Abstract
While photoluminescent graphene quantum dots (GQDs) have long been considered very suitable for bioimaging owing to their protein-like size, super-high photostability and in vivo long-term biosafety, their unique and crucial bioimaging applications in vivo remain unreachable. Herein, we present planted GQDs as an excellent tool for in vivo fluorescent sustainable and multimodality tumor bioimaging in various scenarios. The GQDs were in situ planted in the PEG layer of PEGylated nanoparticles via a bottom-up molecular approach to obtain the NPs-GQDs-PEG nanocomposite. The planted GQDs show more than 4 times prolonged blood circulation and 7~8 times increased tumor accumulation than typical GQDs in vivo. After accessible specificity modification, the multifunctional NPs-GQDs-PEG provides targeted, multimodal molecular imaging for various tumor models in vitro or in vivo. Moreover, the highly photostable GQDs enable long-term, real-time visualization of the local pharmacokinetics of NPs in vivo. Planting GQDs in PEGylated nanomedicine offers a new strategy for broad in vivo biomedical applications of GQDs.
Keywords: Carbon nanomaterials, Fluorescent carbon, Bioimaging, sustainable imaging, Pharmacokinetics, Tumor microenvironment
Table of Contents

The graphene quantum dots (GQDs) were in situ planted in the PEG layer of PEGylated nanoparticles from small molecule pyrene. After accessible specificity modification, the multifunctional NPs-GQDs-PEG provides targeted, multimodal molecular imaging for various tumor models in vitro or in vivo. Moreover, the highly photostable GQDs enable long-term, real-time visualization of the local pharmacokinetics of NPs in vivo.
Introduction
Graphene quantum dots (GQDs) are an emerging class of photoluminescent carbon nanomaterials with a sub-10 nm characteristic size and super-highly photostable and bright fluorescent properties.[1–4] GQDs show much better intrinsic biocompatibility and ease of clearance than typical semiconductor quantum dots (semi-QDs) with necessary surface coating.[5–7] In addition, protein-sized GQDs (dozens of kDa) are more suitable for biological targeting and molecular imaging compared with other quasi-spherical carbon dots (CDs) (hundreds of kDa).[5–6] Moreover, pure GQDs always show low cytotoxicity and excellent long-term biosafety in vivo.[8–12] Recently, GQDs have been successfully explored for in vivo biomedical application potentials, such as magnetic resonance imaging (MRI),[13] distinctive therapeutic platform,[10–11, 14–16] and some preliminary fluorescence imaging.[12, 17–19] From both fundamental and applied perspectives, the highly photostable and bright fluorescence (FL) is one of the most appealing characteristics of GQDs.[5] However, for current fluorescent GQDs, in vivo targeting and multimodal molecular bioimaging are still challenged, and critical fluorescent applications are inaccessible.
Several major hindrances are believed to restrain the broad in vivo bioimaging applications of GQDs significantly. Firstly, the pure GQDs typically exhibit short blood-circulation half-lives and insufficient tumor accumulation,[20] which predominantly affect the effectiveness of GQDs in vivo.[21] Secondly, the regular surface engineering for GQDs, such as stealthy-to-sticky transition and targeting functionalization with binding ligands, would affect the photophysical properties or even significantly quench the photoluminescence.[5–6, 22] Thirdly, the restrictions of post-synthetic engineering would further hamper GQDs from composite with other nanomedicine to achieve multimodal molecular imaging, which is especially needed in precision diagnostics.[23–24] These limitations highlight the need to develop new strategies to formulate, compound, and functionalize GQDs to achieve exclusive bioimaging applications in vivo.
Among various approaches to increase blood circulation and tumor accumulation, surface poly (ethylene glycol) (PEG) grafting is one of the most typical and efficient strategies for nanoparticles (NPs),[25] which could dramatically decrease the undesirable clearance of NPs by the immune system.[26–27] Additionally, therapeutic macromolecules or proteins can generally penetrate the moderately dense PEG layer and load in voids.[28] Herein, we show the strategy for in situ plant GQDs in typical PEGylated NPs to form the multifunctional, targeted nanoprobe (NPC-GQDs-PEG) and then efficiently applied for in vivo bioimaging. The small molecule pyrene (C16H10), a graphene lattice primitive cell, largely penetrated the PEG layer and gradually assembled and grew up in the PEG voids. The planted GQDs naturally inherit the advantages of PEGylation from NPs and show four times increase in blood-circulation half-lives and seven-eight times enhancement of tumor accumulation compared with typical GQDs. Owing to target functionalizing the PEG layer easily, the planted GQDs could specifically label diverse targets (cell surface receptors, cytoskeleton components) in various carcinoma cell lines (breast, liver, and pancreatic) after additional grafting. With the versatility of NPs core, NPC-GQDs-PEG could achieve multimodality MRI and photoacoustic imaging (PAI) simultaneously and fluorescence imaging-guided tumor surgery. Furthermore, we explored the long-term, real-time monitoring of GQDs-planted NPs in vivo and achieved the visualization of local pharmacokinetics (absorption, distribution, metabolism, and excretion) of NPs in the tumor microenvironment and organs for several days. Planting GQDs in PEGylated NPs makes in vivo bioimaging of GQDs more feasible and provides a new strategy for in vivo applications of GQDs.
