Abstract
Pathogenic bacteria produce an elaborate assortment of extracellular and cell-associated bacterial products that enable colonization and establishment of infection within a host. Lipopolysaccharide (LPS) molecules are cell surface factors that are typically known for their protective role against serum-mediated lysis and their endotoxic properties. The most heterogeneous portion of LPS is the O antigen or O polysaccharide, and it is this region which confers serum resistance to the organism. Pseudomonas aeruginosa is capable of concomitantly synthesizing two types of LPS referred to as A band and B band. The A-band LPS contains a conserved O polysaccharide region composed of d-rhamnose (homopolymer), while the B-band O-antigen (heteropolymer) structure varies among the 20 O serotypes of P. aeruginosa. The genes coding for the enzymes that direct the synthesis of these two O antigens are organized into two separate clusters situated at different chromosomal locations. In this review, we summarize the organization of these two gene clusters to discuss how A-band and B-band O antigens are synthesized and assembled by dedicated enzymes. Examples of unique proteins required for both A-band and B-band O-antigen synthesis and for the synthesis of both LPS and alginate are discussed. The recent identification of additional genes within the P. aeruginosa genome that are homologous to those in the A-band and B-band gene clusters are intriguing since some are able to influence O-antigen synthesis. These studies demonstrate that P. aeruginosa represents a unique model system, allowing studies of heteropolymeric and homopolymeric O-antigen synthesis, as well as permitting an examination of the interrelationship of the synthesis of LPS molecules and other virulence determinants.
Lipopolysaccharides (LPS) of gram-negative bacteria are major components of the cell wall. The hydrophobic lipid A component of LPS secures these molecules in the outer membrane, while the core oligosaccharide links the lipid A region to the O antigen or O polysaccharide. The location of these molecules in the outer leaflet of the outer membrane permits interaction of LPS with the external milieu. As a result, early research focused on the role of LPS as a virulence determinant and on its use as a vaccine candidate. Since that time, studies have expanded to include analysis of the chemistry and biosynthesis of the O-antigenic region due to its immunogenicity, serotype specificity, and serum resistance properties.
With the medical and environmental importance of Pseudomonas aeruginosa, major efforts have been directed toward understanding the factors relevant to initial bacterial attachment, evasion of host defenses, and establishment of infection. LPS is one of these factors, and over the last decade remarkable advancements have been made in the fields of P. aeruginosa LPS chemistry and biosynthesis, in particular the progress in the area of O-antigen synthesis and assembly. While the O-antigen synthesis pathways of P. aeruginosa have many properties in common with other characterized LPS systems, unique features of synthesis have been identified. In this review, O-antigen synthesis pathways for the two LPS molecules produced by P. aeruginosa, A band and B band, are discussed in detail, along with future research areas. The role of LPS in the biology and pathogenesis of P. aeruginosa is also examined.
P. AERUGINOSA, THE PATHOGEN
Occurrence of P. aeruginosa Infections
P. aeruginosa typically causes disease only in individuals with impaired host defenses and is thus referred to as an opportunistic pathogen. Such compromised individuals include patients undergoing immunosuppressive therapies (e.g., cancer treatment), those receiving treatment for traumatic skin damage (burn wounds), those with human immunodeficiency virus infections, and those with cystic fibrosis (CF) (78, 107). Recent clinical data indicates P. aeruginosa to be the fourth leading cause of nosocomial infection and the foremost cause of hospital-acquired pneumonia (95). Acquisition of this pathogen within the hospital setting is attributed to contaminated environmental reservoirs (e.g., sinks and respirators), as well as patient-to-patient spread (182).
CF patients in particular are highly susceptible to chronic pulmonary infections with P. aeruginosa. The lung environment of these patients appears to provide a unique niche that promotes chronic microbial colonization. Mutations in the CF transmembrane regulator protein (CFTR) interfere with chloride ion transport in CF patients. This electrolyte imbalance causes dehydration within the lungs and production of a viscous mucous, which significantly impairs mucociliary clearance mechanisms, allowing persistent bacterial colonization (115). Mutations in CFTR are also known to cause undersialylation of epithelial cell surface receptors, which increases P. aeruginosa adherence to host tissue (59, 121). These patients typically experience a progression in pulmonary pathogens, with Haemophilus influenzae and Staphylococcus aureus infections occurring in infants and children and P. aeruginosa and Burkholderia cepacia infections occurring during adolescence and adulthood (186). H. influenzae and S. aureus pulmonary infections are usually controllable with antimicrobial therapies, but effective clearance of these organisms allows subsequent colonization by and chronic establishment of P. aeruginosa within the lungs of these patients (238). More than 80% of CF patients over the age of 26 years are colonized with P. aeruginosa (68). It is these chronic P. aeruginosa respiratory infections which account for most of the pulmonary deterioration and mortality in CF patients, since this organism is usually the only pathogen recovered postmortem from the sputum and lung tissue (67).
Effective antibiotic therapy of P. aeruginosa infections has been problematic, largely due to the high intrinsic resistance of this organism to antimicrobial agents. This resistance is a result of the low permeability of the outer membrane (81), combined with the presence of both β-lactamases (84, 178) and multidrug efflux pumps (119, 183, 184). The outer membrane is thought to reduce the passage of hydrophobic antibiotics due to the highly charged bacterial surface that is stabilized by divalent cations (180). The uptake of small hydrophilic antimicrobial agents, such as β-lactams, occurs via porin proteins, but P. aeruginosa has only 1 to 5% the permeability of Escherichia coli for β-lactams (83). β-Lactam resistance is heightened through the presence of periplasmic β-lactamases, which can be plasmid or chromosomally encoded (80). Recently, various efflux systems (mexAB-oprM [138, 184], mexCD-oprJ [183], and mexEF-oprN [184]) which are able to export structurally unrelated antibiotics, providing P. aeruginosa with multidrug resistance, have been described.
Efforts have been made to gain an in-depth understanding of the above-mentioned resistance mechanisms in an attempt to design more effective therapies for P. aeruginosa infections. One approach has been the coadministration of β-lactams with β-lactamase inhibitors. However, resistance to this treatment combination has also developed (79). There is also the approach of downregulating or inactivating multidrug efflux pumps for enhanced susceptibility to various antibiotics. Interestingly, β-lactamase inhibitors have recently been shown to serve as substrates for the MexAB-OprM pump, which suggests that pump inactivation may enhance the efficacies of β-lactam–β-lactamase inhibitor therapeutic combinations (139). The issue of outer membrane permeability has also been addressed through the coadministration of antibiotic with permeabilizing compounds such as cationic peptides (80, 181). These peptides interact with divalent cation binding sites present on LPS molecules and permeabilize the outer membrane, allowing enhanced antibiotic uptake during this process. In future, one or a combination of these approaches may facilitate improved management of P. aeruginosa infections.
Contribution of Lipopolysaccharide to Host-Bacterium Interactions
LPS molecules are located in the cell wall and thus play an important structural role while mediating interaction with the neighboring environment. The tripartite nature of LPS divides the molecule into a hydrophobic lipid A region, which replaces phospholipids in the outer membrane, a central core oligosaccharide region, and a repeating polysaccharide portion referred to as O antigen or O polysaccharide. The terms “smooth” and “rough” are often used to describe the LPS phenotype. Attachment of the O antigen to core-lipid A results in a smooth LPS phenotype, while core-lipid A lacking O antigen is referred to as rough LPS. The contribution of smooth LPS to virulence has been demonstrated repeatedly by using various animal model systems. A study by Cryz et al. (40) showed that a wild-type strain of P. aeruginosa with smooth LPS was more virulent than were isogenic mutants. In a burned-mouse infection model, the mutant, which has rough LPS, has a 50% lethal dose more than 1,000-fold higher than that of the wild-type strain. This increase in the 50% lethal dose of a strain producing rough LPS demonstrates that O antigen is critical for virulence and that studies directed toward an understanding of LPS biosynthesis are essential. Recent studies by Preston et al. (185) with a mouse cornea infection model and Tang et al. (220) with a neonatal-mouse challenge model confirm that intact smooth LPS is required for P. aeruginosa virulence. In vitro experiments have shown that rough mutants of P. aeruginosa deficient in O-antigen synthesis are sensitive to the killing effects of human serum while wild-type strains with smooth LPS are serum resistant (44, 82). Since the focus of this review centers on LPS biosynthesis in P. aeruginosa, we address the role of lipid A, core oligosaccharide, and O antigen in the pathogenesis of this organism (see below).
When LPS is shed by bacteria into host tissues, it is usually bound by LPS binding protein (136, 222), which is transferred to the CD14 receptor (2395) on macrophages, thereby inducing secretion of cytokines including tumor necrosis factor alpha, interleukin-1 (IL-1), IL-6, IL-8, and IL-10 (150). These cytokines are known markers of inflammatory responses. The lipid A region of LPS, composed of a phosphorylated diglucosamine moiety substituted with fatty acids, is thought to be responsible for most of the biological activities of LPS (also referred to as endotoxin) through the induction of these immunomodulating molecules. Release of these inflammatory mediators enhances host defenses against bacterial infections. Excessive LPS stimulation of the immune system can occur, whereby elevated levels of activated and recruited immune cells results in septic shock and even death (150). P. aeruginosa is one of the top three pathogens responsible for sepsis due to gram-negative bacteria, and LPS from this organism is capable of overstimulating the immune system (62). Interestingly, P. aeruginosa lipid A is less toxic than that of enteric organisms, probably due to the presence of mostly pentaacyl rather than hexaacyl chains (125).
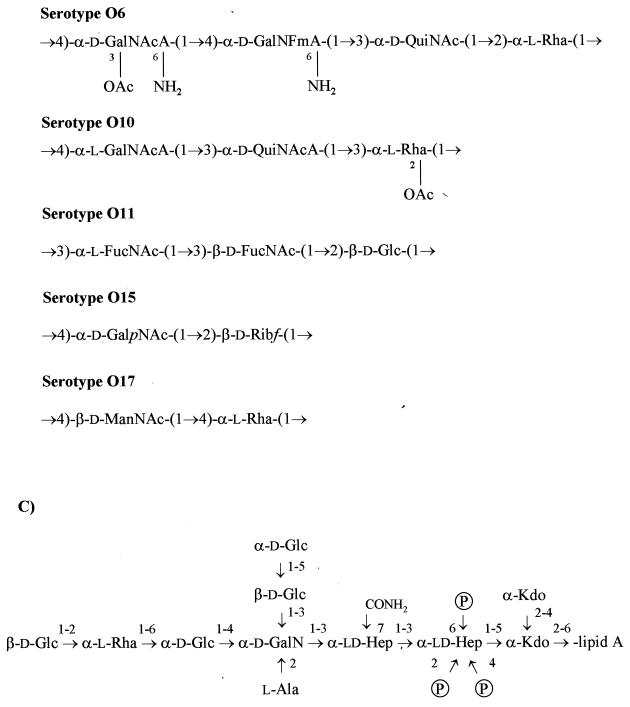
Attached to the lipid A is the core oligosaccharide region of LPS, which can be subdivided into an inner and an outer core. The inner core contains l-glycero-d-manno-heptose and 3-deoxy-d-manno-octulosonic acid (KDO), while the outer core is composed of hexose sugars such as d-glucose (d-Glc). The chemical structure of the P. aeruginosa serotype O5 core oligosaccharide is shown in Fig. 1C. The genetics involved in synthesis of this core region are currently being investigated by our laboratory but are not included in this review (52). The outer core region of P. aeruginosa LPS has recently been proposed to function as a bacterial ligand for association and entry of the organism into corneal cells during the infection stage of keratitis (243). In vitro studies have revealed the terminal d-Glc moiety of the outer core to be the critical residue in binding of the LPS molecule to mouse corneal epithelial cells (243). Recently, Pier et al. (179, 180) assessed the role of CFTR as a receptor for the binding of P. aeruginosa to host epithelial surfaces. In these studies, CFTR-minus epithelial cell lines showed poor ingestion and uptake of P. aeruginosa compared to cell lines transfected with CFTR. These authors have proposed that the presence of CFTR in normal hosts could be a defense mechanism whereby efficient epithelial-cell ingestion of P. aeruginosa, followed by cellular desquamation and swallowing, could facilitate bacterial killing by the digestive system of the host. In this model, the outer core region is believed to be the ligand mediating this association, since strains producing semirough LPS (core plus one O-antigen repeat) and rough LPS (lacking O antigen) with a complete core region were ingested more readily than were those expressing wild-type smooth LPS (180). These data provide evidence demonstrating that LPS variations can affect the uptake of P. aeruginosa by host epithelial cells. These new findings may help elucidate specific host-bacterium interactions and favor the development of therapeutics for chronic P. aeruginosa pulmonary infections in CF patients.
FIG. 1.
Structures of O-antigen and core oligosaccharide P. aeruginosa LPS. (A) A-band O-antigen structure of P. aeruginosa (7). (B) B-band O-antigen structures of P. aeruginosa serotypes O2, O5, O6, O10, O11, O15, O16, O17, O18, and O20 (109–111). The O2, O5, O16, O18, and O20 serotypes compose a cross-reactive serogroup due to similarities in their O-repeat units (130). (C) Core oligosaccharide structure of P. aeruginosa serotype O5 (199). ld-Hep, l-glycero-d-manno-heptose; Kdo, 2-keto-3-deoxy-d-manno-octulosonic acid; CONH2—O-carbamoyl.
As mentioned above, attachment of the O antigen or O polysaccharide to core-lipid A results in the smooth form of LPS. This O-antigenic region is highly immunogenic and elicits a strong antibody response from the host. Resistance to serum is conferred by the presence of O antigen on the cell surface, and the extent of this serum resistance is influenced by O-antigen structure, chain length, and the amount of O antigen substituted on core-lipid A (reviewed in reference 97). P. aeruginosa produces two forms of O antigen, known as A band (homopolymer) and B band (heteropolymer). The A-band O-polysaccharide region is composed of d-rhamnose (d-Rha) residues arranged as trisaccharide repeating units linked α1→2, α1→3, α1→3 (Fig. 1A) (7). The A-band d-rhamnan polysaccharide is composed of approximately 70 d-Rha residues, which is equivalent to 23 repeating units (242). This is shorter than B-band O antigen (composed of ≥50 repeating units [129]), which is thought to mask underlying A-band polysaccharide, since A+B+ strains are not agglutinable with A-band-specific monoclonal antibodies (MAbs) (135). B-band O antigen is composed of di- to pentasaccharide repeating units of various monosaccharides. The composition of the B-band O-antigen trisaccharide repeat of serotype O5 (the serotype of the common laboratory strain PAO1) is di-N-acetylmanosaminuronic acid and N-acetyl-6-deoxygalactose (Fuc2NAc) (Fig. 1B) (109). Several studies have demonstrated that P. aeruginosa LPS confers serum resistance and elicits a protective immune response (40, 63, 82). Recently, a panel of P. aeruginosa LPS-deficient mutants was used to determine that B-band LPS confers serum resistance to the organism while A-band LPS is not protective against serum-mediated lysis (44). Studies have also indicated that while antibodies directed toward B-band LPS are highly protective of neutropenic mice upon challenge with smooth LPS strains, antibodies against A-band LPS are not protective (86).
P. aeruginosa is known to undergo differential expression of some of its virulence factors during infection. These phenotypic changes have for the most part been observed during pulmonary infections in CF patients (reviewed in reference 78). The most dramatic change is the conversion of the organism from a nonmucoid to a mucoid phenotype due to the production of copious amounts of the exopolysaccharide alginate (reviewed in reference 78). This cell surface alteration occurs once P. aeruginosa is well established within the lungs of CF patients and correlates with poor lung function (177). Accompanying the onset of alginate production is initiation of the microcolony mode of growth within the lungs of CF patients, representing a bacterial biofilm composed of cells embedded within an alginate matrix (128). With the emergence of mucoid P. aeruginosa within the lungs, there are cell surface changes with respect to LPS phenotype. Chronic P. aeruginosa isolates from CF patients either lack B-band O antigen entirely or express smaller amounts (70, 82, 131, 177, 182), while the level of A-band O polysaccharide is maintained (135). A study by Lam et al. (135) examined 250 P. aeruginosa isolates from the lungs of CF patients and determined that 68% expressed A-band LPS but none expressed B-band O antigen. Many of these clinical isolates are therefore nontypeable or polytypeable due to the lack of O-antigen polymer or deficiency in high-molecular-weight O-antigen polymer production (131, 177, 182). Such changes in surface properties are problematic for serotyping and epidemiological studies of P. aeruginosa isolates from CF patients (221). Evans et al. (64) demonstrated that 8 of 13 CF patient P. aeruginosa isolates expressing rough LPS could coproduce endogenous smooth LPS along with serotype O11 LPS when transformed with O-antigen genes (wbp, previously rfb) from P. aeruginosa serotype O11. Therefore, in some cases, these rough P. aeruginosa isolates from CF patients have acquired mutations in their wbp region. At present, the mechanisms responsible for these mutations and changes in LPS phenotype are not known.
It is interesting that A band is the LPS molecule selectively maintained on the P. aeruginosa cell surface during chronic CF lung infections. The presence of anti-A-band antibodies within CF patients correlates with both increased duration of P. aeruginosa infections and lower pulmonary function (135). Once P. aeruginosa has colonized the lungs, these LPS modifications are probably beneficial for evasion of host defenses (A band is less immunogenic) and for alteration of susceptibility to antibiotics, since loss of B-band O antigen confers resistance to aminoglycosides (17, 98). A recent study by Asboe et al. (8) examined P. aeruginosa isolates from human immunodeficiency virus-infected patients with respiratory infections. Multiple isolates from single patients infected with the same bacterial strain were shown to become serum sensitive and polyagglutinable/nontypeable. These characteristics, as mentioned above, are associated with a loss of or decrease in B-band O antigen. A-band LPS was not examined in that study. It would be interesting to determine if these respiratory isolates express A-band LPS and if the selection toward B-band deficiency follows a similar mechanism to that of P. aeruginosa pulmonary isolates from CF patients.
Environmental Conditions Influencing Lipopolysaccharide Production
LPS heterogeneity can be achieved through variations in the sugar moieties within the O-antigen repeating unit, the type of stereochemistry (α or β) of the glycosyl linkages within the O-antigen repeat, the addition of noncarbohydrate moieties to the O antigen (i.e., O acetylation), and the proportion of smooth versus rough LPS molecules. Most strains of P. aeruginosa have a capping frequency (core-lipid A molecules substituted with long-chain O antigen) of between 0.2 and 14% (82, 191, 232). Although the regulatory mechanisms governing LPS synthesis and changes in LPS production in P. aeruginosa have not been determined, several studies have been performed to examine the influence of environmental parameters on LPS synthesis. Growth at elevated temperatures decreases B-band O-antigen chain length as the temperature increases (123, 165). Along with this decrease in the level of long-chain O antigen is an increase in the amount of the semirough (SR) B-band LPS (i.e., core-lipid A capped with one O-repeat unit). A study by Kropinski et al. (123) demonstrated that the proportion of SR LPS in P. aeruginosa PAO1 increases from 19.3 to 37.6% when the growth temperature shifts from 15 to 45°C. Interestingly, a recent study by Makin and Beveridge (153) revealed complete loss of B-band O antigen when PAO1 cells were grown at 45°C. The reason for the observed differences between these two studies is not known; however, the data does indicate a correlation between B-band O-antigen synthesis and temperature. The influence of various osmotic conditions on P. aeruginosa LPS synthesis has also been examined. Under conditions of high NaCl, MgCl2, glycerol, and sucrose and low pH and phosphate, there is a decrease in the amount of long-chain B-band O antigen. Both elevated temperature and variation in nutrient levels resulted in modest increases in the chain length of A-band O polysaccharide (153, 165). Future work in this area should be directed at determining how these environmental conditions influence LPS synthesis at the molecular level for a fuller understanding of the regulatory networks controlling the expression of these cell surface molecules.
Formation of Biofilms by P. aeruginosa: Role for Lipopolysaccharide
Biofilm formation has been intensely studied over the years due to its environmental and medical relevance. Bacterial biofilms can develop on solid substrata and generally consist of cells entwined in a protective matrix of extracellular polysaccharides. Establishment of a biofilm requires initial bacterial attachment to a solid surface to allow the growth and development of a mature biofilm. Attachment of P. aeruginosa to surfaces is known to increase transcription of alginate biosynthetic genes, thereby increasing alginate production, which enhances the growth of the biofilm. Transcriptional fusions with a lacZ reporter gene have demonstrated that the transcriptional activities of two alginate structural genes, algD and algC, increase in attached cells compared to planktonic cells (46, 47, 90). To date, regulatory components that are involved in sensing environmental conditions and controlling the transcription of alginate genes from the algC and algD promoters are reasonably well defined (reviewed in references 78 and 162).
The contribution of other polysaccharide molecules (i.e., LPS) to biofilm formation have been less well studied. However, the presence or absence of long-chain LPS polymers and the differences in the chemical nature of LPS molecules influence the physiochemical characteristics of the cell surface. In wild-type P. aeruginosa strains that are nonmucoid and devoid of alginate, the predominant surface polysaccharide is B-band LPS (129). The B-band O polymers are highly anionic and extend beyond the layers of A-band O polysaccharide and outer membrane proteins (98, 129, 154). Makin and Beveridge (154) conducted a study to examine the relationship between cell surface hydrophobicities and relative adhesive properties among P. aeruginosa strains with various LPS phenotypes. They found that A+B+ and A−B+ cells possess the lowest surface hydrophobicity and the lowest surface charge (154). P. aeruginosa cells expressing these predominantly anionic LPS phenotypes (A+B+ and A−B+) adhere to glass more efficiently, implying that electrostatic interactions among the B-band O-antigen polymers play a role in binding (154). In contrast, the ability of the bacteria to adhere to polystyrene was shown to correlate with the relative hydrophobicity of the bacterial cells in the descending order A+B− > A−B− > A+B+ > A−B+ (154). Therefore, changes in the production of either A-band or B-band LPS in P. aeruginosa influence the surface characteristics and probably modify the binding capabilities of these bacteria.
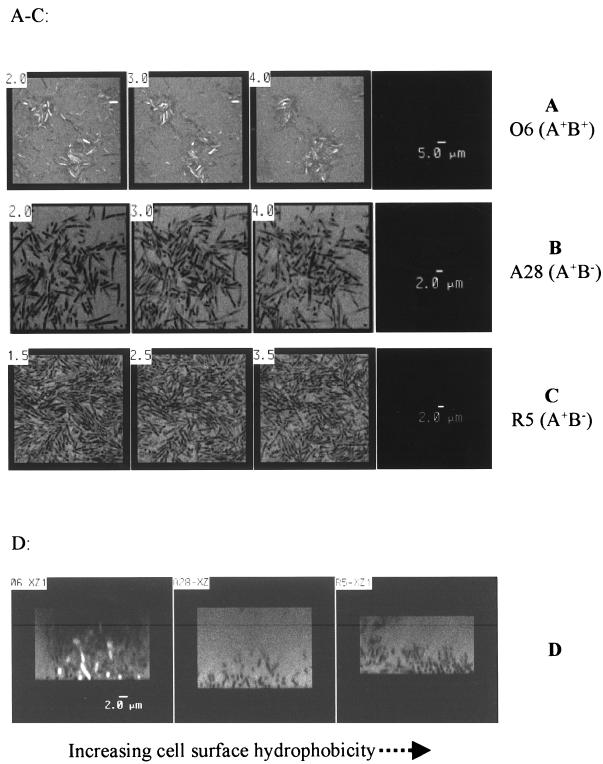
Beveridge et al. (12) recently examined the production of LPS by P. aeruginosa during biofilm formation in vitro and observed that as biofilms mature and thicken, the bacteria undergo changes from an A+B+ LPS phenotype to an A+B− phenotype. This phenotypic change was reversible when cells were removed from the biofilm and allowed to grow in the planktonic mode. Interestingly, a longitudinal study examining P. aeruginosa serial isolates from a number of CF patients with chronic pulmonary infections over a period of several years also showed that the ability of the bacteria to produce B-band LPS was progressively lost over time (135). Thus, there is a need to investigate which form of LPS favors the establishment of biofilms by P. aeruginosa. Such a study was conducted by Flemming et al. (69), who examined a series of P. aeruginosa strains including wild-type serotype O6 (A+B+) and its rough mutant derivatives A28 (A+B−, with a complete core oligosaccharide), R5 (A−B−) (although the A band was detected from whole-cell lysate of this strains by Western immunoblotting, no A-band LPS was produced on the surface of the bacteria due to a deficiency in the LPS core), and Gt700 (A−B−). The LPS structures and other properties of these strains have been characterized previously (44, 159, 160). On the basis of hydrophobic interaction chromatography and a salt aggregation test, the hydrophobic character of these strains was ranked as R5 ≥ A28 > Gt700 > O6. In addition, the anionic characteristics of cell surfaces were determined by electrostatic interaction chromatography and by zeta-potential measurements, ranking the strains as R5 > A28 ≥ Gt700 > O6. Both R5 and A28, which possess more strongly hydrophobic surfaces, demonstrated a significantly greater capacity to form biofilms on stainless steel and glass surfaces. Interestingly, both these adherent mutant strains also have more anionic surfaces based on the zeta-potential measurements (69).
