Abstract
We investigated the regulation of the MexEF-OprN multidrug efflux system of Pseudomonas aeruginosa, which is overexpressed in nfxC-type mutants and confers resistance to quinolones, chloramphenicol and trimethoprim. Sequencing of the DNA region upstream of the mexEF-oprN operon revealed the presence of an open reading frame (ORF) of 304 amino acids encoding a LysR-type transcriptional activator, termed MexT. By using T7-polymerase, a 34-kDa protein was expressed in Escherichia coli from a plasmid carrying the mexT gene. Expression of a mexE::lacZ fusion was 10-fold higher in nfxC-type mutants than in the wild-type strain; however, transcription of mexT as well as the mexT DNA region was unchanged. Located adjacent to mexT but transcribed in opposite direction, the beginning of an ORF termed qrh (quinone oxidoreductase homologue) was identified. Expression of a qrh::lacZ fusion was also found to be activated by MexT. Further, we present evidence for coregulation at the transcriptional and the posttranscriptional level between the MexEF-OprN efflux system and the OprD porin responsible for cross-resistance of nfxC-type mutants to carbapenem antibiotics.
Pseudomonas aeruginosa is a leading cause of hospital acquired infections. Its high intrinsic antibiotic resistance and the ability to develop multidrug resistance pose serious therapeutic problems. For a long time it has been assumed that this elevated intrinsic resistance was mainly due to the low outer membrane permeability of P. aeruginosa which was correlated to the appearance of outer membrane proteins with sizes in the range of 50 kDa. These proteins (OprM, OprJ, and OprN) have now been shown to be part of multidrug efflux systems with broad specificity (13, 25, 26) which catalyze the energy-dependent extrusion of antibiotics such as β-lactams, quinolones, tetracycline, chloramphenicol, macrolides, and trimethoprim. The three efflux systems of P. aeruginosa have similar patterns of genetic organization. The first gene of each operon encodes a periplasmic fusion protein (MexA, MexC, or MexE), the second encodes a cytoplasmic membrane protein (MexB, MexD, or MexF) thought to be the actual efflux pump, and the third gene encodes an outer membrane protein (OprM, OprJ, or OprN). The three proteins are believed to form a channel across the inner and outer membranes. The mexAB-oprM operon is expressed constitutively and contributes to the intrinsic resistance of P. aeruginosa to a variety of toxic substances (14, 15). Transcription of the mexAB-oprM operon is increased in nalB-type mutants (30) due to mutations in the repressor protein MexR (27, 43). The mexCD-oprJ operon (25) is not expressed constitutively but is overexpressed in mutants displaying mutations in nfxB, the gene coding for the transcriptional repressor of this efflux system (24, 35).
The third efflux operon, mexEF-oprN, which confers resistance to quinolones, chloramphenicol, and trimethoprim, is overexpressed in nfxC-type mutants of P. aeruginosa (13). NfxC-type mutants (7) are also cross-resistant to the carbapenem imipenem (3, 8, 18), since they show decreased expression of OprD, an outer membrane protein facilitating the diffusion of basic amino acids, small peptides (40), and several carbapenem antibiotics (39). The mexEF-oprN efflux operon differs from the other efflux systems in that it is positively regulated by a protein belonging to the LysR family of transcriptional activators (13). In the present study, we characterize this activator, called MexT, and show that it is required for the expression of the MexEF-OprN efflux pump. We also demonstrate the involvement of MexT in the regulation of an open reading frame (ORF) adjacent to mexT and present evidence for coregulation at the transcriptional and posttranscriptional levels between the OprD porin and the MexEF-OprN efflux system.
(The MexT sequence and the regulation of the mexEF-oprN operon were presented at the Pseudomonas meeting in Madrid, Spain, in September 1997.)
