Abstract
The mRNA for CspA, a major cold shock protein in Escherichia coli, contains an unusually long (159 bases) 5′ untranslated region (5′-UTR), and its stability has been shown to play a major role in cold shock induction of CspA. The 5′-UTR of the cspA mRNA has a negative effect on its expression at 37°C but has a positive effect upon cold shock. In this report, a series of cspA-lacZ fusions having a 26- to 32-base deletion in the 5′-UTR were constructed to examine the roles of specific regions within the 5′-UTR in cspA expression. It was found that none of the deletion mutations had significant effects on the stability of mRNA at both 37 and 15°C. However, two mutations (Δ56-86 and Δ86-117) caused a substantial increase of β-galactosidase activity at 37°C, indicating that the deleted regions contain a negative cis element(s) for translation. A mutation (Δ2-27) deleting the highly conserved cold box sequence had little effect on cold shock induction of β-galactosidase. Interestingly, three mutations (Δ28-55, Δ86-117, and Δ118-143) caused poor cold shock induction of β-galactosidase. In particular, the Δ118-143 mutation reduced the translation efficiency of the cspA mRNA to less than 10% of that of the wild-type construct. The deleted region contains a 13-base sequence named upstream box (bases 123 to 135), which is highly conserved in cspA, cspB, cspG, and cspI, and is located 11 bases upstream of the Shine-Dalgarno (SD) sequence. The upstream box might be another cis element involved in translation efficiency of the cspA mRNA in addition to the SD sequence and the downstream box sequence. The relationship between the mRNA secondary structure and translation efficiency is discussed.
When a culture of Escherichia coli is shifted from 37 to 15 or 10°C, a number of proteins, called cold shock proteins, are transiently induced during its growth lag period (17, 32, 34). CspA, consisting of 70 amino acid residues, has been identified as a major cold shock protein (12), and its three-dimensional structure has been determined by both X-ray crystallography (28) and nuclear magnetic resonance spectroscopy (9, 24) to consist of a five-antiparallel β-stranded structure. CspA can bind to single-stranded DNA and RNA without high sequence specificity and has been proposed to function as an RNA chaperone at low temperature (16).
In E. coli, nine genes encoding CspA-like proteins, cspA to cspI, have been identified (34). Among them, cspA (12), cspB (18), cspG (23), and cspI (33) are cold shock inducible, and interestingly, cspD is induced during stationary phase and upon nutritional starvation (35). It was proposed that the large CspA family of E. coli may have a function in response to different environmental stresses (34).
Among these cold shock-inducible genes, cspA has been quite extensively investigated for the mechanism of its cold shock induction (34). The cspA promoter is highly active at 37°C, although CspA is hardly detectable at this temperature (8, 21). Even if the cspA promoter is replaced with the lpp promoter, a constitutive promoter for a major outer membrane protein, cspA expression is still cold shock inducible (8), indicating that the cspA induction upon cold shock occurs mainly at the levels of mRNA stability and translation. However, it should be mentioned that the cspA promoter contains an AT-rich upstream element (25) immediately upstream of the −35 region, which is considered to play an important role in efficient transcription initiation at low temperature (8, 11, 21). It has been demonstrated that the cspA mRNA is extremely unstable at 37°C but becomes stable upon cold shock, indicating that the stability of mRNA plays a crucial role in cold shock induction of cspA (3, 8, 10). Furthermore, it was shown that the 14-base downstream box located 12 bases downstream of the translation initiation codon of the cspA mRNA, which is partially complementary to a region, called anti-downstream box, of 16S rRNA (30), plays an important role in efficient translation at low temperature (6, 21). Thus, cspA expression is regulated in a complex manner at the levels of transcription, mRNA stability, and translation.
An important and unique feature of the cspA mRNA is its unusually long 5′ untranslated region (5′-UTR) consisting of 159 bases (31). This feature is also shared with several cold shock genes, which are dramatically induced after temperature downshift, such as cspB (7), cspG (23), and cspI (33). The 5′-UTR is considered to play a crucial role in the cold shock induction of cspA (1, 3, 8, 10, 11, 15, 21). Here, we constructed a series of deletion mutations in the 5′-UTR of cspA and analyzed their effects on cspA expression by examining the amount, stability, and translation efficiency of mRNA. It was found that besides mRNA stability, the 5′-UTR plays an important role in translation efficiency of the cspA mRNA.
MATERIALS AND METHODS
Bacterial strain and media.
E. coli AR137 [MC4100 Δ(malT-ompB) pcnB80] (13) was used and grown in M9-Casamino Acids medium supplemented with d-biotin (50 μg/ml). When necessary, ampicillin was added to a final concentration of 25 μg/ml.
General techniques.
All DNA cloning was carried out according to the method of Sambrook et al. (26). PCR was carried out according to the manufacturer’s instructions (Boehringer) with 25 cycles of amplification steps of 0.5 min at 94°C, 2 min at 50°C, and 1 min at 72°C.
Plasmid construction.