Confocal microscopy was also used to examine the biofilms formed by each of these bacterial strains (134). Both A28 and R5 mutants form luxurious biofilms that spread out evenly on the substratum (glass surface), while the wild-type O6 strain develops a biofilm composed of “microcolonial” clusters of cells that appear to form clumps (Fig. 2). The formation of these clumped biofilms is probably due to specific electrostatic interactions among O-antigen polymers of the LPS molecules. These data imply that the properties of LPS on the surface of P. aeruginosa cells could affect the way in which a biofilm is formed on solid surfaces. Motility due to the presence of flagella generally appears to promote the initial attachment and recolonization in flow systems (50, 120). A recent study by O’Toole and Kolter (174) also demonstrated that flagellum-associated motility is essential for biofilm formation. However, Flemming et al. (69) found that the strongly adherent mutant, R5, lacked flagella and was nonmotile. The presence of flagella did not appear to influence either the hydrophobicity or cell surface anionic character, as seen in the similar rankings of R5 (with no flagella) and A28 (possessing flagella). From these studies, the observations that rough mutants, devoid of A-band and B-band O-antigen polymers, form a more stable biofilm are consistent with the situation for P. aeruginosa, which under certain environmental conditions (i.e., interactions within the CF host) is regulated to produce less high-molecular-weight LPS. At present, the mechanism of LPS regulation is not clearly defined.
FIG. 2.
The architecture of the biofilm formed by P. aeruginosa is influenced by changes in the LPS phenotype. Confocal micrographs of biofilms of fluorescein-stained P. aeruginosa strains grown in glass laminar-flow cells as described by Palmer and Caldwell (175) are shown. After the 9-h growth, the biofilms of the various bacterial strains were approximately 13 to 15 μm thick. (A through C) Optical sections (x-y plane) at 1-μm intervals through biofilms of wild-type strain O6 (A+B+), strain A28 (A+B−, and contains complete core oligosaccharide), and strain R5 (A−B−, contains truncated core oligosaccharide, and A-band LPS is not produced on the surface but accumulated in the bacterial cells), respectively. The characteristics of LPS in the O6 strain and its isogenic mutants have been described by Dasgupta (45). Note the foci of microcolonial growth of wild-type O6 bacterial cells; this mode of growth on a substratum is maintained throughout the biofilm development. The biofilms formed by the mutants, A28 and R5, are evenly spread out, with the most confluent biofilm being formed by the most hydrophobic R5 strain. (D) Optical sections (x-z plane, cross sections) through biofilms of strains O6, A28, and R5 in ascending order of cell surface hydrophobicity (69). The differences in their surface hydrophobicity or hydrophilicity are also revealed by differential fluorescein staining characteristics: cells from both strain A28 and R5 exclude the stain (appeared dark [B and C]) whereas cells of O6 are positively stained (appeared bright [A]). The numbers on the micrographs indicate the distance of the optical sections from the substratum, and the scales of magnification are indicated by the bars. These confocal micrographs were kindly provided by R. Palmer, Center for Environmental Biotechnology, University of Tennessee, Knoxville, Tenn.
In a recent study (134), biofilm formation in the wild-type strain PAO1 (serotype O5; A+B+) and its derived wzm mutant (A−B+; A-band polysaccharide transport mutation [196]) was observed by confocal microscopy. The wild-type PAO1 strain was found to form clumping biofilms similar to those of the O6 strain (data not shown). The A−B+ wzm mutant formed biofilms similar to those of the rough mutants A28 and R5; i.e., the biofilms are evenly spread out on the substratum (reference 134 and data not shown). This nonmicrocolonial biofilm formation by the wzm mutant is unexpected, since the predominant B+ LPS phenotype should provide more hydrophilic charge to this mutant. Instead, the biofilm growth pattern was more widespread and resembled that of B− mutants. Therefore, the physicochemistry with the presence of both A-band and B-band O antigens in wild-type strains appears to promote interactions among polymers of both LPS types. These interactions probably contribute to the cell clumping that leads to microcolony formation in the initial phase of biofilm generation.
GENETICS OF A-BAND O-POLYSACCHARIDE BIOSYNTHESIS
Identification and Organization of A-Band O-Polysaccharide Genes
Several reports described the identification of a neutral polysaccharide composed of d-Rha produced by P. aeruginosa (116, 202, 242). However, it was the study performed by Rivera et al. (191) that distinguished A-band LPS in strain PAO1 as another form of LPS that is separable from the predominant B-band LPS by gel filtration chromatography. They examined fractionated LPS from PAO1 by Western immunoblotting techniques with A-band- and B-band-specific MAbs and by chemical analysis, revealing that this organism coproduces two antigenically and chemically distinct LPS molecules (191, 192). Subsequent immunochemistry studies have demonstrated that 14 of the 20 P. aeruginosa IATS (International Antigenic Typing Scheme) O serotypes produce A-band LPS (42, 135). Although the existence of A band in other gram-negative bacteria has not been studied, reports of LPS bearing the same O-polysaccharide structure as A band have emerged. Organisms expressing these d-rhamnan LPSs include P. syringae pv. morsprunorum C28 (211), P. syringae pv. cerasi 435 (225), B. cepacia (32), and Stenotrophomonas (Xanthomonas) maltophilia (235). Interestingly, the last two bacterial species are among pathogens associated with lung infections in CF patients.
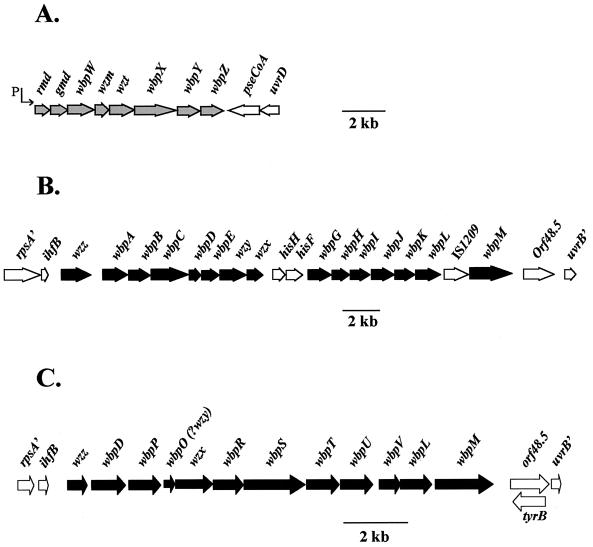
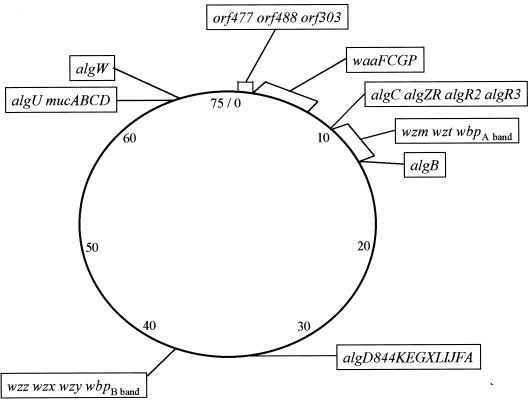
Genes encoding proteins involved in synthesis of a d-rhamnan-containing LPS molecule have been described only for P. aeruginosa. The A-band O-polysaccharide gene cluster was isolated from a P. aeruginosa PAO1 (IATS O5) cosmid library on the basis of its ability to restore A-band LPS synthesis to the A-band deficient mutant rd7513 (142). This cosmid clone, designated pFV3 (containing a 27-kb insert [Fig. 3A]), complemented A-band LPS synthesis in five (O7, O13, O14, O15, and O16) of the six A-band-deficient IATS strains (O7, O12, O13, O14, O15, and O16), indicating that it encoded genes responsible for expression of this cell surface molecule (142). The DNA region responsible for complementation of A-band LPS in rd7513 was localized to a 1.6-kb KpnI fragment, pFV39, through the generation of subclones from pFV3 and the use of transposon mutagenesis (142). Identification of this region led us to focus our sequencing efforts on the 5′ half of the cosmid clone pFV3. Sequence analysis of this 15-kb region revealed eight genes thought to be involved in A-band LPS synthesis (42, 195–197). These LPS genes have been named according to the nomenclature of Reeves et al. (189), whereby wb- represents genes involved in O-antigen biosynthesis and wz- denotes genes involved in O-antigen assembly. Following this system, the P. aeruginosa A-band O-polysaccharide-specific genes have been given the designation wbpA band. The wbpA band genes and the predicted functions of the encoded proteins are listed in Table 1. Lightfoot and Lam (143) used pulsed-field gel electrophoresis and Southern hybridization experiments to map the wbpA band cluster to between 10.5 and 13.3 min on the 75-min map of PAO1 (Fig. 4).
FIG. 3.
Location of A-band and B-band O-antigen genes within their respective cosmid clones. (A) The wbpA band gene cluster isolated from strain PAO1 (serotype O5) is contained on cosmid pFV3. This cluster is composed of eight genes which are probably transcribed from a common ς70-like promoter located upstream of rmd (197). Genes encoding proteins involved in A-band O-polysaccharide synthesis are shown in gray, while non-LPS genes are shown in white. (B) The wbpO5B band gene cluster is found on the cosmid pFV100. (C) wbpO6B band gene cluster. For the B-band clusters, genes encoding proteins involved in B-band O-polysaccharide synthesis for both O5 and O6 are shown in black while non-LPS genes are shown in white.
TABLE 1.
Genes involved in LPS biosynthesis in P. aeruginosa
| Gene function | Gene name
|
Demonstrated or proposed function of gene product and other remarks | Accession no. (reference) | |
|---|---|---|---|---|
| New | Old | |||
| A-band LPS synthesis | rmd | GDP-4-keto-6-deoxy-d-mannose reductase | AF009955 (197) | |
| gmd | gca | GDP-d-mannose dehydratase | U18320 (143) | |
| wbpW | Bifunctional enzyme: phosphomannose isomerase and GDP-mannose pyrophosphorylase | AF009956 (197) | ||
| orf488 | AlgA and WbpW homologue, bifunctional enzyme like WbpW | AF053937 (197) | ||
| wzm | Membrane protein of 2-component ABC transporter | U63722 (196) | ||
| wzt | ATP binding component of ABC transporter | U63723 (196) | ||
| wbpX | Rhamnosyltransferase, transfers α1→2-linked d-Rha to A-band polymer | AF010181 (195) | ||
| wbpY | Rhamnosyltransferase, transfers two α1→3-linked d-Rha to A-band polymer | AF010182 (195) | ||
| wbpZ | Rhamnosyltransferase, transfers a single α1→3-linked d-Rha to the initial A-band acceptor molecule | AF010183 (195) | ||
| wbpL | rfbA | Initial glycosyltransferase, broad specificities: transfers GlcNAc to lipid carrier (for A-band synthesis); transfers Fuc2NAc to lipid carrier (for B-band synthesis) | U17293 (45), U50396 (21, 24, 195) | |
| B-band LPS synthesis | wzz | rol, cld | Modulates O-antigen chain length of B band | U50397 (21, 22) |
| wzy | rfc | O-antigen polymerase for B band | U17294 (51), U50599 (53), U50396 (21), U26685 (38) | |
| wzx | rfbX, wbpF | O-antigen translocase/flippase | U50396 (25) | |
| wbpA | Putative dehydrogenase, required for B-band synthesis | U50396 (21, 26, 27) | ||
| wbpB | Possible oxidoreductase involved in UDP-Fuc2NAc synthesis | U50396 (21) | ||
| wbpC | No role in B-band synthesis, putative O-acetyltransferase | U50396 (20, 21) | ||
| wbpD | Putative UDP-Man2NAc3NAcA 3N-acetyltransferase, required for B-band synthesis | U50396 (20, 21) | ||
| wbpE | Candidate UDP-Man2NAcA aminase | U50396 (21) | ||
| wbpG | Undefined, putative aminotransferase, required for B-band synthesis | U50396 (21) | ||
| wbpH | Putative glycosyltransferase, required for B-band synthesis | U50396 (21, 24) | ||
| wbpI | Putative C2 epimerization of UDP-d-GlcNAc to UDP-d-ManNAc, required for B-band synthesis | U50396 (21) | ||
| wbpJ | Putative glycosytransferase or chaperone, required for B-band synthesis | U50396 (21) | ||
| wbpK | Putative UDP-QuiNAc-4-epimerase for formation of UDP-Fuc2NAc, necessary for B-band synthesis | U50396 (10, 21) | ||
| wbpL | rfbA | Initial transferase for A-band and B-band synthesis (see A-band genes, above) | U17293 (45), U50396 (21, 24, 195) | |
| wbpM | Putative UDP-GlcNAc dehydratase, required for B-band synthesis | U50396 (21, 29) | ||
| Core oligosaccharide synthesis and housekeeping function affecting LPS synthesis | waaC | rfaC | Heptosyltransferase I, inner-core biosynthesis | U70982 (52) |
| waaF | rfaF | Heptosyltransferase II, inner-core biosynthesis | U70983 (52) | |
| algC | Phosphomannose mutase and phosphoglucose mutase activities; essential for alginate synthesis | M60873 (245) | ||
FIG. 4.
The genetic map (75 min) of strain PAO1 illustrates the approximate chromosomal locations of genes involved in synthesis of polysaccharide molecules in P. aeruginosa. Map positions are indicated for genes encoding proteins involved in the synthesis and regulation of alginate (reviewed in reference 78) and in the synthesis of the inner core region (waaFCGP); (52) and A-band and B-band O antigens of the LPS molecule (143). A newly identified locus (orf477 orf488 orf303), which may be involved in synthesis of a unique polysaccharide molecule in P. aeruginosa, is also shown (197). The map is not to scale.
These eight genes are arranged contiguously on pFV3 and are probably transcribed from a promoter found upstream of the first gene, rmd, in this cluster. Three potential promoter sequences have been located upstream of rmd, which correspond to the E. coli ς70 consensus sequence (197). However, two of these putative promoters are nonfunctional on the basis of protein expression experiments with an E. coli background (197). Chromosomal rmd mutants were generated through insertion of a gentamicin resistance (Gmr) cassette within rmd. These mutants no longer express A-band LPS and could be complemented for A-band synthesis by using two subclones that represented the same insert DNA in either orientation (197). This suggested the presence of a functional promoter upstream of rmd; however, the precise transcriptional start site has not yet been determined.
The layout of the wbpA band genes is such that the biosynthetic genes involved in synthesis of the A-band sugar d-Rha (rmd, gmd, and wbpW) are positioned at the beginning of the cluster, followed by those coding for an ATP binding cassette (ABC) transport system (wzm and wzt) and those involved in assembly of the d-rhamnan repeating unit (wbpX, wbpY, and wbpZ) (Fig. 3A). This genetic arrangement is similar to that of the O-antigen gene cluster of E. coli O9a, whereby manC and manB, encoding enzymes required for synthesis of the O9a sugar d-mannose (d-Man), reside at the 5′ end of the cluster and are followed by wzm and wzt, coding for the ABC transport system (106); the 3′ end of the cluster contains genes (wbdA, wbdB, and wbdC) coding for the d-mannosyltransferases that act to assemble the O9a O-repeating unit (105, 106). The wbdEcO9a gene cluster contains one additional gene, wbdD, located between wzt and wbdA, that does not yet have an assigned function for its gene product. The organization of the Yersinia enterocolitica O:3 cluster is somewhat different from that of the wbpA band and wbdEcO9a clusters in that wzm and wzt are positioned among genes encoding the O:3 polysaccharide sugar 6-deoxy-l-altrose (244). The wbb gene arrangement of Klebsiella pneumoniae O1 and O8 and Serratia marcescens O16 is different from the above-mentioned clusters in that wzm and wzt are located at the extreme 5′ ends of their respective gene clusters (35, 102, 218).
The conservation of the A-band O-polysaccharide genes has been determined within the 20 IATS reference strains of P. aeruginosa (42, 195, 196, 197). Southern hybridization analysis has established that all eight genes are present in the 20 IATS strains of P. aeruginosa. However, there are differences in the hybridization profiles of some strains that appear to be serotype specific. The majority of this variation is observed for IATS strains O12, O13, O15, and O16, which do not express A-band LPS (42).
The DNA region on pFV3 flanking the wbpA band cluster has also been examined (195). The 1.5 kb of sequence 5′ to the first A-band gene, rmd, showed no significant similarity to any sequences encoding proteins in the GenBank databases. However, the sequence downstream of the last gene in the cluster, wbpZ, revealed one complete open reading frame (ORF) which coded for a protein of 54 kDa. This ORF is located on the complementary DNA strand and, as shown in Fig. 3A, is transcribed in the opposite direction to that of the A-band gene cluster. A GenBank comparative search of the predicted amino acid sequence encoded by this ORF revealed high homology to coenzyme A transferase proteins from the gram-positive organisms Thermoanaerobacterium thermosaccharolyticum (52.9% identity; GenBank accession no. Z69031) and Clostridium kluyveri (51.3% identity; GenBank accession no. P38946). On the basis of this homology, we have assigned this ORF the designation psecoA. Downstream of psecoA, we identified the 3′ end of a gene whose product displayed homology to the C terminus of a DNA helicase II enzyme (UvrD) from E. coli (66.3% identity; GenBank accession no. P03018). Like psecoA, this P. aeruginosa uvrD homologue is transcribed in the opposite direction to that of the wbpA band cluster. Interestingly, uvrD is also located downstream of the enterobacterial common antigen (ECA) genes wecA to wecG (rfe to rff) at 85 min on the chromosome of E. coli K-12 (166). It is intriguing that another uvr gene homologue was identified during characterization of the B-band LPS gene cluster, wbpB band, in strain PAO1. Burrows et al. (21) identified uvrB to be flanking wbpB band, also at the 3′ end. UvrB is known to function as part of a UvrABC complex that removes thymidine dimers (241). In E. coli, this complex is released from DNA after excision in the presence of UvrD, which is known to unwind DNA duplexes during nucleotide excision and mismatch repair (31, 124, 168). We are curious about whether uvrB and uvrD function as chromosomal markers in other P. aeruginosa serotypes for the B-band and A-band O-antigen biosynthetic gene clusters, respectively.
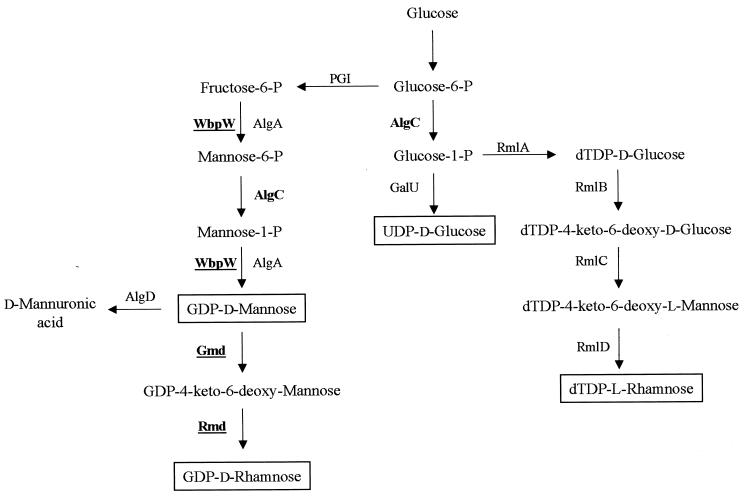
Genes Required for GDP-d-Man and GDP-d-Rha Biosynthesis: Links between A-Band and Alginate
gmd and rmd.
Sugar nucleotide precursors synthesized within the cell cytoplasm are used as donor molecules in the synthesis of cell surface polysaccharides (205). These sugar monomers can be transferred to a lipid carrier molecule, identified as undecaprenol phosphate (Und-P), at the cytoplasmic face of the inner membrane (237). The pathway we have proposed for synthesis of the sugar nucleotide precursor GDP-d-Rha for A-band O-polysaccharide synthesis is illustrated in Fig. 5. This pathway includes GDP-d-Man as an intermediate in GDP-d-Rha synthesis for two reasons. First, early work by Markovitz (156, 157) showed that a soil isolate, Pseudomonas strain GS (ATCC 19241), produced a capsular polysaccharide composed equally of the C-4 epimers d-Rha and d-talomethylose (d-Tal). Crude cell extracts from this bacterium were found to convert GDP-d-Man to GDP-d-Rha and GDP-d-Tal, via a GDP-4-keto-6-deoxy-Man intermediate (157). From this work, Markovitz identified two enzyme fractions from partial column purifications which possessed GDP-d-Man dehydratase and GDP-d-4-keto-6-deoxy-d-Man reductase activity, respectively. Second, Lightfoot and Lam (143) demonstrated that the protein coded for by the A-band gmd gene (formerly called gca) was involved in synthesis of GDP-d-Rha from GDP-d-Man. In that study, supernatants of whole-cell lysates from the A-band mutant rd7513 carrying the gmd gene on a plasmid (pFV39) were incubated with [14C]GDP-d-Man. Data from paper chromatography experiments revealed that rd7513(pFV39) was able to convert GDP-d-Man to a product which migrated at the same rate as the GDP-d-Rha standard. Based on this chromatography data and on the amino acid homology of Gmd to other dehydratase enzymes (42), we propose that Gmd functions as a GDP-d-Man dehydratase converting GDP-d-Man to GDP-4-keto-6-deoxy-d-Man (Fig. 5).
FIG. 5.
Proposed biosynthetic pathways for sugar nucleotide precursors used for synthesis of polysaccharide molecules within P. aeruginosa. GDP-d-Man is a common precursor of both d-mannuronic acid residues of alginate and the d-Rha residues of A-band O polysaccharide. Synthesis of GDP-d-Rha requires WbpW (GMP-PMI), Gmd (GDP-d-Man dehydratase), and Rmd (GDP-4-keto-6-deoxy-d-Man reductase) (42, 197). The d-mannuronic acid residues of alginate are formed through the activities of AlgA (GMP-PMI) (206), AlgC (PMM) (245), and AlgD (GDP-d-Man dehydrogenase) (54). The PMM activity of AlgC is required for GDP-d-Rha synthesis (197, 240), while the phophoglucomutase activity of AlgC is also needed for UDP-d-Glc and TDP-l-Rha for core biosynthesis (39). PGI, phosphoglucose isomerase; GalU, glucose-1-phosphate uridyltransferase; RmlA, glucose-1-phosphate thymidyltransferase; RmlB, dTDP-glucose-4,6-dehydratase; RmlC, dTDP-4-keto-l-rhamnose-3,5-epimerase; RmlD, dTDP-l-rhamnose synthetase.
A second gene in the A-band cluster, rmd, probably encodes the reductase enzyme necessary for converting GDP-4-keto-6-deoxy-d-Man to GDP-d-Rha (Fig. 5). Recently, we have shown that rmd knockout mutants (generated by insertion of a nonpolar gentamicin resistant cassette followed by gene replacement into the wild-type organism) are deficient in A-band LPS synthesis, confirming a role for Rmd in this biosynthetic pathway (197). Protein homology of Rmd reveals similarity to biosynthetic enzymes that modify sugars at the C-4 and C-6 positions. However, no functional homologue of Rmd could be identified within the GenBank databases, probably since this is the first nucleotide sequence report of a reductase required for GDP-d-Rha synthesis. Both Rmd and Gmd contain NAD binding domains (197). The presence of these sites is consistent with the requirement of NAD for both the redox reaction of the dehydratase Gmd and the subsequent reduction of GDP-4-keto-6-deoxy-d-Man by the reductase Rmd (157). A later study by Winkler and Markovitz (234) described the purification from the Pseudomonas strain GS mentioned above of a GDP-4-keto-6-deoxy-d-Man reductase that lacked stereoselectivity. This reductase was found to perform the irreversible reduction of GDP-4-keto-6-deoxy-d-Man to both GDP-d-Rha and GDP-d-Tal, which differ only in the orientation of the C-4 hydroxyl group. Future work on Rmd includes biochemical characterization of the reaction products and determination of the specificity of the reduction reaction. Rmd probably possesses stereoselectivity, producing only GDP-d-Rha, since GDP-d-Tal has not been reported to be a constituent sugar in any of the known polysaccharide molecules of P. aeruginosa. Alternatively, P. aeruginosa may not possess glycosyltransferases capable of recognizing GDP-d-Tal as a substrate.
algC and wbpW/algA/orf477.
Synthesis of the d-mannuronic and l-guluronic acid residues of alginate also proceeds via a GDP-d-Man intermediate (54, 206, 245) (Fig. 5). For this reason, it has previously been suggested that common enzymes may be involved in the formation of the GDP-d-Man precursor for synthesis of both A band and alginate (140). In fact, an alginate enzyme, AlgC, is required for the synthesis of a number of cell surface molecules within P. aeruginosa. AlgC possesses both phosphomannose mutase (PMM) and phosphoglucose mutase (PGM) activities, which are essential for alginate and core oligosaccharide synthesis (39, 245). PMM is required for the conversion of mannose-6-phosphate to mannose-1-phosphate in the synthesis of GDP-d-Man (245) (Fig. 5), while PGM activity is required for conversion of glucose-6-phosphate to glucose-1-phosphate (39). This PGM activity was identified through complementation of the core-deficient mutant AK1012 (39). The cellular pool of glucose-1-phosphate is required for formation of the precursors UDP-d-glucose (d-Glc) and dTDP-l-rhamnose (l-Rha) (Fig. 5), which are presumably the donor sugar nucleotides for the d-Glc and l-Rha residues of the outer core region in P. aeruginosa (Fig. 1C). Synthesis of the O antigen itself may also be affected by an algC mutation in strains that contained d-Glc and/or l-Rha moieties in the O-repeat unit, such as serotype O6 (Fig. 1B).
Interestingly, no gene encoding an enzyme with PMM activity was identified within the A-band gene cluster. This suggested that AlgC might also contribute to the formation of the GDP-d-Man precursor that is converted to GDP-d-Rha for A-band O polysaccharide. To this end, studies involving LPS and colony immunoblots of lysed cells with the A-band-specific MAb N1F10 have shown that the algC mutant AK1012 lacks A band (197, 240). Even though AK1012 expresses a truncated core region that prevents attachment of the A-band O polysaccharide to core-lipid A, detection of intracellular A-band LPS would still be possible if AlgC were not involved in its synthesis. Studies in our laboratory have shown that A-band LPS can be synthesized and maintained on a carrier lipid molecule within the cell cytoplasm when mutations are introduced into wzm and wzt, encoding ABC transport system components (196) (see below). However, the lack of A-band LPS within AK1012 implies that AlgC is also required for synthesis of GDP-d-Rha.