Results
Synthesis and characterization of NPC-GQDs-PEG nanocomposite
Graphene quantum dots (GQDs) were in situ planted into PEGylated nanoparticles to form NPC-GQDs-PEG from graphene-cell molecules pyrene (Fig. 1a). First, the magneto-conjugated polypyrrole polymer (Fe3O4@PPy) nanoparticle core (NPC) was synthesized following an in-situ oxidation polymerization method.[29–30] The NPC is about 40~45 nm in diameter with a polymer shell of about 2~3 nm (Fig. 1b and Fig. S2b). Next, a long chain (10 KDa) amine-PEG reacted with the carboxyl groups of the polymer layer to form PEG grafting layer. Then the 1,3,6-trinitropyrene molecule, a derivative of pyrene (C16H10), was added, penetrated PEG layer voids and in situ formed GQDs under a hydrothermal condensation reaction[31–32] in the mixture of ammonia and hydrazine hydrate solutions (Fig. 1a). After multiple rounds of washing, more than 90% of NPs-GQDs-PEG with high purity can be obtained, which was stable in aqueous colloids for at least three weeks at RT.
Fig. 1. Preparation and characterization of NPC-GQDs-PEG nanoparticles.
a, Schematic process of in situ planting GQDs into PEGylated NPs. b, Representative TEM image of nanoparticle core (NPC). c, Representative AFM image of pure GQDs (insets: height profile along the white line). d, Representative HRTEM image of NPC-GQDs-PEG nanoparticle, the red lines mark single GQDs. e, Raman spectrum of NPC-GQDs-PEG. f, XRD pattern of NPC-GQDs-PEG (insets: pure GQDs). g, Survey XPS spectrum of NPC-GQDs-PEG. h, PL spectra of NPC-GQDs-PEG excited at different wavelengths. i, Arc-lamp photobleaching curves for green-emitting NPC-GQDs-PEG, EGFP, Alex Fluor488 (AF488), and FITC dyes under continuous arc-lamp illumination (10 W cm−2), as plotted as intensity versus normalized total exposure time with an initial emission rate of 1,000 photons/s per molecule. j, Hydrodynamic diameters of nanoparticles of different processes.
The pure GQDs synthesized with the same method have a crystalline structure with a lateral size of 3~4 nm (Fig. S2a) and thickness of 1.2 ± 0.48 nm corresponding to an average of 4 layers atom (Fig. 1c). The obtained NPC-GQDs-PEG nanocomposite has a uniform morphology and diameter of about 50 nm from the scanning electron microscope (SEM) image (Fig. S3). The high-resolution transmission electron microscopy (HRTEM) image depicts the three layers of core-shell structure with crystalline Fe3O4 inner core, amorphous polymer shell, and outermost single-crystalline GQDs (Fig. 1d and Fig. S2c-f). The GQDs layer is about 5~10 nm and consists of 1~3 single-crystalline structure GQDs with a spacing of 0.21 nm matching that of graphene (100) planes. From the Raman spectroscopy (Fig. 1e), the intensity ratio of the ordered G band (1,590 cm−1) to the disordered D band (1,365 cm−1) is about 1.08, indicating the existence of the benzene ring conjugate structure of NPC-GQDs-PEG. The X-ray powder diffraction (XRD) patterns (Fig. 1f) of NPC-GQDs-PEG retain the typical reflection peak of crystalline Fe3O4 cores, the broadened peak (2θ = 20°–25°) of polymer PPy and reflect stacking with a peak near 2θ = 25°–27° which is close to that of pure graphite (3.34 Å). The survey X-ray photoemission spectroscopy (XPS) and the respective high-resolution spectrums (Fig. 1g and Fig. S4) not only reveal the presence of expected Fe-O (532.6 eV), C=C (284.5 eV), C-N (285.8 eV), C-O (286.9 eV) and O-H (531.6 eV) but also show the presence of NH2 (399.9 eV), indicating the GQDs in NPs are functionalized by NH2 which has been approved to locate at the edge sites of GQDs.[31, 33]
The planted GQDs from NPC-GQDs-PEG inherit the bright fluorescence performance of pure GQDs (Fig. S5) and show excitation wavelength-dependent emission with a peak at around 560 nm under the excitation of 490 nm (Fig. 1h). We further compared the photostability of NPC-GQDs-PEG with that of three green-emitting fluorophores FITC, Alexa Fluor 488 (AF488) and EGFP under the same conditions with continuous arc-lamp illumination (10 W cm−2). The time for photobleaching from an initial emission rate of 1,000 photons/molecule down to 500 (t1/2), calculated following the typical method,[34–35] is 135 s, 160 s, and 225 s for FITC, AF488, and EGFP individually (Fig. 1i). However, the NPC-GQDs-PEG exhibits superior photostability and still retains over 90% of the initial performance even after 1500 s illumination (Fig. 1i). Additionally, the NPC-GQDs-PEG nanocomposite inherits the excellent near-infrared (NIR) absorbance properties (800 to 1100 nm) and outstanding super-paramagnetic properties of nanoparticle core (Fig. S6a, b), which are suitable for magnetic resonance imaging (MRI) and photoacoustic imaging (PAI) as shown in Figures S6 c and d. During PEG grafting and GQDs planting, the zeta potential changed from −45 mV of NPC to −29 mV of NPC-PEG (Fig. S7a) and finally increased to −19 mV of NPC-GQDs-PEG and −10 m of NPC-GQDs-PEG/2-DG. Additionally, the hydrodynamic sizes increased from about 64 nm of NPC to 85 nm of NPC-PEG and finally raised to about 95 nm of NPC-GQDs-PEG and 105 nm of NPC-GQDs-PEG/2-DG (Fig. 1j and Fig. S7b). Interestingly, the planted GQDs can gradually release from the PEG nest, and only about 35% remain 7 days after shaking in PBS buffer (Fig. S8). Planting GQDs in the PEGylated NPs dramatically changed the short-term (at least 3~4 days) existence form of GQDs in solution.