Each 5′-UTR deletion was prepared by two-step PCR. A plasmid, pJJG02 (15), which contains the wild-type cspA gene, was used as a template for PCR. For the first step, two independent PCRs were carried out for each mutation. One reaction was done with a combination of primer 67F, 5′-ccttgctagCCGATTAATCATAAATATG-3′ (nucleotides −67 to −49 of cspA), and mutation primer R, which contains a desired deletion mutation and is complementary to the sense cspA sequence, and another reaction was done with primer 4311, 5′-ccggatccagGTTGAACCATTTT-3′ (complementary to nucleotides +186 to +198), and mutation primer F, which also contains the same desired mutation as primer R. In these regions, 5′ tails are shown in lowercase where NheI and BamHI sites are underlined. For the construction of pMM022, pMM023, pMM024, pMM025, pMM026, and pKNJ37, primer D1R, 5′-ACTACACT/TTGATGTGCATTAGC-3′ (complementary to −15 to +1/+28 to +35), and primer D1F, 5′-GCACATCAA/AGTGTAGTAAGGCAA-3′ (−8 to +1/+28 to +42); primer D2R, 5′-CAACGATAA/GCTTTAATGGTCTGT-3′ (complementary to +13 to +27/+56 to +64), and primer D2F, 5′-TAAAGC/TTATCGTTGATACCC-3′ (+22 to +27/+56 to +70); primer D3R, 5′-TAAAGG/CTCTTGAAGGGACTT-3′ (complementary to +41 to +55/+86 to +91), and primer D3F, 5′-TCAAGAG/CCTTTAACGCTTCAAAA-3′ (+49 to +55/+86 to +102); primer D4R, 5′-CGGCGATAT/AATGTGCACTACGAGGG-3′ (complementary to +69 to +85/+118 to +126), and primer D4F, 5′-GCACATT/ATATCGCCGAAAGGC-3′ (+79 to +85/+118 to +132); primer D5R, 5′-TACCTTTAA/GGCGTGCTTTACAGATT-3′ (complementary to +101 to +117/+144 to +152), and primer D5F, 5′-AAAGCACGCC/TTAAAGGTAATACACT-3′ (+108 to +117/+144 to +159); and primer 8064, 5′-TAATTAAG/GATATGGCGTGCTTT-3′ (complementary to +108 to +122/+136 to +143), and primer 8063, 5′-GCCATATC/CTTAATTATTAAAGG-3′ (+115 to +122/+136 to +150), were used, respectively, where the position of each deletion is indicated by a slash. Each set of the first PCR products was mixed, heat denatured, annealed, and extended with Taq DNA polymerase. The resulting products were then amplified again by PCR with primer 67F and primer 4311. The final PCR fragments were digested with NheI and BamHI and inserted into the XbaI-BamHI site of pKM005 (14). For pKNJ37, the PCR fragment was cloned into the XbaI-BamHI site of pRS414X, a pRS414 derivative (29) in which the unique SmaI site has been changed to an XbaI site.
For the construction of pMM007, PCR was carried out with primer 67F and primer 4311 as primers and pJJG02 as a template. The PCR fragment was digested with NheI and BamHI and inserted into the XbaI-BamHI site of pRS414X.
pKNJ38 was constructed as follows: oligonucleotide 8509, 5′-CTAGCCGAA AGGCACAAATTAAGAGGGTATTAATAATGAAAGGGGGAATTCCA- 3′, and oligonucleotide 8510, 5′-AGCTTGGAATTCCCCCTTTCATTATTAATACCCTCTTAATTTGTGCCTTTCGG-3′, were first annealed and then cloned into pKM67 (21) digested with XbaI and HindIII.
The DNA sequences of all the constructs were confirmed by DNA sequencing by the chain-termination method (27).
β-Galactosidase assay.
E. coli AR137 harboring different plasmids was grown at 37°C to mid-log phase in M9-Casamino Acids medium and then transferred to 15°C. The β-galactosidase assay was carried out according to the protocol of Miller (20). The assay was done in duplicate at each time point.
Isolation of RNA and primer extension.
E. coli AR137 harboring different plasmids was grown under the same conditions used for the β-galactosidase assay described above. RNA extraction and primer extension methods were described previously (21). Primer M13-47, 5′-CGCCAGGGTTTTCCCAGTCACGAC-3′, which is complementary to a coding sequence of lacZ, was used. The products were analyzed on a denatured polyacrylamide gel and quantified by using a PhosphorImager (Bio-Rad).
RESULTS
Deletion analysis of the cspA 5′-UTR.
Earlier, we constructed two cspA-lacZ fusions, in which the lacZ gene was transcriptionally fused to cspA at +26 (pKM67) (21) or at +143 (pJJG78) (15) of the cspA mRNA. The β-galactosidase activity of the cells harboring pJJG78 was very low at 37°C and increased about 10-fold at 2 h after temperature downshift to 15°C, whereas the β-galactosidase activity of the cells harboring pKM67 was very high even at 37°C (21), indicating that the region from +26 to +143 in the 5′-UTR of the cspA mRNA plays a crucial role in cspA expression.
In order to further investigate which part of the 5′-UTR is responsible for the positive or negative regulation of cspA expression, we attempted deletion analysis of the 5′-UTR and examined the effects of various deletions on cold shock induction of cspA. For this purpose, a series of 5′-UTR deletion mutants, in which a 26- to 32-base deletion was created at about every 30 bases, were constructed, and the resultant 5′-UTRs were translationally fused to lacZ at the 13th amino acid residue of CspA as described in Materials and Methods. The resultant plasmids, pMM022, pMM023, pMM024, pMM025, and pMM026, contain deletion mutations in the 5′-UTR from +2 to +27, from +28 to +55, from +56 to +86, from +86 to +117, and from +118 to +143, respectively (Fig. 1A). In the case of pMM024, a deletion from +56 to +85 was originally designed, but all the transformants that we analyzed contained an extra base deletion at position +86. Plasmid pMM67 (21), which is the wild-type cspA-lacZ translational fusion construct, was used as a control. E. coli AR137, a pcnB mutant, which is known to maintain pBR322 derivatives in a low copy number (19), was used in order to minimize multicopy effects of the constructed gene on their expression.