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH10B was used for cloning experiments and plasmid propagation. Plasmids pUC119 and pIC20H were used for subcloning and sequencing. Plasmids were conjugated from E. coli S17-1 (36) or by triparental mating with the helper plasmid pRK2013. Transconjugants were selected on M9 minimal medium (16) supplemented with 40 mM citrate as a carbon source or on Luria-Bertani (LB) medium supplemented with ampicillin at 40 μg/ml to counterselect against E. coli. MICs in Mueller-Hinton broth were determined by the microdilution method (11).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| MC1061 | F−araD139 Δ(ara-leu)7696 galE15 galK16 Δ(lac)X74 rpsL thi | Laboratory collection |
| S17-1 | thi pro hsdR recA chr::RP4-2 | 36 |
| DH10B | F−araD139 Δ(ara-leu)7696 galU galK Δ(lac)X74 mcrΔ(mrr-hsdRMS-mcrBC) deoR Φ80dlacZ ΔM15 endA1 nupG recA1 rpsL | Laboratory collection |
| P. aeruginosa | ||
| PAO1 | Wild-type | Laboratory collection |
| PT149 | PAO-7H, PAO1 overproducing MexEF-OprN, selected on ciprofloxacin | 13 |
| PT579 | PAO1 mexT::ΩHg | This study |
| Plasmids | ||
| pUC119 | High-copy-number cloning vector, Apr | 41 |
| pIC20H | High-copy-number cloning vector, Apr | 17 |
| pJQ200mp18 | Mobilizable suicide vector, sacB, Gmr | 28 |
| pWSK29 | Low-copy-number T7 expression vector, Apr | 42 |
| pWNC5 | pWSK29 with 1.5-kbp KpnI-EcoRI fragment expressing mexT under T7 control, Apr | This study |
| pOPN4 | pLAFR3-based cosmid containing 27-kb insert harboring mexE-mexF-oprN, Tcr | 13 |
| pME6001 | Broad-host-range vector, Gmr | D. Haas |
| pMEXT | pNFC8, 1.5-kbp HindIII-EcoRI fragment of pOPN4 cloned in HindIII-EcoRI-cleaved pME6001, Gmr | 13 |
| pMP220 | Promoterless lacZ fusion vector, IncP, Tcr | 37 |
| pNFZ4 | mexE::lacZ fusion on pMP220, Tcr | This study |
| pTZ4 | mexT::lacZ fusion on pMP220, Tcr | This study |
| pQRZ4 | qrh::lacZ fusion on pMP220, Tcr | This study |
| pSE45 | oprD::lacZ transcriptional fusion on pMP220, Tcr | This study |
| pSE50 | oprD::phoA translational fusion on pMP220, Tcr | This study |
Hg, mercury; Tc, tetracycline; Gm, gentamicin; Ap, ampicillin.
DNA sequencing and protein analysis.
DNA sequences were determined from double-stranded templates according to the dideoxy chain termination method (32) with an automatic sequencer (Applied Biosystems model 373A). DNA sequences were processed and analyzed with the PCGENE program (Intelligenetics Inc., Mountain View, Calif.) and the BLAST algorithm (1). Protein alignments were generated by the program CLUSTAL.
Strain and plasmid constructions.
The mexT mutant was constructed by inserting a 2.2-kbp BamHI fragment of pOPN4 (13) carrying the entire mexT gene into the suicide vector pJQ200mp18 (28), yielding pJQN3. MexT was inactivated by inserting the Hgr determinant as a 4.8-kbp BamHI fragment of pHP45ΩHg (6) into the unique BglII site of pJQN3. The resulting construct, called pJQCΩ, was mobilized from strain S17-1 into PAO1. Hgr and Gms colonies were recovered after counterselection on sucrose-containing plates. Three independent colonies were analyzed by Southern blot analysis with a digoxigenin-labeled DNA fragment. All three showed the same banding pattern, in agreement with an integration of the ΩHg cassette into the mexT structural gene. The mexT::ΩHg mutation was then transduced by phage E79tv2 (22) into the nfxC-type mutant PT149. The mexE::lacZ fusion plasmid pNFZ4 was constructed by cloning a 0.5-kbp BglII-EcoRI fragment of cosmid pOPN4 (13) containing the 3′ end of mexT into the promoter probing vector pMP220 (37). To generate the qrh::lacZ fusion plasmid pQRZ4, a 0.5-kbp DNA fragment was amplified from genomic DNA of strain PAO1 with primers procxP1 (5′-CTGCTCGGGGGCCAGGTTCTGAC-3′) and procxM3 (5′-GGTGGGCTCCATGCTGCGTC-3′) by using Pwo polymerase (Boehringer Mannheim). The blunt-ended fragment was cloned into the EcoRV-cleaved vector pIC20H. From the resulting plasmid pPRO1, a BglII-EcoRI fragment was cloned into BglII-EcoRI-cleaved pMP220. The mexT::lacZ fusion plasmid was obtained after HindIII digestion of pQRZ4 and religation. A clone in which the HindIII fragment was in the opposite orientation with respect to that of the lacZ gene as in pQRZ4 was called pTZ4. The transcriptional oprD::lacZ fusion pSE45 was constructed by inserting a 1.3-kbp PCR fragment containing 735 bp upstream of the oprD initiation codon as a BamHI-XbaI fragment into BglII-XbaI-cleaved pMP220. The translational oprD::phoA fusion was constructed by inserting a 941-bp BamHI-HindIII fragment containing the same 735 bp upstream of OprD as in pSE45 and 206 bp of the N-terminal sequence encoding OprD into BamHI-HindIII-cleaved pPHOK102 (31) to yield pSE48. Plasmid pSE50 was obtained by inserting a 3-kbp BamHI-PstI fragment of pSE48 into BglII-PstI-cleaved pMP220. To clone the DNA region upstream of mexT, chromosomal DNA of a P. aeruginosa strain carrying the Gmr suicide plasmid pJQ200mp18 (28) integrated into the oprN gene (unpublished results) was digested with SacI and religated. The ligation mix was used to transform E. coli DH10B, and Gmr clones were selected. The restriction profiles of plasmids from three transformants were analyzed and found to contain the same piece of chromosomal DNA upstream of the mexT gene.
