Abstract
From evolutionary and physiological viewpoints, the Escherichia coli bgl operon is intriguing because its expression is silent (Bgl− phenotype), at least under several laboratory conditions. H-NS, a nucleoid protein, is known as a DNA-binding protein involved in bgl silencing. However, we previously found that bgl expression is still silent in a certain subset of hns mutations, each of which results in a defect in its DNA-binding ability. Based on this fact, we proposed a model in which a postulated DNA-binding protein(s) has an adapter function by interacting with both the cis-acting element of the bgl promoter and the mutated H-NS. To identify such a presumed adapter molecule, we attempted to isolate mutants exhibiting the Bgl+ phenotype in the background of hns60, encoding the mutant H-NS protein lacking the DNA-binding domain by random insertion mutagenesis with the mini-Tn10cam transposon. These isolated mutations were mapped to five loci on the chromosome. Among these loci, three appeared to be leuO, hns, and bglJ, which were previously characterized, while the other two were novel. Genetic analysis revealed that the two insertions are within the rpoS gene and in front of the lrhA gene, respectively. The former encodes the stationary-phase-specific sigma factor, ςS, and the latter encodes a LysR-like DNA-binding protein. It was found that ςS is defective in both types of mutant cells. These results showed that the rpoS function is involved in the mechanism underlying bgl silencing, at least in the hns60 background used in this study. We also examined whether the H-NS homolog StpA has such an adapter function, as was previously proposed. Our results did not support the idea that StpA has an adapter function in the genetic background used.
The Escherichia coli bgl operon is involved in the utilization of β-glucosides, such as salicin and arbutin, as carbon sources (12, 14). Curiously, however, although the bgl operon is intact in the genetic sense (genetically bgl+), its expression is silent in the wild-type background (phenotypically Bgl−) in that the wild-type cells are unable to utilize β-glucosides as sole carbon sources. The mechanism underlying such a silencing of bgl has been a long-standing subject of debate and has been studied extensively, particularly from evolutionary and physiological viewpoints (for a review, see reference 10 and references therein). Recent extensive studies have suggested that for bgl silencing, both the cis-acting element including the bgl promoter region and some trans-acting protein factors are required. In this respect, the DNA element upstream of the bgl promoter was proposed to function in a manner similar to eukaryotic silencer sequences. Schnetz (13) postulated that a region including the bgl promoter is organized into a nucleoprotein structure that prohibits the expression of bgl. The well-known H-NS nucleoid protein was suggested to be a DNA-binding trans-acting factor involved in the formation of the nucleoprotein structure, because expression of the bgl operon is fully derepressed in hns null mutants (2, 21). Several mutant alleles other than hns that affect the Bgl phenotype have been reported. They include gyrA, gyrB, bglJ, and leuO, but their roles in bgl silencing are less evident (3, 6, 19).
In the course of previous studies on the structure-function relationship of H-NS, members of our group realized that bgl expression is still silent in a certain subset of hns mutants (21). The expression of bgl is fully repressed even in cells producing a C-terminally truncated H-NS, which lacks the entire DNA-binding domain. It was thus postulated that the DNA-binding nature of H-NS is not essential for bgl silencing. This led to the hypothesis that a certain DNA-binding protein(s) has an adapter function that allows the H-NS to be properly sited on the cis-acting element of the bgl promoter (21). In the hope of finding an as-yet-unidentified putative adapter, in this study we intensively screened mutants which exhibit the Bgl+ phenotype in a certain genetic background, hns60, specifying a C-terminally truncated H-NS (21). We identified two such mutant alleles, one in rpoS and the other in lrhA. Further analyses of these mutants showed that one of the E. coli sigma factors, ςS, is crucial for bgl silencing in cells producing the C-terminally truncated H-NS. In fact, Free et al. (4) previously addressed a similar issue. They found that bgl silencing caused by a C-terminally truncated H-NS is abolished in an stpA null mutant. Together with the fact that the stpA gene encodes an H-NS homolog, the authors proposed that StpA is the presumed adapter protein. This view is discussed in relation to our findings.