SUMMARY
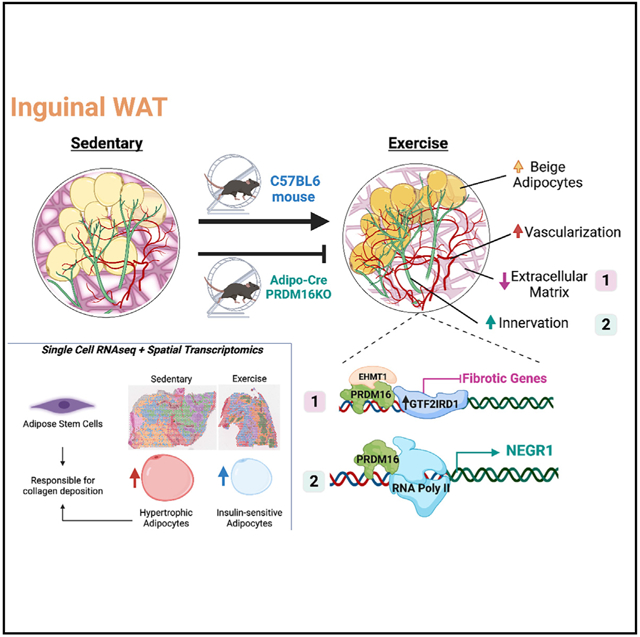
Inguinal white adipose tissue (iWAT) is essential for the beneficial effects of exercise training on metabolic health. The underlying mechanisms for these effects are not fully understood, and here, we test the hypothesis that exercise training results in a more favorable iWAT structural phenotype. Using biochemical, imaging, and multi-omics analyses, we find that 11 days of wheel running in male mice causes profound iWAT remodeling including decreased extracellular matrix (ECM) deposition and increased vascularization and innervation. We identify adipose stem cells as one of the main contributors to training-induced ECM remodeling, show that the PRDM16 transcriptional complex is necessary for iWAT remodeling and beiging, and discover neuronal growth regulator 1 (NEGR1) as a link between PRDM16 and neuritogenesis. Moreover, we find that training causes a shift from hypertrophic to insulin-sensitive adipocyte subpopulations. Exercise training leads to remarkable adaptations to iWAT structure and cell-type composition that can confer beneficial changes in tissue metabolism.
Graphical abstract
In brief
Using multi-omics approaches in conjunction with detailed imaging, Nigro et al. find that exercise training promotes iWAT remodeling through changes in ECM, vascularization, and innervation. Moreover, they identify robust cell-type-specific adaptations in response to exercise that, in combination with structural changes, result in a healthier adipose tissue phenotype.
INTRODUCTION
White adipose tissue (WAT) is a heterogeneous and highly dynamic endocrine organ that is responsive to numerous stimuli and is capable of undergoing remodeling to meet the metabolic and energy demands of the body.1 In obesity, homeostasis of both subcutaneous and visceral WAT depots is impaired, contributing to systemic metabolic disease.2 Pathological expansion of the extracellular matrix (ECM), excessive tissue fibrosis, inflammation, and dysregulated lipid accumulation have been established as important factors in this dysregulation of WAT.2 In contrast to obesity, exercise training can improve systemic metabolic homeostasis, and recent data suggest that some of these beneficial effects of exercise are mediated through adaptations to the subcutaneous inguinal WAT (iWAT).3-6 The underlying molecular and cellular mechanisms mediating the effects of exercise training on iWAT are still poorly understood.
The ECM is a highly dynamic structure that is continuously modified, degraded, and deposited.7 ECM remodeling affects important cellular functions such as adhesion, migration, proliferation, and differentiation and is closely linked with fundamental tissue processes including vascularization8 and innervation.9 Dysregulated ECM remodeling is found in numerous conditions including inflammatory diseases, heart disease, and cancer, and given this clinical relevance, research aimed at understanding the cellular mechanisms of pathological ECM remodeling has become more prominent. ECM remodeling in WAT has been studied mainly in the context of obesity. In WAT, ECM turnover is associated with the formation of new blood vessels, which ensures the healthy expansion of the tissue by preventing tissue hypoxia, fibrosis, and WAT dysfunction.10 There is also a pivotal role of sympathetic innervation in the regulation of WAT physiology, remodeling, and adaptations.11 For example, stimulation of sympathetic innervation in WAT following cold exposure is required for beige cell formation and non-shivering thermogenesis,12,13 which are both critical in maintaining systemic glucose homeostasis. Thus, understanding ECM remodeling and the associated cellular processes of angiogenesis and innervation may facilitate the development of new therapeutic targets for numerous diseases.
In contrast to the investigation of the ECM with obesity, the effect of exercise training on ECM remodeling in iWAT has not been previously studied, and so it is not known if the ECM is essential for the beneficial effects of training on this tissue. Similar to ECM remodeling, there are also no reports investigating the effects of exercise training on iWAT innervation. Exercise training has been shown to increase the expression of the angiogenic factor Vegfa in subcutaneous WAT (scWAT),3,14 suggesting that training promotes iWAT vascularization, although this has not been directly investigated. Thus, while exercise training is known to result in a more favorable metabolic profile in iWAT, it is essential to determine if structural components of iWAT adapt in a beneficial manner to exercise training, as these could provide the basis for novel therapeutic approaches.
Here, we test the hypothesis that exercise training alters ECM deposition and induces innervation and vascularization in iWAT and determine the molecular and cell-type-specific adaptations to iWAT in response to exercise. Using a combination of biochemical, imaging, genetic, and multi-omics approaches including advanced spatial transcriptomics, we find that exercise training dramatically reduces ECM stiffness, increases vascularization, and promotes sympathetic innervation with neuronal refinement in iWAT. Using single-cell transcriptomics, we identify the predominant cell populations that contribute to iWAT ECM physiology and determine how exercise affects collagen deposition by modulating both the cell composition and gene expression profile within specific cell types in iWAT. We also determine that PRDM16 is a regulator of training-induced beiging in iWAT and identify neural growth regulator 1 (NEGR1) as a link between PRDM16 and exercise-induced neuritogenesis. Thus, exercise training is a potent physiological stimulus for beneficial remodeling of iWAT.
RESULTS
Exercise training reduces collagen deposition in iWAT
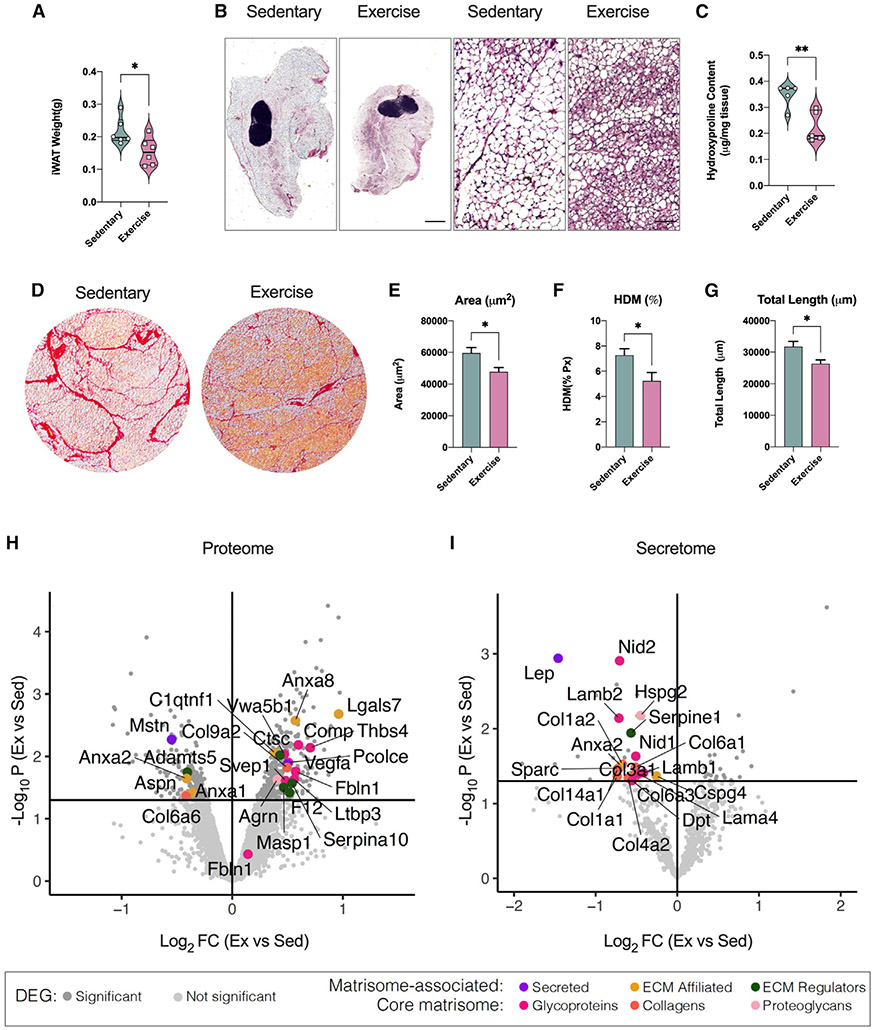
To determine the effects of exercise training on the iWAT ECM, 8-week-old male C57BL/6 mice were fed a standard diet and housed in individual cages with (exercise training) or without (sedentary) voluntary access to a running wheel. Exercise-trained mice ran 6.0 ± 1.6 km/day and had increased food intake compared with sedentary mice (Figure S1A). Sedentary mice increased their body weight by 7% during the 11 days, whereas exercise-trained mice maintained their body weight, although there was no significant difference between the two groups at day 11 (Figure S1B). Exercise training resulted in improved fasting blood glucose, insulin, and homeostatic model assessment for insulin resistance (HOMA-IR) (Figures S1C-S1E). At the tissue level, exercise training reduced fat mass (Figure 1A), and microscopically, it appeared to have a higher number of smaller multi-locular adipocytes compared with sedentary (Figure 1B). To evaluate tissue remodeling, we measured changes in collagen, the main component of the ECM. We found decreased collagen content in trained compared with sedentary iWAT using a hydroxyproline assay (Figure 1C). Sirius red staining also showed thinner and fewer collagen structures in trained iWAT (Figure 1D). Using the TWOMBLI software package,15 we found decreased collagen area in trained iWAT (Figure 1E), reduced “high-density matrix” (HDM) (Figure 1F), and reduced total length of collagen branches compared with sedentary iWAT (Figure 1G). These findings suggest that exercise training causes ECM remodeling through a decrease in iWAT collagen deposition.
Figure 1. Exercise training reduced ECM deposition in iWAT, modulating matrisome-associated and core matrisome gene expression levels.
(A) iWAT mass of male sedentary and trained mice (n = 6/group).
(B) H&E staining images of iWAT from sedentary and trained mice. Scale bar, 1,000 μm (4×) and 50 μm (20×).
(C) Hydroxyproline content in iWAT from sedentary and trained mice (n = 5/group).
(D) Sirius red staining images of iWAT from sedentary and trained mice. Scale bar, 100 μm (10×).
(E–G) Quantification of ECM deposition area (E), percentage of high-density matrix (HDM),.
(F), and total length of fibers (G) in iWAT (n = 4/group; calculated from 8 fields/mouse).
(H) Up- and down-regulated proteins with exercise training detected in iWAT (iWAT proteome analysis). Lines define restriction of log2 fold change (FC) value of 0.5 and −log10 of p value 0.01. Significant proteins are colored based on matrisome subcategories.
(I) Up- and down-regulated proteins with exercise detected in iWAT conditioned media (secretome analysis). Lines define restriction of log2 FC value of 0.5 and −log10 of p value 0.01. Significant proteins are colored based on matrisome subcategories.
Data are presented as mean ± SEM and were compared using unpaired two-tailed Student’s t test. *p < 0.05, and **p < 0.01.
Identification of exercise-modulated ECM genes expressed in iWAT
To determine gene expression changes driving exercise-induced iWAT ECM remodeling, we utilized our previously published microarray dataset generated using iWAT from sedentary mice and 11-day-wheel-trained mice (GEO: GSE68161).3 We performed pathway enrichment analysis using the Matrisome database,16,17 which includes not only all the genes encoding structural ECM components (“core matrisome”) but also the proteins that can interact with and remodel the ECM (“matrisome associated”). We found that 689/837 (82%) and 234/274 (85%) genes expressed in iWAT were associated with matrisome and core matrisome, respectively (Figure S1F). Of these genes, we found that 16% of matrisome-associated and 18% of core matrisome genes were either significantly up-regulated or down-regulated by exercise training (Figure S1F; Table S1), underscoring that there are robust exercise-induced transcriptional changes of ECM genes.
