Abstract
The Tol-peptidoglycan-associated lipoprotein (PAL) system of Escherichia coli is a multiprotein complex of the envelope involved in maintaining outer membrane integrity. PAL and the periplasmic protein TolB, two components of this complex, are interacting with each other, and they have also been reported to interact with OmpA and the major lipoprotein, two proteins interacting with the peptidoglycan. All these interactions suggest a role of the Tol-PAL system in anchoring the outer membrane to the peptidoglycan. Therefore, we were interested in better understanding the interaction between PAL and the peptidoglycan. We designed an in vitro interaction assay based on the property of purified peptidoglycan to be pelleted by ultracentrifugation. Using this assay, we showed that a purified PAL protein interacted in vitro with pure peptidoglycan. A peptide competition experiment further demonstrated that the region from residues 89 to 130 of PAL was sufficient to bind the peptidoglycan. Moreover, the fact that this same region of PAL was also binding to TolB suggested that these two interactions were exclusive. Indeed, the TolB-PAL complex appeared not to be associated with the peptidoglycan. This led us to the conclusion that PAL may exist in two forms in the cell envelope, one bound to TolB and the other bound to the peptidoglycan.
The cell envelope of gram-negative bacteria consists of three layers, the inner membrane, the outer membrane, and the rigid peptidoglycan layer that lies between them in the periplasmic space. Although the components of this envelope have been extensively studied and the three-dimensional structures of some of the proteins have been determined, we still have a limited understanding of their interactions and their assembly mechanism. There is evidence that the peptidoglycan interacts with numerous cell envelope proteins and that these interactions play an important role in the cell envelope organization.
One of the proteins for which such an interaction has been shown is the peptidoglycan-associated lipoprotein (PAL) (28, 29). PAL belongs to the Tol-PAL system, whose proteins are encoded by two operons located at 17 min on the chromosomal map of Escherichia coli (23, 36). The Tol-PAL system consists of several proteins that form two complexes. One, located in the cytoplasmic membrane, consists of the TolA, TolQ, and TolR proteins, which interact with each other by their transmembrane segments (10, 14, 18, 24). The other is linked to the outer membrane and is composed of PAL and the periplasmic protein TolB (4, 17).
The tol-pal genes are involved in maintaining outer membrane integrity (22, 23). Mutations in these genes result in the formation of outer membrane vesicles (3), thus indicating a defect in cell envelope assembly similar to that observed in lpp or ompA mutants (13, 34). Furthermore, Tol proteins have been parasitized to permit group A colicins and single-stranded phage DNA to be transported through the outer membrane (for a review, see reference 21). The Tol-PAL system must play an important role in the envelopes of gram-negative bacteria, since it has been described for Haemophilus influenzae, Pseudomonas aeruginosa, Pseudomonas putida, Actinobacillus pleuropneumoniae, and Helicobacter pylori with similar organizations (9, 12, 32, 33) (GenBank accession no. AE000619). Moreover, homologues of PAL have been found in an even larger number of gram-negative bacteria.
Recently, it has been shown that TolB interacts with Lpp and OmpA and that PAL also interacts with OmpA (7). Furthermore, TolB interacts with trimeric porins of the outer membrane, as does TolA (11, 31). Thus, it appears that the TolB and PAL proteins may be parts of a larger complex involved in anchoring the outer membrane to the peptidoglycan. In this regard, the interaction of PAL with the peptidoglycan should be crucial for the function of the Tol-PAL system.
The interaction of PAL with the peptidoglycan has been characterized by the solubility properties of PAL. In 2% sodium dodecyl sulfate (SDS), PAL is solubilized from an envelope preparation only when heated above 50°C. At 40°C, PAL is not solubilized even when 0.6 M NaCl is added (28, 29). In practice, the interaction of PAL with the peptidoglycan is tested in vivo by incubating whole cell envelopes in Laemmli buffer at 37°C. Under these conditions, PAL is not solubilized. Furthermore, several lipoproteins which are cross-linked to the peptidoglycan by dithiobis(succinimidylpropionate) have been detected, and it was proposed that one of them was PAL (26).
Using PAL-PhoA fusion protein constructs, Lazzaroni and Portalier (23) found that a region located between residues 101 and 116 of PAL was involved in the interaction with the peptidoglycan. In E. coli, this region is localized in a domain also found in OmpA and MotB, two proteins thought to be associated with the peptidoglycan, and in YfiB and YiaD, two lipoproteins with no known function (8). Moreover, a highly conserved sequence (from residues 97 to 114 in PAL) has a strong probability of adopting an α-helical conformation, with the more conserved residues located at one face. It has been proposed that this 18-residue sequence associates with the peptidoglycan (19).