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used in this study are listed in Table 1. The strains were all derivatives of CSH26 [Δ(pro-lac) ara thi] (11). The hns::neo mutation was constructed by replacement of the internal HpaI-PvuII fragment of the hns gene with the neo gene. The recombinant plasmid pTO13 was constructed as follows. A 1.1-kb EcoT22I-DraI fragment encompassing the entire lrhA gene was purified from λ9C2 (8). After treatment with T4 DNA polymerase, the fragment was inserted into the previously blunted BamHI site of pUSI2 (16). The resultant plasmid was designated pTO13.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| BGL1 | Δ(pro-lac) ara thi (λbgl-lacZ | 21 |
| BGL1 hns::neo | BGL1 hns::neo | 21 |
| CU319 | BGL1 zch-3117::Tn10kan | This study |
| CU320 | BGL1 hns60 zch-3117::Tn10kan lrhA83::mini-Tn10cam | This study |
| CU321 | BGL1 lrhA83::mini-Tn10cam | This study |
| TO26 | BGL1 hns::neo stpA::spc | This study |
| TO32 | BGL1 hns60 zch-3117::Tn10kan rpoS23::mini-Tn10cam | This study |
| TO39 | BGL1 rpoS::Tn10 | This study |
| TO40 | BGL1 hns60 zch-3117::Tn10kan rpoS::Tn10 | This study |
| TO41 | BGL1 hns::neo rpoS::Tn10 | This study |
Transposon insertion mutagenesis.
Transposon insertion with mini-Tn10cam was carried out essentially according to the method of Kleckner et al. (7). CSH26 cells were grown in Luria broth at 37°C to the mid-logarithmic phase. A portion of the culture was infected with a lysate of λNK1324 and then plated onto a Luria agar plate containing chloramphenicol (25 μg/ml). The chloramphenicol-resistant (Cmr) transductants were pooled and infected with P1vir phage to prepare a P1 phage lysate, which was stored and used for P1 transduction. Bgl+ transductants were first screened on MacConkey lactose plates containing 0.1% salicin and chloramphenicol (25 μg/ml) and then scored on agar plates containing 0.02% bromothymol blue (as a pH indicator) and 0.5% salicin.
Assaying of β-galactosidase.
Cells were grown at 37°C in TB medium (17) supplemented with 5 mM β-methyl-d-glucoside to induce bgl expression. Ampicillin (50 μg/ml) was added to the medium, if necessary. Assaying of β-galactosidase was performed essentially according to the method of Miller (11).
Immunoblotting analysis.
Total cellular proteins were prepared by precipitation with trichloroacetic acid (final concentration, 5%) and then collected by centrifugation. After a wash with ice-cold acetone, the precipitate was dissolved in 1% (wt/vol) sodium dodecyl sulfate–50 mM Tris-HCl (pH 8)–1 mM EDTA buffer. The protein concentration was accurately determined for each sample with a Micro BCA protein assay reagent kit (Pierce, Rockford, Ill.). Appropriate amounts of total cellular proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting analysis with anti-ςS, anti-CbpA, and anti-H-NS polyclonal antisera.
RESULTS
Isolation of Bgl+ mutants in the hns60 background.
As demonstrated previously (reference 21; see also the introduction), the hns60 allele on the E. coli chromosome specifies a mutant H-NS protein, named H-NSQ92am, which lacks the DNA-binding domain. In this hns60 background, however, expression of bgl is severely repressed (the Bgl− silencing phenotype), as in the case in the wild-type hns background, as mentioned above. In other words, the hns60 allele gives the wild-type phenotype as far as bgl silencing is concerned. Genetic characterization of the hns60 allele not only will provide new insight into the molecular mechanism underlying bgl silencing but also will be advantageous for identifying a presumed adapter molecule (see the introduction). Based on this assumption, strain CU319, carrying both the hns60 allele and the bgl-lacZ operon fusion gene on the chromosome, was mutagenized by transposon insertion mutagenesis with mini-Tn10cam carrying the Cmr gene (7). It should be noted that CU319 shows the Bgl− phenotype (thus also the Lac− phenotype) due to bgl silencing. From 1.2 × 105 Cmr transductants, we isolated 91 candidates exhibiting both the Lac+ and Bgl+ phenotypes by scoring on MacConkey lactose plates containing 0.1% salicin and on agar plates containing salicin and bromothymol blue, respectively. Such double screening should have allowed us to obtain only trans-acting mutations, not cis-acting ones, which somehow affect bgl silencing.
