Abstract
The human complement system plays a crucial role in immune defense. However, its erroneous activation contributes to many serious inflammatory diseases. Since most unwanted complement effector functions result from C5 cleavage into C5a and C5b, development of C5 inhibitors, such as clinically approved monoclonal antibody eculizumab, are of great interest. Here, we developed and characterized two anti-C5 nanobodies, UNbC5-1 and UNbC5-2. Using surface plasmon resonance, we determined a binding affinity of 119.9 pM for UNbC5-1 and 7.7 pM for UNbC5-2. Competition experiments determined that the two nanobodies recognize distinct epitopes on C5. Both nanobodies efficiently interfered with C5 cleavage in a human serum environment, as they prevented red blood cell lysis via membrane attack complexes (C5b-9) and the formation of chemoattractant C5a. The cryo-EM structure of UNbC5-1 and UNbC5-2 in complex with C5 (3.6 Å resolution) revealed that the binding interfaces of UNbC5-1 and UNbC5-2 overlap with known complement inhibitors eculizumab and RaCI3, respectively. UNbC5-1 binds to the MG7 domain of C5, facilitated by a hydrophobic core and polar interactions, and UNbC5-2 interacts with the C5d domain mostly by salt bridges and hydrogen bonds. Interestingly, UNbC5-1 potently binds and inhibits C5 R885H, a genetic variant of C5 that is not recognized by eculizumab. Altogether, we identified and characterized two different, high affinity nanobodies against human C5. Both nanobodies could serve as diagnostic and/or research tools to detect C5 or inhibit C5 cleavage. Furthermore, the residues targeted by UNbC5-1 hold important information for therapeutic inhibition of different polymorphic variants of C5.
Keywords: complement system, innate immunity, nanobody, single-domain antibody (sdAb nanobody), inhibitor, protein structure, structural model, inflammatory diseases
The complement system is an important part of the human innate immune system that is involved in many different biological processes. During embryonal tissue development, it plays a crucial role in cell differentiation and proliferation (1, 2, 3). Later in life, complement is best known for its role in clearing pathogens during infection, but also remains involved in maintaining homeostasis, by removing apoptotic cells and immune complexes (4). Although the complement system is tightly regulated, unwanted complement activation is the root cause of a variety of diseases, including paroxysmal nocturnal hemoglobinuria, atypical hemolytic uremic syndrome, and age-related macular degeneration. Furthermore, erroneous complement activity contributes in more prevalent complex multifactorial diseases like systemic lupus erythematosus, Alzheimer's disease, and coronavirus disease 2019 (5).
The complement system can be activated via three distinct pathways: the classical pathway (CP), the lectin pathway, and the alternative pathway (AP). Together, these pathways result in three major effector functions: [1] opsonization, [2] chemoattraction of immune cells, and [3] direct target cell lysis (6). A central complement component is C5, which circulates in blood at a concentration of ±75 μg/ml (7). C5 is a 190 kDa protein, consisting of two disulfide linked chains (α: 115 kDa, amino acids 678–1676 and β: 75 kDa, amino acids 19–673) (8) and the protein contains a core of eight macroglobulin (MG) domains, and four other domains (“complement C1r/C1s, Uegf, Bmp1” (CUB), C345c, C5d, and C5a) (8, 9). During complement activation, C5 convertases (C4b2bC3b and C3bBbC3b) are formed on the target surface, which cleave native C5 molecules in C5a (8 kDa) and C5b (181 kDa). While C5a is released in the fluid phase, where it functions as a chemoattractant, C5b interacts with complement proteins C6, C7, C8, and multiple copies of C9, forming the membrane attack complex (6).
Since multiple (unwanted) complement effector functions result from the cleavage of C5, many complement inhibitors have been identified and developed to target this particular molecule. Human pathogens have evolved immune evasion proteins to specifically block the cleavage of C5. For example, Staphylococcus aureus expresses staphylococcal superantigen-like protein 7 (SSL7), a protein that potently binds and inhibits cleavage of human C5 (10). Furthermore, ticks (Ornithodoros moubata, Rhipicephalus pulchellus, and Rhipicephalus appendiculatus) contain C5 inhibiting proteins OmCI (11), CirpT (12), and RaCI1, 2, 3 (13), respectively, in their saliva. Moreover, different snake species have evolved a C5-targeting molecule, called cobra venom factor (CVF) (14). This protein structurally resembles complement component C3b and, when bound by Bb, can interact with and cleave human C5 molecules.
In addition to pathogen-derived C5 inhibitors, a wide variety of C5-targeting molecules including monoclonal antibodies have been developed (15, 16). Of these, the best known is the monoclonal antibody eculizumab, that is Food and Drug Administration approved, and which binds to C5 and prevents its cleavage by blocking the interaction of C5 with the C5 convertases (17). After the introduction of eculizumab into the clinic, a genetic polymorphism in the C5 α-chain was identified in a group of poor responders, where a histidine is located in position 885 instead of an arginine (C5 R885H) (18). The structural model of eculizumab and C5 revealed that Arg885 in C5 and Phe101 in eculizumab form a crucial interaction for binding and inhibiting C5 (19, 20, 21).
In this study, we identify and characterize two llama-derived nanobodies that specifically bind and inhibit human complement C5. They bind to C5 with picomolar affinities on two distinct epitopes. Interestingly, one of these nanobodies has an overlapping epitope with eculizumab and is still able to inhibit C5 R885H.
Results
Identification of two C5-targeting nanobodies that interfere with complement
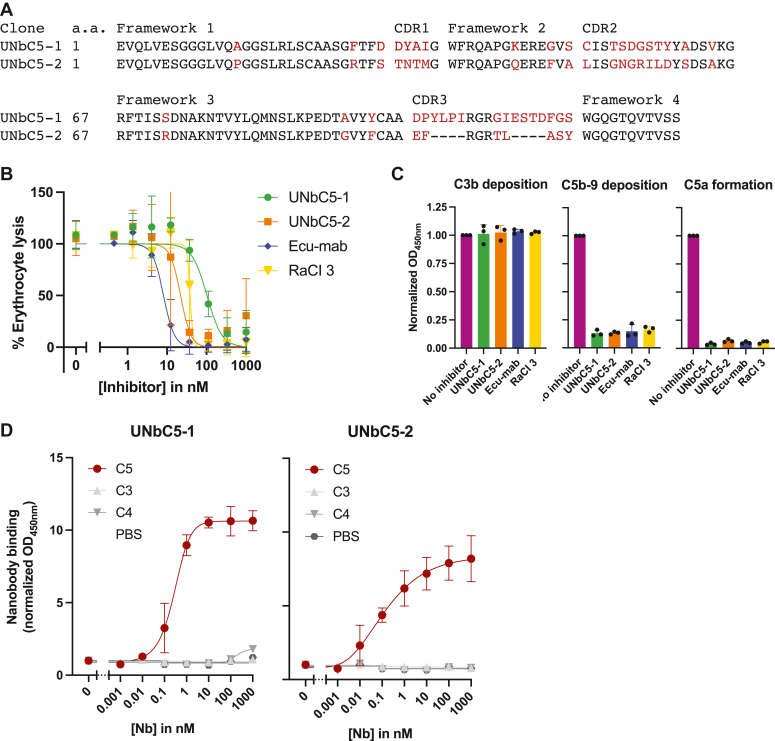
To develop nanobodies targeting C5, llamas were successfully immunized with recombinant human C5 and nanobody phage libraries were generated (Fig. S1, A and B). After two rounds of phage display panning, ∼200 clones were produced in the periplasm of Escherichia coli, and unpurified periplasmic fractions were used to screen for clones that bind C5 in ELISA or inhibit complement activity in a CP hemolysis assay. Based on the nanobody sequences and a CP hemolysis assay used to screen for potent inhibitors (examples shown in Fig. S1C), two clones were selected to further characterize, denoted UNbC5-1 and UNbC5-2. UNbC5-1 (13.5 kDa) and UNbC5-2 (12.9 kDa) are significantly distinct, with only 30% amino acid sequence similarity in their complementarity determining regions (CDR) (Fig. 1A). Next, UNbC5-1 and UNbC5-2 were purified to assess their ability to block complement activity in human serum. Using a CP hemolysis assay (22), we confirmed that UNbC5-1 and UNbC5-2 were potent complement inhibitors (Fig. 1B). Both nanobodies blocked complement-mediated lysis in a dose-dependent manner. As controls we took along in-house produced C5 inhibitors RaCI3 and a monoclonal IgG antibody with the same primary sequence as clinically approved eculizumab (Ecu-mab). The IC50 values of UNbC5-1 and UNbC5-2 are in the same range as that of RaCI3 (Table 1). On a molar basis, Ecu-mab was roughly 3 to 15 times more potent in blocking C5 cleavage than UNbC5-1 and UNbC5-2. This might partially be explained by the fact that Ecu-mab has two antigen-binding domains per molecule and the nanobodies have only one.
Figure 1.
Identification of two C5-targeting nanobodies that interfere with complement.A, protein sequence alignment of UNbC5-1 and UNbC5-2. Frameworks 1 to 4 and CDRs 1 to 3 are indicated. Conserved amino acids are indicated in black and unique amino acids are indicated in red. B, CP mediated hemolysis of antibody-coated sheep erythrocytes incubated with 2.5% normal human serum and a titration of our nanobodies UNbC5-1 and UNbC5-2 and known complement inhibitors RaCI3 and Ecu-mab. The A405 values of the supernatants were measured; the % erythrocyte lysis was calculated using the 0% lysis (buffer) and 100% lysis (milliQ water) control samples. C, CP complement activation on an IgM-coated microtiter plate, incubated with 4% human serum in the presence of buffer (no inhibitor) or 1000 nM inhibitor. Deposition of complement activation products C3b and C5b-9 on the plate was measured using specific anti-C3 and anti-C5b-9 antibodies and HRP-coupled secondary antibodies, at A450. C5a formation was measured by adding the supernatant of the reaction to a microtiter plate coated with C5a capture antibodies, and C5a was detected with a specific C5a detection antibody and HRP-coupled secondary antibodies at A450. Data points were normalized to the maximum levels of C3b and C5b-9 deposition and to the maximum levels of C5a formation when no inhibitor was added. D, nanobody bind to complement protein C5 and not homologs C3 and C4. Microtiter plates were coated with complement proteins and incubated with increasing concentrations of UNbC5-1 and UNbC5-2. Nanobody binding was measured using polyclonal rabbit-anti-VHH QE19 antibodies and donkey-anti-rabbit-HRP antibodies, at an absorbance of 450 nm. Coating with PBS was taken along as a negative control. Data information: (A), sequences were aligned using T-coffee (42). B–D, data represent mean ± SD of three individual experiments (B and D) curves were fitted and IC50 and EC50 values were obtained. CDR, complementarity determining regions; CP, classical pathway; HRP, horseradish peroxide.
Table 1.
IC50 and EC50 values for UNbC5-1, UNbC5-2, Ecu-mab, and RaCI3 for inhibiting complement activity and binding complement C5
| Inhibitor | IC50 (nM) CP | EC50 (nM) C5 WT | EC50 (nM) C5 R885H |
|---|---|---|---|
| UNbC5-1 | 137 ± 83 | 0.38 ± 0.20 | 1.5 ± 0.4 |
| UNbC5-2 | 24 ± 15 | 0.12 ± 0.01 | 22.4 ± 17.0 |
| Ecu-mab | 9 ± 4 | 0.05 ± 0.01 | ND |
| RaCI3 | 42 ± 8 | - | - |
Binding and inhibition curves were fitted using the GraphPad Prism 9.3.0 function “[inhibitor] versus normalized response–variable slope” & “Asymmetrical Sigmoidal, 5PL, X is concentration”. IC50 and EC50 values were calculated for three individual experiments using the fitted curves, and SD were determined.
To confirm that UNbC5-1 and UNbC5-2 block complement at the level of C5, we assessed their capacity to block different steps of the complement reaction. Using an ELISA-based complement activity assay (23), we observed that neither UNbC5-1 nor UNbC5-2 affect the initial steps of complement activation since there is no inhibition of C3b deposition on IgM-coated microtiter plates that were incubated with serum (Fig. 1C). However, UNbC5-1 and UNbC5-2 can prevent the formation of deposited C5b-9 complexes and the release of C5a into the supernatant. Thus, we show that UNbC5-1 and UNbC5-2 block complement activity at the level of C5 cleavage. Consistent with this finding, UNbC5-1 and UNbC5-2 could also prevent complement-mediated hemolysis by the AP (Fig. S1D).
To assess whether UNbC5-1 and UNbC5-2 are indeed specific for C5, we studied the binding to complement proteins C3 and C4, which share sequence and structural homology with C5 (24). In ELISA, UNbC5-1 and UNbC5-2 showed a dose-dependent binding to C5, while no cross-reactivity with C3 or C4 was observed (Fig. 1D). UNbC5-1 and UNbC5-2 bind C5 with EC50 values of 0.38 ± 0.2 and 0.12 ± 0.01 nM, respectively (Table 1). Altogether these data show that UNbC5-1 and UNbC5-2 are C5-specific nanobodies that can interfere with complement activity.
