Abstract
Background
Abnormal methylation of N6-methyladenosine (m6A) is reportedly associated with central nervous system disorders. However, the role of m6A mRNA methylation in unconjugated bilirubin (UCB) neurotoxicity requires further research.
Methods
Rat pheochromocytoma PC12 cells treated with UCB were used as in vitro models. After the PC12 cells were treated with UCB (0, 12, 18, and 24 µM) for 24 h, the total RNA m6A levels were measured using an m6A RNA methylation quantification kit. The expression of m6A demethylases and methyltransferases was detected through western blotting. We determined the m6A mRNA methylation profile in PC12 cells exposed to UCB (0 and 18 µM) for 24 h using methylated RNA immunoprecipitation sequencing (MeRIP-seq).
Results
Compared with the control group, UCB (18 and 24 µM) treatment decreased the expression of the m6A demethylase ALKBH5 and increased the expression of the methyltransferases METTL3 and METTL14, which resulted in an increase in the total m6A levels in PC12 cells. Furthermore, 1533 m6A peaks were significantly elevated and 1331 peaks were reduced in the UCB (18 µM)-treated groups compared with those in the control group. Genes with differential m6A peaks were mainly enriched in protein processing in the endoplasmic reticulum, ubiquitin-mediated proteolysis, cell cycle, and endocytosis. Through combined analysis of the MeRIP-seq and RNA sequencing data, 129 genes with differentially methylated m6A peaks and differentially expressed mRNA levels were identified.
Conclusion
Our study suggests that the modulation of m6A methylation modifications plays a significant role in UCB neurotoxicity.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11033-023-08576-1.
Keywords: N6-methyladenosine, Methylated RNA immunoprecipitation sequencing, Methylation, mRNA profile, Neurotoxicity, Unconjugated bilirubin
Introduction
Hyperbilirubinemia is commonly present in newborn babies. Approximately one in ten neonates develop hyperbilirubinemia in the first 2 weeks of life [1]. Abnormal accumulation of unconjugated bilirubin (UCB) in the blood of newborns can lead to bilirubin crossing the blood–brain barrier and causing brain damage. UCB neurotoxicity can cause irreversible damage to specific regions of the brain, including the globus pallidus, hippocampus, and subthalamic nucleus, and its sequelae mainly include learning disabilities, movement disorders, mental retardation, and cerebral palsy [2]. At present, the potential mechanisms of bilirubin-induced nerve cell damage mainly include oxidative stress [3], apoptosis [4], endoplasmic reticulum (ER) stress [5–7], autophagy [8], and neuroinflammation [9]. However, the fundamentals of bilirubin neurotoxicity are not completely understood and require further elucidation.
Epigenetic modifications play a significant role in UCB neurotoxicity. DNA methylation and histone modifications are also involved in UCB neurotoxicity [10, 11]. DNA methylation may play a critical role in bilirubin-induced neuronal injury. In vitro exposure to high UCB concentrations led to changes in methylation levels of genes in neuronal cells of neonatal Sprague–Dawley rats, and these levels were negatively correlated with gene expression levels [10]. H3K14 acetylation is reportedly involved in bilirubin-induced cerebellar hypoplasia [11]. Currently, the study of epigenetics in bilirubin-induced nerve cell injury is limited to DNA-level epigenomics, whereas the correlation between RNA epigenomics and bilirubin-induced nerve cell injury has not been reported.
N6-methyladenosine (m6A) is one of the most abundant and prevalent chemical modifications, especially in eukaryotic mRNA and long noncoding RNAs [12] m6A is mainly localized in the 3′-untranslated region (UTR) and coding sequence (CDS) of mRNA, and its common sequence is RRACH (R = G or A; H = A, C, or U) [13]. m6A regulates the posttranscriptional processing of mRNAs by affecting splicing, nucleation, degradation, and translation. This modification is regulated by methyltransferases (“writers”) and demethylases (“erasers”). “Writers” are complexes composed of various components, including the RNA methylase m6A methyltransferase 3 (METTL3), METTL14, WTAP, and RBM15, among which the most critical molecule with catalytic active domains is METTL3 [14, 15]. “Erasers” include FTO and the RNA demethylase alkB homolog 5 (ALKBH5), which demethylate m6A residues. Therefore, the combined action of “writers” and “erasers” makes m6A modification a dynamic and reversible process.
In the mammalian brain, m6A modification is essential for cortical neurogenesis [16], cerebellar development [17], and adult neural stem cell proliferation and differentiation [15]. However, abnormal m6A methylation is associated with the pathogenesis of central nervous system (CNS) diseases. For instance, changes in m6A modification are not only involved in chronic CNS diseases, such as Alzheimer’s disease [18], Parkinson’s disease [19], and depression [20], but also in acute brain injuries, such as traumatic brain injury [21] and stroke [22]. However, whether m6A RNA methylation plays a role in bilirubin neurotoxicity remains unknown.
To investigate the effect of m6A RNA methylation in UCB neurotoxicity, we evaluated the changes in the m6A mRNA methylation profile in PC12 cells exposed to UCB using methylated RNA immunoprecipitation (IP) sequencing (MeRIP-seq). We further analyzed the combined MeRIP-seq and RNA sequencing (RNA-seq) data. Herein, we report, for the first time, UCB-induced changes in the transcriptome m6A profile in neural cells and provide a fundamental basis for further studies.
Methods
Cell culture and treatment
PC12 cells (Wuhan Boster Biological Technology, Ltd., Wuhan, China) were isolated from the pheochromocytoma of the rat adrenal medulla. Cells were cultured in Dulbecco’s modified Eagle medium (Gibco, Carlsbad, CA, USA) supplemented with fetal bovine serum (Gibco, Carlsbad, CA, USA; 10%) and penicillin–streptomycin (1%). For the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cell viability assay (F5655; Sigma-Aldrich, St. Louis, MO, USA; Fig. S1), cells were exposed to various concentrations of bilirubin (0, 12, 18, and 24 µM) for 24 h, and the total m6A levels and protein expression of RNA demethylases and methylases were determined. For MeRIP- and RNA-seq, cells were exposed to 0 and 18 µM bilirubin in triplicate to analyze m6A epitranscriptomic and RNA transcriptomic alterations.
Analysis of total m6A levels
Total RNA m6A levels of the bilirubin and control groups were measured using an m6A RNA methylation quantification kit (P-9005; EpiGentek, Farmingdale, NY, USA). Briefly, the RNA-binding reagent was mixed with the total RNA extracted from the cells. Subsequently, the capture and detection antibodies were used to measure the m6A signal, which was enhanced using the enhancement solution. The final absorbance was measured at 450 nm to calculate the amount of m6A as per the manufacturer’s instructions.
