Abstract
Pseudomonas aeruginosa LasB elastase gene (lasB) transcription is controlled by the two-component quorum-sensing system of LasR, and the autoinducer, 3OC12-HSL (N-3-[oxododecanoyl]homoserine lactone). LasR and 3OC12-HSL-mediated lasB activation requires a functional operator sequence (OP1) in the lasB promoter region. Optimal activation of lasB, however, requires a second sequence of 70% identity to OP1, named OP2, located 43 bp upstream of OP1. In this study, we used sequence substitutions and insertion mutations in lasBp-lacZ fusion plasmids to explore the role of OP2 in lasB activation. Our results demonstrate that (i) OP1 and OP2 synergistically mediate lasB activation; (ii) OP2, like OP1, responds to LasR and 3OC12-HSL; and (iii) the putative autoinducer-binding domain of LasR is not required for synergistic activation from OP1 and OP2.
Pseudomonas aeruginosa LasB elastase is a well-characterized exoenzyme that contributes to pathogenesis of the organism in animal models (28). Suggestive of its pathogenic potential is its broad substrate range. LasB elastase acts alone or together with other P. aeruginosa proteases to degrade or inactivate several biologically important substrates, including connective tissues and immune system components (29). Detection of lasB transcript in the sputa of cystic fibrosis patients (44) and of antibodies to elastase in cystic fibrosis patients (1, 23) demonstrates that LasB elastase is produced in the human host.
LasB elastase gene (lasB) transcription depends on a cell density-dependent (quorum-sensing) mechanism of gene activation (35). Bacteria with quorum-sensing systems respond to the attainment of a critical culture density by activating transcription of target genes. Quorum sensing as a regulatory mechanism was first described for the marine bacteria Vibrio harveyi and V. fischeri, in which the attainment of a critical mass of bacteria in the symbiotic host’s light organ results in expression of the bioluminescence (lux) operon (for a review, see reference 11). Quorum-sensing gene activator complexes are composed of two components: a regulatory protein and an N-acylhomoserine lactone (“autoinducer”) molecule. A basal level of autoinducer is constitutively produced and, in the example of V. fischeri, diffuses freely through the cell envelope (22). The intracellular concentration of V. fischeri autoinducer (VAI) is thus dependent upon bacterial cell density. Inside the cell, the autoinducer and regulatory protein form an active complex. The complex recognizes a target gene operator and activates gene transcription (for a review, see reference 11). The first gene of the V. fischeri lux operon codes for autoinducer synthase (luxI). This induction results in a rapid increase in autoinducer concentration that further serves to induce the lux operon, resulting in light emission.
P. aeruginosa quorum-sensing regulatory components are the regulatory protein, LasR, and the autoinducer, 3OC12-HSL (formerly known as PAI-1; N-3-[oxododecanoyl]homoserine lactone) (13, 35, 37). The LasR protein is 30 and 53% identical to V. fischeri regulatory protein (LuxR) at the LuxR autoinducer-binding and DNA-binding domains, respectively (13). The 3OC12-HSL N-acyl side chain is six carbons longer than VAI. Despite structural similarities, the interchangeability of system components in target gene activation is extremely limited (15). In addition, organizational and mechanistic features of the two systems are not identical. In contrast to LuxR and VAI activation of the lux operon, the LasR and 3OC12-HSL complex (LasR/3OC12-HSL) regulated genes are widely separated on the P. aeruginosa chromosome. Two protease genes in addition to lasB are dependent on the presence of LasR/3OC12-HSL: aprA (coding for alkaline protease) and lasA (coding for LasA elastase) (14, 45). Each of these must be independently activated. An additional contrasting feature of the P. aeruginosa system is the active efflux of 3OC12-HSL from the cell, recently reported by Pearson et al. (36), which would limit the intracellular concentration. Thus, cell density and intracellular 3OC12-HSL concentration are not directly proportional in P. aeruginosa.
A sequence upstream of the lasB transcriptional start site, OP1 (−33 to −52 relative to the transcriptional start site; Fig. 1), is a LasR-responsive operator (42). OP1 forms an inverted repeat sequence at 18 of 20 bp and is located immediately upstream of, and may overlap, the lasB promoter sequence. Mutation of OP1 at various positions reduced lasB activity by characteristic amounts (42). Expression of lasB in Escherichia coli was dependent on the presence of LasR and exogenously added 3OC12-HSL, and mutation of OP1 abolished this expression, indicating that OP1 is a LasR-responsive element.
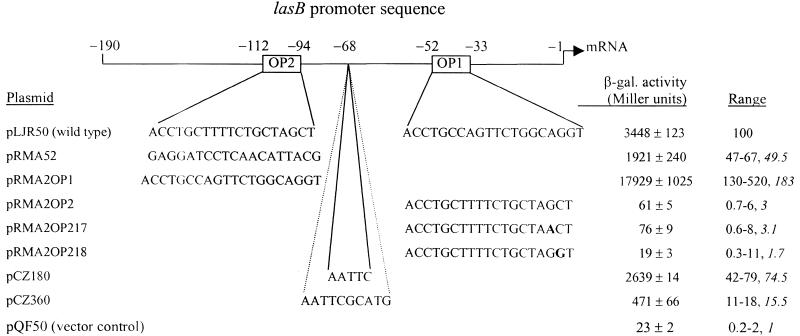
FIG. 1.
Nucleotide sequence of the lasB promoter region (4). Numbering is relative to the lasB transcriptional start site (+1), which is indicated with an arrow. OP1 and OP2 are underlined and labeled accordingly.