UNbC5-1 and UNbC5-2 bind C5 with picomolar affinity and recognize distinct epitopes
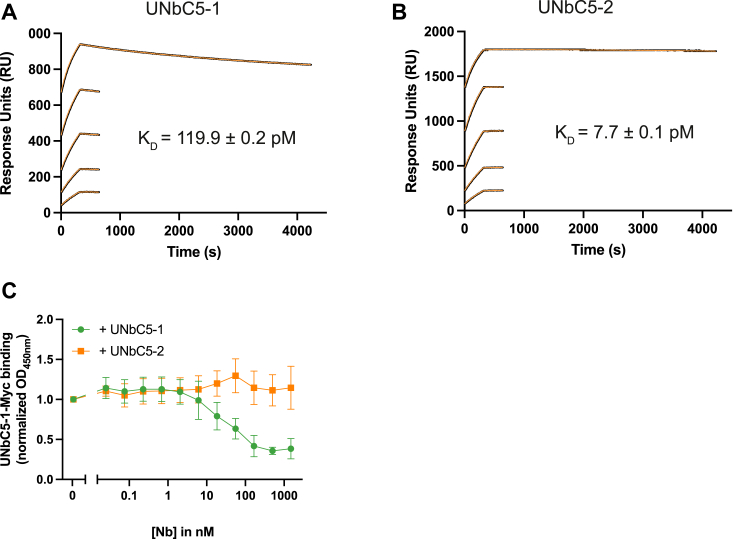
Next, we used surface plasmon resonance (SPR) to determine the affinities of UNbC5-1 and UNbC5-2 for C5. Briefly, biotinylated nanobodies were coupled to streptavidin-coated biosensors, and a kinetic titration with C5 was performed to determine kon rates (Table 2). Next, a dissociation step was performed to measure koff rates and KD values were calculated. UNbC5-1 binds C5 with a KD of 119.9 pM (Fig. 2A), and UNbC5-2 binds C5 with a kD of 7.7 pM (Fig. 2B). As a reference, we took along Ecu-mab and measured a C5 binding affinity of 104 pM, which is in the same range as the published affinity of eculizumab (17.6–120 pM) (Fig. S2) (17, 19, 25). To assess whether the two nanobodies bind overlapping epitopes, we performed competition experiments by ELISA. C5-coated microtiter plates were incubated with Myc-labeled UNbC5-1 (UNbC5-1-Myc) in the presence or absence of unlabeled UNbC5-2. After washing, binding of UNbC5-1-Myc to C5 was quantified. We observed that the presence of UNbC5-2 did not affect binding of UNbC5-1-Myc to C5 (Fig. 2C). As a control, we showed that UNbC5-1 was able to compete with UNbC5-1-Myc. These data indicate that UNbC5-1 and UNbC5-2 can bind C5 simultaneously and not compete for the same epitope.
Table 2.
Binding affinities of UNbC5-1, UNbC5-2 and Ecu-mab for complement C5
| Nanobody | kon (M−1s−1) × 105 | koff (s−1) × 10−6 | KD (pM) |
|---|---|---|---|
| UNbC5-1 | 2.713 ± 0.002 | 3.254 ± 0.06 | 119.9 ± 0.2 |
| UNbC5-2 | 2.937 ± 0.008 | 0.227 ± 0.03 | 7.7 ± 0.1 |
| Ecu-mab | 3.661 ± 0.003 | 3.810 ± 0.10 | 104.0 ± 0.4 |
Nanobody and Ecu-mab affinities were determined by SPR using C5 as analyte and nanobodies and Ecu-mab as a ligand. All measurements were performed in duplicate. Kinetics were determined using Scrubber 2.0 (BioLogic Software).
Figure 2.
UNbC5-1 and UNbC5-2 bind C5 with picomolar affinity and recognize distinct epitopes.A and B, SPR curves with UNbC5-1-biotin (biotinylation method [2]) (A) and UNbC5-2-biotin (biotinylation method [1]) (B) as a ligand and C5 as an analyte at concentrations of 12.5, 6.25, 3.13, 1.56, and 0.78 nM, evaluated over 4000 s. Experimental data are shown in black, and model fit in orange. C, competitive ELISA with C5-coated microtiter plates incubated with 5 nM NbC5-1-Myc and a titration of untagged UNbC5-1 or UNbC5-2. Binding of UNbC5-1-Myc to C5 was measured using anti-Myc antibodies and an HRP-coupled secondary antibody, at A450. Data were normalized on maximum binding of UNbC5-1-Myc, measured when no untagged nanobody was added. Data information: (A and B), experiments were carried out in duplicate as individual experiments at 25 °C. C, data represent mean ± SD of three individual experiments. HRP, horseradish peroxidase; SPR, surface plasmon resonance.
Cryo-EM structures of UNbC5-1 and UNbC5-2 in complex with C5
To obtain structural insights into the nanobody binding sites and inhibitory mechanisms, we determined the structure of both nonoverlapping nanobodies in complex with C5, using single-particle analysis cryo-EM. The micrographs collected showed well-distributed particles in vitreous ice (Fig. S3A). Image processing was performed in CryoSPARC, v3.3/3, where the 2D classification already showed secondary structural features of C5 in complex with both nanobodies (Fig. S3B). 3D classification and refinement of the cryo-EM density map led to an overall map resolution of 3.6 Å, which allowed the subsequent model building of the C5:UNbC5-1:UNbC5-2 structure (Fig. S3C). The structure was built using the previously described C5 crystal structure (3CU7) and AlphaFold generated nanobody models (9, 26). The final refinement of the structure in Phenix 1.20.1 (https://phenix-online.org/version_docs/1.20.1-4487/) showed acceptable model statistics and stereochemistry (Table 3). Fig. S4, A and B show the maps and the resulting models for both C5:nanobody interfaces. Due to a lack of density, the C-terminal C345c domain could not be modeled. A similar flexibility of this C-terminal domain was observed in other C5 cryo-EM structures (9, 12), and the different arrangement of C345c in different crystal structures (9, 10, 13, 19, 27).
Table 3.
Cryo-EM data collection, refinement, and validation statistics
| Data collection and processing | C5:UNbC5-1:UNbC5-2 PDB-8CML, EMDB-16730 |
|---|---|
| Microscope | Talos Arctica |
| Camera | Gatan K2 Summit + GIF |
| Magnification | 130,000 |
| Voltage (kV) | 200 |
| Exposure time frame/total (s) | 0.2/8.0 |
| Number of frames | 40 |
| Electron exposure (e−/Å2) | 54–57 |
| Defocus range (μm) | −0.8 to −2.6 |
| Pixel size (Å) | 1.04 |
| Micrographs (no.) | 1253; 1840 |
| Initial particle images (no.) | 1,483,794 |
| Final particles images (no.) | 193,009 |
| Map Resolution (Å) 0.143 FSC threshold |
3.60 |
| Map resolution range (Å) | 3.1–7.4 |
| Refinement | |
| Model Resolution (Å) 0.143 FSC threshold | 3.60 |
| Map sharpening B factor (Å2) | −120.3 |
| Model composition | |
| Non-hydrogen atoms | 13,594 |
| Protein residues | 1728 |
| Ligands | NAG: 2 |
| B factors (Å2) | |
| Protein | 52.71 |
| Ligands | 50.80 |
| RMS deviations | |
| Bond lengths (Å) | 0.005 |
| Bond angles (o) | 0.842 |
| Validation | |
| MolProbity score | 2.41 |
| Clashscore | 13.01 |
| Rotamer outliers (%) | 2.91 |
| Ramachandran plot | |
| Favored (%) | 93.35 |
| Allowed (%) | 6.01 |
| Outliers (%) | 0.64 |
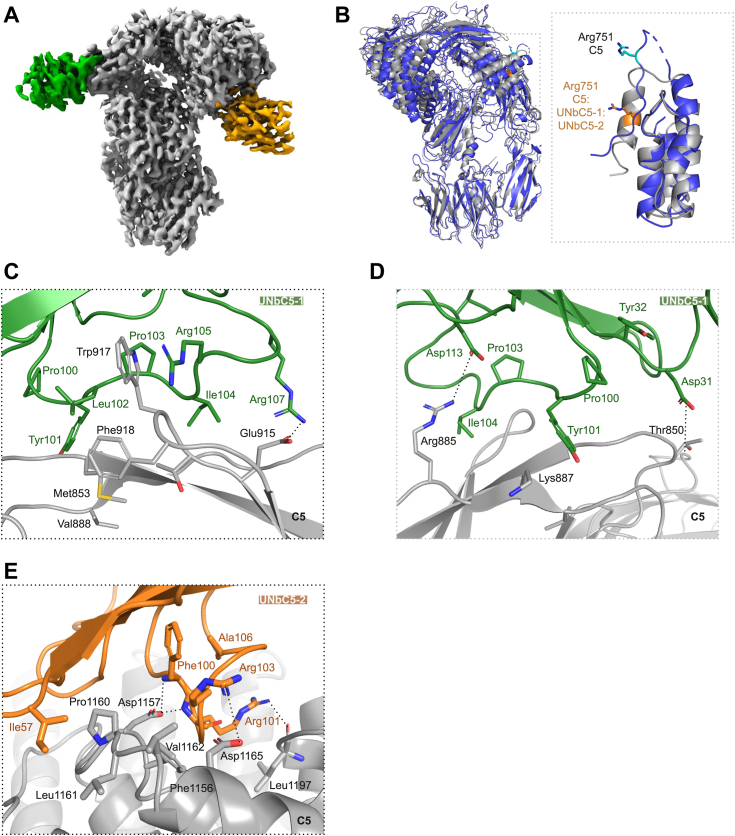
Overall, analysis of the C5 molecule in the C5:UNbC5-1:UNbC5-2 structure revealed interesting differences compared to the C5 crystal structure with Protein Data Bank (PDB) 3CU7 (Fig. 3B, left panel). Most apparent was the different arrangement of the C5a containing the C5 cleavage site, Arg751–Leu752. While residues Asp746–Met754 are disordered in native C5 (PDB 3CU7), they adopt a helical conformation in the C5:UNbC5-1:UNbC5-2 structure, internalizing the cleavage site, Arg751 (Fig. 3B, right panel). Moreover, our C5 structure also slightly deviates from the structure of C5 bound to CVF (PDB 3PVM) (Fig. S5A) (13, 19, 27, 28), with the most apparent differences found in C5a, similar as described above for native C5 (Fig. S5B). Interestingly, the conformation of C5 in complex with our nanobodies is similar to those observed for C5 in complex with the antigen binding fragment (Fab) of eculizumab (eculizumab-Fab) (PDB 5I5K) and OmCI-RaCI3 (PDB 5HCC) (Fig. S5C). In all these structures, the scissile loop in the C5a domain adopts a similar helical conformation (Fig. S5D). In conclusion, the differences between the structure of C5:UNbC5-1:UNbC5-2 and native C5 are similar to the previously described C5 structures in complex with known inhibitors.
Figure 3.
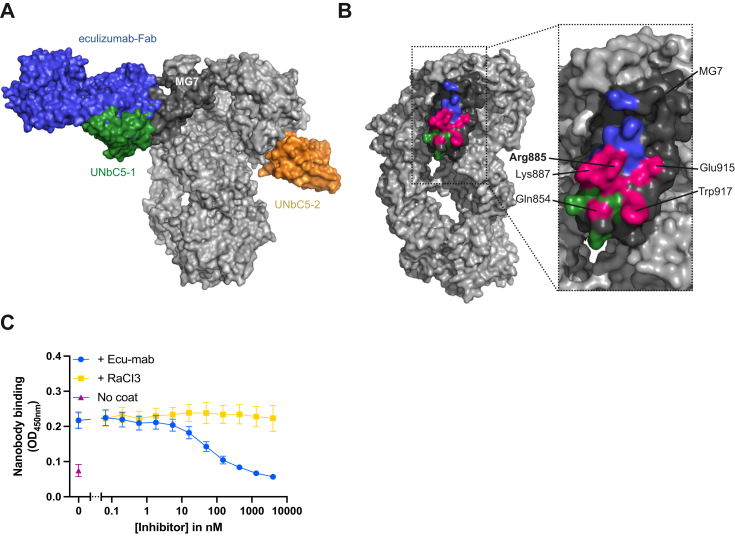
Cryo-EM structures of UNbC5-1 and UNbC5-2 in complex with C5.A, cryo-EM density map of the C5:UNbC5-1:UNbC5-2 complex at 3.6 Å resolution is depicted with C5 colored in gray, UNbC5-1 in green, and UNbC5-2 in orange. B, comparison of the C5 structure excluding the C345c domain (3CU7, blue) to the C5 of the C5:UNbC5-1:UNbC5-2 structure (gray). Left panel shows both structures in cartoon representation, superimposed and aligned by the C5 MG-ring. Domains C5a, C5d, and CUB of the C5:UNbC5-1:UNbC5-2 structure shows a displacement when compared to the C5 structure. Right panel shows a zoom in of the domain C5a reoriented and aligned, both structures with residue R751 showed as sticks in orange for the C5:UNbC5-1:UNbC5-2 structure and in cyan for C5 (3CU7). C–E, zoom in on the C5 interface with nanobodies UNbC5-1 (C and D), and UNbC5-2 (E), in the same colors introduced in panel A. Amino acids that likely contribute to the interface are shown as sticks representation and annotated. Data information: (A), cryo-EM density map visualized in ChimeraX and (B–E) figures produced in PyMOL from refined structure. CUB, complement C1r/C1s, Uegf, Bmp1; MG, macroglobulin.
Binding interfaces
UNbC5-1 and UNbC5-2 bind to the C5 α-chain on opposite domains and with a different epitope–paratope architecture (Fig. 3A). C5:UNbC5-1 displays a binding interface consisting of a hydrophobic core surrounded by polar interactions, whereas the C5:UNbC5-2 interface is mostly formed by salt bridges and hydrogen bonds.