MeRIP-seq and RNA-seq
RNA- and MeRIP-seq experiments were performed by Guangzhou Epigenetic Biotechnology Co., Ltd. (Guangzhou, China). Briefly, total RNA was extracted using the TRIzol™ reagent (Invitrogen, Waltham, MA, USA) for subsequent experiments. Thereafter, mRNA was isolated from the total RNA samples and digested into 100–200-nucleotide fragments. Methylated RNA IP was performed using the Epi™ m6A IP kit (R1804; Epibiotek, Guangzhou, China). The partial nucleotide fragments were used as an input, and both the input and m6A IP samples were fragmented to a size of 150 bp. Illumina NovaSeq 6000 sequencer (Illumina, San Diego, CA, USA) was used to perform the sequencing.
The quality of the mRNA sequencing data was verified using FastQC (v0.11.7), and differential m6A peaks (P < 0.05) between the bilirubin and control groups were analyzed using the exomePeak R package (v2.13.2). Motifs of the m6A peak sequences were determined using HOMER. Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed using the respective databases.
Western blotting
A radioimmunoprecipitation assay lysis buffer (Beyotime, Beijing, China) was supplemented with phenylmethylsulfonyl fluoride (Phygene, Fuzhou, China) and 1% protease inhibitor (MedChemExpress, NJ, USA) to extract the proteins from the PC12 cells. Then, the protein supernatant was mixed with 5× loading buffer and denatured at 95 °C. Proteins of different molecular weights were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to 0.2 μm polyvinylidene fluoride (PVDF) membranes. Next, proteins were blocked with 5% non-fat milk for 60 min. PVDF membranes with protein layers were incubated overnight at 4 °C with primary antibodies against METTL3 (A8307; ABclonal, Wuhan, China) (1:1 000), METTL14 (A8530; ABclonal) (1:1 000), ALKBH5 (ab195377; Abcam, Cambridge, MA, USA) (1:2 000), and FTO (A1438; ABclonal) (1:1 000). The PVDF membranes were then incubated with secondary antibodies (AS014; ABclonal) at 37 °C for 60 min. Image J software (v. 1.53) was used to quantify the grayscale of the protein bands.
Real-time quantitative PCR (RT-qPCR)
The sequences of primers to amplify the regions of the ALKBH5, FTO, METTL3, and METTL14 genes, with the GAPDH gene was as an internal control gene, were designed and synthesized by Sangon Bioengineering (Shanghai) Co., LTD and are shown in Table S1. PC12 cells were treated with bilirubin (0, 12, 18, and 24 µM) for 24 h. After that, RNA was extracted, cDNA was synthesized, and RT-qPCR reaction was conducted using the SYBR Green master mix (TaKaRa Biotech, Tokyo, Japan) based on the instructions from the supplier. Data were analyzed using the 2−ΔCt method.
Statistical analysis
Data were analyzed using the GraphPad Prism V8.0 software (GraphPad Software, San Diego, CA, USA). Data from the western blot analyses are presented as the mean ± standard error of the mean. At least three separate experiments were performed for each study group, and the results were analyzed using one-way analysis of variance. P < 0.05 indicates that the difference is significant.
Results
UCB induced the alteration of total m6A levels in PC12 cells resulting from the changed expression of the m6A methyltransferase and demethylase
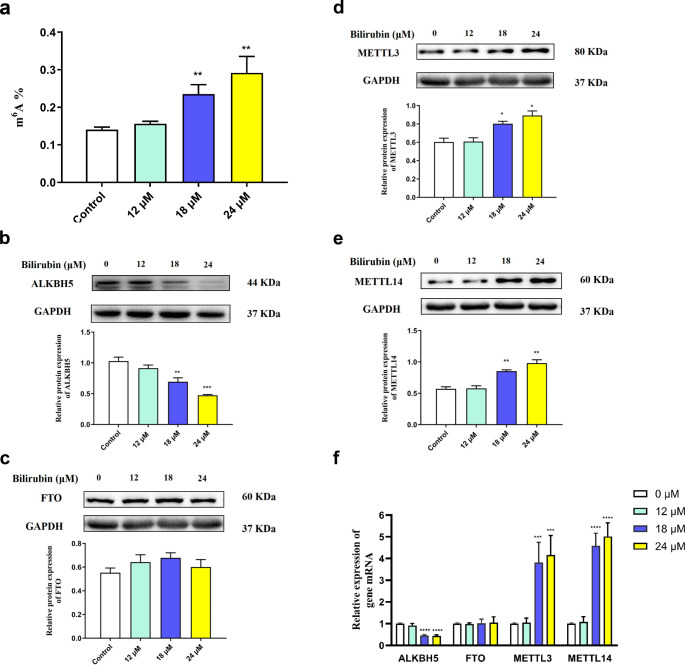
Changes in the total m6A levels were measured in PC12 cells exposed to 12, 18, and 24 µM bilirubin for 24 h. We found that the total m6A levels in the experimental groups treated with 18 and 24 µM bilirubin were significantly higher than those in the control group (P < 0.05) (Fig. 1a). To elucidate the cause of hypermethylation in the bilirubin-treated groups, the levels of m6A methylases and demethylases were determined by performing western blotting. The results showed that the expression of the m6A modified demethylase ALKBH5 (Fig. 1b) significantly decreased and that of the m6A modified methyltransferases METTL3 (Fig. 1c) and METTL14 (Fig. 1d) increased after treatment with 18 or 24 µM bilirubin. However, the expression of the m6A demethylase FTO (Fig. 1e) remained unchanged. These results suggest that bilirubin exposure can promote m6A mRNA hypermethylation in PC12 cells. In addition, compared to that in the control group, the expression of the ALKBH5 mRNA significantly decreased in the PC12 cells when treated with 18 or 24 µM bilirubin, while the expression of the METTL3 and METTL14 mRNA increased (Fig. 1f).
Fig. 1.
Effect of m6A methylation modification in PC12 cells exposed to bilirubin. (a) Total m6A level. (b, c) The protein expression of the RNA demethylases ALKBH5 and FTO. (d, e) The protein expression of the RNA methylases METTL3 and METTL14. (f) The relative expression of ALKBH5, FTO, METTL14, and METTL3 mRNA. Data are presented as the mean ± SD, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.000 compared to the control group
Differential m6A modification profiles in PC12 cells
Based on the results of the total m6A levels and protein expression of RNA demethylases and methylases, PC12 cells were exposed to 0 and 18 µM bilirubin in triplicate to analyze m6A epitranscriptomic alterations through MeRIP- and RNA-seq.