In addition to OP1, a sequence 42 bp upstream of OP1, named OP2 (−94 to −112 relative to the transcriptional start site; Fig. 1), shares 70% identity with OP1. Mutation of OP2 at positions 3 and 5 together reduced lasB induction by 20% (42). These results indicate that all or part of the OP2 sequence is involved in lasB induction.
In this report, we use targeted mutagenesis and sequence replacement to show the extent to which OP2 is involved in lasB induction. We express these mutant lasB upstream regions in the presence or absence of P. aeruginosa regulatory components to show that OP2 is a suboptimal, LasR/3OC12-HSL-responsive sequence. We also show that LasR lacking the putative autoinducer-binding domain elicits synergistic activation from OP1 and OP2, and propose alternative models for OP1- and OP2-mediated lasB activation based on these results.
MATERIALS AND METHODS
Strains and culture conditions.
Strains and plasmids used in this study are listed in Table 1. P. aeruginosa and E. coli used for genetic manipulation were cultured in Luria-Bertani (LB) broth or on LB plates with appropriate antibiotics (100 μg of ampicillin and 30 μg of chloramphenicol per ml for E. coli; 200 μg of carbenicillin per ml for P. aeruginosa), at 37°C. P. aeruginosa harboring plasmids and used for β-galactosidase assays was cultured in tryptic soy broth dialysate (TSBD [33]) with 200 μg of carbenicillin per ml at 37°C, subcultured into TSBD with carbenicillin, and harvested in early stationary phase (optical density at 640 nm [OD640] of ∼2). E. coli MG4 harboring compatible plasmids for β-galactosidase assays were cultured in modified A medium (A medium [27] supplemented with 0.1% yeast extract, 0.4% [vol/vol] glycerol, and 1 mM MgSO4) with appropriate antibiotics at 37°C and subcultured into fresh medium of the same composition. Synthetic 3OC12-HSL was added to the secondary culture to a final concentration of 100 nM where indicated. Due to the exogenous addition of 3OC12-HSL at the time of subculture, samples for β-galactosidase assay were taken in mid-logarithmic-phase growth, at an OD600 of ∼0.4.
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Genotype or descriptiona | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-Blue | recA1 endA1 thi-1 hsdR17 supE44 lac [F′ proAB lacIq Δ(lacZ M15 Tn10 (Tcr)] | Stratagene |
| MG4 | recA1 (argF lacIPOZYA)205 | 25 |
| P. aeruginosa | ||
| PAO1 | Wild type | 18 |
| PAOR1 | ΔlasR::Tcr strain PAO1 derivative | 13 |
| PDO100 | ΔrhlI::Tn501-2 strain PAO1 derivative | 6 |
| PDO111 | rhlR::Tn501-11 strain PAO1 derivative | 6 |
| Plasmids | ||
| pBluescript II KS(+) | General-purpose cloning vector, Apr | Stratagene |
| pQF50 | Broad-host-range transcriptional fusion vector with a promoterless lacZ, Apr | 10 |
| pLJR50 | lasBp-lacZ transcriptional reporter fusion; contains nt +4 to −190 of the lasB promoter region | 46 |
| pRMA52 | pLJR50 harboring random sequence at OP2 location (nt −94 to −112) | This study |
| pRMA2OP1 | pLJR50 harboring OP1 at OP2 location (nt −94 to −112) | This study |
| pRMA2OP2 | pLJR50 harboring OP2 at OP1 location (nt −33 to −52) | This study |
| pRMA2OP217 | Derived from pRMA2OP2; lasBp[nt −35:G→A]-lacZ | This study |
| pRMA2OP218 | Derived from pRMA2OP2; lasBp[nt −34:C→G]-lacZ | This study |
| pCZ180 | pLJR50 harboring a 5-bp insert between −68 and −69 | This study |
| pCZ360 | pLJR50 harboring a 10-bp insert between −68 and −69 | This study |
| pACYC184 | General-purpose cloning vector; Tcr Cmr | 9 |
| pPCS11 | Derived from pACYC184; tacp-lasR | 42 |
| pLJR11 | Derived from pPCS11; lasRΔnt71-478 | This study |
Nucleotides (nt) are numbered relative to the lasB transcriptional start site.
Genetic manipulations.
Transformation of E. coli was carried out by standard methods. Transformation of P. aeruginosa was carried out by the method of Olsen et al. (34). Sequence substitutions and point mutations were generated by oligonucleotide-directed mutagenesis and overlap extension (16). Oligonucleotides used are listed in Table 2. Transcriptional lasBp-lacZ fusion plasmids were generated as described previously (42). Briefly, PCR products were digested with BamHI and HindIII and then ligated into BamHI/HindIII-digested pBluescript II KS(+) (Stratagene, La Jolla, Calif.). Inserts were sequenced for the desired mutation by using a LI-COR automated sequencer (Lincoln, Nebr.). pBluescript II KS(+) harboring an insert of the desired sequence was digested with BamHI and HindIII, and the insert was purified from an agarose gel. The ∼224-bp fragments were ligated into BamHI/HindIII-digested pQF50 to generate lasBp-lacZ transcriptional fusion plasmids.
TABLE 2.
