Abstract
Transposition is a DNA reorganization reaction potentially deleterious for the host. The frequency of transposition is limited by the amount of transposase. Therefore, strict regulation of a transposase is required to keep control over the destructive multiplication of the mobile element. We have shown previously that the expression of the transposase (tnpA) of the Pseudomonas putida PaW85 transposon Tn4652 is positively affected by integration host factor. Here, we present evidence that the amount of the transposase of Tn4652 in P. putida cells is controlled by the transposon-encoded protein (TnpC). Sequence analysis of the 120-amino-acid-long TnpC, coded just downstream of the tnpA gene, showed that it has remarkable similarity to the putative polypeptide encoded by the mercury resistance transposon Tn5041. As determined by quantitative Western blot analysis, the abundance of TnpA was reduced up to 10-fold in the intact tnpC background. In vivo experiments using transcriptional and translational fusions of the tnpA gene and the reporter gene gusA indicated that TnpC operates in the regulation of the transposase of Tn4652 at the post-transcriptional level.
Transposition is a DNA rearrangement process in which a discrete DNA sequence is inserted into a new location in the genome. This reaction is performed by an element-encoded protein called transposase. Mobility of bacterial transposons is strictly regulated to a very low level (10−3 to 10−8 reactions per element per generation [18]) to maintain the balance between their propagation and the potential destructive mutagenic effect on their hosts. The rate of transposition is largely determined by the amount of active transposase. Many of the mechanisms that limit transposase gene expression or transposase protein activity have been described (reviewed in reference 18). These downregulation mechanisms frequently operate coordinately at different levels of transposase expression and help maintain precise control over the amount and activity of transposase in bacteria.
Most of the transposase promoters are weak and often downregulated by transcriptional repressors that may be both transposon-encoded proteins (8, 19, 22) and host factors (13, 21). DNA methylation is also shown to modulate transposase expression in some cases. IS10, IS50, and IS903 carry GATC methylation sites in their transposase promoter regions, and absence of methylation results in increased activity of these promoters (28, 36).
For many transposons, the level of transposase expression is determined by the efficiency of transposase gene translation. Inefficient translation, inhibition of translation by antisense RNA, and programmed translational frameshifting have been described as post-transcriptional mechanisms to regulate transposase expression (7, 9, 31). For example, translation of mRNAs of the transposases of IS10 and IS30 is inhibited by antisense RNAs (2, 31). For synthesis of full-length transposase of several insertion elements, programmed translational frameshifting between the two sequential open reading frames (ORFs) is needed (reviewed in reference 7). Also, transposase stability may be related to control of transposition activity. For instance, IS903 transposase is demonstrated to be sensitive to the Escherichia coli Lon protease (9).
Transposition of several transposons is controlled by regulation of transposase catalytic activity. IS1 and Tn5 modulate transposase catalytic activity with inhibitor proteins coded from the same ORF as the transposase (20, 22). Additionally, many transposases are known to require bacterial host proteins for their activity. Integration host factor (IHF), which is known to alter the conformation of DNA, is the host factor most usually involved in transposition (1, 30, 35). Recently, activity of the transposase of Tn3 was demonstrated to be stimulated by a quite different type of host factor, acyl carrier protein (23).
Pseudomonas putida PaW85 carries transposon Tn4652 in its chromosome. Tn4652 is a 17-kb-long deletion derivative of the toluene degradation xyl gene-carrying transposon Tn4652. Tsuda and Iino (33) have shown that, according to its transposition properties, Tn4652 belongs to the Tn3 family of transposons. We have sequenced the transposase gene tnpA of Tn4652 and shown that transcription from the tnpA promoter is positively affected by IHF (12).
