Abstract
Summary
The phylogenetic signal, frequently used to identify signatures of adaptive evolution or important associations between genes and phenotypes, measures the tendency for recently diverged species to resemble each other more than distantly related species. An example of such a measure is the δ statistic, which uses Shannon entropy to measure the degree of phylogenetic signal between a categorical trait and a phylogeny. In this study, we refined this statistic to account for tree uncertainty, resulting in more accurate assessments of phylogenetic associations. In addition, we provided a more accessible and computationally efficient implementation of the δ statistic that will facilitate its use by the evolutionary community.
Availability and implementation
1 Introduction
The phylogenetic signal measures the tendency for species that have recently diverged to resemble each other more than those that are distantly related (Blomberg and Garland 2002). This signal between phenotypic traits and evolutionary histories helps us to understand the ways in which species evolve and become different.
Recently, we proposed the δ statistic, which is based on the concept of entropy from information theory. It exploits the uncertainty on the ancestral trait’s probability vectors, which can be inferred through maximum likelihood, Bayesian or Approximate Bayesian Computation (ABC) inference, to calculate the degree of phylogenetic signal between a categorical trait and a phylogeny (Borges et al. 2019). This statistic has been applied in various contexts, such as identifying homoplasic sites in SARS-CoV-2 sequences that can hinder phylogenetic reconstruction (Maio et al. 2020) or studying complex social traits, such as the parental care strategies of digger wasps (Field et al. 2020).
A limitation of this statistic is that it ignores common sources of error, and its previous implementation was not optimized for large-scale genomic studies. Here, we address these limitations by providing a new version of this statistic that accounts for tree uncertainty and developing a new Python implementation that will allow its use in large-scale genomic datasets, which are now commonplace in evolutionary biology. We also increased the accessibility and reproducibility of the δ statistic by facilitating its use by the evolutionary community with a web-application.
2 Implementation and usage
The previous δ statistic was implemented in R. Here, we converted it to the Python programming language, using the Numba library (Lam et al. 2015) for faster processing of data items. This new version utilizes the PastML package (Ishikawa et al. 2019), which automatically performs ancestral character reconstructions. The new implementation is on average 12.70 times faster for the standard use of 10 000 iterations (Supplementary Fig. S3). We expect this new implementation will allow applications with genome-scale data. We have also provided a web application that includes a tutorial explaining the usage of the δ statistic and detailed information about the input and output data. This webserver takes on average 10 s per tree (Supplementary Fig. S2) and can be used for smaller-scale applications or for teaching purposes.
3 Application
The previous δ statistic of phylogenetic signal assumed that the given tree is correct (Borges et al. 2019), ignoring possible and likely uncertainties on the topology and branch lengths. To test the impact of tree uncertainty, we used mammalian data retrieved from OrthoMAM (Scornavacca et al. 2019). Our dataset consisted of 1000 protein-coding sequence alignments for 30 mammals. To avoid possible biases due to gene length (i.e. longer genes are expected to estimate more accurate trees), the alignments were trimmed to 1000 base pairs. Our categorical traits consisted of a 2-class (presence and absence of meat in primary diet) and a 3-class (carnivorous, omnivorous, and herbivorous) phenotypes that were defined for the 30 mammalian species based on existing literature (Yahnke et al. 2013) (Supplementary Table S1 and Supplementary Fig. S5). Gene tree estimation was conducted with the Bayesian software RevBayes (Höhna et al. 2016) (further details in Supplementary Text).
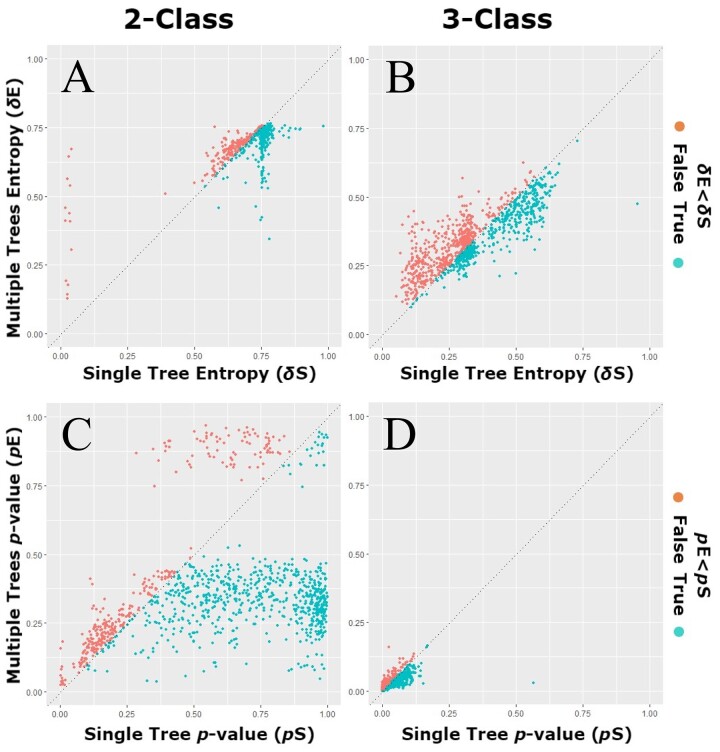
To evaluate the impact of tree uncertainty on the phylogenetic signal, we compared the δ statistic calculated using the previous method (δS) and the extended method proposed here (δE) when accounting for multiple phylogenetic trees. We used 1000 randomly sampled trees from the posterior distribution to average δE. However, we found that δE converges after 840 and 580 trees for the 2- and 3-character traits across all genes (Supplementary Fig. S7). We observed that the two entropies do not have a one-to-one relationship (Fig. 1), with higher δS values having corresponding smaller δE values and vice versa. These discrepancies are related to the overall support of the phylogeny. While δS and δE are similar for trees that have higher posterior clade probabilities, they differ for trees that have overall low support. This result is expected, as for low-supported trees, the maximum a posteriori tree (MAP) might not represent a reasonable gene history. Accounting for distinct posterior-sampled trees thus seems to help recover phylogenetic signal associated with clades that are not present in the MAP.
