhou Science and Technology Fund Project [Grant No. (2020)1Y093] to X.W., Doctoral Science Research Startup Funding of Zunyi Medical University (Grant No.2017-01) to X.W., the National Natural Science Foundation of China (Grant No. 82060620) to Y.Z., Zunyi Science and Technology Fund Project [Grant No. Zunyi Kehe HZ Zi (2021)40] to X.W., Guizhou Science and Technology Program Project (No. Qiankehe Foundation-ZK[2023] General 502) to Y.Z., Zunyi Medical University Postgraduate Research Project Fund Project (Grant No. ZYK88) to X. C. and the Innovation and Entrepreneurship Training Program for Students of Zunyi Medical University (Grant No. ZYDC2020057) to M.T.
Conflicts of interest
There are no conflicts of interest.
Footnotes
Jiarui Liu and Lijia Zhang contributed equally to the writing of this article.
References
- Agundez JA. (2004). Cytochrome P450 gene polymorphism and cancer. Curr Drug Metab 5:211–224. [DOI] [PubMed] [Google Scholar]
- Al-Eitan LN, Rababa’h DM, Alghamdi MA, Khasawneh RH. (2019). Association of CYP gene polymorphisms with breast cancer risk and prognostic factors in the Jordanian population. BMC Med Genet 20:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S, Coombes RC. (2000). Estrogen receptor alpha in human breast cancer: occurrence and significance. J Mammary Gland Biol Neoplasia 5:271–281. [DOI] [PubMed] [Google Scholar]
- An D, Song Z, Yi Y, Zhang Q, Liu J, Zhang Y, et al. (2019). Oroxylin A, a methylated metabolite of baicalein, exhibits a stronger inhibitory effect than baicalein on the CYP1B1-mediated carcinogenic estradiol metabolite formation. Phytother Res 33:1033–1043. [DOI] [PubMed] [Google Scholar]
- Antoniou AC, Easton DF. (2006). Models of genetic susceptibility to breast cancer. Oncogene 25:5898–5905. [DOI] [PubMed] [Google Scholar]
- Anttila S, Raunio H, Hakkola J. (2011). Cytochrome P450-mediated pulmonary metabolism of carcinogens: regulation and cross-talk in lung carcinogenesis. Am J Respir Cell Mol Biol 44:583–590. [DOI] [PubMed] [Google Scholar]
- Apostolou P, Fostira F. (2013). Hereditary breast cancer: the era of new susceptibility genes. Biomed Res Int 2013:747318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barańska A, Dolar-Szczasny J, Kanadys W, Kinik W, Ceglarska D, Religioni U, et al. (2022). Oral contraceptive use and breast cancer risk according to molecular subtypes status: a systematic review and meta-analysis of case-control studies. Cancers 14:574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn HL, Ellsworth DL, Shriver CD, Ellsworth RE. (2015). Role of cytochrome P450 genes in breast cancer etiology and treatment: effects on estrogen biosynthesis, metabolism, and response to endocrine therapy. Cancer Causes Control 26:319–332. [DOI] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. (2012). Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalieri E, Rogan E. (2006). Catechol quinones of estrogens in the initiation of breast, prostate, and other human cancers: keynote lecture. Ann N Y Acad Sci 1089:286–301. [DOI] [PubMed] [Google Scholar]
- Cavalieri EL, Rogan EG. (2016). Depurinating estrogen-DNA adducts, generators of cancer initiation: their minimization leads to cancer prevention. Clin Transl Med 5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons M, Goss P. (2001). Estrogen and the risk of breast cancer. N Engl J Med 344:276–285. [DOI] [PubMed] [Google Scholar]
- Crooke PS, Ritchie MD, Hachey DL, Dawling S, Roodi N, Parl FF. (2006). Estrogens, enzyme variants, and breast cancer: a risk model. Cancer Epidemiol Biomarkers Prev 15:1620–1629. [DOI] [PubMed] [Google Scholar]
- Economopoulos KP, Sergentanis TN. (2010). Three polymorphisms in cytochrome P450 1B1 (CYP1B1) gene and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 122:545–551. [DOI] [PubMed] [Google Scholar]
- Fachal L, Dunning AM. (2015). From candidate gene studies to GWAS and post-GWAS analyses in breast cancer. Curr Opin Genet Dev 30:32–41. [DOI] [PubMed] [Google Scholar]