Matrisome-associated proteins contain ECM regulators, ECM-affiliated proteins, and secreted factors that may interact with core ECM proteins. Figure S1G summarizes the top 10 genes with the greatest absolute fold change for each of these three categories. Exercise training decreased ECM regulators known for their inhibitory role in WAT ECM remodeling (Timp4)18 or their negative effect on the beiging process (Adamts5).19,20 For ECM-affiliated genes, exercise training decreased genes implicated in the development of WAT fibrosis (Galectin-3, Lgals3)21 and increased genes involved in neuronal development (Neuron glial antigen 2; NG2/CSPG4).22,23 A subset of atrisome-associated genes are annotated as secreted factors. Of these, exercise training down-regulated Leptin (Lep), a marker of adipose tissue mass, and S100a8, a mediator of fibrosis and inflammatory response,24 and increased genes involved in the beiging process and adipose tissue innervation (Nrg4 and S100b).25,26
The core matrisome is composed of collagens, ECM glycoproteins, and proteoglycans. Among the genes included in the iWAT core matrisome, 29 genes were significantly down-regulated and 13 genes were significantly up-regulated by exercise training (Figure S1F; Table S1). Matrisome enrichment analysis revealed that out of the 29 down-regulated genes 5 are collagens, 18 are glycoproteins, and 6 are proteoglycans (Figure S1H). To map the exercise-regulated core matrisome genes to the different compartments of the ECM, we generated an iWAT ECM protein-protein interaction (PPI) network by integrating our data with publicly available data27 (Figure S1I). The collagen core components down-regulated by exercise training are mainly located in the basement membrane compartment, while the proteoglycans and glycoproteins are expressed in the interstitial matrix. We validated the effects of exercise training on mRNA expression of selected genes from matrisome-associated and core matrisome using qRT-PCR (Figures S1J and S1K). These results suggest that exercise training induces ECM remodeling through modulation of expression of matrisome-associated and core matrisome genes, which are also known to regulate other tissue processes such as innervation and beiging.
Proteomic profiling revealed iWAT ECM remodeling with exercise training
The total proteome of iWAT from sedentary and trained mice was quantified using high-resolution mass spectrometry. We detected a total of 6,910 proteins, of which 90% were predicted to have an intracellular localization, whereas ~10% were annotated as extracellular proteins. A total of 482 (7%) proteins were differentially expressed between sedentary and trained mice (p <0.05; Table S2), with training up-regulating 300 and down-regulating 182 proteins. Of these, 23 proteins were annotated for matrisome, 17 up-regulated and 6 down-regulated (Figure 1H).
Gene Ontology-Cellular Component (GO-CC) enrichment analysis showed that training led to proteomic changes in all major cellular components in iWAT, with mitochondria being the most affected (Figure S1L). Using the Matrisome database,16,17 we mapped the exercise-regulated proteins and found that exercise increased the matrisome-associated pathways and decreased the core matrisome and collagen-associated pathways (Figure S1M).
To determine the secreted factors involved in iWAT ECM remodeling, we performed a secretome analysis using iWAT conditioned media from sedentary and trained mice. We identified 726 proteins, of which 8% were differentially expressed with exercise training (23 up- and 37 down-regulated), with 50% of the down-regulated proteins being ECM-related genes (Figure 1I; Table S3). No ECM genes were significantly up-regulated in the secretome analysis. Exercise training down-regulated a large number of proteins localized in the extracellular space (Figure S1N) and proteins related to ECM core matrisome (Figure S1O). The most significantly down-regulated core matrisome proteins included LEP, annexin A2 (ANXA2), the glycoprotein SPARC, and the collagen COL1A1 (Figure 1I). Using PPI network analysis, we determined that most of the down-regulated matrisome proteins are located in the basement membrane layer of the ECM (Figure S1P). Overall, the iWAT proteome and secretome analyses corroborate the transcriptomic findings that exercise training down-regulates protein level and secretion of collagen species, mainly found in the basement membrane.
Spatial and single-cell transcriptomics identify main contributors of ECM in iWAT
To identify which adipose tissue cell types are mainly responsible for ECM remodeling, we reanalyzed previously published single-cell RNA sequencing datasets of both adipose stromal-vascular and mature adipocyte fractions6,28 (Figures S2A and S2C). In these data, the down-regulated ECM-secreted proteins that we detected in the multi-omics analysis (Figure 1) were mostly expressed in adipose stem cells (ASCs), preadipocytes, mature adipocytes, and glial and vascular cells (Figures S2A, S2B, and S2D). Since the ASC/preadipocyte cluster showed the greatest consistency in terms of the expression of ECM proteins across the single-cell RNA datasets, we estimated transcription factor (TF) activity in these cells by averaging gene expression of curated targets for each TF. ASCs demonstrated the highest TF activity compared with other cell types, with SMAD3 being one of the TFs with the highest activity within the ASCs (Figure S2E).
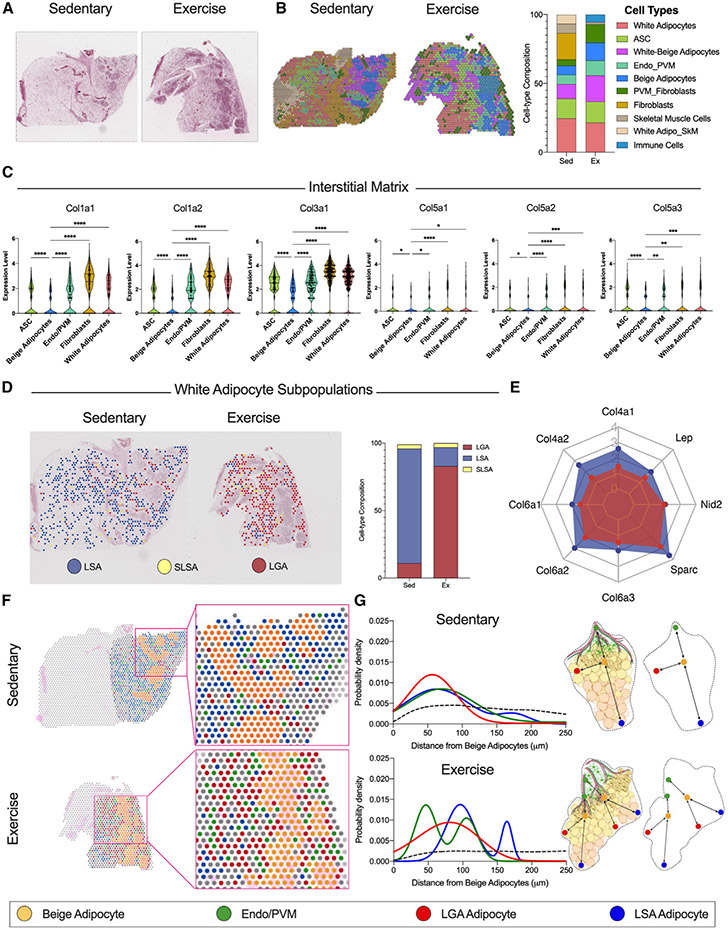
Our next goal was to determine the spatial distribution of the cells contributing to the ECM. This is important because the ECM regulates other spatially confined processes such as innervation and beiging. For this purpose, we performed spatial transcriptomic analysis (Visium Spatial Gene Expression, 10×Genomics) of iWAT from sedentary and trained mice. Each barcoded spot in Visium is ~50 μm in diameter, capturing 1–10 cells/spot based on the detected unique molecular identifier (UMI) (Figure S3A). Exercise-trained iWAT consists of smaller adipocytes and tissue mass compared with sedentary (Figure 1B). Consequently, we observed a smaller number of spots but a larger number of cells per spot in trained iWAT (Figure S3A). We performed spatial pattern clustering (Figure S3B and S3C), scrutinized marker gene expression in the spatially distinct clusters, and annotated the major cell types within each spot (Figure 2B). Spots showing a recognizable cell signature aligned with all the cell clusters visualized in the H&E-stained images (Figures 2A and 2B). Interestingly, exercise training drastically reduced the proportion of fibroblasts while increasing the proportion of beige and white-beige adipocytes and endothelial-perivascular microglia cells (Endo/PVMs). No differences for ASC and white adipocyte proportions were observed (Figure 2B).
Figure 2. Spatial and single-cell transcriptomics identifies main contributors of ECM in iWAT.
(A and B) H&E-stained sections (A) and relative spatial RNA sequencing (RNA-seq) barcoded spot. Scale bar, 2 mm, (B) indicating the cell clusters detected in iWAT from sedentary (left) and exercise (right) mice.
(C) Expression level for interstitial matrix collagens species across 5 selected cell-type clusters.
(D) Spatial distribution maps showing the three white adipocyte subpopulations, LSAs, SLSAs, and LGAs, detected in iWAT from sedentary (left) and exercise (right) mice along with the relative proportion plot.
(E) Basement membrane components gene expression level in LSAs and LGAs. The gene expression level is presented as a scale ranging from zero to four, where four represents the highest expression level.
(F) Spatial distribution maps and relative magnifications showing the selected cell-type clusters in iWAT from sedentary (top) and exercise (bottom) mice: beige adipocyte (yellow), endothelial and perivascular microglia cell (Endo/PVM) (green), LGA (red), and LSA (blue) white adipocyte subpopulations.
(G) Probability density distribution plots showing the distance relative to the beige adipocyte for the LGA, LSA, and Endo/PVM clusters in iWAT from sedentary (top) and exercise (bottom) mice. Dashed black lines indicate the random distribution among the clusters. Summary cartoon explaining the pattern distribution.
Data are presented as mean ± SEM and were compared using one-way ANOVA. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001
By comparing expression of different collagen isoforms across the spatially defined clusters, we found that the spots with a cell signature for fibroblasts, ASCs, Endo/PVMs, and white adipocytes expressed higher levels of major components of the interstitial matrix ECM compartment compared with beige adipocytes (Figure 2C). Further, beige adipocytes expressed lower levels of basement membrane components (Figure S3D). Using our publicly available scWAT single-cell transcriptomics dataset (http://scmetab.mit.edu/), we determined how exercise affects components of the interstitial matrix and basement membrane in the different ASC subpopulations, namely interstitial progenitor cells (IPCs), adipogenesis-regulatory cells (Aregs), and committed preadipocytes (CPs). IPCs, the ASCs with the highest proliferation potential, demonstrate the lowest collagen mRNA levels related to the interstitial matrix and basement membrane. Exercise training decreased the expression of different collagen species in CPs and Aregs only. There was no effect of exercise on ECM components expressed in the IPC subpopulation (Figures S4A and S4B).
Taken together, these data suggest that the mesenchymal stem cell (MSC) compartment consisting of ASCs, in particular CPs and Aregs, preadipocytes, and fibroblasts in iWAT is primarily responsible for collagen synthesis. The reduction of interstitial matrix compartment-related collagens in the trained iWAT was due to a sharp decrease in the proportion of fibroblasts and a concomitant large increase in beige adipocytes, which exhibit the lowest collagen species content.
Exercise training promotes a switch in white adipocyte subpopulations
Beside MSCs, mature adipocytes are a major contributor to collagen synthesis and deposition. The expression of collagens in white adipocytes is directly related to their lipid volume: a larger cell volume results in a thicker basement membrane ECM compartment.29 To test the hypothesis that exercise training directly modulates the expression of collagen species in adipocytes, we annotated the white adipocyte subpopulations using a previously published cell annotation strategy.30 In WAT, there are three distinct white adipocyte subpopulations: lipogenic adipocytes (LGAs), lipid-scavenging adipocytes (LSAs), and stressed LSAs (SLSA). LGAs are the insulin sensitive adipocytes involved in de novo lipogenesis, while LSAs and SLSAs are mainly involved in lipid uptake and transport.30 Interestingly, exercise training shifts the ratio of adipocyte subpopulations from an LSA phenotype toward a significant enrichment of LGAs (Figure 2D). Few SLSAs were detected in both sedentary and trained iWAT. Across both tissues, LGAs showed lower expression of leptin, collagens, and ECM basement membrane components compared with LSAs (Figure 2E).