MexT expression with T7 polymerase.
An E. coli strain harboring the T7 polymerase gene on plasmid pGP1-2 was transformed with the control plasmid pWSK29 or the mexT-carrying plasmid pWNC5. Single transformants were grown at 30°C in LB medium supplemented with the appropriate antibiotics. At an optical density at 600 nm of approximately 0.4, the cultures were shifted for 20 min to 42°C followed by a further 60-min incubation at 37°C. One-milliliter aliquots were centrifuged, and the pellets were resuspended in 1 ml of M9 minimal medium supplemented with all 20 amino acids except methionine and cysteine. 35S-labeled methionine and cysteine (10 μCi) were added, and the mixture was incubated for 2 min in the presence of rifampin at 400 μg/ml. The suspensions were centrifuged, and the pellets were resuspended in 100 μl of sodium dodecyl sulfate loading buffer. The samples were boiled, and 10 μl of each was loaded onto a 12% acrylamide minigel. After migration, the gel was dried and exposed to X-ray film for approximately 18 h.
Determination of β-galactosidase and alkaline phosphatase activities.
Strains were inoculated from −70°C glycerol stocks in LB supplemented with the appropriate antibiotics and grown overnight at 37°C. Cultures were diluted 1:100 in fresh LB without antibiotics. When strains carried two plasmids, antibiotics for the marker on each plasmid were added (gentamicin at 15 μg/ml and tetracycline at 50 μg/ml). β-Galactosidase (21) and alkaline phosphatase (4) activities were determined in triplicate samples at various times during growth.
Nucleotide sequence accession number.
The DNA sequence of mexT and its upstream region have been deposited in the EMBL databank and assigned accession number AJ007825.
RESULTS
Nucleotide sequence of the mexT gene.
The mexE-mexF-oprN operon was previously shown to be located on pOPN4, a pLAFR3-based cosmid clone (13). The DNA region upstream of mexE was sequenced and a single ORF of 912 nucleotides (nt) was identified. This ORF, called mexT, starts 112 nt from the end of the insert of the cosmid and is transcribed in the same direction as the mexE-mexF-oprN operon. A Shine-Dalgarno sequence (GAGGA) was located 6 nt upstream of the initiation codon. The 304-amino-acid sequence of the putative MexT polypeptide was compared to the entries in the Swissprot and EMBL databases by using the BLASTP program (1). Significant homology was found with several members of the LysR family of transcriptional activators (34). The highest amino acid identity (32%) was to NahR (33), the activator of the plasmid-encoded nah operon, specifying genes for the degradation of naphthalene in Pseudomonas putida. The second best alignment (30% amino acid identity) was obtained with NodD, one of the transcriptional activators required for nodulation in Rhizobium spp. Homology was most pronounced toward the N termini of the proteins containing the helix-turn-helix motif.
Mutations leading to overexpression of the mexAB-oprM and the mexCD-oprJ efflux systems have been located in the corresponding regulator genes, mexR (27, 43) and nfxB (24), respectively. The mexT gene was therefore a likely candidate to harbor a mutation responsible for overexpression of the mexEF-oprN operon. The mexT region, encompassing the structural gene and the regulator region, was amplified by PCR from strain PT149 (formerly PAO-7H) overexpressing the MexEF-OprN efflux system and from its parental strain PAO1. However, sequence analysis of these PCR fragments showed no nucleotide changes. The mexT region of two other spontaneous nfxC-type mutants was sequenced. Again, neither of them displayed any nucleotide changes, suggesting that the mutation responsible for the nfxC phenotype was not located in mexT or its regulatory region.
Cloning of the DNA region upstream of mexT.