To determine whether these mutants are previously known ones, such as leuO, hns, and bglJ (2, 6, 19), we carried out a series of P1 mapping analyses using a set of Tn10 insertions, namely, zab-3051::Tn10 for leuO, zci-506::Tn10 for hns, and zji-202::Tn10 for bglJ. The results showed that of the 91 candidate mutations, 83, 1, and 2 appeared to correspond to leuO, hns, and bglJ, respectively. Further P1 mapping analyses with the remaining five mutations, using a set of Tn10 insertions in the entire E. coli chromosome (18), revealed that one mutation (type 1) is linked to cysC95::Tn10 (61 min; 50% linkage) and the others (type 2) are all linked to zfb-223::Tn10 (51 min; 65% linkage). These results suggested that each mutational locus may be novel in terms of bgl silencing.
rpoS and lrhA mutations relieve bgl silencing.
To identify the genes mutagenized in these two types of mini-Tn10cam insertions, each DNA segment encompassing the transposon was first cloned from the chromosome of each representative by using the cam gene (Cmr) as a selectable marker. The results of sequencing analyses of the cloned DNA segments revealed that the transposon is within the rpoS gene in one case (type 1) and upstream of the lrhA open reading frame in the other case (type 2), as shown schematically in Fig. 1. In the latter case, however, the transposon is in the noncoding region between lrhA and its upstream open reading frame (named o405). We assumed that this insertion somehow affects lrhA function, because it is known that the cam gene often exhibits strong promoter activity toward its downstream gene (in this case, lrhA). In any case, we designated these two novel alleles rpoS23::mini-Tn10cam and lrhA83::mini-Tn10cam (referred to herein as rpoS23 and lrhA83, respectively, for clarity).
FIG. 1.
Schematic representations of rpoS23 and lrhA83 mutations on the chromosome. The structures of the chromosomal region encompassing the rpoS gene (A) and the lrhA gene (B) are shown. Polygonal symbols indicate open reading frames as well as their relative directions of transcription. Restriction sites used for the construction of pTO13 are indicated.
Both the rpoS23 and lrhA83 mutants are effective only for hns60, i.e., not for wild-type hns.
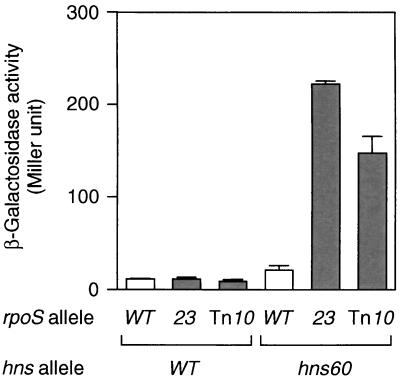
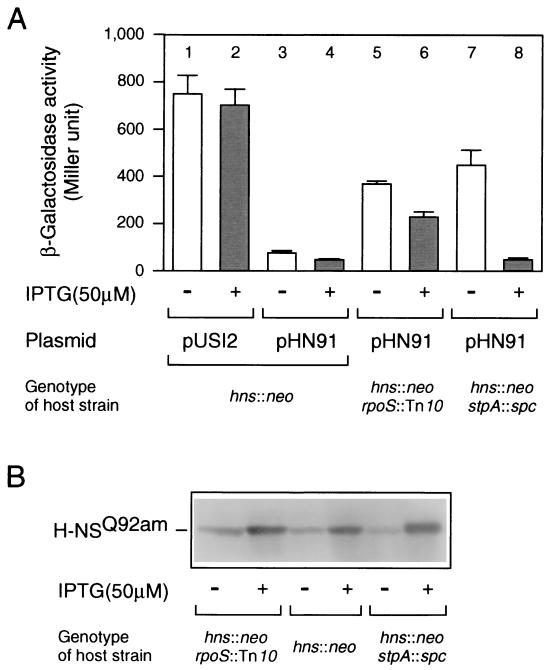
The rpoS gene encodes the ςS subunit of RNA polymerase, which is crucial for transcriptional regulation under several stressful conditions, such as nutrient starvation (9). To clarify the effect of the rpoS23 mutation on bgl silencing, this particular allele was transferred into both the hns+ and hns60 backgrounds, and then the expression of bgl-lacZ was measured in each transductant. As shown in Fig. 2, the β-galactosidase activity expressed by bgl-lacZ was very low in the hns+ background but significantly high in the hns60 background when the rpoS23 allele was introduced. This indicated that the effect of rpoS23 on bgl expression is an event specific to hns60. When another previously characterized