In this study, experiments were carried out to further investigate the interactions between PAL, TolB, and the peptidoglycan and to localize the regions of PAL involved. It is not easy to precisely localize the region of interaction of PAL with the peptidoglycan in vivo because of the different levels of expression of PAL when mutations are introduced. Furthermore, mutations in PAL could also affect the properties of the cell envelope, which would then affect the solubility of the PAL protein. To circumvent this problem, we developed a new in vitro system to assay the interaction between purified PAL protein and purified peptidoglycan. In addition, we used a synthetic peptide covering the region of PAL thought to bind to the peptidoglycan to perform competition analyses. We demonstrated that this region alone is able to interact with the peptidoglycan. Furthermore, this same peptide was able to interact with TolB. This led us to the conclusion that PAL interacts either with TolB or with the peptidoglycan but cannot bind to the two molecules at the same time.
MATERIALS AND METHODS
Purification of PAL, TolB, TolR, and the peptidoglycan.
PAL was purified by exactly the same protocol described for the purification of PAL from H. influenzae (37). We used 1 liter of an E. coli C600 culture at an optical density at 600 nm (OD600) of 1. At the end of the procedure, the buffer was exchanged for 50 mM sodium phosphate buffer (pH 8)–0.08% Triton X-100 by using a PD-10 column (Pharmacia), and the PAL protein was recovered in a 1-ml fraction. The homogeneity of the PAL protein obtained was checked by SDS-polyacrylamide gel electrophoresis (PAGE).
Peptidoglycan was prepared according to the method of Leduc et al. (25). Briefly, cell envelopes corresponding to 1 liter of a culture at an OD600 of 1 were suspended in 10 ml of 9% NaCl, mixed with an equal volume of 8% SDS, and incubated for 30 min at 100°C. After standing at room temperature overnight, the sample was centrifuged at 30°C for 30 min at 400,000 × g (100,000 rpm) in a Beckman TLA-100.4 rotor. The sedimented material was suspended in 1 ml of water, then washed by four cycles of resuspension and recentrifugation in water, and finally resuspended in 0.5 ml of 10 mM sodium phosphate buffer, pH 8.
The purified TolB was a TolBHis derivative described by Bouveret et al. (5). The purification of the TolRIIHis derivative corresponding to the central periplasmic domain of TolR from residues 44 to 117 is described by Journet et al. (18).
Electrophoresis, immunoblotting, and antibodies.
SDS-PAGE and electrotransfer onto nitrocellulose were performed as described previously (20, 35). After transfer, the nitrocellulose membrane was treated with the antisera directed against TolB, PAL, and TolR, as previously described (4, 15, 18, 31).
Peptide synthesis.
The TolR peptide, consisting of residues 113 to 141 of the TolR protein with an additional cysteine at the N terminus (CKDVPYDEIIKALNLLHSAGVKSVGLMTQP), and the PAL peptide, consisting of residues 89 to 130 of the mature PAL protein (DERGTPEYNISLGERRANAVKMYLQGKGVSADQISIVSYGKE), were synthesized according to the method of Barany and Merrifield (1). We used a resin preloaded with 4-hydroxymethyl-phenoxy-methyl-copolystyrene–1% divinylbenzene (0.5 to 0.65 mmol) (Perkin-Elmer, Applied Biosystems Inc.) and an automated synthesizer (model 433A; Perkin-Elmer, Applied Biosystems Inc.). Purification was done with a Beckman high-pressure liquid chromatography apparatus on a Merck C8 reverse-phase column. After lyophilization, the peptides were resuspended in 10 mM sodium phosphate buffer, pH 8. Electrospray mass spectrometry, carried out with a single quad PE-SCIEX API 150ex spectrometer (Perkin-Elmer), and amino acid analyses, performed on a model 6300 Beckman analyzer, showed that synthesis of these two peptides was successful.
In vitro assay for PAL-peptidoglycan binding.
Five micrograms of the purified PAL protein (or TolRIIHis protein), which was first centrifuged for 30 min at 400,000 × g, and 50 μl of purified peptidoglycan were incubated in 10 mM sodium phosphate buffer–150 mM NaCl, pH 8, for 1 h at room temperature (final volume, 100 μl). The mixture was then centrifuged for 30 min at 400,000 × g (Beckman TL100). The supernatant was collected, and the pellet was washed in 500 μl of 10 mM sodium phosphate buffer–500 mM NaCl, pH 8. After identical centrifugation, the wash fraction was collected. Finally, the pellet was resuspended in 40 μl of Laemmli buffer and heated for 10 min at 96°C. After precipitation of the supernatant and wash fractions with 20% trichloroacetic acid, the supernatant, wash, and pellet fractions were analyzed by Western blotting with an anti-PAL antibody.