A gene encoding an enzyme involved in GDP-d-Man biosynthesis was localized within wbpA band. The product of this gene, designated WbpW, is homologous to enzymes functioning as GDP-d-Man pyrophosphorylases (GMP) and phosphomannose isomerases (PMI), including the alginate bifunctional enzyme AlgA, which has both GMP and PMI activities. Through complementation studies with defined E. coli capsule mutants, WbpW was found to possess the same bifunctional enzymatic activity as AlgA, catalyzing two noncontiguous steps in the GDP-d-Man synthesis pathway (197) (Fig. 5). P. aeruginosa wbpW knockout mutants generated by gene replacement in our laboratory continued to produce A-band LPS; however, the amount was significantly reduced from that produced by the parent strain PAO1 (197). Since AlgA has the same enzymatic activity as WbpW, we believed that the level of GDP-d-Man precursor within these wbpW mutants was simply decreased. Elimination of the cellular pool of GDP-d-Man was predicted to occur following the generation of wbpW algA double mutants. This was achieved by using the nonmucoid P. aeruginosa strain FRD1128. This FRD1128 strain was derived from the mucoid CF isolate FRD1 and contains a Tn501 insertion in algX which is polar on algA (34, 169). We used this algA mutant strain as the background for introduction of a chromosomal mutation into wbpW (197). However, these double mutants were still able to produce reduced levels of A-band LPS (197). Provision of algA in trans was sufficient to restore high levels of A-band LPS production to these double mutants, indicating that AlgA can compensate for the absence of WbpW (197). The ability of these wbpW algA double mutants to produce some A-band LPS prompted us to search for other potential wbpW algA homologues within the P. aeruginosa genome of strain PAO1. Using the wbpW nucleotide sequence as a probe, we were able to identify a third homologue, designated orf488 (197). ORF488, along with WbpW and AlgA, contains the highly conserved sugar-1-phosphate binding motif (FVEKP), which is known to be associated with pyrophosphorylase enzymes (163, 197). The existence of ORF488, which probably has GMP and PMI activities, is probably responsible for the A-band LPS synthesis in the wbpW algA double mutants.
Sequence analysis of the region surrounding orf488 revealed two additional ORFs arranged contiguously as orf477, orf488, and orf303 (197). Southern hybridization experiments with a PAO1 PCR product containing these three ORFs demonstrated that all 20 IATS reference strains contained this genetic region (197). Chromosomal mapping places the orf477, orf488, and orf303 region at 0.3 to 0.9 min on the 75-min map of PAO1, as seen in Fig. 4 (197). Based on the amino acid homology of ORF477 to various glycosyltransferases and on its hydrophobicity profile, this enzyme is predicted be an initiating enzyme that adds glucose-1-phosphate to the carrier lipid Und-P. The ORF303 protein is homologous to l-rhamnosyltransferases that are involved in the formation of various cell surface molecules. ORF303 may therefore play a role in catalyzing glycosidic linkages during polysaccharide assembly. Thus, it seems that the wbpW algA homologue orf488 is situated within a gene cluster that may be involved in the formation of another polysaccharide molecule. A study by Kocharova et al. (117) reported the isolation and chemical analysis of a unique polysaccharide molecule, composed of a pentasaccharide repeating unit containing d-Glc, d-Man, and l-Rha, from culture supernatants of P. aeruginosa Fisher immunotype 4 (IATS O1). We postulate that this newly identified gene cluster may be involved in synthesis of this polysaccharide molecule, since the predicted enzyme functions for ORF477, ORF488, and ORF303 correspond to the constituent sugars found in the pentasaccharide repeat. Future work in this area will focus on determining the specific activities of these enzymes, as well as characterization of the DNA regions flanking these three ORFs by using the P. aeruginosa genomic database to identify additional genes that may be involved in synthesis of this molecule.
Of interest is the observation that the pentasaccharide repeating polysaccharide molecule identified by Kocharova et al. (117) was recovered only from cell supernatants of cultures grown for more than 72 h. As mentioned above, our wbpW algA double mutants still synthesize A-band LPS. However, using Western immunoblotting techniques, we could detect only A band from cultures grown for more than 24 h. It is possible that this new gene cluster undergoes upregulation late in the growth cycle, allowing ORF488 to play a compensatory role in the wbpW algA mutants and permitting A-band synthesis to occur. Generation of a mutant lacking orf488, wbpW, and algA will help to prove this hypothesis. Certainly, the existence of these three homologues within P. aeruginosa stresses the need for the GDP-d-Man precursor and leads to the idea of cellular adaptability, with each genetic locus perhaps being regulated under different conditions. For the most part, A-band LPS is known to be expressed constitutively whereas alginate synthesis is highly regulated during growth on solid surfaces and under various nutritional conditions (11, 56, 90). The orf477 orf488 orf303 genetic region is certainly an area deserving of further study to determine the precise polysaccharide molecule that is synthesized, the environmental conditions under which it is produced, and the role that it may play in P. aeruginosa virulence.
Genes Encoding Glycosyltransferases for A-Band Polymer Assembly: WbpL Is Essential for Initiation of Both A-Band and B-Band Synthesis
wbpL.
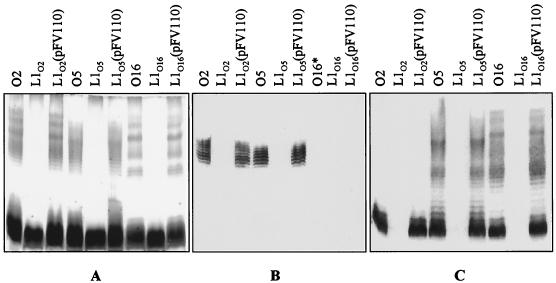
The mechanism of polysaccharide assembly for LPS and capsule synthesis begins through the activities of a glycosyltransferase and Und-P. These enzymes, termed initiating enzymes, contain numerous membrane-associated domains to allow for interactions with the hydrophobic Und-P acceptor. The best-characterized initiating enzymes are WecAEc (Rfe) and WbaPSe (Salmonella enterica), which transfer N-acetylglucosamine-1-phosphate (GlcNAc-1-P) (1, 167) or N-acetylgalactosamine-1-P (GalNAc-1-P) (4) and galactose-1-phosphate (Gal-1-P) (226), respectively, onto Und-P. In our laboratory, an enzyme designated WbpL, which exhibits high homology to glycosyltransferases responsible for initiating polysaccharide synthesis, has been identified within P. aeruginosa (21). The wbpL gene is located within the wbpB band cluster; however, wbpL::Gmr mutations affect both A-band and B-band LPS synthesis (195). No such initiating glycosyltransferase homologue was found with the wbpA band cluster. Figure 6 shows that wbpL mutations abrogate both A-band and B-band synthesis in the cross-reactive serogroups O2, O5, and O16. Based on results from spectroscopic analysis, Fuc2NAc is the first sugar of the serotype O5 repeating unit attached to the core region (198). Since all three serotypes contain a Fuc2NAc moiety in their O-repeat unit (Fig. 1B), WbpL is believed to initiate B-band O-antigen synthesis through the addition of Fuc2NAc-1-P to Und-P (21). Heteropolymeric initiation and synthesis differ from the homopolymeric counterparts, since the former involve the initiating sugar within each O-repeating unit. In the case of B-band O-antigen synthesis, for example, WbpL would be responsible for the addition of Fuc2NAc in the formation of each trisaccharide repeat. In the homopolymers, the initiating “priming sugar” does not form part of the O repeat, since subsequent enzymes act to transfer additional sugar residues processively onto this initial sugar moiety. These differences reflect the cellular location of assembly components for heteropolymeric and homopolymeric O antigens. This is briefly discussed below but is reviewed in detail by Burrows et al. (28).
FIG. 6.
LPS analysis of WbpL mutants in the related serotypes O2, O5, and O16 (195). (A) Silver-stained SDS-PAGE gel; (B and C) Western immunoblots reacted with the A-band-specific MAb N1F10 and the B-band cross-reactive MAb 18-19, respectively. All of the wbpL mutants are deficient in both A-band and B-band LPS, with the parental phenotype being restored through complementation with a subclone containing wbpLO5 (pFV110). Serotype O2 reacts only with MAb 18-19 (C) as a core-plus-one-O-repeat unit, while serotypes O5 and O16 are able to react with the full B-band ladder. The substitution of a guluronic acid residue in the O2 O antigen (Fig. 1B) may be responsible for the formation of an epitope that is not recognized by 18-19 above core-plus-one. ∗, serotype O16 does not express A-band LPS. Reproduced from reference 195 with permission.
As mentioned above, WecAEc is capable of transferring both GlcNAc-1-P and GalNAc-1-P to Und-P. The former serves as the initiating sugar for ECA (167) and heteropolymeric (1) and homopolymeric (106, 190) O-antigen synthesis, while the latter provides initiation for heteropolymeric capsule synthesis (4). WecAEc can therefore initiate the synthesis of structurally distinct molecules. WbpL is similar to WecAEc in being involved in synthesis of the chemically distinct A-band and B-band O antigens. Cross-complementation experiments with WecAEc have shown that this enzyme is able to initiate A-band but not B-band synthesis (195). These data indicate that the initiating sugar for A-band synthesis is probably either GlcNAc-1-P or GalNAc-1-P and that WecAEc is unable to transfer Fuc2NAc-1-P to Und-P for B-band synthesis. Structural analysis is necessary to determine if GlcNAc or GalNAc is in fact the initiating sugar for A band. Regardless of the precise nature of this A-band moiety, WbpL definitively initiates these two LPS molecules with different sugar residues. This phenomenon raises the possibility that WbpL plays a role in the synthesis of other polysaccharide cell surface molecules produced by P. aeruginosa. The enzyme responsible for initiating the synthesis of the mannuronic acid homopolymeric alginate has not yet been identified. It would be of interest to determine whether wbpL::Gmr mutants are capable of alginate production and/or if WbpL is universally required for exopolysaccharide synthesis in P. aeruginosa (194).
wbpX, wbpY, and wbpZ.
Following the initiation event catalyzed by WbpL, homopolymeric biosynthesis proceeds by way of particular glycosyltransferases recognizing acceptor and donor molecules while catalyzing specific glycosidic linkages. The homopolymeric assembly process occurs on Und-P by using the sugar primer GlcNAc provided by the initiating glycosyltransferase at the cytoplasmic face of the inner membrane (230). The process of elongation occurs at the nonreducing terminus of the nascent polysaccharide (228). After assembly or concomitant with this process, the O polysaccharide is transported to the periplasm for attachment to core-lipid A. For the majority of homopolymers characterized thus far, this translocation process occurs via an ABC transport system (16, 106, 155, 218, 244).
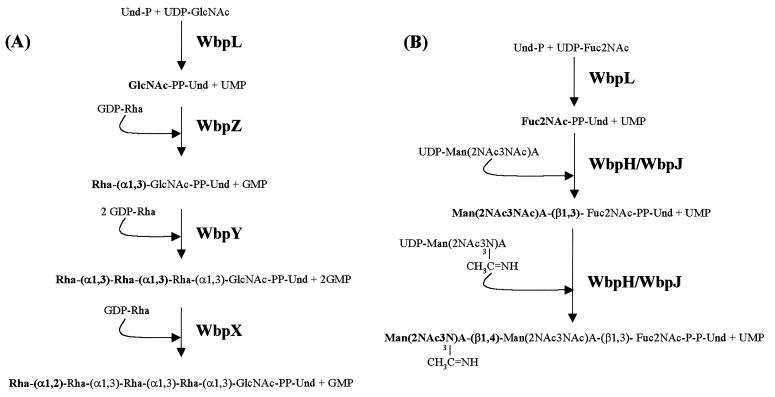
Three genes, wbpX, wbpY, and wbpZ, which encode proteins homologous to glycosyltransferases that assemble various cell surface molecules, were identified at the 3′ end of the wbpA band cluster (Fig. 3A). From these similarity analyses, the three genes were predicted to encode d-rhamnosyltransferases for postinitiation assembly of the A-band O polysaccharide. wbpX::Gmr, wbpY::Gmr, and wbpZ::Gmr PAO1 mutants lacked A-band LPS while maintaining B-band LPS on Western immunoblots (195). These three transferases are therefore dedicated to A-band synthesis, unlike WbpL, which is committed to both LPS synthesis pathways. Interestingly, in trans provision of the wild-type gene in multicopy to each respective mutant restored A-band synthesis, but the length of this A-band polymer differed from that in the parental PAO1 strain by two or three O-repeating units (195). Appropriate intracellular levels of these glycosyltransferases may in fact be necessary to facilitate the expression of a specific polymer phenotype.
These A-band glycosyltransferases exhibited the greatest protein homology to the E. coli O9a mannosyltransferases WbdA, WbdB, and WbdC (105, 106). These E. coli O9a glycosyltransferases serve to assemble a tetrasaccharide d-Man repeat (α1→2, α1→2, α1→3, α1→3) (104–106) composed of linkages similar to A band (α1→2, α1→3, α1→3). Since d-Rha is a 6-deoxy derivative of d-Man, WbpX, WbpY, and WbpZ from P. aeruginosa probably bear a structural resemblance to the E. coli O9a glycosyltransferases that recognize very similar sugar substrates. In vitro transferase studies have shown that WbdC adds the first d-Man residue onto GlcNAc-PP-Und by catalyzing an α1→3 linkage (106). Since WbpZ is 62.1% identical to WbdC, we predicted that WbpZ transfers the first d-Rha residue onto GlcNAc-PP-Und (Fig. 7A). Additional transferase studies with WbdA and WbdB have proven their role in d-mannan assembly; however, the precise linkage assignments for each have not been fully characterized. Preliminary transferase studies by Kido et al. (106) had led to a transferase assignment for WbdA (adds three d-Man α1→2) and WbdB (adds two d-Man α1→3) on the basis of assembling a d-mannan pentasaccharide designated E. coli O9 (α1→2, α1→2, α1→2, α1→3, α1→3). However, recently Kido et al. (104, 105) demonstrated that the E. coli strain used in that study actually contained a tetrasaccharide O repeat (mentioned above), which has been reclassified as O9a. Although the linkages of the pentasaccharide and tetrasaccharide polymers are the same, the absence of one sugar alters the transferase scheme proposed earlier by Kido et al. (106). Further assignment of transferase specificities awaits experimental data from in vitro assays with defined substrates. Isolation and characterization of the true O9 d-mannan transferases, which assemble the pentasaccharide O-repeat unit may provide insight into the assembly mechanisms of both O9 and O9a polymers.
FIG. 7.
Proposed glycosyltransferase mechanism for assembly of A-band (A) and serotype O5 B-band (B) O antigens (195). Note that WbpL participates in initiating both synthesis pathways with, probably, a GlcNAc residue for A band and a Fuc2NAc residue for B band. The specificities of these enzymes remain to proven biochemically. Modified from reference 195.
However, some predictions can be made about the order and specificities of the transferases which mediate assembly reactions of the A-band O polysaccharide based on amino acid homology and the d-rhamnan structure (Fig. 7A) (195). Following addition of the first d-Rha by WbpZ, WbpY may recognize this terminal d-Rha and add two d-Rha moieties α1→3. The terminal α1→3 d-Rha would then serve as an acceptor for WbpX, which adds one d-Rha residue α1→2. Provided that WbpX and WbpY can function in turn, they could continue to extend the A-band O polysaccharide. This assignment requires WbpY to recognize dual acceptors, the α1→2 d-Rha provided by WbpX and the first α1→3 d-Rha moiety added by WbpY. Alternatively, WbpY may possess more than one catalytic domain, which would allow the transfer of two α1→3 d-Rha residues at once and eliminate the need for dual recognition. This transferase scheme is also consistent with the genetic organization of wbpX, wbpY, and wbpZ in the wbpA band cluster, with the genes arranged in the opposite order to which the encoded enzymes act. This is also true for the arrangement of the E. coli O9a transferases encoded by wbdA, wbdB, and wbdC (106). Thus, the transferase which functions first (WbpZ and WbdC) is found last in each of the respective gene clusters. Precise assignment of WbpX, WbpY, and WbpZ in the assembly process of A band requires the development of transferase assays which use chemically defined substrates as acceptor and donor molecules. This area of research will be challenging, since no complete glycosyltransferase mechanism has been unequivocally demonstrated for any homopolymeric LPS O polysaccharides.
A-Band O-Polysaccharide Transport across the Inner Membrane: Evidence for a Wzy (Rfc)-Independent Pathway
wzm and wzt.
Currently, there are two known mechanisms of homopolymeric O-antigen assembly and export across the inner membrane. The more common of the two systems are the ABC transport systems reported for organisms such as E. coli O9a (106), K. pneumoniae O1 and O8 (35, 102), and S. marcescens O16 (218). These systems are composed of two proteins, a hydrophilic ATP-binding component and an integral membrane component, which associate to form paired homodimers (66). The second mechanism has been reported only for S. enterica serovar Borreze O:54 O-polysaccharide synthesis (100). This unique system contains a glycosyltransferase (WbbF) that is believed to couple polymerization and export functions by forming a pore-like structure at its C terminus (100). In either case, these export systems facilitate the translocation of O polysaccharide to the periplasm, allowing its ligation to separately synthesized core-lipid A molecules. Both of these systems proceed independently of an O-antigen polymerase, known as Wzy (discussed below), and are therefore commonly referred to as Wzy-independent pathways (230).
Analysis of the wbpA band cluster revealed two genes, wzm and wzt, which encoded proteins homologous to O-antigen and capsular transport proteins found in other bacterial systems (196). Wzm and Wzt possess specific traits that identify them as components of ABC transport systems. The hydrophobic Wzm protein is an integral membrane protein containing six potential membrane-spanning domains, while the hydrophilic Wzt protein contains a highly conserved ATP binding motif (196). The integral membrane proteins of these systems are believed to form a channel or pore through which the O polysaccharide travels, and hydrolysis of ATP by the hydrophilic component provides the energy for this export process (88, 103). The P. aeruginosa Wzm and Wzt proteins had the highest protein homology to ABC transport components of E. coli O9a (196). As discussed in the glycosyltransferase section (above), O9a O polysaccharide is composed of d-mannan O-repeat units. Since the O9a d-mannan polysaccharide and the A-band d-rhamnan polymer have very similar backbone structures (with d-Rha derived from d-Man), each bacterium would possess comparable export machinery. Similarly, evidence from cross-complementation experiments showed that ABC transporters from K. pneumoniae O1 and O8 (102), and K. pneumoniae O1 and S. marcescens O16 (218) are functionally interchangeable for export of the structurally identical d-galactan I homopolymer.
Chromosomal wzm::Gmr and wzt::Gmr PAO1 mutants maintained the ability to synthesize both A-band and B-band LPS as detected by Western immunoblotting with LPS-specific MAbs (196). The A-band polymer isolated from these mutants, however, was different from that of the PAO1 parent strain. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) showed that A band recovered from both transport mutants had a faster mobility than did the band from PAO1. We predicted that these mutations, which render the transport system inactive, prevent polymer export and subsequent attachment to core-lipid A. The absence of the core-lipid A residues decreases the molecular mass of the LPS molecule, which in turn increases the mobility in SDS-PAGE. Thus, in these mutants, A-band polymer is synthesized but accumulates cytoplasmically on a carrier lipid molecule. These faster-migrating polymers are probably linked to Und-P, as demonstrated by the cleavage of the pyrophosphate bridge during hot-aqueous phenol extraction (256). The phenol lability of this accumulating polymer supported a lack of core-lipid A attachment and indicated that Wzm and Wzt were indeed transport components. The absence of A-band LPS on the cell surface of the wzm and wzt null mutants was corroborated by using immunoelectron microscopy (196). A small amount of A band could be seen within the cell cytoplasm, which supports the notion of intracellular accumulation of the polymer in the absence of a functional transport system.
The presence of these transport components demonstrates that A-band O-polysaccharide export follows the Wzy-independent synthesis pathway, like that of most homopolymers. Mechanistically, this translocation process is believed to be directed via conformational changes in the ATP binding component. Studies on the export system of the E. coli homopolymer K1 capsule have allowed the development of a model whereby the ATP binding protein KpsT associates with the K1 capsule and undergoes a change in conformation upon ATP binding (14, 15). The resulting polymer-KpsT complex is then thought to pass through the membrane at sites defined by KpsM, the integral membrane protein. Upon ATP hydrolysis, KpsT may then return to its previous conformation, which allows deinsertion from the membrane and polymer release to the periplasm. It is not known whether the polymer travels through the membrane attached to Und-P or is transferred to a second, unknown carrier molecule before final ligation to core-lipid A.
Homopolymeric polysaccharide synthesis proceeds independently of two known LPS proteins. Wzy is an O-antigen polymerase that serves to polymerize individual O-antigen units into long-chain O antigen, while Wzz modulates the length of these O-antigen chains. These two proteins are discussed in more detail below, since they both contribute to B-band LPS synthesis. For heteropolymer synthesis, wzy mutations abrogate O-antigen polymerization and result in a phenotype known as SR (core plus one O-repeat unit). No SR mutants have ever been isolated from homopolymeric systems, indicating that these synthesis pathways occur without the activity of Wzy. Support for the Wzz independence for homopolymers stems from studies in enteric systems. The K. pneumoniae O1 d-galactan I homopolymer, for example, has the same chain length distribution in an E. coli K-12 background in the presence and absence of WzzK-12 (230). Similarly, a study by Dodgson et al. (58) revealed the presence of a wzz homologue within E. coli O8 and O9 strains possessing a group IB capsular K antigen. It was determined that this Wzz protein was involved only in regulating the chain length of the heteropolysaccharide capsule and not the homopolymeric polysaccharides O8 and O9 (58, 71). Recently, both Wzy and Wzz have been identified in P. aeruginosa and have been found to have no effect on A-band LPS synthesis, since wzy and wzz null mutants express long-chain A-band polymer with a modality identical to that of the parental strain (22, 51).
An alternative mechanism for controlling the chain length of homopolymers may exist, since they do exhibit strain-specific modal distributions. The incorporation of a 3-O-methyl sugar residue at the terminal position (nonreducing terminus) of the polysaccharide chain may be important in controlling the chain length of these polymers. A 3-O-methyl sugar has been identified as the terminal residue of the homopolymeric O antigens from K. pneumoniae O5 (145), E. coli O8 (94), and Campylobacter fetus serotype B (204). Chemical analyses have also detected 3-O-methyl rhamnose within A-band polysaccharide; however, more definitive studies are still required to determine if it occupies the terminal position (7). The incorporation of these 3-O-methyl sugars would prevent further chain elongation, thereby serving to regulate the length of the homopolymeric O polysaccharide. The mechanism by which this addition occurs and the way in which these 3-O-methyl sugars are synthesized are areas of focus for future research.
GENETICS OF B-BAND O-ANTIGEN BIOSYNTHESIS
Identification and Analysis of B-Band O-Antigen Genes from Serotypes O5, O6, and O11
Isolation of B-band O-antigen gene clusters.
While the chemistry and serology of P. aeruginosa B-band LPS have been studied extensively (reviewed in references 109, 111, and 212), investigation of the genetics underlying its biosynthesis has begun only in the last decade. In our laboratory, we use the IATS (149, 212) designations for 20 reference serotype strains. MAb which differentiate between the first 17 of the 20 IATS serotypes have been generated (132, 133), while the remaining 3 IATS serotypes can be distinguished by using adsorbed polyclonal sera. In a recent review (212), P. aeruginosa B-band LPS was divided into 31 chemotypes that include the 20 IATS serotypes and subtypes (i.e., strains that react with an MAb for a particular IATS serotype but have additional epitopes).
Goldberg et al. (75) and Lightfoot and Lam (143) described the isolation of cosmid clones which contain the gene clusters encoding the B-band LPS of serotypes O11 and O5 respectively. The B-band LPS gene cluster of P. aeruginosa PA103 (serotype O11) was isolated through identification of rough, serum-sensitive P. aeruginosa isolates from CF patients that were complemented to serum resistance by using a cosmid library of PA103 chromosomal DNA (75). In addition, colony immunoblot analysis (with anti-serotype O11 serum) of E. coli HB101 containing the cosmid library was used to identify a single positive clone (75). Analysis of the complementing DNA from both experiments showed that it encoded the synthesis of O antigen that reacted with anti-O11 (Fisher immunotype 2) serum (75). Two of the recombinants reacted with both anti-O11 and anti-O1 (Fisher immunotype 4) serum, probably through complementation of synthesis of the native O antigen (75). These results confirmed the role of B-band LPS in serum resistance.
The serotype O5 B-band gene cluster, wbpB band, was isolated from a cosmid library of P. aeruginosa PAO1 (IATS O5) DNA based on its ability to complement O-antigen synthesis in the B-band-minus mutant, ge6 (143). By using pulsed-field gel electrophoresis and Southern hybridization, wbpB band was mapped to 37 min on the 75-min chromosomal map of PAO1 (Fig. 4). The complementing cosmid, pFV100, is sufficient to encode the biosynthesis of serotype O5 LPS in E. coli HB101, based on reactivity of the recombinant strain with a B-band-specific MAb, MF15-4 (45). Interestingly, attempts to express P. aeruginosa B-band LPS from E. coli K-12 laboratory strains such as DH5α have not been successful (141), suggesting that the core oligosaccharide of E. coli K-12 may not be an appropriate acceptor molecule for P. aeruginosa O antigens. The structure of the core oligosaccharide of HB101, a hybrid of E. coli K-12 and E. coli B, is not known, but it may be more amenable to P. aeruginosa O-antigen attachment than is that of K-12. Alternatively, E. coli HB101, but not E. coli K-12, may have unidentified accessory genes required for expression of P. aeruginosa O antigen in the heterologous host. Attempts to introduce pFV100 into wild-type E. coli B have not been successful, probably due to destruction of the DNA by native restriction-modification systems (23). The identification of appropriate E. coli background strains is vital for further work involving the expression of P. aeruginosa proteins involved in O-antigen synthesis.