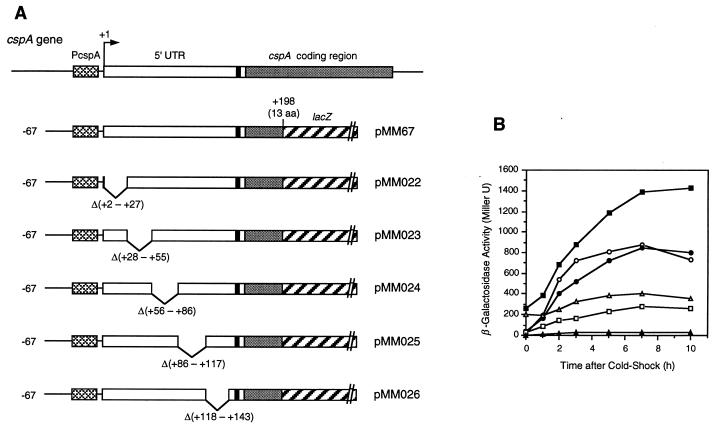
FIG. 1.
Cold shock induction of β-galactosidase. (A) Construction of cspA-lacZ fusions. The wild-type cspA is shown on the top. The cspA-lacZ fusion in each expression plasmid is shown from the 5′ end of the cspA promoter upstream region to lacZ. Nucleotide numbers are given starting from the transcription initiation site as +1, as determined by Tanabe et al. (31). The crosshatched, open, dotted, and diagonally striped bars represent the cspA promoter, its 5′-UTR, the cspA coding region, and the lacZ coding region, respectively. The solid boxes indicate the SD sequence. The positions of deleted regions are shown with nucleotide numbers. (B) Induction patterns of various deletion constructs. At mid-log phase, cultures of E. coli AR137 harboring various plasmids were shifted from 37 to 15°C. Samples were taken at 0, 1, 2, 3, 5, 7, and 10 h after the shift, and β-galactosidase activity was measured. The cspA-lacZ fusions were pMM67 (○), pMM022 (●), pMM023 (□), pMM024 (■), pMM025 (▵), and pMM026 (▴).
Transformed cells were grown in M9-Casamino Acids medium at 37°C, and β-galactosidase activities were measured after temperature downshift from 37 to 15°C. At 37°C (zero time point in Fig. 1B), β-galactosidase activities were 10-fold higher in cells harboring pMM024 (Δ56-86) and pMM025 (Δ86-117) than in cells harboring the wild-type pMM67, while other deletion mutants [pMM022 (Δ2-27), pMM023 (Δ28-55), and pMM026 (Δ118-143)] showed very low β-galactosidase activities. These results suggest that the 5′-UTR from base +56 to +117 is involved in the repression of cspA expression at 37°C. Interestingly, β-galactosidase activity increased almost fivefold with pMM024 (Δ56-86) after temperature downshift, while it increased only less than twofold with pMM025 (Δ86-117), suggesting that the region deleted in pMM025 (Δ86-117) plays an important role in cold shock induction of cspA. Similar to pMM025 (Δ86-117), β-galactosidase activity with pMM023 (Δ28-55) was poorly induced at a low temperature. In particular, the region deleted in pMM026 (Δ118-143) appears to play a crucial role in cspA expression at both high and low temperatures, since β-galactosidase activity was very low at both 37 and 15°C (Fig. 1B). The deletion of the region from base +2 to +27 (pMM022), which contains the cold box sequence involved in cspA autoregulation (15), has little effect on the cold shock induction of cspA (Fig. 1B).
Analysis of cspA-lacZ mRNA.
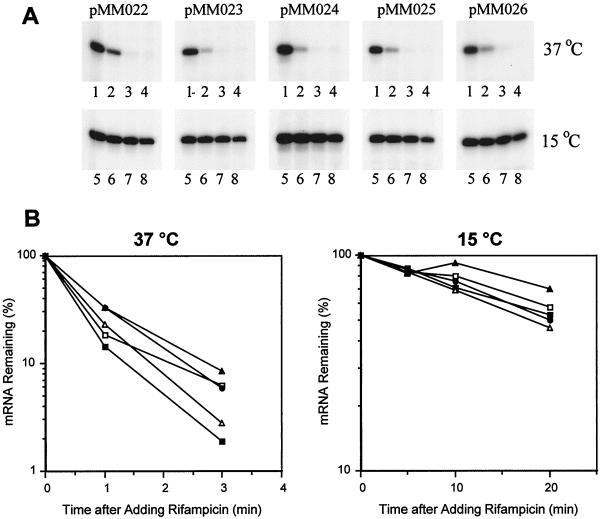
As described above, all deletion mutations, except for pMM022 (Δ2-27), of the 5′-UTR of the cspA mRNA affected cspA-lacZ expression. The cspA promoter is known to be active even at 37°C (8, 11, 21). Since all the deletion constructs have the intact cspA promoter (Fig. 1A), transcription efficiencies of these constructs are likely to be identical. On the other hand, stability of the cspA mRNA is known to be significantly different depending on growth temperatures (1, 3, 8, 10, 11, 21). Therefore, the effect of the deletion mutations on cspA-lacZ expression may be due to different mRNA stabilities of the constructs. To examine this possibility, primer extension analysis was carried out to quantitate the amounts of the cspA-lacZ transcripts for each mutant at different time points after the addition of rifampin at both 37 and 15°C (Fig. 2A). The amounts of transcripts at each time point were estimated with a phosphorimager, and the amounts of mRNA remaining (percentage of the amount at the zero time point) were plotted as shown in Fig. 2B. All the transcripts were unstable at 37°C with their half-lives estimated as being between 30 and 45 s. At 15°C, however, they became very stable with half-lives of between 20 and 40 min. These half-lives are similar to those for the wild-type construct pMM67 obtained previously (21) as well as to those for the wild-type chromosomal cspA (8, 11). It is important to note that in contrast to the similar mRNA half-lives at low temperature, β-galactosidase activities induced at 15°C widely varied among all these constructs as shown in Fig. 1B.