To carry out the competition experiments between PAL and the PAL peptide, two amounts (20 and 100 μg) of the PAL peptide or 250 μg of the TolR peptide, which were first centrifuged at 400,000 × g, were incubated for 1 h with 50 μl of purified peptidoglycan. PAL protein was then added, and the rest of the experiment was performed as described above.
In vivo and in vitro cross-linking with formaldehyde.
In vivo cross-linking experiments with formaldehyde were performed directly on whole cells overexpressing TolB and PAL proteins (strain JC7752 transformed with pBP [4]) for 20 min as described previously (4) and stopped with 10 mM Tris, pH 6.8. Cell envelopes were prepared and treated in 2% SDS–0.1 M NaCl–10% glycerol–10 mM Tris (pH 7.8) at 37°C for 10 min to recover the non-peptidoglycan-associated fraction. Then the pellet was treated in Laemmli loading buffer at 60°C for 10 min to solubilize the peptidoglycan-associated proteins.
For the in vitro experiments, purified TolB (5 μg) and purified PAL (5 μg), in 10 mM sodium phosphate buffer, were mixed in a 15-μl final volume and incubated for 1 h at room temperature. Formaldehyde (1% final concentration) was added, and the mixture was further incubated for 20 min. Cross-linking was stopped by the addition of 10 mM Tris, pH 6.8. The samples were heated at 37°C for 10 min in Laemmli loading buffer and analyzed by SDS-PAGE and Western blotting.
RESULTS
Purified PAL interacts with peptidoglycan in vitro.
To set up a new system allowing detection of the interaction between PAL and the peptidoglycan in vitro, these two molecules were first purified.
The PAL lipoprotein of E. coli was purified by the protocol described by Zlotnick et al. (37). This method, originally used for the purification of the PAL homologue in H. influenzae, consists of several membrane extraction steps with detergents. We followed this method without modification for E. coli C600. After purification, a Coomassie blue-stained gel revealed the presence of three minor contaminants of 7, 32, and 67 kDa (data not shown). The first two may correspond to Lpp and porins, respectively. However, the PAL lipoprotein was more than 95% pure. We obtained approximately 0.5 mg of PAL for 1 liter of culture.
Peptidoglycan was prepared by the protocol described by Leduc et al. (25) (see Materials and Methods). For our assays, we used the property of this purified peptidoglycan to be pelleted by ultracentrifugation at 400,000 × g for 30 min. Therefore, proteins added to the preparation and able to interact with the peptidoglycan should also be pelleted upon ultracentrifugation.
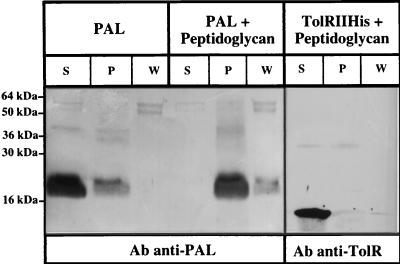
When PAL was incubated with peptidoglycan, the PAL protein pelleted with the peptidoglycan after ultracentrifugation, even after being washed with 0.5 M NaCl (Fig. 1). A negative-control experiment was performed with the purified periplasmic domain of the TolR protein, corresponding to residues 44 to 117 (18). It appeared that this domain of TolR did not pellet with the peptidoglycan after ultracentrifugation (Fig. 1).
FIG. 1.
The purified PAL lipoprotein interacts in vitro with the peptidoglycan. Purified PAL or purified TolRIIHis were incubated with peptidoglycan (+ Peptidoglycan) as described in Materials and Methods. A control incubation was carried out with PAL without peptidoglycan (PAL). After ultracentrifugation and washing as described in Materials and Methods, the Supernatant (S), wash (W), and pellet (P) fractions were loaded on an SDS–15% PAGE gel and electrotransfered to a nitrocellulose membrane, and proteins were detected with antiserum directed against PAL or TolR (Ab anti-PAL or Ab anti-TolR). The positions of the molecular mass markers are indicated on the left.
These in vitro experiments showed that, using purified molecules, it is possible to detect the interaction of PAL with the peptidoglycan. They also showed that the purified PAL was still in a conformation allowing interaction with the peptidoglycan.
The PAL-peptidoglycan interaction is displaced by a peptide composed of residues 89 to 130 of PAL.
The in vitro binding assay described above may be used to determine the regions of a protein involved in this binding. Indeed, peptides binding with the peptidoglycan should compete with the entire protein. We chose to test this hypothesis with a PAL peptide corresponding to residues 89 to 130. This peptide encompasses the region thought to be responsible for the interaction with the peptidoglycan (8, 19, 23) and also several residues which were shown to be involved in TolB and peptidoglycan interaction in vivo (7). Another peptide, corresponding to the C-terminal domain of the TolR protein (from residues 113 to 141), was used as a control.