In 1996, sequencing of the entire O5 cluster was completed by our laboratory (28). More recently, the cloning and sequencing of the B-band O-antigen gene cluster from the clinically relevant IATS serotype O6 was completed by Bélanger et al. (10). The DNA sequence of the O11 cluster has also been determined (48), but only a limited amount of sequence is publicly available (GenBank accession no. U44089). Sequence analysis of the 24.4-kb insert of pFV100, encoding the synthesis of serotype O5 B-band LPS, revealed several genes with homology to those involved in LPS synthesis (21, 45, 51). DNA sequence comparison of the 5′ end of the pFV100 insert with the GenBank database revealed homology in that region to the 3′ halves of wzz genes encoding O-antigen chain length regulators (formerly rol or cld) from a number of gram-negative bacteria (22). The partial P. aeruginosa wzz gene was used as a probe to identify an overlapping cosmid (pFV400) containing the entire O5 wzz gene and adjacent upstream sequences (22). DNA sequence analysis showed no additional sequences upstream of wzz that were involved in O-antigen synthesis, confirming the observation that pFV100 could encode the synthesis of genuine P. aeruginosa O5 O antigen in E. coli HB101. As with the A-band genes, the genes of the B-band O-antigen cluster were named in accordance with the nomenclature proposed by Reeves et al. (189). The wbpB band genes and the predicted function of the encoded proteins are listed in Table 1.
Since the 3′ end of the serotype O5 gene cluster contained sequences conserved across all 20 IATS serotypes (21), a probe from this region was used to identify a 6.5-kb fragment of the serotype O6 B-band LPS gene cluster (10). The remainder of the O6 gene cluster was isolated by long-range PCR, with a downstream primer based on the sequence of the 6.5-kb fragment and an upstream primer based on the highly conserved ihfB (himD) region upstream of the O5 cluster (10).
Organization of the B-band O-antigen gene clusters.
Based on homology and functional studies, the genes that encode proteins involved in assembly of the O antigen in serotype O5 were designated wzx (rfbX, wbpF), wzy (rfc), and wzz (rol, cld) and those involved in biosynthesis and assembly of the nucleotide sugars of the O unit were designated wbpA through wbpN (Fig. 3B). In addition to these ORFs, there are two his genes, hisH and hisF, located between wzx and wbpG and an insertion sequence, IS1209, located between wbpL and wbpM. In the LPS cluster of serotype O6, homologues of wzz, wzx, wbpK, wbpL, wbpM, and wbpN were identified. Mutational analysis has recently shown that WbpN is not involved in O-antigen synthesis, and it has thus been renamed ORF48.5 (10). Other O-antigen-related genes in O6 have been designated wbpO through wbpV (Fig. 3C). The remaining wbp designations, wbpW through wbpZ, have been used to name genes involved in A-band O-antigen synthesis. No complete wzy gene (encoding the O polymerase) could be detected within the O6 cluster, despite the ability of the organism to make long O chains. However, wbpQ, which is approximately 200 bp, has homology to wzy and probably represents a truncated version of that gene (10). Similar truncations of wzy have been detected in the O-antigen clusters of S. enterica groups B and D1 (41).
Southern blot analysis of the 20 IATS reference strains in our laboratory with probes spanning various regions of the O5 B-band LPS gene cluster showed that only wbpM is highly conserved across all 20 P. aeruginosa serotypes. The remainder of the genes in the cluster, up to and including wzz, hybridized only to serotype O5 and the chemically and structurally related serotypes O2, O16, O18, and O20 (21). Limited DNA sequence comparison of various regions of the O-antigen gene clusters (23, 53) indicated that these related serotypes have essentially identical O-antigen clusters. The region 5′ to wzz hybridized to all 20 serotypes of P. aeruginosa, and sequence analysis of this region identified two genes that are highly conserved among bacteria: ihfB, encoding the beta subunit of integration host factor, and rpsA, encoding the S1 ribosomal protein (22). Together, these results show that the related serotypes of the O5 serogroup have very similar LPS gene clusters and that these clusters can be delineated by the conserved genes ihfB at the 5′ end, and wbpM at the 3′ end. Analysis of the serotype O6 B-band O-antigen gene cluster confirmed that it too is flanked by ihfB and wbpM, allowing the 5′ region to be cloned by PCR with a primer based on the O5 ihfB sequence (10). The majority of the remaining genes between these markers are O6 serospecific (10). In serotype O11, orf74.5 (U44089) is identical to wbpM while orf34.5 and orf37.2, which are homologues of wbpL and wbpK, respectively, are less highly conserved. Thus, it appears that the region of the P. aeruginosa chromosome between ihfB and wbpM may be a target site for lateral transfer of O-antigen genes, much like the region between the his and gnd genes of E. coli and S. enterica (13, 173). The presence of wbpM at the 3′ end of each of these three disparate clusters suggests that it is a native and ancestral gene in P. aeruginosa. The wbp genes of serotypes O5 and O6, with the exception of wbpM, have G+C contents at least 10% below the chromosomal average of 67% (10, 21). The higher G+C content of wbpM (∼62%) provides additional evidence that it may represent a more ancestral P. aeruginosa gene.
The identification of non-LPS-related sequences (hisHF and IS1209) in the wbp cluster of serotype O5 is fascinating from an evolutionary standpoint. As mentioned above, enteric O-antigen clusters tend to be linked to the his operon, and we were intrigued to find two contiguous his genes, hisH and hisF, roughly in the centre of the wbp cluster, between wzx and wbpG (Fig. 3B). These genes, which in most organisms are separated by the hisA gene, are unique to those serotypes (O2, O5, O16, O18, and O20) within the O5 serogroup (10). HisH and HisF have the highest protein homology to OrfH3 and OrfH4 of Leptospira borgpetersenii (GenBank accession no. AF078135), the genes for which, as in P. aeruginosa, occur contiguously with no intervening hisA homologue. Interestingly, the L. borgpetersenii his genes are located near the 5′ end of a large cluster of genes thought to be involved in the synthesis of O antigen in that bacterium (annotation of GenBank entry AF078135). OrfH2, encoded by the gene directly upstream of OrfH3 (HisH), has 47% identity to WbpG, which is encoded by the first gene downstream of hisF in P. aeruginosa. Therefore, although there are similarities between the two organisms in this region, their gene orders are different. Inactivation of the wbp hisHF genes does not affect B-band LPS biosynthesis (23). In addition, these genes are probably not required for histidine biosynthesis, since there is a separate set of native his genes on the P. aeruginosa chromosome, unlinked to the B-band O-antigen genes.
The IS1209 element is found between wbpL and wbpM only in serotype O5, and not between the wbpL and wbpM homologues in serotypes O6 and O11. In the latter serotypes, wbpM is probably expressed from the same large transcript as wbpL, while in serotype O5, wbpM is probably expressed from one of several putative outwardly directed promoters within IS1209 (21). In serotype O5, the wbpL gene appears to have been truncated due to insertion of IS1209, since its gene product is only 303 amino acids, compared with 341 amino acids for the corresponding proteins from serotypes O6 and O11 and the closely related homologue, TrsG, from Yersinia enterocolitica O:3 (207). However, this putative truncation does not appear to have affected the function of the protein (10, 195). The IS1209 element, while not present in the O-antigen clusters of serotypes O6 and O11, is nevertheless present on the chromosomes of other P. aeruginosa strains. Of the 20 IATS serotype reference strains, 19 hybridized at high stringency with an IS1209 probe in Southern blot analysis (21). Nucleotide sequence analysis of the 5′ end of a DNA fragment from P. aeruginosa PA103 (serotype O11) showed that a copy of IS1209 is located upstream of exoU (pepA), which encodes a secreted cytotoxic protein (67, 87). IS1209 is related to the gene-activating insertion sequence IS407 of B. cepacia and B. pseudomallei (151, 236) and to similar elements from a number of soil-, plant-, and water-associated bacteria (21). The association of mobile elements with exopolysaccharide gene clusters is not uncommon (61, 89, 213) and may facilitate the lateral transfer of O-antigen genes between strains or species (216). However, the order of genes (wbpK-wbpL-wbpM) at the 3′ ends of the three P. aeruginosa O-antigen clusters examined to date is highly conserved. This level of conservation suggests that the insertion of IS1209 between wbpL and wbpM in serotype O5 may be gratuitous and not related to lateral transfer. Instead, the highly conserved wbpM gene may itself represent a convenient site for recombination between O-antigen gene clusters.
To understand the functions of the proteins encoded within the wbp clusters, it was necessary to have access to information about the chemical nature and structure of the O antigens of serotypes O5 and O6. The structure of the O5 (formerly O:3a,d [112] and then reclassified O:2a,d [111]) O-antigen unit was determined by Knirel et al. (112). The structure of the O5 O antigen is shown in Fig. 1B, along with those of the chemically related serotypes O2 [formerly O:(3a),3c (113); then O:(2a),2 (111)], O16 (formerly O:3a,b [112]; then O:2a,b [111]), O18 (formerly Fisher 7 [109, 148]), and O20 [formerly O:(3a),3d,3f (113); then O:(2a),2d,2f (111)]. These serotypes have closely related O-antigen structures, varying only in the linkage between O units (i.e., serotype O5 is α1→4, while serotype O16 is β1→4), the presence of the C5 epimer of d-mannosaminuronic acid, l-gulosaminuronic acid (in serotypes O2, O18, and O20), or the presence of acetyl substitutions at the C-4 position of Fuc2NAc (serotype O20) (109). The structures of the serotype O6 (and related strains Fisher 1 and Lanyi serotype O4) and O11 O antigens were determined by Knirel and coworkers (57, 114) and are shown in Fig. 1B.
With the structures of the O antigens in hand, one can make predictions about the types and numbers of genes required to encode the synthesis of the polysaccharides. In the following sections, we discuss the biosynthesis of P. aeruginosa B-band O antigens with the well-characterized serotype O5 as a model, comparing it to serotype O6 where applicable. The availability of the majority of the O5 (PAO1) genome sequence since late 1998 has allowed us to examine the chromosome for additional genes involved in LPS synthesis and to look for additional homologues of genes located in the wbp cluster.
Genes Required for Biosynthesis of Hexose Residues of the O-Antigen Units
Synthesis of dideoxy aminohexoses.
Based on their homologies to other LPS and sugar biosynthetic genes, wbpB, wbpK, and wbpM of serotype O5 were predicted to be involved in the biosynthesis of the deoxyaminohexose, UDP-2-N-acetyl-d-fucosamine (UDP-6-deoxy-2-N-acetyl-d-galactosamine [UDP-d-Fuc2NAc]) (21). The precursor sugar for biosynthesis of UDP-d-Fuc2NAc is thought to be UDP-N-acetyl-d-glucosamine (UDP-d-GlcNAc), which is readily available in the cell due to its role in peptidoglycan biosynthesis. The pathway for synthesis of UDP-d-Fuc2NAc from UDP-d-GlcNAc is thought to require an epimerization to generate the galactosyl moiety and a dehydration/oxido-reduction to produce the deoxy form (21). The closest homologue of WbpB is a protein of unknown function from Helicobacter pylori (GenBank accession no. AE000581) (223). Other homologous proteins of WbpB include a number of putative oxidoreductases (21), and WbpB may therefore play that role in UDP-d-Fuc2NAc synthesis. At present, the function of wbpB in P. aeruginosa O5 LPS synthesis has not been confirmed through inactivation of the gene. Despite the presence of a similar deoxy sugar, UDP-6-deoxy-N-acetyl-d-glucosamine (UDP-N-acetyl-d-quinavosamine [UDP-d-QuiNAc]) in the O antigen of serotype O6, no homologue of wbpB could be identified in the B-band gene cluster (10). However, a homologue of wbpB, the Bordetella pertussis gene wlbA, has been identified in the cluster of genes encoding band A lipooligosaccharide, a trisaccharide that contains a methylated Fuc2NAc residue (2). Interestingly, WlbA has been proposed to be a dehydrogenase required for the synthesis of mannosaminuronic acid in B. pertussis (2), a role thought to be played by the essential gene product WbpA in P. aeruginosa O5 (27) (see below). Therefore, the potential role of wbpB in O5 O-antigen synthesis requires further analysis.
WbpK and WbpM both demonstrate homology to dehydratases and epimerases, including the UDP-d-galactose-4-epimerase GalE. Therefore, the specific role played by each of these proteins in UDP-d-Fuc2NAc synthesis is not immediately apparent based on homology. Since wbpM is highly conserved among all 20 IATS serotypes, most of which have either Fuc2NAc or N-acetylgalactosamine (GalNAc) in their O-antigen structures, we previously proposed that wbpM may encode the C-4 epimerase and the less highly conserved wbpK may encode the dehydratase (21). Although a conclusive assignment awaits biochemical analyses of the activities of these proteins, we now have preliminary evidence (discussed below) showing that the reverse may be true (23).
In serotype O6, the gene encoding the C-4 epimerase involved in the conversion of UDP-d-GlcNAc to UDP-d-GalNAc during the formation of UDP-d-GalNAcA has been shown to be WbpP, not WbpM (10). Instead, WbpM, which is essential for both O5 and O6 O-antigen synthesis (21, 10), may be a dehydratase involved in the biosynthesis of d-QuiNAc. A survey of the O antigens of P. aeruginosa shows that most contain either d-QuiNAc or d-FucNAc (109). In O5 and other serotypes, UDP-d-Fuc2NAc may be synthesized from a UDP-d-QuiNAc precursor through the action of a GalE-like C-4 epimerase. In serotype O5, WbpK, which has homology to GalE (21), may function as this epimerase. Based on its ability to partially complement a galE mutant of Salmonella enterica serovar Typhimurium (SL1306), WbpK has weak UDP-glucose-4-epimerase activity (161). Therefore, in O5, WbpK may catalyze the C4 epimerization of UDP-d-QuiNAc to UDP-d-Fuc2NAc. The essential role of WbpK in O5 B-band O-antigen biosynthesis has been demonstrated through generation of wbpK knockout mutants, which no longer produce B-band LPS (10). One caveat to this hypothesis is the presence of a wbpK homologue, wbpV, in serotype O6. The function of wbpV has not been ascertained, but it is essential for O6 O-antigen synthesis (based on mutational analysis) and is not interchangeable with wbpK, based on cross-complementation studies (10).
To demonstrate the role of the highly conserved WbpM in other serotypes, we have generated wbpM::Gmr knockout mutants of serotypes O10, O15, and O17 (Fig. 1B) (29). Mutation of wbpM abrogates O-antigen biosynthesis only in serotype O10, implying that WbpM is not involved in the conversion of UDP-d-GlcNAc to UDP-d-GalNAc (as in O15), as proposed previously, but is probably a dehydratase necessary for the synthesis of UDP-d-QuiNAc and UDP-d-FucNAc (29). These results also show that while WbpM is present in all 20 serotypes, it is not universally required for P. aeruginosa B-band synthesis. We are currently collecting biochemical evidence to support the assignments of function.
WbpM is a representative of a large and growing family of proteins involved in the biosynthesis of bacterial exopolysaccharides such as LPS, lipooligosaccharides, and capsules. These proteins are widely distributed among both gram-negative and gram-positive bacteria (Table 2), many of which are of medical significance. There appear to be at least two distinct lineages of WbpM-like proteins (Table 2) (29). One subfamily, typified by WbpM, contains large (approximately 600-amino-acid) proteins with two distinct domains, with an NAD+/NADP+ binding motif in the C terminus and often a second motif in the N terminus (21). Based on the identification of contiguous but separate ORFs in Vibrio cholerae O139 that were homologous to the N-terminal (ORF10) and C-terminal (ORF11) halves of WbpM, these large proteins were originally postulated to be the result of an ancestral fusion between two shorter proteins (21). However, more recent sequence data showed that the separation of ORF10 and ORF11 of V. cholerae O139 arose from a sequencing error and that there is only a single large ORF10, encoding a 646-amino-acid protein (sequence update April 1998). In the capsular biosynthetic clusters of Staphylococcus aureus serotypes 5 and 8, contiguous large (capD) and short (capE) WbpM homologues are present in the same operon (200, 201) whereas S. aureus serotype 1 has only capD (144).
TABLE 2.
WbpM and its homologues
| Protein name | Organism | Length (amino acids) | Putative function | Accession no. (reference) |
|---|---|---|---|---|
| Subfamily 1a | ||||
| WbpM | Pseudomonas aeruginosa serotype O5 | 665 | Fuc2NAc biosynthesis | U50396 (21) |
| WbpM | Pseudomonas aeruginosa serotype O6 | 665 | Fuc2NAc biosynthesis | AF035937 (10) |
| ORF74.5 | Pseudomonas aeruginosa serotype O11 | 665 | Fuc2NAc biosynthesis | U44089, direct submission |
| TrsG | Yersinia enterocolitica serotype O:3 | 638 | Galactose modification | S51266 (207) |
| WlbL | Bordetella bronchiseptica | 624 | Nucleotide sugar dehydratase/epimerase | AJ007747, direct submission |
| WlbL | Bordetella pertussis | 624 | FucNAcMe biosynthesis | S70683 (2) |
| WlaL | Campylobacter jejuni | 590 | Unknown | Y11648 (74) |
| RfbV | Vibrio cholerae serotype O1 | 621 | Unknown | Y07788 (65) |
| ORF22-30 | Vibrio cholerae serotype O22 | 646 | Unknown | AB012957, direct submission |
| ORF10 | Vibrio cholerae serotype O139 | 646 | Epimerase/dehydratase | U47057 (37) |
| CapD | Staphylococcus aureus serotype 1 | 599 | Type 1 capsule synthesis | U10927 (144) |
| Cap5D | Staphylococcus aureus serotype 5 | 607 | Unknown | U81973 (200) |
| Cap8D | Staphylococcus aureus serotype 8 | 607 | Unknown | U73374 (201) |
| LpsB | Rhizobium etli | 683 | dTDP-glucose-4,6-dehydratase | U56723, direct submission |
| WbiI | Burkholderia pseudomallei | 637 | Epimerase/dehydratase | AF0064070 (55) |
| TP0077 | Treponema pallidum | 538 | Capsular polysaccharide biosynthesis | AE001192 (73) |
| YveM | Bacillus subtilis | 598 | Unknown | Z99121 (126) |
| Subfamily 2b | ||||
| ORF HP0840 | Helicobacter pylori | 333 | Unknown | AE000595 (223) |
| ORF JHP0778 | Helicobacter pylori | 333 | Sugar nucleotide biosynthesis | AE001508 (3) |
| FlaA1 | Caulobacter crescentus | 331 | Unknown | U27301, direct submission |
| Cap5E | Staphylococcus aureus serotype 5 | 342 | Unknown | U81973 (200) |
| Cap8E | Staphylococcus aureus serotype 8 | 342 | Unknown | U73374 (201) |
| Protein D | Methanococcus jannaschii | 333 | Capsular biosynthetic protein | U67549 (18) |
| CapD | Rickettsia prowazekii | 341 | Unknown | AJ235271 (5) |
| KasD | Streptomyces kasugaensis | 329 | NDP-hexose 4,6-dehydratase | AB005901 (92) |
| Gdh | Saccharopolyspora erythraea | 329 | dTDP-d-glucose-4,6-dehydratase | L37354 (146) |
| BbLPS1.16 | Bordetella bronchiseptica | 357 | Nucleotide sugar dehydratase/epimerase | AJ007747, direct submission |
Subfamily 1 contains proteins of approximately 600 amino acids.
Subfamily 2 contains proteins shorter than 360 amino acids.
Although the specific function of these proteins has not yet been demonstrated at the biochemical level, they are functionally homologous (29). A P. aeruginosa O5 wbpM::Gmr mutant, which cannot synthesis B-band O antigen, can be complemented in trans by B. pertussis wlbL (2), S. aureus capD (144), and, weakly, by H. pylori HP0840 (223). The ability of HP0840 to partially complement a wbpM mutation demonstrates that the shorter homologues have activity similar to the larger forms (29). However, the inability of the smaller protein to completely restore O-antigen synthesis in a wbpM::Gmr mutant suggests that the N terminus of the larger proteins is important for full activity. The broad distribution of this protein family among bacteria, many of which are of significance to the medical research community, suggest that they may be of interest as potential therapeutic targets.
Synthesis of uronic acids.
(i) Epimerization.
Two of the three sugars of the O5 O-antigen unit are mannosaminuronic acids, containing either two acetimido groups or one acetimido and one acetaminido group (Fig. 1B). A similar di-N-acetylated mannosaminuronic acid residue is found in the band A trisaccharide of B. pertussis (2). The biosynthetic pathway for these rare sugars is unknown, although likely avenues have been hypothesized (2, 21). These residues are thought to be formed from a UDP-d-GlcNAc precursor that is first epimerized at C-2 to form UDP-d-ManNAc (2, 21). The enzyme thought to perform this step in P. aeruginosa is WbpI, based on its similarity to other UDP-d-GlcNAc-2-epimerases (21). Derivatives of UDP-d-ManNAc are prevalent in bacterial exopolysaccharides, including ECA which is synthesized by a number of enteric species (167). The breadth of this distribution is reflected in the large number of WbpI homologues in the databases. The UDP-d-GlcNAc-2-epimerase involved in ECA biosynthesis, which has recently been given the designation MnaA (for N-acetylmannuronic acid, step A [188a]), has been extensively studied at the biochemical level and can be used as a model for this type of protein. Mutational analysis of MnaA/WbpI has demonstrated that it is essential for B-band LPS biosynthesis (23). A homologous protein, WlbD of B. pertussis, was proposed to be the UDP-d-GlcNAc-2-epimerase in that organism (2). In serotype O6, two of the four residues of the O unit are galactosaminuronic acids. The C-4 epimerase required for conversion of UDP-d-GlcNAc to UDP-d-GalNAc has been identified as WbpP on the basis of its functional homology to VipB, the C-4 epimerase involved in Vi antigen (poly-GalNAcA) biosynthesis in S. enterica sv. Typhi (10, 85).
(ii) Uronic acid formation.
The companion protein of MnaA is MnaB, formerly RffD or WecC (167), for which the P. aeruginosa O5 homologue is WbpA. The MnaB family of proteins includes NAD+-dependent sugar dehydrogenases that generate ManNAcA from ManNAc (188a). WbpA, encoded by the first gene of the putative five-gene operon (wbpA through wbpE), at the 5′ end of the wbp cluster, is essential for O-antigen synthesis, since wbpA::Gmr mutants no longer make B-band LPS (27). WbpA is homologous to the P. aeruginosa sugar dehydrogenase AlgD, which is essential for synthesis of GDP-ManA residues that make up the exopolysaccharide alginate (Fig. 5) (21, 54). During Southern blot analysis of wbpA::Gmr PAO1 mutants with a wbpA probe, an additional DNA fragment homologous to wbpA was identified. A search of the genome sequence revealed that this fragment contained a third sugar dehydrogenase gene, ugd, encoding UDP-glucose dehydrogenase (29). Further genome analysis allowed the identification of a second putative ugd gene, pmrE. In P. aeruginosa, ugd is linked to galU (encoding glucose-1-P uridylyltransferase or UDP-glucose pyrophosphorylase) while pmrE is linked to a cluster of genes homologous to those encoding polymyxin B resistance in S. enterica sv. Typhimurium and E. coli (29). Cross-complementation analyses with the four sugar dehydrogenase genes showed that despite their homology, they were not able to complement heterologous mutants in trans even when present in multiple copies (23, 27).
Although WbpA is essential based on mutational analysis, assignment of this enzyme to a particular step in the pathway for mannuronic acid biosynthesis is not yet possible. While it is homologous to other MnaB proteins, its substrate may be the di-N-acetylated form of mannosamine and not the mono-N-acetylated form of mannosamine generated by MnaA activity. Evidence that WbpA does not act directly on a UDP-ManNAc substrate includes the inability of the E. coli ManNAc dehydrogenase WecC to complement a wbpA::Gmr mutation in trans (23). Interestingly, there is no homologue of WbpA in B. pertussis, and the role of UDP-d-ManNAc dehydrogenase has been assigned to the WbpB homologue, WlbA (2).
In serotype O6, the UDP-GalNAc dehydrogenase WbpO is also encoded by the first gene of the cluster and is preceded by a functional promoter (10). WbpO is functionally homologous (as shown through complementation studies [10]) to VipA, the UDP-GalNAc dehydrogenase forming the GalNAcA molecules of Vi antigen (85), and has been shown by mutation analysis to be essential for O6 O-unit synthesis.
AlgD, the GDP-d-Man dehydrogenase, is also encoded by the first gene of the alginate gene cluster and is operative at the committal step for alginate biosynthesis (54). The location of the sugar dehydrogenase genes wbpA and wbpO at the 5′ ends of disparate O-antigen clusters therefore warrants investigation into possible regulatory phenomena associated with this localization. Inspection of the DNA sequence upstream of wbpA did not reveal any homology to consensus E. coli sigma 70 promoter sequences (29), although such sequences are readily detected in P. aeruginosa. This information, coupled with previous observations that B-band LPS synthesis can be negatively regulated by a number of environmental stimuli, suggests that in P. aeruginosa, B-band O-antigen synthesis is not necessarily constitutive and that closer attention should be paid to the mechanisms of its regulation.
(iii) Amination.
The mannuronic acid residues of the O5 O unit are di-N-acetylated. The N-acetyl group on C-2 is present on the precursor sugar, UDP-GlcNAc, but the second N-acetyl group is added by O-antigen-specific enzymes encoded in the wbp cluster. WbpE is the candidate C-3 aminase, based on its homology to other putative aminases (21) including the B. pertussis protein WlbC, which is thought to catalyze the same reaction. Interesting, one of the WbpE homologues, DegT, was originally identified as a regulatory protein, an assignment that has complicated the annotation of other homologues in this protein family (219). The function of WbpE in O-antigen synthesis has not yet been demonstrated at the biochemical level.