Oligonucleotides used for construction of mutants
| Plasmida | Strandb | Oligonucleotide sequencec | Locationd |
|---|---|---|---|
| Standarde | − | GCATCCATGAAGCTTCGGTGCTTTTCGTGTACCf | +4 to −13 |
| Standard | + | TACGCGGATCCAGGAAAGCGTGCAACTGA | −190 to −171 |
| pRMA52 | − | CGTAATGTTGAGGATCCTCGGCAGCTAGGTTGCC | −94 to −126 |
| pRMA52 | + | GAGGATCCTCAACATTACGATTCCAGCGAAAACATACAG | −112 to −74 |
| pRMA2OP2 | − | AGCTAGCAGAAAAGCAGGTAGCCTTGATTTCGCCGG | −34 to −69 |
| pRMA2OP2 | + | ACCTGCTTTTCTGCTAGCTTTGGCCGCGGGTTCTTTTTGG | −51 to −12 |
| pRMA2OP1 | − | ACCTGCCAGAACTGGCAGGTGGCAGCTAGGTTGCCCCC | −94 to −130 |
| pRMA2OP1 | + | ACCTGCCAGTTCTGGCAGGTATTCCAGCGAAAACATACAG | −113 to −174 |
| pRMA2OP217 | − | CGGCCAAAGTTAGCAGAAAAGCAGGTAGCCTTG | −126 to −159 |
| pRMA2OP217 | + | TTTTCTGCTAACTTTGGCCGCGGG | −45 to −22 |
| pRMA2OP218 | − | CGGCCAAACCTAGCAGAAAAGCAGGTAGCCTTG | −126 to −159 |
| pRMA2OP218 | + | TTTTCTGCTAGGTTTGGCCGCGGG | −45 to −22 |
| pCZ180 | − | CTTGATTTCGCCGAATTCGAAATCTGTATGTTTTCGC | −56 to −87 |
| pCZ180 | + | GAATTCGGCGAAATCAAGGC | −68 to −54 |
| pCZ360 | − | CGCCGAATTCGCATGGAAATCTGTATGTTTTCGC | −64 to −87 |
| pCZ360 | + | CATGCGAATTCGGCGAAATCAAGGC | −68 to −54 |
Plasmid that was constructed by using the respective oligonucleotide.
DNA strand containing the respective oligonucleotide sequence. +, lasB coding strand; −, lasB noncoding strand.
Nucleotide changes are underlined, relevant restriction endonuclease sites are in boldface, and inserted sequences are italicized.
Location of the corresponding sequence within the lasB promoter region relative to the lasB transcriptional start site.
Standard oligonucleotides were used to construct all lasBp-lacZ transcriptional fusion plasmids used in this study.
β-Galactosidase assay.
Assays for β-galactosidase activity were performed according to the method of Miller (27). Three single-colony isolates of each transformation were assayed for strain reproducibility. Cultures were assayed in triplicate, and each experiment was performed at least twice (at least six determinations). While unit activity from each strain varied between experiments, relationships between strains were consistent from experiment to experiment. In addition to unit activity, therefore, β-galactosidase activity levels are also expressed as a range of activity compared to P. aeruginosa PAO1(pLJR50) (parental host harboring the wild-type lasB promoter-lacZ transcriptional fusion, 100%; Table 1) to reflect these relationships.
RESULTS
Effect of OP2 sequence substitutions on lasB expression.
To investigate the contribution of OP2 to lasB expression, a series of OP2 sequence replacements was generated. First, the OP2 sequence (−94 to −112) in pLJR50 was replaced with a random sequence (pRMA52). The replacement sequence maintained the G+C content so as to conserve possible secondary structures (Table 2). In addition, spacing was maintained between OP2 and any upstream P. aeruginosa sequence that may be involved in regulating lasB expression. Replacement of OP2 resulted in a 33 to 53% (median, 50%) decline in P. aeruginosa PAO1 lasB expression (Fig. 2), while mutation of a single critical base pair in OP1, leaving OP2 intact, resulted in a complete absence of lasB expression (42). Thus, OP1 and OP2 together result in a greater induction of lasB than the sum of the individual sequences.
FIG. 2.
Effects of promoter region mutations on lasB activity. P. aeruginosa PAO1 harboring the specified lasBp-lacZ fusion plasmid was cultured and assayed for β-galactosidase activity as described in Materials and Methods. Nucleotides are numbered relative to the lasB transcriptional start site. Level of activity from multiple β-galactosidase assays are expressed as a percentage of PAO1(pLJR50) (100%), followed by the median percentage in italics. Medians were calculated from ≥21 determinations from a total of seven experiments, with PAO1(pLJR50) as an internal control. Nucleotide changes are in boldface.
Based on sequence identity, OP2 may be a minor OP1-like operator. If OP1 and OP2 are operators responding to the same regulatory protein or complex (e.g., LasR/3OC12-HSL), then one might substitute for the other at the other’s location. OP1 was substituted for OP2 at the position of OP2, so that two copies of OP1 were present in the lasBp-lacZ fusion plasmid (pRMA2OP1). PAO1(pRMA2OP1) expressed at 130 to 520% (median, 183%) of the lasB promoter activity from PAO1(pLJR50) (Fig. 2). In addition, OP2 was substituted for OP1 at the position of OP1, so that two copies of OP2 were present in the lasBp-lacZ fusion plasmid (pRMA2OP2). Strain PAO1(pRMA2OP2) expressed at 3% the level of lasB promoter activity of strain PAO1(pLJR50), a slight but still significant induction (P < 0.005; Fig. 2). Together, these results suggest that OP2 is a suboptimal, OP1-like operator.