NPC-GQDs-PEG undergo slower blood clearance and higher tumor accumulation
We first evaluated the intrinsic cytotoxicity of NPC-GQDs-PEG on MCF-7 human breast cancer cells and L929 murine fibroblast cells using a Cell Counting Kit-8 (CCK- 8) assay (Fig. S9). No significant cytotoxicity was observed up to 72 h, even with high concentrations of 150 μg/ml, while exhibiting a slight effect even at 300 μg/ml. We then measured the blood clearance of intravenously injected NPC-GQDs-PEG/2-DG and two surfaces functionalized GQDs (GQDs-OH and GQDs-NH2). The BALB/c mice bearing MCF-7 tumors in three groups (n=4) were intravenously injected with NPC-GQDs-PEG (20 mg kg−1) and pure GQDs (5 mg kg−1), respectively, for the pharmacokinetics examinations. The NPC-GQDs-PEG shows a much slower distribution and later clearance than pure GQDs (Fig. 2a, b, and c). Specifically, the distribution half-life time(tα,1⁄2) of two kinds of GQDs is about 0.4 h, while about 1.46 h for NPC-GQDs-PEG and the elimination half-life time (tβ,1⁄2) is about 4 h for GQDs while about 17 h (17.20 ± 5.16 h) for NPC-GQDs-PEG. Furthermore, the area under the concentration-time (AUC) curve is about 170~180 ID% h g−1 for GQDs (Fig. 2d), while the AUC of NPC-GQDs-PEG surpasses 430 ID% h g−1 36 h after injection (AUC0–36h). The planted GQDs from PEGylated NPs are more persistent in blood circulation than GQDs alone in vivo.
Fig. 2. In vivo blood clearance and biodistribution of NPC-GQDs-PEG.
a-c, Blood clearance kinetics in terms of percent injected dose (ID%) per gram blood for pure GQDs (a-b) or NPC-GQDs-PEG/2-DG (c) in mice after tail-vein injection. d, Quantitative area under the pharmacokinetics curve at three-time points after i.v. injection. e-g, Biodistribution profiles of GQDs (e-f) and NPC-GQDs-PEG (g) in xenograft MCF-7 tumors, as plotted as injected dose (ID%) per gram tumor versus time after i.v. injection. The AUCtumor is the area under the curve. h, Quantitative biodistribution analysis of the probes in organs at different time points. Data are mean ± s.d. of biological replicates (n = 4 mice per group).
It’s widely documented that more extended blood circulation could provide more chances for NPs to reach the tumor through the enhanced permeability and retention (EPR) effect. [36] The concentration profiles of NPC-GQDs-PEG/2-DG and GQDs in tumor times were further investigated (Fig. 2e, f, and g). The area-under-the-curve of the tumor (AUCtumor) is 877.7 ID% h g−1 for NPC-GQDs-PEG while the AUCtumor is just 118.6 ID% h g−1 and 115.9 ID% h g−1 for GQDs (-OH) and GQDs (-NH2) respectively. Following the previously reported method,[20, 37] we further calculated the tumor delivery efficiency of NPs (Fig. S10, Supplementary equations 1–1 and 1–2). The tumor delivery efficiency of NPC-GQDs-PEG is 3.12%, about 7.5 times larger than pure GQDs (0.42% for GQDs-OH, 0.41% for GQDs-NH2) in the xenograft breast cancer model. Further quantitative biodistribution analysis of GQDs in major organs shows the NPC-GQDs-PEG tends to gradually enrich in the liver and spleen partly due to the long blood-circulation half-lives, while the pure GQDs are rapidly excreted through the urinary owing to the ultrasmall size.[38]
In vitro and in vivo targeted, multimodal molecular imaging