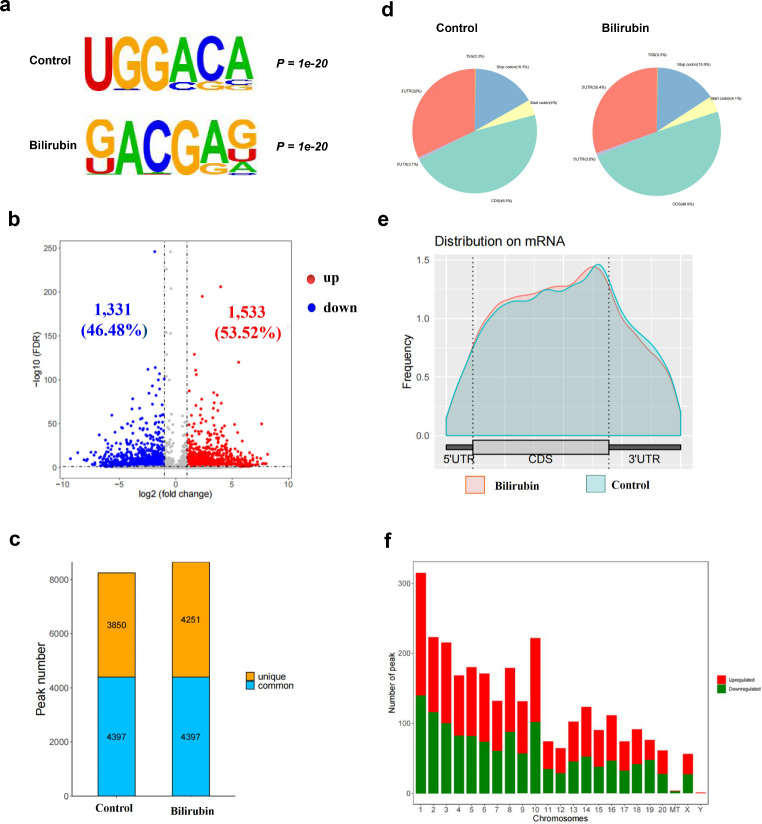
m6A modification was characterized using motif analysis. The consensus motif of the m6A peaks was identified to be G[G]AC[GA] (Fig. 2a), which was consistent with the specific motif of m6A methylation sites. The results suggested that 1533 m6A peaks were significantly elevated and 1331 peaks were reduced in the bilirubin-treated groups compared with those in the control group (fold change ≥ 1 and P < 0.05; Fig. 2b). The m6A methylation level of 10 methylated genes and 10 demethylated genes, listed in Table 1, was significantly altered under treatment with bilirubin in accordance with diff.log2.fc. In the control group, 8247 m6A peaks were recognized, among which 3850 were unique and 4397 were shared with the bilirubin group. Approximately 8600 m6A peaks were identified in the bilirubin group, among which 4251 were unique (Fig. 2c).
Fig. 2.
Differential m6A modification profiles in PC12 cells. (a) The sequence motif of m6A-containing peak regions in the bilirubin and control groups. (b) Volcano plots of differentially methylated mRNAs in the bilirubin group compared with those in the control group. (c) Unique and common peak numbers in the bilirubin and control groups. (d) The proportion of m6A peaks located at the mRNA transcripts. (e) The densities of m6A peaks in the bilirubin and control groups were compared to those in the 5′-UTR, CDS, and 3′-UTR regions. (f) Changes in the m6A peak quantity in different chromosomes after bilirubin treatment
Table 1.
Top 20 genes with altered m6A peaks
| Gene name | chromosome | Peak star | Peak End | lg.p | diff.log2.fc | Regulation |
|---|---|---|---|---|---|---|
| Jak1 | 5 | 119,984,644 | 119,985,175 | -6.01 | -9.32 | down |
| Smad4 | 18 | 69,628,141 | 69,634,083 | -10.1 | -8.68 | down |
| Gpsm2 | 2 | 211,513,403 | 211,513,582 | -2.96 | -8.12 | down |
| Mobp | 8 | 128,841,916 | 128,842,306 | -9.3 | -7.91 | down |
| Sbno1 | 12 | 37,627,019 | 37,628,226 | -2.79 | -7.82 | down |
| Matr3 | 18 | 28,371,575 | 28,371,755 | -12.1 | -7.51 | down |
| Nup155 | 2 | 57,250,191 | 57,251,998 | -6.67 | -7.39 | down |
| Plcg1 | 3 | 156,747,621 | 156,748,564 | -17.8 | -7.24 | down |
| Usp39 | 4 | 100,187,353 | 100,190,182 | -8.38 | -7.23 | down |
| Pygb | 3 | 146,611,545 | 146,612,871 | -6.55 | 7.17 | up |
| Slc38a2 | 7 | 138,089,664 | 138,089,875 | -24.1 | 7.19 | up |
| Ddx1 | 6 | 38,423,066 | 38,423,906 | -13.6 | 7.23 | up |
| Bcar1 | 19 | 43,935,829 | 43,936,009 | -4.68 | 7.29 | up |
| Pdgfrb | 18 | 56,381,091 | 56,381,212 | -2.93 | 7.52 | up |
| Myo9b | 16 | 19,745,273 | 19,746,952 | -14.8 | 7.61 | up |
| Nhlrc2 | 1 | 277,409,493 | 277,409,703 | -4.94 | 7.67 | up |
| Cdk12 | 10 | 86,157,936 | 86,158,116 | -8.62 | 7.81 | up |
| Rspry1 | 19 | 10,812,736 | 10,812,944 | -9.3 | 7.91 | up |
| Zhx3 | 3 | 156,776,897 | 156,777,106 | -13 | 8.01 | up |
| Dhrs9 | 3 | 55,647,252 | 55,647,491 | -3.97 | 8.13 | up |
In general, m6A peaks in the control and bilirubin groups were distributed in the start codon, CDS, stop codon, and 3′-UTR and were mainly located in the CDS near the 3′-UTR. Compared with that in the control group, in the bilirubin group, the distribution of m6A peaks was increased in the CDS of mRNAs and decreased in the 3′-UTRs (Fig. 2d and e). Bilirubin also increased the number of m6A modification peaks in the CDS region.