To explore this hypothesis further, position 17 of the OP2-substituted OP1, which is identical to and corresponds to position 18 of OP1, was mutated to diverge from the OP1-homologous sequence (pRMA2OP217). Independently, position 18 of the OP2-substituted OP1 was mutated to converge with the OP1-homologous sequence (pRMA2OP218). Expression of lasB from these two promoter sequences was surprising: neither mutation dramatically affected expression from OP2 (Fig. 2). We conclude that additional mutations must be generated to establish a consensus sequence between OP1 and OP2.
Effect of insertions on lasB expression.
We sought to determine whether the contribution of OP2 was dependent on helical face in relation to OP1 and the promoter sequence. OP1 and OP2 are separated by 42 bp, or 4.1 turns, and therefore are similarly oriented with respect to helical face. Either 5 or 10 bp (one half-turn or one full turn, respectively) were inserted between OP1 and OP2 upstream of position −68. Surprisingly, insertion of 5 or 10 bp reduced lasB expression by 21 to 58% (median, 25%) and 82 to 89%, respectively (Fig. 2). Since the effect of the 10-bp insertion is greater than that of the random replacement of OP2, these results may reflect a change in activation from OP1 rather than helical face independence of OP2 (see Discussion).
Effect of sequence substitutions on transcript formation.
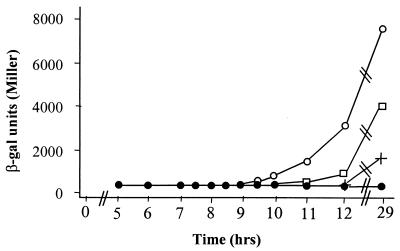
One of three induction scenarios might account for the synergism seen between OP1 and OP2. First, the presence of a second, minor OP1-like operator might allow lasB greater sensitivity to autoinducer concentration, thus resulting in earlier gene activation. Second, OP2 may allow for a longer induction period over the growth cycle. Finally, OP1 and OP2 may together increase the rate of transcript formation, i.e., result in a more efficient promoter. To discriminate between these possibilities, we cultured P. aeruginosa PAO1(pQF50), PAO1(pLJR50), PAO1(pRMA52), and PAO1(pRMA2OP1) and assayed for β-galactosidase activity over the course of the growth cycle. Rates of increase of β-galactosidase activity varied for each of the lasBp-lacZ fusion plasmids, while time of induction and duration of induction remained similar (Fig. 3). These results indicate that OP1 and OP2 increase lasB transcription by increasing the promoter strength rather than by changing the temporal pattern of gene induction.
FIG. 3.
Effect of substituted OP2 on lasB induction over time. Symbols: ●, P. aeruginosa PAO1(pQF50); □, PAO1(pLJR50); +, PAO1(pRMA52); ○, PAO1(pRMA2OP1). Strains were cultured, sampled at the indicated time points, and assayed as described in Materials and Methods.
OP2-mediated lasB induction in regulatory mutant backgrounds.
P. aeruginosa elastase production is dependent on regulatory factors in addition to LasR and 3OC12-HSL. A second system of P. aeruginosa autoinduction has been identified and overlaps in regulating lasB. The complex of RhlR and C4-HSL (formerly known as PAI-2; N-butyrylhomoserine lactone) is required to activate rhamnolipid synthesis (6, 31, 32, 38). A hierarchy of gene activation has been elucidated whereby LasR and 3OC12-HSL partially activate expression of rhlR (RhlR) and rhlI (C4-HSL synthase [24, 39]). In addition to LasR- and 3OC12-HSL-mediated regulation of rhl and lasB, lasB is partially regulated by the rhl system. Mutants of either rhlR or rhlI are deficient not only in rhamnolipid synthesis but also in lasB transcription: rhlI and rhlR backgrounds reduced lasB transcription by 100- and 400-fold, respectively (6). Using pLJR50 (wild-type lasBp-lacZ), we observed decreases of 81 to 96% (median, 87%) and 85 to 88% in rhlI and rhlR backgrounds, respectively (Table 3).
TABLE 3.
Effects of P. aeruginosa regulatory mutant backgrounds on operator-mediated lasB induction
| Plasmid | Mean β-galactosidase activity (Miller units) ± SDa
|
|||
|---|---|---|---|---|
| PAO1 (parent) | PAOR1 (lasR mutant) | PDO100 (rhlI mutant) | PDO111 (rhlR mutant) | |
| pQF50 | 48 ± 3 | 41 ± 1 (1) | 72 ± 4 (2) | 38 ± 1 (1) |
| pLJR50 | 3,203 ± 348 | 30 ± 2 (1) | 135 ± 3 (4–19, 12.5) | 485 ± 13 (12–15) |
| pRMA52 | 1,641 ± 57 | 21 ± 1 (1) | 341 ± 17 (11–24) | 91 ± 11 (6–7) |
| pRMA2OP1 | 4,148 ± 204 | 49 ± 4 (1–2) | 824 ± 35 (22–32, 29) | 425 ± 4 (13–25, 18) |
| pRMA2OP2 | 92 ± 8 | ND | 158 ± 6 (2–23, 3.5) | 62 ± 5 (1–3) |
| pRMA2OP217 | 136 ± 2 | ND | 89 ± 7 (3–4) | 316 ± 24 (7–10) |
| pRMA2OP218 | 127 ± 18 | ND | 48 ± 6 (2–3) | 82 ± 5 (3–7) |
Activity is given in Miller units. Values in parentheses are the ranges of activity expressed as percentages of PAO1(pLJR50) and the median percentages in italics. Medians were calculated from 12 determinations from a total of four independent experiments by using PAO1(pLJR50) as an internal control. ND, not determined.