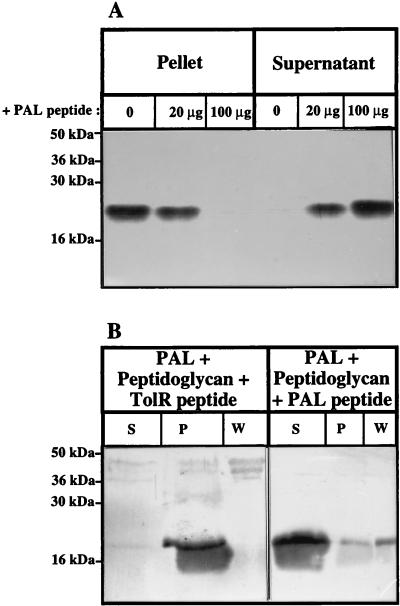
The PAL peptide was added to the PAL-peptidoglycan interaction assay described above at two different concentrations. The peptide prevented the pelleting of the PAL protein with the peptidoglycan in a dose-dependent manner (Fig. 2A). An effect was detected with as little as 20 μg of PAL peptide, representing a 20-fold molar excess compared to the PAL full-length protein (50 μM peptide and 2.5 μM full-length PAL [5 μg]). A control experiment was performed with the peptide corresponding to residues 113 to 141 of the TolR protein. This control peptide did not prevent the pelleting of PAL with the peptidoglycan, even at a concentration higher than the maximum used with the PAL peptide (Fig. 2B). Therefore, the PAL peptide specifically prevented the PAL-peptidoglycan interaction, which suggests that this peptide interacted with the peptidoglycan. The peptide, or part of it, would then adopt a conformation which must be close to the native conformation of this stretch of residues in the entire protein.
FIG. 2.
The PAL peptide competes with PAL for binding to the peptidoglycan. (A) Different amounts of the PAL peptide (0, 20, and 100 μg) were added to the PAL-peptidoglycan binding assay. The different fractions were analyzed by SDS–15% PAGE by immunoblotting with antibodies directed against PAL. (B) One hundred micrograms of the PAL peptide or 250 μg of the TolR peptide was added to the PAL-peptidoglycan binding assay. The different fractions were analyzed on an SDS–15% PAGE gel by immunoblotting with antibodies directed against PAL. The rest of the procedure was as described in the legend to Fig. 1.
The PAL-TolB interaction is displaced by the peptide composed of residues 89 to 130 of PAL.
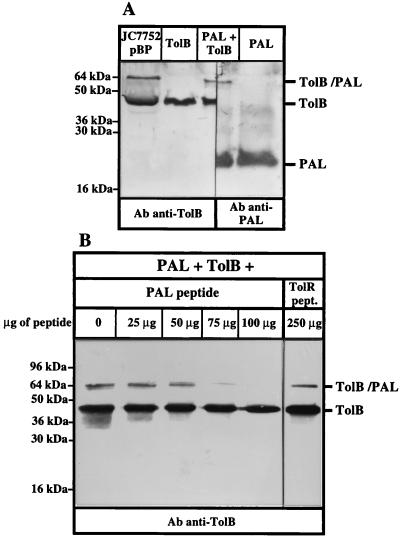
The TolB-PAL interaction has been well characterized in vivo (4, 7). Using purified TolB and PAL proteins, we tested whether this interaction could also be detected in vitro by using cross-linking with formaldehyde. When these two purified proteins were mixed in the presence of 1% formaldehyde and analyzed by Western blotting, a complex of 65 kDa was detected with antibodies directed against TolB and PAL (Fig. 3A). This complex corresponded to the size of the complex obtained in vivo on whole cells. This constitutes the third way to detect the interaction between TolB and PAL, in addition to formaldehyde cross-linking in vivo and coimmunoprecipitation of PAL with TolB (4).
FIG. 3.
The PAL peptide competes with PAL for binding to TolB. (A) Purified PAL and TolBHis proteins can be cross-linked in vitro. Purified PAL and TolBHis proteins were mixed (PAL + TolB) or treated independently with 1% formaldehyde (see Materials and Methods). An in vivo cross-linking was performed as a control to localize the Tol-PAL complex on whole cells overproducing the TolB and PAL proteins (JC7752 pBP). The samples were analyzed by SDS–12% PAGE and by immunoblotting with antibodies directed against TolB and PAL (Ab anti-TolB and Ab anti-PAL). (B) Purified PAL, purified TolBHis, and increasing amounts of the PAL peptide (0 to 100 μg) or 250 μg of the TolR peptide were treated with 1% formaldehyde as described in Materials and Methods. The samples were analyzed by SDS–12% PAGE and by immunoblotting with an antiserum directed against TolB (Ab anti-TolB). The positions of the TolB and PAL proteins, of the TolB-PAL complex, and of the molecular mass markers are indicated.