Functional analysis of differentially m6A methylated mRNAs in PC12 cells
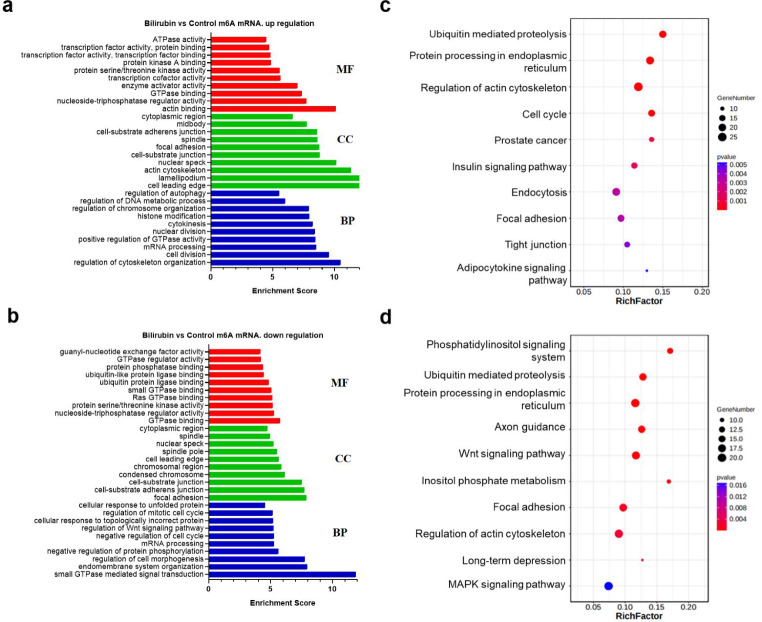
To determine which molecular signaling pathways and functions associated with neurotoxicity were affected by bilirubin, the differentially methylated transcripts were analyzed using GO functional and KEGG pathway analyses. GO enrichment analysis revealed that the genes with differentially m6A methylated peaks were involved in cytokinesis, cell division, mRNA processing, endomembrane system organization, transcription, protein kinase enzymes, and GTPase binding. Further analysis revealed the top 10 significantly enriched biological processes (BPs), cellular components (CCs), and molecular functions (MFs) of the genes with increased m6A peaks, which are shown in Fig. 3a, while GO analysis of genes with decreased m6A peaks is shown in Fig. 3b.
Fig. 3.
Functional analysis of differentially m6A-modified mRNAs in PC12 cells between the bilirubin and control groups. (a) GO terms of genes with upregulated m6A peaks. (b) GO terms of genes with downregulated m6A peaks. (c) KEGG pathway analyses of genes with upregulated m6A peaks. (d) KEGG pathway analyses of genes with downregulated m6A peaks
For the KEGG pathway analysis, we identified 28 and 30 pathways that involved the genes with upregulated and downregulated m6A methylated peaks, respectively. The genes with elevated m6A peaks in the bilirubin group were significantly correlated with ubiquitin-mediated proteolysis, protein processing in the ER, cell cycle, and endocytosis (Fig. 3c). The genes with reduced peaks were significantly related to the phosphatidylinositol signaling system, protein processing in the ER, and the Wnt and mitogen‑activated protein kinase signaling pathways (Fig. 3d).
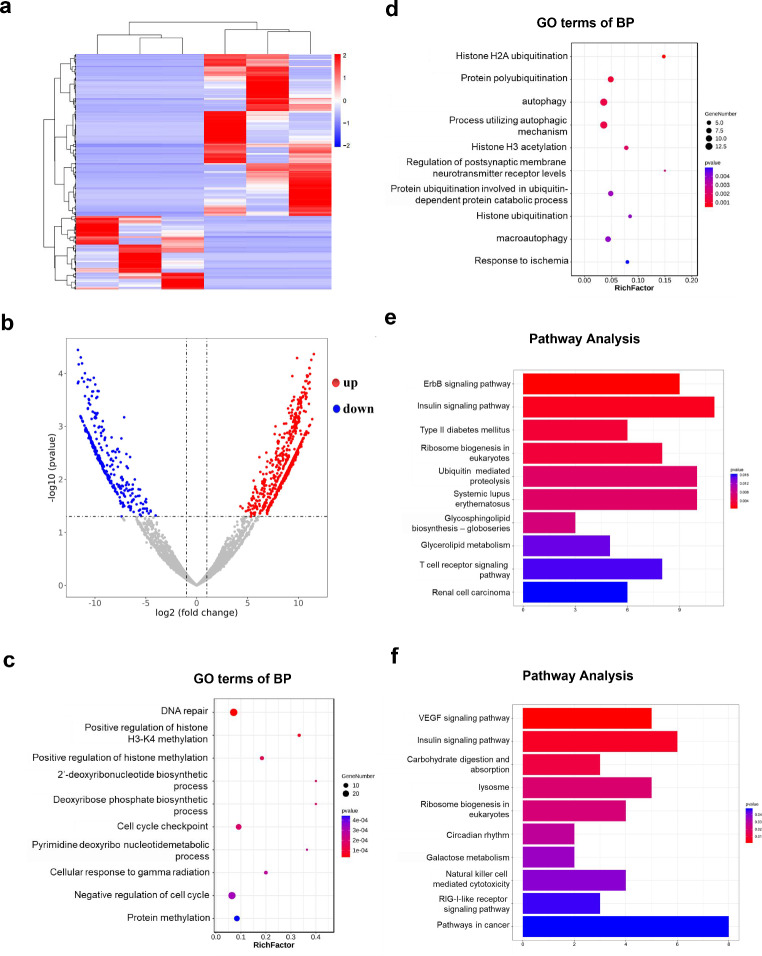
Cluster analysis of differentially expressed mRNAs in PC12 cells
RNA-seq was used to analyze mRNA transcriptional profiles in PC12 cells after bilirubin treatment. Significant differences could be observed in the heat map of mRNA expression patterns between the bilirubin and control groups (Fig. 4a). A volcano map revealed that the expression profiles of 940 mRNAs, including 647 upregulated and 293 downregulated mRNAs, were significantly dysregulated in the bilirubin group compared with those in the control group (fold change > 2 and P < 0.05; Fig. 4b). Subsequently, in order to evaluate what functions these differentially expressed mRNAs play after bilirubin treatment, we performed GO enrichment and KEGG pathway analyses. The results indicated that the significant differentially expressed mRNAs between the bilirubin and control groups (fold change > 2 and P < 0.05) were mainly associated with autophagy, histone H3 acetylation, and DNA repair (Fig. 4c and d). In addition, VEGF signaling pathway, ribosome biogenesis, and lysosomes were found to play a vital role in the bilirubin-treated group (Fig. 4e and f).
Fig. 4.