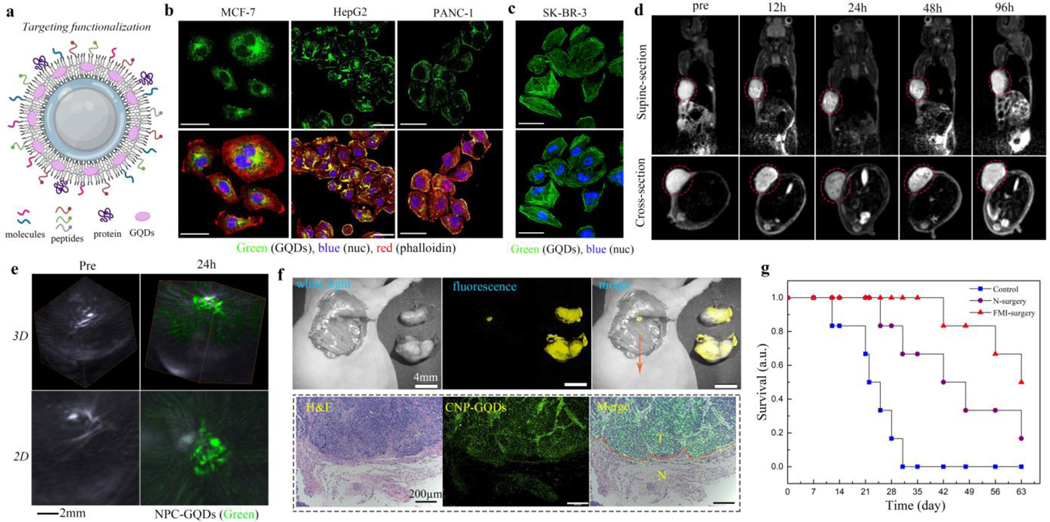
We further explored the potential of NPC-GQDs-PEG for specifically and effectively labeling molecular targets at a subcellular level. The termini of the outermost PEG can be easily conjugated with small molecules, peptides, and proteins using typical EDC/NHS chemistry (Fig. 3a). The 2-Deoxyglucose (2-DG) molecules can significantly increase the NPs endocytosis of some cancer cells due to increased glucose consumption.[30, 39] The NPC-GQDs-PEG/2-DG probes were largely uptake by MCF-7 breast cancer cells after 4 hours of incubation (Fig. 3b). We then attempted the detection of Glypican-3 (GPC-3), a cancer marker overexpressed on the surface of some hepatocellular carcinoma (HCC) cells,[40] with NPC-GQDs-PEG conjugated to GPC-3-binding peptide GP2633. The NPC-GQDs-peptide successfully labeled GPC-3 on the surface of human HepG2 liver cancer cells (Fig. 3b). Similarly, the NPC-GQDs-PEG conjugated to RGD peptide, targeting specifically to plectin-1 cell membrane marker,[41] can also efficiently bind to the surface of plectin 1-positive and human PANC-1 pancreatic cancer cells (Fig. 3b). We then attempted to stain F-actin inside the human SK-BR-3 breast cancer cells using NPC-GQDs-PEG conjugated with phalloidin, which is a highly selective bicyclic peptide used for staining actin filaments. The cytoskeleton F-actin in SK-BR-3 cells was clearly labeled with NPC-GQDs- phalloidin (Fig. 3c). These results indicate that the NPC-GQDs-PEG-based probes can specifically label different types of targets (cell surface cytoskeleton) with varying types of cancer cells (Fig. 3b and Fig. S11).
Fig. 3. In vitro and in vivo targeted tumor imaging based on NPC-GQDs-PEG.
a, Schematic of targeting functionalization for NPC-GQDs-PEG based on different needs. Targeted molecules, peptides, or antibodies can be conjugated in the outer PEG layer. b, Targeting surface or intracellular cancer markers with functionalized NPC-GQDs-PEG: NPC-GQDs-PEG/2-DG for MCF-7 cells, NPC-GQDs-PEG/peptide for HepG2 cells, NPC-GQDs-PEG/RGD for PANC-1 cells. The cytoskeleton was stained with Rhodamine B (red)-phalloidin, and the nucleus (blue) was counterstained with 4’, 6-diamidino-2-phenylindole (DAPI). The bar is 50 μm. c, Targeting cytoskeleton in SK-BR-3 cells with NPC-GQDs-phalloidin (green). The bar is 50 μm. d, Representative T2-MR images of MCF-7 tumor-bearing mice at different time post i.v. Injection, including supine section and cross-section visions. e, 3-dimensional (3D) and 2D PA images of the tumor before and 24 h after i.v. Injection. The bright area is the tumor blood vessels which are rich in hemoglobin. The green signals correspond to the distribution of NPC-GQDs-2-DG. f, Fluorescence imaging-guided intraoperative detection and removal of the microtumor foci from mice bearing MCF-7 tumor. The resected foci tissue section was stained with hematoxylin and eosin stains (H&E), and the green fluorescent signal is from the GQDs. The boundary between the tumor and the normal tissue is marked with a red dashed line. g, Survival curves of mice after different kinds of treatment: no treatment (control), normal surgery (N-surgery), and fluorescence imaging-guided surgery (FMI-surgery). n = 6 mice per group.