Figure 1.
Impact of tree uncertainty on the δ statistic. (A, B) Estimated entropies for the standard (δS) and new (δE) δ statistic in a categorical trait of 2 and 3 classes. (C, D) Probability P of a phylogenetic association for the standard (pS) and new method (pE)
In addition to the entropy comparison, we also computed the probability, P, of finding the empirical entropy value under the standard and new method in their null distributions (ΔS and ΔE, respectively) (Supplementary Table S3), obtained by shuffling the trait vector at the tree tips (Equation 1).
| (1) |
This probability serves as a proxy for the P-value. We observed that the values of P under the new method are on average 1.37 times smaller (2-class: 1.64 times; 3-class: 1.10 times) than in the standard method (Fig. 1 and Supplementary Table S2). This shows that the new method is more likely to identify phylogenetic associations and indicates that accounting for tree uncertainty captures evolutionary signal.
4 Conclusions
The revised δ statistic provides a more accurate discovery of phylogenetic associations, thus delivering a more precise characterization of the genetic players involved in species adaptation. Moreover, our faster and more accessible δ statistic will facilitate its automated use in modern genomic applications, including phylogenetic pipelines with large genomic datasets, or in the classroom for teaching purposes.
Supplementary Material
Contributor Information
Diogo Ribeiro, CIIMAR/CIMAR, Interdisciplinary Centre of Marine and Environmental Research, University of Porto, Terminal de Cruzeiros do Porto de Leixões, 4450-208 Porto, Portugal; Department of Biology, Faculty of Sciences, University of Porto, Rua do Campo Alegre, 4169-007 Porto, Portugal.
Rui Borges, Institut für Populationsgenetik, Vetmeduni Vienna, 1210 Vienna, Austria.
Ana Paula Rocha, CMUP, Centre of Mathematics of the University of Porto, 4169-007 Porto, Portugal; Department of Mathematics, Faculty of Sciences, University of Porto, Rua do Campo Alegre, 4169-007 Porto, Portugal.
Agostinho Antunes, CIIMAR/CIMAR, Interdisciplinary Centre of Marine and Environmental Research, University of Porto, Terminal de Cruzeiros do Porto de Leixões, 4450-208 Porto, Portugal; Department of Biology, Faculty of Sciences, University of Porto, Rua do Campo Alegre, 4169-007 Porto, Portugal.
Supplementary data
Supplementary data are available at Bioinformatics online.
Conflict of interest
None declared.
Funding
This work was supported by the Austrian Science Fund [P34524-B] to R.B.; partially supported by Centre of Mathematics of the University of Porto - Intelligent Systems Associate Laboratory (CMUP-LASI) through the Fundação para a Ciência e a Tecnologia (FCT) project [UIDB/00144/2020] to A.P.R.; and the FCT projects UIDB/04423/2020, UIDP/04423/2020, PTDC/CTA-AMB/31774/2017 [POCI-01-0145-FEDER/031774/2017], and Atlantida [NORTE-01-0145-FEDER-000040] to A.A.
Data availability
The data underlying this article are available at Zenodo (https://doi.org/10.5281/zenodo.7541548).
References
- Blomberg SP, Garland T. Tempo and mode in evolution: phylogenetic inertia, adaptation and comparative methods. J Evol Biol 2002;15:899–910. [Google Scholar]
- Borges R, Machado JP, Gomes C et al. Measuring phylogenetic signal between categorical traits and phylogenies. Bioinformatics 2019;35:1862–9. [DOI] [PubMed] [Google Scholar]
- Field J, Gonzalez-Voyer A, Boulton RA et al. The evolution of parental care strategies in subsocial wasps. Behav Ecol Sociobiol 2020;74:1–12. [Google Scholar]
- Höhna S, Landis MJ, Heath TA et al. RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst Biol 2016;65:726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa SA, Zhukova A, Iwasaki W et al. A fast likelihood method to reconstruct and visualize ancestral scenarios. Mol Biol Evol 2019;36:2069–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SK et al. Numba: a LLVM-based Python JIT compiler. In: Proceedings of the Second Workshop on the LLVM Compiler Infrastructure in HPC, New York, NY, USA: Association for Computing Machinery, 1–6. 2015.
- Maio ND, Walker C, Borges R et al. Issues with SARS-CoV-2 Sequencing Data. Virological.org. 2020. https://virological.org/t/issues-with-sars-cov-2-sequencing-data/473 (accessed on 12 December 2022).
- Scornavacca C, Belkhir K, Lopez J et al. Orthomam v10: scaling-up orthologous coding sequence and exon alignments with more than one hundred mammalian genomes. Mol Biol Evol 2019;36:861–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahnke CJ, Dewey T, Myers P et al. Animal diversity web as a teaching & learning tool to improve research & writing skills in college biology courses. Am Biol Teacher 2013;75:494–8. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available at Zenodo (https://doi.org/10.5281/zenodo.7541548).