Cluster analysis of differentially expressed mRNAs in PC12 cells between the bilirubin and control groups. (a) Heatmap of differentially expressed mRNAs in the bilirubin group compared with the control group. (b) Scatter plots showing the mRNA data (fold changes ≥ 2 and P < 0.05). (c) GO analysis of the upregulated genes in the bilirubin group. (d) GO analysis of the downregulated genes in the bilirubin group. (e) KEGG pathway analyses of the upregulated genes in the bilirubin group. (f) KEGG pathway analysis of the downregulated genes in the bilirubin group
Combined analysis of GO and KEGG pathways with MeRIP- and RNA-seq data
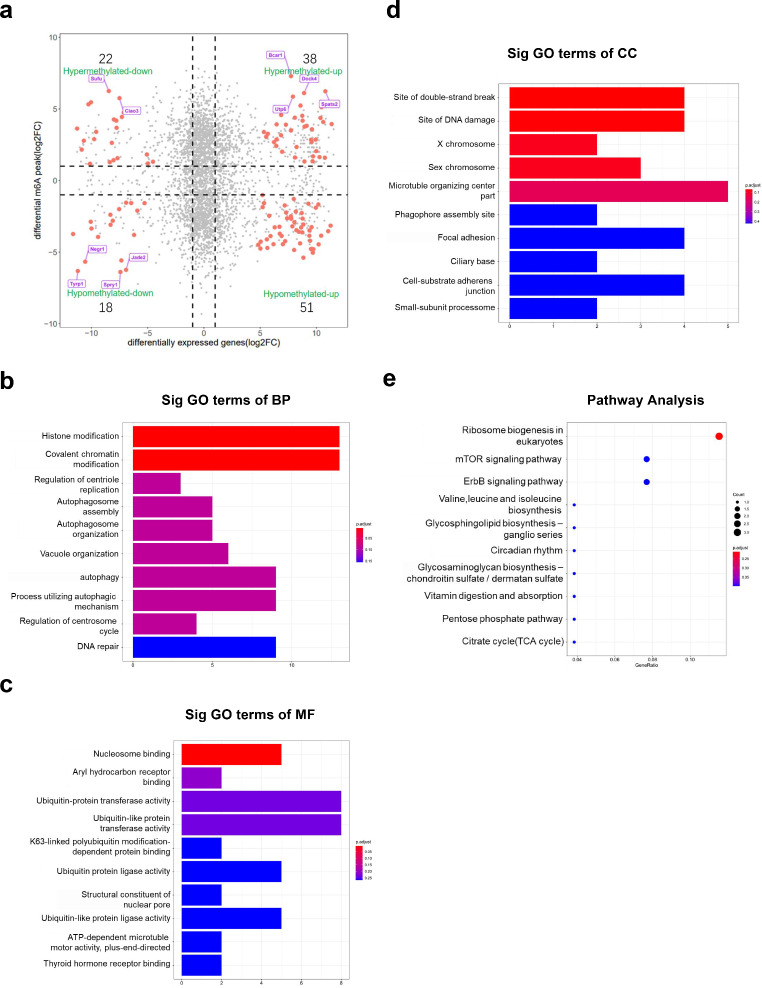
Through a combined analysis of the MeRIP- and RNA-seq results, 129 genes with differentially methylated m6A peaks and differential mRNA levels were divided into four parts, as shown in Fig. 5a. This study showed that 38, 51, 22, and 18 genes were upregulated and hypermethylated, upregulated and hypomethylated, downregulated and hypermethylated, and downregulated and hypomethylated, respectively. GO analysis also revealed that these genes were mainly related to the histone modification pathway (GO term: BP; Fig. 5b), nucleosome adhesion (GO term: MF; Fig. 5c), and protein binding (GO term: CC; Fig. 5d). The KEGG pathway analysis showed that these genes were mainly enriched in ribosome biogenesis in eukaryotes and the mTOR signaling pathway (Fig. 5e).
Fig. 5.
Combined functional analysis of differentially expressed genes between MeRIP-seq and RNA-seq data. (a) Four-quadrant plot showing the genes with differential changes in both m6A modification level and post-transcriptional mRNA level in the bilirubin group compared with the control group. The red dots represent genes with significant differences in m6A modification and mRNA expression between the bilirubin and control groups. (b) GO analysis of BP performed for differentially expressed genes identified through both MeRIP- and RNA-seq analyses. (c) GO analysis of MF of significantly differentially expressed genes identified through both MeRIP- and RNA-seq analyses. (d) GO analysis of CC of significantly differentially expressed genes identified through both MeRIP- and RNA-seq analyses. (e) KEGG pathway analysis of the MeRIP-seq and RNA-seq data
Network construction of major enriched pathway genes in bilirubin-treated PC12 cells
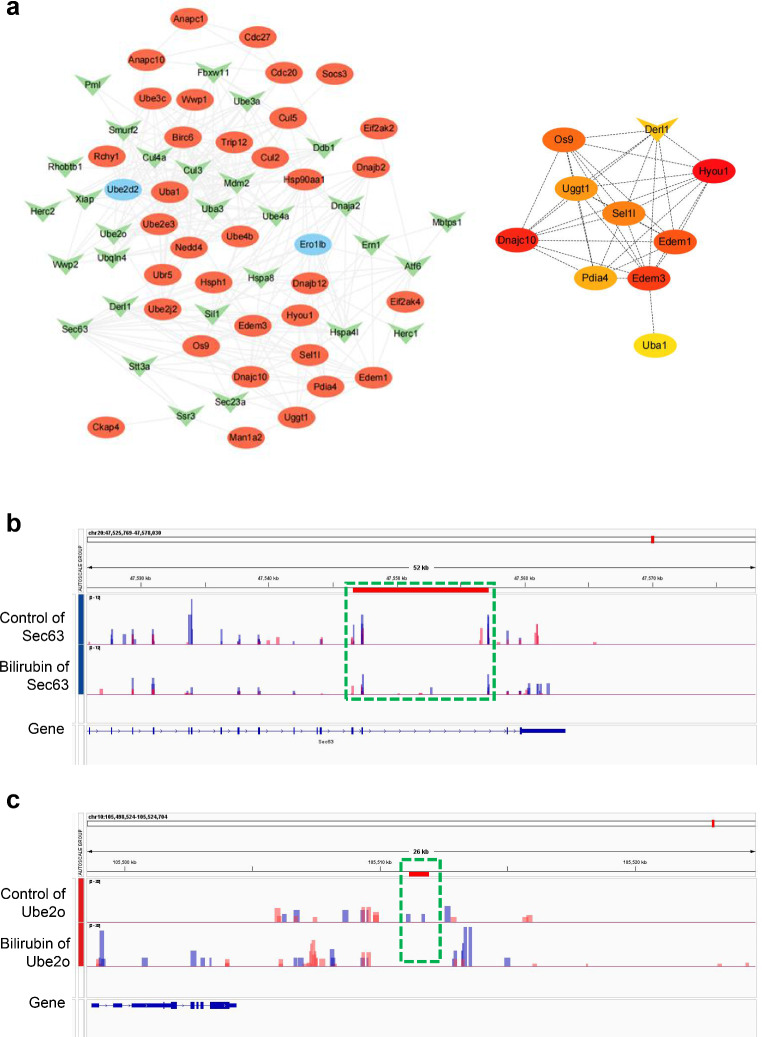
Both upregulated and downregulated m6A-modified genes were enriched in ubiquitin-mediated proteolysis and protein processing in the ER as per KEGG pathway analysis. mRNA interaction networks were constructed using the Cytoscape software to investigate the effect of bilirubin-induced regulation of m6A modification regulation on ubiquitin-mediated proteolysis and protein processing in the ER (Fig. 6a). According to the degree of the differential expression of these m6A-modified genes in the protein processing in the ER and ubiquitin-mediated proteolysis pathways (Tables 2 and 3), Sect. 63 and Ube2o were selected, respectively, and the m6A methylation sites and peak intensities were displayed using the IGV software (Fig. 6b and c).