In this study, we demonstrate that the amount of Tn4652 transposase (TnpA) is downregulated by the Tn4652-encoded protein TnpC. The ORF encoding the 120-amino-acid protein TnpC begins just downstream of tnpA and exhibits striking similarity to an ORF of Tn5041 encoding a putative 120-amino-acid-long polypeptide. In vivo experiments using transcriptional and translational fusions of the tnpA gene and the reporter gene gusA indicate that TnpC interferes with the regulation of TnpA at the post-transcriptional level.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli TG1 (6) was used for the DNA cloning procedures. Bacteria were grown on Luria-Bertani medium (24). Antibiotics were added, with final concentrations as follows: ampicillin, 100 μg/ml for E. coli; carbenicillin, 1,500 μg/ml for P. putida. P. putida was incubated at 30°C. Early-stationary-phase cultures were used for enzyme assays. E. coli was transformed with plasmid DNA as described by Hanahan (11). P. putida was electrotransformed according to the protocol described by Sharma and Schimke (29).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or construction | Source or reference |
|---|---|---|
| E. coli | ||
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′ (traD36 proAB+ lacIqlacZΔM15) | 6 |
| BL21(DE3) | hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) | 32 |
| P. putida | ||
| PaW85 | Tn4652 | 4 |
| PRS2000 | Tn4652 free | 34 |
| Plasmidsa | ||
| pBluescript KS | Cloning vector (Apr) | Stratagene |
| pET19b | Protein expression vector (Apr) | Stratagene |
| pET19-tnpA | tnpA is fused with histidine tag in pET19b | This work |
| pKT240 | Cloning vector (Apr Kmr) | 3 |
| pEST1354 | Plasmid containing Tn4652 upstream of the pheBA operon | 17 |
| pKTtnpA(D/H) | Tn4652 tnpA gene within the 3.2-kb DraI-HindIII fragment cloned into pKT240 | This work (Fig. 3) |
| pKTtnpA(D/P) | Tn4652 tnpA and tnpC genes within the 3.7-kb DraI-PvuI fragment cloned into pKT240 | This work (Fig. 3) |
| pKTtnpA(D/P)* | pKTtnpA(D/P) with the tnpC gene disrupted by frameshift | This work |
| pKTGC/tnpA | pKT240 plus pDEL2-GC promoter region plus the promoterless tnpA gene | This work |
| pKTGC/tnpAC | pKT240 plus pDEL2-GC promoter region plus the promoterless tnpA gene with tnpC | This work |
| pGUS102 | Promoter probe vector containing the gusA gene cloned as a 1.8-kb EcoRI fragment into pBR322 | A. Eriksson |
| pKTGUS | Vector for translational fusions containing the gusA gene without translation initiation codon ATG in pKT240; HindIII restriction site designed at the 5′ end of the gusA is suitable for in-frame cloning | This work |
| pTr1 | PDEL2-GC promoter region plus 42 nt of the coding region of tnpA fused with gusA in pKTGUS | This work (Fig. 5B) |
| pTr2 | PDEL2-GC promoter region plus 546 nt of the coding region of tnpA fused with gusA in pKTGUS | This work (Fig. 5B) |
| pTr3 | PDEL2-GC promoter region plus 1,166 nt of the coding region of tnpA fused with gusA in pKTGUS | This work (Fig. 5B) |
| pKT-ACG | gusA is cloned downstream of tnpC in pKTtnpA(D/P) | This work (Fig. 6B) |
| pKT-AdelCG | ClaI-Cfr10I deletion derivative of pKT-ACG | This work (Fig. 6B) |
| pKT-a1CG | DraI-ClaI deletion derivative of pKT-ACG | This work (Fig. 6B) |
| pKT-a2CG | DraI-NruI deletion derivative of pKT-ACG | This work (Fig. 6B) |
| pKT-CG | DraI-Cfr10I deletion derivative of pKT-ACG | This work (Fig. 6B) |
Oligonucleotides used for construction of the plasmids are described in Materials and Methods.
DNA manipulations.
DNA sequencing was performed with the Sequenase version 2.0 DNA sequencing kit (Amersham). For cloning of the tnpA gene into pET19b, the XbaI and NdeI restriction sites were designed in the 5′ end of tnpA by using oligonucleotide pETtnpA (5′-CCTCTAGA[XbaI]CATATG[NdeI]TGTTCAATGGCATCGGTGG-3′). For amplification and cloning of the PDEL2-GC promoter from plasmid pEST1414 (15), oligonucleotides YrgHind (5′-CCAAAGCTT[HindIII]TGTTTACGATCCAGGC-3′) and AB (5′-GTATGCTTGGCAGTCGT) were used. The ClaI restriction site just flanking the −10 hexamer of the PDEL2-GC promoter was suitable for cloning of tnpA-gusA translational fusions. To design the ClaI restriction site in the 5′ end of the tnpA, oligonucleotide GCtnpA (5′-CTAATCGAT[ClaI]TTTGCCTCGCTTGGGGGAT-3′) was used. For construction of vector pKTGUS for translational fusions, oligonucleotides Gus1 (5′-CTAAAGCTT[HindIII]ACGTCCTGTAGAAACCCCAA-3′) and Gus2 (5′-ACTGATCGTTAAAACTGCCTGG-3′) were used. For construction of translational fusions of tnpA with the reporter gene gusA, oligonucleotide GCtnpA and either oligonucleotide Tr1 (5′-GGTAAGCTT[HindIII]CTGGGCAAGATAGGGTAGGCT-3′), Tr2 (5′-ACCAAGCTT[HindIII]GGCGCTCGAGTCACGACTA), or Tr3 (5′-GAGAAGCTT[HindIII]TCCCGAATCAGGCTGCCAG) were used. For construction of the plasmids pTr1, pTr2, and pTr3 with the tnpC gene, the tnpC under the control of the benzoate-inducible Pi promoter of the pheBA operon (14) was cloned downstream of the tnpA-gusA translational fusions. The tnpC expression cassette was initially designed in pBluescript and was subsequently cloned into plasmids pTr1, pTr2, and pTr3. Inducible expression of the tnpC gene under control of the Pi promoter was tested in plasmid pKTtnpA(D/H) by the ability of TnpC to downregulate TnpA.
For cloning of gusA downstream of tnpC in transcription fusion tnpAC-gusA in plasmid pKT-ACG, an EcoRI restriction site was designed in the 3′ end of tnpC by using oligonucleotide TnpCEco (5′-CCAGAATTC[EcoRI]CCAAGTGCTTACTGTTCGTG-3′).
Overexpression and purification of His-TnpA.