To determine whether a known regulatory protein binds OP2, we assayed lasB promoter activity in various P. aeruginosa regulatory mutant backgrounds. The lasBp-lacZ fusion plasmid pLJR50, pRMA52, pRMA2OP1, pRMA2OP2, or the vector, pQF50, was transformed into strains PDO111 and PDO100 (rhlR and rhlI, respectively), derived from strain PAO1. As in strain PAO1, the OP1 sequence functionally replaced OP2 in each of the rhl strains, although to different extents (Table 3). Likewise, in strain PDO111, expression from pRMA52 was reduced relative to that from pLJR50, a result similar to the pattern of expression in strain PAO1 (Table 3 and Fig. 2). These results indicate that the contribution of RhlR to lasB expression occurs either elsewhere on the lasB regulatory region or indirectly through other regulatory proteins. Interestingly, expression from strain PDO100(pRMA52) was significantly greater than that from strain PDO100(pLJR50) in each experiment. Clearly, the effects of rhlR and rhlI backgrounds on lasB expression are not equivalent.
The same plasmids were transformed into strain PAOR1 (lasR), also derived from P. aeruginosa PAO1. Promoter activity was at background levels from PAOR1(pRMA52), PAOR1(pRMA2OP1), PAOR1(pLJR50), and PAOR1(pRMA2OP2) (Table 3), indicating that LasR is required for the slight induction from pRMA2OP2 seen in strain PAO1 (Fig. 2).
OP2-mediated lasB induction in E. coli in the presence of LasR and 3OC12-HSL.
We next sought to determine whether LasR and 3OC12-HSL induce lasB at OP2 in the absence of other P. aeruginosa factors. Plasmid pLJR50, pRMA52, pRMA2OP1, or pRMA2OP2 was transformed into E. coli MG4 in the presence (pPCS11) or absence (pACYC184) of Ptac-lasR and cultured in the presence or absence of 3OC12-HSL as described in Materials and Methods. As in P. aeruginosa, OP1 functionally replaced OP2 in the presence of LasR and 3OC12-HSL (Table 4). In the presence of lasR and 3OC12-HSL, expression from strains MG4(pRMA52) and MG4(pRMA2OP2) was induced to 5 and 8%, respectively, that from strain MG4(pLJR50) (Table 4). These results indicate that LasR and 3OC12-HSL mediate activation from OP2 as well as from OP1 and in the absence of other P. aeruginosa factors.
TABLE 4.
Expression from operator-substituted lasB in the presence (+) or absence (−) of LasR and 3OC12-HSL
| Plasmid | Mean β-galactosidase activity (Miller units)a ± SD
|
||||
|---|---|---|---|---|---|
| pPCS11 (Ptac-lasR)
|
pLJR11 (Ptac-lasRΔnt71-478)
|
pACYC184 (vector control) (+) | |||
| + | − | + | − | ||
| pQF50 | 0 | ND | ND | ND | 0 |
| pLJR50 | 431 ± 1 | 3 | 24.2 ± 0.4 | 19.8 ± 0.7 | 1 |
| pRMA52 | 23 ± 5 | 2 | 7.6 ± 0.2 | 6.5 ± 0.5 | ND |
| pRMA2OP1 | 332 ± 7 | 3 | 33 ± 2 | 31 ± 2 | ND |
| pRMA2OP2 | 36 ± 1 | 0 | ND | ND | ND |
Units are the average of three determinations. The experiment was repeated twice, with similar results. ND, not determined.
Expression of lasB in the presence of LasRΔaa26-159.
To determine whether synergistic (OP1 and OP2) activation of the lasB promoter requires full-length LasR, a PstI fragment internal to lasR was deleted from pPCS11. This deletion and religation is predicted to code for an in-frame LasRΔaa26-159. This plasmid, pLJR11, was transformed into E. coli MG4 in combination with each of the pQF50-derived lasBp-lacZ fusion plasmids pLJR50, pRMA52, and pRMA2OP1. β-Galactosidase activity was assayed in the presence or absence of 3OC12-HSL. Expression from strain MG4(pLJR11)(pLJR50) was reduced to 6%, in the presence or absence of 3OC12-HSL, that from strain MG4(pPCS11)(pLJR50) (Table 4). This result is consistent with that of K. Tucker’s LasRΔaa26-159 expression vector and may be due to an N-terminus effect on protein folding of the DNA binding domain (47). However, due to low variance, relationships among OP2-substituted promoters expressed in the presence of the truncated protein could be assessed. Expression from MG4(pLJR11)(pRMA52) was reduced by 69% from MG4(pLJR11)(pLJR50), which was restored by substituting OP1 for OP2 (Table 4). These results indicate that the truncated LasR mediates activation from the upstream operator.
DISCUSSION
Our results demonstrate that the contribution of OP2 to lasB activation is sequence specific and that OP1 can substitute for OP2 in lasB activation. With the exception of strain PDO100 (rhlI), replacement of OP2 with random sequence resulted in a decrease in lasB expression (Fig. 2 and Tables 3 and 4). In P. aeruginosa PAO1 (parental), PDO100 (rhlI), and PDO111 (rhlR) and in E. coli in the presence of LasR and 3OC12-HSL, OP1 functionally replaced OP2, resulting in expression equal to or greater than the wild-type sequence (Fig. 2 and Tables 3 and 4). The enhanced activity of strain PAO1(pRMA2OP1) was due to an increase in promoter strength rather than to an earlier induction of expression (Fig. 3).