The interface between C5 and nanobody UNbC5-1 buried a total area of 1535 Å2 (http://www.pymol.org/pymol). UNbC5-1 binds to domain MG7 in C5. This domain is considered a known target for inhibitory molecules for C5 and its homologous complement proteins C4b and C3/C3b. Reported inhibitors that target this domain in C5 are antibody eculizumab (and its derivatives) (19), while nanobodies NbE3 and hC4Nb8 (22, 29) target domain MG7 in C4b, and nanobody hC3Nb1 (30) targets domain MG7 in C3/C3b. In addition, proposed convertase models (Staphylococcal complement inhibitor-convertase (PDB 2WIN), and CVF-C5 (PDB 3PVM)) suggest the involvement of this domain in the convertase–substrate interaction (28, 31). Nanobody UNbC5-1 binds to the connecting loops of the four-stranded antiparallel β-sheet in the MG7 domain in C5 (Fig. 3, C and D). Most of the interface in UNbC5-1 is formed by the extended CDR3, with limited contribution of CDR1 and 2. Residues Tyr101–Ile104, of the CDR3 loop, form a hydrophobic core that interacts with three of C5-MG7 loops with residues Met853, Val888, Trp917, and Phe918. Furthermore, nanobody residues, Arg105 forms a cation-π interaction with residue Trp917 in C5 (Fig. 3C). The paratope architecture is also maintained by residues Tyr32 of CDR1 and Tyr101 of CDR3 that sandwich Phe100 of CDR3, where Tyr101 interacts with C5-MG7 residue Lys887 (Fig. 3D). Additionally, CDR3 residues Arg107 and Asp113 form salt bridges with Glu915 and Arg885 of C5-MG7, respectively (Fig. 3C). Finally, CDR1 helps to stabilize the interface with interactions between CDR1 Asp31 and MG7 Thr850. CDR2 may also contribute to the interface due to its backbone proximity to C5-MG7; nevertheless, side chain interactions cannot be assigned unambiguously.
The C5:UNbC5-2 buried a surface area of 1084 Å2. The C5 binding epitope of UNbC5-2 comprises residues of several α-helices and loops of the C5d domain (Fig. 3E). Similar as the MG7 domain, the C5d domain is targeted by multiple complement inhibitors, including OmCI and RaCI3 (13). The C5 interface is formed by the three CDRs of the nanobody with a major contribution of CDR3. Residue Ile57 of CDR2 interacts with a hydrophobic patch formed by residues Pro1160–Val1162 in C5d. Additionally, some electrostatic interactions are formed between C5d residue Lys1091 and residue Glu99 of CDR3 and between C5d residue Asp1165 with Arg103 from CDR3. Moreover, hydrogen bond interactions are formed between Arg101 from CDR3 and C5d-residue Leu1197 and between C5d residue Asp1157 and the backbones of Pro100 and Arg101 of CDR3. Additionally, the backbone of Asp1157 also interacts with residue Thr33 of CDR1. Other observed interactions are C5d residue Gln1097 and residue Asn54 of CDR2 and stacking of residue Arg103 with Phe1156 of C5d.
The C5-binding interface of UNbC5-2 partly overlaps with RaCI3
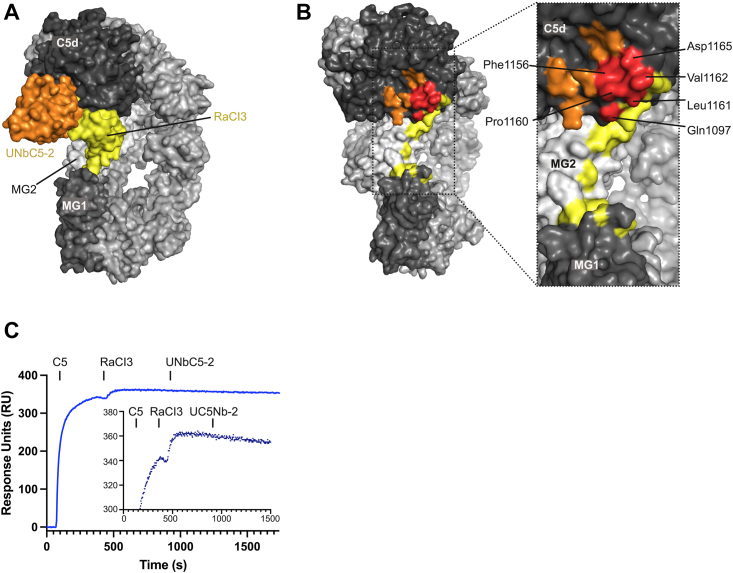
The UNbC5-2 interface in C5 is close to that of RaCI3; therefore, we next compared the structure of C5:UNbC5-1:UNbC5-2 with C5-OmCI-RaCI3 (PDB 5HCC) (Fig. 4A). RaCI3 binds to the cleft formed by domain MG1, MG2, and C5d, interacting with all three domains of C5 (13). A detailed comparison revealed six overlapping residues between the UNbC5-2 and RaCI3 interfaces on C5d (Fig. 4B) (13). In contrast to the C5-RaCI3 interactions, which are mostly driven by extensive van der Waals interactions and hydrogen bonds, the C5:UNbC5-2 interface include hydrophobic interactions with residues Pro1160–Val1162, a hydrogen bond with Gln1097 and a salt bridge with residue Asp1165 of C5d. To investigate whether UNbC5-2 and RaCI3 can bind C5 simultaneously, we performed an SPR assay. Here, we used a Fab domain with the same primary sequence as the Fab domain of eculizumab (Ecu-Fab) as a bait to capture C5 and sequentially injected RaCI3 and UNbC5-2. As expected, we measured association of C5 and RaCI3 (Fig. 4C). Interestingly, there was no increase in signal when UNbC5-2 was added. This indicates that UNbC5-2 and RaCI3 cannot bind C5 together. To conclude, the structural model and SPR assay indicate that the binding epitopes of UNbC5-2 and RaCI3 on C5 partially overlap and that both inhibitors cannot bind C5 simultaneously.
Figure 4.
The C5-binding interface of UNbC5-2 partially overlaps with RaCI3.A, superposition of the C5:UNbC5-1:UNbC5-2 and the C5-RaCI3 complex in surface representation with UNbC5-2 in orange, RaCI3 in yellow, most of the C5 domains in light gray, MG1 in medium gray, and C5d in dark gray. B, left panel shows a 45° rotation view of panel A where footprint of epitopes of UNbC5-2, RaCI3, and their overlapping residues (red) are colored according to panel A. Right panel shows a zoom in of UNbC5-2 and RaCI3 epitopes where overlapping residues are annotated. C, competition SPR between UNbC5-2 and RaCI3. Ecu-Fab was used as a bait followed up by injections of 100 nM C5, RaCI3, and UNbC5-2, at time points 60, 360, and 900 s, respectively. Injection of RaCI3 shows binding to C5 bound to Ecu-Fab. Additionally, sequential injection of UNbC5-2 at 900 s showed no signal increase, denoting competition. Figure also shows a zoom-in of RaCI3 and UNbC5-2 injections from time 0 to 1500 s where response units (RU) 300 to 380 are shown. Data information: (A and B), figures produced in PyMOL. C, experiments performed in duplicate. MG, macroglobulin; SPR, surface plasmon resonance.
UNbC5-1 competes with eculizumab on binding the MG7 domain of C5
A comparative analysis of C5-eculizumab-Fab and C5:UNbC5-1:UNbC5-2, showed an extensive overlap of both the eculizumab–Fab and UNbC5-1 epitope (Fig. 5A), with six overlapping residues (Fig. 5B). The eculizumab-Fab epitope extends through the antiparallel β-strands of the C5-MG7 domain, while the UNbC5-1 epitope is shifted downward and binds part of the β-strands and their connecting loops (19). Eculizumab binds the critical C5 residue Arg885 through a hydrophobic core formed by eculizumab residues Trp33, Phe101 and Trp107 (21). Instead, UNbC5-1 forms a salt bridge with C5-Arg885, at the edge of the epitope interface (Fig. 3D). Both, UNbC5-1 in our structural data and biochemical data with eculizumab (21), show interactions with C5-residues Trp917, Arg885, and Lys887, indicating a binding overlap for both inhibitors (19, 21). To confirm with an in vitro experiment that UNbC5-1 competes with eculizumab for C5 binding, we performed a competition ELISA. Briefly, we coated C5 and measured binding of UNbC5-1 in the presence of a titration of Ecu-mab. As a negative control we added RaCI3 instead of Ecu-mab. Consistent with the structural data, we observed a decrease in UNbC5-1 binding in the presence of Ecu-mab, but not with RaCI3 (Fig. 5C).
Figure 5.
UNbC5-1 competes witheculizumab in binding to the MG7 domain of C5.A, superposition of the C5:UNbC5-1:UNbC5-2 and the C5-eculizumab-Fab complex structure. Surface representation shows epitope overlap between UNbC5-1 and eculizumab-Fab. UNbC5-1 is shown in green, UNbC5-2 in orange, eculizumab-Fab in blue, most domains of C5 in light gray, and domain MG7 in dark gray. B, left panel shows a 45° rotation of panel A. Right panel shows a zoom in of the epitope of UNbC5-1 (green) and eculizumab-Fab (blue) on the MG7 domain (dark gray) with an extensive overlap (pink) of the epitope between the interface of both molecules in C5. The overlapping amino acids for both interfaces are indicated, with residue R885 in bold. C, competition ELISA with C5-coated microtiter plates incubated with 300 nM UNbC5-1 and a titration of Ecu-mab or RaCI3 (5.49–4000 nM, 3-fold). Binding of UNbC5-1 to C5 was measured using polyclonal rabbit-anti-VHH QE19 antibodies and donkey-anti-rabbit-HRP antibodies at A450 nm. Data information: (A and B) figures produced in PyMOL, (C), data represent mean ± SD of four individual experiments. MG, macroglobulin.
UNbC5-1 and UNbC5-2 bind and inhibit the human C5 variant R885H and are not cross-reactive with murine C5
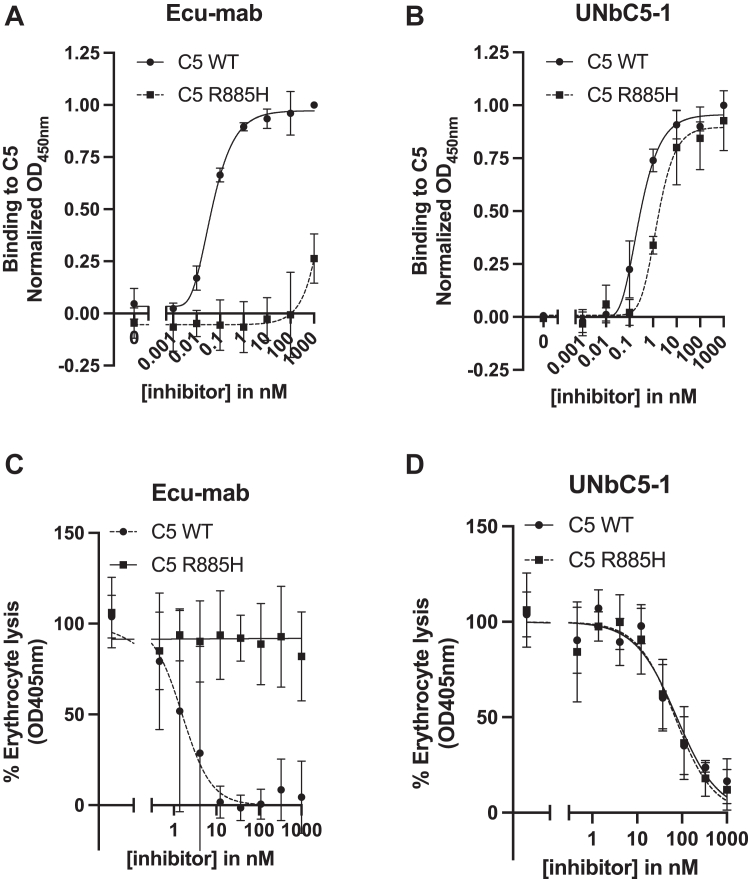
Next, we wondered if UNbC5-1 could recognize the genetic variant C5 R885H (18) that is not targeted by eculizumab. First, we coated microtiter plates with C5 WT and C5 R885H, added UNbC5-1 or Ecu-mab, and measured their binding. As described for eculizumab (18, 20), we observed in ELISA and SPR that Ecu-mab binds C5 R885H poorly (Figs. 6A and S6A). Interestingly, UNbC5-1 binds C5 R885H efficiently (Fig. 6B and Table 1), with a binding affinity of 15.5 nM (Fig. S6B). This affinity is roughly 100 × lower compared to the affinity for C5 WT, which confirms that residue 885 is involved in the binding interface of UNbC5-1 and C5, but is not crucial for its interaction, like it is for eculizumab. Next, we assessed whether UNbC5-1 was also able to prevent complement-mediated erythrocyte lysis with C5 R885H. To assess this, we incubated antibody-coated sheep erythrocytes with C5-depleted serum, replete with physiological concentrations of C5 WT or C5 R885H. Consistent with the binding data, we observed that Ecu-mab failed to inhibit complement activity via C5 R885H (Fig. 6C). On the contrary, UNbC5-1 inhibits complement activity with C5 R885H and C5 WT to a similar extent (Fig. 6D). Furthermore, for UNbC5-2, which binds C5 to an epitope unrelated to amino acid 885, we also observed similar binding efficiencies in ELISA (Fig. S6C). Next, we show that UNbC5-2 inhibits complement-mediated lysis with comparable potencies using C5 R885H and C5 WT (Fig. S6D). Finally, we assessed if UNbC5-1 and UNbC5-2 are cross-reactive with murine C5. Therefore, we performed an AP erythrocyte lysis assay with rabbit erythrocytes and mouse serum. Next to control C5-inhibitors Ecu-mab and RaCI3, we included SSL7, a C5 inhibitor that is described to be cross-reactive with murine C5 (32). UNbC5-1 and UNbC5-2 did not prevent erythrocyte lysis with murine serum (Fig. S6E). As expected SSL7 potently inhibited erythrocyte lysis with murine serum, while RaCI3 only inhibited in the two highest concentrations tested and Ecu-mab did not inhibit at all. A sequence alignment of human and murine C5 confirmed that cross-reactivity would be unlikely, since multiple amino acids involved in the C5:nanobody interfaces are different in C5 from mouse compared to C5 from human (Fig. S6F). Altogether, these data show that UNbC5-1 and UNbC5-2 bind and inhibit human C5 WT and the genetic variant C5 R885H, but are not cross-reactive with murine C5.
Figure 6.