We then tested in vitro the effect of the PAL peptide on this interaction. When increasing amounts of the PAL peptide were added to the TolB-PAL mixture before cross-linking, the formation of the TolB-PAL complex was gradually prevented (Fig. 3B). An effect was detected with as little as 50 μg of PAL peptide, representing a 40-fold molar excess compared to the PAL full-length protein (110 μM peptide and 2.5 μM full-length PAL [5 μg]). The control peptide from the TolR protein described above did not produce this effect. Therefore, the PAL peptide specifically prevented the TolB-PAL interaction. This peptide thus interacted with TolB, confirming that the conformation of the peptide was close to the native one and that residues 89 to 130 of PAL alone can interact with TolB.
The TolB-PAL complex does not interact in vivo with the peptidoglycan.
Our results indicated that the region of PAL spanning residues 89 to 130 interacted with TolB and the peptidoglycan. We wanted to know if these two interactions could take place together or if they were exclusive.
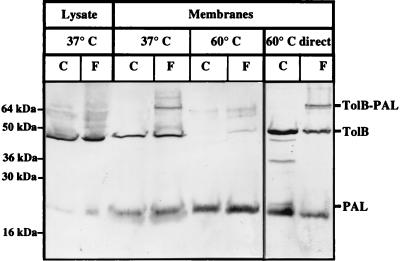
In our previous work, the TolB-PAL complex detected in vivo after cross-linking with formaldehyde was solubilized at 37°C (4). Yet the PAL protein was described as peptidoglycan associated, since it was solubilized in the Laemmli buffer only when heated to at least 50°C (23, 29). Therefore, to better define the solubilization properties of the cross-linked TolB-PAL complex, we performed in vivo cross-linking experiments. After cross-linking whole cells with formaldehyde, envelopes were prepared. The samples were first heated to 37°C and then to 60°C (Fig. 4). The PAL lipoprotein was detected mainly in the 60°C samples, whereas the TolB-PAL complex was detected only in the 37°C sample. We verified that the TolB-PAL complex was not destroyed by treatment at 60°C by incubating the membranes directly at 60°C. Therefore, the TolB-PAL complex is not peptidoglycan associated, suggesting that PAL cannot bind to TolB and to the peptidoglycan at the same time. This is in agreement with the idea that it is the same region, contained in the synthetic peptide, which is responsible for both the interaction with TolB and that with the peptidoglycan.
FIG. 4.
The TolB-PAL complex does not interact in vivo with the peptidoglycan. Whole cells overproducing the TolB and PAL proteins (JC7752 transformed with pBP) were treated with (F) or without (C) 1% formaldehyde, and after the treatment the cell envelopes were prepared and the cell lysate was kept (Lysate). The non-peptidoglycan-associated fraction (37°C) and the peptidoglycan-associated fraction (60°C) were prepared as described in Materials and Methods. In parallel, a sample was treated directly at 60°C (60°C direct). The different fractions (equivalent to 0.3 OD600 unit) were analyzed by SDS–12% PAGE and by immunoblotting with antibodies directed against TolB and PAL. The positions of the TolB and PAL proteins, of the TolB-PAL complex, and of the molecular mass markers are indicated.
DISCUSSION
In this study, we describe a new assay that allowed us to study the binding of PAL to the peptidoglycan in vitro.
We showed that the peptide composed of residues 89 to 130 of PAL competed with a full-length PAL for interaction with the peptidoglycan. To obtain a competition effect, we had to add approximately 20 times as much of the peptide as of the full-length protein. However, we do not know the nature of the sites of interaction in the peptidoglycan, and therefore we do not know the number of these sites. However, since all of the PAL protein binds to the peptidoglycan, it is highly probable that the number of sites in the peptidoglycan is not a limiting factor. Therefore, a 20-fold excess of peptide to obtain a competition suggests that the peptide interaction is efficient and that the peptide is correctly folded. Previous studies have identified mutations that impaired the interaction of PAL with the peptidoglycan (7). Most of them are in the region from residues 89 to 130, but residue substitutions outside this region (positions Gly86, Ala88, and Arg146) were also shown to affect peptidoglycan binding in vivo (7). This suggests that these residues, even if not directly participating in the peptidoglycan interaction, can affect it by changing the conformation of PAL.