Next, we performed a spatial interaction analysis using a previously published pipeline31 to investigate the distance between beige adipocytes and Endo/PVMs, LGAs, and LSAs (Figure 2F). For both sedentary and exercise, we found a clear distribution pattern of these cell populations with beige adipocytes (Figure 2G). We also observed that beige adipocytes were closer to LGAs than to LSAs in both conditions. All cell clusters were equally distributed in the selected region of interest in sedentary iWAT. The interaction analysis model provides a strength score that quantifies the probability of cell proximity between beige adipocytes and the other clusters (probability density). LGAs had the highest probability density to beige adipocytes with an estimated strength score of 8.02, which reflects non-random close proximity (Figures S4C and S4D). In contrast, LSAs were at a greater distance (wide distribution) with an estimated strength score of 5.76 (Figures S4C and S4D). Interestingly, trained iWAT showed two peaks for the probability density. For one peak, there was a reduced distance between beige adipocytes and the Endo/PMV cluster (strength score = 8.26), while the second peak matched the sedentary. The reduced distance peak may be due to the induction of vascularization and innervation in iWAT following training. LSAs also showed a bimodal distribution, but their location is shifted farther from beige adipocytes (Figures 2G, S4C, and S4D). Taken together, these results indicate that exercise training robustly alters the proportion of LSAs to LGAs and modulates cell organization, providing an explanation as to how exercise training affects the basement membrane collagen deposition of the white adipocytes.
Exercise training increases vascularization in iWAT
We next investigated whether the changes in cell-type interactions with exercise training translated to changes in cellular processes such as innervation and vascularization. A significant proportion of the ECM proteins differentially expressed with exercise training in our tissue proteomics and secretome analysis were also annotated in the vascularization and innervation pathways32 (tissue proteome: 46 out of 166; secretome: 11 out of 23) (Figures S5A and S5B). These pathways highlight an important role of ECM proteins in neuronal and vascular remodeling in the context of exercise training.
To determine the effects of exercise training on angiogenesis, we performed immunofluorescence staining of sedentary and trained iWAT using the griffonia simplicifolia lectin (GSA-I) stain, which specifically binds to the blood vessel endothelial cells8,33 (Figure S5C). Analysis of iWAT images using Cell Profiler34 revealed a significantly higher total capillary density (Figure S5D) and capillary-to-adipocyte ratio (Figure S5E) in the trained compared with the sedentary iWAT. This exercise-induced iWAT vascularization was also evident at the gene expression level. Exercise training induced the expression of pro-angiogenic mediators,35 including vascular endothelial growth factor A (Vegfa) and its receptor (Kdr/Vegfr2), the Notch ligand Jagged1 (Jag1), Angiopoietin-1 (Angpt1), and Angiopoietin-2 (Angpt2) (Figures S5F-S5J). These microscopy and gene expression data demonstrate that exercise training increases vascularization in iWAT.
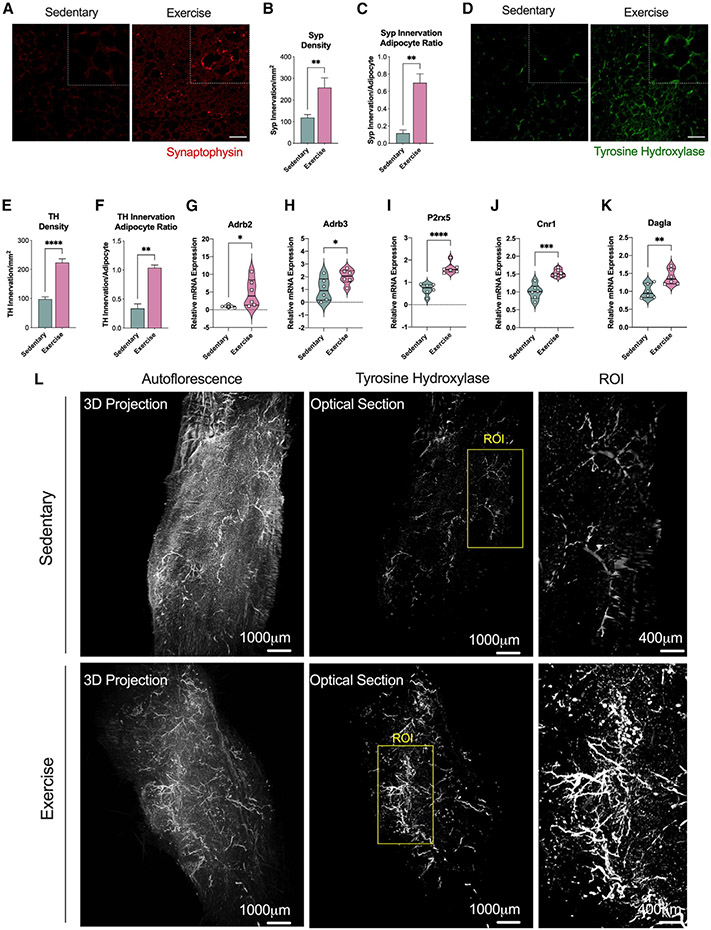
Exercise training increases sympathetic neurite density in iWAT
The effects of exercise training on iWAT innervation were determined with immunofluorescence analysis using two different stains: synaptophysin (Syp), an integral protein on presynaptic vesicles and a pan-marker for neural structures, and tyrosine hydroxylase (Th), a specific marker for sympathetic fibers.13 Exercise training increased the neural projections in iWAT (Figure 3A) with increased synaptophysin density and synaptophysin-to-adipocyte ratio (Figures 3B and 3C) and increased sympathetic innervation as shown in TH-stained images (Figures 3D-3F). We performed “Adipo-Clear,” which is a whole-mount tissue staining that provides a high-resolution volumetric fluorescent imaging of the neural network using TH staining,36 and found a tremendous increase of sympathetic innervation with exercise training compared with sedentary iWAT (Figure 3L). In addition, we clearly observed an increase in neurite development, also known as neuritogenesis (Figure 3L, ROI). We then measured expression of genes known to be involved in iWAT innervation. Exercise led to higher gene expression levels of adrenergic receptors β-2 (Adrb2) and β-3 (Adrb3), as well as the purine receptor P2rx5, a cell surface marker for beige/brown adipocytes that increases upon β3 stimulation37 (Figures 3G-3J). Cannabinoid 1 (Cnr1) and the diacylglycerol lipase α (Dagla), involved in the retrograde endocannabinoid signaling known for its regulatory role in synaptic plasticity, were also found elevated in iWAT following exercise training38 (Figures 3G-3K).
Figure 3. Exercise training increased innervation of iWAT.
(A) Immunofluorescence staining images of iWAT for the pan-innervation marker synaptophysin. Scale bar, 50 μm.
(B and C) Synaptophysin density (B) and synaptophysin innervation per adipocyte ratio (C) in iWAT from sedentary and trained mice (n = 6/group; calculated from 10 fields/mouse).
(D) Immunofluorescence staining images of iWAT for the sympathetic innervation marker TH. Scale bar, 50 μm.
(E and F) TH density (E) and TH innervation per adipocyte ratio (F) in iWAT from sedentary and trained mice (n = 6/group; calculated from 10 fields/mouse).
(G–K) mRNA expression of innervation markers Adrb2 (G), Adrb3 (H), P2rx5 (I), Cnr1 (J), and Dagla (K) in iWAT from sedentary and trained mice (n = 6/group).
(L) Whole-tissue images of iWAT from sedentary (top) and trained (bottom) mice immunolabeled with TH. Maximum intensity projection from a 1,000 μm z stack and high-magnification view of the region of interest (ROI) are shown.
Data are presented as mean ± SEM and were compared using unpaired two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Proteomics data identified up-regulation of proteins involved in the neurovascular remodeling and activation of neural stem cells (VEGFA)39 in self-renewal of neural progenitor cells (i.e., paired related homeobox protein 1 [Prrx1])40,41 and in the clearance of apoptotic neural cells (i.e., scavenger receptor class F member 1 [Scarf1]) (Figure S5K). The down-regulated proteins included the myelin proteolipid protein (Plp1) that is a component of the perineural net (PNN), a lattice-like inflexible structure of the ECM wrapping certain neurons and dendritic processes42 (Figure S5L). Taken together, the microscopic and molecular data underscore the ability of exercise to promote remodeling and refinement of the neuronal compartment in iWAT by promoting neuritogenesis.
Neuronal growth regulator 1 (NEGR1) is an exercise-training-induced cell adhesion molecule involved in neuritogenesis of iWAT
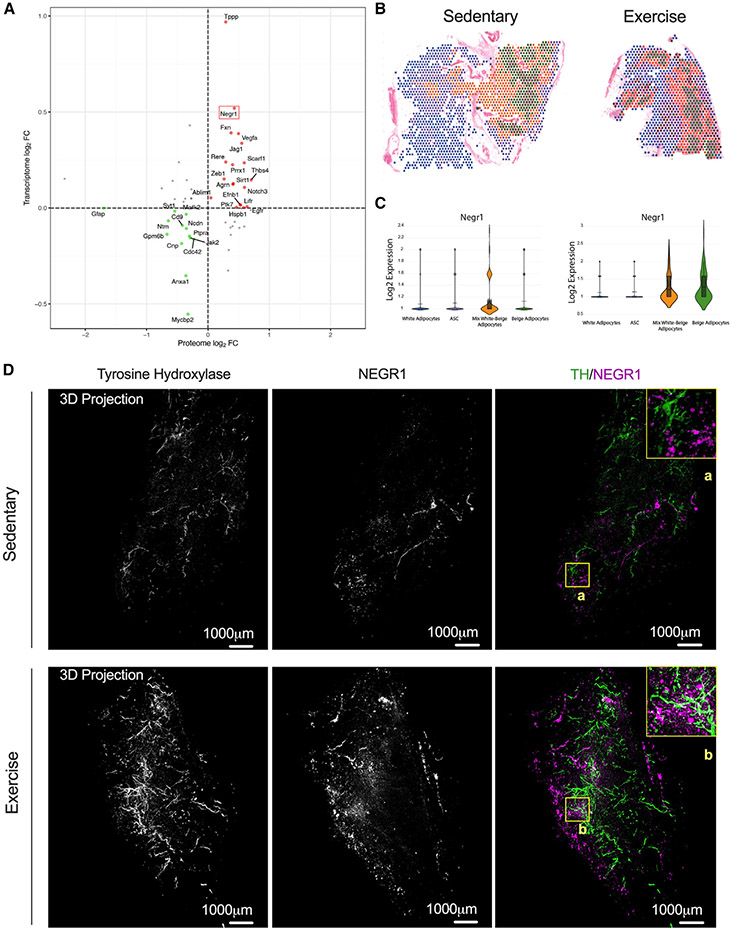
To understand the mechanism by which exercise training promotes neuritogenesis, we used our omics datasets to identify exercise-regulated proteins known to be involved in neurite outgrowth and synapse formation. Correlation analysis of our proteomics and transcriptomics data of all the innervation-related proteins determined NEGR1, a member of the IgLON superfamily of cell adhesion molecules (CAMs), as one of the most up-regulated exercise-induced molecule at both the mRNA and protein levels (Figure 4A). These findings were validated using qPCR and western blot analysis (Figures S6A-S6C). We also confirmed the increase of NEGR1 in a separate cohort of male C57BL/6 mice that underwent voluntary wheel (VW) training for 10 weeks, where training increased NEGR1 under conditions of both chow (20% fat) and high-fat diet (60% fat) (Figure S6D).
Figure 4. NEGR1 is a cell adhesion molecule induced by exercise training in iWAT.
(A) Correlation of RNA and protein expression in iWAT after 11 days of exercise. Negr1 is highlighted with a red box.
(B and C) Visium images (B) and relative individual violin plots (C) showing the Negr1 expression level across the cell clusters detected in iWAT from sedentary (left) and exercise (right).
(D) Whole-tissue images of iWAT from sedentary (top) and trained (bottom) mice immunolabeled with TH (green) and NEGR1 (magenta). Maximum intensity projection from a 1,000 μm z stack and high-magnification view of ROI are shown (A and B).