(iv) N-acetylation.
Following amination of the mannuronic acid residues at C-3, N-acetylation can occur. Inspection of the wbp cluster revealed two contiguous ORFs, wbpC and wbpD, with homology to O-acetyltransferases (OATs), but none with homology to N-acetyltransferases (NATs) (21). Few bacterial NATs other than those acting on aminoglycosides and similar substrates have been identified. Although no NAT genes were identified in the wbp cluster, there is precedent for OATs catalyzing N-acetyltransfer. The enzyme responsible for O-acetylation of the structural polysaccharide, peptidoglycan, in the urinary pathogen Providencia stuartii also has 2′ NAT activity. This enzyme N-acetylates the 2′ position of aminoglycosides including gentamicin, tobramycin, netilimicin, and 6′-N-ethylnetilimicin (187) and is thus responsible for drug resistance as well as peptidoglycan O-acetyl modifications (176). Based on this precedent, we believed that either of the two candidate OAT genes in the wbp cluster could be involved in N-acetylation of the UDP-Man(2NAc3N)A moiety.
Both wbpC and wbpD were inactivated separately with carbenicillin resistance (Cbr) cassettes to assess their specific roles in UDP-Man2NAc3N acetylation. We chose the Cbr cassette over a Gmr cassette for inactivation to avoid introduction of additional acetyltransfer activities into the mutants. Inactivation only of wbpD, not of wbpC, abrogated O5 O-antigen biosynthesis, implicating WbpD as the 3′ NAT for UDP-Man2NAc3NAcA synthesis (20). Lack of WbpC, which is homologous to the O-antigen OAT OafA of S. enterica sv. Typhimurium (208), did not appear to affect O-antigen biosynthesis, consistent with the lack of O-acetyl substitutions in the O5 structure (20). B. pertussis, which also synthesizes UDP-d-Man(2NAc3NAc)A, possesses a homologue of WbpD called WlbB (2) that presumably catalyzes the 3′ NAT reaction in that organism, and it does not contain a homologue of WbpC.
(v) Additional modifications.
In the O5 O unit, the first mannuronic acid residue contains an acetiminido group instead of an acetimido group on C-3 (Fig. 1B). This change is carried by all five serotypes of the O5 serogroup, implying that it occurs prior to the epimerization of this residue in O18 and O20 to l-gulosaminuronic acid. The conversion of an acetyl group to an acetamido group could occur through a simple Schiff’s base reaction; however, the mechanism of this alteration is currently unknown. WbpG, encoded by the O5 cluster, is essential for B-band synthesis (21). WbpG has few homologues in the databases, but it is 65% similar overall to OrfH2 of Leptospira borgpeterensii (GenBank accession no. AF078135) and has limited local similarity to WbrF (formerly ORF2) of the V. cholerae O139 O-antigen gene cluster (37), as well as to NH3-dependent NAD+ synthetases and asparagine synthetases (Table 3). These proteins transfer amino groups by using NH3 or glutamine as substrates. WbpG contains a highly conserved motif also found in these proteins and may therefore be an aminotransferase, forming the C-3 acetiminido group on the first sugar residue in the O5 serogroup O units. Data from Western immunoblot analysis of wbpG mutants with MAb 18-19 (specific for the core-plus-one O repeat; 130) showed that these mutants were not rough, as previously thought, but SR (core plus one O unit). This phenotype, which resembles that of an O polymerase mutant (see below), implies that the presence of the acetiminido group on the terminal residue of the O unit is essential for polymerization of the O antigen (23). Interestingly, wbpS of serotype O6 occupies the same relative position in the O6 gene cluster as does wbpG in O5 (10). WbpS also has limited protein homology to WbrF of V. cholerae O139 but none to WbpG (10, 37, 217). WbpS does not contain the conserved motif shown in Table 3, and its function remains unclear. Serotype O6 and related subtypes contain an amino substituent on the second GalNAcA residue at C-6 (109), but the enzyme required for this addition has not been identified.
TABLE 3.
Conserved motif within WbpG and its homologues
| Protein name | Motifb | Accession no. (reference) |
|---|---|---|
| Pseudomonas aeruginosa WbpG | 67-IIGLSGGVDSSYLAVKVKD-85 | U50396 (21) |
| Leptospira borgpeterensii OrfH2 | 74-ILGISGGVDSSYLAYLAKE-92 | AF078135 (direct submission) |
| Haemophilus ducreyi GMP synthetase, GuaA-1 | 74-ILGLSGGVDSSVTALLLHR-92 | AF057695 (227) |
| Aquifex aeolicus NadE | 314-VLGLSGGIDSSFVACLAVD-330 | AE000715 (49) |
| Archaeglobus fulgidus cons.hyp.a | 23-VIAFSGGVDSSTLAAVCKD-39 | 2650065 (108) |
| Archaeglobus fulgidus fulgidus GuaA-1 | 24-IIALSGGVDSSVCTVLAHK-42 | 2650384 (108) |
| Methanobacterium thermoautotrophicum cons.hyp. | 139-AVALSGGVDSSFSLIAAVK-155 | 2621322 (210) |
| Methanobacterium thermoautotrophicum NadE | 40-VLGLSGGVDSSTVAYLAVN-56 | 2622628 (210) |
| Synechococcus sp. strain NadE | 302-ILGLSGGIDSSLVAAIAVE-318 | 1653472 (99) |
| Rhodobacter capsulatus NadE | 288-VLGLSGGIDSALVAVIAAD-304 | Q03638 (233) |
| Mycobacterium tuberculosis Rv2438c | 423-VIGVSGGLDSTHALIVATH-439 | 1666146 (36) |
| Helicobacter pylori NadE | 29-VYGLSGGLDSAVVGVLCQK-45 | 4154831 (3) |
| Methanococcus jannaschii AsnB | 277-GAFLSGGLDSSTVVGVMRE-293 | 2826362 (18) |
| Bacillus subtilis AsnB | 260-GSFLSGGIDSSFIVSVAKE-276 | 2293165 (137) |
| Mycobacterium tuberculosis AsnB | 281-GAFLSGGIDSTAIAALAIR-297 | Q10374, direct submission |
| Bordetella bronchiseptica amidotransferase | 259-GAYISGGVDSSLVAAMARH-275 | 3451496, direct submission |
| Bordetella bronchiseptica amidotransferase | 267-GAALSGGIDSSAIVCAMRW-285 | 3451502, direct submission |
| Vibrio cholerae WbrF, AsnB | 264-GAFLSGGVDSSTVVGILQS-280 | 2244692 (216); 724317, direct submission; U47057 (37) |
| Consensus | lSGGvDSs |
cons.hyp., conserved hypothetical protein.
The position of the motif within the individual proteins is indicated by the amino acid numbers flanking the sequence.
The first galactosaminuronic acid residue in the O6 O unit contains a formyl, rather than an acetyl, group on N-2. Formyl groups are unusual substituents (109), and the pathway leading to their addition is not yet understood. Serotypes O7 and O8 also carry formyl substituents on N-7 of pseudaminic acid, while this position is unsubstituted in serotype O9 (109).
Lysogeny of serotype O5 with bacteriophage D3 converts the O antigen to an O-acetylated form of the related serotype O16, which differs from O5 in having a β1→4 rather than an α1→4 linkage between O repeats (Fig. 1B; 127). The O-acetyl substitution occurs at C-4 of the Fuc2NAc residue. Similarly, serotype O20, which is also closely related to serotype O5, has an O-acetyl substitution at C-4 of Fuc2NAc (Fig. 1B) (109). Analysis of the bacteriophage D3 genome (122) revealed a gene whose product was homologous to both OafA of S. enterica serovar Typhimurium and WbpC; this gene was designated oac. Complementation experiments showed that the product of this gene was responsible for O acetylation of the O5 O antigen (20). In contrast, WbpC does not appear to be a functional O-antigen OAT, despite its homology to OafA and Oac. The O-acetyl substitutions of the O20 antigen are probably added by an OAT other than WbpC, possibly of bacteriophage origin. Bacteriophages mediate such substitutions during lysogenic conversion of their host, probably to prevent nonproductive adsorption of other phage particles to the O antigen of infected cells (203). The O antigen of serotype O6 is stoichiometrically substituted with O-acetyl groups on C-3 of the GalNAcA residue (109), but the enzyme responsible for this addition has not yet been identified.
The addition of these substituents, whether stoichiometric or otherwise, contributes to the structural diversity of the O antigen. Small changes such as these can have a profound impact on the ability of the organism to infect a host population that has been previously exposed to a related organism. As a case in point, one needs only to contemplate the recent emergence of epidemic strains of V. cholerae O139, which is essentially identical to V. cholerae O1 but with the critical exception of an altered O-antigen/capsule structure (37, 216).
Genes Encoding Glycosyltransferases Required for O-Antigen Unit Assembly
Specific glycosyltransferase proteins are required to form the O-antigen units of P. aeruginosa B-band LPS. B-band LPS is a heteropolymer and follows the typical O-antigen polymerase (Wzy)-dependent pathway of biosynthesis (10, 21, 22, 25, 38, 51, 53, 230). In the Wzy-dependent pathway of synthesis, nonprocessive glycosyltransferase enzymes assemble the heteropolysaccharide O units at the cytoplasmic face on the lipid carrier Und-P (230). Addition of each monosaccharide requires a specific glycosyltransferase that recognizes the donor and acceptor molecules and catalyzes the formation of a glycosidic bond, either α or β, between specific carbons on those molecules.
Three putative glycosyltransferase genes, wbpH, wbpJ, and wbpL, were identified in the O5 wbpB band cluster, located generally in the 3′ region and separated by single intervening ORFs (Fig. 3B). In serotype O6, which synthesizes a tetrasaccharide repeat, four putative glycosyltransferase genes, wbpR, wbpT, wbpU, and wbpL, were identified (Fig. 3C), again in the 3′ region of the cluster. In both cases, the glycosyltransferases are encoded in the opposite order to which they act; i.e., WbpL is the initiating transferase (10, 21, 195).
As discussed earlier, WbpLO5 is required for both A-band and B-band LPS biosynthesis (195). We propose that this is mediated through the addition of GlcNAc-1-P to Und-P to initiate A-band LPS synthesis and the addition of Fuc2NAc-1-P to Und-P to initiate B-band LPS biosynthesis. Similar results were obtained during studies of WbpLO6 (10). However, in the case of serotype O6, it is believed that WbpLO6 transfers QuiNAc-1-P to Und-P to initiate B-band synthesis based on the O6 O-antigen structure (Fig. 1B). Interestingly, cross-complementation of wbpL::Gmr mutants of O5 and O6 showed that WbpLO5 could complement A-band and B-band biosynthesis in the wbpLO6::Gmr strain. In contrast, WbpLO6 complemented A-band biosynthesis to parental levels in a wbpLO5::Gmr mutant but could only weakly complement O5 B-band O-antigen synthesis (10). The O6 enzyme appears to be less efficient in recognizing UDP-d-Fuc2NAc as a donor, while WbpLO5 readily recognizes UDP-d-QuiNAc.
Two of the three residues that make up the O5 trisaccharide O unit are mannosaminuronic acid residues. The putative mannosylaminuronyltransferases, WbpH and WbpJ, are 22.5% identical throughout and are homologous to a number of glycosyltransferases from other bacteria. Particularly significant is the homology of WbpH not only to WbpJ but also throughout its length to the A-band LPS glycosyltransferase, WbpZ (30). As mentioned above, WbpZ is the glycosyltransferase that is thought to add the first d-Rha residue to the GlcNAc-PP-Und primer formed by WbpL (195). The homology between the proteins suggests that they could play similar roles and that WbpH may recognize the initiating Fuc2NAc residue added by WbpL to Und-P (Fig. 7B). In that case, WbpJ could be the third transferase, adding the second Man(2NAc3N)A residue to form the trisaccharide O unit. However, glycosyltransferase genes are generally ordered within exopolysaccharide gene clusters in the opposite order to which they act. In that case, the gene order implies that WbpJ is the second and WbpH is the third glycosyltransferase.
To address the roles of WbpH and WbpJ in O5 O-antigen biosynthesis, the genes were individually inactivated with nonpolar Gmr cassettes and the LPS of the resulting mutants was examined. Inactivation of WbpH abrogated B-band LPS biosynthesis completely, and complementation of the wbpH::Gmr mutant with wbpH in trans led to the restoration of long-chain O-antigen production (24, 30). However, Western immunoblot analysis of the LPS produced by the complemented wbpH::Gmr mutant showed that the structure of the LPS was different from that of the parent. MAb MF15-4, which recognizes a conformational epitope formed by high-molecular-weight O5 O antigen, reacted strongly with LPS isolated from the complemented mutant, while MAb 18-19, specific for the individual and identical trisaccharide O units of serotypes O5 and O16, bound poorly. Taken together, these results suggested that overexpression of WbpH in trans permitted the formation of O antigen that was conformationally similar to the wild type. However, the linkages or types of sugars within the O unit appeared to be altered in such a way that the availability of the epitope for MAb 18-19 was reduced (24, 30). Structural analysis of the LPS from the complemented mutant is necessary to distinguish between these possibilities.
In contrast, a wbpJ::Gmr null mutant produced a greatly reduced amount of O5-like antigen that reacted with the trisaccharide-specific MAb 18-19 (24, 30). This residual LPS was visible as a weak core-plus-one-trisaccharide band on silver-stained SDS-PAGE gels, while a small amount of high-molecular-weight material could be detected on the more sensitive Western immunoblots. Addition of wzy and wzx in trans on a multicopy plasmid slightly increased the amount of core-plus-one material (24). Complementation of the wbpJ::Gmr mutant with wbpJ in trans did not restore wild-type O-antigen synthesis, nor did complementation with constructs containing wbpH and wbpJ, or wbpJ and wbpL. However, a construct containing all three genes complemented the O antigen of the wbpJ::Gmr mutant to a phenotype resembling the parent (24, 30).
Experiments to use spectroscopic methods to determine the structure of the residual O antigen produced by the wbpJ::Gmr mutant, to examine the role of WbpJ and WbpH in O5 O-antigen formation, are under way (30). In light of the protein homology between WbpH and WbpZ and the apparently essential requirement for WbpH but not WbpJ to form an O unit, we propose that WbpH and WbpL only are necessary to form an O5-like O-antigen unit (30). The role of WbpJ is not clear, but it may act to modify the activity of WbpH to generate the specific glycosidic bonds seen within the typical O5 O unit (30). This situation could be analogous to the modulation of the E. coli UDP-d-Glc pyrophosphorylase GalU by its homologue, GalF (158). Although these proteins were originally thought to be isoforms of GalU, Marolda and Valvano (158) showed that GalF does not have pyrophosphorylase activity but, instead, interacts physically with GalU to modify its activity. Along these lines, we are currently investigating the physical interactions between WbpH, WbpJ, and WbpL. It is not yet clear why provision of wbpJ in trans is not able to restore the mutant to the parental phenotype, but the possible involvement of regulatory or stoichiometric effects is under consideration.
Aside from WbpL, there is no homology between the glycosyltransferases that form the O5 and O6 antigens, as would be predicted from their chemical structure. In serotype O6, as mentioned above, WbpLO6 is believed to be the initial transferase of d-QuiNAc, while WbpT and WbpU are believed to transfer the GalNAcA and GalNFmA residues of the O6 unit, respectively (10). WbpT is homologous to galactosyltransferases including WcdD, which is involved in formation of the Vi antigen of S. enterica sv. Typhi, a homopolymer of d-GalNAcA (85). The galactosyltransferase WcdD uses d-GalNAcA as both donor and acceptor, and a similar role is proposed for WbpT, which would add d-GalNAcA to d-GalNFmA in the growing O6 unit (10). Confirmation of WbpT function awaits mutational analysis. Mutants lacking WbpU, which is predicted to be the galactosyltransferase attaching d-GalNFmA to d-QuiNAc, do not synthesize O antigen (10). The similarities of WbpU to galactosyltransferases that add Gal derivatives to GlcNAc-PP-Und strongly implicate it as the second glycosyltransferase. WbpR, the fourth putative glycosyltransferase, is thought to add l-Rha to complete the tetrasaccharide O unit, but its function remains to be ascertained (10).
wbpG: a Potential Control Point for B-Band Lipopolysaccharide Biosynthesis
After isolation of the O5 B-band LPS gene cluster on pFV100 through complementation of mutants lacking B band, we wished to determine which genes were affected in the long-chain O-antigen-deficient Tn5-751 mutants ge6, ge11, and ge12 (141, 143). Through subcloning and complementation experiments, both ge6 and ge12 were found to contain mutations in wbpG (33). ge11 could be complemented only by the entire cosmid, suggesting that it had multiple mutations which were sufficiently distant from one another that they could not be complemented by a single subclone.
Interestingly, analysis of the chromosomal DNA from ge6 and ge12 showed that the sites of transposon insertion were unique and distal to the B-band LPS genes and that the DNA flanking the insertion sites had no homology to known LPS genes (33). Similarly, analysis of the wbpG-containing region from these mutants by Southern blot analysis showed that wbpG from ge6 and ge12 was identical in size to that of the parent PAO1 strain (33). Collectively, these results showed that ge6 and ge12 had acquired spontaneous mutations in wbpG that were unrelated to the Tn5-751 mutation, but the mutations still abrogated B-band LPS synthesis (21). wbpG is the first gene of a putative large operon (wbpG through wbpL) in serotype O5, and it is intriguing that two independently isolated mutants contained lesions in the same gene.
Goldberg et al. (75) showed that addition of the serotype O11 B-band O-antigen gene cluster to various rough phenotypes of P. aeruginosa isolated from CF patients allowed them to produce not only the O11 antigen but in some cases their native O antigen as well. Therefore, the lack of B-band O antigen in CF isolates may arise from the acquisition of spontaneous mutations within the B-band LPS genes. In chronic infections, B-band LPS is frequently lost, and selection for spontaneous B-band-minus strains may be one of the mechanisms underlying this event.
Interestingly, wbpG, which is essential for B-band LPS biosynthesis in serotype O5 (21), has the lowest G+C content (44.5%, which is ca. 22% lower than the chromosomal average) of any gene in the O-antigen cluster. A low G+C content has been proposed to be a mechanism of translational regulation for the Wzy protein (171), and a similar mechanism may be involved in regulation of B-band LPS synthesis through control of the rate of WbpG translation from the wbpG-wbpL transcript. The chromosomal G+C content of P. aeruginosa, ∼67%, is reflected in its preferred usage of codons that are GC rich (i.e., that have G or C in the wobble position) (229). A preponderance of AT-rich codons in the 5′ region of a wbpG-wpbL mRNA may lead to a decreased rate of translation, which in turn could reduce the stability of the large transcript. Inspection of the serotype O6 LPS gene cluster reveals that wbpP, which is preceded by a functional promoter, has the lowest G+C content (46.4%) of the entire O6 cluster (10). Therefore, regulation of wbpP expression may similarly represent a control point for the expression of WbpP, WbpQ, and WzxO6.
Wzy (Rfc)-Dependent Pathway of Synthesis
Effects of Wzx mutations on A-band and B-band LPS biosynthesis.
Once assembled, the O-repeat-PP-Und molecule of heteropolymers is translocated to the periplasmic face of the inner membrane through the action of the hydrophobic protein Wzx (RfbX) (147). Interestingly, all of the reported Wzy-dependent exopolysaccharide clusters encode a Wzx-like protein of approximately 400 to 500 amino acids, with 12 predicted membrane-spanning domains (101). Thus far, there has been only limited analysis of Wzx function, due to a number of technical difficulties. Wzx has proven difficult to express in heterologous systems, probably due to the presence of rare codons with the coding sequence and to the fact that it is an integral membrane protein (25, 152). In enteric bacteria, it has also been difficult to clone wzx and to generate wzx chromosomal mutants (147, 152, 203).
Liu et al. (147) used an S. enterica serogroup B wba deletion mutant (lacking the wzx gene cluster) harboring a Shigella dysenteriae O1 O-antigen cluster containing a transposon insertion in the wzx gene for expression of the S. dysenteriae O1 polysaccharide. Isolation of radiolabelled Und-PP-linked intermediates by butanol extraction and examination of inverted membrane vesicles revealed the apparent accumulation of single S. dysenteriae O1 repeat units linked to Und-P on the cytoplasmic face of the inner membrane (147). Therefore, Wzx is thought to function as a transporter or “flippase” of nascent O units, moving them from the cytoplasmic to the periplasmic face of the inner membrane. Presentation of lipid-linked O units at the periplasmic face by Wzx is thought to precede polymerization and ligation (discussed below).
Homologues of wzx (originally named wbpF in serotype O5) have been identified in both serotypes O5 and O6 and are located approximately in the middle of the O-antigen gene clusters. This arrangement is similar to that of enteric bacteria (188). To demonstrate the role of Wzx in B-band LPS synthesis, chromosomal knockout mutants with mutations in serotype O5 were generated. The frequency of wzx::Gmr mutants that could be isolated was low compared with that of mutants with mutations in other LPS biosynthetic genes, probably reflecting the deleterious nature of the mutation (25, 203). The accumulation of lipid-linked O units may impair the ability of the cell to make other essential polysaccharides, including peptidoglycan, which are built on the same scaffold molecule, Und-P.
Although experiments similar to those performed by Liu et al. (147) cannot readily be replicated in P. aeruginosa due to the rarity of the deoxyhexoses comprising the O unit, it was of interest to observe the effect of a wzx mutation on LPS synthesis. Not surprisingly, the introduction of the wzx::Gmr mutation abrogated B-band LPS biosynthesis (25). More startling was the observation that the majority of the wzx mutants did not produce the homopolymeric O antigen, A-band LPS. The pathways for A-band and B-band biosynthesis had been shown to be separate, as expected based on the homopolymeric and heteropolymeric natures of the respective polysaccharides (22, 38, 51, 196, 230).
The lack of A-band biosynthesis by the majority of wzx mutants was determined to be a temporal phenomenon, since extended growth of the cultures allowed the eventual detection of A-band LPS (25). The effect of the wzx mutation on A-band but not B-band LPS biosynthesis could be alleviated by supplying multiple copies of the gene encoding the initiating glycosyltransferase, WbpL, in trans (25). Therefore, mutation of wzx appeared to have the unexpected effect of reducing the total cellular availability of WbpL in some manner, which in turn delayed the initiation of A-band LPS biosynthesis. Possible explanations for this phenomenon include feedback inhibition of wbpL transcription or WbpL translation by blockage of O-unit translocation, preclusion of WbpL release from a biosynthetic complex due to unsuccessful translocation of the O unit, and interference with initiation of A-band synthesis, perhaps due to the presence of untranslocated B-band O units on Und-P. Whether the effect of the wzx mutation is at the level of transcription, translation, or posttranslation is not yet clear. However, it is intriguing that interruption of the terminal step in O-unit synthesis, translocation, can affect the first step, initiating glycosyltransfer. The unique effects of this mutation on homopolymer synthesis may not have been apparent in enteric wzx mutants, which normally produce a single type of O side chain. There are some strains of E. coli which coproduce a homopolymeric O antigen (O8 or O9) with a heteropolymeric capsular antigen that can be linked to core-lipid A (e.g., K40 [4]). Although a wzx homologue has been identified in the gene cluster for the K40 antigen in such a strain, the effect of its mutation upon O8 synthesis has not been examined (4).
Wzy (Rfc) and Wzz (Rol, Cld) form O-antigen chains of defined length. (i) The O-antigen polymerase, Wzy.
Following Wzx translocation of the O-repeat-PP-Und molecules to the periplasm, they are polymerized into long chains by the O-antigen polymerase, Wzy, via a poorly understood mechanism. Mutants lacking Wzy display an SR LPS phenotype, consisting of core-lipid A capped with one O-repeat unit (172), due to their inability to polymerize O units into chains. Like Wzx, Wzy is an integral membrane protein, with 11 to 13 transmembrane domains (270). The requirement for Wzy in polymerization seems to be restricted to heteropolymeric O antigens (203, 230), as well as other heteropolymeric cell surface polysaccharides such colanic acid (213) and capsules (4).
Although Wzy apparently mediates the formation of a typical glycosidic linkage between O units, it does not display any homology to known glycosyltransferases. In addition, Wzy is active on the periplasmic side of the cytoplasmic membrane, rather than in the cytoplasm (164), unlike most glycosyltransferases. While typical glycosyltransferases form the nascent glycosidic bond at the nonreducing terminus of the acceptor. Wzy forms bonds at the reducing end of the newly translocated O unit. To form the linkage, Wzy transfers the growing polymer to the nascent subunit, a mechanism of synthesis akin to protein and lipid biosynthesis (193). Again, studies of Wzy activity in vitro have been hampered by the inability to express the protein, due to its poor ribosome binding site, its hydrophobicity, and the presence of multiple rare codons within the wzy ORF (43). Primary amino acid sequences are not conserved among Wzy proteins both within and between species, since Wzy recognizes only its cognate O unit, or close approximations there of (53, 203). This lack of homology has made it difficult to identify potential catalytic motifs or active sites in these proteins. The lack of similarity to glycosyltransferases may suggest that despite seemingly incontestable evidence, it is possible that Wzy does not directly catalyze the formation of the glycosidic linkage between O units but acts instead as a scaffold upon which the linkage is formed by another, as yet unidentified, protein. In P. aeruginosa, specific knockout mutation of wzy results in cells elaborating only a single B-band O-antigen unit attached to lipid A-core (51, 53), while A-band LPS biosynthesis remains unaffected. This SR phenotype is consistent with that seen in wzy mutants of other bacteria (203). By using a wzy mutant of P. aeruginosa O5, the point of attachment of the O unit to the core oligosaccharide has recently been identified as an l-Rha residue (198).