rpoS null allele (22) was used for the same analysis, essentially the same result was obtained (Fig. 2, Tn10 bars). In any event, this result suggested that the rpoS gene product is involved in bgl silencing in the hns60 background. We further confirmed this idea through a different approach, as follows. The recombinant plasmid pHN91 carries the hns60 allele under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible tac promoter (20). When the hns60 product, H-NSQ92am, was produced by pHN91 in an hns null mutant (hns::neo), the β-galactosidase activity expressed by bgl-lacZ was markedly reduced (Fig. 3A, lane 4). However, such a result was not observed in an hns::neo rpoS::Tn10 double mutant (Fig. 3A, bar 6). It should be noted that the hns60 product was present in both genetic backgrounds (Fig. 3B). These results also supported the view that rpoS function is required for the bgl silencing caused by H-NSQ92am. It is thus suggested that the rpoS gene product, ςS, may be a presumed adapter molecule, as discussed below.
FIG. 2.
Expression of the bgl operon in rpoS mutants. Cells carrying the rpoS and hns alleles as indicated were grown at 37°C to the mid-logarithmic phase in TB medium containing 5 mM β-methyl-d-glucoside, an inducer for the bgl operon. β-Galactosidase activity expressed by bgl-lacZ was measured by the method of Miller (11). Each value is the mean + standard deviation from four independent assays. WT, wild type.
FIG. 3.
bgl silencing by H-NSQ92am requires rpoS function, but not stpA function. Strains with the indicated mutations and plasmids were grown in either the absence (−) or presence (+) of 50 μM IPTG to induce H-NSQ92am, and then the β-galactosidase activity expressed by bgl-lacZ (A) and the cellular level of H-NSQ92am (B) were measured, as described for Fig. 2. For β-galactosidase activity, each value is the mean + standard deviation from four independent assays.
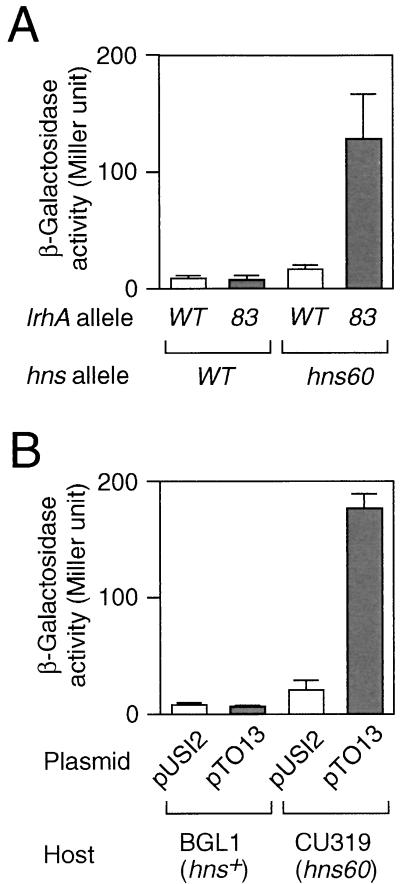
lrhA encodes a DNA-binding protein belonging to the LysR family (lrhA stands for LysR homolog), but its physiological function is not yet clear (1). We examined whether the lrhA83 allele also affects bgl silencing in a manner specific to hns60, as does the rpoS mutant. The results showed that this is the case (Fig. 4A). We then assumed that the effect of lrhA83 is probably due to overproduction of the LrhA protein, as mentioned above. This was also demonstrated to be the case, as follows. The intact lrhA gene was cloned into a multicopy plasmid, pUSI2 (16), to overproduce LrhA under the control of an IPTG-inducible tac promoter. The resultant plasmid, pTO13, was introduced into cells carrying the bgl-lacZ fusion, and then β-galactosidase activity in the presence of IPTG was measured. The results showed that such presumed overproduction of LrhA indeed resulted in the Bgl+ phenotype in the hns60 background (Fig. 4B). Furthermore, consistent with the result with the lrhA83 allele (Fig. 4A), such an event did not occur in the wild-type hns background (Fig. 4B).