For most surgical treatments of cancer, imaging methods such as magnetic resonance imaging (MRI) and photoacoustic imaging (PAI) can provide accurate diagnosis information, and fluorescence molecular imaging (FMI) can define the margins between lesions and normal tissue intraoperatively.[41–42] And the targeted, multifunctional probes are the keys to achieving these. The NPC-GQDs-PEG/2-DG probes were intravenously injected into the BALB/c mice bearing MCF-7 tumors. The MRI signals indicate the dynamic biodistribution of NPC-GQDs-PEG in the whole body and the tumor’s precise location (Fig. 3d and Fig. S12). The PA imaging (Fig. 3e and Fig. S13) signals reflect the massive accumulation and wide distribution of probes in the whole tumor 24 h post-injection, while very weak signals in the surrounding tissues. Many intratumor details and surrounding information are clear at the optimal metabolic point of 24 h post-injection from both MRI and PAI. Based on this comprehensive diagnosis information, FMI-guided surgery was conducted 24 h post-injection of probes. The typical surgical resection was performed by two experienced surgeons to completely remove the tumor tissues at their best using white-light imaging (Fig. S14). However, a small residual focus (<0.3 mm) was easily visualized from the resection bed by detecting the fluorescent signal of probes (Fig. S15 and Fig. 3f upper). We removed the small focus overlying surrounding probe-negative tissue for further histological analysis. Analysis of the hematoxylin and eosin (H&E)-stained tissues confirm that the regions showing fluorescence contained residual tumors left behind after excision, whereas probe-negative regions contained normal tissue (Fig. 3f bottom). Thus, the NPC-GQDs-PEG/2-DG probe can accurately define the margins of tumors and detect residual microtumors. Furthermore, the survival rates of mice (n=6 in each group) in the FMI-guided group (FMI-surgery) are significantly higher than the normal surgery (N-surgery) and control groups (Fig. 3g). Therefore, the FMI guidance of GQDs can tremendously improve the effectiveness of surgery.
In vivo long-term, real-time visualization of local pharmacokinetics of NPs
We further explored the real-time and long-term tracking of GQDs-loaded NPs in the tumor microenvironment, which is believed to be vital for evaluating nanomedicines’ efficacy,[42–43] based on the excellent photostability of GQDs. The local pharmacokinetics of systemically administrated NPs were monitored by fibered confocal fluorescence microscopy with dual-band laser excitation (480 nm and 660 nm) in Balb/c mice bearing xenograft MCF-7 tumors (Fig. 4a and Fig. S16). During tumor imaging, one optical fiber was applied to the tumor surface under the epidermis (S-probe). Then, another optical fiber (D-probe) further penetrated the deep tumor tissue (Fig. 4a). We first assessed the in vivo photostability of NPC-GQDs-PEG/2-DG in tumor tissues under continuously illuminating for 10 min (3~5 mW power) with NPC-AF488-PEG/2-DG as control and Evans blue (EB) as a vascular contrast agent. The fluorescence signal of GQDs remained stable under continuous 10 minutes of illumination with no apparent decay, but only about 20% remained for AF488 after ten minutes of illumination (Fig. 4b and 4c, Fig. S17, S18, and S19). The signal of the EB was also greatly attenuated, but the attenuation amplitude was different owing to the different circulation speeds of the blood.
Fig. 4. Real-time and long-term monitoring of the NPC-GQDs-PEG in local tissues.

a, Schematic illustration of the fibered confocal fluorescence microscopy (FCFM) imaging for tumors and major organs in the abdominal cavity. For imaging tumors, the optical fiber probes were inserted into the superficial (S-probe) and deep tissue (D-probe), respectively. The D-fiber penetrated the abdominal cavity for major organs through a pre-operated small incision in the belly. b, Photostability comparison between NPC-GQDs-PEG/2-DG (green)and NPC-AF488–2-DG (green) in MCF-7 tumor in vivo. Evans blue (red) was used as the vascular contrast agent. The tumor tissues were continuously illuminated for 10 min with a dual-band laser (488 nm and 660 nm) with 3~5 mW power. The scale bar is 50 μm. c, Quantify fluorescence intensity of Evans blue (EB), AF488, and GQDs in tumor under 10 min continuous illumination. d, In vivo monitoring of probes’ accumulation, diffusion, and biodistribution in tumor tissues using FCFM. Green: NPC-GQDs-PEG/2-DG, red: EB. e, Quantify fluorescence intensity of NPC-GQDs-2-DG in superficial and deep tumor tissue at different time points (n=4). f, In vivo monitoring the biodistribution and metabolism of probes in live and kidney using FCFM. g, Quantify fluorescence intensity of NPC-GQDs-PEG/2-DG in living, spleen, and kidney at different time points (n=4).
Time-lapse, sustainable imaging with 10 min continuous illumination for each time point was then performed on the superficial and deep tissues of the tumor in the following three days after intravenous injection of NPC-GQDs-PEG/2-DG (25 mg kg−1). EB injected several hours in advance could not only stain the blood vessels but also penetrate deep tissues. The strong GQDs signals can be found in the vasculature of superficial tissue two hours post-injection, while very weak signals can be found in the deep tissue. From 2 h to 24 h post-injection, NPs gradually exuded from vessels and enriched into surrounding tissues on the surface, with more NPs gradually penetrating the deep tissue simultaneously (Fig. 4d and Supplementary Video 1–4). With the further extension of time, NPs were gradually metabolized from the tumor, but the fluorescence signal of NPs in deep tissue decreased more slowly, which is mainly due to the incompleteness of vasculature and higher tissue pressure. The quantitative fluorescence calculation further confirms these results (Fig. 4e) and exhibits that NPs ‘leave’ the tumor tissue more slowly than ‘enter’ the tumor, especially for the deep tumor tissue. Additionally, real-time and long-term tracking of GQDs-embedded NPs in vivo can be expanded to other organs. A minor surgical opening (3~4 mm wide) was made in the belly to enable the fiber to probe organs in the abdominal cavity (Fig. 4a). After intravenous injection, NPC-GQDs-PEG/2-DG gradually accumulated in the liver and spleen (Fig. 4f, Fig. S20) and reached the maximum at 24 h and 48 h for live and spleen, respectively (Fig. 4g). And the NPs were then gradually clear from the liver and spleen with fluorescent signals obviously decreasing. Interestingly, the signals of GQDs in the kidney had not disappeared with the almost complete clearance of NPC-GQDs-PEG from the blood 48 h and 72 h post-injection (Fig. 4f and 4g). These residual GQDs signals may come from the gradual release of GQDs from NPC-GQDs-PEG over time in vivo. Therefore, the planted GQDs can enable long-term, real-time tracking of the dynamics of systemically administered NPs in tumor microenvironments and major organs.