DNA sequence analysis of the wzy genes from two related serotypes of P. aeruginosa, O2 and O5, showed that the genes are essentially identical (53). In addition, mutation of wzy in either serotype results in an SR phenotype, suggesting there is only a single copy of wzy present in the chromosome (51, 53). This observation is interesting since the linkage between the O units in serotype O2 is β1→4 while the linkage between O units in serotype O5 is α1→4 (109). This linkage is thought to be the result of Wzy activity, and it is not clear how a single protein could catalyze linkages of opposite stereochemistry.
It has previously been reported that lysogeny of serotype O5 by bacteriophage D3 causes seroconversion to an O-acetylated form of serotype O16 (127), which differs from O5 in the presence of a β1→4 instead of an α1→4 linkage between O units. However, inspection of the bacteriophage genome, whose DNA sequence was recently determined (122), revealed no putative O-polymerase genes. Therefore, the activity of other phage proteins probably influences the configuration of the linkage between O units. Similarly, the β configuration of the linkage between O units in serotypes O2 and O16 may arise from the activity of proteins encoded outside the O-antigen gene cluster, possibly of bacteriophage origin, that influence the activity of the Wzy protein. Evidence for this hypothesis includes the observation that D3 DNA hybridizes to chromosomal DNA from serotypes O2 and O16 but not to that from O5 (23).
(ii) The O-antigen chain length regulator, Wzz.
The Wzz protein controls the extent of O-unit polymerization, giving rise to a narrow, strain-specific range of O-antigen chain lengths. This nonrandom distribution of chain lengths can be observed as clusters of LPS bands (consisting of more high-molecular-weight and fewer low-molecular-weight bands), referred to as a modal distribution, after separation of the molecules on SDS-PAGE. In contrast, for bacteria in which the O-antigen chain length is not regulated, the LPS bands on SDS-PAGE are generally those of shorter O-antigen chain length. A decrease in the proportion of high-molecular-weight LPS bands is apparent (nonmodal distribution).
Wzz is predicted to have hydrophobic N and C termini that are embedded in the cytoplasmic membrane, while the large central region of the protein forms a periplasmic loop (171). While its mechanism of action is not yet understood, it is thought to interact with either Wzy or the O-antigen ligase, WaaL (RfaL), to adjust the ratio of polymerization to ligation. Mutants lacking Wzz display a nonmodal distribution of O-chain lengths corresponding to that expected for a process with an equal probability of either polymerization (addition of another O unit) or ligation at each round (9, 76). The O-chain length can vary among strains with different wzz genes but identical O antigens, showing that the activity of the Wzz protein is independent of O-antigen structure (9, 72) and that the O-antigen structure does not determine chain length.
The two theories that have been presented to date to explain the mechanism of this O-chain length modulation are the molecular-clock model of Bastin et al. (9) and the molecular-chaperone model of Morona et al. (171). In the first model, proposed for Salmonella, Wzz is predicted to modulate Wzy between two states, one which favors continued extension of the O polymer and one that favors transfer of the O polymer to WaaL for ligation to core-lipid A. Species-specific O-antigen chain lengths are the result of slight variations in the timing of alteration between one state and the other, resulting from differences conferred by the specific amino acid sequence of the Wzz protein (9, 72).
The second model, proposed by Morona et al. (171) for Shigella, describes Wzz as a molecular chaperone that facilitates the formation of a complex containing a particular ratio of Wzy to WaaL (a ligase). This ratio, which varies with the particular Wzz protein, is thought to determine the frequency of ligation, which in turn specifies the lengths of the polymers. Neither Wzz nor WaaL appears to have specificity for particular O antigens, since they can respectively modulate and ligate a number of different polysaccharides (72). However, results of studies on the capsular antigens of E. coli (K30 and K40), which can be linked to lipid A-core, showed that only core-linked capsular polysaccharide, and not high-molecular-weight LPS-free polysaccharide, could be influenced by the presence or absence of wzz (231). This result lends support to the possibility of interaction between Wzz and WaaL, an interaction that does not occur for high-molecular-weight capsular material (231).
In P. aeruginosa, wzz homologues are localized as the first gene at the 5′ end of both the O5 and O6 O-antigen gene clusters (Fig. 3B). These genes are 44.3% identical to one another, despite the obvious differences in the chemical compositions of the O antigens. The functions of the wzz genes of IATS serotypes O5 and O6 were revealed based on their ability to complement the nonmodal phenotype of a related O6 strain, Fisher 1 (10). The modality conferred by WzzO5 was approximately 12 O units, while that conferred by WzzO6 was approximately 18 O units (10). These results confirm that the modality is a function of the Wzz protein and not of the O-antigen composition.
Mutation of wzz in serotype O5 did not result in a strain with the typical nonmodal O-antigen chain length distribution. Instead, the mutant displayed an O-antigen chain length distribution different from the parental phenotype but still modulated with mostly long chains (22). At the time of the previous report (22), we predicted that there was an additional copy of wzz located elsewhere on the chromosome, whose product was responsible for the observed modulation. Similarly, Stevenson et al. (214) identified a wzz (cld) gene on a plasmid harbored by Shigella flexneri strains that cause reactive arthritis. In S. flexneri, the chromosomal copy of wzz conferred a modality of approximately 15 O units upon the parent strain, while the plasmid-encoded gene product conferred a modality of 90 to 100 O units, unusually long for a Wzz. The biological significance of the extremely long O chains is not known (214).
To determine whether serotype O5 does possess multiple wzz homologues, we used the O5 wzz sequence to probe the genome sequence and detected a second homologue of wzz, which we have designated wzz2 (161). The gene products of wzz and wzz2 are only 20.8% identical, similar to the 20.6% identity between WzzO6 and Wzz2. Wzz2 has the highest protein homology to large Wzz homologues associated with capsule synthesis in K. pneumoniae and Burkholderia solanacerum (6, 91). The wzz2 gene was cloned by PCR and is able to confer modality upon the unmodulated S. dysenteriae O antigen expressed from the rfb deletion E. coli mutant CLM4 (22, 161). Interestingly, the modality conferred by both P. aeruginosa wzz and wzz2 genes in E. coli is similar, in the range of 10 to 12 O units (161). In P. aeruginosa, a knockout mutation of wzz2 abrogates the high-molecular-weight LPS bands of the modal cluster of O5 B band while not affecting the LPS bands of intermediate chain lengths attributed to the activity of wzz (23).
The biological role of O-antigen chain length in conferring resistance to serum killing has been well established, and rough mutants deficient in O-antigen production are easily killed when exposed to human serum. However, the more subtle effects of O-antigen chain length perturbation are only just beginning to be appreciated. Recent studies have shown that the modal distribution of chain lengths, and not the mere presence of long O chains on the cell surface in general, is responsible for serum resistance (19) and infectivity (224). This phenomenon is probably due to the preponderance of shorter O chains in a nonmodal distribution, chain lengths which are incapable of protecting the cell membrane from lethal interactions with serum proteins.
Some intriguing results were reported by Van den Bosch et al. (224), who examined the localization of IcsA, an S. flexneri virulence protein required for intercellular spread in strains with various LPS mutations. In wild-type S. flexneri, IcsA is localized to the pole of the cell, but in wzz mutants with unmodulated O chains, little or no IcsA is produced (224). Other mutations that blocked O-antigen synthesis altogether caused IcsA to be distributed randomly over the cell surface (224). In another study, Iredell et al. (93) showed that mutations affecting O-antigen synthesis prevented export of the type IV pilin subunit, causing it to accumulate in the periplasm. Thus, in addition to the important role of O antigen in preserving the integrity of outer membranes, the control and modulation of O-antigen chain length by Wzz plays a role in the localization of cell surface proteins, including those involved in virulence. P. aeruginosa is among the bacteria that produce type IV pili (118). One of our goals is to examine the fate of P. aeruginosa outer membrane proteins, pili, and flagella, all of which have been implicated in virulence, in various O-antigen mutants including those expressing unmodulated O-chain lengths.
B-band O antigens are Wzy-dependent heteropolymers.
The involvement of the Wzx, Wzy, and Wzz proteins in P. aeruginosa B-band LPS biosynthesis shows that B band is synthesized in the same manner as other heteropolymeric exopolysaccharides and extends the paradigm to the family Pseudomonadaceae. The ability of E. coli Wzz to modify the chain lengths of the amino-rich, acidic, and unusual monosaccharides that make up the O antigens of P. aeruginosa is a testament to the Wzy-dependent synthetic mechanism, based on proteins which recognize one another and not on the chemical structure of the particular polysaccharide. This lack of constraint on polysaccharide composition allows for great versatility in O-antigen structure without the requirement for dedicated proteins to handle its attachment to the cell surface.
Evolution of A-Band and B-Band O-Antigen Gene Clusters
The requirement of a common enzyme, WbpL, for both A-band and B-band LPS synthesis is fascinating with respect to the evolution of these gene clusters in P. aeruginosa. The genes for synthesis of A-band LPS are present in all 20 serotypes, most of which are capable of producing this LPS type. The G+C content of the A-band LPS genes ranges from 64.1 to 68%, similar to the P. aeruginosa chromosomal background of approximately 67%, while that of the B-band LPS genes is at least 10% lower (10, 21, 194). While this difference could imply separate ancestries for the two clusters, the absolute requirement for WbpL in A-band synthesis suggests the gene clusters coevolved. In an analogous system, E. coli O9 and O8 d-Man homopolymers are coexpressed with K30 or K40 heteropolymers, respectively (4, 61). The O8 (direct submission; GenBank accession no. AB010150) and O9a (105) gene clusters have a G+C content of approximately 50%, the same as that of the E. coli chromosomal average, while the K30 and K40 clusters are approximately 31 to 38% G+C (4, 60). While these examples are limited, it will be interesting to see whether other gene clusters encoding the synthesis of homopolymeric polysaccharides conform to the G+C content of the background strain.
Two different theories have been used to explain the reduced G+C contents of heteropolymeric O-antigen gene clusters. The first implicates lateral transfer of the genes from an organism whose G+C content was lower than that of the current host (188, 203). The second, based on analysis of wzy genes, invokes potential translational regulatory mechanisms, since the atypical molar ratio is reflected in the usage of codons atypical for the background (270). The intriguing schism between the G+C contents of interdependent homopolymeric and heteropolymeric O-antigen clusters in the same backgrounds lends credence to the latter theory. However, the reasons why separate mechanisms of regulation should have evolved to control the synthesis of the two types of polysaccharide are not yet clear.
The requirement for WbpL to initiate the synthesis of the common antigen, A-band, implies that wbpL homologues should be present on the chromosome in other strains of P. aeruginosa. The broad substrate specificity of these enzymes for UDP-d-GlcNAc, UDP-d-QuiNAc, and UDP-d-FucNAc and the presence of d-QuiNAc or d-FucNAc in most P. aeruginosa B-band O antigens (109) suggest that homologues of WbpL are likely to be encoded in the B-band gene clusters. The onus to retain a WbpL homologue for initiation of A-band synthesis may explain in part the high degree of gene conservation (“wbpK,” “wbpL,” wbpM) at the 3′ end of the B-band gene clusters. However, despite the conservation of DNA at both the 5′ and 3′ ends of the O-antigen clusters, the genes within a cluster are unique to a particular serotype. The identification of O-antigen genes specific for each serotype may prove useful for molecular epidemiology of rough or nontypeable P. aeruginosa strains.
CONCLUSIONS AND FUTURE DIRECTIONS
From our discussions, it is clear that the A-band and B-band O antigens produced by P. aeruginosa are synthesized by independent mechanisms. The only protein common to both systems is the initiating glycosyltransferase WbpL. Data from our preliminary studies indicate that GlcNAc-1-P and Fuc2NAc-1-P may be the two sugars that WbpL transfers to Und-P for initiation of A-band and B-band LPS synthesis, respectively. The extent of substrate specificities for WbpL and the way in which cellular levels of this enzyme are regulated to accommodate this codependency need to be examined. Recently, there has been speculation that these initiating enzymes, along with glycosyltransferases and biosynthetic enzymes, act to form a membrane-associated complex that allows efficient O-antigen synthesis (101). Future studies addressing possible protein-protein interactions between WbpL and the A-band (WbpX, WbpY, and WbpZ) and B-band (WbpJ and WbpH) glycosyltransferases would be very useful to ascertain complex formation that may occur during O-antigen assembly. Identification of the Und-P binding domain within WbpL would also be valuable, since this is a critical step for initiation of a number of other molecules (e.g., peptidoglycan) by proteins homologous to WbpL.
The housekeeping requirement of AlgC for A-band, core oligosaccharide, and alginate biosynthesis on AlgC is intriguing. The algC gene therefore seems strategically placed distal to the highly regulated algD biosynthetic gene cluster on the chromosome (Fig. 4). Similarly, due to regulatory constraints upon the algD cluster, which contains algA, P. aeruginosa has perhaps maintained functional homologues of AlgA for synthesis of A-band (WbpW) and other unique cell polysaccharides (ORF488 [197]). Similar observations were made in enteric organisms, in which duplicate GDP-d-Man biosynthesis homologues are found within the O-antigen (ManBLPS [PMM activity] and ManCLPS [GMP activity] [4, 96]) and colanic acid (ManBCA, ManCCA [213, 215]) gene clusters. The O-antigen produced by an E. coli O8:K40 strain contains d-Man, while the colanic acid polysaccharide contains l-Fuc, a derivative of GDP-d-Man. As with alginate, colanic acid production has been proven to be environmentally regulated (77). This establishes the need for duplicate enzymes with similar functions within E. coli to allow LPS biosynthesis under various environmental conditions, which lead to the absence of colanic acid synthesis. Interestingly, the gene coding for ManA (PMI activity), like algC, maps outside the O-antigen and colanic acid gene clusters in E. coli and can function independently as a key enzyme for synthesis of both of these cell surface molecules.
Other instances of such enzyme multiplicity are sure to emerge from future analysis of the P. aeruginosa genome sequence. One recent example is the identification of two functional B-band O-antigen chain length regulator homologues, Wzz and Wzz2, within strain PAO1 (22, 161). We are curious to understand the modulation of B-band O-antigen chain length by Wzz and Wzz2. We plan to examine the effect of wzz and wzz2 mutations on serum resistance and the distribution of cell surface proteins in P. aeruginosa, since there is precedence for such effects from studies in other bacteria (19, 93, 224). These studies may help establish a correlation between alterations in B-band O-polymer length during infection and changes in the virulence of the organism.
Many genes encoding proteins involved in synthesis of the B-band O-antigen constituent sugars have been discussed. Biochemical studies to determine their enzymatic properties are under way; however, complexities of the substrates and intermediates of these sugar synthesis pathways make this a formidable task. The conserved nature of WbpM among the P. aeruginosa reference strains and the occurrence of functional homologues in other medically relevant bacteria make WbpM an attractive target for in-depth biochemical studies. Also of interest are the activities of proteins believed to be involved in modification reactions of the O-antigen unit. The presence of such substitutents increases structural diversity among O antigens and influences the immunological properties of LPS molecules (209).
Although preliminary studies have shown that changes in the LPS phenotype affect adherence properties and influence biofilm formation, specific ligands mediating this association have not yet been identified. Future investigations with an extensive panel of defined LPS mutants may help to identify sugar compositions that promote attachment and development of P. aeruginosa biofilms. Transcriptional experiments, similar to those performed for the alginate biosynthetic genes, may also be useful in understanding the regulation of LPS genes during attachment to a substratum. We have discussed various studies in this review that have demonstrated that B-band O antigen is affected by environmental conditions both in vitro and in vivo. A critical point of focus for future work is the identification of the regulatory mechanisms controlling B-band LPS production.
While substantial progress has been achieved in characterizing the pathways for A-band and B-band O-antigen synthesis, many questions remain unanswered. The manner in which these proteins come together within the cell to coordinately synthesize and assemble these O antigens in an ordered manner remains unknown. The process of ligating these O-antigen molecules to core-lipid A is also not yet defined, since no ligase-type protein (WaaL) has been identified within P. aeruginosa. Research focusing on synthesis of the core oligosaccharide molecule is under way, since this region of LPS has recently been implicated in host-bacterium interactions (243). A fuller understanding of the biosynthesis and regulatory networks affecting LPS at the molecular and protein level may assist in the development of a rational therapeutic approach for P. aeruginosa and will undoubtedly contribute to our fundamental knowledge of these cell surface molecules.
ACKNOWLEDGMENTS
This work was supported by grants to J.S.L. from the Canadian Cystic Fibrosis Foundation (CCFF), the Medical Research Council of Canada (grants MT12281 and MT14687), and the Canadian Bacterial Diseases Network (a consortium of the federal Networks of Centres of Excellence program). H.L.R. is a recipient of a CCFF studentship, and L.L.B. is a recipient of a CCFF Kinsmen Fellowship.
We acknowledge our laboratory colleagues past and present who have contributed to the advancement of this field. We thank R. J. Palmer for helpful discussions and collaborative efforts.
ADDENDUM IN PROOF
New information on the P. aeruginosa serotype O11 O-antigen gene cluster became available recently (C. R. Dean, C. V. Franklund, J. D. Retief, M. J. Coyne, Jr., K. Hatano, D. J. Evans, G. B. Pier, and J. B. Goldberg, J. Bacteriol. 181:4275–4284, 1999). The organization of the wbpO11 cluster was very similar to the wbpO5 cluster discussed in this review, and the 5′ region was flanked by the same conserved gene, himD/infB.
REFERENCES
- 1.Alexander D C, Valvano M A. Role of the rfe gene in the biosynthesis of the Escherichia coli O7-specific lipopolysaccharide and other O-specific polysaccharides containing N-acetylglucosamine. J Bacteriol. 1994;176:7079–7084. doi: 10.1128/jb.176.22.7079-7084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen A, Maskell D. The identification, cloning and mutagenesis of a genetic locus required for lipopolysaccharide biosynthesis in Bordetella pertussis. Mol Microbiol. 1996;19:37–52. doi: 10.1046/j.1365-2958.1996.354877.x. [DOI] [PubMed] [Google Scholar]
- 3.Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 4.Amor P A, Whitfield C. Molecular and functional analysis of genes required for expression of group IB K antigens in Escherichia coli: characterization of the his− region containing gene clusters for multiple cell-surface polysaccharides. Mol Microbiol. 1997;26:145–161. doi: 10.1046/j.1365-2958.1997.5631930.x. [DOI] [PubMed] [Google Scholar]
- 5.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Ponten T, Alsmark U C M, Podowski R M, Naeslund A K, Eriksson A S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 6.Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol. 1995;177:1788–1796. doi: 10.1128/jb.177.7.1788-1796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arsenault T L, Huges D W, MacLean D B, Szarek W A, Kropinski A M B, Lam J S. Structural studies on the polysaccharide portion of ‘A-band’ lipopolysaccharide from a mutant (AK14O1) of Pseudomonas aeruginosa strain PAO1. Can J Chem. 1991;69:1273–1280. [Google Scholar]
- 8.Asboe D, Gant V, Aucken H M, Moore D A, Umasankar S, Bingham J S, Kaufmann M E, Pitt T L. Persistence of Pseudomonas aeruginosa strains in respiratory infection in AIDS patients. AIDS. 1998;12:1771–1775. doi: 10.1097/00002030-199814000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Bastin D A, Stevenson G, Brown P K, Haase A, Reeves P R. Repeat unit polysaccharides of bacteria: a model for polymerization resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 10.Bélanger, M., L. L. Burrows, and J. S. Lam. Functional analysis of genes responsible for synthesis of the B-band O-antigen of Pseudomonas aeruginosa serotype O6. Submitted for publication. [DOI] [PubMed]
- 11.Berry A, DeVault J D, Chakrabarty A M. High osmolarity is a signal for enhanced algD transcription in mucoid and nonmucoid Pseudomonas aeruginosa strains. J Bacteriol. 1989;171:2312–2317. doi: 10.1128/jb.171.5.2312-2317.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beveridge T J, Makin S A, Kadurugamuwa J L, Li Z. Interactions between biofilms and the environment. FEMS Microbiol Rev. 1997;20:291–303. doi: 10.1111/j.1574-6976.1997.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 13.Bisercic M, Feutrier J Y, Reeves P R. Nucleotide sequences of the gnd genes from nine natural isolates of Escherichia coli: evidence of intragenic recombination as a contributing factor in the evolution of the polymorphic gnd locus. J Bacteriol. 1991;173:3894–3900. doi: 10.1128/jb.173.12.3894-3900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bliss J M, Silver R P. Coating the surface: a model for expression of capsular polysialic acid in Escherichia coli K1. Mol Microbiol. 1996;21:221–231. doi: 10.1046/j.1365-2958.1996.6461357.x. [DOI] [PubMed] [Google Scholar]
- 15.Bliss J M, Silver R P. Evidence that KpsT, the ATP-binding component of an ATP-binding cassette transporter, is exposed to the periplasm and associates with polymer during translocation of the polysialic acid capsule of Escherichia coli K1. J Bacteriol. 1997;179:1400–1403. doi: 10.1128/jb.179.4.1400-1403.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bronner D, Clarke B R, Whitfield C. Identification of an ATP-binding cassette transport system required for translocation of lipopolysaccharide O-antigen side-chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol Microbiol. 1994;14:505–519. doi: 10.1111/j.1365-2958.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 17.Bryan L E, O’Hara K, Wong S. Lipopolysaccharide changes in impermeability-type aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1984;26:250–255. doi: 10.1128/aac.26.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 19.Burns S M, Hull S I. Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect Immun. 1998;66:4244–4253. doi: 10.1128/iai.66.9.4244-4253.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burrows, L. L., D. Bowdish, A. Kropinski, G. Newton, and J. S. Lam. Analysis of genes involved in acetylation of the O antigens of the O5 serogroup of Pseudomonas aeruginosa. Unpublished data.
- 21.Burrows L L, Charter D F, Lam J S. Molecular characterization of the Pseudomonas aeruginosa serotype O5 (PAO1) B-band lipopolysaccharide gene cluster. Mol Microbiol. 1996;22:481–495. doi: 10.1046/j.1365-2958.1996.1351503.x. [DOI] [PubMed] [Google Scholar]
- 22.Burrows L L, Chow D, Lam J S. Pseudomonas aeruginosa B-band O-antigen chain length is modulated by Wzz (Ro1) J Bacteriol. 1997;179:1482–1489. doi: 10.1128/jb.179.5.1482-1489.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burrows, L. L., and J. S. Lam. 1998. Unpublished data.
- 24.Burrows L L, Lam J S. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Analysis of three glycosyltransferases required for assembly of the Pseudomonas aeruginosa O5 B-band lipopolysaccharide O-antigen unit, abstr. B-400. [Google Scholar]
- 25.Burrows L L, Lam J S. Effect of wzx (rfbX) mutations on A-band and B-band lipopolysaccharide biosynthesis in Pseudomonas aeruginosa serotype O5. J Bacteriol. 1999;181:973–980. doi: 10.1128/jb.181.3.973-980.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burrows, L. L., K. E. Pigeon, R. K. Ernst, S. I. Miller, and J. S. Lam. Role of the AlgD homologues WbpA and Ugd in polysaccharide biosynthesis in Pseudomonas aeruginosa: Ugd is required for aminoarabinose synthesis and polymyxin resistance. Unpublished data.
- 27.Burrows L L, Pigeon K E, Lam J S. Abstracts of the 5th Conference of the International Endotoxin Society 1998. 1998. Analysis of wbpA mutants of Pseudomonas aeruginosa and identification of a wbpA homologue, ugd, in the P. aeruginosa genome, abstr. 88. [Google Scholar]
- 28.Burrows, L. L., H. L. Rocchetta, and J. S. Lam. Assembly pathways for biosynthesis of A-band and B-band lipopolysaccharide in Pseudomonas aeruginosa. In R. Doyle (ed.), Glycomicrobiology, in press. Plenum Press, London, United Kingdom.
- 29.Burrows, L. L., R. Urbanic, and J. S. Lam. Functional conservation of the polysaccharide biosynthetic protein, WbpM, and its homologues in Pseudomonas aeruginosa and other medically significant bacteria. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 30.Burrows, L. L., A. G. Walsh, M. Monteiro, M. Perry, and J. S. Lam. Formation of a serotype O5-like lipopolysaccharide in Pseudomonas aeruginosa continues in the absence of the putative mannosaminuronyltransferase, WbpJ. Unpublished data.