To obtain soluble His-TnpA, E. coli BL21(DE3)(pET19-tnpA) was grown at 22°C in 200 ml of Luria-Bertani medium. Expression of His-TnpA was induced for 3.0 h by adding isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 0.4 mM) when the culture optical density at 590 nanometers reached about 1.0. Cells were pelleted and sonicated in buffer A (100 mM Tris-HCl [pH 7.5], 0.25 mM EDTA, 5 mM β-mercaptoethanol, 1 M NaCl, 0.1% Triton X-100, 10% glycerol). The cell lysate was centrifuged at 15,000 × g for 20 min. Imidazole (100 mM) was added to the supernatant before it was loaded into the Ni2+-iminodiacetic acid-activated chelating Sepharose 6B column previously equilibrated with buffer A. The column was washed with 8 volumes of buffer A supplemented with 100 mM imidazole (pH 6.5). Purified His-TnpA was eluted with buffer A containing 500 mM imidazole. Imidazole and excess salt were removed by dialyzing the eluate against buffer B (75 mM Tris-HCl [pH 7.5], 0.2 mM EDTA, 5 mM β-mercaptoethanol, 200 mM NaCl, 0.1% Triton X-100, 10% glycerol), and the purified protein was stored at −75°C.
Preparation of cell lysates and immunoblotting of TnpA.
Cell lysates were prepared from 30-ml early-stationary-phase cultures. Cells were pelleted and sonicated in 500 μl of 0.5× buffer B. Protein concentration in cleared lysates was estimated as described by Bradford (5). Equal amounts of total protein (40 μg) were used for a Western immunoblotting assay. Proteins were separated by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (BA 85; Schleicher & Schuell). For Western blotting, the membranes were probed with mouse anti-TnpA polyclonal serum diluted 1:5,000, followed by alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (LabAS Ltd., Tartu, Estonia) diluted 1:5,000. The blots were developed with bromochloroindolyl phosphate and nitroblue tetrazolium.
Enzyme assays.
β-Glucuronidase (GUS) activity was assayed by using p-nitrophenyl β-d-glucuronide as the substrate (26). The degradation product of p-nitrophenyl β-d-glucuronide, p-nitrophenol, was detected at 405 nm and GUS-specific activities were measured in nanomoles of p-nitrophenol per minute per optical density unit of cell culture at 590 nm.
Nucleotide sequence accession numbers.
The nucleotide sequences of tnpA and tnpC have been deposited in the EMBL database under the accession no. X83686.
RESULTS
Overexpression and purification of the transposase of Tn4652.
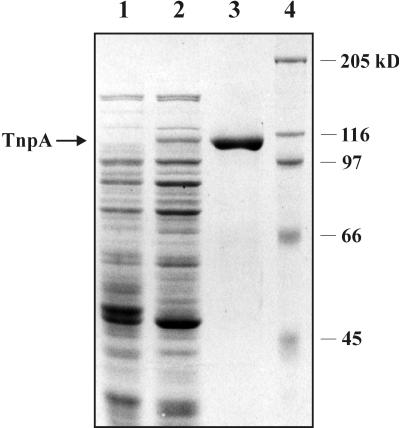
To investigate the regulation of the Tn4652-encoded transposase TnpA, the transposase protein was overexpressed and purified to obtain antibodies against it. Coding sequence of the tnpA gene was fused with N-terminal histidine tag in the protein expression vector pET19b. The His-tagged TnpA was overexpressed in E. coli BL21(DE3) and purified by single-step Ni2+-chelate affinity chromatography. Purification yielded near-homogeneous TnpA protein (Fig. 1, lane 3). The molecular mass of TnpA was estimated to be approximately 114 kDa, which is consistent with the predicted molecular mass of 114.3 kDa suggested by the results of the tnpA gene sequence analysis (12).
FIG. 1.
Sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis demonstrating overexpression and purification of His-tagged TnpA in E. coli BL21(DE3). Lane 1, crude extract from E. coli BL21(DE3)(pET19-tnpA); lane 2, as described for lane 1, but induced with 0.4 mM IPTG; lane 3, purified His-TnpA; lane 4, standard molecular weight markers.
Amount of TnpA is downregulated by the Tn4652-encoded factor.
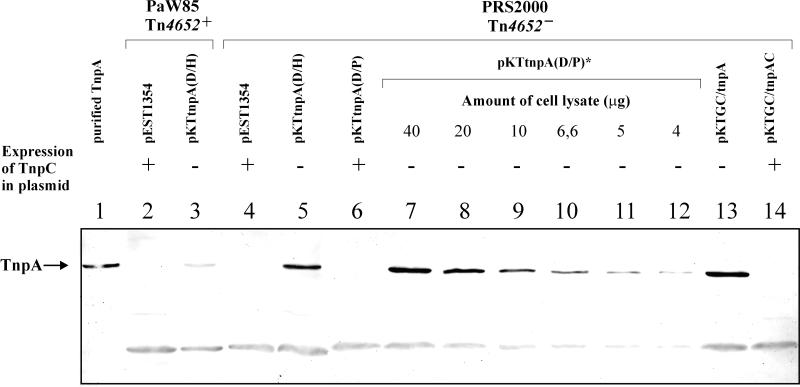
To observe TnpA expression in different genetic backgrounds, we used Western blot analysis with anti-TnpA polyclonal antiserum. We could not detect TnpA in the cell lysate of P. putida PaW85 that carries Tn4652 in its chromosome (data not shown). Similarly, TnpA was not detectable in the cell lysates of P. putida PaW85 and PRS2000 (free of Tn4652) which harbored Tn4652-containing plasmid pEST1354 (17) (Fig. 2, lanes 2 and 4). In order to test whether TnpA expression is downregulated either by some P. putida host factor or by a Tn4652-encoded factor, we generated a subclone of this transposon. The tnpA gene with its native promoter was cloned into the broad-host-range vector plasmid pKT240 to obtain the plasmid pKTtnpA(D/H). This plasmid contained the 3.2-kb fragment of the right arm of Tn4652 from the distal DraI restriction site up to the HindIII site (Fig. 3 and Table 1). Western blot analysis of crude lysates prepared from the cells of P. putida PaW85 and PRS2000 harboring the plasmid pKTtnpA(D/H) allowed detection of the TnpA protein (Fig. 2, lanes 3 and 5). This result pointed to a transposon-encoded regulator of transposase located outside of the DraI-HindIII restriction fragment of Tn4652.