UNbC5-1 binds and inhibits C5 variant R885H.A and B, binding of Ecu-mab (A) and UNbC5-1 (B) to C5 WT and C5 R885H, using C5 WT- or C5 R885H-coated microtiter plates, incubated with increasing concentrations of Ecu-mab or UNbC5-1. Binding was assessed with a monoclonal anti-human-kappa antibody (A) or polyclonal rabbit-anti-VHH QE19 antibodies (B) and donkey-anti-rabbit-HRP secondary antibodies (A and B) at A450. C and D, CP mediated hemolysis of antibody-coated sheep erythrocytes incubated with 2.5% C5-depleted human serum and repleted with physiological concentrations of C5 WT or C5 R885H and a titration of Ecu-mab (C) or UNbC5-1 (D). The A450 values of the supernatants were measured, and the % erythrocyte lysis was calculated using a 0% (buffer) and 100% (milliQ water) control sample. Data information: (A and D), data represent mean ± SD of three individual experiments and curves were fitted. CP, classical pathway.
Discussion
With the identification of UNbC5-1 and UNbC5-2 as specific, high affinity C5 targeting nanobodies, this study adds two new C5 inhibitors to the field. We determined the structure of C5 in complex with both nanobodies UNbC5-1 and UNbC5-2 simultaneously to identify their binding interfaces and potentially get insight into the modes of action. This approach was chosen for experimental efficiency and was performed similarly for C5 in complex with inhibitors CirpT, OmCI, and RaCI1, 2, 3 (12, 13).
For UNbC5-1, the mode of action is likely similar to that of eculizumab, since both the cryo-EM structure and competitive binding experiment reveal overlapping C5-binding sites. For eculizumab, it was suggested that binding to the MG7 domain sterically hinders the interaction between C5 and the C5 convertase, preventing C5 cleavage (19).
For UNbC5-2, the inhibitory mechanism is less evident. Because the binding interface of UNbC5-2 partially overlaps with RaCI3 (13), the inhibitory mechanism could be similar to RaCI. For the RaCI family, it is suggested that binding to C5d (together with OmCI) induces a conformational change in C5 in which the disordered and exposed scissile loop (residues Asp746–Met754) is altered into an ordered α-helix with the cleavage site (Arg751–Leu752) unavailable for C5 cleavage (13, 19, 27, 33). As observed for eculizumab–Fab (derived from mice (19)), OmCI–RaCI3 (derived from ticks (13)) and knob K92 (derived from cows (27)), our llama-derived nanobodies similarly induce a conformational change that leads to the formation of an ordered scissile loop in C5a. The formation of an α-helix is comparable to the conformational change described for small molecule inhibitor (compound 7), which directly interacts with the scissile loop in C5a (34). Since we generated a combined structure with both nanobodies, it is impossible with our data to attribute this proposed mechanism of inhibition to either UNbC5-1, or UNbC5-2, or both. Nevertheless, this structural change in C5 has been suggested to be part of the inhibitory mechanism for the RaCI family, and more recently also for knob K92 (27). This, together with our data, suggests that this partial molecular movement of the α-chain and helical rearrangement of the scissile loop is a common inhibitory mechanism that can be triggered by various C5-inhibitors from different sources and targeting different epitopes across the molecule (12, 19, 27).
Although eculizumab is proven to be an effective therapy in controlling diseases like atypical hemolytic uremic syndrome, a drawback is the therapy resistance in a small proportion of patients with C5 polymorphism (18). This was explained by the fact that the interaction of eculizumab with the Arg885 residue is essential for binding (21). Interestingly, we observed that UNbC5-1 can bind and can inhibit the R885H variant of C5. Even though UNbC5-1 does interact with Arg885, the structure suggests that mutation of Arg885 into a histidine has less impact on the C5:UNbC5-1 interface, because Arg885 interacts mostly with a polar patch (formed by residues Glu915 of MG7 and Arg107 and Asp113 of CDR3 at the edge of the interface) and a His-885 variant could probably still establish a polar interaction. In contrast, Arg885 extends into a hydrophobic pocket in the eculizumab interface surrounded by bulky side chain amino acids such as Trp33, Phe101, and Trp107 where a histidine side chain might be too small to extend inward and interact, therefore disrupting the core interaction of the interface (19, 21). We propose that the detailed binding interface of UNbC5-1 and C5 is a good starting point to further optimize reactivity of UNbC5-1 toward the genetic variant of C5 R885H and obtain more insight in the requirements for efficient C5 inhibition in general.
In the last decades, diverse C5 targeting molecules have been identified and developed, including monoclonal antibodies, single domain antibodies, small molecules, nucleic acid-based therapies, pathogen derived proteins and peptides, but no nanobodies (16). In contrast, nanobodies against other complement components (C1q (35), C3/C3b (30, 36, 37), C4b (22, 29), properdin (38, 39)), and a complement receptor (CRIg (40, 41)) were previously identified. The use of nanobodies as therapeutic molecules has gained increasing interest in many different fields, especially since the successful introduction of the first nanobody in the clinic, caplacizumab (targeting plasma protein von Willebrand factor) (42). Compared to conventional antibodies, nanobodies lack an Fc-tail to activate the immune system, which is favorable when complement inhibition is the goal. At the same time, the lack of an Fc-tail shortens the half-life of nanobodies in circulation while improving tissue penetration. In addition to their therapeutic potential, nanobodies are increasingly used as tools in research and diagnostics because they are easy to produce and label with for example fluorophores (43, 44) or to conjugate to solid matrixes for chromatography (45). Also, the presented C5-specific nanobodies could be developed for specific detection of C5 in complex with biological specimens or for affinity purification of C5 from human plasma. Furthermore, their inhibitory action makes them suited as tools to inhibit the complement system at level of C5 cleavage, thereby studying the role of complement in diseases.
To conclude, in this study we developed and characterized two anti-C5 nanobodies that bind C5 with picomolar affinities and efficiently inhibit complement at the level of C5 cleavage. Both nanobodies can be used to detect C5 and inhibit C5 cleavage for diagnostic and/or research purposes. Finally, both nanobodies have the potential to be further developed for therapeutic purposes, and detailed binding interfaces obtained here could help to further develop high affinity C5 inhibitors that are reactive to different genetic variants of C5.
Experimental procedures
Serum, proteins, and complement inhibitors
Normal human serum was obtained from a pool of healthy donors as previously described (46). C5-depleted serum was obtained from Complement Technology. Complement protein C3 was isolated and purified from freshly obtained human plasma, as described before (47). Complement protein C4 was obtained from Complement Technology. Complement proteins C5 WT (unless stated differently) and C5 R885H and complement inhibitor RaCI3 were recombinantly produced and purified in our lab, using Expi293F cells.
C5 WT and C5 R885H
For C5 expression, we used the pcDNA 3.4 TOPO vector (Thermo Fisher Scientific). This vector was made circular by inserting a multiple cloning site (pcDNA34-MCS) using the pcDNA 3.4 TOPO TA cloning kit (Thermo Fisher Scientific). The XbaI and AgeI restriction sites, adjacent to the original pcDNA34-TOPO site, were used to clone the cystatin(S) signal peptide (coding for MARPLCTLLLLMATLAGALA) with an upstream Kozak (ACCACC) sequence and a downstream NheI and NotI cloning site, followed by a C-terminal sequence coding for a linker (GGGGS), an LPETGG site, and a 6x histidine tag (pcDNA34-Cystatin(S)-Linker-LPETG-HIS vector). The 13 and 15 bp sequences, adjacent to the XbaI and AgeI site and the original pcDNA34-TOPO site, were left intact. Subsequently, the C5 gene (a kind gift from U-Protein Express) without start and stop codon was cloned into this vector, using the NheI and NotI sites (Table 4). For cloning the C5-R885H mutant, we used overlap extension PCR to introduce the R885H mutation using the pcDNA34-Cystatin(S)-C5-Linker-LPETG-HIS vector as a template. Primers used were the following: 5-XbaI: 5′GACCGATCCAGCCTCCGG ACTCTAGAGGATCGAAC and 3-R885H: 5′ GAGCCCTCTACTTTCTGGTGCACACAT TTGGAGGACTTTG combined with 5-R885H: CTCCAAATGTGTGCACCAGAAAG TAGAGGGCTCCTC and 3-AgeI: 5′ GATATCAAACTCATTACTAACCGGTAGGGATC GAAC, with R885H mutations depicted in bold. The final C5-R885H PCR product was cloned into the XbaI and AgeI site of the pcDNA34-MCS vector using Gibson assembly (New England Biolabs). After verification of the correct sequences, the C5 and C5-R885H plasmids were used to transfect Expi293F cells (Thermo Fisher Scientific). Expi293F cells were grown in Expi293 medium (Life Technologies) in culture filter cap Erlenmeyer bottles (Corning) on a rotation platform (125 rotations/min) at 37 °C, 8% CO2. One day before transfection, cells were diluted to 2 × 106 cells/ml. The next day, cells were diluted to 2 × 106 cells/ml using SFM4Transfx-293 medium, containing UltraGlutamine I (VWR International) prior to transfection using PEI HCl MAX (Polysciences). Therefore, 0.5 μg DNA/ml cells, containing 50% empty pcDNA34-MCS “dummy” vector, was added to Opti-MEM (1:10 of total volume; Gibco) and gently mixed. After adding PEI (1 μg/ml) in a PEI/DNA (w/w) ratio of 5:1, the mixture was incubated at room temperature (RT) for 20 min and then added dropwise to the cells while manually rotating the Erlenmeyer culture bottle. After 3 to 5 h, 1 mM valproic acid was added to the transfected cells. After 5 days of expression, the cell supernatant was collected by centrifugation and filtration (0.45 μM). C5 and C5-R885H supernatants were concentrated and buffer exchanged to 25 mM Hepes, 500 mM NaCl, 5 mM benzamidine, pH8.2 using the QuixStand benchtop system (GE HealthCare) and 25 mM imidazole was added before application to a HiTrap Chelating column (GE HealthCare). The column was washed with 50 mM imidazole, before C5 and C5-R885H were eluted using 175 mM imidazole using the ÄKTA pure protein chromatography system (GE HealthCare). C5 and C5-R885H were dialyzed to PBS and stored at −80 °C.
Table 4.