To further investigate the regulation of the mexEF-oprN efflux operon, the DNA region upstream of mexT was cloned by plasmid rescue as described in Materials and Methods. The restriction pattern of three recovered clones was analyzed and found to contain about 10 kbp of chromosomal DNA. The DNA sequence upstream of mexT was determined from one of the plasmids and found to contain the beginning of an ORF transcribed divergently from mexT and located 221 bp from the mexT start codon. The deduced amino acid sequence of the ORF was homologous to a family of quinone oxidoreductases from E. coli (38) and P. aeruginosa (EMBL accession number X85015) as well as to eucaryotic homologues. The intergenic region between mexT and this ORF, called tentatively qrh (quinone oxidoreductase homologue), contained two putative promoter sequences located on the two different DNA strands and overlapping at their −10 regions. The putative mexT (CTGACA-18 bp-GATAAT) and qrh (CTGACA-15 bp-AATAAC) promoter sequences were very similar to the E. coli ς70 consensus sequence (TTGACA-15 to 17 bp-TATAAT).
Examination of the previously sequenced mexE promoter region (13) did not reveal significant homology to the E. coli ς70 consensus sequence. However, the sequence GTATCAC TGTTCGTGATAATCAAAATCTCGTCGTTCGATTAGT was found 58 bp upstream of the mexE start codon. This sequence showed a striking similarity to the sequence of the nod box (9) (NYATCCAYNNYRYRGATGNNNNYNATCNAAACAATCGATTTTA) located upstream of genes regulated by NodD in a Rhizobium spp. MexT is therefore likely to activate mexEF-oprN transcription by binding to the nod-box-like sequence.
Analysis of the mexT gene product and phenotype of a mexT mutant.
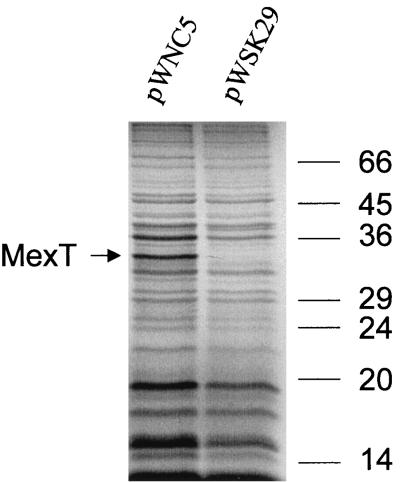
T7 polymerase-directed expression of mexT in E. coli revealed a protein band with a size of 34 kDa (Fig. 1). This size is close to the molecular mass of 33,418 deduced from the nucleotide sequence of mexT, and the labeled protein is therefore likely to correspond to the mexT gene product.
FIG. 1.
Labeling of E. coli cells with [35S]Met and [35S]Cys carrying either the control T7 expression vector pWSK29 or the mexT-carrying plasmid pWNC5. The molecular masses (in kilodaltons) of standard protein markers are indicated on the right.
A PAO1 derivative, called PT579 and inactivated by the insertion of an ΩHg cassette in the coding region of mexT, was constructed. As expected, MICs of the mexT mutant were indistinguishable from those of the wild-type strain (data not shown). The mexT::ΩHg mutation was transferred by transduction into the multidrug-resistant nfxC-type mutant PT149. All of the tested transductants recovered the antibiotic susceptibility profile of the wild-type strain, demonstrating that a functional mexT gene is required for expression of the multidrug resistance phenotype in nfxC-type mutants.
MexT is required for selection of the nfxC phenotype.
Selection of the nfxC phenotype can easily be achieved by plating wild-type PAO1 cells on LB agar containing at least 500 μg of chloramphenicol per ml. To establish the role of MexT in selection of the nfxC phenotype, PAO1 and the mexT::ΩHg mutant PT579 were plated on LB agar plates containing chloramphenicol at 600 μg/ml. While resistant colonies of PAO1 appeared at a frequency of about 10−8, no colonies (<10−10) were obtained with the mexT mutant. Among the 10 colonies derived from PAO1, all presented the nfxC phenotype (data not shown). These results demonstrate the absolute requirement of mexT for the selection of the nfxC antibiotic resistance phenotype. Indeed, PAO1 strains unable to yield nfxC mutants carry mutations in mexT (unpublished result).
Expression of mexE, mexT, and qrh.