Our results indicate that, like OP1, OP2 is a LasR/3OC12-HSL-responsive operator. The OP2 operator in the downstream position mediated a significant level of activation in E. coli in the presence of LasR and 3OC12-HSL (Table 4). However, OP2 is not an efficient substitute for OP1, since the levels of activation from pRMA2OP2 in P. aeruginosa PAO1 and in E. coli in the presence of full-length LasR are 3 and 8%, respectively, that of the nonmutated promoter region (Fig. 2 and Table 4). Interestingly, Stevens and Greenberg (43) also identified a sequence upstream of the lux operator protected by the LuxR DNA-binding domain (LuxRΔN). As with lasB OP2, LuxRΔN interaction at this site was not essential for lux operon transcription.
We reported previously that OP1 functions as a LasR/3OC12-HSL-responsive operator, with its component base pairs exerting characteristic levels of involvement in lasB activation (42). These levels of involvement revealed an asymmetric contribution by each of the operator half-sites, with the promoter-proximal half-site contributing more to lasB activation than the promoter-distal half-site. We speculated that LasR bound to OP1 interacts with RNA polymerase (RNAP) to stabilize the closed complex and facilitate lasB activation. The position of OP1, centered at −42.5 and potentially overlapping the −35 promoter determinant, calls to mind that of the 22-bp inverted repeat CRP galP1 class II promoter operator, which is centered at −41.5 and also overlaps the −35 determinant (5). CRP class II promoters are characterized by interactions between the RNAP α N-terminal domain (αNTD) and the downstream CRP monomer and between the RNAP α C-terminal domain (αCTD) and the upstream CRP monomer (2, 19, 20, 30, 40; for a review, see reference 8). The downstream interaction with αNTD activates transcription, whereas the upstream interaction with αCTD relieves an αCTD inhibitory effect (49, 50).
In addition to promoter and operator sequences, a third DNA element is involved in class II promoter activation. RNAP αCTD contacts an AT-rich sequence upstream of the operator (the UP element [41]), which is essential for relieving αCTD-mediated inhibition (41). In contrast to the overall P. aeruginosa mol% GC of 67 (17), the sequence between OP1 and OP2 is low in G+C content at 25%. Located 4 bp upstream of OP1 is an AT-rich sequence, 5′-AAATCAA-3′ (−63 to −57; Fig. 1). An additional AT-rich sequence of 15 base pairs (−70 to −84; Fig. 1) follows a GC hexamer. One or both of these AT-rich sequences may function as an UP element for RNAP αCTD binding. Based on previous results and sequence analyses, LasR-mediated lasB activation from OP1 may be analogous to CRP class II promoter activation.
The class II-like model of OP1-mediated activation provides a potential explanation of the widely disparate levels of activation seen between the five base pair and the ten base pair insertions (Fig. 2). The insertions are positioned within the GC hexamer, between the two AT-rich sequences (Fig. 1). Neither insertion appears to affect the intrinsic DNA curvature or local bendability (7, 12). Since the 10-bp insertion resulted in lower activation than random-sequence replacement of OP2 (Fig. 2), an impact of this insertion on OP1-mediated activation is probable. The 10-bp insertion may prevent αCTD binding to an OP1-associated UP element, rendering observable the αCTD inhibitory effect. In addition, the 5-bp insert (5′-AATTC-3′) may increase αCTD-UP element binding affinity, counteracting any decrease in OP2 contribution. In vitro binding studies of LasR and RNAP holoenzyme with or without αCTD to the lasB promoter region will be necessary to distinguish effects of UP element alteration from effects of operator spacing and helical orientation.
Several alternative roles for OP2 are worthy of consideration. Protein bound to OP1 and to OP2 may interact in a manner termed pairwise cooperativity (21). In this model, two dimeric proteins bind adjacently and cooperatively to DNA, as in λcI binding to OR1 and OR2 to stimulate transcription at PM (21, 26). However, the length of DNA between OP1 and OP2 is 4.1 helical turns, or ∼140 Å along the axis, while operator spacing of OR1 and OR2 is a single helical turn (26). Multiple helical-turn spacing of operators facilitates loop formation, which is characteristic of promoter repression rather than activation. In addition, the truncated LasR protein as well as the full-length LasR mediated activation at OP2 (Table 4). Activation at OP2 in the absence of a large portion of the N terminus, distal to the DNA binding domain, renders pairwise cooperativity unlikely. However, the moderate and severe reductions in lasB expression from inserting 5 or 10 bp, respectively, between OP1 and OP2 suggest other possibilities (Fig. 2). OP2 may increase the likelihood that LasR will bind to OP1 through one-dimensional diffusion, which is analogous to the sliding mechanism of the lac repressor (3). In this model, OP2 provides an expanded target for LasR; insertion of 5 or 10 bp may reduce the efficiency of LasR tracking to OP1. Alternatively, LasR interacting with OP2 may induce downstream DNA conformational effects that contribute to lasB activation, a phenomenon termed allosteric propagation by Vossen et al. (48). Insertion of 5 or 10 bp may interfere with conformational propagation and reduce the affinity of LasR for OP1. Finally, OP2 may facilitate class I-like promoter activation. This model is based on CRP dimer-mediated activation of the lac promoter (for a review, see reference 8). In this scenario, OP1 would facilitate class II-like activation and OP2 would facilitate class I-like activation, each utilizing an associated UP element. The reality of lasB activation may be a combination of these models. In vitro binding studies are underway to discriminate between these possibilities.