We determined the effects of exercise training on the spatial expression of NEGR1 in a cell-type-specific manner, compared Negr1 expression in the annotated spatial transcriptomics images (Figures 4B and 4C), and acquired high-resolution volumetric fluorescent imaging of a full pad of iWAT stained with TH using Adipo-Clear to distinguish whether training increased NEGR1 expression in neurons or mature adipocytes (Figure 4D). Our spatial transcriptomics data demonstrated that Negr1 is expressed in ASCs and in mature adipocytes (white and beige). Exercise training led to a significant increase in Negr1 expression, and this was mainly observed in the cluster of white/beige and beige adipocytes (Figure 4C). As shown earlier in Figures 2F and 2G, these are the clusters neighboring the nerves and vasculature. To demonstrate that Negr1 expression occurs primarily in differentiated adipocytes, we isolated mature adipocytes and ASCs from sedentary and trained iWAT (Figure S6E). Mature adipocytes were immediately homogenized for protein extraction, while ASCs were cultivated until passage 1 and differentiated into mature adipocytes before mRNA analysis (Figure S6E). We analyzed the mature adipocytes and found an increase of NEGR1 protein with exercise training (Figures S6F and S6G). ASCs from sedentary and trained mice did not exhibit any differences in Negr1 mRNA levels (Figure S6H). In contrast, we observed a significant increase of Negr1 in differentiated adipocytes (Figure S6H). The expression of Negr1 in differentiated adipocytes was also demonstrated using immunofluorescence that showed NEGR1 abundance in cells with lipid droplets (Figure S6I, MERGE). There was higher Negr1 immunofluorescence in the trained iWAT compared with sedentary (Figures 4D and S6I). This increase was predominantly observed in mature adipocytes and not in neurons, as there was no overlap in the fluorescence signaling between neurites and adipocytes (Figure 4D). An animated version of the 3D projection images from the trained mice is shown in Video S1. These data suggest that exercise training induces NEGR1 expression in white/beige adipocyte clusters that are in close proximity to innervated areas of the tissue, indicating a potential role of Negr1 as a neuronal growth regulator at the adipose tissue level.
Next, we performed promoter region analysis to identify potential TFs regulating the expression of Negr1 in iWAT. Using the JASPAR database,43 we found three binding sites for the Pparγ:Rxr heterodimer within the Negr1 promoter region (Figure S7A). This heterodimer is known to be involved in adipogenesis, lipid metabolism, and PRDM16 recruitment,44,45 while PPARγ agonists have been shown to induce a white-to-brown fat conversion through stabilization of the PRDM16 protein.46 To confirm the role of Pparγ in regulating Negr1 expression, we measured Negr1 levels in wild-type (WT) and Pparγ knockout (KO) mouse embryonic fibroblasts (MEFs). Negr1 expression was lower in Pparγ KO MEFs. Moreover, the PPARγ agonist rosiglitazone increased Negr1 levels in the WT MEFs but not in the Pparγ KO cell line (Figure S7B). Rosiglitazone treatment increased Negr1 mRNA expression in mature adipocytes, ASCs, and differentiated adipocytes from sedentary mice, with the greatest degree observed in mature adipocytes47 (Figures S7C-S7E). These results support the in silico promoter region analysis showing that NEGR1 expression is modulated by PPARγ. Given the known effect of PPARγ on PRDM16, we hypothesized that there may be a link between PRDM16 and Negr1 in regulating exercise-training-induced scWAT remodeling and innervation.
To investigate this hypothesis, we isolated mature adipocytes from PRDM16KO mice and detected lower expression of both NEGR1 mRNA and protein compared with WT (Figures S8A-S8C), which supports a potential interplay between NEGR1 and PRDM16. Using previously published PRDM16 ChIP-seq data in both brown adipose tissue and WAT from WT and PRDM16KO mice,48,49 we identified a PRDM16-binding site located within the promoter region of the Negr1 gene (Figure S8D) and found that the promoter region shows an RNA polymerase II (RNA Pol II) peak that is greatly reduced in PRDM16KO. Together, these data suggest that PRDM16 binding promotes NEGR1 transcription (Figure S8D). This finding along with our spatial transcriptomic data shown in Figures 4B and 4D demonstrate that Negr1 expression is restricted in iWAT to sites neighboring the nerves and beige adipocytes, where PRDM16 is crucial for their development.46,50 These findings suggest a link between PRDM16 and NEGR1 that may be responsible for the exercise-induced neurite sympathetic density of iWAT (Figure S8E).
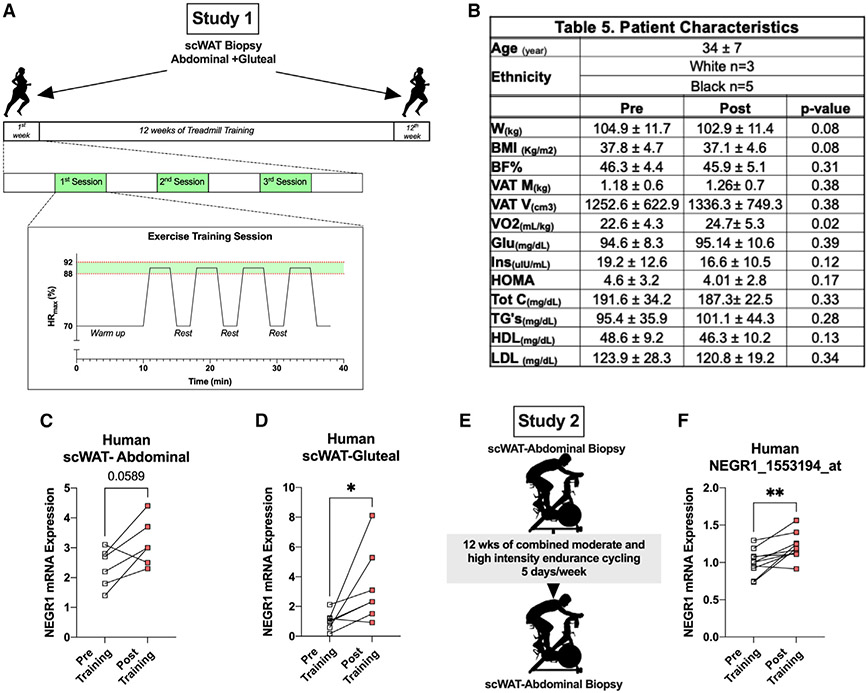
We then evaluated the effects of exercise training on NEGR1 expression in human scWAT collected from two different cohorts before and after supervised exercise training. Study 1 included obese women (n = 8) who performed 12 weeks of treadmill exercise training (Figures 5A and 5B). Training significantly improved maximal oxygen consumption (VO2max), and subjects lost ~2 kg body weight without a significant change in percentage of fat mass (Figure 5B). Exercise training increased NEGR1 mRNA expression in both abdominal and gluteal scWAT (Figures 5C and 5D). Study 2 included lean males (n = 10) who performed 12 weeks of combined moderate- and high-intensity endurance cycling 5 days/week as previously described (Figure 5E).51 Analysis of microarray data showed that exercise training significantly increased NEGR1 levels in abdominal scWAT (GEO: GSE116801) (Figure 5F).5 Our mouse data imply a relationship between Negr1 and beiging, and while exercise-induced beiging in humans is still not fully understood, our results from both these human studies showed an increase in NEGR1 with exercise training regardless of sex, BMI, training period, and fat depot. Taken together, we conclude that NEGR1 is an exercise-induced protein and potential key regulator of neuritogenesis in iWAT.
Figure 5. NEGR1 increased with exercise training in human subcutaneous WAT (scWAT).
(A) Study design to collect scWAT biopsies from obese women (n = 6/8) pre- and post-treadmill exercise training.
(B) Patient characteristics.
(C and D) mRNA expression of NEGR1 in abdominal (C) and gluteal (D) scWAT pre- and post-exercise (n = 6).
(E) Study design to collect abdominal scWAT biopsies from lean men (n = 10) pre- and post-moderate-intensity endurance cycling exercise.
(F) mRNA expression of NEGR1 in abdominal scWAT pre- and post-exercise training (n = 10) from the microarray dataset GEO: GSE116801.
Data are presented as mean ± SEM and were compared using paired Student’s test and two-way ANOVA followed by Tukey’s multiple comparisons t test. *p < 0.05 and **p < 0.01.
Adipo-PRDM16KO mice lack exercise-induced iWAT remodeling
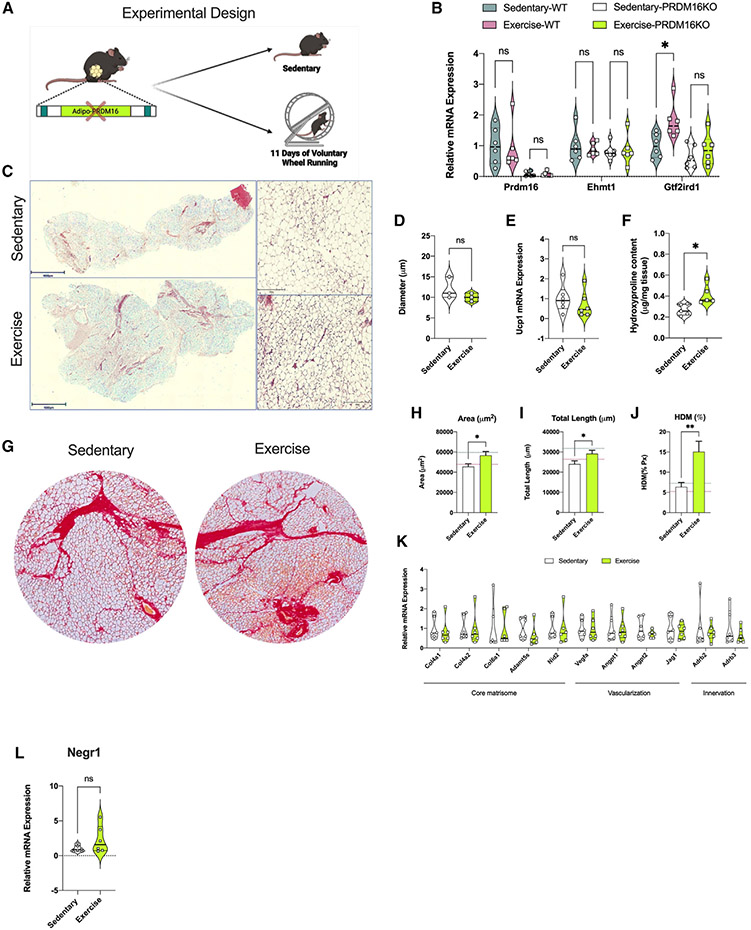
Given the potential relationship of PRDM16 with Negr1 expression in iWAT, the critical role of PRDM16 in tissue remodeling,52 and the paucity of data on the effect of PRDM16 in the exercise-induced iWAT adaptations, we decided to explore the effects of PRDM16 on iWAT remodeling with and without exercise training.52 Prdm16 is a transcriptional regulator that creates a complex with Gtf2ird1 and Ehmt1 to repress expression of pro-fibrotic genes.52,53 Gtf2ird1 is an essential component of this complex and plays a role in inhibiting fibrosis and promoting sympathetic neurite density in scWAT.53-55 We studied both Adipo-cre PRDM16KO and WT mice following 11 days of VW running and found no difference in running distance between genotypes (Figure 6A). In WT mice, exercise training did not alter Prdm16 and Ehmt1 expression levels but did lead to a significant increase in Gtf2ird1 (Figure 6B). We next investigated the phenotype of PRDM16KO and the effects of knocking out Prdm16 on training-induced ECM remodeling in iWAT. Within the PRDM16KO group, body weight was similar between the trained and sedentary mice, while caloric intake was higher in the trained group (Figures S8F and S8G). Trained PRDM16KO mice had lower fasting blood glucose compared with sedentary mice (Figure S8H). In the PRDM16KO mice, exercise training did not result in changes to adipocyte size compared with sedentary (Figures 6C and 6D). Interestingly, lack of PRDM16 prevented an exercise-induced increase in Ucp1 mRNA levels (Figure 6E) and the presence of multi-locular adipocytes. In contrast to trained WT mice (Figure 1C), we found increased collagen deposition in iWAT of trained PRDM16KO mice compared with the sedentary mice as demonstrated using the hydroxyproline assay (Figure 6F) and sirius red staining (Figures 6G-6J). There were no changes in mRNA expression of core matrisome genes or genes involved in vascularization and/or innervation in trained PRDM16KO iWAT (Figure 6K). Staining with TH showed that exercise training in PRDM16KO mice did not result in increased sympathetic neurite density, and this may be linked to a lack of increased NEGR1 mRNA expression (Figures 6L and S8I).
Figure 6. PRDM16 transcriptional complex mediated exercise-induced iWAT remodeling.
(A) Study design used for the PRDM16KO mice (n = 6/group).