FIG. 4.
(A) Expression of the bgl operon in lrhA mutants. Cells carrying the lrhA and hns alleles as indicated were grown and then the β-galactosidase activity was measured, as described for Fig. 2. Each value is the mean + standard deviation from four independent assays. WT, wild type. (B) Effect of overproduction of LrhA on bgl expression. Transformants of the indicated strains with pUSI2 (a control vector) or pTO13 were grown at 37°C to the mid-logarithmic phase in TB medium supplemented with 5 mM β-methyl-d-glucoside, 25 μg of ampicillin per ml, and 100 μM IPTG, and then the β-galactosidase activity was measured as described above.
lrhA83 affects bgl silencing through rpoS function.
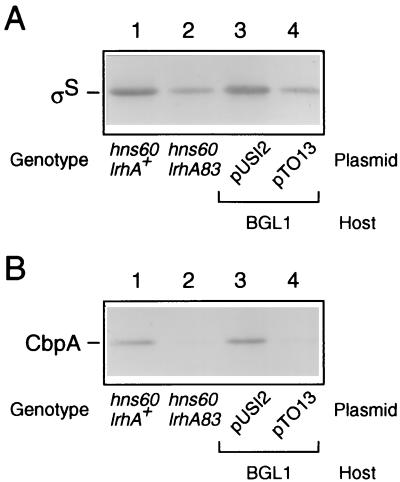
As stated above for the lrhA83 mutant, it is most likely that transposon insertion results in the overproduction of LrhA, thereby yielding the Bgl+ phenotype in the hns60 background. During the course of this study, we came across an intriguing report that the overproduction of LrhA decreases the cellular content of ςS (5). This immediately led us to hypothesize that the Bgl+ phenotype observed in the lrhA83 mutant was due to reduced expression of rpoS. To examine this, the cellular content of ςS was determined by immunoblotting analysis. Indeed, the lrhA83 mutant cells were found to contain a markedly reduced amount of ςS (Fig. 5A; compare lanes 1 and 2). Overproduction of LrhA from plasmid pTO13 caused a similar event in the wild-type lrhA background (Fig. 5A, lanes 3 and 4). These observations are consistent with the conclusion in the previous report (5). To confirm this further, we examined the cellular content of the CbpA protein, whose expression is largely dependent on ςS (22). It was found that the levels of CbpA were markedly reduced in lrhA83 cells and also in LrhA-overproducing cells (Fig. 5B). From these results, we concluded that the lrhA83 mutation affects bgl silencing by modulating the content of ςS in hns60 cells. In other words, we obtained two novel Bgl+ mutations in the hns60 background, both of which impair rpoS function, one directly (rpoS23) and the other indirectly (lrhA83).
FIG. 5.
Effect of LrhA overproduction on cellular content of ςS. Strains CU319 (hns60 lrhA+), CU320 (hns60 lrhA83), BGL1 harboring pUSI2, and BGL1 harboring pTO13 were grown as described for Fig. 2. Total protein samples were prepared at the mid-logarithmic phase, and then immunoblotting analysis of ςS or CbpA was carried out by using 15 or 60 μg of each sample per lane, respectively.
Does StpA function as an adapter molecule?