FIG. 2.
Analysis of mRNA stability. (A) Primer extension analysis of the cells harboring the cspA-lacZ fusions. At mid-log phase, cultures of E. coli AR137 harboring various plasmids were shifted from 37 to 15°C. For measurement of the mRNA stability at 37°C, cultures were shifted back to 37°C after 30 min of incubation at 15°C and rifampin was added to the cultures to a final concentration of 200 μg/ml. RNAs were extracted at 0 (lanes 1), 1 (lanes 2), 3 (lanes 3), and 5 (lanes 4) min after the addition of rifampin. For measurement of the mRNA stability at 15°C, rifampin was added 1 h after the temperature downshift, and then RNAs were extracted at 0 (lanes 5), 5 (lanes 6), 10 (lanes 7), and 20 (lanes 8) min after the addition of rifampin. Primer extension was carried out as described previously (21). (B) Graphic presentation of the results shown in panel A for 37 and 15°C, respectively. The radioactivities of transcripts were measured with a phosphorimager and plotted by using the transcript at the zero time point as 100%. ●, pMM022; □, pMM023; ■, pMM024; ▵, pMM025; and ▴, pMM026.
These results indicate that β-galactosidase activity of each cspA-lacZ construct is not correlated with the stability of mRNA but rather with the amount of mRNA and/or its translation efficiency. Therefore, we next examined the amounts of mRNA for each construct at different time points after cold shock by the primer extension method. The results are shown in Fig. 3A, and the amounts of transcripts were estimated with a phosphorimager. Their relative amounts were calculated by using the amount of the transcript of pMM67 at zero time as 1 (Fig. 3B). At 37°C, the amounts of transcripts for pMM022 (Δ2-27) and pMM024 (Δ56-86) were very similar to that of the wild-type construct pMM67 (Fig. 3B, column 1). It should be noted that β-galactosidase activity of pMM024 (Δ56-86) at 37°C was more than 10 times higher than that of pMM022 (Δ2-27) (Fig. 1B). In the case of pMM023 (Δ28-55), pMM025 (Δ86-117), and pMM026 (Δ118-143), the amounts of transcripts at 37°C are approximately half of that of pMM67. Again, it should be noted that β-galactosidase activity of pMM025 (Δ86-117) was 10 times higher than those of pMM023 (Δ28-55) and pMM026 (Δ118-143) (Fig. 1B). These results indicate that there is no correlation between the amounts of transcripts and the β-galactosidase activities at 37°C. This is consistent with the previous notion that although cspA mRNA was stabilized and accumulated at the nonpermissive temperature in the temperature-sensitive RNase E mutant, CspA was not produced under this condition (8).
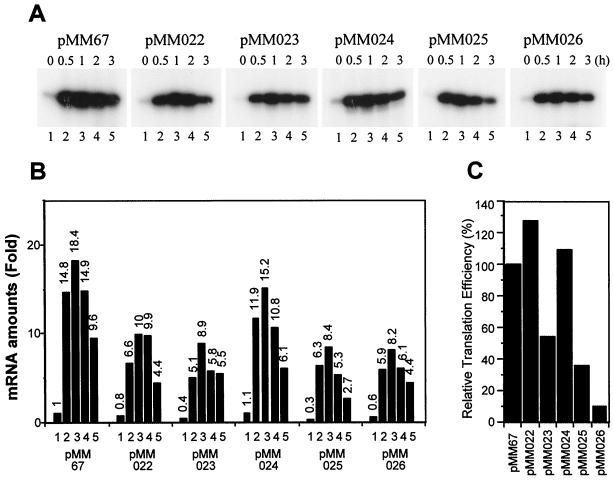
FIG. 3.
Analysis of mRNA level and translational efficiency. (A) Primer extension analysis of the cspA-lacZ fusions. At mid-log phase, cultures of E. coli AR137 harboring various plasmids were shifted from 37 to 15°C. RNAs were prepared from the culture at 37°C (0 h; lanes 1) and at 0.5 (lanes 2), 1 (lanes 3), 2 (lanes 4), and 3 (lanes 5) h after the temperature downshift. Primer extension was carried out as described previously (21). (B) Graphic presentation of the relative amounts of mRNA. Relative mRNA amounts (mean values of two experiments) were calculated from the radioactivities of transcripts shown in panel A with the transcript of pMM67 at 37°C as 1. The relative amount is shown on the top of each column. Columns 1, 0 h; columns 2, after 0.5 h; columns 3, after 1 h; columns 4, after 2 h; and columns 5, after 3 h. (C) Relative translational efficiencies of the cspA-lacZ mRNAs. Translational efficiencies at 15°C were calculated by dividing the increment of β-galactosidase activity during the first 2 h after cold shock by the amount of mRNA with the following formula: [(Gal 2 h) × (OD 2 h) − (Gal 0 h) × (OD 0 h)]/(ave mRNA), where (Gal 0 h) and (Gal 2 h) are β-galactosidase activities at 0 and 2 h after temperature downshift, respectively; (OD 0 h) and (OD 2 h) are the optical densities at 600 nm of the cultures at 0 and 2 h after temperature downshift, respectively; and (ave mRNA) is the average of relative mRNA amounts at 0.5, 1, and 2 h after temperature downshift. Relative translational efficiency of each mRNA was calculated by using the efficiency of mRNA of pMM67 as 100%.