Fig. 6.
The differentially m6A-modified mRNA network in ubiquitin-mediated proteolysis and protein processing in endoplasmic reticulum pathways. (a) The interaction network of differentially m6A-modified mRNA in the ubiquitin-mediated proteolysis pathway and in protein processing of endoplasmic reticulum pathways. The genes shown in red are m6A hypermethylated genes and those shown in green are m6A hypomethylated genes. Genes showed in blue can interact with m6A differentially expressed genes, not m6A differentially expressed genes in MeRIP-seq data. The remaining genes represent the top 10 genes calculated by the number of nodes in the interaction network of differentially m6A-modified mRNA in the ubiquitin-mediated proteolysis pathway and in protein processing of the endoplasmic reticulum pathway. (b, c) Sect. 63 and Ube2o transcripts in the UCB and control groups. Input and IP m6A are displayed as blue and red, respectively
Table 2.
Top 10 altered m6A genes related to protein processing in the ER
| Gene name | chromosome | Peak star | Peak End | lg.p | diff.log2.fc | Regulation |
|---|---|---|---|---|---|---|
| Section 63 | 20 | 47,560,798 | 47,561,068 | -8.69 | -6.67 | down |
| Ero1b | 17 | 90,710,794 | 90,711,154 | -8.62 | -4.85 | down |
| Sec23a | 6 | 80,061,005 | 80,062,850 | -35.8 | -4.56 | down |
| Ssr3 | 2 | 156,012,961 | 156,013,140 | -28.1 | -4.34 | down |
| Dnaja2 | 19 | 22,586,432 | 22,587,544 | -5.97 | -4.17 | down |
| Ern1 | 10 | 94,652,644 | 94,681,854 | -3.69 | -4.08 | down |
| Ube2d3 | 2 | 240,654,769 | 240,655,187 | -5.7 | 3.96 | up |
| Ube4b | 5 | 166,214,129 | 166,215,524 | -11 | 4.25 | up |
| Dnajb2 | 9 | 82,437,551 | 82,439,401 | -9.31 | 4.33 | up |
| Dnajb12 | 20 | 29,455,755 | 29,457,342 | -2.69 | 5.02 | up |
Table 3.
Top 10 altered m6A genes related to ubiquitin-mediated proteolysis
| Gene name | chromosome | Peak star | Peak End | lg.p | diff.log2.fc | Regulation |
|---|---|---|---|---|---|---|
| Ube2o | 10 | 105,511,135 | 105,511,929 | -6.55 | -6.39 | down |
| Herc1 | 8 | 72,116,404 | 72,117,022 | -4.45 | -5.83 | down |
| Ube3a | 1 | 116,648,732 | 116,656,481 | -6.65 | -5.81 | down |
| Pml | 8 | 63,016,204 | 63,018,743 | -2.98 | -4.58 | down |
| Ube4a | 8 | 49,264,827 | 49,269,334 | -11.4 | -4.1 | down |
| Ubr5 | 7 | 76,808,850 | 76,810,827 | -7.92 | 3.71 | up |
| Ube2d3 | 2 | 240,654,769 | 240,655,187 | -5.7 | 3.96 | up |
| Ube4b | 5 | 166,214,129 | 166,215,524 | -11 | 4.25 | up |
| Socs3 | 10 | 106,975,801 | 106,976,040 | -17.7 | 4.68 | up |
| Anapc10 | 19 | 31,815,766 | 31,815,916 | -4.85 | 5.67 | up |
Discussion
In the present study, we first investigated the expression of m6A methylation-modifying enzymes and revealed the m6A landscape of mRNAs in UCB-treated PC12 cells using high-throughput sequencing. PC12 cells are one of the most common models used in neuroscience studies exploring the latent mechanisms of action of neurotoxins [23], including those of bilirubin neurotoxicity [24]. Therefore, PC12 cells were selected as a model to study the potential neurotoxicity of bilirubin in the current study.
We observed that bilirubin treatment caused downregulation of the m6A demethylase ALKBH5 (Fig. 1b) and upregulation of the methyltransferases METTL3 and METTL14 (Fig. 1c and d). This resulted in increased m6A levels and led to an altered m6A expression profile in bilirubin-treated PC12 cells. Stable m6A methylation modifications are crucial in maintaining CNS function. In the mammalian nervous system, m6A methylation participates in various physiological functions, such as neuronal development, synaptic plasticity, learning and memory, addiction and reward learning, and stress responses [25]. Knockdown of FTO in the mouse prefrontal cortex and hippocampus led to enhanced m6A levels and the maintenance of fear memory and loss of learning ability [26, 27]. Specific differential expression of METTL13 and FTO and differential m6A methylation in different brain regions (medial prefrontal cortex and basolateral and central amygdala) were observed in mice during a stress response, which was consistent with the expression of glucocorticoids [28]. ALKBH5 overexpression inhibits cell proliferation via the Wnt/β-catenin signaling pathway, and METTL3 downregulation also reduces cell proliferation and increases apoptosis by inhibiting the Wnt/β-catenin signaling pathway [29]. METTL14 knockdown significantly reduced proliferation and promoted the differentiation of mouse embryonic neural stem cells, thereby inhibiting their self-renewal ability [30]. These results indicate that m6A modification is involved in brain-related pathophysiology. Notably, we also found a specific mRNA m6A methylation profile in PC12 cells exposed to bilirubin, which suggests that mRNA m6A methylation modifications may be involved in bilirubin neurotoxicity.
We detected many differentially methylated mRNAs associated with several significant biological pathways. KEGG pathway analyses illustrated that ER protein processing was regulated by upregulated or downregulated genes with m6A-modified sites in bilirubin-treated PC12 cells. ER stress is closely related to bilirubin neurotoxicity [5–7]. For instance, a transcriptome analysis identified that more than 50 genes were directly involved in ER stress response in SHSY5Y cells with UCB treatment [5]. Bilirubin induced the expression of ER stress-related genes and activated the unfolded protein response, which ultimately caused neurological dysfunction syndrome [6]. UCB upregulated key proteins in ER stress-related pathways and induced mitochondrial dysfunction, which further caused apoptosis of oligodendrocyte progenitor cells, thereby compromising myelination [7]. Recent studies have reported that m6A methylation of mRNAs is involved in ER stress. The m6A methyltransferase WTAP increases ER stress by regulating m6A modification of ATF4 mRNA, thus promoting myocardial ischemia/reperfusion injury [31]. Another m6A methyltransferase, METTL14, accelerates the mRNA degradation of ER stress-related gene CHOP, thereby inhibiting the expression of proapoptotic target genes [32]. METTL14 can promote liver regeneration by mediating m6A modification of polypeptide-processing proteins in the ER [33]. By contrast, accumulation of unfolded or misfolded proteins in the ER lumen induces METTL14 expression by preventing its ubiquitination and degradation, which reveals a crosstalk between ER stress and m6A modification of mRNAs [32]. Based on previous studies and our results, we believe that m6A modification of the ER protein-processing pathway is closely related to bilirubin neurotoxicity and may form a basis for future research.