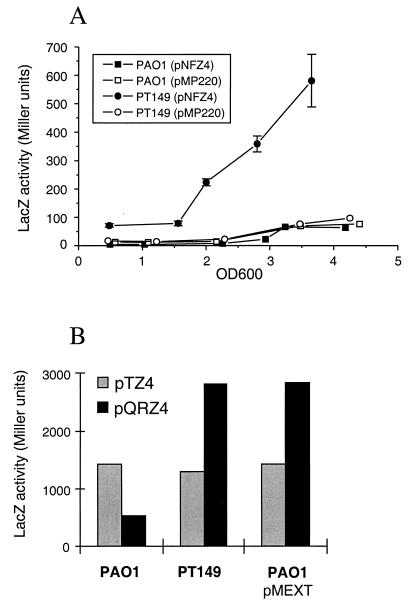
To further study the regulatory circuits of the qrh-mexT-mexEF-oprN DNA region, transcriptional lacZ fusions to mexE (pNFZ4), mexT (pTZ4), and qrh (pQRZ4) were constructed with the low-copy-number IncP derivative pMP220. In PAO1, β-galactosidase activities expressed from the mexE::lacZ fusion were always comparable to those of the control vector pMP220. However, in the nfxC-type mutant PT149 expression of the mexE::lacZ fusion was already higher in the lag phase and further increased during exponential and stationary growth (Fig. 2A). A similar increase in mexE::lacZ expression was found when MexT was introduced in trans on the multicopy plasmid pMEXT (data not shown). These results clearly show a correlation in the level of expression of the mexEF-oprN operon with the antibiotic resistance phenotype. They also suggest that the amount of MexT might be critical to the regulation of the mexEF-oprN operon. Therefore, the mexT::lacZ fusion plasmid pTZ4 was introduced into the wild type and the nfxC mutant PT149. In both strains, the fusion showed similar levels of elevated constitutive expression of β-galactosidase during growth in LB (data not shown), in agreement with a gene displaying a ς70 consensus promoter sequence (Fig. 2B). This result suggests that overexpression of MexEF-OprN in the nfxC-type mutant PT149 was not due to increased levels of mexT transcription.
FIG. 2.
(A) β-Galactosidase activity in P. aeruginosa wild-type strain PAO1 and nfxC-type mutant PT149 carrying either the control plasmid pMP220 or the mexE::lacZ fusion plasmid pNFZ4. Data are the results from one representative experiment. Error bars indicate standard deviations for triplicate determinations of LacZ activity. (B) Expression of mexT::lacZ and qrh::lacZ fusions in wild-type strain PAO1 in the absence or presence of plasmid-encoded copies of MexT and in the nfxC-type mutant PT149. Samples were taken during the mid-exponential growth phase.
Genes controlled by LysR-type activators are often located adjacent to the regulator and transcribed in the opposite direction. We therefore analyzed expression of the qrh::lacZ fusion plasmid pQRZ4. Like that for mexT, qrh expression was found to be constitutive; however, four- to fivefold-higher levels of β-galactosidase were measured in strains PT149 and PAO1 (pMEXT) compared to the wild type (Fig. 2B), suggesting that the qrh gene is also positively regulated by MexT.
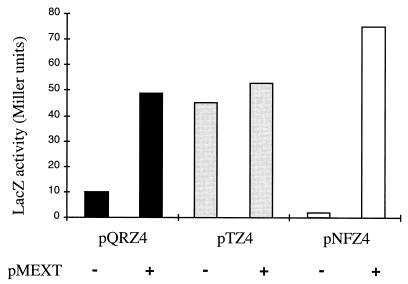
To confirm that plasmid-encoded MexT was sufficient to activate transcription, the lacZ fusions were analyzed in E. coli. In the presence of plasmid pMEXT, expression levels of the mexE::lacZ fusion were increased about 40-fold, while those of the qrh::lacZ fusion were increased fivefold. Plasmid pMEXT had no significant effect on expression of the mexT::lacZ fusion (Fig. 3). These results are in agreement with those found for P. aeruginosa, where plasmid-encoded MexT had a greater effect on mexE than on qrh transcription.
FIG. 3.
Effect of mexT on expression of mexE::lacZ (pNFZ4), mexT::lacZ (pTZ4), and qrh::lacZ (pQRZ4) fusions in E. coli MC1061. The presence and absence of the mexT-carrying plasmid pMEXT are indicated by + and − symbols, respectively.
In order to verify that the observed operon expression was not a singularity of the particular nfxC-type strain PT149, the three lacZ fusions were introduced into four other nfxC type strains as well as into strains overexpressing either the mexAB-oprM (nalB) or the mexCD-oprJ (nfxB) operon. All four nfxC-type strains showed increased expression of the mexE::lacZ fusion (10- to 20-fold) and of the qrh::lacZ fusion (fivefold). In the strains overexpressing either of the two other efflux operons, all three fusions were expressed at levels comparable to those of the wild type (data not shown).
MexT coregulates oprD expression.