(B) mRNA expression of the PRDM16 transcriptional complex genes Prdm16, Ehmt1, and Gtf2ird1 in iWAT from sedentary and trained wild-type (WT) and PRDM16KO mice (n = 6/group).
(C and D) H&E staining images (C) of iWAT from sedentary and trained mice with adipocyte cell size measurement (D) (n = 3/group; calculated from 10 fields/ mouse). Scale bar, 500 μm (4×) and 50 μm (20×), respectively.
(E) mRNA expression of Ucp1 in iWAT from sedentary and trained PRDM16KO mice (n = 6/group).
(F) Hydroxyproline content in iWAT from sedentary and trained PRDM16KO mice (n = 5/group).
(G) Sirius red staining images of iWAT from sedentary (left) and trained (right) PRDM16KO mice. Scale bar, 100 μm (10×).
(H–J) ECM deposition area (H), total length of fibers (I), and percentage of HDM (J) in iWAT of sedentary and trained PRDM16KO mice (n = 4/group; calculated from 8 fields/mouse).
(K) mRNA expression level for core matrisome, vascularization, and innervation markers in iWAT of sedentary and trained PRDM16KO mice (n = 6/group).
(L) mRNA expression of Negr1 in iWAT from sedentary and trained PRDM16KO mice (n = 6/group).
Data are presented as mean ± SEM and were compared using unpaired two-tailed Student’s t test and two-way ANOVA followed by Tukey’s multiple comparisons test. *p < 0.05 and **p < 0.01.
We then used spatial transcriptomics to study transcriptomic changes and changes in the cellular composition and organization of the PRDM16KO iWAT with and without exercise training. We found a total absence of beige adipocytes (Figures 6C, S9A, and S9B) and demonstrated an increased proportion of cells known to be involved in collagen deposition and fibrosis, including fibroblasts in trained PRDM16KO iWAT (Figure S9B). In the PRDM16KO iWAT, both ASCs and mature adipocytes are responsible for the expression of the different collagen species found in the interstitial matrix and basement membrane compartment of ECM (Figures S9C and S9D). A subcluster analysis of the mature adipocytes in the PRDM16KO iWAT showed a similar expression of the different collagen species in LSAs, LGAs, and SLSAs (Figures S9E and S9F). In addition, in the PRDM16KO iWAT, exercise leads to an increased proportion of the LSAs (Figure S9E), in contrast to the results obtained in WT iWAT (Figure 2D). These findings imply an important role of PRDM16 in determining the phenotype of different adipocyte subpopulations. Absence of a functional PRDM16 complex leads to increased collagen deposition, fibrosis, and overall altered ECM remodeling, highlighting the critical role of the PRDM16 in mediating the exercise-induced ECM remodeling.
DISCUSSION
Here, we show that exercise training changes the structural architecture of iWAT and promotes healthy expansion of the tissue by reducing collagen deposition and inducing vascularization and neuritogenesis. ECM is a significant modulator of cell behavior, function, and fate, and structural alterations in ECM have been associated with the development and progression of numerous diseases including cancer and cardiometabolic disease.10,56 Recent studies have demonstrated the important role of adipocyte progenitors in adipose tissue fibrosis in the context of obesity.57 Our results, showing that exercise training favorably regulates ECM remodeling, thereby facilitating healthy expansion of WAT, provides a physiological model for identification of key molecular adaptations, which in turn could become therapeutic targets. As such, our data underscore the critical role of ASCs in ECM reorganization and remodeling in iWAT. Based on our findings, we hypothesize that in ASCs, TGF-β/SMAD3 signaling functions as a master regulator of collagen species that control ECM structure and function. The prominent role of ASCs in mediating exercise- and obesity-induced adaptations has also emerged through our recent multi-tissue, single-cell analysis of exercise and obesity in rodents.6
At the cellular level, we observe that training increases the degree of heterogeneity, changing the proportion of cell types and states, which reflects the “multi-tasking” role of WAT. We demonstrate changes in both the cellular proportion and the molecular signatures of mature adipocytes with a significant increase in the beige adipocyte population. Another intriguing finding is that exercise training promotes a shift toward the insulin-sensitive white adipocytes (LGAs) and reduces the presence of the stressed/hypertrophic adipocytes (LSAs). We show that there is high collagen gene expression in LSAs and that their reduction with exercise training directly reflects the reduction in collagen deposition in trained iWAT. Our data suggest that one of the mechanisms by which exercise training has beneficial effects on systemic metabolism is by promoting a healthier phenotype in white adipocytes, an effect mediated by repressing the LSA subpopulation that plays a prominent role in ECM deposition.
Although exercise-induced vascular and innervation remodeling has been described in skeletal muscle,58 whether this occurs in WAT had not been previously studied. Here, we discover that exercise training results in a robust vascular remodeling and neuronal refinement and we hypothesize that this results in the reconfiguration of WAT to allow for both the metabolic and energy balance requirements of physical activity. According to prior studies, the PRDM16 transcriptional complex, consisting of PRDM16, GTF2IRD1, and EHMT1, is one of the main regulators of neuronal refinement and ECM remodeling in WAT.53,54 Given its important role in tissue remodeling, we investigated the effects of exercise training on the PRDM16 transcriptional complex. Interestingly, we found that exercise training did not increase Prdm16 levels but led to a significant increase in Gtf2ird1, an important component of the PRDM16 transcriptional complex. Thus, the effects of exercise training on beiging are likely mediated through the interaction of different TFs forming a complex and not through an effect to increase Prdm16 levels. Our finding that training failed to alter beiging and mRNA expression of core matrisome, vascularization, and innervation gene markers in iWAT from PRDM16KO mice underscores the important role of this transcriptional complex in mediating the effects of exercise on iWAT. Our spatial transcriptomics data demonstrated the absolute lack of beige adipocytes in the PRDM16KO iWAT as well as the impact of an absent PRDM16 transcription complex on the phenotype of the different mature adipocyte subpopulations. Our data along with recently published data by other groups confirm the critical role of PRDM16 in tissue remodeling, which may be at least partially mediated either through effects on adipocyte transdifferentiation and adipose precursor cells recruitment.52
Studies in the PRDM16KO mice also led us to identify NEGR1 as a potential link between the PRDM16 TF complex and neurite density. NEGR1 is a membrane protein anchored to glycosylphosphatidylinositol that has primarily been studied in the brain, where it mediates neural cell communication and synapse formation.59 NEGR1 has been identified in genome-wide association studies (GWASs) as a gene implicated in obesity and was shown to increase by ~1.8-fold after 12 days of adipocyte differentiation in vitro60 but has not been further studied. We found that NEGR1 is significantly increased in both mouse and human WAT in response to exercise training and is highly expressed in mature adipocytes in proximity to nerve endings, and therefore we hypothesize that NEGR1 is a key regulator of exercise-induced WAT innervation. In addition to neurite outgrowth, we propose that the PDRM16-NEGR1 axis is involved in de novo adipogenesis. We base this hypothesis on the finding that the Pparγ:Rxr TF heterodimer, known to be involved in adipogenesis, has the highest enrichment score at the Negr1 promoter region.
Consistent with the mouse models, our human studies reveal that exercise training increases NEGR1 in scWAT in both males and females. The importance of NEGR1 in metabolic health is also demonstrated by two different GWAS analyses. In one, there was a strong association between NEGR1 common variants and obesity,61-63 and in the second, there was a strong association between a NEGR1 locus variant and moderate-to-vigorous physical activity.64 All these data point to NEGR1 as a novel, exercise-induced molecule expressed in adipocytes that may play a significant role in regulating metabolic health. Future identification of an agonist that increases NEGR1 may prove to be a new target for the treatment of obesity and metabolic disease. In addition, studies of NEGR1 in other tissues may reveal a broader role for NEGR1 in mediating beneficial effects of exercise on neural and metabolic health.
In conclusion, we have discovered that exercise training is a potent stimulus of iWAT remodeling through fundamental changes in ECM, vascularization, and innervation. We identify robust cell-type-specific adaptations in response to exercise training that, in combination with the structural changes, undoubtedly results in a healthier adipose tissue phenotype. Finally, the extensive single-cell, transcriptomic, and proteomic data, in conjunction with detailed imaging and biochemical analyses, provide a resource for identifying novel iWAT factors that can be used to develop therapeutics for the treatment of metabolic disease.
Limitations of the study
Our mouse studies were performed in male mice only. However, we studied the exercise effect on Negr1 in both male and female subjects. We performed spatial transcriptomics using the Visium platform, which can only detect mRNA expression in cell clusters and not individual cells.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Laurie J.
Goodyear (laurie.goodyear@joslin.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository. Mouse and human microarray data were previously published and available in the Gene Expression Omnibus database. Spatial transcriptomic data have been deposited at Mendeley and are publicly available as of the date of publication. The DOI for all the omics are listed in the key resources table. Original western blots (Data S1), and microscopy raw data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-mouse synaptophysin (1:300) | ProteinTech | Cat# 17785-1-AP; RRID: AB_2271365 |
| Mouse anti-mouse tyrosine hydroxylase (1:200) | EMD Millipore | Cat# MAB318; RRID: AB_2201528 |
| Alexa Fluor 488 donkey anti-rabbit (1:400) | Life Technologies | Cat# A21441; RRID: AB_2535859 |
| Alexa Fluor 594 goat anti-rabbit (1:400) | Life Technologies | Cat# A11072; RRID: AB_142057 |
| Alexa Fluor 488 donkey anti-mouse (1:400) | Life Technologies | Cat# A21202; RRID: AB_141607 |
| Alexa Fluor 594 goat anti-mouse (1:400) | Life Technologies | Cat# A11037; RRID: AB_2534095 |
| Alexa Fluor 647 Goat anti-mouse (1:400) | Life Technologies | Cat# A21235; RRID: AB_2535804 |
| Anti-mouse NEGR1 (1:200) | ProteinTech | Cat# 13674-1-AP; RRID: AB_2877969 |
| Biological samples | ||
| Human subcutaneous abdominal and gluteal white adipose tissue | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 1uM insulin | Sigma | Cat# 11376497001 |
| 250nM Dexamethasone | Sigma-Aldrich | Cat# D1756 |
| 1% Pen/Strep | Thermo-Scientific | Cat# 15140122 |
| Direct-zol RNA MiniPrep | Zymo Research | Cat# R2052 |
| Rosiglitazone | Cayman Chemical | Cat# 122320-73-4 |
| High-Capacity Reverse Transcription Kits | Applied Biosystems | Cat# 4368813 |
| Dichloromethane (DCM) | Sigma-Adrich | Cat# 270991 |
| Benzyl ether (DBE) | Sigma-Adrich | Cat# 108014 |
| UltraPure Agarose | Invitrogen | Cat# 16500500 |
| Triton X-100 | Sigma-Adrich | Cat# X100-500ML |
| Tween 20 | Sigma Adrich | Cat# P2287-500ML |
| Critical commercial assays | ||
| Visium Spatial Tissue Optimization Slide & Reagent Kit | 10x Genomics | Cat# PN-1000193 |
| Visium Spatial Gene Expression Solution v1 | 10x Genomics | Cat# PN-1000184 |
| Dual Index Kit TT Set A | 10x Genomics | Cat# PN-1000215 |
| Deposited data | ||
| Raw and processed MS data | This study, ProteomeXchange Data | PXD040894 |
| Raw and processed MS data | This study, ProteomeXchange Data | PXD040894 |
| Visium normalized data | This study, Mendeley Data | https://doi.org/10.17632/r6v7yktxnt.1 |
| Visium raw data and Code for Visium data analyses | This study, Mendeley Data | https://doi.org/10.17632/pktxwdvcbz.1 |
| Proteome and Secretome analysis | Dreyfuss et al.65 | https://github.com/jdreyf/ezlimma |
| Rajbhandari et al.,2019 scRNA-seq data of adipose tissue | Rajbhandari et al.28 | GEO: (GSE133486) |
| Ermont et al.,2022 scRNA-seq data of adipose tissue | Ermont et al.66 | GEO: (GSE176067) |
| Ermont et al.,2022 snRNA-seq data of adipose tissue | Ermont et al.66 | GEO: (GSE176171) |
| Sárvári et al.,2021 snRNA-seq data of adipose tissue | Sárvári et al.30 | GEO: (GSE160729) |
| Takahashi et al.,2019 Human microarray | Takahashi et al.5 | GEO: (GSE116801) |
| Stanford et al.,2015 Mouse microarray | Stanford et al.3 | GEO: (GSE68161) |
| Experimental models: Cell lines | ||
| Primary adipose derived stem cells from mouse iWAT | This study | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: Male C57BL/6NCrl | Charles River Lab | Cat# 027 |
| Mouse: Adipo-PRDM16 KO PRDM16lox/lox | Jackson Laboratory | Cat# 24992 |
| Mouse: Adiponectin-cre | Jackson Laboratory | Cat# 28020 |
| Oligonucleotides | ||
| Total of 50 oligonucleotides | This study | Table S4 |
| Software and algorithms | ||
| CellProfiler 3.0 | Carpenter et al67 | http://cellprofiler.org; RRID: SCR_007358 |
| FIJI macro TWOMBLI | Wershof et al.15 | https://github.com/wershofe/TWOMBLI |
| Fiji v1 | ImageJ | https://imagej.net/software/fiji/; RRID: SCR_002285 |
| Imaris 9.7 | BitPlane | https://imaris.oxinst.com; RRID: SCR_007370 |
| Cytoscape | Shannon et al. 68 | https://cytoscape.org; RRID: SCR_003032 |
| String Database | Szklarczyk et al. 69 | https://string-db.org; RRID: SCR_005223 |
| Metascape | Zhou et al. 32 | https://metascape.org; RRID: SCR_016620 |
| GraphPad Prism v9 | GraphPad | https://www.graphpad.com; RRID: SCR_002798 |
| Loupe Browser version 6.0.0 | 10x Genomics | https://10xgenomics.com; RRID: SCR_018555 |
| R 4.04 | See computational analysis section for R packages used | https://www.r-project.org/about.html |
| Spaceranger-1.3.0 | 10x Genomics | https://www.10xgenomics.com |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All experiments were conducted following NIH guidelines and protocols approved by the IACUC at Joslin Diabetes Center (JDC). 8-week-old male C57BL/6 mice (Charles River Laboratories, MA) were housed at 23°C on a 12/12 h light/dark cycle. Male mice were studied since exercise training in males, but not females, results in beiging adaptations of iWAT. Standard mouse chow diet (9F 5020 Lab Diet, PharmaServ, Inc.) and water were provided ad libitum. For the exercise training experiments, mice were housed with or without a running wheel (24 cm diameter Techniplast) for 11 days. This time period is chosen based on our previous published works showing an optimal training response and maintenance of the adipose tissue depots.3 To generate Adipo-PRDM16 KO PRDM16lox/lox (024992, Jackson Laboratory) mice were crossed with Adiponectin-cre mice (028020, Jackson Laboratory).