After temperature downshift, the amounts of the cspA-lacZ mRNAs dramatically increased in all the constructs, and the induction patterns are shown in Fig. 3. They showed patterns in accumulation of the transcripts very similar to that of the wild-type pMM67, such that the maximal induction was observed at 1 h after temperature downshift. The patterns of mRNA levels were very similar between pMM67 and pMM024 (Δ56-86), while the others also showed a similar induction pattern although the amounts of their mRNAs were approximately half of that of the pMM67 mRNA at each time point. Since the promoter activities of all the deletion constructs are considered to be the same, and in addition their mRNA stabilities were also very similar to that of the wild-type construct (Fig. 2), lower amounts of mRNAs for all the deletion constructs except pMM024 (Δ56-86) are probably due to their lower transcription elongation rate and/or transcription attenuation within the 5′-UTR as reported recently (2). A remarkable finding was that the amounts of mRNA for pMM026 (Δ118-143) accumulated after cold shock were also identical to those for pMM022 (Δ2-27), pMM023 (Δ28-55), and pMM025 (Δ86-117) (Fig. 3B). Nevertheless, cold shock induction of β-galactosidase activity was extremely low for pMM026 (Δ118-143) throughout cold shock treatment (Fig. 1B), indicating that the mRNA for this construct was very poorly translated.
Translational regulation by the 5′-UTR.
Relative translation efficiencies at 15°C were calculated for all the constructs from the increments of β-galactosidase activity during the cold shock and the amounts of mRNA (see the legend to Fig. 3). As shown in Fig. 3C, the translation efficiency of pMM026 (Δ118-143) mRNA was extremely poor and calculated to be 9.5% of that of pMM67 mRNA. Besides pMM026 (Δ118-143), the translation efficiencies of the mRNAs of pMM023 (Δ28-55) and pMM025 (Δ86-117) were relatively low and calculated to be 54 and 36% of that of pMM67 mRNA, respectively. These results clearly indicate that the translation efficiency of mRNA plays an important role in the regulation of cspA expression and that, in particular, the region from base +118 to +143 of the 5′-UTR plays a major role in translation efficiency.
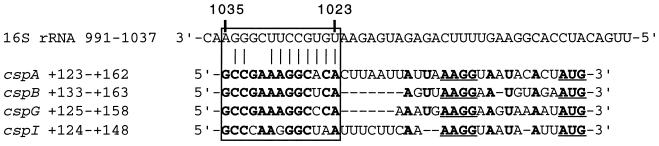
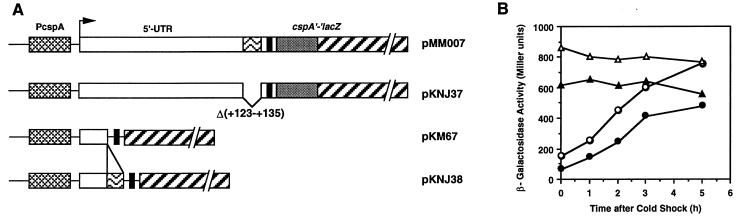
Nucleotide sequence comparison of the 5′-UTRs of the cold-shock-inducible genes, cspA, cspB, cspG, and cspI, reveals that there is a 13-base sequence named upstream box (UB) sequence (from base +123 to +135 of cspA) very well conserved among these genes as shown in Fig. 4. Interestingly, these 13-base sequences are located immediately upstream of the Shine-Dalgarno (SD) sequence and contain a palindromic sequence to form a stable secondary structure (ΔG = −9.5 kcal [Fig. 4, see also Fig. 6]). It is also complementary to the region from base 1023 to 1035 of 16S rRNA (Fig. 4). The significance of these facts will be discussed in the Discussion. In the pMM026 (Δ118-143) mRNA, this UB region has been deleted, which may be the major cause for the poor translation efficiency of the mRNA. In order to characterize the role of the UB sequence in translation efficiency, two new constructs were made; in one construct (pKNJ37), the exact 13-base UB sequence was deleted from the wild-type cspA-lacZ construct (pMM007), and in another (pKNJ38) the 13-base sequence was added at the upstream region of the SD sequence of pKM67 (21) as shown in Fig. 5A. In pKM67, the lacZ gene is fused at base +26, and it has been shown that as a result of the substantial deletion in the 5′-UTR β-galactosidase became expressed even at 37°C without any further induction upon cold shock (21). Note that both pMM007 and pMM67 have the exactly identical insert of cspA in different vectors, pRS414 and pKM005, respectively. Cold shock induction patterns of β-galactosidase activities of these two plasmids (pMM007 and pMM67) are similar (data not shown). Cells were transformed with pKNJ37, pMM007, pKNJ38, and pKM67, and cold shock induction of β-galactosidase activity was examined. Deletion of the UB sequence (pKNJ37) significantly lowered β-galactosidase activity not only at 37°C but also upon cold shock (Fig. 5B). Consistently, when the UB sequence was inserted into pKM67, the constitutive expression of β-galactosidase at 37°C increased by approximately 20% (Fig. 5B). This increment was also kept upon cold shock. These results suggest that the UB sequence may be associated with efficient translation of cspA, although the loss of the UB sequence is unlikely to be the sole defect in pMM026 (Δ118-143).