Several reports have clearly established that in nfxC-type mutants, overexpression of the mexEF-oprN operon is linked to decreased amounts of the OprD porin in the outer membrane (18, 19). Since OprD is the port of entry of the carbapenem imipenem, nfxC-type mutants are cross-resistant to imipenem (3, 8, 18). Whether this cross-resistance is due to transcriptional repression of the oprD gene by MexT was investigated by constructing a pMP220-based lacZ fusion to the oprD gene (pSE45). Twofold-lower LacZ activity was found in the nfxC-type mutant PT149 compared to the wild type (Table 2). Since this weak effect could probably not account for the dramatic decrease in OprD expression in nfxC mutants as determined previously by Western blot analysis (13, 18, 19), we further examined the possibility of posttranscriptional regulation by MexT. A plasmid-encoded oprD::phoA translational fusion (pSE50) was constructed and introduced into PAO1 and PT149. Compared to the wild type, a fivefold decrease was observed in the nfxC-type mutant PT149 (Table 2). Furthermore, a similar fivefold decrease in alkaline phosphatase expression was found when plasmid pMEXT was introduced into PAO1 carrying the oprD::phoA fusion. These results strongly suggest that MexT downregulates oprD expression also at the posttranscriptional level.
TABLE 2.
Effects of nfxC mutation and mexT on transcriptional oprD::lacZ and translational oprD::phoA fusions in P. aeruginosa
| Strain | Plasmid | LacZ or PhoA activitya |
|---|---|---|
| PAO1 | pSE45 (oprD::lacZ) | 2,086 ± 373 |
| PT149(nfxC) | pSE45 | 1,012 ± 31 |
| PAO1 | pSE50 (oprD::phoA) | 10.4 ± 2.1 |
| PT149(nfxC) | pSE50 | 1.8 ± 0.1 |
| PAO1 | pSE50, pME6001 (control plasmid) | 8.7 ± 0.4 |
| PAO1 | pSE50, pMEXT (pME6001::mexT) | 1.8 ± 0.1 |
Results are the means ± standard deviations of triplicate determinations. Samples were taken during the mid-exponential growth phase. Enzymatic activities are expressed in Miller units (LacZ) or alkaline phosphatase units (PhoA).
DISCUSSION
Our results clearly establish MexT as the transcriptional activator of the mexEF-oprN efflux operon and demonstrate its requirement for the expression of the nfxC multidrug resistance phenotype. Furthermore, we found that when the gene is expressed from a multicopy plasmid, mexT on its own is able to activate transcription of a mexE::lacZ fusion in both P. aeruginosa and E. coli. This finding is in agreement with the previous observation that the mexT-carrying plasmid pMEXT (pNFC8) is sufficient to confer a nfxC resistance phenotype to a susceptible PAO1 wild type strain (13). We also show that the mexT DNA region is unchanged in the nfxC-type mutant PT149 and that mexT transcription levels are comparable to those of the wild type. How then can one account for the increased mexEF-oprN transcription in the nfxC mutants? The majority of the LysR-type regulators are synthesized in a nonactive form and become activated upon binding of a cognate effector molecule(s) (34). We therefore assume that in the nfxC-type mutants, the effector molecule of MexT is produced constitutively or in larger amounts than in the wild type, thereby causing permanent activation of MexT and hence overexpression of the MexEF-OprN efflux system. The fact that introduction of additional plasmid-encoded copies of MexT into a wild-type strain also causes increased mexEF-oprN expression can be explained by a shift in the equilibrium between the inactive and active forms of MexT. Such a mechanism has been suggested for the XylS regulator protein of the TOL plasmid pWW0 from P. putida (29), a member of the AraC family of transcriptional activators. Therefore, even in the absence of MexT effector molecules, plasmid-encoded MexT is able to activate mexEF-oprN transcription and to confer a nfxC multidrug resistance phenotype on the susceptible wild type strain.
The MexT homologues NahR and NodD are activated upon binding of salicylate (33) and phenolic plant-derived compounds (20), respectively. Interestingly, MexT not only has significant amino acid similarity to NodD, but the regulatory region of the mexEF-oprN operon also contains a nod box element (9) found upstream of NodD-regulated genes in Rhizobium species. However, neither NodD effectors (flavone, trigonellin, naringenin, and vanilline, etc.) provided at a final concentration of 2 mM nor the NahR inducer salicylate at concentrations of up to 50 mM showed any significant effect on mexEF-oprN transcription (unpublished results). Furthermore, the addition of the MexEF-OprN substrate molecules chloramphenicol, norfloxacin, or trimethoprim at subinhibitory concentrations had no effect on the expression of the mexE::lacZ fusion. Supposing that effector molecules are also substrates of the efflux pump, these results are further evidence that antibiotics are not the natural substrates of the MexEF-OprN efflux pump.