Human studies
Both study protocols were approved by the local Ethical Committees: The Institutional Review Board of East Carolina University for Study 1 and Copenhagen and Frederiksberg (KF 01 289434) for Study 2. Both studies were carried out in compliance with the Declaration of Helsinki and received an exempt status by the Institutional Review Board of Joslin Diabetes Center. All participants provided informed written consent prior to their enrollment in the two studies. Baseline and follow up characteristics of subjects from Study 1 are summarized in Figure 5B. In this study, eight women with mean age 34 ± 7 years of age and BMI 37.8 ± 4.7 kg/m2 performed a 12-week moderate to high intensity exercise training program (Figure 5A). All subjects had to attend the clinical research center of East Carolina University for three supervised exercise sessions per week using the treadmill. Each exercise session began with a 10-min warm up followed by four bouts of high intensity interval training of 5-min duration each during which subjects achieved 88–92% max heart rate. Pre- and post-exercise training subcutaneous WAT biopsies were collected from the abdominal and gluteal area using a biopsy needle. The WAT samples were snap frozen in liquid nitrogen and stored in −80°C until further analysis. In Study 2, as previously described, a total of 10 male subjects with mean age of 30.5 years (95% CI 27–34) and mean BMI of 24.9 kg/m2 (95% CI 23.4–26.3) underwent a 12-week exercise training program which consisted of 60–80 min cycling/day. Biopsies of abdominal scWAT were obtained using a biopsy needle before and after the 12 weeks of training.70 NEGR1 mRNA relative expression was analyzed in human scWAT from the two different studies.
Cell culture
All cell lines used in this study were cultured in high glucose Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS). Preadipocytes and adipocytes were isolated from iWAT of 10-week-old male C57BL6 mice as previously described.4 White preadipocytes were differentiated as described before.4 Briefly, cells were allowed to reach confluency, and treated with induction media supplemented with 2% FBS, 20 nM insulin, 1 nM triiodothyronine (T3), 5 μM dexamethasone, and 0.5 mM isobutylmethylxanthine (IBMX) for 48 h. Then, cells were maintained in differentiation media supplemented with 10% FBS and 1 μM insulin 6 more days. Pparg knockout mouse embryonic fibroblasts (MEFKO) were generously gifted from Dr. Farnaz Shamsi and were cultivated as previously described.71
METHOD DETAILS
Immunocytochemistry and quantification of innervation and vascularization density
Inguinal white adipose tissue (iWAT) was fixed in 10% phosphate-buffered formalin and embedded in paraffin. Immunofluorescence was carried out using rabbit anti-mouse synaptophysin (1:300) (17785-1-AP, Proteintech) and mouse anti-mouse tyrosine hydroxylase (1:200) (MAB318, EMD Millipore) primary antibodies. Primary antibodies were incubated overnight at 4°C. The secondary antibodies used were AlexaFluor 488 donkey anti-rabbit and 594 goat anti-rabbit (A11037, A11072, A11020, A21235, Life Technologies) at a 1:400 dilution, and were incubated for 1 h at room temperature. Images were either captured with an inverted fluorescence microscope (IX51 Olympus) or with a confocal laser scanning microscope (Zeiss LSM 710). Hematoxylin and eosin (H&E) images were acquired with the EVOS M7000 Imaging system (Thermo Fisher System). Synaptophysin, tyrosine hydroxylase and griffonia simplicifolia agglutinin-I were quantified using CellProfiler 3.0 (http://cellprofiler.org). Innervation and vascularization densities were quantified as the number of sprinkles normalized to adipocytes count (mm2/adipocytes).
Measurement of fibrosis and collagen production
Sirius Red staining was used to quantify the extracellular matrix (ECM) and collagen content in white adipose tissue. Briefly, tissues were fixed overnight in 4% paraformaldehyde and processed for standard paraffin embedding. Sections of 3 μm were stained as previously described72 and at least 8 images were captured per slide on an inverted fluorescence microscope (IX51 Olympus) using 10× objective and were analyzed using the FIJI macro TWOMBLI.15 Hydroxyproline content in adipose tissue was measured with a Hydroxyproline Assay Kit (Quickzyme Biosciences).
Whole mount tissue staining and 3D imaging
AdipoClear was performed as previously described.36 Anti-mouse TH (1:200) (MAB318, EMD Millipore) and anti-mouse NEGR1 (1:200) (13674-1-AP, ProteinTech) were used. Whole-tissue iWAT samples were imaged on a light sheet microscope (Ultramiscroscope II, LaVision Biotec) equipped with 1.6× objective lenses and an sCMOs camera (Andor Neo). Images were acquired with the Inspector Pro software (LaVision BioTec). Samples were positioned in an imaging chamber filled with dibenzyl ether (DBE) and ivluminated from one side by the laser light sheet with 488 and 561 nm laser channels. Whole tissue 3D images were analyzed using Fiji v1(ImageJ) and Imaris 9.7 (BitPlane).
Quantitative proteomics of secretome proteins from adipose tissue organ culture
For the secretome analysis, Quantitative Immunoprecipitation (Quant-IP) analysis was performed. Briefly, 20 mg of iWAT was washed three times in PBS 1X and incubated in the M199 media supplemented with 1 μM insulin (11376497001 Sigma), 250nM Dexamethasone (D1756 Sigma-Aldrich) and 1%Pen/Strep (15140122, Thermo-Scientific). After 24hr media was replaced with fresh M199 media plus 1% Pen/Strep. The media was collected after 24hr and immediately stored at −80°C. Debris from the media was removed by centrifugation at 4000 × g. The protein-containing supernatant was mixed with a lysis buffer composed of 50 mM EPPS pH 8, 1 M Urea, 1% SDS, and protease inhibitors (Roche EDTA-free protease inhibitor tablet: 1 mini table per 10 mL). The reduction and alkylation of the tryptic peptides was achieved using 5 mM TCEP, 15 mM Iodoacetamide and 10 mM DTT at 56°C for 30 min. Protein precipitation was done with TCA, precipitate was dissolved 50 mM EPPS. Digestion was performed using Trypsin (1:100) based on protease to protein volume ratio. Each sample was labeled with TMT tags for multiplex proteomics.
Orbitrap-based LC-MS/MS analysis
Peptides were separated using a gradient of 3–36% Buffer B (90% Acetonitrile in 0.1% formic acid) over 120 min. MS analysis was performed using an Orbitrap Fusion™ Tribrid™ Mass Spectrometer (Thermo Scientific, United States) interfaced with an EASY-nLC 1200 System. The following 11-plex data search Sequest parameters were used: Peptide Mass Tolerance = 20 ppm, Fragment Ion Tolerance = 1, Max Internal Cleavage Site = 2, Max differential/Sites = 4. For Reported Quant Parameter: tolerance = 0.003, ms3 = 1, peak picking = max, num isotopes = 2, ms2_isolation_width = 0.7, ms3_isolation_width = 1.2. MS2 spectra were searched using the SEQUEST algorithm against a Uniprot composite database derived from Mouse proteome containing its reversed complement and known contaminants. Peptide spectral matches were filtered to a 1% false discovery rate (FDR) using the target-decoy strategy combined with linear discriminant analysis. The proteins were filtered to a <1% FDR. Proteins were quantified only from peptides with a summed SN threshold of >200 and MS2 isolation specificity of 0.5.
For both secretome and proteome analysis, we used a total of 11 samples including iWAT from cold exposed mice. For the purpose of this study, we are including only data from the sedentary and exercise trained iWATs. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD040894.
Spatial transcriptomics
For spatial transcriptomics we used the 10x Genomics Visium Spatial Gene Expression platform (10x Genomics, Pleasanton, CA, USA). The paraffin-embedded iWAT from sedentary and trained Wild Type and PRDM16KO mice were used for the experiment. Visium Spatial Gene Expression was then carried out according to the manufacturer’s instructions (10x Genomics, Pleasanton, CA, USA) with the following parameters: 20 μm thick sections of adipose tissue were used, H&E staining was carried out by incubating slides for 1 min in isopropanol, 3 min in hematoxylin and 30 s in eosin (1:20 dilution). Permeabilization was carried out for 15 min. All Visium cDNA libraries were indexed, pooled, and sequenced simultaneously on the Illumina NovaSeq6000 platform, supported by the BioMicro Center Core at MIT. In accordance with the Visium protocol, the number of bases sequenced were 28 nt for R1, 120 nt for R2, and 10 nt for each of the indexes.
RNA isolation and quantitative real-time PCR
RNA was isolated from mouse and human adipose tissue using an RNA extraction kit (Direct-zol RNA MiniPrep, Zymo Research). RNA was reverse-transcribed using standard reagents (High-Capacity Reverse Transcription Kits, Applied Biosystems), and cDNA was amplified by RT- PCR. For each gene, mRNA expression was calculated relative to Actb and 18s. Primer sequences used in RT-qPCR are presented in Table S4.