Constructs and sequences used for protein purifications
| # | Description/sequence |
|---|---|
| 1 | pcDNA34-Cystatin(S)-C5-Linker-LPETG-HIS vector sequence from the XbaI up to the AgeI sequence, in which the NheI and NotI site are depicted in gray, the start (ATG) and stop (TAA) codon are underlined and the 13 bp and 15 bp sequences adjacent to the XbaI and AgeI sites are in bold. |
| TCTAGAGGATCGAACCCTTACCACCATGGCAAGACCTCTTTGTACCCTGCTTCTGCTCATGGCGACACTGGCTGGGGCCCTCGCCGCTAGCCAGGAGCAAACATATGTCATTTCAGCACCAAAAATATTCCGTGTTGGAGCATCTGAAAATATTGTGATTCAAGTTTATGGATACACTGAAGCATTTGATGCAACAATCTCTATTAAAAGTTATCCTGATAAAAAATTTAGTTACTCCTCAGGCCATGTTCATTTATCCTCAGAGAATAAATTCCAAAACTCTGCAATCTTAACAATACAACCAAAACAATTGCCTGGAGGACAAAACCCAGTTTCTTATGTGTATTTGGAAGTTGTATCAAAGCATTTTTCAAAATCAAAAAGAATGCCAATAACCTATGACAATGGATTTCTCTTCATTCATACAGACAAACCTGTTTATACTCCAGACCAGTCAGTAAAAGTTAGAGTTTATTCGTTGAATGACGACTTGAAGCCAGCCAAAAGAGAAACTGTCTTAACTTTCATAGATCCTGAAGGATCAGAAGTTGACATGGTAGAAGAAATTGATCATATTGGAATTATCTCTTTTCCTGACTTCAAGATTCCGTCTAATCCTAGATATGGTATGTGGACGATCAAGGCTAAATATAAAGAGGACTTTTCAACAACTGGAACCGCATATTTTGAAGTTAAAGAATATGTCTTGCCACATTTTTCTGTCTCAATCGAGCCCGAATATAATTTCATCGGCTACAAGAACTTTAAGAACTTCGAGATTACCATCAAGGCCAGATACTTCTATAATAAGGTAGTCACTGAGGCTGACGTTTATATCACATTTGGAATAAGAGAAGACTTAAAAGATGATCAAAAAGAAATGATGCAAACAGCAATGCAAAACACAATGTTGATAAATGGAATTGCTCAAGTCACATTTGATTCTGAAACAGCAGTCAAAGAACTGTCATACTACAGTTTAGAAGATTTAAACAACAAGTACCTTTATATTGCTGTAACAGTCATAGAGTCTACAGGTGGATTTTCTGAAGAGGCAGAAATACCTGGCATCAAATATGTCCTCTCTCCCTACAAACTGAATTTGGTTGCTACTCCTCTTTTCCTGAAGCCTGGGATTCCATATCCCATCAAGGTGCAGGTTAAAGATTCGCTTGACCAGTTGGTAGGAGGAGTCCCAGTAACACTGAATGCACAAACAATTGATGTAAACCAAGAGACATCTGACTTGGACCCAAGCAAAAGTGTAACACGTGTTGATGATGGAGTAGCTTCCTTTGTGCTTAATCTCCCATCTGGAGTGACGGTGCTGGAGTTTAATGTCAAAACTGATGCTCCAGATCTTCCAGAAGAAAATCAGGCCAGGGAAGGTTACCGAGCAATAGCATACTCATCTCTCAGCCAAAGTTACCTTTATATTGATTGGACTGATAACCATAAGGCTTTGCTAGTGGGAGAACATCTGAATATTATTGTTACCCCCAAAAGCCCATATATTGACAAAATAACTCACTATAATTACTTGATTTTATCCAAGGGCAAAATTATCCACTTTGGCACGAGGGAGAAATTTTCAGATGCATCTTATCAAAGTATAAACATTCCAGTAACACAGAACATGGTTCCTTCATCCCGACTTCTGGTCTATTACATCGTCACAGGAGAACAGACAGCAGAATTAGTGTCTGATTCAGTCTGGTTAAATATTGAAGAAAAATGTGGCAACCAGCTCCAGGTTCATCTGTCTCCTGATGCAGATGCATATTCTCCAGGCCAAACTGTGTCTCTTAATATGGCAACTGGAATGGATTCCTGGGTGGCATTAGCAGCAGTGGACAGTGCTGTGTATGGAGTCCAAAGAGGAGCCAAAAAGCCCTTGGAAAGAGTATTTCAATTCTTAGAGAAGAGTGATCTGGGCTGTGGGGCAGGTGGTGGCCTCAACAATGCCAATGTGTTCCACCTAGCTGGACTTACCTTCCTCACTAATGCAAATGCAGATGACTCCCAAGAAAATGATGAACCTTGTAAAGAAATTCTCAGGCCAAGAAGAACGCTGCAAAAGAAGATAGAAGAAATAGCTGCTAAATATAAACATTCAGTAGTGAAGAAATGTTGTTACGATGGAGCCTGCGTTAATAATGATGAAACCTGTGAGCAGCGAGCTGCACGGATTAGTTTAGGGCCAAGATGCATCAAAGCTTTCACTGAATGTTGTGTCGTCGCAAGCCAGCTCCGTGCTAATATCTCTCATAAAGACATGCAATTGGGAAGGCTACACATGAAGACCCTGTTACCAGTAAGCAAGCCAGAAATTCGGAGTTATTTTCCAGAAAGCTGGTTGTGGGAAGTTCATCTTGTTCCCAGAAGAAAACAGTTGCAGTTTGCCCTACCTGATTCTCTAACCACCTGGGAAATTCAAGGCGTTGGCATTTCAAACACTGGTATATGTGTTGCTGATACTGTCAAGGCAAAGGTGTTCAAAGATGTCTTCCTGGAAATGAATATACCATATTCTGTTGTACGAGGAGAACAGATCCAATTGAAAGGAACTGTTTACAACTATAGGACTTCTGGGATGCAGTTCTGTGTTAAAATGTCTGCTGTGGAGGGAATCTGCACTTCGGAAAGCCCAGTCATTGATCATCAGGGCACAAAGTCCTCCAAATGTGTGCGCCAGAAAGTAGAGGGCTCCTCCAGTCACTTGGTGACATTCACTGTGCTTCCTCTGGAAATTGGCCTTCACAACATCAATTTTTCACTGGAGACTTGGTTTGGAAAAGAAATCTTAGTAAAAACATTACGAGTGGTGCCAGAAGGTGTCAAAAGGGAAAGCTATTCTGGTGTTACTTTGGACCCTAGGGGTATTTATGGTACCATTAGCAGACGAAAGGAGTTCCCATACAGGATACCCTTAGATTTGGTCCCCAAAACAGAAATCAAAAGGATTTTGAGTGTAAAAGGACTGCTTGTAGGTGAGATCTTGTCTGCAGTTCTAAGTCAGGAAGGCATCAATATCCTAACCCACCTCCCCAAAGGGAGTGCAGAGGCGGAGCTGATGAGCGTTGTCCCAGTATTCTATGTTTTTCACTACCTGGAAACAGGAAATCATTGGAACATTTTTCATTCTGACCCATTAATTGAAAAGCAGAAACTGAAGAAAAAATTAAAAGAAGGGATGTTGAGCATTATGTCCTACAGAAATGCTGACTACTCTTACAGTGTGTGGAAGGGTGGAAGTGCTAGCACTTGGTTAACAGCTTTTGCTTTAAGAGTACTTGGACAAGTAAATAAATACGTAGAGCAGAACCAAAACAGCATCTGCAACAGCTTATTGTGGCTAGTTGAGAATTATCAATTAGATAATGGATCTTTCAAGGAAAATTCACAGTATCAACCAATAAAATTACAGGGTACCTTGCCTGTTGAAGCCCGAGAGAACAGCTTATATCTTACAGCCTTTACTGTGATTGGAATTAGAAAGGCTTTCGATATATGCCCCCTGGTGAAAATCGACACAGCTCTAATTAAAGCTGACAACTTTCTGCTTGAAAATACACTGCCAGCCCAGAGCACCTTTACATTGGCCATTTCTGCGTATGCTCTTTCCCTGGGAGATAAAACTCACCCACAGTTTCGTTCAATTGTTTCAGCTTTGAAGAGAGAAGCTTTGGTTAAAGGTAATCCACCCATTTATCGTTTTTGGAAAGACAATCTTCAGCATAAAGACAGCTCTGTACCTAACACTGGTACGGCACGTATGGTAGAAACAACTGCCTATGCTTTACTCACCAGTCTGAACTTGAAAGACATCAACTACGTGAACCCAGTCATCAAATGGCTATCAGAAGAGCAGAGGTATGGAGGTGGCTTTTATTCAACCCAGGACACAATCAATGCCATTGAGGGCCTGACGGAATATTCACTCCTGGTTAAACAACTCCGCTTGAGTATGGACATCGATGTTTCTTACAAGCATAAAGGTGCCTTACATAATTATAAAATGACAGACAAGAATTTCCTTGGGAGGCCAGTAGAGGTGCTTCTCAATGATGACCTCATTGTCAGTACAGGATTTGGCAGTGGCTTGGCTACAGTACATGTAACAACTGTAGTTCACAAAACCAGTACCTCTGAGGAAGTTTGCAGCTTTTATTTGAAAATCGATACTCAGGATATTGAAGCATCCCACTACAGAGGCTACGGAAACTCTGATTACAAACGCATAGTAGCATGTGCCAGCTACAAGCCCAGCAGGGAAGAATCATCATCTGGCTCCTCTCATGCGGTGATGGACATCTCCTTGCCTACTGGAATCAGTGCAAATGAAGAAGACTTAAAAGCCCTTGTGGAAGGGGTGGATCAACTATTCACTGATTACCAAATCAAAGATGGACATGTTATTCTGCAACTGAATTCGATTCCCTCCAGTGATTTCCTTTGTGTACGATTCCGGATATTTGAACTCTTTGAAGTTGGGTTTCTCAGTCCTGCCACTTTCACAGTGTACGAATACCACAGACCAGATAAACAGTGTACCATGTTTTATAGCACTTCCAATATCAAAATTCAGAAAGTCTGTGAAGGAGCCGCGTGCAAGTGTGTAGAAGCTGATTGTGGGCAAATGCAGGAAGAATTGGATCTGACAATCTCTGCAGAGACAAGAAAACAAACAGCATGTAAACCAGAGATTGCATATGCTTATAAAGTTAGCATCACATCCATCACTGTAGAAAATGTTTTTGTCAAGTACAAGGCAACCCTTCTGGATATCTACAAAACTGGGGAAGCTGTTGCTGAGAAAGACTCTGAGATTACCTTCATTAAAAAGGTAACCTGTACTAACGCTGAGCTGGTAAAAGGAAGACAGTACTTAATTATGGGTAAAGAAGCCCTCCAGATAAAATACAATTTCAGTTTCAGGTACATCTACCCTTTAGATTCCTTGACCTGGATTGAATACTGGCCTAGAGACACAACATGTTCATCGTGTCAAGCATTTTTAGCTAATTTAGATGAATTTGCCGAAGATATCTTTTTAAATGGATGCGCGGCCGCAGGAGGAGGCGGTTCCCTGCCTGAAACGGGCGGTCACCATCACCATCACCATTAAAAGGGTTCGATCCCTACCGGT | |
| 2 | pcDNA34-Cystatin(S)-RacI3-LPETG-HIS vector sequence from the XbaI up to the AgeI sequence, in which the NheI and NotI site are depicted in gray, the start (ATG) and stop (TAA) codon are underlined. |
| TCTAGAGGATCGAACCCTTACCACCATGGCAAGACCTCTTTGTACCCTGCTTCTGCTCATGGCGACACTGGCTGGGGCCCTCGCCGCTAGCAGCGGAGAGTCTCAGTCAATTCAGAGGAAAGGGCAATGTGAGGAAGTCATTTGTCACCGAAAATTGAACCATCTCGGGGAAAGGGTCACCAGCGGATGCCCGACCGGTTGCCTCTGTGTCATAAGGGAGCCAGATAATGTCGATAATGCTAACGGGACATGCTATGCTCTGATGAGCAGTACGACCACGACCACAACAACACCGGACGGCACAACCACCAGTGAGGAGGAGGAAGCGGCCGCACTGCCTGAAACGGGCGGTCACCATCACCATCACCATTAAAAGGGTTCGATCCCTACCGGT | |
| 3 | Ecu-mab VH+ constant region gBlock, with the HAVT20 signal peptide depicted in bold: |
| GATCCAGCCTCCGGACTCTAGAGGATCGAACCCTTGAATTCACCACCATGGCTTGCCCTGGCTTCCTGTGGGCATTGGTAATCAGCACTTGCCTGGAGTTTTCTATGGCTCAGGTACAACTTGTGCAATCTGGGGCTGAAGTTAAAAAACCAGGTGCTAGTGTTAAAGTGAGCTGCAAAGCCAGCGGTTACATCTTCAGTAACTATTGGATACAATGGGTGCGACAAGCGCCCGGACAAGGCTTGGAGTGGATGGGCGAAATTCTGCCCGGAAGCGGTAGCACAGAATACACTGAGAACTTTAAAGATCGAGTAACGATGACCCGAGATACCAGTACTTCTACGGTTTATATGGAACTGTCATCACTGCGCTCTGAAGACACCGCGGTGTATTACTGTGCCAGGTATTTTTTTGGTAGTTCCCCTAACTGGTACTTCGACGTGTGGGGGCAGGGCACGCTGGTTACGGTCAGCTCCGCTAGCACCAAGGGCCCATCGGTCTTCCCCCTGGCGCCCTGCTCCAGGAGCACCTCCGAGAGCACAGCCGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCTGGATTGTATTCTTTGTCATCCGTCGTAACGGTGCCCAGTAGCAATTTCGGCACACAAACATATACTTGCAATGTAGATCATAAGCCAAGCAACACCAAGGTCGATAAGACCGTGGAAAGAAAGTGTTGCGTCGAGTGTCCTCCTTGCCCGGCACCACCCGTGGCGGGTCCCAGTGTCTTCCTGTTCCCACCGAAACCCAAGGATACTCTCATGATATCTCGAACCCCCGAGGTGACATGCGTCGTCGTGGATGTCTCTCAGGAAGACCCGGAGGTCCAATTTAACTGGTATGTTGATGGAGTAGAAGTACATAATGCAAAGACCAAACCACGGGAGGAGCAGTTTAATAGCACCTACAGAGTAGTGAGCGTACTGACGGTGCTTCATCAAGACTGGCTCAACGGAAAAGAATACAAATGTAAAGTGAGTAATAAGGGCCTTCCTAGCTCTATCGAAAAGACGATTTCAAAGGCCAAAGGCCAGCCCAGAGAGCCTCAAGTTTACACGCTTCCGCCATCCCAAGAAGAAATGACAAAGAACCAAGTATCACTCACGTGTTTGGTTAAAGGGTTCTATCCGTCAGACATCGCGGTAGAGTGGGAGTCTAATGGACAGCCTGAGAACAACTATAAAACCACTCCACCCGTTCTCGATTCCGATGGCAGCTTTTTCTTGTACAGTCGCCTGACCGTTGACAAAAGCCGCTGGCAGGAGGGCAACGTGTTCTCCTGTAGTGTGATGCACGAGGCGCTTCACAATCACTACACACAAAAGTCACTTTCTTTGAGCCTGGGCAAATGAACCGGTTAGTAATGAGTTTGATATCTC | |
| 4 | Ecu-mab VL+constant region gBlock, with the HAVT20 signal peptide depicted in bold: |
| GACCGATCCAGCCTCCGGACTCTAGAGGATCGAACCCTTGAATTCACCACCATGGCTTGCCCTGGCTTCCTGTGGGCATTGGTAATCAGCACTTGCCTGGAGTTTTCTATGGCTGATATACAGATGACACAGTCTCCCAGTAGTTTGTCTGCTTCAGTAGGGGATCGAGTCACGATTACATGCGGCGCTTCCGAAAATATCTATGGAGCACTCAACTGGTATCAGCAGAAACCCGGAAAGGCGCCAAAGTTGCTGATTTACGGGGCGACCAACCTGGCTGACGGGGTGCCTAGCAGGTTTAGCGGATCTGGCAGCGGCACAGACTTCACTCTTACAATTTCTTCTCTCCAACCGGAGGACTTCGCTACTTACTACTGCCAAAATGTGCTGAATACGCCATTGACGTTCGGCCAGGGAACCAAGGTCGAGATTAAGCGTACGGTGGCTGCACCATCTGTCTTCATCTTCCCGCCATCTGATGAGCAGTTGAAATCTGGAACTGCCTCTGTTGTGTGCCTGCTGAATAACTTCTATCCCAGAGAGGCCAAAGTACAGTGGAAGGTGGATAACGCCCTCCAATCGGGTAACTCCCAGGAGAGTGTCACAGAGCAGGACAGCAAGGACAGCACCTACAGCCTCAGCAGCACCCTGACGCTGAGCAAAGCAGACTACGAGAAACACAAAGTCTACGCCTGCGAAGTCACCCATCAGGGCCTGAGCTCGCCCGTCACAAAGAGCTTCAACAGGGGAGAGTGTTAGAAGGGTTCGATCCCTACCGGTTAGTAATGAGTTTGATATCTC | |
| 5 | Ecu-Fab VH+constant Fab region gBlock, with the HAVT20 signal peptide depicted in bold: |
| GATCCAGCCTCCGGACTCTAGAGGATCGAACCCTTGAATTCACCACCATGGCTTGCCCTGGCTTCCTGTGGGCATTGGTAATCAGCACTTGCCTGGAGTTTTCTATGGCTCAGGTACAACTTGTGCAATCTGGGGCTGAAGTTAAAAAACCAGGTGCTAGTGTTAAAGTGAGCTGCAAAGCCAGCGGTTACATCTTCAGTAACTATTGGATACAATGGGTGCGACAAGCGCCCGGACAAGGCTTGGAGTGGATGGGCGAAATTCTGCCCGGAAGCGGTAGCACAGAATACACTGAGAACTTTAAAGATCGAGTAACGATGACCCGAGATACCAGTACTTCTACGGTTTATATGGAACTGTCATCACTGCGCTCTGAAGACACCGCGGTGTATTACTGTGCCAGGTATTTTTTTGGTAGTTCCCCTAACTGGTACTTCGACGTGTGGGGGCAGGGCACGCTGGTTACGGTCAGCTCCGCTAGCACCAAGGGCCCATCGGTCTTCCCCCTGGCACCCTCCTCCAAGAGCACCTCTGGGGGCACAGCGGCCCTGGGCTGCCTGGTCAAGGACTACTTCCCCGAACCGGTGACGGTGTCGTGGAACTCAGGCGCCCTGACCAGCGGCGTGCACACCTTCCCGGCTGTCCTACAGTCCTCAGGACTCTACTCCCTCAGCAGCGTGGTGACCGTGCCCTCCAGCAGCTTGGGCACCCAGACCTACATCTGCAACGTGAATCACAAGCCCAGCAACACCAAGGTGGACAAGAAAGTTGAGCCCAAATCTTGTGGAGGTGGAGGCAGTCTGCCGGAGACCGGAGGGCACCATCACCATCACCATTGAAAGGGTTCGATCCCTACCGGTTAGTAATGAGTTTGATATC | |
RaCI 3
For RaCI 3 expression, a RaCI 3 gBlock was cloned into the NheI/NotI site of our pcDNA34-Cystatin(S)-LPETG-HIS vector (Table 4). This vector was cloned as described for C5 WT, with the exception that this pcDNA34 construct lacks the GGGGS linker. After verification of the correct sequence, the plasmid was used to transfect Expi293F cells, as described for the expression of C5 WT and C5 R885H. A total of 1 μg DNA/ml of cells was used. RaCI 3 was isolated using a HISTrap FF column (Cytiva, GE HealthCare) in the ÄKTA pure protein chromatography system (GE HealthCare) according to the manufacturer’s description. Before application to the column, the supernatant was dialyzed against 50 mM Tris, 500 mM NaCl, pH8.0, and 30 mM imidazole was added to reduce aspecific binding. RaCI 3 protein was eluted from the column via a 30 to 250 mM imidazole gradient. Fractions containing protein were collected and dialyzed against 50 mM Tris/300 mM NaCl, pH8.0.