FIG. 2.
Western immunoblot analyses of P. putida PaW85 and PRS2000 cell lysates by using anti-TnpA polyclonal antibodies. Lane 1, purified TnpA protein; lane 2, crude extract from P. putida PaW85(pEST1354); lane 3, crude extract from P. putida PaW85[pKTtnpA(D/H)]; lane 4, crude extract from P. putida PRS2000(pEST1354); lane 5, crude extract from P. putida PRS2000[pKTtnpA(D/H)]; lane 6, crude extract from P. putida PRS2000[pKTtnpA(D/P)]; lanes 7 through 12, gradual dilutions of crude extracts of P. putida PRS2000[pKTtnpA(D/P)*]; lane 13, crude extract from P. putida PRS2000(pKTGC/tnpA); lane 14, crude extract from P. putida PRS2000(pKTGC/tnpAC). The amount of crude lysate was 40 μg per lane except that for lanes 8 to 12, gradual dilutions of cell lysate of P. putida PRS2000[pKTtnpA(D/P)*] were used.
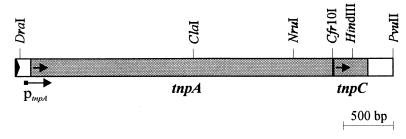
FIG. 3.
Genetic organization of tnpA and tnpC in the right arm of the Tn4652. Right inverted repeat of Tn4652 is marked by a black triangle. Restriction sites relevant to this study are indicated. The arrows indicate the direction of transcription of the tnpA and tnpC genes. The promoter of the tnpA gene is designated ptnpA.
Localization and sequencing of the DNA region of Tn4652 influencing the expression of TnpA.
In order to localize the DNA region that controls the accumulation of TnpA, deletion analysis of Tn4652 was performed. The amount of TnpA was tested in lysates of Tn4652-free P. putida PRS2000 cells carrying plasmids which contained the tnpA gene linked to different regions of Tn4652. Plasmid pKTtnpA(D/P) carried the right arm of DNA of Tn4652 (including also the tnpA gene) from the distal DraI restriction site up to the PvuII site (Fig. 3 and Table 1). Results of the Western blot analysis presented in Fig. 2 show that TnpA was detectable in the cell lysates of bacteria harboring the plasmid pKTtnpA(D/H) (Fig. 2, lane 5), but not in cell lysates of bacteria harboring the plasmid pKTtnpA(D/P) (Fig. 2, lane 6). According to these results, the putative regulator of TnpA was localized just downstream of the tnpA gene, in the DNA region extending to the PvuII site.
Sequence analysis of the DNA region downstream of the tnpA gene revealed a 360-nucleotide (nt)-long ORF starting 8 nt apart from the stop codon of the tnpA (Fig. 3). The predicted protein encoded by this ORF is 120 amino acids long, with a calculated molecular mass of 13.0 kDa. Comparison of the deduced amino acid sequence of the putative regulator (TnpC) of the transposase of Tn4652 with the translated sequences of genes in the EMBL database with the BLAST program revealed homology of TnpC to the putative 120-amino-acid-long polypeptide encoded by the mercury resistance transposon Tn5041 (Fig. 4). Amino acid sequence identity of 52% and similarity of 75% were demonstrated.
FIG. 4.
Alignment of the deduced amino acid sequence of TnpC of Tn4652 with the putative 120-amino-acid-long polypeptide encoded by Tn5041 (16). Identical amino acids are indicated between the two aligned sequences in boldface. Similar amino acids are marked by plus signs.
Intact ORF of tnpC is needed for the downregulation of TnpA.
In order to test whether the TnpC protein indeed acts on the expression of the tnpA gene product, we disrupted the ORF of TnpC in the plasmid pKTtnpA(D/P). The unique HindIII restriction site in tnpC was used to generate a +1 frameshift into the coding sequence of the tnpC gene [plasmid pKTtnpA(D/P)*]. Western blot analysis of the crude lysates prepared from the cells of P. putida PRS2000 harboring either pKTtnpA(D/P) or pKTtnpA(D/P)* demonstrated that in-frame tnpC was needed to decrease the amount of TnpA (Fig. 2, compare lane 6 to lane 7).
However, downregulation of TnpA by TnpC was not complete. We could also detect a small amount of TnpA in the cell lysates of P. putida PRS2000 while intact tnpC was present (not visible in Fig. 2, but seen in overdeveloped filters). To quantify the extent of downregulation of TnpA by TnpC, gradual dilutions of cell lysates of P. putida PRS2000[pKTtnpA(D/P)*] were tested on a Western blot and compared to the amount of TnpA detected in cells containing pKTtnpA(D/P). Four independent measurements with different preparations of cell lysates indicated that the presence of TnpC decreased the abundance of TnpA about 10-fold (Fig. 2, compare lane 6 to lanes 7 through 12).
Testing the effect of TnpC on transcriptional and translational initiation of tnpA.