Discussion
The scheme of in situ planting GQDs from pyrene molecules in the outer PEG grafting layer of nanoparticles provides a new strategy for in vivo bioimaging applications of GQDs. This approach allows GQDs to inherit the rational design and physicochemical properties of PEGylated nanomedicine, which will significantly prolong circulation in the blood and enhance tumor accumulation of GQDs and provide a general, accessible targeting modification platform without damaging the unique structure for GQDs. Additionally, the multi-functionality of nanomedicine makes it possible for multimodal molecular imaging, which complements the intrinsic limitations of GQDs-based fluorescence imaging alone. These provide conditions for the extensive and in-depth applications of GQDs in vivo. Furthermore, owing to the super-high photostability and bright fluorescent property, the planted GQDs provide a sustainable bioimaging method that can long-term, real-time track and visualize the whole pharmacokinetic process of PEGylated NPs in local tissues of tumors and organs. Interestingly, the planted GQDs can also be gradually released from PEGylated NPs and should be cleared from the body, maintaining the excellent long-term biosafety of GQDs.
Reading and understanding many life phenomena and live activities often require sustainable bioimaging with a high spatiotemporal resolution to study the long-term, fast dynamics of fine subcellular activities on a time scale from minutes to days.[35, 44] The typical time-lapse (temporal down-sampling) bioimaging may miss transient but important signals. Therefore, super-highly photostable fluorescent GQDs planted in nanomedicine have great potential to alleviate these undesirable situations in broad applications, such as embryo development,[45] stem cell differentiation trajectories,[46] and imaging-based spatiotemporal single-cell omics.[47] Of course, the current GQDs-planted nanoparticles also have many limitations: one is the limited penetration depth of green fluorescent GQDs, and the other is that the core NPs are not biodegradable in a short time in vivo. However, these can be improved through the optimal choice of core nanomedicine and regulation of near-infrared GQDs in the future. Therefore, such a strategy of promoting strengths and avoiding the weakness of GQDs will broaden the in vivo bioimaging horizons of GQDs.
Experimental Section
Planting GQDs in PEGylated NPs:
1 g pyrene was added to 80 ml HNO3 under refluxing and stirring for 12 h to obtain trinitropyrene and then diluted with deionized water filtered through a 0.22 μm microporous membrane to remove the acid. Meanwhile, 2 ml NPC-PEG solution (1 mg/ml) was dispersed in 30 mL ammonia (0.4 M)/hydrazine hydrate (1.5 M) mixed solvent. The above 1,3,6- trinitropyrene (0.25 g) was then added into 30 ml prepared NPC-PEG (50 μg/ml) ammonia (0.4 M)/hydrazine hydrate (1.5 M) solution. After further sonicated for 30 min, the suspension was transferred to a poly(tetrafluoroethylene) (Teflon)-lined autoclave (40 ml) and heated at 100 °C for five hours. The resulting product was collected with the help of a magnet and purified with repeated centrifugation (20 000 g for 30 min) five times. Finally, the purified NPC-GQDs-PEG were dried at 80 °C for further characterization and property measurement. The pure GQDs-NH2 and GQDs-OH were synthesized following the above procedure except using different reaction mediums: 0.2 M NaOH solution for GQDs-OH while 0.4 M ammonia and 1.5 M hydrazine hydrate for GQDs-NH2.
Animals:
All in vivo experiments were approved by the Animal Care and Use Committee (IACUC) of the Chinese Academy of Medical Sciences Tumor Hospital (#NCC2019A010). Strict adherence to animal care and use protocols was ensured throughout the study. BABL/c nude mice (about five weeks old) were purchased from Beijing Vital River Laboratory Animal Technology Co., China. Mice were implanted with MCF-7 cells (2 × 106, 0.2 ml in H-DMEM culture medium without FBS) subcutaneously near the right side of the axillary. The mice were used when their tumor volumes approached about 60–80 mm3.
Statistical analysis:
The data are presented as means ± standard deviation, as noted in each case. The number of samples, n, are provided. A two-way analysis of variance (ANOVA) was used. P < 0.05 was considered to indicate a statistically significant difference. P < 0.05, P < 0.01, and P < 0.001 are marked with single, double, and triple asterisks, respectively.
Supplementary Material
Acknowledgments
The authors thank the National Natural Science Foundation of Youth Program of China (No. 81901813), the National Natural Science Foundation of China (No. 52072210, 52272278, 52111530230), and the Key R&D projects of social development of Hainan Provincial Department of Science and Technology of China (ZDYF2020137) for their support of funding. In addition, H.Y. and S-H.Y. acknowledge support from the National Institutes of Health in the USA (R01-CA192878).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
Authors have no competing interests.