FIG. 4.
Sequence similarities of cspA, cspB, cspG, and cspI mRNAs around the SD sequence and potential base pairing between cspA mRNA and 16S rRNA. Nucleotide numbers of cspA (31), cspB (7), cspG (23), and cspI (33) mRNA are given starting from the major transcription initiation site as +1. The sequence of 16S rRNA is from the work of Brosius et al. (4). Nucleotides identical in the four csp mRNAs are shown in boldface. The 13-base homologous sequences in cspA, cspB, cspG, and cspI are boxed (the UB). Positions of the SD sequence and the initiation codon are underlined. Potential base pairings between cspA mRNA and 16S rRNA are indicated by vertical lines.
FIG. 6.
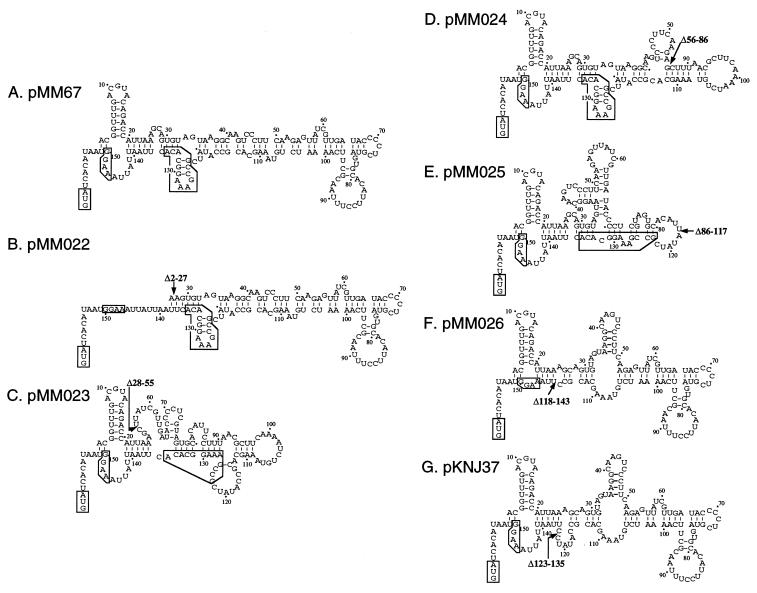
Comparison of the secondary structures of the 5′-UTRs for the deletion constructs. Secondary structures of the 5′-UTR for each deletion construct were predicted with a nucleotide sequence analysis program (DNASIS-Mac; Hitachi Software Engineering Co. Ltd.) based on the method of Zuker and Stieger (36). Nucleotides are numbered as the position in the cspA mRNA starting from the transcription initiation site as +1. The position of the deletion in each mutant is shown by an arrow with the nucleotide numbers of the deleted region. The highly conserved 13-base sequences upstream of the SD sequence designated the UBs are boxed. The initiation codon and the SD sequence are also boxed.
FIG. 5.
Role of the 13-base UB sequence in the cspA 5′-UTR in cspA-lacZ expression. (A) Construction of cspA-lacZ fusions. The constructs are drawn in the same manner as shown in Fig. 1A except for a box with wavy lines, which represents the 13-base UB sequence (5′-GCCGAAAGGCACA-3′) located upstream of the SD sequence. pKNJ37 is identical to pMM007 except for the deletion of the 13-base sequence. In both pMM007 and pKNJ37, cspA was translationally fused to lacZ. pKNJ38 is identical to pKM67 (21) except for the addition of the 13-base sequence by replacing the DNA fragment between XbaI and HindIII with synthesized oligonucleotides as described in Materials and Methods. In both pKM67 and pKNJ38, cspA was transcriptionally fused to lacZ. (B) Cold shock induction of β-galactosidase. The cspA-lacZ fusions were pMM007 (○), pKNJ37 (●), pKNJ38 (▵), and pKM67 (▴).
DISCUSSION
In the present paper, we have attempted to further elucidate the roles of the unusually long 5′-UTR of the cspA mRNA in cspA expression. We made a series of 26- to 32-base deletion mutations encompassing the entire 5′-UTR. These mutated 5′-UTRs were translationally fused to lacZ at the 13th amino acid residue of CspA after the downstream box sequence. At 37°C, pMM022 (Δ2-27), pMM023 (Δ28-55), and pMM026 (Δ118-143) showed β-galactosidase activities similar to the wild-type pMM67, while pMM024 (Δ56-86) and pMM025 (Δ86-117) showed higher β-galactosidase activities than pMM67, indicating that the region from base +56 to +117 is involved in the repression of cspA expression at 37°C to some extent. As shown in Fig. 2 and 3, this repression is not due to the decrease in the amounts and the stability of mRNA. Although it is possible that a factor binding this region might be involved in the repression, a precise mechanism for the repression is unclear at present. Note that the most important fact for the repression of the wild-type cspA expression at 37°C is the extreme instability of the cspA mRNA (3, 8, 10).
Based on the β-galactosidase activities after temperature downshift, mutants can be classified into three classes: class I [pMM022 (Δ2-27) and pMM024 (Δ56-86)], in which β-galactosidase is induced in a fashion similar to that of pMM67; class II [pMM023 (Δ28-55) and pMM025 (Δ86-117)], in which cold shock induction of β-galactosidase is poor; and class III [pMM026 (Δ118-143)], with very low β-galactosidase activities both before and after cold shock. It is worth mentioning that these differences were due to neither the amounts nor the stability of mRNA as evident from Fig. 2 and 3. This supports the notion that the 5′-UTR has another role in translation efficiency in addition to the stability of mRNA.