The stpA gene encodes an H-NS homolog that functions as a multicopy suppressor of hns mutations (15, 24). Free et al. reported previously that the bgl silencing caused by a C-terminally truncated H-NS is relieved in an stpA null strain, and they proposed that StpA is an adapter molecule (4). Surprisingly, we did not succeed in isolating such stpA knockout mutations in our intensive screening. This prompted us to construct a strain carrying both the hns60 allele and an stpA null (stpA::spc) mutation. The double mutant cells were found to be very sick, i.e., they formed very tiny colonies even after prolonged incubation on standard agar plates. Another double mutant carrying stpA::spc and the hns52 (G113D) allele showed the same growth properties as the former one (note that the hns52 allele also specifies an H-NS mutant protein impaired in its DNA-binding ability [21]). In any event, both double mutants exhibited the Bgl− phenotype on MacConkey lactose plates containing 0.1% salicin (data not shown). We then constructed a double mutant carrying both the null alleles (hns::neo and stpA::spc), which was apparently very healthy. We then introduced pHN91, specifying the H-NSQ92am mutant protein, into the hns::neo stpA::spc double mutant in order to determine the effect of the stpA mutation on bgl silencing. The result showed that the stpA mutation has no effect on bgl silencing in the hns60 background, under the H-NSQ92am induction conditions (compare bars 4 and 8 in Fig. 3A). It should be noted that we could not reliably interpret the results from the noninduction conditions because the amounts of the H-NSQ92am protein varied considerably depending on the genetic background.
DISCUSSION
In this study, we demonstrated that rpoS function is essential for bgl silencing by C-terminally truncated H-NS. How ςS functions in the mechanism underlying bgl silencing in the hns60 background is unknown. One explanation is that ςS regulates the expression of an as-yet-unidentified DNA-binding protein(s) that functions as a real adapter molecule. If this is the case, E. coli cells may have redundant genes encoding such adapters or the presumed gene may be essential for cell growth, because in spite of intensive genetic screening, we failed to identify such a single gene, except for rpoS. Alternatively, as we originally expected, ςS itself may be such an adapter molecule that can interact with both the bgl promoter region and H-NSQ92am. ςS may have such a function because it has the ability to bind to both DNA and protein (e.g., certain promoter sequences and RNA polymerase core enzyme). This assumption led us to recall the previous finding that the stability of ςS is somehow affected by H-NS (23). It will be interesting to determine if ςS can bind directly to the H-NS protein.
In this study, we searched for a protein(s) that plays a role in concert with H-NS in the mechanism underlying bgl silencing. Our findings revealed that ςS is crucial for bgl silencing, at least in the hns60 background. However, it should be emphasized that such a ςS function is seemingly dispensable for the bgl silencing observed in the wild-type background. The reason for this is unknown. The molecular mechanisms underlying the bgl silencing caused by the intact and truncated H-NS may be completely different from each other. But this view is unlikely a priori, because the repression mechanisms are equally relieved by certain dominant mutations that cause the overproduction of either LeuO or BglJ, as demonstrated in this and previous studies (6, 19). Rather, we favor the idea that ςS may play a role even in the naturally occurring bgl silencing caused by wild-type H-NS. In this respect, it should be taken into consideration that the C-terminally truncated H-NS is functionally intact with regard to bgl silencing, yet it causes very pleiotropic defects in other aspects of E. coli cell physiology (e.g., proV repression and porin regulation) (21). Thus, the difference between the intact H-NS and C-terminally truncated H-NS may be solely in the thresholds of their sensitivities to the absence of ςS under the growth conditions we used. The ςS content in the hns60 background may be elevated, because H-NS is known as a negative regulator for rpoS expression (23). Such a situation may also be advantageous for ςS to function cooperatively with C-terminally truncated H-NS. Based on this assumption, we are now extensively examining various growth conditions under which ςS affects bgl silencing in the wild-type hns background. In any case, our results suggested that the mechanism underlying bgl silencing is much more complex than originally thought. Whatever the molecular mechanism is, our results indicate that the ςS protein, which is crucial for a wide variety of fundamental cellular processes, is also involved in bgl silencing.
Finally, our result (Fig. 3A) with regard to StpA was not consistent with those of Free et al. (4). As mentioned above, our stpA::spc hns60 double mutant was very unstable. When the cells were spread on appropriate agar plates, a small number of fast-growing colonies appeared among tiny colonies. They exhibited the Bgl+ phenotype, but the results of immunoblotting analysis showed that the hns60 gene product was hardly detected in such fast-growing cells (data not shown). They probably have an additional mutation in the hns60 locus. This result suggests that the double mutant used by Free et al. (4) may have such a secondary mutation. Other explanations, such as the different genetic backgrounds, are also plausible. In addition, we used a bgl-lacZ fusion to directly monitor the transcriptional event, whereas Free et al. measured Bgl enzyme activity. In any case, we failed to demonstrate that the stpA gene is involved in bgl silencing in the hns60 background. This controversial issue remains to be addressed extensively.