The use of purified TolB and PAL proteins provided us a new way of detecting the interaction between these two proteins. Here again, the PAL peptide corresponding to residues 89 to 130 was able to interact with TolB, therefore competing with the full-length lipoprotein for binding to TolB. To obtain a competition effect, we had to add approximately 40 times as much of the peptide as of the full-length protein. Since only a small fraction of TolB and PAL proteins are cross-linked (in vitro as well as in vivo), it is not easy to come to a conclusion regarding these relative amounts. However, we can conclude that at least a fraction of the peptide interacted with TolB. One could wonder why the TolB-peptide complex was not detected by anti-TolB antibodies (cf. Fig. 3B). First, the difference in size between TolB and the complex might be too small to be detected. Second, it is possible that the peptide interacts with TolB without being able to be cross-linked by the formaldehyde but is still sufficient to prevent the full-length PAL protein from binding.
Therefore, we concluded that the PAL peptide comprising residues 89 to 130 was able to interact with TolB and with the peptidoglycan. This, added to the fact that PAL does not bind to TolB and to the peptidoglycan at the same time in vivo, suggests that it is exactly the same region which binds to these two partners.
The two forms of PAL (bound to TolB or bound to the peptidoglycan), as well as the two forms of TolB (free and bound to PAL), must have a physiological significance.
One may speculate, for example, that these different associations play a role in the crossing of the murein sacculus by the colicins. PAL proteins bound to TolB, and not to the peptidoglycan, may define special points in the envelope where the crossing of the peptidoglycan is possible. The colicin could then reach the inner membrane TolA protein, with which they have been shown to interact (2, 6). This hypothesis is consistent with the fact that the TolB-PAL system may constitute contact sites between the inner and outer membranes, allowing the translocation of colicins (16). Although tol mutants are tolerant of colicin action, a pal null mutant is still sensitive to these toxins. However, several studies have demonstrated an indirect involvement of PAL in the translocation of colicins. First, it has been shown that the binding of colicin E3 to TolB prevents the interaction of TolB with PAL (5). Second, point mutations in PAL have been obtained which rendered cells tolerant to colicins (7), perhaps by affecting the step in which TolB is released from the TolB-PAL interaction, when the colicin interacts with TolB.
From another point of view, the shuttling of a protein from a periplasmic and soluble state to a lipoprotein binding state is reminiscent of the shuttling of LolA from a soluble and periplasmic state to a lipoprotein LolB binding state during the transport of lipoproteins to the outer membrane (27). It is therefore very tempting to postulate a role for the Tol-PAL system in transport, including a function of crossing the peptidoglycan where TolB would be the carrier, shuttling between PAL in the outer membrane and the TolQRA proteins in the inner membrane.
Finally, it is still possible that the PAL and TolB proteins also play a role independent of the whole Tol system. Indeed, the C-terminal domain of PAL is present in three other proteins in E. coli, the most important being OmpA (19). It is striking that the periplasmic domain of OmpA is so homologous to that of PAL. It seems that the same protein domain can be anchored to the outer membrane either by a lipid tail or by a protein domain, as is the case for OmpA, whose membrane anchor structure has been determined and shown to be a β-barrel of eight strands (30). One can speculate that the associations of PAL and OmpA with the peptidoglycan are to a certain extent redundant. Furthermore, the finding that TolB, PAL, Lpp, and OmpA may be parts of the same complex points to a role for these proteins in the association of the outer membrane with the underlying peptidoglycan (7).
The new assay system presented in this paper, relying on direct protein-protein or protein-peptidoglycan interactions, will be useful for investigating protein domains and their interactions in more detail. The use of smaller peptides will permit precise determination of the minimal region involved in protein-peptidoglycan binding. In addition, the assay can be used to test the effect of mutations in PAL on its binding behavior. This implies the prior purification of the mutated proteins. However, it seems the most interesting development of our in vitro system, since the study of mutations in PAL in vivo appears to be difficult. Finally, this new assay might also be useful in characterizing the interactions of other membrane components, like OmpA and MotB, with the peptidoglycan.
ACKNOWLEDGMENTS
We thank Jacques Bonicel, who performed the mass spectrometry analyses. We are grateful to Laure Journet for the gift of purified TolRIIHis. We thank Harold Cremer and Pascal Lopez for carefully reading the manuscript.
E. Bouveret was a recipient of an AMN fellowship. This work was supported by the Life Science department and the Mission Physique et Chimie du Vivant of the CNRS, by the INSERM Programme Environnement Santé 96 (EN96C3), and the European Community (BIO4-CT97-2313).