The relative translation efficiencies after temperature downshift for different constructs showed surprisingly significant differences (Fig. 3C), which coincided well with the classification of the constructs at 15°C as described above. The translation efficiencies with pMM022 (Δ2-27) and pMM024 (Δ56-86) (class I) are a little better than that of the wild-type pMM67, those with pMM023 (Δ28-55) and pMM025 (Δ86-117) (class II) are 40 to 50% of that of pMM67, and that with pMM026 (Δ118-143) (class III) is less than 10% of that of pMM67.
In pMM026 (Δ118-143), the deletion mutation is clearly affecting the translation efficiency but not the stability of mRNA. The deleted region was found to contain a 13-base sequence (bases +123 to +135) well conserved in the mRNAs for all the cold shock-inducible CspA family genes, cspA, cspB, cspG, and cspI (Fig. 4). This sequence, designated the UB sequence, may form a distinct secondary structure in both the wild-type pMM67 and class I constructs [pMM022 (Δ2-27) and pMM024 (Δ56-86)] (Fig. 6). Class I constructs showed a translation efficiency similar to that of the wild type. In class II [pMM023 (Δ28-55) and pMM025 (Δ86-117)], which showed 50% of the translation efficiency of the wild type, this secondary structure disappears; however, the predicted secondary structures around the SD sequence in these constructs are still similar to that of the wild-type construct. In contrast, when the UB region is deleted [pMM026 (Δ118-143); class III, which showed a very poor translation efficiency], the SD region forms a more stable secondary structure. This likely prevents recognition of the mRNA by ribosomes, causing a very poor translation efficiency in pMM026 (Δ118-143). Therefore, the UB sequence may function to punctuate the formation of a stable secondary structure immediately upstream of the SD sequence, allowing it to be highly accessible to ribosomes. Alternatively, as the UB sequence is complementary to the 16S rRNA sequence from bases 1023 to 1035 (Fig. 4), it is possible that the UB sequence may be another cis element, which may enhance translation efficiency by forming a duplex with 16S rRNA in addition to the SD sequence and the downstream box (21). Since all the constructs use the identical site of cspA to fuse to lacZ, the observed differences in translation efficiency are considered to be at the level of translation initiation but not at the level of translation elongation.
Consistent with the proposed role of the UB sequence in the translation, the addition of the UB sequence to pKM67 resulted in the increase of β-galactosidase activity by approximately 20%. On the other hand, the deletion of the exact 13-base UB sequence [pKNJ37 (Δ123-135) in Fig. 5] resulted in a 50% reduction of β-galactosidase activity at 37°C and a lower level of induction upon cold shock. Note that the predicted secondary structure surrounding the SD sequence of pKNJ37 (Δ123-135) is the same as that of the wild type (Fig. 6).
In summary, stabilization of the cspA mRNA upon cold shock is prerequisite for CspA production. This stabilization does not require any de novo protein synthesis (5). As presented here, translation efficiency of cspA mRNA turns out to play an important role in cspA expression in addition to the mRNA stabilization. The region from base +118 to +143, containing the UB sequence, of the 5′-UTR was clearly required for the cold shock induction (Fig. 1). However, it is unlikely that a de novo-synthesized activator binds to this region to induce translation upon cold shock, since the cold shock induction of CspA was observed even in the presence of a translation inhibitor such as chloramphenicol (7), although it cannot be ruled out that a preexisting factor might be activated upon cold shock. It is possible that the secondary structure of the 5′-UTR of cspA mRNA at low temperatures, in particular surrounding the SD and UB sequences, might be different from that at 37°C. This structural change might make cspA mRNAs more accessible to ribosomes. It is thus possible that the 5′-UTR of the cspA mRNA might act as an RNA thermometer, as was recently proposed for the rpoH mRNA (22). To know the more precise molecular mechanism of the regulation of cspA expression, determination of the secondary structure of the cspA mRNA and the relationship between the secondary structure and the translation efficiency remain to be addressed. The point mutation analysis in addition to the deletion analysis presented here will give us information on the molecular anatomy of the structure and function of the 5′-UTR of the cspA mRNA.
ACKNOWLEDGMENTS
Kunitoshi Yamanaka and Masanori Mitta contributed equally to this work.
We thank W. Bae, J.-P. Etchegaray, and S. Phadtare for comments.
K.Y. was partly supported by the Uehara Memorial Foundation. This work was supported by a grant from the National Institutes of Health (GM 19043).