Functional analysis of differentially m6A-modified genes after UCB treatment of PC12 cells also showed the enrichment of the ubiquitin-mediated proteolysis in the KEGG pathway. The level of ubiquitinated proteins (Ub-prs) in the blood cells of patients with hyperbilirubinemia is significantly increased [34]. Bilirubin treatment induces the accumulation of Ub-prs in the brain of neonatal rats and inhibits the deubiquitination activity of the proteasome in a dose-dependent manner in vitro [34]. In our study, ubiquitin-mediated proteolysis was the top hypermethylation-affected pathway (Fig. 3c). These results suggested that m6A methylation might play a role in ER stress and that ubiquitin-mediated proteolysis was induced by bilirubin in neuronal cells.
In addition, in the pathways of ubiquitin-mediated proteolysis and protein processing in the ER, we selected the gene with the largest difference in m6A methylated peaks. Finally, we selected Sect. 63 and Ube2o as the target genes for further study. Section 63 plays a key role in the transport of newly synthesized polypeptide precursors into the ER membrane by forming a dimeric complex with Sect. 62 [35]. The low expression of Sect. 63 leads to reduced myelination in both the central and peripheral nervous systems [36]. Ube2o is a member of the ubiquitin-conjugating enzyme family, which plays a vital role in the regulation of protein ubiquitination and cellular functions. Studies have reported that Ube2o has carcinogenic or tumor suppressive roles in human cancers [37–39]. However, whether Sect. 63 or Ube2o is involved in bilirubin neurotoxicity has not been reported and needs to be further elucidated.
Although our study presents some interesting findings and novel insights, it had some limitations. First, this study mainly focused on the m6A modification spectrum. We will further study and reveal the specific molecular mechanism involved in the changes and interactions of the enriched pathways in the m6A modification spectrum. Second, there may be differences between in vivo and in vitro m6A modifications induced by bilirubin in the CNS. In future studies, animal or human samples should be used to investigate changes in m6A modification levels in neonates with bilirubin-induced CNS injury.
Conclusions
The present study confirmed that the profiles of mRNA m6A methylation were significantly altered in bilirubin-induced PC12 cell injury. These results suggested that m6A modification was involved in bilirubin-triggered CNS damage by regulating various pathways, such as ER stress, protease ubiquitination, and cell cycle. Our study provides new insights into the potential epigenetic mechanism of bilirubin neurotoxicity.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors appreciate the technical assistance of Guangzhou Epigenetic Biotechnology Co., Ltd. (Guangzhou, China).
Authors’ contributions
HL, JZ, and SL contributed to conception and design of the study. JZ, CZ, and JL performed experiments and organized the database. GL performed the statistical analysis. JZ and SL wrote the first draft of the manuscript. HL reviewed and edited the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This study was funded by the Natural Science Foundation of Fujian Province (grant number 2020J01327); the Fujian Provincial Health Technology Project (grant number 2020GGB017); the Joint Funds for the Innovation of Science and Technology, Fujian Province (grant number 2020Y9143); the Open Project of the Key Laboratory of Environment and Health of Fujian Medical University (grant number GWGXZD-202002); and the Fujian Maternity and Child Health Hospital (grant number YCXM 20 − 08).
Data availability
The data and materials in the current study are available from the corresponding author upon reasonable request.
Declarations
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jinfu Zhou and Sining Liao contributed equally to this work.
References
- 1.Gamaleldin R, Iskander I, Seoud I, et al. Risk factors for neurotoxicity in newborns with severe neonatal hyperbilirubinemia. Pediatrics. 2011;128:e925–e931. doi: 10.1542/peds.2011-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaplan M, Bromiker R, Hammerman C. Hyperbilirubinemia, hemolysis, and increased bilirubin neurotoxicity. Semin Perinatol. 2014;38:429–437. doi: 10.1053/j.semperi.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Bortolussi G, Codarin E, Antoniali G, et al. Impairment of enzymatic antioxidant defenses is associated with bilirubin-induced neuronal cell death in the cerebellum of Ugt1 KO mice. Cell Death Dis. 2015;6:e1739. doi: 10.1038/cddis.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rauti R, Qaisiya M, Tiribelli C, et al. Bilirubin disrupts calcium homeostasis in neonatal hippocampal neurons: a new pathway of neurotoxicity. Arch Toxicol. 2020;94:845–855. doi: 10.1007/s00204-020-02659-9. [DOI] [PubMed] [Google Scholar]
- 5.Calligaris R, Bellarosa C, Foti R, et al. A transcriptome analysis identifies molecular effectors of unconjugated bilirubin in human neuroblastoma SH-SY5Y cells. BMC Genomics. 2009;10:543. doi: 10.1186/1471-2164-10-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schiavon E, Smalley JL, Newton S, et al. Neuroinflammation and ER-stress are key mechanisms of acute bilirubin toxicity and hearing loss in a mouse model. PLoS ONE. 2018;13:e0201022. doi: 10.1371/journal.pone.0201022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barateiro A, Vaz AR, Silva SL, et al. ER stress, mitochondrial dysfunction and Calpain/JNK activation are involved in Oligodendrocyte Precursor Cell death by Unconjugated Bilirubin. Neuromolecular Med. 2012;14:285–302. doi: 10.1007/s12017-012-8187-9. [DOI] [PubMed] [Google Scholar]
- 8.Qaisiya M, Mardešić P, Pastore B, et al. The activation of autophagy protects neurons and astrocytes against bilirubin-induced cytotoxicity. Neurosci Lett. 2017;661:96–103. doi: 10.1016/j.neulet.2017.09.056. [DOI] [PubMed] [Google Scholar]
- 9.Feng J, Li M, Wei Q, et al. Unconjugated bilirubin induces pyroptosis in cultured rat cortical astrocytes. J Neuroinflammation. 2018;15:23. doi: 10.1186/s12974-018-1064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gou P, Qi X, Yuan R, et al. Tet1-mediated DNA demethylation involves in neuron damage induced by bilirubin in vitro. Toxicol Mech Methods. 2018;28:55–61. doi: 10.1080/15376516.2017.1357775. [DOI] [PubMed] [Google Scholar]