REFERENCES
- 1.Barany G, Merrifield R B. In: The peptide: analysis, synthesis, biology. Gross E, Meinhofer J, editors. Vol. 2. New York, N.Y: Academic Press; 1980. pp. 1–284. [Google Scholar]
- 2.Bénédetti H, Lazdunski C, Lloubès R. Protein import into Escherichia coli: colicins A and E1 interact with a component of their translocation system. EMBO J. 1991;10:1989–1995. doi: 10.1002/j.1460-2075.1991.tb07728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernadac A, Gavioli M, Lazzaroni J C, Raina S, Lloubès R. Escherichia coli mutants form outer membrane vesicles. J Bacteriol. 1998;180:4872–4878. doi: 10.1128/jb.180.18.4872-4878.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouveret E, Dérouiche R, Rigal A, Lloubès R, Lazdunski C, Bénédetti H. Peptidoglycan-associated lipoprotein-TolB interaction. J Biol Chem. 1995;270:11071–11077. doi: 10.1074/jbc.270.19.11071. [DOI] [PubMed] [Google Scholar]
- 5.Bouveret E, Rigal A, Lazdunski C, Bénédetti H. The N-terminal domain of colicin E3 interacts with TolB which is involved in the colicin translocation step. Mol Microbiol. 1997;23:909–920. doi: 10.1046/j.1365-2958.1997.2751640.x. [DOI] [PubMed] [Google Scholar]
- 6.Bouveret E, Rigal A, Lazdunski C, Bénédetti H. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol Microbiol. 1998;27:143–157. doi: 10.1046/j.1365-2958.1998.00667.x. [DOI] [PubMed] [Google Scholar]
- 7.Clavel T, Germon P, Vianney A, Portalier R, Lazzaroni J C. TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol Microbiol. 1998;29:359–367. doi: 10.1046/j.1365-2958.1998.00945.x. [DOI] [PubMed] [Google Scholar]
- 8.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in Gram-positive and Gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 9.Dennis J J, Lafontaine E R, Sokol P A. Identification and characterization of the tolQRA genes of Pseudomonas aeruginosa. J Bacteriol. 1996;178:7059–7068. doi: 10.1128/jb.178.24.7059-7068.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dérouiche R, Bénédetti H, Lazzaroni J C, Lazdunski C, Lloubès R. Protein complex within Escherichia coli inner membrane: TolA N-terminal domain interacts with TolQ and TolR proteins. J Biol Chem. 1995;270:11078–11084. doi: 10.1074/jbc.270.19.11078. [DOI] [PubMed] [Google Scholar]
- 11.Dérouiche R, Gavioli M, Bénédetti H, Prilipov A, Lazdunski C, Lloubès R. TolA central domain interacts with Escherichia coli porins. EMBO J. 1996;15:6408–6415. [PMC free article] [PubMed] [Google Scholar]
- 12.Frey J, Kuhnert P, Villiger L, Nicolet J. Cloning and characterization of an Actinobacillus pleuropneumoniae outer membrane protein belonging to the family of PAL lipoproteins. Res Microbiol. 1996;147:351–361. doi: 10.1016/0923-2508(96)84710-3. [DOI] [PubMed] [Google Scholar]
- 13.Fung J, MacAlister T J, Rothfield L I. Role of murein lipoprotein in morphogenesis of the bacterial division septum: phenotypic similarity of lkyD and lpo mutants. J Bacteriol. 1978;133:1467–1471. doi: 10.1128/jb.133.3.1467-1471.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germon P, Clavel T, Vianney A, Portalier R, Lazzaroni J C. Mutational analysis of the Escherichia coli K-12 TolA N-terminal region and characterization of its TolQ-interacting domain by genetic suppression. J Bacteriol. 1998;180:6433–6439. doi: 10.1128/jb.180.24.6433-6439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorvel J-P, Rigal A, Sarles J, Maroux S. Aminopeptidase N and human blood group A antigenicity along the digestive tract and associated glands in the rabbit. Cell Tissue Res. 1985;239:241–245. doi: 10.1007/BF00214925. [DOI] [PubMed] [Google Scholar]
- 16.Guihard G, Boulanger P, Bénédetti H, Lloubès R, Besnard M, Letellier L. Colicin A and the Tol proteins involved in its translocation are preferentially located in the contact sites between the inner and outer membranes of Escherichia coli cells. J Biol Chem. 1994;269:5874–5880. [PubMed] [Google Scholar]