REFERENCES
- 1.Bae W, Jones P G, Inouye M. CspA, the major cold-shock protein of Escherichia coli, negatively regulates its own gene expression. J Bacteriol. 1997;179:7081–7088. doi: 10.1128/jb.179.22.7081-7088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bae W, Phadtare S, Severinov K, Inouye M. Characterization of Escherichia coli cspE, whose product negatively regulates transcription of cspA, the gene for the major cold shock protein. Mol Microbiol. 1999;31:1429–1441. doi: 10.1046/j.1365-2958.1999.01284.x. [DOI] [PubMed] [Google Scholar]
- 3.Brandi A, Pietroni P, Gualerzi C O, Pon C L. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 4.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etchegaray J P, Inouye M. CspA, CspB, and CspG, major cold shock proteins of Escherichia coli, are induced at low temperature under conditions that completely block protein synthesis. J Bacteriol. 1999;181:1827–1830. doi: 10.1128/jb.181.6.1827-1830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etchegaray J P, Inouye M. Translational enhancement by an element downstream of the initiation codon in Escherichia coli. J Biol Chem. 1999;274:10079–10085. doi: 10.1074/jbc.274.15.10079. [DOI] [PubMed] [Google Scholar]
- 7.Etchegaray J P, Jones P G, Inouye M. Differential thermoregulation of two highly homologous cold-shock genes, cspA and cspB, of Escherichia coli. Genes Cells. 1996;1:171–178. doi: 10.1046/j.1365-2443.1996.d01-231.x. [DOI] [PubMed] [Google Scholar]
- 8.Fang L, Jiang W, Bae W, Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol Microbiol. 1997;23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 9.Feng W, Tejero R, Zimmerman D E, Inouye M, Montelione G T. Solution NMR structure and backbone dynamics of the major cold-shock protein (CspA) from Escherichia coli: evidence for conformational dynamics in the single-stranded RNA-binding site. Biochemistry. 1998;37:10881–10896. doi: 10.1021/bi980269j. [DOI] [PubMed] [Google Scholar]
- 10.Goldenberg D, Azar I, Oppenheim A B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 11.Goldenberg D, Azar I, Oppenheim A B, Brandi A, Pon C L, Gualerzi C O. Role of Escherichia coli cspA promoter sequence and translational apparatus adaptation in the cold shock response. Mol Gen Genet. 1997;256:282–290. doi: 10.1007/s004380050571. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein J, Pollitt N S, Inouye M. Major cold shock proteins of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harlocker S L, Rampersaud A, Yang W-P, Inouye M. Phenotypic revertant mutations of a new OmpR2 mutant (V203Q) of Escherichia coli lie in the envZ gene, which encodes the OmpR kinase. J Bacteriol. 1993;175:1956–1960. doi: 10.1128/jb.175.7.1956-1960.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inouye M. Multipurpose expression cloning vehicles in Escherichia coli. In: Inouye M, editor. Experimental manipulation of gene expression. New York, N.Y: Academic Press, Inc.; 1983. pp. 15–32. [Google Scholar]
- 15.Jiang W, Fang L, Inouye M. The role of 5′-end untranslated region of the mRNA for CspA, the major cold-shock protein of Escherichia coli, in cold-shock adaptation. J Bacteriol. 1996;178:4919–4925. doi: 10.1128/jb.178.16.4919-4925.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 17.Jones P G, VanBogelen R A, Neidhardt F C. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee S J, Xie A, Jiang W, Etchegaray J-P, Jones P G, Inouye M. Family of the major cold-shock protein, CspA (CS7.4) of Escherichia coli, whose members show a high sequence similarity with the eukaryotic Y-box binding proteins. Mol Microbiol. 1994;11:833–839. doi: 10.1111/j.1365-2958.1994.tb00361.x. [DOI] [PubMed] [Google Scholar]
- 19.Lopilato J, Bortuer S, Beckwith J. Mutations in a new chromosomal gene of Escherichia coli K-12, pcnB, reduced plasmid copy number of pBR322 and its derivatives. Mol Gen Genet. 1986;205:285–290. doi: 10.1007/BF00430440. [DOI] [PubMed] [Google Scholar]
- 20.Miller J H. A short course in bacterial genetics—a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 21.Mitta M, Fang L, Inouye M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold shock induction. Mol Microbiol. 1997;26:321–335. doi: 10.1046/j.1365-2958.1997.5771943.x. [DOI] [PubMed] [Google Scholar]
- 22.Morita M T, Tanaka Y, Kodama T S, Kyogoku Y, Yanagi H, Yura T. Translational induction of heat shock transcription factor ς32: evidence for a built-in RNA thermosensor. Genes Dev. 1999;13:655–665. doi: 10.1101/gad.13.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakashima K, Kanamaru K, Mizuno T, Horikoshi K. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J Bacteriol. 1996;178:2994–2997. doi: 10.1128/jb.178.10.2994-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newkirk K, Feng W, Jiang W, Tejero R, Emerson S D, Inouye M, Montelione G T. Solution NMR structure of the major cold shock protein (CspA) from Escherichia coli: identification of a binding epitope for DNA. Proc Natl Acad Sci USA. 1994;91:5114–5118. doi: 10.1073/pnas.91.11.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross W, Gosink K K, Salmon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the a subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schindelin H, Jiang W, Inouye M, Heinemann U. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5119–5123. doi: 10.1073/pnas.91.11.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 30.Sprengart M L, Fuchs E, Porter A G. The downstream box: an efficient and independent translation initiation signal in Escherichia coli. EMBO J. 1996;15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 31.Tanabe H, Goldstein J, Yang M, Inouye M. Identification of the promoter region of the Escherichia coli major cold shock gene, cspA. J Bacteriol. 1992;174:3867–3873. doi: 10.1128/jb.174.12.3867-3873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thieringer H A, Jones P G, Inouye M. Cold shock and adaptation. Bioessays. 1998;20:49–57. doi: 10.1002/(SICI)1521-1878(199801)20:1<49::AID-BIES8>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 33.Wang N, Yamanaka K, Inouye M. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J Bacteriol. 1999;181:1603–1609. doi: 10.1128/jb.181.5.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 35.Yamanaka K, Inouye M. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J Bacteriol. 1997;179:5126–5130. doi: 10.1128/jb.179.16.5126-5130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker M, Stieger P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1982;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]