- Fernandez SV, Russo IH, Russo J. (2006). Estradiol and its metabolites 4-hydroxyestradiol and 2-hydroxyestradiol induce mutations in human breast epithelial cells. Int J Cancer 118:1862–1868. [DOI] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. (2003). Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144:4562–4574. [DOI] [PubMed] [Google Scholar]
- Gajjar K, Martin-Hirsch PL, Martin FL. (2012). CYP1B1 and hormone-induced cancer. Cancer Lett 324:13–30. [DOI] [PubMed] [Google Scholar]
- García-Closas M, Lubin JH. (1999). Power and sample size calculations in case-control studies of gene-environment interactions: comments on different approaches. Am J Epidemiol 149:689–692. [DOI] [PubMed] [Google Scholar]
- García-Closas M, Herbstman J, Schiffman M, Glass A, Dorgan JF. (2002). Relationship between serum hormone concentrations, reproductive history, alcohol consumption and genetic polymorphisms in pre-menopausal women. Int J Cancer 102:172–178. [DOI] [PubMed] [Google Scholar]
- Hanna IH, Dawling S, Roodi N, Guengerich FP, Parl FF. (2000). Cytochrome P450 1B1 (CYP1B1) pharmacogenetics: association of polymorphisms with functional differences in estrogen hydroxylation activity. Cancer Res 60:3440–3444. [PubMed] [Google Scholar]
- Hidaka A, Sasazuki S, Matsuo K, Ito H, Charvat H, Sawada N, et al.; JPHC Study Group (2016). CYP1A1, GSTM1 and GSTT1 genetic polymorphisms and gastric cancer risk among Japanese: a nested case-control study within a large-scale population-based prospective study. Int J Cancer 139:759–768. [DOI] [PubMed] [Google Scholar]
- Hollis PR, Mobley RJ, Bhuju J, Abell AN, Sutter CH, Sutter TR. (2022). CYP1B1 augments the mesenchymal, claudin-low, and chemoresistant phenotypes of triple-negative breast cancer cells. Int J Mol Sci 23:9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Trentham-Dietz A, García-Closas M, Newcomb PA, Titus-Ernstoff L, Hampton JM, et al. (2009). Association of CYP1B1 haplotypes and breast cancer risk in Caucasian women. Cancer Epidemiol Biomarkers Prev 18:1321–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing W, Li L, Zhang X, Wu S, Zhao J, Hou Q, et al. (2020). Genetic profiling of breast cancer with and without preexisting metabolic disease. Transl Oncol 13:245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Hashimoto Y, Wong RK, Mitsui Y, Maekawa S, Chang I, et al. (2018). Influence of lifestyle choices on risks of CYP1B1 polymorphisms for prostate cancer. J Cell Mol Med 22:4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyama R, Wada-Kiyama Y. (2015). Estrogenic endocrine disruptors: molecular mechanisms of action. Environ Int 83:11–40. [DOI] [PubMed] [Google Scholar]
- Li JJ, Weroha SJ, Lingle WL, Papa D, Salisbury JL, Li SA. (2004). Estrogen mediates Aurora-A overexpression, centrosome amplification, chromosomal instability, and breast cancer in female ACI rats. Proc Natl Acad Sci USA 101:18123–18128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Zhu W, Gonzalez FJ. (2017). Potential role of CYP1B1 in the development and treatment of metabolic diseases. Pharmacol Ther 178:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, et al. (2000). Environmental and heritable factors in the causation of cancer – analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343:78–85. [DOI] [PubMed] [Google Scholar]
- Lu Y, Quan C, Chen H, Bo X, Zhang C. (2017). 3DSNP: a database for linking human noncoding SNPs to their three-dimensional interacting genes. Nucleic Acids Res 45:D643–D649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukina SS, Burdennyy AM, Zavarykina TM, Riabchikov DA, Kazubskaya TP, Kruglova MP, et al. (2021). The role of ESR1 gene polymorphic markers in the development of breast cancer and resistance to Tamoxifen therapy. Bull Exp Biol Med 170:350–355. [DOI] [PubMed] [Google Scholar]
- Manikandan P, Nagini S. (2018). Cytochrome P450 structure, function and clinical significance: a review. Curr Drug Targets 19:38–54. [DOI] [PubMed] [Google Scholar]