- 17.Isnard M, Rigal A, Lazzaroni J C, Lazdunski C, Lloubès R. Maturation and localization of the TolB protein required for colicin import. J Bacteriol. 1994;176:6392–6396. doi: 10.1128/jb.176.20.6392-6396.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Journet L, Rigal A, Lazdunski C, Bénédetti H. Role of TolR N-terminal, central, and C-terminal domains in dimerization and interaction with TolA and TolQ. J Bacteriol. 1999;181:4476–4484. doi: 10.1128/jb.181.15.4476-4484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koebnik R. Proposal for peptidoglycan-associating alpha-helical motif in the C-terminal regions of some bacterial cell-surface proteins. Mol Microbiol. 1995;16:1269–1270. doi: 10.1111/j.1365-2958.1995.tb02348.x. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lazdunski C, Bouveret E, Rigal A, Journet L, Lloubès R, Bénédetti H. Colicin import into Escherichia coli cells. J Bacteriol. 1998;180:4993–5002. doi: 10.1128/jb.180.19.4993-5002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazzaroni J C, Portalier R. Genetic and biochemical characterization of periplasmic-leaky mutants of Escherichia coli K-12. J Bacteriol. 1981;145:1351–1358. doi: 10.1128/jb.145.3.1351-1358.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazzaroni J C, Portalier R. The excC gene of Escherichia coli K12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL) Mol Microbiol. 1992;6:735–742. doi: 10.1111/j.1365-2958.1992.tb01523.x. [DOI] [PubMed] [Google Scholar]
- 24.Lazzaroni J C, Vianney A, Popot J L, Bénédetti H, Samatey F, Lazdunski C, Portalier R, Géli V. Transmembrane α-helix interactions are required for the functional assembly of the Escherichia coli Tol complex. J Mol Biol. 1995;246:1–7. doi: 10.1006/jmbi.1994.0058. [DOI] [PubMed] [Google Scholar]
- 25.Leduc M, Joseleau-Petit D, Rothfield L. Interactions of membrane lipoproteins with the murein sacculus of Escherichia coli as shown by chemical cross-linking studies of intact cells. FEMS Microbiol Lett. 1989;60:11–14. doi: 10.1016/0378-1097(89)90068-2. [DOI] [PubMed] [Google Scholar]
- 26.Leduc M, Ishidate K, Shakibai N, Rothfield L. Interactions of Escherichia coli membrane lipoproteins with the murein sacculus. J Bacteriol. 1992;174:7982–7988. doi: 10.1128/jb.174.24.7982-7988.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuyama S, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuno T. A novel peptidoglycan-associated lipoprotein found in the cell envelope of Pseudomonas aeruginosa and Escherichia coli. J Biochem. 1979;86:991–1000. doi: 10.1093/oxfordjournals.jbchem.a132631. [DOI] [PubMed] [Google Scholar]
- 29.Mizuno T. A novel peptidoglycan-associated lipoprotein (Pal) of the Proteus mirabilis outer membrane: characterization of the peptidoglycan associated region of Pal. J Biochem. 1981;91:19–24. doi: 10.1093/oxfordjournals.jbchem.a133675. [DOI] [PubMed] [Google Scholar]
- 30.Pautsch A, Schulz G. Structure of the outer membrane protein A transmembrane domain. Nat Struct Biol. 1998;5:1013–1017. doi: 10.1038/2983. [DOI] [PubMed] [Google Scholar]
- 31.Rigal A, Bouveret E, Lloubès R, Lazdunski C, Bénédetti H. The TolB protein interacts with the porins of Escherichia coli. J Bacteriol. 1997;179:7274–7279. doi: 10.1128/jb.179.23.7274-7279.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Herva J J, Ramos-Gonzalez M I, Ramos J L. The Pseudomonas putida peptidoglycan-associated outer membrane lipoprotein is involved in maintenance of the integrity of the cell envelope. J Bacteriol. 1996;178:1699–1706. doi: 10.1128/jb.178.6.1699-1706.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sen K, Sikkema D J, Murphy T F. Isolation and characterization of the Haemophilus influenzae tolQ, tolR, tolA and tolB genes. Gene. 1996;178:75–81. doi: 10.1016/0378-1119(96)00338-1. [DOI] [PubMed] [Google Scholar]
- 34.Sonntag I, Schwartz H, Hirota Y, Henning U. Cell envelope and cell shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136:280–285. doi: 10.1128/jb.136.1.280-285.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vianney A, Muller M, Clavel J, Lazzaroni J C, Portalier R, Webster R E. Characterization of the tol-pal region of E. coli K-12: translational control of tolR expression by TolQ and identification of a new open reading frame downstream of pal encoding a periplasmic protein. J Bacteriol. 1996;178:4031–4038. doi: 10.1128/jb.178.14.4031-4038.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zlotnick G W, Sanfilippo V T, Mattler J A, Kirkley D H, Boykins R A, Seid R C., Jr Purification and characterization of a peptidoglycan-associated lipoprotein from Haemophilus influenzae. J Biol Chem. 1988;263:9790–9794. [PubMed] [Google Scholar]