- 11.Vianello E, Zampieri S, Marcuzzo T, et al. Histone acetylation as a new mechanism for bilirubin-induced encephalopathy in the Gunn rat. Sci Rep. 2018;8:13690. doi: 10.1038/s41598-018-32106-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv J, Xing L, Zhong X, et al. Role of N6-methyladenosine modification in central nervous system diseases and related therapeutic agents. Biomed Pharmacother. 2023;162:114583. doi: 10.1016/j.biopha.2023.114583. [DOI] [PubMed] [Google Scholar]
- 13.Lu Z, Liu J, Yuan C, et al. m6A mRNA methylation analysis provides novel insights into heat stress responses in the liver tissue of sheep. Genomics. 2021;113:484–492. doi: 10.1016/j.ygeno.2020.09.038. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q, Liang Y, Luo X, et al. N6-methyladenosine RNA modification: a promising regulator in central nervous system injury. Exp Neurol. 2021;345:113829. doi: 10.1016/j.expneurol.2021.113829. [DOI] [PubMed] [Google Scholar]
- 15.Sokpor G, Xie Y, Nguyen HP, Tuoc T. Emerging role of m6 A methylome in Brain Development: implications for neurological Disorders and potential treatment. Front Cell Dev Biol. 2021;9:656849. doi: 10.3389/fcell.2021.656849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yen Y-P, Chen J-A. The m(6)a epitranscriptome on neural development and degeneration. J Biomed Sci. 2021;28:40. doi: 10.1186/s12929-021-00734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du T, Li G, Yang J, Ma K. RNA demethylase Alkbh5 is widely expressed in neurons and decreased during brain development. Brain Res Bull. 2020;163:150–159. doi: 10.1016/j.brainresbull.2020.07.018. [DOI] [PubMed] [Google Scholar]
- 18.Shafik AM, Zhang F, Guo Z, et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021;22:17. doi: 10.1186/s13059-020-02249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang N, Ding C, Zuo Y, et al. N6-methyladenosine and neurological Diseases. Mol Neurobiol. 2022;59:1925–1937. doi: 10.1007/s12035-022-02739-0. [DOI] [PubMed] [Google Scholar]
- 20.Xu Z, Zhu X, Mu S, et al. FTO overexpression expedites wound healing and alleviates depression in burn rats through facilitating keratinocyte migration and angiogenesis via mediating TFPI-2 demethylation. Mol Cell Biochem. 2023 doi: 10.1007/s11010-023-04719-x. [DOI] [PubMed] [Google Scholar]
- 21.Cheng J, Lin L, Yu J, et al. N6-methyladenosine RNA is modified in the rat hippocampus following traumatic brain injury with hypothermia treatment. Front Neurosci. 2023;17:1069640. doi: 10.3389/fnins.2023.1069640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Xi W, Bai Y, et al. FTO-dependent m6A modification of Plpp3 in circSCMH1-regulated vascular repair and functional recovery following stroke. Nat Commun. 2023;14:489. doi: 10.1038/s41467-023-36008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiatrak B, Kubis-Kubiak A, Piwowar A, Barg E (2020) PC12 cell line: cell types, Coating of Culture Vessels, differentiation and other Culture Conditions. 10.3390/cells9040958. Cells 9: [DOI] [PMC free article] [PubMed]
- 24.Mancuso C, Capone C, Ranieri SC, et al. Bilirubin as an endogenous modulator of neurotrophin redox signaling. J Neurosci Res. 2008;86:2235–2249. doi: 10.1002/jnr.21665. [DOI] [PubMed] [Google Scholar]
- 25.Hess ME, Hess S, Meyer KD, et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat Neurosci. 2013;16:1042–1048. doi: 10.1038/nn.3449. [DOI] [PubMed] [Google Scholar]
- 26.Widagdo J, Zhao Q-Y, Kempen M-J, et al. Experience-dependent Accumulation of N6-Methyladenosine in the Prefrontal cortex is Associated with memory processes in mice. J Neurosci. 2016;36:6771–6777. doi: 10.1523/JNEUROSCI.4053-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walters BJ, Mercaldo V, Gillon CJ, et al. The role of the RNA demethylase FTO (Fat Mass and Obesity-Associated) and mRNA methylation in hippocampal memory formation. Neuropsychopharmacology. 2017;42:1502–1510. doi: 10.1038/npp.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engel M, Eggert C, Kaplick PM, et al. The role of m(6)A/m-RNA methylation in stress response regulation. Neuron. 2018;99:389–403e9. doi: 10.1016/j.neuron.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang L, Cao R, Li D, et al. Ethionine-mediated reduction of S-adenosylmethionine is responsible for the neural tube defects in the developing mouse embryo-mediated m6A modification and is involved in neural tube defects via modulating Wnt/β-catenin signaling pathway. Epigenetics Chromatin. 2021;14:52. doi: 10.1186/s13072-021-00426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Li Y, Yue M, et al. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Zhang J, Ma Y, et al. WTAP promotes myocardial ischemia/reperfusion injury by increasing endoplasmic reticulum stress via regulating m(6)a modification of ATF4 mRNA. Aging. 2021;13:11135–11149. doi: 10.18632/aging.202770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei J, Harada BT, Lu D, et al. HRD1-mediated METTL14 degradation regulates m(6)a mRNA modification to suppress ER proteotoxic liver disease. Mol Cell. 2021;81:5052–5065e6. doi: 10.1016/j.molcel.2021.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao X, Shu Y, Chen Y, et al. Mettl14-Mediated m(6)a modification facilitates liver regeneration by maintaining endoplasmic Reticulum Homeostasis. Cell Mol Gastroenterol Hepatol. 2021;12:633–651. doi: 10.1016/j.jcmgh.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang H, Guo M, Liu N, et al. Bilirubin neurotoxicity is associated with proteasome inhibition. Cell Death Dis. 2017;8:e2877. doi: 10.1038/cddis.2017.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung S-J, Kim H (2021) Emerging View on the Molecular Functions of Sect. 62 and Sect. 63 in protein translocation. Int J Mol Sci 22. 10.3390/ijms222312757 [DOI] [PMC free article] [PubMed]
- 36.Monk KR, Voas MG, Franzini-Armstrong C, et al. Mutation of Sect. 63 in zebrafish causes defects in myelinated axons and liver pathology. Dis Model Mech. 2013;6:135–145. doi: 10.1242/dmm.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y, Yang X, Lu Y, et al. UBE2O targets Mxi1 for ubiquitination and degradation to promote lung cancer progression and radioresistance. Cell Death Differ. 2021;28:671–684. doi: 10.1038/s41418-020-00616-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Zhang Z, Li J, et al. The ubiquitin-conjugating enzyme UBE2O modulates c-Maf stability and induces myeloma cell apoptosis. J Hematol Oncol. 2017;10:132. doi: 10.1186/s13045-017-0499-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Z, Liu R, Lu Q, et al. UBE2O promotes hepatocellular carcinoma cell proliferation and invasion by regulating the AMPKα2/mTOR pathway. Int J Med Sci. 2021;18:3749–3758. doi: 10.7150/ijms.63220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials in the current study are available from the corresponding author upon reasonable request.