Ecu-mab and Ecu-Fab
A human IgG2/4 monoclonal antibody with the same primary sequence as clinically approved eculizumab, denoted as Ecu-mab, was produced and purified in our lab. Briefly, the heavy and light chain variable plus constant region amino acid sequences of eculizumab (17) were from patent (WO2017044811A1). gBlocks containing codon optimized sequences of the VH + constant and VL + constant region with an upstream Kozak and HAVT20 signal peptide were cloned into the XbaI/AgeI site of the pcDNA34-MCS vector, using Gibson assembly (New England Biolabs) (Table 4). After verification of the correct sequence, plasmids were used to transfect Expi293F cells (Thermo Fisher Scientific), as described for the expression of C5 WT and C5 R885H, with the only exception that 1 μg DNA/ml of cells was used with a ratio of light chain:heavy chain of 3:2. Ecu–mab was purified using a HiTrap Protein A column (GE HealthCare) and the ÄKTA pure (GE HealthCare), according to the manufacturer’s description. Ecu-mab fractions were pooled and dialyzed against PBS. The Ecu-mab preparation was analyzed by gel filtration on a Superdex 200 10/300 GL column (GE HealthCare) and stored at −20 °C when >95% of Ecu-mab appeared as monomeric antibody. For Ecu-Fab (a Fab domain with the same primary sequence as the Fab domain of eculizumab), the light chain vector was similar as used for Ecu-mab IgG. The heavy chain constant region of Ecu-Fab ends with the amino acid sequence EPKSC, followed by a flexible linker (GGGGS), an LPETG site, and a 6x histidine tag. The gBlock containing the codon optimized sequence of the VH + constant Fab region with an upstream Kozak and HAVT20 signal peptide was cloned into the XbaI/AgeI site of the pcDNA34-MCS vector, using Gibson assembly (New England Biolabs) (Table 4). The expression of Ecu-Fab in Expi293 cells was similar as described for Ecu-mab IgG. The expression supernatant was dialyzed against 50 mM Tris, 500 mM NaCl, pH8.0, and 30 mM imidazole was added before application to a HISTrap column (GE HealthCare). Elution of Ecu-Fab was performed by a gradient of 30 to 250 mM imidazole. Isolated Ecu-Fab was dialyzed against PBS and stored at 4 °C and −80 °C.
Llama immunization, phage library, phage display, and periplasmic fractions
Two llamas (lama glama) were immunized with recombinant human C5 (WT). Phage display libraries were produced according to standard protocols (48). Briefly, complementary DNA was generated from RNA isolated from llama peripheral blood mononuclear cells. Nanobody-encoding genes were cloned into a phagemid pPQ81 vector (a derivative of pHEN1 (49)), and two phage display libraries with estimated sizes of 108 clones/library were generated. To select C5-binding nanobodies, we produced phages and performed two rounds of phage display, according to standard procedures (50, 51), with the antigen immobilized on Nunc Maxisorp plates (VWR 735–0083, Thermo Fisher Scientific) in different concentrations (round 1: 0.5 and 5.0 μg/ml, round 2: 0.1 and 1.0 μg/ml) to have diversity in stringency. Phages (∼1010 CFU/well round one and ∼109 CFU/well round 2) were added for 2 h, at RT and after incubation wells were washed extensively, to remove unbound phages. Phages were eluted with triethanolamine solution (pH > 10), for 15 min, at RT, while shaking, and rescued by infection in E. coli TG1 bacteria. Next, bacteria were plated and ∼200 single colonies were picked to obtain bacterial cultures with one clone/culture. Nanobody expression in the periplasm of E. coli was induced using IPTG for 4 h. Periplasmic fractions were collected via a cycle of freeze and thawing.
Nanobody production and purification
Nanobodies with Myc- and His-tags
For nanobody productions, pPQ81 vectors (derived from pHEN1 (49)) encoding the nanobody genes were transformed into E. coli Rosetta 2 (DE3) BL21 cells (Merck). pPQ81 vectors provided nanobodies with C-terminal Myc- and His-tags (AAASGSLEQKLISEEDLNGAAHHHHHHGAA). Transformed bacteria were inoculated in 2× yeast extract tryptone medium supplemented with 2% glucose and 100 μg/ml ampicillin and grown shaking, at 37 °C, overnight (O/N). The next day cultures were diluted 1:20 in 2× yeast extract tryptone medium supplemented with 0.1% glucose and 100 μg/ml ampicillin and grown at 37 °C, shaking, until an A600 of 0.6 to 0.9 was obtained. Then, nanobody production was induced with 1 mM IPTG and expression was continued for 4 h at 37 °C or O/N at RT under shaking conditions. Next, cultures were spun down for 10 min at 6000g. Pellets were dissolved in PBS and frozen for >30 min at −80 °C or O/N at −20 °C and thawed to release periplasmic fractions in the supernatant. Thawed pellets were centrifuged for 15 min at 6000g. Next, immobilized metal-affinity chromatography was used with ROTI Garose-His/Co beads (Roth) to purify nanobodies via their His-tag. After binding to the beads, nanobodies were eluted with 150 mM imidazole. Subsequently, nanobodies were dialyzed to PBS and concentrations were determined at A280 using the NanoDrop One (Thermo Fisher Scientific). For both nanobodies, production yields were ±5 mg/l bacterial culture. Finally, nanobody size, purity, and concentration were verified using SDS-PAGE (Fig. S7), stained with InstantBlue Safe Coomassie stain (Sigma-Aldrich).
Nanobodies without a Myc-tag
To produce nanobodies without a Myc-tag, nanobody genes were recloned in the pYQ11 vector (previously published as pYQVQ11 (52)), which encodes for a C-terminal C-Direct tag (derived from a FLAG–EPEA tag) containing a free thiol (cysteine) and an EPEA (Glu, Pro, Glu, and Ala) purification tag (C-tag, Thermo Fisher Scientific). Vectors were transformed into yeast cells (Saccharomyces cerevisiae strain VWK18 (53)). For this, expression cells were inoculated in yeast peptone medium supplemented with 2% glucose and grown for 24 h, at 30 °C, shaking. Next, cultures were diluted 1:20 in yeast nitrogen base medium supplemented with 2% glucose and grown at 30 °C under shaking conditions. After 24 h, cultures were diluted 1:10 by adding yeast peptone medium supplemented with 2% glucose and 1% galactose. Nanobody production was continued for >64 h, at 30 °C, shaking, until an A600 of >20 was reached. Next, cells were spun down for 20 min at 6000g and supernatants were collected, filter sterilized (0.45 μm), and stored at 4 °C until purification. Produced nanobodies were purified from yeast supernatants using a CaptureSelect C-tag column (Thermo Fisher Scientific) and an ÄKTA Start (Cytiva). For this, the supernatants were adjusted to neutral pH by adding 10 × PBS. Bound nanobodies were washed with at least five column volumes of PBS and eluted off with 20 mM citrate buffer supplemented with 150 mM NaCl, pH 3.0. Next, samples were neutralized with 100 mM Tris, pH 7.5 and dialyzed to PBS. Protein concentration was determined at A280 using a MultiScan (Thermo Fisher Scientific). Finally, nanobody size, purity, and concentration were confirmed using SDS-PAGE followed by Instant Blue Safe Coomassie staining.
Biotinylated nanobodies, Ecu-mab, and Ecu-Fab
Nanobodies and eculizumab(-Fab) were biotinylated using two methods. [1] Nanobody sequences were modified to contain a C-terminal LPETG-His motif and were subsequently produced and purified in E. coli BL21 Rosetta, like described above. After purification 50 μM LPETG-His nanobody was incubated with 25 μM His-tagged Sortase A7 (His-Tevg-SrtA7) (54) and 1 mM GGGK-Biotin (provided by Louris Feitsma, Medicinal Chemistry, Utrecht University), in 50 mM Tris, supplemented with 300 mM NaCl, for 2 h, at 4 °C. His-Tevg-SrtA7 was modified, expressed in E. coli and purified using its His-tag, in our lab. Next, samples were equilibrated with 20 mM imidazole and run over a HisTrap FF column to remove all His-tagged compounds from the reaction. Subsequently, a 30 kDa Amicon Tube (Merck Millipore) was used to concentrate the biotinylated nanobodies. To remove excess GGGK-biotin, samples were run over a Zeba Spin Desalting column (Thermo Fisher Scientific). [2] Myc-His-tagged nanobodies produced in E. coli and Ecu-mab and Ecu-Fab were randomly biotinylated via N-hydrocysucciminimidyl-labeling. Briefly, nanobodies at a concentration of 50 to 70 μM were incubated for 2 h, at 4 °C, with 20 × molar excess of EZ link NHS-PEG4-Biotin (Thermo Fisher Scientific, 210901BID), in PBS, pH 7.4. Unreacted linker was separated with Bio-Spin 6 columns (Bio-Rad) that were previously equilibrated in PBS. The nanobody concentrations were determined using a Nanodrop One.
Erythrocyte lysis assays
Sheep (Alsever Biotrading) and rabbit blood (kindly provided by Utrecht University, Faculty of Veterinary Medicine) was washed 3 × with PBS and diluted to a 2% erythrocyte suspension in veronal buffered saline (VBS + 145 nM NaCl, pH7.4). For the CP, sheep erythrocytes, preopsonized with 1:2000 polyclonal rabbit-anti-sheep IgM (hemolytic amboceptor (55)) were used, and VBS was supplemented with 0.25 mM MgCl2 and 0.5 mM CaCl2 (VBS++). For the AP, rabbit erythrocytes were used, and VBS was supplemented with 5 mM MgCl2 and 10 mM EGTA (VBS/MgEGTA). Periplasmic fractions (1:5 diluted with PBS) or purified nanobodies (0.45–1000 nM, 3-fold) were incubated for 10 min at RT with normal human serum (CP: 2.5% and AP: 10%) or 2.5% C5-depleted serum and repleted with physiological concentrations of C5 WT or C5 R885H. Next nanobody–serum mixes were added to 2% erythrocyte suspensions, and samples were incubated at 37 °C under shaking conditions (CP: 10 min, AP: 30 min). Afterward plates were spun down for 7 min at 3500 rpm. Supernatants were diluted 1:3 with milliQ water, and hemoglobin release was measured in a flat-bottom plate using an iMark Microplate Reader (Biorad) at 405 nm.
ELISA: coating, incubation times, and development
Unless stated differently, Nunc MaxiSorp plates (VWR 735-0083, Thermo Fisher Scientific) were coated with 2 μg/ml purified protein, diluted in PBS, in a volume of 50 μl. Plates were incubated O/N at 4 °C under still conditions. Next, plates were blocked with 80 μl 4% bovine serum albumin (BSA) in PBS + 0.05% Tween-20 (PBS-T). All following incubation steps were performed in 50 μl 1% BSA PBS-T, shaking, at RT for 1 h. In between all steps, plates were washed 3 × with PBS-T. To develop the ELISAs, tetramethylbenzidine substrate solution of 6 mg/ml, dissolved in dimethyl sulfoxide, was used to activate the enzyme-labeled antibodies, unless stated differently. When a color change was observed, the reaction was stopped with 0.5 M sulfuric acid, and the absorbance was measured at 450 nm using an iMark Microplate Reader.