Quantification of the tnpA-specific mRNA in both tnpC-expressing and tnpC-deficient backgrounds could answer the question of whether TnpC would affect the expression of the tnpA gene product at the transcriptional or at the post-transcriptional level. Since we failed to detect tnpA-specific mRNA in both primer extension and Northern blot analyses, alternative approaches were used to solve this problem. In order to test whether TnpC represses transcription initiation from the tnpA promoter, we replaced the native promoter of the tnpA gene with the constitutive promoter PDEL2-GC described by members of our group previously (15). The promoter of the tnpA gene was earlier localized into the terminal 122-nt DNA region of the right end of Tn4652, and the transcription starting point of the tnpA gene was mapped at 129 nt from the end of this transposon (12). The DNA fragments lacking the terminal 125 nt from the right end of Tn4652 and containing either gene tnpA or tnpAC were fused with the PDEL-GC promoter. The fusions were designed without altering the 5′ end of the tnpA-specific mRNA (Table 1 and Materials and Methods). Obtained plasmids pKTGC/tnpA and pKTGC/tnpAC were introduced into P. putida PRS2000, and Western blot analysis of the cell lysates was performed. Data presented in Fig. 2, lanes 13 and 14, demonstrated that although the promoter of the tnpA gene was replaced with another one, the expression of TnpA was still downregulated by TnpC.
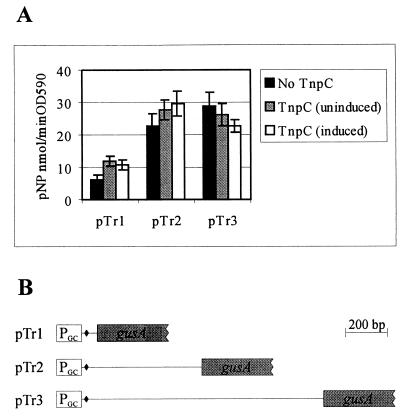
To investigate whether TnpC affects the expression of the tnpA gene at the level of initiation of transcription or translation, we constructed different translational fusions of the 5′ end of the tnpA gene (up to one-third of the gene) with the reporter gene gusA (encodes GUS) (Fig. 5B). Plasmids pTr1, pTr2, and pTr3 contained 42, 546, and 1,143 nt of the coding region of the tnpA gene, respectively, fused with the gusA gene. The control plasmid for translational fusions was designed by substituting transposase start codons (there are two potential ATG start codons of tnpA separated by 6 nt) for ATC in translation fusion plasmid pTr3. The obtained plasmid was introduced into P. putida PRS2000, but no GUS activity was detectable in an enzyme assay using this strain. Thus, this control experiment confirms that translation of the tnpA and gusA fusion starts from the ATG of the tnpA gene. All translational fusions were expressed under the PDEL2-GC promoter (Fig. 5B; Table 1). The tnpC gene, if present, was expressed in the same plasmids under the control of the benzoate-inducible Pi promoter of the pheBA operon (14) (see Materials and Methods). No negative effect of TnpC on GUS activity was observed when expression of these translational fusions was tested in P. putida PRS2000 cells in either the presence or absence of benzoate (Fig. 5A). On the basis of these experiments and considering the results of the promoter change experiment described above, we suggest that TnpC could influence the accumulation of TnpA after either the transcriptional or translational initiation of the tnpA gene.
FIG. 5.
(A) GUS activities measured in P. putida PRS2000 carrying different translational fusion plasmids either together with the tnpC gene or without the tnpC gene. Na-benzoate (10 mM) was used for the induction of tnpC. Data (means ± standard deviations) of at least five independent experiments are presented. (B) Schematic presentation of the translational fusions of the 5′ end of the tnpA gene with the reporter gene gusA. For each fusion, the PDEL2-GC promoter is indicated by an open box, the 5′ region of tnpA is marked by a line, and the translation initiation codon ATG of tnpA is indicated by a black diamond.
Localization of the tnpC promoter region.
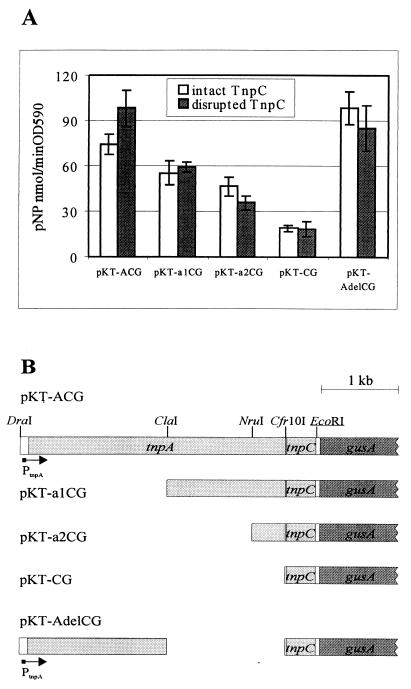
The tnpC gene lies just downstream of the transposase gene tnpA. Thus, the transcription of tnpC could be initiated from its own promoter(s), or it could be cotranscribed with the tnpA gene from the tnpA promoter. In order to measure transcription of the tnpC gene, we constructed the plasmid pKT-ACG that contained the native tnpAC gene cassette and the reporter gene gusA just downstream of the tnpC gene (Fig. 6B; Table 1; Materials and Methods). Additionally, deletion derivatives of pKT-ACG lacking different amounts of the sequence from the 5′ end of tnpA were generated (pKT-a1CG, pKT-a2CG, and pKT-CG) (Fig. 6B). GUS activity in the cells of P. putida PRS2000 harboring these plasmids was estimated (Fig. 6A, white bars). The highest level of GUS activity was detected in P. putida PRS2000 cells carrying the plasmid pKT-ACG (contains the full-length tnpA gene together with its promoter upstream from tnpC). Plasmids pKT-a1CG and pKT-a2CG with deletions from the 5′ sequences of the tnpA gene revealed levels of GUS activity 65 to 75% of that measured in cells carrying pKT-ACG (Fig. 6A). Bacteria containing plasmid pKT-CG (lacks tnpA but harbors all of tnpC), used as a control; showed significantly lower levels of GUS activity. Thus, the estimated GUS activity in our test system represents the sum of the function of the tnpA promoter and the internal promoters of the tnpA gene.