Computational analysis
Proteome and secretome scaled relative intensities were processed as follows. For the tissue secretome, we annotated secreted proteins if the proteins belong to Gene Ontology (GO) terms of either “extracellular region” GO:0005576 or “extracellular space” GO:0005615, or the predicted human secretome from Human Protein Atlas by using three methods for signal peptide prediction. We unbiasedly estimated the empirical sample quality weights in the R package limma (Ritchie et al. 2006). We observed a trend between a protein’s average abundance and its variance and accounted for it in variance estimation using limma-trend (Law et al. 2014). For the proteome, we performed Surrogate Variables Analysis (SVA) to remove unknown batch effects (i.e. the unwanted variation that is independent of the group assignment) using the R package sva. We tested the differential abundance of each protein using limma (Ritchie et al. 2015). Volcano plots were generated using R package ggplot2. The protein-protein interaction analysis was performed using Cytoscape68 open source software integrating the network analysis generated with String Database69 and Metascape.32 Spatial data were aligned to the mm10-2020-A reference using spaceranger-1.3.0 with default parameters for each of the samples. We used the R package Seurat to normalize, identify marker genes, reduce dimensions, cluster, annotate cell types and visualize the spatial data. We integrated our spatial data with the publicly available single-cell datasets using FindTransferAnchors and TransferData functions provided by the Seurat package. For spatial distribution analysis we selected region of interests (ROI) in both sedentary and training using Loupe Browser 6. End/PVM cells, Beige, LGA and LSA adipocytes have been identified as cells/dots expressing Cdh5 and Axl log2FC > 1, Ucp1 log2FC > 4, Pparg and Acaca and Acly log2FC > 1, Nnat and Abcd2 log2FC > 2 respectively. Control plots have been generated in the same ROI selecting cells expressing Car3log2FC > 5. Interaction potential was calculated using the MosaicIA plugin in ImageJ as interaction strength (ε) using parametric Hernquist potential73-75
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are expressed as mean ± SEM. Sample sizes are indicated in the figure legends. All statistical analyses were performed using GraphPad Prism v9. Statistical significance was analyzed by One-way ANOVA and two-way ANOVA followed by Tukey’s multiple comparisons test or unpaired two-tailed Student’s t-test.
Supplementary Material
Highlights.
Exercise training decreases ECM and increases vascularization and innervation in iWAT
Adipose stem cells (ASCs) are major contributors of exercise-induced ECM remodeling
PRDM16 transcriptional complex is necessary for exercise-induced iWAT remodeling
NEGR1 functions as a link between PRDM16 and exercise-induced iWAT innervation
ACKNOWLEDGMENTS
This work was supported by NIH grants R01DK099511 and R01DK101043 (to L.J.G.); K23DK114550 (to R.J.W.M.); T32DK00726042, F32DK12643201,and Joslin DRC Pilot and Feasibility Program award (to M.V.); and the Joslin Diabetes Center DRC (P30 DK36836). The Center for Physical Activity Research (CFAS) is supported by TrygFonden (grant IDs 101390, 20045, and 125132). The Center of Inflammation and Metabolism (CIM) was supported by a grant from the Danish National Research Foundation (DNRF55). We thank Dr. Krithika Ramachandran and Madison M. Columbus for scientific feedback, Drs. Michael R. Blanchard and Mahmoud EL-Rifai for technical assistance with Adipo-Clear image acquisition, and Drs. Hui Pan and Jonathan M. Dreyfuss for statistical analysis support.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112392.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Li S, Gao H, Hasegawa Y, and Lu X (2021). Mini-Review: fight against fibrosis in adipose tissue remodeling. Am. J. Physiol. Endocrinol. Metab 321, E169–E175. 10.1152/ajpendo.00558.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruiz-Ojeda FJ, Méndez-Gutiérrez A, Aguilera CM, and Plaza-Díaz J (2019). Extracellular matrix remodeling of adipose tissue in obesity and metabolic diseases. Int. J. Mol. Sci 20, 4888. 10.3390/ijms20194888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanford KI, Middelbeek RJW, Townsend KL, Lee M-Y, Takahashi H, So K, Hitchcox KM, Markan KR, Hellbach K, Hirshman MF, et al. (2015). A novel role for subcutaneous adipose tissue in exercise-induced improvements in glucose homeostasis. Diabetes 64, 2002–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigro P, Middelbeek RJW, Alves CRR, Rovira-Llopis S, Ramachandran K, Rowland LA, Møller AB, Takahashi H, Alves-Wagner AB, Vamvini M, et al. (2021). Exercise training promotes sex-specific adaptations in mouse inguinal white adipose tissue. Diabetes 70, 1250–1264. 10.2337/db20-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi H, Alves CRR, Stanford KI, Middelbeek RJW, Nigro P, Ryan RE, Xue R, Sakaguchi M, Lynes MD, So K, et al. (2019). TGF-β2 is an exercise-induced adipokine that regulates glucose and fatty acid metabolism. Nat. Metab 1, 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang J, Vamvini M, Nigro P, Ho L-L, Galani K, Alvarez M, Tanigawa Y, Renfro A, Carbone NP, Laakso M, et al. (2022). Single-cell dissection of the obesity-exercise axis in adipose-muscle tissues implies a critical role for mesenchymal stem cells. Cell Metab. 34, 1578–1593.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marcelin G, and Clément K (2021). The multifaceted progenitor fates in healthy or unhealthy adipose tissue during obesity. Rev. Endocr. Metab. Disord 22, 1111–1119. 10.1007/s11154-021-09662-0. [DOI] [PubMed] [Google Scholar]
- 8.Lu P, Takai K, Weaver VM, and Werb Z (2011). Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb. Perspect. Biol 3, a005058. 10.1101/cshperspect.a005058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long KR, and Huttner WB (2019). How the extracellular matrix shapes neural development. Open Biol. 9, 180216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spencer M, Unal R, Zhu B, Rasouli N, McGehee RE Jr., Peterson CA, and Kern PA (2011). Adipose tissue extracellular matrix and vascular abnormalities in obesity and insulin resistance. J. Clin. Endocrinol. Metab 96, E1990–E1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lambert EA, Straznicky NE, Dixon JB, and Lambert GW (2015). Should the sympathetic nervous system be a target to improve cardiometabolic risk in obesity? Am. J. Physiol. Heart Circ. Physiol 309, H244–H258. [DOI] [PubMed] [Google Scholar]
- 12.Bartness TJ, Liu Y, Shrestha YB, and Ryu V (2014). Neural innervation of white adipose tissue and the control of lipolysis. Front. Neuroendocrinol 35, 473–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao Y, Wang H, and Zeng W (2018). Whole-tissue 3D imaging reveals intra-adipose sympathetic plasticity regulated by NGF-TrkA signal in cold-induced beiging. Protein Cell 9, 527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HJ (2018). Exercise training regulates angiogenic gene expression in white adipose tissue. J. Exerc. Rehabil 14, 16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wershof E, Park D, Barry DJ, Jenkins RP, Rullan A, Wilkins A, Schlegelmilch K, Roxanis I, Anderson KI, Bates PA, and Sahai E (2021). A Fiji macro for quantifying pattern in extracellular matrix. Life Sci. Alliance 4, e202000880. 10.26508/lsa.202000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naba A, Clauser KR, Hoersch S, Liu H, Carr SA, and Hynes RO (2012). The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteomics 11. M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, and Hynes RO (2016). The extracellular matrix: tools and insights for the “omics” era. Matrix Biol. 49, 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maquoi E, Munaut C, Colige A, Collen D, and Lijnen HR (2002). Modulation of adipose tissue expression of murine matrix metalloproteinases and their tissue inhibitors with obesity. Diabetes 51, 1093–1101. [DOI] [PubMed] [Google Scholar]
- 19.Bauters D, Cobbaut M, Geys L, Van Lint J, Hemmeryckx B, and Lijnen HR (2017). Loss of ADAMTS5 enhances brown adipose tissue mass and promotes browning of white adipose tissue via CREB signaling. Mol. Metab 6, 715–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bondeson J, Wainwright S, Hughes C, and Caterson B (2008). The regulation of the ADAMTS4 and ADAMTS5 aggrecanases in osteoarthritis: a review. Clin. Exp. Rheumatol 26, 139–145. [PubMed] [Google Scholar]
- 21.Martínez-Martínez E, Calvier L, Rossignol P, Rousseau E, Fernández-Celis A, Jurado-López R, Laville M, Cachofeiro V, and López-Andrés N (2016). Galectin-3 inhibition prevents adipose tissue remodelling in obesity. Int. J. Obes 40, 1034–1038. [DOI] [PubMed] [Google Scholar]
- 22.Sakry D, Neitz A, Singh J, Frischknecht R, Marongiu D, Binamé F, Perera SS, Endres K, Lutz B, Radyushkin K, et al. (2014). Oligodendrocyte precursor cells modulate the neuronal network by activity-dependent ectodomain cleavage of glial NG2. PLoS Biol. 12, e1001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakry D, and Trotter J (2016). The role of the NG2 proteoglycan in OPC and CNS network function. Brain Res. 1638, 161–166. [DOI] [PubMed] [Google Scholar]
- 24.Tammaro A, Florquin S, Brok M, Claessen N, Butter LM, Teske GJD, de Boer OJ, Vogl T, Leemans JC, and Dessing MC (2018). S100A8/A9 promotes parenchymal damage and renal fibrosis in obstructive nephropathy. Clin. Exp. Immunol 193, 361–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng X, Ye M, Resch JM, Jedrychowski MP, Hu B, Lowell BB, Ginty DD, and Spiegelman BM (2019). Innervation of thermogenic adipose tissue via a calsyntenin 3β-S100b axis. Nature 569, 229–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G-X, Zhao X-Y, Meng Z-X, Kern M, Dietrich A, Chen Z, Cozacov Z, Zhou D, Okunade AL, Su X, et al. (2014). The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat. Med 20, 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsutsui K, Machida H, Nakagawa A, Ahn K, Morita R, Sekiguchi K, Miner JH, and Fujiwara H (2021). Mapping the molecular and structural specialization of the skin basement membrane for inter-tissue interactions. Nat. Commun 12, 2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajbhandari P, Arneson D, Hart SK, Ahn IS, Diamante G, Santos LC, Zaghari N, Feng A-C, Thomas BJ, Vergnes L, et al. (2019). Single cell analysis reveals immune cell-adipocyte crosstalk regulating the transcription of thermogenic adipocytes. Elife 8, e49501. 10.7554/eLife.49501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mariman ECM, and Wang P (2010). Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life Sci 67, 1277–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sárvári AK, Van Hauwaert EL, Markussen LK, Gammelmark E, Marcher A-B, Ebbesen MF, Nielsen R, Brewer JR, Madsen JGS, and Mandrup S (2021). Plasticity of epididymal adipose tissue in response to diet-induced obesity at single-nucleus resolution. Cell Metab. 33, 437–453.e5. [DOI] [PubMed] [Google Scholar]
- 31.Shivanandan A, Radenovic A, and Sbalzarini IF (2013). MosaicIA: an ImageJ/Fiji plugin for spatial pattern and interaction analysis. BMC Bioinf. 14, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, and Chanda SK (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun 10, 1523. 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laitinen L (1987). Griffonia simplicifolia lectins bind specifically to endothelial cells and some epithelial cells in mouse tissues. Histochem. J 19, 225–234. [DOI] [PubMed] [Google Scholar]
- 34.Lamprecht MR, Sabatini DM, and Carpenter AE (2007). CellProfiler: free, versatile software for automated biological image analysis. Biotechniques 42, 71–75. [DOI] [PubMed] [Google Scholar]