Immune response ELISA
Wells coated with C5 were incubated with llama serum to measure the presence of C5-binding llama antibodies. Llama serum was diluted in PBS, supplemented with 1% skimmed milk (Marvel). Llama antibodies were detected using 1:2000 primary polyclonal rabbit-anti-VHH antibody QE19 (QVQ Holding BV) and 1:5000 secondary polyclonal donkey-anti-rabbit-HRP antibodies (Jackson ImmunoResearch). ELISA was developed using 3.7 mM O-phenylenediamine dihydrochloride + 50 mM Na2HPO4 2H2O + 25 mM citric acid + 0.03% H2O2 and the reaction was stopped with 0.5 M sulfuric acid.
Binding ELISA
Wells coated with purified complement components C3, C4, C5 WT or C5 R885H were used to assess the binding of our nanobodies and Ecu-mab to different complement proteins. Nanobodies and Ecu-mab were added in a 10-fold titration (0.001–1000 nM). Next, wells containing nanobodies were incubated with 1:2000 primary polyclonal rabbit-anti-VHH antibody QE19 (QVQ Holding BV) and 1:5000 secondary polyclonal donkey-anti-rabbit-HRP antibodies (Jackson ImmunoResearch). To detect Ecu-mab binding, wells were incubated with anti-human-kappa-HRP (Southern Biotech) detection antibodies.
Competition ELISA
Plates coated with purified complement component C5 were used to assess if our nanobodies compete. Five nanomolar UNbC5-1-Myc was incubated with a titration of untagged UNbC5-2 (0.025–1500 nM, 3-fold) for 10 min at RT and subsequently added to the C5 coated wells. Next, wells were incubated with 1:2000 primary monoclonal mouse-anti-Myc-tag clone 9B11 #2274 (Cell Signaling Technologies) followed by 1:5000 secondary monoclonal goat-anti-mouse-PO (Southern Biotech). To assess competition between our nanobodies and Ecu-mab/RaCI3, 300 nM of UNbC5-1 or UNbC5-2 was incubated with a titration of Ecu-mab or RaCI3 (5.49–4000 nM, 3-fold) for 10 min at RT, prior to adding it to the C5 coated wells. Next, nanobody binding was detected using primary polyclonal rabbit-anti-VHH antibody QE19 and 1:5000 secondary polyclonal donkey-anti-rabbit-horseradish peroxidase (HRP) antibodies.
CP complement ELISA
Plates were coated with 3 μg/ml human IgM (Millipore) in 0.1 M sodium carbonate, pH 9.6. Thousand nanomolar inhibitor (UNbC5-1, UNbC5-2, Ecu-mab, or RaCI3) was incubated with 4% normal human serum, added to the IgM coated wells, and incubated for 1 h, at 37 °C, under shaking conditions. To measure C5a formation, 25 μl of the supernatant was diluted with 25 μl 1% BSA PBS-T and added to a Nunc Maxisorp plate coated with 1 μg/ml anti-C5a capture antibody (C5a DuoSet ELISA kit, R&D systems). Next, we detected C5a with 2 μg/ml anti-C5a detection antibody (C5a DuoSet ELISA kit, R&D systems), followed by an incubation of 1:5000 streptavidin-HRP (Southern Biotech). To measure deposition of C3b on the IgM coated plates, 1:10,000 primary anti-C3 WM-1 clone digoxigenin labeled antibodies (Sigma-Aldrich), and 1:8000 secondary anti-digoxigenin-PO antibodies (Roche) were added. To measure deposition of C5b-9 on the IgM coated plates, 1:1000 primary monoclonal mouse-anti-C5b-9 aE11 (produced in our lab, based on (56, 57)) and 1:5000 secondary polyclonal goat-anti-mouse-PO were used.
Surface plasmon resonance
Planar streptavidin coated chips (P-strep, Sens BV) were spotted with biotinylated UNbC5-1 (method [2]), UNbC5-2 (method [1]), and Ecu-mab (method [1]) (25 and 50 nM, in duplicate) under a continuous flow for 1 h using a microspotter (Wasatch). Subsequent SPR experiments were carried out in the IBIS-MX96 (IBIS Technologies) with SPR buffer (20 mM Hepes pH 7.4, 150 mM NaCl, and 0.005% Tween-20). Initial testing showed limited regeneration with different high ionic strength (20 mM Hepes, 2 M NaCl, pH 7.4); ion containing (20 mM Hepes, 150 mM NaCl, 1 M MgCl2, pH 7.4); or low pH regeneration buffers (10 mM glycine, pH 3.0). Therefore, the method of kinetic titration was selected to perform accurate affinity determination. C5 was injected in a series of 14 steps 2-fold dilutions, at concentrations of 0.78, 1.56, 3.13, 6.25, and 12.50 nM, without regeneration on nanobodies or Ecu-mab coated surfaces. Determination of affinities for C5 R885H was performed with the same protocol using concentrations of 25, 50, 100, 200, and 400 nM. The last step of dissociation ran for 60 and 35 min, respectively, for reliable determination of the Koff. Kinetics were determined using Scrubber 2.0 (BioLogic Software, www.biologic.com.au/scrubber.html), where simple bimolecular models were used to fit the data. Due to unsuccessful coating on the chip surface and the instability of RaCI3 and C5, we used Ecu-Fab-biotin (method [2]) as a ligand in the competition assay. C5, RaCI3, and UNbC5-2 were injected sequentially at concentrations of 100 nM, with 5 min association and 4 min dissociation for RaCI3 and UNbC5-2.
Cryo-electron microscopy
Sample preparation and data collection
C5, purchased from Complement Technologies, was diluted to a final concentration of 1 μM and gently mixed with a 1.5 × molar excess of UNbC5-1 and UNbC5-2. The sample was diluted in PBS and incubated on ice for 20 min before freezing on glow discharged R1.2/1.3200 mesh Au holey carbon grids (Quantifoil). The grids were then plunge frozen in ethane using a Vitrobot Mark IV (Thermo Fisher Scientific), at 4 °C. Cryo-EM data were collected on a 200 kV Talos Arctica microscope (Thermo Fisher Scientific) equipped with a K2 summit detector (Gatan) and a post column 20 eV energy filter. Movies were collected in EPU 2 (Thermo Fisher Scientific, https://www.thermofisher.com/nl/en/home/electron-microscopy/products/software-em-3d-vis/epu-software.html) at a magnification of 130,000× with a pixel size of 1.04 Å/pix. Two datasets were collected in two collection sessions with similar settings of 40 frames with a total exposure of 54 and 57 e−/Å2, and a defocus range of −0.8 to 2.6 μm.
Data processing
The datasets of the C5:UNbC5-1:UNbC5-2 complex contained 1461 and 2349 movies (Fig. S3A). Both data sets were processed independently in CryoSPARC, v3.2/3 with the same workflow until 2D classification. Overall, the workflow for each dataset started with patch-based motion correction and patch-based contrast transfer function (CTF) estimation in CryoSPARC, followed by exposure curation that led to a total of 1253 and 1840 exposures, respectively. Then 100 movies of each data set were selected to perform a round of blob picking. Particles were extracted three times binned to 3.14 Å/pix and several rounds of 2D classification were performed to clean and select adequate templates for the CryoSPARC template picker. After template selection, the particles were picked and extracted unbinned with a 320-pixel box, particles of both datasets were merged, and an initial cleanup was performed through 2D classification. A total of 1,483,794 particles from 2D selection were then submitted to generate four initial models (Fig. S3B), that resulted in one good model that was further submitted to two initial model rounds (Fig. S3C). The resulted model with 193,009 particles was further selected for refinement. One round of nonuniform refinement led to a density map with a 3.73 Å resolution, which was further improved to 3.60 Å after local and global CTF refinement followed by a final nonuniform refinement. Global resolution was calculated according to the gold standard Fourier shell correlation, FSC = 0.143 criterion (Fig. S3D). Postprocessing such as sharpening and local resolution was also performed in CryoSPARC (Fig. S3,C and E).
Model building, refinement, and structure figures
To build the model of the C5:UNbC5-1:UNbC5-2 complex, models for the two nanobodies were generated through AlphaFold (26) based on nanobody sequences. The C5 molecule comprising chains α and β of PDB 3CU7 (9) was also used in combination with the nanobody models to rigid-body fit them into the cryo-EM maps using UCSF ChimeraX (58, 59). The model was then iteratively refined using Coot (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/) and Phenix (https://phenix-online.org/version_docs/1.20.1-4487/) real-space refine (60, 61) with geometric restraints. The C345c domain of C5 was not included in the final structures because of the weak density in the map. Figures with protein structures were prepared using PyMOL, as well as the calculation for interface area between C5 and nanobodies (retrieved from: http://www.pymol.org/pymol). Figures with cryo-EM densities were created using UCSF ChimeraX (58, 59) The statistics of the structure come from the refinement in Phenix.
Data analysis and statistical testing
Nanobody sequences were aligned using T-coffee (62). Bar and line graphs were created using GraphPad Prism 9.3.0. Binding and inhibition curves were fitted in GraphPad Prism 9.3.0 using the functions “[inhibitor] versus normalized response–Variable slope” and “Asymmetric Sigmoidal, 5PL, X is concentration”. Fitted curves of three individual experiments were used to calculate IC50 and EC50 values with SD in GraphPad Prism 9.3.0. SPR data were analyzed in Scrubber 2.0 (www.biologic.com.au/scrubber.html) and plotted in GraphPad Prism 9.3.0 (www.graphpad.com).
Data availability
The model coordinates and cryo-EM density maps of the C5 structure in complex with nanobodies UNbC5-1 and UNbC5-2 have been deposited under the following accession numbers 8CML and EMD-16730.
Supporting information
This article contains supporting information (60).
Conflict of interest
E. D. and R. H. are employees of QVQ Holding BV. Other authors declare no conflict of interest with the contents of this article.
Acknowledgments
We thank Wouter Beugelink, Dr Ramon van den Bos and Dr Itziar Serna Martin for their valuable input and scientific advice.
Author contributions
E. M. S., B. W. B., and S. H. M. R. conceptualization; E. M.S. and K. I. D. l. O. B. methodology; E. M. S. and K. I. D. l. O. B. validation; E. M. S., K. I. D. l. O. B., M. R., C. J. C. d. H., F. v. O., D. Y. S., and J. E. v. K. investigation; E. M. S., K. I. D. l. O. B., and S. H. M. R. writing–original draft; E. M. S. and K. I. D. l. O. B. visualization; E. M. S., K. I. D. l. O. B., P. G., and S. H. M. R. funding acquisition; D. A. C. H., E. D., R. H., B. W. B., P. G., and S. H. M. R. supervision; D. A. C. H., R. H., B. W. B., P. G., and S. H. M. R. writing–review and editing.
Funding and additional information
This work was mainly supported by the Netherlands Organization for Scientific Research (NWO) under the TTW Industrial Doctorate (grant agreement no. NWA.ID.17.036 to E. M. S.) and the Consejo Nacional de Ciencia y Tecnología in Mexico (grant agreement No. CVU 604718 to K. I. D. l. O. B.). The project also received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No. 101001937, ERC-ACCENT to SHMR and No. 787241, ERC-C-CLEAR to P. G.).