Interestingly, the increase in expression from pRMA2OP1 over pLJR50 in P. aeruginosa PAO1 was not reproduced in P. aeruginosa PDO111 (rhlR) or in E. coli MG4(pPCS11) (Tables 3 and 4). While strain PDO111 is isogenic with strain PAO1, the role of RhlR in lasB expression is still unclear, and its absence may be limiting lasB expression at a different level of regulation. Alternatively, the increase in OP1 sequence number may require strain- or species-specific concentrations of active LasR/3OC12-HSL in order to maximize expression. These optimal concentrations may be influenced by plasmid copy number or by intracellular binding conditions. The intracellular concentration of 3OC12-HSL may be a limiting factor in active complex formation, particularly if E. coli has an efflux pump functionally homologous to the proposed P. aeruginosa 3OC12-HSL efflux pump (36). Supporting this hypothesis is the autoinducer-independent overexpression from pRMA2OP1 in the presence of the truncated LasR (Table 4).
The disparate effect of the OP2 random-sequence replacement in the two rhl mutant backgrounds was a surprising result (Table 3). The lack of C4-HSL and the lack of RhlR do not result in equivalent phenotypes, suggesting roles independent of each other. The random sequence in pRMA52 may stimulate lasB transcription in an rhlI background. Alternatively, RhlR may bind OP2 in the absence of C4-HSL, resulting in a nonproductive competition with LasR/3OC12-HSL. Replacement of OP2 would then lead to a relaxation of inhibition in strain PDO100 (rhlI), resulting in unexpectedly high activation from pRMA52. Binding by RhlR to OP2 may result in either a less-stable LasR-RNAP complex that prevents promoter isomerization or from formation of an overly stable LasR-RNAP complex that prevents promoter clearance by the polymerase. Binding by RhlR to OP2 would depend on a number of criteria: (i) RhlR has DNA-binding capability in the absence of its cognate autoinducer, (ii) RhlR recognizes OP2, and (iii) OP2-mediated lasB activation is not a generalized protein-DNA binding effect but rather is specific to LasR binding.
ACKNOWLEDGMENTS
We thank the American Lung Association, the North Dakota Agricultural Experiment Station, and North Dakota EPSCoR for supporting this study. Additional support was received through an intramural Grant-in-Aid.
R.M.A. was supported by the American Lung Association Research Grant. We also thank Lou Passador and Barbara H. Iglewski for plasmids and synthetic 3OC12-HSL and Dennis Ohman for use of the P. aeruginosa rhl strains. Finally, we thank Anne Summers, Doug Storey, and Eb Pesci for helpful conversation.
REFERENCES
- 1.Albus A, Saalmann M, Tesch H, Pedersen S S, Doring G. Increased levels of IgG subclasses specific for Pseudomonas aeruginosa exoenzyme and polysaccharide antigens in chronically infected patients with cystic fibrosis. APMIS. 1989;97:1146–1148. doi: 10.1111/j.1699-0463.1989.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 2.Belyaeva T A, Bown J A, Fujita N, Ishihama A, Busby S J W. Location of the C-terminal domain of the RNA polymerase α subunit in different open complexes at the Escherichia coli galactose operon regulatory region. Nucleic Acids Res. 1996;24:2243–2251. doi: 10.1093/nar/24.12.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg O G, Winter R B, von Hippel P H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 4.Bever R A, Iglewski B H. Molecular characterization and nucleotide sequence of the Pseudomonas aeruginosa elastase structural gene. J Bacteriol. 1988;170:4309–4314. doi: 10.1128/jb.170.9.4309-4314.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bingham A H, Ponnambalam S, Chan B, Busby S. Mutations that reduce expression from the P2 promoter of the Escherichia coli galactose operon. Gene. 1986;41:67–74. doi: 10.1016/0378-1119(86)90268-4. [DOI] [PubMed] [Google Scholar]
- 6.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brukner I, Sanchez R, Suck D, Pongor S. Sequence dependent bending propensity of DNA as revealed by DNase I: parameters for trinucleotides. EMBO J. 1995;14:1812–1818. doi: 10.1002/j.1460-2075.1995.tb07169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busby S, Ebright R H. Transcription activation at class II CAP-dependent promoters. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 9.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farinha M A, Kropinski A M. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J Bacteriol. 1990;172:3496–3499. doi: 10.1128/jb.172.6.3496-3499.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua W C, Winans S C, Greenberg E P. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabrielian A, Pongor S. Correlation of intrinsic DNA curvature with DNA property periodicity. FEBS Lett. 1996;393:65–68. doi: 10.1016/0014-5793(96)00855-1. [DOI] [PubMed] [Google Scholar]
- 13.Gambello M J, Iglewski B H. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J Bacteriol. 1991;173:3000–3009. doi: 10.1128/jb.173.9.3000-3009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gambello M J, Kaye S, Iglewski B H. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect Immun. 1993;61:1180–1184. doi: 10.1128/iai.61.4.1180-1184.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray K M, Passador L, Iglewski B H, Greenberg E P. Interchangeability and specificity of components from the quorum-sensing regulatory systems of Vibrio fischeri and Pseudomonas aeruginosa. J Bacteriol. 1994;176:3076–3080. doi: 10.1128/jb.176.10.3076-3080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1988;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 17.Holloway B W, Morgan A F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- 18.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Igarashi K, Hanamura A, Makino K, Aiba H, Mizuno T, Nakata A, Ishihama A. Functional map of the alpha subunit of Escherichia coli RNA polymerase: two modes of transcription activation by positive factors. Proc Natl Acad Sci USA. 1991;88:8958–8962. doi: 10.1073/pnas.88.20.8958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igarashi K, Ishihama A. Bipartite functional map of the E. coli RNA polymerase α subunit: involvement of the C-terminal region in transcription activation by cAMP-CRP. Cell. 1991;65:1015–1022. doi: 10.1016/0092-8674(91)90553-b. [DOI] [PubMed] [Google Scholar]