Data availability
The data supporting this paper’s figures are available upon reasonable request.
References
- [1].Zhu SJ, Song YB, Zhao XH, Shao JR, Zhang JH, Yang B, Nano Res. 2015, 8, 355. [Google Scholar]
- [2].Zheng XT, Ananthanarayanan A, Luo KQ, Chen P, Small 2015, 11, 1620. [DOI] [PubMed] [Google Scholar]
- [3].Yan Y, Gong J, Chen J, Zeng Z, Huang W, Pu K, Liu J, Chen P, Adv. Mater 2019, 31, 1808283. [DOI] [PubMed] [Google Scholar]
- [4].Wu H, Xu H, Shi Y, Yuan T, et al. Chinese J. Chem 2021, 39, 1364. [Google Scholar]
- [5].Dordevic L, Arcudi F, Cacioppo M, Prato M, Nat. Nanotechnol 2022, 17, 112. [DOI] [PubMed] [Google Scholar]
- [6].Younis MR, He G, Lin J, Huang P, Front. Chem 2020, 8, 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Du JJ, Xu N, Fan JL, Sun W, Peng XJ, Small 2019, 15, 1805087. [DOI] [PubMed] [Google Scholar]
- [8].Nurunnabi M, Khatun Z, Huh KM, Park SY, Lee DY, Cho KJ, Lee YK, ACS Nano 2013, 7, 6858. [DOI] [PubMed] [Google Scholar]
- [9].Chong Y, Ma YF, Shen H, Tu XL, Zhou X, Xu JY, Dai JW, Fan SJ, Zhang ZJ, Biomaterials 2014, 35, 5041. [DOI] [PubMed] [Google Scholar]
- [10].Liu HJ, Li CW, Qian Y, Hu L, Fang J, Tong W, Nie RR, Chen QW, Wang H, Biomaterials 2020, 232, 119700. [DOI] [PubMed] [Google Scholar]
- [11].Yang SW, Wang XL, He P, Xu AL, Wang G, Duan JL, Shi YQ, Ding GQ, Small 2021, 17, 2004867. [DOI] [PubMed] [Google Scholar]
- [12].Li S, Su W, Wu H, Yuan T, Yuan C, Liu J, Deng G, et al. Nat. Biomed. Eng 2020, 4, 704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang H, Revia R, Wang K, Kant RJ, Mu QX, Gai Z, Hong KL, Zhang MQ, Adv. Mater 2017, 29, 1605416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li SH, Zhou SX, Li YC, Li XH, Zhu J, Fan LZ, Yang SH, ACS Appl. Mater. Interfaces 2017, 9, 22332. [DOI] [PubMed] [Google Scholar]
- [15].Ge JC, Lan MH, Zhou BJ, Liu WM, Guo L, Wang H, Jia QY, Niu GL, Huang X, Zhou HY, Meng XM, Wang PF, Lee CS, Zhang WJ, Han XD, Nat. Commun 2014, 5, 4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim D, Yoo JM, Hwang H, Lee J, Lee SH, Yun SP, Park MJ, Lee M, Choi S, Kwon SH, Lee S, Kwon SH, Kim S, Park YJ, Kinoshita M, Lee YH, Shin S, Paik SR, Lee SJ, Lee S, Hong BH, Ko HS, Nat. Nanotechnol 2018, 13, 812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu H, Li C, Qian Y, Hu L, Fang J, Tong W, Nie R, Chen Q, Wang H, Biomaterials 2020, 232, 119700. [DOI] [PubMed] [Google Scholar]
- [18].Wang H, Mu QX, Wang K, Revia RA, Yen C, Gu XY, Tian BW, Liu J, Zhang MQ, Appl. Mater. Today 2019, 14, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Li Y, Dong H, Tao Q, Ye C, Yu M, Li J, Zhou H, Yang S, Ding G, Xie X, Biomaterials 2020, 250, 120056. [DOI] [PubMed] [Google Scholar]
- [20].Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW, Nat. Rev. Mater 2016, 1, 1. [Google Scholar]
- [21].Smith BR, Gambhir SS, Chem. Rev 2017, 117, 901. [DOI] [PubMed] [Google Scholar]
- [22].Yamato K, Sekiya R, Suzuki K, Haino T, Angew. Chem. Int. Ed 2019, 58, 9022. [DOI] [PubMed] [Google Scholar]
- [23].Cheng L, Wang XW, Gong F, Liu T, Liu Z, Adv. Mater 2020, 32, 1902333. [DOI] [PubMed] [Google Scholar]
- [24].Chen HM, Zhang WZ, Zhu GZ, Xie J, Chen XY, Nat. Rev. Mater 2017, 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shi JJ, Kantoff PW, Wooster R, Farokhzad OC, Nat. Rev. Cancer 2017, 17, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Blanco E, Shen H, Ferrari M, Nat. Biotechnol 2015, 33, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pelaz B, del Pino P, Maffre P, Hartmann R, Gallego M, Rivera-Fernandez S, de la Fuente JM, Nienhaus GU, Parak WJ, ACS Nano 2015, 9, 6996. [DOI] [PubMed] [Google Scholar]