- McGraw J, Waller D. (2012). Cytochrome P450 variations in different ethnic populations. Expert Opin Drug Metab Toxicol 8:371–382. [DOI] [PubMed] [Google Scholar]
- Min JY, Lee GH, Khanal T, Jin SW, Lee SY, Kim HG, et al. (2022). Upregulation of CYP1B1 by hypoxia is mediated by ERα activation in breast cancer cells. Am J Cancer Res 12:2798–2816. [PMC free article] [PubMed] [Google Scholar]
- Molaei Ramshe S, Ghaedi H, Omrani MD, Geranpayeh L, Alipour B, Ghafouri-Fard S. (2021). Up-regulation of FOXN3-AS1 in invasive ductal carcinoma of breast cancer patients. Heliyon 7:e08179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskhelishvili L, Thompson PA, Kusewitt DF, Wang C, Kadlubar FF. (2001). In situ hybridization and immunohistochemical analysis of cytochrome P450 1B1 expression in human normal tissues. J Histochem Cytochem 49:229–236. [DOI] [PubMed] [Google Scholar]
- Park SA, Na HK, Surh YJ. (2012). Resveratrol suppresses 4-hydroxyestradiol-induced transformation of human breast epithelial cells by blocking IκB kinaseβ-NF-κB signalling. Free Radic Res 46:1051–1057. [DOI] [PubMed] [Google Scholar]
- Park J, Rodriguez JL, O’Brien KM, Nichols HB, Hodgson ME, Weinberg CR, et al. (2021). Health-related quality of life outcomes among breast cancer survivors. Cancer 127:1114–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platet N, Cathiard AM, Gleizes M, Garcia M. (2004). Estrogens and their receptors in breast cancer progression: a dual role in cancer proliferation and invasion. Crit Rev Oncol Hematol 51:55–67. [DOI] [PubMed] [Google Scholar]
- Polimanti R, Piacentini S, Manfellotto D, Fuciarelli M. (2012). Human genetic variation of CYP450 superfamily: analysis of functional diversity in worldwide populations. Pharmacogenomics 13:1951–1960. [DOI] [PubMed] [Google Scholar]
- Qiu J, Du Z, Liu J, Zhou Y, Liang F, Lü Q. (2018). Association between polymorphisms in estrogen metabolism genes and breast cancer development in Chinese women: a prospective case-control study. Medicine (Baltimore) 97:e13337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Hirata H, Majid S, Dahiya R. (2009). Functional significance of cytochrome P450 1B1 in endometrial carcinogenesis. Cancer Res 69:7038–7045. [DOI] [PubMed] [Google Scholar]
- Sarink D, White KK, Loo LWM, Wu AH, Wilkens LR, Le Marchand L, et al. (2022). Racial/ethnic differences in postmenopausal breast cancer risk by hormone receptor status: the multiethnic cohort study. Int J Cancer 150:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling C, Gallicchio L, Miller SR, Langenberg P, Zacur H, Flaws JA. (2007). Genetic polymorphisms, hormone levels, and hot flashes in midlife women. Maturitas 57:120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, et al. (2017). Risk factors and preventions of breast cancer. Int J Biol Sci 13:1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. (2021). Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. (2010). ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia C, Dong X, Li H, Cao M, Sun D, He S, et al. (2022). Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl) 135:584–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye T, Li LL, Peng XM, Li Q. (2021). CYP1B1-AS1 is a novel biomarker in glioblastoma by comprehensive analysis. Dis Markers 2021:8565943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Liu LY, Wang F, Mu K, Yu ZG. (2012). The changes in female physical and childbearing characteristics in China and potential association with risk of breast cancer. BMC Public Health 12:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Hao Z, Zhong Y, Xu Y, Guo M, Zhang B, et al. (2021). Discovery of breast cancer risk genes and establishment of a prediction model based on estrogen metabolism regulation. BMC Cancer 21:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Xie DW, Jin F, Cheng JR, Dai Q, Wen WQ, et al. (2000). Genetic polymorphism of cytochrome P450-1B1 and risk of breast cancer. Cancer Epidemiol Biomarkers Prev 9:147–150. [PubMed] [Google Scholar]