We found that MexT activates transcription not only of the mexEF-oprN efflux operon but also of an adjacent ORF, which we tentatively called qrh. The protein encoded by qrh shows homology to a family of quinone oxidoreductases of eucaryotic and procaryotic origins (38). Whether this gene is involved in the expression of the nfxC phenotype remains to be determined.
Numerous reports have demonstrated a decrease in OprD expression in nfxC-type mutants (12, 13, 18). OprD is a porin which facilitates diffusion of basic amino acids and small peptides and is also the port of entry of carbapenem antibiotics. In addition to MexEF-OprN substrates, nfxC-type strains are therefore cross-resistant to imipenem. Our results with oprD::lacZ and oprD::phoA fusion experiments suggest that the coregulation between oprD and the mexEF-oprN operon is exerted both at the transcriptional and posttranscriptional levels. A 2.5-fold decrease in the expression of a transcriptional oprD::xylE fusion in the presence of MexT has been reported recently by Ochs et al. (23). This result is in agreement with the twofold decrease in expression of our transcriptional oprD::lacZ fusion observed in a nfxC mutant. However, this modest effect seems unlikely to account solely for the almost complete absence of OprD in outer membranes of nfxC mutants (10, 13, 18). Therefore, the observed posttranscriptional effect on oprD expression offers a further explanation. A similar type of coregulation is found in E. coli mar mutants in which increased expression of the AcrAB efflux system is correlated with decreased expression of the porin OmpF (5). This effect is mediated by the antisense micF RNA (2). Alternatively, one can assume that transport of OprD across the cytoplasmic membrane is decreased in nfxC-type mutants by the overexpression of the MexEF-OprN efflux system.
Among the three multidrug efflux systems characterized so far, only the mexAB-oprM system (14) is constitutively expressed. It also displays the broadest substrate specificity and might therefore represent a natural defense mechanism against a variety of harmful substances. The fact that the other two efflux systems are not expressed under normal laboratory conditions and are tightly regulated by their respective regulator protein could suggest a role in more specific tasks, for example in the secretion of cellular metabolites. Identification of their substrates should help to elucidate the physiological role of these efflux pumps.
ACKNOWLEDGMENTS
We thank Mehri Michéa-Hamzehpour and Patrick Plésiat for helpful discussions and Colette Rossier for performing the sequencing. We thank R. Fellay (Novartis, Nyon, Switzerland) for pointing out the homology to the nod box of Rhizobium sp. and for providing chemicals. We are grateful to D. Haas (University of Lausanne, Lausanne, Switzerland) for providing plasmid pME6001.
This work was supported by a grant to T.K. from the Fonds National Suisse pour la Recherche Scientifique.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen J, Delihas N. micF RNA binds to the 5′ end of ompF mRNA and to a protein from Escherichia coli. Biochemistry. 1990;29:9249–9256. doi: 10.1021/bi00491a020. [DOI] [PubMed] [Google Scholar]
- 3.Aubert G, Pozzetto B, Dorche G. Emergence of quinolone-imipenem cross-resistance in Pseudomonas aeruginosa after fluoroquinolone therapy. J Antimicrob Chemother. 1992;29:307–312. doi: 10.1093/jac/29.3.307. [DOI] [PubMed] [Google Scholar]
- 4.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 5.Cohen S P, McMurry L M, Levy S B. marA locus causes decreased expression of OmpF porin in multiple-antibiotic-resistant (Mar) mutants of Escherichia coli. J Bacteriol. 1988;170:5416–5422. doi: 10.1128/jb.170.12.5416-5422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 7.Fukuda H, Hosaka M, Hirai K, Iyobe S. New norfloxacin resistance gene in Pseudomonas aeruginosa PAO. Antimicrob Agents Chemother. 1990;34:1757–1761. doi: 10.1128/aac.34.9.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda H, Hosaka M, Iyobe S, Gotoh N, Nishino T, Hirai K. nfxC-type quinolone resistance in a clinical isolate of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:790–792. doi: 10.1128/AAC.39.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goethals K, Van Montagu M, Holsters M. Conserved motifs in a divergent nod box of Azorhizobium caulinodans ORS571 reveal a common structure in promoters regulated by LysR-type proteins. Proc Natl Acad Sci USA. 1992;89:1646–1650. doi: 10.1073/pnas.89.5.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishii H, Sato K, Hoshino K, Sato M, Yamaguchi A, Sawai T, Osada Y. Active efflux of ofloxacin by a highly quinolone-resistant strain of Proteus vulgaris. J Antimicrob Chemother. 1991;28:827–836. doi: 10.1093/jac/28.6.827. [DOI] [PubMed] [Google Scholar]