ACKNOWLEDGMENTS
We thank Y. Kano (Kyoto Pharmaceutical University) for the kind gift of the stpA::spc allele.
This work was supported by a grant from Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Bongaerts J, Zoske S, Weidner U, Unden G. Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA-N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol. 1995;16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 2.Defez R, De Felice M. Cryptic operon for β-glucoside metabolism in Escherichia coli K12: genetic evidence for a regulatory protein. Genetics. 1981;97:11–25. doi: 10.1093/genetics/97.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiNardo S, Voelkel K A, Sternglanz R, Reynolds A E, Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982;31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 4.Free A, Williams R M, Dorman C J. The StpA protein functions as a molecular adapter to mediate repression of the bgl operon by truncated H-NS in Escherichia coli. J Bacteriol. 1998;180:994–997. doi: 10.1128/jb.180.4.994-997.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson K E, Silhavy T J. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator SprE. J Bacteriol. 1999;181:563–571. doi: 10.1128/jb.181.2.563-571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giel M, Desnoyer M, Lopilato J. A mutation in a new gene, bglJ, activates the bgl operon in Escherichia coli K12. Genetics. 1996;143:627–635. doi: 10.1093/genetics/143.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 8.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 9.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 10.Lopilato J, Wright A. Mechanisms of activation of the cryptic bgl operon. In: Drlica K, Riley M, editors. The bacterial chromosome. Washington, D.C.: American Society for Microbiology; 1990. pp. 439–444. [Google Scholar]
- 11.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 12.Prasad I, Schaefler S. Regulation of the β-glucoside system in Escherichia coli K-12. J Bacteriol. 1974;120:638–650. doi: 10.1128/jb.120.2.638-650.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnetz K. Silencing of Escherichia coli bgl promoter by flanking sequence elements. EMBO J. 1995;14:2545–2550. doi: 10.1002/j.1460-2075.1995.tb07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnetz K, Toloczyki C, Rak B. β-Glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987;169:2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi X, Bennett G N. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J Bacteriol. 1994;176:6769–6775. doi: 10.1128/jb.176.21.6769-6775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibui T, Uchida M, Teranishi Y. A new hybrid promoter and its expression vector in Escherichia coli. Agric Biol Chem. 1988;52:983–988. [Google Scholar]
- 17.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 18.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueguchi C, Ohta T, Seto C, Suzuki T, Mizuno T. The leuO gene product has a latent ability to relieve bgl silencing in Escherichia coli. J Bacteriol. 1998;180:190–193. doi: 10.1128/jb.180.1.190-193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ueguchi C, Seto C, Suzuki T, Mizuno T. Clarification of the dimerization domain and its functional significance for the Escherichia coli nucleoid protein H-NS. J Mol Biol. 1997;274:145–151. doi: 10.1006/jmbi.1997.1381. [DOI] [PubMed] [Google Scholar]
- 21.Ueguchi C, Suzuki T, Yoshida T, Tanaka K, Mizuno T. Systematic mutational analysis revealing the functional domain organization of Escherichia coli nucleoid protein H-NS. J Mol Biol. 1996;263:149–162. doi: 10.1006/jmbi.1996.0566. [DOI] [PubMed] [Google Scholar]
- 22.Yamashino T, Kakeda M, Ueguchi C, Mizuno T. An analogue of the DnaJ molecular chaperone whose expression is controlled by ςS during the stationary phase and phosphate starvation in Escherichia coli. Mol Microbiol. 1994;13:475–483. doi: 10.1111/j.1365-2958.1994.tb00442.x. [DOI] [PubMed] [Google Scholar]
- 23.Yamashino T, Ueguchi C, Mizuno T. Quantitative control of the stationary phase-specific sigma factor, ςS, in Escherichia coli: involvement of the nucleoid protein H-NS. EMBO J. 1995;14:594–602. doi: 10.1002/j.1460-2075.1995.tb07035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang A, Rimsky S, Reaban M E, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]