FIG. 6.
(A) GUS activities measured in P. putida PRS2000 carrying the different transcriptional fusions of the tnpAC region with the reporter gene gusA. Plasmids with disrupted tnpC are marked by asterisks in the text. Data (means ± standard deviations) of at least five independent experiments are presented. pNP, p-nitrophenol; OD590, optical density at 590 nanometers. (B) Schematic depiction of plasmids with transcriptional fusions employed in GUS activity assays. Restriction sites used for construction of deletion derivatives of pKT-ACG are indicated. The EcoRI restriction site in the 3′ end of tnpC is artificial, designed by using oligonucleotide TnpCEco (Materials and Methods). The direction of transcription from the tnpA promoter is indicated by an arrow.
TnpC does not affect transcription elongation of the tnpA gene.
Results of the experiments using the tnpA-gusA translational fusions revealed that TnpC affected TnpA expression after transcriptional initiation of the tnpA gene. To investigate if TnpC operates at the transcription elongation of the tnpA gene, we compared the expression of the reporter gene gusA in plasmids pKT-ACG, pKT-a1CG, and pKT-a2CG (Fig. 6A) and in their TnpC-defective derivatives pKT-AC*G, pKT-a1C*G, and pKT-a2C*G [Fig. 6A; the same strategy employed in the construction of pKTtnpA(D/P)* was used for designing them]. No differences in levels of GUS activity were established in the cells of P. putida PRS2000 harboring the 5′ deletion derivatives of the full-length tnpAC+gusA gene cassette either with intact tnpC or with disrupted tnpC (Fig. 6A). A modest repressive effect of intact tnpC on GUS activity (approximately 25%) appeared in the pKT-ACG-containing cells of P. putida PRS2000 compared to the GUS activity in pKT-AC*G-carrying bacteria (Fig. 6A). To control whether this effect is real and whether it might be obscured by the downstream transcription, plasmids pKT-AdelCG and pKTAdelC*G lacking the second half of the tnpA gene (DNA region between the restriction sites ClaI and Cfr10I) (Fig. 6B) were constructed. GUS activity levels measured in P. putida PRS2000 containing plasmid pKT-AdelCG with either intact or disrupted tnpC were similar (Fig. 6A). Therefore, we suggest that instead of influencing the transcription of the tnpA gene, TnpC affects TnpA expression post-transcriptionally.
DISCUSSION
A high level of transposition activity would be harmful for the host. Therefore, every transposon must have regulatory mechanisms that keep the level of transposition low. Most of these regulatory mechanisms are developed to control the level of active transposase, the protein that carries out the transposition reaction (reviewed in references 7 and 18). Data presented in this paper show that the abundance of the Tn4652 transposase TnpA in P. putida is downregulated by the transposon-encoded protein TnpC.
The amount of the Tn4652 transposase in bacterial cell lysates was monitored by Western blot analysis with polyclonal antibodies against the TnpA protein of Tn4652. The analysis revealed that this protein was not detectable in either Tn4652-containing P. putida PaW85 or Tn4652-free P. putida PRS2000 complemented with Tn4652 in the plasmid pEST1354 (Fig. 2, lanes 2 and 4). However, subcloning of the tnpA gene together with its native promoter allowed us to detect the TnpA protein in both the P. putida PaW85 and PRS2000 backgrounds (Fig. 2, lanes 3 and 5). This indicated that some factor encoded by Tn4652 must be involved in TnpA downregulation. A DNA region affecting the amount of TnpA in bacteria was located just downstream of the tnpA gene, where an ORF encoding a 120-amino-acid-long polypeptide was discovered. Disruption of this ORF demonstrated that the protein encoded by the ORF and named TnpC by us was functioning as a regulator of the TnpA protein (Fig. 2, lanes 6 and 7).
Notably, the level of TnpA was elevated in P. putida PRS2000 compared to the concentration of TnpA in P. putida PaW85 (Fig. 2, lanes 3 and 5). P. putida PaW85 contains Tn4652 in its chromosome. Therefore, we suggest that chromosomally encoded TnpC may act in trans and decrease the amount of plasmid-encoded TnpA. For many transposons encoding both transposase and its inhibitor, it has been shown that transposase can function effectively only in cis but the inhibitor can act in trans as well (22, 27, 31). This mechanism is believed to have evolved to limit the rate of accumulation of transposable elements in the genome (18).