- 11.Jorgensen J H, Ferraro M J, Craig W A, Doern G V, Finegold S M, Fung-Tomc J, Hansen S L, Hindler J, Reller L B, Swenson J M, Tenover F C, Testa R T, Wikler M A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Wayne, Pa: The National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 12.Köhler T, Michea-Hamzehpour M, Epp S F, Pechere J C. Carbapenem activities in Pseudomonas aeruginosa: respective contribution of OprD and efflux systems. Antimicrob Agents Chemother. 1999;43:424–427. doi: 10.1128/aac.43.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köhler T, Michea-Hamzehpour M, Henze U, Gotoh N, Curty L K, Pechère J C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 14.Li X-Z, Nikaido H, Poole K. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X Z, Zhang L, Poole K. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J Bacteriol. 1998;180:2987–2991. doi: 10.1128/jb.180.11.2987-2991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Marsh J L, Erfle M, Wykes E J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984;32:481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- 18.Masuda N, Ohya S. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1992;36:1847–1851. doi: 10.1128/aac.36.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McIver J, Djordjevic M A, Weinman J J, Bender G L, Rolfe B G. Extension of host range of Rhizobium leguminosarum bv. trifolii caused by point mutations in nodD that result in alterations in regulatory function and recognition of inducer molecules. Mol Plant Microbe Interact. 1989;2:97–106. doi: 10.1094/mpmi-2-097. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 22.Morgan A F. Transduction of Pseudomonas aeruginosa with a mutant of bacteriophage E79. J Bacteriol. 1979;139:137–140. doi: 10.1128/jb.139.1.137-140.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochs M M, McCusker M P, Bains M, Hancock R E. Negative regulation of the Pseudomonas aeruginosa outer membrane porin OprD selective for imipenem and basic amino acids. Antimicrob Agents Chemother. 1999;43:1085–1090. doi: 10.1128/aac.43.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okazaki T, Hirai K. Cloning and nucleotide sequence of the Pseudomonas aeruginosa nfxB gene, conferring resistance to new quinolones. FEMS Microbiol Lett. 1992;97:197–202. doi: 10.1016/0378-1097(92)90386-3. [DOI] [PubMed] [Google Scholar]
- 25.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J, Li X Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 26.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 29.Ramos J L, Mermod N, Timmis K N. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987;1:293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- 30.Rella M, Haas D. Resistance of Pseudomonas aeruginosa PAO to nalidixic acid and low levels of beta-lactam antibiotics: mapping of chromosomal genes. Antimicrob Agents Chemother. 1982;22:242–249. doi: 10.1128/aac.22.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez-Quinones F, Hernandez-Alles S, Alberti S, Escriba P V, Benedi V J. A novel plasmid series for in vitro production of phoA translational fusions and its use in the construction of Escherichia coli PhoE::PhoA hybrid proteins. Gene. 1994;151:125–130. doi: 10.1016/0378-1119(94)90642-4. [DOI] [PubMed] [Google Scholar]
- 32.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schell M A. Transcriptional control of the nah and sal hydrocarbon-degradation operons by the nahR gene product. Gene. 1985;36:301–309. doi: 10.1016/0378-1119(85)90185-4. [DOI] [PubMed] [Google Scholar]
- 34.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 35.Shiba T, Ishiguro K, Takemoto N, Koibuchi H, Sugimoto K. Purification and characterization of the Pseudomonas aeruginosa NfxB protein, the negative regulator of the nfxB gene. J Bacteriol. 1995;177:5872–5877. doi: 10.1128/jb.177.20.5872-5877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon R, Priefer U, Pühler A. A broad-host-range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology. 1983;1:784–790. [Google Scholar]
- 37.Spaink H P, Okker R J H, Wijffelman C A, Pees E, Lugtenberg B J J. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- 38.Thorn J M, Barton J D, Dixon N E, Ollis D L, Edwards K J. Crystal structure of Escherichia coli QOR quinone oxidoreductase complexed with NADPH. J Mol Biol. 1995;249:785–799. doi: 10.1006/jmbi.1995.0337. [DOI] [PubMed] [Google Scholar]
- 39.Trias J, Nikaido H. Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1990;34:52–57. doi: 10.1128/aac.34.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trias J, Nikaido H. Protein D2 channel of the Pseudomonas aeruginosa outer membrane has a binding site for basic amino acids and peptides. J Biol Chem. 1990;265:15680–15684. [PubMed] [Google Scholar]
- 41.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 42.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 43.Ziha-Zarifi I, Llanes C, Köhler T, Pechere J C, Plesiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob Agents Chemother. 1998;43:287–291. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]