Reviewed by members of the JBC Editorial Board. Edited by Wolfgang Peti
Supporting information
References
- 1.Reichhardt M.P., Lundin K., Lokki A.I., Recher G., Vuoristo S., Katayama S., et al. Complement in human pre-implantation embryos: attack and defense. Front. Immunol. 2019;10:1–12. doi: 10.3389/fimmu.2019.02234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L., Fukuda N., Shimizu S., Kobayashi H., Tanaka S., Nakamura Y., et al. Role of complement 3 in renin generation during the differentiation of mesenchymal stem cells to smooth muscle cells. Am. J. Physiol. Cell Physiol. 2020;318:C981–C990. doi: 10.1152/ajpcell.00461.2019. [DOI] [PubMed] [Google Scholar]
- 3.Lee J.D., Coulthard L.G., Woodruff T.M. Complement dysregulation in the central nervous system during development and disease. Semin. Immunol. 2019;45 doi: 10.1016/j.smim.2019.101340. [DOI] [PubMed] [Google Scholar]
- 4.Merle N.S., Noe R., Halbwachs-Mecarelli L., Fremeaux-Bacchi V., Roumenina L.T. Complement system part II: role in immunity. Front. Immunol. 2015;6:1–26. doi: 10.3389/fimmu.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garred P., Tenner A.J., Mollnes T.E. Therapeutic targeting of the complement system: from rare diseases to pandemics. Pharmacol. Rev. 2021;73:792–827. doi: 10.1124/pharmrev.120.000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merle N.S., Church S.E., Fremeaux-Bacchi V., Roumenia L.T. Complement system part I – molecular mechanisms of activation and regulation. Front. Immunol. 2015;6:1–30. doi: 10.3389/fimmu.2015.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber R.D., Müller-Eberhard H.J. Assembly of the cytolytic alternative pathway of complement from 11 isolated plasma proteins. J. Exp. Med. 1978;148:1722–1727. doi: 10.1084/jem.148.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laursen N.S., Magnani F., Gottfredsen R.H., Petersen S.V., Andersen G.R. Structure, function and control of complement C5 and its proteolytic fragments. Curr. Mol. Med. 2012;12:1083–1097. doi: 10.2174/156652412802480925. [DOI] [PubMed] [Google Scholar]
- 9.Fredslund F., Laursen N.S., Roversi P., Jenner L., Oliveira C.L.P., Pedersen J.S., et al. Structure of and influence of a tick complement inhibitor on human complement component 5. Nat. Immunol. 2008;9:753–760. doi: 10.1038/ni.1625. [DOI] [PubMed] [Google Scholar]
- 10.Laursen N.S., Gordon N., Hermans S., Lorenz N., Jackson N., Wines B., et al. Structural basis for inhibition of complement C5 by the SSL7 protein from Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 2010;107:3681–3686. doi: 10.1073/pnas.0910565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nunn M.A., Sharma A., Paesen G.C., Adamson S., Lissina O., Willis A.C., et al. Complement inhibitor of C5 activation from the soft tick Ornithodoros moubata. J. Immunol. 2005;174:2084–2091. doi: 10.4049/jimmunol.174.4.2084. [DOI] [PubMed] [Google Scholar]
- 12.Reichhardt M.P., Johnson S., Tang T., Morgan T., Tebeka N., Popitsch N., et al. An inhibitor of complement C5 provides structural insights into activation. Proc. Natl. Acad. Sci. U. S. A. 2020;117:362–370. doi: 10.1073/pnas.1909973116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jore M.M., Johnson S., Sheppard D., Barber N.M., Li Y.I., Nunn M.A., et al. Structural basis for therapeutic inhibition of complement C5. Nat. Struct. Mol. Biol. 2016;23:378–386. doi: 10.1038/nsmb.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel C.W., Fritzinger D.C. Cobra venom factor: structure, function, and humanization for therapeutic complement depletion. Toxicon. 2010;56:1198–1222. doi: 10.1016/j.toxicon.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Harris C.L., Pouw R.B., Kavanagh D., Sun R., Ricklin D. Developments in anti-complement therapy; from disease to clinical trial. Mol. Immunol. 2018;102:89–119. doi: 10.1016/j.molimm.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Harris C.L. Expanding horizons in complement drug discovery: challenges and emerging strategies. Semin. Immunopathol. 2018;40:125–140. doi: 10.1007/s00281-017-0655-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rother R.P., Rollins S.A., Mojcik C.F., Brodsky R.A., Bell L. Discovery and development of the complement inhibitor eculizumab for the treatment of paroxysmal nocturnal hemoglobinuria. Nat. Biotechnol. 2007;25:1256–1264. doi: 10.1038/nbt1344. [DOI] [PubMed] [Google Scholar]
- 18.Nishimura J., Yamamoto M., Hayashi S., Ohyashiki K., Ando K., Brodsky A.L., et al. Genetic variants in C5 and poor response to eculizumab. New Engl. J. Med. 2014;370:632–639. doi: 10.1056/NEJMoa1311084. [DOI] [PubMed] [Google Scholar]
- 19.Schatz-Jakobsen J.A., Zhang Y., Johnson K., Neill A., Sheridan D., Andersen G.R. Structural basis for eculizumab-mediated inhibition of the complement terminal pathway. J. Immunol. 2016;197:337–344. doi: 10.4049/jimmunol.1600280. [DOI] [PubMed] [Google Scholar]
- 20.Brachet G., Bourquard T., Gallay N., Reiter E., Gouilleux-Gruart V., Poupon A., et al. Eculizumab epitope on complement C5: progress towards a better understanding of the mechanism of action. Mol. Immunol. 2016;77:126–131. doi: 10.1016/j.molimm.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 21.Volk A.L., Hu F.J., Berglund M.M., Nordling E., Strömberg P., Uhlen M., et al. Stratification of responders towards eculizumab using a structural epitope mapping strategy. Sci. Rep. 2016;6 doi: 10.1038/srep31365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de la O Becerra K.I., Oosterheert W., van den Bos R.M., Xenaki K.T., Lorent J.H., Ruyken M., et al. Multifaceted activities of seven nanobodies against complement C4b. J. Immunol. 2022;208:2207–2219. doi: 10.4049/jimmunol.2100647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roos A., Bouwman L.H., Munoz J., Zuiverloon T., Faber-Krol M.C., Fallaux-van den Houten F.C., et al. Functional characterization of the lectin pathway of complement in human serum. Mol. Immunol. 2003;39:655–668. doi: 10.1016/s0161-5890(02)00254-7. [DOI] [PubMed] [Google Scholar]
- 24.Muller-Eberhard H.J. The membrane attack complex of complement. Ann. Rev. Immunol. 1986;4:503–528. doi: 10.1146/annurev.iy.04.040186.002443. [DOI] [PubMed] [Google Scholar]
- 25.Parker C. Eculizumab for paroxysmal nocturnal haemoglobinuria. Lancet. 2009;373:759–767. doi: 10.1016/S0140-6736(09)60001-5. [DOI] [PubMed] [Google Scholar]
- 26.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macpherson A., Laabei M., Ahdash Z., Graewert M., James R., Schulze M.S.E.D.D., et al. The allosteric modulation of complement C5 by knob domain peptides. Elife. 2021;10:1–49. doi: 10.7554/eLife.63586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laursen N.S., Andersen K.R., Braren I., Spillner E., Sottrup-Jensen L., Andersen G.R. Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex. EMBO J. 2011;30:606–616. doi: 10.1038/emboj.2010.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarantonello A., Presumey J., Simoni L., Yalcin E., Fox R., Hansen A., et al. An ultrahigh-affinity complement C4b-specific nanobody inhibits in vivo assembly of the classical pathway proconvertase. J. Immunol. 2020;205:1678–1694. doi: 10.4049/jimmunol.2000528. [DOI] [PubMed] [Google Scholar]
- 30.Jensen R.K., Pihl R., Gadeberg T.A.F.F., Jensen J.K., Andersen K.R., Thiel S., et al. A potent complement factor C3–specific nanobody inhibiting multiple functions in the alternative pathway of human and murine complement. J. Biol. Chem. 2018;293:6269–6281. doi: 10.1074/jbc.RA117.001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rooijakkers S.H.M., Wu J., Ruyken M., van Domselaar R., Planken K.L., Tzekou A., et al. Structural and functional implications of the complement convertase stabilized by a staphylococcal inhibitor. Nat. Immunol. 2009;10:721–727. doi: 10.1038/ni.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bestebroer J., Aerts P.C., Rooijakkers S.H.M., Pandey M.K., Köhl J., van Strijp J.A.G., et al. Functional basis for complement evasion by staphylococcal superantigen-like 7. Cell Microbiol. 2010;12:1506–1516. doi: 10.1111/j.1462-5822.2010.01486.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schatz-Jakobsen J.A., Pedersen D.V., Andersen G.R. Structural insight into proteolytic activation and regulation of the complement system. Immunol. Rev. 2016;274:59–73. doi: 10.1111/imr.12465. [DOI] [PubMed] [Google Scholar]
- 34.Nilsson P.H., Johnson C., Quach Q.H., Macpherson A., Durrant O., Pischke S.E., et al. A conformational change of complement C5 is required for thrombin-mediated cleavage, revealed by a Novel ex vivo human whole blood model preserving full thrombin activity. J. Immunol. 2021;207:1641–1651. doi: 10.4049/jimmunol.2001471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laursen N.S., Pedersen D.V., Gytz H., Zarantonello A., Magnus J., Jensen B., et al. Functional and structural characterization of a potent C1q inhibitor targeting the classical pathway of the complement system. Front. Immunol. 2020;11:1–15. doi: 10.3389/fimmu.2020.01504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedersen H., Jensen R.K., Hansen A.G., Gadeberg T.A.F., Thiel S., Laursen N.S., et al. A C3 specific nanobody that blocks all three activation pathways in the human and murine complement system. J. Biol. Chem. 2020;295:8746–8758. doi: 10.1074/jbc.RA119.012339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedersen H., Jensen R.K., Jensen J.M.B., Fox R., Pedersen D.V., Olesen H.G., et al. A complement C3–specific nanobody for modulation of the alternative cascade identifies the C-terminal domain of C3b as functional in C5 convertase activity. J. Immunol. 2020;205:2287–2300. doi: 10.4049/jimmunol.2000752. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen D.V., Rösner T., Hansen A.G., Andersen K.R., Thiel S., Andersen G.R., et al. Recruitment of properdin by bi-specific nanobodies activates the alternative pathway of complement. Mol. Immunol. 2020;124:200–210. doi: 10.1016/j.molimm.2020.06.005. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen D.V., Gadeberg T.A.F., Thomas C., Wang Y., Joram N., Jensen R.K., et al. Structural basis for properdin oligomerization and convertase stimulation in the human complement system. Front. Immunol. 2019;10:1–19. doi: 10.3389/fimmu.2019.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng F., Put S., Bouwens L., Lahoutte T., Matthys P., Muyldermans S., et al. Molecular imaging with macrophage crig-targeting nanobodies for early and preclinical diagnosis in a mouse model of rheumatoid arthritis. J. Nucl. Med. 2014;55:824–829. doi: 10.2967/jnumed.113.130617. [DOI] [PubMed] [Google Scholar]
- 41.Wen Y., Ouyang Z., Schoonooghe S., Luo S., De Baetselier P., Lu W., et al. Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity. Immunobiology. 2017;222:807–813. doi: 10.1016/j.imbio.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 42.Duggan S. Caplacizumab: first global approval. Drugs. 2018;78:1639–1642. doi: 10.1007/s40265-018-0989-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Ineveld R.L., Kleinnijenhuis M., Alieva M., de Blank S., Barrera Roman M., van Vliet E.J., et al. Revealing the spatio-phenotypic patterning of cells in healthy and tumor tissues with mLSR-3D and STAPL-3D. Nat. Biotechnol. 2021;39:1239–1245. doi: 10.1038/s41587-021-00926-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rashidian M., Keliher E.J., Bilate A.M., Duarte J.N., Wojtkiewicz G.R., Jacobsen J.T., et al. Noninvasive imaging of immune responses. Proc. Natl. Acad. Sci. U. S. A. 2015;112:6146–6151. doi: 10.1073/pnas.1502609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pabst T.M., Wendeler M., Wang X., Bezemer S., Hermans P., Hunter A.K. Camelid VHH affinity ligands enable separation of closely related biopharmaceuticals. Biotechnol. J. 2017;12 doi: 10.1002/biot.201600357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berends E.T.M., Dekkers J.F., Nijland R., Kuipers A., Soppe J.A., van Strijp J.A.G., et al. Distinct localization of the complement C5b-9 complex on Gram-positive bacteria. Cell Microbiol. 2013;15:1955–1968. doi: 10.1111/cmi.12170. [DOI] [PubMed] [Google Scholar]
- 47.Berends E.T.M., Gorham R.D., Jr., Ruyken M., Soppe J.A., Orhan H., Aerts P.C., et al. Molecular insights into the surface-specific arrangement of complement C5 convertase enzymes. BMC Biol. 2015;13:1–13. doi: 10.1186/s12915-015-0203-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Haard H.J.W., Bezemer S., Ledeboer A.M., Müller W.H., Boender P.J., Moineau S., et al. Llama antibodies against a lactococcal protein located at the tip of the phage tail prevent phage infection. J. Bacteriol. 2005;187:4531–4541. doi: 10.1128/JB.187.13.4531-4541.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoogenboom H.R., Griffiths A.D., Johnson K.S., Chiswell D.J., Hudson P., Winter G. Multi-subunit proteins on the surface of filamentous phage: methodologies for displaying antibody (Fab) heavy and light chains. Nucleic Acids Res. 1991;19:4133–4137. doi: 10.1093/nar/19.15.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marks J.D., Hoogenboom H.R., Bonnert T.P., Mccafferty J., Griffiths A.D., Winter G. By-passing immunization human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 51.Gangaiah D., Ryan V., Van Hoesel D., Mane S.P., Mckinley E.T., Lakshmanan N., et al. Recombinant limosilactobacillus (Lactobacillus) delivering nanobodies against Clostridium perfringens NetB and alpha toxin confers potential protection from necrotic enteritis. Microbiologyopen. 2022;11 doi: 10.1002/mbo3.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heukers R., Mashayekhi V., Ramirez-Escudero M., De Haard H., Verrips T.C., Van Bergen En Henegouwen P.M.P., et al. Vhh-photosensitizer conjugates for targeted photodynamic therapy of met-overexpressing tumor cells. Antibodies. 2019;8:1–13. doi: 10.3390/antib8020026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gorlani A., Lutje Hulsik D., Adams H., Vriend G., Hermans P., Verrips T. Antibody engineering reveals the important role of J segments in the production efficiency of llama single-domain antibodies in Saccharomyces cerevisiae. Protein Eng. Des. Sel. 2012;25:39–46. doi: 10.1093/protein/gzr057. [DOI] [PubMed] [Google Scholar]
- 54.Jeong H.J., Abhiraman G.C., Story C.M., Ingram J.R., Dougan S.K. Generation of Ca2+-independent sortase A mutants with enhanced activity for protein and cell surface labeling. PLoS One. 2017;12:1–15. doi: 10.1371/journal.pone.0189068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klerx J.P.A.M., Beukelrnan C.J., Dijk H. Van, Willers J.M.N. Microassay for colorimetric estimation of complement activity in Guinea pig, human and mouse serum. J. Immunol. Methods. 1983;63:215–220. doi: 10.1016/0022-1759(83)90425-8. [DOI] [PubMed] [Google Scholar]
- 56.Mollnes T.E., Harboe M., Tschopp J. Monoclonal antibodies recognizing a neoantigen of Poly(C9) detect the human terminal complement complex in tissue and plasma. Scand. J. Immunol. 1985;22:183–195. doi: 10.1111/j.1365-3083.1985.tb01870.x. [DOI] [PubMed] [Google Scholar]
- 57.Bayly-Jones C., Ho B.H.T., Lau C., Leung E.W.W., D’Andrea L., Lupton C.J., et al. The neoepitope of the complement C5b-9 membrane attack complex is formed by proximity of adjacent ancillary regions of C9. Commun. Biol. 2023;6:42. doi: 10.1038/s42003-023-04431-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 61.Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J.M., et al. T-coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:13–17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The model coordinates and cryo-EM density maps of the C5 structure in complex with nanobodies UNbC5-1 and UNbC5-2 have been deposited under the following accession numbers 8CML and EMD-16730.