- 21.Johnson A D, Meyer B J, Ptashne M. Interactions between DNA-bound repressors govern regulation by the λ phage repressor. Proc Natl Acad Sci USA. 1979;76:5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan H B, Greenberg E P. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klinger J D, Strauss D C, Hilton C B, Bass J A. Antibodies to proteases and exotoxin A of Pseudomonas aeruginosa in patients with cystic fibrosis: demonstration by radioimmunoassay. J Infect Dis. 1978;138:49–58. doi: 10.1093/infdis/138.1.49. [DOI] [PubMed] [Google Scholar]
- 24.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 25.Linn T, Ralling G. A versatile multiple- and single-copy vector system for the in vitro construction of transcriptional fusions to lacZ. Plasmid. 1985;14:134–142. doi: 10.1016/0147-619x(85)90073-3. [DOI] [PubMed] [Google Scholar]
- 26.Mao C, Carlson N G, Little J W. Cooperative DNA-protein interactions. J Mol Biol. 1994;235:532–544. doi: 10.1006/jmbi.1994.1011. [DOI] [PubMed] [Google Scholar]
- 27.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. Assay of β-galactosidase; pp. 352–355. [Google Scholar]
- 28.Morihara K, Homma J Y. Pseudomonas proteases. In: Holder I A, editor. Bacterial enzymes and virulence. Boca Raton, Fla: CRC Press, Inc.; 1985. pp. 49–79. [Google Scholar]
- 29.Nicas T I, Iglewski B H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- 30.Niu W, Kim Y, Tau G, Heyduk T, Ebright R H. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ochsner U A, Reiser J. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:6424–6428. doi: 10.1073/pnas.92.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochsner U A, Koch A K, Fiechter A, Reiser J. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:2044–2054. doi: 10.1128/jb.176.7.2044-2054.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohman D E, Sadoff J C, Iglewski B H. Toxin A-deficient mutants of Pseudomonas aeruginosa PA-103: isolation and characterization. Infect Immun. 1980;28:899–908. doi: 10.1128/iai.28.3.899-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olsen R H, Debusscher G, McCombie W R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982;150:60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Passador L, Cook J M, Gambello M J, Rust L, Iglewski B H. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science. 1993;260:1127–1130. doi: 10.1126/science.8493556. [DOI] [PubMed] [Google Scholar]
- 36.Pearson J P, Van Delden C, Iglewski B H. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pearson J P, Passador L, Iglewski B H, Greenberg E P. A second N-acylhomoserine lactone signal produced by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1995;92:1490–1494. doi: 10.1073/pnas.92.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhodius V A, West D M, Webster C L, Busby S J W, Savery N J. Transcription activation at class II CRP-dependent promoters: the role of different activating regions. Nucleic Acids Res. 1997;25:326–332. doi: 10.1093/nar/25.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 42.Rust L, Pesci E C, Iglewski B H. Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. J Bacteriol. 1996;178:1134–1140. doi: 10.1128/jb.178.4.1134-1140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens A M, Greenberg E P. Quorum sensing in Vibrio fischeri: essential elements for activation of the luminescence genes. J Bacteriol. 1997;179:557–562. doi: 10.1128/jb.179.2.557-562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storey D G, Ujack E E, Rabin H R. Population transcript accumulation of Pseudomonas aeruginosa exotoxin A and elastase in sputa from patients with cystic fibrosis. Infect Immun. 1992;60:4687–4694. doi: 10.1128/iai.60.11.4687-4694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toder D S, Gambello M J, Iglewski B H. Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol Microbiol. 1991;5:2003–2010. doi: 10.1111/j.1365-2958.1991.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 46.Toder D S, Ferrell S J, Nezezon J L, Rust L, Iglewski B H. lasA and lasB genes of Pseudomonas aeruginosa: analysis of transcription and gene product activity. Infect Immun. 1994;62:1320–1327. doi: 10.1128/iai.62.4.1320-1327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tucker, K. Personal communication.
- 48.Vossen K M, Stickle D F, Fried M G. The mechanism of CAP-lac repressor binding cooperativity at the E. coli lactose promoter. J Mol Biol. 1996;255:44–54. doi: 10.1006/jmbi.1996.0005. [DOI] [PubMed] [Google Scholar]
- 49.West D, Williams R, Rhodius V, Bell A, Sharma N, Zou C, Fujita N, Ishihama A, Busby S. Interactions between the Escherichia coli cyclic AMP receptor protein and RNA polymerase at class II promoters. Mol Microbiol. 1993;10:789–797. doi: 10.1111/j.1365-2958.1993.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhou Y, Pendergrast P S, Bell A, Williams R, Busby S, Ebright R H. The functional subunit of a dimeric transcription activator protein depends on promoter architecture. EMBO J. 1994;13:4549–4557. doi: 10.1002/j.1460-2075.1994.tb06776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]