- 35.Rudnicki M, Abdifarkosh G, Rezvan O, Nwadozi E, Roudier E, and Haas TL (2018). Female mice have higher angiogenesis in perigonadal adipose tissue than males in response to high-fat diet. Front. Physiol 9, 1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chi J, Crane A, Wu Z, and Cohen P (2018). Adipo-clear: a tissue clearing method for three-dimensional imaging of adipose tissue. J. Vis. Exp 137, e58271. 10.3791/58271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ussar S, Lee KY, Dankel SN, Boucher J, Haering M-F, Kleinridders A, Thomou T, Xue R, Macotela Y, Cypess AM, et al. (2014). ASC-1, PAT2, and P2RX5 are cell surface markers for white, beige, and brown adipocytes. Sci. Transl. Med 6, 247ra103. 10.1126/scitranslmed.3008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castillo PE, Younts TJ, Chávez AE, and Hashimotodani Y (2012). Endocannabinoid signaling and synaptic function. Neuron 76, 70–81. 10.1016/j.neuron.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S-M, Xiao Z-F, Li X, Zhao Y-N, Wu X-M, Han J, Chen B, Li J-Y, Fan C-X, Xu B, et al. (2019). Vascular endothelial growth factor activates neural stem cells through epidermal growth factor receptor signal after spinal cord injury. CNS Neurosci. Ther 25, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shimozaki K, Clemenson GD, and Gage FH (2013). Paired related homeobox protein 1 is a regulator of stemness in adult neural stem/progenitor cells. J. Neurosci 33, 4066–4075. 10.1523/jneurosci.4586-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oshima K, Teo DTW, Senn P, Starlinger V, and Heller S (2007). LIF promotes neurogenesis and maintains neural precursors in cell populations derived from spiral ganglion stem cells. BMC Dev. Biol 7, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heo S, Diering GH, Na CH, Nirujogi RS, Bachman JL, Pandey A, and Huganir RL (2018). Identification of long-lived synaptic proteins by proteomic analysis of synaptosome protein turnover. Proc. Natl. Acad. Sci. USA 115, E3827–E3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Lemma RB, Turchi L, Blanc-Mathieu R, Lucas J, Boddie P, Khan A, Manosalva Pérez N, et al. (2022). Jaspar 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 50, D165–D173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, and Evans RM (2013). PPARγ signaling and metabolism: the good, the bad and the future. Nat. Med 19, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kajimura S (2015). Promoting brown and beige adipocyte biogenesis through the PRDM16 pathway. Int. J. Obes. Suppl 5, S11–S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohno H, Shinoda K, Spiegelman BM, and Kajimura S (2012). PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 15, 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tchkonia T, Giorgadze N, Pirtskhalava T, Tchoukalova Y, Karagiannides I, Forse RA, DePonte M, Stevenson M, Guo W, Han J, et al. (2002). Fat depot origin affects adipogenesis in primary cultured and cloned human preadipocytes. Am. J. Physiol. Regul. Integr. Comp. Physiol 282, R1286–R1296. [DOI] [PubMed] [Google Scholar]
- 48.Harms MJ, Lim H-W, Ho Y, Shapira SN, Ishibashi J, Rajakumari S, Steger DJ, Lazar MA, Won K-J, and Seale P (2015). PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 29, 298–307. 10.1101/gad.252734.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, and Spiegelman BM (2008). Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 22, 1397–1409. 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, and Spiegelman BM (2011). Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Invest 121, 96–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yfanti C, Akerström T, Nielsen S, Nielsen AR, Mounier R, Mortensen OH, Lykkesfeldt J, Rose AJ, Fischer CP, and Pedersen BK (2010). Antioxidant supplementation does not alter endurance training adaptation. Med. Sci. Sports Exerc 42, 1388–1395. 10.1249/mss.0b013e3181cd76be. [DOI] [PubMed] [Google Scholar]
- 52.Wang W, Ishibashi J, Trefely S, Shao M, Cowan AJ, Sakers A, Lim H-W, O’Connor S, Doan MT, Cohen P, et al. (2019). A PRDM16-driven metabolic signal from adipocytes regulates precursor cell fate. Cell Metab. 30, 174–189.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hasegawa Y, Ikeda K, Chen Y, Alba DL, Stifler D, Shinoda K, Hosono T, Maretich P, Yang Y, Ishigaki Y, et al. (2018). Repression of adipose tissue fibrosis through a PRDM16-GTF2IRD1 complex improves systemic glucose homeostasis. Cell Metab. 27, 180–194.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chi J, Wu Z, Choi CHJ, Nguyen L, Tegegne S, Ackerman SE, Crane A, Marchildon F, Tessier-Lavigne M, and Cohen P (2018). Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab. 27, 226–236.e3. [DOI] [PubMed] [Google Scholar]
- 55.Chi J, Lin Z, Barr W, Crane A, Zhu XG, and Cohen P (2021). Early postnatal interactions between beige adipocytes and sympathetic neurites regulate innervation of subcutaneous fat. Elife 10, e64693. 10.7554/eLife.64693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonbol HS (2018). Extracellular matrix remodeling in human disease. J. Microsc. Ultrastruct 6, 123–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shao M, Hepler C, Zhang Q, Shan B, Vishvanath L, Henry GH, Zhao S, An YA, Wu Y, Strand DW, and Gupta RK (2021). Pathologic HIF1α signaling drives adipose progenitor dysfunction in obesity. Cell Stem Cell 28, 685–701.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laughlin MH, and Roseguini B (2008). Mechanisms for exercise training-induced increases in skeletal muscle blood flow capacity: differences with interval sprint training versus aerobic endurance training. J. Physiol. Pharmacol 59, 71–88. [PMC free article] [PubMed] [Google Scholar]
- 59.Joo Y, Kim H, Lee S, and Lee S (2019). Neuronal growth regulator 1-deficient mice show increased adiposity and decreased muscle mass. Int. J. Obes 43, 1769–1782. [DOI] [PubMed] [Google Scholar]
- 60.Bernhard F, Landgraf K, Klöting N, Berthold A, Büttner P, Friebe D, Kiess W, Kovacs P, Blüher M, and Körner A (2013). Functional relevance of genes implicated by obesity genome-wide association study signals for human adipocyte biology. Diabetologia 56, 311–322. [DOI] [PubMed] [Google Scholar]
- 61.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, et al. (2009). Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet 41, 18–24. [DOI] [PubMed] [Google Scholar]
- 62.Willer CJ, Speliotes EK, Loos RJF, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, et al. (2009). Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet 41, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Mägi R, et al. (2010). Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet 42, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klimentidis YC, Raichlen DA, Bea J, Garcia DO, Wineinger NE, Mandarino LJ, Alexander GE, Chen Z, and Going SB (2018). Genome-wide association study of habitual physical activity in over 377,000 UK Biobank participants identifies multiple variants including CADM2 and APOE. Int. J. Obes 42, 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dreyfuss JM, Yuchi Y, Dong X, Efthymiou V, Pan H, Simonson DC, Vernon A, Halperin F, Aryal P, Konkar A, et al. (2021). High-throughput mediation analysis of human proteome and metabolome identifies mediators of post-bariatric surgical diabetes control. Nat. Commun 12, 6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Emont MP, Jacobs C, Essene AL, Pant D, Tenen D, Colleluori G, Di Vincenzo A, Jørgensen AM, Dashti H, Stefek A, et al. (2022).A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yfanti C, Nielsen AR, Akerström T, Nielsen S, Rose AJ, Richter EA, Lykkesfeldt J, Fischer CP, and Pedersen BK (2011). Effect of antioxidant supplementation on insulin sensitivity in response to endurance exercise training. Am. J. Physiol. Endocrinol. Metab 300, E761–E770. [DOI] [PubMed] [Google Scholar]
- 71.Shamsi F, Xue R, Huang TL, Lundh M, Liu Y, Leiria LO, Lynes MD, Kempf E, Wang C-H, Sugimoto S, et al. (2020). FGF6 and FGF9 regulate UCP1 expression independent of brown adipogenesis. Nat. Commun. 11, 1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hemmeryckx B, Bauters D, and Roger H Sirius Red Staining of Murine Tissues V1 (protocols.io.J9rcr56). protocols.io. 10.17504/protocols.io.j9rcr56. [DOI] [Google Scholar]
- 73.Law CW, Chen Y, Shi W, and Smyth GK (2014). voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ritchie ME, Diyagama D, Neilson J, van Laar R, Dobrovic A, Holloway A, and Smyth GK (2006). Empirical array quality weights in the analysis of microarray data. BMC Bioinf. 7, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository. Mouse and human microarray data were previously published and available in the Gene Expression Omnibus database. Spatial transcriptomic data have been deposited at Mendeley and are publicly available as of the date of publication. The DOI for all the omics are listed in the key resources table. Original western blots (Data S1), and microscopy raw data reported in this paper will be shared by the lead contact upon request.
All original code has been deposited at Mendeley and is publicly available as of the date of publication. DOIs are listed in the key resources table.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-mouse synaptophysin (1:300) | ProteinTech | Cat# 17785-1-AP; RRID: AB_2271365 |
| Mouse anti-mouse tyrosine hydroxylase (1:200) | EMD Millipore | Cat# MAB318; RRID: AB_2201528 |
| Alexa Fluor 488 donkey anti-rabbit (1:400) | Life Technologies | Cat# A21441; RRID: AB_2535859 |
| Alexa Fluor 594 goat anti-rabbit (1:400) | Life Technologies | Cat# A11072; RRID: AB_142057 |
| Alexa Fluor 488 donkey anti-mouse (1:400) | Life Technologies | Cat# A21202; RRID: AB_141607 |
| Alexa Fluor 594 goat anti-mouse (1:400) | Life Technologies | Cat# A11037; RRID: AB_2534095 |
| Alexa Fluor 647 Goat anti-mouse (1:400) | Life Technologies | Cat# A21235; RRID: AB_2535804 |
| Anti-mouse NEGR1 (1:200) | ProteinTech | Cat# 13674-1-AP; RRID: AB_2877969 |
| Biological samples | ||
| Human subcutaneous abdominal and gluteal white adipose tissue | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 1uM insulin | Sigma | Cat# 11376497001 |
| 250nM Dexamethasone | Sigma-Aldrich | Cat# D1756 |
| 1% Pen/Strep | Thermo-Scientific | Cat# 15140122 |
| Direct-zol RNA MiniPrep | Zymo Research | Cat# R2052 |
| Rosiglitazone | Cayman Chemical | Cat# 122320-73-4 |
| High-Capacity Reverse Transcription Kits | Applied Biosystems | Cat# 4368813 |
| Dichloromethane (DCM) | Sigma-Adrich | Cat# 270991 |
| Benzyl ether (DBE) | Sigma-Adrich | Cat# 108014 |
| UltraPure Agarose | Invitrogen | Cat# 16500500 |
| Triton X-100 | Sigma-Adrich | Cat# X100-500ML |
| Tween 20 | Sigma Adrich | Cat# P2287-500ML |
| Critical commercial assays | ||
| Visium Spatial Tissue Optimization Slide & Reagent Kit | 10x Genomics | Cat# PN-1000193 |
| Visium Spatial Gene Expression Solution v1 | 10x Genomics | Cat# PN-1000184 |
| Dual Index Kit TT Set A | 10x Genomics | Cat# PN-1000215 |
| Deposited data | ||
| Raw and processed MS data | This study, ProteomeXchange Data | PXD040894 |
| Raw and processed MS data | This study, ProteomeXchange Data | PXD040894 |
| Visium normalized data | This study, Mendeley Data | https://doi.org/10.17632/r6v7yktxnt.1 |
| Visium raw data and Code for Visium data analyses | This study, Mendeley Data | https://doi.org/10.17632/pktxwdvcbz.1 |
| Proteome and Secretome analysis | Dreyfuss et al.65 | https://github.com/jdreyf/ezlimma |
| Rajbhandari et al.,2019 scRNA-seq data of adipose tissue | Rajbhandari et al.28 | GEO: (GSE133486) |
| Ermont et al.,2022 scRNA-seq data of adipose tissue | Ermont et al.66 | GEO: (GSE176067) |
| Ermont et al.,2022 snRNA-seq data of adipose tissue | Ermont et al.66 | GEO: (GSE176171) |
| Sárvári et al.,2021 snRNA-seq data of adipose tissue | Sárvári et al.30 | GEO: (GSE160729) |
| Takahashi et al.,2019 Human microarray | Takahashi et al.5 | GEO: (GSE116801) |
| Stanford et al.,2015 Mouse microarray | Stanford et al.3 | GEO: (GSE68161) |
| Experimental models: Cell lines | ||
| Primary adipose derived stem cells from mouse iWAT | This study | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: Male C57BL/6NCrl | Charles River Lab | Cat# 027 |
| Mouse: Adipo-PRDM16 KO PRDM16lox/lox | Jackson Laboratory | Cat# 24992 |
| Mouse: Adiponectin-cre | Jackson Laboratory | Cat# 28020 |
| Oligonucleotides | ||
| Total of 50 oligonucleotides | This study | Table S4 |
| Software and algorithms | ||
| CellProfiler 3.0 | Carpenter et al67 | http://cellprofiler.org; RRID: SCR_007358 |
| FIJI macro TWOMBLI | Wershof et al.15 | https://github.com/wershofe/TWOMBLI |
| Fiji v1 | ImageJ | https://imagej.net/software/fiji/; RRID: SCR_002285 |
| Imaris 9.7 | BitPlane | https://imaris.oxinst.com; RRID: SCR_007370 |
| Cytoscape | Shannon et al. 68 | https://cytoscape.org; RRID: SCR_003032 |
| String Database | Szklarczyk et al. 69 | https://string-db.org; RRID: SCR_005223 |
| Metascape | Zhou et al. 32 | https://metascape.org; RRID: SCR_016620 |
| GraphPad Prism v9 | GraphPad | https://www.graphpad.com; RRID: SCR_002798 |
| Loupe Browser version 6.0.0 | 10x Genomics | https://10xgenomics.com; RRID: SCR_018555 |
| R 4.04 | See computational analysis section for R packages used | https://www.r-project.org/about.html |
| Spaceranger-1.3.0 | 10x Genomics | https://www.10xgenomics.com |