Investigation of TnpC expression revealed that TnpC is expressed from multiple promoters located inside of the tnpA gene (Fig. 6A). Part of tnpC expression is promoted by the first half of the coding sequence of tnpA and possibly also from the tnpA promoter. However, a larger amount of the transcription of tnpC was initiated from the 3′ terminal half of the tnpA gene. Interestingly, data presented in Fig. 6A showed that when the 3′ terminal half of the tnpA gene was eliminated (plasmid pKT-AdelCG), the GUS activity was about the same as in the case of the full-length tnpAC+gusA cassette (plasmid pKT-ACG). This was approximately twice as high as could be expected on the basis of the simple arithmetical subtraction of downstream promoter activities from the upstream ones. This finding could be interpreted as a diminishing effect of the DNA sequences located in the 3′ terminal half of the tnpA gene on the transcription initiated in the first half of tnpA. Concerning the expression of tnpA, one may speculate that transcription elongation of the tnpA-specific mRNA might be influenced by this region. However, we point out that this silencing effect of the downstream region of tnpA was not related to the intactness of tnpC (Fig. 6A). Therefore, we suspect that besides the TnpC-specific downregulation of TnpA, expression of tnpA could also be influenced by a restraint on the rate of transcription elongation of the transposase gene. Indeed, the transcription elongation rate is not constant and there are multiple examples for retardation of transcription elongation due to certain DNA sequences or the nature of nascent RNA (reviewed in reference 25).
The question about the checkpoint of the TnpC action in the regulation of the concentration of TnpA cannot be answered unambiguously. However, our results support the possibility that TnpC operates in the regulation of the transposase of Tn4652 at the post-transcriptional level. First, it does not interfere with the transcription initiation from the tnpA promoter. Exchanging the tnpA promoter with another one revealed no effect on the ability of TnpC to downregulate expression of TnpA (Fig. 2, lanes 13 and 14). Second, translational fusions of the tnpA gene 5′ end with the reporter gene gusA exhibited no sensitivity to the expression of TnpC (Fig. 5A). Thus, TnpC affected neither the transcriptional nor the translational initiation of the tnpA gene. Third, testing the effect of TnpC on transcription throughout the tnpA gene revealed that transcription elongation was also not altered by TnpC (Fig. 6A). On the basis of these results, we suggest that TnpC functions in regulation of TnpA post-transcriptionally. Moreover, TnpC seems to act after translation initiation, as determined by results obtained from experiments with translational fusions. Herein, it should be noted that it is improbable that translation elongation would be controlled by protein repressors (10). Therefore, it is possible that TnpC acts post-translationally by altering transposase folding and/or transposase stability. However, we cannot exclude the possibility that TnpC is involved in the regulation of tnpA-specific mRNA stability.
Comparison of TnpC with the translated sequences of genes in the EMBL database showed a striking similarity between TnpC and a putative 120-amino-acid-long polypeptide encoded by the mercury resistance transposon Tn5041 (Fig. 4). We have previously shown that TnpA of Tn4652 is very similar to TnpA of Tn5041 (12). Up to now, there are no data about the regulation of TnpA of Tn5041. However, considering the similarity between TnpC of Tn4652 and the putative 120-amino-acid polypeptide of Tn5041, we suggest that a regulatory mechanism similar to that described for TnpC of Tn4652 may also regulate the transposase of Tn5041.
ACKNOWLEDGMENTS
We are grateful to J. Parik for producing mouse anti-TnpA polyclonal serum and to A. Eriksson for kindly providing plasmid pGUS102. We also thank T. Alamäe, L. Kasak, V. Kõiv, and A. Tamm for critically reading the manuscript and for their helpful discussions.
This work was supported by grant no. 2323 from the Estonian Science Foundation.
REFERENCES
- 1.Allison R G, Chaconas G. Role of the A protein-binding sites in the in vitro transposition of Mu DNA. J Biol Chem. 1992;267:19963–19970. [PubMed] [Google Scholar]
- 2.Arini A, Keller M P, Arber W. An antisense RNA in IS30 regulates the translational expression of the transposase. Biol Chem. 1997;378:1421–1431. doi: 10.1515/bchm.1997.378.12.1421. [DOI] [PubMed] [Google Scholar]
- 3.Bagdasarian M M, Amann E, Lurz R, Ruckert B, Bagdasarian M. Activity of the hybrid trp-lac(tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983;26:273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- 4.Bayley S A, Duggleby C J, Worsey M J, Williams P A, Hardy K G, Broda P. Two modes of loss of the TOL function from Pseudomonas putida mt-2. Mol Gen Genet. 1977;154:203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Carter P, Bedoulle H, Winter G. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 1985;13:4431–4443. doi: 10.1093/nar/13.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler M, Fayet O. Translational frameshifting in the control of transposition in bacteria. Mol Microbiol. 1993;7:497–503. doi: 10.1111/j.1365-2958.1993.tb01140.x. [DOI] [PubMed] [Google Scholar]
- 8.Chou J, Lemaux P G, Casadaban M, Cohen S N. Transposition protein of Tn3: identification and characterization of an essential repressor controlled gene product. Nature. 1979;282:801–806. doi: 10.1038/282801a0. [DOI] [PubMed] [Google Scholar]