- 31.Caron P R, Kushner S R, Grossman L. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the UvrABC protein complex. Proc Natl Acad Sci USA. 1985;82:4925–4929. doi: 10.1073/pnas.82.15.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerantola S, Montrozier H. Structural elucidation of two polysaccharides present in the lipopolysaccharide of a clinical isolate of Burkholderia cepacia. Eur J Biochem. 1997;246:360–366. doi: 10.1111/j.1432-1033.1997.00360.x. [DOI] [PubMed] [Google Scholar]
- 33.Charter D F, Matewish M J, Burrows L L, Lam J S. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Characterization of Pseudomonas aeruginosa B-band (O-antigen) lipopolysaccharide mutants of strain PAO1, abstr. B-0427. [Google Scholar]
- 34.Chitnis C E, Ohman D E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol. 1993;8:583–590. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 35.Clarke B R, Bronner D, Keenleyside W J, Severn W B, Richards J C, Whitfield C. Role of Rfe and RfbF in the initiation of biosynthesis of d-galactan I, the lipopolysaccharide O antigen from Klebsiella pneumoniae O1. J Bacteriol. 1995;177:5411–5418. doi: 10.1128/jb.177.19.5411-5418.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver S, Osborne J, Quail M A, Rajandream M A, Rogers J, Rutter S, Seeger K, Skelton S, Squares S, Sqares R, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 37.Comstock L E, Johnson J A, Michalski J M, Morris J G, Jr, Kaper J B. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol Microbiol. 1996;19:815–826. doi: 10.1046/j.1365-2958.1996.407928.x. [DOI] [PubMed] [Google Scholar]
- 38.Coyne M J, Jr, Goldberg J B. Cloning and characterization of the gene (rfc) encoding O-antigen polymerase of Pseudomonas aeruginosa PAO1. Gene. 1995;167:81–86. doi: 10.1016/0378-1119(95)00595-1. [DOI] [PubMed] [Google Scholar]
- 39.Coyne M J, Russell K S, Coyle C L, Goldberg J B. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J Bacteriol. 1994;176:3500–3507. doi: 10.1128/jb.176.12.3500-3507.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cryz S J, Jr, Pitt T L, Furer E, Germanier R. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infect Immun. 1984;44:508–513. doi: 10.1128/iai.44.2.508-513.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Curd H, Liu D, Reeves P R. Relationships among the O-antigen gene clusters of Salmonella enterica groups B, D1, D2, and D3. J Bacteriol. 1998;180:1002–1007. doi: 10.1128/jb.180.4.1002-1007.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Currie H L, Lightfoot J, Lam J S. Prevalence of gca, a gene involved in synthesis of A-band common antigen polysaccharide in Pseudomonas aeruginosa. Clin Diagn Lab Immunol. 1995;2:554–562. doi: 10.1128/cdli.2.5.554-562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniels C, Vindurampulle C, Morona R. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy) Mol Microbiol. 1998;28:1211–1222. doi: 10.1046/j.1365-2958.1998.00884.x. [DOI] [PubMed] [Google Scholar]
- 44.Dasgupta T, de Kievit T R, Masoud H, Altman E, Richards J C, Sadovskaya I, Speert D P, Lam J S. Characterization of lipopolysaccharide-deficient mutants of Pseudomonas aeruginosa derived from serotypes O3, O5, and O6. Infect Immun. 1994;62:809–817. doi: 10.1128/iai.62.3.809-817.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dasgupta T, Lam J S. Identification of rfbA, involved in B-band lipopolysaccharide biosynthesis in Pseudomonas aeruginosa serotype O5. Infect Immun. 1995;63:1674–1680. doi: 10.1128/iai.63.5.1674-1680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davies D G, Chakrabarty A M, Geesey G G. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies D G, Geesey G G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol. 1995;59:1181–1186. doi: 10.1128/aem.61.3.860-867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dean C R, Franklund C V, Retief J D, Coyne M J, Hatano K, Evans D J, Pier G B, Goldberg J B. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Sequence analysis of the O-antigen locus from the serogroup O11 Pseudomonas aeruginosa strain PA103, abstr. D-89. [Google Scholar]
- 49.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olsen G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 50.DeFlaun M F, Marshall B M, Kulle E P, Levy S B. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl Environ Microbiol. 1994;60:2637–2642. doi: 10.1128/aem.60.7.2637-2642.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Kievit T R, Dasgupta T, Schweizer H, Lam J S. Molecular cloning and characterization of the rfc gene of Pseudomonas aeruginosa (serotype O5) Mol Microbiol. 1995;16:565–574. doi: 10.1111/j.1365-2958.1995.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 52.de Kievit T R, Lam J S. Isolation and characterization of two genes, waaC (rfaC) and waaF (rfaF), involved in Pseudomonas aeruginosa serotype O5 inner-core biosynthesis. J Bacteriol. 1997;179:3451–3457. doi: 10.1128/jb.179.11.3451-3457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Kievit T R, Staples T, Lam J S. Pseudomonas aeruginosa rfc genes of serotypes O2 and O5 could complement O-polymerase-deficient semi-rough mutants of either serotype. FEMS Microbiol Lett. 1997;147:251–257. doi: 10.1111/j.1574-6968.1997.tb10250.x. [DOI] [PubMed] [Google Scholar]
- 54.Deretic V, Gill J F, Chakrabarty A M. Gene algD coding for GDP-mannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987;169:351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeShazer D, Brett P J, Woods D E. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol. 1998;30:1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x. [DOI] [PubMed] [Google Scholar]
- 56.DeVault J D, Berry A, Misra T K, Darzins A, Chakrabarty A M. Environmental sensory signals and microbial pathogenesis: Pseudomonas aeruginosa infection in cystic fibrosis. Bio/Technology. 1989;7:352–357. [Google Scholar]
- 57.Dmitriev B A, Knirel Y A, Kocharova N A, Kochetkov N K, Stanislavsky E S, Mashilova G M. Somatic antigens of Pseudomonas aeruginosa. The structure of the polysaccharide chain of Ps. aeruginosa O-serogroup 7 (Lanyi) lipopolysaccharide. Eur J Biochem. 1980;106:643–651. doi: 10.1111/j.1432-1033.1980.tb04612.x. [DOI] [PubMed] [Google Scholar]
- 58.Dodgson C, Amor P, Whitfield C. Distribution of the rol gene encoding the regulator of lipopolysaccharide O-chain length in Escherichia coli and its influence on the expression of group I capsular K antigens. J Bacteriol. 1996;178:1895–1902. doi: 10.1128/jb.178.7.1895-1902.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dosanjh A, Lencer W, Brown D, Ausiello D A, Stow J L. Heterologous expression of delta F508 CFTR results in decreased sialylation of membrane glycoconjugates. Am J Physiol. 1994;266:C360–C366. doi: 10.1152/ajpcell.1994.266.2.C360. [DOI] [PubMed] [Google Scholar]
- 60.Drummelsmith, J. 1999. Personal communication.
- 61.Drummelsmith J, Amor P A, Whitfield C. Polymorphism, duplication, and IS1-mediated rearrangement in the chromosomal his-rfb-gnd region of Escherichia coli strains with group IA and capsular K antigens. J Bacteriol. 1997;179:3232–3238. doi: 10.1128/jb.179.10.3232-3238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dudley M N. Overview of gram-negative sepsis. Am J Hosp Pharm. 1990;47:S3–S6. [PubMed] [Google Scholar]
- 63.Engels W, Endert J, Kamps M A F, van Boven C P A. Role of lipopolysaccharide in opsonization and phagocytosis of Pseudomonas aeruginosa. Infect Immun. 1985;49:182–189. doi: 10.1128/iai.49.1.182-189.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Evans D J, Pier G B, Coyne M J, Jr, Goldberg J B. The rfb locus from Pseudomonas aeruginosa strain PA103 promotes the expression of O antigen by both LPS-rough and LPS-smooth isolates from cystic fibrosis patients. Mol Microbiol. 1994;13:427–434. doi: 10.1111/j.1365-2958.1994.tb00437.x. [DOI] [PubMed] [Google Scholar]
- 65.Fallarino A, Mavrangelos C, Stroeher U H, Manning P A. Identification of additional genes required for O-antigen biosynthesis in Vibrio cholerae O1. J Bacteriol. 1997;179:2147–2153. doi: 10.1128/jb.179.7.2147-2153.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fick R B. Pathogenesis of the Pseudomonas lung lesion in cystic fibrosis. Chest. 1989;96:156–164. doi: 10.1378/chest.96.1.158. [DOI] [PubMed] [Google Scholar]
- 68.Fitzsimmons S C. The changing epidemiology of cystic fibrosis. J Pediatr. 1993;122:1–9. doi: 10.1016/s0022-3476(05)83478-x. [DOI] [PubMed] [Google Scholar]
- 69.Flemming C A, Palmer R J, Jr, Arrage A A, Van de Mei H C, White D C. Cell surface physicochemistry alters biofilm development of Pseudomonas aeruginosa lipopolysaccharide mutants. Biofouling. 1998;13:213–231. [Google Scholar]
- 70.Fomsgaard A, Conrad R S, Galanos C, Shand G H, Høiby N. Comparative immunochemistry of lipopolysaccharides from typable and polyagglutinable Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis. J Clin Microbiol. 1988;26:821–826. doi: 10.1128/jcm.26.5.821-826.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Franco A V, Liu D, Reeves P R. A Wzz (Cld) protein determines the chain length of K lipopolysaccharide in Escherichia coli O8 and O9 strains. J Bacteriol. 1996;178:1903–1907. doi: 10.1128/jb.178.7.1903-1907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franco A V, Liu D, Reeves P R. The wzz (cld) protein in Escherichia coli: amino acid sequence variation determines O-antigen chain length specificity. J Bacteriol. 1998;180:2670–2675. doi: 10.1128/jb.180.10.2670-2675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fraser C M, Norris S J, Weinstock G M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, Sodergren E, Hardham J M, McLeod M P, Salzberg S, Peterson J, Khalak H, Richardson D, Howell J K, Chidambaram M, Utterback T, McDonald L, Artiach P, Bowman C, Cotton M D, Fujii C, Garland S, Hatch B, Horst K, Roberts K, Watthey L, Weidman J, Smith H O, Venter J C. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 74.Fry B N, Korolik V, ten Brinke J A, Pennings M T, Zalm R, Teunis B J, Coloe P J, van der Zeijst B A. The lipopolysaccharide biosynthesis locus of Campylobacter jejuni 81116. Microbiology. 1998;144:2049–2061. doi: 10.1099/00221287-144-8-2049. [DOI] [PubMed] [Google Scholar]
- 75.Goldberg J B, Hatano K, Meluleni G S, Pier G B. Cloning and surface expression of Pseudomonas aeruginosa O antigen in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:10716–10720. doi: 10.1073/pnas.89.22.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Goldman R C, Hunt F. Mechanism of O-antigen distribution in lipopolysaccharide. J Bacteriol. 1990;172:5352–5359. doi: 10.1128/jb.172.9.5352-5359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gottesman S, Stout V. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12. Mol Microbiol. 1991;5:1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x. [DOI] [PubMed] [Google Scholar]
- 78.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hanberger H, Hoffmann M, Lindgren S, Nilsson L E. High incidence of antibiotic resistance among bacteria in 4 intensive care units at a university hospital in Sweden. Scand J Infect Dis. 1997;29:607–614. doi: 10.3109/00365549709035904. [DOI] [PubMed] [Google Scholar]
- 80.Hancock R E W. The bacterial outer membrane as a drug barrier. Trends Microbiol. 1997;5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 81.Hancock R E W. Resistance mechanisms in Pseudomonas aeruginosa and other nonfermentative gram-negative bacteria. Clin Infect Dis. 1998;1:S93–S99. doi: 10.1086/514909. [DOI] [PubMed] [Google Scholar]
- 82.Hancock R E W, Mutharia L M, Chan L, Darveau R P, Speert D P, Pier G B. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypeable strains deficient in lipopolysaccharide O side chains. Infect Immun. 1983;42:170–177. doi: 10.1128/iai.42.1.170-177.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hancock R E W, Speert D P. Antibiotics for Pseudomonas and related infections. In: Dodge J A, Brock D J H, Widdicombe J H, editors. Cystic fibrosis: current topics. New York, N.Y: John Wiley & Sons Inc.; 1996. pp. 245–266. [Google Scholar]
- 84.Hancock R E W, Woodruff W A. Roles of porin and beta-lactamase in beta-lactam resistance of Pseudomonas aeruginosa. Rev Infect Dis. 1988;10:770–775. doi: 10.1093/clinids/10.4.770. [DOI] [PubMed] [Google Scholar]
- 85.Hashimoto Y, Li N, Yokoyama H, Ezaki T. Complete nucleotide sequence and molecular characterization of ViaB region encoding Vi antigen in Salmonella typhi. J Bacteriol. 1993;175:4456–4465. doi: 10.1128/jb.175.14.4456-4465.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hatano K, Goldberg J B, Pier G B. Biologic activities of antibodies to the neutral-polysaccharide component of the Pseudomonas aeruginosa lipopolysaccharide are blocked by O side chains and mucoid exopolysaccharide (alginate) Infect Immun. 1995;63:21–26. doi: 10.1128/iai.63.1.21-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hauser A R, Kang P J, Engel J N. PepA, a secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol Microbiol. 1998;27:807–818. doi: 10.1046/j.1365-2958.1998.00727.x. [DOI] [PubMed] [Google Scholar]
- 88.Higgins C F. ABC transporters from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- 89.Houng H S H, Venkatesan M M. Genetic analysis of Shigella sonnei form I antigen: identification of a novel IS630 as an essential element for the form I antigen expression. Microb Pathog. 1998;25:165–173. doi: 10.1006/mpat.1998.0222. [DOI] [PubMed] [Google Scholar]
- 90.Hoyle B D, Williams L J, Costerton J W. Production of mucoid exopolysaccharide during development of Pseudomonas aeruginosa biofilms. Infect Immun. 1993;61:777–780. doi: 10.1128/iai.61.2.777-780.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang J, Schell M. Molecular characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation at a single promoter. Mol Microbiol. 1995;16:977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 92.Ikeno S, Tsuji T, Higashide K, Kinoshita N, Hamada M, Hori M. A 7.6kb DNA region from Streptomyces kasugaensis M338-M1 includes some genes responsible for kasugamycin biosynthesis. J Antibiot. 1998;51:341–352. doi: 10.7164/antibiotics.51.341. [DOI] [PubMed] [Google Scholar]
- 93.Iredell J R, Stroeher U H, Ward H M, Manning P A. Lipopolysaccharide O-antigen expression and the effect of its absence on virulence in rfb mutants of Vibrio cholerae O1. FEMS Immunol Med Microbiol. 1998;20:45–54. doi: 10.1111/j.1574-695X.1998.tb01110.x. [DOI] [PubMed] [Google Scholar]
- 94.Jansson P-E, Lönngren L, Widmalm G. Structural studies of the O-antigen polysaccharides of Klebsiella pneumoniae O5 and Escherichia coli O8. Carbohydr Res. 1985;145:59–66. doi: 10.1016/s0008-6215(00)90412-9. [DOI] [PubMed] [Google Scholar]
- 95.Jarvis W R, Martone W J. Predominant pathogens in hospital infections. J Antimicrob Chemother. 1992;29:S19–S24. doi: 10.1093/jac/29.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 96.Jiang X-M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the rfb (O antigen) gene cluster of Salmonella serovar typhimurium (strain LT2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 97.Joiner K A. Studies on the mechanism of bacteria resistance to complement-mediated killing and on the mechanism of action of bactericidal antibody. Curr Top Microbiol Immunol. 1985;121:99–133. doi: 10.1007/978-3-642-45604-6_6. [DOI] [PubMed] [Google Scholar]
- 98.Kadurugamuwa J L, Lam J S, Beveridge T J. Interaction of gentamicin with the A band and B band lipopolysaccharides of Pseudomonas aeruginosa and its possible lethal effects. Antimicrob Agents Chemother. 1993;37:715–721. doi: 10.1128/aac.37.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata T. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 100.Keenleyside W J, Whitfield C. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J Biol Chem. 1996;271:28581–28592. doi: 10.1074/jbc.271.45.28581. [DOI] [PubMed] [Google Scholar]
- 101.Keenleyside W J, Whitfield C. Genetics and biosynthesis of lipopolysaccharide O antigens. In: Brade H, Morrison D C, Opal S M, Vogel S N, editors. Endotoxin in health and disease, in press. New York, N.Y: Marcel Dekker, Inc.; 1999. pp. 331–358. [Google Scholar]
- 102.Kelly R F, Whitfield C. Clonally diverse rfb gene clusters are involved in expression of a family of related d-galactan O antigens in Klebsiella species. J Bacteriol. 1996;178:5205–5214. doi: 10.1128/jb.178.17.5205-5214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kerppola R E, Shyamala V K, Klebba P, Ames G F-L. The membrane-bound proteins of periplasmic permeases form a complex. J Biol Chem. 1991;266:9857–9865. [PubMed] [Google Scholar]
- 104.Kido N, Morooka N, Paeng N, Ohtani T, Kobayashi H, Shibata N, Okawa Y, Suzuki S, Sugiyama T, Yokochi T. Production of monoclonal antibody discriminating serological differences in Escherichia coli O9 and O9a polysaccharides. Microbiol Immunol. 1997;41:519–525. doi: 10.1111/j.1348-0421.1997.tb01887.x. [DOI] [PubMed] [Google Scholar]
- 105.Kido N, Sugiyama T, Yokochi T, Kobayashi H, Okawa Y. Synthesis of Escherichia coli O9a polysaccharide requires the participation of two domains of WbdA, a mannosyltransferase encoded within the wb* gene cluster. Mol Microbiol. 1998;27:1213–1221. doi: 10.1046/j.1365-2958.1998.00765.x. [DOI] [PubMed] [Google Scholar]
- 106.Kido N, Torgov V I, Sugiyama T, Uchiya K, Sugihara H, Komatsu T, Kato N, Jann K. Expression of the O9 polysaccharide of Escherichia coli: sequencing of the E. coli O9 rfb gene cluster, characterization of mannosyl transferases, and evidence for an ATP-binding cassette transport system. J Bacteriol. 1995;177:2178–2187. doi: 10.1128/jb.177.8.2178-2187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kielhofner M, Atmar R L, Hamill R J, Musher D M. Life-threatening Pseudomonas aeruginosa infections in patients with human immunodeficiency virus infection. Clin Infect Dis. 1992;14:403–411. doi: 10.1093/clinids/14.2.403. [DOI] [PubMed] [Google Scholar]
- 108.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Peterson S, Reich C I, McNeil L K, Badger J H, Glodek A, Zhou L, Overbeek R, Gocayne J D, Weidman J F, McDonald L, Utterback T, Cotton M D, Spriggs T, Artiach P, Kaine B P, Sykes S M, Sadow P W, D’Andrea K P, Bowman C, Fujii C, Garland S A, Mason T M, Olsen G J, Fraser C M, Smith H O, Woese C R, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1998;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 109.Knirel Y A, Kochetkov N K. The structure of lipopolysaccharides of gram-negative bacteria. III. The structure of O-antigens: a review. Biochemistry. 1994;59:1325–1382. [Google Scholar]
- 110.Knirel Y A, Paramonov N A, Vinogradov E V, Shashkov A S, Dmitriev B A, Kochetkov N K, Kholodkova E V, Stanislavsky E S. Somatic antigens of Pseudomonas aeruginosa. The structure of O-specific polysaccharide chains of lipopolysaccharides of P. aeruginosa O3 (Lanyi), O25 (Wokatsch) and Fisher immunotypes 3 and 7. Eur J Biochem. 1987;167:549–561. doi: 10.1111/j.1432-1033.1987.tb13372.x. [DOI] [PubMed] [Google Scholar]
- 111.Knirel Y A, Vinogradov E V, Kocharova N A, Paramonov N A, Kochetkov N K, Dmitriev B A, Stanislavsky E S, Lanyi B. The structure of O-specific polysaccharides and serological classification of Pseudomonas aeruginosa (a review) Acta Microbiol Hung. 1988;35:3–24. [PubMed] [Google Scholar]
- 112.Knirel Y A, Vinogradov E V, Shashkov A S, Dmitriev B A, Kochetkov N K, Stanislavsky E S, Mashilova G M. Somatic antigens of Pseudomonas aeruginosa. The structure of O-specific polysaccharide chains of P. aeruginosa O:3a,b and O:3a,d lipopolysaccharides. Eur J Biochem. 1982;128:81–90. [PubMed] [Google Scholar]
- 113.Knirel Y A, Vinogradov E V, Shashkov A S, Dmitriev B A, Kochetkov N K, Stanislavsky E S, Mashilova G M. Somatic antigens of Pseudomonas aeruginosa. The structure of the O-specific polysaccharide chains of lipopolysaccharides of P. aeruginosa serogroup O4 (Lanyi) and related serotype O6 (Habs) and immunotype 1 (Fisher) Eur J Biochem. 1985;150:541–550. doi: 10.1111/j.1432-1033.1985.tb09055.x. [DOI] [PubMed] [Google Scholar]
- 114.Knirel Y A, Vinogradov E V, Shashkov A A, Dmitriev B A, Kochetkov N K, Stanislavsky E S, Mashilova G M. Somatic antigens of Pseudomonas aeruginosa. The structure of O-specific polysaccharide chains of P. aeruginosa O:3(a),c and O:3a,d,e lipopolysaccharides. Eur J Biochem. 1983;134:289–297. doi: 10.1111/j.1432-1033.1983.tb07564.x. [DOI] [PubMed] [Google Scholar]
- 115.Koch C, Høiby N. Pathogenesis of cystic fibrosis. Lancet. 1993;341:1065–1069. doi: 10.1016/0140-6736(93)92422-p. [DOI] [PubMed] [Google Scholar]
- 116.Kocharova N A, Knirel Y A, Kochetkov N K, Stanislavsky E S. Characterization of a rhamnan derived from preparations of Pseudomonas aeruginosa lipopolysaccharides. Bioorg Khim. 1988;14:701–703. [PubMed] [Google Scholar]
- 117.Kocharova N A, Knirel Y A, Shashkov A S, Kochetkov N K, Pier G B. Structure of an extracellular cross-reactive polysaccharide from Pseudomonas aeruginosa immunotype 4. J Biol Chem. 1988;263:11291–11295. [PubMed] [Google Scholar]
- 118.Koga T, Ishimoto K, Lory S. Genetic and functional characterization of the gene cluster specifying expression of Pseudomonas aeruginosa pili. Infect Immun. 1993;61:1371–1377. doi: 10.1128/iai.61.4.1371-1377.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Köhler T, Michéa-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechère J-C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 120.Korber D R, Lawrence J R, Sutton B, Caldwell D E. Effect of laminar flow velocity on the kinetics of surface recolonization by Mot+ and Mot−Pseudomonas fluorescens. Microb Ecol. 1989;18:1–9. doi: 10.1007/BF02011692. [DOI] [PubMed] [Google Scholar]
- 121.Krivan H C, Ginsburg V, Roberts D D. Pseudomonas aeruginosa and Pseudomonas cepacia isolated from cystic fibrosis patients bind specifically to gangliotetraosylceramide (asialo GM1) and gangliotriaosylceramide (asialo GM2) Arch Biochem Biophys. 1988;260:493–496. doi: 10.1016/0003-9861(88)90473-0. [DOI] [PubMed] [Google Scholar]
- 122.Kropinski, A. M. 1999. Personal communication.
- 123.Kropinski A M, Lewis V, Berry D. Effect of growth temperature on the lipids, outer membrane proteins, and lipopolysaccharides of P. aeruginosa PAO. J Bacteriol. 1987;169:1960–1966. doi: 10.1128/jb.169.5.1960-1966.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kuemmerle N B, Masker W. Effect of the uvrD mutation on the excision repair. J Bacteriol. 1980;142:535–546. doi: 10.1128/jb.142.2.535-546.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kulshin V A, Zahringer U, Lindner B, Jager K E, Dmitriev B A, Rietschel E T. Structural characterization of the lipid A component of Pseudomonas aeruginosa wild-type and rough mutant lipopolysaccharides. Eur J Biochem. 1991;198:697–704. doi: 10.1111/j.1432-1033.1991.tb16069.x. [DOI] [PubMed] [Google Scholar]
- 126.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 127.Kuzio J, Kropinski A M. O-antigen conversion in Pseudomonas aeruginosa PAO1 by bacteriophage D3. J Bacteriol. 1983;155:203–212. doi: 10.1128/jb.155.1.203-212.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lam J, Chan R, Lam K, Costerton J W. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect Immun. 1980;28:546–556. doi: 10.1128/iai.28.2.546-556.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lam J S, Graham L L, Lightfoot J, Dasgupta T, Beveridge T J. Ultrastructural examination of the lipopolysaccharides of Pseudomonas aeruginosa strains and their isogenic rough mutants by freeze-substitution. J Bacteriol. 1992;174:7159–7167. doi: 10.1128/jb.174.22.7159-7167.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lam J S, Handelsman M Y C, Chivers T R, MacDonald L A. Monoclonal antibodies as probes to examine serotype-specific and cross-reactive epitopes of lipopolysaccharides from serotypes O2, O5, and O16 of Pseudomonas aeruginosa. J Bacteriol. 1992;174:2178–2184. doi: 10.1128/jb.174.7.2178-2184.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lam J S, MacDonald L A, Kropinski A M, Speert D P. Characterization of nontypable strains of Pseudomonas aeruginosa from cystic fibrosis patients by means of monoclonal antibodies and SDS-polyacrylamide gel electrophoresis. Serodiagn Immunother Infect Dis. 1988;2:365–374. [Google Scholar]
- 132.Lam J S, MacDonald L A, Lam M Y. Production of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect Immun. 1987;55:2854–2856. doi: 10.1128/iai.55.11.2854-2856.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lam J S, MacDonald L A, Lam M Y, Duchesne L G, Southam G G. Production and characterization of monoclonal antibodies against serotype strains of Pseudomonas aeruginosa. Infect Immun. 1987;55:1051–1057. doi: 10.1128/iai.55.5.1051-1057.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lam, J. S., and R. Palmer. 1998. Unpublished data.
- 135.Lam M Y C, McGroarty E J, Kropinski A M, MacDonald L A, Pedersen S S, Høiby N, Lam J S. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. J Clin Microbiol. 1989;27:962–967. doi: 10.1128/jcm.27.5.962-967.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lamping N, Hoess A, Yu B, Park T C, Kirschning C J, Pfeil D, Reuter D, Wright S D, Herrmann F, Schumann R R. Effects of site-directed mutagenesis of basic residues (Arg 94, Lys 95, Lys 99) of lipopolysaccharide (LPS)-binding protein on binding and transfer of LPS and subsequent immune cell activation. J Immunol. 1996;157:4648–4656. [PubMed] [Google Scholar]
- 137.Lapidus A, Galleron N, Sorokin A, Ehrlich S D. Sequencing and functional annotation of the Bacillus subtilis genes in the 200 kb rrnB-dnaB region. Microbiology. 1997;143:3431–3441. doi: 10.1099/00221287-143-11-3431. [DOI] [PubMed] [Google Scholar]
- 138.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li X Z, Zhang L, Srikumar R, Poole K. β-Lactamase inhibitors are substrates for the multidrug efflux pumps of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1998;42:399–403. doi: 10.1128/aac.42.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lightfoot J. Molecular cloning of genes involved with expression of lipopolysaccharide in Pseudomonas aeruginosa. Ph.D. thesis. Guelph, Ontario, Canada: University of Guelph; 1993. [Google Scholar]
- 141.Lightfoot, J., and J. S. Lam. 1991. Unpublished data.