- [28].Heyes CD, Kobitski AY, Amirgoulova EV, Nienhaus GU, Journal of Physical Chemistry B 2004, 108, 13387. [Google Scholar]
- [29].Yan H, Zhao LY, Shang WT, Liu ZQ, Xie WS, Qiang C, Xiong ZY, Zhang RR, Li BH, Sun XD, Kang FY, Nano Res. 2017, 10, 704. [Google Scholar]
- [30].Yan H, Shang WT, Sun XD, Zhao LY, Wang JY, Xiong ZY, Yuan J, Zhang RR, Huang QL, Wang K, Li BH, Tian J, Kang FY, Feng SS, Adv. Funct. Mater 2018, 28, 1705710. [Google Scholar]
- [31].Wang L, Wang YL, Xu T, Liao HB, Yao CJ, Liu Y, Li Z, Chen ZW, Pan DY, Sun LT, Wu MH, Nat. Commun 2014, 5, 5357. [DOI] [PubMed] [Google Scholar]
- [32].Xia C, Qiu YR, Xia Y, Zhu P, King G, Zhang X, Wu ZY, Kim JY, Cullen DA, Zheng DX, Li P, Shakouri M, Heredia E, Cui PX, Alshareef HN, Hu YF, Wang HT, Nat. Chem 2021, 13, 887. [DOI] [PubMed] [Google Scholar]
- [33].Wang JY, Yan H, Liu ZQ, Wang ZC, Gao HN, Zhang ZJ, Wang BL, Xu N, Zhang SQ, Liu XJ, Zhang RR, Wang XM, Zhang GF, Zhao LY, Liu K, Sun XD, Nanoscale 2018, 10, 19612. [DOI] [PubMed] [Google Scholar]
- [34].Shaner NC, Lin MZ, McKeown MR, Steinbach PA, Hazelwood KL, Davidson MW, Tsien RY, Nat. Methods 2008, 5, 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hirano M, Ando R, Shimozono S, Sugiyama M, Takeda N, Kurokawa H, Deguchi R, Endo K, Haga K, Takai-Todaka R, Inaura S, Matsumura Y, Hama H, Okada Y, Fujiwara T, Morimoto T, Katayama K, Miyawaki A, Nat. Biotechnol 2022, 40, 1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen SQ, Zhong Y, Fan WF, Xiang JJ, Wang GW, Zhou Q, Wang JQ, Geng Y, Sun R, Zhang Z, Piao Y, Wang JG, Zhuo JY, Cong HL, Jiang HP, Ling J, Li ZC, Yang DD, Yao X, Xu X, Zhou ZX, Tang JB, Shen YQ, Nat. Biomed. Eng 2021, 5, 1019. [DOI] [PubMed] [Google Scholar]
- [37].Chiou WL, J. Pharmacokinet. Biopharm 1978, 6, 539. [DOI] [PubMed] [Google Scholar]
- [38].Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Ipe BI, Bawendi MG, Frangioni JV, Nat. Biotechnol 2007, 25, 1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yuan J, Liu JL, Song Q, Wang D, Xie WS, Yan H, Zhou JF, Wei Y, Sun XD, Zhao LY, ACS Appl. Mater. Interfaces 2016, 8, 24445. [DOI] [PubMed] [Google Scholar]
- [40].Li Y, Zhang J, Gu J, Hu K, Huang S, Conti PS, Wu H, Chen K, Mol. Imaging Biol. 2020, 22, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang Q, Yan H, Jin Y, Wang Z, Huang W, Qiu J, Kang F, Wang K, Zhao X, Tian J, Biomaterials 2018, 183, 173. [DOI] [PubMed] [Google Scholar]
- [42].Ogata G, Ishii Y, Asai K, Sano Y, Nin F, Yoshida T, Higuchi T, Sawamura S, Ota T, Hori K, Maeda K, Komune S, Doi K, Takai M, Findlay I, Kusuhara H, Einaga Y, Hibino H, Nat. Biomed. Eng 2017, 1, 654. [DOI] [PubMed] [Google Scholar]
- [43].Fan Z, Sun LM, Huang YJ, Wang YZ, Zhang MJ, Nat. Nanotechnol 2016, 11, 388. [DOI] [PubMed] [Google Scholar]
- [44].Dong A, He K, Dudok B, Farrell JS, Guan W, Liput DJ, Puhl HL, Cai R, Wang H, Duan J, Albarran E, Ding J, Lovinger DM, Li B, Soltesz I, Li Y, Nat. Biotechnol 2022, 40, 787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Huang Q, Cohen MA, Alsina FC, Devlin G, Garrett A, McKey J, Havlik P, Rakhilin N, Wang E, Xiang K, Mathews P, Wang L, Bock C, Ruthig V, Wang Y, Negrete M, Wong CW, Murthy PKL, Zhang S, Daniel AR, Kirsch DG, Kang Y, Capel B, Asokan A, Silver DL, Jaenisch R, Shen X, Science 2020, 368, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Siebert S, Farrell JA, Cazet JF, Abeykoon Y, Primack AS, Schnitzler CE, Juliano CE, Science 2019, 365, 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lewis SM, Asselin-Labat M-L, Quan N, Berthelet J, Tan X, Wimmer VC, Merino D, Rogers KL, Naik SH, Nat. Methods 2021, 18, 997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting this paper’s figures are available upon reasonable request.