- 9.Derbyshire K M, Grindley N D F. cis preference of the IS903 transposase is mediated by a combination of transposase instability and inefficient translation. Mol Microbiol. 1996;21:1261–1272. doi: 10.1111/j.1365-2958.1996.tb02587.x. [DOI] [PubMed] [Google Scholar]
- 10.Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D. Studies on the transformation of E. coli with plasmids. J Mol Biol. 1983;166:577–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 12.Hõrak R, Kivisaar M. Expression of the transposase gene tnpA of Tn4652 is positively affected by integration host factor. J Bacteriol. 1998;180:2822–2829. doi: 10.1128/jb.180.11.2822-2829.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu S T, Wang H C, Lei G S, Wang S H. Negative regulation of IS2 transposition by the cyclic AMP (cAMP)-cAMP receptor protein complex. J Bacteriol. 1998;180:2682–2688. doi: 10.1128/jb.180.10.2682-2688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasak L, Hõrak R, Nurk A, Talvik K, Kivisaar M. Regulation of the catechol 1,2-dioxygenase- and phenol monooxygenase-encoding pheBA operon in Pseudomonas putida PaW85. J Bacteriol. 1993;175:8038–8042. doi: 10.1128/jb.175.24.8038-8042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasak L, Hõrak R, Kivisaar M. Promoter-creating mutations in Pseudomonas putida: a model system for the study of mutation in starving bacteria. Proc Natl Acad Sci USA. 1997;94:3134–3139. doi: 10.1073/pnas.94.7.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kholodii G Y, Yurieva O V, Gorlenko Z M, Mindlin S Z, Bass I A, Lomovskaya O L, Kopteva A V, Nikiforov V G. Tn5041: a chimeric mercury resistance transposon closely related to the toluene degradative transposon Tn4651. Microbiology. 1997;143:2549–2556. doi: 10.1099/00221287-143-8-2549. [DOI] [PubMed] [Google Scholar]
- 17.Kivisaar M, Hõrak R, Kasak L, Heinaru A, Habicht J. Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid. 1990;24:25–36. doi: 10.1016/0147-619x(90)90022-5. [DOI] [PubMed] [Google Scholar]
- 18.Kleckner N. Regulation of transposition in bacteria. Annu Rev Cell Biol. 1990;6:297–327. doi: 10.1146/annurev.cb.06.110190.001501. [DOI] [PubMed] [Google Scholar]
- 19.Krause H M, Higgins N P. Positive and negative regulation of the Mu operator by Mu repressor and Escherichia coli integration host factor. J Biol Chem. 1986;261:3744–3752. [PubMed] [Google Scholar]
- 20.Krebs M P, Reznikoff W S. Transcriptional and translational sites of IS50. Control of transposase and inhibitor expression. J Mol Biol. 1986;192:781–791. doi: 10.1016/0022-2836(86)90028-8. [DOI] [PubMed] [Google Scholar]
- 21.Kuan C-T, Tessman I. LexA protein of Escherichia coli represses expression of the Tn5 transposase gene. J Bacteriol. 1991;173:6406–6410. doi: 10.1128/jb.173.20.6406-6410.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machida C, Machida Y. Regulation of IS1 transposition by the insA gene product. J Mol Biol. 1989;208:567–574. doi: 10.1016/0022-2836(89)90148-4. [DOI] [PubMed] [Google Scholar]
- 23.Maekawa T, Yanagihara K, Ohtsubo E. Specific nicking at the 3′ ends of the terminal inverted repeat sequences in transposon Tn3 by transposase and an E. coli protein ACP. Genes Cells. 1996;1:1017–1030. doi: 10.1046/j.1365-2443.1996.d01-221.x. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 25.Mooney R A, Artsimovitch I, Landick R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novel G, Didier-Fichet M L, Stoeber F. Inducibility of β-glucuronidase in wild-type and hexuronate-negative mutants of Escherichia coli K-12. J Bacteriol. 1974;120:89–95. doi: 10.1128/jb.120.1.89-95.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reznikoff W S. The Tn5 transposon. Annu Rev Microbiol. 1993;47:945–963. doi: 10.1146/annurev.mi.47.100193.004501. [DOI] [PubMed] [Google Scholar]
- 28.Roberts D, Hoopes B C, McClure W R, Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- 29.Sharma R C, Schimke R T. Preparation of electro-competent E. coli using salt-free growth medium. BioTechniques. 1996;20:42–44. doi: 10.2144/96201bm08. [DOI] [PubMed] [Google Scholar]
- 30.Signon L, Kleckner N. Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favour evolution of new transposons. Genes Dev. 1995;9:1123–1136. doi: 10.1101/gad.9.9.1123. [DOI] [PubMed] [Google Scholar]
- 31.Simons R W, Kleckner N. Translational control of IS10 transposition. Cell. 1983;34:683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- 32.Studier F W, Moffatt B A. Use of bacteriophage T7 polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 33.Tsuda M, Iino T. Genetic analysis of a transposon carrying toluene degrading genes on a TOL plasmid pWW0. Mol Gen Genet. 1987;210:270–276. doi: 10.1007/BF00325693. [DOI] [PubMed] [Google Scholar]
- 34.Wheelis M L, Ornston L N. Genetic control of enzyme induction in β-ketoadipate pathway of Pseudomonas putida: deletion mapping of cat mutations. J Bacteriol. 1972;109:790–795. doi: 10.1128/jb.109.2.790-795.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiater L A, Grindley N D F. γδ transposase and integration host factor bind cooperatively at both ends of γδ. EMBO J. 1988;7:1907–1911. doi: 10.1002/j.1460-2075.1988.tb03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin J C P, Krebs M P, Reznikoff W S. Effect of dam methylation on Tn5 transposition. J Mol Biol. 1988;199:35–45. doi: 10.1016/0022-2836(88)90377-4. [DOI] [PubMed] [Google Scholar]