- 142.Lightfoot J, Lam J S. Molecular cloning of genes involved with expression of A-band lipopolysaccharide, an antigenically conserved form in Pseudomonas aeruginosa. J Bacteriol. 1991;173:5624–5630. doi: 10.1128/jb.173.18.5624-5630.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lightfoot J, Lam J S. Chromosomal mapping, expression and synthesis of lipopolysaccharide in Pseudomonas aeruginosa: a role for guanosine diphospho (GDP)-d-mannose. Mol Microbiol. 1993;8:771–782. doi: 10.1111/j.1365-2958.1993.tb01620.x. [DOI] [PubMed] [Google Scholar]
- 144.Lin W S, Cunneen T, Lee C Y. Sequence analysis and molecular characterization of genes required for the biosynthesis of type 1 capsular polysaccharide in Staphylococcus aureus. J Bacteriol. 1994;176:7005–7016. doi: 10.1128/jb.176.22.7005-7016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lindberg B, Lönngren J, Nimmich W. Structural studies on Klebsiella O group 5 lipopolysaccharides. Acta Chem Scand. 1972;26:2231–2236. doi: 10.3891/acta.chem.scand.26-2231. [DOI] [PubMed] [Google Scholar]
- 146.Linton K J, Jarvis B W, Hutchinson C R. Cloning of the genes encoding thymidine diphosphoglucose 4,6-dehydratase and thymidine diphospho-4-keto-6-deoxyglucose 3,5-epimerase from the erythromycin-producing Saccharopolyspora erythraea. Gene. 1995;153:33–40. doi: 10.1016/0378-1119(94)00809-7. [DOI] [PubMed] [Google Scholar]
- 147.Liu D, Cole R A, Reeves P R. An O-antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J Bacteriol. 1996;178:2102–2107. doi: 10.1128/jb.178.7.2102-2107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu P V. The roles of various fractions of Pseudomonas aeruginosa in its pathogenesis. 3. Identity of the lethal toxins produced in vitro and in vivo. J Infect Dis. 1966;116:481–489. doi: 10.1093/infdis/116.4.481. [DOI] [PubMed] [Google Scholar]
- 149.Liu P V, Matsumoto H, Kusama H, Bergan T. Survey of heat-stable major somatic antigens of Pseudomonas aeruginosa. Int J Syst Bacteriol. 1983;33:256–264. [Google Scholar]
- 150.Lynn W A, Golenbock D T. Lipopolysaccharide antagonists. Immunol Today. 1992;13:271–276. doi: 10.1016/0167-5699(92)90009-V. [DOI] [PubMed] [Google Scholar]
- 151.Mack K, Titball R W. The detection of insertion sequences within the human pathogen Burkholderia pseudomallei which have been identified previously in Burkholderia cepacia. FEMS Microbiol Lett. 1998;162:69–74. doi: 10.1111/j.1574-6968.1998.tb12980.x. [DOI] [PubMed] [Google Scholar]
- 152.Macpherson D F, Manning P A, Morona R. Genetic analysis of the rfbX gene of Shigella flexneri. Gene. 1995;155:9–17. doi: 10.1016/0378-1119(94)00918-i. [DOI] [PubMed] [Google Scholar]
- 153.Makin S A, Beveridge T J. Pseudomonas aeruginosa PAO1 ceases to express serotype-specific lipopolysaccharide at 45°C. J Bacteriol. 1996;178:3350–3352. doi: 10.1128/jb.178.11.3350-3352.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Makin S A, Beveridge T J. The influence of A-band and B-band lipopolysaccharide on the surface characteristics and adhesion of Pseudomonas aeruginosa to surfaces. Microbiology. 1996;142:299–307. doi: 10.1099/13500872-142-2-299. [DOI] [PubMed] [Google Scholar]
- 155.Manning P A, Stroeher U H, Karageorgos L E, Morona R. Putative O-antigen transport genes within the rfb region of Vibrio cholerae O1 are homologous to those for capsule transport. Gene. 1995;158:1–7. doi: 10.1016/0378-1119(95)00124-o. [DOI] [PubMed] [Google Scholar]
- 156.Markovitz A. Isolation of d-talomethylose (6-deoxy-d-talose) and d-rhamnose (6-deoxy-d-mannose) from capsular polysaccharide of a Gram-negative bacterium. J Biol Chem. 1962;237:1767–1775. [PubMed] [Google Scholar]
- 157.Markovitz A. Biosynthesis of guanosine diphosphate d-rhamnose and guanosine diphosphate d-talomethylose from guanosine diphosphate α-d-mannose. J Biol Chem. 1964;239:2091–2098. [PubMed] [Google Scholar]
- 158.Marolda C L, Valvano M A. The GalF protein of Escherichia coli is not a UDP-glucose pyrophosphorylase but interacts with the GalU protein possibly to regulate cellular levels of UDP-glucose. Mol Microbiol. 1996;22:827–840. doi: 10.1046/j.1365-2958.1996.01531.x. [DOI] [PubMed] [Google Scholar]
- 159.Masoud H, Altman E, Richards J C, Lam J S. A general strategy for structural analysis of the oligosaccharide region of lipopolysaccharides. Structure of oligosaccharide component of Pseudomonas aeruginosa IATS serotype O6 mutant R5 rough-type lipopolysaccharide. Biochemistry. 1994;33:10568–10578. doi: 10.1021/bi00201a002. [DOI] [PubMed] [Google Scholar]
- 160.Masoud M, Sadovskaya I, de Kievit T R, Altman E, Richards J C, Lam J S. Structural elucidation of the LPS core region of the O-chain deficient mutant strain A28 from Pseudomonas aeruginosa serotype O6 (IATS) J Bacteriol. 1995;177:6718–6726. doi: 10.1128/jb.177.23.6718-6726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Matewish M, Rocchetta H L, Burrows L L, Rahim R, Pigeon K, Lam J S. Abstracts of the 60th North American Cystic Fibrosis Meeting. 1998. Genomics of Pseudomonas aeruginosa: identification of multiple homologues of proteins required for the biosynthesis of cell-surface polysaccharides, abstr. 374. [Google Scholar]
- 162.May T B, Chakrabarty A M. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 1995;2:151–157. doi: 10.1016/0966-842x(94)90664-5. [DOI] [PubMed] [Google Scholar]
- 163.May T B, Shinabarger D, Boyd A, Chakrabarty A M. Identification of amino acid residues involved in the activity of phosphomannose isomerase-guanosine 5′-diphospho-d-mannose pyrophosphorylase, a bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J Biol Chem. 1994;269:4872–4877. [PubMed] [Google Scholar]
- 164.McGrath B C, Osborn M J. Localization of the terminal steps of O-antigen synthesis in Salmonella typhimurium. J Bacteriol. 1991;173:649–654. doi: 10.1128/jb.173.2.649-654.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McGroarty E J, Rivera M. Growth-dependent alterations in production of serotype-specific and common antigen lipopolysaccharides in Pseudomonas aeruginosa PAO1. Infect Immun. 1990;58:1030–1037. doi: 10.1128/iai.58.4.1030-1037.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Meier U, Mayer H. Genetic location of genes encoding enterobacterial common antigen. J Bacteriol. 1985;163:756–762. doi: 10.1128/jb.163.2.756-762.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P D. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 168.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 169.Monday S R, Schiller N L. Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX. J Bacteriol. 1996;178:625–632. doi: 10.1128/jb.178.3.625-632.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Morona R, van den Bosch L, Manning P A. Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J Bacteriol. 1995;177:1059–1068. doi: 10.1128/jb.177.4.1059-1068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Naide Y, Nikaido H, Makela P H, Wilkinson R G, Stocker B A D. Semi-rough strains of Salmonella. Proc Natl Acad Sci USA. 1965;53:147–153. doi: 10.1073/pnas.53.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nelson K, Selander R K. Intergeneric transfer and recombination of the 6-phosphogluconate dehydrogenase gene (gnd) in enteric bacteria. Proc Natl Acad Sci USA. 1994;91:10227–10231. doi: 10.1073/pnas.91.21.10227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.O’Toole G A, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 175.Palmer R J, Jr, Caldwell D E. A flowcell for the study of plaque removal and regrowth. J Microbiol Methods. 1995;24:171–182. [Google Scholar]
- 176.Payie K G, Clarke A J. Characterization of gentamicin 2′-N-acetyltransferase from Providencia stuartii: its use of peptidoglycan metabolites for acetylation of both aminoglycosides and peptidoglycan. J Bacteriol. 1997;179:4106–4114. doi: 10.1128/jb.179.13.4106-4114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Penketh A, Pitt T, Roberts D, Hodson M E, Batten J C. The relationship of phenotype changes in Pseudomonas aeruginosa to the clinical condition of patients with cystic fibrosis. Am Rev Respir Dis. 1983;127:605–608. doi: 10.1164/arrd.1983.127.5.605. [DOI] [PubMed] [Google Scholar]
- 178.Philippon L N, Naas T, Bouthors A T, Barakett V, Nordmann P. OXA-18, a class D clavulanic acid-inhibited extended-spectrum beta-lactamase from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:2188–2195. doi: 10.1128/aac.41.10.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pier G B, Grout M, Zaidi T S, Goldberg J B. How mutant CFTR may contribute to Pseudomonas aeruginosa infection in cystic fibrosis. Am J Respir Crit Care Med. 1996;154:S175–182. doi: 10.1164/ajrccm/154.4_Pt_2.S175. [DOI] [PubMed] [Google Scholar]
- 180.Pier G B, Grout M, Zaidi T S, Olsen J C, Johnson L G, Yankaskas J R, Goldberg J B. Role of mutant CFTR in hypersusceptibility of cystic fibrosis patients to lung infections. Science. 1996;271:64–67. doi: 10.1126/science.271.5245.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Piers K L, Hancock R E. The interaction of a recombinant cecropin/melittin hybrid peptide with the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1994;12:951–958. doi: 10.1111/j.1365-2958.1994.tb01083.x. [DOI] [PubMed] [Google Scholar]
- 182.Pitt T L. Biology of Pseudomonas aeruginosa in relation to pulmonary infection in cystic fibrosis. J R Soc Med. 1986;79:13–18. [PMC free article] [PubMed] [Google Scholar]
- 183.Poole K, Gotoh N, Tsujimoto H, Zaho Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Nishino T. Overexpression of the mexC-mexD-oprJ operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 184.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Preston M J, Fleiszig S M, Zaidi T S, Goldberg J B, Shortridge V D, Vasil M L, Pier G B. Rapid and sensitive method for evaluating Pseudomonas aeruginosa virulence factors during corneal infections in mice. Infect Immun. 1995;63:3497–3501. doi: 10.1128/iai.63.9.3497-3501.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Quinn J P. Clinical problems posed by multiresistant nonfermenting gram-negative pathogens. Clin Infect Dis. 1998;27:S117–S124. doi: 10.1086/514912. [DOI] [PubMed] [Google Scholar]
- 187.Rather P N, Orosz E, Shaw K J, Hare R, Miller G. Characterization and transcriptional regulation of the 2′-N-acetyltransferase gene from Providencia stuartii. J Bacteriol. 1993;175:6492–6498. doi: 10.1128/jb.175.20.6492-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Reeves P. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- 188a.Reeves, P. Personal communication.
- 189.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 190.Rick P D, Hubbard G L, Barr K. Role of the rfe gene in the synthesis of the O8 antigen in Escherichia coli K-12. J Bacteriol. 1994;176:2877–2884. doi: 10.1128/jb.176.10.2877-2884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Rivera M, Bryan L E, Hancock R E W, McGroarty E J. Heterogeneity of lipopolysaccharides from Pseudomonas aeruginosa: analysis of lipopolysaccharide chain length. J Bacteriol. 1988;170:512–521. doi: 10.1128/jb.170.2.512-521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rivera M, McGroarty E J. Analysis of a common-antigen lipopolysaccharide from Pseudomonas aeruginosa. J Bacteriol. 1989;171:2244–2248. doi: 10.1128/jb.171.4.2244-2248.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Robbins P W, Bray D, Dankert M, Wright A. Direction of chain growth in polysaccharide synthesis. Science. 1967;158:1536–1542. doi: 10.1126/science.158.3808.1536. [DOI] [PubMed] [Google Scholar]
- 194.Rocchetta H L. Molecular analysis of A-band lipopolysaccharide synthesis in Pseudomonas aeruginosa. Ph.D. thesis. Guelph, Ontario, Canada: University of Guelph; 1998. [Google Scholar]
- 195.Rocchetta H L, Burrows L L, Pacan J C, Lam J S. Three rhamnosyltransferases responsible for assembly of the A-band d-rhamnan polysaccharide in Pseudomonas aeruginosa: a fourth transferase, WbpL, is required for the initiation of both A- band and B-band lipopolysaccharide synthesis. Mol Microbiol. 1998;28:1103–1119. doi: 10.1046/j.1365-2958.1998.00871.x. [DOI] [PubMed] [Google Scholar]
- 196.Rocchetta H L, Lam J S. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J Bacteriol. 1997;179:4713–4724. doi: 10.1128/jb.179.15.4713-4724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rocchetta H L, Pacan J C, Lam J S. Synthesis of the A-band polysaccharide sugar d-rhamnose requires Rmd and WbpW: identification of multiple AlgA homologues, WbpW and ORF488, in Pseudomonas aeruginosa. Mol Microbiol. 1998;29:1419–1434. doi: 10.1046/j.1365-2958.1998.01024.x. [DOI] [PubMed] [Google Scholar]
- 198.Sadovskaya, I., E. Altman, J. C. Richards, and J. S. Lam. 1998. Unpublished data.
- 199.Sadovskaya I, Brisson J R, Lam J S, Richards J C, Altman E. Structural elucidation of the lipopolysaccharide core regions of the wild-type strain PAO1 and O-chain-deficient mutant strains AK1401 and AK1012 from Pseudomonas aeruginosa serotype O5. Eur J Biochem. 1998;255:673–684. doi: 10.1046/j.1432-1327.1998.2550673.x. [DOI] [PubMed] [Google Scholar]
- 200.Sau S, Bhasin N, Wann E R, Lee J C, Foster T J, Lee C Y. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology. 1997;143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 201.Sau S, Lee C Y. Cloning of type 8 capsule genes and analysis of gene clusters for the production of different capsular polysaccharides in Staphylococcus aureus. J Bacteriol. 1996;178:2118–2126. doi: 10.1128/jb.178.7.2118-2126.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sawada S T, Kawamura, Masuho Y, Tomibe K. A new common polysaccharide antigen of strains of Pseudomonas aeruginosa detected with a monoclonal antibody. J Infect Dis. 1985;152:1290–1299. doi: 10.1093/infdis/152.6.1290. [DOI] [PubMed] [Google Scholar]
- 203.Schnaitman C A, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Senchenkova S N, Shashkov A S, Knirel Y A, McGovern J J, Moran A P. The O-specific polysaccharide chain of Campylobacter fetus serotype B lipopolysaccharide is a d-rhamnan terminated with 3-O-methyl-d-rhamnose (d-acofriose) Eur J Biochem. 1996;239:434–438. doi: 10.1111/j.1432-1033.1996.0434u.x. [DOI] [PubMed] [Google Scholar]
- 205.Shibaev V L. Biosynthesis of bacterial polysaccharide chains composed of repeating units. Adv Carbohydr Chem Biochem. 1986;44:277–339. doi: 10.1016/s0065-2318(08)60080-3. [DOI] [PubMed] [Google Scholar]
- 206.Shinabarger D, Berry A, May T B, Rothmel R, Fialho A, Chakrabarty A M. Purification and characterization of phosphomannose isomerase-guanosine diphospho-d-mannose pyrophosphorylase—a bifunctional enzyme in the alginate biosynthetic pathway of Pseudomonas aeruginosa. J Biol Chem. 1991;266:2080–2088. [PubMed] [Google Scholar]
- 207.Skurnik M, Venho R, Toivanen P, al-Hendy A. A novel locus of Yersinia enterocolitica serotype O:3 involved in lipopolysaccharide outer core biosynthesis. Mol Microbiol. 1995;17:575–594. doi: 10.1111/j.1365-2958.1995.mmi_17030575.x. [DOI] [PubMed] [Google Scholar]
- 208.Slauch J M, Lee A A, Mahan M J, Mekalanos J J. Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O-antigen: oafA is a member of a family of integral membrane trans-acylases. J Bacteriol. 1996;178:5904–5909. doi: 10.1128/jb.178.20.5904-5909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Slauch J M, Mahan M J, Michetti P, Neutra M R, Mekalanos J J. Acetylation (O-factor 5) affects the structural and immunological properties of Salmonella typhimurium lipopolysaccharide O antigen. Infect Immun. 1995;63:437–441. doi: 10.1128/iai.63.2.437-441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nolling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Smith R W, Zamze S E, Munro S M, Carter K J, Hignett R C. Structure of the sidechain of lipopolysaccharide from Pseudomonas syringae pv morsprunorum C28. Eur J Biochem. 1985;149:73–78. doi: 10.1111/j.1432-1033.1985.tb08895.x. [DOI] [PubMed] [Google Scholar]
- 212.Stanislavsky E S, Lam J S. Pseudomonas aeruginosa antigens as potential vaccines. FEMS Microbiol Rev. 1997;21:243–277. doi: 10.1111/j.1574-6976.1997.tb00353.x. [DOI] [PubMed] [Google Scholar]
- 213.Stevenson G, Andrianopoulos K, Hobbs M, Reeves P R. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Stevenson G, Kessler A, Reeves P R. A plasmid-borne O-antigen chain length determinant and its relationship to other chain length determinants. FEMS Microbiol Lett. 1995;125:23–30. doi: 10.1111/j.1574-6968.1995.tb07330.x. [DOI] [PubMed] [Google Scholar]
- 215.Stevenson G, Lee S J, Romana L K, Reeves P R. The cps gene cluster of Salmonella strain LT2 includes a second mannose pathway: sequence of two genes and relationship to genes in the rfb gene cluster. Mol Gen Genet. 1991;227:173–180. doi: 10.1007/BF00259668. [DOI] [PubMed] [Google Scholar]
- 216.Stroeher U H, Manning P A. Vibrio cholerae serotype O139: swapping genes for surface polysaccharide biosynthesis. Trends Microbiol. 1997;5:178–180. doi: 10.1016/s0966-842x(97)85010-x. [DOI] [PubMed] [Google Scholar]
- 217.Stroeher U H, Parasivam G, Dredge B K, Manning P A. Novel Vibrio cholerae O139 genes involved in lipopolysaccharide biosynthesis. J Bacteriol. 1997;179:2740–2747. doi: 10.1128/jb.179.8.2740-2747.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Szabo M, Bronner D, Whitfield C. Relationships between rfb gene clusters required for biosynthesis of identical d-galactose-containing O antigens in Klebsiella pneumoniae serotype O1 and Serratia marcescens serotype O16. J Bacteriol. 1995;177:1544–1553. doi: 10.1128/jb.177.6.1544-1553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Takagi M, Takada H, Imanaka T. Nucleotide sequence and cloning in Bacillus subtilis of the Bacillus stearothermophilus pleiotropic regulatory gene degT. J Bacteriol. 1990;172:411–418. doi: 10.1128/jb.172.1.411-418.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Tang H B, Dimango E, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.The International Pseudomonas aeruginosa Typing Study Group. A multicenter comparison of methods for typing strains of Pseudomonas aeruginosa predominantly from patients with cystic fibrosis. J Infect Dis. 1994;169:134–142. doi: 10.1093/infdis/169.1.134. [DOI] [PubMed] [Google Scholar]
- 222.Tobias P S, Soldau K, Ulevitch R J. Isolation of a lipopolysaccharide-binding acute phase reactant from rabbit serum. J Exp Med. 1986;164:777–793. doi: 10.1084/jem.164.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Venter J C, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 224.Van den Bosch L, Manning P A, Morona R. Regulation of O-antigen chain length is required for Shigella flexneri virulence. Mol Microbiol. 1997;23:765–775. doi: 10.1046/j.1365-2958.1997.2541625.x. [DOI] [PubMed] [Google Scholar]
- 225.Vinogradov E V, Shashkov A S, Knirel Y A, Zdorovenko G M, Solyanik L P, Gvozdyak R I. Somatic antigens of pseudomonads: structure of the O-specific polysaccharide chain of Pseudomonas syringae pv. syringae (cerasi) 435 lipopolysaccharide. Carbohydr Res. 1991;212:295–299. doi: 10.1016/0008-6215(91)84069-q. [DOI] [PubMed] [Google Scholar]
- 226.Wang L, Liu D, Reeves P R. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J Bacteriol. 1996;178:2598–2604. doi: 10.1128/jb.178.9.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ward C K, Lumbley S R, Latimer J L, Cope L D, Hansen E J. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J Bacteriol. 1998;180:6013–6022. doi: 10.1128/jb.180.22.6013-6022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Weisgerber C, Jann K. Glucosyldiphosphoundecaprenol, the mannose acceptor in the synthesis of the O9 antigen of Escherichia coli. Eur J Biochem. 1982;127:165–168. doi: 10.1111/j.1432-1033.1982.tb06851.x. [DOI] [PubMed] [Google Scholar]
- 229.West S E H, Iglewski B H. Codon usage in Pseudomonas aeruginosa. Nucleic Acids Res. 1988;16:9323–9335. doi: 10.1093/nar/16.19.9323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 231.Whitfield C, Amor P A, Koplin R. Modulation of the surface architecture of Gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol Microbiol. 1997;23:629–638. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]
- 232.Wilkinson S G. Composition and structure of lipopolysaccharides from Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:S941–S949. doi: 10.1093/clinids/5.supplement_5.s941. [DOI] [PubMed] [Google Scholar]
- 233.Willison J C, Tissot G. The Escherichia coli efg gene and the Rhodobacter capsulatus adgA gene code for NH3-dependent NAD synthetase. J Bacteriol. 1994;176:3400–3402. doi: 10.1128/jb.176.11.3400-3402.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Winkler N W, Markovitz A. Guanosine diphosphate-4-keto-d-rhamnose reductase. A non-steroselective enzyme. J Biol Chem. 1971;246:5868–5876. [PubMed] [Google Scholar]
- 235.Winn A M, Wilkinson S G. The O7 antigen of Stenotrophomonas maltophilia is a linear d-rhamnan with a trisaccharide repeating unit that is also present in polymers for some Pseudomonas and Burkholderia species. FEMS Microbiol Lett. 1998;166:57–61. doi: 10.1111/j.1574-6968.1998.tb13183.x. [DOI] [PubMed] [Google Scholar]
- 236.Wood M S, Byrne A, Lessie T G. IS406 and IS407, two gene-activating insertion sequences for Pseudomonas cepacia. Gene. 1991;105:101–105. doi: 10.1016/0378-1119(91)90519-h. [DOI] [PubMed] [Google Scholar]
- 237.Wright A, Dankert M, Fennessey P, Robbins P W. Characterization of a polyisoprenoid compound functional in O-antigen biosynthesis. Proc Natl Acad Sci USA. 1967;57:1798–1803. doi: 10.1073/pnas.57.6.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wright K C. Cystic fibrosis and the pseudomonads. Br J Biomed Sci. 1996;53:140–145. [PubMed] [Google Scholar]
- 239.Wright S D, Ramos R A, Tobias P S, Ulevitch R J, Mathison J C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990;249:1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 240.Ye R W, Zielinski N A, Chakrabarty A M. Purification and characterization of phosphomannomutase/phosphoglucomutase from Pseudomonas aeruginosa involved in biosynthesis of both alginate and lipopolysaccharide. J Bacteriol. 1994;176:4851–4857. doi: 10.1128/jb.176.16.4851-4857.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yeung A T, Mattes W B, Oh E Y, Grossman L. Enzymatic properties of the purified Escherichia coli UvrABC complex. In: Friedberg E C, Bridges B A, editors. Cellular responses to DNA damage. New York, N.Y: Alan R. Liss, Inc.; 1983. pp. 77–168. [Google Scholar]
- 242.Yokota S, Kaya S, Sawada S, Kawamura T, Araki Y, Ito E. Characterization of a polysaccharide component of lipopolysaccharide from Pseudomonas aeruginosaIID 1008 (ATCC 27584) as d-rhamnan. Eur J Biochem. 1987;167:203–209. doi: 10.1111/j.1432-1033.1987.tb13324.x. [DOI] [PubMed] [Google Scholar]
- 243.Zaidi T S, Fleiszig S M J, Preston M J, Goldberg J B, Pier G B. Lipopolysaccharide outer core is a ligand for corneal cell binding and ingestion of Pseudomonas aeruginosa. Investig Ophthalmol Visual Sci. 1996;37:976–986. [PubMed] [Google Scholar]
- 244.Zhang L, Al-Hendy A, Toivanen P, Skurnik M. Genetic organization and sequence of the rfb gene cluster of Yersinia enterocolitica serotype O:3: similarities to the dTDP-l-rhamnose biosynthesis pathway of Salmonella and to the bacterial polysaccharide transport systems. Mol Microbiol. 1993;9:309–321. doi: 10.1111/j.1365-2958.1993.tb01692.x. [DOI] [PubMed] [Google Scholar]
- 245.Zielinski N A, Chakrabarty A M, Berry A. Characterization and regulation of the Pseudomonas aeruginosa algC gene encoding phosphomannomutase. J Biol Chem. 1991;266:9754–9763. [PubMed] [Google Scholar]